SUMMARY
The receptor‐like protein kinases (RLKs) constitute a large and diverse group of proteins controlling numerous plant physiological processes, including development, hormone perception and stress responses. The cysteine‐rich RLKs (CRKs) represent a prominent subfamily of transmembrane‐anchored RLKs. We have identified a putative barley (Hordeum vulgare) CRK gene family member, designated HvCRK1. The mature putative protein comprises 645 amino acids, and includes a putative receptor domain containing two characteristic ‘domain 26 of unknown function’ (duf26) domains in the N‐terminal region, followed by a rather short 17‐amino‐acid transmembrane domain, which includes an AAA motif, two features characteristic of endoplasmic reticulum (ER)‐targeted proteins and, finally, a characteristic putative protein kinase domain in the C‐terminus. The HvCRK1 transcript was isolated from leaves inoculated with the biotrophic fungal pathogen Blumeria graminis f.sp. hordei (Bgh). HvCRK1 transcripts were observed to accumulate transiently following Bgh inoculation of susceptible barley. Transient silencing of HvCRK1 expression in bombarded epidermal cells led to enhanced resistance to Bgh, but did not affect R‐gene‐mediated resistance. Silencing of HvCRK1 phenocopied the effective penetration resistance found in mlo‐resistant barley plants, and the possible link between HvCRK1 and MLO was substantiated by the fact that HvCRK1 induction on Bgh inoculation was dependent on Mlo. Finally, using both experimental and in silico approaches, we demonstrated that HvCRK1 localizes to the ER of barley cells. The negative effect on basal resistance against Bgh and the functional aspects of MLO‐ and ER‐localized HvCRK1 signalling on Bgh inoculation are discussed.
INTRODUCTION
Receptor‐like protein kinases (RLKs) constitute a diverse group of proteins involved in many plant signal transduction pathways regulating numerous plant physiological processes, including growth and development, hormone signalling, transpiration, symbiosis and pathogen defence (Dievart and Clark, 2004; Tichtinsky et al., 2003). A prominent subfamily of RLKs, known as cysteine‐rich RLKs (CRKs), are predicted to be transmembrane receptor proteins and are localized in the plasma membrane (Chen, 2001; Shiu and Bleecker, 2001). CRKs are characterized by the presence of one to four copies of domain 26 of unknown function (duf26), a C–X8–C–X2–C motif in the extracellular receptor region in the N‐terminus and a serine/threonine kinase domain in the C‐terminus (Chen, 2001; Shiu and Bleecker, 2001). The duf26 motifs are conserved domains spread throughout various protein families. According to EMBL‐EBI (http://www.ebi.ac.uk), there are 520 proteins in Viridiplantae (green plants) that possess one or more duf26 domains. Of these, 263 are found in rice (Oryza sativa) and 97 in Arabidopsis (Arabidopsis thaliana), including 46 CRKs (Wrzaczek et al., 2010). It has been suggested that the conserved cysteine residues in the duf26 motif direct the formation of three‐dimensional structures through disulphide bonds (Chen, 2001) and that C–X2–C could be a binding site for transition metal ions (Dykema et al., 1999). Both features may mediate protein–protein interactions important for the activation of CRKs. The expression of CRKs is induced by oxidative stress, pathogen attack and the application of salicylic acid (SA) (Du and Chen, 2000; Wrzaczek et al., 2010), and it has been shown that CRK4, CRK5, CRK13, CRK19 and CRK20 are involved in the regulation of cell death in Arabidopsis (Acharya et al., 2007; Chen et al., 2003, 2004). Furthermore, overexpression of CRK5 and CRK13 in Arabidopsis increases resistance against different pathovars of the bacterial pathogen Pseudomonas syringae (Acharya et al., 2007; Chen et al., 2003).
Plant defence responses are primarily controlled by the transcriptional activation of specific stress genes (Glazebrook, 2001; Singh et al., 2002). Several of these are induced in barley during attack of the biotrophic phytopathogenic fungus Blumeria graminis f.sp. hordei (Bgh), which causes powdery mildew (Collinge et al., 2002; Gregersen et al., 1997). Among these, a CRK gene transcript has also been reported to be induced after Bgh attack (Gregersen and Collinge, 2001). Interestingly, Bgh inoculated onto nonhost Arabidopsis induces the same expression profile of CRKs as both ozone and the bacterial microbe‐associated molecular pattern (MAMP) molecules flagellin (flg22) and hairpin Z (HrpZ) (Wrzaczek et al., 2010), indicating that CRKs in both Arabidopsis and barley play a central role in the mediation of the stress responses driven by apoplastic reactive oxygen species (ROS).
For successful infection, Bgh must overcome at least two levels of defence in barley: first, an early race‐nonspecific basal resistance (i.e. MAMP‐triggered immunity) and, second, a later race‐specific (R‐gene) resistance (i.e. effector‐triggered immunity) involving hypersensitive host cell death, which prevents the establishment of a biotrophic interaction. Both types of resistance involve the accumulation of ROS (Thordal‐Christensen et al., 1997). Basal resistance is exhibited in susceptible as well as R‐gene‐resistant barley cultivars and is associated with cell wall modifications, including the formation of papillae that form in the apoplast in order to prevent Bgh germlings from penetrating the epidermal cell walls (Zeyen et al., 2002). Papilla formation is complex and involves many cellular and biochemical processes, including the early generation of nitric oxide (Prats et al., 2005), hydrogen peroxide (H2O2) (Thordal‐Christensen et al., 1997) and phenolics (Carver et al., 1994), and localized deposition of callose below the attempted site of penetration (Zeyen et al., 2002). If the host plant possesses R genes recognizing the attacking avirulent Bgh isolate, a hypersensitive cell death response is initiated in successfully penetrated epidermal cells and the Bgh biotrophic life cycle is thereby interrupted. However, if the Bgh isolate is virulent, penetrated host epidermal cells survive and Bgh forms an intracellular haustorium that mediates nutrient uptake and supports ectophytic hyphal development.
In barley, recessive loss‐of‐function alleles of the Mlo locus are known to mediate durable and broad‐spectrum resistance towards Bgh as a result of highly effective penetration resistance (Jørgensen, 1994). Papilla formation is reported to occur earlier and is associated with higher levels of ROS and more rapid cross‐linking of phenolics in mlo‐resistant barley compared with susceptible wild‐type Mlo barley (Lyngkjaer et al., 2000). The functional Mlo gene encodes a seven‐transmembrane‐domain G‐coupled receptor protein (Buschges et al., 1997; Devoto et al., 1999). MLO appears to mediate susceptibility to Bgh via a Ca2+‐dependent interaction with calmodulin (Bhat et al., 2005; Kim et al., 2002), and it is considered that MLO is a target used by Bgh to gain entry to the host epidermal cell (Panstruga and Schulze‐Lefert, 2003).
In this article, we present results concerning the functional characterization of HvCRK1, the first CRK isolated from barley. We examined HvCRK1 expression in relation to Bgh attack on barley with or without mlo resistance and after treatment with H2O2 and SA. H2O2 treatment induced the accumulation of HvCRK1 transcripts and we demonstrated that transcript accumulation after Bgh inoculation was dependent on MLO. Furthermore, reduction of HvCRK1 expression by transient‐induced gene silencing (TIGS) resulted in enhanced resistance to Bgh, whereas transient overexpression had no effect on basal and R‐gene resistance. The cellular localization of HvCRK1 was examined and the protein was found to accumulate in the endoplasmic reticulum (ER), which was also supported by an in silico approach. Taken together, these data suggest that HvCRK1 is involved in the negative regulation of basal resistance.
RESULTS
Isolation of the HvCRK1 cDNA and phylogenetic relationship
Using the differential display of mRNA, we identified a transcript encoding a putative barley member of the CRK gene family which accumulated following Bgh inoculation of barley (Gregersen and Collinge, 2001). The full‐length cDNA of 2198 bp was obtained from a cDNA library of Bgh‐inoculated barley by rapid amplification of cDNA ends (RACE). The cDNA included a single 1935‐bp‐long open reading frame, representing a coding region of 645 amino acids (Fig. 1) (EMBL‐EBI Accession: FR717136). SMART analysis of the domain architecture of the deduced primary sequence identified a predicted extracellular region with a 22‐amino‐acid signal peptide and two cysteine‐rich duf26 (C–X8–C–X2–C and C–X10–C–X2–C) domains, a transmembrane region and an acid serine/threonine kinase domain (Fig. 1). The gene product was designated HvCRK1 as it represents the first report of a duf26‐related CRK from barley. The closest homologue in rice was the AK120072 gene which encodes a protein product with 58.3% identity and 71.6% similarity to HvCRK1 (EMBL‐EBI, http://www.ebi.ac.uk). To delineate the evolutionary relationship of HvCRK1 with other annotated plant CRKs, we performed a phylogenetic analysis using the conserved kinase region. RLK families can often be subdivided into clades based on the phylogenetic relationships between members, and genes encoding RLKs with related kinase sequences tend to have similar extracellular domains (Shiu and Bleecker, 2001; Shiu et al., 2004). For this purpose, we used SMART (Schultz et al., 2000) to define the kinase domain sequences of the entire genomic complement of duf26 subfamily RLKs from rice, poplar (Populus trichocarpa) and Arabidopsis. From the analysis, it is clear that the kinase domains cluster in distinct groups according to species, as only one small group contained members from rice, poplar and Arabidopsis. Accordingly, the protein kinase domain sequence in HvCRK1 also seems to be species distinct (Fig. 2).
Figure 1.
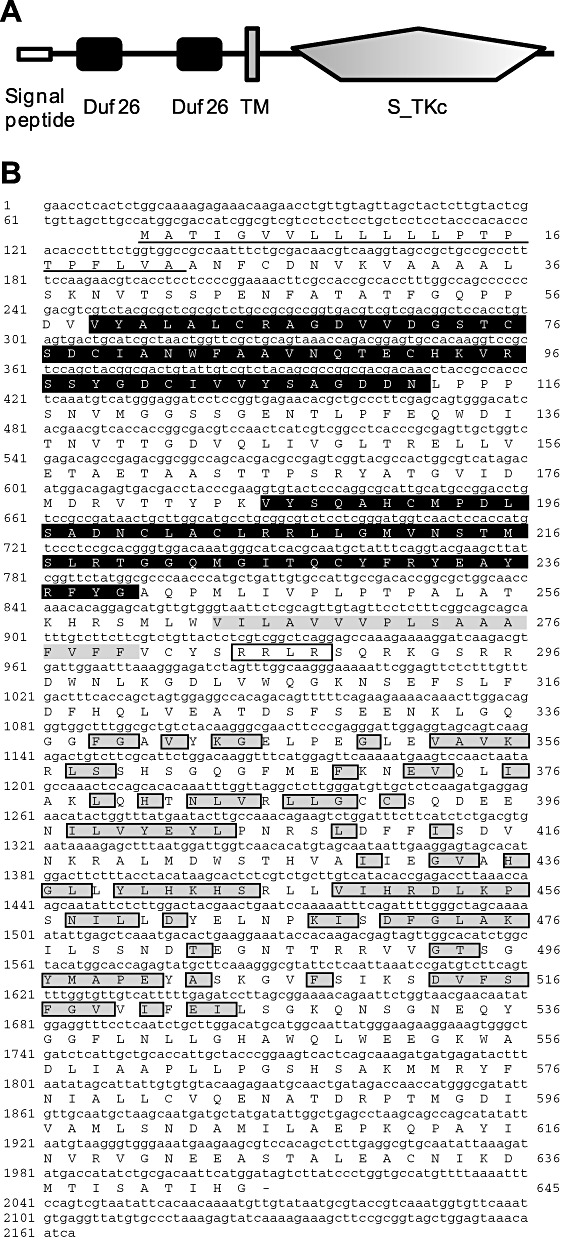
Domain architecture, deduced amino acid sequence and phylogenetic analysis of Hordeum vulgare cysteine‐rich receptor‐like protein kinase (HvCRK1). (A) Schematic diagram of HvCRK1 protein domain architecture showing the signal peptide and two characteristic domain 26 of unknown function (duf26) domains at the N‐terminus, followed by a transmembrane TM domain and a well‐conserved kinase domain. (B) Deduced amino acid sequence of HvCRK1, showing two duf26 domains (highlighted in black), TM domain (highlighted in grey) and the kinase domain (following the TMD). Conserved amino acids in the predicted serine–threonine kinase domain are highlighted in grey boxes (National Center for Biotechnology Information conserved domain search; http://www.ncbi.nlm.nih.gov).
Figure 2.
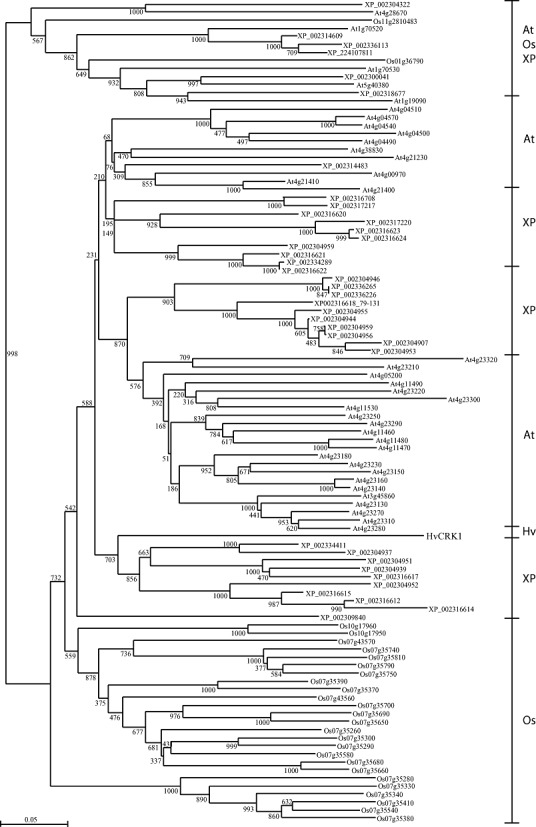
Phylogenetic tree of the predicted amino acid sequence of the kinase domain of Hordeum vulgare cysteine‐rich receptor‐like protein kinase (HvCRK1) and 101 other kinase domains of annotated receptor‐like protein kinase (RLK) genes from Arabidopsis thaliana (At), Hordeum vulgare (Hv), Oryza sativa (Os) and Populus trichocarpa (XP). The kinase domains cluster in species‐distinct groups, except for HvCRK1. These groups are highlighted to the right of the tree. Bootstrap values from 1000 replicates are indicated at each node. Scale bar indicates amino acid substitutions.
Silencing HvCRK1 increases basal resistance in barley towards Bgh
As signal perception in barley mediating the response to Bgh attack involves both early and late recognition, leading to either penetration‐based basal resistance or R‐gene‐mediated race‐specific resistance, involving programmed cell death (PCD), we examined whether the manipulation of HvCRK1 expression influenced basal resistance in compatible interactions and/or PCD in incompatible interactions mediated by Mla9 R‐gene resistance. This was achieved by RNAi gene silencing (TIGS) and overexpression. TIGS was performed using a 536‐bp fragment encoding the extracellular domain of HvCRK1. The HvCRK1 RNAi construct pIPKTA30_CRK1 was generated to produce spliced hairpin double‐stranded RNAs in vivo, and transformed by particle bombardment into barley epidermal cells together with a pUbiGUS reporter plasmid encoding β‐glucuronidase (GUS) (Douchkov et al., 2005). Three days after bombardment, leaf segments were inoculated with either a virulent or avirulent Bgh isolate, and transformed cells were screened for possible effects on the barley–Bgh interaction. In the compatible interaction, epidermal cells transformed with pIPKTA30_CRK1 exhibited a significant decrease in the infection index (Fig. 3A, P < 0.05), i.e. the cells showed increased basal penetration resistance compared with cells carrying the empty vector (pIPKTA30N). In contrast, no effect of the silencing of HvCRK1 was observed in the incompatible interaction, and all Bgh attack was either blocked at the penetration stage or terminated by PCD in the same way as in the empty vector control (Fig. 3A). As a positive control for effects on basal resistance, we included an RNAi construct for Mlo (pIPKTA36), as mutation of this gene provides host resistance to Bgh. In compatible interactions, pIPKTA36‐transformed cells showed a significant reduction (approximately 75%) (Fig. 3A, P < 0.05) in the infection index compared with cells transformed with the empty vector control, as reported previously (Douchkov et al., 2005). No effect of Mlo silencing was observed in the incompatible interaction as a result of mlo resistance or PCD. As a positive control for effects on PCD, we included an RNAi construct expressing the inverted repeat sequence of Mla6, which effectively silences the expression of both the Mla6 and Mla9 R genes because of their high sequence homology (Douchkov et al., 2005; Seeholzer et al., 2010). In the compatible interaction, there was no effect of Mla6 silencing (Fig. 3A), whereas, in the incompatible interaction, we found an increase in the infection of cells transformed with this RNAi construct, indicating successful suppression of R‐gene recognition and subsequent PCD. In an attempt to complement the RNAi studies, we performed overexpression of HvCRK1. The pIPKTA9_CRK1 construct for HvCRK1 overexpression was transformed into barley epidermal cells, together with the reporter plasmid encoding GUS (Douchkov et al., 2005). Four hours after bombardment, leaf segments were inoculated with either a virulent or avirulent Bgh isolate. However, cells transformed with the plasmid overexpressing HvCRK1 did not show any significant changes in infection index compared with leaves transformed with the empty vector (Fig. 3B), irrespective of whether the interaction was compatible or incompatible, indicating that the overexpression of HvCRK1 does not affect basal resistance and PCD. As a positive control for the effects of overexpression, we included the overexpression of a peroxidase gene (TaPrx103) that reduced Bgh infection after overexpression in a compatible interaction, as shown previously (Johrde and Schweizer, 2008). No effect of TaPrx203 overexpression was observed in the incompatible interaction mediated by Mla9 (Fig. 3B). In summary, the silencing of HvCRK1 increased basal resistance, but did not affect PCD, in barley after Bgh attack.
Figure 3.
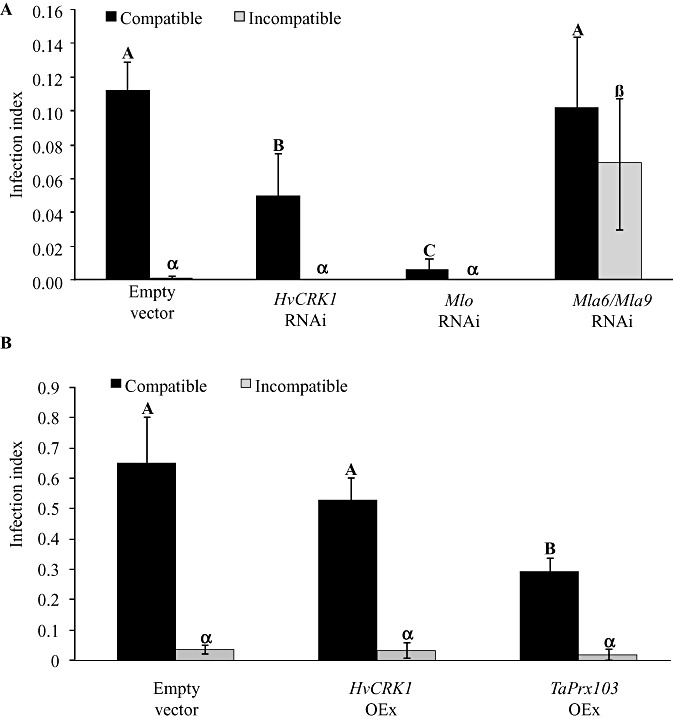
Effects of silencing (RNAi) or overexpression (OEx) of Hordeum vulgare cysteine‐rich receptor‐like protein kinase (HvCRK1) on barley responses to Blumeria graminis f.sp. hordei (Bgh) attack. Bgh infection is given as the infection index, which is the number of haustoria divided by the total number of transformed cells. (A) Bgh infection of barley cells transiently expressing empty vector as control (pIPKTA30N), the RNAi construct for HvCRK1 (pIPKTA30_CRK1), the RNAi construct for Mlo (pIPKTA36) or the RNAi construct for Mla6 and Mla9 (pIPKTA30_Mla6). In the compatible interaction, silencing of HvCRK1 resulted in reduced infection, when compared with empty vector or RNAi against Mla6/Mla9, which have no function in the compatible interaction. RNAi against Mlo gave the expected strong reduction in Bgh infection. In the incompatible interaction, there was no infection as a result of Mla9 R‐gene‐mediated programmed cell death (PCD) of Bgh‐infected cells, except for RNAi against Mla6/Mla9 which silenced the R gene and allowed Bgh infection. (B) Bgh infection of barley cells transiently overexpressing the empty vector as control (pIPKTA9), overexpression of HvCRK1 (pIPKTA9_CRK1) or overexpression of the peroxidase gene TaPrx103 (pWIR3). In the compatible interaction, overexpression of HvCRK1 had no effect on Bgh infection. Overexpression of TaPrx103 gave the expected reduction in Bgh infection. In the incompatible interaction, there was no infection as a result of Mla9 R‐gene‐mediated PCD of Bgh‐infected cells. The infection index is given as the mean ± standard error, and different letters or symbols within each interaction indicate significant differences (n= 5, P= 0.05).
HvCRK1 transcript accumulation in barley leaves after Bgh attack is transient and dependent on MLO
To investigate the spatiotemporal transcript accumulation of HvCRK1 in response to Bgh, we performed a detailed time‐course study using both epidermal cell layer and total leaf extracts of the susceptible barley line P01 following virulent Bgh inoculation. Transcript profiles of Bgh‐inoculated epidermal samples showed that HvCRK1 was strongly expressed, with approximately 50‐fold induction at 6 h after inoculation (hai) and 30‐fold induction at 12 hai, compared with uninoculated plants (Fig. 4A). In our studies, this time‐dependent induction of HvCRK1 expression coincides with the formation of the primary germ tubes by Bgh conidia (6 hai) and the time of Bgh penetration of attacked epidermal cells (12 hai) (data not shown). In total leaf samples (i.e. predominantly mesophyll cells which do not come into direct contact with this fungus), HvCRK1 expression also increased strongly at 6 hai, exhibiting approximately 25‐fold induction, followed by a later induction between 12 and 30 hai, with up to 16‐fold induction (Fig. 4B), compared with uninoculated control plants. The early and transient induction of HvCRK1 transcripts suggests a possible function for HvCRK1 in barley cells during the early recognition of and responses to Bgh.
Figure 4.
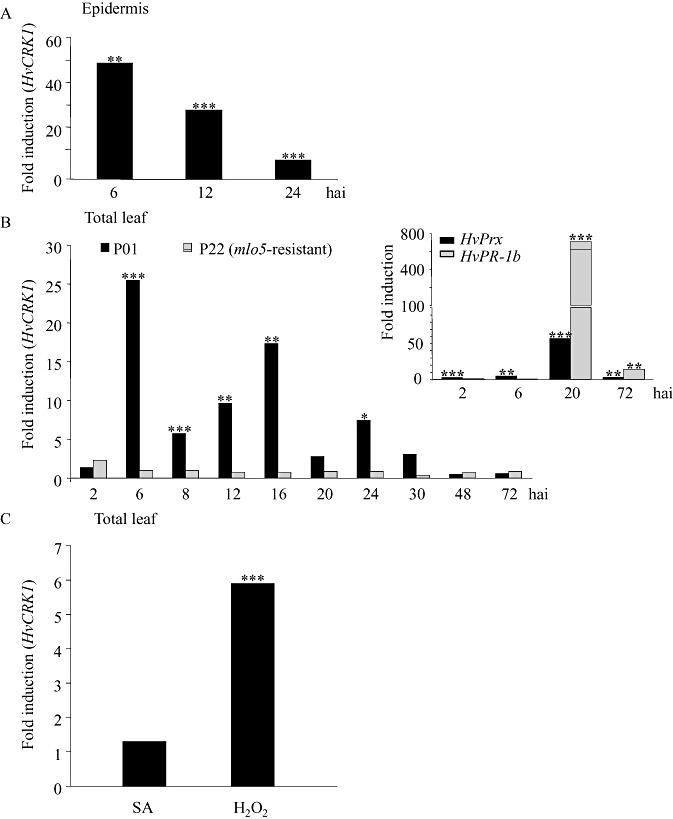
Hordeum vulgare cysteine‐rich receptor‐like protein kinase (HvCRK1) transcript accumulation in the epidermis and total leaves of barley in response to virulent Blumeria graminis f.sp. hordei (Bgh). (A) HvCRK1 transcript accumulation in abaxial epidermal tissue of the barley line P01 at 6, 12 and 24 h after inoculation (hai) of Bgh. (B) HvCRK1 transcript accumulation in total leaves of barley line P01 and the near‐isogenic mlo5‐resistant P22 barley line between 2 and 72 hai. Inset: transcript accumulation of peroxidase (HvPrx) and the pathogenesis‐related protein (HvPR‐1b) gene in the same P22 cDNA. (C) HvCRK1 transcript accumulation in response to 250 µm salicylic acid (SA) or 5 mm hydrogen peroxide (H2O2) 24 h after infiltration into leaves of P01. Values represent the fold induction in Bgh‐inoculated or SA/H2O2‐treated plants. Asterisks indicate significant differences in Bgh‐infected samples compared with uninfected epidermis or leaf samples at the respective time points (n= 5, ***P < 0.001, **P= 0.01–0.001, *P= 0.05–0.01).
Like the barley Mlo gene, HvCRK1 is responsive to Bgh and compromises penetration resistance. To investigate whether HvCRK1 transcript levels are dependent on MLO, we analysed HvCRK1 expression in total leaves in the mlo‐resistant barley line P22 containing the mutant mlo5 allele which is near‐isogenic to P01 (Kølster et al., 1986). Interestingly, the accumulation of the HvCRK1 transcript observed in wild‐type barley cultivar P01 after Bgh attack was completely absent in mlo5‐resistant P22 plants from 6 h onwards. From this time point, we did not observe any induction of HvCRK1 expression (Fig. 4B) in mlo plants. HvCRK1 expression levels in uninoculated P01 and P22 leaves showed no difference (data not shown), suggesting that basal levels of HvCRK1 were unaltered in uninoculated mlo5‐resistant P22. The low expression of HvCRK1 in P22 is unlikely to be caused by a cis‐allelic effect on the introgressed donor genome fragment (linkage drag) in this backcross line because, according to information from flow‐sorted barley chromosome arms (http://webblast.ipk‐gatersleben.de/barley/index.php), it mapped to chromosome 2HL, whereas the introgressed mlo mutation maps to chromosome 4HL.
To ascertain whether there was normal and expected accumulation of defence signalling components in our mlo5‐resistant P22/Bgh inoculation assay, we analysed the early Bgh‐responsive gene (peroxidase) and late Bgh‐induced gene (PR‐1b) (Gregersen et al., 1997) in the same P22 cDNA as above at 2, 6, 20 and 72 hai. The gene encoding peroxidase was induced significantly at all tested time points and PR‐1b was strongly induced at the 20 and 72 hai time points (Fig. 4B), confirming that there was the expected induction of defence signalling components. The differential accumulation patterns of HvCRK1 transcripts in Bgh‐attacked P01 and mlo5‐resistant P22 suggest that the induction of HvCRK1 expression after Bgh attack is dependent on MLO activity in barley.
Effects of SA and H2O2 on HvCRK1 transcript accumulation
As Arabidopsis CRKs are transcriptionally induced by oxidative stress and the application of SA (Wrzaczek et al., 2010), we tested how HvCRK1 transcripts accumulated in whole barley leaves 24 h after infiltration of 5 mm H2O2 or 250 µm SA. Transcripts of HvCRK1 accumulated strongly after H2O2 treatment, but not after SA treatment, suggesting that HvCRK1 is part of a signalling pathway responsive to oxidative stress, but not SA (Fig. 4C).
Subcellular localization of HvCRK1
To study the subcellular localization of HvCRK1 in vivo, we fused the open reading frame of HvCRK1 to the green fluorescent protein (GFP) gene (HvCRK1::GFP) and expressed it under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Using SMART (http://smart.embl‐heidelberg.de/) (Schultz et al., 2000) and PSORT (http://wolfpsort.org/) (Horton et al., 2007), we found that HvCRK1 encoded a potential transmembrane domain (Fig. 1) and had predicted plasma membrane localization. As a result of this prediction, we expected HvCRK1::GFP to accumulate around the plasma membrane. Initially, the fusion protein was transiently expressed in barley epidermal cells after the biolistic delivery of vector DNA, and was analysed by confocal microscopy. HvCRK1::GFP was found to localize in a finely polygonal structured network traversing the cytoplasm, similar to the ER (Fig. 5A; panels I and III). A local concentration of the ER network can be seen to align around the nucleus (Fig. 5A; panel II), in agreement with the actual orientation of this organelle in plant cells. GFP alone exhibited a characteristic diffused cytoplasmic fluorescent staining (Fig. 5A; panels IV and VI) and a dense fluorescent staining in the nucleus (Fig. 5A; panel V). We also included the co‐expression of HvCRK1::GFP and the ER marker YFP::HDEL (Runions et al., 2006), which supported the localization of ER (Fig. S1, see Supporting Information). Although HvCRK1 has a predicted plasma membrane localization, we did not observe any clear indication of the plasma membrane localization of HvCRK1::GFP. Because wounding and a limited number of transformed epidermal cells are major limitations when using biolistic delivery of plasmid DNA for localization studies, we also examined the subcellular localization of HvCRK1 using a more robust Agrobacterium‐mediated transient expression system in Nicotiana benthamiana. Consistent with our observations from barley, a clear cyan fluorescent protein (CFP) fluorescence from the HvCRK1::CFP construct used was observed in the finely polygonal structured ER (Fig. 5B; panel VII), and no clear plasma membrane localization was observed. The gene AHA2 (Liu et al., 2009), a plasma membrane‐associated transporter, was included as a plasma membrane localization control (Fig. S1). These results suggest that our HvCRK1 fusion proteins accumulate mainly in the ER.
Figure 5.
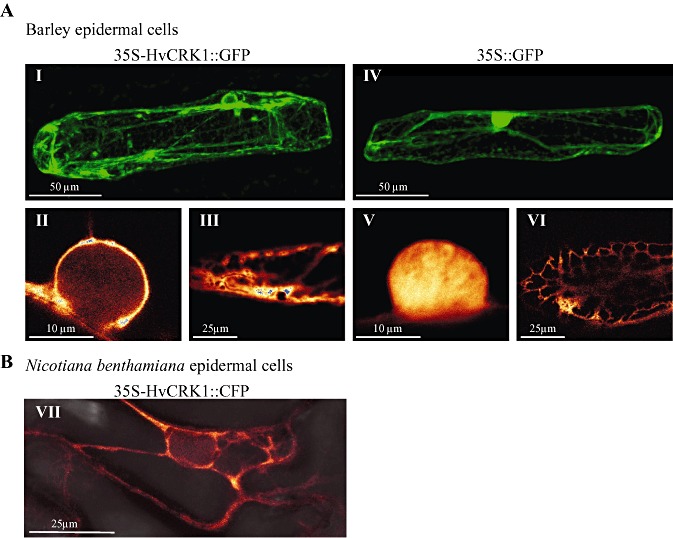
Cellular localization of Hordeum vulgare cysteine‐rich receptor‐like protein kinase (HvCRK1) is mainly associated with the endoplasmic reticulum (ER). (A) HvCRK1::GFP (left column) and green fluorescent protein (GFP) alone (right column), expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter, after biolistic delivery of vector DNA to barley epidermal cells and analysis for GFP fluorescence by confocal microscopy. Panels I and IV show fluorescent (green colour) overall morphology of cells; panels II and V show fluorescent (intensity colour) close‐up images of the nuclei; panels III and VI show fluorescent (intensity colour) images of the central part of transformed cells. HvCRK1::GFP is localized in a finely polygonal structured network traversing the cytoplasm, similar to the ER (I and III), and a local concentration of the network can be observed aligning around the nucleus, like the ER (II). GFP alone exhibited a characteristic diffuse cytoplasmic fluorescent staining (IV and VI) and a dense fluorescent staining in the nucleus (V). (B) Fluorescence (intensity colour) of HvCRK1::CFP Agrobacterium‐transformed Nicotiana benthamiana leaves. HvCRK1::CFP is also localized in a polygonal structured ER‐like network traversing the cytoplasm in N. benthamiana cells (VII). Photographs are maximum intensity images of z‐stack projections.
Arabidopsis AtCRK6 (At4g23140), showing a similar domain structure and sequence homology to HvCRK1 (39% identity and 54% similarity, EMBL‐EBI, http://www.ebi.ac.uk), was shown to be a plasma membrane‐localized protein when transiently expressed under a 35S promoter in N. benthamiana using Agrobacterium transformation (Thomas et al., 2008). However, these authors also showed that single amino acid changes in the transmembrane domain can move the localization from the plasma membrane to the ER (Thomas et al., 2008). Therefore, we compared the transmembrane domains of HvCRK1 and AtCRK6 to predict possible sequence variation that might explain the unexpected subcellular localization of HvCRK1 in N. benthamiana (Table 1). The transmembrane domain of HvCRK1 is predicted to consist of 17 amino acids, whereas AtCRK6 is predicted to comprise 23 amino acids. Five amino acids are identical and five belong to similar hydrophobic groups. The most distinct difference is at positions 17–19, with amino acids leucine–valine–glycine (LVG) in AtCRK6, accounting for more hydrophobicity (3.148), and alanine–alanine–alanine (AAA) in HvCRK1, accounting for less hydrophobicity (2.662). The length and low hydrophobicity of the HvCRK1 transmembrane domain support the observed ER localization (Brandizzi et al., 2002; Szczesna‐Skorupa and Kemper, 2000). ER localization is also supported by the presence of an ER retention signal in the HvCRK1 amino acid sequence. This –R–R–L–R– motif is placed adjacent to the transmembrane region (Fig. 1B).
Table 1.
Length and hydrohobicity of the transmembrane domain (TMD) in Hordeum vulgare cysteine‐rich receptor‐like protein kinase (HvCRK1) and the Arabidopsis homologue AtCRK6 (At4g23140).
| CRK proteins | TMD alignment | TMD length† | Hydrophobicity score‡ | Subcellular localization in Nicotiana benthamiana |
|---|---|---|---|---|
| AtCRK6 | VLVVAVVVLAVLLFIALVGYCFL | 23 | 3.148 | Plasma membrane |
| HvCRK1 | ‐‐‐‐‐‐VILAVVVPLSAAAFVFF *:***:: : *: | 17 | 2.662 | Endoplasmic reticulum |
* = identical amino acid, : = similar amino acid.
DISCUSSION
Barley HvCRK1 is a putative CRK. The closest homologue in rice is the gene AK120072 encoding a CRK protein with 58.3% identity and 71.6% similarity to HvCRK1 (EMBL‐EBI, http://www.ebi.ac.uk). In Arabidopsis, the closest homologues have about 30–35% identity and 50–60% similarity to HvCRK1 and, in general, the Arabidopsis CRKs are much more closely related to each other than to HvCRK1. In both rice and Arabidopsis, duf26‐containing CRKs constitute large families of genes, some of which are known to be induced in response to pathogens and defence‐inducing signals, such as SA and ROS (Dievart and Clark, 2004; Du and Chen, 2000; Shiu and Bleecker, 2001; Wrzaczek et al., 2010). Several of the Arabidopsis CRKs (CRK4, CRK5, CRK13, CRK19 and CRK20) have also been shown to be involved in the regulation of defence reactions and PCD against Pseudomonas syringae, and the overexpression of CRKs leads to increased PCD (Acharya et al., 2007; Chen et al., 2003, 2004).
HvCRK1 regulates basal resistance, but not R‐gene‐mediated PCD
Barley possesses two major types of defence against Bgh. Firstly, an early race‐nonspecific basal resistance which may block Bgh penetration and thereby prevent further fungal infection. However, if this fails and the barley line contains an effective race‐specific R gene, a hypersensitive response leading to PCD will be induced in the next layer of defence in the infected cells preventing the establishment of biotrophy and further fungal growth. The silencing and overexpression studies of HvCRK1 showed that this CRK negatively influenced the level of basal resistance, but did not influence R‐gene‐mediated PCD responses against Bgh, suggesting a role in defence different from that observed for the Arabidopsis CRKs 4, 5, 13, 19 and 20.
Basal resistance in barley against Bgh is characterized by the formation of cell wall appositions, known as papillae, that involve the deposition of callose (a β‐1,3‐glucan polymer) and phenolic compounds on the inner surface of the cell wall at the site of penetration (Zeyen et al., 2002). These physiological changes are mediated by a strong local H2O2 burst which is induced on fungal challenge (Thordal‐Christensen et al., 1997; Zeyen et al., 2002). In Arabidopsis, transcript accumulation of several CRKs is induced by ROS and as a nonhost response to Bgh (Wrzaczek et al., 2010), and, indeed, we observed a strong induction of HvCRK1 following both Bgh attack and the application of H2O2 (Fig. 4), suggesting that the induction of HvCRK1 in response to Bgh in barley could be dependent on H2O2. This is supported by a meta‐analysis of Arabidopsis CRK expression in relation to biotic and abiotic stresses (Wrzaczek et al., 2010), which showed that CRK3, CRK10, CRK11 and CRK19 are responsive to H2O2 and that CRK11, CRK12, CRK13, CRK20, CRK28, CRK29 and CRK36 are responsive to Bgh, indicating that at least the induction of CRK11 in Arabidopsis could be dependent on both Bgh and H2O2.
ROS and SA seem to be connected in a positive feedback loop that amplifies signals leading to defence responses and cell death in Arabidopsis (Overmyer et al., 2003). However, the accumulation of SA could not be detected in barley seedlings after Bgh inoculation in either compatible or incompatible interactions, indicating that defence responses against Bgh do not rely on or induce SA accumulation in barley (Huckelhoven et al., 1999). We also found that treatment with SA did not result in any noticeable changes in HvCRK1 transcript accumulation (Fig. 4).
Finally, we observed no enhanced HvCRK1 transcript accumulation in the mlo5 mutant on Bgh inoculation. The penetration resistance caused by the recessive barley mlo5 allele is physiologically similar to the nonhost penetration resistance in grasses (Zeyen et al., 2002), and it has been shown that one effect of MLO is to negatively regulate the cell wall‐restricted H2O2 burst that is induced by fungal penetration attempts, whereas, in the subtending mesophyll cells, it suppresses a second oxidative burst and cell death (Piffanelli et al., 2002). Although H2O2 can upregulate HvCRK1, the increased H2O2 observed in the fully resistant mlo5 genotype (Piffanelli et al., 2002) could not compensate for the lack of HvCRK1 transcript accumulation, suggesting that an MLO‐dependent factor(s) in addition to H2O2 is important for the upregulation of HvCRK1 transcript levels. More information about the HvCRK1–MLO signalling hierarchy may come from the investigation of HvCRK1 expression in barley lines mutated in the Ror1 and/or Ror2 genes that compromise mlo resistance and make these lines partially susceptible to Bgh. Interestingly, these mutations are also known to cause changes in the accumulation of H2O2 during Bgh attack in mlo‐resistant barley (Huckelhoven et al., 1999).
A reduction in the length and hydrophobicity of the transmembrane domain determines the localization of HvCRK1 to the ER
Results from the subcellular localization of HvCRK1 in vivo indicate that the HvCRK1::GFP fusion protein is targeted primarily to the ER (Fig. 5). However, to rule out any possible effects of wounding responses caused by the biolistic bombardment method in barley, we also investigated the localization of HvCRK1::CFP in tobacco (N. benthamiana) using an Agrobacterium‐mediated transformation system with the same result. Few duf26 proteins have been studied experimentally with respect to their subcellular localization. However, two duf26‐related CRKs, OsRMC (Jiang et al., 2007) and the HvCRK1 homologue, AtCRK6, have been reported to be localized in the plasma membrane (Thomas et al., 2008). Comparing the transmembrane domains of AtCRK6 and HvCRK1, we noticed that the domains differed in length and hydrophobicity (Table 1). It has been postulated that the main characteristic feature determining protein localization to the plasma membrane is the length of the hydrophobic transmembrane domain. As a result, it has been noted that longer and more hydrophobic transmembrane domains tend to be localized to the plasma membrane (Bretscher and Munro, 1993; Szczesna‐Skorupa and Kemper, 2000). Some studies have also indicated specific amino acid sequences in the transmembrane domain as key determinants of extracellular localization (Thomas et al., 2008). In agreement with these observations, it has been shown that the shortening of the transmembrane domain of ATCRK6 from 23 to 18 amino acids results in the retention of the protein in the ER as opposed to its original localization, i.e. plasmodesmata (Thomas et al., 2008). The same study reported that the substitution of the hydrophobic C‐terminal amino acids (LVL) in the domain of PDLP1a (a duf26‐related transmembrane protein) with less hydrophobic amino acids (AAA) resulted in the ER retention of PDLP1a. Based on these previous results, it is plausible that the less hydrophobic distal amino acids (AAA) in the putative transmembrane domain of HvCRK1 might represent specific signals that determine ER retention. In summary, a short transmembrane domain (17 amino acids) and the presence of less hydrophobic C‐terminal amino acids (AAA) may represent key characteristics of HvCRK1 responsible for its apparent ER localization. In addition to the amino acid sequence in the transmembrane domain, both luminal and membrane‐localized ER proteins have been predicted traditionally on the basis of classical C‐terminus‐located retention signals ([KH]DEL) (Teasdale and Jackson, 1996). However, other noncanonical arginine‐based signals directing proteins to the ER have been identified (Michelsen et al., 2007). These trafficking signals conform to the consensus –R–R–X–R, where –X– denotes a large neutral or positively charged amino acid, and have been found in both ion channels and G‐coupled receptors (Ren et al., 2005). Interestingly, analysis of the deduced amino acid sequence of HvCRK1 identified an –R–R–L–R– motif adjacent to the transmembrane region (Fig. 1B). Hence, the predicted subcellular localization using the transmembrane domain as a query is in agreement with our in vivo studies showing HvCRK1 to be localized in the ER. Future studies should focus on the negative defence regulation exerted by ER‐localized HvCRK1.
In summary, HvCRK1 is a putative CRK, which is probably ER‐associated and involved in the negative regulation of basal resistance in barley against Bgh. We argue that this regulation could be dependent on H2O2. However, this regulation occurs in the signalling pathway downstream from MLO function, as HvCRK1 transcripts do not accumulate after Bgh inoculation in mlo‐resistant barley. Overall, this suggests that HvCRK1 could be part of the basal defence regulation controlled by MLO and, through this, a possible direct or indirect target for Bgh‐derived effector proteins.
EXPERIMENTAL PROCEDURES
Plant material and fungal inoculations
The near‐isogenic barley (Hordeum vulgare) lines P01 (susceptible) and P22 (mlo5 resistant) (Kølster et al., 1986) and the cultivars Golden Promise (susceptible) and Roland (carrying Mla9‐mediated resistance) were grown in pots of compost soil in growth chambers (16 h light and approximately 20 °C) for 7 days before the experiments. The fungus Blumeria graminis f.sp. hordei (Bgh) race A6, virulent on P01, was used for gene expression and localization studies. The Bgh isolates 4.8 (avirulent on Mla9) and K1 (virulent on Mla9) were used for RNAi and overexpression experiments. All Bgh isolates were maintained on compatible barley lines by weekly transfer to fresh plants.
Differential display, RACE and cloning of HvCRK1
Differential display was performed as described previously (Jensen et al., 2007). RNA was isolated from abaxial epidermal tissue of barley leaves, 20 h after Bgh inoculation. The reverse transcriptase‐polymerase chain reaction (RT‐PCR) was performed with oligoT12CG and random hexamer primers. Amplification of 5′ and 3′ ends was carried out according to the SMART™ RACE kit (Clontech, Mountain View, CA, USA). Two 5′ cDNA clones were obtained using gene‐specific primers in combination with SMART II oligo primers, and one 3′ cDNA clone was obtained using 3′ CDS primers (SMART™ RACE kit) in combination with the gene‐specific primer. Differential display fragments and RACE products were all cloned into the pGEM‐T Easy vector (Promega, Madison, WI, USA) and sequenced. The overlapping sequences were combined to obtain the 2198‐bp sequence of HvCRK1, which included a single 1935‐bp‐long open reading frame, representing a coding region of 645 amino acids.
RNA extraction and cDNA synthesis
For transcript analyses, 20 abaxial epidermal peels, or the 7‐cm central part of five fully expanded seedling leaves, were prepared for each time point of both control and inoculated plants for each biological replicate. For the analysis of the effects of SA and H2O2, five fully matured seedling leaves of the barley line P01 were infiltrated with 250 µm SA or 5 mm H2O2 and sampled 24 h later. Total RNA was isolated using the RNA mini kit (Qiagen, Valencia, CA, USA) following the manufacturer's instructions. One microgram of purified DNaseI‐treated (Ambion, Austin, TX, USA) quality‐checked RNA was used for cDNA synthesis employing the iScript™ cDNA Synthesis Kit (Bio‐Rad, Hercules, CA, USA) according to the manufacturer's instructions.
Quantitative real‐time PCR analysis
Quantitative real‐time PCR was performed for studies of HvCRK1 expression using the primers RLK729 (5′‐CGCACGGGTGGACAAATG‐3′) and RLK825 (5′‐TCGGCAATGGCACAATCAGC‐3′). Ubiquitin conjugating enzyme 2 (UBC2, AY220735) was used for the normalization of variations in cDNA quantity employing the primers UBC423 (5′‐TCTCGTCCCTGACATTGCCAACAT‐3′) and UBC552 (5′‐TTTCTCGGGACAGCAACACAATCTTCT‐3′). The expression of peroxidase (X58396) and PR‐1b (X74940) was tested in P22 (mlo resistant) using the primers Prx299 (5′‐GGCATGGAACAAAACGCTAT‐3′) and Prx419 (5′‐TGAGTATGTCGGCACAGGAG‐3′), and PR1b158 (5′‐CCGCAGGACTACGTATCACC‐3′) and PR1b282 (5′‐TTGCAGTCGTTGATCCTCTG‐3′), respectively. Relative gene expression differences (R) and statistical significance levels for Bgh‐inoculated samples compared with uninoculated control samples were quantified using rest software (Pfaffl et al., 2002) and expressed as fold induction. C T values included in the analyses were based on five biological replicate measurements, with three technical replicates for each time point and treatment.
Constructs for transient expression studies
For RNAi, a 536‐bp extracellular domain/receptor domain starting at the 66th base pair of HvCRK1 CDS was PCR amplified and cloned into the pENTR™ TOPO vector using the pENTR™ Directional TOPO Cloning Kit (Invitrogen, Carlsbad, CA, USA). PCR was performed using high‐fidelity pfu polymerase (Promega). The RNAi fragment was cloned from the pENTR™ TOPO donor vector into the RNAi destination vector pIPKTA30N as inverted repeats by a single LR recombination reaction using the LR clonase and the Invitrogen Gateway system to generate pIPKTA30_CRK1 (Douchkov et al., 2005). Two other constructs, namely pIPKTA36 (an RNAi construct for silencing Mlo) and pIPKTA30_Mla6 (an RNAi construct for silencing Mla6 and Mla9) (Douchkov et al., 2005), were used as controls. As a result of the high sequence similarity between Mla6 and Mla9 (Seeholzer et al., 2010), pIPKTA30_Mla6 is expected to efficiently silence the expression of both genes. The overexpression construct pIPKTA9_CRK1 was obtained by subcloning a BamHI‐ApaI fragment of the full coding sequence of HvCRK1 under control of the CaMV 35S promoter into the pIPKTA9 vector for overexpression (Dong et al., 2006). The pWIR3 overexpression construct for the peroxidase gene TaPrx103 and empty pIPKTA9 vector (Johrde and Schweizer, 2008) were included as control. An HvCRK1 C‐terminal GFP fusion for studies of subcellular localization in barley was generated in the pIPKTA9 vector by an overlap extension strategy (Dong et al., 2006; Jensen et al., 2007). HvCRK1::CFP fusion proteins for the study of the subcellular localization in N. benthamiana were prepared by amplification of the open reading frame without the stop codon using primers Topo rlk FP (5′‐CACCATGGCGACCATCGGCGT‐3′) and Topo rlk ns RP (5′‐TCCATGAATTGTCGCAGATATGG‐3'). The PCR product was first cloned into the pENTR/D‐TOPO vector (Invitrogen). The resulting plasmids were combined with the destination vector pEarleygate 102 (Earley et al., 2006) in an LR reaction to generate the HvCRK1::CFP fusion construct.
Microprojectile bombardment and challenge inoculation
TIGS and transient overexpression were performed in 7‐day‐old susceptible barley plants of cultivar Golden Promise (mla9) or Roland (Mla9), as described previously (Douchkov et al., 2005; Ihlow et al., 2008). Briefly, leaf segments were bombarded by gold particles that had been coated with either one of our test constructs or one of the control constructs. In addition, pUbiGUS, containing the GUS gene under the control of the maize Ubiquitin promoter, was included and co‐transformed as a reporter for transformed epidermal cells (Schweizer et al., 1999). Leaves were bombarded using the PDS‐1000/He biolistic particle delivery system (Bio‐Rad, Munich, Germany). Leaf segments were inoculated with Bgh, 3 days after bombardment for the RNAi experiments and 4 h after bombardment for the overexpression experiments. After another 40 h of incubation, cells were stained for GUS activity. Leaves were screened for the infection of transformed cells using an automated analysis system (Ihlow et al., 2008). The fraction of successfully formed haustoria (infection index) in GUS‐stained cells was calculated as:
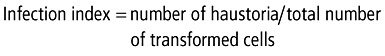 |
and the data were analysed for significance using the Wilcoxon rank‐sum test.
Subcellular localization
The pIPKTA9 plasmid (Dong et al., 2006) harbouring the CaMV 35S promoter was used to express GFP and HvCRK1::GFP transiently in barley epidermal cells, as described previously (Jensen et al., 2007). A total of 10 µg of each expression construct was coated onto 27.5 mg/mL 1‐µm gold particles and transferred into the cells, as described for transient gene silencing experiments. For the subcellular localization of HvCRK1::CFP in N. benthamiana, plants were transformed by the infiltration of Agrobacterium tumefaciens (Liu et al., 2009). Transformed cells were visualized using a Leica TCS SP2/MP confocal laser scanning microscope (Leica Microsystems, Mannheim, Germany).
Bioinformatic tools
Amino acid sequences from poplar, rice and Arabidopsis duf26‐containing RLKs were extracted from RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq/). The batch extraction largely overlapped with the annotation table from Lehti‐Shiu et al. (2009). Kinase domain sequences were extracted and aligned using SMART (http://smart.embl‐heidelberg.de/) (Schultz et al., 2000) and ClustalX (Thompson et al., 1997). A rooted phylogenetic tree was built using NJPlot software (Perriere and Gouy, 1996) using neighbour joining (gap open and gap extension penalties of 10 and 0.2, respectively) with 1000 bootstrap trials. Alignment of the deduced transmembrane domain of HvCRK1 with Arabidopsis CRK (AtCRK6) was carried out using a high‐performance multiple alignment program: Kalign2 (http://msa.sbc.su.se/cgi‐bin/msa.cgi) (Lassmann et al., 2009). PSORT (http://wolfpsort.org/) (Horton et al., 2007) was used to predict the subcellular localization of HvCRK1. The hydrophobicity index of the transmembrane domain was calculated using ProtScale and following the method of Kyte and Doolittle (1982) with a window size of 21 (http://expasy.org/tools/protscale.html).
Accession number
The isolated HvCRK1 cDNA clone described in this article has been deposited in the EMBL database with the following accession number: FR717136
Supporting information
Fig. S1 Cellular localization of Hordeum vulgare cysteine‐rich receptor‐like protein kinase (HvCRK1) is mainly associated with the endoplasmic reticulum (ER). (A) HvCRK1::GFP (top row) and the ER marker YFP::HDEL (Runions et al., 2006) (bottom row), co‐expressed under the control of the CaMV 35S promoter after biolistic delivery of vector DNA to barley epidermal cells, and then analysed for green fluorescent protein/yellow fluorescent protein (GFP/YFP) fluorescence by confocal microscopy. Panels I and III show fluorescent close‐up images of the edge of the transformed cell and panels II and IV show fluorescent images of the central part of the transformed cell. The fluorescence signal patterns appear to be identical, although YFP:HDEL gives a stronger signal than HvCRK1::GFP inside the nucleus. (B) HvCRK1::CFP and the plasma membrane‐associated transporter AtAHA2::CFP (Liu et al., 2009) in Agrobacterium‐transformed Nicotiana benthamiana leaves. Panels I and IV show fluorescent images, panels III and VI show light images and panels II and V show overlay images of the same N. benthamiana cells. HvCRK1::CFP is localized in a polygonal structured ER‐like network traversing the cytoplasm (arrows in II), whereas AtAHA2::CFP is only localized around the cell periphery.
Supporting info item
ACKNOWLEDGEMENTS
This work was supported by the Danish Council for Independent Research, Technology and Production Sciences (projects 23‐03‐0167 and 274‐07‐0173) and the Villum Foundation.
REFERENCES
- Acharya, B.R. , Raina, S. , Maqbool, S.B. , Jagadeeswaran, G. , Mosher, S.L. , Appel, H.M. , Schultz, J.C. , Klessig, D.F. and Raina, R. (2007) Overexpression of CRK13, an Arabidopsis cysteine‐rich receptor‐like kinase, results in enhanced resistance to Pseudomonas syringae . Plant J. 50, 488–499. [DOI] [PubMed] [Google Scholar]
- Bhat, R.A. , Miklis, M. , Schmelzer, E. , Schulze‐Lefert, P. and Panstruga, R. (2005) Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. USA, 102, 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi, F. , Frangne, N. , Marc‐Martin, S. , Hawes, C. , Neuhaus, J.M. and Paris, N. (2002) The destination for single‐pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell, 14, 1077–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher, M.S. and Munro, S. (1993) Cholesterol and the Golgi‐apparatus. Science, 261, 1280–1281. [DOI] [PubMed] [Google Scholar]
- Buschges, R. , Hollricher, K. , Panstruga, R. , Simons, G. , Wolter, M. , Frijters, A. , van Daelen, R. , van der Lee, T. , Diergaarde, P. , Groenendijk, J. , Topsch, S. , Vos, P. , Salamini, F. and Schulze‐Lefert, P. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell, 88, 695–705. [DOI] [PubMed] [Google Scholar]
- Carver, T.L.W. , Zeyen, R.J. , Robbins, M.P. , Vance, C.P. and Boyles, D.A. (1994) Suppression of host cinnamyl alcohol‐dehydrogenase and phenylalanine ammonia‐lyase increases oat epidermal‐cell susceptibility to powdery mildew penetration. Physiol. Mol. Plant Pathol. 44, 243–259. [Google Scholar]
- Chen, K.G. , Du, L.Q. and Chen, Z.X. (2003) Sensitization of defense responses and activation of programmed cell death by a pathogen‐induced receptor‐like protein kinase in Arabidopsis. Plant Mol. Biol. 53, 61–74. [DOI] [PubMed] [Google Scholar]
- Chen, K.G. , Fan, B.F. , Du, L.Q. and Chen, Z.X. (2004) Activation of hypersensitive cell death by pathogen‐induced receptor‐like protein kinases from Arabidopsis. Plant Mol. Biol. 56, 271–283. [DOI] [PubMed] [Google Scholar]
- Chen, Z.X. (2001) A superfamily of proteins with novel cysteine‐rich repeats. Plant Physiol. 126, 473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge, D.B. , Gregersen, P.L. and Thordal‐Christensen, H. (2002) The nature and role of defence response genes in cereals In: The Powdery Mildews: A Comprehensive Treatise (Belanger R.R., Bushnell W.R., Dik A.J. and Carver T.L.W., eds), pp. 146–160. St. Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Devoto, A. , Piffanelli, P. , Nilsson, I. , Wallin, E. , Panstruga, R. , von Heijne, G. and Schulze‐Lefert, P. (1999) Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J. Biol. Chem. 274, 34 993–35 004. [DOI] [PubMed] [Google Scholar]
- Dievart, A. and Clark, S.E. (2004) LRR‐containing receptors regulating plant development and defense. Development, 131, 251–261. [DOI] [PubMed] [Google Scholar]
- Dong, W.B. , Nowara, D. and Schweizer, P. (2006) Protein polyubiquitination plays a role in basal host resistance of barley. Plant Cell, 18, 3321–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchkov, D. , Nowara, D. , Zierold, U. and Schweizer, P. (2005) A high‐throughput gene‐silencing system for the functional assessment of defense‐related genes in barley epidermal cells. Mol. Plant–Microbe Interact. 18, 755–761. [DOI] [PubMed] [Google Scholar]
- Du, L.Q. and Chen, Z.X. (2000) Identification of genes encoding receptor‐like protein kinases as possible targets of pathogen‐ and salicylic acid‐induced WRKY DNA‐binding proteins in Arabidopsis. Plant J. 24, 837–847. [DOI] [PubMed] [Google Scholar]
- Dykema, P.E. , Sipes, P.R. , Marie, A. , Biermann, B.J. , Crowell, D.N. and Randall, S.K. (1999) A new class of proteins capable of binding transition metals. Plant Mol. Biol. 41, 139–150. [DOI] [PubMed] [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K.M. and Pikaard, C.S. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2001) Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr. Opin. Plant Biol. 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Gregersen, P.L. and Collinge, D.B. (2001) Penetration attempts by the powdery mildew fungus into barley leaves are accompanied by increased gene transcript accumulation in the epidermal cell layer. In 5th Congress of the European Foundation for Plant Pathology. J. Plant Pathol. 82, 257–260. Edizioni/ETS. [Google Scholar]
- Gregersen, P.L. , Thordal‐Christensen, H. , Forster, H. and Collinge, D.B. (1997) Differential gene transcript accumulation in barley leaf epidermis and mesophyll in response to attack by Blumeria graminis f.sp. hordei (syn. Erysiphe graminis f.sp. hordei). Physiol. Mol. Plant Pathol. 51, 85–97. [Google Scholar]
- Horton, P. , Park, K.J. , Obayashi, T. , Fujita, N. , Harada, H. , Adams‐Collier, C.J. and Nakai, K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckelhoven, R. , Fodor, J. , Preis, C. and Kogel, K.H. (1999) Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol. 119, 1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihlow, A. , Schweizer, P. and Seiffert, U. (2008) A high‐throughput screening system for barley/powdery mildew interactions based on automated analysis of light micrographs. BMC Plant Biol. 8, Article 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, M.K. , Rung, J.H. , Gregersen, P.L. , Gjetting, T. , Fuglsang, A.T. , Hansen, M. , Joehnk, N. , Lyngkjaer, M.F. and Collinge, D.B. (2007) The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol. Biol. 65, 137–150. [DOI] [PubMed] [Google Scholar]
- Jiang, J.F. , Li, J.H. , Xu, Y.Y. , Han, Y. , Bai, Y. , Zhou, G.X. , Lou, Y.G. , Xu, Z.H. and Chong, K. (2007) RNAi knockdown of Oryza sativa root meander curling gene led to altered root development and coiling which were mediated by jasmonic acid signalling in rice. Plant Cell Environ. 30, 690–699. [DOI] [PubMed] [Google Scholar]
- Johrde, A. and Schweizer, P. (2008) A class III peroxidase specifically expressed in pathogen‐attacked barley epidermis contributes to basal resistance. Mol. Plant Pathol. 9, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen, J.H. (1994) Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 13, 97–119. [Google Scholar]
- Kim, M.C. , Panstruga, R. , Elliott, C. , Muller, J. , Devoto, A. , Yoon, H.W. , Park, H.C. , Cho, M.J. and Schulze‐Lefert, P. (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature, 416, 447–450. [DOI] [PubMed] [Google Scholar]
- Kølster, P. , Munk, L. , Stølen, O. and Løhde, J. (1986) Near‐isogenic barley lines with genes for resistance to powdery mildew. Crop Sci. 26, 903–907. [Google Scholar]
- Kyte, J. and Doolittle, R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Lassmann, T. , Frings, O. and Sonnhammer, E.L.L. (2009) Kalign2: high‐performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 37, 858–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti‐Shiu, M.D. , Zou, C. , Hanada, K. and Shiu, S.H. (2009) Evolutionary history and stress regulation of plant receptor‐like kinase/Pelle genes. Plant Physiol. 150, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Elmore, J.M. , Fuglsang, A.T. , Palmgren, M.G. , Staskawicz, B.J. and Coaker, G. (2009) RIN4 functions with plasma membrane H+‐ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 7, e100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngkjaer, M.F. , Newton, A.C. , Atzema, J.L. and Baker, S.J. (2000) The barley mlo‐gene: an important powdery mildew resistance source. Agronomie, 20, 745–756. [Google Scholar]
- Michelsen, K. , Schmid, V. , Metz, J. , Heusser, K. , Liebel, U. , Schwede, T. , Spang, A. and Schwappach, B. (2007) Novel cargo‐binding site in the beta and delta subunits of coatomer. J. Cell Biol. 179, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer, K. , Brosche, M. and Kangasjarvi, J. (2003) Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 8, 335–342. [DOI] [PubMed] [Google Scholar]
- Panstruga, R. and Schulze‐Lefert, P. (2003) Corruption of host seven‐transmembrane proteins by pathogenic microbes: a common theme in animals and plants? Microb. Infect. 5, 429–437. [DOI] [PubMed] [Google Scholar]
- Perriere, G. and Gouy, M. (1996) WWW‐query: an on‐line retrieval system for biological sequence banks. Biochimie, 78, 364–369. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. , Horgan, G.W. and Dempfle, L. (2002) Relative expression software tool (REST (c)) for group‐wise comparison and statistical analysis of relative expression results in real‐time PCR. Nucleic Acids Res. 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli, P. , Zhou, F.S. , Casais, C. , Orme, J. , Jarosch, B. , Schaffrath, U. , Collins, N.C. , Panstruga, R. and Schulze‐Lefert, P. (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 129, 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats, E. , Mur, L.A.J. , Sanderson, R. and Carver, T.L.W. (2005) Nitric oxide contributes both to papilla‐based resistance and the hypersensitive response in barley attacked by Blumeria graminis f. sp hordei . Mol. Plant Pathol. 6, 65–78. [DOI] [PubMed] [Google Scholar]
- Ren, X.R. , Reiter, E. , Ahn, S. , Kim, J. , Chen, W. and Lefkowitz, R.J. (2005) Different G protein‐coupled receptor kinases govern G protein and beta‐arrestin‐mediated signaling of V2 vasopressin receptor. Proc. Natl. Acad. Sci. USA, 102, 1448–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runions, J. , Brach, T. , Kuhner, S. and Hawes, C. (2006) Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J. Exp. Bot. 57, 43–50. [DOI] [PubMed] [Google Scholar]
- Schultz, J. , Copley, R.R. , Doerks, T. , Ponting, C.P. and Bork, P. (2000) SMART: a web‐based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, P. , Pokorny, J. , Abderhalden, O. and Dudler, R. (1999) A transient assay system for the functional assessment of defense‐related genes in wheat. Mol. Plant–Microbe Interact. 12, 647–654. [Google Scholar]
- Seeholzer, S. , Tsuchimatsu, T. , Jordan, T. , Bieri, S. , Pajonk, S. , Yang, W.X. , Jahoor, A. , Shimizu, K.K. , Keller, B. and Schulze‐Lefert, P. (2010) Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol. Plant–Microbe Interact. 23, 497–509. [DOI] [PubMed] [Google Scholar]
- Shiu, S. , Karlowski, W.M. , Mayer, K.F.X. , Pan, R. , Tzeng, Y. and Li, W. (2004) Comparative analysis of the receptor‐like kinase family in Arabidopsis thaliana and rice. Plant Cell, 2004, 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H. and Bleecker, A.B. (2001) Receptor‐like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA, 98, 10 763–10 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, K.B. , Foley, R.C. and Onate‐Sanchez, L. (2002) Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436. [DOI] [PubMed] [Google Scholar]
- Szczesna‐Skorupa, E. and Kemper, B. (2000) Endoplasmic reticulum retention determinants in the transmembrane and linker domains of cytochrome P4502C1. J. Biol. Chem. 275, 19 409–19 415. [DOI] [PubMed] [Google Scholar]
- Teasdale, R.D. and Jackson, M.R. (1996) Signal‐mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 12, 27–54. [DOI] [PubMed] [Google Scholar]
- Thomas, C.L. , Bayer, E.M. , Ritzenthaler, C. , Fernandez‐Calvino, L. and Maule, A.J. (2008) Specific targeting of a plasmodesmal protein affecting cell‐to‐cell communication. PLoS Biol. 6, 180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D. , Gibson, T.J. , Plewniak, F. , Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z.G. , Wei, Y.D. and Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Tichtinsky, G. , Vanoosthuyse, V. , Cock, J.M. and Gaude, T. (2003) Making inroads into plant receptor kinase signalling pathways. Trends Plant Sci. 8, 231–237. [DOI] [PubMed] [Google Scholar]
- Wrzaczek, M. , Brosche, M. , Salojarvi, J. , Kangasjarvi, S. , Idanheimo, N. , Mersmann, S. , Robatzek, S. , Karpinski, S. , Karpinska, B. and Kangasjarvi, J. (2010) Transcriptional regulation of the CRK/DUF26 group of receptor‐like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 10(95), 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyen, R.J. , Carver, T.L.W. and Lyngkjaer, M.F. (2002) Epidermal cell papillae In: The Powdery Mildews: A Comprehensive Treatise (Belanger R.R., Bushnell W.R., Dik A.J. and Carver T.L.W., eds), pp. 107–125. St. Paul, MN: American Phytopathological Society Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Cellular localization of Hordeum vulgare cysteine‐rich receptor‐like protein kinase (HvCRK1) is mainly associated with the endoplasmic reticulum (ER). (A) HvCRK1::GFP (top row) and the ER marker YFP::HDEL (Runions et al., 2006) (bottom row), co‐expressed under the control of the CaMV 35S promoter after biolistic delivery of vector DNA to barley epidermal cells, and then analysed for green fluorescent protein/yellow fluorescent protein (GFP/YFP) fluorescence by confocal microscopy. Panels I and III show fluorescent close‐up images of the edge of the transformed cell and panels II and IV show fluorescent images of the central part of the transformed cell. The fluorescence signal patterns appear to be identical, although YFP:HDEL gives a stronger signal than HvCRK1::GFP inside the nucleus. (B) HvCRK1::CFP and the plasma membrane‐associated transporter AtAHA2::CFP (Liu et al., 2009) in Agrobacterium‐transformed Nicotiana benthamiana leaves. Panels I and IV show fluorescent images, panels III and VI show light images and panels II and V show overlay images of the same N. benthamiana cells. HvCRK1::CFP is localized in a polygonal structured ER‐like network traversing the cytoplasm (arrows in II), whereas AtAHA2::CFP is only localized around the cell periphery.
Supporting info item


