Summary
Rhynchosporium commune is a haploid fungus causing scald or leaf blotch on barley, other Hordeum spp. and Bromus diandrus.
Taxonomy
Rhynchosporium commune is an anamorphic Ascomycete closely related to the teleomorph Helotiales genera Oculimacula and Pyrenopeziza.
Disease symptoms
Rhynchosporium commune causes scald‐like lesions on leaves, leaf sheaths and ears. Early symptoms are generally pale grey oval lesions. With time, the lesions acquire a dark brown margin with the centre of the lesion remaining pale green or pale brown. Lesions often merge to form large areas around which leaf yellowing is common. Infection frequently occurs in the leaf axil, which can lead to chlorosis and eventual death of the leaf.
Life cycle
Rhynchosporium commune is seed borne, but the importance of this phase of the disease is not fully understood. Debris from previous crops and volunteers, infected from the stubble from previous crops, are considered to be the most important sources of the disease. Autumn‐sown crops can become infected very soon after sowing. Secondary spread of disease occurs mainly through splash dispersal of conidia from infected leaves. Rainfall at the stem extension growth stage is the major environmental factor in epidemic development.
Detection and quantification
Rhynchosporium commune produces unique beak‐shaped, one‐septate spores both on leaves and in culture. The development of a specific polymerase chain reaction (PCR) and, more recently, quantitative PCR (qPCR) has allowed the identification of asymptomatic infection in seeds and during the growing season.
Disease control
The main measure for the control of R. commune is the use of fungicides with different modes of action, in combination with the use of resistant cultivars. However, this is constantly under review because of the ability of the pathogen to adapt to host plant resistance and to develop fungicide resistance.
Introduction
The fungal pathogen Rhynchosporium commune causes one of the most destructive diseases of barley (Hordeum vulgare L.), scald or leaf blotch (Fig. 1A,B), especially in areas with cool temperate climates. Yield losses ranging from 10% to 45% have been reported (Brown, 1985; Shipton et al., 1974). Grain quality can also be affected, leading to discounted prices for quality uses, such as malting. Rhynchosporium commune has been one of the major threats to barley production for over a century. It is present in all barley‐growing areas from northern and central Europe to the Middle East, Central Asia, North and South Africa, the Americas, Australia and New Zealand (Brunner et al., 2007; Robbertse et al., 2001; Shipton et al., 1974; von Korff et al., 2004) (Fig. 2). In the UK, R. commune is presently causing a national yield loss after treatment worth £4.8 million (at £100 per tonne) (Home‐Grown Cereals Authority, 2011).
Figure 1.
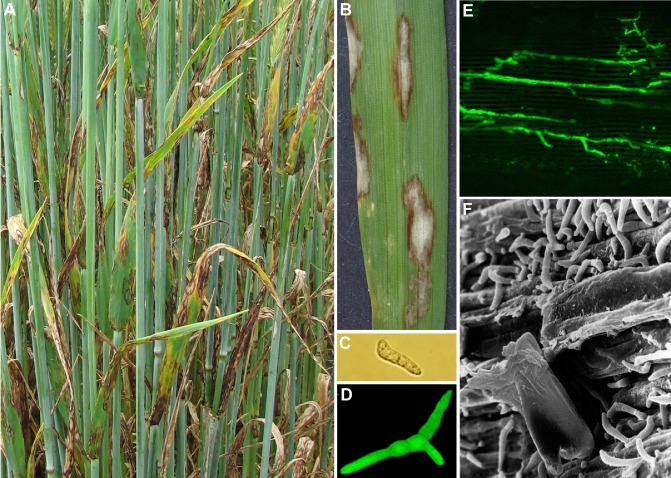
(A) Symptoms on a highly susceptible spring barley cultivar Optic, 2009. (B) Typical lesions with dark brown borders on a susceptible barley leaf caused by Rhynchosporium commune. (C) Typical R. commune conidium, strain L2A. (D) Green fluorescent protein (GFP)‐tagged R. commune strain UK7 conidium with three germination tubes. (E) Hyphal network of GFP‐tagged R. commune strain UK7 in the epidermis of barley cultivar Ingrid 7 days post‐inoculation (dpi). (F) Rhynchosporium commune sporulation on a susceptible barley leaf.
Figure 2.
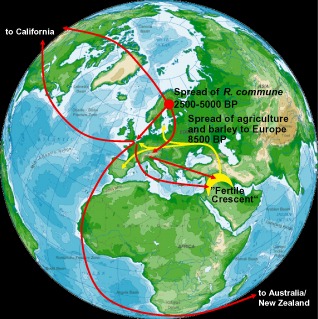
The proposed origin of Rhynchosporium commune in northern Europe and its subsequent spread southwards and to northern America and Australia. Red arrows indicate migration routes of R. commune around the world. Yellow arrows are migration routes of Neolithic farmers into Europe (modified from Brunner et al., 2007).
The control of R. commune by the use of resistant plant cultivars, fungicides or cultural practices has not proven to be sustainable (Shipton et al., 1974; Xi et al., 2000). The fungal population can change rapidly, thereby defeating new barley resistance genes and fungicides after just several seasons of their widespread commercial use (Newton et al., 2001; Oxley et al., 2003). Therefore, the development of sustainable management strategies relies on an improved understanding of R. commune biology and its interactions with the barley host and fungicides. This article presents an overview of Rhynchosporium taxonomy, host range, epidemiology, population variability, strategies for integrated control of the disease and advances in our understanding of its biology and interactions with its host plants.
Taxonomy, Pathogen Evolution and Host Range
Rhynchosporium isolated from rye in the Netherlands was first described by Oudemans in 1897 as Marsonia secalis Oud. (Oudemans, 1897). In the same year, Frank (1897) referred to a disease of barley and rye in Germany caused by the same fungus. In 1901, Heinsen reclassified the fungus in the new genus Rhynchosporium, because of its typical beak‐shaped, one‐septate spores (Fig. 1C,F), naming it Rhynchosporium graminicola Heinsen (Heinsen, 1901). The occurrence of Rhynchosporium in Britain was first recorded in 1919 by Cotton (Brooks, 1928). At about the same time, Davis (1919, 1921) in the USA renamed the fungus Rhynchosporium secalis (Oud.) J.J. Davis in compliance with the International Rules of Nomenclature. Rhynchosporium secalis remained generally accepted as the pathogen infecting barley, rye, triticale and other grasses, including Agropyron spp., Hordeum spp. and Bromus diandrus, for almost a century.
Over the years, numerous attempts have been made to characterize the host specialization of Rhynchosporium isolates through pathogenicity studies (Zaffarano et al., 2011). Using a population genetics approach and restriction fragment length polymorphism (RFLP) markers, Zaffarano et al. (2006) found evidence for host specialization in populations of Rhynchosporium originating from different host species. In 2008, they demonstrated host specialization of rye‐, barley‐ and Agropyron‐infecting Rhynchosporium isolates by cross‐infection studies (Zaffarano et al., 2008). Later, based on phylogenetic analyses of multilocus DNA sequence data from Rhynchosporium isolates originating from different hosts, they resolved the monophyletic groups into three species according to their respective hosts (Zaffarano et al., 2011). As R. secalis was first described on rye, this name is retained for fungal isolates infecting rye and triticale. Rhynchosporium isolates infecting barley and other Hordeum spp., as well as B. diandrus, now belong to a distinct species, R. commune. Isolates infecting Agropyron spp. represent a species called R. agropyri. Analyses by Zaffarano et al. (2008) also suggested that the barley‐, rye‐ and Agropyron‐adapted Rhynchosporium species did not originate from each other, but rather from a common unknown ancestor. Another member of the Rhynchosporium species complex, R. orthosporum, had previously been isolated from cocksfoot, Dactylis glomerata (Caldwell, 1937). This species is morphologically different from the other Rhynchosporium species as it lacks the typical beak‐shaped conidia.
Rhynchosporium nomenclature and classification provide little information on its relatedness to other fungi. Comparison of the internal transcribed spacer (ITS) regions of the ribosomal DNA and mating‐type DNA sequences revealed that R. commune is an anamorphic Ascomycete closely related to the helotialean plant pathogens Pyrenopeziza brassicae and Oculimacula yallundae (Foster and Fitt, 2003; Goodwin, 2002). Interestingly, the ITS sequence of R. commune differs by only 3.19% from that of O. yallundae, but by 6.06% from that of R. orthosporum. This close relationship was unexpected and could not have been deduced from morphological traits. The anamorph of Oculimacula, Helgardia, previously belonged to Ramulispora (Crous et al., 2003). Ramulispora produces five‐ to seven‐celled conidia (Wiese, 1987) that may be branched (Robbertse et al., 1995), whereas conidia from R. commune are two‐celled and unbranched (Fig. 1C,F) (Caldwell, 1937). Based on ITS analysis, it was predicted that, if a teleomorph of R. commune exists, it would be a species of Oculimacula.
Infection Biology and Epidemiology
Rhynchosporium is a polycyclic pathogen with several generations of spores developing during the crop growing season. Primary inoculum probably originates from crop debris or infected seeds (Fig. 3). Secondary spread occurs through splash‐dispersed conidia from infected leaves (Fitt et al., 1989; Zhan et al., 2008). Rainfall at the growth stage of stem extension, usually in April in the UK, is the major environmental factor in epidemic development (Atkins et al., 2010). Rhynchosporium commune can infect any part of the leaf and produce spots or blotches of irregular shape (Fig. 1B) (Brooks, 1928). As a result of the tendency for water retention between the auricle and the stem, lesions are also often found there (Brooks, 1928).
Figure 3.
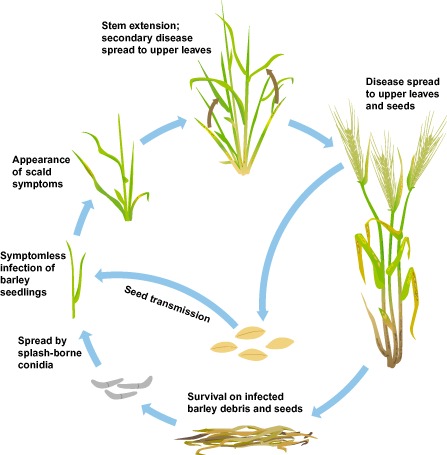
Rhynchosporium commune development during the barley growing season.
Infection of barley ears can result in severe grain infection (Skoropad, 1959). Rhynchosporium commune can be transmitted by seeds and seed dust remaining on the soil surface (Fig. 3) (Reed, 1957). Infection in seeds can be seen as a typical lesion at the base of the awn and show a dark brown margin with a light centre (Skoropad, 1959). However, seed infection can remain symptomless (Lee et al., 2001a, b), which implies that visible analysis of the seeds may not always be accurate when determining seed quality (Kay and Owen, 1973; Skoropad, 1959). Seedlings grown from infected seeds were found to have symptoms at the tip of the coleoptile 4–6 days after emergence, or remained symptomless (Habgood, 1971; Ozoe, 1956; Skoropad, 1959). On average, 2% of grain in diseased crops can be infected, which can result in up to 85% of subsequent seedlings being infected (Skoropad, 1959). Although splash dispersal of R. commune conidia contributes to the short‐distance spread in the field (McDonald et al., 1999; Shipton et al., 1974), transport of infected seeds may be responsible for the long‐distance dispersal of inoculum in general, as well as the spread of new physiological races (Lee et al., 2001b; Ozoe, 1956).
In humid conditions, conidia germinate on the leaf surface (Fig. 1D), producing hyphae that penetrate the cuticle directly above epidermal cells. Subsequent fungal growth is confined to the subcuticular region of the epidermis (Fig. 1E) (Jones and Ayres, 1974; Lehnackers and Knogge, 1990; Thirugnanasambandam et al., 2011; Xi et al., 2000). During the early stages of subcuticular growth, thin hyphae with broadly spaced septae tend to grow along the anticlinal cell walls (Fig. 1E), including those of the stomatal guard cells, but no growth of mycelium through the stomata has been observed (Ayesu‐Offei and Clare, 1970; Lehnackers and Knogge, 1990; Thirugnanasambandam et al., 2011). Prior to sporulation, thicker fungal hyphae with closely spaced septae align themselves parallel to the leaf surface (Ayesu‐Offei and Clare, 1970; Horbach et al., 2011; Lehnackers and Knogge, 1990) and eventually form a dense subcuticular stroma. New R. commune conidia are produced on conidiophores, which erupt through the leaf cuticle (Fig. 1F) in apparently healthy leaf regions (Davis and Fitt, 1990; Davis et al., 1994; Howlett and Cooke, 1987, 1992; Lehnackers and Knogge, 1990). In addition, sporulation occurs in the lesion areas (Lehnackers and Knogge, 1990).
The development of R. commune is characterized by a long phase of asymptomatic growth between penetration and occurrence of the typical disease symptoms, necrotic lesions with dark brown margins (Fig. 1B) (Davis and Fitt, 1990; Lehnackers and Knogge, 1990). Indeed, several generations of the pathogen may occur before symptoms appear. Although the disease is spread from the lower to upper leaves by rain splash (Fig. 3) (Fitt et al., 1986), sometimes severe symptoms appear on the upper leaves of the crop, which previously exhibited little visible signs of disease (Shaw, 1987). Thus, severe epidemics, resulting in considerable yield loss, can occur in crops with an initially low level of disease which did not justify a fungicide application (Jenkins and Jemmett, 1967). During the asymptomatic phase, collapsing epidermal cells represent the earliest microscopically visible evidence of disease. This phase ends with the appearance of the typical scald lesions, which are caused by the collapse of mesophyll cells beneath extensive fungal mycelia (Lehnackers and Knogge, 1990; Thirugnanasambandam et al., 2011; Zhan et al., 2008). The greyish colour in the middle of the blotch is caused by the formation of spores on the surface (Fig. 1B). The size of the lesion can vary as a function of environmental conditions and cultivar resistance. The lesions may merge and destroy the entire leaf (Fig. 1A,B).
Originally R. commune was considered to be a necrotroph, because of the necrotic lesions it produces. Unlike true necrotrophs, which often have a wide host range (Lucas, 1998) and trigger host cell death in order to feed exclusively on dead or dying host tissue, R. commune is restricted to barley, other Hordeum spp. and B. diandrus. During an extended asymptomatic phase, the fungus is presumed to acquire nutrients biotrophically from leaky host cells (Davis et al., 1994; Jones and Ayres, 1972). The lifestyle of R. commune can be better described by one of the contemporary meanings of the term hemibiotrophy (Luttrell, 1974; Oliver and Ipcho, 2004; Zhan et al., 2008). It is applied to species such as Cladosporium fulvum, Mycosphaerella graminicola and P. brassicae, which have an extended (4–14 days) asymptomatic phase, followed by the increasing development of plant tissue damage (Oliver and Ipcho, 2004). Unlike true biotrophic plant pathogens, R. commune, C. fulvum, M. graminicola and P. brassicae do not produce haustoria, specific feeding structures, which invaginate the plant cell membrane, thus forming an intimate association with the host (Oliver and Ipcho, 2004). Unlike M. graminicola, which appears to benefit nutritionally from triggering host programmed cell death (PCD) (Keon et al., 2007), there is no evidence of R. commune benefiting from the collapse of epidermal and, later, mesophyll cells as, by that time, the front of the infection has moved away from the necrotized part of the leaf (A. Avrova, unpublished data; S. Kirsten and W. Knogge, unpublished data).
Pathogen Variation
Rhynchosporium commune is a diverse pathogen with a high potential to evolve relatively quickly (Brown, 1985; Burdon et al., 1994; Jorgensen and Smedegaard‐Petersen, 1995; McDermott et al., 1989; McDonald et al., 1999) and adapt to changes in environmental conditions and host resistance (Habgood, 1973; Jackson and Webster, 1976; Xi et al., 2003; Zhang et al., 1992). Rhynchosporium commune isolates, even those originating from the same lesion, often differ in colony colour and morphology, shape and size of conidia, sporulation and germination rates, pathogenicity, virulence, response to nutritional conditions, fungicide resistance and molecular profile (Ali et al., 1976; Brown, 1985; Ceoloni, 1980; Goodwin et al., 1994; Habgood, 1973; Hansen and Magnus, 1973; Kari and Griffiths, 1993; Newman, 1985; Newton et al., 2001; Owen, 1963; Salamati and Tronsmo, 1997; Schein, 1958; Williams et al., 2003; Williams and Owen, 1975).
Zaffarano et al. (2006) suggested that gene flow is common at the local level, whereas it is low between regions on the same continent, and rare between continents. Around 75% of the total genetic diversity on a continent and about 40% of the worldwide RFLP variation in R. commune were found in a 1‐m2 sampling area (Salamati et al., 2000; Zaffarano et al., 2006). Almost 60% of the total genetic variation was found in a single barley field (Zaffarano et al., 2006). Intriguingly, the most variable R. commune populations are found in Scandinavia, rather than the Middle East, which is considered to be the centre of origin of its barley host (Zaffarano et al., 2006). Rhynchosporium commune may have originated in northern Europe at 2500–5000 bp following a host switch, most probably from a wild grass onto cultivated barley, shortly after barley was introduced into northern Europe (Brunner et al., 2007; Zaffarano et al., 2006). Rhynchosporium commune subsequently spread southwards into already established European barley‐growing areas (Fig. 2).
Several reasons proposed to explain the high genetic diversity include the large population size (McDermott et al., 1989), frequency‐dependent selection (Goodwin et al., 1993; McDermott et al., 1989), spontaneous mutation (Goodwin et al., 1994; Williams et al., 2003), gene flow (Goodwin et al., 1994), sexual reproduction (McDonald et al., 1999; Salamati et al., 2000) and asexual recombination (Forgan et al., 2007; Goodwin et al., 1994; Newman and Owen, 1985; Newton, 1989; Williams et al., 2003).
Although genetic diversity within R. commune populations is high, the use of molecular markers has shown it to be lower (Newton et al., 2001; Salamati et al., 2000; Zaffarano et al., 2006) than that of the highly variable wheat pathogen M. graminicola (Linde et al., 2003). Unlike M. graminicola, which regularly reproduces sexually, the sexual stage of R. commune has not been identified, although the finding of nearly perfect gametic equilibrium in most populations suggests that the fungus is capable of sexual reproduction (Linde et al., 2003; Salamati et al., 2000). Analysis of multilocus associations, genotype diversity and mating locus allele frequencies has suggested that sexual recombination is occurring in most of the populations (Foster and Fitt, 2003; Linde et al., 2003; Zaffarano et al., 2006). Both mating alleles were frequently found in the same lesion or leaf, providing opportunities for isolates carrying opposite mating alleles to interact and reproduce sexually (Linde et al., 2003).
Disease Control
Agronomic practices
Infected straw provides a reservoir of inoculum for splash dispersal (Fig. 3) when weather conditions favour the development of R. commune infection (Fitt et al., 1987). Rhynchosporium commune can survive on straw for about 1 year, depending on the ambient conditions, but cannot oversummer in straw left in the open field or buried in soil (Ozoe, 1956; Skoropad, 1959). The germination rate of conidia from dead leaves in spring is also affected by the duration of the freezing period and the number of alternating wet and dry periods in the autumn and spring (Skoropad, 1966). The viability of R. commune conidia may also be affected by naturally occurring bacteria in the soil (Newton et al., 2004a).
The combination of continuous barley cultivation and reduced tillage leads to the accumulation of crop debris in the field and, with it, to a buildup of inoculum (Elen, 2002). Over 40 years ago, Hansen and Magnus reported an increase in scald that might have been caused by the shift from crop rotation to continuous barley cultivation (Hansen and Magnus, 1969). More recently, reduced tillage and continuous spring barley cultivation have led to an increase in the occurrence of Rhynchosporium in the Nordic countries (Arvidsson, 1998; Rasmussen, 1984). Crop rotation, or even a 1‐year interruption with oats, is effective in controlling the occurrence of the disease on barley (Elen, 2002). Similarly, commonly used stubble management practices, such as grazing, reduce the amount of R. commune inoculum available for subsequent disease development (Mayfield and Clare, 1984).
Chemical control and fungicide resistance
Fungicides are widely used to protect crops as they can provide very high levels of disease control. Foliar fungicides are used on most barley crops in Europe. However, the long‐term effectiveness of fungicides depends on the ability of pathogens to evolve fungicide resistance.
During the 1970s and 1980s, R. commune was effectively controlled by the application of the methyl benzimidazole carbamates (MBCs) and demethylation inhibitors (DMIs; ‘triazoles’), alone or in mixtures. Since the first detection of resistance to MBC fungicides in the early 1990s, the frequency of resistant isolates has increased rapidly (Kendall et al., 1994; Taggart et al., 1998, 1999). Resistance to MBCs is now widespread in R. commune populations in the UK (Locke and Phillips, 1995; Taggart et al., 1999). It is mediated by mutation in a single gene, β‐tubulin (Wheeler et al., 1995), and is also associated with decreased pathogenicity (Kendall et al., 1993).
In contrast, resistance to triazole fungicides has evolved more slowly because of the polygenic nature of this resistance, and may involve several mechanisms (Cooke et al., 2004; Zhan et al., 2005, 2006). Nevertheless, increasing resistance to fungicides, such as flusilazole and epoxiconazole, has been reported in the UK (Oxley et al., 2003). Rhynchosporium commune resistance to triazole fungicides is not associated with a fitness penalty (Kendall et al., 1993). Exposure to flusilazole, tebuconazole and epoxiconazole can result in a 10‐fold decrease in the sensitivity of the R. commune population to these fungicides (Cooke et al., 2004; Robbertse et al., 2001), indicating erosion in their effectiveness. Although there is cross‐resistance between the different triazoles, no cross‐resistance between the imidazole and triazole DMIs has been found (Kendall et al., 1993).
Despite the partial loss of DMI efficacy in some parts of the UK and Europe, DMIs remain one of the most important fungicide groups for the control of barley diseases (Walters et al., 2012). However, it is recommended that they be used mixed with other fungicides with a different mode of action. The recommended mixing partners are the ‘quinone outside inhibitors’ (QoIs; strobilurins) or anilinopyrimidines, and the newer succinate dehydrogenase inhibitor (SDHI) fungicides. Rhynchosporium commune resistance to QoI fungicides was also reported by the Fungicide Resistance Action Group (FRAG) during 2008 in northern France (Walters et al., 2012). Similar to other fungal pathogens of barley, complete resistance of R. commune to all QoI fungicides is the result of a single point mutation in the cytochrome b gene (Sierotzki et al., 2000). However, no further R. commune isolates resistant to QoI fungicides have been reported since then anywhere in Europe (Walters et al., 2012). Therefore, although resistance is expected to develop in the future, the current levels of QoI resistance remain very low and have not affected R. commune disease control when following the recommendations for resistance management.
Pathogens will continue to develop fungicide resistance as long as a selection pressure is applied. Therefore, an integrated crop protection (ICP) system needs to be implemented to slow down the loss of effective fungicides. The most effective ICP system currently adopted includes the application of the appropriate dose at the correct time and the mixing of fungicides with different modes of action, in combination with the use of resistant cultivars.
Host Resistance
The deployment of host resistance is the most sustainable method of protecting barley from pests and pathogens, including Rhynchosporium. However, the genetic basis of such resistance must ensure its durability and avoid a disproportionate cost to the plant, resulting in yield loss. The deployment of resistant cultivars in combination with the monitoring of pathogen populations can lead to a reduction in the number of pesticide applications and prolong the lifespan of individual resistance genes.
Qualitative/major gene resistance
Active nonhost resistance (NHR) of plants to potential pathogens is based on the recognition of race‐nonspecific, microbe‐associated molecular patterns (MAMPs) by pattern recognition receptors (PRRs) present in the plant cell membrane. Race‐specific resistance arises after successful suppression of NHR by a pathogen. It involves major plant resistance (R) genes, which directly or indirectly recognize the products of certain pathogen effector genes, termed avirulence (Avr) genes. This triggers a qualitative resistance response called effector‐triggered immunity (ETI) (Jones and Dangl, 2006). A pathogen population usually consists of several races, or pathotypes, with different alleles at Avr gene loci. Likewise, plants may possess several Avr gene‐associated R genes/alleles, which allow the recognition of individual pathogen races (Knogge, 1996).
In barley, several major R genes against R. commune have been described (Goodwin et al., 1990; Habgood and Hayes, 1971; Shipton et al., 1974). As a result of their insufficient and partly confusing description in the literature, Bjornstad et al. (2002) proposed a new nomenclature for R genes against R. commune. It takes into account the fact that several previously described distinct R genes are actually alleles of the same R gene. They listed seven different R genes [Rrs1 (11 alleles), Rrs2 (two alleles), Rrs3, Rrs4 (two alleles), Rrs12, Rrs13 and Rrs14], as well as four unconfirmed R genes (Rh5, rh8, Rh10 and rh11). The first seven R genes have been located on a consensus bin‐map (Zhan et al., 2008). Although, none of the R genes against R. commune have been cloned to date, the recent sequencing of the barley genome should aid in their identification.
Quantitative/partial resistance
Quantitative resistance, also termed horizontal, partial or race‐nonspecific resistance, is based on multiple genes with partial effects, which may control different mechanisms (Poland et al., 2009). Quantitative resistance may affect different stages in the life cycle of R. commune. It can influence the development of scald epidemics in barley crops by decreasing the leaf area affected by lesions (Williams and Owen, 1975) or by affecting sporulation (Kari and Griffiths, 1993; Xue and Hall, 1991).
The level of quantitative resistance is greatly influenced by site and season through genotype‐by‐environment interactions (Kari and Griffiths, 1993). Therefore, it is well suited to being studied by the mapping of quantitative trait loci (QTLs) (Zhan et al., 2008). Several QTL clusters for resistance to R. commune have been mapped to all barley chromosomes, except chromosome 5H, which interestingly also lacks a major R gene (Schweizer and Stein, 2011; Zhan et al., 2008). The complex genetic nature of quantitative resistance has led to the assumption that this form of resistance will be more durable than major gene‐mediated resistance (Walters et al., 2012).
According to the UK Recommended List of Cereal Cultivars (http://www.hgca.com), winter barley cultivars have, on average, much higher partial resistance to R. commune (Zhan et al., 2008). This difference is even more pronounced if spring barley cultivars are winter sown alongside true winter cultivars (Newton et al., 2004b). The gene pools of the two barley types are largely separate. It is possible that winter barley cultivars have more actively selected resistance to R. commune because they are routinely exposed to the pathogen (Zhan et al., 2008). A recent QTL mapping study of R. commune resistance in a winter barley × spring barley cross demonstrated the resistance of the winter parent to be independent of genes controlling seasonal growth habit (Looseley et al., 2011). However, some of the differences between winter and spring types may be attributable to linkage or pleiotropic effects of cold tolerance or vernalization genes in winter cultivars. Field trials in Scotland have suggested that ratings for spring barley give a more accurate indication of cultivar resistance, whereas the resistance of winter barley is overestimated (Oxley et al., 2003).
There are other types of ‘resistance’, such as ‘disease escape’, which involve no biochemical recognition between the barley cultivar and R. commune strain. This can be associated with cultivar height, maturity or canopy structure, which limits the upward spread of splash‐dispersed R. commune conidia (Bingham et al., 2008; Walters et al., 2012). Early stem elongation, for example, could decrease the spread of late epidemics. Other potentially useful constitutive plant defence traits that may influence disease escape are the repellent action of the leaf surface and hairs, and leaf topography, which reduces pathogen attachment and/or spread or inhibits spore germination (Walters et al., 2012).
Molecular Aspects of the Host–Pathogen Interaction
Rhynchosporium commune is characterized by its unusual development inside host leaf tissues. As a result of the subcuticular growth of the mycelia, the epidermal cell walls separate fungal hyphae from host plasma membranes. This places secreted fungal molecules (proteins, secondary metabolites) into the centre of interest to explain how the fungus communicates with the host and manipulates the plant physiology in its favour. After detecting the presence of toxic compounds in fungal culture filtrates (Ayesu‐Offei and Clare, 1971), early molecular studies targeted a family of oligoglucosides of 1,2‐propanediol. These rhynchosporosides, some of which were originally believed to be host selective (Auriol et al., 1978), caused necrotic lesions in detached leaves. Another compound with necrotic activity, the isocoumarin (+)‐orthosporin, was isolated from R. orthosporum by Ichihara et al. (1989). It remains to be shown, however, whether these compounds or additional products of fungal polyketide synthases (C. Wenzel and W. Knogge, unpublished data) play a role during pathogenesis. Furthermore, glycosphingolipids were isolated from membranes of R. commune (Sakaki et al., 2001). These compounds are structurally related to cerebrosides A, B and C from Magnaporthe grisea and other fungi, which have been shown to induce defence reactions in rice based on precise structural requirements (Koga et al., 1998; Umemura et al., 2000). Again, it is not known whether these lipids are relevant to the interaction of R. commune with its host plant.
Recent years have witnessed a dramatic increase in the number of characterized secreted effector proteins from several plant‐pathogenic bacteria, oomycetes and fungi (Desveaux et al., 2006; Ellis et al., 2009; Hogenhout et al., 2009; Ma and Guttman, 2008; Schornack et al., 2009; Stergiopoulos and de Wit, 2009; Stukenbrock and McDonald, 2009; Tyler, 2009). A small family of necrosis‐inducing small secreted proteins (NIP1, NIP2, NIP3) was purified from culture filtrates of R. commune as early as 1991 (Wevelsiep et al., 1991). On injection into leaves of barley and other grasses, these structurally unrelated proteins cause necrosis resembling the disease symptoms. Mature NIP1 contains 60 amino acids (10 cysteines), NIP2 93 amino acids (six cysteines) and NIP3 98 amino acids (eight cysteines). In contrast with NIP1 and NIP2, NIP3 is post‐translationally processed; the protein carries a carbohydrate moiety near the N‐terminus and appears to be proteolytically shortened at the C‐terminus (Kirsten et al., 2012; Wevelsiep et al., 1993). No biochemical activity is known for NIP2 to date, whereas NIP1 and NIP3 have been shown to stimulate the plant plasma membrane‐localized H+‐ATPase, the enzyme controlling the electrochemical gradient at the plant plasma membrane (Wevelsiep et al., 1993). Modulation of this gradient may affect the regulation of essential membrane transport processes, such as nutrient export (Elmore and Coaker, 2011). Alternatively, apoplastic acidification may serve to optimize the conditions for enzymatic degradation of the plant cell wall, as has been discussed for several phytopathogenic fungi, such as Sclerotinia sclerotiorum and Botrytis cinerea (Prusky and Yakoby, 2003).
NIP1 has a dual role in plant disease and resistance (van't Slot and Knogge, 2002). In addition to its necrotizing pathogenicity‐associated activity, it is the product of one of the first Avr genes identified in phytopathogenic fungi, AvrRrs1 (Rohe et al., 1995). The protein induces the expression of plant defence genes (PR1, PR5, PR9, PR10) specifically in barley cultivars carrying the R gene Rrs1 (Hahn et al., 1993; Steiner‐Lange et al., 2003). However, in contrast with the interaction between Avr and R genes in most plant–pathogen interactions, a visible hypersensitive response (HR) is not triggered by NIP1 (Fig. 4) (Hahn et al., 1993) and generally does not appear to occur in this pathosystem. Nevertheless, small necrotic flecks have been recorded on leaves of some resistant cultivars inoculated with certain isolates of R. commune (Bjornstad et al., 2002).
Figure 4.

The effect of NIP1 demonstrated on the Rrs1 cultivar ‘Turk’ 21 days post‐inoculation (dpi) with NIP1‐expressing Rhynchosporium commune wild‐type (WT) strain UK7 (top panel) and the NIP1 deletion mutant UK7ΔNIP1 (bottom panel).
Natural selection drives pathogens to avoid recognition by R proteins. This can be achieved by losing either the expression or the function of an effector with no apparent cost to pathogen fitness. Strong selective pressures, together with the simple genetic architecture of major gene resistance, mean that this process can be rapid. Both of these strategies have been deployed by R. commune to enable fungal growth in the presence of the Rrs1 gene (Houston and Ashworth, 1957; Rohe et al., 1995), as revealed by the analysis of several hundred isolates collected worldwide. In 45% of the isolates, the NIP1 gene is absent (Schürch et al., 2004). Likewise, deletion of NIP1 from R. commune wild‐type strain UK7 produced a mutant virulent on the Rrs1 cultivar ‘Turk’ (Fig. 4). In addition, several structural variants exist, some of which attenuate or prevent Rrs1‐based recognition by the host plant, but at the same time abolish the necrotic and ATPase‐stimulating activity (Fiegen and Knogge, 2002).
On NIP1 expression in a heterologous system (Gierlich et al., 1999), the solution structure of the molecule was solved (van't Slot et al., 2003) and binding studies were carried out (van't Slot et al., 2007). A single class of binding site with identical binding characteristics was found in membranes from barley genotypes, irrespective of the presence or absence of Rrs1, as well as from other cereals, but not from Arabidopsis thaliana. Interestingly, protein variants that were inactive as Avr factors showed similar binding affinities to the highly active NIP1, suggesting that the Rrs1 gene does not encode the NIP1 receptor and the binding of NIP1 to its target is not sufficient for recognition by Rrs1.
NIP2 and NIP3 also show some degree of structural variation (D. Croll and B. A. McDonald, unpublished data). In contrast with NIP1, however, both genes occur in almost all (NIP2 in 92%, NIP3 in 99.6%) of the analysed R. commune isolates, indicating the importance of both proteins for the fungus (Schürch et al., 2004). NIP2 and NIP3 are transcribed during fungal development in susceptible host leaves, whereas NIP1 transcripts are already abundant in spores (Kirsten et al., 2012). When the fungal biomass starts to increase drastically in leaves of susceptible plants several days post‐inoculation, biosynthesis of the three proteins decreases rapidly. This suggests that they are functionally important during the earlier stages of the interaction, when fungal hyphae spread along the leaf blade before the development of a dense subcuticular stroma. Deletion of a single NIP gene resulted in a host genotype‐dependent reduction in pathogenicity of the fungal mutants (Kirsten et al., 2012). This suggests that the proteins contribute quantitatively to fungal pathogenicity, i.e. their combined activities lead to stronger fungal growth during infection. Alternatively, they may be involved in recognition events, which increase the plant defence response in a quantitative manner (Tao et al., 2003). The plant recognition factors involved may be encoded at quantitative resistance loci, thus turning recognized proteins into Avr effectors in quantitative disease resistance.
Dna‐Mediated Transformation
Rhynchosporium commune was first transformed to hygromycin‐B and phleomycin resistance using polyethylene glycol (PEG)/CaCl2 treatment of protoplasts (Rohe et al., 1996). Transformation frequencies varied from 59 to 493 transformants per 10 μg of DNA and 5 × 107 protoplasts. The antibiotic‐resistant phenotype appeared to be stable under selective and nonselective conditions for several generations. Co‐transformation using the Escherichia coli uidA gene encoding β‐glucuronidase (GUS) under the control of the Aspergillus nidulans promoter and terminator sequences on a nonselectable plasmid occurred at frequencies of up to 66%.
More recently, Agrobacterium tumefaciens‐mediated transformation (ATMT) was used to generate R. commune transformants expressing the green fluorescent protein (GFP) or DsRed fluorescent protein (Kirsten et al., 2011; Linsell et al., 2011; Thirugnanasambandam et al., 2011). These transformants were utilized to compare R. commune growth using confocal microscopy in both the susceptible and resistant cultivars. In addition, GFP‐tagged transformants allowed the quantification of fungal development in planta using pattern recognition software (Baum et al., 2011), as well as fungal growth and the impact of growth inhibitors ex planta (Kirsten et al., 2011).
Like most fungal plant pathogens, R. commune is haploid. This allows the use of gene‐specific deletions via homologous recombination to elucidate gene function. Through protoplast transformation or ATMT, deletion mutants were generated to characterize the function of the effector genes NIP1, NIP2 and NIP3 (Kirsten et al., 2012), as well as several other fungal genes (W. Knogge, unpublished data). Although ATMT is a more efficient transformation system, it requires the construction of plasmids by cloning, which can be time consuming. In contrast, protoplast transformation can be used with the split marker approach to generate deletion mutants in fungi (Catlett et al., 2003).
Conclusions and Future Perspectives
Recent advances in next‐generation sequencing (NGS) technologies have enabled the sequencing of the genomes from strains of all Rhynchosporium species, as well as the transcriptomes from different (including early) developmental stages of R. commune during the interaction with its barley host (W. Knogge and A. Avrova, unpublished data). The available sequence information has allowed the identification of numerous additional pathogen effector proteins, followed by their functional characterization, in order to understand fungal virulence mechanisms. It is important to understand redundancy within such pathogen effectors. Redundant effectors, such as NIP1 (AvrRrs1), are known to be readily lost or modified by the pathogen, resulting in a lower durability of the host R genes recognizing these effectors (Houston and Ashworth, 1957; Rohe et al., 1995). Therefore, breeding should aim to target the introgression of R genes recognizing pathogen effectors which are nonredundant and therefore essential for pathogenicity. As a result of the pressure on the pathogen to preserve the function of these effectors, they are likely to remain more conserved in pathogen populations.
The transient expression of pathogen genes in Nicotiana benthamiana using A. tumefaciens (agroinfiltration) or virus (Potato virus X, PVX) has revolutionized solanaceous pathogen genomics. It has enabled the discovery and functional profiling of late blight R genes and Avr genes at an unprecedented rate (Vleeshouwers et al., 2011). This technology promises to accelerate the engineering of late blight‐resistant potato varieties (Vleeshouwers et al., 2011). Modification of the recently developed Agrobacterium delivery system for the Barley stripe mosaic virus (BSMV) (Yuan et al., 2011) to allow the in planta expression of small secreted fungal proteins in cereals (K. Kanyuka et al., unpublished data) can greatly facilitate the screening of extensive collections of barley germplasm. It can lead to the identification of novel sources of resistance to R. commune, as well as other fungal pathogens of cereals, which can be used in breeding. The BSMV‐mediated expression system will also allow the characterization of resistance already present in current barley breeding populations. Functional characterization of pathogen effectors can assist in predicting the durability of individual R proteins recognizing these effectors. This will have a direct impact on disease resistance breeding programmes by providing the rapid identification of effective resistance sources, and the implementation of resistance in the field.
Finally, the sequence information on the genomes of all four Rhynchosporium species can be exploited through comparative genomics techniques to unravel the molecular basis of fungal speciation and host specialization. It will allow the identification of conserved, as well as species‐specific, effectors involved in host‐specific interaction. It will also help to answer questions about the evolution of the genus Rhynchosporium.
Acknowledgements
We thank Cavan Convery for help with the drawing of Fig. 3. AA was supported by the Scottish Government Rural and Environment Science and Analytical Services (RESAS). WK was supported by the Leibniz Association.
References
- Ali, S.M. , Mayfield, A.H. and Clare, B.G. (1976) Pathogenicity of 203 isolates of Rhynchosporium secalis on 21 barley cultivars. Physiol. Plant Pathol. 9, 135–143. [Google Scholar]
- Arvidsson, J. (1998) Effects of cultivation depth in reduced tillage on soil physical properties, crop yield and plant pathogens. Eur. J. Agron. 9, 79–85. [Google Scholar]
- Atkins, S.D. , Fitt, B.D.L. , Fraaije, B. , Harvey, S. , Lynott, J. and Newton, A.C. (2010) The epidemiological importance of asymptomatic infection of winter barley by Rhynchosporium secalis and its consequences for crop protection and breeding. Proc. Crop Prot. Northern Britain , 81–86.
- Auriol, P. , Strobel, G. , Pio Beltran, J. and Gray, G. (1978) Rhynchosporoside, a host‐selective toxin produced by Rhynchosporium secalis, the causal agent of scald disease of barley. Proc. Natl. Acad. Sci. USA, 75, 4339–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayesu‐Offei, E.N. and Clare, B.G. (1970) Processes in the infection of barley leaves by Rhynchosporium secalis . Aust. J. Biol. Sci. 23, 299–307. [Google Scholar]
- Ayesu‐Offei, E.N. and Clare, B.G. (1971) Symptoms of scald disease induced by toxic metabolites of Rhynchosporium secalis . Aust. J. Biol. Sci. 24, 169–174. [DOI] [PubMed] [Google Scholar]
- Baum, T. , Navarro‐Quezada, A. , Knogge, W. , Douchkov, D. , Schweizer, P. and Seiffert, U. (2011) HyphArea—automated analysis of spatiotemporal fungal patterns. J. Plant Physiol. 168, 72–78. [DOI] [PubMed] [Google Scholar]
- Bingham, I.J. , Hoad, S.P. , Newton, A.C. and Thomas, W.T.B. (2008) Avoidance and tolerance of foliar disease in barley: opportunities for improvement. Proc. Crop Prot. Northern Britain , 139–144.
- Bjornstad, A. , Patil, V. , Tekauz, A. , Maroy, A.G. , Skinnes, H. , Jensen, A. , Magnus, H. and MacKey, J. (2002) Resistance to scald (Rhynchosporium secalis) in barley (Hordeum vulgare) studied by near‐isogenic lines: I. Markers and differential isolates. Phytopathology, 92, 710–720. [DOI] [PubMed] [Google Scholar]
- Brooks, F.T. (1928) Observations on Rhynchosporium secalis (Oud.) Davis, leaf blotch of barley and rye. New Phytol. 27, 215–219. [Google Scholar]
- Brown, J.S. (1985) Pathogenic variation among isolates of Rhynchosporium secalis from cultivated barley growing in Victoria, Australia. Euphytica, 34, 129–133. [Google Scholar]
- Brunner, P.C. , Schürch, S. and McDonald, B.A. (2007) The origin and colonization history of the barley scald pathogen Rhynchosporium secalis . J. Evol. Biol. 20, 1311–1321. [DOI] [PubMed] [Google Scholar]
- Burdon, J.J. , Abbott, D.C. , Brown, A.H.D. and Brown, J.S. (1994) Genetic structure of the scald pathogen (Rhynchosporium secalis) in South East Australia: implications for control strategies. Aust. J. Agric. Res. 45, 1445–1454. [Google Scholar]
- Caldwell, R.M. (1937) Rhynchosporium scald of barley, rye, and other grasses. J. Agric. Res. 55, 175–198. [Google Scholar]
- Catlett, N.L. , Lee, B.‐N. , Yoder, O.C. and Turgeon, B.G. (2003) Split‐marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. News, 50, 9–11. [Google Scholar]
- Ceoloni, C. (1980) Race differentiation and search for sources of resistance to Rhynchosporium secalis in barley in Italy. Euphytica, 29, 547–553. [Google Scholar]
- Cooke, L.R. , Locke, T. , Lockley, K.D. , Phillips, A. , Sadiq, M.D.S. , Coll, R. , Black, L. , Taggart, P.J. and Mercer, P.C. (2004) The effect of fungicide programmes based on epoxiconazole on the control and DMI sensitivity of Rhynchosporium secalis in winter barley. Crop Prot. 23, 393–406. [Google Scholar]
- Crous, P.W. , Groenewald, J.Z.E. and Gams, W. (2003) Eyespot of cereals revisited: ITS phylogeny reveals new species relationships. Eur. J. Plant Pathol. 109, 841–850. [Google Scholar]
- Davis, H. and Fitt, B.D. (1990) Symptomless infection of Rhynchosporium secalis on leaves of winter barley. Mycol. Res. 94, 557–560. [Google Scholar]
- Davis, H. , Fitt, B.D.L. and Evans, R.L. (1994) Atypical, green leaf blotch lesions on barley leaves infected by Rhynchosporium secalis (Oud.) Davis. New Phytol. 127, 139–145. [DOI] [PubMed] [Google Scholar]
- Davis, J.J. (1919) Notes on parasitic fungi in Wisconsin. VI. Trans. Wis. Acad. Sci. Arts. Lett. 19, 705–727. [Google Scholar]
- Davis, J.J. (1921) Notes of parasitic fungi in Wisconsin. Trans. Wis. Acad. Sci. Arts. Lett. 20, 413–431. [Google Scholar]
- Desveaux, D. , Singer, A.U. and Dangl, J.L. (2006) Type III effector proteins: doppelgangers of bacterial virulence. Curr. Opin. Plant Biol. 9, 376–382. [DOI] [PubMed] [Google Scholar]
- Elen, O. (2002) Plant protection in spring cereal production with reduced tillage. III. Cereal diseases. Crop Prot. 21, 195–201. [Google Scholar]
- Ellis, J.G. , Rafiqi, M. , Gan, P. , Chakrabarti, A. and Dodds, P.N. (2009) Recent progress in discovery and functional analysis of effector proteins of fungal and oomycete plant pathogens. Curr. Opin. Plant Biol. 12, 399–405. [DOI] [PubMed] [Google Scholar]
- Elmore, J.M. and Coaker, G. (2011) The role of the plasma membrane H+‐ATPase in plant microbe interactions. Mol. Plant, 4, 416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegen, M. and Knogge, W. (2002) Amino acid alterations in isoforms of the effector protein NIP1 from Rhynchosporium secalis have similar effects on its avirulence‐ and virulence‐associated activities on barley. Physiol. Mol. Plant Pathol. 61, 299–302. [Google Scholar]
- Fitt, B.D.L. , Creighton, N.F. , Lacey, M.E. and McCartney, H.A. (1986) Effects of rainfall intensity and duration on dispersal of Rhynchosporium secalis conidia from infected barley leaves. Trans. Br. Mycol. Soc. 86, 611–618. [Google Scholar]
- Fitt, B.D.L. , Gregory, P.H. , Todd, A.D. , McCartney, H.A. and Macdonald, O.C. (1987) Spore dispersal and plant disease gradients: a comparison between 2 empirical models. J. Phytopathol. Phytopathol. Z. 118, 227–242. [Google Scholar]
- Fitt, B.D.L. , McCartney, H.A. and Walklate, P.J. (1989) The role of rain in dispersal of pathogen inoculum. Annu. Rev. Phytopathol. 27, 241–270. [Google Scholar]
- Forgan, A.H. , Knogge, W. and Anderson, P.A. (2007) Asexual genetic exchange in the barley pathogen Rhynchosporium secalis . Phytopathology, 97, 650–654. [DOI] [PubMed] [Google Scholar]
- Foster, S.J. and Fitt, B.D.L. (2003) Isolation and characterisation of the mating‐type (MAT) locus from Rhynchosporium secalis . Curr. Genet. 44, 277–286. [DOI] [PubMed] [Google Scholar]
- Frank, A.B. (1897) Über die Zerstörung der Gerste durch einen neuen Getreidepilz. Wochenschr. Brauerei, 42, 518–520. [Google Scholar]
- Gierlich, A. , van't Slot, K.A.E. , Li, V.M. , Marie, C. , Hermann, H. and Knogge, W. (1999) Heterologous expression of the avirulence gene product, NIP1, from the barley pathogen Rhynchosporium secalis . Protein Expr. Purif. 17, 64–73. [DOI] [PubMed] [Google Scholar]
- Goodwin, S.B. (2002) The barley scald pathogen Rhynchosporium secalis is closely related to the discomycetes Tapesia and Pyrenopeziza . Mycol. Res. 106, 645–654. [Google Scholar]
- Goodwin, S.B. , Allard, R.W. and Webster, R.K. (1990) A nomenclature for Rhynchosporium secalis pathotypes. Phytopathology, 80, 1330–1336. [Google Scholar]
- Goodwin, S.B. , Saghai Maroof, M.A. , Allard, R.W. and Webster, R.K. (1993) Isozyme variation within and among populations of Rhynchosporium secalis in Europe, Australia, and the United States. Mycol. Res. 97, 49–58. [Google Scholar]
- Goodwin, S.B. , Webster, R.K. and Allard, R.W. (1994) Evidence for mutation and migration as sources of genetic variation in populations of Rhynchosporium secalis . Phytopathology, 84, 1047–1053. [Google Scholar]
- Habgood, R.M. (1971) The transmission of Rhynchosporium secalis by infected barley seed. Plant Pathol. 20, 80–81. [Google Scholar]
- Habgood, R.M. (1973) Variation in Rhynchosporium secalis . Trans. Br. Mycol. Soc. 61, 41–47. [Google Scholar]
- Habgood, R.M. and Hayes, J.D. (1971) The inheritance of resistance to Rhynchosporium secalis in barley. Heredity, 27, 25–37. [Google Scholar]
- Hahn, M. , Jüngling, S. and Knogge, W. (1993) Cultivar‐specific elicitation of barley defense reactions by the phytotoxic peptide NIP1 from Rhynchosporium secalis . Mol. Plant–Microbe Interact. 6, 745–754. [DOI] [PubMed] [Google Scholar]
- Hansen, L.R. and Magnus, H.A. (1969) Leaf spot fungi on barley in Norway. Forskn. Fors. Landbr. 20, 95–105. [Google Scholar]
- Hansen, L.R. and Magnus, H.A. (1973) Virulence spectrum of Rhynchosporium secalis in Norway and sources or resistance in barley. Phytopathol. Z. 76, 303–313. [Google Scholar]
- Heinsen, E. (1901) Beobachtungen über den neuen Getreidepilz Rhynchosporium graminicola . Jahrb. Hamburg Wiss. Anstalt. 18, 43–55. [Google Scholar]
- Hogenhout, S.A. , Van der Hoorn, R.A.L. , Terauchi, R. and Kamoun, S. (2009) Emerging concepts in effector biology of plant‐associated organisms. Mol. Plant–Microbe Interact. 22, 115–122. [DOI] [PubMed] [Google Scholar]
- Home‐Grown Cereals Authority (HGCA) (2011) The HGCA Barley Disease Management Guide. Stoneleigh Park, Warwickshire: HGCA. [Google Scholar]
- Horbach, R. , Navarro‐Quesada, A.R. , Knogge, W. and Deising, H.B. (2011) When and how to kill a plant cell: infection strategies of plant pathogenic fungi. J. Plant Physiol. 168, 51–62. [DOI] [PubMed] [Google Scholar]
- Houston, B.R. and Ashworth, L.J. (1957) Newly determined races of the barley scald fungus in California. Phytopathology, 47, 525. [Google Scholar]
- Howlett, S.G. and Cooke, B.M. (1987) Scanning electron microscopy of sporulation in Rhynchosporium secalis . Trans. Br. Mycol. Soc. 88, 547–549. [Google Scholar]
- Howlett, S.G. and Cooke, B.M. (1992) More scanning electron micrographs of conidial production in Rhynchosporium secalis on barley. Mycologist, 6, 16–17. [Google Scholar]
- Ichihara, A. , Hashimoto, M. , Hirai, T. , Takeda, I. , Sasamura, Y. , Sakamura, S. , Sato, R. and Tajimi, A. (1989) Structure, synthesis, and stereochemistry of (+)‐orthosporin, a phytotoxic metabolite of Rhynchosporium orthosporum . Chem. Lett. 8, 1495–1498. [Google Scholar]
- Jackson, L.F. and Webster, R.K. (1976) Race differentiation, distribution, and frequency of Rhynchosporium secalis in California. Phytopathology, 66, 719–725. [Google Scholar]
- Jenkins, J.E.E. and Jemmett, J.L. (1967) Barley leaf blotch. Natl. Agric. Advis. Serv. Q. Rev. 75, 127–132. [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones, P. and Ayres, P.G. (1972) The nutrition of the subcuticular mycelium of Rhynchosporium secalis (barley leaf blotch): permeability changes induced in the host. Physiol. Plant Pathol. 2, 383–392. [Google Scholar]
- Jones, P. and Ayres, P.G. (1974) Rhynchosporium leaf blotch of barley studied during the subcuticular phase by electron microscopy. Physiol. Plant Pathol. 4, 229–233. [Google Scholar]
- Jorgensen, H.J.L. and Smedegaard‐Petersen, V. (1995) Pathogenic variation of Rhynchosporium secalis in Denmark and sources of resistance in barley. Plant Dis. 79, 297–301. [Google Scholar]
- Kari, A.G. and Griffiths, E. (1993) Components of partial resistance of barley to Rhynchosporium secalis: use of seedling tests to predict field resistance. Ann. Appl. Biol. 123, 545–561. [Google Scholar]
- Kay, J.G. and Owen, H. (1973) Transmission of Rhynchosporium secalis on barley grain. Trans. Br. Mycol. Soc. 60, 405–411. [Google Scholar]
- Kendall, S. , Hollomon, D.W. , Ishii, H. and Heaney, S.P. (1994) Characterization of benzimidazole‐resistant strains of Rhynchosporium secalis . Pestic. Sci. 40, 175–181. [Google Scholar]
- Kendall, S.J. , Hollomon, D.W. , Cooke, L.R. and Jones, D.R. (1993) Changes in sensitivity to DMI fungicides in Rhynchosporium secalis . Crop Prot. 12, 357–362. [Google Scholar]
- Keon, J. , Antoniw, J. , Carzaniga, R. , Deller, S. , Ward, J.L. , Baker, J.M. , Beale, M.H. , Hammond‐Kosack, K. and Rudd, J.J. (2007) Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant–Microbe Interact. 20, 178–193. [DOI] [PubMed] [Google Scholar]
- Kirsten, S. , Siersleben, S. and Knogge, W. (2011) A GFP‐based assay to quantify the impact of effectors on the ex planta development of the slowly growing barley pathogen Rhynchosporium commune . Mycologia, 103, 1019–1027. [DOI] [PubMed] [Google Scholar]
- Kirsten, S. , Navarro‐Quezada, A. , Penselin, D. , Wenzel, C. , Matern, A. , Leitner, A. , Baum, T. , Seiffert, U. and Knogge, W. (2012) Necrosis‐inducing proteins of Rhynchosporium commune, effcetors in quantitative disease resistance. Mol. Plant–Microbe Interact. in press. [DOI] [PubMed] [Google Scholar]
- Knogge, W. (1996) Molecular basis of specificity in plant/fungus interactions. Eur. J. Plant Pathol. 102, 807–816. [Google Scholar]
- Koga, J. , Yamauchi, T. , Shimura, M. , Ogawa, N. , Oshima, K. , Umemura, K. , Kikuchi, M. and Ogasawara, N. (1998) Cerebrosides A and C, sphingolipid elicitors of hypersensitive cell death and phytoalexin accumulation in rice plants. J. Biol. Chem. 273, 31 985–31 991. [DOI] [PubMed] [Google Scholar]
- von Korff, M. , Udupa, S.M. , Yahyaoui, A. and Baum, M. (2004) Genetic variation among Rhynchosporium secalis populations of West Asia and North Africa as revealed by RAPD and AFLP analysis. J. Phytopathol. 152, 106–113. [Google Scholar]
- Lee, H.K. , Tewari, J.P. and Turkington, T.K. (2001a) Symptomless infection of barley seed by Rhynchosporium secalis . Can. J. Plant Pathol. Rev. Can. Phytopathol. 23, 315–317. [Google Scholar]
- Lee, H.K. , Tewari, J.P. and Turkington, T.K. (2001b) A PCR‐based assay to detect Rhynchosporium secalis in barley seed. Plant Dis. 85, 220–225. [DOI] [PubMed] [Google Scholar]
- Lehnackers, H. and Knogge, W. (1990) Cytological studies on the infection of barley cultivars with known resistance genotypes by Rhynchosporium secalis . Can. J. Bot. 68, 1953–1961. [Google Scholar]
- Linde, C.C. , Zala, M. , Ceccarelli, S. and McDonald, B.A. (2003) Further evidence for sexual reproduction in Rhynchosporium secalis based on distribution and frequency of mating‐type alleles. Fungal Genet. Biol. 40, 115–125. [DOI] [PubMed] [Google Scholar]
- Linsell, K.J. , Keiper, F.J. , Forgan, A. and Oldach, K.H. (2011) New insights into the infection process of Rhynchosporium secalis in barley using GFP. Fungal Genet. Biol. 48, 124–131. [DOI] [PubMed] [Google Scholar]
- Locke, T. and Phillips, A.N. (1995) The occurrence of carbendazim resistance in Rhynchosporium secalis on winter barley in England and Wales in 1992 and 1993. Plant Pathol. 44, 294–300. [Google Scholar]
- Looseley, M.E. , Newton, A.C. , Atkins, S.D. , Fitt, B.D.L. , Fraije, B. , Thomas, W.T.B. , Keith, R. , Lynott, J. and Harrap, D. (2011) Genetic basis of control of Rhynchosporium secalis infection and symptom expression in barley. Euphytica 184, 47–56. [Google Scholar]
- Lucas, J.A. (1998) Plant Pathology and Plant Pathogens. Oxford: Blackwell. [Google Scholar]
- Luttrell, E.S. (1974) Parasitism of fungi on vascular plants. Mycologia, 66, 1–15. [Google Scholar]
- Ma, W.B. and Guttman, D.S. (2008) Evolution of prokaryotic and eukaryotic virulence effectors. Curr. Opin. Plant Biol. 11, 412–419. [DOI] [PubMed] [Google Scholar]
- Mayfield, A.H. and Clare, B.G. (1984) Survival over summer of Rhynchosporium secalis in host debris in the field. Aust. J. Agric. Res. 35, 789–797. [Google Scholar]
- McDermott, J.M. , McDonald, B.A. , Allard, R.W. and Webster, R.K. (1989) Genetic variability for pathogenicity, isozyme, ribosomal DNA and colony color variants in populations of Rhynchosporium secalis . Genetics, 122, 561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, B.A. , Zhan, J. and Burdon, J.J. (1999) Genetic structure of Rhynchosporium secalis in Australia. Phytopathology, 89, 639–645. [DOI] [PubMed] [Google Scholar]
- Newman, P.L. (1985) Variation amongst isozymes of Rhynchosporium secalis . Plant Pathol. 34, 329–337. [Google Scholar]
- Newman, P.L. and Owen, H. (1985) Evidence of asexual recombination in Rhynchosporium secalis . Plant Pathol. 34, 338–340. [Google Scholar]
- Newton, A.C. (1989) Somatic recombination in Rhynchosporium secalis . Plant Pathol. 38, 71–74. [Google Scholar]
- Newton, A.C. , Searle, J. , Guy, D.C. , Hackett, C.A. and Cooke, D.E.L. (2001) Variability in pathotype, aggressiveness, RAPD profile, and rDNA ITS1 sequences of UK isolates of Rhynchosporium secalis . Z. Pflanzenkr. Pflanzenschutz J. Plant Dis. Prot. 108, 446–458. [Google Scholar]
- Newton, A.C. , Toth, I.K. , Neave, P. and Hyman, L.J. (2004a) Bacterial inoculum from a previous crop affects fungal disease development on subsequent nonhost crops. New Phytol. 163, 133–138. [DOI] [PubMed] [Google Scholar]
- Newton, A.C. , Swanston, J.S. and Guy, D.C. (2004b) Enhanced durability and utility of genes for resistance by deployment in cultivar mixtures In: Biology of Plant–Microbe Interactions, Volume 4 (Tikhonovich I., Lugtenberg B. and Provorov N., eds), pp. 240–243. St Petersburg: International Society for Molecular Plant‐Microbe Interactions. [Google Scholar]
- Oliver, R.P. and Ipcho, S.V.S. (2004) Arabidopsis pathology breathes new life into the necrotrophs‐vs‐biotrophs classification of fungal pathogens. Mol. Plant Pathol. 5, 347–352. [DOI] [PubMed] [Google Scholar]
- Oudemans, C.A.J. (1897) Observations mycologiques. Konink. Akad. Wetensch. Amsterdam, 6, 86–92. [Google Scholar]
- Owen, H. (1963) Physiologic races of Rhynchosporium secalis on cultivated barley. Trans. Br. Mycol. Soc. 46, 604–608. [Google Scholar]
- Oxley, S.J.P. , Cooke, L.R. , Black, L. , Hunter, A. and Mercer, P.C. (2003) Management of Rhynchosporium in different barley varieties and cropping systems. Home‐Grown Cereals Authority, Project Report 315. London: Home‐Grown Cereals Authority. [Google Scholar]
- Ozoe, S. (1956) Studies on the Rhynchosporium scald of barley and its control. Shimane Prefect. Agric. Inst. Bull. 1, 1–122. [Google Scholar]
- Poland, J.A. , Balint‐Kurti, P.J. , Wisser, R.J. , Pratt, R.C. and Nelson, R.J. (2009) Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14, 21–29. [DOI] [PubMed] [Google Scholar]
- Prusky, D. and Yakoby, N. (2003) Pathogenic fungi: leading or led by ambient pH? Mol. Plant Pathol. 4, 509–516. [DOI] [PubMed] [Google Scholar]
- Rasmussen, K.J. (1984) Methods of soil tillage for spring barley on coarse sandy silts. Tidsskr. Planteavl. 88, 443–452. [Google Scholar]
- Reed, H.E. (1957) Studies of barley scald. Bull. Tennessee Univ. Agric. Exp. Station, 268, 1–43. [Google Scholar]
- Robbertse, B. , Campbell, G.F. and Crous, P.W. (1995) Revision of Pseudocercosporella‐like species causing eyespot disease of wheat. S. Afr. J. Bot. 61, 43–48. [Google Scholar]
- Robbertse, B. , van der Rijst, M. , van Aarde, I.M.R. , Lennox, C. and Crous, P.W. (2001) DMI sensitivity and cross‐resistance patterns of Rhynchosporium secalis isolates from South Africa. Crop Prot. 20, 97–102. [Google Scholar]
- Rohe, M. , Gierlich, A. , Hermann, H. , Hahn, M. , Schmidt, B. , Rosahl, S. and Knogge, W. (1995) The race‐specific elicitor, NIP1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J. 14, 4168–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohe, M. , Searle, J. , Newton, A.C. and Knogge, W. (1996) Transformation of the plant pathogenic fungus, Rhynchosporium secalis . Curr. Genet. 29, 587–590. [DOI] [PubMed] [Google Scholar]
- Sakaki, T. , Zähringer, U. , Warnecke, D.C. , Fahl, A. , Knogge, W. and Heinz, E. (2001) Sterol glycosides and cerebrosides accumulate in Pichia pastoris, Rhynchosporium secalis and other fungi under normal conditions or under heat shock and ethanol stress. Yeast, 18, 679–695. [DOI] [PubMed] [Google Scholar]
- Salamati, S. and Tronsmo, A.M. (1997) Pathogenicity of Rhynchosporium secalis isolates from Norway on 30 cultivars of barley. Plant Pathol. 46, 416–424. [Google Scholar]
- Salamati, S. , Zhan, J. , Burdon, J.J. and McDonald, B.A. (2000) The genetic structure of field populations of Rhynchosporium secalis from three continents suggests moderate gene flow and regular recombination. Phytopathology, 90, 901–908. [DOI] [PubMed] [Google Scholar]
- Schein, R.D. (1958) Pathogenic specialization in Rhynchosporium secalis . Phytopathology, 48, 477–480. [Google Scholar]
- Schornack, S. , Huitema, E. , Cano, L.M. , Bozkurt, T.O. , van Oliva, R., Damme, M. , Schwizer, S. , Raffaele, S. , Chaparro‐Garcia, A. , Farrer, R. , Segretin, M.E. , Bos, J. , Haas, B.J. , Zody, M.C. , Nusbaum, C. , Win, J. , Thines, M. and Kamoun, S. (2009) Ten things to know about oomycete effectors. Mol. Plant Pathol. 10, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch, S. , Linde, C.C. , Knogge, W. , Jackson, L.F. and McDonald, B.A. (2004) Molecular population genetic analysis differentiates two virulence mechanisms of the fungal avirulence gene NIP1 . Mol. Plant–Microbe Interact. 17, 1114–1125. [DOI] [PubMed] [Google Scholar]
- Schweizer, P. and Stein, N. (2011) Large‐scale data integration reveals colocalization of gene functional groups with meta‐QTL for multiple disease resistance in barley. Mol. Plant–Microbe Interact. 24, 1492–1501. [DOI] [PubMed] [Google Scholar]
- Shaw, M.W. (1987) Assessment of upper movement of rain splash using a fluorescent tracer method and its application to the epidemiology of cereal pathogens. Plant Pathol. 36, 201–213. [Google Scholar]
- Shipton, W.A. , Boyd, W.J.R. and Ali, S.M. (1974) Scald of barley. Rev. Plant Pathol. 53, 839–861. [Google Scholar]
- Sierotzki, H. , Wullschleger, J. and Gisi, U. (2000) Point mutation in cytochrome b gene conferring resistance to strobilurin fungicides in Erysiphe graminis f. sp. tritici field isolates. Pestic. Biochem. Physiol. 68, 107–112. [Google Scholar]
- Skoropad, W.P. (1959) Seed and seedling infection of barley by Rhynchosporium secalis . Phytopathology, 49, 623–626. [Google Scholar]
- Skoropad, W.P. (1966) Sporulating potential of Rhynchosporium secalis on naturally infected leaves of barley. Can. J. Plant Sci. 46, 243–247. [Google Scholar]
- van't Slot, K.A.E. and Knogge, W. (2002) A dual role of microbial pathogen‐derived proteins in plant disease and resistance. Crit. Rev. Plant Sci. 21, 229–271. [Google Scholar]
- van't Slot, K.A.E. , van den Burg, H.A. , Kloks, C.P.A.M. , Hilbers, C.W. , Knogge, W. and Papavoine, C.H.M. (2003) Solution structure of the plant disease resistance‐triggering protein NIP1 from the fungus Rhynchosporium secalis shows a novel β‐sheet fold. J. Biol. Chem. 278, 45 730–45 736. [DOI] [PubMed] [Google Scholar]
- van't Slot, K.A.E. , Gierlich, A. and Knogge, W. (2007) A single binding site mediates resistance‐ and disease‐associated activities of the effector protein NIP1 from the barley pathogen Rhynchosporium secalis . Plant Physiol. 144, 1654–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner‐Lange, S. , Fischer, A. , Böttcher, A. , Rouhara, I. , Liedgens, H. , Schmelzer, E. and Knogge, W. (2003) Differential defense reactions in leaf tissues of barley in response to infection by Rhynchosporium secalis and to treatment with a fungal avirulence gene product. Mol. Plant–Microbe Interact. 16, 893–902. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. and de Wit, P.J.G.M. (2009) Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Stukenbrock, E.H. and McDonald, B.A. (2009) Population genetics of fungal and oomycete effectors involved in gene‐for‐gene interactions. Mol. Plant–Microbe Interact. 22, 371–380. [DOI] [PubMed] [Google Scholar]
- Taggart, P.J. , Cooke, L.R. , Mercer, P.C. and Shaw, M.W. (1998) Effects of fungicides used to control Rhynchosporium secalis where benzimidazole resistance is present. Crop Prot. 17, 727–734. [Google Scholar]
- Taggart, P.J. , Locke, T. , Phillips, A.N. , Pask, N. , Hollomon, D.W. , Kendall, S.J. , Cooke, L.R. and Mercer, P.C. (1999) Benzimidazole resistance in Rhynchosporium secalis and its effect on barley leaf blotch control in the UK. Crop Prot. 18, 239–243. [Google Scholar]
- Tao, Y. , Xie, Z.Y. , Chen, W.Q. , Glazebrook, J. , Chang, H.S. , Han, B. , Zhu, T. , Zou, G.Z. and Katagiri, F. (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae . Plant Cell, 15, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirugnanasambandam, A. , Wright, K.M. , Atkins, S.D. , Whisson, S.C. and Newton, A.C. (2011) Infection of Rrs1 barley by an incompatible race of the fungus Rhynchosporium secalis expressing the green fluorescent protein. Plant Pathol. 60, 513–521. [Google Scholar]
- Tyler, B.M. (2009) Entering and breaking: virulence effector proteins of oomycete plant pathogens. Cell. Microbiol. 11, 13–20. [DOI] [PubMed] [Google Scholar]
- Umemura, K. , Ogawa, N. , Yamauchi, T. , Iwata, M. , Shimura, M. and Koga, J. (2000) Cerebroside elicitors found in diverse phytopathogens activate defense responses in rice plants. Plant Cell Physiol. 41, 676–683. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A. , Raffaele, S. , Vossen, J.H. , Champouret, N. , Oliva, R. , Segretin, M.E. , Rietman, H. , Cano, L.M. , Lokossou, A. , Kessel, G. , Pel, M.A. and Kamoun, S. (2011) Understanding and exploiting late blight resistance in the age of effectors. Annu. Rev. Phytopathol. 49, 507–531. [DOI] [PubMed] [Google Scholar]
- Walters, D.R. , Avrova, A. , Bingham, I.J. , Burnett, F.J. , Fountaine, J. , Havis, N.D. , Hoad, S.P. , Hughes, G. , Looseley, M. , Oxley, S.J.P. , Renwick, A. , Topp, C.F.E. and Newton, A.C. (2012) Control of foliar diseases in barley: towards an integrated approach. Eur. J. Plant Pathol. 133, 33–73. [Google Scholar]
- Wevelsiep, L. , Kogel, K.‐H. and Knogge, W. (1991) Purification and characterization of peptides from Rhynchosporium secalis inducing necrosis in barley. Physiol. Mol. Plant Pathol. 39, 471–482. [Google Scholar]
- Wevelsiep, L. , Rüpping, E. and Knogge, W. (1993) Stimulation of barley plasmalemma H+‐ATPase by phytotoxic peptides from the fungal pathogen Rhynchosporium secalis . Plant Physiol. 101, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, J.E. , Kendall, S.J. , Butters, J. , Hollomon, D.W. and Hall, L. (1995) Using allele‐specific oligonucleotide probes to characterize benzimidazole resistance in Rhynchosporium secalis . Pestic. Sci. 43, 201–209. [Google Scholar]
- Wiese, M.V. (1987) Compendium of Wheat Diseases, 2nd edn 112 pp. St. Paul, MN: APS Press. [Google Scholar]
- Williams, K. , Donnellan, S. , Smyl, C. , Scott, L. and Wallwork, H. (2003) Molecular variation in Rhynchosporium secalis isolates obtained from hotspots. Australas. Plant Pathol. 32, 257–262. [Google Scholar]
- Williams, R.J. and Owen, H. (1975) Susceptibility of barley cultivars to leaf blotch and aggressiveness of Rhynchosporium secalis races. Trans. Br. Mycol. Soc. 65, 109–114. [Google Scholar]
- Xi, K. , Burnett, P.A. , Tewari, J.P. , Chen, M.H. , Turkington, T.K. and Helm, J.H. (2000) Histopathological study of barley cultivars resistant and susceptible to Rhynchosporium secalis . Phytopathology, 90, 94–102. [DOI] [PubMed] [Google Scholar]
- Xi, K.N. , Turkington, T. , Meadus, J. , Helm, J. and Tewari, J. (2003) Dynamics of Rhynchosporium secalis pathotypes in relation to barley cultivar resistance. Mycol. Res. 107, 1485–1492. [DOI] [PubMed] [Google Scholar]
- Xue, G. and Hall, R. (1991) Components of parasitic fitness in Rhynchosporium secalis and quantitative resistance to scald in barley as determined with a dome inoculation chamber. Can. J. Plant Pathol. Rev. Can. Phytopathol. 13, 19–25. [Google Scholar]
- Yuan, C. , Li, C. , Yan, L. , Jackson, A.O. , Liu, Z. , Han, C. , Yu, J. and Li, D. (2011) A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE , 6, e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffarano, P.L. , McDonald, B.A. , Zala, M. and Linde, C.C. (2006) Global hierarchical gene diversity analysis suggests the Fertile Crescent is not the center of origin of the barley scald pathogen Rhynchosporium secalis . Phytopathology, 96, 941–950. [DOI] [PubMed] [Google Scholar]
- Zaffarano, P.L. , McDonald, B.A. and Linde, C.C. (2008) Rapid speciation following recent host shifts in the plant pathogenic fungus Rhynchosporium . Evolution, 62, 1418–1436. [DOI] [PubMed] [Google Scholar]
- Zaffarano, P.L. , McDonald, B.A. and Linde, C.C. (2011) Two new species of Rhynchosporium . Mycologia, 103, 195–202. [DOI] [PubMed] [Google Scholar]
- Zhan, J. , Linde, C.C. , Jurgens, T. , Merz, U. , Steinebrunner, F. and McDonald, B.A. (2005) Variation for neutral markers is correlated with variation for quantitative traits in the plant pathogenic fungus Mycosphaerella graminicola . Mol. Ecol. 14, 2683–2693. [DOI] [PubMed] [Google Scholar]
- Zhan, J. , Stefanato, F.L. and McDonald, B.A. (2006) Selection for increased cyproconazole tolerance in Mycosphaerella graminicola through local adaptation and in response to host resistance. Mol. Plant Pathol. 7, 259–268. [DOI] [PubMed] [Google Scholar]
- Zhan, J. , Fitt, B.D.L. , Pinnschmidt, H.O. , Oxley, S.J.P. and Newton, A.C. (2008) Resistance, epidemiology and sustainable management of Rhynchosporium secalis populations on barley. Plant Pathol. 57, 1–14. [Google Scholar]
- Zhang, Q. , Webster, R.K. , Crandall, B.A. , Jackson, L.F. and Saghai Maroof, M.A. (1992) Race composition and pathogenicity associations of Rhynchosporium secalis in California. Phytopathology, 82, 798–803. [Google Scholar]


