Summary
Sclerotinia sclerotiorum causes a devastating disease in oilseed rape (Brassica napus) resulting in a tremendous yield loss worldwide. Studies on various host–pathogen interactions have shown that plant WRKY transcription factors are essential for defence. For the B. napus–S. sclerotiorum interaction, little direct evidence has been found with regard to the biological roles of specific WRKY genes in host resistance. In this study, we isolated a B. napus WRKY gene, BnWRKY33, and found that the gene is highly responsive to S. sclerotiorum infection. Transgenic B. napus plants overexpressing BnWRKY33 showed markedly enhanced resistance to S. sclerotiorum, constitutive activation of the expression of BnPR1 and BnPDF1.2, and inhibition of H2O2 accumulation in response to pathogen infection. Further, we isolated a mitogen‐activated protein (MAP) kinase substrate gene, BnMKS1, and found that not only can BnWRKY33 interact with BnMKS1, which can also interact with BnMPK4, using the yeast two‐hybrid assay, consistent with their collective nuclear localization, but also BnWRKY33, BnMKS1 and BnMPK4 are substantially and synergistically expressed in response to S. sclerotiorum infection. In contrast, the three genes showed differential expression in response to phytohormone treatments. Together, these results suggest that BnWRKY33 plays an important role in B. napus defence to S. sclerotiorum, which is most probably associated with the activation of the salicylic acid (SA)‐ and jasmonic acid (JA)‐mediated defence response and inhibition of H2O2 accumulation, and we propose a potential mechanism in which BnMPK4–BnMKS1–BnWRKY33 exist in a nuclear localized complex to regulate resistance to S. sclerotiorum in oilseed rape.
Keywords: Brassica napus, disease resistance, Sclerotinia sclerotiorum, WRKY33 transcription factor
Introduction
Oilseed rape (Brassica napus) is an agriculturally important oilseed crop. Its contribution to global oilseed production is considerable: approximately 64 million metric tonnes (MMT) were produced worldwide in 2012, with China producing about 14 MMT and Canada, the European Union, India and Australia being the other major contributors [Statistics Division of the Food and Agriculture Organization (FAOSTAT) data 2012, http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567#ancor]. In oilseed rape farming, Sclerotinia disease, caused by Sclerotinia sclerotiorum, is one of the most devastating diseases worldwide. It causes rotting of leaves, stems and pods, resulting in a tremendous loss in seed yield: for example, 10%–20% of yield losses every year, and up to 80% in severely infected fields in China (Wu et al., 2013). No immune or highly resistant germplasm in B. napus has been reported to date, and few genetic sources of host resistance to the pathogen are available to breeders (Liu S et al., 2005). Disease management depends heavily on the application of fungicides to the crop, but this may cause environmental contamination, increase farming costs and may be ineffective because of the difficulties associated with the application of sprays to thick canopies and the lack of suitable forecasting methods to enable the timely application of fungicides. Attempts have been made to engineer disease resistance in important crop plants. However, the molecular mechanisms of host defence to S. sclerotiorum remain poorly understood in the B. napus–S. sclerotiorum interaction, which restricts the engineering of resistance by transgenic approaches.
Plant defence responses include the transcriptional control of the expression of stress‐responsive genes (Chen and Chen, 2002; Chen et al., 2002; Maleck et al., 2000; Mysore et al., 2002), including a number of transcription factors (TFs) whose abundance is altered as a result of pathogen challenge. Plant WRKY TFs, proteins containing WRKY zinc‐finger motifs, regulate many plant defence responses to diverse biotic and abiotic stresses (Chen and Chen, 2002; Eulgem and Somssich, 2007). The WRKY domain is defined by the conserved amino acid sequence WRKYGQK, and WRKY proteins can be classified into three groups according to the number of WRKY domains and the characteristic of their zinc‐finger‐like motif (Eulgem et al., 2000). There is a large body of indirect evidence implicating plant WRKY proteins in plant defence responses to pathogens. For example, in Arabidopsis thaliana, it has been observed that 49 AtWRKY genes are regulated by Pseudomonas syringae or treatment with salicylic acid (SA), a defence signalling molecule (Chen and Chen, 2002; Dong et al., 2003). In Brassica napus, the transcript abundance of 13 tested BnWRKY genes is responsive to S. sclerotiorum and one or more hormones, including SA, jasmonic acid (JA) and abscisic acid (ABA) (Yang et al., 2009).
Transgenic approaches have provided further direct evidence that plant WRKY genes are involved in plant defence. In A. thaliana, transient expression of AtWRKY29 in leaves leads to reduced disease symptoms (Asai et al., 2002), and overexpression of AtWRKY18 and AtWRKY70 results in increased resistance to virulent pathogens (Chen and Chen, 2002; Li et al., 2004). In addition, the recessive RRS1‐R gene RRS1, which encodes the AtWRKY52 protein, confers resistance to the bacterial pathogen Ralstonia solanacearum (Deslandes et al., 2002; Lahaye, 2002). In particular, a study has shown that AtWRKY33 is required for resistance to Botrytis cinerea (Zheng et al., 2006). More recently, the overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis has been shown to enhance resistance to S. sclerotiorum (Chen et al., 2013).
In rice, plants overexpressing OsWRKY13 showed enhanced resistance to both bacterial Xanthomonas oryzae pv. oryzae (Xoo) and fungal Magnaporthe grisea infections (Qiu D et al., 2007, 2008). Similarly, overexpression of OsWRKY53 and OsWRKY45 led to enhanced resistance to M. grisea (Chujo et al., 2007; Shimono et al., 2007). Transgenic rice plants overexpressing OsWRKY03 and OsWRKY71 showed enhanced resistance to Xoo (Chujo et al., 2008; Liu XQ et al., 2005). In contrast, overexpression of OsWRKY62 compromised Xa21‐mediated resistance to Xoo (Peng et al., 2008). In tobacco, virus‐induced silencing of three NtWRKY genes compromises N gene‐mediated resistance to Tobacco mosaic virus (Liu et al., 2004).
Despite the obvious importance of plant WRKYs in various host–pathogen interactions, there have been no reports to date investigating the biological functions of WRKY TFs directly involved in host defence in the B. napus–S. sclerotiorum interaction, one of the most important host–pathogen interactions, by a transgenic approach. In Arabidopsis, AtWRKY33 plays an important role in resistance to Botrytis cinerea, a necrotrophic fungus closely related to S. sclerotiorum (Zheng et al., 2006), and AtWRKY33 has been found to interact with AtMKS1, an AtMPK4 substrate (Andreasson et al., 2005). Further, a previous study has shown that B. napus plants overexpressing BnMPK4, a B. napus homologous gene of AtMPK4, show enhanced resistance to S. sclerotiorum (Wang et al., 2009). Therefore, we investigated the involvement of BnWRKY33 in defence responses against S. sclerotiorum infection. We found that B. napus transgenic lines overexpressing BnWRKY33 showed enhanced resistance to S. sclerotiorum, and that both BnMPK4 and BnWRKY33 interact with BnMKS1. Mitogen‐activated protein (MAP) kinase signalling plays a central role in signal transduction mechanisms, linking upstream receptors and downstream targets and leading to the rapid activation of defence responses on recognition of invading pathogens. Our data suggest a potential mechanism in which BnMPK4–BnMKS1–BnWRKY33 exist in a nuclear localized complex to regulate resistance to the pathogen in oilseed rape.
Results
Sequence analysis of BnWRKY33 and its response to S. sclerotiorum infection in oilseed rape
A full‐length cDNA was cloned from a cDNA library of B. napus cv. Zhongshuang9 by a homology cloning approach, was consequently identified as a homologous gene of AtWRKY33 and was designated as BnWRKY33 (GenBank accession no. KF712488). BnWRKY33 encodes 483 deduced amino acid residues with a calculated molecular mass of 53 kDa and a predicted pI of 8.37.
The deduced amino acid sequence of BnWRKY33 was used in a blastp search of the National Center for Biotechnology Information (NCBI) database. As shown in Fig. S3 (see Supporting Information), alignment of the 10 top‐scoring matches with the BnWRKY33 sequence demonstrated that all 11 proteins are highly similar and that BnWRKY33 belongs to group 1 of the WRKY superfamily of plant TFs based on the presence of two WRKY domains (Eulgem et al., 2000). The two WRKY domains with their Cx4Cx22/23HxH zinc‐finger motifs are highly conserved in these sequences and the C‐terminal WRKY domain contains a consensus aspartic acid (Asp) residue at the same position, which may be a critical residue for interaction with proteins containing a conserved VQ motif (Cheng et al., 2012). In contrast, at the relatively conserved position of N‐terminal regions, these proteins contain four to five serine–proline (Ser‐Pro) residues that are potential MAP kinase phosphorylation sites (Liu and Zhang, 2004; Sharrocks et al., 2000). In addition, as expected from TFs, all these proteins contain highly conserved putative nuclear localization signals (NLSs) predicted by the psort ii program.
In order to determine whether BnWRKY33 functions in defence responses to S. sclerotiorum, we used the B. napus cv. Zhongshuang9, with higher resistance to S. sclerotiorum, to detect the expression of BnWRKY33 in response to pathogen infection, employing quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The results showed that BnWRKY33 transcription was induced rapidly and strongly by pathogen infection (Fig. 1). In particular, the transcript level of BnWRKY33 increased by an average of nine‐fold over noninoculated controls at 24 and 48 h after pathogen inoculation, indicating that BnWRKY33 is highly responsive to S. sclerotiorum in B. napus. These primary results led us to conduct further studies on BnWRKY33.
Figure 1.
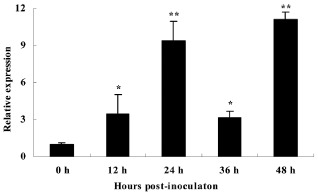
Response of BnWRKY33 to Sclerotinia sclerotiorum infection. Relative expression levels of BnWRKY33 in Brassica napus were determined by real‐time quantitative polymerase chain reaction at 0, 12, 24, 36 and 48 h post S. sclerotiorum inoculation. Values are means of three replicates. The error bars show the standard deviation. The significances of the gene expression differences between each time point and the 0‐h time point are indicated: **(Student's t‐test, P > 0.01) or *(Student's t‐test, P > 0.05).
Transgenic plants overexpressing BnWRKY33 show enhanced resistance to S. sclerotiorum
To further functionally characterize BnWRKY33, we overexpressed the gene in B. napus and investigated its possible function in resistance to S. sclerotiorum. The full‐length WRKY33 cDNA was cloned behind the cauliflower mosaic virus (CaMV) 35S promoter and transformed into B. napus plants (Fig. 2a). Hygromycin and PCR were used to screen BnWRKY33 transgenic lines. Three independent transgenic lines (6, 8 and 10) showed much higher levels of expression of the BnWRKY33 gene than the untransformed control by semi‐quantitative RT‐PCR analysis (Fig. 2b), and were used for further analysis.
Figure 2.
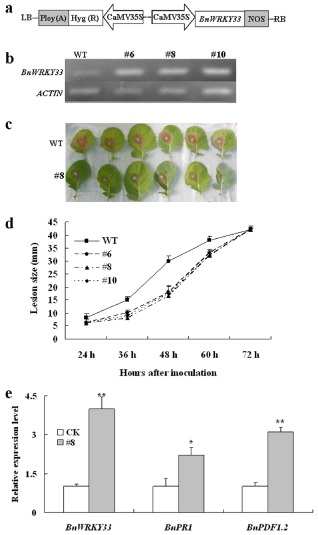
Characterization of BnWRKY33‐overexpressing lines. WT, untransformed wild‐type control; #6, #8 and #10, three independent BnWRKY33 transgenic T2 lines. (a) Diagram of the plasmids used in this study. CaMV35S, cauliflower mosaic virus 35S promoter; NOS, terminator. (b) Validation of BnWRKY33‐overexpressing lines at transcription levels revealed by reverse transcription‐polymerase chain reaction (RT‐PCR). (c) Disease responses of inoculated plants at 48 h post‐inoculation (hpi). Photographs were taken of leaves from three plants of WT and three hygromycin‐ and PCR‐positive plants of line 8. (d) Disease progression is shown from 24 to 72 hpi. Error bars indicate standard deviations. Differences in susceptibility between WT and the transgenic lines were significant (P < 0.05) from 36 to 60 hpi. (e) Relative expression levels of BnWRKY33, BnPDF1.2 and BnPR1 were quantified by real‐time qPCR. Values are means of three replicates. The error bars show the standard deviation. The significances of the gene expression differences between line 8 and WT (CK) are indicated: **(Student's t‐test, P > 0.01) or *(Student's t‐test, P > 0.05).
To test the effect of the transgene on resistance to S. sclerotiorum, T2 generation transgenic plants were selected by hygromycin screening and confirmed by PCR for the presence of the BnWRKY33 transgene. Leaves from these PCR‐positive plants and their untransformed controls at the six‐true‐leaf stage were used for inoculation with S. sclerotiorum mycelial plugs. The results indicated that BnWRKY33‐overexpressing plants developed less severe disease symptoms and sustained less tissue damage than untransformed controls (Fig. 2c). Investigation of disease progression showed that soft‐rotting necrosis occurred as early as 24 h post‐inoculation (hpi), and the lesion size differences between the transgenic plants and untransformed controls became apparent at 36 hpi and up to a maximum at 48 hpi [Figs 2d and S4 (see Supporting Information)], suggesting that transgenic plants did not support rapid lesion expansion when compared with the untransformed controls. The difference in the rate of lesion expansion suggests that BnWRKY33‐mediated defences appear to inhibit or delay the spread of the pathogen. Ultimately, disease symptoms were limited to leaves that had been inoculated and fungal spread virtually stopped after 72 hpi. These results show that overexpression of BnWRKY33 triggers plant resistance to S. sclerotiorum infection.
To investigate whether overexpression of BnWRKY33 leads to expression changes in defence marker genes, we detected the transcript abundance of BnPDF1.2 and BnPR1 using quantitative RT‐PCR. Although expression levels of BnWRKY33 in the transgenic plants were significantly elevated by about four‐fold, the levels of BnPR1 and BnPDF1.2 increased by about 2.5‐fold and 3.3‐fold, respectively (Fig. 2e). These results show that overexpression of BnWRKY33 in B. napus leads to activation of BnPR1 and BnPDF1.2 in the absence of induction by chemicals or pathogen treatment. The two genes PDF1.2 and PR1 are considered as marker genes for the JA‐mediated and SA‐mediated defence pathways, respectively (Brodersen et al., 2006; Penninckx et al., 1996; Thomma et al., 1998). Thus, our results suggest that BnWRKY33 overexpression may activate the SA‐ and JA‐mediated defence responses.
Transgenic plants overexpressing BnWRKY33 inhibit H2O2 accumulation in response to S. sclerotiorum infection
The generation of reactive oxygen species (ROS, e.g. O2 – and H2O2) is well documented as one of the earliest and most universal responses of plants to biotic stress, and H2O2 in particular, as a major component of ROS, is involved in a broad range of plant physiological processes, including disease defence. To further understand the resistance mechanism of transgenic plants overexpressing BnWRKY33, we collected inoculated leaves of transgenic plants and untransformed wild‐type (WT) controls at time points of 0, 6, 12, 18 and 24 hpi and stained them with 3,3‐diaminobenzidine (DAB) to visualize H2O2 accumulation. As shown in Fig. 3, a red–brown precipitate in inoculated leaves occurred in the untransformed WT control as early as 6 hpi but, in BnWRKY33 transgenic plants, a red–brown precipitate occurred at 18 hpi, suggesting that the overexpression of BnWRKY33 delayed the production of H2O2. The results demonstrate that transgenic plants overexpressing BnWRKY33 show inhibited H2O2 accumulation in response to S. sclerotiorum infection.
Figure 3.
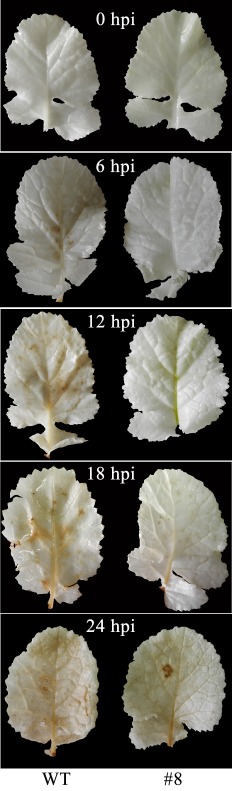
Transgenic plants overexpressing BnWRKY33 inhibit H2O2 accumulation in response to Sclerotinia sclerotiorum infection. In situ detection of H2O2 was performed using 3,3‐diaminobenzidine staining in the untransformed wild‐type (WT) control and the transgenic line 8 at 0, 6, 12, 18 and 24 h post‐inoculation (hpi).
Isolation of BnMKS1 from B. napus and its sequence analysis
A previous study has demonstrated that B. napus plants overexpressing BnMPK4 show enhanced resistance to S. sclerotiorum (Wang et al., 2009). In Arabidopsis, AtWRKY33 was found to interact with AtMKS1, a substrate of AtMPK4, and a homologue of BnMPK4. Thus, we investigated whether BnWRKY33 is capable of interacting with BnMKS1, which may, in turn, interact with BnMPK4. For B. napus, however, there is no information on the sequence of BnMKS1. To continue our studies, we first set out to clone the gene BnMKS1.
Brassica napus originated from a spontaneous hybridization of Brassica rapa L. (syn. campestris; AA, 2n = 20) and Brassica oleracea L. (CC, 2n = 18). Here, based on the MKS1 homologous sequence from B. oleracea, a full‐length cDNA was cloned from a cDNA library of B. napus by PCR, consequently identified as the homologous gene of AtMKS1 and designated as BnMKS1 (GenBank accession no. KF712487). BnMKS1 encodes 217 deduced amino acid residues with a calculated molecular mass of 23 kDa and a predicted pI of 6.29.
The deduced amino acid sequence of BnMKS1 was used in a blastp search of the NCBI database. As shown in Fig. S5 (see Supporting Information), alignment of the 11 top‐scoring matches with the BnMKS1 sequence showed that these sequences contain four conserved regions (domains I, II, III and IV). In the highly conserved domain II region, there is a conserved short FxxxVQxLTG motif, indicating that these proteins belong to members of the VQ protein family. The function of the VQ motif is unknown, but loss‐of‐function mutants and/or overexpressing lines for VQ genes are altered in their tolerance to abiotic stress or resistance to pathogen infection, suggesting that members of this protein family play important roles in responses to environmental conditions (Cheng et al., 2012). As would be expected from MAP kinase substrates, these proteins contain numerous Ser/tryptophan (Trp)‐Pro residues, the potential MAP kinase phosphorylation sites. BnMKS1 contains 10 Ser‐Pro sites, one each in domains I and II, two each in domains III and IV and three in the C‐terminal region. psort prediction showed that the protein sequence of BnMKS1 lacks putative NLSs and other predicted subcellular targeting sequences.
Interaction of BnMKS1, BnWRKY33 and BnMPK4
We used a Gal4 transcription activation‐based yeast two‐hybrid system to investigate the interaction of BnMKS1, BnWRKY33 and BnMPK4. We first fused whole BnWRKY33 protein with the DNA binding domain (BD) of Gal4 (BD‐W33), but found that the fusion protein itself had transcription‐activating activity and activated reporter genes in yeast cells. Thus, we fused Gal4 BD with whole BnMKS1 protein and did not find activation of reporter genes when the fusion protein was expressed in yeast cells. Then, we fused BnWRKY33 and BnMPK4, respectively, with the Gal4 activation domain (AD) in the vector pGADT7. Double transformed cells were selected on –Leu/–Trp dropout medium (medium lacking leucine and tryptophan), and interaction of proteins was checked on high‐stringency –Ade/–His/–Leu/–Trp dropout medium (medium lacking adenine, histidine, leucine and tryptophan) with β‐galactosidase. The results showed that BnMKS1 was capable of interacting with both BnWRKY33 and BnMPK4 (Fig. 4a). We also fused Gal4 BD with whole BnMPK4 protein to investigate the interaction between BnMPK4 and BnWRKY33, but found that they cannot interact (data not shown). The results suggest that BnMPK4–BnMKS1–BnWRKY33 exist in a complex to regulate the plant defence response.
Figure 4.
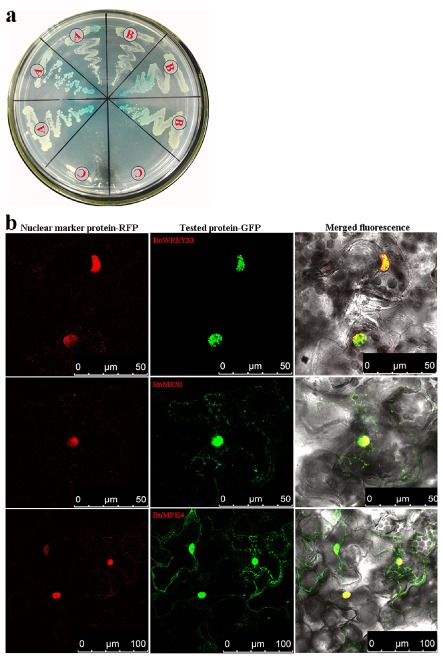
Interaction and subcellular localization of BnWRKY33, BnMKS1 and BnMPK4 proteins. (a) Interaction of BnWRKY33, BnMKS1 and BnMPK4 in yeast cells. The Gal4 DNA binding domain (BD)‐BnMKS1 fusion bait vectors were co‐transformed with the activation domain (AD)‐BnWRKY33 or BnMPK4 fusion prey vectors into yeast cells, and the transformant cells were assayed for LacZ reporter gene expression on high‐stringency (SD/–Ade/–His/–Leu/–Trp) plates: A, pGBKT7‐BnMKS1 + pGDAT7‐BnWRKY33; B, pGBKT7‐BnMKS1 + pGDAT7‐BnMPK4; C, BnMKS1 (pGBKT7‐BnMKS1) and SV40 large T‐antigen (pGADT7‐T) co‐transformants were used as negative controls. (b) Subcellular localization of BnWRKY33, BnMKS1 and BnMPK4. In planta localization in Nicotiana benthamiana leaves of protein‐green fluorescent protein (GFP), nuclear marker protein‐red fluorescent protein (RFP) and merged fluorescence from RFP and GFP.
Subcellular localization of BnWRKY33, BnMKS1 and BnMPK4
As BnWRKY33, BnMKS1 and BnMPK4 exist in a complex, the three proteins should co‐localize. Further, the function of BnWRKY33, as a TF, normally requires that it is localized in the nucleus, although TFs targeting chloroplasts, mitochondria or endoplasmic reticulum (ER) have also been identified (Schwacke et al., 2007). Thus, we verified the possible nuclear localization of the three proteins using a tobacco transient expression system.
Three constructs expressing gene fusions between the coding region of BnWRKY33, BnMKS1 or BnMPK4 and a sequence encoding an enhanced green fluorescent protein (eGFP) were produced. Each of the three constructs, together with another construct expressing a gene fusion of the red fluorescent protein (RFP) gene and a nuclear‐localized marker gene, was transiently co‐transferred by injection of transformed Agrobacterium tumefaciens cells into tobacco leaves. Five days after injection, GFP and RFP fluorescence in leaf cells was associated with regions of nuclei autofluorescence, and co‐transformation of each of the three genes and the marker gene led to yellow fluorescence (Fig. 4b). Our results indicate that BnWRKY33, BnMKS1 and BnMPK4 are primarily located in the nucleus, which is consistent with their ability to interact in vivo. Unexpectedly, BnMPK4 and BnMKS1, but not BnWRKY33, also appear to be located in the cytomembrane to some extent (Fig. 4b), suggesting that both BnMPK4 and BnMKS1 are involved in other plant physiological processes without the involvement of BnWRKY33.
Expression of BnWRKY33, BnMKS1 and BnMPK4 during the activation of plant defence responses
We also compared the expression of BnWRKY33, BnMKS1 and BnMPK4 in response to S. sclerotiorum infection in B. napus. As shown in Fig. 5, the three genes were induced by the pathogen and showed similar expression profiles. In particular, the expression of the three genes was substantially and synergistically enhanced at 24 and 48 hpi.
Figure 5.
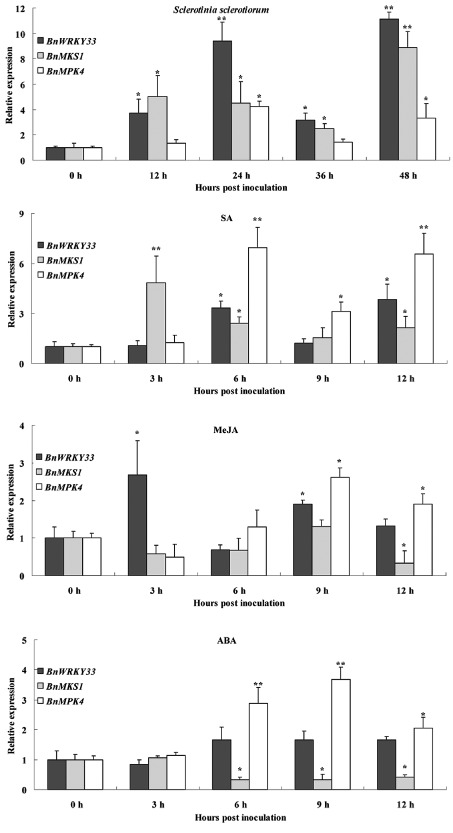
Expression of BnWRKY33, BnMKS1 and BnMPK4 during activation of plant defence responses. Expression of BnWRKY33, BnMKS1 and BnMPK4 in response to Sclerotinia sclerotiorum and treatment with chemicals. Plants were inoculated or treated with the various chemicals as described in Experimental procedures. SA, salicylic acid; MeJA, methyl jasmonate; ABA, abscisic acid. Values are means of three replicates. Error bars indicate standard deviations. The significances of the gene expression differences between each time point and the 0‐h time point are indicated: **(Student's t‐test, P > 0.01) or *(Student's t‐test, P > 0.05).
In contrast, in response to treatments with SA and methyl jasmonate (MeJA), two important defence signalling molecules that exhibit antagonistic interactions in their expression of mediated defence genes, the three genes showed different expression profiles, although their expression was up‐regulated together at certain time points. Interestingly, although the expression levels of BnWRKY33 during treatment with MeJA were elevated significantly by more than an average of 2.6‐fold over noninoculated controls at 3 h, the levels of BnMKS1 at the corresponding time points increased by more than 4.8‐fold during treatment with SA (Fig. 5). In addition, for the gene BnMPK4, involved in the repression of SA signalling (Wang et al., 2009), SA appeared to be more effective than MeJA in the activation of its expression. Its induction peak was at 3 hpi with 6.9‐fold greater expression than in non‐inoculated controls with SA treatment, whereas, with MeJA treatment, the peak was at 9 h with 2.6‐fold greater expression. During treatment with ABA, another defence signalling molecule that antagonizes JA in the mediation of defence gene expression (Anderson et al., 2004), the BnMPK4 gene was induced strongly at 6–9 h after ABA treatment. The expression of BnWRKY33 increased modestly within 6–12 h after ABA treatment. In contrast, the expression of BnMKS1 was reduced persistently by 0.33‐fold at 6–12 h after ABA treatment.
Discussion
In Arabidopsis and other plants, loss‐of‐function and gain‐of‐function studies have demonstrated that the WRKY genes are involved in plant defence against pathogens, but little direct evidence has been found for the biological roles of specific WRKY genes from B. napus in defence against the pathogen S. sclerotiorum. In this study, our results provide new data indicating that BnWRKY33 plays an important role in resistance to S. sclerotiorum in B. napus. Not only is the expression of BnMPK4 increased strongly (up to 11‐fold) by S. sclerotiorum infection, but it is also induced by SA and JA, two phytohormones associated with defence against the pathogen in oilseed rape (Wang et al., 2009). Further, overexpression of BnWRKY33 in transgenic plants significantly enhances the resistance to S. sclerotiorum. These new data, which indicate enhanced resistance to necrotrophic S. sclerotiorum in oilseed rape plants, are complementary to the positive effects observed in a study on the AtWRKY33 gene, which is required for resistance to two other necrotrophic fungi, Alternaria brassicicola and Botrytis cinerea, in Arabidopsis plants (Zheng et al., 2006). We observed that BnWRKY33 transgenic plants showed enhanced expression of PR1 and PDF1.2, suggesting the activation of both SA and JA signalling. These observations are similar to those reported for AtWRKY75, which plays a role in defence against S. sclerotiorum in Arabidopsis plants (Chen et al., 2013). It has been shown that SA‐ and JA‐mediated defence responses are involved in defence against S. sclerotiorum in B. napus (Wang et al., 2012), suggesting that the activation of the two signalling pathways may explain, at least in part, the positive effect of BnWRKY33 in B. napus resistance to S. sclerotiorum.
Overexpression of BnWRKY33 may involve redox control to enhance resistance to the necrotrophic pathogen S. sclerotiorum at the early stage. Previous studies have shown that a decrease in H2O2 levels in the host reduces the susceptibility to necrotrophic Botrytis cinerea (Govrin and Levine, 2000), and the disease severity of the pathogen increases when spraying various crops with H2O2 (Elad, 1992). In this study, BnWRKY33 transgenic plants showed inhibition of H2O2 accumulation in response to S. sclerotiorum infection and exhibited enhanced resistance to the pathogen. This is consistent with results showing that exposure of B. napus plants to H2O2 solution dramatically decreases the resistance to S. sclerotiorum (Wang et al., 2009). In the case of BnMPK4, BnMPK4 transgenic plants with decreased H2O2 levels exhibited enhanced resistance to S. sclerotiorum (Wang et al., 2009). Likewise, the Atmpk4 mutant with increased H2O2 levels (Nakagami et al., 2006) exhibits increased susceptibility to necrotrophic Alternaria brassicicola (Brodersen et al., 2006). The oxidative burst, the controlled release of O2 – and H2O2, is one of the earliest and most universal responses observed in plants following pathogen challenge (Apel and Hirt, 2004). This response occurs in both compatible and incompatible responses, and a resulting frequent outcome is programmed cell death (PCD), commonly known as the hypersensitive response (HR), which may deprive biotrophic pathogens of a food source, but may be beneficial to necrotrophic pathogens (Kim et al., 2008). For example, in biotrophic pathogen–host interactions, an increase in H2O2 can, in general, lead to enhanced host resistance. By contrast, in necrotrophic pathogen–host interactions, the cell death driven by ROS does not seem to enhance host resistance. Further, host cell death inhibition leads to markedly enhanced resistance to necrotrophic S. sclerotiorum in transgenic tobacco plants expressing negative regulators of mammalian apoptosis (Dickman et al., 2001); when ROS induction is inhibited, apoptotic‐like cell death induced by oxalic acid does not occur and the PCD response is required for disease development (Kim et al., 2008). More recently, it has been shown that the control of cell death governs the outcome of the S. sclerotiorum–plant interaction (Kabbage et al., 2013) and, once infection is established, the necrotrophic S. sclerotiorum induces the generation of plant ROS, leading to PCD of host tissue, the result of which is of direct benefit to the pathogen (Williams et al., 2011). Thus, taking these data together, it is suggested that the inhibition of H2O2 accumulation in transgenic plants overexpressing BnWRKY33 is another defence mechanism against necrotrophic S. sclerotiorum.
The positive role of BnWRKY33 in B. napus defence against S. sclerotiorum is also consistent with the possible regulation of BnWRKY33 by BnMPK4, a positive regulator of resistance to the pathogen (Wang et al., 2009). The C‐terminal WRKY domain of BnWRKY33, a member of group 1, contains a consensus Asp residue, a potential residue for interaction with VQ proteins (Cheng et al., 2012), such as BnMKS1, which contains a conserved VQ motif, representing the core of a protein–protein interaction domain (Cheng et al., 2012; Morikawa et al., 2002), referred to here as domain II. Simultaneously, in the N‐terminal region of BnMKS1 and throughout the sequence of BnWRKY33, there are four Ser‐Pro sites and 10 Ser‐Pro sites, respectively, indicating that the two proteins may be phosphorylated by MAP kinases, such as BnMPK4. Further, consistent with the collective nuclear localization of BnWRKY33, BnMKS1 and BnMPK4, yeast two‐hybrid analysis showed that BnWRKY33 can interact with BnMKS1, which, in turn, interacts with BnMPK4, and that BnMPK4 and BnWRKY33 do not interact directly with each other; this is in good agreement with the results of studies on their homologues in Arabidopsis (Qiu JL et al., 2008; Zheng et al., 2006). This could imply that BnMPK4 and BnWRKY33 exist in a complex that depends on BnMKS1, and that BnMKS1 may function as an adaptor linking BnMPK4 activity to the TF BnWRKY‐regulated defence responses. The interactions may also explain the nuclear localization of BnMKS1, which lacks predicted NLSs. Furthermore, the three genes BnWRKY33, BnMKS1 and BnMPK4 were synergistically expressed in response to S. sclerotiorum infection. Thus, these results indicate that BnWRKY33 functions downstream of BnMPK4 via BnMKS1 to regulate S. sclerotiorum defence responses in oilseed rape.
Qiu JL et al. (2008) have shown the existence of MKS1–WRKY33 complexes both before and after phosphatase treatments of MKS1, and that phospho‐mimics, non‐phosphorylatable and wild‐type forms of MKS1 bind WRKY33 equally well in yeast, suggesting that the complex exists independently of MKS1 phosphorylation. Interestingly, MPK4 also interacts equally well with these forms of MKS1 in yeast. Thus, the interaction may not involve MKS1 phosphorylation. One possible explanation could be that other unknown factors are involved in the interaction. The precise mechanism remains to be determined.
Sclerotinia sclerotiorum is considered to be one of the most damaging pathogens; it is capable of causing disease on at least 408 described species of plant from 278 genera in 75 families, including important crops, such as cotton, tomato, sunflower, dry bean, soybean and oilseed rape (Boland and Hall, 1994). The expression of plant WRKY genes associated with pathogen infection has been investigated in Arabidopsis and B. napus. In Arabidopsis plants, AtWRKY28 and AtWRKY75 were up‐regulated on challenge with oxalic acid, an important pathogenicity determinant of S. sclerotiorum. Moreover, overexpression of AtWRKY28 or AtWRKY75 in Arabidopsis enhances host resistance to the pathogen (Chen et al., 2013). In B. napus, Yang et al. (2009) investigated the expression profiles of 16 selected WRKY genes during the plant defence response and showed that 10 genes, including BnWRKY33, were induced after infection with S. sclerotiorum in the susceptible B. napus cv. Westar using qPCR. Using a B. napus oligonucleotide microarray, Zhao et al. (2007) investigated the patterns of gene expression in B. napus infected by S. sclerotiorum, and showed that BnWRKY33 was induced in a partially resistant cv. ZY821. These results are consistent with our observations in cv. Zhongshuang 9 with higher resistance than cv. ZY821 (Wang et al., 2004). The biphasic response of BnWRKY33 expression to S. sclerotiorum infection can be explained by the recent observation that defence against the pathogen in oilseed rape is associated with the sequential activation of SA and JA signalling (Wang et al., 2012), as the expression of BnWRKY33 can be induced by both SA and JA. Consequently, we further identified its role in the resistance to S. sclerotiorum through a transgenic approach in B. napus. The results from our and previous studies suggest that many WRKY genes are involved in host resistance to S. sclerotiorum.
It is interesting that BnMKS1 was induced rapidly and strongly at the earlier time point (3 h) after treatment with SA, whereas BnWRKY33 was induced by JA at the same time point. Further, the expression differences between BnMKS1 and BnWRKY33 were more obvious during treatment with ABA, another plant hormone showing antagonistic interactions with JA in defence gene expression. The expression of BnMKS1 was significantly down‐regulated by ABA, whereas BnWRKY33, together with BnMPK4, were induced at the corresponding time point. It has been shown that the SA‐mediated signalling pathway provides protection from biotrophic fungi, oomycetes and bacteria, such as Erysiphe orontii, Hyaloperonospora (formerly Peronospora) parasitica and Pseudomonas syringae, whereas the JA‐mediated signalling pathway activates defence responses against many necrotrophic fungi, such as Alternaria brassicicola and Botrytis cinerea (Glazebrook, 2005; Penninckx et al., 1996; Thomma et al., 1998). The role of ABA seems to depend not only on the specific interaction, but also on the pathogen's lifestyle (Adie et al., 2007). Thus, the different induction of BnWRKY33 and BnMKS1 by these defence signalling molecules suggests that the two genes are subject to different defence signal regulation, as BnMKS1, but not BnWRKY33, is also located in the cytomembrane (Fig. 4b). Indeed, in contrast with the resistance role of AtWRKY33 in defence against B. cinerea in Arabidopsis, constitutive expression of AtMKS1 confers susceptibility to the necrotrophic pathogen infection (Fiil and Petersen, 2011; Petersen et al., 2010) and loss‐of‐function Atmks1 mutants exhibit increased susceptibility to biotrophic P. syringae (Petersen et al., 2010).
In summary, this study demonstrates that transgenic oilseed rape overexpressing BnWRKY33 shows significantly enhanced resistance to S. sclerotiorum, which is most probably associated with the activation of the SA‐ and JA‐mediated defence responses and the inhibition of H2O2 accumulation. A potential mechanism is proposed in which BnMPK4–BnMKS1–BnWRKY33 exist in a nuclear localized complex to regulate resistance to S. sclerotiorum in oilseed rape. Data from this study may help to shed light on the mechanisms underlying the resistance to S. sclerotiorum and may provide critical clues to help curb crop diseases caused by S. sclerotiorum.
Experimental Procedures
Plant and fungal materials
The B. napus cultivar Zhongshuang9 was used in this study. Plants were grown in a plant growth room under the following growth conditions: 20 ± 2 °C with a photoperiod of 16 h light and 8 h dark at a light intensity of 44 μmol/m2/s and 60%–90% relative humidity. Fresh sclerotia of the fungus S. sclerotiorum, collected from oilseed rape stems in the field in Zhenjiang, China, were germinated to produce hyphal inoculum on potato dextrose agar (PDA).
Isolation of BnWRKY33 and BnMKS1 cDNA
Total RNA from B. napus leaf tissues was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). After removal of genomic DNA contamination by DNase I (TaKaRa, Dalian, China), 200 ng of poly(A)+ mRNA was converted into cDNA by MMLV Reverse Transcriptase (Promega, Madison, WI, USA). The cDNA template was used for PCR analysis subsequently. For the BnWRKY33 cDNA, a full‐length cDNA was obtained using the primers 5′‐ATGGATCCGTCGGAGTCTTT‐3′ and 5′‐TTATCCCCAAATATGACTGAACAA‐3′ according to the sequence of a B. napus WRKY33 homologue (Yang et al., 2009). BnMKS1 cDNA was acquired using the primers 5′‐ATGGCTGCTTCTTCCCTTC‐3′ and 5′‐TCAAGACAAAAACGAATCAAAG‐3′ based on the MKS1 homologue sequence from B. oleracea (Andreasson et al., 2007). The PCR products were inserted into pMD19‐T simple vector (TaKaRa) and then sequenced (Invitrogen, Shanghai, China). Subcellular localization of the deduced polypeptides was predicted by wolf psort (http://wolfpsort.org) (Horton et al., 2007). The molecular mass and theoretical pI (isoelectric point) of the polypeptides were determined using the Compute pI/MW tool (http://web.expasy.org) (Wilkins et al., 1999). Multiple‐aligned sequences were determined by ClustalX (Thompson et al., 1997), and GeneDoc was used to manually edit the results.
Plasmid construction for transgenic plant generation
To construct a vector for the constitutive expression of BnWRKY33, the vector pCAMBIA1300‐35S‐Nos was generated by inserting two fragments from the vector pEGAD containing the CaMV 35S promoter and CaMV Nos terminator into the EcoRI/KpnI and BamHI/HindIII sites of pCAMBIA1300, respectively. The PCR primers were 5′‐gaattcTTAATTAAGAGCTCGCATGCC‐3′ and 5′‐ggtaccGTCCCCGTGTTCTCTCCAA‐3′ for the insertion of the CaMV 35S promoter. The primers were 5′‐ggatccGAATTTCCCCGATCGTTCAA′ and 5′‐aagcttGATCTAGTAACATAGATGACACCGC‐3′ for the insertion of the CaMV Nos terminator. pCAMBIA1300‐35S‐Nos contains a hygromycin‐resistant gene in its T‐DNA region for selection of transgenic plants by hygromycin. Then, a 1.473‐kb full‐length BnWRKY33 cDNA was PCR amplified from its cDNA clone with the primers 5′‐ggtaccATGGCTGCTTCTTCCCTTC‐3′ and 5′‐ggatccTCAAGACAAAAACGAATCAAAG‐3′, and inserted into the KpnI/BamHI sites of pCAMBIA1300‐35S‐Nos, creating a BnWRKY33‐expressing vector 1300‐35S‐W33‐NOS. The inserted sequences were confirmed by restriction enzyme digestion and sequencing, and the 1300‐35S‐W33‐NOS vector was transformed into Agrobacterium tumefaciens (LBA4404). The plants were then grown in a protected field in Zhenjiang, China, and transformed by in planta Agrobacterium‐mediated transformation according to the procedure described by Wang et al. (2009). Three independent transformants overexpressing BnWRKY33 were examined as described in the Results section. Hygromycin‐resistant T2 generation plants were identified by PCR from the progeny of the primary transformants.
Plant inoculation
Mycelia of S. sclerotiorum were cultured on PDA. Agar plugs (3 mm in diameter) were excised from the edges of growing colonies and upended onto the adaxial surface of plant leaves of seedlings at the six‐leaf stage. Three independent T2 lines were screened with hygromycin (100 mg/L), and leaves of hygromycin‐resistant plants were collected for PCR detection. The PCR‐positive plants were used for inoculation. Inoculation of the detached leaves was performed as described previously (Dong et al., 2008). The experiment was in a randomized complete block design and was repeated three times, with 16 leaves from eight plants used in each replicate. Twenty hours after inoculation and at intervals thereafter, the lesion size was determined as the diameter of the lesion after S. sclerotiorum infection.
H2O2 detection by DAB staining
Leaves were soaked in DAB solution (1 mg/mL, pH 3.8) for 8 h in the dark at room temperature and then placed in acetic acid–glycerol–ethanol (1 : 1 : 1, v/v/v) and boiled for 5 min in a water bath. Subsequently, the solution was exchanged and the leaves were maintained in 60% glycerol. DAB was polymerized locally in the presence of H2O2, giving a visible brown stain.
Chemical treatments
Detached leaves were treated with SA (Mallinckrodt Baker, Deventer, the Netherlands) by dipping the leaves into a solution of 0.015% (v/v) Silwet L77 (Van Meeuwen Chemicals BV, Weesp, the Netherlands) containing 100 μm SA, as described by Leon‐Reyes et al. (2010). Control treatments were dipped in a solution containing 0.015% (v/v) Silwet L77. The MeJA solution was diluted to 100 μm and used to treat detached leaves as described by Thomma et al. (2000). Treatments without MeJA were used as controls.
RNA extraction, cDNA synthesis, semi‐quantitative reverse transcription and quantitative real‐time PCR (qRT‐PCR)
Total RNA was isolated using Trizol reagent. Total RNA (2 μg) was treated with RNase‐free DNase (Invitrogen, Carlsbad, CA, USA), and first‐strand cDNA was synthesized with oligo‐dT using a first‐strand cDNA Synthesis Kit (Invitrogen). For semi‐quantitative reverse transcription, B. napus BnACTIN was used as reference gene for internal control. For qRT‐PCR, BnACTIN and BnUBC21 (ubiquitin‐conjugating enzyme 21) were used as reference genes. The expression of the target genes and reference genes in plants was analysed by qRT‐PCR using SYBR green real‐time PCR master mix in an Eppendorf Mastercycler ep Realplex2 PCR system. Quantification of the threshold cycle (Ct) values in quantitative PCR analysis was achieved using the 2[–ΔΔC(t)] method (Livak and Schmittgen, 2001). Primers for BnACTIN (AF111812) were 5′‐GCTGACCGTATGAGCAAAG‐3′ and 5′‐AAGATGGATGGACCCGAC‐3′; primers for BnUBC21 (EV086936) were 5′‐CCTCTGCAGCCTCCTCAAGT‐3′ and 5′‐CATATCTCCCCTGTCTTGAAATGC‐3′; primers for BnWRKY33 were 5′‐AGAGGACGGTTACAACTGGAGAAA‐3′ and 5′‐TGTCGGACAGCTTGGGAAAG‐3′; primers for BnPDF1.2 (AY884023) were 5′‐CATCACCCTTCTCTTCGCTGC‐3′ and 5′‐ATGTCCCACTTGACCTCTCGC‐3′; primers for BnPR1 (AY623008) were 5′‐ATGCCAACGCTCACAACCA‐3′ and 5′‐CACGGGACCTACGCCTACT‐3′; primers for BnMKS1 were 5′‐GGAAGAAGCAGCCGAGT‐3′ and 5′‐GCGAGAACATCCCACCT‐3′; primers for BnMPK4 (EU581868) were 5′‐GCACGAAAGGATTGGCTAC‐3′ and 5′‐CGATGGGACGAAGAGGAG‐3′. These primer sets were tested by dissociation curve analysis and verified for the absence of nonspecific amplification (Fig. S1, see Supporting Information), and the efficiencies (E) of the primer‐specific PCR amplifications were tested (Fig. S2, see Supporting Information).
Yeast two‐hybrid assays
A yeast two‐hybrid assay was performed using the GAL4‐based two‐hybrid system adopted from Clontech (Clontech, Changping District, Beijing, China). BnMKS1 was PCR amplified from its cDNA clone with the gene‐specific primers 5′‐CGgaattcATGGATCCGTCGGAGTCT‐3′ and 5′‐AActgcagTTATCCCCAAATATGACTGAAC‐3′, and inserted into the EcoRI/PstI sites of pGBKT7, creating a pGBKT7‐BnMKS1 fusion construct. BnWRKY33 was cloned into the EcoRI/XhoI sites of pGDAT7 using PCR primers 5′‐CGgaattcATGGCTGCTTCTTCCCTT‐3′ and 5′‐AActcgagcTCAAGACAAAAACGAATCAAAG‐3′, creating a pGDAT7‐BnWRKY33 fusion construct. Likewise, the pGDAT7‐BnWMPK4 fusion construct was generated using PCR primers 5′‐CGgaattcATGTCGGCGGAGAACTGT‐3′ and 5′‐AActcgagcttaCTGAGGATTGAACTTGACTGTTT‐3′. The pGBKT7‐BnMKS1 construct was transformed into AH109 cells on selective medium (minimal SD base supplemented with 2% glucose and the required dropout solution, –Trp in this case), and it was verified that they did not autonomously activate reporter genes. Singly transformed strains were used as a yeast stock in which to transform the pGDAT7‐protein constructs. Double transformed cells were selected on –Leu/–Trp auxotrophy medium and the interaction of proteins was checked on high‐stringency –Ade/–His/–Leu/–Trp auxotrophy medium with β‐galactosidase. BnMKS1 (pGBKT7‐BnMKS1) and SV40 large T‐antigen (pGADT7‐T) co‐transformants were used as negative controls.
Subcellular localization of BnWRKY33, BnMKS1 and BnMPK4
The BnWRKY33, BnMKS1 and BnMPK4 genes were cloned into the destination vector pK7FWG2.0 to create a C‐terminal GFP fusion (Karimi et al., 2002) using the Gateway LR recombinase (Invitrogen, China). The nuclear marker gene INDEHISCENT (IND) was cloned into pK7FWG2.0 to create a C‐terminal RFP fusion (Tan et al., 2009). These constructs were introduced into Agrobacterium tumefaciens (LBA4404).
Agrobacterium infiltration into Nicotiana benthamiana leaves was performed as described previously (Wood et al., 2009). Cells of Agrobacterium tumefaciens strain were cultured at 28 °C for 2 days on Luria–Bertani (LB) agar medium containing 50 μg/mL kanamycin and 2.5 μg/mL tetracycline. The recombinant agrobacteria were grown in 10 mL LB liquid medium supplemented with appropriate antibiotics at 28 °C, and then harvested by centrifugation. The cell pellet was resuspended in buffer [10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES), pH 5.6, 10 mm MgCl2 and 150 μm acetosyringone], adjusted to a final optical density at 600 nm (OD600) of 0.6, and incubated for 3 h at room temperature before inoculation. Nicotiana benthamiana leaves were hand infiltrated using a needleless 1‐mL syringe with the transformed Agrobacterium mixed with Agrobacterium containing the gene‐silencing suppressor p19 (Voinnet et al., 2003). Inoculated plants were incubated at 26 °C in a growth chamber for 1–2 days. Five days after transformation, fluorescence was monitored by confocal microscopy. Fluorescence emission was at 510–540 nm for GFP and 580–584 nm for RFP, and excitation was at 488 nm for GFP and 543 nm for RFP.
Statistical analysis
Statistical analysis was performed using the SAS program (SAS Institute Inc., Cary, NC, USA). The data relating to lesion size were subjected to one‐way analysis of variance followed by a comparison of the means according to a significant difference test at P < 0.05. Using ΔCt values (target – reference), pairwise comparisons relating to PCR were conducted according to Student's t‐test at P < 0.05 or P < 0.01 under the assumption that variances are unequal.
Supporting information
Fig. S1 Specificity of quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) amplicons. (a) 1% agarose gel electrophoresis showing amplification of a single product of the expected size for all tested genes. M represents DL2000 DNA marker. (b) Dissociation curves with single peaks generated for all amplicons.
Fig. S2 The efficiencies (E) of the primer‐specific polymerase chain reaction (PCR) amplifications.
Fig. S3 BnWRKY33 and homologues. The amino acid sequence of BnWRKY33 was aligned with those of the 10 closest matching proteins from a blast search. Identical amino acids are shown in black boxes, and similar amino acids are shown in grey boxes. The WRKYGQK peptide stretch is shown in pink. The zinc‐finger‐like motifs in the two WRKY domains are shown in blue. Putative nuclear localization signals are shown in yellow. Putative mitogen‐activated protein (MAP) kinase phosphorylation sites (S/P) are shown in green. Critical residues for interaction with VQ proteins are shown in brown. Species abbreviations are as follows: Bn, Brassica napus; Bo, Brassica oleracea; Cr, Capsella rubella; ThUn‐protein, Thellungiella halophila unnamed protein; AtPu‐protein, Arabidopsis thaliana putative WRKY33; Al, Arabidopsis lyrata; Cr, Capsella rubella; RcTF, Ricinus communis transcription factor; Jc, Jatropha curcas.
Fig. S4 Disease progression of Sclerotinia sclerotiorum in BnWRKY33‐overexpressing lines and the untransformed wild‐type control. Photographs were taken of leaves from three wild‐type plants and three hygromycin‐ and PCR‐positive plants of line 8. WT, untransformed wild‐type control; #8, BnWRKY33 transgenic T2 line; hpi, h post‐inoculation.
Fig. S5 BnMKS1 and homologues. The amino acid sequence of BnMKS1 was aligned with those of the 10 closest matching proteins from a blast search. Identical amino acids are shown in black boxes, and similar amino acids are shown in grey boxes. Putative domains I, II, III and IV are underlined in the consensus. The FxxxVQxLTG motif is shown in pink. Putative mitogen‐activated protein (MAP) kinase phosphorylation sites (S/P) are shown in green. Species abbreviations are as follows: Bn, Brassica napus; Bo, Brassica oleracea; At, Arabidopsis thaliana; AlHy‐protein, Arabidopsis lyrata hypothetical protein; CrHy‐protein, Capsella rubella hypothetical protein; TcPu‐MKS1, Theobroma cacao putative MKS1; Gm, Glycine max; PtPr‐protein, Populus trichocarpa predicted protein; Ca, Cicer arietinum; Mt, Medicago truncatula; Fv, Fragaria vesca; RcPu‐MKS1, Ricinus communis putative MKS1.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (No. 31071672 and 31271760), a China Postdoctoral Science Foundation‐funded project (2011M500873 and 2013T60507), the Natural Science Fund for Colleges and Universities in Jiangsu Province of China (10KJB210001), the Jiangsu Province Postdoctoral Science Foundation Funded Project (1102130C) and Jiangsu University (09JDG061).
References
- Adie, B.A. , Perez‐Perez, J. , Perez‐Perez, M.M. , Godoy, M. , Sanchez‐Serrano, J.J. , Schmelz, E.A. and Solano, R. (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell, 19, 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.P. , Badruzsaufari, E. , Schenk, P.M. , Manners, J.M. , Desmond, O.J. , Ehlert, C. , Maclean, D.J. , Ebert, P.R. and Kazan, K. (2004) Antagonistic interaction between abscisic acid and jasmonate‐ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell, 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson, E. , Jenkins, T. , Brodersen, P. , Thorgrimsen, S. , Petersen, N.H. , Zhu, S. , Qiu, J.L. , Micheelsen, P. , Rocher, A. , Petersen, M. , Newman, M.A. , Nielsen, H.B. , Hirt, H. , Somssich, I. , Mattsson, O. and Mundy, J. (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24, 2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson, E. , Brodersen, P. , Jenkins, T. , Mundy, J. , Petersen, N.H. , Thorgrimsen, S. and Rocher, A. (2007) Plant disease resistance and SAR regulator protein. United States Patent No.: 2007/0271623 A1.
- Apel, K. and Hirt, H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.L. , Gomez‐Gomez, L. , Boller, T. , Ausubel, F.M. and Sheen, J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Boland, G.J. and Hall, R. (1994) Index of plant hosts of Sclerotinia sclerotiorum . Can. J. Plant Pathol. 16, 93–108. [Google Scholar]
- Brodersen, P. , Petersen, M. , Bjorn Nielsen, H. , Zhu, S. , Newman, M.A. , Shokat, K.M. , Rietz, S. , Parker, J. and Mundy, J. (2006) Arabidopsis MAP kinase 4 regulates salicylic acid‐ and jasmonic acid/ethylene‐dependent responses via EDS1 and PAD4. Plant J. 47, 532–546. [DOI] [PubMed] [Google Scholar]
- Chen, C. and Chen, Z. (2002) Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen‐induced Arabidopsis transcription factor. Plant Physiol. 129, 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Provart, N.J. , Glazebrook, J. , Katagiri, F. , Chang, H.S. , Eulgem, T. , Mauch, F. , Luan, S. , Zou, G. , Whitham, S.A. , Budworth, P.R. , Tao, Y. , Xie, Z. , Chen, X. , Lam, S. , Kreps, J.A. , Harper, J.F. , Si‐Ammour, A. , Mauch‐Mani, B. , Heinlein, M. , Kobayashi, K. , Hohn, T. , Dangl, J.L. , Wang, X. and Zhu, T. (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell, 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Liu, J. , Lin, G. , Wang, A. , Wang, Z. and Lu, G. (2013) Overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum . Plant Cell Rep. 32, 1589–1599. doi: 10.1007/s00299-013-1469-3. [DOI] [PubMed] [Google Scholar]
- Cheng, Y. , Zhou, Y. , Yang, Y. , Chi, Y.J. , Zhou, J. , Chen, J.Y. , Wang, F. , Fan, B. , Shi, K. , Zhou, Y.H. , Yu, J.Q. and Chen, Z. (2012) Structural and functional analysis of VQ motif‐containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 159, 810–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo, T. , Takai, R. , Akimoto‐Tomiyama, C. , Ando, S. , Minami, E. , Nagamura, Y. , Kaku, H. , Shibuya, N. , Yasuda, M. , Nakashita, H. , Umemura, K. , Okada, A. , Okada, K. , Nojiri, H. and Yamane, H. (2007) Involvement of the elicitor‐induced gene OsWRKY53 in the expression of defense‐related genes in rice. Biochim. Biophys. Acta, 1769, 497–505. [DOI] [PubMed] [Google Scholar]
- Chujo, T. , Kato, T. , Yamada, K. , Takai, R. , Akimoto‐Tomiyama, C. , Minami, E. , Nagamura, Y. , Shibuya, N. , Yasuda, M. , Nakashita, H. , Umemura, K. , Okada, A. , Okada, K. , Nojiri, H. and Yamane, H. (2008) Characterization of an elicitor‐induced rice WRKY gene, OsWRKY71. Biosci. Biotechnol. Biochem. 72, 240–245. [DOI] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Theulieres, F. , Hirsch, J. , Feng, D.X. , Bittner‐Eddy, P. , Beynon, J. and Marco, Y. (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1‐R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA, 99, 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman, M.B. , Park, Y.K. , Oltersdorf, T. , Li, W. , Clemente, T. and French, R. (2001) Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc. Natl. Acad. Sci. USA, 98, 6957–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, J. , Chen, C. and Chen, Z. (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. [DOI] [PubMed] [Google Scholar]
- Dong, X. , Ji, R. , Guo, X. , Foster, S.J. , Chen, H. , Dong, C. , Liu, Y. , Hu, Q. and Liu, S. (2008) Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta, 228, 331–340. [DOI] [PubMed] [Google Scholar]
- Elad, Y. (1992) The use of antioxidants (free radical scavengers) to control grey mould (Botrytis cinerea) and white mould (Sclerotinia sclerotiorum) in various crops. Plant Pathol. 41, 417–426. [Google Scholar]
- Eulgem, T. and Somssich, I.E. (2007) Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. , Rushton, P.J. , Robatzek, S. and Somssich, I.E. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Fiil, B.K. and Petersen, M. (2011) Constitutive expression of MKS1 confers susceptibility to Botrytis cinerea infection independent of PAD3 expression. Plant Signal. Behav. 6, 1425–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Govrin, E.M. and Levine, A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea . Curr. Biol. 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Horton, P. , Park, K.J. , Obayashi, T. , Fujita, N. , Harada, H. , Adams‐Collier, C.J. and Nakai, K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage, M. , Williams, B. and Dickman, M.B. (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum . PLoS Pathog. 9, e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M. , Inze, D. and Depicker, A. (2002) GATEWAY vectors for Agrobacterium‐mediated plant transformation. Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kim, K.S. , Min, J.‐Y. and Dickman, M.B. (2008) Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant–Microbe Interact. 21, 605–612. [DOI] [PubMed] [Google Scholar]
- Lahaye, T. (2002) The Arabidopsis RRS1‐R disease resistance gene—uncovering the plant's nucleus as the new battlefield of plant defense? Trends Plant Sci. 7, 425–427. [DOI] [PubMed] [Google Scholar]
- Leon‐Reyes, A. , Van der Does, D. , De Lange, E.S. , Delker, C. , Wasternack, C. , Va Wees, S.C. , Ritsema, T. and Pieterse, C.M. (2010) Salicylate‐mediated suppression of jasmonate‐responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta, 232, 1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Brader, G. and Palva, E.T. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate‐mediated and salicylate‐mediated signals in plant defense. Plant Cell, 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Wang, H. , Zhang, J. , Fitt, B.D. , Xu, Z. , Evans, N. , Liu, Y. , Yang, W. and Guo, X. (2005) In vitro mutation and selection of doubled‐haploid Brassica napus lines with improved resistance to Sclerotinia sclerotiorum . Plant Cell Rep. 24, 133–144. [DOI] [PubMed] [Google Scholar]
- Liu, X.Q. , Bai, X.Q. , Qian, Q. , Wang, X.J. , Chen, M.S. and Chu, C.C. (2005) OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res. 15, 593–603. [DOI] [PubMed] [Google Scholar]
- Liu, Y. and Zhang, S. (2004) Phosphorylation of 1‐aminocyclopropane‐1‐carboxylic acid synthase by MPK6, a stress‐responsive mitogen‐activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell, 16, 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. and Dinesh‐Kumar, S.P. (2004) Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N‐mediated resistance to tobacco mosaic virus. Plant J. 38, 800–809. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(–Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Maleck, K. , Levine, A. , Eulgem, T. , Morgan, A. , Schmid, J. , Lawton, K.A. , Dangl, J.L. and Dietrich, R.A. (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Morikawa, K. , Shiina, T. , Murakami, S. and Toyoshima, Y. (2002) Novel nuclear‐encoded proteins interacting with a plastid sigma factor, Sig1, in Arabidopsis thaliana . FEBS Lett. 514, 300–304. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S. , Crasta, O.R. , Tuori, R.P. , Folkerts, O. , Swirsky, P.B. and Martin, G.B. (2002) Comprehensive transcript profiling of Pto‐ and Prf‐mediated host defense responses to infection by Pseudomonas syringae pv. tomato . Plant J. 32, 299–315. [DOI] [PubMed] [Google Scholar]
- Nakagami, H. , Soukupová, H. , Schikora, A. , Zársk, V. and Hirt, H. (2006) A mitogen‐activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281, 38 697–38 704. [DOI] [PubMed] [Google Scholar]
- Peng, Y. , Bartley, L.E. , Chen, X. , Dardick, C. , Chern, M. , Ruan, R. , Canlas, P.E. and Ronald, P.C. (2008) OsWRKY62 is a negative regulator of basal and Xa21‐mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant, 1, 446–458. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A. , Eggermont, K. , Terras, F.R. , Thomma, B.P. , De Samblanx, G.W. , Buchala, A. , Metraux, J. , Manners, J.M. and Broekaert, W.F. (1996) Pathogen‐induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid‐independent pathway. Plant Cell, 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, K. , Qiu, J.L. , Lutje, J. , Fiil, B.K. , Hansen, S. , Mundy, J. and Petersen, M. (2010) Arabidopsis MKS1 is involved in basal immunity and requires an intact N‐terminal domain for proper function. PLoS ONE, 5, e14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, D. , Xiao, J. , Ding, X. , Xiong, M. , Cai, M. , Cao, Y. , Li, X. , Xu, C. and Wang, S. (2007) OsWRKY13 mediates rice disease resistance by regulating defense‐related genes in salicylate‐ and jasmonate‐dependent signaling. Mol. Plant–Microbe Interact. 20, 492–499. [DOI] [PubMed] [Google Scholar]
- Qiu, D. , Xiao, J. , Xie, W. , Liu, H. , Li, X. , Xiong, L. and Wang, S. (2008) Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol. Plant, 1, 538–551. [DOI] [PubMed] [Google Scholar]
- Qiu, J.L. , Fiil, B.K. , Petersen, K. , Nielsen, H.B. , Botanga, C.J. , Thorgrimsen, S. , Palma, K. , Suarez‐Rodriguez, M.C. , Sandbech‐Clausen, S. , Lichota, J. , Brodersen, P. , Grasser, K.D. , Mattsson, O. , Glazebrook, J. , Mundy, J. and Petersen, M. (2008) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke, R. , Fischer, K. , Ketelsen, B. , Krupinska, K. and Krause, K. (2007) Comparative survey of plastid and mitochondrial targeting properties of transcription factors in Arabidopsis and rice. Mol. Genet. Genomics, 277, 631–646. [DOI] [PubMed] [Google Scholar]
- Sharrocks, A.D. , Yang, S.H. and Galanis, A. (2000) Docking domains and substrate‐specificity determination for MAP kinases. Trends Biochem. Sci. 25, 448–453. [DOI] [PubMed] [Google Scholar]
- Shimono, M. , Sugano, S. , Nakayama, A. , Jiang, C.J. , Ono, K. , Toki, S. and Takatsuji, H. (2007) Rice WRKY45 plays a crucial role in benzothiadiazole‐inducible blast resistance. Plant Cell, 19, 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, X. , Zhang, L. and Xia, Z. (2009) Molecular cloning and characterization of a putative BnHEC3 gene in oilseed rape (Brassica napus). Int. J. Biol. 1, 71–77. [Google Scholar]
- Thomma, B.P. , Eggermont, K. , Penninckx, I.A. , Mauch‐Mani, B. , Vogelsang, R. , Cammue, B.P. and Broekaert, W.F. (1998) Separate jasmonate‐dependent and salicylate‐dependent defense‐response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA, 95, 15 107–15 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H. , Eggermont, K. , Broekaert, W.F. and Cammue, B.P.A. (2000) Disease development of several fungi on Arabidopsis can be reduced by treatment with methyl jasmonate. Plant Physiol. Biochem. 38, 421–427. [Google Scholar]
- Thompson, J.D. , Gibson, T.J. , Plewniak, F. , Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O. , Rivas, S. , Mestre, P. and Baulcombe, D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Zheng, Y. , Wang, X. and Yang, Q. (2004) Breeding of the Brassica napus cultivar Zhongshuang 9 with high‐resistance to Sclerotinia sclerotiorum and dynamics of its important defense enzyme activity. Sci. Agric. Sin. 37, 23–28. [Google Scholar]
- Wang, Z. , Mao, H. , Dong, C. , Ji, R. , Cai, L. , Fu, H. and Liu, S. (2009) Overexpression of Brassica napus MPK4 enhances resistance to Sclerotinia sclerotiorum in oilseed rape. Mol. Plant–Microbe Interact. 22, 235–244. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Tan, X. , Zhang, Z. , Gu, S. , Li, G. and Shi, H. (2012) Defense to Sclerotinia sclerotiorum in oilseed rape is associated with the sequential activations of salicylic acid signaling and jasmonic acid signaling. Plant Sci. 184, 75–82. [DOI] [PubMed] [Google Scholar]
- Wilkins, M.R. , Gasteiger, E. , Bairoch, A. , Sanchez, J.C. , Williams, K.L. , Appel, R.D. and Hochstrasser, D.F. (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. [DOI] [PubMed] [Google Scholar]
- Williams, B. , Kabbage, M. , Kim, H.‐J. , Britt, R. and Dickman, M.B. (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7, e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, C.C. , Petrie, J.R. , Shrestha, P. , Mansour, M.P. , Nichols, P.D. , Green, A.G. and Singh, S.P. (2009) A leaf‐based assay using interchangeable design principles to rapidly assemble multistep recombinant pathways. Plant Biotechnol. J. 7, 914–924. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Cai, G. , Tu, J. , Li, L. , Liu, S. , Luo, X. , Zhou, L. , Fan, C. and Zhou, Y. (2013) Identification of QTLs for resistance to Sclerotinia stem rot and BnaC.IGMT5.a as a candidate gene of the major resistant QTL SRC6 in Brassica napus . PLoS ONE, 8, e67740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. , Jiang, Y. , Rahman, M.H. , Deyholos, M.K. and Kav, N.N. (2009) Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 9, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Wang, J. , An, L. , Doerge, R.W. , Chen, Z.J. , Grau, C.R. , Meng, J. and Osborn, T.C. (2007) Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus . Planta, 227, 13–24. [DOI] [PubMed] [Google Scholar]
- Zheng, Z. , Qamar, S.A. , Chen, Z. and Mengiste, T. (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Specificity of quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) amplicons. (a) 1% agarose gel electrophoresis showing amplification of a single product of the expected size for all tested genes. M represents DL2000 DNA marker. (b) Dissociation curves with single peaks generated for all amplicons.
Fig. S2 The efficiencies (E) of the primer‐specific polymerase chain reaction (PCR) amplifications.
Fig. S3 BnWRKY33 and homologues. The amino acid sequence of BnWRKY33 was aligned with those of the 10 closest matching proteins from a blast search. Identical amino acids are shown in black boxes, and similar amino acids are shown in grey boxes. The WRKYGQK peptide stretch is shown in pink. The zinc‐finger‐like motifs in the two WRKY domains are shown in blue. Putative nuclear localization signals are shown in yellow. Putative mitogen‐activated protein (MAP) kinase phosphorylation sites (S/P) are shown in green. Critical residues for interaction with VQ proteins are shown in brown. Species abbreviations are as follows: Bn, Brassica napus; Bo, Brassica oleracea; Cr, Capsella rubella; ThUn‐protein, Thellungiella halophila unnamed protein; AtPu‐protein, Arabidopsis thaliana putative WRKY33; Al, Arabidopsis lyrata; Cr, Capsella rubella; RcTF, Ricinus communis transcription factor; Jc, Jatropha curcas.
Fig. S4 Disease progression of Sclerotinia sclerotiorum in BnWRKY33‐overexpressing lines and the untransformed wild‐type control. Photographs were taken of leaves from three wild‐type plants and three hygromycin‐ and PCR‐positive plants of line 8. WT, untransformed wild‐type control; #8, BnWRKY33 transgenic T2 line; hpi, h post‐inoculation.
Fig. S5 BnMKS1 and homologues. The amino acid sequence of BnMKS1 was aligned with those of the 10 closest matching proteins from a blast search. Identical amino acids are shown in black boxes, and similar amino acids are shown in grey boxes. Putative domains I, II, III and IV are underlined in the consensus. The FxxxVQxLTG motif is shown in pink. Putative mitogen‐activated protein (MAP) kinase phosphorylation sites (S/P) are shown in green. Species abbreviations are as follows: Bn, Brassica napus; Bo, Brassica oleracea; At, Arabidopsis thaliana; AlHy‐protein, Arabidopsis lyrata hypothetical protein; CrHy‐protein, Capsella rubella hypothetical protein; TcPu‐MKS1, Theobroma cacao putative MKS1; Gm, Glycine max; PtPr‐protein, Populus trichocarpa predicted protein; Ca, Cicer arietinum; Mt, Medicago truncatula; Fv, Fragaria vesca; RcPu‐MKS1, Ricinus communis putative MKS1.


