Summary
Fungal pathogens continue to pose a significant threat to crop production and food supply. The early stages of plant–fungus interactions are mostly mediated by microbe‐associated molecular pattern (MAMP) molecules, perceived by plant pattern recognition receptors (PRRs). Currently, the identified fungal MAMP molecules include chitin, chitosan, β‐glucans, elicitins and ergosterol. Although the molecular battles between host plants and infecting fungal phytopathogens have been studied extensively, many aspects still need to be investigated to obtain a holistic understanding of the intrinsic mechanisms, which is paramount in combating fungal plant diseases. Here, an overview is given of the most recent findings concerning an ‘orphan’ fungal MAMP molecule, ergosterol, and we present what is currently known from a synopsis of different genes, proteins and metabolites found to play key roles in induced immune responses in plant–fungus interactions. Clearly, integrative investigations are still needed to provide a comprehensive systems‐based understanding of the dynamics associated with molecular mechanisms in plant–ergosterol interactions and associated host responses.
Keywords: ergosterol, fungal pathogens, genomics, MAMPs, metabolomics, plant–fungus interactions, proteomics
Apocalypse Now: Fungal Phytopathogen Warfare
Global losses of economically important crop plants as a result of the destructive interaction of virulent fungi and their respective elicitors/effectors with plant hosts have always been a major concern to food yield and security, as well as to the maintenance of biodiversity within ecosystems (Fisher et al., 2012; Sharma et al., 2011). Despite this fact, there is a substantial short‐fall in information on specific plant–fungus interactions when compared with the vast number of investigations into plant–bacterium interactions. Thus, as a result of the increase in emerging infectious diseases (EIDs) during the 21st century, research in this regard has intensified worldwide (Pennisi, 2010). Indeed, it is estimated that up to 70% of global EIDs affecting all kingdoms of life are caused either directly or indirectly by fungal pathogens (Fisher et al., 2012).
The up to 125 million tonne loss in the five most economically important food crops (potatoes, maize, rice, wheat and soybean) represents the potentially devastating current‐day impact of fungal pathogens on both food security and economic stability. This contributes greatly to the promotion of poverty and financial loss within global agricultural sectors. Losses occurring as a result of maize, rice and wheat diseases exclusively total nearly $60 billion per year; thus, if the spread of these diseases and those affecting other plants can be managed more effectively, up to 600 million people could be fed through increased food yield (Fisher et al., 2012). This showcases the extensive impact of fungal pathogens on modern food security, and thus the integrity of agricultural industries must be maintained effectively to curb crop losses as much as possible (Fisher et al., 2012). Thus, an understanding of the molecular mechanisms involved in plant–fungus interactions is of paramount importance, as this would open up the possibility to develop novel, more effective and sustainable strategies to control or eradicate fungal diseases in plants. To set the terrain for the current combat scene, a patrolled mission on general background information and plant–fungus interactions will first be surveyed before delving into specific plant–ergosterol interactions.
Innate Immunity: The Central Command Unit to Coordinate Plant Defence
The continuing biological war between plants and various microbial pathogens, as with all co‐evolving organisms, for survival of the fittest, is inescapable (Bittel and Robatzek, 2007). To defend themselves against pathogens, plants utilise a protection system (passive resistance) that involves an array of structural barriers and pre‐formed antimicrobial phytoanticipins to prevent/attenuate invasion by potential attackers (Pritchard and Birch, 2011; Schwessinger and Zipfel, 2008). However, this protection system can be successfully overcome by numerous phytopathogens to ultimately cause infection and disease. Lacking an adaptive immune system, plants may then initiate dynamic innate immune defences that are activated once danger signals have been perceived.
The molecular battles occurring between host plants and their infecting phytopathogens have been fascinating fields of research for decades (Jones and Dangl, 2006; Pieterse et al., 2009). Since the 1940s, scientists have been trying to unravel the complex molecular mechanisms that comprise the plant innate immune system (Flor, 1942; Thomma et al., 2011). The notion of these immune responses and their interactions with the pathogens causing plant diseases is far more complex, and has been revised and re‐formulated many times. The conception of the initial gene‐for‐gene resistance hypothesis (Flor, 1942; Keen, 1990), the more specific characterisation and grouping of various pathogenic signature molecules into different classes of elicitors and effectors (Bent and Mackey, 2007; Boller and Felix, 2009), the coinage of the interrelated ‘zig‐zag hypothesis’ terms microbe/pathogen‐associated molecular pattern (M/PAMP)‐triggered immunity (M/PTI) and effector‐triggered immunity (ETI) (Jones and Dangl, 2006), and the concepts of systemic acquired resistance (SAR) (Van der Biezen and Jones, 1998) and induced systemic resistance (ISR) have all contributed greatly to our understanding and elucidation of this multicomponent immune system of plants (Fig. 1). It should be noted that, from here onwards, the term MAMP is preferred to PAMP so as to be more inclusive. Until recently, these concepts and pathways have largely been treated as separate entities and studied in an isolated sense; however, recent reports have suggested a more intricate interplay and continuity between defence pathways, which ultimately culminate in a single, complex interconnected signalling network deployed by the plant as necessary (Boller and Felix, 2009; Katagiri and Tsuda, 2010; Qi et al., 2011; Thomma et al., 2011; Vidhyasekaran, 2014).
Figure 1.
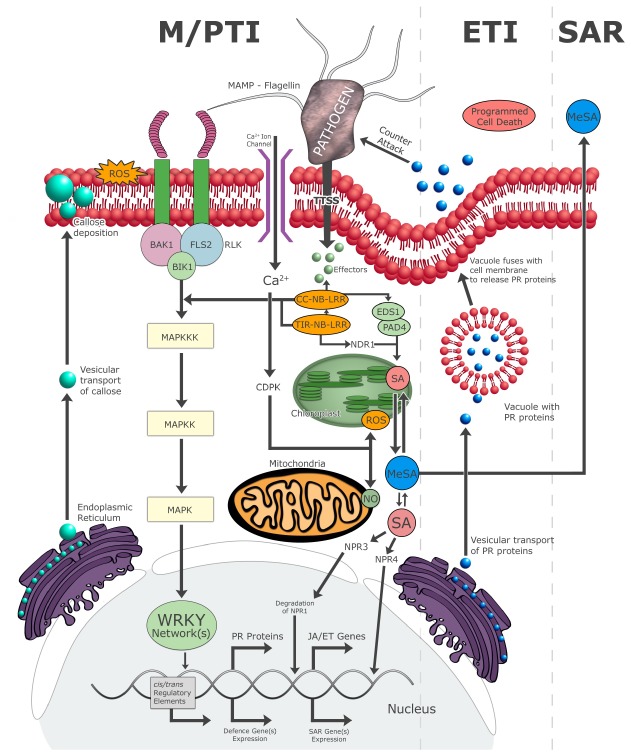
Diagram illustrating the interplay between plant innate immune responses and cellular events with regard to microbe/pathogen‐associated molecular pattern (M/PAMP)‐triggered immunity (M/PTI), effector‐triggered immunity (ETI) and the concomitant hypersensitive response (HR)/programmed cell death (PCD), and systemic acquired resistance (SAR) (adapted from Muthamilarasan and Prasad, 2013). BAK1, brassinosteroid insensitive 1 (BRI1)‐associated kinase; BIK1, Botrytis‐induced kinase 1; CC‐NB‐LRR, coiled‐coil nucleotide‐binding leucine‐rich‐repeat; CDPK, calcium‐dependent protein kinase; EDS1, enhanced disease susceptibility 1; ET, ethylene; FLS2, flagellin‐sensitive 2; JA, jasmonic acid; MAPK(K/K), mitogen‐activated protein kinase (kinase/kinase); MeSA, methyl‐SA; NPR, nonexpressor of PR gene; PAD4, phytoalexin deficient 4; PR, pathogenesis‐related; RLK, receptor‐like kinase; ROS, reactive oxygen species; SA, salicylic acid; TIR, Toll and Interleukin 1 transmembrane receptor; TTSS, type III secretion system.
The basic concept of immunity and host resistance remains largely the same. In order for the successful establishment of a phytopathogen within a potential host plant, initial avoidance and traversing of the host's pre‐formed defence responses are essential (Deepak et al., 2006; Fan and Doerner, 2012; Gonzalez‐Lamothe et al., 2009; Madala et al., 2011; Sanabria et al., 2010). During the process certain conserved molecular signatures/motifs present in a particular class of microbes (MAMPs) may be specifically recognised by dedicated host receptors (pattern recognition receptors or PRRs), thereby initiating inducible defence response cascades (Beck et al., 2012; Henry et al., 2012; Sanabria et al., 2010; Zipfel, 2008,2009), resulting in immunity through MTI activation (Jones and Dangl, 2006). It should also be noted that wounding or pathogen attack may lead to the production of damage‐associated molecular patterns (DAMPs), which are categorised and recognised in a similar fashion to MAMPs. From here, the paradigm of MTI and ETI, or the zig‐zag hypothesis of plant–pathogen interactions and plant defences, originated (Fig. 1). In this regard, MAMPs can be described as general elicitors of the basal defence response. Bacterial MAMPs include flagellin (Zipfel, 2008), EF‐Tu (elongation factor‐thermounstable) (Postel and Kemmerling, 2009), LPS (lipopolysaccharide) from Gram‐negative bacteria (Gerber and Dubery, 2004; Gerber et al., 2006) and peptidoglycan from Gram‐positive bacteria (Nürnberger et al., 2004), whereas fungal MAMPs include chitin (Granado et al., 1995), chitosan (Amborabé et al., 2008), xylanase (Hou et al., 2013), elicitin proteins (Yang et al., 2012), cerato‐platanin proteins (Frías et al., 2013) and the sterol ergosterol (Rossard et al., 2010).
Attack and Defence: Fungus Versus Plant
Induced responses ultimately require the activation of many complex cascades, and one of the initial responses to ensue following MAMP perception and host infection is transcriptional reprogramming of the host cell transcriptome (Vidhyasekaran, 2014), with downstream protein expression, signal transduction and the production of numerous secondary metabolites, including phytoalexins. Although the molecular mechanisms of these induced immune responses are not yet fully understood, the general plant defence reaction can be simplified into interconnected steps: following the perception of the non‐self/altered‐self/damaged‐self entities (Sanabria et al., 2010), the early cellular events include changes in plasma membrane permeability, the influx of Ca2+ and H+ and efflux of Cl− and K+, pH changes, plasma membrane depolarisation at the immediate site of infection, generation of reactive oxygen and nitrogen species (ROS and RNS), ion‐induced signalling, mitogen‐activated protein (MAP) kinase activation and accumulation of pathogenesis‐related (PR) proteins (Fig. 1) (Almagro et al., 2009; Arasimowicz and Floryszakwieczorek, 2007; Lecourieux et al., 2006; Ma, 2011; Piater et al., 2004; Vatsa et al., 2011a, b). The molecules of these cellular events can even circulate to distal parts of the plant, giving rise to SAR (Mishina and Zeier, 2007) and an associated increase in the levels of phytohormones, thereby effectively controlling the spread of the pathogen through the priming of uninfected plant parts and destruction of infected plant tissues.
However, many aspects of the molecular mechanisms involved in plant–pathogen interactions are still unknown. Transcriptomic‐ and proteomic‐based studies have provided tremendous knowledge about the plant immune response, but with certain limitations. Metabolomics is now being suggested as the ultimate level of post‐genomic analyses, as it provides a comprehensive picture of the molecular organisation of an organism under defined biological conditions, thus reflecting both transcriptional and post‐transcriptional regulation (Allwood et al., 2008, 2010; Tugizimana et al., 2013). Plant–microbe interactions can thus be described as a co‐evolving molecular war between pathogen and host, with specialised weaponry available to both partners that can be activated and used as required, and the continuous competition between the opponents leads to successful infection by the pathogen or effective resistance in the host.
Ergosterol: Betraying The Presence of Fungal Attackers
Ergosterol, a 5,7‐diene oxysterol, is the most predominant sterol found in fungal cell membranes (Mohd As'wad et al., 2011; Zhao et al., 2005) and is of particular interest as a MAMP because of its potential to activate lipid‐based signalling events. Ergosterol biosynthesis has never been reported in plants and is thus recognised as ‘non‐self’ by a plant cell, thereby falling into the lipophilic class of biotic elicitors (Angelova et al., 2006; Granado et al., 1995; Sanabria et al., 2010). Although in planta determination of ergosterol can be used to estimate the level of fungal infection (Mohd As'wad et al., 2011), i.e. as a fungal marker as documented in cereal crops, including barley and corn (Dohnal et al., 2010), to date, this MAMP has not received as much attention as would be expected.
Sterols are part of the vast family of isoprenoid compounds, and display both chemical complexity and remarkable functional diversity in living organisms (Liu and Nes, 2009). Sterols that are present in the plasma membranes of eukaryotic cells are essential for the organisation and function of these structures, with ergosterol being the major component in lower eukaryotes, whereas cholesterol is found in higher eukaryotes (Hsueh et al., 2005; Xu et al., 2001), and sitosterol is the most abundant phytosterol (Roche et al., 2008). These steroids differ structurally, with ergosterol having two additional double bonds (at positions C7–C8 and C22–C23) and a methyl group at C24 of the side‐chain (Fig. 2). Such features are essential for functionality and appear relatively late during ergosterol biosynthesis (Liu and Nes, 2009; Naumowicz et al., 2011). These structural differences also presumably allow the plant cell to recognise ergosterol as a ‘non‐self’, foreign MAMP (Fig. 3).
Figure 2.
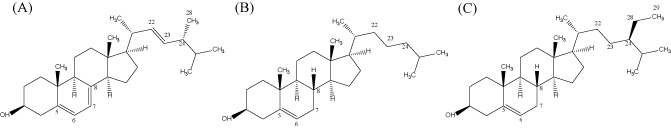
Chemical structures of ergosterol (A), cholesterol (B) and sitosterol (C), indicating structural similarities and differences. Compared with cholesterol, ergosterol has unsaturation at positions C7–C8 and an additional methyl group (C28) at the C24 position, whereas sitosterol has an additional ethyl group (C28–C29) at the C24 position.
Figure 3.
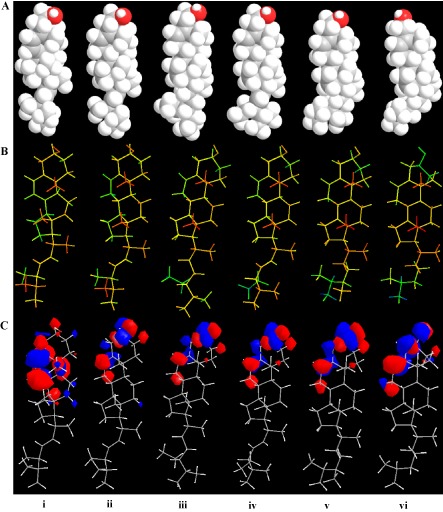
Spacefilling (A), stereochemical stick (B) and frontier molecular orbital (FMO) (C) models for ergosterol (i), brassicasterol (ii), sitosterol (iii), stigmasterol (iv), campesterol (v) and cholesterol (vi). In (A), lone electron pairs (white circles) are indicated on the OH (red) groups of all sterols. In (B), the depth of colour indicates the progression of stereochemistry as planar (yellow/orange), forward (orange/red) and backward (green/blue) projections. In (C), the FMO surfaces (colour‐coded wavefunctions: red, positive; blue, negative) visually represent the various stable electron distributions of a molecule and, according to frontier orbital theory, the shapes and symmetries are crucial in predicting the reactivity of a species, and the stereochemical and regiochemical outcome of a chemical reaction.
Unfortunately, the mechanism of ergosterol recognition by plant cells is not known thus far. Scientists have hypothesised that plants either possess an ergosterol receptor/exploit an alternative MAMP receptor, or that ergosterol uptake leads to perturbations of a lipid raft structure because of the ability of this sterol to form very stable microdomains (Amborabé et al., 2003; Xu et al., 2001). Although the signal transduction pathway induced after recognition has not been fully elucidated to date, researchers have shown that ergosterol acts as a MAMP molecule in tobacco and tomato plants, resulting in an MTI response.
Soldiers at The Look‐Out Posts: Proteins Involved in Perception of Fungal Attackers
Structural constituents of fungal cell membranes and walls induce the same types of basal innate immunity as other well‐studied elicitors, such as LPS and flagellin (Iriti and Faoro, 2007; Zipfel et al., 2004). Fungal MAMPs include ergosterol, β‐glucans, chitin/chitosan, ethylene‐inducing xylanase and the very recently reported cerato‐platanin BcSpL1 from Botrytis cinerea, whereas secreted elicitin proteins, such as cryptogein, have been reported from oomycetes, such as Phytophthora (Amborabé et al., 2008; Frías et al., 2013; Shimizu et al., 2010; Zipfel, 2009).
The initiation of an immune response involves the binding of a MAMP or effector to either a transmembrane PRR or an intracellular receptor (Thomma et al., 2011), with the PRRs being classified into two classes, namely receptor‐like kinases (RLKs) and receptor‐like proteins (RLPs) (Monaghan and Zipfel, 2012; Yang et al., 2012). To date, only a few plant PRRs for fungal MAMPs have been identified. The best‐described include those that perceive chitin, namely chitin‐elicited receptor kinase 1 (CERK1) and chitin elicitor binding protein (CEBiP) (Zipfel, 2009), the extracellular domains of which consist of leucine‐rich repeats (LRRs) and are more specifically denoted as eLRRs (Mazzotta and Kemmerling, 2011; Yang et al., 2012). The CERK1/CEBiP receptor's activity and interaction with chitin have been shown in vitro, and seem to be responsible for conferring some resistance to fungi in plants. Furthermore, knockouts of OsCERK1 in rice consequently reduced the capacity of the plant to perceive and adequately defend itself against fungal pathogens (Shimizu et al., 2010; Yang et al., 2012), whereas a very recent communication reported on CERK1‐independent fungal resistance in Arabidopsis (Narusaka et al., 2013). Further research is, however, required to definitively establish the roles played by these two important receptors in fungal pathogen perception and defence with regard to chitin and various chito‐oligosaccharides. The ethylene‐induced 22‐kDa xylanase protein from Trichoderma viride, however, has been shown to be a prominent fungal elicitor, and has been demonstrated to stimulate ethylene production during the defence response. Two PPRs in tomato cells have also been identified, and fall into the LRR‐RLP class of PRRs, namely LeEIX1 and LeEIX2. Only LeEIX2 is capable of inducing a defence response, whereas LeEIX1 assists LeEIX2 and aids endocytosis of the chitin‐bound receptors (Mazzotta and Kemmerling, 2011).
Sounding the Alarm: Transduction of Ergosterol‐Triggered Signals
On binding of an elicitor to a receptor, a signalling cascade is activated and, in turn, results in the activation and expression of multiple defence‐related genes, with the corresponding proteins translated from their mRNA counterparts being the key players in many signal transduction pathways that activate the initial defence responses when the plant perceives MAMPs (Sanabria et al., 2012).
Akin to most plant–fungus interactions, signal transduction events specifically set in motion following the perception of ergosterol are characterised by changes in plasma membrane potential (Amborabé et al., 2003; Rossard et al., 2006), changes in the fluxes of H+ ions (Amborabé et al., 2003; Granado et al., 1995; Kasparovsky et al., 2003; Rossard et al., 2010), rapid transient growth medium alkalinisation (Amborabé et al., 2003; Granado et al., 1995), mobilisation of internal calcium stores, activation of an oxidative burst and production of ROS (Kasparovsky et al., 2003), involving protein kinases and phospholipases, namely protein kinase C (PKC) and phospholipase A2 (PLA2) (Kasparovsky et al., 2004).
Regulatory enzymes are an essential part of these signal transduction pathways, and the most prominent participants are protein kinases and phosphatases, which act in concert to activate and deactivate, respectively, protein kinase components in signal transduction during plant–pathogen interactions (Gerber and Dubery, 2004; Gerber et al., 2006; Xing et al., 2002; Zhu and Li, 2013). Furthermore, these cascades have been found to be a converging point at the start of the defence‐signalling network (Angelova et al., 2006; He et al., 2007; Peck et al., 2001; Zhang and Klessig, 2001).
Enter the Omics in Plant–Fungus Interactions for Setting the Crosshairs on Ergosterol
The outcome of plant–fungus interactions relies on the complex interplay of numerous molecular signals, and both parties have elaborate pathways employed specifically in perception, recognition and infection/defence mechanisms. As defence and immune mechanisms against fungal MAMPs and, in particular, ergosterol are poorly understood relative to their bacterial counterparts, the challenge in current research is to use modern ‘‐omic’ technologies to unravel and reveal the cellular events and signal transduction pathways that are activated in plants following the perception of fungal pathogens or MAMPs (Ferreira et al., 2006).
Although numerous plant defence responses to fungal pathogens have already been studied and characterised, these have, to a large extent, been rather narrowly focused, and more recent strategies seem to favour bio‐analytical techniques as well as more complete, comprehensive experimental designs. For this reason, it is important to consider all‐inclusive research approaches that focus on ‘‐omics’ techniques, including genomics, transcriptomics, proteomics and metabolomics, thereby providing a greater wealth of potential knowledge that can be gained from a particular investigation (Lederberg and McCray, 2001).
Activating the defensome: transcription factors (TFs) and regulatory proteins
The activation and increased production of specific enzymes, hormones and metabolites in the host following pathogen perception are regarded as vital components of the ensuing host response. However, the importance of assessing gene regulation following host invasion and pathogenic challenge should not be neglected in order to more completely verify and evaluate the interplay between the two parties in such complex immune response cascades. As such, a large number of genes have also been identified as possible candidates that code for key proteins involved in a wide variety of plant–pathogen interactions. Many of these genes have been positively identified as being involved either directly or indirectly in the pathogen perception of plants.
To date, numerous studies have focused specifically on genomic/transcriptomic findings and, in particular, on TFs and gene regulatory elements that play commanding roles in transcriptional reprogramming of the host cell transcriptome, thereby initiating and potentiating the increased expression of defence biomolecules. It seems that some of these early defence responses are mediated by the expression of numerous WRKY TFs, which contain a domain having a highly conserved WRKYGQK sequence, as well as a novel zinc finger motif (Eulgem et al., 2000). These TFs have been shown to be involved in plant–fungus interactions. For example, overexpression of AtWRKY80 in Arabidopsis leads to the activation of numerous defence responses, whereas AtWRKY8 and AtWRKY70 have been shown to control cross‐talk between jasmonic acid (JA)‐ and salicylic acid (SA)‐dependent defence response pathways and exhibit increased resistance to Botrytis infection (Ishihama et al., 2011). Additional investigations on WRKY70 knockout lines of Arabidopsis have also shown increased susceptibility to infection by B. cinerea, thereby indicating the importance of this class of TFs in plant–fungus defences (Chen et al., 2010; Knoth et al., 2007; Li et al., 2006).
Furthermore, in a recent study conducted by Peng et al. (2012), it was found that the OsWRKY30 gene was involved in specific defence responses in rice (Oryza sativa) following various treatments, including treatment of the host with JA and methyl jasmonate, as well as SA and the fungal rice pathogens, Rhizoctonia solani and Magnaporthe grisea. OsWRKY30 showed increased transcript accumulation post‐JA and SA treatment, and overexpression of this gene also led to the activation of numerous defence genes involved in the JA synthesis and defence‐signalling pathways, including LOX (lipoxygenase), AOS2 (allene oxide synthase 2), PR‐3 and PR‐10, and increased JA accumulation following fungal pathogen infection.
Research conducted on AtWRKY62 has also found a link between its activated expression through both SA‐ and JA‐dependent signalling, with NPR‐1 (Nonexpressor of pathogenesis‐related gene 1) being involved in its activity (Pandey and Somssich, 2009). Unfortunately, these putative pathways have not yet been well studied, and AtWRKY62 involvement in general immune responses is not clearly understood. Yet, other studies have shown the involvement of closely related WRKYs 18, 40 and 60 in Arabidopsis plant resistance to various pathogens. These three WRKYs seem to be interlinked, and may provide functional redundancy in both fungal and bacterial defence responses (Pandey and Somssich, 2009). AtWRKY33 appears to be involved in defence against necrotrophic pathogens, and its putative interactions with WRKY3 and WRKY4 genes, and with WRKY48, suggest functional redundancy regarding these responses. Interactions of some of the more common, well‐studied WRKY genes are summarised in Fig. 4 with regard to fungal MAMP perception in plants. It is clear that our understanding of the interconnected responses of individual members of the complex plant WRKY network is still not complete, but the large amount of research currently being conducted on this class of TFs is sure to provide further insights into plant defence responses in the future (Pandey and Somssich, 2009).
Figure 4.
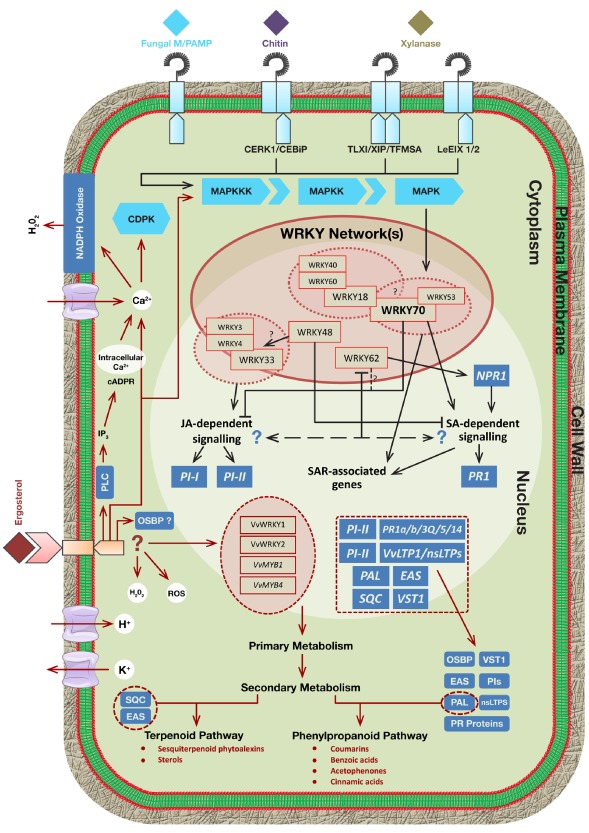
Summary of all known/putative mechanisms and players of important fungal microbe/pathogen‐associated molecular pattern (M/PAMP) perception, and those influenced by ergosterol. The structures of the known fungal pattern recognition receptor (PRR) complexes (those representing the recognition of chitin and xylanase) have been included and indicate downstream/intracellular mechanisms activated by their perception at the cell membrane. These mechanisms can be extended to provide a suitable hypothesis for ergosterol perception. Ergosterol has been shown to trigger both Ca2+ influx and release of intracellular Ca2+ stores, which has been observed with the perception of other fungal M/PAMPs by the plant immune system. The hypothesised receptor, based on the assumption that ergosterol may be perceived in the same manner as other M/PAMPs, is indicated in maroon on the left. A possible intracellular receptor, oxysterol‐binding protein (OSBP), is included. The known metabolic pathways, and their resultant metabolites, that are involved in the defence response following ergosterol treatment are shown (maroon arrows). In addition, putative interactions between some well‐studied WRKY transcription factors are also represented within the nucleus, creating an intricate WRKY network activated through various upstream interactions by MAPK‐based signalling. Dotted lines indicate putative/unestablished reactions, and question marks represent unknown mechanisms of interaction. Three important metabolic enzymes, which contribute towards the production of specific ergosterol‐induced metabolites, are SQC, EAS and PAL. These are circled by dotted lines in the lower portion of the figure. It is important to note that the genes, proteins and metabolites implicated in this figure have resulted from studies in tobacco and grape plants. cADPR, cyclic ADP‐ribose; CDPK, calcium‐dependent protein kinase; CeBiP, chitin elicitor binding protein; CERK1, chitin elicitor receptor kinase; EAS, epi‐aristolochene synthase; H2O2, hydrogen peroxide; IP3, inositol 3‐phosphate; JA, jasmonic acid; LeEIX, Lycopersicon esculentum ethylene‐induced xylanase; MAPK, mitogen‐activated protein kinase; MAPKK, MAP kinase kinase; MAPKKK, MAP kinase kinase kinase; NPR1, Nonexpressor of pathogenesis‐related gene 1; nsLTP, nonspecific lipid‐transfer protein; MYB, myeloblastosis; OSBP, oxysterol‐binding protein; PAL, phenylalanine ammonia lyase; PLC, phospholipase C; PI, proteinase inhibitor; PR, pathogenesis‐related; ROS, reactive oxygen species; SA, salicylic acid; SAR, systemic acquired resistance; SQC, sesquiterpenoid cyclase; TFs, transcription factors; TFMSA, trifluoromethanesulphonic acid; TLXI, thaumatin‐like xylanase inhibitor; VST1, Vitis stilbene synthase 1; Vv, Vitis vinifera; XIP, xylanase inhibitor protein.
Key transcripts that have proven to be useful in the study of plant responses towards ergosterol are the type‐I lipid‐transfer proteins (LTPs). In a study conducted in 2006 on grapevine, Vitis vinifera, it was found that VvLTP1 exhibited activated expression following ergosterol treatment and, subsequent to intensive studies of the gene's promoter region, several cis‐acting regulatory elements were identified that may be involved in general and ergosterol‐specific plant defence response activation of LTP genes (Laquitaine et al., 2006). The various regulatory elements identified within the promoter region of the gene were tested for activation by deletional analysis of the promoter region linked to β‐glucuronidase (GUS) as a reporter gene (Laquitaine et al., 2006). Following ergosterol elicitation, it was postulated that one W‐Box, EIRE (ethylene‐induced response element), as well as a further identified W‐Box, may be required for VvLTP1 gene activation initiated specifically by ergosterol (Laquitaine et al., 2006). Furthermore, expression of VvMYB4 and VvWRKY1 genes coding for TFs associated specifically with binding to W‐Box and MYB‐box cis‐acting promoter elements (Stracke et al., 2001; Ülker and Somssich, 2004) increased the expression of VvLTP1 to greater than 10‐fold in vitro, whereas VvMYB2 increased in expression by approximately eightfold. In addition, following ergosterol elicitation, a 23‐fold increase in the expression of VvWRKY1 was noted within 5 h post‐treatment, and a near fivefold increase in VvLTP1 expression within 24 h post‐treatment, specifically, thus highlighting the involvement of all the above in defence‐related VvLTP1 activation initiated specifically by ergosterol (Laquitaine et al., 2006). Moreover, it was found that the priming of grapevine with ergosterol resulted in enhanced resistance to B. cinerea and the up‐regulated expression of VST1 (coding for the defence‐related enzyme stilbene synthase). This gene is responsible for increased phytoalexin production and plays an active role in defence against stilbene‐sensitive pathogens (Coutos‐Thévenot et al., 2001), and was also discovered, together with the accumulation of the phytoalexin resveratrol, following ergosterol elicitation (Laquitaine et al., 2006).
Positioning the infantry: the PR proteins
In addition to the elucidation of early plant defence responses, some studies have also been conducted on ergosterol's involvement in the activation of specific defence genes and thus subsequent protein counterparts. It has been well documented that a large arsenal of defence‐related genes is expressed and the most prominent are the PR genes, responsible for the production of PR proteins (Van Loon et al., 2006). PR proteins are one of the most important aspects of immune response mechanisms and have been grouped into 17 established families (Naseri et al., 2012; Sels et al., 2008), and have been classified according to structure, function and biological activity (refer to Table 1). It should be noted that Table 1 only summarises the main PR protein family groups and their antifungal functions, but there are also subfamilies that are distinguished by particular properties. In addition, not all of the PR protein families have been identified for one individual plant species and, in some cases, are not present at all. As such, a brief description of the individual PR proteins identified specifically in ergosterol responses is given.
Table 1.
Summary of classified pathogenesis‐related (PR) proteins in plant systems, and their general antimicrobial and antifungal activities
| Family | Original discovery | Common name | Function | References |
|---|---|---|---|---|
| PR‐1 | Tobacco PR‐1a | PR‐1 | Unknown, possible antifungal activity | Asensio et al. (2004); Elvira et al. (2008); Zhu et al. (2012) |
| PR‐2 | Tobacco PR‐2 | β‐1,3‐Glucanases | Cell wall degradation | Ebrahim et al. (2011); Vigers et al. (1991) |
| PR‐3 | Tobacco P, Q | Chitinases | Enzymatic degradation of chitin fragments in fungal cell walls | Ebrahim et al. (2011); Sharma et al. (2011); Torres et al. (2012) |
| PR‐4 | Tobacco ‘R’ | Chitin‐binding proteins | Assist with chitin hydrolysis by chitinase | Borad and Sriram (2008); Li et al. (2010) |
| PR‐5 | Tobacco S | Thaumatin‐like proteins (TLPs), permatin, osmotin | Permeabilise fungal and other cell membranes, inhibit hyphal growth | Borad and Sriram (2008); Grenier and Asselin (1990); Naseri et al. (2012); Van Loon and Van Strien (1999) |
| PR‐6 | Tomato inhibitor I | Proteinase inhibitors | Inhibition of fungal enzymes, prevent chitin synthesis | Van Loon and Van Strien (1999) |
| PR‐7 | Tomato P69 | Endoproteinases | Endoproteinase activity | Van Loon et al. (2006) |
| PR‐8 | Cucumber chitinase | Class III endochitinases, glycosyl hydrolases | Lysozyme activity | Edreva (2005); Van Loon and Van Strien (1999); Wanderley‐Nogueira et al. (2012) |
| PR‐9 | Tobacco | Peroxidases | Involved in enzymatic cross‐linking of cell wall components | Edreva (2005); Li et al. (2010) |
| PR‐10 | Parsley ‘PR1’ | Ribonucleases/ribosome inactivating protein | Enzymatic cleavage of viral RNA | Borad and Sriram (2008); Li et al. (2010); Soh et al. (2011) |
| PR‐11 | Tobacco class V chitinase | Chitinases | Fungal cell wall degradation | Edreva (2005) |
| PR‐12 | Radish Rs‐AFP3 | Lipid‐transfer protein (LTP), defensins | Transfer phospholipids between membranes, fungal membrane permeabilisation | Kader (1996); Sels et al. (2008) |
| PR‐13 | Arabidopsis THI2.1 | Thionins | Permeabilise cell membranes | Sels et al. (2008); Van Loon and Van Strien (1999) |
| PR‐14 | Barley LTP4 | LTPs, nonspecific (ns)LTPs |
Involved in cutin synthesis Transport of sterol molecules across cell membranes and possible ergosterol binding (nsLTPs in rice) |
Blilou et al. (2000); Cheng et al. (2004); Van Loon and Van Strien (1999) |
| PR‐15 | Barley OxOa | Oxalate oxidases, germins | Unknown | Bernier and Berna (2001); Zhang et al. (1995); Zhou et al. (1998) |
| PR‐16 | Barley OxOLP | Germin‐like proteins | Unknown | Van Loon et al. (2006) |
| PR‐17 | Tobacco PRp27 | Basic secretory proteins | Unknown | Van Loon et al. (2006) |
One study in particular focused on specific expressional changes and accumulation of transcripts of a number of genes coding for PR proteins, which may be distinctively grouped according to their physical and chemical properties, cellular localisation and purported functions in plant defence responses in tobacco following ergosterol elicitation (Lochman and Mikes, 2006). On investigation of the specific defence mechanisms following ergosterol perception, it was found that PR‐1, PR‐3 and PR‐5 were expressed (Lochman and Mikes, 2006), as well as the PR‐14 [nonspecific (ns)LTPs] family (Laquitaine et al., 2006), in accordance with increased gene transcript levels.
Five separate PR protein coding genes (PR1a, PR1b, PR1 basic, PR3Q and PR5), together with the genes PI‐I and PI‐II coding for proteinase inhibitors, were also investigated in Nicotiana tabacum following elicitation with nanomolar concentrations of ergosterol. It was found that transcripts of both PR1a and PR1b rapidly accumulated and relative expression increased to greater than 60‐ and 10‐fold, respectively, for these candidates over a period of 48 h following ergosterol infiltration. Furthermore, transcript levels of both PR3Q and PR5 genes also showed increases in relative gene expression of greater than fourfold over a 24‐h period, with PR3Q expression increasing to more than 10‐fold within 48 h post‐elicitation (Lochman and Mikes, 2006).
It was interesting to note that transient increases in relative expression of PI‐I and PI‐II genes were observed following leaf infiltration with three separate sterols (cholesterol, sitosterol and stigmasterol) employed as controls, as well as with water, with maximum expression occurring at 24 h post‐elicitation. This was expected as the up‐regulated expression of proteinase inhibitors has been strongly linked to plant wounding, stress responses and JA application among others, as would be evoked through leaf infiltration (Choi et al., 2000; Linthorst et al., 1993). However, this transient increase in the relative expression of PI‐I and PI‐II genes was not seen following ergosterol infiltration, thereby suggesting a potential suppression of these genes and thus the potential downstream pathways that may be affected (Lochman and Mikes, 2006). From all the analysed relative expression changes in the genes investigated, it was thus postulated that ergosterol may function to increase the expression of certain key defence‐related genes, specifically those involved in or affected by the SA signalling pathway, and that ergosterol may mediate possible cross‐talk between SA and JA signalling pathways, leading to the inactivation of the JA signalling pathway in tobacco (Lochman and Mikes, 2006).
nsLTPs are small, basic, cysteine‐rich proteins whose function in plants has been a matter of intense debate since their initial discovery in the 1970s (Kader, 1975). Potential roles have been postulated as to their involvement in intracellular phospholipid trafficking (Kader, 1975), somatic embryogenesis and plant developmental mechanisms (Coutos‐Thévenot et al., 1993; Fleming et al., 1992), transport of structural molecules, including cutin monomers, in plants during cuticle assembly (Meijer et al., 1993; Sterk et al., 1991) and, more recently, putative roles in numerous plant defence responses (Blilou et al., 2000; Carvalho et al., 2001). In a 2003 study utilising grape cell suspensions, the changes in expression of certain PR‐14 genes coding for several nsLTPs were studied in response to various elicitor molecules, including both ergosterol and a protein fraction purified from B. cinerea. Three full‐length nsLTP cDNA clones were initially identified in two grape cultivars and the full‐length cDNA clone of 41B‐nsLTP was subsequently employed as a probe for Northern blot analyses of PR‐14 (nsLTP) gene expression status in the cell cultures following elicitor induction (Gomès et al., 2003). Ergosterol and the uncharacterised purified B. cinerea protein fraction drastically increased the hybridisation signals following elicitor induction, but only minor expression increases were noted following treatment with cholesterol and sitosterol. Moreover, SA had a negligible effect on gene expression status, whereas JA showed a small but significant increase in gene induction (Gomès et al., 2003). To further assess the expression kinetics in response to ergosterol and the B. cinera fraction, these two elicitors were studied on cell suspensions, and it was found that nsLTP transcripts increased at 5 h post‐ergosterol treatment (150‐fold) and 10 h post‐B. cinerea proteinaceous elicitor treatment (190‐fold) (Gomès et al., 2003).
Deploying the chemical defence units: the metabolomic squadrons
In order to deal with a large number of biotic and abiotic stressors, plants have evolutionarily developed a metabolic network that is highly diverse and complex when compared with that of other organisms. The main challenge of metabolomics studies, however, is the enormous complexity and diversity of the metabolome (Allwood et al., 2010; Dunn and Ellis, 2005; Sumner et al., 2003), and it should be noted that the application of metabolomics to the investigation of plant–pathogen interactions is still in its infancy (Okazaki and Saito, 2012; Tugizimana et al., 2013).
The chemical defence arsenal of plants can include pre‐formed antimicrobial phytoanticipins, described as ‘low‐molecular‐weight’ antimicrobial compounds that are present in plants before challenge by microorganisms, or are produced after infection solely from pre‐existing precursors (Bourgaud et al., 2001; Dixon, 2001; Essenberg, 2001; Grayer and Kokubun, 2001; Mert‐Türk, 2002). In contrast, phytoalexins are host‐synthesised, low‐molecular‐weight compounds with protective properties (antimicrobial, photoprotective, structure stabilising and signalling), the de novo biosynthesis and accumulation of which are induced in plants following biotic or abiotic stress/attack, i.e. the production of phytoalexins requires the de novo activation and expression of the enzymes involved in their respective biosynthetic pathways (Edreva et al., 2008; Mert‐Türk, 2002; Pedras and Ahiahonu, 2005). Phytoalexins include a wide spectrum of compounds that form structurally and functionally diverse metabolite families, such as stilbenes, coumarins, sesquiterpenes and polyketides, as well as other phenylpropanoid and flavonoid derivatives (Hammerschmidt, 1999; Smith, 1996). More than 300 phytoalexins have now been characterised from the members of more than 20 families. Generally, plants of a particular family produce phytoalexins that are structurally related (Dixon, 2001; Harborne, 2000).
Various studies have reported the antifungal activity of phytoalexins against numerous phytopathogenic fungi, also revealing a complex relationship between phytoalexin accumulation and disease resistance that is greatly pathogen dependent (Ahuja et al., 2012; Mert‐Türk, 2002; Tierens et al., 2001). The work of Huffaker et al. (2011) describes maize‐produced acidic sesquiterpenoid phytoalexins, zealexins, which significantly inhibited the growth of fungal pathogens at physiologically relevant conditions. Furthermore, deficiency in phytoalexin production has been shown to enhance the susceptibility of plants to phytopathogenic fungal infection (Glazebrook et al., 1997; Thomma et al., 1999). However, in their continuous adaptation and co‐evolution with plants, some phytopathogenic fungi have acquired efficient abilities to overcome host chemical defences through enzyme‐mediated detoxification of phytoalexins and phytoanticipins (Pedras and Ahiahonu, 2005; Schmidt and Panstruga, 2011). It has been shown, for instance, that the phytopathogenic fungus, Gibberella pulicaris, a major cause of dry‐rot of stored potatoes (Solanum tuberosum), has the ability to detoxify the sesquiterpene phytoalexins, rishitin and lubimin, produced by potato by enzyme‐mediated epoxidation, dehydrogenation and cyclisation (Desjardins et al., 1992; Gardner et al., 1994; Pedras and Ahiahonu, 2005).
The perception of ergosterol results in the activation of a number of defence genes and the production of defence‐related secondary metabolites (Kasparovsky et al., 2003, 2004; Lochman and Mikes, 2006). Lochman and Mikes (2006) also demonstrated that the treatment of tobacco cells with nanomolar concentrations of ergosterol led to the de novo gene expression of PAL genes (coding for phenylalanine ammonia lyase, a key defence‐related enzyme responsible for the initiation of the phenylpropanoid pathway that ultimately leads to the production of SA and other phenolics). Furthermore, 5‐epi‐aristolochene synthase and sesquiterpene synthase (important enzymes related to the synthesis of various sesquiterpenoid phytoalexins) were responsive towards ergosterol (Lochman and Mikes, 2006; Rossard et al., 2010).
Although it is known that ergosterol elicits the synthesis of phytoalexins, such as resveratrol (grapevine) and capsidiol (potato and tobacco) (Kasparovsky et al., 2003; Robert et al., 2001, 2002), the work of Tugizimana et al. (2012) is the only study reported to have investigated the effect of ergosterol on plant secondary metabolism from a metabolomic perspective. Five sesquiterpenoid phytoalexins (capsidiol, lubimin, phytuberin, rishitin and solavetivone) were identified/annotated, indicating ergosterol‐induced activation of the terpenoid pathway. Moreover, the study indicated that the perception of ergosterol, acting as a ‘non‐self’ M/PAMP molecule, induces significant and dynamic metabolomic alterations in tobacco cell suspensions. These changes included variation in the levels of the constitutively expressed metabolites and the production of new metabolites (Tugizimana et al., 2014).
Potential Ergosterol Sensor Proteins as Host Sentinels
Although modern approaches with regard to plant–fungus interactions have certainly yielded significant insights to date, no clear model pertaining to ergosterol perception and the subsequent signalling cascade has yet been established (Fig. 4).
Data presented by Vega and Boland (1988) first suggested the existence of binding sites in plants for sterols (endogenous sitosterol and stigmasterol), although both the biological role and specificity with respect to ‘self’ and ‘non‐self’ recognition remained to be examined. Plant cells are presumably able to recognise ergosterol as ‘non‐self’ (Sanabria et al., 2010) because of the distinguishing structural features of the molecule (Figs 2 and 3). Given that ergosterol has been considered as a MAMP molecule (Nürnberger et al., 2004), it would be rational to assume that there exists a receptor for perception. Granado et al. (1995) reported a stimulus–response perception system for ergosterol in tomato cells. Here, subnanomolar concentrations of ergosterol purified from Cladosporium fulvum spore extracts were found to be able to elicit extracellular alkalinisation with a selectivity and sensitivity that resembled steroid hormone perception in animals. None of the endogenous plant sterols that were evaluated (sitosterol, stigmasterol and campesterol) exhibited similar activity and, based on these observations, the authors postulated that plants possess ergosterol receptors for the detection of fungi in plant–pathogen or plant–symbiont interactions, thereby adding to the concept of receptor‐mediated sterol perception. A similar postulation with hydrogen peroxide‐associated signalling was made a year later by Kauss and Jeblick (1996).
Later studies confirmed specific ergosterol perception in comparative investigations with other sterols (sitosterol, stigmasterol, campesterol and cholesterol) by plants (Rossard et al., 2010; Vatsa et al., 2011a). The former noted that, if Beta vulgaris leaf cells were first exposed to ergosterol, a refractory state resulted which was characterised by a change in pH of the extracellular environment. Following a second application of ergosterol after a 2‐h period, no secondary pH changes were observed. However, after a first application with one of the other sterols or chitosan, cells responded to a subsequent application of ergosterol. This is indicative that ergosterol perception does not interfere with that of the other tested sterols and chitosan, and vice versa, and could imply that plants possess a specific receptor for ergosterol. These results pointed to an ergosterol‐sensitive recognition mechanism in Beta vulgaris leaf cells that shares many of the properties of well‐described recognition systems for MAMPs, such as flg22, elf, chitin and LPS (Boller and Felix, 2009; Gerber and Dubery, 2004), including transient increases in [Ca2+]cyt, modification of proton fluxes, an oxidative burst and activation of MAPKs corresponding to SIPK and WIPK, which was also the case observed with cryptogein treatment of N. tabacum cells (Vatsa et al., 2011a). In the latter study pertaining to the structural analogues of ergosterol (cholesterol, sitosterol, stigmasterol and campesterol), only campesterol exhibited slight activity to trigger a rise in calcium concentration and a very low oxidative burst, suggesting that there are some shared but, more so, differential/specific structural features (Fig. 3) that are recognised by the perception system for ergosterol.
Osman et al. (2001) reported on the decrease in defence responses (Ca2+ signalling, extracellular alkalinisation and SAR) in tobacco following treatments with elicitin (cryptogein) mutated in its ability to bind sterols. From this study, it is very apparent that, in order for elicitins to trigger biological responses, the elicitors need to be sterol loaded. As oomycetes, such as Phytophthora, do not synthesise their own sterols, these would most likely be acquired from the plasma membranes of the host. Moreover, sterol loading of the elicitin was shown to be essential and a prerequisite for binding to high‐affinity sites. As such, defence responses thus appear to be dependent on perception only if the elicitin is sterol loaded. Interestingly, however, Trigos et al. (2005) reported on the identification of ergosterol in Phytophthora dreschsleri, a soil‐borne phytopathogen. The authors noted that this is an unusual metabolite for this oomycete member and is, to date, the only such documentation in this regard.
Research conducted by Avrova et al. (2004) showed the specific up‐regulation of an oxysterol‐binding protein‐coding gene (OSBP) in potato following infection of plants with Phytophthora infestans. In mammalian systems, OSBP activity is divided between the endoplasmic reticulum and Golgi apparatus, where it imparts sterol‐dependent regulation of phosphoinositide and sphingolipid pathways. It has been hypothesised further that OSBPs specifically bind to, and interact with, molecules such as ergosterol, and possibly play a role in vesicle trafficking in fungi and fungal perception in mammals and plants (Im et al., 2005). Furthermore, phytosterols may undergo oxidation and/or oxygenation in the presence of ROS (Burlini et al., 2011), thereby generating altered molecular patterns within the sterols (i.e. oxysterols) and ultimately resulting in defence responses. This, coupled with research conducted by Madala et al. (2012), which showed the specific up‐regulation of an OSBP gene in Arabidopsis thaliana following LPS elicitation, makes OSBP an attractive target to study for specific pathways of fungal perception in plants, and the potential elucidation of convergent defence cascades employed in the recognition and defence against numerous phytopathogens.
Therefore, the potential/hypothesised perception mechanism(s) for ergosterol (Fig. 4) might involve a membrane‐localised receptor‐like kinase(s) (analogous to the brassinosteroid receptor that binds steroid hormones; Hou et al., 2013; Yang et al., 2012) binding to/interacting with intracellular oxysterol‐binding proteins, or localised perturbations of membrane structures [e.g. changes in the detergent‐resistant microdomain (DRM) composition of lipid rafts and the activation of DRM‐associated proteins, membrane depolarisation, opening of calcium channels, etc.]. In support of the latter, it has been reported that RLKs are over‐represented in plant DRMs (Zappel and Panstruga, 2008), and the same has been reported for flagellin, cryptogein and chitin elicitation in plants (Cacas et al., 2012).
A Battle Won?
As part of their innate immune system, it would appear that plants have evolved a system for the recognition of exogenous sterols that probably targets the fungal‐specific ergosterol. It is clear that, although modern approaches regarding plant–fungus interactions have certainly yielded significant insights to date, no clear model pertaining to ergosterol perception and the subsequent signalling cascades has yet been established. Further investigations of ergosterol as a MAMP, its recognition by plant cells and its ability to trigger MTI are still needed. The use of large‐scale quantifiable tools, such as ‘‐omics’ approaches (e.g. genomics, transcriptomics, proteomics and metabolomics), will certainly provide comprehensive insights into the understanding of ergosterol–plant interactions. The discovery of a receptor for ergosterol would add impetus to this important area of research and generate renewed interest in its application in plant biotechnology and crop protection.
Acknowledgements
The South African National Research Foundation (NRF) and the University of Johannesburg are acknowledged for financial support.
References
- Ahuja, I. , Kissen, R. and Bones, M.A. (2012) Phytoalexins in defense against pathogens. Trends Plant Sci. 17, 73–90. [DOI] [PubMed] [Google Scholar]
- Allwood, J.W. , Ellis, D. and Goodacre, R. (2008) Metabolomic technologies and their application to the study of plants and plant–host interactions. Physiol. Plant. 132, 117–135. [DOI] [PubMed] [Google Scholar]
- Allwood, J.W. , Clarke, A. , Goodacre, R. and Mur, L.A.J. (2010) Dual metabolomics: a novel approach to understanding plant–pathogen interactions. Phytochemistry, 71, 590–597. [DOI] [PubMed] [Google Scholar]
- Almagro, L. , Gómez Ros, L.V. , Belchi‐Navarro, S. , Bru, R. , Ros Barceló, A. and Pedreño, M. (2009) Class III peroxidases in plant defence reactions. J. Exp. Bot. 60, 377–390. [DOI] [PubMed] [Google Scholar]
- Amborabé, B.‐E. , Rossard, S. , Pérault, J.M. and Roblin, G. (2003) Specific perception of ergosterol by plant cells. C. R. Biol. 326, 363–370. [DOI] [PubMed] [Google Scholar]
- Amborabé, B.‐E. , Bonmort, J. , Fleurat‐Lessard, P. and Roblin, G. (2008) Early events induced by chitosan on plant cells. J. Exp. Bot. 59, 2317–2324. [DOI] [PubMed] [Google Scholar]
- Angelova, Z. , Georgiev, S. and Roos, W. (2006) Elicitation of plants. Biotechnol. Biotechnol. Equip. 20, 72–83. [Google Scholar]
- Arasimowicz, M. and Floryszakwieczorek, J. (2007) Nitric oxide as a bioactive signalling molecule in plant stress responses. Plant Sci. 172, 876–887. [Google Scholar]
- Asensio, T. , Crespo, J.F. , Sanchez‐Monge, R. , Lopez‐Torrejon, G. , Somoza, M.L. , Rodriguez, J. and Salcedo, G. (2004) Novel plant pathogenesis‐related protein family involved in food allergy. J. Allergy Clin. Immunol. 114, 896–899. [DOI] [PubMed] [Google Scholar]
- Avrova, A.O. , Taleb, N. , Rokka, V.M. , Heilbronn, J. , Campbell, E. , Hein, I. , Gilroy, E.M. , Cardle, L. , Bradshaw, J.E. , Stewart, H.E. , Fakim, Y.J. , Loake, G. and Birch, P.R.J. (2004) Potato oxysterol binding protein and cathepsin B are rapidly up‐regulated in independent defence pathways that distinguish R‐gene‐mediated and field resistance to Phytophthora infestans . Mol. Plant Pathol. 5, 45–56. [DOI] [PubMed] [Google Scholar]
- Beck, M. , Heard, W. , Mbengue, M. and Robatzek, S. (2012) The INs and OUTs of pattern recognition receptors at the cell surface. Curr. Opin. Plant Biol. 15, 367–374. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. and Mackey, D. (2007) Elicitors, effectors and R genes: the new paradigm and lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. [DOI] [PubMed] [Google Scholar]
- Bernier, F. and Berna, A. (2001) Germins and germin‐like proteins: plant do‐all proteins. But what do they do exactly? Plant Physiol. Biochem. 39, 545–554. [Google Scholar]
- Bittel, P. and Robatzek, S. (2007) Microbe‐associated molecular patterns (MAMPs) probe plant immunity. Curr. Opin. Plant Biol. 10, 335–341. [DOI] [PubMed] [Google Scholar]
- Blilou, I. , Ocampo, J.A. and Garcia‐Garrido, J.M. (2000) Induction of LTP (Lipid transfer protein) and PAL (Phenylalanine ammonia‐lyase) gene expression in rice roots colonised by the arbuscular mycorrhizal fungus Glomus mosseae . J. Exp. Bot. 51, 1969–1977. [DOI] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Borad, V. and Sriram, S. (2008) Pathogenesis‐related proteins for the plant protection. Asian J. Exp. Sci. 22, 189–196. [Google Scholar]
- Bourgaud, F. , Gravot, A. , Milesi, S. and Gontier, E. (2001) Production of plant secondary metabolites: a historical perspective. Plant Sci. 161, 839–851. [Google Scholar]
- Burlini, N. , Iriti, M. , Daghetti, A. , Faoro, F. , Ruggiero, A. and Bernasconi, S. (2011) Benzothiadiazole (BTH) activates sterol pathway and affects vitamin D3 metabolism in Solanum malacoxylon cell cultures. Plant Cell Rep. 30, 2131–2141. [DOI] [PubMed] [Google Scholar]
- Cacas, J.‐L. , Furt, F. , Le Guédard, M. , Schmitter, J.‐M. , Buré, C. , Gerbeau‐Pissot, P. , Moreau, P. , Bessoule, J.‐J. , Simon‐Plas, F. and Mongrand, S. (2012) Lipids of plant membrane rafts. Prog. Lipid Res. 51, 272–299. [DOI] [PubMed] [Google Scholar]
- Carvalho, A.O. , Machado, O.L.T. , Cunha, M.A. , Santos, I.S. and Gomes, V.M. (2001) Antimicrobial peptides and immunolocalisation of an LTP in Vigna unguiculata seeds. Plant Physiol. Biochem. 39, 137–146. [Google Scholar]
- Chen, L. , Hamada, S. , Fujiwara, M. , Zhu, T. , Thao, N.P. , Wong, H.L. , Krishna, P. , Ueda, T. , Kaku, H. , Shibuya, N. , Kawasaki, T. and Shimamoto, K. (2010) The Hop/Sti1‐Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe 7, 185–196. [DOI] [PubMed] [Google Scholar]
- Cheng, C.S. , Samuel, D. , Liu, Y.J. , Shyu, J.C. , Lai, S.M. , Lin, K.F. and Lyu, P.C. (2004) Binding mechanism of nonspecific lipid transfer proteins and their role in plant defence. Biochemistry, 43, 13 628–13 636. [DOI] [PubMed] [Google Scholar]
- Choi, D. , Park, J.A. , Seo, Y.S. , Chun, Y.J. and Kim, W.T. (2000) Structure and stress‐related expression of two cDNAs encoding proteinase inhibitor II of Nicotiana glutinosa L. Biochim. Biophys. Acta, 1492, 211–215. [DOI] [PubMed] [Google Scholar]
- Coutos‐Thévenot, P. , Jouenne, T. , Maës, O. , Guerbette, F. , Grosbois, M. , Le Caer, J.P. , Boulay, M. , Deloire, A. , Kader, J.C. and Guern, J. (1993) Four 9‐kDa proteins excreted by somatic embryos of grapevine are isoforms of lipid‐transfer proteins. Eur. J. Biochem. 217, 885–889. [DOI] [PubMed] [Google Scholar]
- Coutos‐Thévenot, P. , Poinssot, B. , Bonomelli, A. , Yean, H. , Breda, C. , Buffard, D. , Esnault, R. , Hain, R. and Boulay, M. (2001) In vitro tolerance to Botrytis cinerea of grapevine 41B rootstock in transgenic plants expressing the stilbene synthase Vst1 gene under the control of a pathogen‐inducible PR‐10 promoter. J. Exp. Bot. 52, 901–910. [DOI] [PubMed] [Google Scholar]
- Deepak, S.A. , Ishii, H. and Park, P. (2006) Acibenzolar‐S‐methyl primes cell wall strengthening genes and reactive oxygen species forming/scavenging enzymes in cucumber after fungal pathogen attack. Physiol. Mol. Plant Pathol. 69, 52–61. [Google Scholar]
- Desjardins, E.A. , Gardener, W.H. and Weltring, M.K. (1992) Detoxification of sesquiterpene phytoalexins by Gibberella pulicaris (Fusarium sambucinum) and its importance for virulence on potato tubers. J. Ind. Microbiol. 9, 201–211. [Google Scholar]
- Dixon, R.A. (2001) Natural products and plant disease resistance. Nature, 411, 843–847. [DOI] [PubMed] [Google Scholar]
- Dohnal, V. , Jezkova, A. , Pavlikova, L. , Musilek, K. , Jun, D. and Kuca, K. (2010) Fluctuation in the ergosterol and deoxynivalenol content in barley and malt during malting process. Anal. Bioanal. Chem. 397, 109–114. [DOI] [PubMed] [Google Scholar]
- Dunn, W.B. and Ellis, D.I. (2005) Metabolomics: current analytical platforms and methodologies. Trends Anal. Chem. 24, 285–294. [Google Scholar]
- Ebrahim, S. , Usha, K. and Singh, B. (2011) Pathogenesis‐related (PR) proteins?: chitinase and β‐1,3‐glucanase in defence mechanism against malformation in mango (Mangifera indica L). Sci. Hort. 130, 847–852. [Google Scholar]
- Edreva, A. (2005) Pathogenesis‐related proteins?: research progress in the last 15 years. Gen. Appl. Plant Physiol. 31, 105–124. [Google Scholar]
- Edreva, A. , Velikova, V. , Tsonev, T. , Dagnon, S. , Gürel, A. , Aktaş, L. and Gesheva, E. (2008) Stress‐protective role of secondary metabolites: diversity of functions and mechanisms. Gen. Appl. Plant Physiol. 34, 67–78. [Google Scholar]
- Elvira, M.I. , Galdeano, M.M. , Gilardi, P. , García‐Luque, I. and Serra, M.T. (2008) Proteomic analysis of pathogenesis‐related proteins (PRs) induced by compatible and incompatible interactions of pepper mild mottle virus (PMMoV) in Capsicum chinense L3 plants. J. Exp. Bot. 59, 1253–1265. [DOI] [PubMed] [Google Scholar]
- Essenberg, M. (2001) Prospects for strengthening plant defences through phytoalexin engineering. Physiol. Mol. Plant Pathol. 59, 71–81. [Google Scholar]
- Eulgem, T. , Rushton, P.J. , Robatzek, S. and Somssich, I.E. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Fan, J. and Doerner, P. (2012) Genetic and molecular basis of nonhost disease resistance: complex, yes; silver bullet, no. Curr. Opin. Plant Biol. 15, 1–7. [DOI] [PubMed] [Google Scholar]
- Ferreira, R.B. , Monteiro, S. , Freitas, R. , Santos, C.N. , Chen, Z. , Batista, L.M. , Duarte, J. , Borges, A. and Teixeira, A.R. (2006) Fungal pathogens: the battle for plant infection. Crit. Rev. Plant Sci. 25, 505–524. [Google Scholar]
- Fisher, M.C. , Henk, D.A. , Briggs, C.J. , Brownstein, J.S. , Madoff, L.C. , McCraw, S.L. and Gurr, S.J. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature, 484, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, A.J. , Mandel, T. , Hoffman, S. , Sterk, P. , De Vries, S.C. and Kuhlemeier, C. (1992) Expression pattern of a putative lipid transfer protein gene within the shoot apex. Plant J. 2, 855–862. [PubMed] [Google Scholar]
- Flor, H.H. (1942) Inheritance of pathogenicity in Melampsora lini . Phytopathology, 32, 653–669. [Google Scholar]
- Frías, M. , Brito, N. and Gonzáles, C. (2013) The Botrytis cinerea cerato‐platanin BcSpI1 is a potent inducer of systemic acquired resistance (SAR) in tobacco and generates a wave of salicylic acid expanding from the site of application. Mol. Plant Pathol. 14, 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, W.H. , Desjardins, E.A. , McCormick, P.S. and Weisleder, D. (1994) Detoxification of the potato phytoalexin rishitin by Gibberella pulicaris . Phytochemistry, 37, 1001–1005. [Google Scholar]
- Gerber, I.B. and Dubery, I.A. (2004) Protein phosphorylation in Nicotiana tabacum cells in response to perception of lipopolysaccharides from Burkholderia cepacia . Phytochemistry, 65, 2957–2966. [DOI] [PubMed] [Google Scholar]
- Gerber, I.B. , Laukens, K. , Witters, E. and Dubery, I.A. (2006) Identification of lipopolysaccharide‐responsive phosphoproteins in Nicotiana tabacum cells. Plant Physiol. Biochem. 44, 369–379. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. , Zook, M. , Mert, F. , Kagan, I. , Rogers, E.E. , Crute, R.I. , Holub, B.E. , Hammerschmidt, R. and Ausubel, M.F. (1997) Phytoalexin‐deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics, 146, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomès, E. , Sagot, E. , Gaillard, C. , Laquitaine, L. , Poinsot, B. , Sanejouand, H.Y. , Delrot, S. and Coutos‐Thévenot, P. (2003) Nonspecific lipid‐transfer protein genes expression in grape (Vitis sp.) cells in response to fungal elicitor treatments. Mol. Plant–Microbe Interact. 16, 456–464. [DOI] [PubMed] [Google Scholar]
- Gonzalez‐Lamothe, R. , Mitchell, G. , Gattuso, M. , Diarra, M.S. , Malouin, F. and Bouarab, K. (2009) Plant antimicrobial agents and their effects on plant and human pathogens. Int. J. Mol. Sci. 10, 3400–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granado, J. , Felix, G. and Boller, T. (1995) Perception of fungal sterols in plants: subnanomolar concentrations of ergosterol elicit extracellular alkalinisation in tomato cells. Plant Physiol. 107, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayer, R.J. and Kokubun, T. (2001) Plant–fungal interactions: the search for phytoalexins and other antifungal compounds from higher plants. Phytochemistry, 56, 253–263. [DOI] [PubMed] [Google Scholar]
- Grenier, J. and Asselin, A. (1990) Some pathogenesis‐related proteins are chitosanases with lytic activity against fungal spores. Mol. Plant–Microbe Interact. 3, 401–407. [Google Scholar]
- Hammerschmidt, R. (1999) Phytoalexins: what have we learned after 60 years? Annu. Rev. Phytopathol. 37, 285–306. [DOI] [PubMed] [Google Scholar]
- Harborne, J.B. (2000) Arsenal for survival: secondary plant products. Taxon, 49, 435–449. [Google Scholar]
- He, P. , Shan, L. and Sheen, J. (2007) Elicitation and suppression of microbe‐associated molecular pattern‐triggered immunity in plant–microbe interactions. Cell. Microbiol. 9, 1385–1396. [DOI] [PubMed] [Google Scholar]
- Henry, G. , Thonart, P. and Ongena, M. (2012) PAMPs, MAMPs, DAMPs and others: an update of the diversity of plant immunity elicitors. Biotechnol., Agron., Soc. Environ. 16, 257–268. [Google Scholar]
- Hou, S. , Zhang, C. , Yang, Y. and Wu, D. (2013) Recent advances in plant immunity: recognition, signaling, response and evolution. Biol. Plant. 57, 11–25. [Google Scholar]
- Hsueh, Y.‐W. , Gilbert, K. , Trandum, C. , Zuckermann, M. and Thewalt, J. (2005) The effect of ergosterol on dipalmitoylphosphatidylcholine bilayers: a deuterium NMR and calorimetric study. Biophys. J. 88, 1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker, A. , Kaplan, F. , Vaughan, M.M. , Dafoe, J.N. , Ni, X. , Rocca, R.J. , Alborn, T.H. , Teal, E.A.P. and Schmelz, A.E. (2011) Novel acidic sesquiterpenoids constitute a dominant class of pathogen‐induced phytoalexins in maize. Plant Physiol. 156, 2082–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im, Y.J. , Raychaudhuri, S. , Prinz, W.A. and Hurley, J.H. (2005) Structural mechanism for sterol sensing and transport by OSBP‐related proteins. Nature, 437, 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriti, M. and Faoro, F. (2007) Review of innate and specific immunity in plants and animals. Mycopathologia, 164, 57–64. [DOI] [PubMed] [Google Scholar]
- Ishihama, N. , Yamada, R. , Yoshioka, M. , Katou, S. and Yoshioka, H. (2011) Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defence response. Plant Cell, 23, 1153–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kader, J.C. (1975) Proteins and the intracellular exchange of lipids: stimulation of phospholipids exchange between mitochondria and microsomal fractions by protein isolated from potato tuber. Biochim. Biophys. Acta, 380, 31–44. [PubMed] [Google Scholar]
- Kader, J.C. (1996) Lipid‐transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 627–654. [DOI] [PubMed] [Google Scholar]
- Kasparovsky, T. , Milat, M.L. , Humbert, C. , Blein, J.P. , Havel, L. and Mikes, V. (2003) Elicitation of tobacco cells with ergosterol activates a signal pathway including mobilisation of internal calcium. Plant Physiol. Biochem. 41, 495–501. [Google Scholar]
- Kasparovsky, T. , Blein, J.P. and Mikes, V. (2004) Ergosterol elicits an oxidative burst in tobacco cells via phospholipase A2 and protein kinase C signal pathway. Plant Physiol. Biochem. 42, 429–435. [DOI] [PubMed] [Google Scholar]
- Katagiri, F. and Tsuda, K. (2010) Understanding the plant immune system. Mol. Plant–Microbe Interact. 23, 1531–1536. [DOI] [PubMed] [Google Scholar]
- Kauss, H. and Jeblick, W. (1996) Influence of salicylic acid on the induction of competence for H2O2 elicitation (comparison of ergosterol with other elicitors). Plant Physiol. 111, 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen, N.T. (1990) Gene‐for‐gene complementarity in plant–pathogen interactions. Annu. Rev. Genet. 24, 447–463. [DOI] [PubMed] [Google Scholar]
- Knoth, C. , Ringler, J. , Dangl, J.L. and Euglem, T. (2007) Arabidopsis WRKY70 is required for full RPP4‐mediated disease resistance and basal defence against Hyaloperonospora parasitica . Mol. Plant–Microbe Interact. 20, 120–128. [DOI] [PubMed] [Google Scholar]
- Laquitaine, L. , Gomès, E. , Françios, J. , Marchive, C. , Pascal, S. , Hamdi, S. , Atanassova, R. , Delrot, S. and Coutos‐Thévenot, P. (2006) Molecular basis of ergosterol‐induced protection of grape against Botrytis cinerea: induction of Type‐I LTP promoter activity, WRK, and stilbene synthase gene expression. Mol. Plant–Microbe Interact. 19, 1103–1112. [DOI] [PubMed] [Google Scholar]
- Lecourieux, D. , Ranjeva, R. and Pugin, A. (2006) Calcium in plant defence‐signalling pathways. New Phytol. 171, 249–269. [DOI] [PubMed] [Google Scholar]
- Lederberg, J. and McCray, A.T. (2001) Ome sweet Omics – A genealogical treasury of words. Scientist, 15, 8. [Google Scholar]
- Li, J. , Brader, G. , Kariola, T. and Palva, E.T. (2006) WRKY 70 modulates the selection of signalling pathways in plant defense. Plant J. 46, 477–491. [DOI] [PubMed] [Google Scholar]
- Li, X. , Xia, B. , Jiang, Y. , Wu, Q. , Wang, C. , He, L. , Peng, F. and Wang, R. (2010) A new pathogenesis‐related protein, LrPR4, from Lycoris radiata, and its antifungal activity against Magnaporthe grisea . Mol. Biol. Rep. 37, 995–1001. [DOI] [PubMed] [Google Scholar]
- Linthorst, H.J. , Brederode, F.T. , van der Does, C. and Bol, J.F. (1993) Tobacco proteinase inhibitor I genes are locally, but not systemically induced by stress. Plant Mol. Biol. 21, 985–992. [DOI] [PubMed] [Google Scholar]
- Liu, J. and Nes, W.D. (2009) Steroidal triterpenes: design of substrate‐based inhibitors of ergosterol and sitosterol synthesis. Molecules, 14, 4690–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochman, J. and Mikes, V. (2006) Ergosterol treatment leads to the expression of a specific set of defence‐related genes in tobacco. Plant Mol. Biol. 62, 43–51. [DOI] [PubMed] [Google Scholar]
- Ma, W. (2011) Roles of Ca2+ and cyclic nucleotide gated channel in plant innate immunity. Plant Sci. 181, 342–346. [DOI] [PubMed] [Google Scholar]
- Madala, N.E. , Molinaro, A. and Dubery, I.A. (2011) Deciphering the structural and biological properties of the lipid‐A moiety of lipopolysaccharides from Burkholderia cepacia strain ASP B 2D, in Arabidopsis thaliana . Glycobiology, 21, 184–194. [DOI] [PubMed] [Google Scholar]
- Madala, N.E. , Molinaro, A. and Dubery, I.A. (2012) Distinct carbohydrate and lipid‐based molecular patterns within lipopolysaccharides from Burkholderia cepacia contribute to defence‐associated differential gene expression in Arabidopsis thaliana . Innate Immun. 18, 140–154. [DOI] [PubMed] [Google Scholar]
- Mazzotta, S. and Kemmerling, B. (2011) Pattern recognition in plant innate immunity. J. Plant Pathol. 93, 7–17. [Google Scholar]
- Meijer, E.A. , De Vries, S.C. , Sterk, P. , Gadella, D.W. , Writz, K.W. and Hendricks, T. (1993) Characterisation of a nonspecific lipid‐transfer protein EP2 from carrot (Daucus carota L.). Mol. Cell. Biochem. 123, 159–166. [DOI] [PubMed] [Google Scholar]
- Mert‐Türk, F. (2002) Phytoalexins: defence or just a response to stress? J. Cell Mol. Biol. 1, 1–6. [Google Scholar]
- Mishina, T.E. and Zeier, J. (2007) Pathogen‐associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50, 500–513. [DOI] [PubMed] [Google Scholar]
- Mohd As'wad, A.W. , Sariah, M. , Paterson, R.R.M. , Zainal Abidin, M.A. and Lima, N. (2011) Ergosterol analyses of oil palm seedlings and plants infected with Ganoderma . Crop Prot. 30, 1438–1442. [Google Scholar]
- Monaghan, J. and Zipfel, C. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. [DOI] [PubMed] [Google Scholar]
- Muthamilarasan, M. and Prasad, M. (2013) Plant innate immunity: an updated insight into defence mechanism. J. Biosci. 38, 1–17. [DOI] [PubMed] [Google Scholar]
- Narusaka, Y. , Shinya, T. , Narusaka, M. , Motoyama, N. , Shimada, H. , Murakami, K. and Shibuya, N. (2013) Presence of LYM2 dependent but CERK1 independent disease resistance in Arabidopsis . Plant Signal. Behav. 8, e25345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseri, G. , Sohani, M.M. , Pourmassalehgou, A. and Allahi, S. (2012) In planta transformation of rice (Oryza sativa) using thaumatin‐like protein gene for enhancing resistance to sheath blight. J. Biotechnol. 11, 7885–7893. [Google Scholar]
- Naumowicz, M. , Petelska, A.D. and Figaszewski, Z.A. (2011) Chronopotentiometric studies of phosphatidylcholine bilayers modified by ergosterol. Steroids, 76, 967–973. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T. , Brunner, F. , Kemmerling, B. and Piater, L. (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Okazaki, Y. and Saito, K. (2012) Recent advances of metabolomics in plant biotechnology. Plant Biotechnol. Rep. 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, H. , Vauthrin, S. , Mikes, V. , Milat, M.‐A. , Panabières, F. , Marais, A. , Brunie, S. , Maume, B. , Ponchet, M. and Blein, J.‐P. (2001) Mediation of elicitin activity on tobacco is assumed by elicitor–sterol complexes. Mol. Biol. Cell 12, 2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S.P. and Somssich, I.E. (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck, S.C. , Nühse, T.S. , Hess, D. , Iglesias, A. , Meins, F. and Boller, T. (2001) Directed proteomics identifies a plant‐specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell, 13, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedras, M.S.C. and Ahiahonu, P.W.K. (2005) Metabolism and detoxification of phytoalexins and analogs by phytopathogenic fungi. Phytochemistry, 66, 391–411. [DOI] [PubMed] [Google Scholar]
- Peng, X. , Hu, Y. , Tang, X. , Zhou, P. , Deng, X. , Wang, H. and Guo, Z. (2012) Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta, 236, 1485–1498. [DOI] [PubMed] [Google Scholar]
- Pennisi, E. (2010) Armed and dangerous. Science, 327, 804–805. [DOI] [PubMed] [Google Scholar]
- Piater, L.A. , Nürnberger, T. and Dubery, I.A. (2004) Identification of a lipopolysaccharide responsive erk‐like MAP kinase in tobacco leaf tissue. Mol. Plant Pathol. 5, 331–341. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Leon‐Reyes, A. , Van der Ent, S. and Van Wees, S.C.M. (2009) Networking by small‐molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Postel, S. and Kemmerling, B. (2009) Plant systems for recognition of pathogen‐associated molecular patterns. Semin. Cell Dev. Biol. 20, 1025–1031. [DOI] [PubMed] [Google Scholar]
- Pritchard, L. and Birch, P. (2011) A systems biology perspective on plant–microbe interactions: biochemical and structural targets of pathogen effectors. Plant Sci. 180, 584–603. [DOI] [PubMed] [Google Scholar]
- Qi, Y. , Tsuda, K. , Glazebrook, J. and Katagiri, F. (2011) Physical association of pattern‐triggered immunity (PTI) and effector‐triggered immunity (ETI) immune receptors in Arabidopsis. Mol. Plant Pathol. 12, 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, N. , Ferran, J. , Breda, C. , Coutos‐Thevenot, P. , Boulay, M. , Buffard, D. and Esnault, R. (2001) Molecular characterisation of the incompatible interaction of Vitis vinifera leaves with Pseudomonas syringae pv. pisi: expression of genes coding for stilbene synthase and class 10 PR‐Protein. Eur. J. Plant Pathol. 107, 249–261. [Google Scholar]
- Robert, N. , Roche, K. , Lebeau, Y. , Breda, C. , Bouley, M. , Esnault, R. and Buffard, D. (2002) Expression of grapevine chitinase genes in berries and leaves infected by fungal and bacterial pathogens. Plant Sci. 162, 389–400. [Google Scholar]
- Roche, Y. , Gerbeau‐Pissot, P. , Buhot, B. , Thomas, D. , Bonneau, L. , Gresti, J. , Mongrand, S. , Perrier‐Cornet, J.‐M. and Simon‐Plas, F. (2008) Depletion of phytosterols from the plant plasma membrane provides evidence for disruption of lipid rafts. FASEB J. 22, 3980–3991. [DOI] [PubMed] [Google Scholar]
- Rossard, S. , Luini, E. , Pérault, J.M. , Bonmort, J. and Roblin, G. (2006) Early changes in membrane permeability, production of oxidative burst and modification of PAL activity induced by ergosterol in cotyledons of Mimosa pudica . J. Exp. Bot. 57, 1245–1252. [DOI] [PubMed] [Google Scholar]
- Rossard, S. , Roblin, G. and Atanassova, R. (2010) Ergosterol triggers characteristic elicitation steps in Beta vulgaris leaf tissues. J. Exp. Bot. 61, 1807–1816. [DOI] [PubMed] [Google Scholar]
- Sanabria, N.M. , Huang, J.‐C. and Dubery, I.A. (2010) Self/nonself perception in plants in innate immunity and defence. Self/Nonself Immun. Recog. 1, 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabria, N.M. , van Heerden, H. and Dubery, I.A. (2012) Molecular characterization and regulation of a Nicotiana tabacum S‐domain receptor‐like kinase gene induced during an early rapid response to lipopolysaccharides. Gene, 501, 39–48. [DOI] [PubMed] [Google Scholar]
- Schmidt, S.M. and Panstruga, R. (2011) Pathogenomics of fungal plant parasites: what have we learnt about pathogenesis? Curr. Opin. Plant Biol. 14, 392–399. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B. and Zipfel, C. (2008) News from the frontline: recent insights into PAMP‐triggered immunity in plants. Curr. Opin. Plant Biol. 11, 389–395. [DOI] [PubMed] [Google Scholar]
- Sels, J. , Mathys, J. , De Coninck, B.M.A. , Cammue, B.P.A. and De Bolle, M.F.C. (2008) Plant pathogenesis‐related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950. [DOI] [PubMed] [Google Scholar]
- Sharma, N. , Sharma, K.P. , Gaur, R.K. and Gupta, V.K. (2011) Role of chitinase in plant defence. Asian J. Biochem. 6, 29–37. [Google Scholar]
- Shimizu, T. , Nakano, T. , Takamizawa, D. , Desaki, Y. , Ishii‐Minami, N. , Nishizawa, Y. , Minami, E. , Okada, K. , Yamane, H. , Kaku, H. and Shibuya, N. (2010) Two LysM receptor molecules, CEBiP and OsCERK1 cooperatively regulate chitin elicitor signal in rice. Plant J. 64, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.J. (1996) Accumulation of phytoalexins: defence mechanism and stimulus response system. New Phytol. 132, 1–45. [DOI] [PubMed] [Google Scholar]
- Soh, H.C. , Park, A.R. , Park, S. , Back, K. , Yoon, J.B. , Park, H.G. and Kim, Y.S. (2011) Comparative analysis of pathogenesis‐related protein 10 (PR10) genes between fungal resistant and susceptible peppers. Eur. J. Plant Pathol. 132, 37–48. [Google Scholar]
- Sterk, P. , Booij, H. , Schellekens, G.A. , Van Kammen, A. and De Vries, S.C. (1991) Cell‐specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell, 3, 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R. , Werber, M. and Weisshaar, B. (2001) The R2R3‐MYB gene family in Arabidopsis thaliana . Curr. Opin. Plant Biol. 4, 447–456. [DOI] [PubMed] [Google Scholar]
- Sumner, L.W. , Mendes, P. and Dixon, R.A. (2003) Plant metabolomics: large‐scale phytochemistry in the functional genomics era. Phytochemistry, 62, 817–836. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H. , Nelissen, I. , Eggermont, K. and Broekaert, W.F. (1999) Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to fungus Alternaria brassicicola . Plant J. 19, 163–171. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H. , Nürnberger, T. and Joosten, M.H.A.J. (2011) Of PAMPs and effectors: the blurred PTI–ETI dichotomy. Plant Cell, 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierens, K.F.M. , Thomma, B.P.H.J. , Brouwer, M. , Schmidt, J. , Kistner, K. , Porzel, A. , Mauch‐Mani, M. , Cammue, B.P.A. and Broekaert, W.F. (2001) Study of the role of antimicrobial glucosinolate‐derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol. 125, 1688–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, J.M. , Calderón, H. , Rodríguez‐Arango, E. , Morales, J.G. and Arango, R. (2012) Differential induction of pathogenesis‐related proteins in banana in response to Mycosphaerella fijiensis infection. Eur. J. Plant Pathol. 133, 887–898. [Google Scholar]
- Trigos, Á. , Castellanos‐Onorio, O. , Salinas, A. and Yáñez‐Morales, M.J. (2005) Ergosterol from Phytophthora drechsleri, an unusual metabolite of a member of this genus. Mycopathologia, 159, 469–471. [DOI] [PubMed] [Google Scholar]
- Tugizimana, F. , Steenkamp, P.A. , Piater, L.A. and Dubery, I.A. (2012) Ergosterol‐induced sesquiterpenoid synthesis in tobacco cells. Molecules, 17, 1698–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugizimana, F. , Piater, L.A. and Dubery, I.A. (2013) Plant metabolomics: a new frontier in phytochemical analysis. S. Afr. J. Sci. 109, 1–11. [Google Scholar]
- Tugizimana, F. , Steenkamp, P.A. , Piater, L.A. and Dubery, I.A. (2014) Multi‐platform metabolomic analyses of ergosterol‐induced dynamic changes in Nicotiana tabacum cells. PLoS ONE, 9, e87846. doi: 10:1371/journal.pone.0087846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker, B. and Somssich, I.E. (2004) WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498. [DOI] [PubMed] [Google Scholar]
- Van der Biezen, E.A. and Jones, J.D. (1998) Plant disease‐resistance proteins and the gene‐for‐gene concept. Trends Biochem. Sci. 23, 454–456. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C. and Van Strien, E.A. (1999) The families of pathogenesis‐related proteins, their activities, and comparative analysis of PR‐1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. [Google Scholar]
- Van Loon, L.C. , Rep, M. and Pieterse, C.M.J. (2006) Significance of inducible defence‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Vatsa, P. , Chiltz, A. , Luini, E. , Vandelle, E. , Pugin, A. and Roblin, G. (2011a) Cytosolic calcium rises and related events in ergosterol‐treated Nicotiana cells. Plant Physiol. Biochem. 49, 764–773. [DOI] [PubMed] [Google Scholar]
- Vatsa, P. , Chiltz, A. , Bourque, S. , Wendehenne, D. , Garcia‐Brugger, A. and Pugin, A. (2011b) Involvement of putative glutamate receptors in plant defence signaling and NO production. Biochimie, 93, 2095–2101. [DOI] [PubMed] [Google Scholar]
- Vega, M.A. and Boland, R.L. (1988) Presence of sterol‐binding sites in the cytosol of French‐bean (Phaseolus vulgaris) roots. Biochem. J. 250, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidhyasekaran, P. (2014) PAMP signals in plant innate immunity: signal perception and transduction In: Signaling and Communication in Plants (Baluška F., ed.), pp. 17–115, 228. Dordrecht, Heidelberg, New York, London: Springer. [Google Scholar]
- Vigers, A.J. , Roberts, W.K. and Selitrennikoff, C.P. (1991) A new family of plant antifungal proteins. Mol. Plant–Microbe Interact. 4, 315–323. [DOI] [PubMed] [Google Scholar]
- Wanderley‐Nogueira, A.C. , Belarmino, L.C. , Soares‐Cavalcanti, N.D.M. , Bezerra‐Neto, J.P. , Kido, E.A. , Pandolfi, V. and Abdelnoor, R.V. (2012) An overall evaluation of the Resistance (R) and Pathogenesis‐Related (PR) superfamilies in soybean, as compared with Medicago and Arabidopsis . Genet. Mol. Biol. 35, 260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, T. , Ouellet, T. and Miki, B. (2002) Towards genomic and proteomic studies of protein phosphorylation in plant–pathogen interactions. Trends Plant Sci. 5, 224–230. [DOI] [PubMed] [Google Scholar]
- Xu, X. , Bittman, R. , Duportail, G. , Heissler, D. , Vilcheze, C. and London, E. (2001) Effect of the structure of natural sterols and sphingolipids on the formation of ordered sphingolipid/sterol domains (rafts): comparison of cholesterol to plant, fungal, and disease‐associated sterols and comparison of sphingomyelin, cerebrosides, and ceramide. J. Biol. Chem. 276, 33 540–33 546. [DOI] [PubMed] [Google Scholar]
- Yang, X. , Deng, F. and Ramonell, K.M. (2012) Receptor‐like kinases and receptor‐like proteins: keys to pathogen recognition and defense signalling in plant innate immunity. Front. Biol. 7, 155–166. [Google Scholar]
- Zappel, N.F. and Panstruga, R. (2008) Heterogeneity and lateral compartmentalization of plant plasma membrane. Curr. Opin. Plant Biol. 11, 632–640. [DOI] [PubMed] [Google Scholar]
- Zhang, S. and Klessig, D.F. (2001) MAPK cascades in plant defence signaling. Trends Plant Sci. 6, 520–527. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Collinge, D.B. and Thordal‐Christensen, H. (1995) Germin‐like oxalate oxidase, a H2O2‐producing enzyme, accumulates in barley attacked by the powdery mildew fungus. Plant J. 8, 139–145. [Google Scholar]
- Zhao, X.R. , Lin, Q. and Brookes, P.C. (2005) Does soil ergosterol concentration provide a reliable estimate of soil fungal biomass? Soil Biol. Biochem. 37, 311–317. [Google Scholar]
- Zhou, F. , Zhang, Z. , Gregersen, P.L. , Mikkelsen, J.D. , de Neergaard, E. , Collinge, D. and Thordal‐Christensen, H. (1998) Molecular characterization of the oxalate oxidase involved in the response of barley to the powdery mildew fungus. Plant Physiol. 117, 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, F. , Xu, M. , Wang, S. , Jia, S. , Zhang, P. , Lin, H. and Xi, D. (2012) Prokaryotic expression of pathogenesis‐related protein 1 gene from Nicotiana benthamiana: antifungal activity and preparation of its polyclonal antibody. Biotechnol. Lett. 34, 919–924. [DOI] [PubMed] [Google Scholar]
- Zhu, L. and Li, N. (2013) Quantitation, networking, and function of protein phosphorylation in plant cells. Science, 3, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. (2008) Pattern‐recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20, 10–16. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. (2009) Early molecular events in PAMP‐triggered immunity. Curr. Opin. Plant Biol. 12, 414–420. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D.G. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]


