Summary
Chromobacterium sp. strain C61 displays antifungal activities in vitro and has been used successfully for the biocontrol of plant diseases under field conditions. In this study, the roles of extracellular chitinase and an antifungal compound produced by strain C61 were investigated to elucidate their contributions to biological control activity. The bacterium possessed a locus chi54 encoding an extracellular chitinase, and mutation of chi54 eliminated chitinase production. Production of the extracellular enzyme and expression of the chi54 transcript were increased in the wild‐type strain when chitin was added to the culture medium. In vitro assays showed that purified chitinase inhibited spore germination of multiple pathogens. However, the in planta biocontrol activity of filtrates of cultures grown in the presence of chitin was lower than that of filtrates grown without chitin, indicating that correlation between chitinase and biocontrol activity was lacking. The analysis of C61 culture filtrates revealed an antifungal cyclic lipopeptide, chromobactomycin, whose structure contained a unique nonameric peptide ring. The purified chromobactomycin inhibited the growth of several phytopathogenic fungi in vitro, and plant application significantly reduced disease severity for several pathogens. Furthermore, the production of chromobactomycin was reduced in cultures amended with chitin. These data suggest that the production of both the extracellular chitinase Chi54 and the newly identified antibiotic chromobactomycin can contribute, in an interconnected way, to the suppression of plant disease by Chromobacterium sp. strain C61.
Introduction
Diverse rhizobacteria promote plant growth and/or suppress plant diseases (Lugtenberg and Kamlikova, 2009). The mechanisms involved in plant disease suppression are mediated by a variety of secreted products thought to contribute to antibiosis, competition and the induction of host resistance. The large majority of studies of individual biocontrol agents highlight only a single mode of action, despite the recognition that the most successful agents probably work at the field scale through multiple mechanisms (Kim YC et al., 2011). The contributions of different mechanisms, expressed by a single agent, to the emergent property of biological control remain relatively understudied.
Lytic enzymes that degrade fungal cell walls, particularly extracellular chitinases, contribute to pathogen suppression (Dahiya et al., 2006; Kobayashi et al., 2002). Chitinase‐producing bacterial strains are studied because the enzyme (EC 3.2.1.14) hydrolyses the β‐1,4‐linkage of chitin, a polymer widely distributed in phytopathogenic fungal cell walls (Flach et al., 1992). Chitinases are grouped into families depending on the similarities of the amino acids in their catalytic domain (Cohen‐Kupiec and Chet, 1998). Plant chitinases and chitinases from Streptomyces sp. belong to family 19, which function as endochitinases, have lysozyme activity and are antifungal (Watanabe et al., 1999). Most bacterial chitinases are classified into family 18 and show less antifungal activity than that of family 19 chitinases (Dahiya et al., 2006). However, studies indicate that some biocontrol‐active rhizobacteria produce family 18 chitinases that inhibit the growth of plant pathogenic fungi (Arora et al., 2007; Kobayashi et al., 2002; Li et al., 2008).
Antibiotics and other metabolites produced by different biocontrol bacteria also suppress phytopathogenic fungi, both directly and indirectly. Pyrrolnitrin, pyoluteorin, 2,4‐diacetylphloroglucinol (DAPG) and phenazines are implicated in biocontrol mediated by fluorescent pseudomonads (Dubuis et al., 2007). Cyclic lipopeptides (CLPs) are produced by many bacterial genera and can show lytic and growth‐inhibiting properties against various plant pathogenic pathogens (Raaijmakers et al., 2010). For example, CLPs produced by biocontrol Bacillus species, including fengycins and iturin, are antifungal against Fusarium graminearum (Wang et al., 2007), Colletotrichum dematium (Hiradate et al., 2002) and Magnaporthe grisea (Tendulkar et al., 2007). However, compounds such as DAPG and CLPs can be multifunctional, affecting both pathogens and plant hosts in a variety of ways, e.g. affecting host defence status and altering rooting patterns (Kim YC et al., 2011).
We have identified a root‐colonizing Chromobacterium sp. C61 as a useful biocontrol agent using chitinase production as a criterion for selecting and testing biocontrol bacteria (Kim et al., 2008). Formulations of Chromobacterium sp. C61 were used in combination with other culture methods to control Alternaria blight and anthracnose of ginseng (Kim et al., 2010) and Phytophthora blight of pepper (Kim et al., 2008) in the field. We presumed that the chitinase activity of C61 accounted for its potent ability to inhibit Rhizoctonia solani growth (Park et al., 2005). The major extracellular chitinase from the bacterium had a molecular weight of 54 kDa and a pI of 8.7 (Park et al., 2007). Our recent draft genome sequence analysis of Chromobacterium sp. C61 identified a single chitinase gene (chi54) (Kim HJ et al., 2011), and site‐directed mutagenesis of this gene was performed to identify the key amino acids involved in chitinase activity. For example, replacing the tyrosine at amino acid 218 with a serine in the Chi54 protein enhances chitinase activity (Park et al., 2007).
In this study, we examined the role of the major chitinase and a newly identified antifungal compound in the biological control of different plant diseases by Chromobacterium sp. strain C61. Our results show that a novel CLP secreted by Chromobacterium sp. strain C61, and an extracellular chitinase, contribute in an interconnected way to both in vitro antifungal and in planta biocontrol activities.
Results
Biocontrol activities of the Chromobacterium sp. C61 chitinase mutants
Strong chitinase production by the wild‐type strain was confirmed using sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE), followed by staining for activity and Western analysis using a polyclonal antibody that detects the C61 secreted chitinase (Fig. 1). Insertional inactivation of a chi54 locus in mutant C61Chi54M was verified by polymerase chain reaction (PCR) analysis of the sequence of the mutated gene (data not shown). This mutant did not grow on chitin as a sole carbon source, and Chi54 activity was not detected by activity staining or antibody recognition (Fig. 1). A mutant producing a more active chitinase, C61Chi54TS, as a result of a site‐directed mutation that replaced a serine residue with a tyrosine (Park et al., 2007), showed greater activity than the wild‐type (Fig. 1C).
Figure 1.
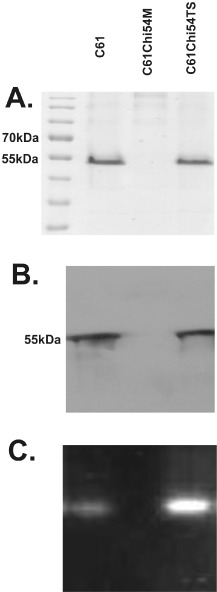
Comparison of chitinase activity from the chi54 mutant (C61Chi54M) and the mutant (C6IChi54TS) compared with the wild‐type (C61) strain of Chromobacterium sp. C61. Coomassie blue staining (A), Western blot (B) and chitinase activity staining (C) of proteins from C61 separated by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE). Proteins (20 μg) were partially purified from culture filtrates of the wild‐type and mutant strains (C61, C61Chi54M and C61Chi54TS) grown in Luria–Bertani (LB) broth containing 0.2% colloidal chitin.
Culture filtrates of the wild‐type Chromobacterium sp. C61 and the mutant C61Chi54TS both produced significant in planta biocontrol activity against rice blast, tomato leaf blight and wheat leaf rust (P < 0.05 for each; Fig. 2). In contrast, the filtrates from the C61Chi54M mutant showed reduced activity (Fig. 2). Thus, the biological control activity of C61 can be attributed, in part, to the levels of extracellular chitinase activity produced.
Figure 2.
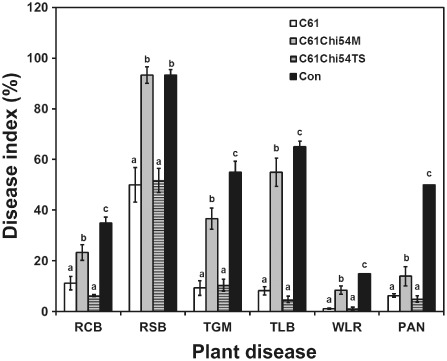
In planta biocontrol efficacies of the extracellular products of Chromobacterium sp. C61 and its chitinase mutants. Intact plants were sprayed with cell‐free bacterial cultures of the C61 wild‐type(C61), C61Chi54M and C61Chi54TS (1:3 dilution), and water was used as the negative control treatment. The disease index resulting from each pathogen was based on the areas of the infected lesion. PAN, red pepper anthracnose; RCB, rice blast; RSB, rice sheath blight; TGM, tomato gray mould; TLB, tomato late blight; WLR, wheat leaf rust. Data are means ± standard deviations of three independent experiments with nine plants per treatment. Bars labelled with different letters for each pathosystem have significantly different (P < 0.05) levels of disease based on Duncan's multiple range test.
Effect of chitin on chitinase expression, and in vitro and in planta biocontrol of C61
Transcriptional analysis identified chi54 transcripts in the stationary phase in wild‐type cells grown in the presence of colloidal chitin (Fig. 3A). Extracellular chitinase was detected only when the wild‐type and C61Chi54TS mutant were grown with chitin and, as expected, there was no detectable extracellular chitinase activity with the C61Chi54M mutant (Fig. 3B). However, in vitro, antifungal activity against the fungal plant pathogens R. solani and Botrytis cinerea by the wild‐type Chromobacterium sp. C61 was higher on potato dextrose agar (PDA) without chitin (Fig. 3C), indicating that chitinase induction reduced the overall antibiotic‐like activity on the plates. In addition, no significant difference was observed between the extent of antagonism between the C61Chi54TS construct and wild‐type C61, although the C61Chi54M mutant was less antagonistic.
Figure 3.
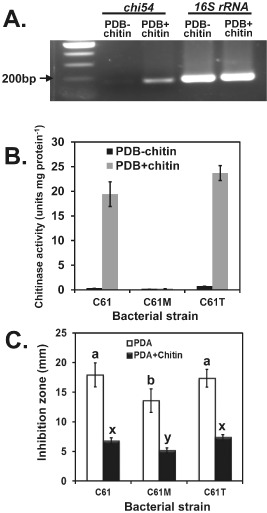
Effect of chitin amendment on the production of extracellular chitinase activity and antifungal activities of Chromobacterium sp. C61. (A) Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of the accumulation of transcripts from the chi54 gene in Chromobacterium sp. C61. Total RNA was isolated from wild‐type cells grown to the stationary phase [optical density at 600 nm (OD 600 nm) > 2.4] in potato dextrose broth (PDB) with or without chitin. RT‐PCRs were halted at the end of 24 cycles and the PCR products were loaded onto 1.5% agarose gel and visualized with ethidium bromide staining. Three independent experiments were conducted with similar results, and the data shown are from one experiment. (B) Chitinase activities of extracellular proteins partially purified from the growth of chitinase mutants and wild‐type strains in PDB with and without colloidal chitin. (C) Antifungal activity against Rhizoctonia solani produced by inocula of Chromobacterium sp. C61, C61Chi54M and C61Chi54TS on potato dextrose agar (PDA) or a chitin‐amended PDA plate. Data with different letters indicate that inhibition was significantly different (P < 0.05) based on Duncan's multiple range test.
To test the potential of the chitinase enzyme to inhibit fungal growth, the antifungal activity of purified Chi54 was tested in vitro. After incubation for 10 h, spore germination from most tested fungal pathogens was decreased significantly (P < 0.05) with 2 and 0.2 mg/mL purified Chi54. No antagonism for Fusarium oxysporum, Colletotrichum gloeosporioides or B. cinerea was observed below 0.2 mg/mL (Fig. S1, see Supporting Information). Germination of Cladosporium spharospermum was the most sensitive to Chi54 treatment, and conidial germination of F. oxysporum was the most resistant.
Subsequent in planta bioassays using cell‐free extracts of Chromobacterium sp. C61 supported the differential effects seen in vitro. For example, foliar application of the cell‐free supernatant of C61 grown in potato dextrose broth (PDB) significantly reduced the symptoms of rice blast, rice sheath blight, tomato gray mould, tomato leaf blight and wheat leaf rust (Fig. 4). However, control levels declined when the growth medium was amended with chitin (Fig. 4). These data indicated that components other than Chi54 secreted by C61 contributed to its in vitro and in planta biocontrol activities.
Figure 4.
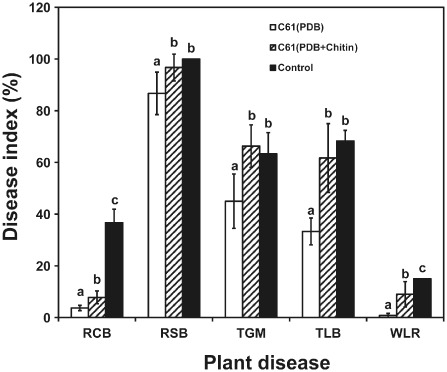
In planta biocontrol efficacies of cell‐free culture filtrates of Chromobacterium sp. C61 grown in the presence or absence of chitin. Applications were made by spraying intact plants with cell‐free culture filtrates of C61 (1:3 dilution) grown in potato dextrose broth (PDB) or PDB + chitin; water was used as the negative control treatment. The disease index for each pathogen was based on the areas of the infected lesion. RCB, rice blast; RSB, rice sheath blight; TGM, tomato gray mould; TLB, tomato late blight; WLR, wheat leaf rust. Data are means ± standard deviations of three independent experiments with nine plants per treatment. Bars with different letters for each pathosystem have significantly different (P < 0.05) levels of disease based on Duncan's multiple range test.
Identification of the antifungal compound produced by C61
A new antifungal metabolite from wild‐type C61 was extracted with ethyl acetate from culture medium. High‐performance liquid chromatography (HPLC) analyses of these extracts from cultures of the wild‐type indicated that a product (elution time, 8.5 min) had high antifungal activity (Fig. S2, see Supporting Information). Its structure was analysed using two‐dimensional 1H–1H correlation spectroscopy (2D 1H–1H COSY), 1H–1H rotating frame nuclear Overhauser effect spectroscopy (1H–1H ROESY), 1H–1H total correlation spectroscopy (1H–1H TOCSY), 1H–13C heteronuclear multiple‐quantum correlation spectroscopy (1H–13C HMQC), 1H–13C heteronuclear multiple‐bond correlation spectroscopy (1H–13C HMBC), distortionless enhancement by polarization transfer (DEPT) and 13C spectroscopy in methanol‐d 4 and dimethylsulphoxide (DMSO)‐d 6. The 1H and 13C resonance assignments of the isolated compound are presented in Tables S1 and S2 (see Supporting Information). Proton resonances for most aliphatic hydrocarbon chain groups and all amino acid residues were assigned, using the nuclear magnetic resonance (NMR) cross peaks, to structures in the main and side chains. We named the isolated compound chromobactomycin based on the novelty of its structure, which contained one β‐hydroxymyristate (HMS) group and nine amino acids (Fig. 5).
Figure 5.
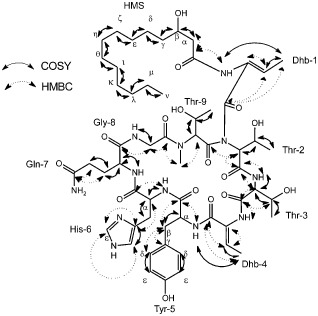
Chemical structure correlations by correlation spectroscopy (COSY) and heteronuclear multiple‐bond correlation spectroscopy (HMBC) of chromobactomycin. Solid arrows and dotted arrows refer to 1H–1H and 1H–13C correlations, respectively. Dhb, dihydroxybutyric acid; Gln, glutamine; Gly, glycine; His, histidine; HMS, β‐hydroxymyristate; Thr, threonine; Tyr, tyrosine.
Time‐of‐flight mass spectrometry (TOF MS) analysis of chromobactomycin detected an (M + H)+ peak at m/z 1196.6 with the predicted formula C57H86N12O16 (Fig. S4, see Supporting Information). A cyclic peptide was formed with a connection between the carboxyl group of Thr‐2 and the amine group of Thr‐9. Additional data for the structural analysis are presented in Figs 3, 4, 5, 6 (see Supporting Information). The MS3 spectra of the compound yielded a fragment ion peak at m/z 868.4, which represented the loss of HMS and dihydroxybutyric acid (Dhb). A second fragmentation produced a mass at m/z 568.3, suggesting the loss of a glutamine–glycine–threonine moiety. Two main ions were detected (m/z 387.2 and 304.1) in a third fragmentation, and represented the loss of two threonines from the structure with m/z 568.3 and the loss of Dhb UT from the peak at m/z 387.2. The proposed structure for chromobactomycin, based on the NMR and MS analyses, is shown in Fig. 5.
Figure 6.
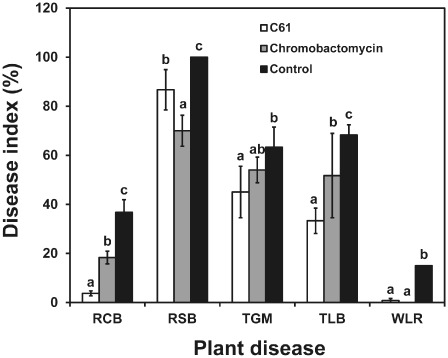
In planta biocontrol efficacies of purified chromobactomycin or cell‐free culture filtrates of Chromobacterium sp. C61. Applications were made by spraying intact plants with cell‐free culture filtrates of C61 (1:3 dilution) and the partially purified chromobactomycin (50 μg/mL); water was used as the negative control treatment. The disease index for each pathogen was based on the areas of the infected lesion. RCB, rice blast; RSB, rice sheath blight; TGM, tomato gray mould; TLB, tomato late blight; WLR, wheat leaf rust. Data are means ± standard deviations of three independent experiments with nine plants per treatment. Bars with different letters for each pathosystem have significantly different (P < 0.05) levels of disease based on Duncan's multiple range test.
Biocontrol activity of the purified chromobactomycin
Chromobactomycin, purified by preparative HPLC, when added to filter discs at 400 μg/disc, inhibited significantly the mycelial growth of R. solani, Pyricularia grisea, B. cinerea, Alternaria longipes and C. gloeosporioides in vitro. However, neither Phytophthora capsici nor F. oxysporum were inhibited in these assays (Table 1). There was no antibacterial activity against pv. oryzae (KACC10384), Xanthomonas campestris pv. oryzae, Pseudomonas syringae pv. tomato DC3000 or Bacillus subtilis QST713 (Table 1). Foliar applications of the partially purified chromobactomycin (50 μg/mL) reduced disease incidence in four pathosystems (Fig. 6).
Table 1.
In vitro activity of chromobactomycin against plant pathogenic fungi and bacteria
| Microorganism | Inhibition zone (mm) |
|---|---|
| Rhizoctonia solani (KACC40111) | 6.0a |
| Pyricularia grisea (KACC40439) | 6.0 |
| Fusarium oxysporum (KACC40183) | –b |
| Phytophthora capsici (KACC40157) | – |
| Botrytis cinerea (KACC40573) | 7.0 |
| Alternaria longipes (KACC40028) | 5.0 |
| Colletotrichum gloeosporioides (KACC40003) | 5.5 |
| Xanthomonas campestris pv. oryzae | – |
| Pseudomonas syringae pv. tomato DC3000 | – |
| Bacillus subtilis QST713 | – |
Inhibition zone (mm) between the fungal mycelia or bacterial spreading and the edge of the paper disc treated with chromobactomycin (400 μg/disc). Data are the means of two independent experiments.
No inhibition was detected.
The cell‐free culture filtrate of the C61 strain had a surface tension of 72.7 mN/mL, which was similar to that of the water control. However, the surface tension‐purified chromobactomycin was surface active, 42 mN/mL at 400 μg/μL. This finding indicates that the chromobactomycin produced by the C61 strain possessed weak surfactant activity.
Although the presence of chitin in the growth medium increased chitinase production, chromobactomycin production was reduced and correlated with lower in vitro biocontrol activity (Table 2). Based on the mass intensity value, chromobactin levels in the wild‐type [(326.67 ± 24.63) × 104] and C61Chi54TS mutant [(303.67 ± 14.81) × 104] samples were higher than that for the C61Chi54M mutant [(22.71 ± 9.18) × 104]. One possibility for these differences in CLP levels could be variations in growth rate. Although the growth of the Chi54 mutants was similar to that of the wild‐type in PDB or Luria–Bertani (LB) broth without chitin, growth of the C61Chi54M mutant had a lag phase of 6–9 h in PDB broth containing chitin (Fig. S11, see Supporting Information).
Table 2.
Effect of a colloidal chitin amendment on the production of extracellular chitinase, production of chromobactomycin and in vitro antifungal activity of the Chromobacterium sp. C61
| Strain | Growth condition | Chitinase activity (U/mL/min)a | Concentration of chromobactomycin (mg/L)b | Inhibition zone (mm)c | |
|---|---|---|---|---|---|
| Rhizoctonia solani | Botrytis cinerea | ||||
| C61 | –Chitin | 20.9 ± 0.7 | 88.2 ± 13.9 | 10.7 ± 0.6* | 11.8 ± 1.1* |
| C61 | +Chitin | 59.8 ± 7.4* | 60.8 ± 19.5 | 6.3 ± 0.6 | 7.5 ± 0.5 |
Wild‐type and mutants were grown in potato dextrose broth (PDB) with or without chitin for 3 days prior to the determination of chitinase activity using the fluorescent substrate 4‐methylumbelliferyl‐β‐d‐N,N‐diacetylchitobioside in the culture filtrates. One unit (U) of chitinase activity was defined as the release of one micromole of 4‐methylumbelliferone per minute per millilitre of culture filtrate.
The concentration of chromobactomycin in the culture filtrates of wild‐type and mutants was determined by high‐performance liquid chromatography (HPLC) using purified chromobactomycin as the authentic standard.
Inhibited fungal growth of R. solani and B. cinerea by Chromobacterium sp. C61 was determined on potato dextrose agar (PDA) plates with or without colloidal chitin amendments.
Data are from three independent experiments and are represented as means and standard deviations. Asterisk indicates a significant difference between growth medium based on Student's t‐test at P < 0.01.
Discussion
Because the suppression of plant disease results from multiple activities of biocontrol bacteria, it is important to determine which activities contribute the most to biological control under various conditions (Kim YC et al., 2011). Here, we demonstrate that the bacterium Chromobacterium sp. C61 produces and secretes both a chitinase and a novel CLP, which can contribute to biological control (Figs 2 and 5, respectively). Both lytic enzymes and CLP have been associated with biocontrol activity expressed by various microorganisms. Chitin is found in the cell walls of true fungi and in insect exoskeletons (Gooday, 1990), and amendment of soils with chitin has been correlated with increased populations of chitinolytic microorganisms (Wang et al., 2002). However, this is the first demonstration that both compounds actually contribute to the in vitro and in planta biocontrol activity noted by a single strain, i.e. C61.
Interestingly, the production of these two biocontrol‐related compounds by C61 grown in liquid medium seems to be interconnected in some way. Specifically, the presence of added chitin induces an increase in Chi54 production (Fig. 3) and a concominant decrease in chromobactomycin. At this point, it is not clear how the production and/or secretion of the two products are regulated. Recently, we found that a Tn5 mutant in C61, which lacked antifungal activity, had a transposon insertion in a sequence identified as a luxI‐encoding homoserine lactone synthase (H.J. Kim et al., unpublished results). Thus, it may be that Chi54 and chromobactomycin production are under quorum sensing control. Quorum sensing has been shown to regulate the expression of a major chitinase gene in Chromobacterium violaceum (Stauff and Bassler, 2011). Moreover, CLPs in other pseudomonads are also regulated by quorum sensing (Raaijmakers et al., 2006). Consequently, the fact that growth of the mutant on chitin‐amended medium demonstrated a long lag phase would correlate with reduced chromobactomycin levels, because of a delay in reaching a cell density threshold for quorum sensing to occur. Another possibility is that chromobactomycin production in the C61 strain is affected by metabolites from chitinase degradation, such as N‐acetyl‐glucosamine.
Although positively regulated by chitin (Monreal and Reese, 1969), negative regulation in the presence of glucose and N‐acetyl‐glucosamine in Serratia marcescens has been reported (Watanabe et al., 1997). Consequently, it seems that both environmental and nutrient conditions could be playing a role in the coordination of chitinase and CLP production in C61, as reviewed for other microbes by Fernando et al. (2005).
The CLP chromobactomycin from Chromobacterium sp. C61 contained an unusual amino acid, Dhb (2‐amino‐2‐butenoic acid), which has been reported previously for microcystin heptapeptides (Sano and Kaya, 1995, 1998). Dhb is also present in other important CLPs in the tolaasin and syringomycin produced by pseudomonads (Raaijmakers et al., 2006). Dhb‐1 connects the HMS and the cyclic peptide, possibly involving a thiotemplate mechanism, as discussed previously by Christiansen et al. (2008). The nonameric peptide of chromobactomycin is unique among the diverse CLP antibiotics produced by other bacteria (Ongena et al., 2007; Raaijmakers et al., 2006). Future research on chromobactomycin should involve larger scale fermentation and preparative extracts, so that the lethal dose 50% (LD50) for various pathogens can be accurately determined and the nature of the resistance of some phytopathogens to this chemical can be evaluated.
Interestingly, CLPs play diverse roles in the ecology of plant beneficial bacteria (Raaijmakers et al., 2010). Many CLPs have biosurfactant activity and thus are important in colony spread, biofilm formation and root colonization on plant surfaces (Ongena and Jacques, 2007). The induction of systemic resistance against plant diseases has also been documented for particular CLPs (Jourdan et al., 2009; Ongena et al., 2007; Raaijmakers et al., 2010). Their antifungal activities are varied: orfamide and putisolvins inhibit zoospore motility and cause zoospore lysis, but have no effect on mycelial growth (de Bruijin et al., 2007; Kruijt et al., 2009). Weak biosurfactant activity was observed for chromobactomycin. Currently, we are examining the anticancer properties of chromobactomycin as reported for the CLP from Chromobacterium violaceum (Cheng et al., 2007). Studies of mutants impaired in the biosynthesis genes for chromobactomycin in C61 will be needed to confirm the relative importance of the antibiotic to chitinase activity in the biocontrol potential of this strain. The draft genome sequence of Chromobacterium sp. C61 indicated that a potential CLP operon exists (Kim HJ et al., 2011). Because biocontrol is multifactorial in nature (Kim YC et al., 2011), it will be interesting to investigate how the environmental factors affect the metabolome required for biocontrol. This work will contribute to the maximization of the utilization of biocontrol agents under field conditions.
Experimental Procedures
Bacterial strains and growth conditions
The chitinolytic Chromobacterium strain C61, isolated previously from Korean soil (Park et al., 2005), was grown on nutrient broth (NB; Difco, Detroit, MI, USA) or LB medium (Sambrook et al., 1989) containing 50 μg/mL ampicillin and 0.2% colloidal chitin at 28 °C for routine culturing. Colloidal chitin was prepared as described previously (Park et al., 2007). Escherichia coli strains used in the genetic manipulations to obtain Chromobacterium sp. mutants were grown at 37 °C on LB medium (Sambrook et al., 1989). The medium was supplemented with kanamycin (50 μg/mL) for the growth of strains containing the plasmids pRL648 (Elhai and Wolk, 1988) and pCRII (Invitrogen Corporation, Carlsbad, CA, USA), with spectinomycin (50 μg/mL) for growth of a helper plasmid pRK2073 and tetracycline (12.5 μg/mL) for growth of the marker exchange vector pCPP54 (Miller et al., 1997). All strains were stored in 15% glycerol at −80 °C. The bacterial strains used to assess fungal antagonism were grown to an optical density at 600 nm (OD600nm) in LB medium, centrifuged and the cells were suspended in sterile water to 1 × 108 colony‐forming units (CFU)/mL.
Construction of Chromobacterium sp. C61 chitinase mutants
The cloning and subcloning procedures involved in the generation of the mutants were performed as described in Sambrook et al. (1989) and Ausubel et al. (1989). Plasmid DNA was isolated using an AccuPrep plasmid extraction kit (Bioneer Inc., Daejeon, South Korea), and genomic DNA was isolated by the cetyltrimethylammonium bromide (CTAB)‐NaCl method (Sambrook et al., 1989). Restriction enzymes and modified enzymes were purchased from New England Biolabs (Ipswich, MA, USA).
A mutant defective in chi54 gene expression was generated. The chi54 gene harboured in plasmid pChi54 (Park et al., 2007) was disrupted by the insertion of a 0.9‐kb BamHI‐digested fragment containing a kanamycin resistance gene from pRL648 into the unique BamHI site within the gene (Elhai and Wolk, 1988). The interrupted chi54 gene was exchanged with the chromosomal wild‐type copy using the pCPP54 exchange vector, as described previously (Miller et al., 1997). Putative chitinase‐deficient mutants were selected on plate medium containing 5% sucrose based on their resistance to kanamycin and their sensitivity to tetracycline.
A mutant, C61T, producing a more active chitinase, was generated by substitution of a tyrosine for a serine by modification of the chi54 gene sequence (Park et al., 2007). Assessment of chitinase production involved the transfer of aliquots (5 μL) of the bacterial suspension onto 0.1% colloidal chitin–50% LB agar plates.
PCR analysis, using DNA extracted employing the standard CTAB method (Ausubel et al., 1989), was performed with chi54‐specific primers (chi54F, 5′‐GAACGCGTGCTGTTGTAAGA‐3′; chi54R, 5′‐GAATTGAGGAGGTCCGAATG‐3′) with Bioneer PCR premix to check for the presence of the interrupted gene in the chi54 mutant and altered sequence in the T218S overactive mutant.
Chi54 transcript analysis
Chromobacterim sp. C61 cells were harvested from PDB cultures with or without 0.5% colloidal chitin at OD600nm = 0.7 for mid‐log phase cells. RNA was isolated using Trizol™ in accordance with the manufacturer's manual (Invitrogen Corporation). Reverse transcription (RT)‐PCR was conducted using the QuantiTect SYBR Green Reverse Transcription‐PCR kit (Qiagen, Valencia, CA, USA). Two specific primers, forward (5′‐AGAACAAGCGCCGCTACTAC‐3′) and reverse (5′‐AGAACAAGCGCCGCTACTAC‐3′), for the chi54 gene, and specific primers for the 16S rRNA gene of Chromobacterium species, forward (5′‐CCCACTGCCTCCCGTAAGGA‐3′) and reverse (5′‐TGGCTCAGAACGAACGCTGG‐3′), were used. A 25‐μL mixture was incubated for 30 min at 50 °C for reverse transcription. Quantitative RT‐PCR was performed using a Rotor‐Gene 3000 Real Time Cycler machine (Corbett Research Inc., Sydney, Australia) for 40 cycles, with denaturation at 94 °C for 15 s, annealing at 55 °C for 30 s and extension at 72 °C for 30 s. Quantitative RT‐PCRs were halted at the end of 24 cycles and the PCR products were loaded onto 1.5% agarose gels and visualized with ethidium bromide staining.
Assessment of chitinase activity
Chromobacterium sp. strains were grown in PDB with or without 0.2% colloidal chitin at 28 °C for 48 h with agitation. Cells were removed from the cultures by centrifugation and chitinase activity was assessed in the culture supernatant. Enzyme activity was assayed by measuring the hydrolysis of 4‐methylumbelliferyl‐β‐d‐N,N‐diacetylchitobioside (Sigma, St. Louis, MO, USA) in a reaction mixture (100 μL) containing 0.1 mm substrate and enzyme in 100 mm KH2PO4 buffer, pH 7.0. After incubation at 37 °C for 10 min, the reaction was stopped by the addition of 0.1 mL of 0.2 m Na2CO3, and the formation of chromogen was measured at an excitation wavelength of 360 nm and an emission wavelength of 440 nm in a Bio‐Tek FLX‐8000 fluorometer (Winooski, VT, USA). One unit of chitinase activity was expressed as micromoles of liberated 4‐methylumbelliferone per minute per millilitre of culture supernatant or per milligram of purified chitinase. Activity was also assessed by observing the extent of the clearing zones on LB plates containing 0.2% colloidal chitin after transfer of test sample aliquots.
Partial purification of the extracellular proteins involved adjusting the supernatant to 50% ammonium sulphate with stirring at 4 °C for 24 h. The precipitated proteins were harvested by centrifugation at 7000 g for 30 min, and were then dissolved in 1 mL of 20 mm sodium acetate (pH 6.0) before storage at 20 °C. The protein concentration of each sample was determined by measuring the absorbance at 595 nm using the protein assay dye reagent method as described by the manufacturer (Bio‐Rad Laboratories, San Francisco, CA, USA). The extracellular chitinase production from Chromobacterium sp. C61 wild‐type and chitinase mutant was determined according to the method of Park et al. (2007). Briefly, the same amount of protein (20 μg), partially purified from the culture filtrates, was loaded onto SDS‐PAGE gels. The proteins on the SDS‐PAGE gels were stained with Coomassie blue for Western analysis, and their molecular weights were determined by comparison with low range SDS‐PAGE standards (Bio‐Rad Laboratories). The proteins were transferred to negatively charged nylon membranes (Roche Applied Science, Mannheim, Germany) by electroblotting and incubated at 4 °C for 24 h with a polyclonal antibody prepared previously against the purified Chi54 chitinase (Park et al., 2005). After washing twice with TBS‐T buffer (20 mm Tris‐HCl, pH 7, 100 mm NaCl, 0.5% Tween‐20), the membrane was incubated with rabbit secondary antibody (Sigma) at room temperature for 1 h, followed by washing twice for 15 min with TBS‐T and air drying. The chitinase bands were detected by exposing the membrane to Western Lighting™ Plus‐ECL Enhanced Chemiluminescence Substrate (PerkinElmer Inc., San Jose, CA, USA). In addition, the same amount of protein (20 μg), partially purified from the culture filtrates, was loaded onto SDS‐PAGE gels, the proteins were separated and bands with chitinolytic activity were detected on the gels using 4‐methylumbelliferyl N‐acetyl‐β‐d‐glucosamine [4‐MU‐(GlcNAc)2].
Antifungal activity of the Chromobacterium sp. strain and the extracellular product
R. solani (KACC40111) and B. cinerea (KACC40573) were obtained from the Korea Agriculture Type Culture Collection Center (Suwon, South Korea). The pathogens were grown on PDA (Difco) at 28 °C. Suspensions of the Chromobacterium sp. C61 strains were spotted onto PDA plates prepared with or without colloidal chitin to measure antifungal activity, and agar discs (0.5 cm in diameter) bearing mycelia of each pathogen were placed in the centre of the plate. The antagonistic activities of the strains were evaluated by determining the extent of inhibited fungal mycelial growth compared with control plates lacking any bacterial inoculum. Cell‐free filtrates were prepared from cultures grown on PDB containing 0.2% colloidal chitin for 3 days at room temperature with 200 r/min agitation to measure antifungal activities. An aliquot of the filtrate was boiled for 5 min to test the resilience of antifungal activity to heat. Both the control and heat‐treated culture filtrates were transferred onto PDA plates to assess the inhibition of R. solani and B. cinerea growth, and onto plates with colloidal chitin to visualize chitinase activity.
Growth studies of the chi54 mutant
Growth of the chi54 mutant was compared with that of the wild‐type in PDB with or without amendment with 0.2% colloidal chitin. The bacterial strains were grown at 25 °C with shaking at 200 r/min in PDB (Difco) for 24 h, and adjusted to OD600 nm = 0.1. Further growth was determined from serial dilutions by measuring CFU/mL growing on LB agar plates supplemented with appropriate antibiotics: 50 μg/mL ampicillin for the wild‐type and 50 μg/mL kanamycin for the Chi54 mutants.
Assessment of in vitro antifungal activity of the purified Chi54
Inhibition of the fungal spore germination of Chromobacterium sp. C61 Chi54 by chitinase was examined using Chi54 generated from an E. coli strain containing the chi54 gene in an expression vector, as described previously (Park et al., 2007). Four fungal pathogens, C. gloeosporioides (KACC4003), B. cinerea (KACC40573), Cladosporium spharospermum (KACC43548) and F. oxysporum (KACC40183), were obtained from the Korea Agriculture Type Culture Collection Center. Conidial suspensions (1 × 105 conidia/mL) of the fungal pathogens were prepared in sterile water from fungal cultures grown on PDA plates. Equal volumes of Chi54 (0.02, 0.2, 2 mg/mL) in 10 mm sodium acetate buffer (pH 6.0) or the buffer alone, and each fungal conidial suspension (1 × 103 conidia/mL), were mixed in a tube and incubated at 28 °C for 10 h. Germination of the fungal spores was observed under phase contrast microscopy with samples applied to a haemocytometer. The experiment was performed in duplicate.
Isolation of antifungal compounds
C61 was grown in 50 L LB medium with shaking at 27 °C for 36 h. Cells were removed by centrifugation at 10 000 g for 20 min and the supernatant was extracted twice with the same volume of ethyl acetate. The ethyl acetate extracts were dried in an evaporator at 45 °C and dissolved in a mixture of ethyl acetate–methanol (90:10, v/v). The extracts were loaded onto SPE (Supelco, Bellefonte, PA, USA) silica gel cartridge columns conditioned with an ethyl acetate–methanol (90:10, v/v) solvent system. The methanol concentration was increased by 10% to 100% for each elution step. The eluates were collected and concentrated to dryness in a rotary evaporator at 45 °C.
After measurement of the antifungal activities of the eluates, as described below, the active fractions were dissolved in methanol and injected into a prep‐HPLC system equipped with a Dionex P680 model dual pump and a Dionex PDA‐100 model photodiode array detector (Germering, Bayern, Germany) at 210 nm. The HPLC column was a μ‐Bondapak C‐18 stainless steel column (7.8 × 300 mm2; film thickness, 10 μm). The solvent system was 85% (v/v) aqueous methanol, supplemented with 0.1% trifluoroacetic acid, and the flow rate was 2.0 mL/min. Each peak detected by HPLC analysis was collected and subjected to antifungal bioassays. All solvents used were of HPLC grade and were purchased from Fisher Scientific (Pittsburgh, PA, USA).
Instrumental analysis
A Varian model 500MS ion trap liquid chromatography (LC) mass spectrometer, connected to a 212‐LC Binary LC system (Varian, Palo Alto, CA, USA) with a Prostar 335 photodiode array detector at 210, 260 and 280 nm, was employed to identify the isolated antifungal compound in the cultures. The HPLC column was a Chromsep C18 stainless steel column (150 × 2.0 mm2). The mobile phase consisted of acetonitrile and ultrapure water containing 0.1% (v/v) formic acid, where the acetonitrile concentration was increased from 10% to 90% (v/v) over 30 min at a flow rate of 0.2 mL/min. The HPLC system was interfaced to an LC‐MS2 system equipped with electrospray ionization (ESI) interface in positive ion mode. The drying gas temperature and pressure were 300 °C and 20 psi, respectively. The capillary voltage was 80 V. Mass spectra were obtained in the m/z range 100–1500.
Mass spectra were obtained from a hybrid ion‐trap TOF mass spectrometer (Shimadzu, Tokyo, Japan) equipped with an ESI source in positive ion mode at a mass resolution of 10 000 full width at half‐maximum to calculate the exact mass. The analytical conditions were as follows: scan range, m/z 700–1500; spray voltage, +4.50 kV; detector voltage, 1.70 kV; skimmer voltage, 8.0 V; pressure of TOF region, 2.2 × 10−4 Pa; temperature, 40 °C; ion source temperature, 200 °C; trap cooling gas (Ar) flow rate, 94 mL/min; ion trap pressure, 1.8 × 102 Pa; collision gas (Ar) flow rate, 43 mL/min; ion accumulation time, 10 ms; precursor ion selected width, 3.0 m/z units; selected time, 50 ms; collision‐induced dissociation collision time, 30 ms; collision energy, 100%; qz = 0.251. MSn data were acquired using the targeted function. The Shimadzu Composition Formula Predictor was also used to verify identifications. A Varian model Unity INFINITY plus 125‐MHz NMR spectrometer and INOVA 500‐MHz NMR spectrometer were used for the 13C and 1H analyses, respectively.
Determination of biosurfactant activity
Biosurfactant activity in cell‐free culture medium was measured using a DuNouy model 3010 tensiometer (DuNouy, Tokyo, Japan), as described previously (Kim et al., 2007), and by the drop collapse assay using 10W‐40 Penzoil (Penzoil Products Co., Houston, TX, USA), as described by Burch et al. (2010). The bacterial supernatants were prepared by centrifugation at 12 000 g for 10 min, and filtered through a 0.2‐μm Millipore filter (Billerica, MA, USA) to remove residual bacterial cells. For surface tension measurement, the instrument was calibrated with air and water, giving a reading of 72.75 mN/mL. The surface tension measurement and drop collapse assay were conducted in three independent experiments with three replicates per sample.
Measurement of in vitro antifungal activity
Wild‐type and mutant strain cells were grown in PDB with or without 0.2% colloidal chitin for 3 days to measure extracellular chromobactomycin production from C61. Chromobactomycin was extracted into ethyl acetate, fractionated by HPLC as described above and used in methanolic solutions for antifungal assays. The phytopathogenic fungal pathogens were A. longipes (KACC40028), R. solani (KACC40111), P. grisea (KACC40439), F. oxysporum (KACC40183), Ph. capsici (KACC40157), B. cinerea (KACC40573) and Colletotrichum gloeosporioides (KACC4003), each obtained from the Korea Agriculture Type Culture Collection Center. These pathogens were grown on PDA (Difco) at 28 °C. Xanthomonas campestris pv. oryzae, Ps. syringae pv. tomato DC3000 and Ba. subtilis QST718 were streaked onto fresh LB agar plates and incubated for 2 days at room temperature to measure the antibacterial activities of the purified chromobactomycin. The bacterial cells were harvested, resuspended in sterile water, adjusted to OD600nm = 0.1 and 1 mL of the bacterial suspension was spread onto fresh LB agar plates. The CLP dissolved in methanol was added to sterile paper discs (8 mm in diameter; Advantec, Toyo Roshi Kaisha, Japan), dried and placed on the LB agar plates which were spread with the bacterial suspensions. The plates were incubated at 27 °C, and clear zones around the paper discs were assessed after 3 days.
The Chromobacterium sp. C61 strain inoculum was transferred to the edges of PDA plates to measure antifungal activity. Assays were also performed to measure mycelial growth inhibition on PDA plates with paper discs loaded with the purified materials. Methanol was used as a negative control. Sterilized paper discs (8 mm in diameter; Advantec) were treated with the isolated compound (400 μg/disc) dissolved in methanol. The paper discs were dried and placed at a distance of 2.5 cm from the pathogens provided as PDA plugs (5 mm in diameter) covered with growing fungal mycelia. The plates were incubated at 27 °C, and fungal growth was assessed after 5 days.
Plant disease suppression biocontrol assays
The biocontrol activity of Chromobacterium sp. strain C61 was tested in planta against six different plant pathogens (Kim et al., 2001), including Magnaporthe oryzae and R. solani on rice plants, B. cinerea and Ph. infestans on tomato plants, Puccinia recondita on wheat plants and Colletotrichum coccodes on red pepper plants. The in vivo antifungal bioassays were conducted as described previously (Kim et al., 2001). Briefly, plants were grown in vinyl pots in a glasshouse at 25 ± 5 °C for 1–4 weeks. The plants were sprayed with 1:3 dilutions of cell‐free culture filtrates of 3‐day‐old C61, C61Chi54M and C61Chi54TS grown in PDB with and without chitin, or the purified chromobactomycin (50 μg/mL) with Tween‐20 as a wetting agent. Distilled water with Tween‐20 was used as the negative control. The treated plant seedlings were inoculated with spores or mycelial suspensions from one of the six plant pathogens after 24 h, as described previously (Kim et al., 2001). Disease symptoms were rated 3–7 days after inoculation, depending on the pathogen. The pots were arranged in a randomized complete‐block design, with three replicates per treatment, and each replicate consisted of nine plants. The disease index was determined by measuring the percentage of the infected leaf area. The experiment was conducted three times.
Statistical analysis
Data were subjected to analysis of variance using IBM SPSS Statistics version 19 (IBM Corporation, Somers, NY, USA). The differential effects of the bacterial treatments on disease severity were determined by Duncan's multiple range test (P < 0.05). The effect of a chitin amendment in the growth medium on the production of chitinase was evaluated using Student's t‐test (P < 0.01).
Supporting information
Fig. S1 Effect of the purified Chi54 protein on the germination rate of phytopathogenic fungal spores. Fungal spores of Colletotrichum gloeosporioides (Cg), Botrytis cinerea (Bc), Cladosporium spharospermum (Cs) and Fusarium oxysporum (Fo) were treated with different concentrations of the purified Chi54 protein in 10 mm sodium acetate buffer (pH 6.0) or the buffer without the Chi54 protein. The germination rate was measured by counting the number of fungal spores that germinated versus the total number of spores using a haemocytometer under a phase contrast microscope 10 h after incubation at 28 °C. The data are the means of two independent experiments and the vertical bars indicate the standard deviations. Different letters indicate significant differences between fungal pathogens according to Duncan's multiple range test (P < 0.05).
Fig. S2 High‐performance liquid chromatograms of ethyl acetate‐extracted compounds from cell‐free filtrates of Chromobacterium sp. C61. The solvent system was 85% (v/v) aqueous methanol supplemented with 0.1% trifluoroacetic acid at 2.0 mL/min, and the peaks were detected at 210 nm. The chromobactomycin peak from Chromobacterium sp. C61 is shown with an arrow, and this material was used to obtain the time‐of‐flight mass spectrum provided.
Fig. S3 Ion trap‐time‐of‐flight mass spectrometry (IT‐TOF MS), MS2 and MS3 data of the isolated antifungal compound.
Fig. S4 Correlation spectra of dihydroxybutyric acid (Dhb) in methanol‐d 6 of the isolated compound.
Fig. S5 Heteronuclear multiple‐bond correlation spectra of dihydroxybutyric acid (Dhb) and Tyr‐5 in methanol‐d 6 of the isolated compound.
Fig. S6 Heteronuclear multiple‐quantum correlation spectra of β‐hydroxymyristate in methanol‐d 6 of the isolated compound.
Fig. S7 Heteronuclear multiple‐bond correlation spectra in methanol‐d 6 identifying all carbonyl groups in the amino acid residues of the isolated compound.
Fig. S8 Heteronuclear multiple‐bond correlation spectra of Tyr‐5 in methanol‐d 6 of the isolated compound.
Fig. S9 Connectivities observed in correlation and heteronuclear multiple‐bond correlation spectra, leading to sequence assignment of the isolated compound.
Fig. S10 The proposed modifications of chromobactomycin structure identified by tandem mass spectrometry (MS2). Dhb, dihydroxybutyric acid; Gln, glutamine; Gly, glycine; His, histidine; HMS, β‐hydroxymyristate; 3‐HTDA, 3‐hydroxytetradecanoic acid; Thr, threonine; Tyr, tyrosine.
Fig. S11 Effects of the chi54 mutations on aerobic growth in the presence of chitin. Cultures were grown aerobically in potato dextrose broth (PDB) with 0.2% chitin, and the growth of bacterial strains was determined by measuring the colony‐forming units/mL (CFU/mL) of serial diluted cultures. Bacterial populations were assessed at each time point using analysis of variance (ANOVA), and mean CFU/mL values were compared according to Duncan's multiple range test (*P < 0.05 and **P < 0.01). The data are the means of three independent experiments and vertical bars indicate standard deviations.
Table S1 1H chemical shift assignments of chromobactomycin in methanol‐d 4 (A) and dimethylsulphoxide (DMSO)‐d 6 (B).
Table S2 13C chemical shift assignments of chromobactomycin in methanol‐d 4 (A) and dimethylsulphoxide (DMSO)‐d 6 (B).
Acknowledgements
This study was supported by a grant from the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries (311019‐03), Ministry for Food, Agriculture, Forest, and Fisheries and partially supported by the World Class University project of the National Research Foundation of Korea (grant no. R32‐2009‐000‐20047‐0).
References
- Arora, N.K. , Kim, M.J. , Kang, S.C. and Maheshwari, D.K. (2007) Role of chitinase and β‐1,3‐glucanase activities produced by a fluorescent pseudomonad and in vitro inhibition of Phytophthora capsici and Rhizoctonia solani . Can. J. Microbiol. 53, 207–212. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M. , Brent, R. , Kingston, R.E. , Moore, D.D. , Seidman, J.G. , Smith, J.A. and Struhl, K. (1989) Current Protocols in Molecular Biology. New York: Wiley. [Google Scholar]
- de Bruijin, I. , de Kock, M.J.D. , Yang, M. , de Waad, P. , van Beek, T.A. and Raaijmakers, J.M. (2007) Genome‐based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol. 63, 417–428. [DOI] [PubMed] [Google Scholar]
- Burch, A.Y. , Shimada, B.K. , Browne, P.J. and Lindow, S.E. (2010) Novel high‐throughput detection method to assess bacterial surfactant production. Appl. Environ. Microbiol. 75, 5363–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y.‐Q. , Yang, M. and Matter, A.M. (2007) Characterization of a gene cluster responsible for the biosynthesis of anticancer agent FK228 in Chromobacterium violaceum No. 968. Appl. Environ. Microbiol. 73, 3460–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen, G. , Yoshida, W.Y. , Blom, J.F. , Portmann, C. , Gademann, K. , Hemscheide, T. and Kurmayer, R. (2008) Isolation and structure determination of two microcystins and sequence comparison of the McyABC adenylation domains in Planktothrix species. J. Nat. Prod. 71, 1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen‐Kupiec, R. and Chet, I. (1998) The molecular biology of chitin digestion. Curr. Opin. Biotechnol. 9, 270–277. [DOI] [PubMed] [Google Scholar]
- Dahiya, N. , Tewari, R. and Hoondai, G.S. (2006) Biotechnological aspects of chitinolytic enzymes: a review. Appl. Microbiol. Biotechnol. 71, 773–782. [DOI] [PubMed] [Google Scholar]
- Dubuis, C. , Keel, C. and Haas, D. (2007) Dialogues of root‐colonizing biocontrol pseudomonads. Eur. J. Plant Pathol. 119, 311–328. [Google Scholar]
- Elhai, J. and Wolk, C.P. (1988) A versatile class of positive‐selection vectors based on the nonviability of palindrome‐containing plasmids that allows cloning into long polylinkers. Gene, 68, 119–138. [DOI] [PubMed] [Google Scholar]
- Fernando, W.G.D. , Nakkereran, S. and Zhang, Y. (2005) Biosynthesis of antibiotics by PGPR and its relation in biocontrol of plant diseases In: PGPR; Biocontrol and Biofertilizer (Siddigui Z.A., ed.), pp. 67–109. Dordrecht, The Netherlands: Springer. [Google Scholar]
- Flach, J. , Pilet, P.E. and Jolles, P. (1992) What's new in chitinase research? Experientia 48, 701–716. [DOI] [PubMed] [Google Scholar]
- Gooday, G.W. (1990) The ecology of chitin degradation. Adv. Microb. Ecol. 11, 387–430. [Google Scholar]
- Hiradate, S. , Yoshida, S. , Sugie, H. , Yada, H. and Fujii, Y. (2002) Mulberry anthracnose antagonists (iturins) produced by Bacillus amyloliquefaciens RC‐2. Phytochemistry, 61, 693–698. [DOI] [PubMed] [Google Scholar]
- Jourdan, E. , Henry, G. , Duby, F. , Dommes, J. , Barthelemy, J.P. , Thonart, P. and Ongena, M. (2009) Insights into the defense‐related events occurring in plant cells following perception of surfactin‐type lipopeptide from Bacillus subtilis . Mol. Plant–Microbe Interact. 22, 456–468. [DOI] [PubMed] [Google Scholar]
- Kim, H.J. , Park, J.Y. , Han, S.H. , Lee, J.H. , Rong, X. , McSpadden Gardener, M.B. , Park, S.K. and Kim, Y.C. (2011) Draft genome sequence of the biocontrol bacterium Chromobacterium sp. strain C‐61. J. Bacteriol. 193, 6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.‐C. , Choi, G.J. , Park, J.‐H. , Kim, H.T. and Cho, K.Y. (2001) Activity against plant pathogenic fungi of phomalactone isolated from Nigrospora sphaerica . Pest Manag. Sci. 57, 554–559. [DOI] [PubMed] [Google Scholar]
- Kim, S.K. , Kim, S.R. , Choi, M.S. , Park, C.E. , Kim, Y.C. , Kim, K.Y. , Whang, K.S. , Oh, K.‐T. and Kim, I.S. (2007) Soybean oil‐degrading bacterial cultures as a potential for control of green peach aphids (Myzus Persicae). J. Microbiol. Biotechnol. 17, 1700–1703. [PubMed] [Google Scholar]
- Kim, Y.C. , Jung, H. , Kim, K.Y. and Park, S.K. (2008) An effective biocontrol bioformulation against Phytophthora blight of pepper using growth mixtures of combined chitinolytic bacteria under different field conditions. Eur. J. Plant Pathol. 120, 373–382. [Google Scholar]
- Kim, Y.C. , Lee, J.H. , Bae, Y.‐S. , Sohn, B.‐K. and Park, S.K. (2010) Development of effective environmentally‐friendly approaches to control Alternaria blight and anthracnose diseases of Korean ginseng. Eur. J. Plant Pathol. 127, 443–450. [Google Scholar]
- Kim, Y.C. , Leveau, J. , McSpadden Gardener, B.B. , Pierson, E.A. , Pierson, L.S. III and Ryu, C.‐M. (2011) The multifactorial basis for plant health promotion by plant‐associated bacteria. Appl. Environ. Microbiol. 77, 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, D.Y. , Reedy, R.M. , Bick, J. and Oudemans, P.V. (2002) Characterization of a chitinase gene from Stenotrophomonas maltophilia strain 34S1 and its involvement in biological control. Appl. Environ. Microbiol. 68, 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijt, M. , Tran, H. and Raaijmakers, J.M. (2009) Functional, genetic and chemical characterization of biosurfactants produced by plant growth‐promoting Pseudomonas putida 267. J. Appl. Microbiol. 107, 546–556. [DOI] [PubMed] [Google Scholar]
- Li, J.‐G. , Jiang, Z.‐Q. , Xu, L.‐P. , Sun, F.‐F. and Guo, J.‐H. (2008) Characterization of chitinase secreted by Bacillus cereus strain CH2 and evaluation of its efficacy against Verticillium wilt of eggplant. BioControl, 53, 931–944. [Google Scholar]
- Lugtenberg, B. and Kamlikova, F. (2009) Plant growth promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–546. [DOI] [PubMed] [Google Scholar]
- Miller, C.D. , Kim, Y.C. and Anderson, A.J. (1997) Cloning and mutational analysis of the gene for the stationary‐phase inducible catalase (catC) from Pseudomonas putida . J. Bacteriol. 179, 5241–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monreal, J. and Reese, E.T. (1969) The chitinase of Serratia marcescens . Can. J. Microbiol. 15, 689–696. [DOI] [PubMed] [Google Scholar]
- Ongena, M. and Jacques, P. (2007) Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. [DOI] [PubMed] [Google Scholar]
- Ongena, M. , Jourdan, E. , Adam, A. , Paquot, M. , Brans, A. , Joris, B. , Arpigny, J.‐L. and Thonart, P. (2007) Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. [DOI] [PubMed] [Google Scholar]
- Park, S.K. , Lee, M.C. and Harman, G.E. (2005) The biocontrol activity of Chromobacterium sp. strain C61 against Rhizoctonia solani depends on the productive ability of chitinase. Plant Pathol. J. 21, 275–282. [Google Scholar]
- Park, S.K. , Kim, C.W. , Kim, H. , Jung, J.S. and Harman, G.E. (2007) Cloning and high‐level production of a chitinase from Chromobacterium sp. and the role of conserved or nonconserved residues on its catalytic activity. Appl. Microbiol. Biotechnol. 74, 791–804. [DOI] [PubMed] [Google Scholar]
- Raaijmakers, J.M. , de Bruijin, I. and Kock, M.J.D. (2006) Cyclic lipopeptide production by plant‐associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Mol. Plant–Microbe Interact. 19, 699–710. [DOI] [PubMed] [Google Scholar]
- Raaijmakers, J.M. , de Bruijin, I. , Nybroe, O. and Ongena, M. (2010) Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34, 1037–1062. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Frithsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sano, T. and Kaya, K. (1995) A 2‐amino‐2‐butenoic acid (Dhb)‐containing microcystin isolated from Oscillatoria agardhii . Tetrahedron Lett. 36, 8603–8606. [Google Scholar]
- Sano, T. and Kaya, K. (1998) Two new (E)‐2‐amino‐butenoic acid (Dhb)‐containing microcystin isolated from Oscillatoria agardhii . Tetrahedron, 54, 463–470. [Google Scholar]
- Stauff, D.L. and Bassler, B.L. (2011) Quorum sensing in Chromobacterium violaceum: DNA recognition and gene regulation by the CviR receptor. J. Bacteriol. 193, 3871–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tendulkar, S.R. , Saikumari, Y.K. , Patel, V. , Raghotama, S. , Munshi, T.K. , Balaram, P. and Chattoo, B.B. (2007) Isolation, purification, and characterization of an antifungal molecule produced by Bacillus licheniformis BC98, and its effect on phytopathogen Magnaporthe grisea . J. Appl. Microbiol. 103, 2331–2339. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Liu, J. , Chen, H. and Yao, J. (2007) Characterization of Fusarium graminearum inhibitory lipopeptide from Bacillus subtilis IB. Appl. Microbiol. Biotechnol. 76, 889–894. [DOI] [PubMed] [Google Scholar]
- Wang, S.L. , Yen, Y.H. , Tsiao, W.J. , Chang, W.T. and Wang, C.L. (2002) Production of antimicrobial compounds by Monascus purpureus CCRC31499 using shrimp and crab shell powder as a carbon source. Enzyme Microb. Technol. 31, 337–344. [Google Scholar]
- Watanabe, T. , Kimura, K. , Sumiya, T. , Nikaidou, N. , Suzuki, K. , Suzuki, M. , Taiyoji, M. , Ferrer, S. and Regue, M. (1997) Genetic analysis of the chitinase system of Serratia marcescens 217. J. Bacteriol. 179, 7111–7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T. , Kanai, R. , Kawase, T. , Tanabe, T. , Mitsutomi, M. , Sakuda, S. and Miyashita, K. (1999) Family of chitinases of Streptomyces species: characterization and distribution. Microbiology, 145, 3353–3363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Effect of the purified Chi54 protein on the germination rate of phytopathogenic fungal spores. Fungal spores of Colletotrichum gloeosporioides (Cg), Botrytis cinerea (Bc), Cladosporium spharospermum (Cs) and Fusarium oxysporum (Fo) were treated with different concentrations of the purified Chi54 protein in 10 mm sodium acetate buffer (pH 6.0) or the buffer without the Chi54 protein. The germination rate was measured by counting the number of fungal spores that germinated versus the total number of spores using a haemocytometer under a phase contrast microscope 10 h after incubation at 28 °C. The data are the means of two independent experiments and the vertical bars indicate the standard deviations. Different letters indicate significant differences between fungal pathogens according to Duncan's multiple range test (P < 0.05).
Fig. S2 High‐performance liquid chromatograms of ethyl acetate‐extracted compounds from cell‐free filtrates of Chromobacterium sp. C61. The solvent system was 85% (v/v) aqueous methanol supplemented with 0.1% trifluoroacetic acid at 2.0 mL/min, and the peaks were detected at 210 nm. The chromobactomycin peak from Chromobacterium sp. C61 is shown with an arrow, and this material was used to obtain the time‐of‐flight mass spectrum provided.
Fig. S3 Ion trap‐time‐of‐flight mass spectrometry (IT‐TOF MS), MS2 and MS3 data of the isolated antifungal compound.
Fig. S4 Correlation spectra of dihydroxybutyric acid (Dhb) in methanol‐d 6 of the isolated compound.
Fig. S5 Heteronuclear multiple‐bond correlation spectra of dihydroxybutyric acid (Dhb) and Tyr‐5 in methanol‐d 6 of the isolated compound.
Fig. S6 Heteronuclear multiple‐quantum correlation spectra of β‐hydroxymyristate in methanol‐d 6 of the isolated compound.
Fig. S7 Heteronuclear multiple‐bond correlation spectra in methanol‐d 6 identifying all carbonyl groups in the amino acid residues of the isolated compound.
Fig. S8 Heteronuclear multiple‐bond correlation spectra of Tyr‐5 in methanol‐d 6 of the isolated compound.
Fig. S9 Connectivities observed in correlation and heteronuclear multiple‐bond correlation spectra, leading to sequence assignment of the isolated compound.
Fig. S10 The proposed modifications of chromobactomycin structure identified by tandem mass spectrometry (MS2). Dhb, dihydroxybutyric acid; Gln, glutamine; Gly, glycine; His, histidine; HMS, β‐hydroxymyristate; 3‐HTDA, 3‐hydroxytetradecanoic acid; Thr, threonine; Tyr, tyrosine.
Fig. S11 Effects of the chi54 mutations on aerobic growth in the presence of chitin. Cultures were grown aerobically in potato dextrose broth (PDB) with 0.2% chitin, and the growth of bacterial strains was determined by measuring the colony‐forming units/mL (CFU/mL) of serial diluted cultures. Bacterial populations were assessed at each time point using analysis of variance (ANOVA), and mean CFU/mL values were compared according to Duncan's multiple range test (*P < 0.05 and **P < 0.01). The data are the means of three independent experiments and vertical bars indicate standard deviations.
Table S1 1H chemical shift assignments of chromobactomycin in methanol‐d 4 (A) and dimethylsulphoxide (DMSO)‐d 6 (B).
Table S2 13C chemical shift assignments of chromobactomycin in methanol‐d 4 (A) and dimethylsulphoxide (DMSO)‐d 6 (B).


