Summary
Pseudomonas savastanoi pv. savastanoi is the causal agent of olive (Olea europaea) knot disease and an unorthodox member of the P. syringae complex, causing aerial tumours instead of the foliar necroses and cankers characteristic of most members of this complex. Olive knot is present wherever olive is grown; although losses are difficult to assess, it is assumed that olive knot is one of the most important diseases of the olive crop. The last century witnessed a large number of scientific articles describing the biology, epidemiology and control of this pathogen. However, most P. savastanoi pv. savastanoi strains are highly recalcitrant to genetic manipulation, which has effectively prevented the pathogen from benefitting from the scientific progress in molecular biology that has elevated the foliar pathogens of the P. syringae complex to supermodels. A number of studies in recent years have made significant advances in the biology, ecology and genetics of P. savastanoi pv. savastanoi, paving the way for the molecular dissection of its interaction with other nonpathogenic bacteria and their woody hosts. The selection of a genetically pliable model strain was soon followed by the development of rapid methods for virulence assessment with micropropagated olive plants and the analysis of cellular interactions with the plant host. The generation of a draft genome of strain NCPPB 3335 and the closed sequence of its three native plasmids has allowed for functional and comparative genomic analyses for the identification of its pathogenicity gene complement. This includes 34 putative type III effector genes and genomic regions, shared with other pathogens of woody hosts, which encode metabolic pathways associated with the degradation of lignin‐derived compounds. Now, the time is right to explore the molecular basis of the P. savastanoi pv. savastanoi–olive interaction and to obtain insights into why some pathovars like it necrotic and why some like it knot.
Synonyms
Pseudomonas syringae pv. savastanoi.
Taxonomy
Kingdom Bacteria; Phylum Proteobacteria; Class Gammaproteobacteria; Family Pseudomonadaceae; Genus Pseudomonas; included in genomospecies 2 together with at least P. amygdali, P. ficuserectae, P. meliae and 16 other pathovars from the P. syringae complex (aesculi, ciccaronei, dendropanacis, eriobotryae, glycinea, hibisci, mellea, mori, myricae, phaseolicola, photiniae, sesami, tabaci, ulmi and certain strains of lachrymans and morsprunorum); when a formal proposal is made for the unification of these bacteria, the species name P. amygdali would take priority over P. savastanoi.
Microbiological properties
Gram‐negative rods, 0.4–0.8 × 1.0–3.0 μm, aerobic. Motile by one to four polar flagella, rather slow growing, optimal temperatures for growth of 25–30 °C; oxidase negative, arginine dihydrolase negative; elicits the hypersensitive response on tobacco; most isolates are fluorescent and levan negative, although some isolates are nonfluorescent and levan positive.
Host range
P. savastanoi pv. savastanoi causes tumours in cultivated and wild olive and ash (Fraxinus excelsior). Although strains from olive have been reported to infect oleander (Nerium oleander), this is generally not the case; however, strains of P. savastanoi pv. nerii can infect olive. Pathovars fraxini and nerii are differentiated from pathovar savastanoi mostly in their host range, and were not formally recognized until 1996. Literature before about 1996 generally names strains of the three pathovars as P. syringae ssp. savastanoi or P. savastanoi ssp. savastanoi, contributing to confusion on the host range and biological properties.
Disease symptoms
Symptoms of infected trees include hyperplastic growths (tumorous galls or knots) on the stems and branches of the host plant and, occasionally, on leaves and fruits.
Epidemiology
The pathogen can survive and multiply on aerial plant surfaces, as well as in knots, from where it can be dispersed by rain, wind, insects and human activities, entering the plant through wounds. Populations are very unevenly distributed in the plant, and suffer drastic fluctuations throughout the year, with maximum numbers of bacteria occurring during rainy and warm months. Populations of P. savastanoi pv. savastanoi are normally associated with nonpathogenic bacteria, both epiphytically and endophytically, and have been demonstrated to form mutualistic consortia with Erwinia toletana and Pantoea agglomerans, which could result in increased bacterial populations and disease symptoms.
Disease control
Based on preventive measures, mostly sanitary and cultural practices. Integrated control programmes benefit from regular applications of copper formulations, which should be maintained for at least a few years for maximum benefit. Olive cultivars vary in their susceptibility to olive knot, but there are no known cultivars with full resistance to the pathogen.
Useful websites
http://www.pseudomonas‐syringae.org/; http://genome.ppws.vt.edu/cgi‐bin/MLST/home.pl; ASAP access to the P. savastanoi pv. savastanoi NCPPB 3335 genome sequence https://asap.ahabs.wisc.edu/asap/logon.php.
Introduction
Pseudomonas syringae is an economically important pathogen and one of the most relevant models for the study of plant–microbe interactions (e.g. Mansfield, 2009; Mansfield et al., 2012). The species is currently a taxonomic conundrum and has been pulled together with P. amygdali, P. avellanae, P. cannabina, P. caricapapayae, P. ficuserectae, P. meliae, P. savastanoi, P. tremae and P. viridiflava into a group designated as the P. syringae complex, which could correspond to at least nine different species (Gardan et al., 1999; Parkinson et al., 2011; Young, 2010).
Pathovars of the P. syringae complex generally exploit the plant apoplast as a parasitic niche and cause foliar necrosis in diverse plant hosts, with a minority of strains causing other types of symptoms, such as vascular diseases on woody plants (Agrios, 2005). Remarkable exceptions include a few pathovars producing aerial tumours in woody plants, such as P. savastanoi pv. savastanoi. Pseudomonas savastanoi pv. savastanoi is the causal agent of olive (Olea europaea) knot disease, whose symptoms include hyperplastic growths (tumorous galls or knots) on the stems and branches of the host plant and, occasionally, on leaves and fruits (Fig. 1). Olive knot is present worldwide, wherever olive is grown, and is considered to be one of the most important diseases of olive (CMI, 1987; Quesada et al., 2010a; Young, 2004). Diverse research groups worldwide have made substantial contributions towards the understanding of the biology, epidemiology and control of this pathogen; however, most strains of P. savastanoi pv. savastanoi are highly recalcitrant to genetic manipulation (Pérez‐Martínez et al., 2007), which has significantly slowed down their molecular analysis.
Figure 1.
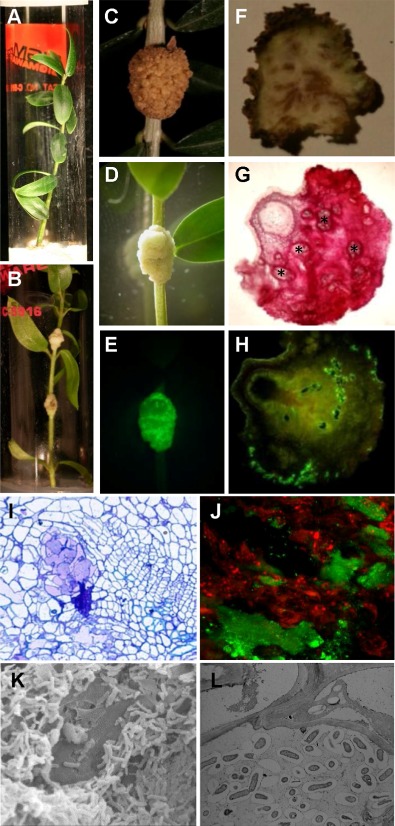
Symptoms produced by Pseudomonas savastanoi pv. savastanoi NCPPB 3335 in olive plants and pathogen visualization within knots. In vitro micropropagated olive plants not inoculated (A) and inoculated (B). (C) Knot induced on a 2‐year‐old olive plant 90 days post‐inoculation (dpi). Real‐time monitoring of green fluorescent protein (GFP)‐tagged P. savastanoi infection of a young micropropagated olive plant at 30 dpi (D) and complementary epifluorescence microscopy image (E). (F) Cross‐section of the knot exposed in (C) showing necrosis associated with infection of the stem. (G) Cross section of a 30‐dpi knot, stained with methylene blue–picrofuchsin; asterisks indicate newly formed bundles of xylem vessels. (H) Transverse section of a knot, induced by GFP‐tagged NCPPB 3335, showing GFP emission within the lumen of xylem vessels, in the internal cavities and at the periphery of the tumour tissue. (I) Semithin cross‐section of a knot stained with toluidine blue. Stained primary and secondary walls show dark and light blue in colour, respectively. (J) Scanning confocal electron microscopy (SCLM) image of a knot induced by GFP‐tagged NCPPB 3335. (K) Scanning electron micrograph showing a group of rod‐shaped P. savastanoi cells. (L) Transmission electron micrograph of ultrathin section of a knot showing pathogen cells colonizing the intercellular spaces of the host tissue.
The growing availability of microbial genomes has promoted a new research era in the field of plant–microbe interactions, leading to the identification of potentially comprehensive repertoires of putative virulence genes and the emergence of unified models of interaction between prototypical pathogens and plant hosts (Lindeberg et al., 2008; Mansfield, 2009; Schneider and Collmer, 2010). Extensive recent research efforts have focused on Pseudomonas diseases of herbaceous plants, with knowledge on the virulence and pathogenicity determinants specific for the infection of woody plants, including those of tumour‐inducing strains, lagging far behind. The selection of strain P. savastanoi pv. savastanoi NCPPB 3335 as a research model (Pérez‐Martínez et al., 2007) has opened up the way for the application of high‐throughput molecular tools to the analysis of the molecular basis of bacterial adaptation to woody hosts.
Taxonomy and Population Biology
Despite significant advances in molecular phylogeny and taxonomy, the nomenclature and classification of P. savastanoi pv. savastanoi are still a source of confusion. This bacterium is part of the P. syringae complex, encompassing at least 60 pathovars and several other Pseudomonas species (Bull et al., 2010; Young, 2010). A study limited to a few taxa formally classified pathovars glycinea, phaseolicola and savastanoi into the new species P. savastanoi (Gardan et al., 1992), to which pathovars fraxini, nerii and retacarpa were later added (Bull et al., 2010). DNA–DNA hybridization distributed P. syringae into at least nine separate genomospecies (Gardan et al., 1999; Young, 2010). Pseudomonas savastanoi pv. savastanoi was included in genomospecies 2, together with 16 other P. savastanoi–P. syringae pathovars (see ‘Summary’) and the species P. amygdali, P. ficuserectae and P. meliae; when genomospecies 2 is formally named, however, the species should be designated P. amygdali and not P. savastanoi (Gardan et al., 1999). Multilocus sequence analyses have shown that P. savastanoi pv. savastanoi NCPPB 3335 is evolutionarily closer to P. syringae pathovars aesculi 2250 and NCPPB 3681, tabaci ATCC 11528 and phaseolicola 1448A (genomospecies 2) than to P. syringae pv. tomato DC3000 (genomospecies 3) or P. syringae pv. syringae B728a (genomospecies 1) (Fig. S1, see Supporting Information) (Parkinson et al., 2011; Sarkar and Guttman, 2004). These studies support the genomospecies 2 grouping and indicate that it might encompass at least nine further pathovars (broussonetiae, castaneae, cerasicola, cunninghamiae, daphniphylli, fraxini, nerii, rhaphiolepidis and retacarpa) plus P. tremae (Parkinson et al., 2011; Sarkar and Guttman, 2004). Therefore, what name should be used for this bacterium? Although P. savastanoi is being widely used in the literature, the P. syringae designation helps to avoid the false idea that this pathogen is a different species from, for example, P. syringae pv. tabaci.
Natural isolates of P. savastanoi pv. savastanoi are heterogeneous, both phenotypically and genotypically (Table 1), although they tend to generate clonal populations in colonized areas (e.g. Quesada et al., 2008; Sisto et al., 2007). There is an important variation in virulence, with strains showing either low, intermediate or, most commonly, high virulence to diverse olive cultivars (Penyalver et al., 2006), and also variation in the size and morphology of tumours in artificial inoculations (Pérez‐Martínez et al., 2007). Certain isolates in central Italy are nonfluorescent and produce levan, in contrast with the majority of other isolates (Marchi et al., 2005). Amplified fragment length polymorphism (AFLP) data cluster these levan‐positive isolates separately from most of the common levan‐negative isolates. Arbitrarily primed polymerase chain reaction (PCR) (Krid et al., 2009; Scortichini et al., 2004), AFLP (Sisto et al., 2007) and typing with IS53 (Quesada et al., 2008) have revealed high levels of polymorphism; in addition, AFLP clearly differentiates pathovar savastanoi from pathovars fraxini and nerii. In general, genetic variability associates with geographical origin, with strains from the same area having a closer genetic relationship than those from different areas (Krid et al., 2009; Matas et al., 2009; Quesada et al., 2008; Sisto et al., 2007), suggesting a preference for clonal colonization of olive orchards; indeed, the spread of bacteria from inoculated to noninoculated trees in an olive orchard, where they produce tumours, has been documented in less than 1 year (Quesada et al., 2010a). Typing with IS53 revealed higher diversity than any of the other techniques, and could be used to track strains in the environment, because many strains display unique patterns (Quesada et al., 2008).
Table 1.
Phenotypic and genetic differences among selected pathovars of Pseudomonas savastanoi a
| P. savastanoi pv. | Plant hostb | Genomic location of hormone biosynthesis genesc | |||||
|---|---|---|---|---|---|---|---|
| Ash | Oleander | Olive | Spanish broom | iaaMH | iaaL | ptz | |
| fraxini | c | – | c | – | nd | uk | uk |
| nerii | K | K | K | – | P | P | P |
| savastanoi | K | – | K | – | Ch | Ch | Ch |
| retacarpa | – | – | – | K | uk | uk | uk |
Ash, Fraxinus excelsior; oleander, Nerium oleander; olive, Olea europaea; Spanish broom, Retama sphaerocarpa; c, cankers accompanied by wart‐like excrescences; K, knots; –, no visible symptoms.
Symbols indicate that the gene is located in the chromosome (Ch) or in plasmids (P) in at least 70% of the strains examined; nd; not detected; uk, unknown. Genes iaaMH and iaaL are involved in the biosynthesis of indoleacetic acid and indoleacetic acid lysine, respectively, whereas ptz codes for an isopentenyl transferase, involved in the biosynthesis of cytokinins.
Epidemiology and Control
Pseudomonas savastanoi pv. savastanoi does not survive for long in soil, and is normally found as an epiphyte and also endophytically, being able to migrate to produce secondary knots in new wounds (Ercolani, 1978; Penyalver et al., 2006; Quesada et al., 2007). Epiphytic life allows the build‐up of populations for plant colonization and also fosters the interaction with other microbial communities in the phyllosphere. The pathogen is usually introduced to new areas through infected plant material. The bacterium can survive and multiply as a saprophyte on plant surfaces (Ercolani, 1978; Quesada et al., 2007), as well as inside knots, from where it can be disseminated by rain, wind‐blown aerosols, insects and cultural practices, such as pruning. It enters the plant through any type of wound, such as leaf scars or those caused by pruning, harvesting, frost and hail. The presence of knots in even a single tree normally leads to the rapid infection of the whole orchard because the pathogen is very rapidly and efficiently disseminated, with significant colonization of healthy trees in as little as 1 year (Quesada et al., 2010a). The size of P. savastanoi pv. savastanoi populations is highly variable, even by several orders of magnitude between different leaves of the same shoot (Quesada et al., 2007), with the highest populations occurring in rainy months with moderate temperatures (10–20 °C).
A plethora of nonpathogenic bacterial species is found colonizing olive leaves or closely associated with knots produced by P. savastanoi pv. savastanoi, and the sizes of their populations are often positively correlated (Ercolani, 1978; Marchi et al., 2006; Moretti et al., 2011; Ouzari et al., 2008; Quesada et al., 2007; Rojas et al., 2004). Several of these species can synthesize large amounts of indoleacetic acid (IAA), which may favour the proliferation of the pathogen and colonization of the plant (Cimmino et al., 2006; Marchi et al., 2006; Ouzari et al., 2008). Pantoea agglomerans is the species most frequently found to be associated with P. savastanoi pv. savastanoi populations, its growth being stimulated in the presence of active populations of the pathogen (Marchi et al., 2006; Quesada et al., 2007). Their interaction is not completely understood, and can apparently lead to either an increase in virulence or a decrease in pathogen populations (Hosni et al., 2011; Marchi et al., 2006). As described below (see ‘Other virulence factors’), a recent study has demonstrated that both Erwinia toletana and P. agglomerans can form stable communities in planta (Hosni et al., 2011).
The literature is elusive with regard to the crop losses caused by olive knot, which greatly depend on the geographical location and olive cultivar, although it is generally accepted that it is one of the most important diseases affecting the olive crop (Young, 2004). Tree vigour, growth and yield can be moderately or severely reduced, as can the size and quality of the fruits (Quesada et al., 2010a; Schroth et al., 1973). Olive knot cannot be eradicated once it is established in a tree or orchard, and therefore its control is based on preventive measures, mostly sanitary and cultural practices (Quesada et al., 2010a, b; Young, 2004). Methods should aim to avoid the introduction and dissemination of the pathogen, for instance by using certified pathogen‐tested trees and rootstocks to start new olive groves (EPPO, 2006), by minimizing the wounding of trees and by reducing epiphytic populations of the pathogen. Detection and diagnosis of the pathogen can be performed using diverse rapid and highly sensitive PCR methodologies (see Table S1, see Supporting Information), some of which allow the differentiation of pathovars fraxini, nerii and savastanoi. Pivotal to efficient disease management is a carefully planned and executed pruning, which should always start with healthy trees and be avoided in wet weather. Chemical control with copper compounds has been traditionally used in both nurseries and the field (Teviotdale and Krueger, 2004; Young, 2004). An extensive and systematic study (Quesada et al., 2010b) reported a significant reduction in pathogen populations from the very first application of copper compounds, either copper oxychloride or cuprocalcic sulphate plus mancozeb. Nevertheless, treatments should be part of an appropriate integrated control programme that includes the regular application of two copper treatments per year. This schedule produced the greatest difference with respect to the untreated control in the third year, after five copper treatments, resulting in a significant reduction in the average number of knots per plant (Quesada et al., 2010b). Conversely, acibenzolar‐S‐methyl treatments did not result in a significant reduction in disease symptoms.
Reduction of host susceptibility, including the use of resistant cultivars, is the most effective method for the integrated control of plant diseases; unfortunately, there are no known olive cultivars that are completely resistant to the pathogen. Early comparative studies showed a considerable degree of phenotypic variation among olive cultivars, ranging from high susceptibility to a certain resistance (reviewed in Young, 2004). A larger assay evaluated the effect on symptom development of diverse variables—cultivar, plant age, development of secondary knots, inoculum dose and strain virulence—and proposed a standardized method to assess cultivar susceptibility (Penyalver et al., 2006). These authors demonstrated large differences in disease response with small variations in the inoculum dose, which might explain the discrepancies in cultivar assessment among different studies, and classified 29 cultivars in three categories of high, medium and low susceptibility to the pathogen.
Pseudomonas Savastanoi pv. Savastanoi: Life Inside the Knot
Pseudomonas savastanoi pathogenicity and virulence are generally tested on 1–3‐year‐old olive plants (Glass and Kosuge, 1988; Hosni et al., 2011; Iacobellis et al., 1994; Penyalver et al., 2006; Pérez‐Martínez et al., 2007; Sisto et al., 2004). Apart from the space required, this often results in a large variability in the size and number of knots that develop. In vitro techniques have been widely used to study the pathogenicity and virulence of animal bacterial pathogens and can also be conveniently applied in plant pathology. Several techniques have been described for the micropropagation of a vast number of fruit trees, including several olive varieties, facilitating the mass production of clonal and disease‐free plants that can easily be maintained under controlled conditions in growth chambers. The use of in vitro micropropagated olive plants has been established as a fast and inexpensive method to study the pathogenicity and virulence of P. savastanoi strains isolated from olive and oleander knots (Rodríguez‐Moreno et al., 2008). As observed previously with older olive plants, symptom development in micropropagated olive plants is highly dependent on both the olive variety and the strain. Nevertheless, histological modifications observed in in vitro olive plants after infection by P. savastanoi pv. savastanoi strains (Marchi et al., 2009; Rodríguez‐Moreno et al., 2008, 2009) are very similar to those in older olive plants (Smith, 1920; Surico, 1977; Temsah et al., 2008), further confirming the suitability of this model system.
Tagging of P. savastanoi pv. savastanoi NCPPB 3335 with the green fluorescent protein (GFP), in combination with the use of in vitro olive plants and epifluorescence microscopy, allows the real‐time monitoring of disease development at the whole‐tumour level, as well as the monitoring of bacterial localization inside knots at the single‐cell level by scanning confocal electron microscopy. In addition, scanning and transmission electron microscopy can be used for detailed ultrastructural analysis of tumour histology, as well as for the visualization of the P. savastanoi pv. savastanoi lifestyle within knot tissues (Fig. 1) (Rodríguez‐Moreno et al., 2009). A combination of these microscopy techniques was used for the in vivo analysis of P. savastanoi pv. savastanoi NCPPB 3335 mutants affected in virulence (Bardaji et al., 2011; Pérez‐Martínez et al., 2010).
Genomic Insights into P. Savastanoi pv. Savastanoi Pathogenicity and Virulence
In this section, we review how the recent sequencing of the P. savastanoi pv. savastanoi NCPPB 3335 draft genome, and the complete sequence of its three‐plasmid complement, has allowed the identification of the virulence gene complement of this tumour‐inducing pathogen of woody hosts (Bardaji et al., 2011; Rodríguez‐Palenzuela et al., 2010).
Phytohormones
In P. savastanoi, IAA is synthesized from tryptophan in two steps catalysed by the products of the genes iaaM (tryptophan monooxygenase) and iaaH (indoleacetamide hydrolase) (Comai and Kosuge, 1982; Palm et al., 1989). In addition, P. savastanoi pv. nerii (oleander isolates) also converts IAA to IAA‐lysine through the action of the iaaL gene (Glass and Kosuge, 1988), which is also present in most P. syringae complex pathovars (Glickmann et al., 1998). Although P. savastanoi pv. savastanoi strains contain two iaaL alleles (Matas et al., 2009), IAA‐lysine has not been detected in culture filtrates of P. savastanoi strains isolated from olive (Evidente et al., 1986; Glass and Kosuge, 1988). Two chromosomally encoded iaaM, iaaH and iaaL alleles were also found in the genome of P. savastanoi pv. savastanoi NCPPB 3335; however, the iaaM‐2 and iaaH‐1 alleles appeared to be pseudogenes (Rodríguez‐Palenzuela et al., 2010). Resequencing of these two loci has recently confirmed that, in fact, iaaM‐2 is a pseudogene, whereas iaaH‐1 encodes a complete coding sequence (CDS).
Gene iaaL is widely distributed within the P. syringae complex (Glickmann et al., 1998), and its phylogeny (Fig. 2) is largely congruent with the phylogeny deduced from housekeeping genes (Fig. S1), suggesting that iaaL is ancestral to the complex. However, clustering of iaaL from P. syringae pv. oryzae 1_6 (genomospecies 4) with genomospecies 2 (Fig. 2) provides evidence of horizontal transfer. This is not surprising because iaaL is often found in several copies and located in plasmids (Glickmann et al., 1998; Matas et al., 2009), although the transfer appears to preferentially occur within the P. syringae complex (not shown). Conversely, highly conserved iaaMH alleles are present in only a handful of P. syringae complex strains (Table S2, see Supporting Information) (Glickmann et al., 1998); nevertheless, diverse pathovars contain CDSs (Baltrus et al., 2011) whose deduced products show very low identity to those of iaaMH (e.g. PSPTO_0518/PSPTO_4204; 29.3%/29.7% amino acid identity), but high identity with putative monooxygenase and amidase genes common in the P. syringae complex (e.g. 99%/89% amino acid identity with PSA3335_4651/PSA3335_4172 from NCPPB 3335), and whose role in IAA biosynthesis has not been demonstrated. The limited data available also suggest the horizontal transfer of iaaMH within the P. syringae complex, which is also less related to the corresponding genes of other organisms (Table S2).
Figure 2.

Unrooted neighbour‐joining (NJ) tree of iaaL nucleotide sequences from strains of the Pseudomonas syringae complex. (See Fig. S1 for methodology and Table S3 for accession numbers.) Only the pathovar name and strain designation are shown; all strains belong to P. syringae, except P. cannabina pv. alisalensis ES4326, which was previously designated as P. syringae pv. maculicola.
Genes for phytohormone biosynthesis have a disparate genomic localization in different tumour‐inducing strains of P. savastanoi (Table 1), with those for the biosynthesis of cytokinins (CKs) preferentially located in plasmids of the pPT23A family in P. savastanoi pv. savastanoi (Macdonald et al., 1986; Pérez‐Martínez et al., 2008; Silverstone et al., 1993). The ptz gene, encoding an isopentenyl transferase and characterized by a low G + C content (43.4% G + C), was found in a potential genomic island located in plasmid pPsv48A of P. savastanoi pv. savastanoi NCPPB 3335 (Bardaji et al., 2011). Knots induced in olive plants by P. savastanoi strains cured of plasmids containing ptz are smaller (Bardaji et al., 2011; Iacobellis et al., 1994; Rodríguez‐Moreno et al., 2008) and show a lower presence of spiral vessels (Bardaji et al., 2011), than those induced by wild‐type strains. Another gene putatively involved in the biosynthesis of CKs, gene ipt, encoding a putative isopentenyl‐diphosphate delta‐isomerase, was found in plasmid pPsv48C of P. savastanoi pv. savastanoi NCPPB 3335; however, its role in virulence has not been tested, as derivatives lacking pPsv48C are not yet available (Bardaji et al., 2011).
Apparently, P. savastanoi pv. savastanoi does not belong to the group of 2‐oxoglutarate‐dependent ethylene producers, a pathway dependent on gene efe in several P. syringae pathovars (Weingart et al., 1999). First, no homology to an efe probe was found by hybridization analysis of 32 different P. savastanoi pv. savastanoi plasmids (Pérez‐Martínez et al., 2008). Second, proteins homologous to ethylene‐forming enzymes from P. syringae pv. phaseolicola, pv. glycinea and pv. pisi have not been found in the P. savastanoi pv. savastanoi NCPPB 3335 genome (Rodríguez‐Palenzuela et al., 2010).
Type III secretion system (T3SS) and effectors
Cluster analysis of HrpS protein sequences (Inoue and Takikawa, 2006) has shown that P. savastanoi pv. savastanoi NCPPB 3335 belongs to group I, which comprises exclusively proteins from P. syringae pathovars from genomospecies 2 (Gardan et al., 1999). In relation to HrpA, P. savastanoi pv. savastanoi NCPPB 3335 contains an hrpA2 gene, which is highly similar to those of P. syringae pathovars phaseolicola, glycinea and tabaci (Pérez‐Martínez et al., 2010).
In agreement with Sisto et al. (2004), a T3SS mutant of strain NCPPB 3335 was also unable to multiply in olive tissues and induce the formation of knots in woody olive plants. Interestingly, tumours induced by the T3SS mutant on young micropropagated olive plants did not show the necrosis and internal open cavities observed in knots induced by the wild‐type strain (Pérez‐Martínez et al., 2010).
Bioinformatic analysis of the P. savastanoi pv. savastanoi NCPPB 3335 genome sequence (Rodríguez‐Palenzuela et al., 2010) has allowed a prediction of hop genes, including 19 putative T3SS effectors with amino acid identities of 65%–80% to previously described effectors. In addition, a further 11 candidate genes do not share sequence similarity with known effectors (Rodríguez‐Palenzuela et al., 2010). A later revision of this genome sequence identified four new candidate effectors: AvrPto1, HopAT1′, HopAZ1 and HopF4 (Hops Database, http://www.pseudomonas‐syringae.org/home.html) (Fig. 3). Furthermore, sequencing of the three‐plasmid complement of this strain revealed that two of the T3SS effector genes are plasmid encoded: hopAF1 (plasmid pPsv48A) and hopAO1 (plasmid pPsv48B) (Bardaji et al., 2011). Figure 3 shows an updated and corrected comparison of the T3SS effector gene complements of P. savastanoi pv. savastanoi NCPPB 3335 and other sequenced plant‐pathogenic pseudomonads. Translocation analysis of the T3SS effector repertoire of P. savastanoi pv. savastanoi NCPPB 3335 is currently in progress.
Figure 3.

Updated and corrected comparison of the type III effector gene complements of Pseudomonas savastanoi pv. savastanoi (Psv) NCPPB 3335 and other sequenced plant‐pathogenic pseudomonads. Pph, P. syringae pv. phaseolicola; Pta, P. syringae pv. tabaci. Gene hopAF1‐2, plasmid encoded in NCPPB 3335, shows 73%–74% amino acid identity with hopAF1 from Psy B728a, Pph 1448A and Pto DC3000. Psv NCPPB 3335 effectors included in the Hop database (http://www.pseudomonas‐syringae.org/pst_func_gen2.htm) are indicated in bold type. #Plasmid‐encoded gene; asterisks indicate putative pseudogenes; hop genes truncated by a frameshift or a premature stop codon are indicated by a single quotation mark (Lindeberg et al., 2008).
Other virulence factors
The pathogenicity of P. savastanoi pv. savastanoi in olive critically depends on quorum sensing (QS) regulation. The QS system of P. savastanoi pv. savastanoi strain DAPP‐PG 722 consists of a luxI homologue (pssI) and a luxR homologue (pssR) (Hosni et al., 2011). However, the lack of signal production in a pssI mutant of this pathogen has been shown to be complemented in planta by the presence of wild‐type E. toletana, a nonpathogenic bacterium that is very often found to be associated with the olive knot pathogen (Hosni et al., 2011). Erwinia toletana produces the same N‐acyl‐homoserine lactone molecules as P. savastanoi pv. savastanoi; moreover, populations of E. toletana significantly decline over time after inoculation in olive tissues, but increase on co‐inoculation with a strain of P. savastanoi pv. savastanoi. This relationship is mutualistic, because the populations of P. savastanoi pv. savastanoi also increase significantly when the pathogen is co‐inoculated with E. toletana; in addition, the knot size also increases, reflecting an increase in virulence (Hosni et al., 2011). The mechanism underlying this relationship is not fully clear, but it appears to result, at least in part, from the sharing of QS signalling mediated by N‐acyl‐homoserine lactones.
Other known virulence determinants in plant‐pathogenic Pseudomonas include phytotoxins, cell wall‐degrading hydrolytic enzymes, extracellular polysaccharides, iron uptake systems, resistance to plant‐derived antimicrobials, adhesion, and the general processes of motility and chemotaxis. Annotation of the P. savastanoi pv. savastanoi NCPPB 3335 draft genome revealed the existence of 551 genes potentially involved in several processes that could contribute to virulence, most of which are conserved in P. syringae pv. phaseolicola 1448A. However, the subset of P. savastanoi pv. savastanoi NCPPB 3335‐specific genes (not found in 1448A), includes a cellulase, a pectate lyase and a putative filamentous haemagglutinin (Rodríguez‐Palenzuela et al., 2010). Genes for levansucrase, the enzyme responsible for the biosynthesis of the exopolysaccharide levan, are found in the genome of all sequenced P. syringae strains, although their numbers vary from three to one (O'Brien et al., 2011). Only a single levansucrase‐coding gene (PSA3335_2033) was identified in P. savastanoi pv. savastanoi NCPPB 3335 (Rodríguez‐Palenzuela et al., 2010), probably because P. savastanoi pv. savastanoi strains are, in general, levan negative, whereas the P. syringae pathovars of LOPAT subgroup 1a are all levan positive (Lelliott and Stead, 1987). The relevance of all these putative virulence factors in P. savastanoi has not been reported to date.
Metabolic Versatility and Adaptation to Woody Hosts
Pseudomonas syringae pathovars are nutritionally specialized for growth in the plant environment relative to nonpathogenic pseudomonads (Rico et al., 2011). Biolog GN2 MicroPlate Technology (Bochner et al., 2001) has revealed that the carbon utilization profiles of five different P. savastanoi pv. savastanoi strains, including NCPPB 3335, are almost identical. However, comparative analysis with previously reported data for P. syringae pathovars and nonpathogenic pseudomonads (Mithani et al., 2011; Rico and Preston, 2008) shows that the metabolic activities of P. savastanoi pv. savastanoi are more similar to those shown by P. syringae pv. tabaci ATCC 11528, P. syringae pv. tomato DC3000 and P. syringae pv. syringae B728a than to those observed for P. syringae pv. phaseolicola 1448A (Fig. 4), despite the fact that both strains ATCC 11528 and 1448A cluster together with P. savastanoi pv. savastanoi NCPPB 3335 by multilocus sequence analysis of housekeeping genes (Group 3, Fig. S1). Thus, nutritional divergence does not mirror phylogenetic divergence, possibly as a result of host‐specific features or pathogen evolutionary history.
Figure 4.
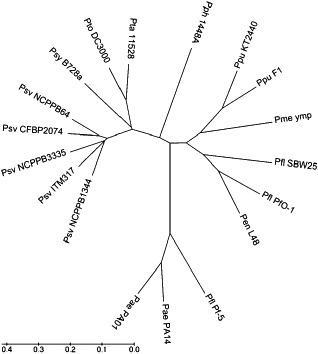
Unrooted unweighted pair group method with arithmetic mean (UPGMA) tree based on nutrient utilization data of Pseudomonas savastanoi pv. savastanoi (Psv) and other pseudomonads. Metabolic activities of Psv strains, tested using Biolog GN2 plates, were compared with carbon utilization data reported for P. syringae strains and nonplant pathogenic species of Pseudomonas (Rico and Preston, 2008). The tree was constructed using MEGA5 (Tamura et al., 2011) and is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the dendrogram. Distances were computed using the maximum composite likelihood method and are in units of the number of substrate utilizations per site. P. syringae pathovars: syringae, Psy; tomato, Pto; tabaci, Pta; phaseolicola, Pph. Pseudomonas putida, Ppu; P. entomophila, Pme; P. fluorescens, Pfl; P. aeruginosa, Pae.
The production of phenolic compounds, which provide a natural defence against pathogen attack, is greatly increased in olive knots induced by P. savastanoi pv. savastanoi (Cayuela et al., 2006), suggesting that bacterial resistance to phenols could be of paramount importance in pathogenicity. The P. savastanoi pv. savastanoi NCPPB 3335 genome (Rodríguez‐Palenzuela et al., 2010) encodes a region of about 15 kb, named VR8 (60.1% G + C), which is absent in all sequenced P. syringae strains infecting herbaceous plants, but shared with P. syringae pathovars infecting woody hosts, such as aesculi (Green et al., 2010), morsprunorum and actinidiae (Fig. 5), which are pathogenic to chestnut, cherry and kiwi, respectively. Among other genes encoded in this region, the antABC and catBCA operons are involved in the degradation of anthranilate and catechol, respectively, and could offer a selective advantage for growth in woody hosts. Indeed, the antABC cluster is homologous to the anthranilate degradation genes found on plasmid pCAR1 of Pseudomonas resinovorans (Nojiri et al., 2002; Urata et al., 2004), a bacterium commonly found in the lubricating oils of wood mills. Other metabolic pathways involving the cat and/or ant genes included in the KEGG Pathway Database (http://www.genome.jp/kegg/pathway.html) are those related to the degradation of benzoate, fluorobenzoate, toluene, chlorocyclohexane and chlorobenzene. In P. savastanoi pv. savastanoi NCPPB 3335 and all other strains encoding VR8, the genetic content and chromosomal location of this region are identical (Fig. 5). However, genetic elements suggesting its possible acquisition by horizontal transfer were not found bordering VR8 in P. savastanoi pv. savastanoi NCPPB 3335 (Rodríguez‐Palenzuela et al., 2010).
Figure 5.
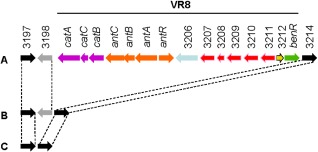
Schematic map of variable region 8 (VR8) in the genomes of Pseudomonas savastanoi pv. savastanoi NCPPB 3335 and other sequenced P. syringae pathovars. (A) Pseudomonas savastanoi pv. savastanoi NCPPB 3335, P. syringae pathovars aesculi strains 2250 and NCPPB 3681, morsprunorum MAFF302280 and actinidiae MAFF302091. (B) Pseudomonas syringae pathovars tabaci ATCC 11528, mori 301020, phaseolicola 1448A, glycinea race 4, lachrymans MAFF302278 and japonica MAFF301072. (C) Pseudomonas syringae pathovars syringae B728a, tomato DC3000 and oryzae 1_6. Black and grey arrows indicate genes flanking VR8 in P. savastanoi pv. savastanoi NCPPB 3335 which are present (PSA3335_3197 and PSA3335_3214) or not (PSA3335_3198), respectively, in the genome of all the strains analysed. Pink and orange arrows indicate genes involved in the catabolism of catechol (catBCA) and anthranilate (antABC and antR), respectively. PSA3335_3197, outer membrane protein; PSA3335_3198, ribosomal‐protein‐S5p‐alanine acetyltransferase (rimJ); PSA3335_3206, aerotaxis receptor; PSA3335_3207, nitrilotriacetate monooxygenase component B, flavin reductase; PSA3335_3208, protein involved in meta‐pathway of phenol degradation; PSA3335_3209, putative oxygenase subunit; PSA3335_3210, short‐chain alcohol dehydrogenase/reductase; PSA3335_3211, dienelactone hydrolase; PSA3335_3212, hypothetical protein; PSA3335_3214, voltage‐dependent potassium channel protein.
Plasmid Genetics and Biology
Plasmids are the main agents in the horizontal exchange of DNA amongst bacteria, and the P. syringae complex contains a significant horizontal gene pool distributed in diverse native plasmids (Jackson et al., 2011). Strains of P. savastanoi pv. savastanoi usually contain one to six plasmids (around 10 to >100 kb) (Murillo and Keen, 1994; Pérez‐Martínez et al., 2008). Most of these belong to the pPT23A‐like family of plasmids (PFP), characterized for sharing a highly conserved replication module (Gibbon et al., 1999), although strains might contain from zero to four non‐PFP plasmids. As usual in the P. syringae complex, plasmid profiles are highly variable and often strain specific, offering a simple method of strain tracking (Pérez‐Martínez et al., 2007, 2008). Nevertheless, plasmid profiles of P. syringae complex strains are dynamic and often change in response to repeated subculture or interaction with the host (e.g. Lovell et al., 2011).
PFP plasmids carry a panoply of genes involved in pathogenicity, virulence and adaptation to the environment, such as genes for T3SS effectors, type IV secretion systems, phytotoxins, phytohormones and resistance to antibiotics and heavy metals, as well as an array of insertion sequences (Sundin, 2007). Similar types of genes have been found in 32 native plasmids from 10 P. savastanoi pv. savastanoi strains using a macroarray containing 135 different genes, albeit with a limited presence of rulAB genes for UV radiation tolerance. This could be significant because rulAB genes often appear to control the expression of integrases, and are predicted to facilitate the dispersal of associated T3SS effector genes (Jackson et al., 2011). Native plasmids from P. savastanoi pv. savastanoi contain diverse virulence genes carried indistinctly by PFP and non‐PFP plasmids (Pérez‐Martínez et al., 2008), although PFP plasmids have been traditionally recognized as the main, or the only, repository of valuable genes in the P. syringae complex. At least eight T3SS effector genes (Jackson et al., 2002; Pérez‐Martínez et al., 2008) are frequently found on P. savastanoi pv. savastanoi plasmids. Other relevant virulence genes are those involved in the biosynthesis of phytohormones, which were the first plasmid‐borne pathogenicity genes found in Pseudomonas spp. (Comai and Kosuge, 1980). Unlike pathovar nerii, most strains of pathovar savastanoi carry chromosomal copies of genes for the biosynthesis of IAA and CKs (Table 1).
The complete sequences of the three‐PFP plasmid complement of strain NCPPB 3335 (pPsv48A, 78 kb; pPsv48B, 45 kb; pPsv48C, 42 kb) (Bardaji et al., 2011) contain 152 predicted CDSs; the majority (38 CDSs) have been annotated as hypothetical proteins, followed by 37 CDSs involved in DNA metabolism, including plasmid replication and maintenance. Each of the plasmids contain at least one putative toxin–antitoxin system, involved in plasmid maintenance, which is probably why we could not obtain derivatives cured of the three plasmids (Bardaji et al., 2011). The three plasmids contain seven putative virulence genes, five of which are putative type III effectors preceded by an Hrp‐box: pPsv48A contains a chimeric copy of gene hopAF1, included in the transposon effector ISPsy30, plus three copies of a large CDS found in many plant‐associated proteobacteria, whereas pPsv48B contains gene hopAO1 (avrPphD2). In addition, two genes putatively involved in CK biosynthesis, ptz (PSPSV_A0024) and ipt (PSPSV_C0024), are also found in plasmids A and C, respectively.
Plasmids are very plastic and dynamic molecules, facilitating the exchange of sequences among them and with the chromosome (Jackson et al., 2011; Ma et al., 2007; Sundin, 2007). This is illustrated by plasmids pPsv48B and pPsv48C, which probably arose from a duplication event because their replication gene, repA, is 98.6% identical. However, they only share around 10 kb with at least 80% identity, implying they participate in an active exchange of DNA. Indeed, pPsv48B contains a complete type IVA secretion system and a well‐conserved origin of conjugational transfer, suggesting that it might be conjugative; in addition, pPsv48C also contains an origin of transfer and could be mobilizable by pPsv48B. Although, in principle, plasmids can be transferred to very distant organisms, they tend to propagate within a specific host clade (Jackson et al., 2011). A phylogenetic analysis of the repA gene from diverse PFP plasmids, and of other genes carried by them, indicate that they are actively exchanging DNA and moving amongst P. syringae complex pathovars (Ma et al., 2007).
The role of native plasmids in the life cycle of P. savastanoi pv. savastanoi has not been assessed in detail because of the difficulties of genetic manipulation and plasmid curing. Nevertheless, in diverse strains of P. savastanoi pv. savastanoi and pv. nerii, certain native plasmids are essential for the expression of wild‐type symptoms, to reach high population densities in planta and for competitive fitness, all of which are related to the presence in these plasmids of genes for IAA and/or CK biosynthesis (Bardaji et al., 2011; Iacobellis et al., 1994; Rodríguez‐Moreno et al., 2008; Silverstone et al., 1993). As these effects are very drastic, they could conceivably have obscured more subtle roles in the pathogenic process of other plasmid‐borne genes; however, the current availability of genetically tractable strains and plasmid sequences will facilitate a more detailed analysis of their potential role.
Future Prospects
Diseases of woody plants caused by pathovars of the P. syringae complex are of major concern in fruit‐producing areas and nurseries worldwide, and result in considerable economic losses (Kennelly et al., 2007). Undoubtedly, advances in the understanding of diseases caused by P. syringae pathovars on herbaceous plants, including the model plant Arabidopsis, are relevant to our understanding of fruit tree diseases, and vice versa. However, there is a pressing need for appropriate research model systems facilitating the identification and analysis of specific determinants involved in bacterial interactions with trees and shrubs. A series of studies in recent years have made significant advances in the biology, ecology, genetics and genomics of P. savastanoi pv. savastanoi, which has emerged as a powerful and uniquely valuable model for the study of the molecular basis of disease production and tumour formation in woody hosts. Analysis of the P. savastanoi–olive interaction, and comparison with the model systems of herbaceous plants, can provide insights into the interactions of other bacterial pathogens with woody hosts and address relevant unresolved questions, such as: What is the role of the T3SS system and its effectors during infection of woody tissues? Are there differences in the metabolic network required by bacterial pathogens for survival in woody hosts and herbaceous hosts? What virulence determinants are singularly required for infection of woody tissues? What factors are involved in tumour induction by P. savastanoi and what evolutionary advantage derives from producing them instead of necroses? To what degree do bacterial consortia influence disease incidence and severity, and can they be targeted for disease control? What traits govern host specificity in P. savastanoi pathovars? Comparative genomics among P. syringae and P. savastanoi pathovars is generating workable hypotheses to critically investigate these questions. However, a great deal of research remains to establish genome‐wide approaches that will allow the functional characterization of bacterial interactions with woody hosts and to develop effective control strategies for Pseudomonas diseases. Genetic dissection of the P. savastanoi pv. savastanoi–olive pathosystem is technically very challenging and requires the analysis of the always unfriendly woody plants but, as Osgood wisely summarized in the delightful Billy Wilder film, ‘Well, nobody's perfect’.
Supporting information
Fig. S1 Evolutionary relationships of Pseudomonas savastanoi pv. savastanoi and selected P. syringae pathovars. The tree was constructed by multilocus sequence analysis using a concatenated dataset (exactly 12 000 nucleotides) of acnB, fruK, gapA, gltA, gyrB, pgi, recA and rpoD genes. Phylogenetic groups 1, 2, 3 and 4 (Sarkar and Guttman, 2004; Studholme, 2011) correspond to genomospecies (Gsp) 3, 1, 2 and 4 (Gardan et al., 1999), respectively. Sequence alignment using Muscle; determination of the optimal nucleotide substitution model and phylogenetic tree construction were performed using MEGA5 (Tamura et al., 2011); all positions containing gaps and missing data were eliminated using the option of complete deletion. Bootstrap values (1000 repetitions) are shown on the branches. Similar or identical topologies were obtained by maximum likelihood. The scale bar represents nucleotide substitutions per site.
Table S1 Primers used for the detection of Pseudomonas savastanoi pv. savastanoi.
Table S2 Comparison of the deduced products of iaaM‐1 (PSA3335_1475) and iaaH‐1 (PSA3335_1476), from Pseudomonas savastanoi pv. savastanoi NCPPB 3335, with their homologues in selected organismsa.
Table S3 Accession numbers and coordinates of the nucleotide sequences used for the construction of the neighbour‐joining tree shown in Fig. 2.
Acknowledgements
This work was supported by the Spanish Plan Nacional I+D+i grants AGL2008‐05311‐C02‐01, AGL2008‐05311‐C02‐02, AGL2011‐30343‐C02‐01 and AGL2011‐30343‐C02‐02 (Ministerio de Economía y Competitividad), co‐financed by Fondo Europeo de Desarrollo Regional (FEDER), and by grant P08‐CVI‐03475 from the Junta de Andalucía, Spain (http://www.juntadeandalucia.es). IMM and IMA were supported by the Ramón Areces Foundation (Spain) and by an FPU fellowship from the Ministerio de Economía y Competitividad (Spain), respectively. We thank L. Rodríguez‐Moreno for confocal and electron microscopy images and T. Osinga for help with the English language.
References
- Agrios, G.N. (2005) Plant Pathology. San Diego, CA: Elsevier Academic Press. [Google Scholar]
- Baltrus, D.A. , Nishimura, M.T. , Romanchuk, A. , Chang, J.H. , Mukhtar, M.S. , Cherkis, K. , Roach, J. , Grant, S.R. , Jones, C.D. and Dangl, J.L. (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 7, e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardaji, L. , Pérez‐Martínez, I. , Rodríguez‐Moreno, L. , Rodríguez‐Palenzuela, P. , Sundin, G.W. , Ramos, C. and Murillo, J. (2011) Sequence and role in virulence of the three plasmid complement of the model tumor‐inducing bacterium Pseudomonas savastanoi pv. savastanoi NCPPB 3335. PLoS ONE, 6, e25705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner, B.R. , Gadzinski, P. and Panomitros, E. (2001) Phenotype microarrays for high‐throughput phenotypic testing and assay of gene function. Genome Res. 11, 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, C.T. , De Boer, S.H. , Denny, T.P. , Firrao, G. , Fischer‐Le Saux, M. , Saddler, G.S. , Scortichini, M. , Stead, D.E. and Takikawa, Y. (2010) Comprehensive list of names of plant pathogenic bacteria, 1980–2007. J. Plant Pathol. 92, 551–592. [Google Scholar]
- Cayuela, J.A. , Rada, M. , Rios, J.J. , Albi, T. and Guinda, A. (2006) Changes in phenolic composition induced by Pseudomonas savastanoi pv. savastanoi infection in olive tree: presence of large amounts of verbascoside in nodules of tuberculosis disease. J. Agric. Food Chem. 54, 5363–5368. [DOI] [PubMed] [Google Scholar]
- Cimmino, A. , Andolfi, A. , Marchi, G. , Surico, G. and Evidente, A. (2006) Phytohormone production by strains of Pantoea agglomerans from knots on olive plants caused by Pseudomonas savastanoi pv. savastanoi . Phytopathol. Mediterr. 45, 247–252. [Google Scholar]
- CMI (1987) Pseudomonas syringae pv. savastanoi (E.F. Smith) Young, Dye & Wilkie CMI Distribution Maps of Plant Diseases, Map 135, 4th edn. Wallingford, Oxon.: CAB International. [Google Scholar]
- Comai, L. and Kosuge, T. (1980) Involvement of plasmid deoxyribonucleic acid in indoleacetic acid synthesis in Pseudomonas savastanoi . J. Bacteriol. 143, 950–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai, L. and Kosuge, T. (1982) Cloning and characterization of iaaM, a virulence determinant of Pseudomonas savastanoi . J. Bacteriol. 149, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPPO (2006) Pathogen‐tested olive trees and rootstocks. EPPO Bull. 36, 77–83. [Google Scholar]
- Ercolani, G.L. (1978) Pseudomonas savastanoi and other bacteria colonizing surface of olive leaves in the field. J. Gen. Microbiol. 109, 245–257. [Google Scholar]
- Evidente, A. , Surico, G. , Iacobellis, N.S. and Randazzo, G. (1986) a‐N‐acetyl‐indole‐3‐acetyl‐e‐L‐lysine: a metabolite of indole‐3‐acetic‐acid from Pseudomonas syringae pv. savastanoi . Phytochemistry, 25, 125–128. [Google Scholar]
- Gardan, L. , Bollet, C. , Abu Ghorrah, M. , Grimont, F. and Grimont, P.A.D. (1992) DNA relatedness among the pathovar strains of Pseudomonas syringae subsp. savastanoi Janse (1982) and proposal of Pseudomonas savastanoi sp. nov. Int. J. Syst. Bacteriol. 42, 606–612. [Google Scholar]
- Gardan, L. , Shafik, H. , Belouin, S. , Broch, R. , Grimont, F. and Grimont, P.A.D. (1999) DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959). Int. J. Syst. Bacteriol. 49, 469–478. [DOI] [PubMed] [Google Scholar]
- Gibbon, M.J. , Sesma, A. , Canal, A. , Wood, J.R. , Hidalgo, E. , Brown, J. , Vivian, A. and Murillo, J. (1999) Replication regions from plant‐pathogenic Pseudomonas syringae plasmids are similar to ColE2‐related replicons. Microbiology, 145, 325–334. [DOI] [PubMed] [Google Scholar]
- Glass, N.L. and Kosuge, T. (1988) Role of indoleacetic acid lysine synthetase in regulation of indoleacetic‐acid pool size and virulence of Pseudomonas syringae subsp. savastanoi . J. Bacteriol. 170, 2367–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickmann, E. , Gardan, L. , Jacquet, S. , Hussain, S. , Elasri, M. , Petit, A. and Dessaux, Y. (1998) Auxin production is a common feature of most pathovars of Pseudomonas syringae . Mol. Plant–Microbe Interact. 11, 156–162. [DOI] [PubMed] [Google Scholar]
- Green, S. , Studholme, D.J. , Laue, B.E. , Dorati, F. , Lovell, H. , Arnold, D. , Cottrell, J.E. , Bridgett, S. , Blaxter, M. , Huitema, E. , Thwaites, R. , Sharp, P.M. , Jackson, R.W. and Kamoun, S. (2010) Comparative genome analysis provides insights into the evolution and adaptation of Pseudomonas syringae pv. aesculi on Aesculus hippocastanum . PLoS ONE, 5, e10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosni, T. , Moretti, C. , Devescovi, G. , Suárez‐Moreno, Z.R. , Fatmi, M.B. , Guarnaccia, C. , Pongor, S. , Onofri, A. , Buonaurio, R. and Venturi, V. (2011) Sharing of quorum‐sensing signals and role of interspecies communities in a bacterial plant disease. ISME J. 5, 1857–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobellis, N.S. , Sisto, A. , Surico, G. , Evidente, A. and DiMaio, E. (1994) Pathogenicity of Pseudomonas syringae subsp. savastanoi mutants defective in phytohormone production. J. Phytopathol. 140, 238–248. [Google Scholar]
- Iacobellis, N.S. , Caponero, A. and Evidente, A. (1998) Characterization of Pseudomonas syringae ssp. savastanoi strains isolated from ash. Plant Pathol. 47, 73–83. [Google Scholar]
- Inoue, Y. and Takikawa, Y. (2006) The hrpZ and hrpA genes are variable, and useful for grouping Pseudomonas syringae bacteria. J. Gen. Plant Pathol. 72, 26–33. [Google Scholar]
- Jackson, R.W. , Mansfield, J.W. , Ammouneh, H. , Dutton, L.C. , Wharton, B. , Ortiz‐Barredo, A. , Arnold, D.L. , Tsiamis, G. , Sesma, A. , Butcher, D. , Boch, J. , Kim, Y.J. , Martin, G.B. , Tegli, S. , Murillo, J. and Vivian, A. (2002) Location and activity of members of a family of virPphA homologues in pathovars of Pseudomonas syringae and P. savastanoi . Mol. Plant Pathol. 3, 205–216. [DOI] [PubMed] [Google Scholar]
- Jackson, R.W. , Vinatzer, B. , Arnold, D.L. , Dorus, S. and Murillo, J. (2011) The influence of the accessory genome on bacterial pathogen evolution. Mob. Genet. Elem. 1, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse, J.D. (1982) Pseudomonas syringae subsp. savastanoi (ex Smith) subsp. nov., nom. rev., the bacterium causing excrescences on Oleaceae and Nerium oleander L. Int. J. Syst. Bacteriol. 32, 166–169. [Google Scholar]
- Kennelly, M.M. , de Cazorla, F.M., Vicente, A. , Ramos, C. and Sundin, G.W. (2007) Pseudomonas syringae diseases of fruit trees. Progress toward understanding and control. Plant Dis. 91, 4–17. [DOI] [PubMed] [Google Scholar]
- Krid, S. , Rhouma, A. , Quesada, J.M. , Penyalver, R. and Gargouri, A. (2009) Delineation of Pseudomonas savastanoi pv. savastanoi strains isolated in Tunisia by random‐amplified polymorphic DNA analysis. J. Appl. Microbiol. 106, 886–894. [DOI] [PubMed] [Google Scholar]
- Lelliott, R.A. and Stead, D.E. (1987) Methods for the Diagnosis of Bacterial Diseases of Plants. Oxford: Blackwell Scientific Publications. [Google Scholar]
- Lindeberg, M. , Stavrinides, J. , Chang, J.H. , Alfano, J.R. , Collmer, A. , Dangl, J.L. , Greenberg, J.T. , Mansfield, J.W. and Guttman, D.S. (2005) Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen. Pseudomonas syringae. Mol Plant Microbe Interact. 18, 275–282. [DOI] [PubMed] [Google Scholar]
- Lindeberg, M. , Myers, C.R. , Collmer, A. and Schneider, D.J. (2008) Roadmap to new virulence determinants in Pseudomonas syringae: insights from comparative genomics and genome organization. Mol. Plant–Microbe Interact. 21, 685–700. [DOI] [PubMed] [Google Scholar]
- Lovell, H.C. , Jackson, R.W. , Mansfield, J.W. , Godfrey, S.A.C. , Hancock, J.T. , Desikan, R. and Arnold, D.L. (2011) In planta conditions induce genomic changes in Pseudomonas syringae pv. phaseolicola. Mol. Plant Pathol. 12, 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Z. , Smith, J.J. , Zhao, Y. , Jackson, R.W. , Arnold, D.L. , Murillo, J. and Sundin, G.W. (2007) Phylogenetic analysis of the pPT23A plasmid family of Pseudomonas syringae . Appl. Environ. Microbiol. 73, 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald, E.M.S. , Powell, G.K. , Regier, D.A. , Glass, N.L. , Roberto, F. , Kosuge, T. and Morris, R.O. (1986) Secretion of zeatin, ribosylzeatin, and ribosyl‐1′′‐methylzeatin by Pseudomonas savastanoi: plasmid‐coded cytokinin biosynthesis. Plant Physiol. 82, 742–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, J.W. (2009) From bacterial avirulence genes to effector functions via the hrp delivery system: an overview of 25 years of progress in our understanding of plant innate immunity. Mol. Plant Pathol. 10, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. , Citovsky, V. , Sriariyanum, M. , Ronald, P. , Dow, M.A.X. , Verdier, V. , Beer, S.V. , Machado, M.A. , Toth, I.A.N. , Salmond, G. and Foster, G. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614 – 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchi, G. , Viti, C. , Giovannetti, L. and Surico, G. (2005) Spread of levan‐positive populations of Pseudomonas savastanoi pv. savastanoi, the causal agent of olive knot, in central Italy. Eur. J. Plant Pathol. 112, 101–112. [Google Scholar]
- Marchi, G. , Sisto, A. , Cimmino, A. , Andolfi, A. , Cipriani, M.G. , Evidente, A. and Surico, G. (2006) Interaction between Pseudomonas savastanoi pv. savastanoi and Pantoea agglomerans in olive knots. Plant Pathol. 55, 614–624. [Google Scholar]
- Marchi, G. , Mori, B. , Pollacci, P. , Mencuccini, M. and Surico, G. (2009) Systemic spread of Pseudomonas savastanoi pv. savastanoi in olive explants. Plant Pathol. 58, 152–158. [Google Scholar]
- Matas, I.M. , Pérez‐Martínez, I. , Quesada, J.M. , Rodriguez‐Hervá, J.J. , Penyalver, R. and Ramos, C. (2009) Pseudomonas savastanoi pv. savastanoi contains two iaaL paralogs, one of which exhibits a variable number of a trinucleotide (TAC) tandem repeat. Appl. Environ. Microbiol. 75, 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithani, A. , Hein, J. and Preston, G.M. (2011) Comparative analysis of metabolic networks provides insight into the evolution of plant pathogenic and nonpathogenic lifestyles in Pseudomonas . Mol. Biol. Evol. 28, 483–499. [DOI] [PubMed] [Google Scholar]
- Moretti, C. , Hosni, T. , Vandemeulebroecke, K. , Brady, C. , De Vos, P. , Buonaurio, R. and Cleenwerck, I. (2011) Erwinia oleae sp. nov., isolated from olive knots caused by Pseudomonas savastanoi pv. savastanoi . Int. J. Syst. Evol. Microbiol. 61, 2745–2752. [DOI] [PubMed] [Google Scholar]
- Murillo, J. and Keen, N.T. (1994) Two native plasmids of Pseudomonas syringae pathovar tomato strain PT23 share a large amount of repeated DNA, including replication sequences. Mol. Microbiol. 12, 941–950. [DOI] [PubMed] [Google Scholar]
- Nojiri, H. , Maeda, K. , Sekiguchi, H. , Urata, M. , Shintani, M. , Yoshida, T. , Habe, H. and Omori, T. (2002) Organization and transcriptional characterization of catechol degradation genes involved in carbazole degradation by Pseudomonas resinovorans strain CA10. Biosci. Biotechnol. Biochem. 66, 897–901. [DOI] [PubMed] [Google Scholar]
- O'Brien, H.E. , Thakur, S. and Guttman, D.S. (2011) Evolution of plant pathogenesis in Pseudomonas syringae: a genomics perspective. Annu. Rev. Phytopathol. 49, 269–289. [DOI] [PubMed] [Google Scholar]
- Ouzari, H. , Khsairi, A. , Raddadi, N. , Jaoua, L. , Hassen, A. , Zarrouk, M. , Daffonchio, D. and Boudabous, A. (2008) Diversity of auxin‐producing bacteria associated to Pseudomonas savastanoi‐induced olive knots. J. Basic Microbiol. 48, 370–377. [DOI] [PubMed] [Google Scholar]
- Palm, C.J. , Gaffney, T. and Kosuge, T. (1989) Cotranscription of genes encoding indoleacetic‐acid production in Pseudomonas syringae subsp. savastanoi. J. Bacteriol. 171, 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson, N. , Bryant, R. , Bew, J. and Elphinstone, J. (2011) Rapid phylogenetic identification of members of the Pseudomonas syringae species complex using the rpoD locus. Plant Pathol. 60, 338–344. [Google Scholar]
- Penyalver, R. , García, A. , Ferrer, A. , Bertolini, E. , Quesada, J.M. , Salcedo, C.I. , Piquer, J. , Pérez‐Panadés, J. , Carbonell, E.A. , del Río, C. , Caballero, J.M. and López, M.M. (2006) Factors affecting Pseudomonas savastanoi pv. savastanoi plant inoculations and their use for evaluation of olive cultivar susceptibility. Phytopathology, 96, 313–319. [DOI] [PubMed] [Google Scholar]
- Pérez‐Martínez, I. , Rodríguez‐Moreno, L. , Matas, I.M. and Ramos, C. (2007) Strain selection and improvement of gene transfer for genetic manipulation of Pseudomonas savastanoi isolated from olive knots. Res. Microbiol. 158, 60–69. [DOI] [PubMed] [Google Scholar]
- Pérez‐Martínez, I. , Zhao, Y. , Murillo, J. , Sundin, G.W. and Ramos, C. (2008) Global genomic analysis of Pseudomonas savastanoi pv. savastanoi plasmids. J. Bacteriol. 190, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Martínez, I. , Rodríguez‐Moreno, L. , Lambertsen, L. , Matas, I.M. , Murillo, J. , Tegli, S. , Jiménez, A.J. and Ramos, C. (2010) Fate of a Pseudomonas savastanoi pv. savastanoi type III secretion system mutant in olive plants (Olea europaea L.). Appl. Environ. Microbiol. 76, 3611–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada, J.M. , García, A. , Bertolini, E. , López, M.M. and Penyalver, R. (2007) Recovery of Pseudomonas savastanoi pv. savastanoi from symptomless shoots of naturally infected olive trees. Int. Microbiol. 10, 77–84. [PubMed] [Google Scholar]
- Quesada, J.M. , Pérez‐Martínez, I. , Ramos, C. , López, M.M. and Penyalver, R. (2008) IS53: an insertion element for molecular typing of Pseudomonas savastanoi pv. savastanoi . Res. Microbiol. 159, 207–215. [DOI] [PubMed] [Google Scholar]
- Quesada, J.M. , Penyalver, R. , Pérez‐Panadés, J. , Salcedo, C.I. , Carbonell, E.A. and López, M.M. (2010a) Dissemination of Pseudomonas savastanoi pv. savastanoi populations and subsequent appearance of olive knot disease. Plant Pathol. 59, 262–269. [Google Scholar]
- Quesada, J.M. , Penyalver, R. , Pérez‐Panadés, J. , Salcedo, C.I. , Carbonell, E.A. and López, M.M. (2010b) Comparison of chemical treatments for reducing epiphytic Pseudomonas savastanoi pv. savastanoi populations and for improving subsequent control of olive knot disease. Crop Prot. 29, 1413–1420. [Google Scholar]
- Rico, A. and Preston, G.M. (2008) Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast‐induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol. Plant–Microbe Interact. 21, 269–282. [DOI] [PubMed] [Google Scholar]
- Rico, A. , McCraw, S.L. and Preston, G.M. (2011) The metabolic interface between Pseudomonas syringae and plant cells. Curr. Opin. Microbiol. 14, 31–38. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Moreno, L. , Barceló‐Muñoz, A. and Ramos, C. (2008) In vitro analysis of the interaction of Pseudomonas savastanoi pvs. savastanoi and nerii with micropropagated olive plants. Phytopathology, 98, 815–822. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Moreno, L. , Jiménez, A.J. and Ramos, C. (2009) Endopathogenic lifestyle of Pseudomonas savastanoi pv. savastanoi in olive knots. Microb. Biotechnol. 2, 476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Palenzuela, P. , Matas, I. , Murillo, J. , López‐Solanilla, E. , Bardaji, L. , Pérez‐Martínez, I. , Rodríguez‐Moskera, M.E. , Penyalver, R. , López, M.M. , Quesada, J.M. , Biehl, B.S. , Perna, N.T. , Glasner, J.D. , Cabot, E.L. , Neeno‐Eckwall, E. and Ramos, C. (2010) Annotation and overview of the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 draft genome reveals the virulence gene complement of a tumour‐inducing pathogen of woody hosts. Environ. Microbiol. 12, 1604–1620. [DOI] [PubMed] [Google Scholar]
- Rojas, A.M. , García de los Rios, J.E. , Saux, M.F.‐L. , Jimenez, P. , Reche, P. , Bonneau, S. , Sutra, L. , Mathieu‐Daudé, F. and McClelland, M. (2004) Erwinia toletana sp. nov., associated with Pseudomonas savastanoi‐induced tree knots. Int. J. Syst. Evol. Microbiol. 54, 2217–2222. [DOI] [PubMed] [Google Scholar]
- Sarkar, S.F. and Guttman, D.S. (2004) Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70, 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, D.J. and Collmer, A. (2010) Studying plant–pathogen interactions in the genomics era: beyond molecular Koch's postulates to systems biology. Annu. Rev. Phytopathol. 48, 457–479. [DOI] [PubMed] [Google Scholar]
- Schroth, M.N. , Osgood, J.W. and Miller, T.D. (1973) Quantitative assessment of the effect of the olive knot disease on olive yield and quality. Phytopathology, 63, 1064–1065. [Google Scholar]
- Scortichini, M. , Rossi, M.P. and Salerno, M. (2004) Relationship of genetic structure of Pseudomonas savastanoi pv. savastanoi populations from Italian olive trees and patterns of host genetic diversity. Plant Pathol. 53, 491–497. [Google Scholar]
- Silverstone, S.E. , Gilchrist, D.G. , Bostock, R.M. and Kosuge, T. (1993) The 73‐kb pIAA plasmid increases competitive fitness of Pseudomonas syringae subspecies savastanoi in oleander. Can. J. Microbiol. 39, 659–664. [DOI] [PubMed] [Google Scholar]
- Sisto, A. , Cipriani, M.G. and Morea, M. (2004) Knot formation caused by Pseudomonas syringae subsp. savastanoi on olive plants is hrp‐dependent. Phytopathology, 94, 484–489. [DOI] [PubMed] [Google Scholar]
- Sisto, A. , Cipriani, M.G. , Tegli, S. , Cerboneschi, M. , Stea, G. and Santilli, E. (2007) Genetic characterization by fluorescent AFLP of Pseudomonas savastanoi pv. savastanoi strains isolated from different host species. Plant Pathol. 56, 366–372. [Google Scholar]
- Smith, E.F. (1920) Bacterial Diseases of Plants. Philadelphia, PA: W.B. Saunders Company. [Google Scholar]
- Studholme, D.J. (2011) Application of high‐throughput genome sequencing to intrapathovar variation in Pseudomonas syringae . Mol. Plant Pathol. 12, 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin, G.W. (2007) Genomic insights into the contribution of phytopathogenic bacterial plasmids to the evolutionary history of their hosts. Annu. Rev. Phytopathol. 45, 129–151. [DOI] [PubMed] [Google Scholar]
- Surico, G. (1977) Osservazioni istologiche sui tubercoli della ‘rogna’ dell'Olivo. Phytopathol. Mediterr. 16, 109–125. [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temsah, M. , Hanna, L. and Saad, A.T. (2008) Anatomical pathogenesis of Pseudomonas savastanoi on olive and genesis of knots. J. Plant Pathol. 90, 225–232. [Google Scholar]
- Teviotdale, B.L. and Krueger, W.H. (2004) Effects of timing of copper sprays, defoliation, rainfall, and inoculum concentration on incidence of olive knot disease. Plant Dis. 88, 131–135. [DOI] [PubMed] [Google Scholar]
- Urata, M. , Miyakoshi, M. , Kai, S. , Maeda, K. , Habe, H. , Omori, T. , Yamane, H. and Nojiri, H. (2004) Transcriptional regulation of the ant operon, encoding two‐component anthranilate 1,2‐dioxygenase, on the carbazole‐degradative plasmid pCAR1 of Pseudomonas resinovorans strain CA10. J. Bacteriol. 186, 6815–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingart, H. , Völksch, B. and Ullrich, M.S. (1999) Comparison of ethylene production by Pseudomonas syringae and Ralstonia solanacearum . Phytopathology, 89, 360–365. [DOI] [PubMed] [Google Scholar]
- Young, J.M. (2004) Olive knot and its pathogens. Australas. Plant Pathol. 33, 33–39. [Google Scholar]
- Young, J.M. (2010) Taxonomy of Pseudomonas syringae . J. Plant Pathol. 92, S5–S14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Evolutionary relationships of Pseudomonas savastanoi pv. savastanoi and selected P. syringae pathovars. The tree was constructed by multilocus sequence analysis using a concatenated dataset (exactly 12 000 nucleotides) of acnB, fruK, gapA, gltA, gyrB, pgi, recA and rpoD genes. Phylogenetic groups 1, 2, 3 and 4 (Sarkar and Guttman, 2004; Studholme, 2011) correspond to genomospecies (Gsp) 3, 1, 2 and 4 (Gardan et al., 1999), respectively. Sequence alignment using Muscle; determination of the optimal nucleotide substitution model and phylogenetic tree construction were performed using MEGA5 (Tamura et al., 2011); all positions containing gaps and missing data were eliminated using the option of complete deletion. Bootstrap values (1000 repetitions) are shown on the branches. Similar or identical topologies were obtained by maximum likelihood. The scale bar represents nucleotide substitutions per site.
Table S1 Primers used for the detection of Pseudomonas savastanoi pv. savastanoi.
Table S2 Comparison of the deduced products of iaaM‐1 (PSA3335_1475) and iaaH‐1 (PSA3335_1476), from Pseudomonas savastanoi pv. savastanoi NCPPB 3335, with their homologues in selected organismsa.
Table S3 Accession numbers and coordinates of the nucleotide sequences used for the construction of the neighbour‐joining tree shown in Fig. 2.


