Summary
In the Potyvirus genus, the P1 protein is the first N‐terminal product processed from the viral polyprotein, followed by the helper‐component proteinase (HCPro). In silencing suppression patch assays, we found that Potato virus Y (PVY) HCPro expressed from a P1‐HCPro sequence increased the accumulation of a reporter gene, whereas protein expressed from an HCPro sequence did not, even with P1 supplied in trans. This enhancing effect of P1 has been noted in other potyviruses, but has remained unexplained. We analysed the accumulation of PVY HCPro in infiltrated tissues and found that it was higher when expressed from P1‐HCPro than from HCPro sequences. Co‐expression of heterologous suppressors increased the steady‐state level of mRNA expressed from the HCPro sequence, but not that of protein. This suggests that, in the absence of P1 upstream, either HCPro acquires a conformation that affects negatively its activity or stability, or that its translation is reduced. To test these options, we purified HCPro expressed in the presence or absence of upstream P1, and found no difference in purification pattern and final soluble state. By contrast, alteration of the Kozak context in the HCPro mRNA sequence to favour translation increased partially suppressor accumulation and activity. Furthermore, protein activity was not lower than in protein expressed from P1‐HCPro sequences. Thus, a direct role for P1 on HCPro suppressor activity or stability, by influencing its conformation during translation, can be excluded. However, P1 could still have an indirect effect favouring HCPro accumulation. Our data highlight the relevance of cis‐acting translational elements in the heterologous expression of HCPro.
Introduction
Members of the genus Potyvirus are plus‐sense RNA viruses that express a single polyprotein which undergoes proteolytic cleavage to generate the final products, although a small essential gene expresses via translational frameshifting (Chung et al., 2008). Extensive work to understand the proteolytic activities and processing of the potyviral polyprotein was carried out in the late 1980s and early 1990s, mainly on Tobacco etch virus (TEV) and Plum pox virus (PPV) using in vitro translation systems. Three proteases are involved in the processing of the polyprotein: (i) the P1 protein, a serine‐type protease that detaches itself at its C‐terminus from the adjacent helper‐component proteinase (HCPro) (Verchot and Carrington, 1995a; Verchot et al., 1991); (ii) HCPro, a papain‐like protease that detaches itself at its C‐terminus from the polyprotein (Carrington and Herndon, 1992); and (iii) the small nuclear inclusion body protein (NIa), which targets the remaining polyprotein cleavage sites (Carrington et al., 1988; García et al., 1989).
The first N‐terminal product from the polyprotein corresponds to the P1 protein, followed by HCPro. Early studies implicated the P1 protein in viral genome amplification (Verchot and Carrington, 1995b), an effect that operates in trans (Verchot and Carrington, 1995b), whereas its protease activity does not (Verchot et al., 1991). Mutations that prevented P1 from detaching from HCPro severely affected the viability of the virus, but this was restored by adding an NIa protease site at the P1–HCPro boundary, indicating that detachment of P1 from HCPro rather than P1 activity is essential to virus infectivity. P1 is, in fact, dispensable, but virus accumulation and movement are then severely diminished (Verchot and Carrington, 1995a).
HCPro, by contrast, is essential in the potyvirus infection cycle. In addition to being a protease and essential for the horizontal transmission of these viruses by vectors, it enhances the pathogenicity of other viruses, such as potex‐, cucumo‐ and tobamoviruses (Pruss et al., 1997), and suppresses gene silencing defences; it has been demonstrated that Potato virus Y (PVY) HCPro alone, expressed transgenically, can reverse the virus‐induced systemic silenced state of reporter transgenes, and that TEV HCPro alone, expressed from a virus vector, can prevent the systemic silencing of a transgene (Anandalakshmi et al., 1998; Brigneti et al., 1998). However, HCPros from TEV, PVY, PPV and Potato virus A (PVA), expressed from P1‐HCPro sequences, prevent the local silencing of reporter genes in agroinfiltration patch assays (Canto and Palukaitis, 2002; Johansen and Carrington, 2001; Rajamäki et al., 2005; Valli et al., 2006).
The mode by which HCPro interferes with the host antiviral gene silencing defences is not yet fully understood. However, it is known that PVY HCPro can interact in vitro with long (250 nucleotides) nucleic acids (Maia and Bernardi, 1996; Urcuqui‐Inchima et al., 2000), and TEV and Zucchini yellow mosaic virus (ZYMV) HCPros can interact in vitro with synthetic double‐stranded (ds) small RNAs (Mérai et al., 2006; Shiboleth et al., 2007). This binding in the case of Papaya ringspot virus HCPro was found to be temperature dependent (Mangrauthia et al., 2009). Binding in vitro to synthetic small RNAs of hexa‐histidine‐tagged TEV HCPro, purified from virus‐infected plants, was also found to be dependent on both small RNA size and the presence or absence of overhangs, and was enhanced by the addition of Drosophila embryo or Arabidopsis thaliana extracts (Lakatos et al., 2006). Turnip mosaic virus (TuMV) HCPro has been shown to interfere with the biogenesis and action of microRNAs, although no direct binding to these small RNAs was observed (Chapman et al., 2004). In all of these cases, the binding to small RNAs occurred at protein : RNA molar ratios much higher than the 2 : 1 characterized in the P19 and 2b suppressors of Tomato bushy stunt virus (TBSV) and Cucumber mosaic virus (CMV) (González et al., 2012; Vargasson et al., 2003), making it unlikely that HCPro interferes with the antiviral silencing response by the sequestration of small RNAs.
Interactions of HCPro with host proteins involved in gene silencing processes have so far only been reported between ZYMV HCPro and the RNA methyltransferase HEN‐1 in vitro, whose activity was inhibited (Jamous et al., 2011). However, HCPro interacts with host proteins that intervene in processes other than gene silencing, e.g. a calmodulin‐related protein (rgs‐CaM) (Anandalakshmi et al., 2000), which is able to bind and inhibit the activities of dsRNA‐binding viral suppressors, as well as to direct their degradation through the autophagy pathway (Nakahara et al., 2012), and the A. thaliana transcription factor RAV2, whose expression appears to be required for HCPro suppressor activity (Endres et al., 2010). HCPro also binds components of the proteasome, a structure potentially involved in antiviral defence (Ballut et al., 2005; Dielen et al., 2011; Jin et al., 2007a), translation initiation factors (Ala‐Poikela et al., 2011) and chloroplast factors (Cheng et al., 2008; Jin et al., 2007b).
With regard to its conformation, HCPro is a cytoplasmic protein of around 50 kDa with three domains: the N‐terminal domain, associated with aphid transmission (Blanc et al., 1997; Canto et al., 1995a) and interaction with proteasomal units (Jin et al., 2007a); the central domain, associated with the suppression of silencing function (Shiboleth et al., 2007); and a C‐terminal domain containing its protease activity (Carrington and Herndon, 1992). In plants and in vitro, HCPro has been shown to self‐interact and to form soluble aggregates (Plisson et al., 2003; Ruíz‐Ferrer et al., 2005; Thornbury et al., 1985; Urcuqui‐Inchima et al., 1999; Zheng et al., 2011), which could have functional relevance. In addition to self‐interaction, HCPro has been shown to interact with other viral components, although whether it binds to the P1 protein in vivo remains unclear (Merits et al., 1999; Zilian and Maiss, 2011).
From early studies, it is known that the presence of P1 upstream of the HCPro sequence increases the activity of the latter, both as a pathogenicity enhancer and as a suppressor of gene silencing when expressed from heterologous systems, such as T‐DNAs transiently or constitutively, or viral vectors: for example, the presence of P1 and of the viral 5' nontranslated region upstream of a TEV HCPro sequence expressed from a Potato virus X (PVX) virus vector strongly enhanced the stability and accumulation of the minus‐strand RNA of the vector when compared with PVX expressing HCPro alone (Pruss et al., 1997). An enhancing effect of transgenically expressed P1 on the efficiency of TEV HCPro suppression of the virus‐induced gene silencing (VIGS) of a transgene reporter has also been observed (Anandalakshmi et al., 1998). Furthermore, local suppression of the silencing of a transiently expressed reporter by PPV HCPro in agroinfiltration patch assays occurred only if the latter was expressed as P1‐HCPro, rather than as HCPro alone (Valli et al., 2006). Similarly, the total absence of P1 or some insertions in the PVA P1 cistron resulted in reduced accumulation of HCPro when expressed by agroinfiltration from a P1‐HCPro sequence, and affected its suppressor of silencing activity on a β‐glucuronidase reporter. A role for P1 as a stabilizer of PVA HCPro, allowing strong suppression of silencing and high RNA levels during transient expression, was therefore hypothesized (Rajamäki et al., 2005). This accumulated experimental evidence on the enhancing effects in cis of P1 on HCPro activity (Anandalakshmi et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998; Pruss et al., 1997; Rajamäki et al., 2005; Valli et al., 2006) has caused many researchers to use P1‐HCPro rather than HCPro in their experimental studies. However, the reasons for these enhancing effects of P1 on HCPro accumulation and activity in these diverse experimental systems have remained largely unexplained.
We have investigated the enhancing effect on the accumulation and biological activity of PVY HCPro of the upstream presence in cis of the P1 protein in agroinfiltration patch assays. We have also studied how nucleotide positions around the initiation codon influence the translatability, accumulation and activity of HCPro when expressed in the absence of P1. We show that PVY HCPro lack of suppressor activity when expressed in the absence of P1 upstream can be partially compensated for by enhanced protein translation in a more favourable Kozak translation context. Furthermore, we show a correlation between suppressor activity and its accumulation, and exclude a role for P1 on either HCPro suppressor activity or stability by functioning in its conformational maturation during translation. Our results do not rule out some indirect contribution of P1 to HCPro accumulation, and highlight the relevance of cis‐acting translational elements in the heterologous expression of HCPro in plants.
Results
The presence upstream in cis of the viral P1 protein provides strong local suppression of silencing activity to PVY HCPro in patch assays
PVY HCPro expressed transiently by agroinfiltration from a 35S promoter‐driven P1‐HCPro sequence in a binary vector (construct P1‐HCPro; Table 1) suppressed the partial silencing of a co‐infiltrated green fluorescent protein (GFP) reporter construct, leading to increased GFP‐derived fluorescence in the infiltrated patch under an ultraviolet (UV) lamp (Fig. 1A, both leaves, top left vs. right patches, and top Western blot panel). HCPro tagged with a methionine plus six histidines at its N‐terminus, expressed from a P1‐6x‐HCPro sequence (construct P1‐6x‐HCPro; Table 1), was also able to induce a similar increase in fluorescence (Fig. 1A, top vs. bottom left patches and top Western blot panel). Thus, the addition of the tag did not affect the local suppressor of silencing activity of PVY HCPro. The 6 × histidine‐tagged HCPro had a similar size to the native protein, around 50 kDa, indicating that proteolytic self‐cleavage by P1 had not been affected (Fig. 1A, middle and bottom Western blot panels). We therefore used 6 × histidine‐tagged HCPro in this work instead of HCPro because of its convenience with regard to serological detection and the purification from plants.
Table 1.
Representation of the N‐terminal amino acid sequences and the nucleotides around the translation initiation codon in the helper‐component proteinase (HCPro) constructs used in this work
| Construct | Amino acid sequence at protein N‐terminus | Suppressor activity | Kozak plant consensus AACAAUGGC |
|---|---|---|---|
| P1‐HCPro | Gln‐Phe/Ser‐Asn‐ | +++ | AUCAAUGGC |
| P1‐6x‐HCPro | Gln‐Phe/Ser‐Ala‐Ser‐Met‐6 × His‐Ser‐Asn‐ | +++ | AUCAAUGGC |
| HCPro | Met‐Ser‐Asn‐ | –+ | AUCCAUGUC |
| 6x‐HCPro | Met‐6 × His‐Ser‐Asn‐ | –+ | AUCCAUGCA |
| 6x‐HCPro (Ala) | Met‐Ala‐6 × His‐Ser‐Asn‐ | ++ | AUCCAUGGC |
The first column shows the binary construct names. The second column shows, in italic, the last amino acids from P1, the catalytic separation site indicated as ‘/’ and any extra amino acids that remain attached to HCPro; the original amino acid sequences appear in roman. Proteins expressed from construct HCPro or P1‐HCPro only differ in that there is an additional N‐terminal methionine (Met) at the N‐terminus of the former; HCPro expressed from construct 6x‐HCPro or P1‐6x‐HCPro differs in that there is a serine‐alanine‐serine triplet (Ser‐Ala‐Ser) at the N‐terminus of the latter. Construct 6x‐HCPro (Ala) contains an added alanine between the starting methionine and the 6 × histidines. The suppressor activities in patch assays of each construct are shown in the next column (+++, ++ and + refer to strong, intermediate and weak/no suppressor activity, respectively). The nucleotide sequences flanking the AUG translation initiation codon in the corresponding mRNAs, with regard to the consensus found in plants (Lützke et al., 1987), are shown in the last column.
Figure 1.
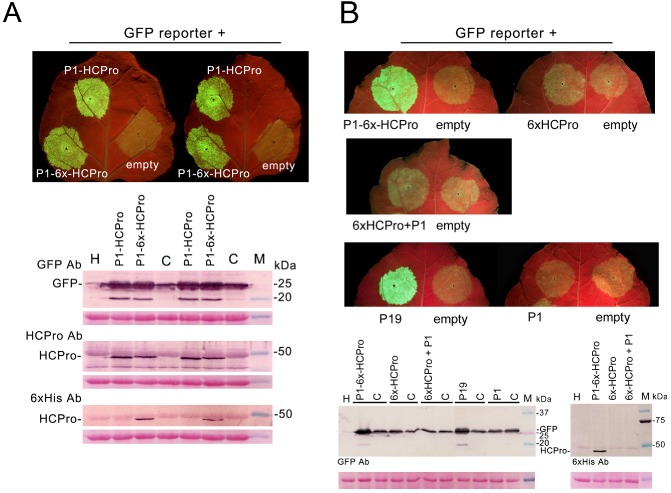
Assessment of the suppressor of silencing activity of the helper‐component proteinase (HCPro) constructs in agroinfiltration patch assays in Nicotiana benthamiana leaves. (A) Addition of an N‐terminal 6 × histidine tag to the HCPro protein does not affect its suppressor of silencing activity: a binary vector expressing a green fluorescent protein (GFP) reporter was co‐infiltrated together with the empty binary vector pROK2 (right side of both leaves), together with a binary vector expressing HCPro from a P1‐HCPro sequence (construct P1‐HCPro; top patch on left side of leaves) or 6 × histidine‐tagged (6x) HCPro from a P1‐6x‐HCPro sequence (construct P1‐6x‐HCPro; bottom patch on left side of leaves). The increase in GFP‐derived fluorescence under UV light was similar in both cases. The amounts of GFP protein and of suppressor detected by Western blotting using antibodies against GFP, HCPro and histidine tags were also similar (top, middle and bottom Western blot panels, respectively). (B) In the absence of P1 upstream in cis, 6 × histidine‐tagged HCPro expressed from a 6x‐HCPro sequence (construct 6x‐HCPro) barely suppressed the silencing of the GFP reporter when compared with that shown by construct P1‐6x‐HCPro (top leaf panel, compare right vs. left leaf, and left Western blot panel). The addition of P1 in trans from a different binary vector (construct P1) did not alter this lack of suppressor activity (middle leaf panel, and left Western blot membrane). However, the suppressor activity of construct P1‐6x‐HCPro was comparable with that shown by the Tomato bushy stunt virus P19 protein, a strong suppressor of gene silencing (left leaf in top leaf panel vs. left leaf in bottom leaf panel, and left Western blot membrane), whereas P1 protein on its own did not show any suppressor activity in this type of assay (bottom leaf panel, right leaf). The Western blot panel to the right shows the accumulation of HCPro in the same samples. Western blot analyses of infiltrated tissue were made at 4 days post‐agroinfiltration. In both Western blot panels, lane H denotes healthy plant extract, used as negative control, and lane M shows the molecular weight markers. The bottom panels below the Western blots in (A) and (B) show the Ponceau S‐stained membranes after blotting, as controls of loading. Lanes labelled C refer to the GFP plus empty binary vector controls co‐infiltrated on the right side of the corresponding leaves.
In contrast with HCPro expressed from construct P1‐6x‐HCPro (Fig. 1B, top left leaf, and left Western blot panel), HCPro expressed from a 6x‐HCPro sequence that lacked the upstream P1 sequence (construct 6x‐HCPro; Table 1) failed to efficiently suppress the silencing of the GFP reporter (Fig. 1B, top right leaf, left vs. right patches, and left Western blot panel), and the same occurred with HCPro expressed from an HCPro sequence (data not shown). Co‐expression of P1 in trans from a different binary vector, together with construct 6x‐HCPro, failed to suppress the silencing of the GFP reporter (Fig. 1B, middle leaf, left vs. right patches, and left Western blot panel), as did P1 expressed alone (Fig. 1B, bottom right leaf, and left Western blot panel). For comparison, the suppression activity of the P19 protein is shown (Fig. 1B, bottom left leaf). Interestingly, HCPro could only be detected serologically in patches infiltrated with construct P1‐6x‐HCPro, but not in those infiltrated with construct 6x‐HCPro (Fig. 1B, right Western blot panel), alone or together with P1 expressed in trans.
In agroinfiltrated patches, steady‐state levels of mRNAs transcribed from construct 6x‐HCPro were several‐fold lower than those transcribed from construct P1‐6x‐HCPro [Fig. 2A, top Northern blot panels, and middle quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) charts, second vs. fourth lanes]. This is probably caused by their partial targeted degradation by the host gene silencing defences, as happens to any T‐DNA‐expressed gene in the absence of an efficient suppressor (Canto and Palukaitis, 2002; Johansen and Carrington, 2001). This would also explain the lack of accumulation of HCPro expressed from construct 6x‐HCPro (Fig. 1B, right Western blot panel). This was indeed the case, as co‐expression of the heterologous viral suppressors 2b from CMV or P19 from TBSV led to a several‐fold increase in construct 6x‐HCPro mRNA levels in the infiltrated patches, approaching those of construct P1‐6x‐HCPro (Fig. 2A, top Northern blot panels, and middle qRT‐PCR charts, fifth and sixth vs. second lane). Surprisingly, HCPro was serologically detected when expressed from construct P1‐6x‐HCPro, but hardly or not at all when expressed from construct 6x‐HCPro, even when silencing of the latter mRNA was prevented by the 2b or P19 suppressor (Fig. 2A, bottom Western blot panels, second vs. fifth to sixth lanes). These data suggest that, in this latter case, either a conformational alteration negatively affects the stability and/or activity of HCPro, or its translation is negatively affected.
Figure 2.
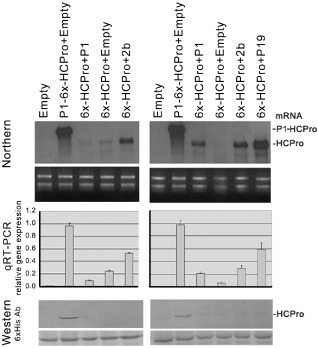
Quantification of the accumulation of helper‐component proteinase (HCPro) expressed from construct P1‐6x‐HCPro and from construct 6x‐HCPro and of the steady‐state levels of their mRNAs in the presence or absence of heterologous suppressors of silencing in agroinfiltrated patches of Nicotiana benthamiana leaves, in two independent experiments (left and right sets of panels, respectively). In each experiment, patches were infiltrated with empty vector (first lane), construct P1‐6x‐HCPro plus empty vector (second lane) and construct 6x‐HCPro plus vectors expressing: P1, empty vector, the Cucumber mosaic virus 2b protein suppressor of silencing and (in the second experiment only) the Tomato bushy stunt P19 suppressor of silencing (third, fourth and fifth lanes, respectively). Analyses of infiltrated tissue were made at 4 days post‐agroinfiltration. HCPro mRNA accumulation in infiltrated tissues was visualized by Northern blot (top panel), and also quantified by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) (middle panel). Protein accumulation was visualized by Western blot (bottom panel). The panels below the Northern blots show the total nucleic acid samples stained with ethidium bromide in separate gels, as controls of equal loading. The panels below the Western blots show the Ponceau S‐stained membranes after blotting, as controls of loading.
HCPro expressed from either 6x‐HCPro or P1‐6x‐HCPro sequences does not display a difference in its purification properties or soluble state
To test whether structural differences exist between HCPros expressed from construct P1‐6x‐HCPro and construct 6x‐HCPro, which might explain the differences observed in protein steady‐state levels and in the respective suppressor activities (Fig. 1B), as well as the lack of enhancing effect of heterologous suppressors on the accumulation of HCPro expressed from construct 6x‐HCPro (Fig. 2), we undertook the purification from plants of protein expressed from both constructs. This allowed us to assess their behaviour during the differential fractionation, precipitation, concentration and nitrilotriacetic acid resin‐binding steps, and also their final soluble states. To do this, the P1‐6x‐HCPro and 6x‐HCPro sequences from constructs P1‐6x‐HCPro and 6x‐HCPro were transferred to PVX vectors for their expression from a subgenomic RNA and large‐scale protein expression and purification from Nicotiana benthamiana plants. Expression of either 6x‐HCPro or P1‐6x‐HCPro from the chimeric PVX vector increased the severity of virus infection symptoms in N. benthamiana plants in both cases. However, this did not translate into increased virus accumulation (Fig. S1, see Supporting Information).
In both cases, protein was expressed successfully, although HCPro expressed from the subgenomic viral RNA containing the 6x‐HCPro sequence accumulated to around 10%–20% of the levels of HCPro expressed from a viral mRNA containing the P1‐6x‐HCPro sequence (Fig. 3A, compare lanes 3 vs. 8 from the left). Despite this difference, both proteins were successfully isolated to near purity using their 6 × histidine tags (Fig. 3B). No differences could be discerned in protein behaviour during the different purification steps (Fig. 3B, top vs. bottom Coomassie‐stained gels and Western blot panels). Purified proteins were then subjected to size fractionation by high‐performance liquid chromatography (HPLC) and their elution profiles were analysed by Western blot. We found that, in both cases, purified soluble HCPro eluted from the column in similar profiles, with the peaks eluting at fractions that could be consistent with tetrameric forms (Fig. 3C). Therefore, no differences were apparent in vitro between HCPros expressed from either P1‐6x‐HCPro or 6x‐HCPro sequences during their purification, or in their soluble aggregated states.
Figure 3.
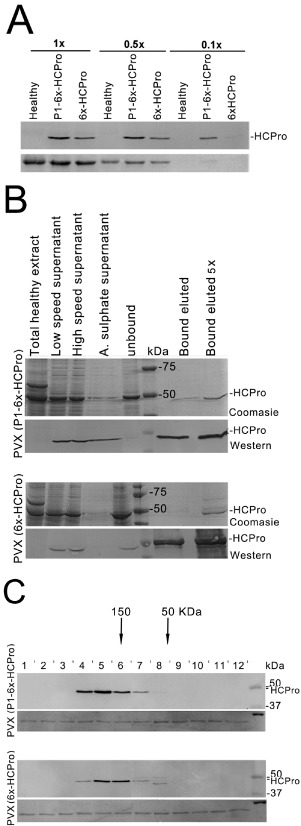
Expression of helper‐component proteinase (HCPro) from either P1‐6x‐HCPro or 6x‐HCPro sequences in subgenomic RNAs from Potato virus X (PVX)‐based vectors (constructs PVX‐P1‐6x‐HCPro and PVX‐6x‐HCPro, respectively), and their purification from infected plant tissue. Tissue from 15 infected plants (approximately 100 g) was pooled for extraction and protein analysis by Western blot and protein purification. (A) The accumulation of HCPro derived from the PVX vector containing a P1‐6x‐HCPro or 6x‐HCPro sequence in infected leaf tissues was assessed by Western blot. Numbers (1×, 0.5× and 0.1×) indicate the relative dilution of each extract sample. HCPro expressed from the latter sequence accumulated at levels that were two‐ to ten‐fold lower than those expressed from the former sequence. (B) Purification of HCPros from plants infected by either PVX construct was achieved successfully in both cases, with similar patterns of protein enrichment at each purification step. In each case, the top panels show the Coomassie blue‐stained sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) fractionation of the protein sample. The bottom panels show the detection of HCPro in the purified samples by Western blot. Each lane was loaded with 7.5 μL of each step of the purification process indicated above the lane. (C) Fractionation of the purified HCPros by high‐performance liquid chromatography (HPLC), followed by Western blot analysis of the eluted fractions, shows that both displayed a similar soluble aggregated pattern. The elution peaks of known size marker proteins are indicated by arrows. HPLC‐eluted fractions of 100 μL were added to 0.5 μg of bovine serum albumin (BSA) as carrier before precipitation with acetone prior to Western blot analysis (middle panels), and the panels below the Western blots show the BSA band in the Ponceau S‐stained membranes after the blot transfer as controls of loading and recovery after acetone precipitation.
Improved translatability results in increased HCPro accumulation and suppressor activity
The respective N‐termini of the HCPros expressed from constructs P1‐HCPro, P1‐6x‐HCPro, HCPro and 6x‐HCPro are shown in Table 1, as well as their local suppressor of silencing activity in patch assays and the sequences upstream and downstream of the AUG translation initiation codons in their mRNAs. Kozak motifs were less favourable in the HCPro and 6x‐HCPro constructs, encoding proteins without suppressor activity, than in P1‐HCPro and P1‐6x‐HCPro constructs, relative to the consensus published for plants. In plant mRNAs, the most frequent two nucleotides after the AUG initiation codon correspond to G and C, present in 85% and 77% of all plant mRNAs, respectively. This results in an alanine after the starting methionine (Lützke et al., 1987). In constructs 6x‐HCPro and HCPro, these two nucleotides correspond to CA and TC, respectively. It could be possible that, despite the increase in the levels of HCPro mRNA induced by heterologous suppressors (Fig. 2A), the corresponding proteins failed to increase because of poor ribosomal affinity for the initiation codon, negatively affecting accumulation and overall suppressor activity. To test this, we added a GCA (encoding alanine) after the AUG initiation codon [construct 6x‐HCPro (Ala)] to create a Kozak context favourable for translation, comparable with that found in the native P1‐HCPro or in construct P1‐6x‐HCPro. The new construct showed increased protein accumulation, when compared with the undetectable levels found in the case of construct 6x‐HCPro (Fig. 4A, top panel). Densitometric analysis of protein bands in Western blot of total protein from the infiltrated patches showed that HCPro accumulation was over 30% of that found in patches infiltrated with construct P1‐6x‐HCPro (Fig. 4A, top panel, sixth and eighth vs. second lane from the left in Western blot). Steady‐state levels of the corresponding mRNAs were also approximately 50% higher than those found in the case of construct 6x‐HCPro (Fig. 4A, bottom panel, sixth and eighth vs. fourth lane from the left in the qRT‐PCR chart), but still half of the levels found in the case of construct P1‐6x‐HCPro (Fig. 4A, bottom panel, sixth and eighth vs. second lane from the left in the qRT‐PCR chart). Interestingly, co‐expression of the heterologous suppressor P19 failed to increase the level of translated HCPro further (Fig. 4A, middle panel, seventh and ninth vs. sixth and eighth lanes from the left in Western blot), despite the fact that, in all cases, it actually increased the HCPro mRNA levels further. Indeed, the protein levels fell slightly (Fig. 4A, top panel, seventh and ninth vs. sixth and eight lanes from the left). This could be caused by competition between HCPro‐ and P19‐encoding mRNAs for the cellular translational machinery.
Figure 4.
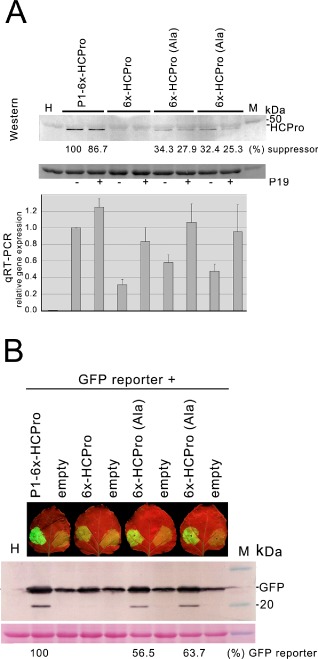
The effects of alterations in the Kozak context on helper‐component proteinase (HCPro) translatability, accumulation and activity. An alanine was introduced between the starting methionine and the 6 × histidine tag encoded by construct 6x‐HCPro [Met‐Ala‐6xHis; construct 6x‐HCPro (Ala)] to assess whether an improved Kozak context sequence (from ATCCATGCA to ATCCATGGC) would increase the transient accumulation of HCPro and its suppressor activity. (A) Quantification of the steady‐state accumulation of protein expressed from construct 6x‐HCPro (Ala) vs. that from either construct 6x‐HCPro or P1‐6x‐HCPro in agroinfiltrated patches (top Western blot panel), and of their corresponding mRNAs [bottom quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) chart] Patches were infiltrated with empty vector (first lane), construct P1‐6x‐HCPro (second and third lanes), construct 6x‐HCPro (fourth and fifth lanes) and construct P1‐6x‐HCPro (Ala) (sixth to ninth lanes), together with either empty vector (lanes labelled ‘–’) or a binary vector expressing P19 (lanes labelled ‘+’). Each lane corresponds to a different patch. Western blot analyses of infiltrated tissue were made at 4 days post‐agroinfiltration. A densitometric analysis of the HCPro bands is shown below the Western blot. Data are given as a percentage of protein relative to that found in the patch in which construct P1‐6x‐HCPro plus the empty vector were co‐infiltrated (100%). The addition of alanine increased the levels of suppressor protein, from undetectable to over 30% of that found in the patch co‐infiltrated with construct P1‐6x‐HCPro plus empty vector. (B) Quantification of the ability of the same constructs to suppress the silencing of a co‐infiltrated green fluorescent protein (GFP) reporter. A densitometric analysis of the reporter bands is shown below the Western blot. Data are given as a percentage of protein detected relative to that found in the patch in which the GFP reporter and construct P1‐6x‐HCPro were co‐infiltrated (a value of 100%). The extra alanine increased the accumulation of the GFP reporter in the infiltrated patches to 56%–63% of that found in the patch co‐infiltrated with construct P1‐6x‐HCPro plus empty vector. In both (A) and (B), lane M shows molecular weight markers, and the small panels below the Western blots show the membranes stained with Ponceau‐S as controls of loading.
We then tested the comparative suppressor of silencing activity of HCPro expressed from construct 6x‐HCPro (Ala) on a co‐infiltrated GFP reporter in patch assays. Under the UV lamp, a phenotype of strong suppression of the silencing of the reporter could be seen (Fig. 4B, top panel, left vs. right infiltrated patches in the corresponding leaves). Increased GFP protein levels were also apparent in Western blot analysis of total proteins from the infiltrated patches. Densitometric analysis of Western blot bands showed that GFP accumulation caused by HCPro expressed from construct 6x‐HCPro (Ala) was around 60% of that induced by HCPro expressed from construct P1‐6x‐HCPro (Fig. 4B, middle panel).
Discussion
From early studies, it is known that, in heterologous expression systems, suppression of silencing activity by HCPros produced by viruses in the genus Potyvirus is stronger if the whole 5' coding region of the polyprotein, rather than HCPro alone, is expressed (fragments P1‐HCPro, sometimes P1‐HCPro‐P3; Anandalakshmi et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998; Pruss et al., 1997; Rajamäki et al., 2005; Valli et al., 2006). These experiments assessed the suppression of silencing of a reporter gene during transient expression from agroinfiltrated T‐DNAs, the reversal of systemic VIGS in transgenic plants and the increase in the accumulation of heterologous viruses in either mixed infections or viral chimeras that expressed the protein. However, the reasons for these enhancing effects of P1 on HCPro accumulation and activity, or, alternatively, the lack of these enhancing effects in the absence of P1 in cis in these diverse experimental systems, have remained largely unexplained. However, it is also known that P1 does not have suppression of silencing activity in these expression systems (Anandalakshmi et al., 1998; Brigneti et al., 1998; Rajamäki et al., 2005; Valli et al., 2006). The enhancing effect therefore appears to be limited to P1 expressed from the same mRNA as HCPro, as part of a polyprotein. To explain the auxiliary effects of P1 on HCPro suppressor activity, Rajamäki et al. (2005) hypothesized a stabilizing effect of P1 on HCPro that would lead to increased accumulation and activity. The way in which this stabilization takes place was not explained. One possibility is that P1 expressed together with HCPro as a polyprotein could be required for correct post‐translational maturation of HCPro, in order to acquire suppressor of silencing activity. In the absence of P1 upstream, an inadequate maturation/conformation could cause HCPro to exhibit either reduced suppressor activity or reduced stability that would eventually lead to reduced suppressor activity. Another view of stability could be that P1 could prevent HCPro from being degraded by host processes, such as the proteasome, autophagy through interaction with rgs‐CaM, RAV2 or other hypothetical mechanisms. Although, for unknown reasons, this would only happen when P1 was expressed in cis.
In our patch assay system, we also found that PVY HCPro displayed suppressor of silencing activity on a reporter only in the presence of P1 upstream in cis (Fig. 1B). Protein analysis showed that a lack of suppressor activity correlated with a lack of HCPro accumulation (Figs 1B and 2). This could be caused by the partial silencing known to affect any gene expressed from agroinfiltrated T‐DNAs in the absence of a functional suppressor of silencing (Canto and Palukaitis, 2002; Johansen and Carrington, 2001). However, co‐expression of strong heterologous suppressors of silencing, such as 2b or P19, failed to increase the steady‐state levels of HCPro, even though the levels of their transcript mRNAs increased substantially (Fig. 2). This excludes the possibility that HCPro expressed in the absence of P1 upstream exhibits a conformational alteration that negatively affects its suppressor activity, but not its stability. Therefore, as proposed by Rajamäki et al. (2005), a lack of stability that leads to small amounts of protein and therefore little or no suppressor activity would be an option. To investigate this possibility, we purified HCPro expressed from either the 6x‐P1‐HCPro or 6x‐HCPro sequence in PVX vectors. Although the expression from chimeric PVX vectors increased, in both cases, the severity of virus infection symptoms in N. benthamiana plants, this did not translate into increased virus accumulation (Fig. S1), in agreement with observations in previous studies on the synergistic effects of HCPro (González‐Jara et al., 2005). This happened despite the fact that the levels of HCPro in infected plants in the former case were five to ten times higher than in the latter (Fig. 3A). Therefore, a greater accumulation of HCPro, when expressed from a P1‐6x‐HCPro sequence, did not have a stronger synergistic effect on the PVX chimera than when expressed from a 6x‐HCPro sequence. In addition, the presence of P1 did not apparently have any effect on PVX. The differences in HCPro accumulation are, however, similar to those found by Pruss et al. (1997) in PVX expressing either TEV P1‐HCPro or HCPro alone.
Despite the differences in starting protein levels, we successfully purified both HCPros, which displayed similar behaviour during a process that involved differential fractionation, precipitation, concentration and nitrilotriacetic acid resin‐binding steps (Fig. 3B). Size fractionation of the purified samples showed similar oligomerization patterns (Fig. 3C). Therefore, we found no differences in protein stability in vitro. The serological detection of 6x‐HCPro expressed from a PVX chimera (albeit at low levels) vs. its nondetection when expressed transiently from an agroinfiltrated T‐DNA could be caused by the transient nature of the latter, with expression kinetics different from those of a virus vector, with protein steady‐state accumulation falling below the antibody detection threshold.
On the other hand, when expressed in the context of either its own virus, or as part of a polyprotein that undergoes cleavage in heterologous systems, the first amino acid of HCPro is a serine in the vast majority of the members of the genus Potyvirus (http://www.dpvweb.net/potycleavage/species.html). For this reason, when HCPro is expressed as a single gene, at least a codon for methionine must be added 5' to the HCPro sequence to enable translation, and the Kozak context sequence would be different. The context of expression of HCPro is thus not the same when expressed as a single gene or as part of a polyprotein (Table 1). In plants, the consensus Kozak sequence differs from that of animals, both qualitatively and quantitatively, with nucleotides +4 and +5 (relative to the AUG +1 to +3 initiation codon) modulating initiation codon selection in plants (Lützke et al., 1987). In plant mRNAs, the most frequent nucleotides at these two consecutive positions correspond to G and C (85% and 77% frequency, respectively), which result in an alanine after the initial methionine (Lützke et al., 1987). This is the case for the PVY polyprotein and its P1 protein. Our PVY constructs P1‐HCPro and P1‐6x‐HCPro thus contain a GC after the AUG initiation codon (Table 1), whereas, in constructs HCPro and 6x‐HCPro, UC (from the first serine) and CA (from the first histidine) after their respective initiation codons (Table 1) are much less common in plants, and perhaps would not favour the efficient binding of the ribosome to initiate translation. In support of this hypothesis, we found that, even if mRNA levels were comparable, HCPro expressed from construct 6x‐HCPro barely accumulated in comparison with protein expressed from a 6x‐P1‐HCPro sequence (Fig. 2). We tested the possibility that translation was being negatively affected by a poor Kozak context by adding a GCA after the AUG to improve the Kozak context [construct 6x‐HCPro (Ala); Table 1]. We found that, in patch assays, HCPro expressed from construct 6x‐HCPro (Ala) showed both increased protein accumulation and suppressor activity on a GFP reporter, although the protein levels were lower than those of HCPro expressed from construct P1‐6x‐HCPro (Fig. 4). Quantitative analysis of the steady‐state levels of suppressor expressed from constructs P1‐6x‐HCPro, 6x‐HCPro and 6x‐HCPro (Ala and of the GFP reporter) (Fig. 4) showed a clear correlation between this suppressor accumulation and activity. However, the possibility that differences in the accumulation levels of the different HCPro proteins could be caused partly by differences in protein stability derived from the different N‐termini, rather than only by differences in translation efficiency from the mRNAs, cannot be ruled out completely.
Together, our data rule out the possibility that a faulty post‐translational maturation of protein expressed in the absence of upstream P1 abolishes HCPro activity. In addition, as we found no differences in protein stability in vitro, P1 does not appear to be required during translation for HCPro to acquire stability. By contrast, our experiments showed that the Kozak context is relevant to HCPro expression and the steady‐state levels acquired by HCPro in patch assays. We also demonstrated that HCPro expressed in the absence of P1 does not display less suppressor activity than HCPro expressed in its presence, when the relative accumulation levels are taken into account (Fig. 4). However, because the levels of suppressor were lower when expressed alone than when expressed together with P1 as a polyprotein (c. 30%), even after improving the Kozak context, and the same applied to the activity on a GFP reporter in patch assays (c. 50%), our data do not exclude a potential role of cis‐P1 in further increasing the accumulation of HCPro by interfering with its degradation by an, as yet unidentified, host process, or by preventing a proportion of HCPro molecules from adopting a structure during translation that facilitates their degradation, such that they are not available to perform their functions.
Experimental Procedures
Plasmid constructs
For transient expression in plants, proteins were cloned into pROK2‐based binary vectors. Cloning of the P1HCPro sequence from PVY into pROK2 has been described previously (Canto and Palukaitis, 2002). To generate constructs P1 and HCPro, P1 and HCPro sequences were each amplified by PCR with appropriate oligonucleotides, and cloned into pROK2 after digestion of fragments and vector with BamHI and SacI, respectively. The nucleotide sequences immediately 5' and 3' of the translation start codon ATG are shown in Table 1. To add an N‐terminal tag of methionine plus six histidines (Met‐6 × His; ATG‐CAT‐CAC‐CAT‐CAC‐CAT‐CAC) to HCPro in the HCPro construct, the sequences corresponding to the tag, plus the first 5' 330 nucleotides of HCPro, were amplified by PCR with appropriate 5' oligonucleotides that encoded the Met‐6 × His sequence and a 3' oligonucleotide (nucleotides 319–339). The PCR fragment was digested with NheI and XhoI (a unique site present at nucleotide 330 of the HCPro sequence) and inserted into the above‐mentioned construct, linearized with XbaI‐XhoI in place of the original sequence, thus generating construct 6x‐HCPro. To insert the Met‐6 × His sequence between the P1 and HCPro sequences, the P1 sequence was amplified by PCR with an appropriate 5' oligonucleotide and a 3' oligonucleotide complementary to the end of the P1 sequence plus a serine codon and an NheI site [CAGTTT(P1)AGC(serine)GCTAGC(NheI site; proline‐serine)]. In addition, a sequence from construct 6x‐HCPro was amplified by PCR using a 5' oligonucleotide encoding the Met‐6 × His tag sequence with an added NheI site upstream, and an appropriate 3' oligonucleotide. Both PCR fragments were then cleaned, digested with NheI and ligated in vitro. The ligation product was used to amplify a PCR fusion fragment using the P1 5' oligonucleotide and the 3' oligonucleotide at the HCPro encoding sequence (nucleotides 319–339). The fusion fragment thus obtained contained a P1‐(Ser‐Pro‐Ser‐Met‐6 × His‐Met)‐HCPro sequence at the fusion sites. The fusion PCR product was digested with BamHI and XhoI, and cloned into equally linearized construct P1‐HCPro, generating construct P1‐6x‐HCPro. For large‐scale purification of HCPro, the 6x‐HCPro and P1‐6x‐HCPro sequences were amplified by PCR with appropriate oligonucleotides and cloned into a binary vector pgR 107 expressing infectious PVX (Lu et al., 2003) linearized with ClaI and SmaI. To obtain HCPro with an N‐terminal methionine‐alanine‐six histidines tag (Met‐Ala‐6 × His; ATG‐GCA‐CAT‐CAC‐CAT‐CAC‐CAT‐CAC), a fragment was amplified from construct 6x‐HCPro by PCR with a 5' oligonucleotide that encoded this Met‐Ala‐6 × His tag sequence and an appropriate 3' oligonucleotide in the HCPro sequence (nucleotides 319–339). The PCR fragment was digested with NheI‐XhoI and inserted into XbaI‐XhoI‐linearized construct 6x‐HCPro, to generate construct 6x‐HCPro (Ala). Binary vectors expressing TBSV P19 and CMV 2b have been described previously (Canto et al., 2006; González et al., 2010, respectively).
Transient expression of genes in plants
For transient expression assays (patch assays), binary vectors were transferred to electrocompetent Agrobacterium tumefaciens C58C1 derived from a single colony, to prevent bacterial variability. Cultures were grown to exponential phase in Luria–Bertani medium with antibiotics at 28 °C. For infiltration, each bacterial culture was diluted to a final optical density of 0.2 at 600 nm. Different cultures harbouring different T‐DNAs were then combined, and infiltration of the mixtures was performed as described by Canto and Palukaitis (2002).
Local suppression of silencing in agroinfiltration patch assays
A free GFP reporter gene expressed from a binary vector under the control of the 35S promoter was expressed transiently in an N. benthamiana leaf, either co‐infiltrated with the empty binary vector pROK2, or with another vector expressing a protein to be tested for suppression of silencing activity. To determine the levels of fluorescence derived from the transiently expressed free eGFP, leaves were illuminated at 4–6 days post‐infiltration with a Blak Ray® long‐wave UV lamp (UVP, Upland, CA, USA), as described by González et al. (2010).
Immunoblot detection of proteins and analysis
Total protein from infiltrated tissue discs was extracted with a pestle and mortar in extraction buffer [0.1 m Tris‐HCl, pH 8, 10 mm ethylenediaminetetraacetic acid (EDTA), 0.1 m LiCl, 1% β‐mercaptoethanol and 1% sodium dodecylsulphate (SDS)] (6 μL/mg of fresh tissue), and the samples were boiled and fractionated by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) in 10% (for HCPro detection) or 15% (for GFP detection) gels. Each lane was loaded with 7.5 μL of extract sample. Gels were wet blotted in tris‐glycine buffer onto Hybond‐P poly(vinylidene difluoride) (PVDF) membranes (GE Healthcare, Amersham, Buckinghamshire, UK). For immunological detection of GFP, a rabbit GFP polyclonal antiserum by G. Cowan (James Hutton Institute, Dundee, UK) was used. For detection of 6x‐HCPro, a mouse monoclonal antiserum to six histidines was used (Sigma‐Aldrich, St Louis, MO, USA). A rabbit polyclonal antiserum against PVY HCPro was used in the Western blot shown in Fig. 1A (Canto et al., 1995b). For the detection of PVX by Western blot, a commercial rabbit antibody was used (No. 070375/500; Loewe Biochemica GmbH, Sauerlach, Germany). Blotted proteins were detected using commercial secondary antibodies and SigmaFast™ 5‐bromo‐4‐chloro‐3‐indolyl‐phosphate/nitroblue tetrazolium (BCIP/NBT) substrate tablets (Sigma‐Aldrich). Comparative protein densitometric analysis of blotted proteins was made with the Quantity One 4.6.3 1‐D analysis software (Bio‐Rad Laboratories, Hercules, CA, USA).
RNA isolation from plants and Northern blot and qRT‐PCR analyses
For the Northern blot and qRT‐PCR analysis shown in Fig. 2, total RNA was extracted from 50 mg of leaves using a Plant RNA Mini Kit (Omega, Norcross, GA, USA) and resuspended in 50 μL of water. For Northern blot analysis, 7.5‐μL samples were fractionated in 1% denaturing agarose gels, and transferred to nylon membranes (Hybond‐N+; GE Healthcare), as described by Canto and Palukaitis (2002). HCPro mRNAs were detected using a digoxigenin‐labelled RNA probe to the whole PVY HCPro sequence, partially hydrolysed with 5 vol of 0.1 m NaOH for 5 min at 37 °C, and following the manufacturer's instructions for the substrate (Roche Diagnostics GmbH, Mannheim, Germany). One‐step qRT‐PCR was performed in a Rotor‐GeneQ real‐time PCR detection system (Qiagen GmbH, Hilden, Germany) using total RNA preparations treated with TURBO DNA‐free kit (Ambion, Life Technologies Corporation, Carlsbad, CA, USA). The assay was performed using 15 μL of a reaction mix containing 7.5 μL of Brilliant III Ultra‐Fast qRT‐PCR Master Mix (Agilent, Santa Clara, CA, USA), 1.8 μL of RNAase‐free water, 0.75 μL of reverse transcriptase, 0.15 μL of 100 mm dithiothreitol, 0.3 μm of each primer and 15 ng of total RNA. All reactions were performed in triplicate and their averaged values were obtained. qRT‐PCR was carried out at 50 °C for 10 min, 95 °C for 3 min and for 40 cycles of 95 °C for 10 s and 60 °C for 20 s. PCRs were performed, recorded and analysed using the Rotor‐Gene Q series Software. Synthesis of cDNA products of approximately 150 base pairs (bp) in length was verified by 2% agarose gel electrophoresis and direct sequencing, and by melting curve analysis containing a single melt curve peak using the Rotor‐Gene Q series software. 18S rRNAs were used for normalization because of their similar expression levels across all different agroinfiltrations, and their PCR amplification efficiencies. The following primers were used: forward primer Q‐PCR‐HcF2 (5'‐CCAGGAGTCAGCAGAAAATG) and reverse primer Q‐PCR‐HcR2 (5'‐GGTGCTTTTTAGTTGGTGGATAG) for the amplification of PVY HcPro, and forward primer 18S‐F (5'‐GCCCGTTGCTGCGATGATTC) and reverse primer 18S‐R (5'‐GCTGCCTTCCTTGGATGTGG) for the amplification of 18S rRNA.
For the Northern blot analysis of positive‐sense PVX genomic and subgenomic RNAs shown in Fig. S1, total nucleic acids from infected plants were extracted from 2 g of individual plants using 6 mL of extraction buffer (0.1 m Tris‐HCl, pH 8, 10 mm EDTA, 0.1 m LiCl, 1% β‐mercaptoethanol and 1% SDS), followed by two phenol–chloroform extractions, ethanol precipitation and resuspension in 2 mL of water. Samples of 7.5 μL were fractionated in 1% denaturing agarose gels, wet blotted to nylon membranes and detected with a digoxigenin‐labelled probe to the viral CP (González‐Jara et al., 2004), following the manufacturer's instructions for the substrate (Roche Diagnostics GmbH).
Purification of HCPro from plants and in vitro analysis
A protocol was set up based on modifications to three previous methods (Blanc et al., 1999; Ruíz‐Ferrer et al., 2005; Sasaya et al., 2000). Briefly, leaves infected with PVX expressing 6x‐HCPro (∼100 g), showing fully developed symptoms 10 days after inoculation, were homogenized in 350 mL of chilled 100 mm Tris‐HCl (pH 8.0), 20 mm Mg2SO4, 500 mm NaCl, 0.5 mm ethyleneglycol‐bis(2‐aminoethylether)‐N,N,N',N′‐tetraacetic acid (EGTA) buffer supplemented with 0.2% Na2SO3, 0.1% polyvinylpyrrolidone and 5 mm β‐mercaptoethanol. After filtration, centrifugation and ultracentrifugation, precipitation with 40% (NH4)2SO4 was carried out, discarding the supernatant and recovering the protein by resuspension of the pellet. The 6x‐HCPro molecule was retained in nitrilotriacetic acid resin (Qiagen GmbH), packed on a polypropylene column and eluted with 100 mm Tris‐HCl (pH 8.0), 20 mm Mg2SO4, 500 mm NaCl and 400 mm EGTA. Purified 6x‐HCPro was concentrated finally using Centrifugal filter units (Millipore, Billerica, MA, USA) and stored at −80 °C until use.
For HPLC fractionation analysis, purified protein samples were subjected to overnight dialysis against column buffer (100 mm Tris‐HCl, pH 7.5, 20 mm Mg2SO4, 50 mm NaCl, 1 mm dithiothreitol) and concentrated to 10 μg/mL using an Amicon Ultra‐4 centrifugal device minicolumn (Millipore). Samples eluted by HPLC were collected in 100‐μL aliquots, combined with 0.5 μg bovine serum albumin (BSA) as a carrier and precipitated with four volumes of acetone, before SDS‐PAGE plus Western blot analysis (one aliquot per Western blot well). Proteins of known size (monoclonal antibody IgG1 and fragment Fab) were also fractionated by HPLC to estimate the size of the eluted HCPro.
Supporting information
Fig. S1 Assessment of the effect of the expression of 6x‐HCPro or P1‐6x‐HCPro from a Potato virus X (PVX) vector on virus infection symptoms and accumulation. (A) At 7 days post‐inoculation (dpi). Infection with the PVX vector induced mosaic, vein clearing and curling in systemically infected leaves when compared with healthy plants (bottom vs. top left plants). Expression of either 6x‐HCPro or P1‐6x‐HCPro from the PVX vector resulted in similar symptoms in both cases, stronger than those induced by the PVX vector, with more severe curling in the systemically infected leaves (bottom and top right plants vs. bottom left plant). (B) Despite these differences in the severity of infection symptoms, the steady‐state levels of virus were similar in the three cases [top Western blot panel against viral coat protein (CP); numbers (1× and 0.2×) indicate the relative dilution of the sample extract]. Differential HCPro accumulation was also confirmed (bottom Western blot panel against the 6 × histidine tag). The panels below the Western blots show the Ponceau S‐stained membranes after blotting, as controls of loading. (C) The accumulation of viral genomic and CP subgenomic RNAs for the three viruses was also visualized by Northern blot with a probe against the viral CP (top panel), which recognizes all viral RNAs, and a probe against HCPro (bottom panel), which recognizes the full‐length genomic and HCPro‐containing subgenomics: triple‐gene‐block (TGB) subgenomic, and the HCPro subgenomic proper, but not the CP. TGB and HCPro subgenomics failed to resolve as sharp bands. The panels below the Northern blots show the total nucleic acid samples stained with ethidium bromide in separate gels, as controls of equal loading.
Acknowledgements
This work was supported by grant AGL2008‐03482 from the Spanish Ministry of Innovation and Science and by a joint grant between the Spanish Ministry of Economy and Competitivity (AC1/2009‐0855) to T. Canto and the Department of Science and Technology of the Government of India (DST/INT/SPAIN/P‐9/2009) to S. Praveen. N. Sahana was funded by a grant from the Indian Agricultural Research Institute, New Delhi. The authors thank Professor Peter Palukaitis for critical reading of the manuscript.
References
- Ala‐Poikela, M. , Goytia, E. , Halkonen, T. , Rajamaki, M.‐L. and Valkonen, J.P.T. (2011) Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors sIF(iso)4E and eIF4E and contains a 4E binding motif. J. Virol. 85, 6784–6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandalakshmi, R. , Pruss, G.J. , Ge, X. , Marathe, R. , Mallory, A.C. , Smith, T.H. and Vance, V.B. (1998) A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA, 95, 13 079–13 084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandalakshmi, R. , Marathe, R. , Ge, X. , Herr, J.J.M. , Mau, C. , Mallory, A. , Pruss, G. , Bowman, L. and Vance, V.B. (2000) A calmodulin‐related protein that suppresses porstranscriptional gene silencing in plants. Science, 290, 142–144. [DOI] [PubMed] [Google Scholar]
- Ballut, L. , Drucker, M. , Pugniére, M. , Cambon, F. , Blanc, S. , Roquet, F. , Candresse, T. , Schmid, H.‐P. , Nicolas, P. , Le Gall, O. and Badaoui, S. (2005) HcPro, a multifunctional protein encoded by a plant RNA virus, targets the 20S proteasome and affects its enzymatic activities. J. Gen. Virol. 88, 2595–2603. [DOI] [PubMed] [Google Scholar]
- Blanc, S. , López‐Moya, J.‐J. , Wang, R. , García‐Lampasona, S. , Thornbury, D.‐W. and Pirone, T.P. (1997) A specific interaction between coat protein and helper component correlates with aphid transmission of a potyvirus. Virology, 231, 141–147. [DOI] [PubMed] [Google Scholar]
- Blanc, S. , Dolja, V.V. , Llave, C. and Pirone, T.P. (1999) Histidine‐tagging and purification of Tobacco etch potyvirus helper component protein. J. Virol. Methods, 77, 11–15. [DOI] [PubMed] [Google Scholar]
- Brigneti, G. , Voinnet, O. , Li, W.X. , Ding, S.W. and Baulcombe, D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana . EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Canto, T. and Palukaitis, P. (2002) Generation of siRNAs by T‐DNA sequences does not require active transcription or homology to sequences in the plant. Mol. Plant–Microbe Interact. 15, 1137–1146. [DOI] [PubMed] [Google Scholar]
- Canto, T. , López‐Moya, J.J. , Serra‐Yoldi, M.T. , Díaz‐Ruíz, J.R. and López‐Abella, D. (1995a) Different helper component mutations associated with lack of aphid transmissibility in two isolates of Potato virus Y . Phytopathology, 85, 1519–1524. [Google Scholar]
- Canto, T. , Ellis, P. , Bowler, G. and López‐Abella, D. (1995b) Production of monoclonal antibodies to Potato virus Y helper component‐protease and their use for strain differentiation. Plant Dis. 79, 234–237. [Google Scholar]
- Canto, T. , Uhrig, J. , Swanson, M. , Wright, K. and MacFarlane, S. (2006) Translocation of Tomato bushy stunt virus P19 protein into the nucleus by ALY proteins compromises its suppressor of silencing activity. J. Virol. 80, 9064–9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C. and Herndon, K.L. (1992) Characterization of the potyviral HC‐Pro autoproteolytic cleavage site. Virology, 187, 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C. , Cary, S.M. and Dougherty, W.G. (1988) Mutational analysis of Tobacco etch virus polyprotein processing: cis‐ and trans‐ proteolytic activities of polyproteins containing the 49‐kDa proteinase. J. Virol. 62, 2313–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, E.J. , Prokhnevsky, A.I. , Gopinath, K. , Dolja, V.V. and Carrington, J. (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y.‐Q. , Liu, Z.‐M. , Xu, J. , Zhou, T. , Wang, M. , Chen, Y.‐T. , Li, H.‐F. and Fan, Z.‐F. (2008) HC‐Pro protein of sugar cane mosaic virus interacts specifically with maize ferredoxin‐5 in vitro and in planta . J. Gen. Virol. 89, 2046–2054. [DOI] [PubMed] [Google Scholar]
- Chung, B.Y.‐W. , Miller, W.A. , Atkins, J.F. and Firth, A.E. (2008) An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA, 105, 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dielen, A.‐S. , Sassaki, F.T. , Walter, J. , Michon, T. , Menard, G. , Pagny, G. , Krause‐Sakate, R. , Maia, I. , Badaoui, S. , Le Gall, O. , Candresse, T. and German‐Retama, S. (2011) The 20S proteasome α5 subunit of Arabidopsis thaliana carries an RNase activity and interacts in planta with the Lettuce mosaic potyvirus HcPro protein. Mol. Plant Pathol. 12, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres, M.W. , Gregory, B.D. , Gao, Z. , Foreman, A.W. , Mlotshwa, S. , Ge, X. , Pruss, G.J. , Ecker, J.R. , Bowman, L.H. and Vance, V. (2010) Two plant viral suppressors of silencing require the ethylene‐inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathog. 6, e1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García, J.A. , Riechmann, J.L. and Laín, S. (1989) Proteolytic activity of the Plum pox potyvirus NIa‐like protein in Escherichia coli . Virology, 170, 362–369. [DOI] [PubMed] [Google Scholar]
- González, I. , Martínez, L. , Rakitina, D. , Lewsey, M.G. , Atienzo, F.A. , Llave, C. , Kalinina, N. , Carr, J.P. , Palukaitis, P. and Canto, T. (2010) Cucumber Mosaic Virus 2b protein subcellular targets and interactions: their significance to its RNA silencing suppressor activity. Mol. Plant–Microbe Interact. 23, 294–303. [DOI] [PubMed] [Google Scholar]
- González, I. , Rakitina, D. , Semashko, M. , Taliansky, M. , Praveen, S. , Palukaitis, P. , Carr, J.P. , Kalinina, N. and Canto, T. (2012) RNA binding is more relevant to the suppression of silencing function of Cucumber mosaic virus 2b protein than nuclear localization. RNA, 18, 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Jara, P. , Tenllado, F. , Martínez‐García, B. , Atencio, F.A. , Barajas, D. , Vargas, M. , Díaz‐Ruíz, J. and Díaz‐Ruíz, J.R. (2004) Host‐dependent differences during synergistic infection by potyviruses with potato virus X. Mol. Plant Pathol. 5, 29–35. [DOI] [PubMed] [Google Scholar]
- González‐Jara, P. , Atencio, F.A. , Martínez‐García, B. , Barajas, D. , Tenllado, F. and Día‐Ruíz, J.R. (2005) A single amino acid mutation in the Plum pox virus Helper component‐proteinase gene abolishes both synergistic and RNA silencing suppression activities. Virology, 95, 894–901. [DOI] [PubMed] [Google Scholar]
- Jamous, R.M. , Boonrod, K. , Fuellgrabe, M.W. , Ali‐Shtayeh, M.S. , Krczal, G. and Wassenegger, M. (2011) The helper component‐proteinase of the Zucchini yellow mosaic virus inhibits the Hua Enhancer 1 methyltransferase activity in vitro . J. Gen. Virol. 92, 2222–2226. [DOI] [PubMed] [Google Scholar]
- Jin, Y. , Ma, D. , Dong, J. , Jin, J. , Li, D. , Deng, C. and Wang, T. (2007a) HC‐Pro protein of Potato virus Y can interact with three Arabidopsis thaliana 20S proteasome subunits in planta. J. Virol. 81, 12 881–12 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y. , Ma, D. , Dong, J. , Li, D. , Deng, C. , Jin, J. and Wang, T. (2007b) The HC‐Pro protein of Potato virus Y interacts with NtMinD of tobacco. Mol. Plant–Microbe Interact. 20, 1505–1511. [DOI] [PubMed] [Google Scholar]
- Johansen, L.K. and Carrington, J.C. (2001) Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium‐mediated transient expression system. Plant Physiol. 126, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D. and Carrington, J.C. (1998) A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell, 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Lakatos, L. , Csorba, T. , Pantaleo, V. , Chapman, E.J. , Carrington, J.C. , Liu, Y. , P., Dolja, V.V. , Calvino, L.F. , López‐Moya, J.J. and Burgyán, J. (2006) Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25, 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Malcuit, I. , Moffett, P. , Ruíz, M.T. , Peart, J. , Wu, A.J. , Rathjen, J.P. , Bendahmane, A. , Day, L. and Baulcombe, D. (2003) High throughput virus‐induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lützke, H.A. , Chow, K.C. , Mickel, F.S. , Moss, K.A. , Kern, H.F. and Scheele, G.A. (1987) Selection of AUG initiation codons differs in plants and animals. EMBO J. 6, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia, I.G. and Bernardi, F. (1996) Nucleic acid‐binding properties of a bacterially expressed potato virus Y helper component proteinase. J. Gen. Virol. 77, 869–877. [DOI] [PubMed] [Google Scholar]
- Mangrauthia, S.K. , Singh Shakya, V.P. , Jain, R.K. and Praveen, S. (2009) Ambient temperature perception in papaya for Papaya ringspot virus interaction. Virus Genes, 38, 429–434. [DOI] [PubMed] [Google Scholar]
- Mérai, Z. , Kerényi, Z. , Kertész, S. , Magda, M. , Lakatos, L. and Silhavy, D. (2006) Double‐stranded RNA binding may be a general plant RNA viral strategy to suppress silencing. J. Virol. 80, 5747–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merits, A. , Guo, D. , Järvekülg, L. and Saarma, M. (1999) Biochemical and genetic evidence for interactions between potato A potyvirus‐encoded proteins P1 and P3 and proteins of the putative replication complex. Virology, 263, 15–22. [DOI] [PubMed] [Google Scholar]
- Nakahara, K.S. , Masuta, C. , Yamada, S. , Shimura, H. , Kashihara, Y. , Wada, T.S. , Meguro, A. , Goto, K. , Tadamura, K. , Sueda, K. , Sekiguchi, T. , Shao, J. , Itchoda, N. , Matsumara, T. , Igarashi, M. , Ito, K. , Carthew, R.W. and Uyeda, I. (2012) Tobacco calmodulin‐like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA, 109, 10 113–10 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plisson, C. , Drucker, M. , Blanc, S. , German‐Retama, S. , Le Gall, O. , Thomas, C. and Bron, P. (2003) Structural characterization of HC‐Pro, a plant virus multifunctional protein. J. Biol. Chem. 278, 23 753–23 761. [DOI] [PubMed] [Google Scholar]
- Pruss, G. , Ge, X. , Shi, X.M. , Carrington, J.C. and Vance, V.B. (1997) Plant viral synergism: the potyviral genome encodes a broad‐range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell, 9, 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamäki, M.‐L. , Kelloniemi, J. , Alminaite, A. , Kekarainen, T. , Rabenstein, F. and Valkonen, J.P.T. (2005) A novel insertion site inside the potyvirus P1 cistron allows expression of heterologous proteins and suggests some P1 functions. Virology, 342, 88–101. [DOI] [PubMed] [Google Scholar]
- Ruíz‐Ferrer, V. , Boskovic, J. , Alfonso, C. , Rivas, G. , Llorca, O. , López‐Abella, D. and López‐Moya, J.J. (2005) Structural analysis of Tobacco etch potyvirus HC‐Pro oligomers involved in aphid transmission. J. Virol. 79, 3758–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaya, T. , Torrance, L. , Cowan, G. and Ziegler, A. (2000) Aphid transmission studies using helper component proteins of Potato virus Y expressed from a vector derived from Potato virus X . J. Gen. Virol. 81, 1115–1119. [DOI] [PubMed] [Google Scholar]
- Shiboleth, Y.M. , Haronsky, E. , Leibman, D. , Arazi, T. , Wassenegger, M. , Whitham, S.A. , Gaba, V. and Gal‐On, A. (2007) The conserved FRNK box in HC‐Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J. Virol. 81, 13 135–13 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornbury, D.W. , Hellmann, G.M. , Rhoads, R.E. and Pirone, T.P. (1985) Purification and characterization of potyvirus helper component. Virology, 144, 260–267. [DOI] [PubMed] [Google Scholar]
- Urcuqui‐Inchima, S. , Walter, J. , Drugeon, G. , German‐Retama, S. , Haenni, A.‐L. , Candresse, T. , Bernardi, F. and Le Gall, O. (1999) Potyvirus helper component‐proteinase self‐interaction in the yeast two‐hybrid system and delineation of the interaction domain involved. Virology, 258, 95–99. [DOI] [PubMed] [Google Scholar]
- Urcuqui‐Inchima, S. , Maia, I.G. , Arruda, P. , Haenni, A.‐L. and Bernardi, F. (2000) Deletion mapping of the potyviral helper component‐proteinase reveals two regions involved in RNA binding. Virology, 268, 104–111. [DOI] [PubMed] [Google Scholar]
- Valli, A. , Martín‐Hernández, A.M. , López‐Moya, J.J. and García, J.A. (2006) RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the Family Potyviridae that lacks the cysteine protease HCPro. J. Virol. 80, 10055–10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargasson, J.M. , Szittya, G. , Burgyan, J. and Hall, T.M. (2003) Size selective recognition of siRNAs by an RNA silencing suppressor. Cell, 115, 799–811. [DOI] [PubMed] [Google Scholar]
- Verchot, J. and Carrington, J.C. (1995a) Debilitation of plant potyvirus infectivity by P1 proteinase‐inactivating mutations and restoration by second‐site modifications. J. Virol. 69, 1582–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot, J. and Carrington, J.C. (1995b) Evidence that the potyvirus P1 protein functions as an accessory factor for genome amplification. J. Virol. 69, 3668–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot, J. , Koonin, E.V. and Carrington, J.C. (1991) The 35‐kDa protein from the N‐terminus of the potyviral polyprotein functions as a third virus‐encoded proteinase. Virology, 185, 527–535. [DOI] [PubMed] [Google Scholar]
- Zheng, H. , Yan, F. , Lu, Y. , Sun, L. , Lin, L. , Cai, L. , Hou, M. and Chen, J. (2011) Mapping the self‐interacting domains of TuMV HC‐Pro and the subcellular localization of the protein. Virus Genes, 42, 110–116. [DOI] [PubMed] [Google Scholar]
- Zilian, E. and Maiss, E. (2011) Detection of plum pox potyviral protein–protein interactions in planta using an optimized mRFP‐based bimolecular fluorescence complementation system. J. Gen. Virol. 92, 2711–2723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Assessment of the effect of the expression of 6x‐HCPro or P1‐6x‐HCPro from a Potato virus X (PVX) vector on virus infection symptoms and accumulation. (A) At 7 days post‐inoculation (dpi). Infection with the PVX vector induced mosaic, vein clearing and curling in systemically infected leaves when compared with healthy plants (bottom vs. top left plants). Expression of either 6x‐HCPro or P1‐6x‐HCPro from the PVX vector resulted in similar symptoms in both cases, stronger than those induced by the PVX vector, with more severe curling in the systemically infected leaves (bottom and top right plants vs. bottom left plant). (B) Despite these differences in the severity of infection symptoms, the steady‐state levels of virus were similar in the three cases [top Western blot panel against viral coat protein (CP); numbers (1× and 0.2×) indicate the relative dilution of the sample extract]. Differential HCPro accumulation was also confirmed (bottom Western blot panel against the 6 × histidine tag). The panels below the Western blots show the Ponceau S‐stained membranes after blotting, as controls of loading. (C) The accumulation of viral genomic and CP subgenomic RNAs for the three viruses was also visualized by Northern blot with a probe against the viral CP (top panel), which recognizes all viral RNAs, and a probe against HCPro (bottom panel), which recognizes the full‐length genomic and HCPro‐containing subgenomics: triple‐gene‐block (TGB) subgenomic, and the HCPro subgenomic proper, but not the CP. TGB and HCPro subgenomics failed to resolve as sharp bands. The panels below the Northern blots show the total nucleic acid samples stained with ethidium bromide in separate gels, as controls of equal loading.


