SUMMARY
Atf1‐homologous basic region leucine zipper (bZIP) transcription factors are known to act downstream of the stress‐activated mitogen‐activated protein kinase (SAPK) cascade in mammals, as well as in several fungi; they regulate the transcription of genes involved in the general stress response. Functional analyses of BcAtf1 in Botrytis cinerea show that it is also connected to the SAPK BcSak1, as it shares several stress response target genes. However, Δbcatf1 mutants are not hypersensitive to osmotic or oxidative stress, as are Δbcsak1 mutants. Both BcSak1 and BcAtf1 are regulators of differentiation, but their roles in these processes are almost inverse as, in contrast with Δbcsak1, Δbcatf1 mutants are significantly impaired in conidia production and do not differentiate any sclerotia. They show extremely vigorous growth in axenic culture, with a thick layer of aerial hyphae and a marked increase in colonization efficiency on different host plants and tissues. In addition, the sensitivity to cell wall‐interfering agents is increased strongly. Microarray analyses demonstrate that the loss of BcAtf1 leads to extensive transcriptional changes: apart from stress response genes, the expression of a broad set of genes, probably involved in primary metabolism, cell wall synthesis and development, is affected by BcAtf1. Unexpectedly, BcAtf1 also controls secondary metabolism: the mutant contains significantly elevated levels of phytotoxins. These data indicate that BcAtf1 controls a diversity of cellular processes and has broad regulatory functions.
INTRODUCTION
Homologues of the basic region leucine zipper (bZIP) activating transcription factor Atf1 are players in diverse cellular processes and normally act downstream of the stress‐activated mitogen‐activated protein kinase (SAPK) cascade. This regulatory cascade is conserved in eukaryotic cells and has been analysed in detail in mammals, where the SAPK, JNK (c‐Jun N‐terminal kinase) and HOG (high‐osmolarity glycerol response) homologue p38 activates ATF2 (van Dam et al., 1995; Gupta et al., 1995; Livingstone et al., 1995), as well as in fission yeast, whose SAPK Sty1 controls Atf1 (Takeda et al., 1995). In fungi, the Atf1 protein from fission yeast has been analysed extensively. Activated and stabilized via phosphorylation by Sty1, the transcription factor regulates genes involved in general stress responses, such as the catalase gene ctt1 (Wilkinson et al., 1996). Atf1 is also involved in sexual development, the entry into the stationary phase, conjugation and meiosis (Shiozaki and Russell, 1996, Takeda et al., 1995; Watanabe and Yamamoto, 1996), where it modulates meiotic recombination and heterochromatin formation (Kon et al., 1997). Atf1 can form heterodimers frequently with the bZIP factor Pcr1 (Kon et al., 1997); both interact in the transcriptional regulation of stress response genes (Sansóet al., 2008). In addition, Atf1 plays a role in cell cycle control (Ors et al., 2009) and is a target for the ubiquitin‐proteasome system, indicating that this association is disrupted by the stress‐induced phosphorylation of Atf1 (Lawrence et al., 2009).
The diverse functions and regulatory mechanism of the Atf1 protein are reflected by its heterogeneous roles in filamentous fungi. The atf1 knockout in Neurospora crassa results in mutants impaired in asexual reproduction and reveals a role of Atf1 in the regulation of genes involved in germination and in the circadian rhythm (Yamashita et al., 2008), whereas inactivation of atfA in Aspergillus nidulans does not affect asexual reproduction per se. However, ΔatfA conidia are highly susceptible to oxidative and heat stress (Hagiwara et al., 2008); AtfA regulates a set of oxidative and osmotic stress‐responsive genes (Balázs et al., 2010). Recently, it has been demonstrated that AtfA interacts physically with SakA (Lara‐Rojas et al., 2011). The orthologue CpTf1 from the rye pathogen Claviceps purpurea does not control growth or sporulation in axenic culture, but regulates the transcription of the stress responsive catalase CpCat1 and of genes not directly involved in stress responses. Its deletion reduces fungal virulence significantly (Nathues et al., 2004; E. Nathues and P. Tudzynski, unpublished data). Furthermore, in the rice pathogen Magnaporthe oryzae, the orthologue Moatf1 is involved in the oxidative stress response and is important for full virulence (Guo et al., 2010). In this work, the role of the Atf1 homologue BcAtf1 was investigated in the ascomycete Botrytis cinerea, a broad host range necrotrophic phytopathogen (recently reviewed by Choquer et al., 2007; van Kan, 2006; Tudzynski and Kokkelink, 2009). In this fungus, BcAtf1 is obviously not exclusively controlled by the SAPK cascade. We show here that the phenotype of Δbcatf1 differs significantly from that of Δbcsak1 (Segmüller et al., 2007). We present evidence that BcAtf1 affects several signalling and differentiation processes, as well as secondary metabolism. Microarray analyses were used to clarify the signalling network controlled by BcAtf1.
RESULTS
Identification and deletion of the bcatf1 gene in B. cinerea
The bcatf1 open reading frame (ORF) was not automatically annotated in the B05.10 sequence (http://www.broadinstitute.org/annotation/genome/botrytis_cinerea), but was obtained by heterologous polymerase chain reaction (PCR) with primer pairs based on the DNA sequence of known atf1‐homologous genes from different filamentous fungi considering the B. cinerea codon usage (primer 27−30, data not shown). The ORF identified consists of 1789 bp with two introns of 174 and 49 bp [confirmed by reverse transcription (RT)‐PCR]. The first intron belongs to the group of noncanonical GC‐AG introns (Rep et al., 2006). The BcAtf1 protein (521 amino acids) contains the conserved BRLZ domain (smart00338) (Marchler‐Bauer et al., 2005). Conservation of this domain is quite high in different ascomycetes [71.4% identity to A. nidulans AtfA (Hagiwara et al., 2008) and 77.6% identity to Schizosaccharomyces pombe Atf1 (Takeda et al., 1995)], as well as in the mammalian ATF2 (44.9% identity to Homo sapiens ATF2; Maekawa et al., 1989) (Fig. 1). However, comparison of the full‐length protein sequences revealed limited similarity to these transcription factors (30.7% identity to A. nidulans AtfA, 21.7% identity to S. pombe Atf1 and 17.7% identity to H. sapiens ATF2; data not shown). In the sequence of another strain of B. cinerea that has been published recently (T4 strain; Amselem et al., 2011), the bcatf1 ORF is split into two predicted ORFs, BofuT4_P101510.1 and BofuT4_P101520.1, respectively (http://urgi.versailles.inra.fr/Species/Botrytis/).
Figure 1.

Protein alignment of the basic region leucine zipper (bZIP) domain of BcAtf1 and bZIP sequences of (hypothetical) Atf1 proteins (ClustalW). [Aspergillus nidulans AtfA (AY166595.1), Claviceps purpurea CpTf1 (AJ428492.1), Gibberella zeae (XM_390318.1), Neurospora crassa (AL451017.1), Sclerotinia sclerotiorum (XM_001593436.1), Schizosaccharomyces pombe (NM_001021546.1), Homo sapiens (BC026175.1)]. Residues that match the consensus sequence exactly are shaded (black).
To create a bcatf1 deletion strain for functional analysis, a replacement approach was used: the ORF of bcatf1 in the wild‐type (WT) strain B05.10 was replaced by a nourseothricin resistance cassette (Fig. S1A, see Supporting Information). Homologous integration was analysed for several transformants via diagnostic PCR. Single spore isolation resulted in homokaryotic strains of deletion mutants that were screened for the absence of the bcatf1 WT allele by PCR (Fig. S1B). Southern blot analysis confirmed homologous integration into the bcatf1 locus in the genomes of the transformants, Δbcatf1_B and Δbcatf1_L, respectively, without further ectopic integration of the replacement cassette (Fig. S1C). Both independent mutants showed the same phenotypes.
BcAtf1 is important for normal vegetative growth and differentiation
Δbcatf1 mutants have a distinct phenotype in axenic culture (see Fig. 2). Although WT grows only on the agar surface on complete medium (CM) after 3 days under day/night conditions, Δbcatf1 develops more aerial hyphae than WT, resulting in a fluffy phenotype (Fig. 2A, panel a). Microscopic analysis revealed elongated hyphae of the mutant compared with WT, but no differences in the distribution of septa (Fig. S2, see Supporting Information).
Figure 2.
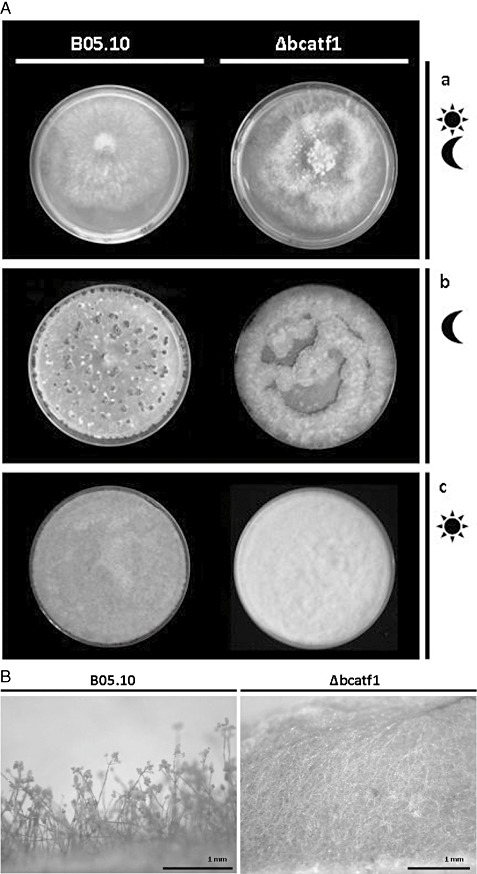
Differentiation of Δbcatf1 under standard cultivation conditions. (A) Growth test. (a) Growth under day/night conditions (3 days, B5 + 2% glucose + 1 g/L yeast extract): wild‐type (WT, B05.10) shows flat growth on top of the medium; Δbcatf1 shows the production of aerial hyphae. (b) Growth in permanent darkness (2 weeks, B5 + 2% glucose + 1 g/L yeast extract): WT shows sclerotia formation; Δbcatf1 shows the formation of conidia. (c) Growth in permanent light [complex medium (potato dextrose agar + bean)]: WT shows the production of large amounts of conidia (black colour); Δbcatf1 shows almost no production of conidia. (B) Cross‐section of fungal mycelium of Δbcatf1 in comparison with B05.10 (complete medium plates after 5 days). WT starts to differentiate conidiophores containing conidia, whereas Δbcatf1 only produces an abundance of aerial hyphae.
Under permanent light on CM, WT produced conidia profusely after 6 days, whereas the deletion mutant was severely affected with regard to conidia formation (Fig. 2A, panel c). As shown in Fig. 2B, WT started to differentiate conidiophores and conidia after 5 days, whereas Δbcatf1 only produced abundant aerial hyphae. After incubation in continuous darkness for 2 weeks on CM, WT formed sclerotia, the fungal survival structures that are also a prerequisite for sexual reproduction. However, Δbcatf1 produced conidia and failed to produce sclerotia under these conditions, indicating that the light‐dependent developmental processes are disturbed (Fig. 2A, panel b). The complementation of Δbcatf1 restored the WT phenotype (Fig. S8, see Supporting Information).
Assessment of the production of conidia by Δbcatf1 was performed under standard growth conditions [2 weeks on B5 + 2% glucose + 1 g/L yeast extract (B5/Glc/ye), day/night switch]. Δbcatf1 produced only 2.5% of the conidia produced by WT (Fig. S3, see Supporting Information). The germination rates of conidia of WT and the mutant were comparable, also under oxidative stress conditions (10 mm H2O2) and on hydrophobic surfaces (data not shown).
BcATF1 has limited impact on stress tolerance
As BcAtf1 is a potential component of the SAPK cascade, Δbcatf1 was analysed for the typical characteristics of Δbcsak1. The tolerance of Δbcatf1 was tested by growth under different conditions of stress. The data are summarized in Fig. 3. Osmotic stress was triggered by both sugars and salts. On CM plus xylose, sorbitol, saccharose and fructose (1 m and 2 m), no significant growth differences between WT and Δbcatf1 could be detected (data not shown). However, glucose, as well as KCl, inhibited the growth rates of the deletion mutant slightly, whereas the high osmotic pressure induced by sorbitol or NaCl did not cause significant growth repression of Δbcatf1. Therefore, there is no clear evidence for a significant impact of BcAtf1 on the general osmotic stress response. Two other major features of Δbcsak1 were not shared by Δbcatf1: Δbcatf1 and the WT strain showed comparable sensitivity to oxidative stress (Fig. S4, see Supporting Information) and to the fungicides fludioxonil and iprodione (Fig. 3). However, cell wall stress caused by Congo red and calcofluor white resulted in reduced growth of Δbcatf1 (Fig. 3), which has also been observed in Δbcsak1 (Liu et al., 2011). Calcofluor white and Congo red act by binding to nascent chitin chains, thereby inhibiting the assembly enzymes that connect chitin to 1,3‐glucan and 1,6‐glucan, resulting in a weakened cell wall (Bulawa, 1993; Ram and Klis, 2006). Therefore, the loss of BcAtf1 seems to lead to a fragile cell wall assembly system that is much more susceptible to interfering compounds. In the presence of the detergent sodium dodecylsulphate (SDS), which attacks cellular membranes, the growth rates of WT and Δbcatf1 were comparable.
Figure 3.
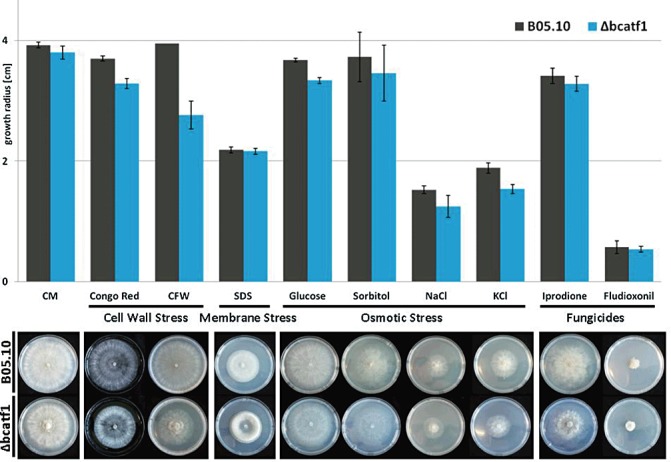
Growth in the presence of various stressors. Growth of Δbcatf1 and the wild‐type (WT) was tested on stress‐inducing media (3 days post‐inoculation). Complete medium was supplemented with Congo red (2 mg/mL), calcofluor white (CFW) (2 mg/mL), sodium dodecylsulphate (SDS) (0.02%), glucose (1 m), sorbitol (1 m), NaCl (1 m), KCl (1 m), iprodione (0.1 µg/mL) and fludioxonil (0.01 µg/mL). (A) Bar chart (growth radius in centimetres, mean value of three biological replicates, standard error is indicated). (B) Representative plates.
BcATF1 controls in planta growth
The infection cycle of B. cinerea can be divided into defined steps: adhesion of conidia, germination, appressoria formation, penetration of the host plant, killing of tissue and formation of primary lesions, expansion via tissue maceration and sporulation (reviewed in van Kan, 2006). During lesion expansion, fungal hyphae grow inside the host, as well as on the surface of the host, until the whole tissue is macerated (see WT infection; Fig. 4). Δbcatf1 displayed a quite different growth behaviour in planta: it grew inside the bean leaf tissue, but also broke through the abaxial epidermis and differentiated aerial hyphae on both adaxial and abaxial surfaces of the leaf. This almost uncontrolled growth is comparable with the mutant's vigorous growth in axenic culture. One week after infection, WT began to sporulate on its host (closed infection cycle), but there were no conidia visible with Δbcatf1 (Fig. 4). Similar results were obtained when conidia solutions were dropped onto cucumber fruits: Δbcatf1 produced an abundance of aerial hyphae and grew not only on the host surface, but also through the receptacle that forms the cucumber carpel within the pericarp of the fruit, whereas WT hardly crossed the epidermis, as shown in transverse sections of the infection sites (Fig. 4B). Complementation of Δbcatf1 restored the WT phenotype (Fig. S9, see Supporting Information).
Figure 4.

Pathogenicity assay of Δbcatf1 on different host plants and tissues in comparison with B05.10. Agar plugs were placed on leaves of Phaseolus vulgaris (A), as well as on fruits of organic cucumber (B). dpi, days post‐infection.
BcAtf1 is targeted by the SAPK cascade, but has important SAPK‐independent features
In fungi, Atf1‐like factors are generally considered to be downstream targets of the SAPK cascade. However, the corresponding B. cinerea mutants (Δbcatf1 vs. Δbcsak1) show strikingly different phenotypes, indicating different or even antagonistic functions (Table 1). Δbcsak1 forms no conidia and more sclerotia than WT; Δbcatf1 produces some conidia (especially abundant in the dark), but no sclerotia. They also act differently on exposure to stress. Unlike Δbcatf1, Δbcsak1 is more resistant than WT to the fungicide iprodione (Segmüller et al., 2007; Fig. 3), and is highly sensitive to H2O2 and NaCl (Fig. S4). In planta, deletion of bcsak1 results in a penetration defect of the fungus, whereas Δbcatf1 is more aggressive than WT. These data indicate that BcSak1 and BcAtf1 have different roles in fungal development, stress tolerance and virulence. However, Northern data suggest a significant upstream influence of the SAPK cascade, as shown in Fig. 5. BcSak1 and BcAtf1 share a set of target genes expressed in response to oxidative stress (10 mm H2O2). These targets include genes encoding for catalase B (BC1G_12856), the opsin Bop1 (BC1G_02456), the DNA damage response protein DDR48 (BC1G_10423), a mannitol dehydrogenase (BC1G_09259) and a G‐β‐like protein (BC1G_10054) (Fig. 5), which were mainly identified via a BcSak1 macroarray approach (J. Heller et al., unpublished results). Except for the G‐β‐like protein, regulation of the expression of these genes by oxidative stress conditions is lost in Δbcsak1 and Δbcatf1. Therefore, the expression of these genes is dependent on both the SAPK cascade and the Atf1 transcription factor, probably as a result of a direct regulation of BcAtf1 by BcSak1, as shown recently in A. nidulans (Lara‐Rojas et al., 2011). Another set of BcSak1 target genes, however, is not controlled by BcAtf1. These include prominent redox regulation genes encoding thioredoxin and thioredoxin reductase, glutathione peroxidase and glutaredoxin (see Fig. 5). The expression of the G‐β‐like protein transcript is regulated by both the mitogen‐activated protein kinase and the transcription factor. In this case, the G‐β‐like protein gene is repressed under standard growth conditions, but induced under conditions of oxidative stress, in both mutants, underlining its concerted regulation via SAPK and BcAtf1. With the exception of the catalase B gene, regulation of all SAPK–BcAtf1 target genes is independent of the most prominent transcriptional regulator of the oxidative stress response, Bap1 (Temme and Tudzynski, 2009; Fig. 5). BcAtf1 itself is induced at transcript level after exposure to H2O2 independent of SAPK (Segmüller, 2007).
Table 1.
Phenotypic differences of Δbcsak1 and Δbcatf1.
| Δbcsak1 | Δbcatf1 | |
|---|---|---|
| Conidiogenesis | No conidia | Impaired conidia production |
| Sclerotia formation | Enhanced sclerotia formation | No sclerotia |
| Pathogenesis | Penetration defect | Enhanced growth and invasion |
| Resistance to iprodione | Yes | No |
| Sensitivity to H2O2 | Yes | No |
| Sensitivity to NaCl | Yes | No |
Figure 5.
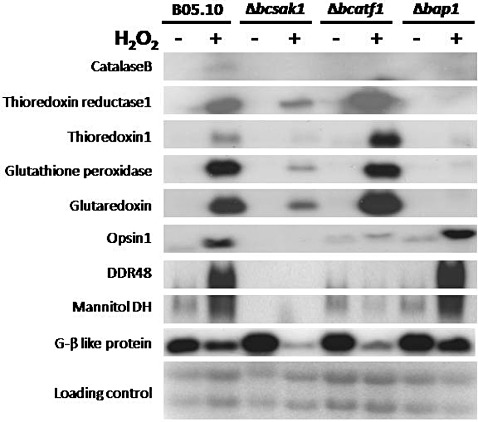
H2O2‐dependent gene expression in B05.10 and Δbcsak1, Δbcatf1 and Δbap1. Cultivation was performed in CD medium with 10 mm H2O2 (+) or without H2O2 (–) for 30 min (see Materials and methods for further information). Loading control: rRNA.
Loss of bcatf1 has genome‐wide consequences
To obtain a broader view of the impact of BcAtf1 on the transcript profile of B. cinerea, microarray analyses were performed using RNA of WT and Δbcatf1_L grown for 5 days under day/night standard conditions on cellophane. At this time, the mutant began to produce aerial hyphae, allowing a direct comparison of the normal WT growth and the fluffy Δbcatf1 phenotype. Two biological repeats for microarray analyses were performed; a third experiment as biological repeat was used for confirmation by Northern hybridization (for the full dataset, see Table S2 in Supporting Information). ArrayStar analysis of the data revealed 511 ORFs which were significantly differentially expressed in Δbcatf1. The amount of ORFs up‐regulated in Δbcatf1 was slightly higher (278 ORFs) than the number of predicted genes that were repressed in this strain (233 ORFs). These genes were classified according to their known function following the FunCat annotation scheme (Ruepp et al., 2004). Figure 6A presents the microarray results in a bar chart, and a selected set of Northern data is shown in Fig. 6B. Genes involved in metabolic processes were strongly derepressed in Δbcatf1, probably inducing a hyperactivation of fungal metabolism. In this group, and in the category ‘development’, there was a relatively high number of ORFs whose products were probably involved in fungal cell wall biosynthesis and modification, such as a chitin deacetylase, glucanases and a 1,3‐glucan synthase, indicating an incessant development of the fungal cell wall. This could explain the excrescent growth and overproduction of aerial hyphae of Δbcatf1 relative to WT. The up‐regulated genes categorized to ‘cell rescue, defence and virulence’ include many genes responsible for plant cell wall degradation, including a cutinase, xylanase and endopolygalacturonase 4, which are induced even in axenic culture. As the growth in planta concurs with the growth on agar plates, this emphasizes the aberrant uncontrolled growth of the mutant. Δbcatf1 probably senses little of its environment because it does not grow towards nutrient sources. This can also explain its aggressive growth on host plants. The number of ‘signal transduction’ genes regulated by the bcatf1 deletion is limited and mainly includes transcription factors. As these are not bZIP transcription factors, a regulatory function of potential dimerization partners of BcAtf1 cannot be substantiated by these data. The group of genes encoding for transporters is strongly deregulated in Δbcatf1. MFS transporters, including the BMR1 transporter and an ABC transporter, but also sugar permeases and an amino acid permease, are transcriptionally activated in Δbcatf1. However, even more genes of this category encoding multidrug resistance factors, as well as putative sulphate, phosphate and uracil permeases, are down‐regulated in the deletion strain, indicating a total perturbation of transport mechanisms in Δbcatf1. Genes involved in secondary metabolism were merged into an additional category of this microarray approach because of their high abundance and conspicuously high induction according to the data obtained: 26 predicted ORFs involved in the secondary metabolism of B. cinerea were significantly up‐regulated in Δbcatf1. These genes include key enzymes of secondary metabolite production, among them known genes of phytotoxin biosynthesis.
Figure 6.
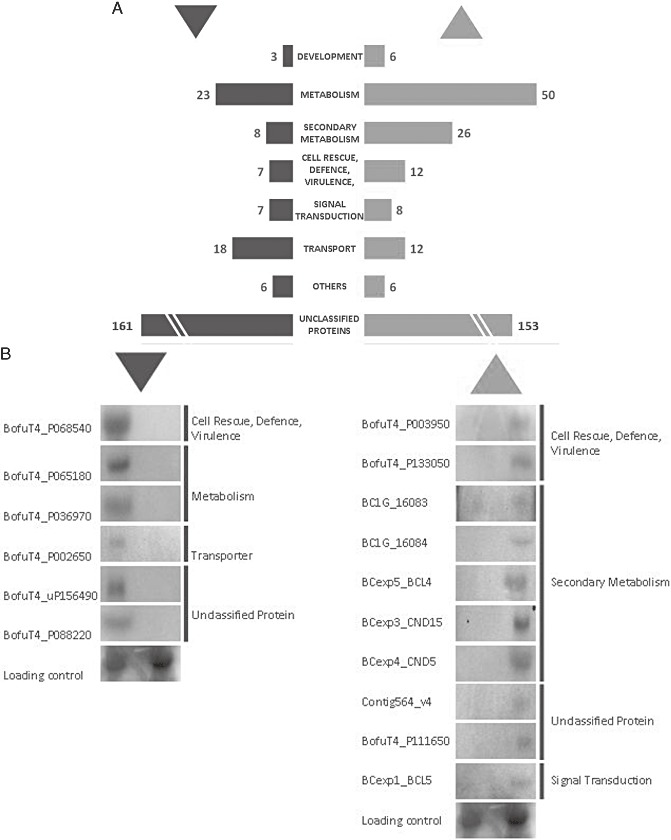
Gene expression profile of Δbcatf1. (A) Bar chart presents genes significantly up‐regulated (▴) and down‐regulated (▾) in Δbcatf1 compared with B05.10. (B) Northern blot experiments with BcAtf1 target genes. Cultivation was performed under the conditions as described for microarray probes. See Table S2 for detailed gene descriptions. Loading control: actinA.
BcAtf1 controls secondary metabolite production
Botrytis cinerea secretes a spectrum of phytotoxic compounds, including the sesquiterpene botrydial and the polyketide botcinic acid, as the best‐studied representatives so far. Botrydial causes necrotic lesions on many host plants in a light‐dependent manner and can be detected in axenic culture as well as in planta (Collado et al., 2007; Colmenares et al., 2002); botcinic acid and related botcinins have phytotoxic and antifungal activity (Tani et al., 2005). Genes for both botrydial and botcinic acid biosynthesis are clustered (Dalmais et al., 2011; Siewers et al., 2005), and are by far the most up‐regulated genes in Δbcatf1 in this experiment. The botcinic acid gene cluster organization is still not completely understood. However, most ORFs potentially involved in botcinic acid biosynthesis have been identified (Dalmais et al., 2011), and nearly all of these genes have been found in the BcAtf1 microarray approach as up‐regulated ORFs in Δbcatf1. Botrytis cinerea is known to produce the plant hormone abscisic acid (ABA) in axenic culture (Siewers et al., 2004). The genes encoding for enzymes involved in ABA biosynthesis are located in the immediate vicinity. Three of the four genes identified for ABA production (bcaba1, bcaba2 and bcaba4) are highly up‐regulated in Δbcatf1. In the case of botrydial, the three key enzymes of the corresponding gene cluster, namely the P450 monooxygenase BcBot1 (Siewers et al., 2004), the sesquiterpene synthase BcBot2 and the p450 monooxygenase BcBot3 (Pinedo et al., 2008), are highly up‐regulated in the Δbcatf1 strain under conditions that are not known to enhance botrydial production. These genes, previously described as CND5, CND15 and CND11, respectively, were identified as calcineurin‐dependent genes in a macroarray approach: the botrydial genes are co‐regulated by the Gα protein Bcg1, as well as by calcineurin (Schumacher et al., 2008a; Viaud et al., 2003). To further investigate the regulatory function of BcAtf1 in this network, expression of the genes encoding the P450 monooxygenase BcBot1/CND5 and the sesquiterpene synthase BcBot2/CND15 was determined under botrydial production condition (see Materials and methods) with and without the addition of the exogenous calcineurin inhibitor cyclosporin A (CsA). The results (Fig. S5, see Supporting Information) confirm the repressing function of BcAtf1 on the expression of the botrydial cluster genes, but also show that it is partly overruled by calcineurin. The expression of bcbot1 and bcbot2 was significantly higher in Δbcatf1 than in WT, but was also reduced, although not completely abolished, by the addition of CsA to the culture (Fig. S5A). These experiments were performed under standard day/night conditions. As the formation of botrydial is light dependent, a parallel experiment was performed in continuous darkness (Fig. S5B). As expected, the botrydial genes were not induced in WT under these conditions, whereas Δbcatf1 showed reduced, but constitutive, expression of bcbot1 and bcbot2, independent of CsA addition.
Chemical analyses confirm that the amounts of phytotoxins formed in axenic culture correlate with the expression data (Table 2; Fig. S6, see Supporting Information): botrydial, botryendial, which have strong phytotoxic activity in planta (Collado et al., 2007; Colmenares et al., 2002), and botcinin A are significantly elevated in the deletion strain. These data underline the function of BcAtf1 as a repressor of genes regulating secondary metabolism in B. cinerea. To test whether this regulatory effect is achieved by direct binding of BcAtf1 at the promoters of the target genes, yeast one‐hybrid analyses were performed. The predicted DNA‐binding domain of BcAtf1 was cloned in a haploid yeast strain in frame with the Gal4 activator domain, whereas promoters of a representative set of regulated genes were cloned in another haploid strain upstream of a histidine reporter gene (HIS3; see Materials and methods). These genes were CutA, BcBOT1 and three genes that are part of the botcinic acid biosynthesis gene cluster (BcPKS6, BC1G_15841.1 and BC1G_16085.1; BC1G_02936.1; Dalmais et al., 2011). As shown in Fig. S7 (see Supporting Information), mating of the haploid strains did not allow the recovery of diploid yeasts in which the BcAtf1‐binding domain activates the expression of the HIS3 gene. These results suggest that there is no physical interaction between the BcAtf1 protein and the tested promoters, which strongly suggests an indirect/global effect of this transcription factor on the CutA gene and the expression of genes involved in the biosynthesis of phytotoxins.
Table 2.
Metabolites isolated (mg) from B05.10 and Δbcatf1 (for information on their structures, see Fig. S6).
| Compound | B05.10 | Δbcatf1 | Δbcatf1‐2 | Phytotoxicity in planta |
|---|---|---|---|---|
| Botrydial (1) | 7 | 13 | 10 | Strong |
| Dihydrobotrydial (2) | 6 | 1.5 | 2 | Low |
| Botryendial (3) | 1 | 9.6 | 7 | Strong |
| Botryenalol (4) | — | — | 9 | Low |
| Botrydienalol (5) | — | — | 5 | Low |
| β‐Acetoxy‐9β,10β‐dihydroxyprobotriane (6) | <1 | 1 | — | — |
| Botcinic acid (7) | 7 | 4 | — | Strong |
| Botcinin A (8) | 5 | 22 | 20 | Strong |
| Botrylactone (9) | <1 | 4 | 10 | — |
DISCUSSION
We have shown that the loss of the B. cinerea bZIP transcription factor BcAtf1 has significant phenotypical consequences, emphasizing the diverse functions of the BcAtf1 protein. The transcription factor is an important player for fungal differentiation processes such as hyphal development, conidia and sclerotia formation, and for ‘balanced’ infection of host tissue. In addition, it acts as a repressor of secondary metabolite production (Fig. 7) and is at least partly controlled by the mitogen‐activated protein kinase BcSak1.
Figure 7.
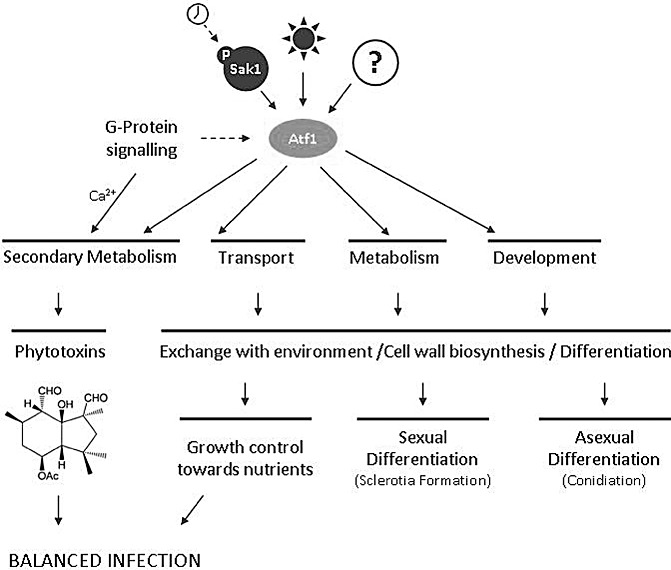
BcAtf1 controls various differentiation processes and phytotoxin production. BcAtf1 affects hyphal morphology, sexual and asexual differentiation and secondary metabolism in Botrytis cinerea, presumably as a result of various signals, such as circadian clock‐induced activation of the stress‐activated mitogen‐activated protein kinase (SAPK) BcSak1, and subsequent BcAtf1 activation, light‐induced regulatory systems, heterotrimeric G‐protein signalling and unknown regulators. Its regulatory function on phytotoxin production, as well as on hyphal growth control, seems to be essential for optimal balanced growth on host plants. Arrows indicate both the activating and repressing influence of BcAtf1 on downstream processes. Chemical structure: botrydial (representative secondary metabolite). Broken arrows: hypothetical signal transduction. See text for further information.
In addition to the striking phenotype of the mutant's fluffy mycelium, loss of bcatf1 results in a strongly reduced production of conidia. The role of the Atf1 protein in conidia development has been described previously in other filamentous fungi, such as N. crassa and Aspergillus oryzae (Sakamoto et al., 2009; Yamashita et al., 2008). In N. crassa, conidiation has been shown to be induced by several environmental signals (Davis, 2000; Springer, 1993). Light regulation of conidiation has been described via the activity of the white collar complex (Lauter et al., 1997; Linden and Macino, 1997), which regulates the fluffy gene transcription, encoding a major regulator of conidiation in response to blue light (Olmedo et al., 2010). In addition, conidiation is controlled by an endogenous circadian clock (Brunner and Ka'ldi, 2008; Dunlap and Loros, 2004; Heintzen and Liu, 2007; Tan et al., 2004), which is connected to SAPK signalling (de Paula et al., 2008). Regulation of Atf1 via the Os‐2 mitogen‐activated protein kinase from N. crassa associated with the circadian clock has been proposed (Yamashita et al., 2008). As general elementary cellular mechanisms regulate the asexual reproduction in N. crassa in a complex concerted manner, the same processes may also exist in B. cinerea. In A. nidulans, initiation of conidiation requires both the inhibition of G‐protein signalling and the activation of development‐specific functions (reviewed in Yu, 2006). Activation of this G‐protein signalling stimulates hyphal proliferation, which, in turn, represses conidiation (Hicks et al., 1997). Constitutive activation of FadA (Gα subunit) signalling causes uncontrolled accumulation of hyphal mass and the absence of sporulation, resulting in a fluffy autolytic phenotype (Mah and Yu, 2006; Yu et al., 1996). This phenotype seems to resemble Δbcatf1, indicating that there may be a connection between BcAtf1 and cyclic adenosine monophosphate (cAMP) signalling. As Δbcgb1 (Gβ subunit) from B. cinerea produces aerial hyphae, but strongly reduced amounts of conidia (J. Schumacher, unpublished data, WWU Muenster), similar to Δbcatf1, BcAtf1 and cAMP signalling may control differentiation and conidia formation in B. cinerea in a concerted manner.
A link between BcAtf1 and light perception is assumed, as the mutant forms conidia, but no sclerotia, in the dark. This indicates a perturbation of asexual and sexual differentiation processes in Δbcatf1, probably as a result of deregulated or missing light sensors. One of these light sensors could be the opsin Bop1, which was shown to be activated by BcAtf1 under stress conditions. Opsins are retinal‐binding proteins, with seven transmembrane helices, capable of absorbing light. However, their function in fungi is unclear (reviewed in Purschwitz et al., 2006).
A concerted role of the SAPK BcSak1 and the transcription factor BcAtf1 has been explored in B. cinerea, as this regulatory cascade has been analysed extensively in S. pombe previously (Shiozaki and Russell, 1996; Wilkinson et al., 1996). In addition to a large number of independently regulated genes, a set of BcSak1–BcAtf1 target genes has been identified to be identically controlled in both mutants after exposure to H2O2. This effect of oxidative stress on both proteins is conserved in yeasts and filamentous fungi (Hagiwara et al., 2009; Wilkinson et al., 1996). Joint target genes of BcSak1 and BcAtf1 include the stress responsive protein DDR48 (BC1G_10423), as well as a mannitol dehydrogenase (BC1G_09259). This protein has been connected to the stress response in fungi (Ruijter et al., 2003). The function of the light receptor protein opsin Bop1 (BC1G_02456) and a G‐β‐like protein (BC1G_10054) remain to be elucidated under these cultivation conditions. However, as these genes show the same regulation pattern in both Δbcsak1 and Δbcatf1, they are obviously controlled via a BcSak1–BcAtf1 cascade. The situation is different for the catalase B (BC1G_12856) encoding gene, as its activation is not only dependent on BcSak1 and BcAtf1, but also on the SAPK‐independent Bap1 transcription factor, the major regulator of the oxidative stress response in B. cinerea (Temme and Tudzynski, 2009). The expression of oxidative stress response target genes is controlled by both Atf1 and Pap1 in an H2O2 dose‐dependent way in fission yeast (Quinn et al., 2002). In B. cinerea, catalase B expression probably requires the recruitment of both transcription factors to its promoter region under all tested H2O2 concentrations. In N. crassa, some SAPK (Os‐2) target gene expression is also dependent on Atf1, but the phenotypes of both the SAPK and transcription factor knock‐out mutants differ greatly, as in B. cinerea (Yamashita et al., 2008). The data presented here also indicate that the BcAtf1 protein is a downstream component of the SAPK BcSak1 in B. cinerea. Its transcriptional induction under oxidative stress, however, is independent of SAPK, providing evidence for protein–protein interaction, rather than a transcriptional regulation, of BcSak1 on BcAtf1. However, BcAtf1 is obviously not the major, or only, transcription factor regulated by the mitogen‐activated protein kinase. Another putative transcriptional regulator, BcReg1, which has been identified recently, is a good candidate for control by BcSak1. The corresponding deletion mutant shows some phenotypic similarities to Δbcsak1. A direct interaction of BcReg1 and BcSak1 remains to be shown (Michielse et al., 2011). As Δbcatf1 only shares little or even opposite phenotypic features to Δbcsak1, it is evident that BcAtf1 is not only activated, but might also be functionally repressed, by BcSak1. This new finding substantiates the growing evidence that highly conserved signalling components have different ‘wiring’ in different fungi. Further factors must be involved in the regulation of BcAtf1.
Microarray analysis of the Δbcatf1 transcriptome has contributed to the understanding of its global gene expression control. Deregulation of developmental and metabolic target genes explains, to some extent, the phenotype of Δbcatf1, causing its uncontrolled enhanced growth in vitro and in vivo. The transcriptional profile of Δbcatf1 has uncovered a deregulation of several genes involved in fungal cell wall biosynthesis and modification, respectively. This probably leads to a permanent restructuring of the cell wall during hyphal growth. Aberrant cell wall assembly may also cause the sensitivity of the mutant to agents such as calcofluor white. Transport mechanisms seem to be strongly deregulated in Δbcatf1, which may result in deficient sensing of the environment. For example, the fungus may not recognize nutrient sources, or nutrients may not be transported effectively. Nutrients are not only a source of energy, but also regulators of metabolism, growth and development. The adaptation to changing nutritional conditions is mediated by a variety of signalling pathways, including those that initiate the sensing of nutrients directly at the plasma membrane (Holsbeeks et al., 2004). A deregulation of transporter gene expression, as observed in Δbcatf1, may therefore result in the deregulation of metabolism, growth and development. It may explain why the fungus fails to grow in a directed manner towards nutrient sources provided by the solid medium, but differentiates aerial hyphae.
The growth pattern is also visible on different host plants, where the mutants grow through the tissue and differentiate white aerial hyphae on both sides of the leaf. Analysis of the biotrophic fungus C. purpurea revealed significant amounts of H2O2 in Δcptf1‐infected rye epidermal tissues that prevented further growth of the mutant, probably as a result of an induction of the host's oxidative burst caused by Δcptf1 (Nathues et al., 2004). This defence reaction is never observed during C. purpurea WT infection, as the fungus infects its host in a symptomless manner. Similar host reactions may be caused by infection with Δbcatf1, but a stronger induction of the oxidative burst does not stop pathogen advance, but probably enhances the infection of this necrotrophic fungus, as observed in the Δbcatf1–host interaction. Although host invasion by Δbcatf1 seems to be more aggressive than WT infection, in the final analysis it is inefficient, because the mutant hardly produces any conidia after total host tissue maceration. In addition to its generally invasive growth behaviour, the mutant's aggressive expansion on the host may also be explained by its enhanced production of secondary metabolites, such as botrydial and botcinic acid, as the inactivation of biosynthesis genes indicated that both toxins have a role in plant tissue colonization (Dalmais et al., 2011; Siewers et al., 2005). Thus, the protein is a repressor of these secondary metabolites in B. cinerea. In contrast, Δbcsak1 shows reduced secondary metabolite production (J. Heller et al., unpublished results). This effect may be caused by a direct transcriptional regulation of corresponding gene clusters via the transcriptional regulator bcreg1 (Michielse et al., 2011). Such a direct regulatory effect of Atf1 proteins on secondary metabolism has not been shown previously. Recently, studies in A. parasiticus have postulated an influence of a related protein, AtfB, on secondary metabolism, because of the binding of this transcription factor on promoters of aflatoxin cluster genes (Roze et al., 2011). In B. cinerea, we found no evidence for a direct binding of BcAtf1 at promoters of genes involved in toxin biosynthesis, as strongly suggested by the yeast one‐hybrid analyses reported here. BcAtf1 clearly is part of the global regulatory network controlling the secondary metabolism in B. cinerea, together with the Gα protein Bcg1, as well as calcineurin and its downstream transcription factor BcCrz1 (2008a, 2008b; Viaud et al., 2003). This latter regulatory mechanism is not overruled in Δbcatf1, as inhibition of calcineurin negatively affects botrydial cluster gene expression, even in Δbcatf1. Interestingly, the activating function of calcineurin on botrydial cluster gene transcription in day/night rhythm does not exist in total darkness. In the dark, only in Δbcatf1, but not in WT, the transcription of botrydial cluster genes was detected and their transcription was independent of calcineurin control (Fig. S5). Thus BcAtf1 represses the activation of cluster gene transcription in light and dark, whereas calcineurin activation of these genes only occurs in the light. In the absence of BcAtf1, its repressing function is abolished.
Various novel functions of the Atf1 transcription factor in B. cinerea as a global regulator are summarized in Fig. 7. This work presents evidence that BcAtf1 is not only activated by the SAPK cascade, but shows phenotypic features which indicate BcSak1‐independent regulation, or even a repressing function of the mitogen‐activated protein kinase on BcAtf1 activity, depending on the environmental conditions. This might explain the inverse influence of both proteins on sclerotia formation, pathogenicity and secondary metabolism. BcAtf1 does not only control the stress response, but plays a major role in general differentiation processes and secondary metabolite production. The latter effect has not been demonstrated in other fungi to date. As the deletion of bcatf1 increases secondary metabolite production, this repressing function may be conserved in other fungi, and may even be used for the overproduction of certain compounds. In B. cinerea, this provides further evidence for a connection between controlled secondary metabolite production and optimal balanced growth on host plants.
EXPERIMENTAL PROCEDURES
Fungal and bacterial strains
Botrytis cinerea Pers. Fr. [teleomorph Botryotinia fuckeliana (de Bary) Whetz] strain B05.10 (Quidde et al., 1999) was used as recipient strain for the transformation experiments, and as a WT control. For comparative expression studies, mutant strains of B05.10 bcsak1 (Segmüller et al., 2007) and bap1 (Temme and Tudzynski, 2009) were used. Subcloning was performed in strain TOP10F′ (Invitrogen, Darmstadt, Germany).
Media and culture conditions
Botrytis cinerea strains were grown on CM (Pontecorvo et al., 1953), B5/Glc/ye (B5 + 2% glucose + 1 g/L yeast extract) or potato dextrose agar (PDA; containing 10% bean leaves) at 20 °C under near‐UV for conidiation.
‘Stress’ cultivation
For axenic culture shift experiments and subsequent RNA isolation, cultivation was performed according to Temme and Tudzynski (2009).
CsA inhibition assay
Cultivation was performed according to Viaud et al. (2003); 20 µg/mL of CsA were added to the liquid media. Cultivation was performed in the dark.
Phytotoxin analysis
Phytotoxin analysis was performed as described by Michielse et al. (2011). Strains were grown on malt agar medium (20 g/L d‐glucose, 20 g/L malt extract, 20 g/L agar, 1 g/L peptone, pH 6.5–7.0) at 25 °C for 10 days. Semi‐preparative high‐performance liquid chromatography (HPLC) afforded compounds 1–9 from the WT strain and mutants of B. cinerea (Table 2; Fig. S6).
Standard molecular methods
Fungal genomic DNA was isolated as described by Cenis (1992). Plasmid DNA was isolated using a plasmid DNA preparation kit (Genomed, Bad Oeynhausen, Germany).
Southern and Northern blot analyses
Southern blot analysis was performed according to Sambrook et al. (1989). For Northern analysis, RNA was isolated from mycelial samples using the Trizol method (Invitrogen), as described previously (Temme and Tudzynski, 2009)
Conditions for microarray probes
Fungal strains (B05.10 and Δbcatf1) were cultivated on CM plates with cellophane for 5 days under day/night light conditions. Three experiments were performed.
Microarray analyses
Botrytis cinerea four‐plex arrays (Roche NimbleGen Systems, Inc., Madison, WI, USA) were used in this work. These microarrays comprise 72 000 60‐mer probe sets. The probe sets were designed based on B. cinerea predicted genes from strains T4 and B05.10, and consist of 21 200 gene models with about three probes/gene, including 12 164 common T4‐B05.10 ORFs, 4072 T4‐specific ORFs and 3218 B05.10‐specific ORFs, as well as 1734 expressed sequence tag (EST) unisequences and 12 experimental genes. More information on the array design can be obtained from Amselem et al. (2011). Microarray hybridization, data acquisition, summarization and normalization were conducted by NimbleGen. The normalized data from the probe sets of the four arrays hybridized with fungal probes (two biological repetitions for each strain) were further analysed using ArrayStar microarray analysis software (DNAStar, Inc., Madison, WI, USA). By performing a moderate t‐test with false discovery rate (FDR) multiple‐test correction (Benjamini and Hochberg, 1995), 511 ORFs and ESTs were selected with ≥ 95% confidence as being differentially expressed. Selected B. cinerea ORFs were examined by blastx against the National Center for Biotechnology Information (NCBI) nonredundant protein sequence database. Proteins were classified according to their known function or according to the FunCat annotation scheme (Ruepp et al., 2004).
DNA sequencing
DNA sequencing was performed with the Big Dye Terminator v3.1 sequencing kit (Applied Biosystems, Frankfurt, Germany). After PCR, samples were purified by column chromatography and sequenced in an ABI Prism capillary sequencer (model 3730). For sequence analysis and the construction of phylogenetic trees, DNAStar (DNAStar, Inc.) was used.
RT‐PCR analyses
RT‐PCR analyses were performed for the confirmation of introns in bcatf1 using the primer pair 1/2 (Table 1), as described in Temme and Tudzynski (2009).
Yeast one‐hybrid analysis
The binding domains of BcAtf1 and CreA were predicted using supfam search tools (http://supfam.cs.bris.ac.uk). Gene fragments encoding predicted domains were amplified by RT‐PCR and cloned into the pACTIIst expression vector containing the LEU2 gene as selection marker (kindly provided by M. Fromont, Institute Pasteur, Paris, France; Fromont‐Racine et al., 1997). Primers were designed so that the coding sequence was in frame with the Gal4 activation domain of pACTIIst. For bcatf1, primers (13/14) included NcoI and BamHI restriction sites, respectively. For creA, primers were 15 and 16. PCR products were cloned between the NcoI and BamHI sites of pACTIIst, and used to transform the yeast strain Y187 (Clontech, Heidelberg, Germany) employing the LiAc method described by Gietz et al. (1992). Transformants were recovered on selective media (–leucine). Promoters of the B. cinerea genes BC1G_02936.1 (CutA), BC1G_15841.1, BC1G_16085.1, BcPKS6 and BcBOT1 were cloned into the integration vector pINT1_HIS3NB which contains the HIS3 reporter gene (kindly provided by P. F. B. Ouwerkerk, Leiden University, Leiden, The Netherlands; Meijer et al., 1998). Primers to amplify promoter regions of these genes included a NotI (forward primer) and a SpeI (reverse primer) restriction site (underlined), respectively (see Table S1). PCR products were cloned between the SpeI and NotI restriction sites of pINT1‐HIS3NB upstream of the HIS3 gene. Plasmids were digested with NcoI and SacI to recover fragments, including the promoter, the HIS3 gene, the APT1 gene (confers resistance to G418) and the PDC6 gene flanking regions. Fragments were transformed into yeast CG1945 (Clontech). Homologous recombination events at the PDC6 locus were selected with G418 (Ouwerkerk and Meijer, 2001). Yeast mating was performed by spreading onto two independent plates: the MATα haploid yeasts harbouring the B. cinerea DNA‐binding domains on horizontal lines, and the MATα haploid strains harbouring the promoters on vertical lines. After velvet replication of the two plates on a single plate, diploids were obtained at the intersections between the lines. Then, the plate was newly replicated on selection medium (–leucine, –histidine) with 5 mm 3‐amino‐1,2,4,triazole (3‐AT) (Sigma, Sigma‐Aldrich, Munich, Germany) and incubated at 30 °C for 10 days. Diploids able to grow on this medium are positive one‐hybrid interactants.
Replacement vector construction
For the deletion of bcatf1, the 5′ and 3′ flanking regions of the gene were amplified and cloned into the nourseothricin resistance vector pNR1 (Malonek et al., 2004) (Fig. S1). The replacement fragment was excised with SacI/XhoI and used to transform strain B05.10.
Transformation of B. cinerea
Protocols for protoplast formation and transformation were adapted from ten Have et al. (1998) as described by Schulze Gronover et al. (2001). Δbcatf1 was complemented via transformation with the bcatf1 gene, including its native promoter and terminator sequences.
Pathogenicity assays
Botrytis cinerea strains for the standard pathogenicity test, as described by Klimpel et al. (2002), were inoculated on primary leaves of Phaseolus vulgaris L. genotype N90598 (originating from J. D. Kelly, Michigan State University, East Lansing, MI, USA). In addition, cucumber fruits were used for pathogenicity studies and incubated under the same conditions, and the infection process was monitored and documented photographically.
Quantification of conidia formation
Conidia were harvested from agar plates at 2 weeks post‐inoculation, filtered and counted (see Fig. S3).
Germination assay
The germination assays were performed as described by Doehlemann et al. (2006).
Microscopic analyses
To study hyphal morphology, a Leica DMRBE epifluorescence microscope (Wetzlar, Germany), with a PixelFly digital camera (PCO Computer Optics GmbH, Kelheim, Germany) and filter set A (BP 340 to 380, RKP 400, LP 430), was used.
Supporting information
Fig. S1 (A) Replacement strategy for deletion of bcatf1. The flanking regions of bcatf1 were amplified via polymerase chain reaction (PCR). Modified primer (9/10 and 11/12; see Table S1 in Supporting Information) integrated specific restriction sites (SacI/SacII and Apa/XhoI, respectively) for endonucleases into the flanking regions, allowing for directed cloning of these fragments into the pNR1 replacement vector. OliC, promoter from Aspergillus nidulans; nat1, nourseothricin acetyltransferase (Streptomyces noursei); T‐tub, tubulin terminator from Botrytis cinerea. A linear fragment obtained by digestion of pNR1_Δbcatf1 with SacI and ApaI was used for the transformation of B. cinerea wild‐type strain B05.10. Primers used for amplification of the flanking regions: 3, BcAtf1_F1_for/9; 4, BcAtf1_F1_rev/10; 5, BcAtf1_F2_for/11; 6, BcAtf1_F2_rev/12. (B) Verification of homologous integration in the bcatf1 locus and control of homokaryotic deletion strains (B.9, N3, N.6, L.11). Diagnostic PCR was performed using primers 1 (BcAtf1_probe_for/3) and 2 (BcAtf1_probe_rev/4) for amplification of the wild‐type fragment (0.5 kb). Primers 7 (BcAtf1_for control 2/5) and 9 (pOliR/7) (1.8 kb) were used to amplify the DNA fragment F1 consisting of the flanking region 1 upstream of bcatf1 and the first part of the resistance cassette promoter region. Primers 8 (BcAtf1_F2_rev_control/6) and 10 (T‐tub2/8) (0.9 kb) were used to amplify a fragment F2 with the end of the resistance cassette terminator and the flanking region 2 downstream of bcatf1. Amplification of these DNA fragments should only be possible in cases of homologous integration of the replacement fragment into the bcatf1 gene locus. (C) Southern blot analysis of Δbcatf1 strains. Genomic DNA was digested with HindIII and separated via agarose gel electrophoresis. Flanking region 2 used for the replacement cassette served as probe. The F2 probe hybridized with the wild‐type 1.8‐kb HindIII fragment, as well as with the 1.1‐kb fragment of the Δbcatf1 mutants. Marker: DNA ladder mix (Fermentas, St. Leon‐Roth, Germany).
Fig. S2 Hyphal morphology of Δbcatf1 in comparison with the wild‐type B05.10. The mutant's hyphae are elongated in comparison with B05.10 hyphae, but show regular normal septation. Scale bar, 100 μm. The strains were grown for 2 days on CM‐overlaid microscope slides and stained with calcofluor white. After 2–4 days of incubation in a humid chamber at 20 °C, the colonies were incubated for 5 min in 1% (w/v) calcofluor white solution and then washed with water.
Fig. S3 Quantification of conidiospore production of Δbcatf1. Test tubes contain the quantified conidiospore solutions. Conidiospores were harvested from agar plates and spore suspensions of each strain were diluted in H2O to 5 × 105 spores/mL; 4 × 104 spores were plated on CM (three plates/strain) and incubated at 18 °C for 2 weeks under permanent near‐UV light to enhance conidiospore formation. After 2 weeks, conidiospores were floated off the plates in a defined volume of 15 mL H2Obidest, filtered over a Nytex membrane and washed twice with H2Obidest. Spores were resuspended in 20 mL of H2Obidest. For spore quantification, dilution series were counted.
Fig. S4 Growth in the presence of oxidative stressors. Growth of the Δbcatf1 strain, Δbcsak1 strain and wild‐type B05.10 on different stress‐inducing media after 3 days. CM was supplemented with H2O2 (20 mM), menadione (500 μM) and NaCl (1 M).
Fig. S5 Expression of botrydial cluster genes in Δbcatf1 strain and B05.10. Activation of bcbot1/CND5 and bcbot2/CND15, respectively, is dependent on calcineurin, as its inhibitor, cyclosporin A (CsA), inhibits their expression. A strikingly enhanced expression of these cluster genes can be observed in the bcatf1 deletion strain, which is reduced with the addition of CsA under day/night growth conditions (A). There is no expression of the cluster genes visible in the wild‐type and only a slight CsA‐independent expression in Δbcatf1 when cultures are grown in the dark (B). Loading control: rRNA.
Fig. S6 Isolated toxins from parental and mutant strains.
Fig. S7 Physical interaction between DNA‐binding domain of transcription factors and promoters revealed by yeast one‐hybrid assay. Haploid yeast strains harbouring the Botrytis cinerea DNA‐binding domains of the transcription factors ATF1 or CreA in frame with the Gal4 activator domain were grown on horizontal lines, whereas haploid strains harbouring the promoters of the regulated genes upstream of a HIS3 reporter gene were grown on vertical lines. One medium without histidine, with the growth of diploids (large colonies) at the intersections, indicates positive one‐hybrid interactants. The positive control CreA binds to all five promoters, whereas ATF1 does not bind to any of the tested promoters.
Fig. S8 Differentiation of Δbcatf1, the wild‐type B05.10 and the complementation strain cΔbcatf1 under standard cultivation conditions. Growth test. (A) After 3 days on complete medium (B5 + 2% glucose + 1 g/L yeast extract), the wild‐type (WT) and complementation strain cΔbcatf1 grow flat on top of the medium, whereas the mutant produces aerial hyphae. (B) Under permanent light on complex medium [potato dextrose agar (PDA) + bean], the WT and complementation strain cΔbcatf1 produce large amounts of conidiospores causing the black colour of the mycelium. The Δbcatf1 strain hardly produces any spores. (C) After 2 weeks in permanent darkness (B5 + 2% glucose + 1 g/L yeast extract), the WT and complementation strain cΔbcatf1 form sclerotia, whereas there is no formation of sclerotia, but only of conidiospores, in the Δbcatf1 strain.
Fig. S9 Pathogenicity assay of Δbcatf1, the wild‐type B05.10 and complementation strain cΔbcatf1. Agar plaques were placed on leaves of Phaseolus vulgaris. Photographs were taken 2 days post‐infection.
Table S1 Primers used in this work (sequence: 5′ → 3′).
Table S2 Gene expression profile of Δbcatf1. +/−, up‐/down‐regulated genes in Δbcatf1.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank Brian Williamson for critical reading of the manuscript, Eva Nathues and Julia Schumacher for sharing data prior to publication, Klaus B. Tenberge for technical advice and the German Research Foundation (DFG, project Tu/50‐14 and Graduate School 1409 ‘Molecular Interactions of Pathogens with Biotic and Abiotic Surfaces’) for financial support. IGC is grateful to Ministry of Science and Innovation Spain for financial support AGL2009‐13359‐C02‐01. The yeast one‐hybrid experiments were funded by the Institute National de la Recherche Agronomique, Sante ′de Plantes et Environnement Department.
REFERENCES
- Amselem, J. , Cuomo, C. , van Kan, J. , Viaud, M. , Benito, E. , Couloux, A. , Coutinho, P.M. , de Vries, R.P. , Dyer, P.S. , Fillinger, S. , Fournier, E. , Gout, L. , Hahn, M. , Kohn, L. , Lapalu, N. , Plummer, K.M. , Pradier, J.M. , Quevillon, E. , Sharon, A. , Simon, A. , ten Have, A. , Tudzynski, B. , Tudzynski, P. , Wincker, P. , Andrew, M. , Beever, R. , Anthouard, V. , Beffa, R. , Benoit, I. , Bouzid, O. , Brault, B. , Chen, Z. , Choquer, M. , Collemare, J. , Cotton, P. , Danchin, E.G. , Da Silva, C. , Gautier, A. , Giraud, C. , Giraud, T. , Gonzalez, C. , Grossetete, S. , Güldener, U. , Henrissat, B. , Howlett, B.J. , Kodira, C. , Kretschmer, M. , Lappartient, A. , Leroch, M. , Levis, C. , Mauceli, E. , Neuvéglise, C. , Oeser, B. , Pearson, M. , Poulain, J. , Poussereau, N. , Quesneville, H. , Rascle, C. , Schumacher, J. , Ségurens, B. , Sexton, A. , Sirven, C. , Soanes, D.M. , Talbot, N.J. , Templeton, M. , Yandava, C. , Yarden, O. , Zeng, Q. , Rollins, J.A. , Lebrun, M.A. and Dickman, M.B. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genet. 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázs, A. , Pócsi, I. , Hamari, Z. , Leiter, E. , Emri, T. , Miskei, M. , Oláh, J. , Tóth, V. , Hegedus, N. , Prade, R.A. , Molnár, M. and Pócsi, I. (2010) AtfA bZIP‐type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans . Mol. Genet. Genomics, 283, 289–303. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300. [Google Scholar]
- Brunner, M. and Ka'ldi, K. (2008) Interlocked feedback loops of the circadian clock of Neurospora crassa . Mol. Microbiol. 68, 255–262. [DOI] [PubMed] [Google Scholar]
- Bulawa, C.E. (1993) Genetics and molecular biology of chitin synthesis in fungi. Annu. Rev. Microbiol. 47, 505–534. [DOI] [PubMed] [Google Scholar]
- Cenis, J.L. (1992) Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 20, 2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquer, M. , Fournier, E. , Kunz, C. , Levis, C. , Pradier, J.M. , Simon, A. and Viaud, M. (2007) Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 277, 1–10. [DOI] [PubMed] [Google Scholar]
- Collado, I.G. , Sánchez, A.J. and Hanson, J.R. (2007) Fungal terpene metabolites: biosynthetic relationships and the control of the phytopathogenic fungus Botrytis cinerea . Nat. Prod. Rep. 24, 674–686. [DOI] [PubMed] [Google Scholar]
- Colmenares, A.J. , Aleu, J. , Durán‐Patrón, R. , Collado, I.G. and Hernández‐Galán, R. (2002) The putative role of botrydial and related metabolites in the infection mechanism of Botrytis cinerea . J. Chem. Ecol. 28, 997–1005. [DOI] [PubMed] [Google Scholar]
- Dalmais, B. , Schumacher, J. , Moraga, J. , Le Pêcheur, P. , Tudzynski, B. , Collado, I.G. and Viaud, M. (2011) The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Mol. Plant Pathol. 12, 564–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam, H. , Wilhelm, D. , Herr, I. , Steffen, A. , Herrlich, P. and Angel, P. (1995) ATF‐2 is preferentially activated by stress‐activated protein kinases to mediate c‐jun induction in response to genotoxic agents. EMBO J. 14, 1798–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R.H. (2000) Neurospora. Contributions of a Model Organism. Oxford: Oxford University Press. [Google Scholar]
- Doehlemann, G. , Berndt, P. and Hahn, M. (2006) Different signaling pathways involving a G‐alpha protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol. Microbiol. 59, 821–836. [DOI] [PubMed] [Google Scholar]
- Dunlap, J.C. and Loros, J.J. (2004) The Neurospora circadian system. J. Biol. Rhythms, 19, 414–424. [DOI] [PubMed] [Google Scholar]
- Fromont‐Racine, M. , Rain, J.C. and Legrain, P. (1997) Toward a functional analysis of the yeast genome through exhaustive two‐hybrid screens. Nat. Genet. 16, 277–282. [DOI] [PubMed] [Google Scholar]
- Gietz, D. , St Jean, A. , Woods, R.A. and Schiestl, R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M. , Guo, W. , Chen, Y. , Dong, S. , Zhang, X. , Zhang, H. , Song, W. , Wang, W. , Wang, Q. , Lv, R. , Zhang, Z. , Wang, Y. and Zheng, X. (2010) The basic leucine zipper transcription factor Moatf1 mediates oxidative stress response and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae . Mol. Plant–Microbe Interact. 23, 1058–1063. [DOI] [PubMed] [Google Scholar]
- Gupta, S. , Campbell, D. , Dérijard, B. and Davis, R.J. (1995) Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science, 267, 389–393. [DOI] [PubMed] [Google Scholar]
- Hagiwara, D. , Asano, Y. , Yamashino, T. and Mizuno, T. (2008) Characterization of bZip‐type transcription factor AtfA with reference to stress responses of conidia of Aspergillus nidulans . Biosci. Biotechnol. Biochem. 72, 2756–2760. [DOI] [PubMed] [Google Scholar]
- Hagiwara, D. , Asano, Y. , Marui, J. , Yoshimi, A. , Mizuno, T. and Abe, K. (2009) Transcriptional profiling for Aspergillus nidulans HogA MAPK signaling pathway in response to fludioxonil and osmotic stress. Fungal Genet. Biol. 46, 868–878. [DOI] [PubMed] [Google Scholar]
- ten Have, A. , Mulder, W. , Visser, J. and van Kan, J.A.L. (1998) The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea . Mol. Plant–Microbe Interact. 11, 1009–1016. [DOI] [PubMed] [Google Scholar]
- Heintzen, C. and Liu, Y. (2007) The Neurospora crassa circadian clock. Adv. Genet. 58, 25–66. [DOI] [PubMed] [Google Scholar]
- Hicks, J.K. , Yu, J.H. , Keller, N.P. and Adams, T.H. (1997) Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G protein dependent signaling pathway. EMBO J. 16, 4916–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsbeeks, I. , Lagatie, O. , Van Nuland, A. , Van de Velde, S. and Thevelein, J.M. (2004) The eukaryotic plasma membrane as a nutrient‐sensing device. Trends Biochem. Sci. 29, 556–564. [DOI] [PubMed] [Google Scholar]
- van Kan, J.A.L. (2006) Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11, 247–253. [DOI] [PubMed] [Google Scholar]
- Klimpel, A. , Schulze Gronover, C. , Williamson, B. , Stewart, J.A. and Tudzynsk, B. (2002) The adenylate cyclase (BAC) in Botrytis cinerea is required for full pathogenicity. Mol. Plant Pathol. 3, 439–450. [DOI] [PubMed] [Google Scholar]
- Kon, N.M. , Krawchuk, D. , Warren, B.G. , Smith, G.R. and Wahls, W.P. (1997) Transcription factor Mts1/Mts2(Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe . Proc. Natl. Acad. Sci. USA, 94, 13 765–13 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara‐Rojas, F. , Sanchez, O. , Kawasaki, L. and Aguirre, J. (2011) Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol. Microbiol. 80, 436–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter, F.R. , Yamashiro, C.T. and Yanofsky, C. (1997) Light stimulation of conidiation in Neurospora crassa: studies with the wild‐type strain and mutants wc‐1, wc‐2 and acon‐2. J. Photochem. Photobiol. B 37, 203–211. [Google Scholar]
- Lawrence, C.L. , Jones, N. and Wilkinson, C.R. (2009) Stress‐induced phosphorylation of S. pombe Atf1 abrogates its interaction with F box protein Fbh1. Curr. Biol. 19, 1907–1911. [DOI] [PubMed] [Google Scholar]
- Linden, H. and Macino, G. (1997) White collar 2, a partner in blue‐light signal transduction, controlling expression of light‐regulated genes in Neurospora crassa . EMBO J. 16, 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Soulié, M.C. , Perrino, C. and Fillinger, S. (2011) The osmosensing signal transduction pathway from Botrytis cinerea regulates cell wall integrity and MAP kinase pathways control melanin biosynthesis with influence of light. Fungal Genet. Biol. 48, 377–387. [DOI] [PubMed] [Google Scholar]
- Livingstone, C. , Patel, G. and Jones, N. (1995) ATF‐2 contains a phosphorylation‐dependent transcriptional activation domain. EMBO J. 14, 1785–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa, T. , Sakura, H. , Kanei‐Ishii, C. , Sudo, T. , Yoshimura, T. , Fujisawa, J. , Yoshida, M. and Ishii, S. (1989) Leucine zipper structure of the protein CRE‐BP1 binding to the cyclic AMP response element in brain. EMBO J. 8, 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah, J.H. and Yu, J.H. (2006) Upstream and downstream regulation of asexual development in Aspergillus fumigatus . Eukaryot. Cell, 5, 1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malonek, S. , Rojas, M.C. , Hedden, P. , Gaskin, P. , Hopkins, P. and Tudzynski, B. (2004) The NADPH:cytochrome P450 reductase gene from Gibberella fujikuroi is essential for gibberellin biosynthesis. J. Biol. Chem., 279, 25 075–25 084. [DOI] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , Anderson, J.B. , Derbyshire, M.K. , DeWeese‐Scott, C. , Gonzales, N.R. , Gwadz, M. , Hao, L. , He, S. , Hurwitz, D.I. , Jackson, J.D. , Ke, Z. , Krylov, D. , Lanczycki, C.J. , Liebert, C.A. , Liu, C. , Lu, F. , Lu, S. , Marchler, G.H. , Mullokandov, M. , Song, J.S. , Thanki, N. , Yamashita, R.A. , Yin, J.J. and Zhang Dbryant, S.H. (2005) CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33, 192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, A.H. , Ouwerkerk, P.B. and Hoge, J.H. (1998) Vectors for transcription factor cloning and target site identification by means of genetic selection in yeast. Yeast, 14, 1407–1415. [DOI] [PubMed] [Google Scholar]
- Michielse, C.B. , Becker, M. , Heller, J. , Moraga, J.R. , Collado, I.G. and Tudzynski, P. (2011) The Botrytis cinerea REG1 protein, a putative transcriptional regulator, is required for pathogenicity, conidiogenesis and for the production of secondary metabolites. Mol. Plant–Microbe Interact. 24, 1074–1085. [DOI] [PubMed] [Google Scholar]
- Nathues, E. , Joshi, S. , Tenberge, K.B. , von den Driesch, M. , Oeser, B. , Bäumer, N. , Mihlan, M. and Tudzynski, P. (2004) CPTF1, a CREB‐like transcription factor, is involved in the oxidative stress response in the phytopathogen Claviceps purpurea and modulates ROS level in its host Secale cereale . Mol. Plant–Microbe Interact. 17, 383–393. [DOI] [PubMed] [Google Scholar]
- Olmedo, M. , Ruger‐Herreros, C. and Corrochano, L.M. (2010) Regulation by blue light of the fluffy gene encoding a major regulator of conidiation in Neurospora crassa . Genetics, 184, 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ors, A. , Grimaldi, M. , Kimata, Y. , Wilkinson, C.R. , Jones, N. and Yamano, H. (2009) The transcription factor Atf1 binds and activates the APC/C ubiquitin ligase in fission yeast. J. Biol. Chem. 284, 23 989–23 994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwerkerk, P.B. and Meijer, A.H. (2001) Yeast one‐hybrid screening for DNA protein interactions. Curr. Protoc. Mol. Biol. 12, 1–22. [DOI] [PubMed] [Google Scholar]
- de Paula, R.M. , Lamb, T.M. , Bennett, L. and Bell‐Pedersen, D. (2008) A connection between MAPK pathways and circadian clocks. Cell Cycle, 7, 2630–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinedo, C. , Wang, C.M. , Pradier, J.M. , Dalmais, B. , Choquer, M. , Le Pêcheur, P. , Morgant, G. , Collado, I.G. , Cane, D.E. and Viaud, M. (2008) Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea . ACS Chem. Biol. 3, 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo, G.V. , Roper, J.A. , Hummonns, L.M. , MacDonald, K.D. and Buften, A.W.J. (1953) The genetics of Aspergillus nidulans . Adv. Genet. 5, 141–238. [DOI] [PubMed] [Google Scholar]
- Purschwitz, J. , Muller, S. , Kastner, C. and Fischer, R. (2006) Seeing the rainbow: light sensing in fungi. Curr. Opin. Microbiol. 9, 566–571. [DOI] [PubMed] [Google Scholar]
- Quidde, T. , Büttner, P. and Tudzynski, P. (1999) Evidence for three different specific saponin‐detoxifying activities in Botrytis cinerea and cloning of a gene coding for a putative avenacinase. Eur. J. Plant Pathol. 105, 273–283. [Google Scholar]
- Quinn, J. , Findlay, V.J. , Dawson, K. , Millar, J.B. , Jones, N. , Morgan, B.A. and Toone, W.M. (2002) Distinct regulatory proteins control the graded transcriptional response to increasing H(2)O(2) levels in fission yeast Schizosaccharomyces pombe . Mol. Biol. Cell, 13, 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram, A.F. and Klis, F. (2006) Model organisms identification of fungal cell wall mutants using susceptibility assays based on calcofluor white and Congo red. Nat. Protoc. 1, 2253–2256. [DOI] [PubMed] [Google Scholar]
- Rep, M. , Duyvesteijn, R.G. , Gale, L. , Usgaard, T. , Cornelissen, B.J. , Ma, L.J. and Ward, T.J. (2006) The presence of GC‐AG introns in Neurospora crassa and other euascomycetes determined from analyses of complete genomes: implications for automated gene prediction. Genomics, 87, 338–347. [DOI] [PubMed] [Google Scholar]
- Roze, L.V. , Chanda, A. , Wee, J. , Awad, D. and Linz, J.E. (2011) Stress‐related transcription factor AtfB integrates secondary metabolism with oxidative stress response in aspergilli. J. Biol. Chem. 286, 35 137–35 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp, A. , Zollner, A. , Maier, D. , Albermann, K. , Hani, J. , Mokrejs, M. , Tetko, I. , Güldener, U. , Mannhaupt, G. , Münsterkötter, M. and Mewes, H.W. (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32, 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter, G.J. , Bax, M. , Pate, L.H. , Flitter, S.J. , van de Vondervoort, P.J. , de Vries, R.P. , vanKuyk, P.A. and Visser, J. (2003) Mannitol is required for stress tolerance in Aspergillus niger conidiospores. Eukaryot. Cell, 2, 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K. , Iwashita, K. , Yamada, O. , Kobayashi, K. , Mizuno, A. , Akita, O. , Mikami, S. , Shimoi, H. and Gomi, K. (2009) Aspergillus oryzae atfA controls conidial germination and stress tolerance. Fungal Genet. Biol. 46, 887–897. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sansó, M. , Gogol, M. , Ayté, J. , Seidel, C. and Hidalgo, E. (2008) Transcription factors Pcr1 and Atf1 have distinct roles in stress‐ and Sty1‐dependent gene regulation. Eukaryot. Cell, 7, 826–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze Gronover, C. , Kasulke, D. , Tudzynski, P. and Tudzynski, B. (2001) The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea . Mol. Plant–Microbe Interact. 14, 1293–1302. [DOI] [PubMed] [Google Scholar]
- Schumacher, J. , Viaud, M. , Simon, A. and Tudzynski, B. (2008a) The Ga subunit BCG1, the phospholipase C (BcPLC1) and the calcineurin phosphatase co‐ordinately regulate gene expression in the grey mould fungus Botrytis cinerea . Mol. Microbiol. 67, 1027–1050. [DOI] [PubMed] [Google Scholar]
- Schumacher, J. , de Larrinoa, I.F. and Tudzynski, B. (2008b) Calcineurin‐responsive zinc finger transcription factor CRZ1 of Botrytis cinerea is required for growth, development and full virulence on bean plants. Eukaryot. Cell, 7, 584–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segmüller, N. (2007) Molecular mechanisms for scavenging and production of reactive oxygen species in the phytopathogenic fungus Botrytis cinerea . PhD Thesis, Westf. Wilhelms Universität, Münster.
- Segmüller, N. , Ellendorf, U. , Tudzynski, B. and Tudzynski, P. (2007) BcSAK1, a stress‐activated MAP kinase is involved in vegetative differentiation and pathogenicity in Botrytis cinerea . Eukaryot. Cell, 6, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki, K. and Russell, P. (1996) Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10, 2276–2288. [DOI] [PubMed] [Google Scholar]
- Siewers, V. , Smedsgaard, J. and Tudzynski, P. (2004) The P450 monooxygenase BcABA1 is essential for abscisic acid biosynthesis in Botrytis cinerea . Appl. Environ. Microbiol. 70, 3868–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewers, V. , Viaud, M. , Jimenez‐Teja, D. , Collado, I.G. , Schulze Gronover, C. , Tudzynski, B. and Tudzynski, P. (2005) Functional analysis of the cytochrome P450 monooxygenase gene bcbot1 of Botrytis cinerea indicates that botrydial is a strain‐specific virulence factor. Mol. Plant–Microbe Interact. 18, 602–612. [DOI] [PubMed] [Google Scholar]
- Springer, M.L. (1993) Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa . BioEssays, 15, 365–374. [DOI] [PubMed] [Google Scholar]
- Takeda, T. , Toda, T. , Kominami, K. , Kohnosu, A. , Yanagida, M. and Jones, N. (1995) Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14, 6193–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Y. , Merrow, M. and Roenneberg, T. (2004) Photoperiodism in Neurospora crassa . J. Biol. Rhythms, 19, 135–143. [DOI] [PubMed] [Google Scholar]
- Tani, H. , Koshino, H. , Sakuno, E. and Nakajima, H. (2005) Botcinins A, B, C, and D, metabolites produced by Botrytis cinerea, and their antifungal activity against Magnaporthe grisea, a pathogen of rice blast disease. J. Nat. Prod. 68, 1768–1772. [DOI] [PubMed] [Google Scholar]
- Temme, N. and Tudzynski, P. (2009) Does Botrytis cinerea ignore H(2)O(2)‐induced oxidative stress during infection? Characterization of Botrytis activator protein 1. Mol. Plant–Microbe Interact. 22, 987–998. [DOI] [PubMed] [Google Scholar]
- Tudzynski, P. and Kokkelink, L. (2009) Botrytis cinerea: molecular aspects of a necrotrophic life‐style In: The Mykota, Vol. V Plant Relationships, 2nd edn. (Deising H., ed.), pp. 29–50. Heidelberg, New York: Springer. [Google Scholar]
- Viaud, M. , Brunet‐Simon, A. , Brygoo, Y. , Pradier, J.M. and Levis, C. (2003) Cyclophilin A and calcineurin functions investigated by gene inactivation, cyclosporin A inhibition and cDNA arrays approaches in the phytopathogenic fungus Botrytis cinerea . Mol. Microbiol. 50, 1451–1465. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y. and Yamamoto, M. (1996) Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol. Cell. Biol. 16, 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, M.G. , Samuels, M. , Takeda, T. , Toone, W.M. , Shieh, J.C. , Toda, T. , Millar, J.B. and Jones, N. (1996) The Atf1 transcription factor is a target for the Sty1 stress‐activated MAP kinase pathway in fission yeast. Genes Dev. 10, 2289–2301. [DOI] [PubMed] [Google Scholar]
- Yamashita, K. , Shiozawa, A. , Watanabe, S. , Fukumori, F. , Kimura, M. and Fujimura, M. (2008) ATF‐1 transcription factor regulates the expression of ccg‐1 and cat‐1 genes in response to fludioxonil under OS‐2 MAP kinase in Neurospora crassa . Fungal Genet. Biol. 45, 1562–1569. [DOI] [PubMed] [Google Scholar]
- Yu, J.H. (2006) Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans . J. Microbiol. 44, 145–154. [PubMed] [Google Scholar]
- Yu, J.H. , Wieser, J. and Adams, T.H. (1996) The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15, 5184–5190. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 (A) Replacement strategy for deletion of bcatf1. The flanking regions of bcatf1 were amplified via polymerase chain reaction (PCR). Modified primer (9/10 and 11/12; see Table S1 in Supporting Information) integrated specific restriction sites (SacI/SacII and Apa/XhoI, respectively) for endonucleases into the flanking regions, allowing for directed cloning of these fragments into the pNR1 replacement vector. OliC, promoter from Aspergillus nidulans; nat1, nourseothricin acetyltransferase (Streptomyces noursei); T‐tub, tubulin terminator from Botrytis cinerea. A linear fragment obtained by digestion of pNR1_Δbcatf1 with SacI and ApaI was used for the transformation of B. cinerea wild‐type strain B05.10. Primers used for amplification of the flanking regions: 3, BcAtf1_F1_for/9; 4, BcAtf1_F1_rev/10; 5, BcAtf1_F2_for/11; 6, BcAtf1_F2_rev/12. (B) Verification of homologous integration in the bcatf1 locus and control of homokaryotic deletion strains (B.9, N3, N.6, L.11). Diagnostic PCR was performed using primers 1 (BcAtf1_probe_for/3) and 2 (BcAtf1_probe_rev/4) for amplification of the wild‐type fragment (0.5 kb). Primers 7 (BcAtf1_for control 2/5) and 9 (pOliR/7) (1.8 kb) were used to amplify the DNA fragment F1 consisting of the flanking region 1 upstream of bcatf1 and the first part of the resistance cassette promoter region. Primers 8 (BcAtf1_F2_rev_control/6) and 10 (T‐tub2/8) (0.9 kb) were used to amplify a fragment F2 with the end of the resistance cassette terminator and the flanking region 2 downstream of bcatf1. Amplification of these DNA fragments should only be possible in cases of homologous integration of the replacement fragment into the bcatf1 gene locus. (C) Southern blot analysis of Δbcatf1 strains. Genomic DNA was digested with HindIII and separated via agarose gel electrophoresis. Flanking region 2 used for the replacement cassette served as probe. The F2 probe hybridized with the wild‐type 1.8‐kb HindIII fragment, as well as with the 1.1‐kb fragment of the Δbcatf1 mutants. Marker: DNA ladder mix (Fermentas, St. Leon‐Roth, Germany).
Fig. S2 Hyphal morphology of Δbcatf1 in comparison with the wild‐type B05.10. The mutant's hyphae are elongated in comparison with B05.10 hyphae, but show regular normal septation. Scale bar, 100 μm. The strains were grown for 2 days on CM‐overlaid microscope slides and stained with calcofluor white. After 2–4 days of incubation in a humid chamber at 20 °C, the colonies were incubated for 5 min in 1% (w/v) calcofluor white solution and then washed with water.
Fig. S3 Quantification of conidiospore production of Δbcatf1. Test tubes contain the quantified conidiospore solutions. Conidiospores were harvested from agar plates and spore suspensions of each strain were diluted in H2O to 5 × 105 spores/mL; 4 × 104 spores were plated on CM (three plates/strain) and incubated at 18 °C for 2 weeks under permanent near‐UV light to enhance conidiospore formation. After 2 weeks, conidiospores were floated off the plates in a defined volume of 15 mL H2Obidest, filtered over a Nytex membrane and washed twice with H2Obidest. Spores were resuspended in 20 mL of H2Obidest. For spore quantification, dilution series were counted.
Fig. S4 Growth in the presence of oxidative stressors. Growth of the Δbcatf1 strain, Δbcsak1 strain and wild‐type B05.10 on different stress‐inducing media after 3 days. CM was supplemented with H2O2 (20 mM), menadione (500 μM) and NaCl (1 M).
Fig. S5 Expression of botrydial cluster genes in Δbcatf1 strain and B05.10. Activation of bcbot1/CND5 and bcbot2/CND15, respectively, is dependent on calcineurin, as its inhibitor, cyclosporin A (CsA), inhibits their expression. A strikingly enhanced expression of these cluster genes can be observed in the bcatf1 deletion strain, which is reduced with the addition of CsA under day/night growth conditions (A). There is no expression of the cluster genes visible in the wild‐type and only a slight CsA‐independent expression in Δbcatf1 when cultures are grown in the dark (B). Loading control: rRNA.
Fig. S6 Isolated toxins from parental and mutant strains.
Fig. S7 Physical interaction between DNA‐binding domain of transcription factors and promoters revealed by yeast one‐hybrid assay. Haploid yeast strains harbouring the Botrytis cinerea DNA‐binding domains of the transcription factors ATF1 or CreA in frame with the Gal4 activator domain were grown on horizontal lines, whereas haploid strains harbouring the promoters of the regulated genes upstream of a HIS3 reporter gene were grown on vertical lines. One medium without histidine, with the growth of diploids (large colonies) at the intersections, indicates positive one‐hybrid interactants. The positive control CreA binds to all five promoters, whereas ATF1 does not bind to any of the tested promoters.
Fig. S8 Differentiation of Δbcatf1, the wild‐type B05.10 and the complementation strain cΔbcatf1 under standard cultivation conditions. Growth test. (A) After 3 days on complete medium (B5 + 2% glucose + 1 g/L yeast extract), the wild‐type (WT) and complementation strain cΔbcatf1 grow flat on top of the medium, whereas the mutant produces aerial hyphae. (B) Under permanent light on complex medium [potato dextrose agar (PDA) + bean], the WT and complementation strain cΔbcatf1 produce large amounts of conidiospores causing the black colour of the mycelium. The Δbcatf1 strain hardly produces any spores. (C) After 2 weeks in permanent darkness (B5 + 2% glucose + 1 g/L yeast extract), the WT and complementation strain cΔbcatf1 form sclerotia, whereas there is no formation of sclerotia, but only of conidiospores, in the Δbcatf1 strain.
Fig. S9 Pathogenicity assay of Δbcatf1, the wild‐type B05.10 and complementation strain cΔbcatf1. Agar plaques were placed on leaves of Phaseolus vulgaris. Photographs were taken 2 days post‐infection.
Table S1 Primers used in this work (sequence: 5′ → 3′).
Table S2 Gene expression profile of Δbcatf1. +/−, up‐/down‐regulated genes in Δbcatf1.
Supporting info item
Supporting info item


