SUMMARY
MicroRNAs (miRNAs) regulate the abundance of target mRNAs by guiding cleavage at sequence complementary regions. In this study, artificial miRNAs (amiRNAs) targeting conserved motifs of the L (replicase) gene of Watermelon silver mottle virus (WSMoV) were constructed using Arabidopsis pre‐miRNA159a as the backbone. The constructs included six single amiRNAs targeting motifs A, B1, B2, C, D of E, and two triple amiRNAs targeting motifs AB1E or B2DC. Processing of pre‐amiRNAs was confirmed by agro‐infiltration, and transgenic Nicotiana benthamiana plants expressing each amiRNA were generated. Single amiRNA transgenic lines expressing amiR‐LB2 or amiR‐LD showed resistance to WSMoV by delaying symptom development. Triple amiRNA lines expressing amiR‐LB2, amiR‐LD and amiR‐LC provided complete resistance against WSMoV, with no indication of infection 28 days after inoculation. Resistance levels were positively correlated with amiRNA expression levels in these single and triple amiRNA lines. The triple amiR‐LAB1E line did not provide resistance to WSMoV. Similarly, the poorly expressed amiR‐LC and amiR‐LE lines did not provide resistance to WSMoV. The amiR‐LA‐ and amiR‐LB1‐expressing lines were susceptible to WSMoV, and their additional susceptibility to the heterologous Turnip mosaic virus harbouring individual target sequences indicated that these two amiRNAs have no effect in vivo. Transgenic lines expressing amiR‐LB2 exhibited delayed symptoms after challenge with Peanut bud necrosis virus having a single mismatch in the target site. Overall, our results indicate that two amiRNAs, amiR‐LB2 and amiR‐LD, of the six designed amiRNAs confer moderate resistance against WSMoV, and the triple construct including the two amiRNAs provides complete resistance.
INTRODUCTION
RNA silencing is a mechanism in which a small RNA targets a complementary region of an mRNA resulting in cleavage and subsequent degradation (Baulcombe, 2004, 2005) or translational inhibition of the mRNA (Aukerman and Sakai, 2003; Brodersen et al., 2008). Small RNAs involved in the RNA silencing processes include microRNAs (miRNAs) and small interfering RNAs (siRNAs), whereas long double‐stranded RNAs and hairpin precursor RNAs are required to serve as substrates for Dicer‐like (DCL) protein to produce miRNAs or siRNAs (Bartel, 2004; Carrington and Ambros, 2003; Kim, 2005; Zamore and Haley, 2005). These small RNAs are then loaded into RNA‐induced silencing complexes (RISCs) containing an ARGONAUTE protein. The small RNA‐programmed RISC cleaves an mRNA at a complementary sequence site defined by the antisense small RNA, and the cleaved target mRNA is then degraded (Khvorova et al., 2003; Llave et al., 2002; Schwarz et al., 2003; Vaucheret et al., 2004). Endogenous miRNAs are processed from noncoding RNA precursors that form double‐stranded hairpin structures. The stem region of the hairpin is processed into a 21‐nucleotide mature miRNA duplex. The miRNA strand complementary to its target gene is then recognized by the RISC that guides the miRNA to degrade the target mRNA (Bartel, 2004).
The expression of specific miRNAs on already known miRNA precursor backbones has been proven to be able to induce highly specific gene silencing in Arabidopsis thaliana (Park et al., 2009; Schwab et al., 2006), tobacco (Alvarez et al., 2006), tomato (Alvarez et al., 2006), rice (Warthmann et al., 2008), moss (Khraiwesh et al., 2008) and alga (Molnar et al., 2009). Moreover, the application of artificial miRNA (amiRNA) targeting the gene silencing suppressor gene of plus‐strand RNA virus can generate virus resistance (Ai et al., 2011; Duan et al., 2008; Niu et al., 2006; Qu et al., 2007). The expression of amiRNA in a 273‐nucleotide A. thaliana pre‐miR159a backbone conferred virus resistance on Arabidopsis (Niu et al., 2006), tobacco (Ai et al., 2011) and tomato (Zhang et al., 2011). In addition, pre‐amiR159‐P69, targeting the gene silencing suppressor of Turnip yellow mosaic virus (TYMV), can be processed by Nicotiana benthamiana to produce functional amiR159‐P69 to confer resistance against the recombinant Turnip mosaic virus (TuMV) GP69 harbouring the amiR159‐P69 targeting sequence (Lin et al., 2009). Furthermore, amiRNAs directed against highly accessible target sites confer effective resistance to Cucumber mosaic virus (CMV) (Duan et al., 2008).
In engineering resistance against plant viruses, the amiRNA strategy has several advantages over transgene‐homology‐dependent gene silencing. The amiRNAs have minimal off‐target effects, as the amiRNA sequences are much shorter than the viral genomic sequences used for homology‐dependent gene silencing. If the plant genomic sequence is known, precise antiviral amiRNAs lacking complementary host target sequences can be designed (Garcia and Simon‐Mateo, 2006). Moreover, the environmental biosafety concerns of viral sequences that might complement or recombine with nontarget viruses do not apply to amiRNAs (Garcia and Simon‐Mateo, 2006).
The family Bunyaviridae includes five genera: Orthobunyavirus, Phlebovirus, Hantavirus, Nairovirus and Tospovirus (Fauguet et al., 2005). Watermelon silver mottle virus (WSMoV) is a member of the plant‐infecting genus Tospovirus (Fauguet et al., 2005), and is transmitted by Thrips palmi Karny in a persistent manner (Chen et al., 1990). WSMoV limits the production of cucurbits in Eastern Asia (Chen et al., 1990; Iwaki et al., 1984; Yeh et al., 1992). Virions of WSMoV are enveloped quasispherical particles containing three single‐stranded nucleic acid segments, denoted as L, M and S RNAs (Fauguet et al., 2005; Yeh et al., 1992), in a negative‐sense or ambisense organization. The complete WSMoV genomic sequence has been determined (Chu and Yeh, 1998; Chu et al., 2001; Yeh et al., 1996). The L RNA of WSMoV is of negative polarity, encoding a putative RNA‐dependent RNA polymerase (RdRp, 331.8 kDa) in the viral complementary strand (Chu et al., 2001). Comparison of the RdRp protein encoded by L RNA of WSMoV with those of other tospoviruses has revealed a highly conserved region containing five RdRp motifs (Chu et al., 2001), which are essential for virus replication.
In this study, amiRNAs targeting five conserved motifs of the L strand of WSMoV were expressed in N. benthamiana plants to generate transgenic resistance against tospoviruses. Our results indicated that two amiRNAs, amiR‐LB2 and amiR‐LD, of the six designed amiRNAs were effective against WSMoV. Moreover, an additive effect was obtained when these two amiRNAs were expressed in a triple amiRNA construct. In addition, the levels of the virus resistance were positively correlated with the amiRNA expression levels. This is the first report of the management of negative‐sense plant RNA viruses using the amiRNA strategy targeting the highly conserved motifs of the viral replicase gene.
RESULTS
Construction of amiRNAs targeting L RNA of WSMoV
Six amiRNAs containing sequences complementary to nucleotides 4157–4137, 4412–4392, 4451–4431, 4537–4517, 4661–4641 and 4709–4689 of the WSMoV L viral (v) strand (negative‐sense strand) were designed (Fig. 1). These amiRNA sequences replaced the original 21‐nucleotide miR159a sequence in the pre‐miRNA159a backbone. The six pBA‐pre‐amiRNA vectors were named pBA‐pre‐amiR‐LA, pBA‐pre‐amiR‐LB1, pBA‐pre‐amiR‐LB2, pBA‐pre‐amiR‐LC, pBA‐pre‐amiR‐LD and pBA‐pre‐amiR‐LE (Fig. 2A).
Figure 1.
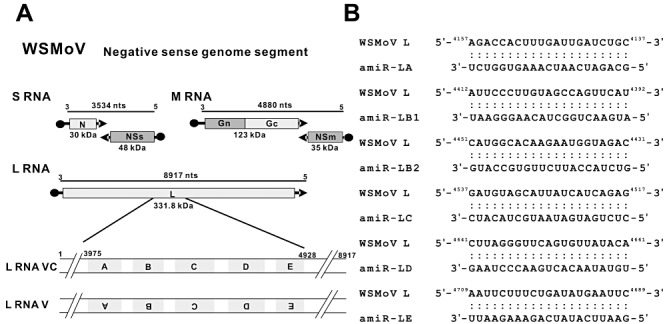
Construction of artificial microRNAs (amiRNAs) targeting conserved motifs of the L (replicase) gene of Watermelon silver mottle virus (WSMoV). (A) The conserved motifs A, B, C, D and E are shown on L RNA. (B) Six amiRNAs (amiR‐LA, amiR‐LB1, amiR‐LB2, amiR‐LC, amiR‐LD and amiR‐LE) targeting individual motifs of L RNA viral strand were designed. The sequences targeted by individual amiRNAs are shown in base pairs.
Figure 2.
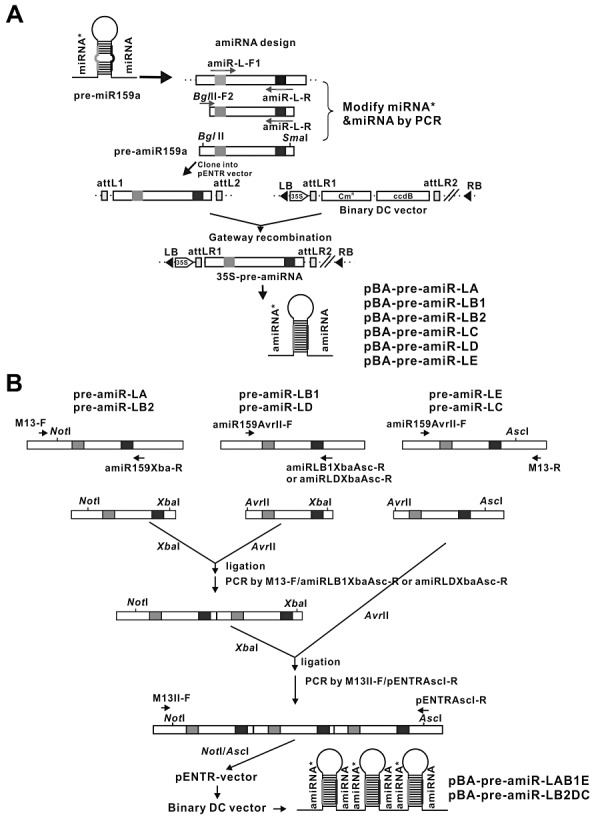
Construction of artificial microRNAs (amiRNAs) targeting the conserved motifs of the tospoviral replicase sequence in the pre‐miR159a backbone. (A) Construction of single amiRNAs. MicroRNA159a (miR159a) processed from the pre‐miRNA159a backbone is presented as a hairpin. Forward primer amiR‐L‐F1 and reverse primer amiR‐L‐R, containing the complementary sequence to an individual amiRNA, were used to replace the miRNA159a and miRNA159a* sequences with an amiRNA targeting the A, B1, B2, C, D or E conserved motif of the L gene (replicase) of Watermelon silver mottle virus by first polymerase chain reaction (PCR). BglII‐F2 primer containing a BglII site and R primer containing a SmaI site were used in nested PCR. The pre‐amiRNA159a was digested with BglII/SmaI, subcloned into the pENTR vector and placed downstream of the 35S promoter in a binary destination vector (destination cassette, DC) by the Gateway recombination system. (B) Strategy for triple amiRNA construction. The primer pair M13‐F/amiR159Xba‐R was used to amplify amiR‐LA (or amiR‐LB2). The primer pairs amiR159AvrII‐F/amiLB1XbaAscI‐R and amiR159AvrII‐F/amiLDXbaAscI‐R were used to amplify amiR‐LB1 and amiR‐LD, respectively. The primer pair amiR159AvrII‐F/M13‐R was used to amplify amiR‐LE or amiR‐LC. From the amplified products, the amiR‐LA or amiR‐LB2 fragment was digested by XbaI and ligated to AvrII‐digested amiR‐LB1 or amiR‐LD fragment, respectively. After ligation, the double amiRNA segment was amplified by the primer pair M13‐F/amiLB1XbaAscI‐R or M13‐F/amiLDXbaAscI‐R. The fragment was digested by XbaI and ligated to AvrII‐digested amiR‐LE or amiR‐LC fragment. The triple amiRNA segment was amplified by M13II‐F/pENTRAscI. The triple amiRNA fragment was released from the amplified product by NotI/AscI digestion and subcloned into the pENTR vector to generate pENTR‐amiR‐LAB1E and pENTR‐amiR‐LB2DC, and then into binary DC vector by Gateway recombination to form triple amiRNA constructs pre‐amiR‐LAB1E and pre‐amiR‐LB2DC.
Furthermore, two triple amiRNA precursors were constructed by separately ligating pre‐amiR‐LA, pre‐amiR‐LB1 and pre‐amiR‐LE, or pre‐amiR‐LB2, pre‐amiR‐LD and pre‐amiR‐LC, in the pENTR vector and placed downstream of the 35S cauliflower mosaic virus (CaMV) promoter in the binary vector. These two triple amiRNA constructs were named pBA‐pre‐amiR‐LAB1E and pBA‐pre‐amiR‐LB2DC (Fig. 2B). All six individual constructs and the two triple constructs, which contained a 35S CaMV promoter (Fig. 2), were used to transform plants of N. benthamiana.
Transient expression of amiRNAs using agro‐infiltration
Transient expression of amiRNA constructs in N. benthamiana plants was verified by agro‐infiltration. The detection of high amiRNA expression levels indicated that all the amiRNAs can be successfully processed from the respective constructs. Transient expression of individual amiRNA constructs was used as a positive control to examine the expression of the corresponding amiRNA in transgenic plants (Fig. 3A–H).
Figure 3.
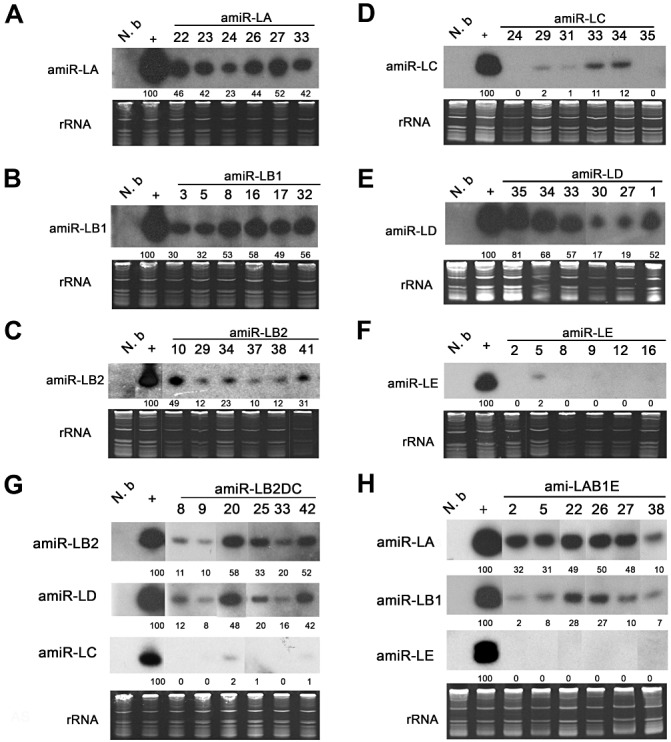
Expression of artificial microRNAs (amiRNAs) in transgenic Nicotiana benthamiana plants. (A–H) Detection of amiRNA expression in T1 plants of different lines carrying amiR‐LA, amiR‐LB1, amiR‐LB2, amiR‐LC, amiR‐LD, amiR‐LE, amiR‐LAB1E and amiR‐LB2DC constructs (10 plants as one sample for each line). Total RNAs extracted from N. benthamiana leaves agro‐infiltrated with individual constructs were used as positive controls (+). rRNA was used as loading control. Line numbers are indicated on the top. The amiRNA expression levels in agro‐infiltrated plants were arbitrarily set as 100 for each construct. The numbers on the panels represent the percentage amiRNA expression levels.
Expression of amiRNAs in transgenic plants
At least 30 independent T0 transgenic lines of each construct were obtained and confirmed by polymerase chain reaction (PCR). Northern hybridization analysis showed different amiRNA expression levels in the T0 plants of pre‐amiR‐LA, pre‐amiR‐LB1, pre‐amiR‐LB2 and pre‐amiR‐LD transgenic lines (data not shown). However, relatively lower or undetectable levels of amiRNA expression were detected in most pre‐amiR‐LC and pre‐amiR‐LE transgenic lines. For each construct, T1 progenies from six independent lines with different levels of amiRNA expression were selected for further assay (Fig. 3). On the basis of the expression levels, numerical values were assigned for each transgenic line as follows. The amiRNA expression level in agro‐infiltrated plants was arbitrarily set to 100 and different expression levels from each construct were graded as: >30 = high (+++), 20–29 = moderate (++), 10–19 = low (+), 0–9 = very low (+/−) and 0 = no (−) expression (Table 2 and Fig. 3A–H).
Table 2.
Expression of artificial microRNAs (amiRNAs) and resistance evaluation of selected transgenic Nicotiana benthamiana lines (T1 plants) after challenge with Watermelon silver mottle virus (WSMoV) and segregation of T1 plants on MSBar medium.
| Construct | Line | Expression level of amiRNA # | Infected/total inoculated plants† at | Segregation of T1 plants‡ | Prediction of copy numbers | ||||
|---|---|---|---|---|---|---|---|---|---|
| 14 dpi | 28 dpi | ||||||||
| Expt 1 + Expt 2 | Expt 1 + Expt 2 | BarR | BarS | ||||||
| I/T | R (%) | I/T | R (%) | ||||||
| amiR‐LA | 22 | +++ | 12/12 | 0 | 12/12 | 0 | 114 | 34 | 1 |
| 23 | +++ | 12/12 | 0 | 12/12 | 0 | 162 | 47 | 1 | |
| 24 | ++ | 12/12 | 0 | 12/12 | 0 | 111 | 6 | >1 | |
| 26 | +++ | 11/12 | 8 | 11/12 | 8 | 103 | 9 | >1 | |
| 27 | +++ | 11/12 | 8 | 11/12 | 8 | 86 | 39 | 1 | |
| 33 | +++ | 12/12 | 0 | 12/12 | 0 | 124 | 37 | 1 | |
| amiR‐LB1 | 3 | +++ | 12/12 | 0 | 12/12 | 0 | 121 | 39 | 1 |
| 5 | +++ | 12/12 | 0 | 12/12 | 0 | 89 | 27 | 1 | |
| 8 | +++ | 12/12 | 0 | 12/12 | 0 | 140 | 7 | >1 | |
| 16 | +++ | 12/12 | 0 | 12/12 | 0 | 48 | 4 | >1 | |
| 17 | +++ | 11/12 | 8 | 11/12 | 8 | 98 | 39 | 1 | |
| 32 | +++ | 12/12 | 0 | 12/12 | 0 | 92 | 28 | 1 | |
| amiR‐LB2 | 10 | +++ | 2/12 | 83 | 7/12 | 42 | 139 | 5 | >1 |
| 29 | + | 6/12 | 50 | 12/12 | 0 | 101 | 51 | 1 | |
| 34 | ++ | 6/12 | 50 | 10/12 | 17 | 118 | 44 | 1 | |
| 37 | + | 5/12 | 58 | 12/12 | 0 | 114 | 40 | 1 | |
| 38 | + | 6/12 | 50 | 11/12 | 8 | 134 | 5 | >1 | |
| 41 | +++ | 3/12 | 75 | 7/12 | 42 | 85 | 2 | >1 | |
| amiR‐LC | 24 | − | 12/12 | 0 | 12/12 | 0 | 86 | 30 | 1 |
| 29 | +/− | 12/12 | 0 | 12/12 | 0 | 54 | 31 | 1 | |
| 31 | +/− | 12/12 | 0 | 12/12 | 0 | 94 | 34 | 1 | |
| 33 | + | 12/12 | 0 | 12/12 | 0 | 117 | 9 | >1 | |
| 34 | + | 12/12 | 0 | 12/12 | 0 | 100 | 31 | 1 | |
| 35 | − | 12/12 | 0 | 12/12 | 0 | 101 | 46 | 1 | |
| amiR‐LD | 1 | +++ | 3/12 | 75 | 12/12 | 0 | 84 | 3 | >1 |
| 27 | + | 11/12 | 8 | 12/12 | 0 | 93 | 47 | 1 | |
| 30 | + | 7/12 | 42 | 12/12 | 0 | 52 | 33 | 1 | |
| 33 | +++ | 3/12 | 75 | 11/12 | 8 | 150 | 3 | >1 | |
| 34 | +++ | 6/12 | 50 | 11/12 | 8 | 138 | 11 | >1 | |
| 35 | +++ | 5/12 | 58 | 11/12 | 8 | 103 | 26 | 1 | |
| amiR‐LE | 2 | − | 12/12 | 0 | 12/12 | 0 | 92 | 31 | 1 |
| 5 | +/− | 12/12 | 0 | 12/12 | 0 | 170 | 0 | >1 | |
| 8 | − | 12/12 | 0 | 12/12 | 0 | 123 | 34 | 1 | |
| 9 | − | 12/12 | 0 | 12/12 | 0 | 115 | 36 | 1 | |
| 12 | − | 12/12 | 0 | 12/12 | 0 | 128 | 39 | 1 | |
| 16 | − | 12/12 | 0 | 12/12 | 0 | 70 | 6 | >1 | |
| amiR‐LAB1E | 2 | +++ | 11/12 | 8 | 11/12 | 8 | 134 | 14 | >1 |
| 5 | +++ | 11/12 | 8 | 11/12 | 8 | 163 | 15 | >1 | |
| 22 | +++ | 12/12 | 0 | 12/12 | 0 | 26 | 13 | 1 | |
| 26 | +++ | 12/12 | 0 | 12/12 | 0 | 137 | 25 | >1 | |
| 27 | +++ | 12/12 | 0 | 12/12 | 0 | 87 | 19 | 1 | |
| 38 | + | 12/12 | 0 | 12/12 | 0 | 94 | 6 | >1 | |
| amiR‐LB2DC | 8 | + | 3/12 | 75 | 6/12 | 50 | 77 | 24 | 1 |
| 9 | + | 3/12 | 75 | 7/12 | 42 | 145 | 47 | 1 | |
| 20 | +++ | 0/12 | 100 | 0/12 | 100 | 139 | 56 | 1 | |
| 25 | +++ | 0/12 | 100 | 0/12 | 100 | 116 | 3 | >1 | |
| 33 | ++ | 0/12 | 100 | 1/12 | 92 | 154 | 10 | >1 | |
| 42 | +++ | 0/12 | 100 | 0/12 | 100 | 145 | 1 | >1 | |
| NT | 12/12 | 0 | 12/12 | 0 | 0 | 150 | 0 | ||
amiRNA expression levels were determined by Northern hybridization (Fig. 3). Expression levels in transgenic plants were graded as >30 = high (+++), 20–29 = moderate (++), 10–19 = low (+), 0–9 = very low (+/−) and 0 = no (−) expression, in proportion to the expression level in agro‐infiltrated plants, arbitrarily set as 100. ‘+’ symbols reflect the situation of first amiRNA expression in the lines of the triple constructs.
For each amiRNA construct, six independent transgenic lines were challenged with WSMoV, and analysed for resistance to WSMoV for 28 days in two independent experiments. Nontransgenic (NT) plants were used as controls. Six T1 plants of each line were tested for WSMoV infection. I/T, the nominator (I) indicates the number of infected plants [symptomatic, enzyme‐linked immunosorbent assay (ELISA) positive] and the denominator (T) indicates the total number of inoculated plants. R, number of symptomless plants with resistance presented as a percentage of the total inoculated plants.
The segregation of T1 progenies was analysed on MSBar medium. BarR, number of T1 plants that survived on MSBar medium; BarS, number of T1 plants that perished on MSBar medium.
Relatively lower or undetectable levels of amiR‐LC and amiR‐LE were seen in all the T0 lines and the T1 progenies of transgenic lines carrying the corresponding single amiRNAs (Fig. 3D,F) or triple amiRNAs (Fig. 3G,H), in contrast with the higher expression levels of the other four amiRNAs (amiR‐LA, amiR‐LB1, amiR‐LB2 and amiR‐LD), in either single (Fig. 3A–C,E) or triple constructs (Fig. 3G,H). Transient expression of the amiRNAs detected by agro‐infiltration, used as a positive control (Fig. 3A–H), indicated that the lower level of amiRNA expression of certain amiRNAs was not a result of the probe.
Bioassay for WSMoV resistance in transgenic N. benthamiana plants
The T1 plants analysed (Fig. 3) were used for WSMoV inoculation to evaluate their transgenic resistance. Nontransgenic (NT) plants of N. benthamiana and six T1 plants from individual selected transgenic lines carrying individual amiRNA constructs were mechanically challenged with WSMoV in two independent experiments. Plants with symptoms were confirmed by enzyme‐linked immunosorbent assay (ELISA) and the percentage resistances to WSMoV are summarized in Table 2. At 7 days post‐inoculation (dpi), typical disease symptoms were observed in all NT plants, and in 92%–100% plants of the 12 T1 progenies from the lines expressing amiR‐LA, amiR‐LB1, amiR‐LC, amiR‐LE or amiR‐LAB1E. No significant delay in symptom development was noticed in these lines. By contrast, at 14 dpi, the infected plants of amiR‐LB2 (10, 29, 34, 37, 38 and 41) and amiR‐LD (1, 30, 33, 34 and 35) lines displayed moderate resistance to WSMoV, with resistance levels ranging between 42% and 83%, as calculated from the numbers of plants without symptoms. Eleven T1 plants of amiR‐LD line 27, expressing a relatively lower level of amiRNA, were susceptible to WSMoV at 14 dpi. However, the symptoms were milder than those on the WSMoV‐infected NT plants (Fig. 4A, i and ii). Strikingly, all the plants of the four triple amiR‐LB2DC lines (20, 25, 33 and 42) displayed 100% resistance to WSMoV at 14 dpi (Table 1). Figure 4A (iii) shows a representative symptomless systemic leaf of the line amiR‐LB2DC‐20.
Figure 4.
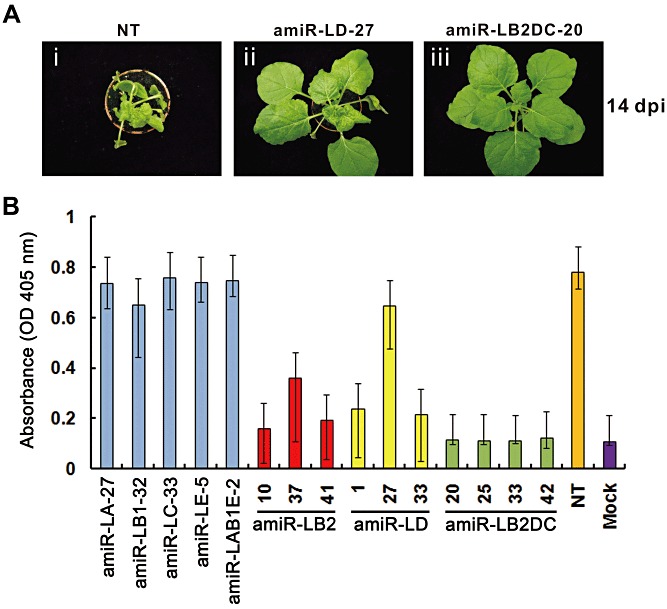
Symptoms and virus accumulation detected by enzyme‐linked immunosorbent assay (ELISA) of different transgenic Nicotiana benthamiana lines, 14 days post‐inoculation with Watermelon silver mottle virus (WSMoV). (A) Typical symptoms were observed on a nontransgenic (NT) plant (i), mild mosaic symptoms on a T1 plant of the amiR‐LD‐27 line (ii) and no symptoms on a T1 plant of the amiR‐LD‐20 line (iii). (B) WSMoV was detected by indirect ELISA with six plants/line. One representative line was used for pre‐amiR‐LA, pre‐amiR‐LB1, pre‐amiR‐LC, pre‐amiR‐LE and pre‐amiR‐LAB1E constructs, three lines were used for pre‐amiR‐LB2 and pre‐amiR‐LD, and four lines were used for pre‐amiR‐LB2DC transgenic plants. WSMoV‐infected NT and mock‐inoculated plants (Mock) of N. benthamiana were used as positive and negative controls, respectively.
Table 1.
Nucleotide sequences of the primers used in this study.
| Primer name | Sequence |
|---|---|
| amiR‐LA‐F1 | 5′‐GACGATGGAAGAGACCACTTTGATTGATCTGCCATGAGTTGAGCAGG‐3′ |
| amiR‐LA‐R | 5′‐TTGACCCGGGATGAGACCACTTTGATTGATCTGCGAAGAGTAAAAGCC‐3′ |
| amiR‐LB1‐F1 | 5′‐GACGATGGAAGATTCCCTTGTAGCCAGTTCATCATGAGTTGAGCAGG‐3′ |
| amiR‐LB1‐R | 5′‐TTGACCCGGGATGATTCCCTTGTAGCCAGTTCATGAAGAGTAAAAGCC‐3′ |
| amiR‐LB2‐F1 | 5′‐GACGATGGAAGCATGGCACAAGAATGGTAGACCATGAGTTGAGCAGG‐3′ |
| amiR‐LB2‐R | 5′‐TTGACCCGGGATGCATGGCACAAGAATGGTAGACGAAGAGTAAAAGCC‐3′ |
| amiR‐LC‐F1 | 5′‐GACGATGGAAGGATGTAGCATTATCATCAGAGCATGAGTTGAGCAGG‐3′ |
| amiR‐LC‐R | 5′‐TTGACCCGGGATGGATGTAGCATTATCATCAGAGGAAGAGTAAAAGCC‐3′ |
| amiR‐LD‐F1 | 5′‐GACGATGGAAGCTTAGGGTTCAGTGTTATACACATGAGTTGAGCAGG‐3′ |
| amiR‐LD‐R | 5′‐TTGACCCGGGATGCTTAGGGTTCAGTGTTATACAGAAGAGTAAAAGCC‐3′ |
| amiR‐LE‐F1 | 5′‐GACGATGGAAGAATTCTTTCTGATATGAATTCCATGAGTTGAGCAGG‐3′ |
| amiR‐LE‐R | 5′‐TTGACCCGGGATGAATTCTTTCTGATATGAATTCGAAGAGTAAAAGCC‐3′ |
| BgIll‐F2 | 5′‐TCGATAGATCTTGATCTGACGATGGAAG‐3′ |
| M13‐F | 5′‐GTTTTCCCAGTCACGAC‐3′ |
| M13II‐F | 5′‐GTAAAACGACGGCCAG‐3′ |
| amiR159Xba‐R | 5′‐GCGGCGCGCCCTCTAGATGACCCGGGATG‐3′ |
| amiR159AvrII‐F | 5′‐GGGTTTCCTAGGACAGTTTGCTTATGTCGGATCC‐3′ |
| amiRLB1XbaAsc‐R | 5′‐GCGGCGCGCCCTCTAGATGACCCGGGATGATTCCC‐3′ |
| amiRLDXbaAsc‐R | 5′‐GCGGCGCGCCCTCTAGATGACCCGGGATGCTTA‐3′ |
| M13‐R | 5′‐CAGGAAACAGCTATGAC‐3′ |
| pENTRAscI‐R | 5′‐CTTTGTACAAGAAAGCTGGGTCGGCGCG‐3′ |
| GFPNcoI‐F | 5′‐CCACGCCCATGGTGAGCAAGGGCGA‐3′ |
| TuGFP‐LA‐R1 | 5′‐AAGTGGTCTCCTTGTACAGCTCGTCCATGCC‐3′ |
| TuGFP‐LA‐R2 | 5′‐CAGCTAGCCCGCAGATCAATCAAAGTGGTCTCCTTGTACAGC‐3′ |
| TuGFP‐LB1‐R1 | 5′‐CAAGGGAATCCTTGTACAGCTCGTCCATGCC‐3′ |
| TuGFP‐LB1‐R2 | 5′‐CAGCTAGCCCATGAACTGGCTACAAGGGAATCCTTGTACAGC‐3′ |
| TuGFP‐LD‐R1 | 5′‐AACCCTAAGCTTGTACAGCTCGTCCATGCC‐3′ |
| TuGFP‐LD‐R2 | 5′‐CAGCTAGCTGTATAACACTGAACCCTAAGCTTGTACAGC‐3′ |
The 21 nucleotides representing the complementary sequences to individual artificial microRNAs (amiRNAs) in the specific primers for the construction of each amiRNA, and the 21 nucleotides representing individual amiRNAs for the construction of recombinant Turnip mosaic virus (TuMV), are in bold italic. The 3′ end (bold) of the common primer BglII‐F2 overlaps the 5′ portion (bold) of the amiR‐L‐F1 primer. The cloning sites of BglII and XmaI, in primers BglII‐F2 and amiR‐L‐R, respectively, are in italic.
In ELISA, WSMoV nucleocapsid protein (NP) was undetectable in all the inoculated plants of four of the six amiR‐LB2DC lines (20, 25, 33 and 42) and the plants were symptomless. Similarly, the WSMoV NP‐undetectable plants of amiR‐LB2 and amiR‐LD lines were also symptomless. By contrast, the NT control plants and those from amiR‐LA, amiR‐LB1, amiR‐LC, amiR‐LE and amiR‐LAB1E lines, which accumulated high levels of WSMoV NP, displayed symptoms. Figure 4B shows the average ELISA readings for representative lines of each construct. At 28 dpi, 58% of plants expressed symptoms in higher amiR‐LB2‐expressing lines (10 and 41), whereas most of the plants of amiR‐LD lines showed symptoms. By contrast, fewer plants of the amiR‐LB2DC lines (8, 9 and 33) developed symptoms and no plants with symptoms were found in amiR‐LB2DC lines (20, 25 and 42) (Table 2). All WSMoV‐resistant plants monitored for an additional month remained symptomless and ELISA negative. Thus, our results indicate that the triple amiR‐LB2DC lines expressing functional amiR‐LB2 and amiR‐LD confer much higher levels of resistance to WSMoV infection than the lines expressing single amiR‐LB2 or amiR‐LD.
In summary, transgenic lines expressing amiR‐LB2 or amiR‐LD conferred moderate resistance to WSMoV, but virus resistance was not observed in transgenic lines expressing amiR‐LA or amiR‐LB1. The expression levels of amiR‐LC and amiR‐LE were low, and no transgenic resistance was observed in any transgenic lines carrying these amiRNAs. Consistent with the results of single amiRNA‐LA and amiRNA‐LB1, most amiR‐LAB1E lines did not show significant levels of resistance to WSMoV. Strikingly, the triple amiR‐LB2DC line conferred complete resistance to WSMoV, apparently an additive effect of amiR‐LB2 and amiR‐LD.
To address the possibility that the amiRNAs might target endogenous mRNAs, we searched for potential targets for our amiRNAs in the Institute for Genomic Research (TIGR) N. benthamiana gene database at http://bioinfo3.noble.org/miRNA/miRU.htm. In TIGR N. benthamiana Gene Index 1, we found three mismatches in the pairing of the N. benthamiana transcript TC2429 with amiR‐LC, which did not produce any resistant line. No other putative targets were identified for the other five amiRNAs. However, we cannot exclude the possibility that genes that have not yet been identified could be potential targets, although all transgenic lines showed a normal phenotype.
Segregation analysis of T1 progenies
All the T1 progeny seeds were cultured on MSBar medium [Murashige and Skoog (MS) medium containing 10 mg/L glufosinate ammonium] for segregation assay (Table 2). The 3:1 segregation ratio of T1 progenies revealed that the T‐DNA was inserted at a single locus in several lines, including several resistant lines (Table 2). However, the results also imply multiple copies of T‐DNA in several susceptible or resistant lines (Table 2).
Resistance levels are positively correlated with the expression levels of amiRNA
Transgenic lines expressing different levels of amiR‐LB2 (Fig. 3C) or amiR‐LD (Fig. 3E) showed variable levels of resistance to WSMoV (Table 2). To determine whether the resistance levels correlated with amiRNA expression levels, Northern hybridization analysis was performed (with 10 T1 plants/line) for three amiR‐LD lines (1, 27 and 33) and three amiR‐LB2DC lines (8, 9 and 20) (Fig. 5A). In the three amiR‐LD lines tested, line 27, with a lower amiR‐LD expression level, was more susceptible to WSMoV than lines 1 and 33, with relatively higher amiR‐LD expression levels (Fig. 5B, top panels). Similar results were also obtained for the amiR‐LB2DC lines (Fig. 5B, bottom panels). Our results show that the resistance level is correlated with the expression levels of amiRNAs (Fig. 5).
Figure 5.
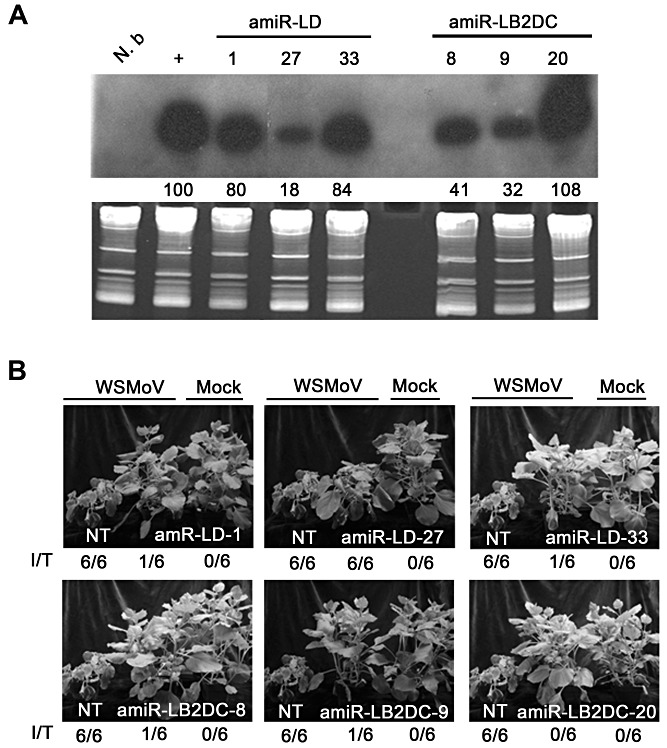
Correlation of the levels of resistance to Watermelon silver mottle virus (WSMoV) of artificial microRNA (amiRNA) transgenic Nicotiana benthamiana plants with the expression levels of amiRNA. (A) Expression levels of amiRNA before inoculation of amiR‐LD (1, 27 and 33) and amiR‐LB2DC (8, 9 and 20) transgenic lines. The relative expression levels of amiRNAs in transgenic lines were compared with the expression level in agro‐infiltrated tissue, arbitrarily set as 100. (B) Symptoms at 14 days after WSMoV inoculation. In each section of (B), a WSMoV‐inoculated transgenic plant (centre) is flanked by a WSMoV‐infected nontransgenic (NT) control plant (left) and a mock‐inoculated transgenic plant (right). I/T (as defined in Table 2) represents the results at 14 days post‐inoculation.
Transgenic lines conferring resistance to Tospovirus with one mismatch in the target sequence
To test the specificity of amiRNAs for virus resistance, the T1 plants of nine WSMoV‐resistant lines, amiR‐LB2 (10, 37 and 41), amiR‐LD (1, 33 and 35) and amiR‐LB2DC (20, 33 and 42) lines, and five WSMoV‐susceptible lines (amiR‐LA‐27, amiR‐LB1‐32, amiR‐LAB1E‐5, amiR‐LAB1E‐22 and amiR‐LAB1E‐26) were challenged with Peanut bud necrosis virus (PBNV), Watermelon bud necrosis virus (WBNV), Capsicum chlorosis virus (CaCV), Calla lily chlorotic spot virus (CCSV) and Tomato spotted wilt virus (TSWV) (three plants/line). Alignment of the amiR‐LB2‐ and amiR‐LD‐targeted sequences in these five tospoviruses (Fig. 6A, top and bottom panels, respectively) revealed the presence of two to three mismatches in the amiR‐LB2‐targeted sequences of WBNV, CaCV, CCSV and TSWV, and two to four mismatches in the amiR‐LD‐targeted sequences of PBNV, WBNV, CaCV, CCSV and TSWV, when G:U pairing was allowed. However, only one mismatch was found in the amiR‐LB2‐targeted sequence of PBNV. All the plants of the tested amiR‐LB2 lines were susceptible to CaCV, CCSV, TSWV (Fig. 6) and WBNV (data not shown). All three plants of each transgenic line expressing amiR‐LB2 (lines amiR‐LB2‐10, amiR‐LB2‐41, amiR‐LB2DC‐20, amiR‐LB2DC‐33 and amiR‐LB2DC‐42) exhibited 2–3 days of delayed symptoms, when inoculated with PBNV (Fig. 6B, ii), in comparison with the NT control.
Figure 6.
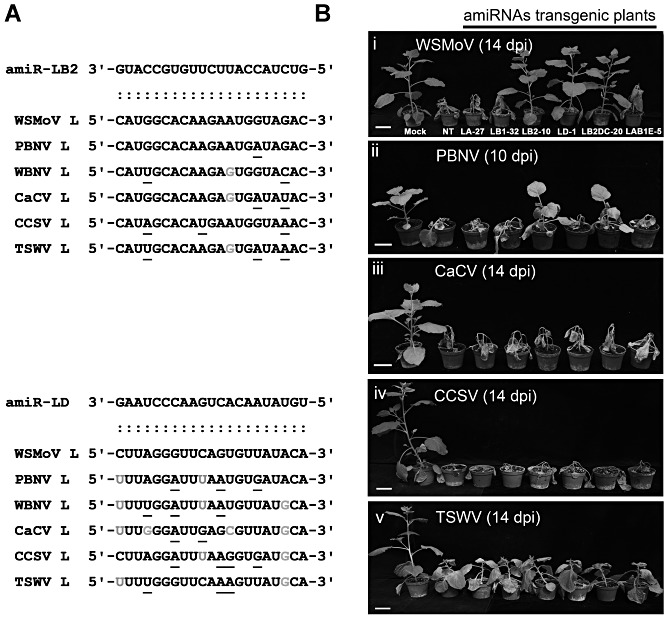
Infectivity assay of artificial microRNA (amiRNA) transgenic Nicotiana benthamiana plants challenged with different serogroup tospoviruses. (A) Alignment of amiR‐LB2 and amiR‐LD with the targeting sequences of five Watermelon silver mottle virus (WSMoV) serogroup viruses [WSMoV, Peanut bud necrosis virus (PBNV), Watermelon bud necrosis virus (WBNV), Capsicum chlorosis virus (CaCV) and Calla lily chlorotic spot virus (CCSV)] and Tomato spotted wilt virus (TSWV). The underlined letters indicate nucleotide mismatches and the grey letters indicate G:U paring between viral RNA and amiRNA. The sequences of L RNAs of WSMoV (accession number NC_003832), TSWV (NC_002052), PBNV (NC_003614), CaCV (NC_008302) and CCSV (FJ822962) were obtained from the National Center for Biotechnology Information (NCBI) GenBank database. The L RNA of WBNV was sequenced by T‐CC in our laboratory. (B) The symptoms of T1 plants 14 days post‐inoculation (dpi) with WSMoV (i), CaCV (iii), CCSV (iv) and TSWV (v), and 10 dpi with PBNV (ii). Three T1 plants from each line were used for challenge inoculation.
Susceptibility of transgenic lines expressing amiR‐LA or amiR‐LB1 to heterologous TuMV carrying the target sequences
To test the biological function of amiRNA‐LA and amiRNA‐LB1, which did not provide resistance to homologous WSMoV, we further used heterologous TuMV recombinants harbouring the 21‐nucleotide amiR‐LA‐ or amiR‐LB1‐targeted sequence (TuMV‐GLA or TuMV‐GLB1, respectively) to challenge T1 plants of two amiR‐LA‐expressing lines (22 and 27) and two amiR‐LB1‐expressing lines (16 and 32) (Fig. 7A). TuMV‐GLD carrying the amiR‐LD targeted sequence was also constructed for challenge inoculation of two WSMoV‐resistant lines: amiR‐LD‐33 and amiR‐LD‐35 (Fig. 7A). All T1 plants of the amiR‐LA‐22, amiR‐LA‐27, amiR‐LB1‐16 and amiR‐LB1‐32 lines, which showed mosaic symptoms at 5 dpi with TuMV‐GLA or TuMV‐GLB1, were severely retarded in growth, as observed at 10 dpi (Fig. 7B). All plants of the amiR‐LD‐33 and amiR‐LD‐35 lines did not show any symptoms at 5 dpi with TuMV‐GLD; however, 30%–50% showed symptoms at 10 dpi (Fig. 7B). All the NT control plants and transgenic lines showed severe symptoms after inoculation with TuMV‐GFP. When compared with the NT control, as detected by ELISA using TuMV CP antiserum, similar high levels of virus accumulation were noticed in symptomatic plants of transgenic lines carrying amiR‐LA or amiR‐LB1 at 10 dpi, whereas significantly lower levels of virus accumulation were found in plants of amiR‐LD transgenic lines (Fig. 7C). Our results imply that amiR‐LA and amiR‐LB1 also have no biological function, when their target sequences are present in TuMV recombinants.
Figure 7.
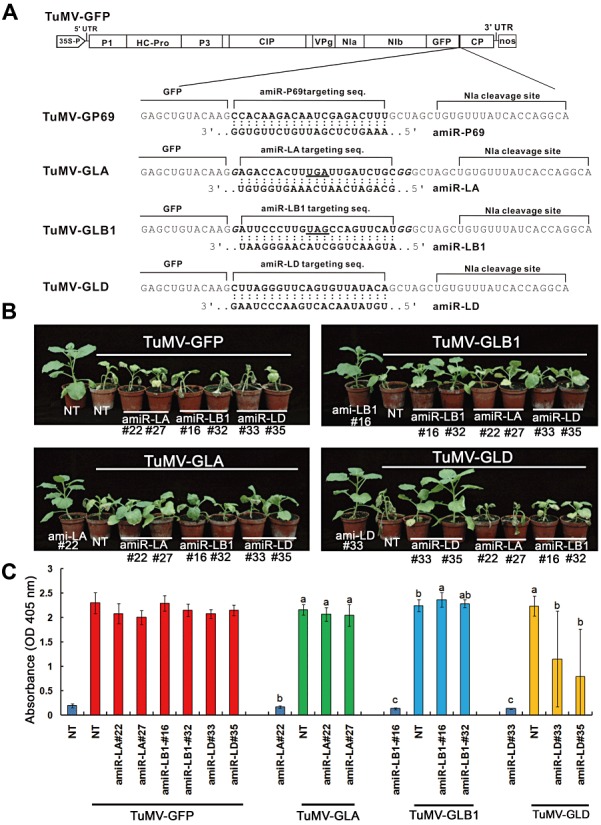
Evaluation of transgenic resistance of Nicotiana benthamiana plants expressing amiR‐LA, amiR‐LB or amiR‐LD by challenge with recombinant Turnip mosaic viruses (TuMV) carrying the corresponding artificial microRNA (amiRNA) target sequences. (A) Construction of infectious clones of TuMV recombinants. The 21‐nucleotide P69 targeting sequence of TuMV‐GP69 was replaced with the amiR‐LA, amiR‐LB1 or amiR‐LD targeting sequence to generate TuMV‐GLA, TuMV‐GLB1 or TuMV‐GLD, respectively. The additional three G nucleotides (italic) in TuMV‐GLA or TuMV‐GLB1 were used to avoid stop codons (UGA or UAG, underlined) and frameshifts of the viral reading frame. (B) Symptoms of transgenic lines expressing amiR‐LA (22 and 27), amiR‐LB (16 and 32) or amiR‐LD (33 and 35) at 10 days post‐inoculation with TuMV‐GFP, TuMV‐GLA, TuMV‐GLB1 or TuMV‐GLD. NT, nontransgenic N. benthamiana plants. (C) Virus accumulation was detected by indirect enzyme‐linked immunosorbent assay (ELISA). For each line, 10 plants were used for challenge inoculation. TuMV‐GFP‐infected NT and mock‐inoculated plants were used as positive and negative controls, respectively. Crowning of two adjacent bars of the histograms with the same letter is indicative of a nonsignificant difference, according to Fisher's least‐significant difference (LSD) test (P < 0.05) (SAS Institute Inc., Cary, NC, USA).
DISCUSSION
Several approaches have been used to manage the deadly diseases caused by thrip‐borne tospoviruses in agricultural crops. Amongst these strategies, the transgenic approach has been proven to be efficacious and to provide durable resistance against these viruses (1995, 1997; Rudolph et al., 2003; Schwach et al., 2004). Current transgenic strategies for plant RNA viruses focus on the expression of long pathogen‐related double‐stranded RNAs (dsRNAs) or hairpin RNAs to confer virus resistance in plants (Bucher et al., 2006). These dsRNAs are processed by Dicer into functional siRNAs, which mediate the cleavage of viral RNA transcripts, leading to resistance. In most investigations, the resistance level is correlated directly with siRNA expression levels (Kalantidis et al., 2002). In this study, we utilized the amiRNA strategy to generate resistance against WSMoV, a thrips‐borne negative‐sense RNA plant virus. We chose five different nucleotide motifs of WSMoV L negative‐sense viral RNA as targets for amiRNA‐mediated antiviral silencing, as these motifs were found to be highly conserved in WSMoV serogroup viruses (Chu et al., 2001). As these five highly conserved nucleotide motifs encode amino acids essential for the function of RdRp in virus replication, their key roles in virus survival would restrict these regions of the viral RNAs from being mutated, which, in turn, would minimize the possibility of the virus being able to evade the amiRNA surveillance mechanism by mutations.
In this study, we generated transgenic N. benthamiana plants expressing single or triple amiRNAs targeting the conserved motifs of the WSMoV L RNA viral strand. All of these pre‐amiRNAs were efficiently processed to the mature forms in plants of N. benthamiana, when expressed transiently. Qu et al. (2007) have reported that the expression of an amiRNA targeting the silencing suppressor 2b of CMV can efficiently inhibit 2b gene expression and suppressor function in transient expression assays and confer effective resistance to CMV infection. However, in our investigation, the resistance levels varied among the constructs. Amongst the transgenic lines expressing six individual amiRNAs, amiR‐LB2 and amiR‐LD lines were moderately resistant to WSMoV. The expression levels of amiR‐LC and amiR‐LE transgenic lines were low, and no transgenic resistance was seen in transgenic lines carrying either of these two constructs. Although high expression levels of these two amiRNAs were detected by agro‐infiltration, we failed to obtain transgenic lines with comparable levels of expression. It is possible that these amiRNAs might be sensitive to exonuclease in transgenic plants during the long process of regeneration. Furthermore, the relative amiR‐LC and amiR‐LE expression levels seem to be lower than those of other amiRNAs in the agro‐infiltration assay. It is possible that the secondary structure in and around the constructed amiRNA may affect the efficiency of the processing of these two amiRNAs.
Comparison of our results with those of previous studies showed that the amiR‐LB2‐ and amiR‐LD‐expressing N. benthamiana lines (T1 progenies) exhibited lower levels of resistance to WSMoV than those provided by amiR159‐P69‐ and amiR159‐HC‐Pro‐expressing Arabidopsis lines to TYMV and TuMV (Niu et al., 2006). Arabidopsis pre‐miRNA159a backbone‐carried amiRNA may express more efficiently in the homologous host Arabidopsis than the heterologous tobacco, the pre‐miRNA 159a of which has its own species‐specific variations. Thus, to control a virus in a particular crop by the amiRNA approach, the cognate miRNA precursor backbone may need to be derived from the crop or a closely related species to ensure effective amiRNA processing and consequent virus resistance. In addition, amiR159‐P69‐expressing N. benthamiana lines (T2 or T3 progenies) were highly resistant to TuMV‐GP69. This may be a result of the segregation of T‐DNA in T1 progenies in our study, which mostly comprised heterozygous combinations of the amiRNA inserts and was different from the durable amiRNA expression in T2 or T3 progenies in the amiR159‐P69 lines (Lin et al., 2009).
In the present transgenic N. benthamiana lines expressing amiRNAs targeting conserved motifs of the L gene of WSMoV, the levels of expression of amiRNA were positively correlated with the degrees of virus resistance. However, the resistance levels were not perfectly correlated with the copy numbers of T‐DNAs. Hence, lower levels of amiRNA expression and, consequently, weaker degrees of resistance in some transgenic lines may be a result of possible co‐suppression of amiRNA transgenes resulting from multiple copy integration. The T1 segregation results also indicated that the amiRNA expression levels were not perfectly positively correlated with the copy numbers of T‐DNA in the transgenic lines. However, the resistance of all individuals of the T2 generation plants of lines amiR‐LB2DC‐20, as those of the T1 generation, indicated that the stable transgenic resistance was faithfully transferred from the T1 to T2 generation.
Previous reports have shown that not all siRNA species against a given mRNA target are equally effective in mediating cleavage. The efficacy of RNA silencing depends not only on the nature of siRNAs, but also on the local structures of the target mRNAs (Luo and Chang, 2004; Overhoff et al., 2005; Schubert et al., 2005). As target recognition in less structured areas influences siRNA‐RISC catalysis (Duan et al., 2008), we propose that the susceptibility observed in the amiR‐LA‐ and amiR‐LB1‐expressing transgenic lines may be twofold. First, the rigid secondary structure of L RNA may have restricted access of the amiRNA‐programmed RISC to enter the target site for cleavage. Second, the synthetic amiRNAs may not be loaded into the RISC and, as a result, the target cannot be cleaved. To address these possibilities, we used the heterologous virus TuMV (Lin et al., 2009), harbouring the 21‐nucleotide amiR‐LA or amiR‐LB1 targeting site, to challenge amiR‐LA or amiR‐LB1 transgenic lines. Our results revealed that the amiR‐LA‐ and amiR‐LB1‐expressing lines were susceptible to either homologous WSMoV or heterologous TuMV harbouring the target sequences. Hence, we assume that the lack of biological function of amiR‐LA and amiR‐LB1 in transgenic plants might be a result of the failure in loading into RISC, and probably not caused by the secondary structure of the target site of A and B motifs in L RNA of WSMoV. Further investigations are needed to clarify this issue.
Simon‐Mateo and Garcia (2006) have reported the possibility that amiRNA‐mediated virus resistance may be overcome by the emergence of virus variants with mutations in the miRNA target sequence. To enhance the resistance level and to provide durable broad‐spectrum resistance against different viruses, the multiple amiRNA strategy can be used, in which several amiRNAs can be expressed under a single promoter (Duan et al., 2008; Niu et al., 2006). In this study, we generated transgenic tobacco expressing three different amiRNAs of amiR‐LB2DC and amiR‐LAB1E constructs. We found that amiR‐LB2DC lines expressing triple amiRNAs, including the effective amiR‐LB2 and amiR‐LD, provided enhanced levels of robust resistance against WSMoV, in comparison with the lines expressing the corresponding individual amiRNAs. Niu et al. (2006) have reported that double amiRNA (amiR159‐HC‐Pro and amiR159‐P69)‐expressing transgenic Arabidopsis provides resistance against two heterologous viruses, TuMV and TYMV. Recently, Ai et al. (2011) have reported that transgenic Nicotiana tabacum expressing amiRNAs targeting HC‐Pro and p25 from A. thaliana pre‐miR159a show highly specific resistance against Potato virus Y and Potato virus X. Thus, transgenic plants expressing three different amiRNAs are likely to provide a more durable virus resistance, as the possibility for the evolution of strains with mutations at three different conserved motifs is remote. However, in this study, neither amiR‐LA nor amiR‐LB1 from transgenic lines expressing triple amiR‐LAB1E exhibited any additive effects, probably because each single amiRNA was ineffective in conferring resistance. The consistent reduction in the expression of amiRNAs in the second position in both triple amiRNA constructs might be a result of unfavourable pre‐amiRNA secondary structures causing physical or steric hindrance to the processing of the second amiRNA.
When transgenic lines were tested with different serogroups of tospoviruses to assess possible broad‐spectrum resistance, only the lines expressing amiR‐LB2 showed a 2–3‐day delay in symptom development after challenge inoculation with PBNV, whereas all the other transgenic lines were susceptible. In a systematic study of the nucleotide positions required for amiRNA‐mediated resistance, Lin et al. (2009) identified positions 3–6, 9 and 12 of amiRNA as being critical. Chimeric heterologous viruses TuMV‐GP69 with mutations at these sites were pathogenic on 82% of amiR159‐P69 plants. Our data on PBNV are consistent with this observation, as there is one mismatch on the sixth position from the 5′ end of amiR‐LB2 within the targeting site. All of the WSMoV‐resistant transgenic lines were susceptible to four other tospovirus species, in which two to four mismatches existed in the targeting sites. This result is not surprising, as even two mutations within the 21‐nucleotide sequence will break down the resistance conferred by amiRNA‐mediated silencing (Lin et al., 2009).
In this study, amiR‐LB2 and amiR‐LD transgenic lines targeting highly conserved regions of the negative strand L RNA conferred resistance to WSMoV. The L conserved motif in RdRp of WSMoV is required for biological function, especially in virus replication. We believe that the amiRNA mediates cleavage of the negative strand (viral strand) and, consequently, no positive strand synthesis will take place for viral translation and replication. This is the first report of the management of negative‐sense plant RNA viruses using the amiRNA strategy. In future, we aim to combine the triple amiRNAs targeting three different regions of the L gene with the amiRNA targeting the common epitope of the gene silencing suppressor NSs (Chen et al., 2006) to confer broad‐spectrum resistance against different serogroups of Asia‐type tospoviruses. In addition, we wish to extend this amiRNA approach to the engineering of crop plants for resistance against different plant viruses, which is important for sustainable agriculture.
EXPERIMENTAL PROCEDURES
Construction of amiRNAs targeting conserved motifs of the RdRp of WSMoV
A 273‐bp fragment containing the entire sequence of the A. thaliana miR159a was cloned into the pENTR vector (Invitrogen, Carlsbad, CA, USA) to obtain pENTR‐pre‐miR159a (Niu et al., 2006). The pre‐miR159a was modified into synthetic sequences targeting the highly conserved motifs A, B, C, D and E of WSMoV L RNA (Chu et al., 2001) by two rounds of PCR (Fig. 1). Plasmid pENTR‐pre‐miR159a (as the template) and six pairs of forward and reverse primers, i.e. amiR‐L (A, B1, B2, C, D or E)‐F1 and amiR‐L (A, B1, B2, C, D or E)‐R (Table 1), were used in the first PCR. The forward primers contained sequences complementary (bold italic) to mature amiRNAs (amiR‐LA, amiR‐LB1, amiR‐LB2, amiR‐LC, amiR‐LD and amiR‐LE). The reverse primers contained an XmaI site (italic) and the sequences complementary (bold italic) to mature amiRNAs (Table 1).
In the second PCR, the product of the first PCR was used as the template with the primers BglII‐F2 (5′‐TCGATAGATCTTGATCTGACGATGGAAG‐3′) and amiR‐L (A, B1, B2, C, D or E)‐R to amplify a 223‐bp DNA fragment. The common primer BglII‐F2 contains a common sequence of amiR‐L (A, B1, B2, C, D or E)‐F1 primers (shown in bold) and a BglII site (italic) (Table 1). After BglII/XmaI digestions, the individual DNA fragments were cloned into BglII/XmaI‐digested pENTR‐pre‐miR159a vector to generate six different constructs in the pENTR vector. The Gateway system (Invitrogen) procedure was followed to transfer these pre‐amiRNAs to the plant binary Gateway destination vector pBA‐DC‐HA to generate six pBA‐pre‐amiRNA vectors, expressed from a 35S promoter (Fig. 2A).
Construction of triple amiRNAs
To generate the triple amiRNA constructs, several primers were designed (Table 1). Using pENTR‐pre‐amiR‐LA or pENTR‐pre‐amiR‐LB2 as the template, the PCR fragment containing amiR‐LA or amiR‐LB2 with an XbaI site was amplified by PCR with the primer pair M13‐F/amiR159Xba‐R, separately (Table 1). Using pENTR‐pre‐amiR‐LB1 or pENTR‐pre‐amiR‐LD as the template, and primer pair amiR159AvrII‐F/amiRLB1XbaAsc‐R or amiR159AvrII‐F/amiRLDXbaAsc‐R, the fragment containing amiR‐LB1 or amiR‐LD, both with an AvrII site at the 5′ end and an XbaI site at the 3′ end, was amplified. This PCR product was digested with AvrII, ligated to the previous XbaI‐digested PCR product (Fig. 2B) and used as the template for a second PCR with primer pair M13‐F/amiRLB1XbaAsc‐R or M13‐F/amiLDXbaAsc‐R (Table 1). The primer pair amiR159AvrII‐F/M13‐R was used to amplify the fragment containing amiR‐LE or amiR‐LC with an AvrII site at the 5′ end and an AscI site at the 3′ end of pENTR‐pre‐amiR‐LE or pENTR‐pre‐amiR‐LC. The amplified product was digested with AvrII, ligated to the XbaI‐digested second PCR product (Fig. 2B) and used as the template for the third PCR with primer pair M13II‐F/pENTRAscI‐R. The final PCR product was separately digested with NotI/AscI and cloned into NotI/AscI‐digested pENTR‐pre‐miR159a vector to generate pENTR‐pre‐amiR‐LAB1E and pENTR‐pre‐amiR‐LB2DC. The precursors containing individual triple amiRNAs were placed downstream of the 35S promoter of the binary vector pBA‐DC‐myc (Niu et al., 2006) using Gateway recombination to generate pBA‐pre‐amiR‐LAB1E and pBA‐pre‐amiR‐LB2DC (Fig. 2B).
Transient expression by agro‐infiltration of N. benthamiana
Eight constructs, pBA‐pre‐amiR‐L (A, B1, B2, C, D and E), pBA‐pre‐amiR‐LAB1E and pBA‐pre‐amiR‐LB2DC, were transferred to Agrobacterium tumefaciens ABI strain and used for the agro‐infiltration of N. benthamiana leaves. Two days after infiltration, total RNA was extracted from the infiltrated leaves using Trizol reagent (Invitrogen) for further analysis.
Transformation of N. benthamiana plants
Agro‐infiltrated N. benthamiana leaves were surface sterilized with 70% ethanol for 1 min, followed by 0.6% bleach with 0.01% Tween‐20 for 10 min, and rinsed with sterile water for 3 min three times. Leaf discs were cultured on MSBar selection medium containing Murashige and Skoog (MS) salts, 1 mg/L 6‐benzylaminopurine (BAP), 0.1 mg/L α‐naphthaleneacetic acid (NAA), 300 mg/L carbenicillin and 10 mg/L glufosinate ammonium for the selection of transformed cells. The transformed cells were placed in MSBar medium for shoot regeneration for 2 months. Developed shoots were excised and cultured on MS medium containing 0.15 mg/L NAA for rooting. Individual shoots were micropropagated on the same medium to generate three to five plants for preliminary tests. Rooted shoots were transplanted onto Florobella potting compost–sand mix (3:1) in a growth chamber. T1 seeds were collected from self‐pollinated T0 plants grown in a temperature‐controlled (23–28 °C) glasshouse. T1 progenies that survived on MSBar medium after germination were used for further assays. The segregation of T1 progenies was calculated on MSBar medium.
Northern hybridization
Total RNA was extracted from leaves of a T0 plant of individual transgenic lines, 10 T1 progeny from each line and NT control plants using Trizol reagent (Invitrogen). Fifteen micrograms of total RNA were resolved on a 15% polyacrylamide/1 × TBE [8.9 mm tris(hydroxymethyl)aminomethane (Tris), 8.9 mm boric acid, 20 mm ethylenediaminetetraacetic acid (EDTA)]/8 m urea gel and blotted onto a Hybond‐N+ membrane (GE Healthcare, Buckinghamshire, UK). DNA oligonucleotide with the exact complementary sequence to amiRNAs was end‐labelled with γ‐32P‐ATP using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA) to generate probes. Hybridization was carried out in ULTRAHyb‐Oligo solution (Ambion Inc., Austin, TX, USA), according to the manufacturer's directions, and signals were detected by autoradiography at −80 °C. In each case, the probe contained the exact antisense sequence of the target amiRNA.
Viruses and challenge inoculation of transgenic plants
An isolate of WSMoV and an isolate of CCSV were collected from watermelon (Yeh et al., 1992) and calla lily (Chen et al., 2005), respectively, in Taiwan. A high‐temperature‐recovered isolate (HT‐1) of CaCV from gloxinia (Hsu et al., 2000) was obtained from H. T. Hsu (US Department of Agriculture, Beltsville, MD, USA). WBNV (Jain et al., 1998) and PBNV (Satyanarayana et al., 1996) were provided by R. Premanand (Mahyco Research Centre, Maharashtra, India). TSWV NY isolate was provided by R. Provvidenti (New York State Experiment Station, Geneva, NY, USA). All of these viruses were used as inocula to evaluate transgenic resistance under glasshouse conditions. Leaves from infected N. benthamiana were ground in 10 mm potassium phosphate buffer (pH 7.0) containing 10 mm sodium sulphite, and the extract was mechanically introduced into N. benthamiana leaves. After 1 week, infected tissues were extracted in 100 volumes (v/w) of the same buffer. T1 plants of different transgenic N. benthamiana lines expressing each of the amiRNA constructs were collected after culturing on MS medium with glufosinate ammonium, and were grown in a glasshouse for 2 weeks (five‐ to six‐leaf stage). Plants were dusted with 600‐mesh carborundum on the three top fully expanded leaves and then mechanically rubbed with individual inoculum. NT N. benthamiana plants were used as controls. All inoculated plants were kept in a glasshouse (23–28 °C) and symptom development was monitored daily for 6 weeks.
Indirect ELISA
Leaf tissues (10 mg) from different systemic leaves of each plant challenged with WSMoV/TuMV recombinants were collected by punching six discs (0.5 cm in diameter) at 14/10 dpi. The accumulation of WSMoV or TuMV recombinants was assessed by indirect ELISA using the antiserum to the WSMoV N protein (Chu et al., 2001) or the antiserum to TuMV (Chen et al., 2003). Goat anti‐rabbit immunoglobulin G conjugated with alkaline phosphatase (KPL, Inc., Gaithersburg, MD, USA) was used as the secondary antibody, and p‐nitrophenyl phosphate (Sigma‐Aldrich Corporation, MO, USA) was used as the substrate for signal detection at 405 nm employing a VERSAmax Tunable Microplate Reader (Molecular Devices, Sunnyvale, CA, USA), 30 min after the addition of the substrate.
Construction of recombinant viruses of TuMV‐GLA, TuMV‐GLB1 and TuMV‐GLD containing amiRNA‐targeted sequences
TuMV‐GP69 in vivo infectious clone was generated from p35STuMV‐GFP comprising a 35S promoter and the full‐length cDNA of TuMV‐GFP (Lin et al., 2009). The 21‐nucleotide P69 targeting sequences of TuMV‐GP69 were replaced by amiR‐LA, amiR‐LB1 or amiR‐LD by two rounds of PCR using the primers listed in Table 1. The primer pairs GFPNcoI‐F/TuGFP‐LA‐R1, GFPNcoI‐F/TuGFP‐LB1‐R1 and GFPNcoI‐F/TuGFP‐LD‐R1 were used for the first PCR. The amplified DNA fragments were used as the templates for the second PCR with primer pairs GFPNcoI‐F/TuGFP‐LA‐R2, GFPNcoI‐F/TuGFP‐LB1‐R2 and GFPNcoI‐F/TuGFP‐LB1‐R2. The amplified products after NcoI/XhoI digestion were cloned into NcoI/XhoI‐digested TuMV‐GP69 to generate infectious clones TuMV‐GLA, TuMV‐GLB1 and TuMV‐GLD which contained amiR‐LA‐, amiR‐LB1‐ and amiR‐LD‐targeted sequences, respectively.
Challenge inoculation with recombinant viruses
To evaluate the efficiency of amiR‐LA‐, amiR‐LB1‐ and amiR‐LD‐mediated resistances to recombinant viruses TuMV‐GLA, TuMV‐GLB1 and TuMV‐GLD, virus challenge inoculation was performed as described by Lin et al. (2009) with minor modifications. All recombinant viruses were propagated from infectious DNAs (1 µg) mechanically applied onto the leaves of plants of Chenopodium quinoa Willd. At 7 dpi, viruses isolated from single lesions were transferred to the plants of the systemic host N. benthamiana for amplification. Nicotiana benthamiana leaves with infection symptoms (5 dpi) were used as the sources of inocula (20‐fold dilution in 0.01 m phosphate buffer, pH 7.0, v/w) for challenge inoculation of NT N. benthamiana plants and transgenic lines expressing amiR‐LA (22 and 27), amiR‐LB (16 and 32) or amiR‐LD (33 and 35). The experiments were repeated twice with 10 T1 plants/line. NT and transgenic lines were also challenged with TuMV‐GFP as controls. TuMV recombinant virus infections (10 dpi) were confirmed with TuMV antiserum (Chen et al., 2003) by ELISA.
ACKNOWLEDGEMENTS
This study was supported by the project NSC 97–2313‐B‐005–036‐MY3 from the National Science Council, Taiwan.
REFERENCES
- Ai, T. , Zhang, L. , Gao, Z. , Zhu, C.X. and Guo, X. (2011) Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol. 13, 304–316. [DOI] [PubMed] [Google Scholar]
- Alvarez, J.P. , Pekker, I. , Goldshmidt, A. , Blum, E. , Amsellem, Z. and Eshed, Y. (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell, 18, 1134–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman, M.J. and Sakai, H. (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2‐like target genes. Plant Cell, 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2005) RNA silencing. Trends Biochem. Sci. 30, 290–293. [DOI] [PubMed] [Google Scholar]
- Brodersen, P. , Sakvarelidze‐Achard, L. , Bruun‐Rasmussen, M. , Dunoyer, P. , Yamamoto, Y.Y. , Sieburth, L. and Voinnet, O. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science, 320, 1185–1190. [DOI] [PubMed] [Google Scholar]
- Bucher, E. , Lohuis, D. , van Poppel, P.M. , Geerts‐Dimitriadou, C. , Goldbach, R. and Prins, M. (2006) Multiple virus resistance at a high frequency using a single transgene construct. J. Gen. Virol. 87, 3697–3701. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. and Ambros, V. (2003) Role of microRNAs in plant and animal development. Science, 301, 336–338. [DOI] [PubMed] [Google Scholar]
- Chen, C.C. , Shy, J.F. and Yeh, S.D. (1990) Thrips transmission of Tomato spotted wilt virus from watermelon. Plant Prot. Bull. 32, 331–332. [Google Scholar]
- Chen, C.C. , Chao, C.H. , Chen, C.C. , Yeh, S.D. , Tsai, H.T. and Chang, C.A. (2003) Identification of Turnip mosaic virus isolates causing yellow stripe and spot on calla lily. Plant Dis. 87, 901–905. [DOI] [PubMed] [Google Scholar]
- Chen, C.C. , Chen, T.C. , Lin, Y.H. , Yeh, S.D. and Hsu, H.T. (2005) A chlorotic spot disease on calla lilies (Zantedeschia spp.) is caused by a tospovirus serologically but distantly related to Watermelon silver mottle virus . Plant Dis. 89, 440–445. [DOI] [PubMed] [Google Scholar]
- Chen, T.C. , Huang, C.W. , Kuo, Y.W. , Liu, F.L. , Yuan, C.H. , Hsu, H.T. and Yeh, S.D. (2006) Identification of common epitopes on a conserved region of NSs proteins among tospoviruses of Watermelon silver mottle virus serogroup. Phytopathology, 96, 1296–1304. [DOI] [PubMed] [Google Scholar]
- Chu, F.H. and Yeh, S.D. (1998) Comparison of ambisense mRNA of Watermelon silver mottle virus with other tospoviruses. Phytopathology, 88, 351–358. [DOI] [PubMed] [Google Scholar]
- Chu, F.H. , Chao, C.H. , Chung, M.H. , Chen, C.C. and Yeh, S.D. (2001) Completion of the genome sequence of Watermelon silver mottle virus and utilization of degenerate primers for detecting tospoviruses in five serogroups. Phytopathology, 91, 361–368. [DOI] [PubMed] [Google Scholar]
- Duan, C.G. , Wang, C.H. , Fang, R.X. and Guo, H.S. (2008) Artificial MicroRNAs highly accessible to targets confer efficient virus resistance in plants. J. Virol. 82, 11 084–11 095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauguet, C.M. , Mayo, M.A. , Maniloff, J. , Maniloof, J. , Desselberger, U. and Ball, L.A. (2005) Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses, p. 1259 London: Academic Press. [Google Scholar]
- Garcia, J.A. and Simon‐Mateo, C. (2006) A micropunch against plant viruses. Nat. Biotechnol. 24, 1358–1359. [DOI] [PubMed] [Google Scholar]
- Hsu, H.T. , Ueng, P.P. , Chu, F.H. , Ye, Z. and Yeh, S.D. (2000) Serological and molecular characterization of a high temperature‐recovered virus belonging to Tospovirus serogroup IV. J. Gen. Plant Pathol. 66, 167–175. [Google Scholar]
- Iwaki, M. , Honda, Y. , Hanada, K. and Tochihara, H. (1984) Silver mottle disease of watermelon caused by Tomato spotted wilt virus . Plant Dis. 68, 1006–1008. [Google Scholar]
- Jain, R.K. , Pappu, H.R. , Pappu, S.S. , Reddy, M.K. and Vani, A. (1998) Watermelon bud necrosis tospovirus is a distinct virus species belonging to serogroup IV. Arch. Virol. 143, 1637–1644. [DOI] [PubMed] [Google Scholar]
- Kalantidis, K. , Psaradakis, S. , Tabler, M. and Tsagris, M. (2002) The occurrence of CMV‐specific short RNAs in transgenic tobacco expressing virus‐derived double‐stranded RNA is indicative of resistance to the virus. Mol. Plant–Microbe Interact. 15, 826–833. [DOI] [PubMed] [Google Scholar]
- Khraiwesh, B. , Ossowski, S. , Weigel, D. , Reski, R. and Frank, W. (2008) Specific gene silencing by artificial MicroRNAs in Physcomitrella patens: an alternative to targeted gene knockouts. Plant Physiol. 148, 684–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova, A. , Reynolds, A. and Jayasena, S.D. (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell, 115, 209–216. [DOI] [PubMed] [Google Scholar]
- Kim, V.N. (2005) MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6, 376–385. [DOI] [PubMed] [Google Scholar]
- Lin, S.S. , Wu, H.W. , Elena, S.F. , Chen, K.C. , Niu, Q.W. , Yeh, S.D. , Chen, C.C. and Chua, N.H. (2009) Molecular evolution of a viral non‐coding sequence under the selective pressure of amiRNA‐mediated silencing. PLoS Pathog. 5, e1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C. , Kasschau, K.D. , Rector, M.A. and Carrington, J.C. (2002) Endogenous and silencing‐associated small RNAs in plants. Plant Cell, 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, K.Q. and Chang, D.C. (2004) The gene‐silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochem. Biophys. Res. Commun. 318, 303–310. [DOI] [PubMed] [Google Scholar]
- Molnar, A. , Bassett, A. , Thuenemann, E. , Schwach, F. , Karkare, S. , Ossowski, S. , Weigel, D. and Baulcombe, D. (2009) Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii . Plant J. 58, 165–174. [DOI] [PubMed] [Google Scholar]
- Niu, Q.W. , Lin, S.S. , Reyes, J.L. , Chen, K.C. , Wu, H.W. , Yeh, S.D. and Chua, N.H. (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24, 1420–1428. [DOI] [PubMed] [Google Scholar]
- Overhoff, M. , Alken, M. , Far, R.K. , Lemaitre, M. , Lebleu, B. , Sczakiel, G. and Robbins, I. (2005) Local RNA target structure influences siRNA efficacy: a systematic global analysis. J. Mol. Biol. 348, 871–881. [DOI] [PubMed] [Google Scholar]
- Park, W. , Zhai, J. and Lee, J.Y. (2009) Highly efficient gene silencing using perfect complementary artificial miRNA targeting AP1 or heteromeric artificial miRNA targeting AP1 and CAL genes. Plant Cell Rep. 28, 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins, M. , de Haan, P. , Luyten, R. , van Veller, M. , van Grinsven, M.Q. and Goldbach, R. (1995) Broad resistance to tospoviruses in transgenic tobacco plants expressing three tospoviral nucleoprotein gene sequences. Mol. Plant–Microbe Interact. 8, 85–91. [DOI] [PubMed] [Google Scholar]
- Prins, M. , Kikkert, M. , Ismayadi, C. , de Graauw, W. , de Haan, P. and Goldbach, R. (1997) Characterization of RNA‐mediated resistance to tomato spotted wilt virus in transgenic tobacco plants expressing NS(M) gene sequences. Plant Mol. Biol. 33, 235–243. [DOI] [PubMed] [Google Scholar]
- Qu, J. , Ye, J. and Fang, R. (2007) Artificial microRNA‐mediated virus resistance in plants. J. Virol. 81, 6690–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, C. , Schreier, P.H. and Uhrig, J.F. (2003) Peptide‐mediated broad‐spectrum plant resistance to tospoviruses. Proc. Natl. Acad. Sci. USA, 100, 4429–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana, T. , Mitchell, S.E. , Reddy, D.V. , Brown, S. , Kresovich, S. , Jarret, R. , Naidu, R.A. and Demski, J.W. (1996) Peanut bud necrosis tospovirus S RNA: complete nucleotide sequence, genome organization and homology to other tospoviruses. Arch. Virol. 141, 85–98. [DOI] [PubMed] [Google Scholar]
- Schubert, S. , Grunweller, A. , Erdmann, V.A. and Kurreck, J. (2005) Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions. J. Mol. Biol. 348, 883–893. [DOI] [PubMed] [Google Scholar]
- Schwab, R. , Ossowski, S. , Riester, M. , Warthmann, N. and Weigel, D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell, 18, 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach, F. , Adam, G. and Heinze, C. (2004) Expression of a modified nucleocapsid‐protein of Tomato spotted wilt virus (TSWV) confers resistance against TSWV and Groundnut ringspot virus (GRSV) by blocking systemic spread. Mol. Plant Pathol. 5, 309–316. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S. , Hutvagner, G. , Du, T. , Xu, Z. , Aronin, N. and Zamore, P.D. (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell, 115, 199–208. [DOI] [PubMed] [Google Scholar]
- Simon‐Mateo, C. and Garcia, J.A. (2006) MicroRNA‐guided processing impairs Plum pox virus replication, but the virus readily evolves to escape this silencing mechanism. J. Virol. 80, 2429–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H. , Vazquez, F. , Crete, P. and Bartel, D.P. (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warthmann, N. , Chen, H. , Ossowski, S. , Weigel, D. and Herve, P. (2008) Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE, 3, e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, S.D. , Lin, Y.C. , Cheng, Y.H. , Jih, C.L. , Chen, M.J. and Chen, C.C. (1992) Identification of tomato spotted wilt‐like virus infecting watermelon in Taiwan. Plant Dis. 76, 835–840. [Google Scholar]
- Yeh, S.D. , Sun, I.J. , Ho, H.M. and Chang, T.F. (1996) Molecular cloning and nucleotide sequence analysis of the S RNA of Watermelon silver mottle tospovirus . Acta Hortic. 431, 244–260. [Google Scholar]
- Zamore, P.D. and Haley, B. (2005) Ribo‐gnome: the big world of small RNAs. Science, 309, 1519–1524. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Li, H. , Zhang, J. , Zhang, C. , Gong, P. , Ziaf, K. , Xiao, F. and Ye, Z. (2011) Expression of artificial microRNAs in tomato confers efficient and stable virus resistance in a cell‐autonomous manner. Transgenic Res. 20, 569–581. [DOI] [PubMed] [Google Scholar]


