SUMMARY
Ascochyta and Phoma are fungal genera containing several important plant pathogenic species. These genera are morphologically similar, and recent molecular studies performed to unravel their phylogeny have resulted in the establishment of several new genera within the newly erected Didymellaceae family. An analysis of the structure of fungal mating‐type genes can contribute to a better understanding of the taxonomic relationships of these plant pathogens, and may shed some light on their evolution and on differences in sexual strategy and pathogenicity. We analysed the mating‐type loci of phylogenetically closely related Ascochyta and Phoma species (Phoma clematidina, Didymella vitalbina, Didymella clematidis, Peyronellaea pinodes and Peyronellaea pinodella) that co‐occur on the same hosts, either on Clematis or Pisum. The results confirm that the mating‐type genes provide the information to distinguish between the homothallic Pey. pinodes (formerly Ascochyta pinodes) and the heterothallic Pey. pinodella (formerly Phoma pinodella), and indicate the close phylogenetic relationship between these two species that are part of the disease complex responsible for Ascochyta blight on pea. Furthermore, our analysis of the mating‐type genes of the fungal species responsible for causing wilt of Clematis sp. revealed that the heterothallic D. vitalbina (Phoma anamorph) is more closely related to the homothallic D. clematidis (Ascochyta anamorph) than to the heterothallic P. clematidina. Finally, our results indicate that homothallism in D. clematidis resulted from a single crossover between MAT1‐1 and MAT1‐2 sequences of heterothallic ancestors, whereas a single crossover event followed by an inversion of a fused MAT1/2 locus resulted in homothallism in Pey. pinodes.
INTRODUCTION
Ascochyta and Phoma are important fungal genera with a world‐wide distribution. The genus Ascochyta harbours pathogens occurring on a broad range of plants, and is responsible for significant economic losses of peas, beans and forage legumes. The genus Phoma includes multiple plant pathogens, saprobes and even human pathogens (Aveskamp et al., 2008). These two genera are morphologically very similar (Boerema, 1997; Boerema and Bollen, 1975; Brewer and Boerema, 1965; Wollenweber and Hochapfel, 1936), and the criteria for their delimitation are based on conidiogenesis and the percentage of septate conidia produced (Boerema and Bollen, 1975). However, the distinctive features of conidiogenesis can only be observed using scanning electron microscopy, and conidiogenesis of the genus Ascochyta has been disputed (Buchanan, 1987; Punithalingam, 1979). As Phoma species produce predominantly aseptate conidia and Ascochyta species mainly septate conidia, the ratio of aseptate to septate conidia produced on artificial media is used as a practical criterion for identification. However, this trait is highly variable for Phoma, and therefore this criterion is also of limited value for accurate identification (Onfroy et al., 1999). As a result of these problems in correct identification, multiple synonyms of Phoma and Ascochyta species are known, e.g. A. clematidina, P. clematidina, A. pinodella, P. pinodella, A. argillacea, P. argillacea, A. caricae‐papayae and P. caricae‐papayae (Boerema et al., 2004).
Recent molecular studies of Phoma spp. (2008, 2009a, 2009b, 2010; 2009, 2010), aimed at unravelling the phylogeny of these genera, have resulted in the establishment of several new genera within the newly erected Didymellaceae family, including both Ascochyta and Phoma species (de Gruyter et al., 2009). In 2010, Phoma pinodella and Didymella pinodes (Ascochyta anamorph) were described in the newly erected genus Peyronellaea as Pey. pinodella and Pey. pinodes (Aveskamp et al., 2010). These fungi, together with Ascochyta pisi and the recently described Phoma koolunga (Davidson et al., 2009), constitute the disease complex causing Ascochyta blight of pea, one of the most important diseases affecting field peas in the world. Although the results of multiple studies have confirmed that there are differences between the species, these studies have also shown that the relationship between Pey. pinodes and Pey. pinodella is much closer than that between either of these species and A. pisi (Barve et al., 2003; Chilvers et al., 2009; Faris‐Mokaiesh et al., 1996; Fatehi et al., 2003; Peever, 2007) or P. koolunga (Davidson et al., 2009). The same situation was observed within a recently characterized disease complex occurring on Clematis. A multigene phylogeny performed on fungal strains isolated from wilting Clematis revealed three distinct clades among strains formerly identified as Phoma clematidina. The sexual fungi Didymella vitalbina (Phoma anamorph) and Didymella clematidis (Ascochyta anamorph) were recognized as being the cause of wilt of different wild Clematis spp., next to the previously recognized asexual P. clematidina, which seems to occur on Clematis hybrids (Woudenberg et al., 2009). Here, the relationship between D. vitalbina (Phoma anamorph) and D. clematidis (Ascochyta anamorph) is much closer than that between either of these species and P. clematidina. These results again indicate the need for taxonomic clarification and correct identification.
Although the internal transcribed spacer (ITS) is the region of the genome most commonly used as a genetic marker in identification and phylogeny studies to date (Begerow et al., 2010; Seifert, 2009), it has been shown that mating‐type loci, as well as other rapidly evolving loci, may provide much better resolution among closely related taxa (Barve et al., 2003; Peever et al., 2007). Mating‐type genes evolve more quickly than other regions of the genome, but are highly conserved within species, making them useful for the phylogenetic analysis of closely related species.
In all heterothallic filamentous ascomycetes studied to date, sexual reproduction is controlled by a single regulatory locus (the mating‐type or MAT locus). In heterothallic (outbreeding) species, the mating‐type locus contains one of two forms of dissimilar sequence. These sequences are named idiomorphs as the alternative versions of the mating‐type locus are completely dissimilar, but are located at the same chromosomal location within the genome (Metzenberg and Glass, 1990). By convention, mating‐type idiomorphs of complementary isolates are termed MAT1‐1 and MAT1‐2 (Turgeon and Yoder, 2000). In contrast, homothallic fungi that do not require the presence of a complementary isolate to complete a sexual cycle contain both mating‐type genes (either physically linked or unlinked) in a single genome. Characteristic for MAT1‐1 isolates is the presence of a gene (MAT1‐1‐1) within the MAT1‐1 mating‐type locus that encodes a protein with an α‐domain and, for MAT1‐2 isolates, the presence of a gene (MAT1‐2‐1) within the MAT1‐2 mating‐type locus that encodes a protein containing a high‐mobility group (HMG) domain (Coppin et al., 1997).
In 1953, the homothallic nature of Pey. pinodes was confirmed by mono‐ascospore cultures (Baumann, 1953). The sexual state of Pey. pinodella has also been described in the past (Bowen et al., 1997) and, as single ascospore‐derived cultures failed to produce pseudothecia, it was assumed to be heterothallic. On the basis of the presence in pure cultures of both the teleomorph and anamorph state, D. clematidis was predicted to be a homothallic fungus, whereas no information was available about the sexual state of P. clematidina and D. vitalbina (Woudenberg et al., 2009). For none of these fungi molecular data on the underlying genetic basis of these mating systems were available. We aimed to use molecular data to confirm the sexual state of Pey. pinodes, Pey. pinodella and D. clematidis and to determine the sexual state of P. clematidina and D. vitalbina. Furthermore, using a phylogenetic analysis based on parts of the mating‐type loci, we aimed to confirm the taxonomy and species boundaries. Finally, by genomic comparison and a study of the structural organization of the mating‐type loci, we sought to obtain a better understanding of the evolution of homo‐ and heterothallism in these closely related and co‐occurring Phoma and Ascochyta species. In this article, we describe the cloning, characterization and genomic comparison of the complete mating‐type loci of P. clematidina, D. vitalbina, D. clematidis, Pey. pinodes, Pey. pinodella and P. herbarum (the type species of Phoma).
RESULTS
Cloning of mating‐type loci
Full‐length mating‐type loci of the examined species were cloned by a combination of methods. Initially, an attempt was made to amplify conserved parts of the MAT1‐1‐1 and MAT1‐2‐1 mating‐type genes using primers previously employed for the amplification of the α‐box (MAT1‐1‐1) of Leptosphaeria maculans and the HMG‐box (MAT1‐2‐1) of Ascochyta rabiei (Barve et al., 2003; Cozijnsen and Howlett, 2003). On the cloned partial MAT sequences, several subsequent chromosome walking steps in both the upstream and downstream directions were performed to obtain the whole mating‐type loci. Only the entire MAT1‐2 of P. herbarum and P. clematidina could thus be generated. On the basis of these MAT1‐2 loci and the sequences of five reference MAT loci, new primers directed against DNA sequences flanking the idiomorphs were designed (1, 2). The use of these ‘idiomorph primers’ resulted in the amplification of the entire MAT1‐1 of P. clematidina and D. vitalbina, and both MAT1‐1 and MAT1‐2 of Pey. pinodella. The sequences of all newly obtained mating‐type loci were used to design Phoma‐specific MAT1‐1 and MAT1‐2 primers (Table 1). For those species still lacking sequence information about the full‐length mating‐type locus (D. clematidis, Pey. pinodes and D. vitalbina), the fragments obtained after polymerase chain reaction (PCR) with the Phoma‐specific primers were used as a starting point for a new chromosome walking procedure. Finally, the Phoma‐specific primers were used on all isolates under investigation (Table 3). According to the Phoma‐specific PCRs, seven P. clematidina strains were MAT1‐1 positive and three were MAT1‐2 positive, of the 12 tested Pey. pinodella strains five contained the MAT1‐1 and seven the MAT1‐2 idiomorph, and of the seven tested D. vitalbina strains four were MAT1‐1 and two were MAT1‐2; the amplification of MAT sequences from one D. vitalbina strain remained unsuccessful. These results indicate the heterothallic nature of P. clematidina, Pey. pinodella and D. vitalbina. However, all Pey. pinodes strains and the D. clematidis strain gave positive results with both Phoma‐specific MAT1‐1 and MAT1‐2 primers, thus indicating the homothallic nature of these species. Finally, no P. herbarum strains gave positive results with MAT1‐1 primers, but six of the 15 strains tested were MAT1‐2 positive.
Table 1.
Primers designed for MAT idiomorph polymerase chain reaction (PCR) and Phoma‐specific high‐mobility group (HMG) motif and partial MAT1 PCR.
| Name | Nucleotide sequence (5′–3′) | Used for |
|---|---|---|
| MATidio_Fd | GGRAGRATIGCIGAYTGGAARGG | MAT idiomorph |
| MATidio_Rv | TGGIGITGYGGIACKTTYATYTGG | |
| HMGF_Phoma | CGYCCRATGAAYTGCTGGAT | HMG motif |
| HMGR_Phoma | CRGGCTTRCGAGGRSWRTACTT | |
| MAT1F2_Phoma | CTGGAATIGCWGRCATGGC | Partial MAT1 |
| MAT1R2_Phoma | TGTCGCTTYGYICGTCGC |
Table 2.
GenBank numbers from control strains.
| Name | MAT1‐1‐1 | MAT1‐2‐1 | ITS | ||
|---|---|---|---|---|---|
| Didymella lentis | DQ341314 | DQ341315 | Chérif et al. (2006) | DQ383953 | Peever et al. (2007) |
| Didymella rabiei | DQ341313 | DQ341312 | Barve et al. (2003) | DQ383949 | Peever et al. (2007) |
| Pyrenophora graminae | DQ823079 | DQ823080 | Rau et al. (2007) | – | |
| Pyrenophora teres | AY950585 | AY950586 | Rau et al. (2005) | – | |
| Leptosphaeria maculans | AY174048 | AY174049 | Cozijnsen and Howlett (2003) | – |
Table 3.
Isolates used in phylogenetic analyses.
| Collection number* | Mating type† | Host | Origin | GenBank no. |
|---|---|---|---|---|
| ITS‡ | ||||
| Didymella clematidis | ||||
| CBS 123705, PD 97/13460–1, ICMP 13664 | 1/2 | Clematis ligusticifolia | USA | FJ515593 |
| Didymella. vitalbina | ||||
| CBS 454.64 | ? | Clematis vitalba | France | FJ515605 |
| CBS 911.87 | 1 | Clematis vitalba | Germany | FJ515592 |
| PD 75/294 | 1 | Clematis sp. | Unknown | FJ515596 |
| CBS 123707, PD 97/13460–2, ICMP 13664 | 1 | Clematis vitalba | Switzerland | FJ515595 |
| PD 04373904‐2B | 2 | Clematis vitalba | Netherlands | FJ515603 |
| CBS 123706, PD 04373904–5 | 2 | Clematis vitalba | Netherlands | FJ515594 |
| PD 04417700–3 | 1 | Clematis vitalba | Netherlands | FJ515604 |
| Peyronellaea pinodella | ||||
| CBS 116.28 | 2 | Unknown | Unknown | JF810508 |
| CBS 108.31 | 2 | Unknown | USA | JF810509 |
| CBS 110.32, MUCL 292 | 1 | Medicago sativa | Netherlands | EU167565 |
| CBS 351.34, MUCL 9927, MUCL 18217 | 2 | Unknown | Unknown | JF810510 |
| CBS 107.46 | 1 | Pisum sativum | Netherlands | JF810511 |
| CBS 108.46 | 1 | Pisum sativum | Netherlands | JF810512 |
| CBS 403.65, PD 57/90, IMI 116998 | 1 | Unknown | Unknown | JF810513 |
| CBS 531.66 | 2 | Trifolium pratense | USA | FJ427052 |
| CBS 317.90, PD 77/948 | 1 | Trifolium sp. | Netherlands | JF810514 |
| CBS 318.90, PD 81/729 | 2 | Pisum sativum | Netherlands | FJ427051 |
| CBS 319.90, PD 84/207 | 2 | Beta vulgaris var. rubra | Netherlands | JF810515 |
| CBS 133.92 | 2 | Glycine soja | Hungary | JF810516 |
| Peyronellaea pinodes | ||||
| CBS 206.28 | 1/2 | Pisum sp. | Unknown | JF810517 |
| CBS 249.47 | 1/2 | Pisum sativum | Scotland | JF810518 |
| CBS 250.47 | 1/2 | Pisum sativum | Netherlands | JF810519 |
| CBS 251.47 | 1/2 | Pisum sativum | Netherlands | JF810520 |
| CBS 252.47 | 1/2 | Pisum sativum | Netherlands | JF810521 |
| CBS 329.51 | 1/2 | Pisum sp. | Germany | JF810522 |
| CBS 235.55 | 1/2 | Pisum sp. | Unknown | GU237805 |
| CBS 525.77 | 1/2 | Pisum sativum | Belgium | GU237883 |
| CBS 159.78 | 1/2 | Pisum sativum | Iraq | GU237786 |
| CBS 374.84, PD 79/674 | 1/2 | Pisum sativum | Netherlands | JF810523 |
| Phoma clematidina | ||||
| CBS 201.49 | 2 | Clematis sp. | Netherlands | FJ426991 |
| CBS 195.64 | 1 | Clematis jackmannii | Netherlands | FJ426990 |
| CBS 102.66 | 2 | Clematis sp. | England | FJ426988 |
| CBS 520.66, PD 64/657 | 2 | Selaginella sp. | Netherlands | FJ426992 |
| CBS 108.79, PD 78/522 | 1 | Clematis sp. | Netherlands | FJ426989 |
| PD 80/683 | 1 | Clematis sp. | Netherlands | FJ515597 |
| PD 91/1865 | 1 | Clematis sp. | Netherlands | FJ515598 |
| PD 95/895 | 1 | Clematis sp. | Netherlands | FJ515599 |
| PD 97/12061 | 1 | Clematis‘Purple spider’ | Netherlands | FJ515600 |
| PD 97/12062 | 1 | Clematis‘New Dawn’ | Netherlands | FJ515601 |
| Phoma herbarum | ||||
| CBS 276.37, PD 92/332, MUCL 9920 | ? | Wood pulp | Sweden | JF810524 |
| CBS 368.61 | 2 | Ulmus sp. | Netherlands | JF810525 |
| CBS 369.61 | ? | Ulmus sp. | Netherlands | JF810526 |
| CBS 370.61 | 2 | Ulmus sp. | Netherlands | JF810527 |
| CBS 567.63, ATCC 15053, MUCL 9889 | ? | Malus sylvestris | USA | JF810528 |
| CBS 615.75, PD 73/665, IMI 199779 | 2 | Rosa multiflora | Netherlands | FJ427022 |
| CBS 502.91, PD 86/276 | ? | Nerium sp. | Netherlands | GU237874 |
| CBS 503.91, PD 87/499 | 2 | Thuja sp. | Netherlands | JF810529 |
| CBS 829.97 | ? | Ornithogenic soil | Antarctica | JF810530 |
| CBS 830.97 | ? | Soil from foot of glacier | Antarctica | JF810531 |
| CBS 100953 | ? | Soil near glacier | Antarctica | JF810532 |
| CBS 101145, ATCC 12569, IMI 049948 | ? | White lead paint | UK | AY293803 |
| PD 85/930 | 2 | Streptocarpus sp. | Unknown | JF810533 |
| PD 87/652 | ? | Soil | Netherlands | JF810534 |
| PD 90/1454 | 2 | Lycopersicon esculentum | Netherlands | JF810535 |
Bold numbers indicate strains for which the mating‐type locus is fully sequenced. ATCC, American Type Culture Collection, Manassas, VA, USA; CBS, CBS Fungal Biodiversity Centre, Utrecht, the Netherlands; ICMP, International Collection of Micro‐organisms from Plants, Auckland, New Zealand; IMI, International Mycological Institute, CABI‐Bioscience, Egham, Surrey, UK; MUCL, (agro)industrial fungi and yeast collection of the Belgian Co‐ordinated Collections of Micro‐organisms (BCCM), Louvain‐la‐Neuve, Belgium; PD, Dutch Plant Protection Service, Wageningen, the Netherlands.
Isolates marked as mating‐type 1 were positive in polymerase chain reaction (PCR) with the Phoma‐specific α‐box (MAT1‐1‐1) primers; isolates marked as mating‐type 2 were positive in PCR with the Phoma‐specific HMG‐motif (MAT1‐2‐1) primers; isolates positive with both Phoma‐specific PCRs are marked with ‘1/2’ and isolates in which both Phoma‐specific PCRs were negative are marked with ‘?’. Numbers in bold indicate isolates with a positive result in the initial α‐box (MAT1‐1‐1) PCR or HMG motif (MAT1‐2‐1) PCR.
Bold numbers indicate sequences determined in this study.
Structural organization of heterothallic mating‐type loci
The P. clematidina MAT1‐1 and MAT1‐2 sequences obtained resulted in the assembly of 3.6 and 3.5 kb of sequence, respectively. blast2 analysis of these sequences indicated that, in the MAT1‐1 and MAT1‐2 isolates, 1.8 kb of MAT1‐1 and 2.1 kb of MAT1‐2 were dissimilar and thus belonged to the idiomorphs (Metzenberg and Glass, 1990). Similar analysis of the D. vitalbina MAT1‐1 and MAT1‐2 sequences (3 and 3.1 kb, respectively) obtained identified a MAT1‐1 idiomorph of 1.9 kb and a MAT1‐2 idiomorph of 2.2 kb. The sequenced mating‐type loci of the heterothallic Pey. pinodella (2.6 kb of MAT1‐1 and 3.1 kb of MAT1‐2) revealed a MAT1‐1 idiomorph of 2.4 kb and a MAT1‐2 idiomorph of 2.8 kb. As indicated above, the amplification of mating‐type sequences from P. herbarum was only successful for 40% of the isolates tested and only MAT1‐2 sequences were obtained. Chromosome walking resulted in the generation of 4.3 kb of P. herbarum MAT1‐2 sequences. blast2 analyses of the P. herbarum MAT1‐2 sequences against the MAT1‐1 sequences of P. clematidina, D. vitalbina and Pey. pinodella suggested that the P. herbarum MAT1‐2 idiomorph had a length of 3.2 kb (Table 4).
Table 4.
Idiomorph lengths of the heterothallic Phoma clematidina, Didymella vitalbina, Peyronellaea pinodella and Phoma herbarum.
| Species | MAT1‐1 * | MAT1‐2 |
|---|---|---|
| Phoma clematidina | 1.8 | 2.1 |
| Didymella vitalbina | 1.9 | 2.2 |
| Peyronellaea pinodella | 2.4 | 2.8 |
| Phoma herbarum | – | 3.2 |
Length in kb.
Upstream of the idiomorphs of all of these species, (part of) an additional open reading frame (ORF) was identified with highest similarity to ORF1, an ORF found near the idiomorph of other loculoascomycetes e.g. Ascochyta lentis, A. rabiei (Chérif et al., 2006), Pyrenophora teres and Pyrenophora graminea (Rau et al., 2007). Furthermore, part of an ORF similar to a DNA lyase was found downstream of the idiomorphs (Fig. 1a). DNA lyases are found more often near the MAT loci of other fungi (Arzanlou et al., 2010; Waalwijk et al., 2002).
Figure 1.
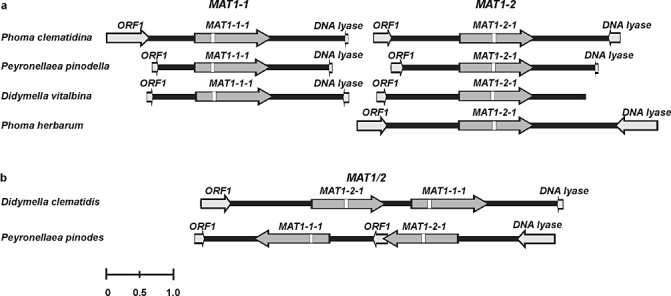
Schematic representation of the organization of mating‐type loci of the heterothallic Phoma clematidina, P. herbarum, Peyronellaea pinodella and Didymella vitalbina (a) and of the homothallic Pey. pinodes and D. clematidis (b). Mating‐type genes are indicated by dark grey arrows, and other predicted (partial) gene models by light grey arrows. The positions of predicted introns are marked by white boxes. The marker at the bottom indicates a size of 1 kb.
Structural organization of homothallic mating‐type loci
Chromosome walking along the genomic DNA of the homothallic species Pey. pinodes and D. clematidis resulted in the generation of 5.3 and 5.4 kb of sequence, respectively. The D. clematidis MAT1/2 mating‐type locus contained a MAT1‐1‐1 located downstream on the same strand as the MAT1‐2‐1 gene. Similar to the situation in the heterothallic fungi, upstream of the D. clematidis mating‐type locus was an ORF with homology to ORF1, and a partial ORF with homology to a DNA lyase was detected downstream of the mating‐type locus (Fig. 1b).
The situation in Pey. pinodes, the other homothallic species examined, was completely different. This locus also contained an intact MAT1‐1‐1 downstream on the same strand as MAT1‐2‐1. Interestingly, in between MAT1‐2‐1 and MAT1‐1‐1, an additional copy of ORF1 was found. Finally, the orientation of this (MAT1‐1‐1/ORF1/MAT1‐2‐1) fusion locus was inverted compared with the situation in the other species (Fig. 1b).
Characterization of mating‐type genes
An analysis of all the mating‐type genes indicated a high similarity between the different species. The length of the predicted MAT1‐1‐1 genes varied between 1120 and 1148 nucleotides. All genes contained a single predicted intron varying between 46 and 56 nucleotides (Table 5). The length of the predicted MAT1‐2‐1 genes varied between 1094 and 1115 nucleotides, and all genes contained a single intron of 55 or 56 nucleotides (Table 5) The predicted intron in MAT1‐1‐1 was located within sequences encoding the characteristic α‐domain motif. This position matches exactly an intron position found in all dothideomycetous MAT1‐1‐1 genes examined. Similarly, the predicted intron in MAT1‐2‐1 was located within the HMG domain encoding sequence and at a position perfectly conserved in dothideomycetous MAT1‐2‐1 genes (data not shown) (Arzanlou et al., 2010; Barve et al., 2003; Groenewald et al., 2006; Stergiopoulos et al., 2007; Turgeon et al., 1993). In addition, the sizes of the deduced MAT1‐1‐1 (varying between 357 and 364 amino acids) and MAT1‐2‐1 (varying between 345 and 352 amino acids) proteins were highly similar and within the range observed for other dothideomycetous MAT1‐1‐1 and MAT1‐2‐1 proteins (data not shown).
Table 5.
Comparison of the gene organization of MAT1‐1‐1 and MAT1‐2‐1 in idiomorphs of Phoma clematidina, Didymella vitalbina, Didymella clematidis, Peyronellaea pinodella, Peyronellaea pinodes and Phoma herbarum.
| Species | Mating type | Gene size (nt) | Intron size (nt) | Mat1‐1‐1 * | Mat1‐2‐1 * | ||||
|---|---|---|---|---|---|---|---|---|---|
| Intron position | Protein size (aa) | Gene size (nt) | Intron size (nt) | Intron position | Protein size (aa) | ||||
| Phoma clematidina | Heterothallic | 1143 | 48 | 245–292 | 364 | 1115 | 56 | 494–549 | 352 |
| Didymella vitalbina | Heterothallic | 1146 | 54 | 245–298 | 363 | 1106 | 56 | 484–540 | 349 |
| Didymella clematidis | Homothallic | 1148 | 56 | 245–300 | 363 | 1094 | 56 | 484–540 | 345 |
| Peyronellaea pinodella | Heterothallic | 1120 | 46 | 245–290 | 357 | 1108 | 55 | 488–542 | 350 |
| Peyronellaea pinodes | Homothallic | 1120 | 46 | 245–290 | 357 | 1111 | 55 | 488–542 | 351 |
| Phoma herbarum | Heterothallic | 1111 | 55 | 494–548 | 351 | ||||
aa, amino acids; nt, nucleotides.
Pairwise comparisons of the deduced protein sequences indicated that MAT1‐1‐1 and MAT1‐2‐1 of the heterothallic Pey. pinodella were more similar to those of the homothallic Pey. pinodes (91.9% and 92.6% identity, respectively) than to those of another heterothallic species. The same was observed for MAT1‐1‐1 and MAT1‐2‐1 of the heterothallic D. vitalbina and the homothallic D. clematidis, with 87.9% and 83.9% identity.
Phylogenetic analysis
On the basis of the publication of Aveskamp et al. (2010), Didymella urticicola (GenBank accession number GU237761) was used as outgroup in the ITS phylogeny. The ITS alignment contained 58 strains, including the outgroup strain, and had a total length of 455 characters, 13 of which were parsimony uninformative and 26 were parsimony informative. The heuristic search resulted in two most parsimonious trees [tree length, 50 steps; consistency index (CI) = 0.860; retention index (RI) = 0.983; rescaled consistency index (RC) = 0.845]. The Bayesian analysis resulted in 6202 trees from which the 50% majority rule consensus tree and posterior probabilities were calculated. In both phylogenetic analyses, five well‐supported clades were obtained dividing P. herbarum, P. clematidina, D. vitalbina and D. clematidis, but the Pey. pinodella and Pey. pinodes strains all clustered in one clade with 99% bootstrap support/0.99 posterior probability (Fig. 2).
Figure 2.
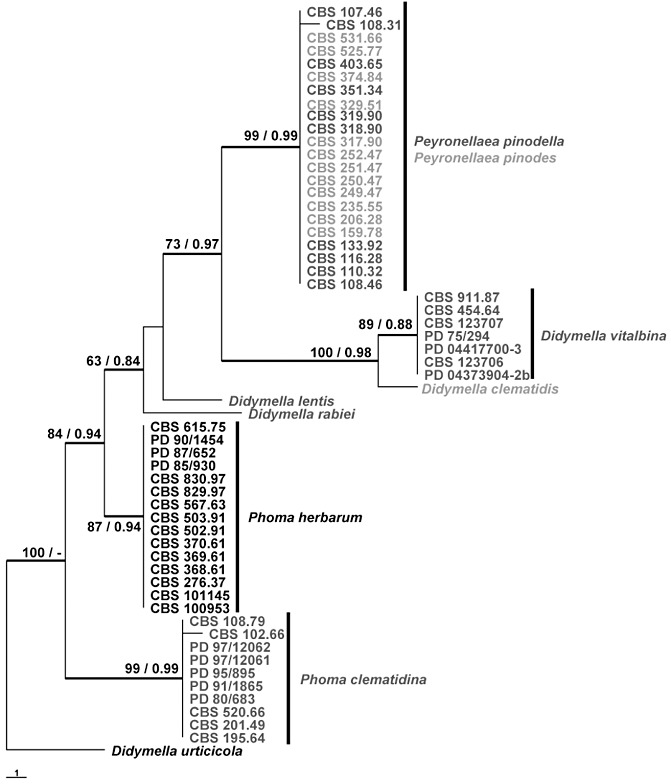
One of the two most parsimonious trees obtained from a heuristic search of the internal transcribed spacer (ITS) sequence alignment. The scale bar shows one change, strict consensus branches are thickened and bootstrap support values from 1000 bootstrap replicates (BS) and posterior probabilities (PP) are shown at the nodes. BS/PP values below 60/0.6 are omitted. Confirmed heterothallic species are indicated in dark grey and confirmed homothallic species in light grey. The tree is rooted with Didymella urticicola GU237761. CBS, Centraalbureau voor Schimmelcultures, Utrecht, the Netherlands; PD, Dutch National Reference Centre of the Plant Protection Service, Wageningen, the Netherlands.
For the phylogeny based on the MAT1‐1‐1 and MAT1‐2‐1 alignments, part of the known sequences of Pyrenophora teres and Pyr. graminae (Table 2) were used as outgroup. The alignments consisted of 31/33 strains, respectively, including the outgroup strains, and had a total length of 405/235 characters, seven/six of which were parsimony uninformative and 122/130 were parsimony informative. The search resulted in four most parsimonious trees (tree length, 208 steps; CI = 0.803; RI = 0.948; RC = 0.761) for the partial MAT1‐1‐1 alignment and in one most parsimonious tree (tree length, 272 steps; CI = 0.721; RI = 0.929; RC = 0.670) for the MAT1‐2‐1 alignment. The Bayesian analyses resulted in 2702 and 2962 trees, respectively, from which the 50% majority rule consensus tree and posterior probabilities were calculated (Fig. 3). Both analyses indicated that all species used clustered in well‐supported clades, with Pey. pinodella close to Pey. pinodes and D. vitalbina close to D. clematidis.
Figure 3.
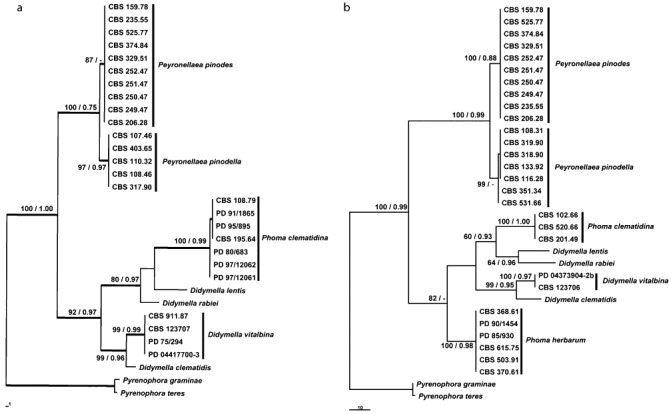
Most parsimonious trees obtained from a heuristic search of the partial MAT1‐1‐1 (a) and MAT1‐2‐1 (b) sequence alignments. The scale bars show the changes; bootstrap support values from 1000 bootstrap replicates (BS) and posterior probabilities (PP) are shown at the nodes. BS/PP values below 60/0.6 are omitted. The trees are rooted with Pyrenophora teres and Pyrenophora graminaea. The first of four most parsimonious trees; the strict consensus branches are thickened. CBS, Centraalbureau voor Schimmelcultures, Utrecht, the Netherlands; PD, Dutch National Reference Centre of the Plant Protection Service, Wageningen, the Netherlands.
Similarities between heterothallic and homothallic mating‐type loci
The phylogenetic analyses, as well as the pairwise comparisons of the deduced MAT1‐1‐1 and MAT1‐2‐1 proteins, indicated that the heterothallic Pey. pinodella was closely related to the homothallic Pey. pinodes. Similarly, the heterothallic D. vitalbina was closely related to the homothallic D. clematidis. This was also shown by blast2 pairwise alignment of the sequenced MAT1‐1 and MAT1‐2 regions of P. clematidina, Pey. pinodella, D. vitalbina and P. herbarum to the mating‐type loci of Pey. pinodes and D. clematidis. A graphical analysis of the pairwise alignments with highest levels of similarity, Pey. pinodella versus Pey. pinodes and D. vitalbina versus D. clematidis, is shown in Fig. 4. This analysis shows that the D. clematidis MAT1/2 locus appears to consist of a fusion between MAT1‐1 and MAT1‐2 sequences of D. vitalbina. The 5′ region of the D. clematidis MAT1/2 locus is highly similar to the entire MAT1‐2 idiomorph of D. vitalbina, whereas the 3′ region of the D. clematidis MAT1/2 locus is highly similar to large parts of the D. vitalbina MAT1‐1 idiomorph. At the MAT1/2 fusion junction, a small stretch of sequence identity between MAT1‐1 and MAT1‐2 could be identified (Fig. 4a).
Figure 4.
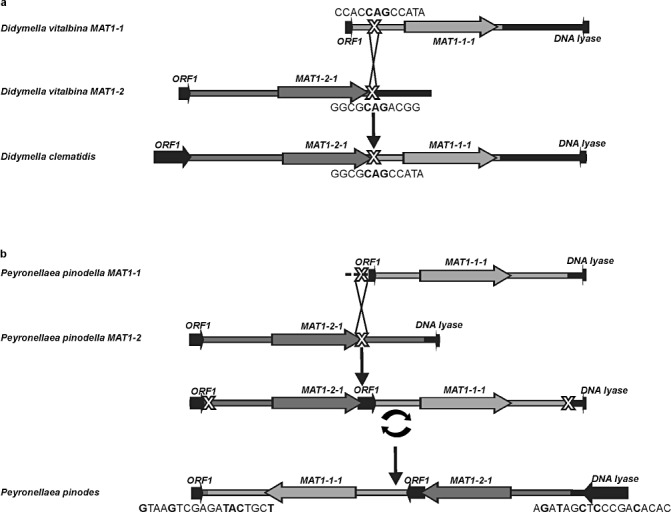
Schematic overview of the sequence similarities between the mating‐type loci of the heterothallic Didymella vitalbina and homothallic D. clematidis (a) and between the heterothallic Peyronellaea pinodella and homothallic Pey. pinodes (b). Also depicted are the putative recombination events responsible for the evolution of the homothallic species from heterothallic ancestors. Predicted gene models are indicated by arrows. Putative recombination spots are marked with a large ‘X’ and the putative inversion event is marked by a circular arrow. Sequences of the predicted junctions are indicated below the figure. All MAT1‐1‐derived areas are indicated in light grey; all MAT1‐2‐derived areas are indicated in dark grey; nonidiomorphic areas are indicated in black; sequences at fusion junctions that are shared between MAT1‐1 and MAT1‐2 are indicated in bold black.
The MAT1/2 locus of Pey. pinodes seemingly consists of an inverted fusion product between MAT1‐1 and MAT1‐2 sequences of Pey. pinodella. Within the MAT1/2 fusion product, sequences highly related to ORF1, located outside the Pey. pinodella MAT loci, can also be found. The 5′ region of the Pey. pinodes MAT1/2 locus is highly similar to the entire inverted MAT1‐1 idiomorph of Pey. pinodella, and the 3′ region of the Pey. pinodes MAT1/2 locus is highly similar to most of the inverted Pey. pinodella MAT1‐2 idiomorph (Fig. 4b). The position of the MAT1/2 fusion junction could not be identified. However, upstream of MAT1‐2‐1 and downstream of MAT1‐1‐1, the boundaries of the inversion could be identified. Interestingly, the sequence found at the 3′ boundary is highly similar to the sequence found at the 5′ boundary when reverse complemented. In total, 10 of 18 nucleotides and eight of the first 11 positions are shared between the two motifs (Fig. 5a).
Figure 5.

Sequence analysis of inversion junctions in Peyronellaea pinodes. (a) Alignment of the 5′ junction of the putative inversion in Pey. pinodes to the reverse complemented (RC) sequence of the 3′ junction. The putative motif is indicated as a shaded box and identical nucleotides are marked in bold and have a larger font size. (b) Alignment of the 5′ and 3′Pey. pinodes junctions to Peyronellaea pinodella MAT1‐1 and MAT1‐2 sequences. The sequences of the 3′ junctions are reverse complemented (RC) for better comparison of the putative motif (shaded box), as also indicated in (a). Identical nucleotides are indicated in bold and nucleotides shared by all three sequences have a larger font size.
DISCUSSION
Recently, molecular and morphological studies were initiated to unravel the phylogeny of the anamorphic genus Phoma and associated genera, such as Ascochyta. This resulted in the establishment of a new family containing several Phoma and Ascochyta species (de Gruyter et al., 2009), the characterization of a disease complex occurring on Clematis sp. (Woudenberg et al., 2009) and the renaming of several species belonging to the disease complex causing Ascochyta blight (Aveskamp et al., 2010). These molecular analyses were based on sequences commonly used for fungal characterization, e.g. ITS, actin, β‐tubulin. However, the use of mating‐type genes in studies aimed at the elucidation of the phylogeny and species boundaries might provide better resolution. The mating‐type genes are more variable than these barcode genes and evolve at a faster rate (Turgeon, 1998). Moreover, in addition to high inter‐species variation, they generally exhibit low intra‐species variation and therefore can be used to sort out species relationships in taxon‐rich complexes. For example, the study of mating‐type genes has helped to solve the phylogenetic relationship of the net and spot forms of Pyrenophora teres (2005, 2007), the phylogeny of Ascochyta spp. associated with legumes (Barve et al., 2003), the relationship between oat‐ and wheat‐infecting Phaeosphaeria avenaria (Ueng et al., 2003) and the recognition of species within the Fusarium graminearum complex (O'Donnell et al., 2004).
Our work, as well as that performed previously by others, has also clearly demonstrated the benefits of using mating‐type sequences to determine species boundaries. Peyronellaea pinodella and Pey. pinodes could not be distinguished from each other on the basis of ITS sequences. However, the phylogeny based on MAT1‐1‐1 and MAT1‐2‐1 was capable of distinguishing between these two species (Fig. 3) (Barve et al., 2003), thus confirming the morphologically based characterization of the two species. This also demonstrates the risk associated with identification solely on the basis of molecular characters without real relevance for the lifestyle or biology of a species, e.g. ITS. This is especially relevant when studying/identifying quarantine organisms that are part of species complexes. Moreover, it also illustrates the enduring importance of classical morphology‐based taxonomy.
Mating‐type analysis can also provide a means to elucidate the sexual strategy of fungal plant pathogens. This is important as the reproductive strategy can influence the success of control (breeding) measurements. Many fungal species are classified as asexual within anamorphic genera because of the lack of a characterized teleomorph. Studies on mating‐type loci of presumed asexual fungi have revealed that many of these species possess MAT genes, which may even be expressed (Kerenyi et al., 2004; Paoletti et al., 2005; Yun et al., 2000). This indicates that a lack of (obvious) sexual recombination is not a result of the absence of basal elements controlling the sexual reproductive machinery (Sharon et al., 1996). The fact that approximately one‐fifth of all fungi still have no described sexual stage can be explained by the possibility that mating only occurs under specific environmental conditions and/or only within a time frame that exceeds normal laboratory crossing experiments (O'Gorman et al., 2008). In the case of cryptic heterothallic fungi, it can also be explained by the absence of one of the compatible mating types as a result of geographical separation, as has been reported for populations of Didymella (Ascochyta) rabiei (Barve et al., 2003; Kaiser and Kusmenoglu, 1997).
Our comparisons of the mating‐type genes did not only distinguish between Pey. pinodella and Pey. pinodes, but also confirmed morphological studies, indicating that both species exhibit a completely different sexual strategy. The sexual state of Pey. pinodella has been described previously (Bowen et al., 1997) as heterothallic. In 1953, the homothallic nature of Pey. pinodes was confirmed by mono‐ascospore cultures (Baumann, 1953). Our molecular studies confirm these conclusions. In addition, the predicted homothallic nature of D. clematidis, exhibiting both the teleomorph and anamorph states in pure culture (Woudenberg et al., 2009), is confirmed. On the basis of our molecular studies, we predict that P. clematidina and D. vitalbina are heterothallic species.
Amplification of the mating‐type sequences from P. herbarum was only successful in six of the 15 isolates tested. None of the PCRs aimed at the amplification of MAT1‐1‐1 sequences were successful and only MAT1‐2‐1 sequences were obtained. The MAT1‐2 mating‐type locus obtained exhibited the typical characteristics and organization observed for the heterothallic P. clematidina, Pey. pinodella and D. vitalbina (Fig. 1). The fact that the amplification of MAT1‐1‐1 sequences remained unsuccessful despite the close phylogenetic relationship with the other examined species (Fig. 2) suggests that MAT1‐1‐1 sequences in P. herbarum have either become absent or corrupted. Thus, these data suggest that P. herbarum might originally have been a functionally heterothallic fungus that lost its capacity for sexual reproduction and is now a genetically heterothallic, but functionally asexual, fungus. This is also supported by multilocus sequence analysis suggesting a clonal nature of P. herbarum isolates (M.M. Aveskamp, unpublished data).
A major question in fungal biology is whether homothallism has arisen from heterothallism or the other way round. Population genetics models have suggested that evolution from heterothallism to homothallism is the most likely scenario (Nauta and Hoekstra, 1992). Phylogenetic analyses and analyses of the MAT structure in Cochliobolus and Stemphylium species have shown that, in these species, heterothallism is indeed the ancestral state (Inderbitzin et al., 2005; Yun et al., 1999). In the genus Aspergillus, the ancestral state is still under debate. The predominance of known homothallic species over known heterothallic species, phylogenetic analyses and comparative genomics suggest that homothallism is the ancestral state in this genus (Galagan et al., 2005; Geiser et al., 1998; Varga et al., 2000). However, recent analysis of the Neosartorya fischeri MAT loci has suggested a heterothallic ancestral state (Rydholm et al., 2007).
In our study, the structural organization of the mating‐type loci from all heterothallic species was conserved and similar to the organization of other loculoascomycetes (Chérif et al., 2006; 2005, 2007). In the two examined homothallic species, the mating‐type locus contained both MAT1‐1‐1 and MAT1‐2‐1 (Fig. 1), whereas the genomic boundaries of the homothallic mating‐type loci were identical to the flanking regions of the heterothallic MAT loci. Moreover, phylogenetic analysis showed that the homothallic mating‐type genes did not cluster together, but clustered with heterothallic mating‐type genes. All of these data strongly support the idea of an independent evolution of homothallism from a heterothallic ancestral state.
The organization of the mating‐type locus of the homothallic D. clematidis and the observed sequence similarity with the mating‐type loci of the related heterothallic D. vitalbina could well be explained by a single crossover event between MAT1‐1 and MAT1‐2 sequences of heterothallic ancestors. The presence of a small stretch of sequence identity between MAT1‐1 and MAT1‐2 at the exact position of the MAT1/2 fusion junction strongly suggests that the D. clematidis MAT1/2 fusion locus originated from such a crossover event (Fig. 4a). The organization of the D. clematidis MAT1/2 mating‐type locus is very similar to the organization seen within homothallic Crivellia. In addition, in that species, the organization was explained by a crossover between a small stretch of identity shared between the mating‐type loci of heterothallic ancestors (Inderbitzin et al., 2006). Moreover, the existence of several homothallic Cochliobolus sp. with MAT loci containing a (partial) MAT1‐1‐1 fused to a (partial) MAT1‐2‐1 has been explained by such recombination events (Yun et al., 1999)
The organization of the MAT locus of the homothallic Pey. pinodes is more complicated (Fig. 1) and, to some extent, resembles the organization observed in the homothallic Cochliobolus kusanoi (Yun et al., 1999). In C. kusanoi, part of a sequence, normally found downstream of the MAT loci in heterothallic species, is located between the mating‐type genes. Similarly, in Pey. pinodes, ORF1, found upstream of the MAT locus in heterothallic species, is located in between MAT1‐1‐1 and MAT1‐2‐1 (1, 4). We propose that the organization of the MAT1/2 locus of Pey. pinodes arose by a single crossover event followed by an inversion. The putative initial recombination event probably occurred between sequences located at the 3′ end of MAT1‐2‐1 and sequences found within ORF1 upstream of the MAT1‐1 idiomorph. A subsequent inversion of this fused MAT1‐2/ORF1/MAT1‐1 region could then have resulted in the observed Pey. pinodes MAT1/2 organization. In contrast with D. clematidis, the position of the proposed initial recombination event (the MAT1/2 fusion junction) could not be identified. This is because no sequence information about the putative crossover site in ORF1 of Pey. pinodella was available. However, the junctions resulting from the proposed inversion events could clearly be identified (4, 5). The motif at the 5′ boundary of the proposed inversion is highly similar to the reverse complemented motif found at the 3′ boundary of the inversion. This suggests that, in an ancestor of Pey. pinodes, these regions of similarity formed a loop structure, resulting in crossover and subsequent inversion of the fused MAT1/2 locus. Sequences resembling these motifs were also identified in both the MAT1‐1 and MAT1‐2 regions of Pey. pinodella (Fig. 5b), thus strengthening the hypothesis that Pey. pinodes originated from heterothallic ancestors, such as Pey. pinodella, after a single crossover event followed by an inversion. The proposed evolution of the homothallic D. clematidis and Pey. pinodes from heterothallic ancestors either resembling or identical to D. vitalbina and Pey. pinodella, is depicted in Fig. 4.
As mentioned previously, the phylogenetic analyses show that the mating‐type genes of the heterothallic species Pey. pinodella and D. vitalbina are more related to the mating‐type genes of the homothallic species Pey. pinodes and D. clematidis, respectively, than to the mating‐type genes of other heterothallic phylogenetically related species. This observation suggests a common evolutionary history between Pey. pinodella/Pey. pinodes, on the one hand, and D. vitalbina/D. clematidis on the other. Interestingly, in both cases, the heterothallic species and the closely related homothallic species share the same host. Peyronellaea pinodella and Pey. pinodes are part of the Ascochyta blight complex on pea, whereas D. vitalbina and D. clematidis are both pathogens on Clematis spp. It has been shown that the presence of multiple closely related species on the same host can correlate with great evolutionary dynamics acting at mating‐type loci (Arzanlou et al., 2010) with potential implications for speciation. The occurrence of recombinations as described above at MAT loci can lead to reproductive isolation between homothallic and heterothallic isolates, and result in the establishment of new species. Additionally, the co‐occurrence of species on a single host could also lead to close physical interactions and potentially even to the exchange of genetic material through inter‐ and intraspecies mating, hybridization or anastomosis. This could also result in the establishment of novel species. Therefore, we postulate that the co‐occurrence of multiple (related) species in both time and space on a single host (species complex) can be both the cause and consequence of (multiple) speciation events.
EXPERIMENTAL PROCEDURES
Isolates, culture and DNA extraction
All isolates used in this study are listed in Table 3. Strains of the species associated with the disease complex on Clematis, D. clematidis, D. vitalbina and P. clematidina, and the closely related species causing Ascochyta blight of pea, Pey. pinodella and Pey. pinodes, together with the type species of Phoma, P. herbarum, were selected. Freeze–dried strains were obtained from the culture collections of the Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands and the Dutch National Reference Centre of the Plant Protection Service (PD), Wageningen, the Netherlands, and were revived in 2 mL of malt/peptone (50%/50%) liquid medium. Subsequently, the cultures were transferred and maintained on oatmeal agar (OA) (Crous et al., 2009). DNA extractions were performed using an Ultraclean Microbial DNA Isolation Kit (Mobio Laboratories, Carlsbad, CA, USA), according to the manufacturer's instructions. All DNA extracts were diluted 10 times in MilliQ water and stored at 4 °C before use.
Isolation of partial MAT sequences
The primers HMG‐L and HMG‐R, originally used for the amplification of the HMG motif of A. rabiei (teleomorph: Didymella rabiei) (Barve et al., 2003), as well as the primers 1 and 2 described for the amplification of the α‐box of L. maculans (anamorph: Phoma lingam) (Cozijnsen and Howlett, 2003), were used in an attempt to amplify part of the mating‐type genes of the isolates presented in Table 3. The PCR mixture contained 0.5 µL of diluted genomic DNA, 2 µm of each primer, 0.5 U of BIOTAQ DNA polymerase (Bioline, Luckenwalde, Germany), 0.1 mm deoxynucleoside triphosphates (dNTPs), 2.5 mm MgCl2 and 1 × NH4 reaction buffer (Bioline). Conditions for the amplification were an initial denaturation step of 5 min at 94 °C, followed by 40 cycles of 45 s at 94 °C, 45 s at 55 °C and 45 s at 72 °C, and a final elongation step of 7 min at 72 °C. The PCR products were visualized by electrophoresis and sequenced in both directions using PCR primers and the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), following the manufacturer's instructions.
Chromosome walking, idiomorph PCR, specific HMG domain and MAT‐1 PCR
The partial MAT sequences obtained were aligned in Bionumerics v4.60 (Applied Maths, Sint‐Martens‐Latem, Belgium). Primer3 v0.4.0 (Rozen and Skaletsky, 2000) and the program Vector NTI Advanced 10 (Invitrogen, Carlsbad, CA, USA) were used to create additional primers that were subsequently employed to determine additional sequences up‐ and downstream of the partial MAT sequences, and, finally, the complete MAT idiomorph. This chromosome walking was performed using the DNA Walking Speedup Kit (Seegene Inc., Rockville, MD, USA), according to the manufacturer's instructions. The products obtained by chromosome walking were ligated into pGEM®‐T easy vector (Promega, Madison, WI, USA) according to the manufacturer's instructions. The vector with insert was transformed into JM109‐competent Escherichia coli cells (Promega). Recombinants were analysed by colony PCR with the universal M13F and M13R primers. The colony PCR mixture contained 1.5 µL of liquid colony, 0.2 µm of each primer, 0.4 U BIOTAQ DNA polymerase (Bioline), 0.03 mm dNTPs, 1.5 mm MgCl2 and 1 × NH4 reaction buffer (Bioline). Conditions for the amplification were an initial denaturation step of 5 min at 94 °C, followed by 35 cycles of 20 s at 94 °C, 20 s at 55 °C and 100 s at 72 °C, and a final elongation step of 7 min at 72 °C. The PCR products were visualized on gels and sequenced as described above using M13F and M13R universal primers.
The multiple sequence products obtained by the chromosome walking procedure were assembled and edited in Bionumerics v4.60 (Applied Maths). blastx analysis of the assembled sequences against the National Center for Biotechnology Information (NCBI) nonredundant protein database (Altschul et al., 1997) was used to predict the end of the idiomorphs.
New primers were designed on the basis of the flanking regions of the idiomorphs and five reference sequences (Table 2), and employed in an attempt to amplify directly the entire idiomorph of the species used in this study (Table 1). The idiomorph PCR mixture contained 1.0 µL of diluted DNA, 0.2 µm of each primer, 1 U BIOTAQ DNA polymerase (Bioline), 0.1 mm dNTPs, 2 mm MgCl2 and 1 × NH4 reaction buffer (Bioline). Conditions for the amplification were an initial denaturation step of 5 min at 94 °C, followed by 35 cycles of 45 s at 94 °C, 45 s at 58 °C and 3 min at 72 °C, and a final elongation step of 10 min at 72 °C. The products were cloned as described above and subsequently fully sequenced.
Based on the newly sequenced idiomorphs, Phoma‐specific MAT1 and HMG motif primers were designed (Table 1). The PCR mixture in this Phoma‐specific PCR contained 1 µL of diluted genomic DNA, 0.2 µm of each primer, 0.5 U BIOTAQ DNA polymerase (Bioline), 0.06 mm dNTPs, 2.4 mm MgCl2 and 1 × NH4 reaction buffer (Bioline). Conditions for the amplification were an initial denaturation step of 5 min at 94 °C, followed by 10 cycles of 30 s at 94 °C and 30 s at 65 °C with a decrease of 0.7 °C every cycle, and 30 s at 72 °C, followed by 30 cycles with an annealing temperature of 58 °C, and a final elongation step of 7 min at 72 °C.
For the species for which no MAT1‐1 and/or MAT1‐2 idiomorphs had been obtained, the newly obtained HMG motif or partial MAT1‐1 PCR sequences were used to perform additional chromosome walking as described above.
The gene‐finding software fgenesh (Softberry Inc., Mount Kisco, NY, USA) was used to predict the gene structure of all mating‐type genes. Lasergene Seqbuilder v7.2.1(DNASTAR Inc., Madison, WI, USA) software was used to make a graphical representation of the structural organization of the mating genes.
Phylogenetic analysis
Phylogenetic analyses were performed on parts of the ITS1, ITS2 and 5.8S rRNA gene (ITS), and on parts of the MAT1‐1‐1 and MAT1‐2‐1 sequences obtained using the Phoma‐specific primers. The primers V9G (de Hoog and Gerrits van den Ende, 1998) and ITS4 (White et al., 1990) were used for the amplification of the ITS region, as described previously (Woudenberg et al., 2009). The multiple sequence alignments were made with mafft v6.850b (http://mafft.cbrc.jp/alignment/server/index.html), using the L‐INS‐i setting (Katoh et al., 2005). Phylogenetic analyses of the sequence data consisted of a parsimony analysis conducted in paup v4.0b10 (Swofford, 2003) and a Bayesian analysis conducted with MrBayes v3.1.2 (Huelsenbeck and Ronquist, 2001). In the parsimony analyses, the heuristic search option with 100 random taxa additions was used, with tree bisection and reconstruction (TBR) as the branch‐swapping algorithm. Alignment gaps were treated as missing. The robustness of the parsimony tree was evaluated by 1000 bootstrap replicates (Hillis and Bull, 1993). The Bayesian analyses were run with a GTR model with gamma‐distributed rate variation for the ITS alignment and an HKY model with gamma‐distributed rate variation for the MAT1‐1‐1 and MAT1‐2‐1 alignments; models were selected using Findmodel (http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html). Further settings included a ‘temperature’ value of 0.05, five million generations and a sample frequency of 100. The run was automatically stopped as soon as the average standard deviation of split frequencies dropped below 0.01. The resulting trees were printed with Treeview v1.6.6 (Page, 1996)
Nucleotide sequence accession numbers
All sequences generated were deposited in GenBank. ITS sequences were deposited with accession numbers JF810508–JF810535. MAT1‐1 mating‐type loci of P. clematidina, Pey. pinodella and D. vitalbina were deposited with accession numbers JF815528, JF815529 and JF815527, respectively. The sequences of the MAT1‐2 mating‐type loci of P. clematidina, Pey. pinodella, D. vitalbina and P. herbarum were deposited with accession numbers JF815530, JF815531, JF815532 and JF815526, respectively. The sequences of the mating‐type loci of Pey. pinodes and D. clematidis were deposited with accession numbers JF815533 and JF815534, respectively.
ACKNOWLEDGEMENTS
This research was financially supported by the Dutch Ministry of Agriculture, Nature and Food Quality through an endowment of the Fonds Economische Structuurversterking programme ‘Versterking Infrasctructuur Plantgezondheid’. The authors would like to thank J. Z. Groenewald (CBS‐KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands) for guidance in the phylogenetic analyses and two anonymous reviewers for critical reading of the manuscript and helpful comments.
REFERENCES
- Altschul, S.F. , Madden, T.L. , Schaffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzanlou, M. , Crous, P.W. and Zwiers, L.‐H. (2010) Evolutionary dynamics of mating‐type loci of Mycosphaerella spp. occurring on banana. Eukaryot. Cell, 9, 164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aveskamp, M.M. , De Gruyter, J. and Crous, P.W. (2008) Biology and recent developments in the systematics of Phoma, a complex genus of major quarantine significance. Fungal Divers. 31, 1–18. [Google Scholar]
- Aveskamp, M.M. , Verkley, G.J.M. , de Gruyter, J. , Murace, M.A. , Perello, A. , Woudenberg, J.H.C. , Groenewald, J.Z. and Crous, P.W. (2009a) DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia, 101, 363–382. [DOI] [PubMed] [Google Scholar]
- Aveskamp, M.M. , Woudenberg, J.H.C. , De Gruyter, J. , Turco, E. , Groenewald, J.Z. and Crous, P.W. (2009b) Development of taxon‐specific sequence characterized amplified region (SCAR) markers based on actin sequences and DNA amplification fingerprinting (DAF): a case study in the Phoma exigua species complex. Mol. Plant Pathol. 10, 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aveskamp, M.M. , de Gruyter, J. , Woudenberg, J.H.C. , Verkley, G.J.M. and Crous, P.W. (2010) Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Stud. Mycol. 65, 1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barve, M.P. , Arie, T. , Salimath, S.S. , Muehlbauer, F.J. and Peever, T.L. (2003) Cloning and characterization of the mating type (MAT) locus from Ascochyta rabiei (teleomorph: Didymella rabiei) and a MAT phylogeny of legume‐associated Ascochyta spp. Fungal Genet. Biol. 39, 151–167. [DOI] [PubMed] [Google Scholar]
- Baumann, G. (1953) Untersuchungen zur Biologie van Mycosphaerella pinodes (Berk. et. Blox.) Stone. Kühn-Archiv. 67, 305–383. [Google Scholar]
- Begerow, D. , Nilsson, H. , Unterseher, M. and Maier, W. (2010) Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl. Microbiol. Biotechnol. 87, 99–108. [DOI] [PubMed] [Google Scholar]
- Boerema, G.H. (1997) Contributions towards a monograph of Phoma (coelomycetes). 5. Subdivision of the genus in sections. Mycotaxon, 64, 321–333. [Google Scholar]
- Boerema, G.H. and Bollen, G.J. (1975) Conidiogenesis and conidial septation as differentiating criteria between Phoma and Ascochyta . Persoonia, 8, 111–144. [Google Scholar]
- Boerema, G.H. , De Gruyter, J. , Noordeloos, M.E. and Hamers, M.E.C. (2004) Phoma Identification Manual. Differentiation of Specific and Infra‐Specific Taxa in Culture. Wallingford, CT: CABI Publishing. [Google Scholar]
- Bowen, J.K. , Lewis, B.G. and Matthews, P. (1997) Discovery of the teleomorph of Phoma medicaginis var. pinodella in culture. Mycol. Res. 101, 80–84. [Google Scholar]
- Brewer, J.G. and Boerema, G.H. (1965) Electron microscope observations on the development of pycnidiospores in Phoma and Ascochyta spp. Proc. K. Ned. Akad. Wet. C, 68, 86–97. [Google Scholar]
- Buchanan, P.K. (1987) A reappraisal of Ascochytula and Ascochytella (coelomycetes) In: Mycological Papers, Vol. 156, pp. 1–83. Kew, Surrey: CABI Publishing. [Google Scholar]
- Chérif, M. , Chilvers, M. , Akamatsu, H. , Peever, T.L. and Kaiser, W. (2006) Cloning of the mating type locus from Ascochyta lentis (teleomorph: Didymella lentis) and development of a multiplex PCR mating assay for Ascochyta species. Curr. Genet. 50, 203–215. [DOI] [PubMed] [Google Scholar]
- Chilvers, M.I. , Rogers, J.D. , Dugan, F.M. , Stewart, J.E. , Chen, W. and Peever, T.L. (2009) Didymella pisi sp. nov., the teleomorph of Ascochyta pisi . Mycol. Res. 113, 391–400. [DOI] [PubMed] [Google Scholar]
- Coppin, E. , Debuchy, R. , Arnaise, S. and Picard, M. (1997) Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61, 411–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozijnsen, A.J. and Howlett, B.J. (2003) Characterisation of the mating‐type locus of the plant pathogenic ascomycete Leptosphaeria maculans . Curr. Genet. 43, 351–357. [DOI] [PubMed] [Google Scholar]
- Crous P.W., Verkley G.J.M., Groenewald J.Z. and Samson R.A. (eds) (2009) CBS Laboratory Manual Series 1: Fungal Biodiversity. Utrecht: CBS‐KNAW Fungal Biodiversity Centre. [Google Scholar]
- Davidson, J.A. , Hartley, D. , Priest, M. , Krysinska‐Kaczmarek, M. , Herdina, McKay, A. and Scott, E.S. (2009) A new species of Phoma causes ascochyta blight symptoms on field peas (Pisum sativum) in South Australia. Mycologia, 101, 120–128. [DOI] [PubMed] [Google Scholar]
- Faris‐Mokaiesh, S. , Boccara, M. , Denis, J. , Derrien, A. and Spire, D. (1996) Differentiation of the ‘Ascochyta complex’ fungi of pea by biochemical and molecular markers. Curr. Genet. 29, 182–190. [DOI] [PubMed] [Google Scholar]
- Fatehi, J. , Bridge, P.D. and Punithalingam, E. (2003) Molecular relatedness within the ‘Ascochyta pinodes‐complex’. Mycopathologia, 156, 317–327. [DOI] [PubMed] [Google Scholar]
- Galagan, J.E. , Calvo, S.E. , Cuomo, C. , Ma, L.‐J. , Wortman, J.R. , Batzoglou, S. , Lee, S.‐I. , Basturkmen, M. , Spevak, C.C. , Clutterbuck, J. , Kapitonov, V. , Jurka, J. , Scazzocchio, C. , Farman, M. , Butler, J. , Purcell, S. , Harris, S. , Braus, G.H. , Draht, O. , Busch, S. , D'Enfert, C. , Bouchier, C. , Goldman, G.H. , Bell‐Pedersen, D. , Griffiths‐Jones, S. , Doonan, J.H. , Yu, J. , Vienken, K. , Pain, A. , Freitag, M. , Selker, E.U. , Archer, D.B. , Penalva, M.A. , Oakley, B.R. , Momany, M. , Tanaka, T. , Kumagai, T. , Asai, K. , Machida, M. , Nierman, W.C. , Denning, D.W. , Caddick, M. , Hynes, M. , Paoletti, M. , Fischer, R. , Miller, B. , Dyer, P. , Sachs, M.S. , Osmani, S.A. and Birren, B.W. (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae . Nature, 438, 1105–1115. [DOI] [PubMed] [Google Scholar]
- Geiser, D.M. , Frisvad, J.C. and Taylor, J.W. (1998) Evolutionary relationships in Aspergillus section Fumigati inferred from partial β‐tubulin and hydrophobin DNA sequences. Mycologia, 90, 831–845. [Google Scholar]
- Groenewald, M. , Groenewald, J.Z. , Harrington, T.C. , Abeln, E.C.A. and Crous, P.W. (2006) Mating type gene analysis in apparently asexual Cercospora species is suggestive of cryptic sex. Fungal Genet. Biol. 43, 813–825. [DOI] [PubMed] [Google Scholar]
- de Gruyter, J. , Aveskamp, M.M. , Woudenberg, J.H.C. , Verkley, G.J.M. , Groenewald, J.Z. and Crous, P.W. (2009) Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycol. Res. 113, 508–519. [DOI] [PubMed] [Google Scholar]
- de Gruyter, J. , Woudenberg, J.H.C. , Aveskamp, M.M. , Verkley, G.J.M. , Groenewald, J.Z. and Crous, P.W. (2010) Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma . Mycologia, 102, 1066–1081. [DOI] [PubMed] [Google Scholar]
- Hillis, D.M. and Bull, J.J. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42, 182–192. [Google Scholar]
- de Hoog, G.S. and Gerrits van den Ende, A.H. (1998) Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses, 41, 183–189. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J.P. and Ronquist, F. (2001) MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755. [DOI] [PubMed] [Google Scholar]
- Inderbitzin, P. , Harkness, J. , Turgeon, B.G. and Berbee, M.L. (2005) Lateral transfer of mating system in Stemphylium . Proc. Natl. Acad. Sci. USA, 102, 11 390–11 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderbitzin, P. , Shoemaker, R.A. , O'Neill, N.R. , Turgeon, B.G. and Berbee, M.L. (2006) Systematics and mating systems of two fungal pathogens of opium poppy: the heterothallic Crivellia papaveracea with a Brachycladium penicillatum asexual state and a homothallic species with a Brachycladium papaveris asexual state. Can. J. Bot. 84, 1304–1326. [Google Scholar]
- Kaiser, W.J. and Kusmenoglu, I. (1997) Distribution of mating types and the teleomorph of Ascochyta rabiei on chickpea in Turkey. Plant Dis. 81, 1284–1287. [DOI] [PubMed] [Google Scholar]
- Katoh, K. , Kuma, K.‐I. , Toh, H. and Miyata, T. (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerenyi, Z. , Moretti, A. , Waalwijk, C. , Olah, B. and Hornok, L. (2004) Mating type sequences in asexually reproducing Fusarium species. Appl. Environ. Microbiol. 70, 4419–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg, R.L. and Glass, N.L. (1990) Mating type and mating strategies in Neurospora . Bioessays, 12, 53–59. [DOI] [PubMed] [Google Scholar]
- Nauta, M.J. and Hoekstra, R.F. (1992) Evolution of reproductive systems in filamentous ascomycetes. 1. Evolution of mating types. Heredity, 68, 405–410. [DOI] [PubMed] [Google Scholar]
- O'Donnell, K. , Ward, T.J. , Geiser, D.M. , Kistler, H.C. and Aoki, T. (2004) Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 41, 600–623. [DOI] [PubMed] [Google Scholar]
- O'Gorman, C.M. , Fuller, H.T. and Dyer, P.P.S. (2008) Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus . Nature, 457, 471–474. [DOI] [PubMed] [Google Scholar]
- Onfroy, C. , Tivoli, B. , Corbière, R. and Bouznad, Z. (1999) Cultural, molecular and pathogenic variability of Mycosphaerella pinodes and Phoma medicaginis var. pinodella isolates from dried pea (Pisum sativum) in France. Plant Pathol. 48, 218–229. [Google Scholar]
- Page, R.D.M. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Paoletti, M. , Rydholm, C. , Schwier, E.U. , Anderson, M.J. , Szakacs, G. , Lutzoni, F. , Debeaupuis, J.‐P. , Latge, J.‐P. , Denning, D.W. and Dyer, P.S. (2005) Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus . Curr. Biol. 15, 1242–1248. [DOI] [PubMed] [Google Scholar]
- Peever, T. (2007) Role of host specificity in the speciation of Ascochyta pathogens of cool season food legumes. Eur. J. Plant Pathol. 119, 119–126. [Google Scholar]
- Peever, T.L. , Barve, M.P. and Stone, L.J. (2007) Evolutionary relationships among Ascochyta species infecting wild and cultivated hosts in the legume tribes Cicereae and Vicieae. Mycologia, 99, 59–77. [DOI] [PubMed] [Google Scholar]
- Punithalingam, E. (1979) Graminicolous Ascochyta species In: Mycological Papers, Vol. 142, pp. 1–214. Kew, Surrey: CABI Publishing. [Google Scholar]
- Rau, D. , Maier, F.J. , Papa, R. , Brown, A.H. , Balmas, V. , Saba, E. , Schaefer, W. and Attene, G. (2005) Isolation and characterization of the mating‐type locus of the barley pathogen Pyrenophora teres and frequencies of mating‐type idiomorphs within and among fungal populations collected from barley landraces. Genome, 48, 855–869. [DOI] [PubMed] [Google Scholar]
- Rau, D. , Attene, G. , Brown, A. , Nanni, L. , Maier, F. , Balmas, V. , Saba, E. , Schäfer, W. and Papa, R. (2007) Phylogeny and evolution of mating‐type genes from Pyrenophora teres, the causal agent of barley ‘net blotch’ disease. Curr. Genet. 51, 377–392. [DOI] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H.J. (2000) Primer 3 on the WWW for general users and for biologist programmers In: Bioinformatics Methods and Protocols: Methods in Molecular Biology (Krawetz S. and Misener S., eds), pp. 365–386. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Rydholm, C. , Dyer, P.S. and Lutzoni, F. (2007) DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating‐type loci of the self‐compatible ascomycete mold Neosartorya fischeri . Eukaryot. Cell, 6, 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert, K.A. (2009) Progress towards DNA barcoding of fungi. Mol. Ecol. Resour. 9, 83–89. [DOI] [PubMed] [Google Scholar]
- Sharon, A. , Yamaguchi, K. , Christiansen, S. , Horwitz, B.A. , Yoder, O.C. and Turgeon, B.G. (1996) An asexual fungus has the potential for sexual development. Mol. Gen. Genet. 251, 60–68. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. , Groenewald, M. , Staats, M. , Lindhout, P. , Crous, P.W. and De Wit, P.J.G.M. (2007) Mating‐type genes and the genetic structure of a world‐wide collection of the tomato pathogen Cladosporium fulvum . Fungal Genet. Biol. 44, 415–429. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2003) PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Turgeon, B.G. (1998) Application of mating type gene technology to problems in fungal biology. Annu. Rev. Phytopathol. 36, 115–137. [DOI] [PubMed] [Google Scholar]
- Turgeon, B.G. and Yoder, O.C. (2000) Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 31, 1–5. [DOI] [PubMed] [Google Scholar]
- Turgeon, B.G. , Bohlmann, H. , Ciuffetti, L.M. , Christiansen, S.K. , Yang, G. , Schafer, W. and Yoder, O.C. (1993) Cloning and analysis of the mating‐type genes from Cochliobolus heterostrophus . Mol. Gen. Genet. 238, 270–284. [DOI] [PubMed] [Google Scholar]
- Ueng, P.P. , Dai, Q. , Cui, K. , Czembor, P.C. , Cunfer, B.M. , Tsang, H. , Arseniuk, E. and Bergstrom, G. (2003) Sequence diversity of mating‐type genes in Phaeosphaeria avenaria . Curr. Genet. 43, 121–130. [DOI] [PubMed] [Google Scholar]
- Varga, J. , Vida, Z. , Toth, B. , Debets, F. and Horie, Y. (2000) Phylogenetic analysis of newly described Neosartorya species. Anton. Leeuw. Int. J. G. 77, 235–239. [DOI] [PubMed] [Google Scholar]
- Waalwijk, C. , Mendes, O. , Verstappen, E.C.P. , de Waard, M.A. and Kema, G.H.J. (2002) Isolation and characterization of the mating‐type idiomorphs from the wheat septoria leaf blotch fungus Mycosphaerella graminicola . Fungal Genet. Biol. 35, 277–286. [DOI] [PubMed] [Google Scholar]
- White, T.J. , Bruns, T. , Lee, S. and Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: PCR Protocols: A Guide to Methods and Applications (Innis M.A., Gelfand D.H., Sninsky J.J. and White T.J., eds), pp. 315–322. San Diego, CA: Academic Press. [Google Scholar]
- Wollenweber, H.W. and Hochapfel, H. (1936) Beiträge zur Kenntnis parasitärer und saprophytotischer Pilze. I. Phomopsis, Dendrophoma, Phoma und Ascochyta und ihre Beziehung zur Fruchtfäule. Z. Parasitenkd. 8, 561–605. [Google Scholar]
- Woudenberg, J.H.C. , Aveskamp, M.M. , de Gruyter, J. , Spiers, A.G. and Crous, P.W. (2009) Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia, 22, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, S.H. , Berbee, M.L. , Yoder, O.C. and Turgeon, B.G. (1999) Evolution of the fungal self‐fertile reproductive life style from self‐sterile ancestors. Proc. Natl. Acad. Sci. USA, 96, 5592–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, S.‐H. , Arie, T. , Kaneko, I. , Yoder, O.C. and Turgeon, B.G. (2000) Molecular organization of mating type loci in heterothallic, homothallic, and asexual Gibberella/Fusarium species. Fungal Genet. Biol. 31, 7–20. [DOI] [PubMed] [Google Scholar]


