Summary
Many pathogenic bacteria have evolved a type III secretion system (T3SS) to successfully invade their host. This extracellular apparatus allows the translocation of proteins, called type III effectors (T3Es), directly into the host cells. T3Es are virulence factors that have been shown to interfere with the host's immunity or to provide nutrients from the host to the bacteria. The Gram‐negative bacterium Ralstonia solanacearum is a worldwide major crop pest whose virulence strongly relies on the T3SS. In R. solanacearum, transcriptional regulation has been extensively studied. However, very few data are available concerning the role played by type III‐associated regulators, such as type III chaperones and T3SS control proteins. Here, we characterized HpaP, a putative type III secretion substrate specificity switch (T3S4) protein of R. solanacearum which is not secreted by the bacterium or translocated in the plant cells. HpaP self‐interacts and interacts with the PopP1 T3E. HpaP modulates the secretion of early (HrpY pilin) and late (AvrA and PopP1 T3Es) type III substrates. HpaP is dispensable for the translocation of T3Es into the host cells. Finally, we identified two regions of five amino acids in the T3S4 domain that are essential for efficient PopP1 secretion and for HpaP's role in virulence on tomato and Arabidopsis thaliana, but not required for HpaP–HpaP and HpaP–PopP1 interactions. Taken together, our results indicate that HpaP is a putative R. solanacearum T3S4 protein important for full pathogenicity on several hosts, acting as a helper for PopP1 secretion, and repressing AvrA and HrpY secretion.
Keywords: bacterial wilt, pathogenicity, secretion, translocation, T3Es (type III effectors), T3S4 (type III secretion substrate specificity switch), T3SS (type III secretion system).
Introduction
The translocation of pathogenicity factors into host cells is a paradigm of parasite–host interactions (He et al., 2004; Hueck, 1998). As documented for numerous bacterial pathogens, pathogenicity proteins, also called type III effectors (T3Es), are delivered from the bacterial cytoplasm into host cells via a molecular syringe called the type III secretion system (T3SS). In plant pathogens, the T3SS proteins are encoded by 23–29 genes, named the hrp gene cluster, required for the hypersensitive response and pathogenicity of the bacteria (He et al., 2004). Among these genes, four subgroups can be distinguished: (i) core component genes with 6–10 non‐conserved genes and nine hrc genes conserved among animal and plant pathogenic bacteria (hrc, for hrp conserved); (ii) two to four transcriptional regulators; (iii) genes encoding for extracellular proteins (pilin, translocators and harpins); and (iv) two to four genes encoding for secretion regulators (Tampakaki et al., 2010).
In recent years, many efforts have been made to elucidate T3E functions of plant pathogenic bacteria (Block et al., 2008; Canonne and Rivas, 2012; McCann and Guttman, 2008), whereas only limited information on T3SS post‐translational regulations is available. Recent reviews have focused more specifically on the involvement of type III secretion chaperones or type III‐associated proteins (Büttner, 2012; Büttner and Bonas, 2009; Büttner and He, 2009; Lohou et al., 2006). Type III chaperones were first characterized as being involved in the transcription or secretion of T3Es in animal pathogenic bacteria (Feldman and Cornelis, 2003; Parsot et al., 2003). Indeed, T3Es are produced in the cytoplasm, where they need to be stabilized and unfolded into a competent state to facilitate their secretion/translocation into the host (Parsot et al., 2003). Type III chaperones are small, acidic, leucine‐rich proteins. They can interact with T3Es (class IA or IB chaperones) or with proteins that allow the insertion of the hrp pilus into the cell membrane, i.e. the translocators (class II chaperones) (Parsot et al., 2003). The secretion of T3Es is also controlled by proteins of the YscP/FliK family, containing a T3S4 (type III secretion substrate specificity switch) domain (Büttner, 2012; Tampakaki et al., 2010). These T3S4 proteins are key players in orchestrating the translocation of T3Es. They are related to the YscP/FliK family, as the concept of substrate specificity switching by T3SS emerged after the characterization of the FliK protein in the flagellar T3SS (Patterson‐Delafield et al., 1973) and of the YscP protein in Yersinia species (Agrain et al., 2005; Edqvist et al., 2003; Journet et al., 2003). A major role of FliK and YscP proteins is to control flagellar hook and needle lengths, respectively, and to regulate the secretion of late substrates (Journet et al., 2003; Minamino et al., 1999). In plant pathogenic bacteria, the YscP/FliK family members characterized so far are HpaC from Xanthomonas campestris pv. vesicatoria (Büttner et al., 2006) (called HpaCXcv hereafter) and HrpP from Pseudomonas syringae pv. tomato (Morello and Collmer, 2009) (called HrpPPst hereafter). HpaCXcv is not secreted and has been demonstrated to be important for the secretion of T3Es and translocon proteins (Lorenz et al., 2008). HrpPPst is a T3SS substrate translocated into plant cells and is essential for the function of the T3SS (Morello and Collmer, 2009). The differences observed in their functions could be a result of the fact that these two plant pathogenic bacteria have a different type of T3SS (Tampakaki et al., 2010). Ralstonia solanacearum has a similar (Hrp2) T3SS to Xanthomonas spp. (Büttner, 2012; Tampakaki et al., 2010), but, to date, the role of putative T3S4 proteins in R. solanacearum has not been described. Lorenz et al. (2008) reported that HpaP (for hrp‐associated P) in R. solanacearum is 27% identical to HpaCXcv, and Agrain et al. (2005) defined 18% sequence identity over 90 amino acids between HpaP and YscP from Yersinia pestis. R. solanacearum, the causal agent of bacterial wilt, is a root pathogen that affects more than 200 plant species (Genin and Denny, 2012; Hayward, 2000). As for many Gram‐negative bacteria, the major role in R. solanacearum pathogenicity is attributed to the T3SS because corresponding mutants cannot provoke the hypersensitive response (HR), the rapid cell death of plant cells directly at the inoculation site, in resistant plants or symptoms in susceptible host plants (hrp– phenotype). R. solanacearum has one of the largest T3E repertoires (74 effectors in the GMI1000 strain) among other Gram‐negative plant pathogenic bacteria (Poueymiro and Genin, 2009). In this large repertoire, several T3Es have been shown to trigger plant immune responses (or effector‐triggered immunity, ETI). This is the case for PopP1 and PopP2, two members of the YopJ/Avrxv family, on Petunia hybrida ecotype St‐40 (Arlat et al., 1994; Lavie et al., 2002) and Nicotiana species (Poueymiro et al., 2009) for PopP1, and on Arabidopsis thaliana ecotype Nd‐1 for PopP2 (Deslandes et al., 1998). AvrA, another T3E, is also required for ETI on Nicotiana species (Poueymiro et al., 2009). Despite the huge number of candidate virulence factors, to date, only a few T3Es have been identified as being required for the pathogenicity of R. solanacearum (Angot et al., 2006; Cunnac et al., 2004; Remigi et al., 2011; Sole et al., 2012).
The present work constitutes the first functional study of HpaP, a putative T3S4 protein, in R. solanacearum. The hpaP mutant has been shown to be impaired in both the induction of HR on tobacco plants and disease symptoms on tomato plants (van Gijsegem et al., 2002). Here, we characterize the involvement of HpaP in the control of the secretion/translocation of T3Es. Our data suggest that this regulation involves direct protein–protein interactions. Finally, we show that HpaP is important for the full pathogenicity of R. solanacearum on several host plants, and that two conserved sequences of five amino acids in the T3S4 domain of HpaP are required for the establishment of the disease.
Results
HpaP is a putative T3S4 protein conserved in the R. solanacearum species complex
The hpaP gene from R. solanacearum GMI1000 (NP_522423.1) is part of the conserved hrp gene cluster, and is localized in an operon together with hrcV and hrcU (Fig. 1a). HpaP is conserved among all the sequenced strains of R. solanacearum, although the overall protein similarity among strains appears to be lower than for HrcV and HrcU (Fig. 1a). Phylogenetic analysis of hpaP genes reveals that hpaP phylogeny matches perfectly with the species phylogeny separating the four phylotypes into three distinctive clades (Peeters et al., 2013) (Fig. 1b). This indicates that hpaP was probably acquired early before the diversification that led to the actual organization of the R. solanacearum species complex into four phylotypes.
Figure 1.
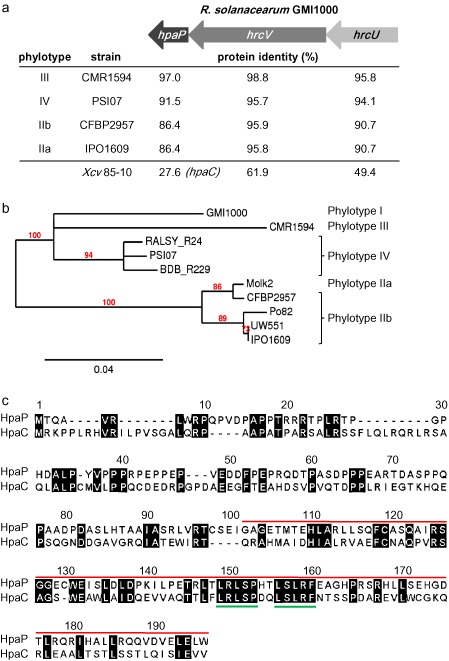
HpaP is a putative type III secretion substrate specificity switch (T3S4) protein conserved in the Ralstonia solanacearum species complex. (a) Protein identity between HpaP, HrcV and HrcU from R. solanacearum GMI1000 and homologues from strains representative of the R. solanacearum species complex diversity or from Xanthomonas campestris pv. vesicatoria (Xcv) 85‐10. Sequences were compared using SIAS (http://imed.med.ucm.es/Tools/sias.html). Arrows represent the hrcU‐hrcV‐hpaP operon. (b) Unrooted phylogenetic tree of hpaP based on sequence similarities using the maximum likelihood principle. Ten sequenced strains belonging to the four phylotypes of the R. solanacearum species complex were included. Branch support values were calculated using the approximate likelihood ratio test (aLRT) and are shown only if >70%. Branch lengths indicate sequence divergence. (c) Amino acid sequence alignment of HpaP from R. solanacearum GMI1000 (GenBank accession: CAB58249) and its homologue HpaCXcv 85‐10 (GenBank accession: CAJ22055). Sequences were aligned using MAFFT version 7 (http://mafft.cbrc.jp/alignment/server). Conserved residues are shaded in black. Numbers refer to the HpaP sequence. The red line indicates the localization of the T3S4 domain as described for HpaCXcv. Green lines underline the two deletions made in this study, corresponding to hpaP::hpaPΔ149–153 and hpaP::hpaP Δ156–160 mutants.
We looked for amino acid conservation between HpaP and other proteins of the YscP/FliK family, which have a T3S4 domain in their C‐terminus part. No significant identity could be uncovered between the HpaP protein of GMI1000 and homologues from Hrp1 T3SS plant‐pathogenic bacteria, such as HrpPPst (Morello and Collmer, 2009) or known T3S4 proteins from animal pathogenic bacteria (i.e. Yersina enterocolitica YscP) (Edqvist et al., 2003; Journet et al., 2003). Significant protein identity was identified only for homologues in Xanthomonas species, i.e. 31.0% for X. oryzae pv. oryzae MAFF311018 and 29.8% for X. axonopodis pv. citri 306. HpaP from R. solanacearum GMI1000 also shares low identity (27.6%) with the characterized T3S4 protein HpaCXcv 85‐10 (Lorenz et al., 2008; Schulz and Büttner, 2011) (Fig. 1a). A comparative genomic analysis shows that the organization of loci containing the hpaP/hpaCXcv genes is collinear. An alignment between HpaP and HpaCXcv using the MAFFT program (Katoh et al., 2002) highlighted that homology between these two proteins is mainly located in the C‐terminal region, which has been described to harbour the T3S4 domain (Lorenz et al., 2008) (Fig. 1c).
HpaP is a virulence factor on both A. thaliana and tomato plants
The hpaP mutation did not affect the growth of the bacterium in complete or minimal media (data not shown). A mutant strain complemented by the wild‐type allele was generated using a plasmid derived from a recently published construction allowing stable insertion in a permissive chromosomal site (Monteiro et al., 2012). We then root inoculated susceptible host plants, A. thaliana ecotype Col‐0 and Solanum lycopersicum cv. Marmande VR, with the GMI1000 strain, the hpaP mutant and the complemented strain, and the symptoms were scored daily. As shown previously (van Gijsegem et al., 2002), the hpaP mutant was hypovirulent on tomato. We validated this result by showing that this reduced virulence on tomato could be complemented by an ectopic copy of the hpaP gene (Fig. S1, see Supporting Information). We also showed that the hpaP mutant is strongly reduced in aggressiveness on A. thaliana, and this was also confirmed by complementation (Fig. 2).
Figure 2.
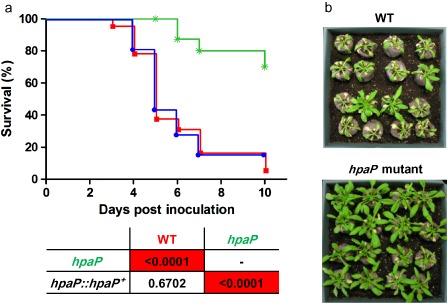
The hpaP mutant is strongly reduced in virulence on Arabidopsis thaliana. (a) Kaplan–Meier survival analysis of A. thaliana plants inoculated with Ralstonia solanacearum wild‐type strain (red squares), hpaP mutant (green stars) and the complemented strain hpaP::hpaP + (blue circles). Each strain was root inoculated on at least 16 A. thaliana plants. P values from Gehan–Breslow–Wilcoxon tests are shown below the graph. Red boxes indicate P < 0.01. Data were pooled from three independent experiments. (b) Representative photographs taken 6 days post‐inoculation. WT, wild‐type.
HpaP is not secreted or translocated via the T3SS and is able to self‐interact
As the T3S4 protein HrpPPst is translocated into plant cells (Morello and Collmer, 2009), we assayed the secretion and translocation of HpaP. First, the presence of HpaP‐HA was monitored after 8, 12 and 18 h of growth using a T3SS‐inducing minimal medium (MM) (Guéneron et al., 2000) (see Experimental procedures). Immunoblotting of the cell pellet and culture supernatant extracts using anti‐haemagglutinin (HA) antibody revealed that HpaP‐HA was produced in bacterial cells at all the sampling times, but not secreted in these conditions (Fig. 3a). PopP2 secretion was monitored as a positive secretion control to check that HpaP‐HA does not lead to an altered type III secretion in this strain (Fig. 3a). In order to verify whether HpaP could be translocated in planta, we infiltrated Nicotiana tabacum leaves using the wild‐type strain and a hrcV mutant (a T3SS impaired mutant) carrying the HpaP‐CyaA′ fusion protein, as described by Cunnac et al. (2004). We checked that the GMI1000 strain carrying HpaP‐CyaA′ was able to induce an HR on tobacco plants to make sure that the T3SS was still functional in this strain (data not shown). Plant leaf material was sampled 7 h after infiltration of the recombinant R. solanacearum strains and extracts were assayed for cyclic adenosine monophosphate (cAMP) quantification (Fig. 3b). Four independent biological replicates were made and no significant difference (paired t‐test, P = 0.13) was detected between the wild‐type strain and the hrcV mutant, showing that HpaP is not translocated via the T3SS.
Figure 3.
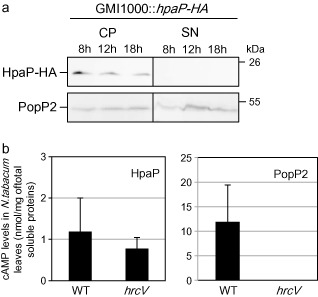
HpaP is not secreted in vitro or translocated in planta. (a) After 8, 12 and 18 h of culture in secretion medium, Ralstonia solanacearum wild‐type strain cell pellet (CP) and culture supernatant (SN) were analysed by immunoblotting. HpaP protein (22 kDa) was detected using anti‐haemagglutinin (anti‐HA) antibody. PopP2 was used as a positive control of the functional secretion of the strain used. This Western blot is representative of three independent replicates. (b) Translocation assay of HpaP and PopP2 proteins in Nicotiana tabacum leaves using HpaP‐CyaA′ and PopP2‐CyaA′ (positive control) fusion proteins. Cyclic adenosine monophosphate (cAMP) levels were detected using cAMP Biotrak competitive enzyme immunoassay. Four independent biological replicates were made. WT, wild‐type.
We then investigated the ability of HpaP to self‐interact, in a similar manner to that which was described for the HpaCXcv T3S4 protein (Büttner et al., 2006). We performed in vitro glutathione S‐transferase (GST) pull‐down assays using the HpaP full‐length protein. The GST‐HpaP fusion protein was expressed in Escherichia coli and immobilized on glutathione sepharose before incubation with an E. coli lysate expressing 6His‐HpaP (His, histidine). Eluates containing bound proteins were then analysed using GST and 6His‐specific antibodies. We were able to detect 6His‐HpaP in the presence of GST‐HpaP, but not with GST alone, indicating that HpaP is able to self‐interact, in dimeric or oligomeric forms (Fig. 4).
Figure 4.
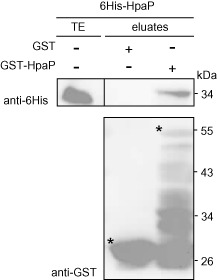
HpaP is able to self‐interact in vitro. Glutathione S‐transferase (GST) and GST‐HpaP were immobilized on glutathione sepharose and incubated with an Escherichia coli lysate containing 6His‐HpaP (His, histidine). Total cell lysates (TE) and eluted proteins (eluates) were analysed using antibodies directed against GST and the 6His epitope. Bands corresponding to GST and GST fusion proteins are marked by asterisks; lower bands represent degradation products. The experiment was repeated three times with similar results.
HpaP modulates the secretion of PopP1, AvrA and HrpY, but is dispensable for effector translocation in plant cells
In order to assay the potential role of HpaP in regulating the secretion/translocation of T3Es, we first analysed the secretion of several T3Es in the wild‐type and hpaP mutant strains. The T3SS‐defective mutant hrcV was used as a control (Table 1; Figs 5 and S2, see Supporting Information). Four previously well‐characterized R. solanacearum T3Es were selected for the assay: PopP1 (Lavie et al., 2002; Poueymiro et al., 2009), PopP2 (Deslandes et al., 2003), AvrA (Poueymiro et al., 2009; Turner et al., 2009) and GALA7 (Angot et al., 2006). Strains were grown in secretion medium for 8, 12 and 18 h, and cell pellets and culture supernatants were harvested at each time point before analysing the protein extracts by immunoblotting using specific anti‐T3E antibodies (see Experimental procedures). All the T3Es were detected in the cell pellet extracts without significant variation along the sampling time course and among bacterial strains. This indicates that HpaP does not seem to play a role in the production of these T3Es. The four T3Es were detected in the culture supernatant of the wild‐type strain and of the hpaP mutant (but not in the hrcV mutant). A quantification of the Western blot signal, followed by a statistical analysis (analysis of variance, ANOVA), showed no significant difference in secretion between the wild‐type strain and the hpaP mutant for GALA7 (P = 0.55) or PopP2 (P = 0.37). However, these analyses revealed that AvrA and PopP1 were more (P = 0.026) and less (P = 0.004) secreted, respectively, in the hpaP mutant compared with the wild‐type strain. As a control, the decrease in PopP1 secretion was complemented in the hpaP::hpaP + strain (Fig. S3, see Supporting Information). We also quantified the secretion of the R. solanacearum HrpY pilus protein, and observed that the HrpY signal was increased in the hpaP mutant cell pellets (P = 10−4) (Fig. S2) and supernatants (P = 0.009) compared with the wild‐type strain. This enhanced secretion of HrpY in the hpaP mutant significantly increased over time (P = 0.004) (Fig. 5 and Fig. S2), and therefore suggested that T3E secretion modulation in the hpaP mutant was not caused by a lack of type III pilin assembly.
Table 1.
Strains and plasmids used in this study
| Relevant characteristics* | Source | |
|---|---|---|
| Plasmids used for cloning | ||
| pENTR/SD/D‐TOPO | Gateway™ entry vector, Shine–Dalgarno sequence, Kmr | Invitrogen (Carlsbad, CA, USA) |
| pDONR207 | Gateway™ entry vector, Gmr Cmr | Invitrogen (Carlsbad, CA, USA) |
| pGEX‐GW | Gateway™ expression vector, N‐terminal GST fusion, Apr | Invitrogen (Carlsbad, CA, USA) |
| pTH19‐GW | Gateway™ expression vector, N‐terminal 6His tag, Apr | Invitrogen (Carlsbad, CA, USA) |
| pNP78 | pENTR/SD/D‐TOPO containing KQGS* between AttL1 and AttL2 sites, Kmr | This study |
| pGBG |
Matchmaker™ pGBKT7 yeast expression vector made Gateway™ compatible, N‐terminal BD fusion, Kmr, Cmr |
L. Deslandes (unpublished) |
| pGAD |
Matchmaker™ pGADT7 yeast expression vector made Gateway™ compatible, N‐terminal AD fusion, Apr, Cmr |
L. Deslandes (unpublished) |
| pLBy10 | pDONR207 derivative carrying hpaP, Gmr | This study |
| pMP17 | pDONR207 derivative carrying AvrA, Gmr | This study |
| pNP200 | pDONR207 derivative carrying GALA7, Gmr | This study |
| pDL154 | pLBy10 derivative carrying hpaP deletion codons 149–153, Gmr | This study |
| pDL152 | pLBy10 derivative carrying hpaP deletion codons 156–160, Gmr | This study |
| pNP260 | pRCG derivative (Monteiro et al., 2012) with the GALA7 promoter followed by a Gateway™ destination cassette and a single HA epitope tag, Gmr Kmr | This study |
| pNP329 | pRCG derivative (Monteiro et al., 2012) with the GALA7 promoter followed by a Gateway™ destination cassette and a triple HA epitope tag, Gmr Kmr | This study |
| pSC205 | pET26B(+) derivative carrying a Gateway cassette, Kmr | This study |
| pMT1 | pSC205 derivative carrying CyaA 4–1197 N‐terminal part, Kmr | This study |
| pCM351 | Allelic exchange vector, Apr, Gmr, Tcr | Marx and Lidstrom (2002) |
| pAM5 | pLAFR3 carrying 2‐kb fragment containing hrpB, Tcr | Guéneron et al. (2000) |
| Final vectors introduced in Ralstonia solanacearum by natural transformation | ||
| pGG18 | pCM351 carrying upstream and downstream DNA fragments of the hpaP gene | This study |
| pGGL7 | pNP260 derivative carrying hpaP, Gmr Kmr | This study |
| pDL185 | pNP329 derivative carrying hpaP deletion codons 149–153, Gmr | This study |
| pDL186 | pNP329 derivative carrying hpaP deletion codons 156–160, Gmr | This study |
| pMT2 | pMT1 derivative carrying hpaP, Kmr | This study |
| pMT3 | pMT1 derivative carrying PopP1, Kmr | This study |
| pMT4 | pMT1 derivative carrying PopP2, Kmr | This study |
| pMT5 | pMT1 derivative carrying AvrA, Kmr | This study |
| pMT6 | pMT1 derivative carrying GALA7, Kmr | This study |
| Final vectors used for yeast two‐hybrid assays | ||
| pDL49 | pGAD derivative carrying hpaP, Apr | This study |
| pDL158 | pGAD derivative carrying hpaP deletion codons 149–153, Apr | This study |
| pDL159 | pGAD derivative carrying hpaP deletion codons 156–160, Apr | This study |
| pLD1 | pGBG derivative carrying PopP1, Kmr | This study |
| pLD2 | pGBG derivative carrying PopP2, Kmr | This study |
| pDL2 | pGBG derivative carrying AvrA, Kmr | This study |
| pNP210 | pGBG derivative carrying GALA7, Kmr | This study |
| Final vectors used for pull‐down assays | ||
| pDL188 | pGEX‐GW derivative carrying KQGS*, Apr | This study |
| pDL189 | pGEX‐GW derivative carrying hpaP, Apr | This study |
| pDL171 | pGEX‐GW derivative carrying hpaP deletion codons 149–153, Apr | This study |
| pDL172 | pGEX‐GW derivative carrying hpaP deletion codons 156–160, Apr | This study |
| pDL190 | pTH19‐GW derivative carrying hpaP, Apr | This study |
| pDL191 | pTH19‐GW derivative carrying PopP1, Apr | This study |
| Yeast strain | ||
| AH109 | Matchmaker™ yeast strain | BD Biosciences, Palo Alto, CA, USA |
| Escherichia coli strain | ||
| BL21(DE3) | F– ompT gal dcm lon hsdSB(rB – mB –) | Invitrogen, Life Technologies, Carlsbad, CA, USA |
| R. solanacearum strains | ||
| GMI1000 | Wild‐type strain | Salanoubat et al. (2002) |
| GMI1415 | hpaP::Tn5B20 mutant, Kmr | Gough et al. (1993) |
| GMI1694 | hrcV ::Ω mutant, Spr | Cunnac et al. (2004) |
| GRS747 | GMI1000ΔhpaP mutant, Gmr | This study |
*Apr, Cmr, Gmr, Kmr, Tcr and Spr, resistances to ampicillin, chloramphenicol, gentamycin, kanamycin, tetracycline and spectinomycin, respectively; AD, activation domain; BD, binding domain; GST, glutathione S‐transferase; His, histidine.
Figure 5.
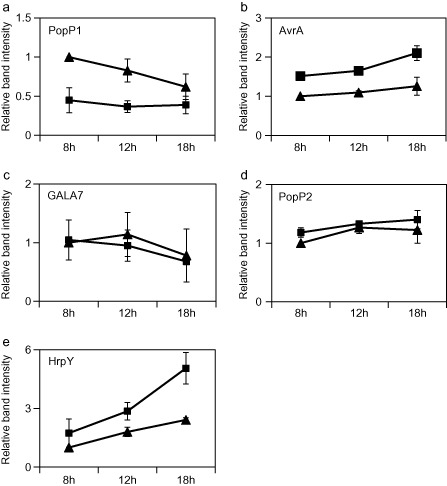
HpaP modulates the secretion of AvrA, PopP1 and HrpY. Ralstonia solanacearum GMI1000 wild‐type strain (triangles) and hpaP mutant (squares) were cultivated in secretion medium. Culture supernatants were harvested after 8, 12 and 18 h of culture and analysed by immunoblotting. PopP1 (a), AvrA (b), GALA7 (c) and PopP2 (d) type III effectors, and HrpY pilin (e), were detected using their respective antibodies for three independent biological replicates. Band intensities for all conditions were quantified using GeneTools (Syngene, Cambridge, UK). For each protein, the band intensity corresponding to the wild‐type strain after 8 h of culture was used as an internal reference. For other time points of the wild‐type and of the hpaP mutant, ratios of band intensities compared with this reference were calculated. Means and standard errors at each time point were plotted on the graphs and correspond to the measurement of three independent biological replicates.
We then looked for the in planta translocation of AvrA, GALA7, PopP1 and PopP2 by infiltrating N. tabacum leaves with the wild‐type strain and hpaP and hrcV mutants, each expressing T3E‐CyaA′ fusion proteins (see Experimental procedures). In order to monitor comparative translocation assays, all T3E‐CyaA′ fusion proteins were integrated by single homologous recombination at each T3E locus, resulting in reporter fusions expressed by the cognate T3E promoters. In planta cAMP levels were measured in four independent biological replicates (Table 2). Considering that the amount of cAMP produced by the T3E‐CyaA′ fusion proteins in the four replicates could vary by more than 50%, as reported previously (Schechter et al., 2004), we calculated the ratio of cAMP production in the two mutants versus the wild‐type strain after normalizing each cAMP quantification within each biological replicate as a percentage of the wild‐type cAMP production (Table 2). All four T3Es are translocated in planta as they were detected at a significantly higher level in the wild‐type and hpaP mutant compared with the hrcV mutant, indicating that their translocation in planta was not dependent on HpaP. Ratios of translocated proteins in the wild‐type compared with the hpaP mutant were calculated. No difference was detected between the wild‐type and hpaP mutant, except for PopP1, whose translocation was reduced for all replicates, with an average translocation of 48% in the hpaP mutant compared with the wild‐type (Table 2, in bold).
Table 2.
Translocation in planta is not dependent on HpaP. Translocation assay of type III effectors (T3Es) AvrA, GALA7, PopP1 and PopP2 in Nicotiana tabacum leaves using T3E‐CyaA′ fusion proteins in the wild‐type strain (WT), hpaP mutant and hrcV mutant (a type III secretion system‐defective mutant). Cyclic adenosine monophosphate (cAMP) levels (nmol cAMP/mg of total soluble proteins) were measured using the cAMP Biotrak competitive enzyme immunoassay in four independent biological replicates. Average ratios and standard errors of translocated protein in the wild‐type compared with the hpaP mutant were calculated for the four replicates
| WT | hrcV/WT | hpaP/WT | |
|---|---|---|---|
| AvrA | 100 | 0.04 ± 0.03 | 183 ± 149 |
| GALA7 | 100 | 0.09 ± 0.07 | 126 ± 132 |
| PopP1 | 100 | 0.03 ± 0.02 | 48 ± 28 |
| PopP2 | 100 | 1 ± 1.5 | 104 ± 63 |
HpaP interacts specifically with PopP1
We then examined whether HpaP could physically interact with the T3Es analysed in the secretion/translocation studies. Yeast two‐hybrid assays using the binding domain (BD) and activation domain (AD) of the transcription factor Gal4 were performed. In the case of an interaction, cells were able to grow on a selective MM lacking histidine (synthetic dropout medium lacking leucine, tryptophan and histidine, SD/–Leu/–Trp/–His). When required, we added to the selective medium the minimal concentration of 3‐amino‐1,2,4‐triazole (3‐AT) (see Experimental procedures) required to suppress the interactions between baits (BD‐T3E) and the prey AD‐T‐antigen used as a negative control. Among the four T3Es tested, HpaP was found to interact only with PopP1 (Fig. 6a). To confirm this interaction, a GST‐HpaP fusion protein was expressed in E. coli and immobilized on glutathione sepharose before incubation with an extract from E. coli expressing 6His‐PopP1. Bound proteins were eluted and analysed using anti‐GST and anti‐6His antibodies. 6His‐PopP1 was detected only in the presence of GST‐HpaP and not with GST alone, confirming the interaction between HpaP and PopP1 (Fig. 6b).
Figure 6.
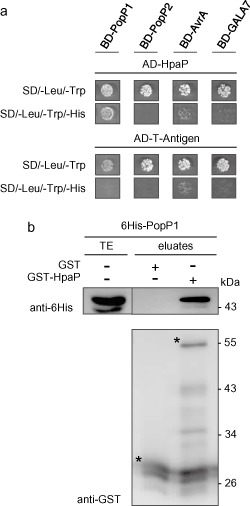
HpaP interacts specifically with the type III effector (T3E) PopP1. (a) Interaction studies between HpaP and several T3Es. Yeast cells were co‐transformed by AD‐HpaP and BD‐PopP1, BD‐PopP2, BD‐AvrA or BD‐GALA7. Double transformation and interaction were tested by plating yeasts on synthetic dropout medium lacking leucine and tryptophan (SD/–Leu/–Trp) and synthetic dropout medium lacking leucine, tryptophan and histidine (SD/–Leu/–Trp/–His), respectively. 3‐Aminotriazole (3‐AT) was added to suppress autoactivation when necessary. Three biological replicates were performed giving the same results. (b) In vitro validation of the interaction between HpaP and PopP1. Glutathione S‐transferase (GST) and GST‐HpaP were immobilized on glutathione sepharose and incubated with an Escherichia coli lysate containing 6His‐PopP1. Total cell lysates (TE) and eluted proteins (eluates) were analysed using antibodies directed against GST and the 6His epitope. Bands corresponding to GST and GST fusion proteins are marked by asterisks; lower bands represent degradation products. Experiments were repeated twice with similar results. AD, activation domain; BD, binding domain; His, histidine.
Two deletions of five amino acids in the T3S4 domain of HpaP do not abolish self‐interaction or interaction with PopP1, but are essential for HpaP function
In order to characterize the contribution of the T3S4 domain in HpaP function, we generated two deletions of five amino acids in the most conserved regions between HpaP and HpaCXcv (Schulz and Büttner, 2011) (Fig. 1c). We analysed the involvement of these regions in the ability of HpaP to self‐interact or to interact with PopP1. GST pull‐down assays were performed using the two HpaP derivatives (GST‐HpaPΔ149–153 and GST‐HpaPΔ156–160) employing anti‐GST and anti‐6His antibodies. Equal amounts of 6His‐HpaP were detected with either HpaP or the deleted versions (Fig. 7a), indicating that these conserved amino acids in the T3S4 domain of HpaP are dispensable for its self‐interaction. We then examined whether the T3S4 deletion derivatives of HpaP were still able to bind to PopP1 in the yeast two‐hybrid assay. Cells were co‐transformed with BD‐PopP1 and AD‐HpaPΔ149–153 or AD‐HpaPΔ156–160, and plated on the different minimal media. AD‐HpaP derivatives required 3‐AT to suppress the interactions with the negative control bait BD‐p53. We detected an interaction between PopP1 and the two HpaP deleted versions, indicating that the two deletions of five amino acids did not alter the capacity of HpaP to bind PopP1 (Fig. 7b). We also tested whether the two deletions designed in the HpaP T3S4 domain had an impact on PopP1 secretion. The hapP mutant was complemented with the two hpaP T3S4 deletion derivatives (strains hpaP::hpaPΔ149–153 and hpaP::hpaPΔ156–160) using the same stable chromosomal insertion system (see Experimental procedures). We checked that the mutated proteins were indeed produced in the complemented strains at the same level as in the wild‐type construct (Fig. S4, see Supporting Information). Interestingly, secretion of PopP1, but not PopP2, was highly reduced at all the time points tested in the supernatants of both strains carrying hpaP deletion derivatives compared with the wild‐type version (Fig. 8a). Finally, these two strains were assayed for their pathogenicity on tomato and A. thaliana. Whatever the host, hpaP::hpaPΔ149–153 and hpaP::hpaPΔ156–160 strains behaved similarly to the hpaP mutant on tomato (Fig. 8b) and on A. thaliana (Fig. 8c), showing a dramatic decrease in their aggressiveness compared with the wild‐type (P < 0.0001); this suggests that both regions of five amino acids in the putative C‐terminal T3S4 domain of HpaP are involved in the efficient secretion of PopP1, and are required for R. solanacearum pathogenicity.
Figure 7.
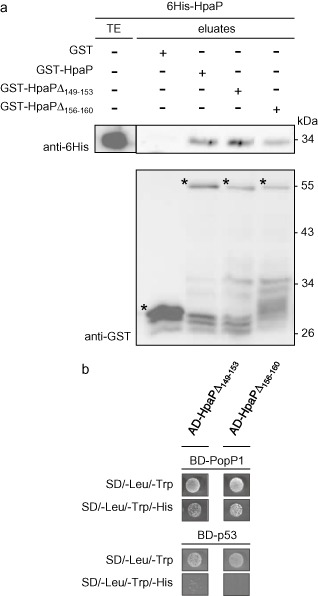
Interaction studies with HpaP deletion derivatives mutated in the type III secretion substrate specificity switch (T3S4) domain. (a) Deletions in the T3S4 domain do not abolish the self‐interaction of HpaP. Immobilized glutathione S‐transferase (GST), GST‐HpaP, GST‐HpaPΔ149–153 and GST‐HpaPΔ156–160 were incubated with 6His‐HpaP. Total cell lysates (TE) and eluted proteins (eluates) were analysed using antibodies directed against GST and the 6His epitope. Bands corresponding to GST and GST fusion proteins are marked by asterisks; lower bands represent degradation products. (b) Deletions in the T3S4 domain do not abolish interaction between HpaP and the type III effector PopP1. Yeast cells were co‐transformed by BD‐PopP1 and either AD‐HpaPΔ149–153 or AD‐HpaPΔ156–160. Double transformation and interaction were tested by plating yeasts on synthetic dropout medium lacking leucine and tryptophan (SD/–Leu/–Trp) and synthetic dropout medium lacking leucine, tryptophan and histidine (SD/–Leu/–Trp/–His), respectively. 3‐Aminotriazole (3‐AT) was added to suppress autoactivation. Experiments in (a) and (b) were repeated twice with similar results. AD, activation domain; BD, binding domain; His, histidine.
Figure 8.
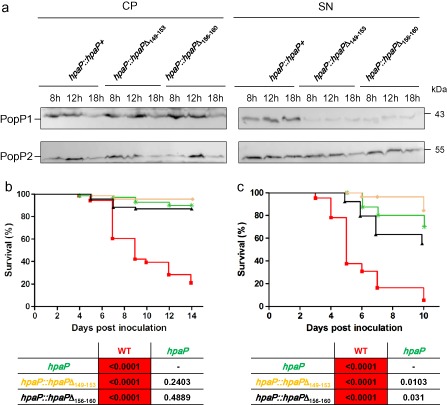
hpaPΔ149–153 and hpaPΔ156–160 mutants behaved similarly to the hpaP mutant. (a) PopP1 is specifically less secreted in both hpaP deletion derivative mutants (hpaP::hpaPΔ149–153 and hpaP::hpaPΔ156–160) compared with the wild‐type version (hpaP::hpaP +). Ralstonia solanacearum strains were cultivated in secretion medium, cell pellets (CP) and culture supernatants (SN) were harvested after 8, 12 and 18 h of culture and analysed by immunoblotting. Effector proteins PopP1 (43 kDa) and PopP2 (53 kDa) were detected using their respective antibodies. Two independent biological replicates were made with similar results. Both hpaP deletion mutants are strongly reduced in virulence on tomato (b) and Arabidopsis thaliana (c). Kaplan–Meier survival analysis of plants inoculated with R solanacearum wild‐type strain (WT) (red squares), hpaP mutant (green stars) and the deletion mutants hpaP::hpaPΔ149–153 (yellow diamonds) and hpaP::hpaPΔ156–160 (black triangles). Each strain was root inoculated on 24 tomato and at least 16 A. thaliana plants. P values from Gehan–Breslow–Wilcoxon tests are shown below each graph. Red boxes indicate P < 0.01. Data were pooled from three independent experiments.
Discussion
T3SS‐dependent pathogenesis of R. solanacearum appears to be extremely complex, notably as a result of the large repertoire of T3Es of the bacterium and the functional redundancy (Remigi et al., 2011; Sole et al., 2012). The study of the key proteins putatively involved in the regulatory mechanisms linked to the secretion/translocation of T3Es is therefore an attractive way to identify sets of T3Es potentially required for disease establishment. Little work is available on such type III regulators in R. solanacearum, as only the putative class IB chaperone, HpaB, was demonstrated to be required for the translocation of 66 T3Es in strain RS1000 (Mukaihara et al., 2010), leading to an hrp– phenotype of the hpaB mutant (Mukaihara et al., 2004). HpaP was originally described as an hrp‐associated protein involved in R. solanacearum pathogenicity towards tomato plants (van Gijsegem et al., 2002; Gough et al., 1993). HpaP does not have the characteristics of a chaperone sensu stricto as described by Parsot et al. (2003). Indeed, HpaP is larger (22 kDa) than most chaperones (c. 15 kDa), and is unusually rich in proline residues (Gough et al., 1993). In this study, we validated the strong impact of the hpaP mutation on the aggressiveness of R. solanacearum on tomato plants, and also highlighted the effect of the same mutant on A. thaliana. As we showed that HpaP is not itself secreted or translocated into plant cells, and modulates the secretion of type III substrates, we hypothesize that HpaP has an indirect role in the contribution to the pathogenicity of R. solanacearum towards tomato and A. thaliana. We also showed that HpaP could self‐interact, as its homologue HpaCXcv (Büttner et al., 2006). Interestingly, Büttner et al. (2006) showed that HpaCXcv could interact with a Xanthomonas chaperone, HpaBXcv, and that both proteins could form homo‐ and hetero‐oligomeric complexes promoting the secretion of the same set of T3Es. As a hpaBXcv orthologue exists in R. solanacearum, also named hpaB, it would be worth investigating whether a functional HpaP–HpaB interaction also occurs in R. solanacearum.
The production of the HrpY pilin of R. solanacearum was found to be more abundant in the supernatant of an hpaP mutant. However, this variation in the pilin subunit secretion does not lead to a hrp– phenotype, whereas it does so for two YscP/FliK‐related proteins, HrpPPst and Yersinia spp. YscP. The latter controls the needle length of the T3SS, preventing effector secretion until the needle is completely formed (Agrain et al., 2005; Journet et al., 2003). Thus, the yscP mutant produces needles of indeterminate length and is unable to secrete T3Es efficiently (Edqvist et al., 2003; Journet et al., 2003). The hrpPPst mutant is strongly affected in its pathogenicity, as no effectors are secreted and only a very small amount of the pilin subunit is secreted (Morello and Collmer, 2009). In our study, the putative over‐secretion of HrpY by the hpaP mutant does not lead to a general over‐secretion of the type III substrates. Indeed, our data show that some T3Es do not seem to be deregulated by the hpaP mutant in their secretion/translocation (PopP2 and GALA7), whereas the secretion/translocation of other T3Es appears to be modulated (PopP1 and AvrA). We observed that PopP1 and AvrA were less and more secreted, respectively, in an hpaP mutant compared with the wild‐type strain (Fig. 5). Although the over‐secretion of AvrA apparently does not lead to an increased translocation in planta, the quantitative lower secretion of PopP1 in the hpaP mutant correlates with a lower translocation in planta (Table 2). It is tempting to speculate that this decreased secretion/translocation of PopP1 results from a regulation of PopP1 delivery through a direct HpaP–PopP1 interaction (Fig. 6). This regulation appears to be specific to PopP1, as neither differential secretion/translocation nor direct interaction was identified with PopP2, which belongs to the same YopJ/AvrRxv family. How direct or indirect interactions of HpaP with PopP1 and AvrA, respectively, promote their differential secretion remains to be addressed. To date, no link between secretion/translocation control by a protein of the YscP/FliK family and a direct interaction with T3Es could be inferred. Büttner et al. (2006) showed that secretion was abolished for four X. campestris pv. vesicatoria T3Es (AvrBs3, XopC, XopJ and XopF1), and HpaCXcv has been shown to be specifically involved in XopJ and XopF1 translocation, whereas AvrBs3 and XopC translocation is HpaCXcv independent. Direct interaction studies have also shown that HpaCXcv is able to bind to AvrBs3 and XopF1, two T3Es with different translocation behaviours. Secretion of the HrpEXcv pilin was not affected in the absence of HpaCXcv(Büttner et al., 2006). Lorenz et al. (2008) postulated a type III secretion hierarchy in X. campestris pv. vesicatoria between early (HrpB2, a protein required for pilus assembly) and later (translocon and T3E proteins) T3SS substrates. Our finding that HrpY is significantly over‐produced in the cell pellets in the absence of HpaP suggests a control of HpaP on HrpY production or stability. HrpY is also over‐secreted in the hpaP mutant, with a regular increase over time. This could be linked to a switch model, where HpaP acts temporally by inhibiting HrpY secretion at later time points when the pilus is formed. HpaP therefore appears to control the secretion of at least three T3SS substrates, including early (HrpY) and late (AvrA and PopP1) substrates.
To date, we still do not know whether there is an order in the delivery of R. solanacearum T3Es, but this study shows that HpaP is a key player in pathogenicity on diverse hosts, potentially through the quantitative regulation of T3E delivery into plant cells. It also suggests that T3S4 homologue proteins have developed different modes of action, which is also supported by their relatively low level of conserved identity at the protein level compared with T3SS structural components (Fig. 1). The fact that HpaP and HpaCxcv proteins are not secreted indicates that they do not act as molecular ruler proteins, as do T3S4 proteins from animal pathogenic bacteria. It is noteworthy that the highest identity between both proteins is located in the C‐terminal part, defined as the putative T3S4 domain. This domain was initially described in animal pathogenic bacteria as required to switch substrate specificity for T3E export when T3SS assembly is completed (Agrain et al., 2005). This study identified two sequences of five amino acids in the T3S4 domain of HpaP in R. solanacearum required for pathogenicity. Indeed, although the HpaP deletion derivatives are still able to interact with the HpaP wild‐type protein or with PopP1 (Fig. 7), we demonstrated that both hpaP deletion derivatives cannot complement the hpaP disruption mutant phenotype for efficient PopP1 secretion or for disease establishment on tomato plants or A. thaliana (Fig. 8). Büttner et al. (2006) showed that the hpaCXcv mutant displayed reduced disease symptom formation and reduced HR on susceptible and resistant pepper, respectively. Moreover, Schulz and Büttner (2011) demonstrated that the same two sequences of five amino acids were involved in these reduced responses. Thus, HpaP and HpaCxcv proteins contribute significantly to bacterial pathogenicity on their respective hosts, and the conserved sequences in the putative T3S4 domains appear to be essential in this process. This work also shows that the HpaP T3S4 sequences have distinct functional roles, being dispensable for the HpaP–PopP1 interaction, but necessary for HpaP‐dependent PopP1 secretion. We have not yet been able to connect the plant phenotypes observed with the hpaP‐deleted mutants with its control over T3E translocation. Our hypothesis is that, beyond PopP1, HpaP could control the delivery of other T3Es, collectively resulting in the strong hypoaggressive phenotype of the hpaP mutant. Thus, to advance further in the functional characterization of HpaP, a global comparison of the secretomes of the wild‐type strain and the hpaP mutant could help to identify the sets of T3Es whose secretion/translocation is controlled in part by HpaP, and their collective involvement for disease establishment.
Experimental Procedures
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this work are listed in Table 1. Escherichia coli strains were grown at 37 °C in Luria–Bertani medium (Ausubel et al., 1989). Ralstonia solanacearum strains were grown in complete B medium or MM, as described by Poueymiro et al. (2009). When needed, antibiotics were added at the following final concentrations (mg/L): kanamycin (25), gentamycin (10), ampicillin (50) and chloramphenicol (25) for E. coli; kanamycin (50), spectinomycin (40), gentamycin (10) and tetracycline (10) for R. solanacearum. The plasmids used in this study were constructed by Gateway technology (Invitrogen, Carlsbad, CA, USA) following the instructions of the manufacturer.
Phylogenetic analysis
Construction of the phylogenetic tree was based on sequence similarities using the maximum likelihood principle (Guindon et al., 2009) via the phylogeny.fr platform (Dereeper et al., 2008). Sequences were first aligned using the MAFFT program and edited using GBlocks (Castresana, 2000). We selected 10 sequenced strains that belonged to all phylotypes: the reference strain GMI1000 (Salanoubat et al., 2002) from phylotype I, Molk2 (C. Boucher and S. Genin, unpublished data) from phylotype IIa; IPO1609 (Guidot et al., 2009), CFBP2957 (Remenant et al., 2010), UW551 (Gabriel et al., 2006) and Po82 (Xu et al., 2011) from phylotype IIb; CMR1594 (Remenant et al., 2010) from phylotype III; and PSI07 (Remenant et al., 2010), Blood Disease Bacterium (BDB) strain R229 and Ralstonia syzygii strain R24 (Remenant et al., 2011) from phylotype IV.
Creation of a hpaP deletion strain, hpaP and hpaPΔT3S4 complementation constructs
A hpaP deletion mutant was generated through a double recombination event using the pCM351 cre‐lox vector base (Marx and Lidstrom, 2002) carrying upstream and downstream DNA fragments of the hpaP gene and named pGG18 (primers in Table S1, see Supporting Information). This construct was used to transform strain GMI1000 through natural transformation as described by Gonzalez et al. (2011), thus resulting in the ΔhpaP strain GRS747.
The GMI1415 hpaP mutant was complemented with either a wild‐type hpaP copy or two independent T3S4 domain deletion variants. The hpaP wild‐type sequence contained in the Gateway™ entry vector pLBy10 was cloned into pNP260. Linearized plasmid was then integrated at a defined bacterial chromosome site (Monteiro et al., 2012) into the naturally competent hpaP mutant, as described by Gonzalez et al. (2011) (hpaP::hpaP +). To generate the hpaP T3S4 deletion derivatives, pLBy10 was employed as a template for polymerase chain reaction (PCR) amplification to generate deletions using the QuikChange II XL Site‐Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). Independent deletions were introduced between amino acids 149–153 and 156–160 (primers in Table S1). Chromosomal insertions of the hpaP deletion derivatives cloned into pNP329 were integrated into the hpaP mutant (hpaP::hpaPΔ149–153 and hpaP::hpaPΔ156–160) by natural transformation as described above.
Plant assays and statistical analysis
Twenty‐four tomato plants (Solanum lycopersicum L. cultivar Super Marmande VR) were inoculated with 40 mL of a bacterial suspension containing 108 bacteria/mL, as described by Gonzalez et al. (2011). For the A. thaliana Col‐0 ecotype, 16 plants were inoculated with 108 bacteria/mL, as described by Deslandes et al. (1998), without cutting the roots. Symptom appearance was scored daily and independently for each plant, using a macroscopic scale describing the observed wilting: 0, no wilting; 1, 25% of leaves wilted; 2, 50%; 3, 75%; 4, complete wilting. For subsequent analysis, the data were transformed into a binary index: 0, less or equal to 50% leaves wilted; 1, >50% wilted leaves. The disease developments of the two given strains were compared using Kaplan–Meier survival analysis (Bland and Altman, 1998), with the Gehan–Breslow–Wilcoxon method to compute the P value to test the null hypothesis of identical survival experience of the tested strains. A P value lower than 0.01 was considered to be significant. Each pathogenicity test was repeated three times, and statistical analyses were performed with Prism version 5.0 (GraphPad Software, San Diego, CA, USA).
In vitro secretion assays and immunoblot analysis
GMI1000 wild‐type strain, GMI1694 hrcV mutant and GMI1415 hpaP mutant, carrying the pAM5 plasmid (Guéneron et al., 2000) that leads to a higher transcriptional activity of T3SS‐regulated genes (Table 1), were first cultivated in B medium in overnight cultures, pelleted for 5 min at 2700 g and then resuspended in MM at an optical density at 600 nm (OD600nm) of 0.2. MM bacteria cultures were then harvested after 8, 12 and 18 h of culture, set at the same concentration and pelleted for 10 min at 4300 g. Pelleted cells and supernatants were then treated as described by Poueymiro et al. (2009). To compare the secretion of T3Es and HrpY pilin between strains, equal amounts of protein were loaded onto a sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel, followed by electrophoresis and transfer to Protran BA85 nitrocellulose membranes (Whatman, Maidstone, UK). Antibodies used for Western blotting were AvrA (Poueymiro et al., 2009), PopP1 (Lavie et al., 2002) and PopP2 (courtesy of L. Deslandes, INRA/CNRS, Castanet Tolosan, France). The GALA7 polyclonal rabbit antibody was generated by a classical immunization protocol (GeneCust Europe, Dudelange, Luxembourg) with the full‐length GALA7 protein purified from E. coli as a 6His‐tagged protein. By using R. solanacearum single gala mutants and E. coli over‐expressing recombinant GALA6 and GALA7, it was shown that this antibody is specific to GALA7 and cannot recognize its closest paralogue GALA6. GALA7 antibody was used at 1:10 000 dilution. Goat anti‐rabbit antibody conjugated with horseradish peroxidase was used as secondary antibody (1:50 000) (Santa Cruz Biotechnology, Santa Cruz, CA, USA). At least three independent biological replicates were made. The same experiments were performed using hpaP::hpaP +, hpaP::hpaPΔ149–153 and hpaP::hpaPΔ156–160 carrying the pAM5 plasmid. The experiment was repeated twice.
Adenylate cyclase (CyaA) translocation assay
Translocation assays were carried out mostly as described by Poueymiro et al. (2009). In order to allow the insertion of the N‐terminal part of the CyaA gene (CyaA′ = CyaA 4–1197) at the locus of each protein studied, CyaA′ was amplified (primers in Table S1) and cloned into pSC205 plasmid. hpaP and the T3Es studied (AvrA, GALA7, PopP1 and PopP2) were then integrated into pMT1. HpaP‐CyaA′ and T3E‐CyaA′ constructs were integrated into naturally competent strains (GMI1000 wild‐type strain, GMI1694 hrcV mutant and GRS747 hpaP mutant). Bacteria carrying the protein‐CyaA′ fusion were grown overnight at 28 °C in B medium supplemented with half doses of antibiotics, pelleted for 5 min at 2700 g and resuspended in sterile water at OD600nm = 0.1. Bacteria were infiltrated into N. tabacum cv. Bottom Special leaves, tissues were harvested after 7 h and frozen in liquid nitrogen. cAMP dosage was carried out using an Amersham cAMP Biotrak Enzyme immunoassay (EIA) kit by GE Healthcare (Little Chalfont, Buckinghamshire, UK). Four independent biological replicates were performed for each protein translocation tested.
Yeast two‐hybrid analysis
Yeast two‐hybrid analysis was performed using a Matchmaker GAL4 Two‐Hybrid System 3 (Clontech, Mountain View, CA, USA) according to the manufacturer's instructions. The generation of plasmids carrying BD or AD fusions was performed by Gateway™ LR recombination using pGBG and pGAD. Saccharomyces cerevisiae strain AH109 was co‐transformed with BD and AD fusion‐containing vectors and plated onto low‐stringency minimal SD medium (lacking leucine and tryptophan) to select for the presence of both plasmids. Double transformants were tested for interaction by spotting six successive 10‐fold dilutions onto minimal SD/–Leu/–Trp/–His medium. 3‐AT was added to suppress autoactivation by the bait protein when necessary (0.5, 1, 2 and 5 mm 3‐AT with GALA7, HpaPΔ derivatives, PopP2 and AvrA, respectively). Co‐transformation of these two plasmids with our constructs was also performed to verify the specificity of interaction. Each interaction test was performed on four different colonies. Two to three biological repetitions were performed for each interaction tested.
GST pull‐down assays
GST and GST fusion proteins were synthesized in E. coli BL21(DE3). Bacterial cells from 50‐mL cultures were resuspended in phosphate‐buffered saline (PBS), pH 7.4, and sonicated 10 times for 15 s at 4 °C by an Analog Sonifier Cell Disruptor (Branson, Danbury, CT, USA). Insoluble cell debris was removed by centrifugation at 16 000 g for 20 min at 4 °C, and soluble GST and GST fusion proteins were immobilized on a glutathione sepharose matrix for 1 h at room temperature. Unbound proteins were removed by washing twice with cold PBS containing 0.1% Triton X‐100. Glutathione sepharose matrix was then incubated with E. coli lysates containing the 6His fusion proteins for 1 h at room temperature. Unbound proteins were removed by washing four times with cold PBS containing 0.1% Triton X‐100, and bound proteins were eluted twice with 10 mm of reduced glutathione for 15 min at room temperature. Two microlitres of input protein lysate and 20 μL of eluted proteins were analysed by 10% SDS‐PAGE and immunoblotting using anti‐GST and anti‐6His horseradish peroxidase‐labelled antibodies. Antibody reactions were visualized by enhanced chemiluminescence (GE Healthcare). Experiments were repeated at least twice.
Supporting information
Fig. S1 The hpaP mutant is strongly reduced in virulence on tomato. Kaplan–Meier survival analysis of tomato plants inoculated with Ralstonia solanacearum wild‐type strain (red squares), hpaP mutant (green stars) and the complemented strain hpaP::hpaP + (blue circles). Each strain was root inoculated on 24 tomato plants. The P values from Gehan–Breslow–Wilcoxon tests are shown below the graph. Red boxes indicate P < 0.01. Data were pooled from three independent experiments.
Fig. S2 HpaP modulates the secretion of AvrA and PopP1. Ralstonia solanacearum wild‐type strain (WT), hpaP and hrcV mutants were cultivated in secretion medium, cell pellets (CP) and culture supernatants (SN) were harvested after 8, 12 and 18 h of culture and analysed by immunoblotting. Effector proteins PopP1 (43 kDa), AvrA (26 kDa), PopP2 (53 kDa), GALA7 (68 kDa) and type III secretion system pilin HrpY (14 kDa) were detected using their respective antibodies. Type III effectors (T3Es) and HrpY were visualized in the CP and not SN of the hrcV mutant (a type III secretion system‐defective mutant). Three independent biological replicates were made. Western blots of one representative experiment are presented here.
Fig. S3 Complementation of hpaP mutation for the secretion of PopP1. Ralstonia solanacearum wild‐type strain (WT), hpaP mutant and complemented strain (hpaP::hpaP +) were cultivated in secretion medium for 8 h. Culture supernatant (SN) was harvested and effector proteins PopP1 (43 kDa) and PopP2 (53 kDa) were analysed by immunoblotting using anti‐PopP1 and anti‐PopP2 antibodies.
Fig. S4 Control of the production of the HpaP wild‐type protein and of two HpaP deletion derivatives (HpaPΔ 149–153 and HpaPΔ 156–160). Ralstonia solanacearum wild‐type strain (WT) and hpaP mutants expressing the hpaP wild‐type version (hpaP +) or hpaP deletion derivatives (hpaPΔ 149–153 and hpaPΔ 156–160) were cultivated in secretion medium for 8 h. Cell pellet (CP) extracts were harvested and HpaP proteins (26 kDa) were detected by immunoblotting using anti‐haemagglutinin (anti‐HA) antibody.
Table S1 Primers used in this study.
Acknowledgements
We thank members of our laboratory: Lenaïck Belliot for the pLBy10 construct, Sébastien Cunnac for the pSC205 plasmid, Marie Poueymiro for the pMP17 plasmid and Guillaume Gacon‐Labat for pGG18. We thank Laurent Deslandes for providing yeast two‐hybrid vectors and for the generous gift of the PopP2 antibody. We thank Jean Luc Pariente, Serge Bosc and Claudette Icher for tomato and tobacco plant preparation. We thank Amandine Gastebois and Sébastien Cunnac for advice on translocation assays, and Marie Françoise Jardinaud for advice on statistical analyses. We also thank Susana Rivas, Mathilde Fagard (INRA, Versailles) and Laurent Deslandes for helpful discussions. D.L. was funded by a grant from the French Ministry of National Education and Research. This work was supported by a French Agence Nationale de la Recherche grant (ANR‐2010‐JCJC‐1710‐01) to F.V. This work was supported by the French Laboratory of Excellence project ‘TULIP’ (ANR‐10‐LABX‐41; ANR‐11‐IDEX‐0002‐02).
References
- Agrain, C. , Callebaut, I. , Journet, L. , Sorg, I. , Paroz, C. , Mota, L.J. and Cornelis, G.R. (2005) Characterization of a Type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica . Mol. Microbiol. 56, 54–67. [DOI] [PubMed] [Google Scholar]
- Angot, A. , Peeters, N. , Lechner, E. , Vailleau, F. , Baud, C. , Gentzbittel, L. , Sartorel, E. , Genschik, P. , Boucher, C. and Genin, S. (2006) Ralstonia solanacearum requires F‐box‐like domain‐containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. USA, 103, 14 620–14 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlat, M. , van Gijsegem, F. , Huet, J.C. , Pernollet, J.C. and Boucher, C.A. (1994) PopA1, a protein which induces a hypersensitivity‐like response on specific Petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum . EMBO J. 13, 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M. , Brent, R. , Kingston, R.E. , Moore, D.D. , Seidman, J.G. , Smith, J.A. and Struhl, K. (1989) Current Protocols in Molecular Biology. New York: Greene Publishing Associates & Wiley Interscience. [Google Scholar]
- Bland, J.M. and Altman, D.G. (1998) Survival probabilities (the Kaplan–Meier method). Br. Med. J. 317, 1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block, A. , Li, G. , Fu, Z.Q. and Alfano, J.R. (2008) Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 11, 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. (2012) Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant‐ and animal‐pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. and Bonas, U. (2009) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. [DOI] [PubMed] [Google Scholar]
- Büttner, D. and He, S.Y. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. , Lorenz, C. , Weber, E. and Bonas, U. (2006) Targeting of two effector protein classes to the type III secretion system by a HpaC‐ and HpaB‐dependent protein complex from Xanthomonas campestris pv. vesicatoria . Mol. Microbiol. 59, 513–527. [DOI] [PubMed] [Google Scholar]
- Canonne, J. and Rivas, S. (2012) Bacterial effectors target the plant cell nucleus to subvert host transcription. Plant Signal. Behav. 7, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana, J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. [DOI] [PubMed] [Google Scholar]
- Cunnac, S. , Occhialini, A. , Barberis, P. , Boucher, C. and Genin, S. (2004) Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128. [DOI] [PubMed] [Google Scholar]
- Dereeper, A. , Guignon, V. , Blanc, G. , Audic, S. , Buffet, S. , Chevenet, F. , Dufayard, J.F. , Guindon, S. , Lefort, V. , Lescot, M. , Claverie, J.M. and Gascuel, O. (2008) Phylogeny.fr: robust phylogenetic analysis for the non‐specialist. Nucleic Acids Res. 36, W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L. , Pileur, F. , Liaubet, L. , Camut, S. , Can, C. , Williams, K. , Holub, E. , Beynon, J. , Arlat, M. and Marco, Y. (1998) Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen Ralstonia solanacearum . Mol. Plant–Microbe Interact. 11, 659–667. [DOI] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Peeters, N. , Feng, D.X. , Khounlotham, M. , Boucher, C. , Somssich, I. , Genin, S. and Marco, Y. (2003) Physical interaction between RRS1‐R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA, 100, 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist, P.J. , Olsson, J. , Lavander, M. , Sundberg, L. , Forsberg, A. , Wolf‐Watz, H. and Lloyd, S.A. (2003) YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J. Bacteriol. 185, 2259–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M.F. and Cornelis, G.R. (2003) The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol. Lett. 219, 151–158. [DOI] [PubMed] [Google Scholar]
- Gabriel, D.W. , Allen, C. , Schell, M. , Denny, T.P. , Greenberg, J.T. , Duan, Y.P. , Flores‐Cruz, Z. , Huang, Q. , Clifford, J.M. , Presting, G. , Gonzalez, E.T. , Reddy, J. , Elphinstone, J. , Swanson, J. , Yao, J. , Mulholland, V. , Liu, L. , Farmerie, W. , Patnaikuni, M. , Balogh, B. , Norman, D. , Alvarez, A. , Castillo, J.A. , Jones, J. , Saddler, G. , Walunas, T. , Zhukov, A. and Mikhailova, N. (2006) Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol. Plant Microbe Interact. 19, 69–79. [DOI] [PubMed] [Google Scholar]
- Genin, S. and Denny, T.P. (2012) Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. [DOI] [PubMed] [Google Scholar]
- van Gijsegem, F. , Vasse, J. , De Rycke, R. , Castello, P. and Boucher, C. (2002) Genetic dissection of Ralstonia solanacearum hrp gene cluster reveals that the HrpV and HrpX proteins are required for Hrp pilus assembly. Mol. Microbiol. 44, 935–946. [DOI] [PubMed] [Google Scholar]
- Gonzalez, A. , Plener, L. , Restrepo, S. , Boucher, C. and Genin, S. (2011) Detection and functional characterization of a large genomic deletion resulting in decreased pathogenicity in Ralstonia solanacearum race 3 biovar 2 strains. Environ. Microbiol. 13, 3172–3185. [DOI] [PubMed] [Google Scholar]
- Gough, C.L. , Genin, S. , Lopes, V. and Boucher, C.A. (1993) Homology between the HrpO protein of Pseudomonas solanacearum and bacterial proteins implicated in a signal peptide‐independent secretion mechanism. Mol. Gen. Genet. 239, 378–392. [DOI] [PubMed] [Google Scholar]
- Guéneron, M. , Timmers, A.C. , Boucher, C. and Arlat, M. (2000) Two novel proteins, PopB, which has functional nuclear localization signals, and PopC, which has a large leucine‐rich repeat domain, are secreted through the hrp‐secretion apparatus of Ralstonia solanacearum . Mol. Microbiol. 36, 261–277. [DOI] [PubMed] [Google Scholar]
- Guidot, A. , Elbaz, M. , Carrere, S. , Siri, M.I. , Pianzzola, M.J. , Prior, P. and Boucher, C. (2009) Specific genes from the potato brown rot strains of Ralstonia solanacearum and their potential use for strain detection. Phytopathology, 99, 1105–1112. [DOI] [PubMed] [Google Scholar]
- Guindon, S. , Delsuc, F. , Dufayard, J.F. and Gascuel, O. (2009) Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537, 113–137. [DOI] [PubMed] [Google Scholar]
- Hayward, A.C. (2000) Ralstonia Solanacearum In: Encyclopaedia of Microbiology, Volume 4 (Lederberg J., ed.), pp. 32–42. San Diego, CA: Academic ; Press. [Google Scholar]
- He, S.Y. , Nomura, K. and Whittam, T.S. (2004) Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta, 1694, 181–206. [DOI] [PubMed] [Google Scholar]
- Hueck, C.J. (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet, L. , Agrain, C. , Broz, P. and Cornelis, G.R. (2003) The needle length of bacterial injectisomes is determined by a molecular ruler. Science, 302, 1757–1760. [DOI] [PubMed] [Google Scholar]
- Katoh, K. , Misawa, K. , Kuma, K. and Miyata, T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie, M. , Shillington, E. , Eguiluz, C. , Grimsley, N. and Boucher, C. (2002) PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host‐specificity factor and modulates aggressiveness of Ralstonia solanacearum . Mol. Plant–Microbe Interact. 15, 1058–1068. [DOI] [PubMed] [Google Scholar]
- Lohou, D. , Lonjon, F. , Genin, S. and Vailleau, F. (2013) Type III chaperones & Co in bacterial plant pathogens: a set of specialized bodyguards mediating effector delivery. Front Plant Sci. 4, e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, C. , Schulz, S. , Wolsch, T. , Rossier, O. , Bonas, U. and Büttner, D. (2008) HpaC controls substrate specificity of the Xanthomonas type III secretion system. PLoS Pathog. 4, e1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx, C.J. and Lidstrom, M.E. (2002) Broad‐host‐range cre‐lox system for antibiotic marker recycling in gram‐negative bacteria. Biotechniques, 33, 1062–1067. [DOI] [PubMed] [Google Scholar]
- McCann, H.C. and Guttman, D.S. (2008) Evolution of the type III secretion system and its effectors in plant–microbe interactions. New Phytol. 177, 33–47. [DOI] [PubMed] [Google Scholar]
- Minamino, T. , Gonzalez‐Pedrajo, B. , Yamaguchi, K. , Aizawa, S.I. and Macnab, R.M. (1999) FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 34, 295–304. [DOI] [PubMed] [Google Scholar]
- Monteiro, F. , Sole, M. , van Dijk, I. and Valls, M. (2012) A chromosomal insertion toolbox for promoter probing, mutant complementation, and pathogenicity studies in Ralstonia solanacearum . Mol. Plant–Microbe Interact. 25, 557–568. [DOI] [PubMed] [Google Scholar]
- Morello, J.E. and Collmer, A. (2009) Pseudomonas syringae HrpP is a type III secretion substrate specificity switch domain protein that is translocated into plant cells but functions atypically for a substrate‐switching protein. J. Bacteriol. 191, 3120–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaihara, T. , Tamura, N. , Murata, Y. and Iwabuchi, M. (2004) Genetic screening of Hrp type III‐related pathogenicity genes controlled by the HrpB transcriptional activator in Ralstonia solanacearum . Mol. Microbiol. 54, 863–875. [DOI] [PubMed] [Google Scholar]
- Mukaihara, T. , Tamura, N. and Iwabuchi, M. (2010) Genome‐wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant–Microbe Interact. 23, 251–262. [DOI] [PubMed] [Google Scholar]
- Parsot, C. , Hamiaux, C. and Page, A.L. (2003) The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6, 7–14. [DOI] [PubMed] [Google Scholar]
- Patterson‐Delafield, J. , Martinez, R.J. , Stocker, B.A. and Yamaguchi, S. (1973) A new fla gene in Salmonella typhimurium—flaR—and its mutant phenotype‐superhooks. Arch. Mikrobiol. 90, 107–120. [DOI] [PubMed] [Google Scholar]
- Peeters, N. , Guidot, A. , Vailleau, F. and Valls, M. (2013) Ralstonia solanacearum, a widespread bacterial plant pathogen in the post‐genomic era. Mol. Plant Pathol. 14, 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro, M. and Genin, S. (2009) Secreted proteins from Ralstonia solanacearum: a hundred tricks to kill a plant. Curr. Opin. Microbiol. 12, 44–52. [DOI] [PubMed] [Google Scholar]
- Poueymiro, M. , Cunnac, S. , Barberis, P. , Deslandes, L. , Peeters, N. , Cazale‐Noel, A.C. , Boucher, C. and Genin, S. (2009) Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host‐range specificity on tobacco. Mol. Plant–Microbe Interact. 22, 538–550. [DOI] [PubMed] [Google Scholar]
- Remenant, B. , Coupat‐Goutaland, B. , Guidot, A. , Cellier, G. , Wicker, E. , Allen, C. , Fegan, M. , Pruvost, O. , Elbaz, M. , Calteau, A. , Salvignol, G. , Mornico, D. , Mangenot, S. , Barbe, V. , Medigue, C. and Prior, P. (2010) Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics, 11, 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenant, B. , de Cambiaire, J.C. , Cellier, G. , Jacobs, J.M. , Mangenot, S. , Barbe, V. , Lajus, A. , Vallenet, D. , Medigue, C. , Fegan, M. , Allen, C. and Prior, P. (2011) Ralstonia syzygii, the blood disease bacterium and some Asian R. solanacearum strains form a single genomic species despite divergent lifestyles. PLoS ONE, 6, e24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigi, P. , Anisimova, M. , Guidot, A. , Genin, S. and Peeters, N. (2011) Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 192, 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanoubat, M. , Genin, S. , Artiguenave, F. , Gouzy, J. , Mangenot, S. , Arlat, M. , Billault, A. , Brottier, P. , Camus, J.C. , Cattolico, L. , Chandler, M. , Choisne, N. , Claudel‐Renard, C. , Cunnac, S. , Demange, N. , Gaspin, C. , Lavie, M. , Moisan, A. , Robert, C. , Saurin, W. , Schiex, T. , Siguier, P. , Thebault, P. , Whalen, M. , Wincker, P. , Levy, M. , Weissenbach, J. and Boucher, C.A. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum . Nature, 415, 497–502. [DOI] [PubMed] [Google Scholar]
- Schechter, L.M. , Roberts, K.A. , Jamir, Y. , Alfano, J.R. and Collmer, A. (2004) Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186, 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, S. and Büttner, D. (2011) Functional characterization of the type III secretion substrate specificity switch protein HpaC from Xanthomonas campestris pv. vesicatoria . Infect. Immun. 79, 2998–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole, M. , Popa, C. , Mith, O. , Sohn, K.H. , Jones, J.D. , Deslandes, L. and Valls, M. (2012) The awr gene family encodes a novel class of Ralstonia solanacearum type III effectors displaying virulence and avirulence activities. Mol. Plant–Microbe Interact. 25, 941–953. [DOI] [PubMed] [Google Scholar]
- Tampakaki, A.P. , Skandalis, N. , Gazi, A.D. , Bastaki, M.N. , Sarris, P.F. , Charova, S.N. , Kokkinidis, M. and Panopoulos, N.J. (2010) Playing the ‘Harp’: evolution of our understanding of hrp/hrc genes. Annu. Rev. Phytopathol. 48, 347–370. [DOI] [PubMed] [Google Scholar]
- Turner, M. , Jauneau, A. , Genin, S. , Tavella, M.J. , Vailleau, F. , Gentzbittel, L. and Jardinaud, M.F. (2009) Dissection of bacterial wilt on Medicago truncatula revealed two type III secretion system effectors acting on root infection process and disease development. Plant Physiol. 150, 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Zheng, H.J. , Liu, L. , Pan, Z.C. , Prior, P. , Tang, B. , Xu, J.S. , Zhang, H. , Tian, Q. , Zhang, L.Q. and Feng, J. (2011) Complete genome sequence of the plant pathogen Ralstonia solanacearum strain Po82. J. Bacteriol. 193, 4261–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The hpaP mutant is strongly reduced in virulence on tomato. Kaplan–Meier survival analysis of tomato plants inoculated with Ralstonia solanacearum wild‐type strain (red squares), hpaP mutant (green stars) and the complemented strain hpaP::hpaP + (blue circles). Each strain was root inoculated on 24 tomato plants. The P values from Gehan–Breslow–Wilcoxon tests are shown below the graph. Red boxes indicate P < 0.01. Data were pooled from three independent experiments.
Fig. S2 HpaP modulates the secretion of AvrA and PopP1. Ralstonia solanacearum wild‐type strain (WT), hpaP and hrcV mutants were cultivated in secretion medium, cell pellets (CP) and culture supernatants (SN) were harvested after 8, 12 and 18 h of culture and analysed by immunoblotting. Effector proteins PopP1 (43 kDa), AvrA (26 kDa), PopP2 (53 kDa), GALA7 (68 kDa) and type III secretion system pilin HrpY (14 kDa) were detected using their respective antibodies. Type III effectors (T3Es) and HrpY were visualized in the CP and not SN of the hrcV mutant (a type III secretion system‐defective mutant). Three independent biological replicates were made. Western blots of one representative experiment are presented here.
Fig. S3 Complementation of hpaP mutation for the secretion of PopP1. Ralstonia solanacearum wild‐type strain (WT), hpaP mutant and complemented strain (hpaP::hpaP +) were cultivated in secretion medium for 8 h. Culture supernatant (SN) was harvested and effector proteins PopP1 (43 kDa) and PopP2 (53 kDa) were analysed by immunoblotting using anti‐PopP1 and anti‐PopP2 antibodies.
Fig. S4 Control of the production of the HpaP wild‐type protein and of two HpaP deletion derivatives (HpaPΔ 149–153 and HpaPΔ 156–160). Ralstonia solanacearum wild‐type strain (WT) and hpaP mutants expressing the hpaP wild‐type version (hpaP +) or hpaP deletion derivatives (hpaPΔ 149–153 and hpaPΔ 156–160) were cultivated in secretion medium for 8 h. Cell pellet (CP) extracts were harvested and HpaP proteins (26 kDa) were detected by immunoblotting using anti‐haemagglutinin (anti‐HA) antibody.
Table S1 Primers used in this study.


