Summary
Resistance to Soybean mosaic virus (SMV) in soybean is conferred by three dominant genes: Rsv1, Rsv3 and Rsv4. Over the years, scientists in the USA have utilized a set of standard pathotypes, SMV‐G1 to SMV‐G7, to study interaction with Rsv‐genotype soybeans. However, these pathotypes were isolated from a collection of imported soybean germplasm over 30 years ago. In this study, 35 SMV field isolates collected in recent years from 11 states were evaluated for gain of virulence on soybean genotypes containing individual Rsv genes. All isolates were avirulent on L78‐379 (Rsv1), whereas 19 were virulent on L29 (Rsv3). On PI88788 (Rsv4), 14 of 15 isolates tested were virulent; however, only one was capable of systemically infecting all of the inoculated V94‐5152 (Rsv4). Nevertheless, virulent variants from 11 other field isolates were rapidly selected on initial inoculation onto V94‐5152 (Rsv4). The P3 cistrons of the original isolates and their variants on Rsv4‐genotype soybeans were sequenced. Analysis showed that virulence on PI88788 (Rsv4) was not associated, in general, with selection of any new amino acid, whereas Q1033K and G1054R substitutions were consistently selected on V94‐5152 (Rsv4). The role of Q1033K and G1054R substitutions, individually or in combination, in virulence on V94‐5152 (Rsv4) was confirmed on reconstruction in the P3 cistron of avirulent SMV‐N, followed by biolistic inoculation. Collectively, our data demonstrate that SMV has evolved virulence towards Rsv3 and Rsv4, but not Rsv1, in the USA. Furthermore, they confirm that SMV virulence determinants on V94‐5152 (Rsv4) reside on P3.
Introduction
Three dominant resistance (R) genes, Rsv1, Rsv3 and Rsv4, confer resistance in soybean [Glycine max (L.)] to Soybean mosaic virus (SMV) (Saghai Maroof et al., 2008a). However, it is unknown whether these genes are present in the elite soybean cultivars grown in the USA. These R genes were discovered when a collection of imported soybean germplasm was screened against a set of standard SMV pathotypes in the USA (Buzzell and Tu, 1989; Cho and Goodman, 1979; Kiihl and Hartwig, 1979; Ma et al., 1995, 2002). These pathotypes, SMV‐G1–SMV‐G7, represented 98 isolates obtained from seeds in the United States Department of Agriculture (USDA) soybean germplasm collection, originated from at least 17 countries, and had been grown in glasshouse or field plots more than three decades ago. The identification of these pathotypes was based solely on phenotypic reactions on Rsv1 allele‐containing soybean differential genotypes (Buzzell and Tu, 1984; Cho and Goodman, 1979). As the soybean accessions from which these pathotypes were isolated were all of foreign origin, it is probable that SMV‐G1–SMV‐G7 did not evolve in the USA, but in the countries of origin (Cho and Goodman, 1979; Jain et al., 1992). As such, they may not be true representatives of current SMV isolates in soybean fields in North America. It should be noted that SMV is seed and aphid borne in soybean in nature (Domier et al., 2007) and, similar to any other RNA virus, it is versatile (Chowda‐Reddy et al., 2011; Hajimorad et al., 2003, 2008, 2011; Seo et al., 2011). Hence, as a result of selection pressures imposed by different soybean cultivars or means of transmission via seeds or aphids under field conditions, SMV variants are expected to emerge and or change in prevalence over time.
The resistance phenotype expressed by soybean genotypes containing Rsv1, Rsv3 or Rsv4 differs on mechanical inoculation with SMV pathotypes G1–G7. Rsv1, identified originally in PI96983 (Kiihl and Hartwig, 1979), confers symptomless resistance (extreme resistance, ER) against SMV‐G1–SMV‐G6, whereas Rsv3 conditions ER against SMV‐G5–SMV‐G7 (Buzzell and Tu, 1989; Chen et al., 1994; Cho and Goodman, 1979; Gunduz et al., 2002). SMV‐G7 causes stem‐tip necrosis in Rsv1‐genotype soybeans, whereas SMV‐G1–SMV‐G4 provoke systemic necrosis in Rsv3‐genotype soybeans (Buzzell and Tu, 1989; Cho and Goodman, 1979; Gunduz et al., 2000, 2002; Hajimorad et al., 2003). Interestingly, Rsv4 in V94‐5125 initially exhibited broad resistance to all SMV pathotypes G1–G7 (Buss et al., 1997; Ma et al., 1995). This finding was promising as it gave the impression that durable and broad resistance against SMV in soybean could be developed by utilizing Rsv4. However, in a follow‐up study by Ma et al. (2002), a susceptibility response to SMV‐G1 was observed in a soybean genotype harbouring an Rsv4 allele at 21 days post‐inoculation (dpi). This response was designated as late susceptibility (LS) because of disease symptom appearance at 21 dpi rather than 7 dpi, which is commonly observed in most SMV‐susceptible cultivars. Gunduz et al. (2004) also noticed LS in a different soybean genotype containing an Rsv4 allele from PI88788 in the context of heterozygosity when mechanically inoculated with SMV‐G1.
The underlying mechanisms of resistance conferred by Rsv genes against SMV also differ. Resistance mediated by Rsv1 and Rsv3 involves active defence pathways, whereas that of Rsv4 appears to be passive and interferes with cell‐to‐cell and systemic movements (Gunduz et al., 2004; Hajimorad and Hill, 2001; Zhang et al., 2009). A strong candidate gene for Rsv1 belonging to the coiled‐coil, nucleotide‐binding, leucine‐rich repeat class of disease R genes has been identified, and it appears that Rsv3 also belongs to the same class of R genes (Hayes et al., 2004; Suh et al., 2011). However, Rsv4 is not genetically similar to Rsv1 or Rsv3 and appears to belong to a novel class of disease R genes (Saghai Maroof et al., 2010).
SMV, a single‐stranded, positive‐sense RNA virus, is a species within the genus Potyvirus belonging to the family Potyviridae. Its genome, approximately 10 kb, contains a long open reading frame (ORF) and a small overlapping ORF, known as ‘pipo’ (Adams et al., 2005; Chung et al., 2008; Wen and Hajimorad, 2010). On expression, the resultant single large polypeptide is processed post‐translationally by three viral‐encoded proteinases to yield a number of multifunctional proteins, including helper‐component proteinase (HC‐Pro), P3 and cytoplasmic inclusion (CI) proteins (Urcuqui‐Inchima et al., 2001). The small ORF embedded in the P3 cistron (i.e. pipo) has the potential to encode PIPO in the +2 frame in relation to the polyprotein ORF, which participates in virus movement (Vijayapalani et al., 2012; Wei et al., 2010; Wen and Hajimorad, 2010).
The resistance‐breaking determinants of SMV on Rsv‐genotype soybeans have been characterized to various extents. The virulence determinants on Rsv1‐genotype soybeans have been mapped to HC‐Pro and P3 by three different experimental approaches, and it has been shown that concurrent non‐synonymous mutations in both cistrons are essential, and sufficient, for conversion of avirulence to virulence (Eggenberger et al., 2008; Hajimorad et al., 2008, 2011; Wen et al., 2011). The determinants for virulence on Rsv3‐genotype soybeans have been mapped to the CI protein by two independent research groups, and it has been shown that a single amino acid substitution, at a minimum, converts avirulence to virulence (Seo et al., 2009a; Zhang et al., 2009). However, the virulence determinants on V94‐5152 (Rsv4) have been mapped to P3 via comparative genomics of naturally occurring virulent and avirulent SMVs, in which amino acids at the polyprotein positions 1033 (lysine) and 1054 (arginine) were found to be crucial for virulence (Chowda‐Reddy et al., 2011).
Our objectives in this study were as follows: (i) to determine whether North American isolates of SMV have evolved virulence towards soybean genotypes containing Rsv1, Rsv3 or Rsv4; and (ii) to determine whether virulence on Rsv4‐genotype soybeans is reliant on specific amino acids at polypeptide positions 1033 and 1054. In this article, we consider any SMV collected from an open soybean field in the USA as an American isolate irrespective of whether it was obtained from elite soybean cultivars grown in farmers’ fields or from breeding lines, including germplasm, grown in agricultural experimental stations.
Results
Revisiting virulence of the standard SMV pathotypes on Rsv1‐, Rsv3‐ and Rsv4‐genotype soybeans
As expected, SMV pathotypes G1–G7 were all capable of systemic infection in Williams (rsv) and Essex (rsv) (Table 1). However, only SMV‐G7 and its derivative variant, SMV‐G7d, were virulent on L78‐379 (Rsv1) (Buzzell and Tu, 1984; Cho and Goodman, 1979; Eggenberger et al., 2008; Hajimorad et al., 2003). In agreement with our previous report (Hajimorad et al., 2003), and contrary to the reports by Buzzell and Tu (1984) and Chen et al. (1994), both subcultures of SMV‐G7a were incapable of infecting L78‐379 (Rsv1) (Table 1). On L29 (Rsv3), SMV pathotypes G1, G2 (N), G4 and G7a were all virulent (Table 1). However, SMV‐G3 was avirulent on L29 (Rsv3), irrespective of whether wild‐type or progeny viruses derived from the molecularly cloned genome served as inoculum sources (Table 1). On V94‐5152 (Rsv4), SMV‐G1, SMV‐G4 and SMV‐G7a were virulent on initial inoculation and caused systemic infection in at least one plant as early as 14 dpi. When sap extract from SMV‐G7a‐infected V94‐5152 (Rsv4) was inoculated on additional plants, the majority became infected (Table 1). On PI88788 (Rsv4), SMV‐G1–SMV‐G4 and SMV‐G7a were all virulent on initial inoculation and some induced systemic mosaic in the majority of plants as early as 10 dpi (Table 1). However, SMV‐G5–SMV‐G7 and SMV‐G7d were all avirulent (Table 1). The virulence of SMV‐G1 on PI88788 (Rsv4) is in agreement with a previous report (Gunduz et al., 2004). Interestingly, when sap extract from the infected PI88788 (Rsv4) was inoculated onto V94‐5152 (Rsv4), SMV‐G1 and SMV‐G4, but not SMV‐G2 (N), were virulent as well (Table 1). These observations suggest that a number of standard SMV pathotypes are capable of infecting Rsv4‐genotype soybeans; however, the strength of resistance to SMV in V94‐5152 (Rsv4) differs from that of PI88788 (Rsv4).
Table 1.
Responses of soybean genotypes to mechanical inoculation with standard Soybean mosaic virus (SMV) pathotypes
| SMV pathotypea | Soybean genotype | |||||
|---|---|---|---|---|---|---|
| Williams (rsv) | L78‐379 (Rsv1) | L29 (Rsv3) | Essex (rsv) | V94‐5152 | PI88788 | |
| (Rsv4) | ||||||
| G1 | 21b, c/22 | 0/11 | 7/9 | 9/9 | 1b/28 [2b/8]d | 19b/21 |
| G2(N) | 17/17 | 0/10 | 9/10 | 9/9 | 0/16 [0/78] | 13b/20 (26b/30)e |
| G3f | 25b/28 | 0/15 | 0/31 | 13/14 | 0/41 [1b/15] | 6b/19 |
| G4 | 13b/14 | 0/11 | 10/10 | 14/14 | 4b/29 [5b/7] | 13b/13 |
| G5 | 26/26 | 0/12 | 0/8 | 9/10 | 0/31 | 0/12 |
| G6 | 17/17 | 0/9 | 0/10 | 11/11 | 0/32 | 0/14 |
| G7 | 18/18 | 9/16 | 0/9 | 7/9 | 0/26 | 0/27 |
| G7ag | 19b/19 | 0/12 | 8/8 | 7/7 | 14/38b (39b/41) | 23b/47 |
| G7d | 19/19 | 11/11 | 0/10 | 10/10 | 0/25 | 0/27 |
Progeny viruses derived from biolistically inoculated Williams82 (rsv) with molecularly cloned genomes of SMV‐N (pSMV‐N), SMV‐G7 (pSMV‐G7) and SMV‐G7d (pSMV‐G7d) serving as the initial inoculum. SMV‐N served as a representative isolate of the G2 pathotype group (Jain et al., 1992). For SMV‐G3, progeny viruses derived from the molecularly cloned genome (pSMV‐G3) also served as the initial inoculum. For all the other SMV pathotypes, wild‐type viruses in Williams (rsv) served as the initial inoculum.
The P3 cistron was reverse transcriptase‐polymerase chain reaction (RT‐PCR) amplified from systemically infected leaves of one of the infected plants and sequenced.
Total number of plants systemically infected/total plants inoculated on primary leaves. The inoculated plants were evaluated at 21 days post‐inoculation by antigen‐coated indirect enzyme‐linked immunosorbent assay (IELISA). Irrespective of disease symptom expression, all the plants were assayed individually for the presence of SMV in non‐inoculated leaves by IELISA.
Numbers shown in brackets indicate total number of systemically infected plants/total number of plants inoculated using sap extract from an infected PI88788 (Rsv4) plant.
Numbers shown in parentheses indicate total number of systemically infected plants/total number of plants inoculated using sap extract from an infected plant of the same genotype. Thus, these represent second passage of the virus in the same soybean genotype.
Data are combined results of inoculation using wild‐type SMV‐G3 or virus progenies derived from the infectious cDNA clone.
The data are combined results for SMV‐G7a (ATCC PV‐624) and SMV‐G7a (ATCC PV‐724) subcultures.
Virulence of American isolates of SMV on L78‐379 (Rsv1) and L29 (Rsv3)
The inoculation of 35 American isolates on L78‐379 (Rsv1) showed that none had evolved virulence (Table 2). By contrast, 19 of 35 isolates had evolved virulence towards L29 (Rsv3). The presence of SMV in each of the infected plants was confirmed by antigen‐coated indirect enzyme‐linked immunosorbent assay (IELISA) (Table 2). Virulence towards L29 (Rsv3) was present among isolates collected from all states, except those from Arkansas and Ohio (Table 2).
Table 2.
Responses of soybean genotypes to mechanical inoculation with American isolates of Soybean mosaic virus (SMV)
| Isolatea | Soybean genotype | |||||
|---|---|---|---|---|---|---|
| Williams (rsv) | L78‐379 (Rsv1) | L29 (Rsv3) | Essex (rsv) | V94‐5152 | PI88788 | |
| (Rsv4) | ||||||
| AL1 | 17b c/17 | 0/9 | 8/8 | 10/10 | 1b/15 (18b/19)d | 11b/11 |
| AL2 | 17/17 | 0/10 | 9/9 | 10/10 | 0/14 | NDe |
| AL3 | 17b/18 | 0/10 | 9/10 | 9/9 | 1b/15 (13b/13) | 13b/20 (12b/12) |
| AL4 | 19/20 | 0/9 | 9/9 | 9/9 | 0/15 | ND |
| AL5 | 18/18 | 0/9 | 7/7 | 9/9 | 0/14 | ND |
| AL6 | 18/19 | 0/11 | 8/8 | 8/9 | 0/17 | ND |
| ArE1 | 21/21 | 0/10 | 0/10 | 11/11 | 0/18 | ND |
| ArE2 | 19/19 | 0/11 | 0/9 | 10/10 | 0/14 | ND |
| ArE5 | 20b/20 | 0/12 | 0/9 | 11/11 | 1b/15 (14b/14) | 0/23 [11b/11]f |
| ArG2 | 21/21 | 0/10 | 0/9 | 11/11 | 0/14 | ND |
| IA | 18b/19 | 0/12 | 9/9 | 5/5 | 1b/16 (16b/17) | 15b/15 |
| IL3 | 19b/19 | 0/11 | 11/11 | 11/11 | 13b/13 (14b/14) | 11b/11 |
| IL4 | 16b/16 | 0/10 | 8/8 | 8/8 | 1b/29 [7b/9] | 7b/10 |
| IL5 | 19/19 | 0/10 | 3/7 | 8/8 | 0/12 [3b/9] | 14b/14 |
| KY | 20b/20 | 0/16 | 12/12 | 9/9 | 1b/22 [9b/10] | 10b/11 |
| MN | 17b/17 | 0/11 | 9/9 | 10/10 | 3b/11 (22b/23) | 10b/12 |
| MS4 | 40b/43 | 0/35 | 17/17 | 18/19 | 1b/46 (17b/19) | 16b/30 |
| MS9 | 12/18 | 0/10 | 0/10 | 7/7 | 0/14 | ND |
| MS10 | 22b/22 | 0/8 | 10/10 | 9/9 | 1b/14 (15b/15) | 12b/14 |
| MS11 | 24/25 | 0/9 | 0/19 | 11/11 | 0/16 | ND |
| MS12 | 24/24 | 0/10 | 0/16 | 9/10 | 0/16 | ND |
| MS13 | 18/18 | 0/9 | 9/10 | 7/7 | 0/15 | ND |
| OH | 20b/20 | 0/18 | 0/10 | 11/11 | 1b/11 (14b/15) | 8b/24 (24b/25) |
| TNP | 20/20 | 0/14 | 11/11 | 10/10 | 0/13 [11b/15] | 14b/15 |
| TN2 | 39b/39 | 0/21 | 17/17 | 18/18 | 6b/25 (29b/30) | 17b/23 |
| TN3 | 13/14 | 0/10 | 0/9 | 9/9 | 0/15 | ND |
| TN4 | 15/15 | 0/11 | 0/12 | 8/9 | 0/16 | ND |
| TN5 | 17/17 | 0/9 | 0/9 | 9/9 | 0/16 | ND |
| TN6 | 13/13 | 0/10 | 0/9 | 10/10 | 0/14 | ND |
| TN7 | 15/15 | 0/11 | 0/11 | 7/9 | 0/16 | ND |
| TN8 | 16/16 | 0/10 | 0/10 | 9/10 | 0/15 | ND |
| TN9 | 12/13 | 0/10 | 0/11 | 6/8 | 0/15 | ND |
| TN10 | 14/14 | 0/9 | 0/9 | 9/9 | 0/16 | ND |
| VA | 37/37 | 0/19 | 23/23 | 20/20 | 0/35 [7b/9] | 30b/31 |
| WI | 17/19 | 0/14 | 9/11 | 10/10 | 0/27 | ND |
Isolates originated from Alabama (AL1–AL6), Arkansas (ArE1, ArE2, ArE5, ArG2), Iowa (IA), Illinois (IL3–IL5), Kentucky (KY), Minnesota (MN), Mississippi (MS4, MS9–MS13), Ohio (OH), Tennessee (TNP, TN2–TN10), Virginia (VA) and Wisconsin (WI). All isolates were originally inoculated mechanically to Williams (rsv), systemically infected leaf tissues were harvested, frozen in the presence of liquid nitrogen and stored at −80 °C until served as the initial inoculum source.
The P3 cistron from one of the infected plants was reverse transcriptase‐polymerase chain reaction (RT‐PCR) amplified and directly sequenced.
Total number of plants systemically infected/total number inoculated on primary leaves with sap extract from infected Williams (rsv). The inoculated plants were evaluated at 21 days post‐inoculation by indirect enzyme‐linked immunosorbent assay (IELISA). Irrespective of disease symptoms, all the inoculated plants were assayed individually for the presence of SMV in non‐inoculated leaves.
Numbers shown in parentheses indicate total number of systemically infected plants/total number of plants inoculated with sap extract from an infected plant of the same soybean genotype. Thus, these represent second passage of the virus in the same soybean genotype.
Not done.
Numbers shown in brackets indicate total number of systemically infected plants/total number of inoculated plants when sap extract from PI88788 (Rsv4) was inoculated on V94‐5152 (Rsv4), and vice versa.
Virulence of American isolates on Rsv4‐genotype soybeans
SMV‐IL3 was the only American isolate that infected systemically all of the inoculated V94‐5152 (Rsv4) plants in the initial inoculation. The presence of SMV‐IL3 in the systemically infected leaves was confirmed immunologically as early as 10 dpi (Table 2; Fig. 1). In addition, 11 other American isolates were capable of systemically infecting V94‐5152 (Rsv4), but mostly with very low infection rates (Table 2). Isolates AL1, AL3, ArE5, IA, IL4, KY, MS4, MS10 and OH each infected systemically a single V94‐5152 (Rsv4) plant, whereas MN and TN2 infected three of 11 and six of 25 inoculated plants, respectively (Table 2). Regardless, when sap extract from the systemically infected leaves of one of the infected V94‐5152 (Rsv4) plants was inoculated again onto V94‐5152 (Rsv4), almost all became systemically infected (Table 2).
Figure 1.

Detection of Soybean mosaic virus (SMV) by immunoprinting. (A) Immunoprints of non‐inoculated trifoliate leaflets from V94‐5152 (Rsv4) inoculated on primary leaves with SMV‐IL3 or buffer (Mock) and harvested at different days post‐inoculation (dpi). (B) Phenotypes of the trifoliate leaflets prior to immunoprinting.
In contrast with the response of V94‐5152 (Rsv4), 14 of 15 American isolates inoculated onto PI88788 (Rsv4) were virulent and systemically infected the majority of the inoculated plants (Table 2). ArE5 was the only isolate that failed to infect PI88788 (Rsv4) when sap from Williams served as the inoculum (Table 2). However, when sap extract from an infected V94‐5152 (Rsv4) plant was used to inoculate PI88788 (Rsv4) plants, they all became systemically infected (Table 2). Conversely, when sap extracts from systemically infected leaves of PI88788 (Rsv4) were used to inoculate V94‐5152 (Rsv4), IL5, TNP and VA isolates, which exhibited avirulence phenotypes in the initial inoculation, gained virulence (Table 2).
Comparison of resistance phenotype to SMV in V94‐5152 (Rsv4) versus PI88788 (Rsv4)
The standard SMV pathotypes, as well as the American isolates, widely exhibited virulence on PI88788 (Rsv4), but not on V94‐5152 (Rsv4) (Tables 1 and 2). This observation led us to hypothesize that the phenotype of resistance mediated by Rsv4 differs in these two soybean genotypes. To test this hypothesis, SMV‐N‐GUS was biolistically inoculated onto attached primary leaves of V94‐5152 (Rsv4) and PI88788 (Rsv4). Both genotypes allowed virus replication in the inoculated leaves, evident by β‐glucuronidase (GUS) expression (Fig. 2). However, unlike GUS expression in the inoculated leaves of Essex (rsv), which extended to the veins, the infected foci in the inoculated leaves of PI88788 (Rsv4) and V94‐5152 (Rsv4) remained restricted (Fig. 2). Regardless, systemic GUS expression was detected in the non‐inoculated leaves of PI88788 (Rsv4), albeit in a few small foci of infected cells, but not in V94‐5152 (Rsv4) (Fig. 2). Under similar conditions, SMV‐N‐GUS caused extensive systemic infection in non‐inoculated leaves of Essex (rsv), evident by efficient GUS expression (Fig. 2). These observations indicate that Rsv4‐mediated resistance affects local and systemic movements of SMV, in agreement with the findings by Gunduz et al. (2004). Furthermore, they confirm that the strength of resistance to SMV differs between V94‐5152 (Rsv4) and PI88788 (Rsv4).
Figure 2.
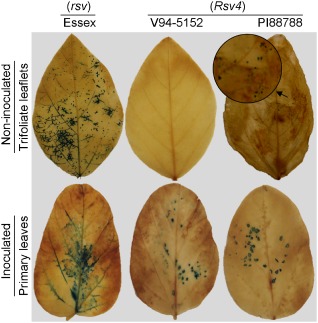
β‐Glucuronidase (GUS) expression in soybean leaf tissues following biolistic inoculation with Soybean mosaic virus (SMV)‐N‐GUS. Attached primary leaves were inoculated and tissues were harvested for GUS expression analysis at 21 days post‐inoculation. It should be noted that SMV‐N‐GUS was capable of limited systemic infection in PI88788 (Rsv4), but not in V94‐5152 (Rsv4). The arrow points to small foci of infected cells expressing GUS, which are displayed in the enlarged insert.
Association of 1033K or 1054R with virulence on Rsv4‐genotype soybeans
Chowda‐Reddy et al. (2011) identified 1033K and 1054R, located on P3, as SMV virulence determinants on V94‐5152 (Rsv4). To determine whether the virulence of American isolates as well as the standard SMV pathotypes on V94‐5152 (Rsv4) can be attributed to these determinants, the P3 cistrons of all the virulent isolates on Rsv4‐genotype soybeans were analysed. The predicated P3 amino acid sequences showed that the virulent variant on V94‐5152 (Rsv4) had either lysine at polypeptide position 1033 (isolates ArE5, IA, OH, G3) or arginine at 1054 (isolates AL1, AL3, IL3, IL4, IL5, KY, MN, MS4, MS10, TNP, TN2, VA, G1, G4, G7a) (Table 3). It should be noted that the virulence of SMV‐G7a (PV‐724) on V94‐5152 (Rsv4) was associated with both Q1033R and G1054R substitutions, and both were retained in the second passage (Table 3). The presence of 1033K or 1054R in P3 of virulent variants from V94‐5152 (Rsv4) was irrespective of the source of the inoculum [i.e. sap extracts from Williams (rsv) or PI88788 (Rsv4)]. The nucleotide sequences of the P3 cistrons of virulent viruses from the second passage on V94‐5152 (Rsv4) were also determined. Pair‐wise comparison of the deduced amino acid sequences derived from the first and second passages on V94‐5152 (Rsv4) showed no difference (Table 3). This observation indicates that, once 1033K or 1054R was selected, it became fixed and remained stable.
Table 3.
Amino acids at polypeptide positions 1033 and 1054 of Soybean mosaic virus (SMV) isolates deduced from nucleotide sequences of the P3 cistrons recovered from various soybean genotypesa
| Isolate | 1033 | 1054 | ||||
|---|---|---|---|---|---|---|
| Soybean genotype | Soybean genotype | |||||
| Williams | V94‐5152 | PI88788 | Williams | V94‐5152 | PI88788 | |
| (rsv) | (Rsv4) | (Rsv4) | (rsv) | (Rsv4) | (Rsv4) | |
| AL1 | Glnb | Gln (Gln) | Gln | Gly | Arg (Arg) | Gly |
| AL3 | Gln | Gln (Gln) | Gln (Gln) | Gly | Arg (Arg) | Gly (Gly) |
| ArE5 | Gln | Lys (Lys) | [Lys] | Ser | Ser (Ser) | [Ser] |
| IA | Gln | Lys (Lys) | Gln | Gly | Gly (Gly) | Gly |
| IL3 | Gln | Gln (Gln) | Gln | Arg | Arg (Arg) | Arg |
| IL4 | Gln | Gln [Gln] | Gln | Gly | Arg [Arg] | Gly |
| IL5 | NDc | [Gln] | Gln | ND | [Arg] | Gly |
| KY | Gln | Gln [Gln] | Gln | Gly | Arg [Arg] | Gly |
| MN | Gln | Gln (Gln) | Gln | Gly | Arg (Arg) | Gly |
| MS4 | Gln | Gln (Gln) | Gln | Gly | Arg (Arg) | Gly |
| MS10 | Gln | Gln (Gln) | Gln | Gly | Arg (Arg) | Gly |
| OH | Gln | Lys (Lys) | Lys (Lys) | Ser | Ser (Ser) | Ser (Ser) |
| TNP | ND | [Gln] | Gln | ND | [Arg] | Gly |
| TN2 | Gln | Gln (Gln) | Gln | Gly | Arg (Arg) | Gly |
| VA | ND | [Gln] | Gln | ND | [Arg] | Gly |
| G1 | Gln | Gln [Gln] | Gln | Gly | Arg [Arg] | Gly |
| G2 (N)d | Gln | NAe | Gln (Gln) | Gly | NA | Gly (Gly) |
| G3 | Gln | [Lys] | Gln | Ser | [Ser] | Ser |
| G4 | Gln | Gln [Gln] | Gln | Gly | Arg [Arg] | Gly |
| G7a (PV‐624) | Gln | Gln (Gln) | Gln | Arg | Arg (Arg) | Arg |
| G7a (PV‐724) | Gln | Arg (Arg) | Gln | Gly | Arg (Arg) | Gly |
Positions of the amino acids are based on the genome of SMV‐N (GenBank Accession Number D00507).
Amino acids shown outside of parentheses were identified from systemically infected leaves of Williams (rsv) or from other genotypes when sap extracts from Williams (rsv) served as the inoculum. Those in parentheses are from systemically infected leaves when sap extracts from infected tissues of the same soybean genotype were used as the inoculum (second passage in the same genotype). Those shown in brackets were identified in the systemically infected leaves when sap extract from PI88788 (Rsv4) was inoculated on V94‐5152 (Rsv4), and vice versa.
Not done.
Amino acids shown for SMV‐N in Williams (rsv) were deduced from the published nucleotide sequences.
Not applicable.
Interestingly, P3 of the majority of virulent American isolates from PI88788 (Rsv4), including those of standard SMV pathotypes G1–G4, did not contain lysine or arginine at polypeptide positions 1033 or 1054, respectively (Table 3). Exceptions were SMV isolates ArE5, IL3, OH and SMV pathotype G7a (ATCC PV‐624), which contained lysine or arginine at polypeptide positions 1033 or 1054, respectively (Table 3). We used two subcultures of SMV‐G7a deposited in the American Type Culture Collection (ATCC) by independent researchers and maintained in two different laboratories. Surprisingly, SMV‐G7a (ATCC PV‐724) contained glycine, but not arginine at position 1054. Both SMV‐G7a isolates encode glutamine at polyprotein position 1033.
To determine whether 1033K and 1054R were present in the initial field isolates, we obtained the nucleotide sequences of P3 cistrons of all virulent American isolates, as well as the standard SMV pathotypes, from the infected Williams (rsv) tissues, which served as the initial inoculum source. Pair‐wise comparison of the P3 sequences derived from Williams (rsv) with those of the same isolates from V94‐5152 (Rsv4) showed that none, except SMV‐IL3 and SMV‐G7a (ATCC PV‐624), contained 1033K or 1054R in the initial inoculum (Table 3). Interestingly, the nucleotide sequences of the P3 cistrons of all isolates from PI88788 (Rsv4) were identical to those amplified from Williams (rsv), except those of SMV‐ArE5 and SMV‐OH (Table 3). It should be noted that additional passages of SMV‐AL3 and SMV‐G2 (N) in PI88788 (Rsv4) did not affect the residues at either the 1033 or 1054 positions (Table 3). These observations suggest that lysine or arginine at polypeptide positions 1033 or 1054, respectively, are associated mostly with virulence on V94‐5152 (Rsv4) and to a limited extent on PI88788 (Rsv4).
Impact of amino acid substitutions at polypeptide positions 1033 or 1054 on the virulence of SMV‐N on Rsv4‐genotype soybeans
SMV‐N was avirulent on V94‐5152 (Rsv4), but virulent on PI88788 (Rsv4), irrespective of whether biolistic inoculation with the infectious cDNA clone or mechanical inoculation with progeny viruses replicated in Williams (rsv) was used (Table 1; Fig. 2). SMV‐N‐derived progeny viruses from PI88788 (Rsv4), unlike those of other standard pathotypes or American isolates, were also avirulent on V94‐5152 (Rsv4) (Table 1). The inability of SMV‐N to adapt experimentally to V94‐5152 could be a result of its unique genetic background that influences its evolvability (Montarry et al., 2011). To determine whether Q1033K or G1054R substitution is sufficient to confer virulence to avirulent SMV‐N on V94‐5152 (Rsv4), SMV‐NQ1033K, SMV‐NG1054R and SMV‐NQ1033K+G1054R were constructed and biolistically inoculated onto attached primary leaves of Rsv4‐genotype soybeans. All SMV‐N‐derived P3 mutants infected systemically 75%–100% of both Rsv4‐genotype soybeans as early as 7–10 dpi (Table 4; Fig. 3). Symptoms induced by SMV‐NQ1033K+G1054R on V94‐5152 (Rsv4) were more severe than those caused by the other two single mutant viruses (Fig. 3). Apparently, the combination of Q1033K and G1054R has a synergistic or additive effect on virulence, as systemic symptoms appeared more rapidly and the infected plants became severely stunted. Regardless, the P3 cistrons derived from progeny viruses in Rsv4‐genotype soybeans were identical to those of the parental plasmids, except those of progenies derived from SMV‐NG1054R in V94‐5152 (Rsv4), in which infection was associated with a sequence polymorphism at nucleotide position 3265 (Table 4).
Table 4.
Responses of soybean genotypes to biolistic inoculation with Soybean mosaic virus (SMV)‐N or its derivative P3 mutants
| Virus | Soybean genotype | ||
|---|---|---|---|
| Essex (rsv) | V94‐5152 (Rsv4) | PI88788 (Rsv4) | |
| SMV‐N | 4b/4 | 0/4 | 4a c/4 |
| SMV‐NQ1033K | 4/4 | 3a/4 | 4a/4 |
| SMV‐NG1054R | 4/4 | 3a/4 | 3a/4 |
| SMV‐NQ1033K+G1054R | 4/4 | 6a/6 | 4a/4 |
The P3 cistron from systemically infected leaves of one of the plants was reverse transcriptase‐polymerase chain reaction (RT‐PCR) amplified and sequenced.
Total number of plants infected systemically/total number inoculated biolistically on primary leaves with the cDNA clones. The inoculated plants were evaluated at 21 days post‐inoculation by symptomatology and indirect enzyme‐linked immunosorbent assay (IELISA).
Infection of V94‐5152 (Rsv4) with SMV‐NG1054R was associated with a sequence polymorphism at position 3265, where both GTA and GCA codons encoding valine and alanine, respectively, were present at polyprotein position 1045. No newly emerged in planta substitution was observed in the P3 cistrons of progenies derived from any of the other mutant viruses in the two Rsv4‐genotype soybeans.
Figure 3.

Virulence of Soybean mosaic virus‐N and its P3‐derivative mutants on Rsv4‐genotype soybeans following biolistic inoculation with cDNA clones. Attached primary leaves were targeted for inoculation and plants were photographed at 21 days post‐inoculation. The inserts display enlarged leaflets from systemically infected trifoliate leaves.
SMV‐N and its P3‐derived mutants also caused systemic infection in 75%–100% of biolistically inoculated PI88788; however, the symptoms induced by the mutants appeared more rapidly, as early as 7–10 dpi (Table 4; Fig. 3). The P3 cistrons of progeny viruses lacked any in planta‐generated new amino acid substitutions. These observations further confirm that SMV‐N is intrinsically virulent on PI88788 (Rsv4). Nevertheless, Q1033K or G1054R substitution enhanced SMV‐N movement and the severity of symptoms in PI88788 (Rsv4) (Fig. 3). Together, these observations demonstrate that the avirulence function of SMV‐N on V94‐5152 (Rsv4) resides on P3, and a Q1033K or G1054R substitution is essential and sufficient to convert avrSMV‐N to virSMV‐N.
Discussion
Interactions of SMV with various soybean genotypes containing Rsv1, Rsv3 or Rsv4 have been studied extensively in the USA (Saghai Maroof et al., 2008a). However, a set of standard SMV pathotypes isolated from imported soybean germplasm three decades ago, which had probably evolved elsewhere in the world, was utilized in those studies (Cho and Goodman, 1979; Jain et al., 1992). Since identification, these SMV pathotypes have been maintained at various laboratories and subjected to propagation in different soybean genotypes. Replication of SMV, similar to other RNA viruses, is prone to a high mutation rate which affects the heterogeneity of the viral population (Drake and Holland, 1999; Hajimorad et al., 2003, 2008). Furthermore, shifting viruses between different hosts also affects quasispecies cloud size (Schneider and Roossinck, 2001). Thus, although standard SMV pathotypes have served scientists well in revealing the genetics of resistance mediated by Rsv genes and their inheritance, they do not truly represent the current American field isolates with regard to the evolution of virulence towards Rsv‐genotype soybeans. As a result of the absence of publicly available information on the deployment of Rsv genes in the elite soybean cultivars grown in the USA, it is unknown whether SMV is directly subject to positive selection pressure imposed by any of these R genes.
It is not surprising that all the American field isolates in this study were avirulent on L78‐379 (Rsv1). It has been well established that the gain of virulence by an avirulent SMV on Rsv1‐genotype soybeans requires mutations in two cistrons with a combination of at least three simultaneous amino acid substitutions: two in HC‐Pro and one in P3, or two in P3 and one in HC‐Pro (Eggenberger et al., 2008; Hajimorad et al., 2008, 2011). Elsewhere in the world, virulence of SMV on Rsv1‐genotype soybeans has been reported only from South Korea (Choi et al., 2005); however, the resistance‐breaking determinants of these virulent isolates have not been identified. Interestingly, the Korean isolates were obtained from soybean cultivars containing Rsv1 or one of its alleles (Choi et al., 2005). Thus, the prevalence of the Korean virulent isolates is probably a consequence of direct selection pressure imposed by Rsv1.
Contrary to the lack of virulence on L78‐379 (Rsv1), the majority of the American isolates were virulent on L29 (Rsv3). SMV field isolates from Canada and South Korea have also evolved widespread virulence towards Rsv3 (Choi et al., 2005; Seo et al., 2009b; Viel et al., 2009). The determinants for virulence on L29 (Rsv3) have been mapped to the CI protein (Seo et al., 2009a; Zhang et al., 2009). This SMV‐encoded protein plays a crucial role in replication and movement (Urcuqui‐Inchima et al., 2001). However, SMV isolates show a high level of diversity in the CI cistron (Seo et al., 2009b). It appears that the cost of fitness of resistance‐breaking virulent isolates on Rsv3‐genotype soybeans is not also high, as SMV‐N, a virulent strain, is highly pathogenic on rsv3‐genotype soybeans as well (Seo et al., 2009a; Zhang et al., 2009). Perhaps a combination of these two factors has contributed to the prevalence of naturally occurring virulent SMV isolates on Rsv3‐genotype soybeans. However, one cannot exclude the possibility that one of the Rsv3 alleles is also present in the elite soybean cultivars grown which imposes positive selection pressure on SMV evolution towards virulence.
It has been shown previously that the Rsv4 allele in PI88788 (Rsv4) expresses LS in the context of heterozygosity to the standard SMV pathotype G1 (Gunduz et al., 2004), whereas V94‐5152 (Rsv4) displays broad resistance to all the standard SMV pathotypes (Gunduz et al., 2002). In this study, we showed that SMV pathotypes G1–G4 and G7a, as well as 14 of the 15 American isolates, were also virulent on PI88788 (Rsv4). Interestingly, P3 of all virulent isolates, with the exceptions of ArE5 and OH, from infected PI88788 (Rsv4) showed no amino acid substitution when compared with P3 of the same isolates from Williams (rsv). Direct biolistic inoculation of SMV‐N‐GUS or SMV‐N also revealed virulence on PI88788 (Rsv4) with no amino acid substitution in P3. However, SMV‐N‐GUS was inefficient in moving systemically in PI88788 (Rsv4), probably because of the negative impact of fusion to GUS (German‐Retana et al., 2000). This is mainly because biolistic inoculation of SMV‐N resulted in efficient systemic infection. These observations indicate that the Rsv4‐mediated resistance mechanism against SMV that operates in PI88788 (Rsv4) is not as robust as that of V94‐5152 (Rsv4) in preventing systemic movement. This could be a result of the genetic differences between the Rsv4 alleles in these two soybean genotypes. It should be noted that the strength of resistance to SMV also varies among soybean genotypes containing different Rsv1 alleles (Chen et al., 1994; Cho and Goodman, 1979; Hayes et al., 2004). However, it is equally possible that the difference in the strength of resistance is a reflection of difference(s) in the genetic background of PI88788 (Rsv4) and V94‐5152 (Rsv4). We have also noticed elsewhere that the strength of resistance mediated by Rsv1 alleles in PI96983 and L78‐379 differs as well (Hajimorad et al., 2003, 2008, 2011). These observations support the notion that other host factors probably influence R‐mediated resistance responses (Banerjee et al., 2001).
Despite weakness in the strength of resistance in PI88788 (Rsv4), the observation that systemic symptoms induced by SMV‐N appeared between 21 and 28 dpi, whereas those of SMV‐N‐derived P3 mutants occurred at about 7–10 dpi, indicates the presence of some interference with the systemic movement of the wild‐type virus. Moreover, the inability of SMV‐ArE5 and SMV‐OH to infect systemically PI88788 (Rsv4), unless they contained the Q1033K substitution, also suggests that this soybean genotype confers strong strain‐specific resistance to SMV. The observation that the replication of SMV in PI88788 (Rsv4) somehow enhanced virulence on V94‐5152 (Rsv4) further supports the existence of some level of positive selection pressure in this genotype. However, virulent variants were not represented in the consensus sequences of P3 cistrons of viral populations recovered from PI88788 (Rsv4). P3 cistrons of virulent isolates on PI88788 (Rsv4) in the second passage also remained identical to those recovered in the first passage. In a study published elsewhere, we observed that replication of an avirulent SMV in L800 (3gG2), a recombinant soybean inbred line containing a genetic element from the complex Rsv1 locus, also enhanced the evolution of virulence towards PI96983 (Rsv1) (Hajimorad et al., 2011).
In contrast with PI88788 (Rsv4), resistance to systemic movement of SMV in V94‐5152 (Rsv4) is robust. Nevertheless, 12 of the 35 American isolates caused systemic infection in V94‐5152 (Rsv4); however, the majority infected only a single plant. Without exception, P3 of all virulent variants on V94‐5152 (Rsv4) contained R1033K or G1054R substitutions. The P3 sequences of SMV‐IL3 and SMV‐G7a (ATCC PV‐624) were unique, as they were identical when recovered from Williams (rsv), PI88788 (Rsv4) or V94‐5152 (Rsv4). This suggests that SMV‐IL3 and SMV‐G7a (ATCC PV‐624) evolved virulence elsewhere, where 1054R was selected and became fixed. SMV‐IL3 was isolated from breeding plots (Domier et al., 2003), whereas SMV‐G7a has been retained in laboratories since its identification (Buzzell and Tu, 1984). It is likely that the virulence of SMV‐G7a (ATCC PV‐624) has evolved experimentally as a result of inadvertent exposures to Rsv4‐genotype soybeans during maintenance. This is mainly because SMV‐G7a (ATCC PV‐724), which was maintained in another laboratory, contained 1054G.
Regardless, it appears that the virulent variants derived from 11 other American isolates were selected under our experimental conditions because of strong Rsv4‐mediated selection pressure in V94‐5152. However, the origin of the mutations essential for gain of virulence is unknown. It is possible that such variants existed as a small subpopulation in each of the field isolates, but were rapidly selected in V94‐5152 (Rsv4). It is equally possible that, on transfer of each of these isolates to Williams (rsv), such mutations emerged in this host, but remained as a small subpopulation because of a lack of selection pressure. In our previous study on SMV/Rsv1 pathosystems, we also noticed that replication of an avirulent SMV, or its derivative chimeras, in Williams82 (rsv) enhanced the emergence of virulent variants on Rsv1‐genotype soybeans (Hajimorad et al., 2008, 2011). This was despite the absence of any detectable mutation in the consensus nucleotide sequences of such viral populations from Williams82 (rsv) (Hajimorad et al., 2008). However, one cannot exclude the possibility that, in the current study, the virulent variants emerged in the primary leaves of V94‐5152 (Rsv4). Inoculation of SMV‐N‐GUS showed that the resistance mechanism mediated by the Rsv4 allele in V94‐5152 (Rsv4) allows for replication at the site of inoculation.
Unlike SMV field isolates, progeny viruses derived from cDNA clones of SMV‐N, SMV‐G7 and SMV‐G7d failed to adapt experimentally to V94‐5152 (Rsv4). Our numerous attempts to adapt SMV‐N to V94‐5152 (Rsv4), using sap extracts from the infected PI88788 (Rsv4), also failed. It is probable that there is greater heterogeneity in viral populations of the field isolates when compared with those derived from molecularly cloned genomes. Complexity in viral populations of field isolates has been demonstrated for another potyvirus (Maliogka et al., 2012). Thus, it is probable that variants with 1033K or 1054R evolved under field conditions, but were selected under our experimental conditions because of the presence of a strong positive selection pressure. The observation that the virulence of some of the isolates on V94‐5152 (Rsv4) was enhanced on replication in PI88788 (Rsv4) probably indicates that this Rsv4‐genotype soybean amplified an existing subpopulation consisting of virulent variants, but not to a detectable level in the consensus sequences.
In conclusion, SMV field isolates were detected with virulence on Rsv3‐ and Rsv4‐ genotype soybeans, but not on Rsv1‐genotype soybeans, in the USA. It appears that one of the determinants for virulence on V94‐5152 (Rsv4) (1054R) has already been fixed in the P3 cistron of SMV‐IL3 and one of the subcultures of the SMV standard pathotype G7a (ATCC PV‐624). The only other naturally occurring virulent isolate of SMV on V94‐5152 (Rsv4) has been reported from Canada, which encodes 1033K (Chowda‐Reddy et al., 2011). Scanning the full‐length P3 cistrons of more than 75 SMV isolates available in GenBank for the presence of 1033K or 1054R showed that Chinese strains C13, C16 and 4469‐4 also harbour 1054R (Accession Numbers GQ491075, GQ491078 and HM590055, respectively) (Yang et al., 2011); however, the virulence of these isolates on V94‐5152 (Rsv4) is unknown. Regardless, it appears that substitution at position 1054, relevant to virulence on V94‐5152 (Rsv4), is more frequent than at 1033. American field isolates that initially lacked 1033K or 1054R were capable of rapid adaptation to V94‐5152 (Rsv4), in which G1054R rather than R1033K was selected in P3 of the majority. Reconstruction of Q1033K or G1054R in avirulent SMV‐N showed that each of these substitutions is essential and sufficient to confer virulence on V94‐5152 (Rsv4). In general, it seems that Rsv4‐mediated resistance against SMV is not strong; hence, it will probably not be durable if deployed alone. The durability of resistance mediated by R genes against plant viruses has been attributed to a number of mutations required for gain of virulence (Harrison, 2002). To develop durable and broad resistance to SMV in soybean, stacking the three Rsv genes together seems to be the best approach (Saghai Maroof et al., 2008b; Shi et al., 2009).
Experimental Procedures
Viruses
Subcultures of the standard SMV pathotypes G1, G3–G6 and G7a (ATCC PV‐624 and PV‐724) were obtained from J. H. Hill (Iowa State University, Ames, IA, USA) and L.L. Domier (University of Illinois, Urbana, IL, USA). Progeny viruses from biolistically inoculated Williams82 (rsv) derived from pSMV‐N, pSMV‐G7 and pSMV‐G7d (GenBank Accession Numbers D00507, AY216010 and AY216987, respectively) (Hajimorad et al., 2003; Wang et al., 2006) served as sources for the standard SMV pathotypes G2, G7 and G7d. In addition to wild‐type SMV‐G3, progeny viruses from biolistically inoculated Williams82 (rsv) with pSMV‐G3 (Kanematsu et al., 2001) also served as inoculum. pSMV‐N‐GUS has been described previously (Wang et al., 2006).
SMV field isolates from Alabama, Arkansas, Illinois, Iowa, Kentucky, Minnesota, Mississippi, Ohio, Virginia and Wisconsin were provided by J. F. Murphy (Auburn University, Auburn, AL, USA), I. E. Tzanetakis (University of Arkansas, Fayetteville, AK, USA), L. L. Domier, J. H. Hill, S. A. Ghabrial (University of Kentucky, Lexington, KY, USA), B. E. Lockhart (University of Minnesota, St. Paul, MN, USA), S. Sabanadzovic (Mississippi State University, Mississippi State, MS, USA), M. G. Redinbaugh (Ohio State University, Columbus, OH, USA), S. A. Tolin (Virginia Tech, Blacksburg, VA, USA) and C. R. Grau (University of Wisconsin, Madison, WI, USA), respectively. SMV isolates from Tennessee were collected from soybean grown at the East Tennessee Research and Education Center (Fajolu et al., 2010). All field isolates were obtained from systemically infected leaves of naturally infected soybeans, except for WI from Wisconsin which was recovered from newly emerged seedlings when seeds from a breeding line produced in Wisconsin were grown in a growth chamber. The infected leaves originated from either elite soybean cultivars or breeding lines, including soybean germplasm, grown in experimental plots. All isolates were mechanically transferred to Williams (rsv) and the infected tissues were stored at −80 °C. None of the isolates subjected to single local lesion cloning or sap extract from systemically infected leaves of Williams (rsv) served as the initial inoculum.
Soybean genotypes
Williams (rsv), Williams82 (rsv) and Essex (rsv), all universally susceptible to SMV, L78‐379 (Rsv1), a near isoline of Williams containing the Rsv1 allele derived from PI96983 (Bernard et al., 1991), L29, a Williams isoline containing Rsv3 derived from Hardee (Gunduz et al., 2000), V94‐5152, an Essex isoline containing the Rsv4 allele from PI486355 (Buss et al., 1997), and PI88788, containing an allelic R gene to SMV at the Rsv4 locus in V94‐5152 (Gunduz et al., 2004), were used in this study.
SMV inoculation and detection
Plants were inoculated mechanically or biolistically. For mechanical inoculation, sap extract in 50 mm phosphate buffer, pH 7.0, was rub inoculated onto carborundum‐dusted (600‐mesh) primary leaves (Hajimorad and Hill, 2001). For biolistic inoculation, infectious cDNA clones were inoculated onto fully expanded attached primary leaves, as described previously (Hajimorad et al., 2003, 2008). The inoculated plants were maintained in a growth chamber operating at 22 °C with a photoperiod of 16 h until evaluation for infection based on symptoms and indexing of non‐inoculated leaves by IELISA or reverse transcriptase‐polymerase chain reaction (RT‐PCR) (Hajimorad et al., 2008; Malapi‐Nelson et al., 2009). Each of the inoculated plants, irrespective of symptom expression, was assayed individually for the presence of SMV by IELISA. Histochemical assay of GUS expression was performed according to Jefferson (1987). SMV was also detected by immunoprinting of leaf tissues to nitrocellulose membrane with polyclonal antibodies against SMV coat protein, using Fast Red TR/Naphthol AS‐MX as the precipitin substrate, according to the manufacturer's instructions (Sigma‐Aldrich, St. Louis, MO, USA).
Construction of SMV‐N‐derived P3 mutants by site‐directed mutagenesis
To generate pSMV‐NQ1033K, primer N‐3296a (Table S1, see Supporting Information) served as the mutagenic primer. PCR‐derived amplicons were synthesized in the presence of primers N‐3296a and SMV‐2271s, and pSMV‐N served as the template. The amplicons were digested with KpnI and SpeI and ligated into pSMV‐N to generate pSMV‐NQ1033K. To construct pSMV‐NG1054R and pSMV‐NQ1033K+G1054R, primer N‐3221s served as the mutagenic primer (Table S1). PCR products were synthesized in the presence of primers N‐3892a and N‐3221s (Table S1), and pSMV‐N served as the template. Subsequently, the amplicons were digested with SpeI and SalI and ligated into pSMV‐N and pSMV‐NQ1033K to generate pSMV‐NG1054R and pSMV‐NQ1033K+G1054R, respectively. The presence of the introduced mutation(s) and the absence of any unwanted PCR‐generated substitution in each of the cDNA clones were verified by sequencing the entire amplified regions. The infectivity of the mutants was tested by biolistic inoculation. The stability of the introduced mutations in the progeny viruses was verified following RT‐PCR amplification of the P3 cistron from systemically infected leaf tissues, followed by analysis.
RNA extraction, RT‐PCR and sequencing
Total RNA was isolated from systemically infected leaves using an RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). To generate nucleotide sequences of the P3 cistron, total RNA was reverse transcribed using 10‐mer random primers in the presence of SuperScript reverse transcriptase III (Invitrogen, Carlsbad, CA, USA), as instructed by the manufacturer. Primers SMV‐2030s and SMV‐3910a (Table S1) were used for PCR amplification of the P3 cistron in the presence of Ex Taq polymerase (Takara Bio, Madison, WI, USA), and RT‐generated products served as the template. The amplicons were purified with a QIAquick‐PCR Purification Kit (Qiagen) and sequenced using primers SMV‐2271s and SMV‐2919s (Table S1). Sequencing was performed at The University of Tennessee DNA Sequencing Facility and sequences were edited, analysed and aligned using Vector NTI (Lu and Moriyama, 2004). The full‐length nucleotide sequences of the P3 cistrons of virulent variants of the following pathotypes or field isolates from V94‐5154 (Rsv4) were deposited in GenBank under the following Accession Numbers; SMV‐G1 (JQ915063), SMV‐G4 (JQ915064), SMV‐G7a (ATCC PV‐624) (JQ915065), SMV‐AL1 (JQ915066), SMV‐AL3 (JQ915067), SMV‐ArE5 (JQ915068), SMV‐IA (HQ845726), SMV‐IL3 (HQ845727), SMV‐IL4 (JQ915069), SMV‐IL5 (HQ845728), SMV‐MN (HQ845729), SMV‐MS4 (HQ845730), SMV‐MS10 (HQ845731), SMV‐OH (HQ845732), SMV‐TN2 (JQ915070) and SMV‐VA (HQ845733). Near full‐length sequences (short of 21 and 12 nucleotides at the 5′ and 3′ ends, respectively) of a virulent variant of SMV‐KY and SMV‐TNP on V94‐5152 (Rsv4), determined by bidirectional sequencing of overlapping RT‐PCR fragments and using a primer walking approach, are also deposited in GenBank under Accession Numbers HQ845736 and HQ845735, respectively.
Supporting information
Table S1 Sequences of oligonucleotide primers used for site‐directed mutagenesis, reverse transcriptase‐polymerase chain reaction (RT‐PCR) and sequencing.
Acknowledgements
We are grateful to Drs L. L. Domier, S. A. Ghabrial, C. R. Grau, J. H. Hill, B. E. Lockhart, J. F. Murphy, M. G. Redinbaugh, S. Sabanadzovic, S. A. Tolin and I. E. Tzanetakis for providing SMV isolates. We would also like to thank Drs J. H. Hill and A. L. Eggenberger (Iowa State University, Ames, IA, USA) for providing a copy of pSMV‐N‐GUS, and P. Chen (University of Arkansas, Fayetteville, AK, USA), R. L. Nelson (USDA, Agricultural Research Services, Urbana, IL, USA) and M. A. Saghai Maroof (Virginia Tech, Blacksburg, VA, USA) for providing seeds of various soybean genotypes. This project was funded in part by the University of Tennessee College of Agricultural Sciences and Natural Resources, the Tennessee Agricultural Experimental Station, Knoxville, TN, USA, North Central Soybean Research Program and the Tennessee Soybean Promotion Board Program.
References
- Adams, M.J. , Antoniw, J.F. and Fauquet, C.M. (2005) Molecular criteria for genus and species discrimination within the family Potyviridae . Arch. Virol. 150, 459–479. [DOI] [PubMed] [Google Scholar]
- Banerjee, D. , Zhang, X. and Bent, A.F. (2001) The leucine‐rich‐repeat domain can determine effective interaction between RPS2 and other host factors in Arabidopsis RPS2‐mediated disease resistance. Genetics 158, 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, R.L. , Nelson, R.L. and Creemens, C.R. (1991) USDA soybean genetic collection: isoline collection. Soybean Genet. Newsl. 18, 27–57. [Google Scholar]
- Buss, G.R. , Ma, G. , Chen, P. and Tolin, S.A. (1997) Registration of V94‐5152 soybean germplasm resistant to soybean mosaic potyvirus. Crop Sci. 37, 1987–1988. [Google Scholar]
- Buzzell, R.I. and Tu, J.C. (1984) Inheritance of soybean resistance to soybean mosaic virus. J. Hered. 75, 82. [Google Scholar]
- Buzzell, R.I. and Tu, J.C. (1989) Inheritance of a soybean stem‐tip necrosis reaction to soybean mosaic virus. J. Hered. 80, 400–401. [Google Scholar]
- Chen, P. , Buss, G.R. , Roane, C.W. and Tolin, S.A. (1994) Inheritance in soybean of resistant and necrotic reactions to soybean mosaic virus strains. Crop Sci. 34, 414–422. [Google Scholar]
- Cho, E.‐K. and Goodman, R.M. (1979) Strains of soybean mosaic virus: classification based on virulence in resistant soybean cultivars. Phytopathology, 69, 467–470. [Google Scholar]
- Choi, B.K. , Koo, J.M. , Ahn, H.J. , Yum, H.J. , Choi, C.W. , Ryu, K.H. , Chen, P. and Tolin, S.A. (2005) Emergence of Rsv‐resistance breaking Soybean mosaic virus isolates from Korean soybean cultivars. Virus Res. 112, 42–51. [DOI] [PubMed] [Google Scholar]
- Chowda‐Reddy, R.V. , Sun, H. , Chen, H. , Poysa, V. , Ling, H. , Gijzen, M. and Wang, A. (2011) Mutations in the P3 protein of Soybean mosaic virus G2 isolates determine virulence on Rsv4‐genotype soybean. Mol. Plant–Microbe Interact. 24, 37–43. [DOI] [PubMed] [Google Scholar]
- Chung, B.Y.‐W. , Miller, W.A. , Atkins, J.F. and Firth, A.E. (2008) An overlapping essential gene in the potyviridae. Proc. Natl. Acad. Sci. USA, 105, 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier, L.L. , Latorre, I.J. , Steinlage, T.A. , McCoppin, N. and Hartman, G.L. (2003) Variability and transmission by Aphis glycines of North American and Asian Soybean mosaic virus isolates. Arch. Virol. 148, 1925–1941. [DOI] [PubMed] [Google Scholar]
- Domier, L.L. , Steinlage, T.A. , Hobbs, H.A. , Wang, Y. , Herrera‐Rodriguez, G. , Haudenshield, J.S. , McCoppin, N.K. and Hartman, G.L. (2007) Similarities in seed and aphid transmission among Soybean mosaic virus isolates. Plant Dis. 91, 546–550. [DOI] [PubMed] [Google Scholar]
- Drake, J.W. and Holland, J.J. (1999) Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA, 96, 13 910–13 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenberger, A.L. , Hajimorad, M.R. and Hill, J.H. (2008) Gain of virulence on Rsv1‐genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC‐Pro. Mol. Plant–Microbe Interact. 21, 931–936. [DOI] [PubMed] [Google Scholar]
- Fajolu, O.L. , Wen, R.‐H. and Hajimorad, M.R. (2010) Occurrence of Alfalfa mosaic virus in soybean in Tennessee. Plant Dis. 94, 1505. [DOI] [PubMed] [Google Scholar]
- German‐Retana, S. , Candresse, T. , Alias, E. , Delbos, R.‐P. and Le Gal, O. (2000) Effects of green fluorescent protein or β‐glucuronidase tagging on the accumulation and pathogenicity of a resistance‐breaking Lettuce mosaic virus isolate in susceptible and resistant lettuce cultivars. Mol. Plant–Microbe Interact. 13, 316–324. [DOI] [PubMed] [Google Scholar]
- Gunduz, I. , Buss, G.R. , Ma, G. , Chen, P. and Tolin, S.A. (2000) Genetic analysis of resistance to Soybean mosaic virus in OX670 and Harosoy soybean. Crop Sci. 41, 1785–1791. [Google Scholar]
- Gunduz, I. , Buss, G.R. , Chen, P. and Tolin, S.A. (2002) Characterization of SMV resistance genes in Tousan 140 and Hourei soybean. Crop Sci. 42, 90–95. [DOI] [PubMed] [Google Scholar]
- Gunduz, I. , Buss, G.R. , Chen, P. and Tolin, S.A. (2004) Genetic and phenotypic analysis of Soybean mosaic virus resistance in PI88788 soybean. Phytopathology, 94, 687–692. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. and Hill, J.H. (2001) Rsv1‐mediated resistance against Soybean mosaic virus‐N is hypersensitive response‐independent at inoculation site, but has the potential to initiate a hypersensitive response‐like mechanism. Mol. Plant–Microbe Interact. 14, 587–598. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2003) Evolution of Soybean mosaic virus‐G7 molecularly cloned genome in Rsv1‐genotype soybean results in emergence of a mutant capable of evading Rsv1‐mediated recognition. Virology, 314, 497–509. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2008) Adaptation of Soybean mosaic virus avirulent chimeras containing P3 sequences from virulent strains to Rsv1‐genotype soybeans is mediated by mutations in HC‐Pro. Mol. Plant–Microbe Interact. 21, 937–946. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Wen, R.‐H. , Eggenberger, A.L. , Hill, J.H. and Saghai Maroof, M.A. (2011) Experimental adaptation of an RNA virus mimics natural evolution. J. Virol. 85, 2557–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, B.D. (2002) Virus variation in relation to resistance‐breaking in plants. Euphytica, 124, 181–192. [Google Scholar]
- Hayes, A.J. , Jeong, S.C. , Gore, M.A. , Yu, Y.G. , Buss, G.R. , Tolin, S.A. and Saghai Maroof, M.A. (2004) Recombination within a nucleotide‐binding‐site/leucine‐rich‐repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics, 166, 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, R.K. , McKern, N.M. , Tolin, S.A. , Hill, J.H. , Barnett, O.W. , Tosic, M. , Ford, R.E. , Beachy, R.N. , Yu, M.H. , Ward, C.W. and Shukla, D.D. (1992) Confirmation that fourteen potyvirus isolates from soybean are strains of one virus by comparing coat protein peptide profiles. Phytopathology, 82, 294–299. [Google Scholar]
- Jefferson, R.A. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Kanematsu, S. , Eggenberger, A.L. and Hill, J.H. (2001) Construction of an infectious cDNA clone of soybean mosaic virus G3. Jpn. J. Phytopathol. 67, 155. [Google Scholar]
- Kiihl, R.A.S. and Hartwig, E.E. (1979) Inheritance of reaction to soybean mosaic virus in soybeans. Crop Sci. 19, 372–375. [Google Scholar]
- Lu, G. and Moriyama, E.N. (2004) Vector NTI, a balanced all‐in‐one sequence analysis suite. Brief. Bioinform. 5, 378–388. [DOI] [PubMed] [Google Scholar]
- Ma, G. , Chen, P. , Buss, G.R. and Tolin, S.A. (1995) Genetic characteristics of two genes for resistance to soybean mosaic virus in PI486355 soybean. Theor. Appl. Genet. 91, 907–914. [DOI] [PubMed] [Google Scholar]
- Ma, G. , Chen, P. , Buss, G.R. and Tolin, S.A. (2002) Complementary action of two independent dominant genes in Columbia soybean for resistance to soybean mosaic virus. J. Hered. 93, 179–184. [DOI] [PubMed] [Google Scholar]
- Malapi‐Nelson, M. , Wen, R.‐H. , Ownley, B.H. and Hajimorad, M.R. (2009) Co‐infection of soybean with Soybean mosaic virus and Alfalfa mosaic virus results in disease synergism and alteration in accumulation level of both viruses. Plant Dis. 93, 1259–1264. [DOI] [PubMed] [Google Scholar]
- Maliogka, V.I. , Salvador, B. , Carbonell, A. , Saenz, P. , Leon, D.S. , Oliveros, J.C. , Delgadillo, M.O. , Garcia, J.A. and Simon‐Mateo, C. (2012) Virus variants with differences in the P1 protein coexist in a Plum pox virus population and display particular host‐dependent pathogenicity features. Mol. Plant Pathol. doi: 10.1111/J.1364-3703.2012.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarry, J. , Doumayrou, J. , Simon, V. and Moury, B. (2011) Genetic background matters: a plant–virus gene‐for‐gene interaction is strongly influenced by genetic contexts. Mol. Plant Pathol. 12, 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai Maroof, M.A. , Tucker, D.M. and Tolin, S.A. (2008a) Genomics of viral–soybean interactions In: Genetics and Genomics of Soybean (Stacey G., ed.), pp. 293–319. New York: Springer. [Google Scholar]
- Saghai Maroof, M.A. , Jeong, S.C. , Gunduz, I. , Tucker, D.M. , Buss, G.R. and Tolin, S.A. (2008b) Pyramiding of soybean mosaic virus resistance genes by marker‐assisted selection. Crop Sci. 48, 517–526. [Google Scholar]
- Saghai Maroof, M.A. , Tucker, D.M. , Skoneczka, J.A. , Bowman, B.C. , Tripathy, S. and Tolin, S.A. (2010) Fine mapping and candidate gene discovery of the Soybean mosaic virus resistance gene, Rsv4 . Plant Genome, 3, 14–22. [Google Scholar]
- Schneider, W. and Roossinck, M.J. (2001) Genetic diversity in RNA virus quasispecies is controlled by host–virus interactions. J. Virol. 75, 6566–6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.‐K. , Lee, S.‐H. and Kim, K.‐H. (2009a) Strain‐specific cylindrical inclusion protein of Soybean mosaic virus elicits extreme resistance and a lethal systemic hypersensitive response in two resistant soybean cultivars. Mol. Plant–Microbe Interact. 22, 1151–1159. [DOI] [PubMed] [Google Scholar]
- Seo, J.‐K. , Ohshima, K. , Lee, H.‐G. , Son, M. , Choi, H.‐S. , Lee, S.‐H. , Sohn, S.‐H. and Kim, K.‐H. (2009b) Molecular variability and genetic structure of the population of Soybean mosaic virus based on the analysis of complete genome sequences. Virology, 393, 91–103. [DOI] [PubMed] [Google Scholar]
- Seo, J.‐K. , Sohn, S.‐H. and Kim, K.‐H. (2011) A single amino acid change in HC‐Pro of soybean mosaic virus alters symptom expression in a soybean cultivar carrying Rsv1 and Rsv3 . Arch. Virol. 156, 135–141. [DOI] [PubMed] [Google Scholar]
- Shi, A. , Chen, P. , Li, D. , Zheng, C. , Zhang, B. and Hou, A. (2009) Pyramiding multiple genes for resistance to soybean mosaic virus in soybean using molecular markers. Mol. Breed. 23, 113–124. [Google Scholar]
- Suh, S.J. , Bowman, B.C. , Jeong, N. , Yang, K. , Kastl, C. , Tolin, S.A. , Saghai Maroof, M.A. and Jeong, S.‐C. (2011) The Rsv3 locus conferring resistance to Soybean mosaic virus is associated with a cluster of coiled coil nucleotide‐binding leucine‐rich repeat genes. Plant Genome, 4, 55–64. [Google Scholar]
- Urcuqui‐Inchima, S. , Haenni, A.L. and Bernardi, F. (2001) Potyvirus proteins: a wealth of functions. Virus Res. 74, 157–175. [DOI] [PubMed] [Google Scholar]
- Viel, C. , Ide, C. , Cui, X. , Wang, A. , Farsi, M. , Michelutti, R. and Stromvik, M. (2009) Isolation, partial sequencing, and phylogenetic analyses of Soybean mosaic virus (SMV) in Ontario and Quebec. Can. J. Plant Pathol. 31, 108–113. [Google Scholar]
- Vijayapalani, P. , Maeshima, M. , Nagasaki‐Takekuchi, N. and Miller, W.A. (2012) Interaction of the trans‐frame potyvirus protein P3N‐PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog. 8, e1002639. doi: 10.1371/journal.ppat.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Eggenberger, A. , Hill, J. and Bogdanove, A.J. (2006) Pseudomonas syringae effector avrB confers soybean cultivar‐specific avirulence on Soybean mosaic virus adapted for transgene expression but effector avrPto does not. Mol. Plant–Microbe Interact. 19, 304–312. [DOI] [PubMed] [Google Scholar]
- Wei, T. , Zhang, C. , Hong, J. , Xiong, R. , Kasschau, K.D. , Zhou, X. , Carrington, J.C. and Wang, A. (2010) Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N‐PIPO. PLoS Pathog. 6, e1000962. doi: 10.1371/journal.ppat.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, R.‐H. and Hajimorad, M.R. (2010) Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology, 400, 1–7. [DOI] [PubMed] [Google Scholar]
- Wen, R.‐H. , Saghai Maroof, M.A. and Hajimorad, M.R. (2011) Amino acid changes in P3, and not the overlapping pipo‐encoded protein, determine virulence of Soybean mosaic virus on functionally immune Rsv1‐genotype soybean. Mol. Plant Pathol. 12, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Gong, J. , Li, H. , Li, C. , Wang, D. , Li, K. and Zhi, H. (2011) Identification of a novel Soybean mosaic virus isolate in China that contains a unique 5′ terminus sharing high sequence homology with Bean common mosaic virus . Virus Res. 157, 13–18. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Hajimorad, M.R. , Eggenberger, A.L. , Tsang, S. , Whitham, S.A. and Hill, J.H. (2009) Cytoplasmic inclusion of Soybean mosaic virus serves as a virulence determinant on Rsv3‐genotype soybean and a symptom determinant. Virology, 391, 240–248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Sequences of oligonucleotide primers used for site‐directed mutagenesis, reverse transcriptase‐polymerase chain reaction (RT‐PCR) and sequencing.


