Summary
Various plant viruses ubiquitously mediate the induction of miR168, resulting in the control of ARGONAUTE 1 (AGO1), which is the pivotal component of the microRNA (miRNA) regulation pathway and can also exhibit antiviral function. Here, we demonstrate that miR168‐driven control of AGO1 can persist for a long time in virus‐infected plants and can be an important component of symptom development. We also show that infection of RNA viruses belonging to various genera is associated with the transcriptional induction of the MIR168 precursor gene. Moreover, in a transient expression study, we reveal that different unrelated viral suppressors of RNA silencing (VSRs) are responsible for the enhanced accumulation of miR168. The induction of miR168 accumulation is an early function of VSRs and this activity is associated with the control of the endogenous AGO1 protein level. The common ability of unrelated VSRs to induce the miR168 level implies that this activity might be a component of the host defence suppression in plant–virus interactions.
Introduction
RNA silencing is a conserved regulatory mechanism controlling endogenous gene expression and also serving as an antiviral defence system (Ding, 2010; Ding and Voinnet, 2007; Mlotshwa et al., 2005). Virus infection in plants triggers the production of virus‐specific small interfering RNAs (siRNAs) by the RNA silencing machinery, which are then incorporated into the RNA‐induced silencing complex (RISC), inducing the sequence‐specific degradation of the complementary viral RNAs (Pantaleo et al., 2007). However, as a counteracting strategy, plant viruses have evolved to encode potent suppressor proteins (viral suppressors of RNA silencing; VSRs) to disable the RNA silencing‐based host defence system (Burgyan, 2008; Burgyan and Havelda, 2011; Mlotshwa et al., 2008).
The ARGONAUTE (AGO) proteins are the executor components of RISCs, bringing about the degradation or translational inhibition of target RNA specified by the incorporated small RNA (Mallory and Vaucheret, 2010). The role of AGO1 as the key constituent of the major antiviral RISC is supported by the following observations: (i) AGO1 recruits virus‐specific siRNAs (Zhang et al., 2006); (ii) virus‐specific siRNAs of Cymbidum ringspot virus (CymRSV) co‐fractionate with large protein complexes that contain AGO1 and probably correspond to RISCs (Csorba et al., 2010; Pantaleo et al., 2007); (iii) several VSRs are able to interact directly with AGO1 inhibiting its antiviral functions (Azevedo et al., 2010; Baumberger et al., 2007; Bortolamiol et al., 2007; Csorba et al., 2010; Giner et al., 2010; Pazhouhandeh et al., 2006; Zhang et al., 2006); (iv) ago1 hypomorphic mutants are more susceptible to viral infections (Morel et al., 2002; Qu et al., 2008). In addition to AGO1, other AGO proteins can be involved in RNA silencing‐based defence reactions (Alvarado and Scholthof, 2011; Harvey et al., 2011; Jaubert et al., 2011; Scholthof et al., 2011; Wang et al., 2011). AGO1 also plays a role, as the key RISC component, in the microRNA (miRNA) pathway regulating target RNAs through cleavage or translational inhibition (Mallory and Bouche, 2008; Mallory and Vaucheret, 2010; Mallory et al., 2008). The level of AGO1 is controlled by the action of an miRNA (miR168) (Rhoades et al., 2002; Vaucheret et al., 2004); however, fine regulation of the AGO1 level requires additional factors, such as the action of AGO1‐derived siRNAs on AGO1 mRNA (Mallory and Vaucheret, 2009), AGO1 mediated post‐transcriptional stabilization of miR168 and the co‐regulated expression of the AGO1 and MIR168 genes (Vaucheret et al., 2006). Recent studies have described the increased expression of miR168 and AGO1 mRNA in virus‐infected plants (Csorba et al., 2007; Havelda et al., 2008; Lang et al., 2011; Varallyay et al., 2010; Zhang et al., 2006). In line with the vital role of AGO1 in RNA silencing‐based defence, we have shown previously that the induction of AGO1 mRNA expression is a potential host defence mechanism enhancing the concentration of the main executor molecule, whereas the induction of miR168 and the subsequent translational control of AGO1 protein accumulation is a viral counter defence strategy (Varallyay et al., 2010). Using a transient expression system and virus infection studies, it was revealed that the p19 VSR of CymRSV is responsible for the induction of miR168 and the subsequent regulation of the AGO1 level (Varallyay et al., 2010). However, the potential role of AGO1 level control in symptom development and the factors responsible for miR168 induction in various unrelated virus infections were not investigated.
In this study, we reveal that the control of AGO1 accumulation in Nicotiana benthamiana is mediated by several viruses belonging to different genera. We demonstrate that, during the infection process of different viruses, the transcriptional activity of the MIR168a gene is induced in Arabidopsis thaliana. Using a transient protein expression assay, we show that VSRs of different virus genera are responsible for the induced accumulation of miR168, which is associated with the control of the AGO1 level. The observation that distinct and unrelated viral VSRs are able to induce miR168 accumulation indicates that the miR168‐driven control of AGO1 may be a component of viral invasion strategies in several plant–virus interactions.
Results
Virus infection‐induced AGO1 down‐regulation can persist in virus‐infected plants
Crucifer‐infecting Tobamovirus (crTMV) infection in A. thaliana, similar to CymRSV‐infected N. benthamiana, induces the over‐accumulation of miR168 and mediates the subsequent control of AGO1 protein (Varallyay et al., 2010). Moreover, the activity of crTMV p122 VSR, similar to p19 VSR, is based on siRNA binding (Csorba et al., 2007). We investigated the longevity of AGO1 down‐regulation in crTMV‐infected plants at 21 °C, as the lack of early necrotic symptoms in crTMV‐infected A. thaliana made it possible to follow the infection process over a longer period of time. RNA and protein samples were taken at different time points [7, 11, 17, 21, 25 and 28 days post‐inoculation (dpi)] from the symptomatic upper leaves and analysed for the presence of miR168, AGO1 mRNA and AGO1 protein (Fig. 1A). We found that AGO1 protein was markedly down‐regulated at the later time points, indicating that this phenomenon is present throughout the virus invasion process. Moreover, we also detected increased levels of miR168 and AGO1 mRNA at all time points (Fig. 1A). Longer exposure of the Northern blot revealed the robustness of the crTMV infection‐mediated up‐regulation of AGO1 mRNA relative to its ubiquitous basal expression in mock‐inoculated plants. The appearance of two AGO1‐specific bands is a typical feature of AGO1 mRNA expression in plants (Mallory and Vaucheret, 2009; Varallyay et al., 2010). As a control, we used the conservative miR159 directing the cleavage of the mRNA of the MYB family member transcription factor, which binds to the promoter of the floral meristem identity gene LEAFY, playing an important role in floral development (Achard et al., 2004). The level of miR159 displayed moderate changes in the samples during the period of the experiment, but showed characteristically increased accumulation in virus‐infected samples, as described previously (Csorba et al., 2007). The persistent deficiency of AGO1 protein and the potential sequestration of miRNAs by p122 in crTMV‐infected A. thaliana may alter the expression of miRNA‐controlled mRNAs. To test this hypothesis, we investigated the expression level of auxin response factor (ARF) mRNAs, which are known to be regulated by miRNA‐mediated cleavage (Mallory et al., 2005). ARF10, ARF16 and ARF17 are targeted by miR160, whereas miR167 mediates the cleavage of ARF6 and ARF8 mRNAs. These genes control diverse developmental responses to auxin signals, including cell elongation, division and differentiation, in both roots and shoots (Rhoades et al., 2002). Enhanced expression of ARF10, ARF17 and ARF6 mRNAs was observed in virus‐infected samples in spite of increased miR160 and miR167 levels at 21 dpi (Fig. 1B). To further investigate our findings, we tested the miRNA sequestration ability of p122 VSR and the incorporation of miR168 into high‐molecular‐weight complexes by size separation gel filtration experiments at late time points (17 dpi) of virus infection (Fig. S1, see Supporting Information). We revealed that, in addition to the down‐regulation of AGO1, the p122 VSR is able to efficiently bind miRNAs (miR168, miR159 and also miR160). The majority of miRNAs are preferentially bound by p122 VSR, resulting in a further decrease in the concentration of active RISCs. Moreover, in spite of the extreme efficiency of p122 VSR binding to miR168, a significant amount of the miR168 excess is able to incorporate into higher molecular weight protein complexes, suggesting an active production of miR168 in virus‐infected plants (Fig. S1). These data imply that persistent AGO1 protein deficiency and the efficient miRNA sequestration mediated by p122 can induce the altered expression of miRNA‐regulated mRNAs, which could be a component of the development of virus infection‐induced disease symptoms.
Figure 1.
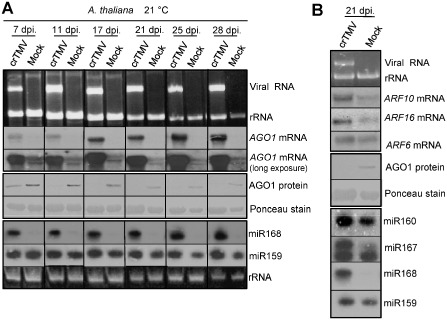
Virus infection‐induced ARGONAUTE 1 (AGO1) down‐regulation can persist in virus‐infected plants. (A) Systemically infected leaves of crucifer‐infecting Tobamovirus (crTMV)‐inoculated Arabidopsis thaliana plants were homogenized and used for RNA and protein extractions. The corresponding samples were used for the detection of AGO1 mRNA, AGO1 and miR168 expression at different time points. Top panels: Northern blot analyses of virus‐infected and mock‐inoculated plants for AGO1 mRNA at 21 °C. Ethidium bromide‐stained agarose gel was also used as loading control. Middle panels: Western blot of total protein extract for AGO1 (A . thaliana). Ponceau‐stained membrane was used as loading control. Bottom panels: Small RNA Northern blot analyses using miR168‐ and miR159‐specific locked nucleic acid (LNA) probes. Relative gel loadings are also indicated by ethidium bromide staining of ribosomal RNAs (rRNA). (B) Northern blot analyses of virus‐infected and mock‐inoculated plants for ARF10, ARF16 and ARF6 mRNAs. Ethidium bromide‐stained membrane was used as loading control (top panels). Western blot of total protein extracts for AGO1 (A . thaliana). Ponceau‐stained membrane was used as loading control (middle panels). Small RNA Northern blot analyses using miR160‐, miR167‐, miR168‐ and miR159‐specific LNA probes (bottom panels). Relative gel loadings are also indicated by ethidium bromide staining of ribosomal RNAs (rRNA).
Control of AGO1 level is a universal phenomenon in virus infections
The induction of miR168 and the subsequent down‐regulation of the AGO1 protein level have been demonstrated for Tombusvirus–N. benthamiana, crTMV–A. thaliana and Turnip crinkle virus (TCV)–A. thaliana systems (Varallyay et al., 2010). To test how ubiquitous is this regulation process, we further investigated this phenomenon by infecting N. benthamiana plants with crTMV, Tobacco etch virus (TEV), TCV and both N. benthamiana and A. thaliana with Cucumber mosaic virus (CMV; Fny strain) (Fig. 2). These viruses belong to different virus genera including Tobamo‐, Poty‐, Carmo‐ and Cucumoviruses. Samples were taken at an early stage of the infection process after the development of disease symptoms on the first systemically infected leaves; these were then homogenized and divided into two parts for total RNA extractions and protein analyses. At the investigated time points, high levels of viral RNA accumulation were detected, except for TEV infection, in ethidium bromide‐stained gels (Fig. 2A). In TEV infection, the viral RNA level was below the detection limit in the extract, but the robust induction of miR168 and the down‐regulation of AGO1 protein indicate an efficient virus infection process. The induction of AGO1 mRNA was evident in TCV‐ and CMV‐infected plants; however, no significant alteration was observed during TEV infection (Fig. 2B), which is probably a specific feature of this interaction. In crTMV‐infected N. benthamiana (Fig. 2B), we detected AGO1 mRNA down‐regulation, which is the opposite of what we experienced in crTMV‐infected A. thaliana (Fig. 1A). This phenomenon is not attributable to the viral accumulation level as crTMV accumulates to a high level in N. benthamiana (Fig. 2A). crTMV infection‐induced down‐regulation of AGO1 mRNA possibly results from the general inhibitory effect (shut‐off) of crTMV infection on the level of N. benthamiana host mRNAs (Havelda et al., 2008) and the fast development of severe necrotic symptoms. The accumulation of AGO1 protein in the corresponding samples was efficiently down‐regulated in TEV‐ and CMV‐infected plants (Fig. 2C). TCV infection in A. thaliana induces efficient down‐regulation of AGO1 protein (Varallyay et al., 2010); however, here we showed that TCV infection in N. benthamiana is associated with less efficient control of the AGO1 protein level, similar to that of Tombusvirus infections (Varallyay et al., 2010), keeping AGO1 at the wild‐type level in spite of the induced expression of AGO1 mRNA (Fig. 2C). We found that all of the viruses used induced the expression of miR168 (Fig. 2D). These data indicate that, although the control of the AGO1 level is ubiquitous in different host–virus interactions, its efficiency varies depending on the particular host–virus combinations.
Figure 2.
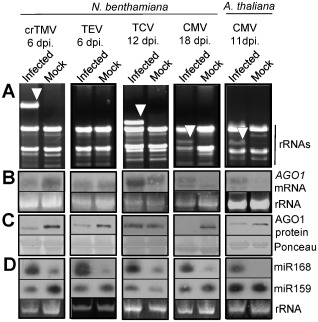
Control of ARGONAUTE 1 (AGO1) level is a universal phenomenon in virus infection. Systemically infected leaves of crucifer‐infecting Tobamovirus (crTMV)‐, Tobacco etch virus (TEV)‐, Turnip crinkle virus (TCV)‐ and Cucumber mosaic virus (CMV)‐inoculated Nicotiana benthamiana and CMV‐inoculated Arabidopsis thaliana plants were homogenized and used for RNA and protein extractions. The samples were used for the detection of AGO1 mRNA, AGO1 and miR168 expression. (A) Relative virus accumulation visualized by ethidium bromide‐stained RNA samples from mock‐inoculated and virus‐infected plants separated on agarose gel. Arrowhead indicates the accumulation of viral RNAs. (B) Northern blot analyses of virus‐infected and mock‐inoculated plants for AGO1 mRNA at 21 °C. Ethidium bromide‐stained membranes were used as loading control. (C) Western blot of total protein extracts for N . benthamiana AGO1 or A. thaliana AGO1 accumulation. Ponceau‐stained membrane was used as loading control. (D) Small RNA Northern blot analyses using miR168‐ and miR159‐specific locked nucleic acid (LNA) probes. Relative gel loadings are also indicated by ethidium bromide staining of ribosomal RNAs (rRNA).
Virus infection induces MIR168a expression in A. thaliana
In further experiments, we intended to investigate how virus infections can lead to an increased miR168 level. The efficient control of AGO1 accumulation in virus‐infected plants suggested that miR168 over‐accumulation is not only the result of the potential stabilizing effect of certain viral VSRs, but is actively mediated during the virus infection process. To test whether virus infections can induce the expression of miR168 precursors, we investigated the effect of TCV, CMV and crTMV infection on the expression of MIR168a and MIR168b genes in the well‐characterized A. thaliana system. We used transgenic A. thaliana plants expressing the β‐glucuronidase (GUS) reporter gene under the control of the promoter of either the MIR168a or MIR168b gene (Vaucheret, 2009) in these experiments. TCV, CMV and crTMV infections in A. thaliana are associated with miR168 induction and the subsequent control of AGO1 (Varallyay et al., 2010; Figs 1 and 2). Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analyses of RNA samples extracted from TCV‐, CMV‐ and crTMV‐infected transgenic plants revealed that expression of the GUS mRNA was induced in pMIR168a:GUS (Fig. 3A), but not in pMIR168b:GUS (Fig. 3B), transgenic plants. Moreover, quantitative RT‐PCR analyses of the endogenous MIR168a and MIR168b precursors also showed the virus infection‐induced expression of MIR168a, but not the MIR168b gene (Fig. 3A,B). These findings indicate that virus infections are able to specifically enhance the transcriptional activity of the MIR168a precursor, contributing to the over‐accumulation of mature miR168. These data are in line with our previous observation, where the over‐accumulation of an intermediate processing product of MIR168a precursor RNA was observed in TCV‐, CMV‐ and crTMV‐infected plants (Varallyay et al., 2010). To further test the specific induction of the MIR168a gene, we investigated the accumulation of MIR168a and MIR168b precursor processing products and found that, in line with quantitative RT‐PCR data, only MIR168a processing products showed increased accumulation (Fig. 3C). In contrast, the level of intermediate product (about 80 nucleotides long) associated with the processing of the MIR168b precursor showed no alteration in virus‐infected plants (Fig. 3C). Our data show that the induction of MIR168a activity is less efficient in TCV infection than in CMV and crTMV infections. Altogether, these data show that TCV, CMV and crTMV infections are able to induce the transcriptional activity of the MIR168a gene in A. thaliana.
Figure 3.
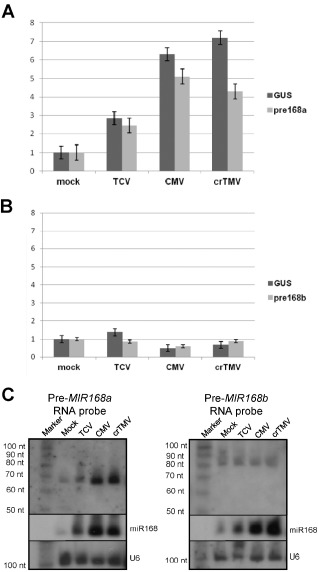
Virus infection‐mediated induction of MIR 168a expression in A rabidopsis thaliana. RNA extracts from Turnip crinkle virus (TCV)‐, Cucumber mosaic virus (CMV)‐ and crucifer‐infecting Tobamovirus (crTMV)‐infected and mock‐inoculated transgenic A. thaliana [expressing β‐glucuronidase (GUS) reporter under the control of promoters of MIR168a and MIR168b genes] were subjected to quantitative reverse transcription‐polymerase chain reaction (RT‐PCR). Relative expression changes are shown (using cyclophilin as endogenous control) for GUS and MIR168a precursor [MIR168a::GUS (A)] and for GUS and MIR168b precursor [MIR168b::GUS (B)] transgenic plants. (C) A rabidopsis thaliana total RNA samples of virus‐ and mock‐inoculated leaves were separated on polyacrylamide gel and used for Northern blot analyses using RNA probe complementary to MIR168a (Pre‐MIR168a) or MIR168b (Pre‐MIR168b) precursors. The membrane was stripped and used for subsequent hybridizations with locked nucleic acid (LNA) probe specific to mature miR168 and DNA probe detecting U6 snRNA (loading control).
Unrelated VSRs of various plant viruses induce miR168 accumulation and control the AGO1 protein level
Next, we wanted to reveal which viral proteins are responsible for the induction of the miR168 level and the subsequent AGO1 level control in different plant–virus interactions. Virus infections are often associated with alterations in endogenous miRNA levels (Bazzini et al., 2007; Csorba et al., 2007). In addition, in transgenic studies, it has been shown that VSRs of plant viruses are responsible for the alteration of endogenous miRNA and miRNA* levels, and also induce changes in target mRNA levels (Chapman et al., 2004; Dunoyer et al., 2004; Kasschau et al., 2003; Mallory et al., 2002; Mlotshwa et al., 2005; Zhang et al., 2006). The ability of VSRs to interfere with miRNA expression prompted us to investigate the role of different unrelated VSRs in the miR168‐driven control of AGO1. Transgenic expression of plant VSRs is a powerful system to understand the molecular action of these molecules; however, it may not reveal all functions of VSRs as they are continuously present during the development of the transgenic lines, often at a low level because of their deteriorating effects. Moreover, the transgenic approach, because of the continuous presence of VSRs, does not distinguish between the early and late effects of VSRs. To avoid the potential long‐term effects of the transgenic expression of VSRs, we used an Agrobacterium‐mediated transient expression system to test the ability of different VSRs for the specific induction of miR168 accumulation. Using this system, we introduced a high level of VSR protein for a short period of time into fully developed leaf cells in order to mimic the early phase of the virus infection process. Previously, using the Agrobacterium‐mediated transient expression system, we demonstrated that the p19 VSR of Tombusviruses is able to enhance the miR168 level and down‐regulate the level of endogenous AGO1 protein (Varallyay et al., 2010). To test whether different unrelated VSRs are also able to control the AGO1 protein level via miR168 induction, N. benthamiana leaves were infiltrated with Agrobacterium harbouring viral suppressor constructs. Leaf samples were collected at 4 dpi, homogenized and divided into two parts for protein and RNA analyses, ensuring the comparability of the samples. Different viral suppressors [p122 of crTMV (Csorba et al., 2007), p19 of CymRSV (Vargason et al., 2003), p38 of TCV (Merai et al., 2005), HcPro of TEV (Lozsa et al., 2008; Merai et al., 2005), 2b of CMV (Zhang et al., 2006)] were used. We found that, in all cases, the transient expression of various VSRs induced the over‐accumulation of miR168 compared with the control infiltrations [empty or green fluorescent protein (GFP)‐expressing binary vector], suggesting that the VSRs investigated in this study are able to modulate the accumulation of miR168 (Fig. 4A). As a control, we also investigated the expression of other miRNAs. miR171 targets mRNAs encoding the GRAS domain or SCARECROW‐like proteins involved in radial patterning in roots, signalling by light and gibberellin (Rhoades et al., 2002). miR165 mediates the control of mRNAs encoding HD‐Zip transcription factors, including Phabulosa (PHB) and Phavoluta (PHV), involved in axillary meristem initiation and leaf development (Yao et al., 2009). miR164 regulates mRNAs coding for NAC domain‐containing proteins, such as Cup‐Shaped Cotyledon 2 (CUC2), which is required for shoot apical meristem formation (Hasson et al., 2011). These miRNAs in our system displayed either no or only a moderate increase in p122‐ and HcPro‐infiltrated samples (Fig. 4A). In line with our previous results, demonstrating that TCV infection is associated with less efficient MIR168a induction, we found that the p38 VSR of TCV has the weakest effect on miR168 induction (Fig. 4A). To test the biological relevance of the observed miR168 accumulation in VSR‐expressing samples, the level of endogenous AGO1 protein was investigated. We found that the AGO1 protein level was down‐regulated in all samples expressing VSR constructs, compared with the control infiltrations (Fig. 4B). All of the VSRs used were shown to have RNA‐binding capacity to some extent (Burgyan and Havelda, 2011), raising the possibility that the RNA‐binding ability of other viral proteins may also induce the accumulation of miR168. To investigate this hypothesis and to test whether the infiltrated VSR constructs express appropriately in the infiltrate patches, we repeated the experiments using the GFP reporter construct to measure the activity of VSRs and Tombusvirus coat protein (CP)‐expressing construct as a control (Fig. S2, see Supporting Information). We found that all of the VSRs used expressed at the appropriate level and worked efficiently, blocking the silencing processes against GFP. Moreover, the expression of CP did not result in the specific induction of miR168, implying that affinity of CP to RNA molecules alone is not sufficient for miR168 induction (Fig. S2). Altogether, these data indicate that miR168 induction is an immediate and specific function of the investigated VSRs, and this activity of VSRs is associated with the down‐regulation of the endogenous AGO1 protein.
Figure 4.
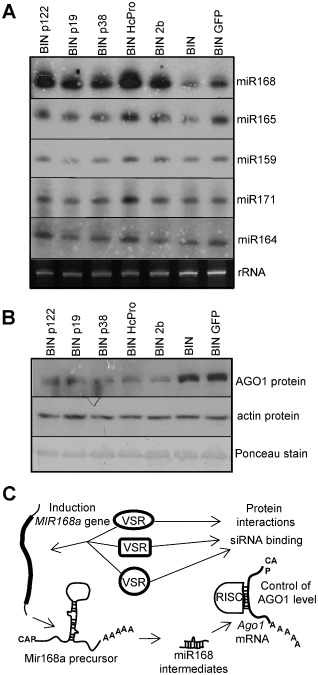
Viral suppressors of RNA silencing (VSRs) of various plant viruses are able to induce miR168 accumulation and control ARGONAUTE 1 (AGO1) level. Total RNA and protein samples were extracted from the leaves infiltrated with A grobacterium suspension expressing p122 of crucifer‐infecting Tobamovirus (crTMV) (BINp122), p19 of Cymbidum ringspot virus (CymRV) (BIN p19), p38 of Turnip crinkle virus (TCV) (BIN p38), HcPro of Tobacco etch virus (TEV) (BIN HcPro), 2b of Cucumber mosaic virus (CMV) (BIN 2b), empty BIN vector (BIN) and 35S‐GFP (BIN GFP). GFP, green fluorescent protein. (A) Total RNA extracts were separated on polyacrylamide gel and used for Northern blot analyses for subsequent hybridizations with locked nucleic acid (LNA) probes specific to mature miR168, miR165, miR159, miR171 and miR164. Relative gel loadings are also indicated by ethidium bromide staining of ribosomal RNAs (rRNA). (B) Western blot analyses of total protein extracts for AGO1 accumulation using AGO1‐1 (Nicotiana benthamiana) antibody. Equal loading was verified by detection of actin protein accumulation and by Ponceau staining of the membrane after Western blotting. (C) Schematic model of miR168‐driven virus infection‐mediated control of AGO1 protein accumulation. RISC, RNA‐induced silencing complex.
Discussion
The various VSRs described so far are able to target different stages of the RNA silencing‐based host defence pathway, such as viral RNA recognition, dicing, RISC formation, amplification and RNA targeting (Burgyan and Havelda, 2011; Mlotshwa et al., 2008). Recent insights into the detailed molecular mechanisms and functions of VSRs have begun to reveal their potential multiple actions in disabling the RNA silencing‐based host defence system. Cucumovirus 2b VSR has been shown to be able to bind siRNAs in a size‐dependent manner (Chen et al., 2008) and also to physically interact with AGO1, inhibiting its slicing activity (Zhang et al., 2006). Similarly, TCV p38 VSR has been shown to bind to double‐stranded viral RNAs in a size‐independent manner (Merai et al., 2005), and this protein interacts, through two GW repeats, with AGO1 (Azevedo et al., 2010). The induction of miR168 is a widespread feature of plant virus infections (Csorba et al., 2007; Havelda et al., 2008; Lang et al., 2011; Varallyay et al., 2010; Zhang et al., 2006). Moreover, it has been shown recently that p19 VSR, in addition to its viral siRNA sequestering ability, is responsible for the induction of miR168 and the subsequent control of AGO1 accumulation (Varallyay et al., 2010). In this work, we tested the regulation of the AGO1 level in different plants infected with viruses belonging to various genera, and we observed that the control of AGO1 over‐accumulation, although with different efficiency, exists in all of the investigated plant–virus interactions. The ubiquitous presence of miR168 induction and the associated suppression of AGO1 accumulation in virus‐infected plants imply that control of the potential AGO1‐containing antiviral RISC is a general strategy of plant viruses. Previously, it has been demonstrated that the elimination of the miRNA recognition site from AGO1 mRNA results in the inhibition of miR168‐mediated regulation of AGO1 in crTMV‐infected A. thaliana, indicating direct connection between miR168 induction and AGO1 down‐regulation (Varallyay et al., 2010). Here, we showed that crTMV infection induced persistent and efficient down‐regulation of AGO1 protein in the host plant. This finding implies that the persistent AGO1 protein deficiency can interfere with the appropriate regulation of the miRNA pathway, contributing to the development of symptoms. However, the miRNA sequestering ability of crTMV p122 VSR (Csorba et al., 2007) can also be responsible for the enhanced accumulation of miRNA‐targeted mRNAs and disease symptom development in infected A. thaliana. Our data show that the AGO1 down‐regulation and miRNA sequestering persist at late time points of the infection process, potentially contributing to symptom development by disturbing the miRNA pathway. However, we cannot exclude that, at these late stages of virus infection, other factors can also contribute to the altered regulation of endogenous genes, thus influencing symptom development. For example, it has been described that Tobamovirus infection can reprogramme the auxin response, indicating that ARF levels might be partly affected by virus‐induced stress responses (Padmanabhan et al., 2008). Moreover, the transgenic expression of various VSRs revealed the central role of miR167‐targeted ARF8 in the manifestation of developmental aberrations also in Turnip mosaic virus infection‐induced pathogenesis (Jay et al., 2011). We identified the increased activity of MIR168a, but not the MIR168b precursor gene, in crTMV‐, TCV‐ and CMV‐infected A. thaliana plants. This observation can reveal the source of the excess of miR168 in infected A. thaliana. These data indicate that miR168 over‐accumulation is not only the result of the stabilizing effect of VSRs but, at least partly, is actively mediated by the increased transcription of the MIR168a gene. We have shown previously that the induction of miR168 is mediated by the p19 VSR of Tombusviruses (Varallyay et al., 2010). In virus‐infected plants, miR168 induction is generally associated with the enhanced accumulation of AGO1 mRNA. The paradoxical co‐expression of MIR168a and AGO1 genes is also present in healthy plants, and is suggested to be a part of the refined regulatory network of AGO1 protein level regulation (Vaucheret et al., 2006). In our previous work, we demonstrated that the induction of the AGO1 mRNA level is part of the host defence reaction against virus infection, whereas the induction of miR168 is mediated mainly by the viral p19 VSR. Infection with a p19 VSR‐deficient virus resulted in the elimination of miR168 induction and was accompanied by the enhanced accumulation of AGO1 mRNA and AGO1 protein (Varallyay et al., 2010). These data indicate the potential role of translational inhibition in miR168‐mediated AGO1 regulation. In crTMV‐infected zll‐3 (ago10) mutants, we showed that AGO1 repression was inhibited in spite of the induction of miR168 and AGO1 mRNA, indicating that AGO1 mRNA is under translational control. Our data suggest that virus infection‐induced miR168 is sorted mainly into ZWILLE/PINHEAD/AGO10, which exerts its activity at the level of translational repression (Burgyan and Havelda, 2011; Varallyay et al., 2010).
Our main aim was to identify the factors responsible for miR168 induction in other plant–virus interactions. We hypothesized that different VSRs may also possess a similar function to p19 VSR. Using a transient assay, which mimics the virus infection process, we revealed that four different VSRs (p122, p38, HcPro and 2b) were also able to induce miR168. This action of VSRs was an early function as, at the investigated time point (4 dpi), the robust induction of miR168 was not accompanied by significant changes in the level of other miRNAs. More importantly, similar to virus infections, the transiently expressed VSR‐mediated miR168 over‐accumulation was associated with the down‐regulation of the AGO1 protein, indicating its biological relevance. Our findings suggest that VSR‐mediated miR168‐driven control of AGO1 may be an additional mechanism to alleviate the action of RNA silencing, thereby facilitating efficient viral replication and invasion by reducing the effect of antiviral RISC. According to this model (Fig. 4C), various VSRs, possessing different main silencing suppression functions, such as siRNA binding or specific protein interaction with the components of the silencing machinery, are able to induce the transcriptional activity of the MIR168a gene, resulting in miR168‐driven control of AGO1 protein accumulation during the virus infection process. The efficiency of this mechanism varies, but was found to be present in all plant–virus interactions investigated in this study. However, the mode of action of these VSRs to modulate the activity of the MIR168a gene remains to be answered. All of the investigated VSRs show certain RNA‐binding capacities, although, for 2b and p38 VSRs, other specific protein–protein functions were also described (Azevedo et al., 2010; Zhang et al., 2006). It is possible that common RNA‐binding ability may be the factor responsible indirectly for the induction of the expression level of miR168 in the infected plants. However, it cannot be excluded that other specific characteristics of the investigated VSRs are involved in the regulation of the miR168 level. Our observations suggest that several distinct and parallel silencing activities of the same VSR protein, attacking different components of the silencing pathway simultaneously, are necessary to efficiently inhibit the RNA silencing‐based host defence.
Experimental Procedures
Plant materials, viruses
Arabidopsis thaliana (Columbia) and N. benthamiana were used as systemic hosts. Plants were grown in soil under normal growth conditions and were infected by viruses by the sap inoculation method using purified virions or purified total RNA. Virus‐infected plants were grown in a Fitotron at 21 °C. TCV, CMV strain Fny, crTMV and TEV were used for infections.
Plasmids and primers
Arabidopsis thaliana AGO1, ARF10, ARF16, ARF6, MIR168a precursor, MIR168b precursor and GUS, and N. benthamiana AGO1‐1 genes were amplified from cDNAs produced by a Revert‐aid First‐strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) from total RNA extract using the oligos detailed here. An 820‐bp fragment of A. thaliana AGO1 (NM_103737.3, 2349–3169) and an 821‐bp fragment of N. benthamiana AGO1‐1 (DQ321488.1, 2187–3008) were amplified with the use of the following oligonucleotides: Ago1s, 5′‐AAATGTGAAGGTTGGAGGAAG; Ago1as, 5′‐GATGTCTCTGGCTCCATGTA. For ARF6 of A. thaliana, a 428‐bp fragment (NM_001036038.2, 3275–3703) was amplified using ARF6s (CGACGTTCTTCTCCTCGGCG) and ARF6as (GTATTACGAGTCGACAGAGGG). For ARF10 of A. thaliana, a 557‐bp fragment (NM_128394.4, 1436–1993) was amplified using ARF10s (CTCCGGCGATGTTTCTATCG) and ARF10as (CGTTGTCGCCACCAATATCC). For ARF16 of A. thaliana, a 408‐bp fragment (NM_119154.4, 1685–2093) was amplified using ARF16s (GACAATGGGAACAACACCATGC) and ARF16as (GGCTCCTGATGCATCCCTATAGAG). For the miR168a precursor, a 223‐bp fragment (ATCHRIV50 chromosome 4, 162 659–162 881) was amplified using Pre168aS (TTCCCGGGTCATATCCCTGTCTAAAGGG) and Pre168aAS (TTGGATCCGCGTAGAAATCTTCCAGATC). For the miR168b precursor, a 227‐bp fragment (AB020744.1, 40214–40441) was amplified using Pre168bS (TTCCCGGGTGGTATCTGTTTTCTCCTCC) and Pre168bAS (TTGGATTCCGTGTCAGATCTGAAAGTTGG), and cloned into the pGEMT Easy vector.
RNA isolation and Northern blotting
Total RNA was extracted from virus‐infected, systemically symptomatic or infiltrated leaves. Homogenized plant material (0.1 g) was resuspended in 400 μL of extraction buffer [0.1 m glycine–NaOH, pH 9.0, 100 mm NaCl, 10 mm ethylenediaminetetraacetic acid (EDTA), 2% sodium dodecylsulphate and 1% sodium lauroylsarcosine] and divided into two aliquots. One part (300 μL) was used for RNA extraction with the phenol–chloroform method, and the other part was used for protein extraction. For Northern blot analyses of mRNAs, 8–10 μg of total RNA were separated on formaldehyde–1.2% agarose gels and blotted to Nytran N membrane (Schleicher & Schuell, Dassel, Germany). PCR products were used to produce radiolabelled random probes with a Decalabel DNA Labeling Kit (Fermentas) for endogenous mRNA detection. Membranes were hybridized using the Perfect Hyb buffer (Sigma‐Aldrich, St. Louis, MO, USA) according to the manufacturer's instructions. For small RNA Northern blot analyses, the total RNA samples (8–10 μg) were fractionated on denaturing 12% polyacrylamide gels containing 8 m urea and transferred to Nytran N membrane (Schleicher & Schuell). Membranes were probed with radiolabelled locked nucleic acid (LNA) oligonucleotide probes (Exiqon, Vedbaek, Denmark), complementary to the mature miRNAs, as described by Varallyay et al. (2008). miR168a and miR168b precursor intermediates were detected with minus‐strand‐specific RNA probe.
Real‐time RT‐PCR
For quantitative real‐time RT‐PCR analyses, total RNA was further purified with TRI Reagent (Sigma) and treated with DN‐ase (Fermentas). Total RNA (1 μg) was reverse transcribed with an ABI High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) using a random hexamer primer, according to the manufacturer's instructions. Real‐time PCRs were performed with the Power SYBR GREEN PCR Master Mix (Applied Biosystems). A two‐step PCR protocol was used with denaturation at 95 °C and annealing/elongation at 60 °C. Primers for amplification were as follows: AthCPHqrtS (GGCGAGAAAGGAATGGGAAA) and AthCPHqrtAS (GTTCTTGGCGGTGAAATCA) (amplifying a 111‐bp fragment of cyclophilin; AK228231.1179‐289); GUS1222qrts (TGGAGTATTGCCAACGAACC) and GUS1348qrtas (CGCAGAACATTACATTGACGCG) (amplifying 126 bp of GUS; S69414.1, 1222–1348); premiR168aqrt165s (TGATAGTAGAGTCTCACCATCGG) and premiR168aqrt273as (CCCTGCTCACAAACCAATAAAGG) (amplifying 108 nucleotides of premiR168a; EU549055, 165–273); and premiR168bqrt68s (CCGATTCAGTTGATACAAGACG) and premiR168bqrt195as (GCGATGATTGTTAAAGTTACCG) (amplifying 127 nucleotides of premiR168b; EU549069, 68–195). Gene expression ratios were estimated from C T values using the ΔΔC T approximation. Cyclophilin (CPH) was used to normalize the cDNA input. Standard deviations were calculated based on triplicates in two different independent plant samples. In all cases, differences in mRNA accumulation ratios showed the same tendency of variation, although individual ratios were not necessarily identical in the two experiments. Relative concentrations of GUS mRNA, MIR168a and MIR168b precursor RNAs were calculated in the virus‐infected samples relative to mock samples, using CPH as a reference gene, as its concentration is constant during virus infection (Havelda et al., 2008).
Agrobacterium infiltration
Agrobacterium tumefaciens C58C1/pBIN61 or A. tumefaciens AGL1/pGreen0029 harbouring the appropriate plasmid [pBINp19 (CymRSV), pBIN (Empty), pBIN‐GFP, pBINp122 (crTMV;Flag tagged), pBINp38 (TCV), pBINHcPro (TEV), pBIN2b (CMV FNy)] was infiltrated into leaves of young N. benthamiana plants (five plants per construction) according to the method described previously (Vargason et al., 2003). The infiltrated leaves containing the same individual constructs were pooled and used for Northern and Western blotting.
Protein analysis
For protein analyses, 0.1 g of plant material was collected (from systemically virus‐infected or agroinfiltrated leaves) and homogenized in an ice‐cold mortar in 400 μL of extraction buffer (0.1 m glycine–NaOH, pH 9.0, 100 mm NaCl, 10 mm EDTA, 2% sodium dodecylsulphate and 1% sodium lauroylsarcosine); 100 μL of 2 × Laemmli buffer was added to 100 μL of the homogenized sample and used for protein analysis. The remaining sample was complemented with 300 μL of extraction buffer and further processed for RNA extraction. This method ensures the comparability of protein and RNA samples. Protein samples were treated as described previously (Csorba et al., 2007), and 20–30 μL of sample were resolved on 8% sodium dodecylsulphate‐polyacrylamide gel electrophoresis, blotted to poly(vinylidene difluoride) (PVDF) transfer membrane (Hybond‐P; GE Healthcare, Freiburg, Germany) and subjected to Western blot analysis. Membranes were blocked using 5% nonfat dry milk in phosphate‐buffered saline (PBS) containing 0.05% Tween 20 (PBST) for 30 min. The target proteins were detected using anti‐N. benthamiana AGO1‐1 (1 : 1000; the custom antibody against N. benthamiana AGO1‐1 protein) (Varallyay et al., 2010), A. thaliana anti‐AGO1 [Agrisera (AS09527); 1 : 2500] and anti‐actin [Sigma (A0480); 1 : 5000]. After washing the membranes in PBST, secondary peroxidase‐conjugated anti‐rabbit or anti‐mouse antibodies were added depending on the particular antibody. The signals were visualized by a chemiluminescence (ECL kit; Amersham) according to the manufacturer's instructions. The membranes were stained with Ponceau stain to check protein loading.
Supporting information
Fig. S1 MicroRNA (miRNA) sequestration by p122 viral suppressor of RNA silencing (VSR) and miR168 incorporation into high‐molecular‐weight complexes at the late stage of crucifer‐infecting Tobamovirus (crTMV) infection.
Fig. S2 Effect of Tombusvirus coat protein (CP) expression on miR168 induction and analysis of the biological activity of various viral suppressors of RNA silencing (VSRs) in agroinfiltrated leaves.
Acknowledgements
We thank Carly Turnbull, György Szittya, József Burgyán, Daniel Silhavy and Ferenc Marincs for critical reading and helpful comments on the manuscript. We also thank Herve Vaucheret for generously providing transgenic plants expressing GUS reporter under the control of promoters of MIR168 genes. This research was supported by grants from the Hungarian Scientific Fund (OTKA K78351 and PD78049) and COST FA0806.
References
- Achard, P. , Herr, A. , Baulcombe, D.C. and Harberd, N.P. (2004) Modulation of floral development by a gibberellin‐regulated microRNA. Development, 131, 3357–3365. [DOI] [PubMed] [Google Scholar]
- Alvarado, V.Y. and Scholthof, H.B. (2011) AGO2: a new argonaute compromising plant virus accumulation. Front. Plant Sci. 2, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo, J. , Garcia, D. , Pontier, D. , Ohnesorge, S. , Yu, A. , Garcia, S. , Braun, L. , Bergdoll, M. , Hakimi, M.A. , Lagrange, T. and Voinnet, O. (2010) Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen‐encoded GW repeat protein. Genes Dev. 24, 904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N. , Tsai, C.H. , Lie, M. , Havecker, E. and Baulcombe, D.C. (2007) The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17, 1609–1614. [DOI] [PubMed] [Google Scholar]
- Bazzini, A.A. , Hopp, H.E. , Beachy, R.N. and Asurmendi, S. (2007) Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc. Natl. Acad. Sci. USA, 104, 12 157–12 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolamiol, D. , Pazhouhandeh, M. , Marrocco, K. , Genschik, P. and Ziegler‐Graff, V. (2007) The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 17, 1615–1621. [DOI] [PubMed] [Google Scholar]
- Burgyan, J. (2008) Role of silencing suppressor proteins. Methods Mol. Biol. 451, 69–79. [DOI] [PubMed] [Google Scholar]
- Burgyan, J. and Havelda, Z. (2011) Viral suppressors of RNA silencing. Trends Plant Sci. 16, 265–272. [DOI] [PubMed] [Google Scholar]
- Chapman, E.J. , Prokhnevsky, A.I. , Gopinath, K. , Dolja, V.V. and Carrington, J.C. (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.Y. , Yang, J. , Lin, C. and Yuan, Y.A. (2008) Structural basis for RNA‐silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep. 9, 754–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba, T. , Bovi, A. , Dalmay, T. and Burgyan, J. (2007) The p122 subunit of Tobacco Mosaic Virus replicase is a potent silencing suppressor and compromises both small interfering RNA‐ and microRNA‐mediated pathways. J. Virol. 81, 11 768–11 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba, T. , Lozsa, R. , Hutvagner, G. and Burgyan, J. (2010) Polerovirus protein P0 prevents the assembly of small RNA‐containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 62, 463–472. [DOI] [PubMed] [Google Scholar]
- Ding, S.W. (2010) RNA‐based antiviral immunity. Nat. Rev. Immunol. 10, 632–644. [DOI] [PubMed] [Google Scholar]
- Ding, S.W. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer, P. , Lecellier, C.H. , Parizotto, E.A. , Himber, C. and Voinnet, O. (2004) Probing the microRNA and small interfering RNA pathways with virus‐encoded suppressors of RNA silencing. Plant Cell, 16, 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Giner, A. , Lakatos, L. , Garcia‐Chapa, M. , Lopez‐Moya, J.J. and Burgyan, J. (2010) Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. Plos Pathog. 6, e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, J.J. , Lewsey, M.G. , Patel, K. , Westwood, J. , Heimstadt, S. , Carr, J.P. and Baulcombe, D.C. (2011) An antiviral defense role of AGO2 in plants. PLoS ONE, 6, e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson, A. , Plessis, A. , Blein, T. , Adroher, B. , Grigg, S. , Tsiantis, M. , Boudaoud, A. , Damerval C. and Laufs, P. (2011) Evolution and diverse roles of the CUP‐SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell, 23, 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelda, Z. , Varallyay, E. , Valoczi, A. and Burgyan, J. (2008) Plant virus infection‐induced persistent host gene downregulation in systemically infected leaves. Plant J. 55, 278–288. [DOI] [PubMed] [Google Scholar]
- Jaubert, M. , Bhattacharjee, S. , Mello, A.F. , Perry, K.L. and Moffett, P. (2011) ARGONAUTE2 mediates RNA‐silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 156, 1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay, F. , Wang, Y. , Yu, A. , Taconnat, L. , Pelletier, S. , Colot, V. , Renou, J.P. and Voinnet, O. (2011) Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis. PLoS Pathog. 7, e1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D. , Xie, Z. , Allen, E. , Llave, C. , Chapman, E.J. , Krizan, K.A. and Carrington, J.C. (2003) P1/HC‐Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell, 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Lang, Q. , Jin, C. , Lai, L. , Feng, J. , Chen, S. and Chen, J. (2011) Tobacco microRNAs prediction and their expression infected with Cucumber mosaic virus and Potato virus X. Mol. Biol. Rep. 38, 1523–1531. [DOI] [PubMed] [Google Scholar]
- Lozsa, R. , Csorba, T. , Lakatos, L. and Burgyan, J. (2008) Inhibition of 3′ modification of small RNAs in virus‐infected plants requires spatial and temporal co‐expression of small RNAs and viral silencing‐suppressor proteins. Nucleic Acids Res. 36, 4099–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A. and Vaucheret, H. (2010) Form, function, and regulation of ARGONAUTE proteins. Plant Cell, 22, 3879–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C. and Bouche, N. (2008) MicroRNA‐directed regulation: to cleave or not to cleave. Trends Plant Sci. 13, 359–367. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C. and Vaucheret, H. (2009) ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 10, 521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C. , Reinhart, B.J. , Bartel, D. , Vance, V.B. and Bowman, L.H. (2002) A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro‐RNAs in tobacco. Proc. Natl. Acad. Sci. USA, 99, 15 228–15 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C. , Bartel, D.P. and Bartel, B. (2005) MicroRNA‐directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell, 17, 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C. , Elmayan, T. and Vaucheret, H. (2008) MicroRNA maturation and action—the expanding roles of ARGONAUTEs. Curr. Opin. Plant Biol. 11, 560–566. [DOI] [PubMed] [Google Scholar]
- Merai, Z. , Kerenyi, Z. , Molnar, A. , Barta, E. , Valoczi, A. , Bisztray, G. , Havelda, Z. , Burgyan, J. and Silhavy, D. (2005) Aureusvirus P14 is an efficient RNA silencing suppressor that binds double‐stranded RNAs without size specificity. J. Virol. 79, 7217–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa, S. , Schauer, S.E. , Smith, T.H. , Mallory, A.C. , Herr, J.M., Jr , Roth, B. , Merchant, D.S. , Ray, A. , Bowman, L.H. and Vance, V.B. (2005) Ectopic DICER‐LIKE1 expression in P1/HC‐Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell, 17, 2873–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa, S. , Pruss, G.J. and Vance, V. (2008) Small RNAs in viral infection and host defense. Trends Plant Sci. 13, 375–382. [DOI] [PubMed] [Google Scholar]
- Morel, J.B. , Godon, C. , Mourrain, P. , Beclin, C. , Boutet, S. , Feuerbach, F. , Proux, F. and Vaucheret, H. (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post‐transcriptional gene silencing and virus resistance. Plant Cell, 14, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan, M.S. , Kramer, S.R. , Wang, X. and Culver, J.N. (2008) Tobacco mosaic virus replicase–auxin/indole acetic acid protein interactions: reprogramming the auxin response pathway to enhance virus infection. J. Virol. 82, 2477–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo, V. , Szittya, G. and Burgyan, J. (2007) Molecular bases of viral RNA targeting by viral small interfering RNA‐programmed RISC. J. Virol. 81, 3797–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh, M. , Dieterle, M. , Marrocco, K. , Lechner, E. , Berry, B. , Brault, V. , Hemmer, O. , Kretsch, T. , Richards, K.E. , Genschik, P. and Ziegler‐Graff V. (2006) F‐box‐like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA, 103, 1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F. , Ye, X. and Morris, T.J. (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4‐initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. USA, 105, 14 732–14 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W. , Reinhart, B.J. , Lim, L.P. , Burge, C.B. , Bartel, B. and Bartel, D. (2002) Prediction of plant microRNA targets. Cell, 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Scholthof, H.B. , Alvarado, V.Y. , Vega‐Arreguin, J.C. , Ciomperlik, J. , Odokonyero, D. , Brosseau, C. , Jaubert, M. , Zamora, A. and Moffett P. (2011) Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana . Plant Physiol. 156, 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varallyay, E. , Burgyan, J. and Havelda, Z. (2008) MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc. 3, 190–196. [DOI] [PubMed] [Google Scholar]
- Varallyay, E. , Valoczi, A. , Agyi, A. , Burgyan, J. and Havelda, Z. (2010) Plant virus‐mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 29, 3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason, J.M. , Szittya, G. , Burgyan, J. and Tanaka Hall, T.M. (2003) Size selective recognition of siRNA by an RNA silencing suppressor. Cell, 115, 799–811. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H. (2009) AGO1 homeostasis involves differential production of 21‐nt and 22‐nt miR168 species by MIR168a and MIR168b. PLoS ONE, 4, e6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H. , Vazquez, F. , Crete, P. and Bartel, D.P. (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H. , Mallory, A.C. and Bartel, D.P. (2006) AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell, 22, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.B. , Jovel, J. , Udomporn, P. , Wang, Y. , Wu, Q. , Li, W.X. , Gasciolli, V. , Vaucheret H. and Ding S.W. (2011) The 21‐nucleotide, but not 22‐nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana . Plant Cell, 23, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X. , Wang, H. , Li, H. , Yuan, Z. , Li, F. , Yang, L. and Huang, H. (2009) Two types of cis‐acting elements control the abaxial epidermis‐specific transcription of the MIR165a and MIR166a genes. FEBS Lett. 583, 3711–3717. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Yuan, Y.R. , Pei, Y. , Lin, S.S. , Tuschl, T. , Patel, D.J. and Chua, N.H. (2006) Cucumber mosaic virus‐encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 MicroRNA (miRNA) sequestration by p122 viral suppressor of RNA silencing (VSR) and miR168 incorporation into high‐molecular‐weight complexes at the late stage of crucifer‐infecting Tobamovirus (crTMV) infection.
Fig. S2 Effect of Tombusvirus coat protein (CP) expression on miR168 induction and analysis of the biological activity of various viral suppressors of RNA silencing (VSRs) in agroinfiltrated leaves.


