Summary
Background
Xanthomonas campestris pv. campestris (Xcc) (Pammel) Dowson is a Gram‐negative bacterium that causes black rot, the most important disease of vegetable brassica crops worldwide. Intensive molecular investigation of Xcc is gaining momentum and several whole genome sequences are available.
Taxonomy
Bacteria; Phylum Proteobacteria; Class Gammaproteobacteria; Order Xanthomonadales; Family Xanthomonadacea; Genus Xanthomonas; Species X. campestris.
Host range and symptoms
Xcc can cause disease in a large number of species of Brassicaceae (ex‐Cruciferae), including economically important vegetable Brassica crops and a number of other cruciferous crops, ornamentals and weeds, including the model plant Arabidopsis thaliana. Black rot is a systemic vascular disease. Typical disease symptoms include V‐shaped yellow lesions starting from the leaf margins and blackening of the veins.
Race structure, pathogenesis and epidemiology
Collections of Xcc isolates have been differentiated into physiological races based on the response of several brassica species lines. Black rot is a seed‐borne disease. The disease is favoured by warm, humid conditions and can spread rapidly from rain dispersal and irrigation water.
Disease control
The control of black rot is difficult and relies on the use of pathogen‐free planting material and the elimination of other potential inoculum sources (infected crop debris and cruciferous weeds). Major gene resistance is very rare in B. oleracea (brassica C genome). Resistance is more readily available in other species, including potentially useful sources of broad‐spectrum resistance in B. rapa and B. carinata (A and BC genomes, respectively) and in the wild relative A. thaliana.
Genome
The reference genomes of three isolates have been released. The genome consists of a single chromosome of approximately 5 100 000 bp, with a GC content of approximately 65% and an average predicted number of coding DNA sequences (CDS) of 4308.
Important genes identified
Three different secretion systems have been identified and studied in Xcc. The gene clusters xps and xcs encode a type II secretion system and xps genes have been linked to pathogenicity. The role of the type IV secretion system in pathogenicity is still uncertain. The hrp gene cluster encodes a type III secretion system that is associated with pathogenicity. An inventory of candidate effector genes has been assembled based on homology with known effectors. A range of other genes have been associated with virulence and pathogenicity, including the rpf, gum and wxc genes involved in the regulation of the synthesis of extracellular degrading enzymes, xanthan gum and lipopolysaccharides.
Useful website
Introduction
The genus Xanthomonas includes economically important pathogenic bacteria that are generally associated with plants (Hayward, 1993; Vauterin et al., 1990). The taxonomy of this genus was initially determined according to host preference (typically the host of origin) and, consequently, a large number of species and pathovars have been defined (Burkholder, 1957). Morphological and other physiological and biochemical characters were subsequently used to classify the Xanthomonas isolates into eight phenotypic groups (Van Den Mooter and Swings, 1990). The Xanthomonas species were later reclassified on the basis of DNA–DNA hybridization, leading to X. campestris being restricted to comprise only the vascular pathogen X. campestris pv. campestris (Pammel) Dowson (Xcc), which causes black rot of brassica species, and additional pathovars that cause vascular or leaf spot diseases in cruciferous hosts, including X. campestris pv. aberrans (Knösel) Dye, armoraciae (McCullock) Dye, barbareae (Burkholder) Dye, incanae (Kendrick & Baker) Dye and raphani (White) Dye (Vauterin et al., 1995).
Debate continues with regard to what constitutes different pathovars. For example, some authors, such as Alvarez et al. (1994), have considered that X. campestris pv. raphani, a pathovar originally described by White (1930), which has a broad range of hosts within the Brassicaceae and Solanaceae, and X. campestris pv. armoraciae, described one year earlier by McCulloch (1929) as a leaf spot disease of horse radish, are synonymous. Other authors, such as Tamura et al. (1994) and Vicente et al. (2006), have considered them to be distinct pathovars with a different host range. Other X. campestris pathovars have received less attention. Some of these pathovars, such as X. campestris pv. aberrans, may not be distinct from Xcc (Fargier and Manceau, 2007; Fargier et al., 2011; Vicente et al., 2001). Fargier and Manceau (2007) considered that the species can be restricted to three pathovars (campestris, raphani and incanae), but some isolates from ornamental crucifers, which are currently identified as pv. campestris or incanae, may still belong to distinct pathovars (Vicente et al., 2006).
The Disease
Black rot was first described by Garman (1894) as a disease of cabbage in Kentucky, USA. He isolated two types of bacteria from diseased plants, but could not determine which type of bacterium was causing the disease. In Iowa, USA, Pammel (1895a, b) observed a similar disease in rutabaga and turnip, and showed that the disease was caused by a bacterium (named Bacillus campestris) with yellow pigmented colonies in culture. Reports from Wisconsin also attributed the disease of turnips and cabbage to the yellow bacterium (Russell, 1898; Smith, 1898). Since then, the disease has been identified in all continents wherever Brassicaceae crops are grown (Bradbury, 1986), and is considered to be the most important disease of vegetable brassica crops worldwide (Williams, 1980).
Brassica oleracea (including cabbage, cauliflower, broccoli, Brussels sprouts and kale) is economically the most important host of Xcc. However, the disease also occurs in other brassica crops, radish, ornamental crucifers and related weed species (Bradbury, 1986). Some accessions of Arabidopsis thaliana, the model plant for molecular plant research, are also susceptible when inoculated with Xcc.
Life Cycle
Black rot is primarily a seed‐borne disease (Cook et al., 1952). However, the disease can also be transmitted in infected transplants, infested soil, crop residues and carry‐over in related weed species (Schaad and Alvarez, 1993; Walker, 1953). Schaad and White (1974) and Dane and Shaw (1996) showed that Xcc can survive in the soil, independent from the host, for approximately 40 days in winter and 20 days in summer. The results of Arias et al. (2000) showed that high soil matric potential (saturated soils) can reduce the survival of the pathogen. The pathogen can survive longer in soil within plant tissues than as free living cells. Kocks and Zadoks (1996) showed that crop residues in fresh (2 weeks) refuse piles are more effective in spreading the disease than older (4 months) piles. In some conditions, cruciferous weeds can survive all year round and can provide potential carry‐over inoculum for the crops (Schaad and Dianese, 1981). Arias et al. (2000) showed that epiphytic survival of the bacteria on the phylloplane is dependent on the plant species, as bacteria survived for 48 days on cabbage, mustard and lettuce, but only for 9 days on rice. In some cases, infected crops have also been shown to provide inoculum for the weeds (Dane and Shaw, 1996), and one study has indicated that weeds do not play an important role in the dissemination of black rot (Schaad and Thaveechai, 1983).
The bacteria can disperse over short distances via wind, insects, aerosols, irrigation water, rain, farm equipment and workers. Commercial vegetable brassica crops are raised from transplants. In plant nurseries that produce module‐raised transplants, the overhead irrigation system can increase significantly the dissemination of the bacteria, and can subsequently lead to a high level of disease in the field; changing the irrigation method can therefore limit the spread of the disease (Roberts et al., 2007).
The bacteria generally enter the plant through hydathodes on the leaf margins, when droplets of guttation contaminated with bacteria are reabsorbed into the leaf (Russell, 1898). This mode of entry is dependent on a combination of environmental, biological and mechanical factors (Meier, 1934). In contrast, stomata generally do not appear to be important for Xcc infection, because the disease generally does not spread into surrounding tissues, even though the bacteria can enter the plant through the stomata and produce small dark spots (Cook et al., 1952). This suggests that vascular movement of bacteria is essential for disease development. The bacteria can also enter the plant through wounds caused by machinery, insects, animals, rain, irrigation and wind.
The typical symptom of black rot is the formation of V‐shaped, chlorotic yellow lesions with vertices towards the middle vein of the leaves (Fig. 1a,b) and darkened veins that result from bacterial movement in the vascular system. The affected tissues can become necrotic, and leaves can fall prematurely; systemic infections can cause stunted growth (Fig. 1c) and the death of young plants. Secondary infection by other bacterial species can also contribute to further development of severe rotting of vegetable tissue. The infection is often latent when temperatures are low, as the bacteria can persist in the vascular system without producing symptoms and, when the temperature rises, the typical symptoms become evident (Cook et al., 1952; Schaad, 1982; Walker, 1953).
Figure 1.
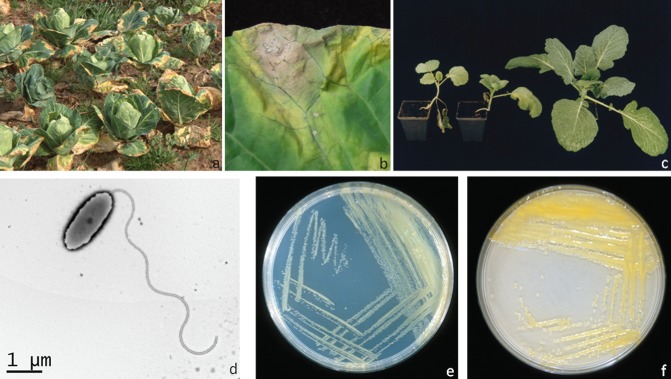
(a) Symptoms of black rot on a cabbage field. (b) Typical black rot V‐shaped lesion on a cabbage leaf. (c) Two plants of Savoy cabbage with symptoms of systemic infection following inoculation of Xanthomonas campestris pv. campestris, and a healthy control plant. (d) Electron microscopy image of a X. campestris pv. campestris rod‐shaped cell showing a single polar flagellum. (e) Xanthomonas campestris pv. campestris culture growing on King's medium B. (f) Xanthomonas campestris pv. campestris culture growing on Yeast Dextrose Calcium Carbonate medium.
In general, Xcc thrives as a severe disease agent in warm, humid climates and, consequently, is most serious in tropical, subtropical and humid continental regions (Williams, 1980). Given the global distribution of Xcc, black rot will become an increasingly important disease constraint favoured by climate change in more northern latitudes of vegetable production, including the warmer regions of Europe, such as the southern regions of the UK.
Black Rot Control
Sanitation and management practices, including crop rotation, weed control and the use of assayed clean seed, can provide significant control of the disease (Schaad and Alvarez, 1993). For example, black rot was a minor disease in the most important production areas in the USA during certain decades, probably because growers followed the recommended practices, including the use of tested, disinfected seed and rotation of seedbeds. However, there was a resurgence of the disease during the 1970s, probably associated with the use of F1 hybrid seed produced in areas in which the disease was endemic (Williams, 1980).
Standard seed testing methods have been developed (Roberts and Koenraadt, 2006). The tolerance for reliable disease control through seed testing needs to be adjusted according to the system of production, e.g. the number of seeds tested should be higher for transplants raised with overhead irrigation than for direct‐drilled crops (Roberts et al., 2007). Seed treatments, including hot water, antibiotics, antibiotics and sodium hypochlorite, hydrogen peroxide and hot acidified cupric acetate or zinc sulphate, are available, but no treatment is totally effective. Several methods can be used to reduce the spread of the disease during transplant raising, including the use of web and flow irrigation systems instead of overhead irrigation and of chlorine dioxide in the irrigation water (Krauthausen et al., 2011).
The development and use of black rot‐resistant cultivars have long been recognized as important methods of control, but, in practice, have had only limited success (Taylor et al., 2002). Natural variation and the inheritance of black rot resistance have been studied in several brassica species and, so far, no disease resistance gene has been cloned. Most studies have focused on B. oleracea (representing the C genome of brassicas), and a limited number of sources of resistance have been identified, including the cabbage cultivar Early Fuji and the cabbage accession PI 436606 (cv. Heh Yeh da Ping Tou) (Camargo et al., 1995; Dickson and Hunter, 1987; Hunter et al., 1987; Taylor et al., 2002; Vicente et al., 2002; Williams et al., 1972). Badger Inbred‐16, a line derived from Early Fuji, contains quantitative trait loci (QTLs) for black rot resistance which have been genetically mapped (Camargo et al., 1995).
The most common and potentially useful sources of black rot resistance occur in the A and B genomes of brassica species, and a number of sources of resistance have been identified in the different species containing these genomes (Bain, 1952; Taylor et al., 2002; Westman et al., 1999). The inheritance of major gene resistance has been studied in the diploid B. rapa (A genome) and in the tetraploids B. carinata (BC genome) and B. napus (AC genome) (Guo et al., 1991; Ignatov et al., 2000; Vicente et al., 2002). A single dominant race‐specific gene has been mapped to the A genome in B. napus (Vicente et al., 2002), and QTLs that control resistance to at least two of the most prevalent races of Xcc have been mapped in a Chinese cabbage accession of B. rapa (Soengas et al., 2007).
Genes present in the brassica A and B genomes could potentially provide durable black rot control, especially if strong race‐specific genes (matching the most prevalent races) could be combined in a genetic background of race‐nonspecific genes (e.g. providing quantitative resistance). To achieve this aim, genes from the wild relative A. thaliana could potentially be easier and quicker to characterize molecularly, and either be used directly in transgenic brassica crops, or facilitate the identification and interspecific transfer of homologous black rot resistance genes from A or B genome sources into vegetable crops. Interestingly, most A. thaliana accessions are resistant to one or more races of Xcc, and more than half exhibit broad‐spectrum resistance to all major races of the pathogen (described below), suggesting that this wild relative of brassica crops could indeed provide useful sources of durable black rot resistance (Holub, 2007). Tsuji et al. (1991) showed that the resistance to an Xcc isolate in the accession Columbia is controlled by a single dominant gene/locus. In addition, Buell and Somerville (1997) described a monogenic and a digenic resistance mechanism in this accession, and mapped the three genes involved. Plant mutants impaired in resistance to Xcc have been isolated and a gene involved in the establishment of the hypersensitive response (HR) and defence response has been identified and mapped (Lummerzheim et al., 2004). However, although A. thaliana and Xcc provided one of the earliest experimental models for the investigation of the interactions of A. thaliana with a major crop pathogen (Simpson and Johnson, 1990), the molecular basis of natural variation in black rot resistance is largely unexplored in this pathosystem.
Pathogen Identification and Detection
The bacterium Xanthomonas campestris pv. campestris (Pammel) Dowson is a Gram‐negative rod, that occurs mostly alone or in pairs and is usually motile by means of a single polar flagellum (Fig. 1d). Most strains form yellow, mucoid, glistening colonies (Fig. 1e,f). The yellow pigments, xanthomonadins (mono‐ or dibromo‐arylpolyene structures), and the exopolysaccharide xanthan, responsible for the mucoid or viscous cultures, are typical of the genus (Vauterin et al., 1995), although the existence of atypical pigmented isolates has been reported (Poplawsky and Chun, 1995).
The taxonomy of the genus was mainly based on the hosts of origin and the phenotypic characteristics until the early 1990s. A detailed study of the phenotypic characteristics of the genus was conducted by Van den Mooter and Swings (1990). Vauterin et al. (1995) later reclassified the genus on the basis of DNA–DNA hybridization studies. In the new classification, the species X. campestris was restricted to strains that cause disease in Brassicaceae (Cruciferae) plants (including X. campestris pv. aberrans, armoraciae, barbarea, campestris, incanae, raphani and, possibly, plantaginis). The reclassification is mainly supported by data obtained through other molecular techniques, including amplified fragment length polymorphism (AFLP) and polymerase chain reaction (PCR) fingerprinting (Rademaker et al., 2000), but there has been some discussion on the shifts in the classification of some groups of isolates (Schaad et al., 2000; Vauterin et al., 2000).
The DNA–DNA hybridization technique is not suitable for the routine identification of new pathogen isolates, and so other molecular methods have been developed. Studies of the 16S rRNA gene (Hauben et al., 1997; Moore et al., 1997a, b) and the 16S–23S intergenic region (Gonçalves and Rosato, 2002) can generally only be used to identify the strains at the genus level. Simões et al. (2007) differentiated species of Xanthomonas by PCR‐restriction fragment length polymorphism of the genes rpfB and atpD involved in the regulation of pathogenicity factors and the synthesis of ATP.
Methods based on DNA sequencing have become more popular as the cost of sequencing has decreased. The sequencing of genes that encode conserved proteins involved in essential cell processes and collectively constitute the ‘core genome’ has been developed for the identification of pathogens. Parkinson et al. (2007, 2009) have shown that sequences of DNA gyrase subunit B (gyrB) can be used as an identification tool at the genus, species and, possibly, pathovar level of Xanthomonas; this method does not have sufficient resolution to differentiate isolates within each pathovar. Young et al. (2008) showed that multilocus sequence analysis (MLSA) based on the partial sequences of four genes, chaperone protein dnaK (dnaK), tonB‐dependent receptor (fyuA), gyrB and RNA polymerase sigma factor (rpoD), can differentiate X. campestris from other species and can possibly also detect differences between some isolates of the same species. The results of MLSA of eight genes, ATP synthase β chain (atpD), dnaK, elongation factor P (efP), glutamine synthetase I (glnA), gyrB, rpoD, triosephosphate isomerase (tpiA) and fyuA, showed high genetic diversity, with Xcc isolates forming two distinct groups; the results also support the existence of two other related pathovars (raphani and incanae) (Fargier et al., 2011).
The identification of Xcc at the pathovar level is generally based on the isolation of the pathogen using semi‐selective media. The currently used protocol for the detection of the pathogen in seeds uses Fieldhouse‐Sasser and mCS20ABN media (Koenraadt et al., 2005; Roberts and Koenraadt, 2006). The morphology of the cultures is generally then checked in subcultures on media such as Yeast Dextrose Calcium Carbonate. Classic bacteriological tests, carbon source metabolic fingerprinting (Biolog, Hayward, CA, USA) (Poplawsky and Chun, 1995), fatty acid analysis (MIDI, Newark, DE, USA) (Massomo et al., 2003) and serological tests using polyclonal or monoclonal antibodies (Alvarez et al., 1994; Franken, 1992) have been used to speed up the identification of the organisms. All of these methods rely on the availability of databases with the results obtained with representative isolates of different species and pathovars, but frequently problems with the standard isolates used (e.g. misidentification) can complicate the interpretation of new results. The inoculation of susceptible brassica seedlings is still the most reliable method, as it provides the ultimate confirmation of the identification of the pathovar (Roberts and Koenraadt, 2006). However, all of these methods are time consuming and inadequate for high‐throughput screening.
Several molecular methods have been used for the identification and characterization of the molecular diversity of Xcc and related pathovars. Rademaker et al. (2005) used PCR primers that amplified repetitive sequences dispersed across bacterial genomes to generate a method to distinguish DNA ‘fingerprinting’ of isolates. Several studies have demonstrated that rep‐PCR (using REP, ERIC and BOX primers) can differentiate isolates at the species, pathovar and intrapathovar level of X. campestris (Rademaker et al., 2005; Vicente et al., 2006). Nevertheless, the comparison of gel profiles and the standardization of the method between laboratories are still difficult to achieve (Parkinson et al., 2007). A DNA probe was developed for the detection of Xcc, but, although the method worked for infected leaves, it was generally not sufficiently sensitive to detect the pathogen in seeds (Shih et al., 2000).
The hrp (hypersensitive response and pathogenicity) genes encode type III secretion systems (T3SSs) (see section on Secretion systems). This gene cluster is involved in plant–pathogen interactions, the growth and development of symptoms in plants and is largely conserved; therefore, these genes are good candidates for molecular diagnostics of different species or pathovars. Berg et al. (2006) and Zaccardelli et al. (2007) developed PCR methods using primers that amplify part of the hrpF gene and the hrcC secretin‐like gene, respectively. These methods allowed the identification of a range of Xcc isolates, but were also positive for isolates of the closely related pathovars aberrans, armoraciae, raphani, barbarea and incanae.
In the near future, the comparison of whole genome sequences might constitute the basis for the classification and identification of X. campestris, and PCR methods with primers related to pathogenicity genes might become part of the routine protocol for the identification of Xcc. Presently, for the molecular identification to the genus or species level, MLSA has the advantage of being a cheaper, more flexible technique to compare bacteria, and is a practical, easier alternative to hybridization studies and full sequencing. It is possible that different sets of housekeeping genes need to be selected to target variation between and within different species (Young et al., 2008), but, as the number of genes increases, the analysis of results becomes more complicated. At the pathovar level, primers based on hrp or other genes linked to pathogenicity might partially substitute the need to test susceptible plants to confirm the identification.
Pathogen Races
A race structure for Xcc was first proposed by Kamoun et al. (1992). The authors described five races (numbered 0–4) based on the reaction of different brassica species (Table 1). Vicente et al. (1998) and Ignatov et al. (1998b) have subsequently shown that race 1 can be subdivided into two or three races on the basis of their reaction on several accessions of B. oleracea and B. carinata. A revised race classification was proposed by Vicente et al. (2001) based on a much larger collection of isolates (Tables 2 and 3). Three races (1, 2 and 4) were retained from Kamoun et al. (1992); however, no isolate was found that matched race 3, and so this race was dropped from the new race classification. Three variant classes were identified amongst the previous race 1 isolates based on the reactions of two B. oleracea accessions and an accession of B. carinata: a new race 1 that refers to the most commonly found variant, a new race 3 to accommodate a rare variant represented by the type strain of Xcc (ATCC33913; NCPPB 528) and an additional race 5 for three non‐UK isolates, including an isolate previously included in X. campestris pv. aberrans (Vicente et al., 2001). It was proposed that race 0 should be reassigned to a new race 6 to avoid the implication that these isolates lacked avirulence genes; although these isolates are pathogenic in all the differentials currently used, partial resistance to this race has been observed in brassica accessions (J. D. Taylor et al., unpublished data; Horticulture Research International, Warwick, UK). Race 2 is only represented by a single isolate (HRI 3849A), which was used in the earliest molecular investigations of black rot resistance in A. thaliana (Buell and Somerville, 1997; Kamoun and Kado, 1990; Tsuji et al., 1991) (Table 3). More recently, race 7 has been added by Jensen et al. (2007, 2010) and Fargier and Manceau (2007). In addition, Fargier and Manceau (2007) included races 8 and 9 for the classification of isolates that have a narrow host range in the differential cultivars (Tables 2 and 3).
Table 1.
Differentiation of Xanthomonas campestris pv. campestris races according to Kamoun et al. (1992)
| Differential cultivar | Race | ||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |
| Early Jersey Wakefield (Brassica oleracea) | + | + | + | + | + |
| Just Right Turnip, Tokyo Turnip Hybrid (Brassica rapa) | + | + | + | − | − |
| Seven Top Turnip (B. rapa) | + | + | − | + | − |
| Florida Broad Leaf India Mustard (Brassica juncea) | + | − | + | − | − |
+, compatible interaction (susceptibility); −, incompatible interaction (resistance).
Table 2.
Postulated gene‐for‐gene model to explain the relationship between Brassica lines and races of Xanthomonas campestris pv. campestris (adapted from Vicente et al., 2001, Fargier and Manceau, 2007 and Jensen et al., 2010)
| Differential cultivar or accession | Race/avirulence gene (A) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ||||||
| A1 | A1 | A1? | A1 | A1 | ||||||||||
| A2 | A2 | |||||||||||||
| A3 | A3 | A3 | A3 | A3 | ||||||||||
| A4 | A4 | |||||||||||||
| Resistance gene (R) | A5 | |||||||||||||
| Wirosa F1 (Brassica oleracea) | + | (+)/+ | + | + | + | + | + | + | + | |||||
| Just Right Hybrid Turnip (Brassica rapa) | R4 | + | + | + | − | + | + | + | + | − | ||||
| COB60 (Brassica napus) | R2? | R4 | + | (+) | + | − | + | + | + | nt | nt | |||
| Seven Top Turnip (B. rapa) | R2 | R4 | + | − | +,v | −,v | +,v | + | + | − | − | |||
| PIC1 (Brassica carinata) | R1 | R4? | ‐ | (+) | − | − | + | + | + | − | − | |||
| FBLM2 (Brassica juncea) | R1 | R4? | R5 | ‐ | + | − | − | (+) | + | − | − | − | ||
| Miracle F1 (B. oleracea) | R2? | R3 | + | −/(+) | −/(+) | + | − | + | + | − | − | |||
| SxD1 (B. oleracea) | R3 | + | −/(+) | − | + | − | (+)/+ | + | nt | nt | ||||
+, compatible interaction (susceptibility); −, incompatible interaction (resistance); (+), weakly pathogenic; nt, not tested; v, variable; ? indicates that the gene might be present, although it might not be necessary to explain the interaction.
Table 3.
Race type strains of Xanthomonas campestris pv. campestris (adapted from Vicente et al., 2001 and Fargier and Manceau, 2007)
| HRI/UW isolate number | Other collections (number) | Host | Country (year) | Race |
|---|---|---|---|---|
| 3811 | P. Williams (PHW1205) | Brassica oleracea | USA | 1 |
| 3849A | C.I. Kado (2D520) | B. oleracea var. botrytis | USA | 2 |
| 5212 | NCPPB (528), CFBP (524)1, LMG (568), ATCC (33913) | B. oleracea var. gemmifera | UK (1957) | 3 |
| 1279A | – | B. oleracea var. capitata | UK (1984) | 4 |
| 3880 | NCPPB (2986) | B. oleracea var. capitata | Australia (1975) | 5 |
| 6181 | – | Brassica rapa | Portugal (1996) | 6 |
| – | CFBP (4953) | B. oleracea var. botrytis | Belgium (1999) | 7 |
| 8450A | B. D. Jensen (N47) | B. oleracea var. capitata | Nepal (2001) | 7 |
| – | CFBP (1124), LMG (8032) | B. oleracea var. botrytis | France (1967) | 8 |
| 3961 | NCPPB (1145), LMG (8004), CFBP (6650) | B. oleracea var. botrytis | UK (1958) | 9 |
CFBP, Collection Française de Bactéries Phytopathogènes, INRA, France; HRI/UW, The University of Warwick (ex‐Warwick HRI), Wellesbourne, UK; LMG, Laboratorium voor Microbiology, Gent, Belgium; NCPPB, National Collection of Plant Pathogenic Bacteria, Sand Hutton, UK.
Interestingly, the type strain (ATCC33913; NCPPB528) can cause atypical blight symptoms in brassicas (Alvarez et al., 1994; Chen et al., 1994) characterized by dark necrotic lesions with limited chlorosis, and, in this respect, is not typical of the majority of Xcc isolates. These symptoms have also been observed in B. oleracea accessions inoculated with isolates of races 5 and 6. The ability to elicit blight symptoms may be under genetic control (Chen et al., 1994), but may also be influenced by environmental conditions, in particular temperature.
Homozygous brassica lines (i.e. inbred or doubled haploid lines) provide the best means for reproducible, routine identification of Xcc races, and seed stocks can be readily regenerated by researchers or public stock centres. However, the current host differential set includes, for example, the cultivar Seven Top Turnip, which exhibits variable reactions (possibly as a result of a genetic mixture from open pollination in the commercial seed production). F1 hybrids are also included, which are impossible for researchers to regenerate, and may have limited availability in future years depending on the commercial success of each of these cultivars. To overcome part of these constraints, doubled haploids from several accessions of B. oleracea, B. napus, B. carinata and B. juncea were produced at the University of Warwick, Warwick HRI (now part of the School of Life Sciences), to replace the previous differential lines described by Vicente et al. (2001). These include doubled haploid lines that replace Cobra, PI199947, Florida Broad Leaf Mustard and Miracle F1.
Gene‐for‐gene interactions can be used to explain the relationship between bacterial isolates and differential lines. The proposed gene‐for‐gene model presented in Table 2 is based on the interaction of at least five matching gene pairs. The genes that confer resistance to the most important races (1 and 4) are designated R1 and R4. The model allows for the possible inclusion of additional gene pairs if new races and differentials are identified. In general, the model was constructed in a manner that reflects the origin of the allotetraploid brassica species (Nagaharu, 1935): R1 originates from the B genome, R3 from the C genome and R4 from the A genome. The proposed model needs to be supported by genetic and molecular data from both the host and the pathogen to be fully validated. In the case of the host, results of crosses made to establish the inheritance of resistance to some of the races indicate that R1, R3 and R4 are single dominant genes (Vicente et al., 2002).
A simpler gene‐for‐gene model has been proposed by He et al. (2007) based on the interactions between Xcc isolates and cultivars of Brassica (B. juncea, B. oleracea, B. rapa), radish (Raphanus sativus) and pepper (Capsicum annuum) (Table 4). It is possible that some of the resistance/avirulence genes proposed by the authors correspond to genes included in Table 2: the gene Rc1 might correspond to R1, Rc2 to R3 and Rc3 to R4.
Table 4.
Postulated gene‐for‐gene model to explain the relationship between Brassica spp., radish and pepper lines and isolates of Xanthomonas campestris pv. campestris (adapted from He et al., 2007)
| Differential cultivar or line | Avirulence gene (A) | |||||||
|---|---|---|---|---|---|---|---|---|
| xop AH (avrXccC) | ||||||||
| . | avrRc2 * | |||||||
| . | xopE (avrXccE1)1 | |||||||
| . | avrBs1 | |||||||
| Resistance gene (R) | ||||||||
| Guangtou (Brassica juncea var. megarrhiza) | Rc1 | − | + | + | ||||
| Jingfeng‐1 (Brassica oleracea) and Huaye (Raphanus sativus var. longipinnatus) | Rc2 | + | − | + | ||||
| Zhongbai‐83 (Brassica rapa var. pekinensis) | Rc3 | + | + | − | ||||
| ECW10R (Capsicum annuum v. latum) | Bs1 | + | + | + | HR | |||
*This gene has not yet been identified.
+, compatible interaction (susceptibility); −, incompatible interaction (resistance); HR, hypersensitive response.
Worldwide, races 1 and 4 are predominant, but their relative frequencies in B. oleracea crops appear to vary with geographical region. For example, race 1 appears to be more common than race 4 in the UK, whereas race 4 has been shown to be the predominant race in Portugal (Vicente, 2004), northwestern Spain (Lema et al., 2012) and some East African countries, such as Tanzania and Uganda (Mulema et al., 2012). Other races are generally rare, but may be more common in other host species that are less frequently surveyed. Races 2 and 6 were absent in a collection of isolates from Japan and Russia (Ignatov et al., 1998a). Nepal and northwest Spain seem to have diverse populations of Xcc, with five different races identified in B. oleracea crop plants (Jensen et al., 2010; Lema et al., 2012). The low frequency of race 3 worldwide may be a result of the extensive use of cultivars that are resistant to this race. However, this may not have been the case 50 years ago when the type strain of Xcc (ATCC33913, NCPPB528) was collected in 1957. Race 5 is also rare, but was recently found in Nepal (Jensen et al., 2010) and in a field in northwest Spain (Lema et al., 2012). Race 6 was not found in the UK, but this may have been a result of the preponderance of isolates obtained from B. oleracea. In Portugal and northwest Spain, race 6 was common in turnip (Lema et al., 2012; Vicente, 2004).
The gene‐for‐gene model and the availability of defined ‘race type strains’ should assist in the selection and evaluation of plant material for breeding programmes and may be the basis for molecular studies. Disease resistance screening should be performed with isolates that represent the pathogenic variation of Xcc, and therefore should at least include the major races 1 and 4. In addition, isolates of race 6 should be useful to detect potential race‐nonspecific resistance. The monitoring of the frequency and distribution of races worldwide is essential to the development of effective strategies for the breeding of black rot‐resistant cultivars. Future brassica crops will benefit from the combination of major genes that confer strong resistance to the most common races of the pathogen (R1 and R4) and, if possible, race‐nonspecific genes that could confer quantitative resistance to all known races.
The Pathogen Genome
Three isolates of Xcc have been fully sequenced, including the type strain ATCC33913 (NCPPB528; LMG568; ICMP13; DSM3586) (da Silva et al., 2002), isolate 8004 (a spontaneous rifampicin‐resistant mutant from NCPPB1145) (Qian et al., 2005) and the industrial high‐xanthan‐producing isolate B100 (Vorhöelter et al., 2008). The type strain has been identified as race 3 (Vicente et al., 2001), isolate 8004 has been assigned to race 9 (Fargier and Manceau, 2007) and B100 belongs to race 1 (J. G. Vicente, unpublished results). The isolate 8004 has featured in several studies of phytopathological properties, such as the secretion of extracellular enzymes and exopolysaccharides (Tang et al., 1991), cell–cell signalling and biofilm formation (Torres et al., 2007). Further information about the Xcc genomes is summarized in Table 5. An isolate of X. campestris pv. raphani (756C), a closely related pathogen that causes a nonvascular, leaf spot pathogen on Brassicaceae and other hosts such as tomato, has also been sequenced recently (Bogdanove et al., 2011). These X. campestris genomes comprise circular chromosomes of approximately 5 000 000 base pairs (bp), have a high G + C content and do not carry plasmids.
Table 5.
Xanthomonas campestris pv. campestris strains that have been completely sequenced and features of the genome (adapted from Vorhöelter et al., 2008)
| Isolate | ATCC 33913 | 8004 | B100 |
|---|---|---|---|
| Other designations | NCPPB (528), HRI5212, LGM 568, CFBP 5241 | Rifampicin‐resistant laboratory strain derived from NCPPB 1145, CFBP 6650 | |
| Host of origin | Brussels sprouts (Brassica oleracea var. gemmifera) | Cauliflower (B. oleracea var. botrytis) | |
| Country of origin | UK | UK | |
| Year of isolation | 1957 | 1958 | |
| Race | 3 | 9 | 1 |
| Size (bp) | 5 076 187 | 5 148 708 | 5 079 002 |
| G + C content (%) | 65.0 | 64.9 | 65.0 |
| CDS—predicted number | 4181 | 4273 | 4471 |
| CDS—function assigned | 2708 | 2671 | 2878 |
| Ribosomal RNA operons | 2 | 2 | 2 |
| Transfer RNAs | 54 | 54 | 54 |
| Insertion sequence elements | 109 | 115 | 59 |
| Reference | da Silva et al. (2002) | Qian et al. (2005) | Vorhöelter et al. (2008) |
CDS, coding DNA sequence; CFBP, Collection Française de Bactéries Phytopathogènes, INRA, France; HRI, School of Life Sciences (ex‐Warwick HRI), University of Warwick, Wellesbourne, UK; LMG, Laboratorium voor Microbiology, Gent, Belgium; NCPPB, National Collection of Plant Pathogenic Bacteria, Sand Hutton, UK.
The comparison of the sequences of Xcc type strain ATCC33913 and 8004 indicated that significant genomic‐scale rearrangement across the replication axis between two IS1478 elements and the loss and acquisition of blocks of genes, rather than point mutations, constitute the main genetic variation between the strains (Qian et al., 2005). The sequence of B100 is extensively collinear to the strain 8004, but differs from the type strain ATCC33913 by the inversion of a large chromosomal fragment (Vorhöelter et al., 2008). This may indicate that the strains 8004 and B100 have originated via recent recombination events (Qian et al., 2005).
He et al. (2007) have constructed a whole genome microarray of the Xcc strain 8004 and have used this resource to study the genetic diversity and host specificity of this pathogen by array‐based comparative hybridization with genomes of 18 pathogenic isolates collected from different host plants and various geographical regions of China. A core set of 3405 conserved coding sequences was identified and 730 coding sequences that were absent or highly divergent amongst the isolates. Of the 304 proven or postulated pathogenicity genes currently known in Xanthomonas, 258 were conserved and 46 were highly divergent amongst the isolates.
These current reference genomes of Xcc are useful for genome‐wide comparisons with publicly available genomes from other Xanthomonas species, including strain 306 of X. axonopodis pv. citri (da Silva et al., 2002), which represents the citrus canker pathogen, strain 85‐10 of X. axonopodis pv. vesicatoria (Thieme et al., 2005), which represents the bacterial spot pathogen which specifically attacks pepper (and not tomato), strains of the rice bacterial blight pathogen X. oryzae pv. oryzae, KACC10331 (Lee et al., 2005), MAFF 311018 (Ochiai et al., 2005) and PXO99A (Salzberg et al., 2008), strain BLS256 of X. oryzae pv. oryzicola (Bogdanove et al., 2011), strain GPE PC73 of X. albilineans (Pieretti et al., 2009) and strains from X. vasicola pv. vasculorum and musacearum, the cause of banana wilt (Studholme et al., 2010). For example, Lu et al. (2008) compared six gene clusters associated with pathogenicity across genomes of eight Xanthomonas isolates representing vascular and nonvascular diseases of rice, brassicas, pepper and citrus. Interestingly, plasmid DNA was only observed in the reference isolates of X. axonopodis pv. citri and pv. vesicatoria.
Secretion Systems
Reference genomes of Xcc have begun to reveal an extensive inventory of genes that may be required for plant associations on the basis of homology with known genes from other Xanthomonas or Pseudomonas species. Most importantly, genome‐wide comparisons have revealed three secretion systems (types II, III and IV) in Xcc that have been associated with pathogenic bacteria of plants or animals.
The type II secretion system (T2SS) mediates the transport of proteins into the extracellular space. This system can secrete plant cell wall‐degrading enzymes, including cellulose, polygalacturonase, xylanase and protease. Two classes of T2SS have been identified in Xanthomonas: xps and xcs. The xps cluster is present in all Xanthomonas genomes currently sequenced and in other genera (e.g. Xylella), whereas the xcs genes are only present in some of the Xanthomonas genomes, including Xcc and X. campestris pv. raphani (Lu et al., 2008). Qian et al. (2005) obtained six pathogenicity‐deficient mutants (xpsD, xpsE, xpsF, xpsK, xpsL and xpsM) related to the xps system, whereas no pathogenicity‐deficient mutants were found on the 12 annotated genes related to the xcs system; therefore, the xps genes are related to pathogenicity, whereas the xcs genes may have other roles not essential for pathogenicity or may be nonfunctional.
The mutation of DsbB, a protein involved in disulphide bond formation in the periplasm of Xcc, reduced virulence, cell mobility and bacterial growth in planta and resulted in ineffective T2SS and T3SS (Jiang et al., 2008b).
The type IV secretion system (T4SS) is important for the release of macromolecules, and this system is used by Gram‐negative bacteria to translocate protein and DNA substrates across the cell envelope into target cells (Souza et al., 2011). However, the role of T4SS in the pathogenicity of Xanthomonas is uncertain. The T4SS carried in the race 9 isolate 8004 is encoded mainly by genes in the virD4 and the virB cluster that consists of nine open reading frames (ORFs). Qian et al. (2005) identified a mutated gene encoding the channel‐forming protein VirB8, and concluded that the T4SS contributes to the virulence of the pathogen. However, He et al. (2007) have shown that deletion of virD4 and the virB cluster of strain 8004 does not affect the virulence of the pathogen.
The T3SS is generally thought to be essential for pathogenicity and for the initiation of disease in susceptible host plants, and can be involved in the elicitation of cell death and other defence responses in resistant plants. The T3SS is encoded by the hrp cluster of genes and translocates effector proteins directly into the plant cells; it is also possible that effectors can exit the bacterial phytopathogens via the Hrp pathways (Lindgren, 1997). Individual genes have been named hrp, hrp‐conserved (hrc) or hrp‐associated (hpa). Most hrp gene sequences are only found in Xanthomonas and related genera, whereas hrc genes are conserved in many plant and animal pathogens. In Xcc, the hrp cluster consists of 26 genes extending from hpa2 to hrpF (da Silva et al., 2002). The Xcc genomes do not contain hpa3 and the N‐terminal region of xopF1, although these genes are present in other Xanthomonas species, including X. campestris pv. raphani (Lu et al., 2008). Four mutants of genes encoding the T3SS machinery (hrcJ, hrcU, hrcV, hrpE) and one encoding a regulatory protein (hrpG) were nonpathogenic (Qian et al., 2005). Mutations in the regulator hrpG of strain 8004 can make all the hrp genes express constitutively and enhance the intensity of the HR reaction in pepper (Jiang et al., 2006).
Wei et al. (2007) showed that hpaR, a putative marR family transcriptional regulator, is essential for the pathogenicity of strain 8004 on cabbage, is required for the HR response on the nonhost pepper and is under the positive control of the two key hrp gene regulators HrpG and HrpX. The GntR family is a frequent group of helix–turn–helix transcriptional regulators in bacteria. One of the six GntR regulators, HpaR1, positively and negatively affects the expression of HR and pathogenicity hrp genes that encode the type III secretion via hrpG (An et al., 2011).
Several genes that are regulated in an HrpX‐dependent manner possess a consensus nucleotide sequence TTCGB–N15–TTCGB (B can be base C, G or T), which has been termed the plant‐inducible‐promoter box, or PIP box. The detection of PIP boxes provides a refined tool for the identification of candidate genes in the hrpX regulon, as well as effector protein genes of the type III pathway. For example, 17 putative PIP box sequences have been described in the promoter regions of Xcc ATCC33913 (da Silva et al., 2002) and 56 genes have been predicted to have PIP boxes in strain 8004 (Jiang et al., 2009).
Type III Effector Genes
There has been considerable research effort to identify and understand type III effector function in Xanthomonas species. White et al. (2009) reviewed in detail the type III effectors from Xanthomonas and classified them into 39 different groups (called Xop) reflecting sequence similarity. From these, 21 appear to be present in the Xcc strain ATCC33913. Nine effectors, considered to be core effectors, are present in almost all Xanthomonas isolates, with the exception of X. albilineans and, in some cases, X. campestris pv. raphani; other effectors are found in a more limited number of species/isolates (Bogdanove et al., 2011; Ryan et al., 2011). From the extensive list of genes encoding putative T3SS proteins in Xcc and X. campestris pv. raphani, summarized in Table 6, effort has begun to identify potential effector proteins (which are important for pathogenicity in a susceptible host) and avirulence determinants (which elicit defence in a resistant host). Effector proteins are generally thought to be involved in the suppression of the host defence system or in the alteration of host metabolism to favour nutrient uptake of the pathogen. However, experimental verification of effector function can be difficult. For example, deletion of individual effector genes can often result in little or no reduction in pathogenicity because of the presence of alternative functional (redundant) effector proteins (Cunnac et al., 2009). Some T3SS effectors may activate defences (e.g. rapid host cell death) when infiltrated into plant tissue, and thus indicate a potential role as avirulence (Avr) proteins in a resistant host (Flor, 1971; Keen, 1990).
Table 6.
Occurrence, features and function of putative and proven effector/avirulence genes of Xanthomonas campestris pv. campestris
| Effector class | Synonym | Gene ID in ATCC33913 | Gene ID in 8004 | Gene ID in B100 | Gene ID in Xcr 756C | PIP box | Features and function on pathogenicity |
|---|---|---|---|---|---|---|---|
| Core effectors* | |||||||
| AvrBs2 | XCC0052 | XC 0052 | xcc‐b100 0057 | No | Yes | Glycerophosphoryl diester phosphodiesterase | |
| Avirulence in Brassica juncea, B. carinata and B. oleracea and HR on pepper (Ignatov et al., 2002) or no effect (Castañeda et al., 2005) | |||||||
| XopK | XCC2899 | XC 1210 | xcc‐b100 1254 | No | Unknown | ||
| XopL | XopLR | XCC4186 | XC 4273 | xcc‐b100 4400 | No | Yes | Leucine‐rich protein |
| Reduced virulence on radish (Jiang et al., 2009) | |||||||
| XopN | XCC0231 | XC 0241 | xcc‐b100 0253 | No | ARM/HEAT repeat | ||
| Reduced virulence on radish (Jiang et al., 2008a) | |||||||
| XopP | XCC1247 | XC 2994 | xcc‐b100 3057 | XCA_1500 | Yes | Unknown | |
| Reduced virulence on radish (Jiang et al., 2009) | |||||||
| XopQ | XCC1072 | XC 3177 | xcc‐b100 3274 | No | Yes | Putative inosine–uridine nucleotide N‐ribohydrolase | |
| Reduced virulence on radish (Jiang et al., 2009) | |||||||
| XopR | XCC0258 | XC 0268 | xcc‐b100 0280 | XCA_4254 | Unknown | ||
| XopX | XCC0529 | XC 0541 | xcc‐b100 0558 and 0559 | No | Methionine‐rich protein | ||
| XCC0530 | XC 0542 | ||||||
| XopZ | XCC1975 | XC 2210 | xcc‐b100 2274 | No | Unknown | ||
| Other effectors† | |||||||
| AvrBs1 | XCC2100 | XC 2081 | xcc‐b100 2396 | No | No | HR on pepper (He et al., 2007) | |
| XopD | XCC2896 | XC 1213 | xcc‐b100 1256 | No | SUMO cysteine protease (C48 family), EAR motif, DNA binding, nuclear localization(Canonne et al., 2012) | ||
| XopE | AvrXccE1 | XCC1629 | XC 2602 | No | No | Yes | Putative transglutaminase |
| Avirulence on B. rapa (He et al., 2007) | |||||||
| XopF | XCC1218 | XC 3024 | xcc‐b100 3087 | XCA_1470 | Unknown | ||
| XopG | XCC3258 | XC 0967 | xcc‐b100 2655 | No | M27 family peptidase (Clostridium toxin) | ||
| XopH | AvrBs1.1 | XCC2099 | XC 2082 | xcc‐b100 2395 | No | No | Putative tyrosine phosphatase |
| XopJ | AvrXccB | XCC3731 | XC 3802 | No | No | Yes | Putative cysteine protease (C55 family) or serine/threonine acetyltransferase. Ubiquitin‐like protease |
| No effect on pathogenicity (Jiang et al., 2009) | |||||||
| XopAC | AvrAC | XCC2565 | XC 1553 | xcc‐b100 1596 | XCA_2914 | Yes | Leucine‐rich protein |
| Avirulence recognized in vascular tissues of Arabidopsis (Xu et al., 2008) | |||||||
| XopAD | No | No | No | XCA_1464 | SKWP repeat protein | ||
| XopAG | XCC3600 | XC 0563 | xcc‐b100 0580 | No | Unknown | ||
| XopAH | AvrXccC | XCC2109 | XC 2004 | xcc‐b100 2071 | No | Yes | Dual function. Avirulence in B. juncea and B. rapa; virulence in B. oleracea (Castañeda et al., 2005; He et al., 2007; Wang et al., 2007) |
| XopAL1 | AvrPhpE | XCC1246 | XC 2995 | xcc‐b100 3058 | XCA_1499 | Yes | Reduced virulence on radish (Jiang et al., 2009) |
| XopAL2 | Downstream XCC3574 | Downstream XC_3916 | xcc‐b100 0616 | No | Unknown | ||
| XopAM | XopR1 | XCC1089 | XC 3160 | xcc‐b100 3256 | No | Yes | Reduced virulence on radish (Jiang et al., 2009) |
| XopAT | No | No | No | XCA_1464a | Yes | Unknown. No similarity with other known proteins (Bogdanove et al., 2011) | |
| XopA | XCC1240 | XC 3002 | xcc‐b100 3065 | XCA_1492 | Harpin, may not be a t3e | ||
| HpaA | XCC1224 | XC 3018 | xcc‐b100 3081 | XCA_1476 | T3 secretion control protein, may not be a t3e | ||
| HrpW | XCC1219 | XC 3023 | xcc‐b100 3086 | XCA_1471 | Pectate lyase, may not be a t3e | ||
| AvrXccA1 | AvrXca | XCC4229 | XC 4318 | xcc‐b100 4450 | XCA_4581 | No | May not be a t3e. Virulence of Xcr in Arabidopsis (Parker et al., 1993) |
| AvrXccA2 | XCC2396 | XC 1716 | xcc‐b100 1770 | XCA_2696 | No | Unknown, may not be a t3e |
Adapted from da Silva et al. (2002), White et al. (2009), Jiang et al. (2009) and Ryan et al. (2011).
*Nine core effectors are found in most Xanthomonas spp., with the exception of X. albilineans and, in some cases, X. campestris pv. raphani (Xcr).
†Other effectors are found in a more limited number of Xanthomonas spp. isolates.
t3e, Type III effector.
Xu et al. (2006) showed that the mutagenesis of eight candidate genes from strain 8004 had no affect on pathogenicity in Chinese radish and cabbage. However, Xu et al. (2006) and He et al. (2007) showed that mutants of avrBs1 lost the ability to elicit an HR in the pepper line ECW10R that contains the corresponding R gene Bs1 (Table 4). The AvrBs1 effector is responsible for the elicitation of HR on pepper dependent on T3SS (Xu et al., 2006). However Castañeda et al. (2005) did not detect an HR variation between the avrBs1 mutants and the wild‐type 528T. He et al. (2007) suggested that it is possible that the function of avrBs1 is redundantly encoded in the Xcc type strain and that the expression is different in the two strains.
Ignatov et al. (2002) reported that an avrBs2 homologue, designated avrRxc1/3, determined the avirulence of Xcc 512/2 (a natural mutant strain of a race 3 isolate) on brassica plants with the B genome, including B. juncea cv. Florida Mustard and B. carinata line PI199947, and on some B. oleracea cultivars (including the lines SR1/3, Badger Inbred‐16/2 and cv. Miracle F1). This gene was also responsible for a mesophyllic HR on leaves of Florida Mustard and nonhost pepper plants carrying Bs2. Xu et al. (2006) showed that avrBs2 is required for full virulence of strain 8004 on Chinese radish and cabbage. In contrast, Castañeda et al. (2005) showed that the deletion of the entire putative avrBs2 homologue in the type strain had no effect on the interaction with Florida Mustard and pepper. The strains employed by Ignatov et al. (2002) are probably derived from the type strain used by da Silva et al. (2002) and Castañeda et al. (2005); therefore, it is difficult to reconcile these results and the role of the avrBs2 homologue on pathogenicity/race specificity still needs to be clarified.
Genes from the AvrBs3 family have been found in a number of Xanthomonas species and pathovars, and in Ralstonia solanacearum (White et al., 2009). This large family of closely related T3 effectors constitutes the TAL effector family. All members of the family have a common structure with tandem repeats of a sequence of amino acids, typically of 34 amino acids in the central part of the proteins, nuclear localization signals (NLSs) and an acidic activation domain (AD); the avirulence and virulence specificities of an AvrBs3 member are dependent on the number and order of the repeats and the NLSs and AD (Boch and Bonas, 2010; Gurlebeck et al., 2006).
Interestingly, the three Xcc strains sequenced do not contain genes from the avrBs3/pthA family, whereas three members of this family have been identified in an isolate named as X. campestris pv. armoraciae. These three genes were designated hax2, hax3 and hax4 (homologue of avrBs3 in Xanthomonas) (Kay et al., 2005). The three Hax proteins are translocated via the T3SS. The Hax3 and Hax4 proteins have the typical structure of the AvrBs3‐like effectors, with 34 amino acid repeats in the central part, but Hax2 has 35 amino acid repeats that contain an additional proline residue. The three hax genes have an additive effect on disease symptoms in radish, with hax2 having the strongest influence, and two hax genes (hax3 and hax4) have a Bs4‐dependent avirulence activity in tomato (Kay et al., 2005).
Transconjugants of Chinese isolates containing avrXccE1 (xopE) became avirulent on Chinese cabbage (B. rapa) cv. Zhongbai‐83 (He et al., 2007) (Table 4). In contrast, Castañeda et al. (2005) did not notice any effect on pathogenicity on the plants tested, but it is possible that the lines tested by these authors did not possess the R gene responsive to avrXccE1.
Jiang et al. (2008a) showed that XopN from strain 8004 (designated XopXccN) is required for full virulence. A mutant of the strain 8004 with an insertion in xopXccN was significantly weaker than strain 8004 when inoculated on Chinese radish (Raphanus sativus var. radiculus) cv. Manshenhong. The expression of xopXccN is regulated by the key hrp regulators HrpG and HrpX.
The protein sequence of XopD has been identified in strain B100 (Canonne et al., 2012). This type III effector targets the Arabidopsis transcription factor MYB30, possibly as part of a virulence strategy that allows Xcc to suppress the plant cell responses to infection (Canonne et al., 2011).
Mutagenesis of the eight candidate effector genes from type strain ATCC33913 (syn. 528T) had no effect on pathogenicity when the mutants were used to inoculate six crucifer species, and none of the mutated genes singly or in any combination affected the nonhost HR elicited by the type strain in pepper. However, insertion or deletion mutants in a locus of a gene designated avrXccFM (XopAH) became virulent on Florida Mustard, and therefore changed the race specificity of the isolate (Castañeda et al., 2005). The T3SS effector AvrXccC of strain 8004, which belongs to the AvrB effector family of Xanthomonas, has been shown to have a dual effect: this effector is required for full bacterial virulence in a susceptible B. oleracea cv. Jingfeng 1 and for avirulence in a B. napiformis L.H. Bailey [syn. B. juncea (L.) Czern. var. napiformis (Pailleux & Bois) Kitam.] accession (not specified) (Wang et al., 2007). The avrXccC gene was expressed in the race 2 strain HRI3849A (which causes symptoms on the mustard accession used by these authors) and the resulting strain was avirulent. He et al. (2007) also showed that an avrXccC mutant of 8004 became pathogenic on mustard (B. juncea var. megarrhiza Tsen et Lee) cv. Guangtou and Chinese cabbage (B. rapa ssp. pekinensis) cv. Zhongbai‐83. The avrXccC gene of strain 8004, which confers avirulence on some mustard cultivars, is identical to avrXccFM of strain ATCC33913 (syn. 528T), which confers avirulence on Florida Mustard (He et al., 2007). Wang et al. (2007) showed that AvrXccC is anchored to the plant plasma membrane and its avirulence function for host recognition depends on its location. The expression of avrXccC is hrpG/hrpX dependent.
Xcc also contains genes encoding leucine‐rich repeat (LRR) proteins. These motifs are commonly involved in protein–protein interactions and are found in several classes of plant disease resistance genes. Xu et al. (2008) showed that one of these genes, xopAC, named avrACXcc8004, encodes a protein containing LRRs; this gene is a type III effector that appears to be restricted to strains of X. campestris. The A. thaliana accession Col‐0 was resistant to the wild‐type 8004 strain and to an avrACXcc8004 mutant when the leaf mesophyll was infiltrated with bacterial suspensions, but the Col‐0 ecotype became susceptible to the mutant when the bacterium was introduced into the vascular system by piercing the central vein of the leaves, indicating that the product of avrACXcc8004 is recognized in vascular tissues and this gene might be related to the ability to colonize the xylem of a host.
Some avr/effector genes have also been identified in closely related pathovars, including isolates identified as X. campestris pv. armoraciae and raphani (possibly synonymous). Parker et al. (1993) cloned an avirulence gene of an isolate identified as X. campestris pv. raphani (isolate 1067). This isolate was avirulent in all the A. thaliana accessions tested by the authors. When the avrXca gene was transferred to Xcc strain 8004, it strongly reduced symptom development and bacterial growth in Columbia plants, but did not affect virulence to Brassica plants. The avrXca gene encodes a protein of 67 kDa that has no homology with known sequences and confers avirulence in a number of A. thaliana accessions, except one (‘Kas‐1’). The avirulence phenotype is not Hrp dependent and the interaction with A. thaliana did not lead to a characteristic HR, indicating that this may not be a type III effector. More recently, Corbett et al. (2005) have characterized a virulence factor, designated Svx, of E. carotovora ssp. atroseptica which shows homology to AvrXca. This protein is secreted by the type II secretion apparatus and the transcription of the svx gene is regulated by quorum sensing; the function of this protein is unknown, but the authors consider that it may play a role in the pathogenicity of E. carotovora pv. atroseptica.
So far, there is some evidence that 12 of the putative effector genes from Xcc are phenotypically functional in a very restricted number of isolates. The avirulence genes presented in the model in Table 2 have not been proven and linked to the effectors of Xcc (Table 6), although it is likely that A1 might correspond to XopAH and A4 to XopE. Some putative effectors could be pseudogenes or poorly expressed genes in the isolates studied, or the genes might have small effects on pathogenicity or fitness that have not been detected in the assays (Castañeda et al., 2005). Further research into the putative effectors using other isolates and hosts might still show that they have a function in pathogenicity and/or race specificity. It is also possible that some effectors were once pathogenicity genes that determined the host range of the pathogen; the function of some of these effectors could be gratuitous or even detrimental, but their structure might allow rapid adaptive selection for pathogenic function (Gabriel, 1999).
Other Virulence and Pathogenicity Factors
The surface structure and appendages of phytopathogenic bacteria are important for the attachment, colonization and infection of the host. Type IV pili may contribute to bacterial pathogenesis by affecting surface motility, microcolony and biofilm formation, adhesion, immune evasion and cell signalling (Craig et al., 2004). The comparison of genomes from isolates ATCC33913 and 8004 showed that at least 26 genes related to the pili assembly are highly conserved, and mutations in two of the assembly genes (pilB and pilC) have reduced virulence (Qian et al., 2005).
Many phytopathogenic bacteria produce a large number of factors that might be essential or contribute to cause disease. The bacteria from the genus Xanthomonas typically produce yellow, membrane‐bound pigments, called xanthomonadins. These pigments have a role in the maintenance of the ecological fitness of the bacteria, protecting the cells against photooxidative stress. Xanthomonadin biosynthesis is encoded by the pig cluster of genes. Poplawsky and Chun (1998) have shown that the mutation of pigB results in a reduction in epiphytic survival and decrease in infection via the hydathodes. The biosynthesis of xanthomonadin is regulated by 3‐hydroxybenzoic acid, a diffusible factor (DF) that is associated with a range of biological functions; DF‐deficient mutants are nonpigmented, impaired in survival ability and less virulent (He et al., 2011).
Xcc produces a range of extracellular enzymes (including proteases, pectinases and endoglucanase). The extracellular enzymes are capable of degrading the plant cell components and may be required to overcome plant defence responses, to allow bacteria to move into uncolonized plant tissues and to mobilize plant polymers for nutritional purposes (Torres et al., 2007).
In Xcc, the synthesis of extracellular degrading enzymes and exopolysaccharides is regulated by products of the rpf genes (rpfABFCHGDIE) (da Silva et al., 2002). The rpf gene cluster of Xcc regulates the genes involved in the synthesis of extracellular hydrolytic enzymes and extracellular polysaccharides (EPSs) (including xanthan) and plays a role in motility, toxin production, oxidative stress resistance, aerobic respiration and biofilm formation (Dow et al., 2003; Wilson et al., 1998). A diffusible signal factor (DSF) responsible for cell–cell signalling is involved in the transcriptional control of these genes (Barber et al., 1997). Some genes under the control of the rpf/DSF system are required for the first stages of endophytic colonization (Gudesblat et al., 2009). Genes that encode proteins involved in the synthesis or degradation of cyclic‐di‐guanosine‐3′,5′‐monophosphate (cyclic‐di‐GMP), a second messenger involved in the regulation of cellular functions, have been shown to play significant roles in virulence to plants. Complex regulatory networks possibly allow the bacteria to adapt to different environments and to modulate virulence factors (Ryan et al., 2007). The expression levels of proteases and endoglucanases are reduced when the rpfI gene is inactivated in Xcc, suggesting that rpfI may have a function in the extensive tissue degeneration that is characteristic of black rot (Dow et al., 2000). RpfF is involved in the synthesis of DSF, and a two‐component system comprising RpfG and the complex sensor kinase RpfC is involved in sensing and responding to DSF (Ryan et al., 2010). Mutants deficient in RpfG, a phosphodiesterase that degrades cyclic‐di‐GMP, or the elevation of cyclic‐di‐GMP via the overexpression of a GGDEF domain protein WspR, caused the aggregation of cells, reduction in mobility and decrease in production of virulence factors, extracellular enzymes and exopolysaccharides (Hsiao et al., 2011). Mutations of rpfF or rpfC also lead to a reduction in the synthesis of virulence factors. The cell–cell interactions are highly regulated and dynamic; the interactions involving HD‐GYP and GGDEF domain proteins are dependent on DSF signalling, influence the localized expression of cyclic‐di‐GMP and mediate virulence (Ryan et al., 2007).
The Mips (macrophage infectivity potentiators) genes encode proteins reported as virulence factors in human pathogens. Zang et al. (2007) showed that a mip‐like gene of Xcc is involved in pathogenicity through an effect on the production of exopolysaccharides and on the activity of extracellular proteases.
EPSs play an important role in the pathogenicity and virulence of many bacteria, both in terms of direct interactions with host cells and in conferring protection against hostile environments (Coplin and Cook, 1990). The most important EPS secreted by Xcc is xanthan gum, an EPS that has many industrial applications. Xanthan is a complex EPS with a cellulosic backbone and trisaccharide side chains of two mannose and one glucuronate residues that are attached to every second glucose in the backbone (Becker et al., 1998; Jansson et al., 1975; Vorhöelter et al., 2008; Yun et al., 2006). Yun et al. (2006) have shown that a xanthan minus mutant strain and a mutant strain that produces truncated xanthan fail to cause disease in A. thaliana and Nicotiana benthamiana, and induce the deposition of callose in these plants; pretreatment of the plant with xanthan restores the pathogenicity of both strains. The results indicate that xanthan induces susceptibility to Xcc by suppressing callose deposition. The bacterial cyclic β‐(1,2)‐glucan also has an effect on plant defence suppression and callose deposition, but, unlike xanthan, which acts locally, cyclic glucan acts systemically as part of a counter‐defensive strategy that might facilitate the spread of the pathogen in the plants (Rigano et al., 2007).
Xanthan has a broad range of applications in food and nonfood products from the oil, pharmaceutical, cosmetic, paper, paint and textile industries, and is mainly used as a thickening, stabilizing, gelling and emulsifying agent (Becker et al., 1998). The biosynthesis of xanthan production and the genes that encode for the enzymes involved have been studied (Ielpi et al., 1993; Becker et al., 1998). Xanthan synthesis is encoded by twelve gum genes (gumBCDEFGHIJKLM), which are located in a single gene cluster of 12 kb that is mainly expressed as an operon from a promoter upstream of the first gene, gumB (Vojnov et al., 2001). The genes required for the synthesis of xanthan and its nucleotide sugar precursors are highly conserved among the three sequenced strains (ATCC33913, 8004 and B100), but are significantly different when compared with the other sequenced Xanthomonas; a model for the biosynthesis of xanthan based on the genome annotation has been proposed (Vorhöelter et al., 2008). The disruption of the rmlA, xanA and gumK genes, which are responsible for the biosynthesis of intermediates for xanthan production, can lead to a loss of pathogenicity (Qian et al., 2005).
The lipopolysaccharide (LPS) is an essential component of the outer membrane of Gram‐negative bacteria, and is an important virulence factor in Xanthomonas (Dow et al., 1995; He et al., 2007; Patil et al., 2007). Over 20 genes for LPS synthesis have been characterized in Xcc, including xanAB, rmlABCB, rfaXY, lpsIJ and 15 genes that constitute the wxc cluster (Vorholter et al., 2001). The wxc gene cluster is involved in the synthesis of the LPS O‐antigen, the most variable part of LPS. The wxc genes are highly divergent between Xcc strains, indicating that different isolates might produce varied LPSs (He et al., 2007). The LPS gene cluster of Xcc B100 is significantly different from those of 8004 and ATCC33913, but is almost identical to the LPS cluster of X. campestris pv. raphani 756C. This could indicate that the LPS cluster of B100 was acquired recently via horizontal gene transfer from X. campestris pv. raphani 756C or, more likely, that the X. campestris pv. raphani strain diverged recently from an Xcc with a B100‐type LPS locus (Patil et al., 2007). Qian et al. (2005) showed that the disruption of a number of wxc genes resulted in a significant reduction in virulence.
Future Perspectives
Research into Xcc and closely related pathovars has now reached the genomic age, although it still lags behind the progress made from the investigation of Pseudomonas pathogens, such as P. syringae pv. tomato and maculicola. Our understanding of Xcc is increasing rapidly through functional and comparative genomic studies, and we are starting to understand the role of some of the key genes involved in pathogenicity. Nevertheless, there are still many areas that require further work, including the study of the mode of entry of the pathogen, such as comparisons between the vascular pathogen Xcc, which generally penetrates the host via the hydathodes, and the nonvascular pathogen X. campestris pv. raphani, which generally penetrates the host through stomata. The effect of the environment and genetic factors in determining the preferred mode of entry of these pathogens is still under‐studied. The importance of epiphytic survival and factors that contribute to it are also not well understood.
Recent studies have shown the role of certain effector proteins, but most have still not been characterized and their molecular targets and function are still unknown. Most of the effector/avirulence genes in the postulated gene‐for‐gene model have not yet been identified molecularly.
With the advances in sequencing, additional sequences of different Xcc isolates, including multiple isolates from each race and additional sequences of isolates of closely related pathovars, should soon be available. The comparison of multiple sequences should then provide more clues on the important genes that determine pathogenicity, including effectors; the presence and absence of particular genes and the variability between isolates will provide clues on the role and evolution of these genes. Functional analysis of the genes can then confirm their role in the plant–pathogen interactions.
On the host side, a number of disease resistance genes in brassicas and A. thaliana have been postulated, some have been mapped, but none have been cloned. The effort to identify more disease resistance genes should be continued, and some of the most important crops could be improved with the incorporation of disease resistance genes from closely related species via interspecific crosses and embryo rescue, or via transformation. It is also possible that the engineering of the promoters of known R genes to recognize multiple effectors could contribute to the development of broad‐spectrum disease‐resistant crops (Ryan et al., 2011). The control of the disease could be improved by pyramiding disease resistance genes and/or by using multiple lines with different gene combinations that could be mixed and balanced to match the avirulence genes present in the bacterial population (Pink and Puddephat, 1999).
The application of functional genomics and proteomics to bacteria in planta to identify virulence factors, and the application of functional genomics and proteomics to both resistant and susceptible host plants inoculated with Xcc, will provide key information on the interaction between the bacteria and the hosts. Research on the diversity of Xcc, pathogenicity factors and evolution, together with host–pathogen interaction studies, should lead to improvements in the prevention and control of the black rot of crucifers.
Acknowledgements
The authors thank Drs J. D. Taylor, S. J. Roberts, P. Hand and D.A.C. Pink for their support and work on black rot, Vânia Passo and Joseph Mulema for discussions and work on effector sequences of Xcc and X. campestris pv. raphani, and Carol Evered for taking electron microscopy photographs of Xcc. We also thank F. J. Vorhöelter for sending us strain B100 for race typing. Our work on black rot and leaf spot pathogens performed at Wellesbourne, UK, since 1996 has been funded by the Department for the Environment, Food and Rural Affairs of the UK (DEFRA), the Biotechnology and Biological Sciences Research Council (BBSRC), the UK Department for International Development (DFID) and grants from the Portuguese Foundation for Science and Technology (FCT).
References
- Alvarez, A.M. , Benedict, A.A. , Mizumoto, C.Y. , Hunter, J.E. and Gabriel, D.W. (1994) Serological, pathological and genetic diversity among strains of Xanthomonas campestris infecting crucifers. Phytopathology, 84, 1449–1457. [Google Scholar]
- An, S.Q. , Lu, G.T. , Su, H.Z. , Li, R.F. , He, Y.Q. , Jiang, B.L. , Tang, D.J. and Tang, J.L. (2011) Systematic mutagenesis of all predicted GntR genes in Xanthomonas campestris pv. campestris reveals a gntR family transcriptional regulator controlling hypersensitive response and virulence. Mol. Plant–Microbe Interact. 24, 1027–1039. [DOI] [PubMed] [Google Scholar]
- Arias, R.S. , Nelson, S.C. and Alvarez, A.M. (2000) Effect of soil‐matric potential and phylloplanes of rotation‐crops on the survival of a bioluminescent Xanthomonas campestris pv. campestris . Eur. J. Plant Pathol. 106, 109–116. [Google Scholar]
- Bain, D.C. (1952) Reaction of Brassica seedlings to black rot. Phytopathology, 42, 497–500. [Google Scholar]
- Barber, C.E. , Tang, J.L. , Feng, J.X. , Pan, M.Q. , Wilson, T.J.G. , Slater, H. , Dow, J.M. , Williams, P. and Daniels, M.J. (1997) A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24, 555–566. [DOI] [PubMed] [Google Scholar]
- Becker, A. , Katzen, F. , Puhler, A. and Ielpi, L. (1998) Xanthan gum biosynthesis and application: a biochemical/genetic perspective. Appl. Microbiol. Biotechnol. 50, 145–152. [DOI] [PubMed] [Google Scholar]
- Berg, T. , Tesoriero, L. and Hailstones, D.L. (2006) A multiplex real‐time PCR assay for detection of Xanthomonas campestris from brassicas. Lett. Appl. Microbiol. 42, 624–630. [DOI] [PubMed] [Google Scholar]
- Boch, J. and Bonas, U. (2010) Xanthomonas AvrBs3 family‐type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Koebnik, R. , Lu, H. , Furutani, A. , Angiuoli, S.V. , Patil, P.B. , Van Sluys, M.A. , Ryan, R.P. , Meyer, D.F. , Han, S.W. , Aparna, G. , Rajaram, M. , Delcher, A.L. , Phillippy, A.M. , Puiu, D. , Schatz, M.C. , Shumway, M. , Sommer, D.D. , Trapnell, C. , Benahmed, F. , Dimitrov, G. , Madupu, R. , Radune, D. , Sullivan, S. , Jha, G. , Ishihara, H. , Lee, S.W. , Pandey, A. , Sharma, V. , Sriariyanun, M. , Szurek, B. , Vera‐Cruz, C.M. , Dorman, K.S. , Ronald, P.C. , Verdier, V. , Dow, J.M. , Sonti, R.V. , Tsuge, S. , Brendel, V.P. , Rabinowicz, P.D. , Leach, J.E. , White, F.F. and Salzberg, S.L. (2011) Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J. Bacteriol. 193, 5450–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury, J.F. (1986) Guide to Plant Pathogenic Bacteria. Slough: CAB International. [Google Scholar]
- Buell, C.R. and Somerville, S.C. (1997) Use of Arabidopsis recombinant inbred lines reveals a monogenic and a novel digenic resistance mechanism to Xanthomonas campestris pv campestris . Plant J. 12, 21–29. [DOI] [PubMed] [Google Scholar]
- Burkholder, W.H. (1957) Genus II. Xanthomonas Dowson 1939 In: Bergey's Manual of Determinative Bacteriology, 7th edn. (Breed R.S., Murray E.G.D. and Smith N.R., eds), pp. 152–183. Baltimore, MD: Williams & Wilkins Co. [Google Scholar]
- Camargo, L.E.A. , Williams, P.H. and Osborn, T.C. (1995) Mapping of quantitative trait loci controlling resistance of Brassica oleracea to Xanthomonas campestris pv. campestris in the field and greenhouse. Phytopathology, 85, 1296–1300. [Google Scholar]
- Canonne, J. , Marino, D. , Jauneau, A. , Pouzet, C. , Briere, C. , Roby, D. and Rivas, S. (2011) The Xanthomonas type III effector XopD targets the Arabidopsis transcription factor MYB30 to suppress plant defense. Plant Cell, 23, 3498–3511. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Canonne, J. , Pichereaux, C. , Marino, D. , Roby, D. , Rossignol, M. and Rivas, S. (2012) Identification of the protein sequence of the type III effector XopD from the B100 strain of Xanthomonas campestris pv campestris . Plant Signal. Behav. 7, 184–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda, A. , Reddy, J.D. , El‐Yacoubi, B. and Gabriel, D.W. (2005) Mutagenesis of all eight avr genes in Xanthomonas camplestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol. Plant–Microbe Interact. 18, 1306–1317. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Roberts, P.D. and Gabriel, D.W. (1994) Effects of a virulence locus from Xanthomonas campestris 528T on pathovar status and ability to elicit blight symptoms on crucifers. Phytopathology, 84, 1458–1465. [Google Scholar]
- Cook, A.A. , Walker, J.C. and Larson, R.H. (1952) Studies on the disease cycle of black rot of crucifers. Phytopathology, 42, 162–167. [Google Scholar]
- Coplin, D.L. and Cook, D. (1990) Molecular genetics of extracellular polysaccharide biosynthesis in vascular phytopathogenic bacteria. Mol. Plant–Microbe Interact. 3, 271–279. [DOI] [PubMed] [Google Scholar]
- Corbett, M. , Virtue, S. , Bell, K. , Birch, P. , Burr, T. , Hyman, L. , Lilley, K. , Poock, S. , Toth, I. and Salmond, G. (2005) Identification of a new quorum‐sensing‐control led virulence factor in Erwinia carotovora subsp. atroseptica secreted via the type II targeting pathway. Mol. Plant–Microbe Interact. 18, 334–342. [DOI] [PubMed] [Google Scholar]
- Craig, L. , Pique, M.E. and Tainer, J.A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378. [DOI] [PubMed] [Google Scholar]
- Cunnac, S. , Lindeberg, M. and Collmer, A. (2009) Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr. Opin. Microbiol. 12, 53–60. [DOI] [PubMed] [Google Scholar]
- Dane, F. and Shaw, J.J. (1996) Survival and persistence of bioluminescent Xanthomonas campestris pv. campestris on host and non‐host plants in the field environment. J. Appl. Bacteriol. 80, 73–80. [Google Scholar]
- Dickson, M.D. and Hunter, J.E. (1987) Inheritance of resistance in cabbage seedlings to black rot. HortScience, 22, 108–109. [Google Scholar]
- Dow, J.M. , Osbourn, A.E. , Wilson, T.J.G. and Daniels, M.J. (1995) A locus determining pathogenicity of Xanthomonas campestris is involved in lipopolysaccharide biosynthesis. Mol. Plant–Microbe Interact. 8, 768–777. [DOI] [PubMed] [Google Scholar]
- Dow, J.M. , Feng, J.X. , Barber, C.E. , Tang, J.L. and Daniels, M.J. (2000) Novel genes involved in the regulation of pathogenicity factor production within the rpf gene cluster of Xanthomonas campestris . Microbiology, 146, 885–891. [DOI] [PubMed] [Google Scholar]
- Dow, J.M. , Crossman, L. , Findlay, K. , He, Y.Q. , Feng, J.X. and Tang, J.L. (2003) Biofilm dispersal in Xanthomonas campestris is controlled by cell–cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA, 100, 10995–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargier, E. and Manceau, C. (2007) Pathogenicity assays restrict the species Xanthomonas campestris into three pathovars and reveal nine races within X. campestris pv. campestris . Plant Pathol. 56, 805–818. [Google Scholar]
- Fargier, E. , Saux, M.F.L. and Manceau, C. (2011) A multilocus sequence analysis of Xanthomonas campestris reveals a complex structure within crucifer‐attacking pathovars of this species. Syst. Appl. Microbiol. 34, 156–165. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971) Current status of gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Franken, A. (1992) Application of polyclonal and monoclonal antibodies for the detection of Xanthomonas campestris pv. campestris in crucifer seeds using immunofluorescence microscopy. Neth. J. Plant Pathol. 98, 95–106. [Google Scholar]
- Gabriel, D.W. (1999) Why do pathogens carry avirulence genes? Physiol. Mol. Plant Pathol. 55, 205–214. [Google Scholar]
- Garman, H. (1894) A bacterial disease of cabbage. Kentucky Agr. Exp. Stn. Rep. 3, 43–46. [Google Scholar]
- Gonçalves, E.R. and Rosato, Y.B. (2002) Phylogenetic analysis of Xanthomonas species based upon 16S–23S rDNA intergenic spacer sequences. Int. J. Syst. Evol. Microbiol. 52, 355–361. [DOI] [PubMed] [Google Scholar]
- Gudesblat, G.E. , Torres, P.S. and Vojnov, A.A. (2009) Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell‐to‐cell signal‐regulated virulence factor. Plant Physiol. 149, 1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H. , Dickson, M.H. and Hunter, J.E. (1991) Brassica napus sources of resistance to black rot in crucifers and inheritance of resistance. Hortscience, 26, 1545–1547. [Google Scholar]
- Gurlebeck, D. , Thieme, F. and Bonas, U. (2006) Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163, 233–255. [DOI] [PubMed] [Google Scholar]
- Hauben, L. , Vauterin, L. , Swings, J. and Moore, E.R.B. (1997) Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int. J. Syst. Bacteriol. 47, 328–335. [DOI] [PubMed] [Google Scholar]
- Hayward, A.C. (1993) The hosts of Xanthomonas In: Xanthomonas (Swings J.G. and Civerolo E.L., eds), pp. 1–119. London: Chapman & Hall. [Google Scholar]
- He, Y.Q. , Zhang, L. , Jiang, B.L. , Zhang, Z.C. , Xu, R.Q. , Tang, D.J. , Qin, J. , Jiang, W. , Zhang, X. , Liao, J. , Cao, J.R. , Zhang, S.S. , Wei, M.L. , Liang, X.X. , Lu, G.T. , Feng, J.X. , Chen, B. , Cheng, J. and Tang, J.L. (2007) Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris . Genome Biol. 8, R218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y.W. , Wu, J. , Zhou, L. , Yang, F. , He, Y.O. , Jiang, B.L. , Bai, L.Q. , Xu, Y.Q. , Deng, Z.X. , Tang, J.L. and Zhang, L.H. (2011) Xanthomonas campestris diffusible factor is 3‐hydroxybenzoic acid and is associated with xanthomonadin biosynthesis, cell viability, antioxidant activity, and systemic invasion. Mol. Plant–Microbe Interact. 24, 948–957. [DOI] [PubMed] [Google Scholar]
- Holub, E.B. (2007) Natural variation in innate immunity of a pioneer species. Curr. Opin. Plant Biol. 10, 415–424. [DOI] [PubMed] [Google Scholar]
- Hsiao, Y.M. , Liu, Y.F. , Fang, M.C. and Song, W.L. (2011) XCC2731, a GGDEF domain protein in Xanthomonas campestris, is involved in bacterial attachment and is positively regulated by Clp. Microbiol. Res. 166, 548–565. [DOI] [PubMed] [Google Scholar]
- Hunter, J.E. , Dickson, M.H. and Ludwig, J.W. (1987) Source of resistance to black rot of cabbage expressed in seedlings and adult plants. Plant Dis. 71, 263–266. [Google Scholar]
- Ielpi, L. , Couso, R.O. and Dankert, M.A. (1993) Sequential assembly and polymerization of the polyprenol‐linked pentasaccharide repeating unit of the xanthan polysaccharide in Xanthomonas campestris . J. Bacteriol. 175, 2490–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatov, A. , Hida, K. and Kuginuki, Y. (1998a) Black rot of crucifers and sources of resistance in Brassica crops. JARQ Japan Agric. Res. Q. 32, 167–172. [Google Scholar]
- Ignatov, A. , Kuginuki, Y. and Hida, K. (1998b) Race‐specific reaction of resistance to black rot in Brassica oleracea . Eur. J. Plant Pathol. 104, 821–827. [Google Scholar]
- Ignatov, A. , Kuginuki, Y. and Hidam, K. (2000) Distribution and inheritance of race‐specific resistance to Xanthomonas campestris pv. campestris in Brassica rapa and B. napus . J. Russ. Phytopathol. Soc. 1, 89–94. [Google Scholar]
- Ignatov, A.N. , Monakhos, G.F. , Djalilov, F.S. and Pozmogova, G.V. (2002) Avirulence gene from Xanthomonas campestris pv. campestris homologous to the avrBs2 locus is recognized in race‐specific reaction by two different resistance genes in brassicas. Russ. J. Genet. 38, 1404–1410. [PubMed] [Google Scholar]
- Jansson, P.E , Kenne, L. and Lindberg, B. (1975) Structure of extracellular polysaccharide from Xanthomonas campestris . Carbohydr. Res. 45, 275–282. [DOI] [PubMed] [Google Scholar]
- Jensen, B.D. , Vicente, J.G. , Manandhar, H.K. and Roberts, S.J. (2007) An increasing risk for bacterial plant diseases in Northern Europe? Occurrence and diversity of Xanthomonas campestris pv. campestris in vegetable Brassica fields in Nepal and identification of a new race. In: Plant Biotech Denmark . Copenhagen, Denmark: Faculty of Life Sciences, University of Copenhagen. [DOI] [PubMed] [Google Scholar]
- Jensen, B.D. , Vicente, J.G. , Manandhar, H.K. and Roberts, S.J. (2010) Occurrence and diversity of Xanthomonas campestris pv. campestris in vegetable brassica fields in Nepal. Plant Dis. 94, 298–305. [DOI] [PubMed] [Google Scholar]
- Jiang, B. , Xu, R.Q. , Li, X.Z. , Wei, H.Y. , Bai, F. , Hu, X. , He, Y.Q. and Tang, J.L. (2006) Construction and characterization of a hrpG mutant rendering constitutive expression of hrp genes in Xanthomonas campestris pv. campestris . Prog. Nat. Sci. 16, 480–485. [Google Scholar]
- Jiang, B.L. , He, Y.Q. , Cen, W.J. , Wei, H.Y. , Jiang, G.F. , Jiang, W. , Hang, X.H. , Feng, J.X. , Lu, G.T. , Tang, D.H. and Tang, J.L. (2008a) The type III secretion effector xopXccN of Xanthomonas campestris pv. campestris is required for full virulence. Res. Microbiol. 159, 216–220. [DOI] [PubMed] [Google Scholar]
- Jiang, B.L. , Liu, J. , Chen, L.F. , Ge, Y.Y. , Hang, X.H. , He, Y.Q. , Tang, D.J. , Lu, G.T. and Tang, J.L. (2008b) DsbB is required for the pathogenesis process of Xanthomonas campestris pv. campestris . Mol. Plant–Microbe Interact. 21, 1036–1045. [DOI] [PubMed] [Google Scholar]
- Jiang, W. , Jiang, B.L. , Xu, R.Q. , Huang, J.D. , Wei, H.Y. , Jiang, G.F. , Cen, W.J. , Liu, J. , Ge, Y.Y. , Li, G.H. , Su, L.L. , Hang, X.H. , Tang, D.J. , Lu, G.T. , Feng, J.X. , He, Y.Q. and Tang, J.L. (2009) Identification of six type III effector genes with the PIP box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity. Mol. Plant–Microbe Interact. 22, 1401–1411. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. and Kado, C.I. (1990) A plant‐inducible gene of Xanthomonas campestris pv. campestris encodes an exocellular component required for growth in the host and hypersensitivity on nonhosts. J. Bacteriol. 172, 5165–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. , Kamdar, H.V. , Tola, E. and Kado, C.I. (1992) Incompatible interactions between crucifers and Xanthomonas campestris involve a vascular hypersensitive response: role of the hrpX locus. Mol. Plant–Microbe Interact. 5, 22–33. [Google Scholar]
- Kay, S. , Boch, J. and Bonas, U. (2005) Characterization of AvrBs3‐like effectors from a Brassicaceae pathogen reveals virulence and avirulence activities and a protein with a novel repeat architecture. Mol. Plant–Microbe Interact. 18, 838–848. [DOI] [PubMed] [Google Scholar]
- Keen, N.T. (1990) Gene‐for‐gene complementarity in plant–pathogen interactions. Annu. Rev. Genet. 24, 447–463. [DOI] [PubMed] [Google Scholar]
- Kocks, C.G. and Zadoks, J.C. (1996) Cabbage refuse piles as sources of inoculum for black rot epidemics. Plant Dis. 80, 789–792. [Google Scholar]
- Koenraadt, H. , Van Bilsen, J. and Roberts, S.J. (2005) Comparative test of four semi‐selective agar media for the detection of Xanthomonas campestris pv. campestris in brassica seeds. Seed Sci. Technol. 33, 115–125. [Google Scholar]
- Krauthausen, H.J. , Laun, N. and Wohanka, W. (2011) Methods to reduce the spread of the black rot pathogen, Xanthomonas campestris pv. campestris, in brassica transplants. J. Plant Dis. Prot. 118, 7–16. [Google Scholar]
- Lee, B.M. , Park, Y.J. , Park, D.S. , Kang, H.W. , Kim, J.G. , Song, E.S. , Park, I.C. , Yoon, U.H. , Hahn, J.H. , Koo, B.S. , Lee, G.B. , Kim, H. , Park, H.S. , Yoon, K.O. , Kim, J.H. , Jung, C. , Koh, N.H. , Seo, J.S. and Go, S.J. (2005) The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lema, M. , Cartea, M.E. , Sotelo, T. , Velasco, P. and Soengas, P. (2012) Discrimination of Xanthomonas campestris pv. campestris races among strains from northwestern Spain by Brassica spp. genotypes and rep‐PCR. Eur. J. Plant Pathol. 133, 159–169. [Google Scholar]
- Lindgren, P.B. (1997) The role of hrp genes during plant–bacterial interactions. Annu. Rev. Phytopathol. 35, 129–152. [DOI] [PubMed] [Google Scholar]
- Lu, H. , Patil, P. , Van Sluys, M.‐A. , White, F.F. , Ryan, R.P. , Dow, J.M. , Rabinowicz, P. , Salzberg, S.L. , Leach, J.E. , Sonti, R. , Brendel, V. and Bogdanove, A.J. (2008) Acquisition and evolution of plant pathogenesis‐associated gene clusters and candidate determinants of tissue‐specificity in Xanthomonas . PLoS ONE, 3, e3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lummerzheim, M. , Kroj, T. , Ferreira, M. , Tronchet, M. , Godard, F. , Van Montagu, M. and Roby, D. (2004) An Arabidopsis mutant with altered hypersensitive response to Xanthomonas campestris pv. campestris, hxc1, displays a complex pathophenotype. Mol. Plant Pathol. 5, 453–464. [DOI] [PubMed] [Google Scholar]
- Massomo, S.M.S. , Nielsen, H. , Mabagala, R.B. , Mansfeld‐Giese, K. , Hockenhull, J. and Mortensen, C.N. (2003) Identification and characterisation of Xanthomonas campestris pv. campestris strains from Tanzania by pathogenicity tests, Biolog, rep‐PCR and fatty acid methyl ester analysis. Eur. J. Plant Pathol. 109, 775–789. [Google Scholar]
- McCulloch, L. (1929) A bacterial leaf spot of horse‐radish caused by Bacterium campestris var. armoraciae, n. var. J. Agric. Res. 38, 269–287. [Google Scholar]
- Meier, D. (1934) A cytological study of the early infection stages of the black rot of cabbage. Bull. Torrey Bot. Club, 61, 173–190. [Google Scholar]
- Moore, E.R.B. , Kruger, A.S. , Hauben, L. , Seal, S.E. , Daniels, M.J. , De Baere, R. , De Wachter, R. , Timmis, K.N. and Swings, J. (1997a) 16S rRNA gene sequence analyses and inter‐ and intrageneric relationships of Xanthomonas species and Stenotrophomonas maltophilia . FEMS Microbiol. Lett. 155, 231–231. [DOI] [PubMed] [Google Scholar]
- Moore, E.R.B. , Kruger, A.S. , Hauben, L. , Seal, S.E. , DeBaere, R. , DeWachter, R. , Timmis, K.N. and Swings, J. (1997b) 16S rRNA gene sequence analyses and inter‐ and intrageneric relationships of Xanthomonas species and Stenotrophomonas maltophilia . FEMS Microbiol. Lett. 151, 145–153. [DOI] [PubMed] [Google Scholar]
- Mulema, J.K. , Vicente, J.G. , Pink, D.A.C. , Jackson, A. , Chacha, D.O. , Wasilwa, L. , Kinyua, Z.M. , Karanja, D.K. , Holub, E.B. and Hand, P. (2012) Characterisation of isolates that cause black rot of crucifers in East Africa. Eur. J. Plant Pathol. 133, 427–438. [Google Scholar]
- Nagaharu, U. (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7, 389–452. [Google Scholar]
- Ochiai, H. , Inoue, V. , Takeya, M. , Sasaki, A. and Kaku, H. (2005) Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. JARQ Japan Agric. Res. Q. 39, 275–287. [Google Scholar]
- Pammel, L.H. (1895a) Bacteriosis of rutabaga (Bacillus campestris n sp.). Am. Mon. Microsc. J. 16, 145–151. [Google Scholar]
- Pammel, L.H. (1895b) Bacteriosis of rutabaga (Bacillus campestris n. sp.). Iowa State Coll. Agric. Exp. Stn. Bull. 27, 130–134. [Google Scholar]
- Parker, J.E. , Barber, C.E. , Fan, M.J. and Daniels, M.J. (1993) Interaction of Xanthomonas campestris with Arabidopsis thaliana: characterization of a gene from X. c. pv. raphani that confers avirulence to most A. thaliana accessions. Mol. Plant–Microbe Interact. 6, 216–224. [DOI] [PubMed] [Google Scholar]
- Parkinson, N. , Aritua, V. , Heeney, J. , Cowie, C. , Bew, J. and Stead, D. (2007) Phylogenetic analysis of Xanthomonas species by comparison of partial gyrase B gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2881–2887. [DOI] [PubMed] [Google Scholar]
- Parkinson, N. , Cowie, C. , Heeney, J. and Stead, D. (2009) Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int. J. Syst. Evol. Microbiol. 59, 264–274. [DOI] [PubMed] [Google Scholar]
- Patil, P.B. , Bogdanove, A.J. and Sonti, R.V. (2007) The role of horizontal transfer in the evolution of a highly variable lipopolysaccharide biosynthesis locus in xanthomonads that infect rice, citrus and crucifers. BMC Evol. Biol. 7, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti, I. , Royer, M. , Barbe, V. , Carrere, S. , Koebnik, R. , Cociancich, S. , Couloux, A. , Darrasse, A. , Gouzy, J. , Jacques, M.A. , Lauber, E. , Manceau, C. , Mangenot, S. , Poussier, S. , Segurens, B. , Szurek, B. , Verdier, V. , Arlat, M. and Rott, P. (2009) The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem‐limited Xanthomonadaceae . BMC Genomics, 10, 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink, D. and Puddephat, I. (1999) Deployment of disease resistance genes by plant transformation—a ‘mix and match’ approach. Trends Plant Sci. 4, 71–75. [DOI] [PubMed] [Google Scholar]
- Poplawsky, A.R. and Chun, W. (1995) Strains of Xanthomonas campestris pv. campestris with atypical pigmentation isolated from commercial crucifer seeds. Plant Dis. 79, 1021–1024. [Google Scholar]
- Poplawsky, A.R. and Chun, W. (1998) Xanthomonas campestris pv. campestris requires a functional pigB for epiphytic survival and host infection. Mol. Plant–Microbe Interact. 11, 466–475. [DOI] [PubMed] [Google Scholar]
- Qian, W. , Jia, Y.T. , Ren, S.X. , He, Y.Q. , Feng, J.X. , Lu, L.F. , Sun, Q.H. , Ying, G. , Tang, D.J. , Tang, H. , Wu, W. , Hao, P. , Wang, L.F. , Jiang, B.L. , Zeng, S.Y. , Gu, W.Y. , Lu, G. , Rong, L. , Tian, Y.C. , Yao, Z.J. , Fu, G. , Chen, B.S. , Fang, R.X. , Qiang, B.Q. , Chen, Z. , Zhao, G.P. , Tang, J.L. and He, C.Z. (2005) Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris . Genome Res. 15, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademaker, J.L.W. , Hoste, B. , Louws, F.J. , Kersters, K. , Swings, J. , Vauterin, L. , de Vauterin, P. and Bruijn, F.J. (2000) Comparison of AFLP and rep‐PCR genomic fingerprinting with DNA–DNA homology studies: Xanthomonas as a model system. Int. J. Syst. Evol. Microbiol. 50, 665–677. [DOI] [PubMed] [Google Scholar]
- Rademaker, J.L.W. , Louws, F.J. , Schultz, M.H. , Rossbach, U. , Vauterin, L. , de Swings, J. and Bruijn, F.J. (2005) A comprehensive species to strain taxonomic framework for Xanthomonas . Phytopathology, 95, 1098–1111. [DOI] [PubMed] [Google Scholar]
- Rigano, L.A. , Payette, C. , Brouillard, G. , Marano, M.R. , Abramowicz, L. , Torres, P.S. , Yun, M. , Castagnaro, A.P. , El Oirdi, M. , Dufour, V. , Malamud, F. , Dow, J.M. , Bouarab, K. and Vojnov, A. (2007) Bacterial cyclic beta‐(1,2)‐glucan acts in systemic suppression of plant immune responses. Plant Cell, 19, 2077–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, S.J. and Koenraadt, H. (2006) 7‐019: detection of Xanthomonas campestris pv. campestris on Brassica spp. In: International Rules for Seed Testing , pp. 1–16. Bassersdorf: International Seed Testing Association (ISTA). [Google Scholar]
- Roberts, S.J. , Brough, J. and Hunter, P.J. (2007) Modelling the spread of Xanthomonas campestris pv. campestris in module‐raised brassica transplants. Plant Pathol. 56, 391–401. [Google Scholar]
- Russell, H.L. (1898) A bacterial rot of cabbage and allied plants. Wis. Agric. Exp. Stn. Bull. 65, 1–39. [Google Scholar]
- Ryan, R.P. , Fouhy, Y. , Lucey, J.F. , Jiang, B.L. , He, Y.Q. , Feng, J.X. , Tang, J.L. and Dow, J.M. (2007) Cyclic di‐GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris . Mol. Microbiol. 63, 429–442. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. , McCarthy, Y. , Andrade, M. , Farah, C.S. , Armitage, J.P. and Dow, J.M. (2010) Cell–cell signal‐dependent dynamic interactions between HD‐GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris . Proc. Natl. Acad. Sci. USA, 107, 5989–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, R.P. , Vorholter, F.J. , Potnis, N. , Jones, J.B. , Van Sluys, M.A. , Bogdanove, A.J. and Dow, J.M. (2011) Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nat. Rev. Microbiol. 9, 344–355. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L. , Sommer, D.D. , Schatz, M.C. , Phillippy, A.M. , Rabinowicz, P.D. , Tsuge, S. , Furutani, A. , Ochiai, H. , Delcher, A.L. , Kelley, D. , Madupu, R. , Puiu, D. , Radune, D. , Shumway, M. , Trapnell, C. , Aparna, G. , Jha, G. , Pandey, A. , Patil, P.B. , Ishihara, H. , Meyer, D.F. , Szurek, B. , Verdier, V. , Koebnik, R. , Dow, J.M. , Ryan, R.P. , Hirata, H. , Tsuyumu, S. , Lee, S.W. , Ronald, P.C. , Sonti, R.V. , Van Sluys, M.A. , Leach, J.E. , White, F.F. and Bogdanove, A.J. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics, 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, N.W. (1982) Detection of seedborne bacterial plant pathogens. Plant Dis. 66, 885–890. [Google Scholar]
- Schaad, N.W. and Alvarez, A. (1993) Xanthomonas campestris pv. campestris: cause of black rot of crucifers In: Xanthomonas (Swings J.G. and Civerolo E.L., eds), pp. 51–55. London: Chapman & Hall. [Google Scholar]
- Schaad, N.W. and Dianese, J.C. (1981) Cruciferous weeds as sources of inoculum of Xanthomonas campestris in black rot of crucifers. Phytopathology, 71, 1215–1220. [Google Scholar]
- Schaad, N.W. and Thaveechai, N. (1983) Black rot of crucifers in Thailand. Plant Dis. 67, 1231–1234. [Google Scholar]
- Schaad, N.W. and White, W.C. (1974) Survival of Xanthomonas campestris in soil. Phytopathology, 64, 1518–1520. [Google Scholar]
- Schaad, N.W. , Vidaver, A.K. , Lacy, G.H. , Rudolph, K. and Jones, J.B. (2000) Evaluation of proposed amended names of several pseudomonads and xanthomonads and recommendations. Phytopathology, 90, 208–213. [DOI] [PubMed] [Google Scholar]
- Shih, H. , Lin, Y. , Huang, H. , Tzeng, K. , Hsu, S. , Shih, H.D. , Lin, Y.C. , Huang, H.C. , Tzeng, K.C. and Hsu, S.T. (2000) A DNA probe for identification of Xanthomonas campestris pv. campestris, the causal organism of black rot of crucifers in Taiwan. Bot. Bull. Acad. Sin. 41, 113–120. [Google Scholar]
- da Silva, A.C.R. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , do Alves, L.M.C., Amaral, A.M. , Bertolini, M.C. , Camargo, L.E.A. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Clapina, L.P. , Cicarelli, R.M.B. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J.S. , Ferreira, R.C.C. , Ferro, M.I.T. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G.M. , Lemos, M.V.F. , Locali, E.C. , Machado, M.A. , Madeira, A. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F.M. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T.M. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A.D. , de Silva, C., Souza, R.F. , Spinola, L.A.F. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I.D. , dos Santos, M.T. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Simões, T.H.N. , Gonçalves, E.R. , Rosato, Y.B. and Mehta, A. (2007) Differentiation of Xanthomonas species by PCR‐RFLP of rpfB and atpD genes. FEMS Microbiol. Lett. 271, 33–39. [DOI] [PubMed] [Google Scholar]
- Simpson, R.B. and Johnson, L.J. (1990) Arabidopsis thaliana as a host for Xanthomonas campestris pv. campestris . Mol. Plant–Microbe Interact. 3, 233–237. [Google Scholar]
- Smith, E.F. (1898) The black rot of the cabbage. US Dep. Agric. Farmer's Bull. 68, 1–21. [Google Scholar]
- Soengas, P. , Hand, P. , Vicente, J.G. , Pole, J.M. and Pink, D.A.C. (2007) Identification of quantitative trait loci for resistance to Xanthomonas campestris pv. campestris in Brassica rapa . Theor. Appl. Genet. 114, 637–645. [DOI] [PubMed] [Google Scholar]
- Souza, D.P. , Andrade, M.O. , Alvarez‐Martinez, C.E. , Arantes, G.M. , Farah, C.S. and Salinas, R.K. (2011) A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PLoS Pathog. 7, e1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme, D.J. , Kemen, E. , MacLean, D. , Schornack, S. , Aritua, V. , Thwaites, R. , Grant, M. , Smith, J. and Jones, J.D.G. (2010) Genome‐wide sequencing data reveals virulence factors implicated in banana Xanthomonas wilt. FEMS Microbiol. Lett. 310, 182–192. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Takikawa, Y. , Tsuyumu, S. and Goto, M. (1994) Bacterial spot of crucifers caused by Xanthomonas campestris pv. raphani . Ann. Phytopathol. Soc. Jpn. 60, 281–287. [Google Scholar]
- Tang, J.L. , Liu, Y.N. , Barber, C.E. , Dow, J.M. , Wootton, J.C. and Daniels, M.J. (1991) Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris . Mol. Gen. Genet. 226, 409–417. [DOI] [PubMed] [Google Scholar]
- Taylor, J.D. , Conway, J. , Roberts, S.J. , Astley, D. and Vicente, J.G. (2002) Sources and origin of resistance to Xanthomonas campestris pv. campestris in Brassica genomes. Phytopathology, 92, 105–111. [DOI] [PubMed] [Google Scholar]
- Thieme, F. , Koebnik, R. , Bekel, T. , Berger, C. , Boch, J. , Buttner, D. , Caldana, C. , Gaigalat, L. , Goesmann, A. , Kay, S. , Kirchner, O. , Lanz, C. , Linke, B. , McHardy, A.C. , Meyer, F. , Mittenhuber, G. , Nies, D.H. , Niesbach‐Klosgen, U. , Patschkowski, T. , Ruckert, C. , Rupp, O. , Schneiker, S. , Schuster, S.C. , Vorholter, F.J. , Weber, E. , Puhler, A. , Bonas, U. , Bartels, D. and Kaiser, O. (2005) Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187, 7254–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, P.S. , Malamud, F. , Rigano, L.A. , Russo, D.M. , Marano, M.R. , Castagnaro, A.P. , Zorreguieta, A. , Bouarab, K. , Dow, J.M. and Vojnov, A.A. (2007) Controlled synthesis of the DSF cell–cell signal is required for biofilm formation and virulence in Xanthomonas campestris . Environ. Microbiol. 9, 2101–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, J. , Somerville, S.C. and Hammerschmidt, R. (1991) Identification of a gene in Arabidopsis thaliana that controls resistance to Xanthomonas campestris pv. campestris . Physiol. Mol. Plant Pathol. 38, 57–65. [Google Scholar]
- Van Den Mooter, M. and Swings, J. (1990) Numerical analysis of 295 phenotypic features of 266 Xanthomonas strains and related strains and an improved taxonomy of the genus. Int. J. Syst. Bacteriol. 40, 348–369. [DOI] [PubMed] [Google Scholar]
- Vauterin, L. , Swings, J. , Kersters, K. , Gillis, M. , Mew, T.W. , Schroth, M.N. , Palleroni, N.J. , Hildebrand, D.C. , Stead, D.E. , Civerolo, E.L. , Hayward, A.C. , Maraite, H. , Stall, R.E. , Vidaver, A.K. and Bradbury, J.F. (1990) Towards an improved taxonomy of Xanthomonas . Int. J. Syst. Bacteriol. 40, 312–316. [Google Scholar]
- Vauterin, L. , Hoste, B. , Kersters, K. and Swings, J. (1995) Reclassification of Xanthomonas . Int. J. Syst. Bacteriol. 45, 472–489. [Google Scholar]
- Vauterin, L. , Rademaker, J. and Swings, J. (2000) Synopsis on the taxonomy of the genus Xanthomonas . Phytopathology, 90, 677–682. [DOI] [PubMed] [Google Scholar]
- Vicente, J.G. (2004) A Podridao Negra Das Cruciferas (Lopes G., ed). Alcobaca: COTHN Centro Operativo e Tecnologico Hortofruticola. [Google Scholar]
- Vicente, J.G. , Ignatov, A. , Conway, J. , Roberts, S.J. and Taylor, J.D. (1998) Development of an improved Brassica differential series for the identification of races of Xanthomonas campestris pv. campestris . In: 7th International Congress of Plant Pathology , Edinburgh, p. 2.2.71.
- Vicente, J.G. , Conway, J. , Roberts, S.J. and Taylor, J.D. (2001) Identification and origin of Xanthomonas campestris pv. campestris races and related pathovars. Phytopathology, 91, 492–499. [DOI] [PubMed] [Google Scholar]
- Vicente, J.G. , Taylor, J.D. , Sharpe, A.G. , Parkin, I.A.P. , Lydiate, D.J. and King, G.J. (2002) Inheritance of race‐specific resistance to Xanthomonas campestris pv. campestris in Brassica genomes. Phytopathology, 92, 1134–1141. [DOI] [PubMed] [Google Scholar]
- Vicente, J.G. , Everett, B. and Roberts, S.J. (2006) Identification of isolates that cause a leaf spot disease of brassicas as Xanthomonas campestris pv. raphani and pathogenic and genetic comparison with related pathovars. Phytopathology, 96, 735–745. [DOI] [PubMed] [Google Scholar]
- Vojnov, A.A. , Slater, H. , Daniels, M.J. and Dow, J.M. (2001) Expression of the gum operon directing xanthan biosynthesis in Xanthomonas campestris and its regulation in planta. Mol. Plant–Microbe Interact. 14, 768–774. [DOI] [PubMed] [Google Scholar]
- Vorhöelter, F.J. , Schneiker, S. , Goesmann, A. , Krause, L. , Bekel, T. , Kaiser, O. , Linke, B. , Patschkowski, T. , Rueckert, C. , Schmid, J. , Sidhu, V.K. , Sieber, V. , Tauch, A. , Watt, S.A. , Weisshaar, B. , Becker, A. , Niehaus, K. and Puehler, A. (2008) The genome of Xanthomonas campestris pv. campestris B 100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 134, 33–45. [DOI] [PubMed] [Google Scholar]
- Vorholter, F.J. , Niehaus, K. and Puhler, A. (2001) Lipopolysaccharide biosynthesis in Xanthomonas campestris pv. campestris: a cluster of 15 genes is involved in the biosynthesis of the LPS O‐antigen and the LPS core. Mol. Genet. Genomics, 266, 79–95. [DOI] [PubMed] [Google Scholar]
- Walker, J.C. (1953) Cauliflower, cabbage, and others. In: Plant Diseases. The Yearbook of Agriculture 1953 , pp. 425–430. Washington: US Department of Agriculture. [Google Scholar]
- Wang, L.F. , Tang, X.Y. and He, C.Z. (2007) The bifunctional effector AvrXccC of Xanthomonas campestris pv. campestris requires plasma membrane‐anchoring for host recognition. Mol. Plant Pathol. 8, 491–501. [DOI] [PubMed] [Google Scholar]
- Wei, K. , Tang, D.J. , He, Y.Q. , Feng, J.X. , Jiang, B.L. , Lu, G.T. , Chen, B.S. and Tang, J.L. (2007) hpaR, a putative marR family transcriptional regulator, is positively controlled by HrpG and HrpX and involved in the pathogenesis, hypersensitive response, and extracellular protease production of Xanthomonas campestris pathovar campestris . J. Bacteriol. 189, 2055–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westman, A.L. , Kresovich, S. and Dickson, M.H. (1999) Regional variation in Brassica nigra and other weedy crucifers for disease reaction to Alternaria brassicicola and Xanthomonas campestris pv. campestris . Euphytica, 106, 253–259. [Google Scholar]
- White, F.F. , Potnis, N. , Jones, J.B. and Koebnik, R. (2009) The type III effectors of Xanthomonas . Mol. Plant Pathol. 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, H.E. (1930) Bacterial spot of radish and turnip. Phytopathology, 20, 653–662. [Google Scholar]
- Williams, P.H. (1980) Black rot: a continuing threat to world crucifers. Plant Dis. 64, 736–742. [Google Scholar]
- Williams, P.H. , Staub, T. and Sutton, J.C. (1972) Inheritance of resistance in cabbage to black rot. Phytopathology, 62, 247–252. [Google Scholar]
- Wilson, T.J.G. , Bertrand, N. , Tang, J.L. , Feng, J.X. , Pan, M.Q. , Barber, C.E. , Dow, J.M. and Daniels, M.J. (1998) The rpfA gene of Xanthomonas campestris pathovar campestris, which is involved in the regulation of pathogenicity factor production, encodes an aconitase. Mol. Microbiol. 28, 961–970. [DOI] [PubMed] [Google Scholar]
- Xu, R.Q. , Li, X.Z. , Wei, H.Y. , Jiang, B. , Li, K. , He, Y.Q. , Feng, J.X. and Tang, J.L. (2006) Regulation of eight avr genes by hrpG and hrpX in Xanthomonas campestris pv. campestris and their role in pathogenicity. Prog. Nat. Sci. 16, 1288–1294. [Google Scholar]
- Xu, R.Q. , Blanvillain, S. , Feng, J.X. , Jiang, B.L. , Li, X.Z. , Wei, H.Y. , Kroj, T. , Lauber, E. , Roby, D. , Chen, B. , He, Y.Q. , Lu, G.T. , Tang, D.J. , Vasse, J. , Arlat, M. and Tang, J.L. (2008) AvrAC(Xcc8004), a type III effector with a leucine‐rich repeat domain from Xanthomonas campestris pathovar campestris confers avirulence in vascular tissues of Arabidopsis thaliana ecotype Col‐0. J. Bacteriol. 190, 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.M. , Park, D.C. , Shearman, H.M. and Fargier, E. (2008) A multilocus sequence analysis of the genus Xanthomonas . Syst. Appl. Microbiol. 31, 366–377. [DOI] [PubMed] [Google Scholar]
- Yun, M.H. , Torres, P.S. , El Oirdi, M. , Rigano, L.A. , Gonzalez‐Lamothe, R. , Marano, M.R. , Castagnaro, A.P. , Dankert, M.A. , Bouarab, K. and Vojnov, A. (2006) Xanthan induces plant susceptibility by suppressing callose deposition. Plant Physiol. 141, 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccardelli, M. , Campanile, F. , Spasiano, A. and Merighi, M. (2007) Detection and identification of the crucifer pathogen, Xanthomonas campestris pv. campestris, by PCR amplification of the conserved Hrp/type III secretion system gene hrcC . Eur. J. Plant Pathol. 118, 299–306. [Google Scholar]
- Zang, N. , Tang, D.J. , Wei, M.L. , He, Y.Q. , Chen, B.S. , Feng, J.X. , Xu, J. , Gan, Y.Q. , Jiang, B.L. and Tang, J.L. (2007) Requirement of a mip‐like gene for virulence in the phytopathogenic bacterium Xanthomonas campestris pv. campestris . Mol. Plant–Microbe Interact. 20, 21–30. [DOI] [PubMed] [Google Scholar]


