Summary
Victorin, the host‐selective toxin produced by the fungus Cochliobolus victoriae, induces programmed cell death (PCD) in victorin‐sensitive oat lines with characteristic features of animal apoptosis, such as mitochondrial permeability transition, chromatin condensation, nuclear DNA laddering and rRNA/mRNA degradation. In this study, we characterized a calcium‐binding protein, namely AsALG‐2, which might have a role in the victorin‐induced PCD. AsALG‐2 is homologous to the Apoptosis‐Linked Gene ALG‐2 identified in mammalian cells. Northern blot analysis revealed that the accumulation of AsALG‐2 transcripts increased during victorin‐induced PCD, but not during necrotic cell death. Salicylic acid, chitosan and chitin strongly activated the expression of general defence response genes, such as PR‐10; however, neither induced cell death nor the accumulation of AsALG‐2 mRNA. Pharmacological studies indicated that victorin‐induced DNA laddering and AsALG‐2 expression were regulated through similar pathways. The calcium channel blocker, nifedipine, moderately inhibited the accumulation of AsALG‐2 mRNA during cell death. Trifluoperazine (calmodulin antagonist) and K252a (serine‐threonine kinase inhibitor) reduced the victorin‐induced phytoalexin accumulation, but did not prevent the victorin‐induced DNA laddering or accumulation of AsALG‐2 mRNA. Taken together, our investigations suggest that there is a calcium‐mediated signalling pathway in animal and plant PCD in common.
Introduction
Host cell death occurs during many, but not all, interactions between plants and the pathogens that infect them. This cell death can be associated with disease resistance or susceptibility, dependent on the lifestyle of the pathogen (Greenberg and Yao, 2004). Under physiological conditions, the animal apoptotic response is elicited by specialized molecular networks comprising multiple signalling pathways, and appears to be pivotal for the shaping of lymphocyte repertoires and prevention of autoimmune diseases (Hünig and Schimpl, 1997). The suicide signal can be received and transferred into the cell through death receptors that initiate signalling cascades, which shortly lead to cell demise (Chinnaiyan and Dixit, 1997).
The process of programmed cell death (PCD) is essential to ensure a robust defence response against invading pathogens (Greenberg, 1996), and the activation of rapid cell death during the hypersensitive response (HR) can protect plants from at least some pathogens. However, excessive PCD compromises plant fitness (Yao and Greenberg, 2006). Recently, studies have indicated that cell death associated with various diseases might be a genetically programmed process (Greenberg, 1996). For example, Arabidopsis genes called ACD2 (accelerated cell death) and LSD1 (lesions simulating disease) activate the HR and multiple defences in the absence of a pathogen (Greenberg et al., 1994). Loss of the LSD1 and ACD1 genes leads to runaway cell death, and ACD2 modulates the extent of PCD triggered by P. syringae and protoporphyrin IX treatment (Yao and Greenberg, 2006). The product of the Tasseoseed2 (TS2) gene, which is required for cell death during sex determination, generates a steroid‐like molecule which might function as a signal to provoke a cell suicide programme (DeLong et al., 1993). AGD2 (aberrant growth and death2) has been shown to function in cell death and/or growth control, as well as the defence response, similar to that described for the function of NFkB (Song et al., 2004a, b).
In animal cells, many genes involved in apoptosis have been cloned and characterized. Recent findings have revealed the details of the mechanisms by which Bcl‐2 and its homologous proteins, such as Bcl‐XL, suppress cell death (Reed, 1996). The Caenorhabditis elegans cell death gene ced‐3 has been proposed to act as a cysteine protease in the initiation of PCD in C. elegans as well as in mammals (Miura et al., 1993). Several proteins involved in apoptosis have been cloned in mouse, such as ALG‐2 and ALG‐3 (Vito et al., 1996), and DAD1 (defender against apoptotic cell death) (Tanaka et al., 2001).
In plants, it is intriguing that toxin‐induced cell death might occur by a mechanism that shares some common features with apoptosis in animal cells (Ryerson and Heath, 1996) Recent studies have reported that At‐DAD1 and At‐DAD2, the two Arabidopsis thaliana homologues of DAD1, can suppress the onset of DNA fragmentation in A. thaliana, supporting the involvement of the endoplasmic reticulum in this form of plant PCD (Danon et al., 2004). This may suggest that plants and invertebrates share a DAD1‐mediated pathway to suppress PCD (Apte et al., 1995).
Using the differential display method, we isolated a phytotoxin‐induced oat cDNA homologous to the apoptosis‐linked gene‐2 (ALG‐2), which we named AsALG‐2 (Jang et al., 2002; Lacanà et al., 1997; Vito et al., 1996). Homologues of ALG‐2 have also been found in Drosophila (Tsuda et al., 2006), Suberites domuncula (Wiens et al., 2004), Lubomirskia baicalensis (Wiens et al., 2006), Xenopus tropicalis (2002), yeast (Chen and Sytkowski, 2005) and Homo sapiens (Satoh et al., 2002a, b). In this study, we report the molecular characterization of AsALG‐2 and discuss the correlation between the transcriptional activation of the AsALG‐2 gene and PCD in oat.
Results
Cloning of an ALG‐2‐like gene from oat leaves treated with the host‐specific toxin, victorin
To isolate oat genes involved in PCD, we treated victorin‐sensitive oat leaves (Iowa × 469) with different concentrations of victorin: at 0.25 ng/mL, at which victorin induces phytoalexin accumulation or activates general defence response genes (Mayama et al., 1986; Tada et al., 2001; Yang et al., 2004), and, at 5 ng/mL, at which victorin causes apoptotic cell death (Kusaka et al., 2004; Tada et al., 2001; Yao et al., 2002). We also treated the victorin‐insensitive oat line Iowa × 424 with victorin as a control. Reverse transcription‐polymerase chain reaction (RT‐PCR) was performed to amplify cDNAs from the victorin‐treated oat leaves using an RNAspectra™ Red Fluorescent mRNA differential Display System (GenHunter®, Nashville, TN, USA). In parallel with the occurrence of cell death, many differentially displayed cDNAs appeared in the victorin‐sensitive oat leaves treated with 5 ng/mL victorin, in comparison with those in other treatment combinations where no cell death was induced (data not shown).
To identify the differentially expressed cDNAs, we isolated 123 fragments and subjected them to sequencing analysis. We designated one of the clones AsALG‐2 (abbreviation for Avena sativa apoptosis‐linked gene 2), which has a calcium (Ca2+)‐binding motif and shows sequence similarity with apoptosis‐linked gene 2 (ALG‐2) identified in animals. To clone the full‐length cDNA of AsALG‐2, we performed 5'‐rapid amplification of cDNA ends (5'‐RACE), and the nucleotide sequence of the resulting clone was determined. AsALG‐2 encoded a polypeptide of 371 amino acid residues. The deduced amino acid sequence had 40% identity and about 62% similarity with ALG‐2 of Strongylocentrotus purpuratus (accession number XP_787000) and also had considerable similarities to ALG‐2 sequences in Bombyx mori (accession number NP_0010104187), Gallus gallus (accession number NP_419075), Homo sapiens (accession number NP_037364), Lubomirskia baicalensis (accession number CAJ12143) and Mus musculus (accession number NP_035181). AsALG‐2 is also highly homologous to the Ca2+‐binding proteins derived from the genome sequences of Oryza sativa, A. thaliana and Pisum sativum. Similar to the ALG‐2 proteins in animal cells, AsALG‐2 contains two Ca2+‐binding motifs similar to the canonical EF‐hand (Fig. 1; Vito et al., 1996). Based on the high degree of similarity, ALG‐2 proteins seem to be conserved during evolution.
Figure 1.

Inferred polypeptide sequence of the AsALG‐2 (Avena sativa apoptosis‐linked gene 2) protein. The two calcium‐binding consensus sequences are shown in bold.
AsALG‐2 expression is up‐regulated by PCD inducer
To understand how AsALG‐2 works in the apoptotic cell death pathway, we examined its transcriptional regulation under different conditions. Peeled oat leaves were treated with 5 ng/mL victorin and 20 μm calcium ionophore, which have been reported to induce apoptotic cell death (Tada et al., 2001). In victorin‐sensitive oat leaves treated with 5 ng/mL victorin, AsALG‐2 mRNA was detected as a single 1.5‐kb transcript as early as 2 h after treatment (data not shown). The accumulation of AsALG‐2 mRNA increased rapidly at 4 h, reached a maximum at 6 h and decreased thereafter (Fig. 2). When treated with a low concentration of victorin (0.25 ng/mL), which induced the accumulation of phytoalexin, AsALG‐2 mRNA was also detectable at a low level (data not shown). The AsALG‐2 mRNA was not detectable in healthy leaves, in victorin‐sensitive oat (Iowa × 469) leaves treated with water or in victorin‐insensitive oat (Iowa × 424) leaves treated with a high concentration of victorin (data not shown). The accumulation of AsALG‐2 mRNA was also strongly induced by another cell death inducer, calcium ionophore, and the pattern of induction was similar to that induced by victorin (Fig. 2). These results suggest that AsALG‐2 responds to a signal(s) of victorin or calcium ionophore during apoptosis. Previous studies have indicated that almost 100% of cells in the first layer coming into direct contact with victorin solution are killed 6 h after treatment (Yao et al., 2001). However, AsALG‐2 mRNA was detectable even at 24 h after victorin treatment; therefore, AsALG‐2 may be expressed in oat cells that are not in direct contact with victorin.
Figure 2.
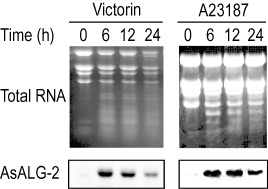
Expression of AsALG‐2 (Avena sativa apoptosis‐linked gene 2) during apoptotic cell death induced by the different apoptotic cell death inducers victorin and calcium ionophore. Primary leaf segments of the victorin‐sensitive oat line cv. Iowa × 469 were treated with victorin (5 ng/mL) and calcium ionophore (20 µm). Total RNAs were extracted from the treated leaf segments, separated on a denatured agarose gel and transferred onto a nylon membrane. Blots were hybridized with the cDNA probe of AsALG‐2.
The accumulation of AsALG‐2 is not mediated by non‐PCD triggers
To address whether the expression of the AsALG‐2 gene is induced in association with apoptotic cell death, we examined transcript levels of AsALG‐2 in oat cells treated with necrotic cell death inducers and elicitors that invoke a disease resistance response, but have very little activity for the induction of apoptotic cell death. We have shown previously that treatment with copper sulphate at 30 mm and heat shock (100 °C for 5 s) induces necrosis, but not apoptotic cell death, in oat cells (Hoat et al., 2006). Interestingly, no significant increase in AsALG‐2 mRNA accumulation was detected in CuSO4‐ or heat shock‐induced necrotic cell death (data not shown), suggesting that AsALG‐2 is not involved in the signalling pathway of necrosis.
Previous studies have indicated that the treatment of oat leaves with salicylic acid, chitosan and chitin at low concentrations does not induce apoptotic cell death, but activates general defence response genes (Yang et al., 2004). To investigate whether AsALG‐2 expression is up‐regulated under conditions in which the general defence response, but not apoptotic cell death, occurs, oat leaves were treated with salicylic acid, chitosan and chitin at concentrations of 0.5 μm, 0.1 mg/mL and 0.5 mg/mL, respectively, for the indicated times. Northern analysis showed that the mRNA expression of PR10, a marker gene of the general defence response, was strongly induced by all the treatments tested. However, interestingly, none of the elicitors, except victorin, induced the expression of AsALG‐2 mRNA (Fig. 3).
Figure 3.
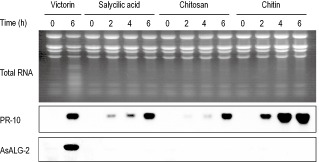
Effects of nonapoptotic inducers on the accumulation of AsALG‐2 (Avena sativa apoptosis‐linked gene 2) mRNA. Peeled oat leaf segments were treated with victorin and other nonapoptosis inducers (salicylic acid, 0.5 μm; chitosan, 0.1 mg/mL; chitin, 0.5 mg/mL) for the times indicated above the lanes. Total RNAs were extracted from the leaf segments and analysed by agarose gel electrophoresis, followed by ethidium bromide staining and Northern analysis with cDNA probes for PR‐10 and AsALG‐2.
Taken together, our investigation indicated that AsALG‐2 appeared to be specifically induced during apoptotic cell death but not necrosis and thus, not to be involved in phytoalexin biosynthesis or general defence response. It should be noted, however, that we did not detect AsALG‐2 mRNA in oat leaves infected with an incompatible race of the crown rust fungus (data not shown), where HR occurred. This could be explained by that relatively small number of HR cells in the infected leaves made AsALG‐2 mRNA accumulation below a detectable level or that victorin‐induced cell death and HR cell death were regulated by discrete pathways at least in part.
Signalling pathways involved in the induction of AsALG‐2 gene expression by victorin
To investigate the signalling pathways involved in the induction of AsALG‐2 mRNA during victorin‐induced cell death, we applied a pharmacological approach. Electrolyte leakage can be induced in oat by non‐cell death inducers, such as chitosan and chitin. In our hands, the level of leakage was not very well correlated with the extent of cell death assayed by fluorescein diacetate staining (data not shown). We used terminal deoxynucleotidyl transferase‐mediated uridine triphosphate nick end labelling (TUNEL) assay for the detection of cell death and for the analysis of the effect of the various pharmacological agents on the induction of cell death, DNA laddering and rRNA degradation (Hoat et al., 2006; Tada et al., 2001; Yao et al., 2002). The inhibitors used in this study were 5 mm nifedipine as a Ca2+ channel blocker, 5 μm cycloheximide as a de novo protein synthesis inhibitor, 100 μg/mL sodium dodecylsulphate (SOD) and 100 μm N‐acetylcysteine (NAC) as radical scavengers, 2 mm diphenyleneiodonium (DPI) as a NADPH (nicotinamide adenine dinucleotide phosphate, reduced form) oxidase–NO synthesis inhibitor, 150 μg/mL aprotinin as a serine protease inhibitor, 20 μm pepstatin A as an aspartic protease inhibitor and 500 μm E‐64 as a cysteine protease inhibitor. The results indicated that none of the pharmacological agents used inhibit victorin‐induced cell death (Tada et al., 2001). The addition of nifedipine, a Ca2+ channel blocker, to the test solution suppressed victorin‐induced DNA laddering (Tada et al., 2001), victorin‐induced hydroxyanthranilate N‐hydroxycinnamoyltransferase (AsHHT) and caffeoyl‐CoA 3‐O‐methyltransferase (AsCCoAOMT) (Yang et al., 2004) and inhibited the accumulation of AsALG‐2 mRNA (Fig. 4). These results suggest that Ca2+ plays a key role in many signalling pathways in oat, such as phytoalexin accumulation (Yang et al., 2004), DNA laddering (Tada et al., 2001), rRNA/mRNA degradation (Hoat et al., 2006) and AsALG‐2 mRNA induction.
Figure 4.
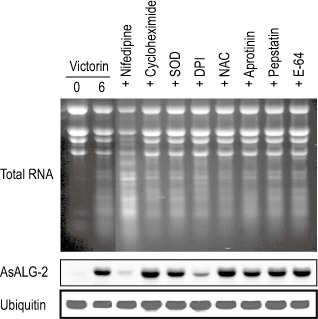
Effects of various inhibitors on the expression of AsALG‐2 (Avena sativa apoptosis‐linked gene 2) during victorin‐induced apoptotic cell death. Peeled oat leaf segments were treated with victorin and victorin plus the following inhibitors: 5 mm nifedipine as a calcium channel blocker; 5 μm cycloheximide as a de novo protein synthesis inhibitor; 100 μg/mL sodium dodecylsulphate (SOD) as a radical scavenger; 100 μm diphenyleneiodonium (DPI) as a NADPH (nicotinamide adenine dinucleotide phosphate, reduced form) oxidase–NO synthesis inhibitor; 2 mm N‐acetylcysteine (NAC) as a radical scavenger; 150 μg/mL aprotinin as a serine protease inhibitor; 20 μm pepstatin A as an aspartic protease inhibitor; and 500 μm E‐64 as a cysteine protease inhibitor. Total RNA was extracted from the leaf segments 6 h after treatment and analysed by agarose gel electrophoresis, followed by ethidium bromide staining and Northern analysis with cDNA probes for AsALG‐2.
Trifluoperazine and K252a have been reported to moderately reduce the victorin‐induced phytoalexin (Tada et al., 2001; Yang et al., 2004), but do not prevent the victorin‐induced DNA laddering in oats (Hoat et al., 2006; Tada et al., 2001). Interestingly, in parallel with victorin‐induced DNA laddering, none of the inhibitors suppressed the accumulation of AsALG‐2 mRNA (data not shown), suggesting that the phytoalexin biosynthesis and victorin‐induced cell death were, at least in part, regulated discrete pathways, and protein kinase and calmodulin were involved only in the former pathway. Moreover, none of the inhibitors, such as cycloheximide, NAC, pepstatin and E‐64, which show partial inhibition of victorin‐induced DNA laddering, but not RNA degradation (Hoat et al., 2006; Tada et al., 2001), were effective for the inhibition of AsALG‐2 expression (Fig. 4). Notably, DPI, a NADPH oxidase–NO synthesis inhibitor strongly suppressed the level of victorin‐induced accumulation of AsALG‐2 mRNA. Therefore, NADPH oxidase‐mediated reactive oxygen species (ROS) were probably involved in the AsALG‐2 induction pathway. Taken together, the above results suggest that the signalling pathways of victorin‐induced DNA laddering, rRNA degradation and accumulation of AsALG‐2 are involved in victorin‐induced cell death, mediated by different pathways with some overlapping upstream signals.
Discussion
AsALG‐2 is a Ca 2+‐dependent protein
In animal systems, ALG‐2 is a 22‐kDa Ca2+‐binding protein belonging to the penta‐EF (PEF)‐hand protein family, including peflin, sorcin and grancalcin, as well as the large and small subunits of calpains (Kitaura et al. 1999; Maki et al., 1997), which contain the Ca2+‐binding helix–loop–helix structure (Lo et al., 1999; Vito et al., 1996). Peflin has features common to other PEF proteins (dimerization and translocation to membranes) and may modulate the function of ALG‐2 in Ca2+ signalling (Kitaura et al., 2001). ALG‐2 is associated with several proteins, including annexin VII, annexin XI and Alix/AIP1 (ALG‐2‐interacting protein X/apoptosis‐linked gene‐2‐interacting protein), in a Ca2+‐dependent manner (Satoh et al., 2002a,b). Alix/AIP1, a protein that interacts with ALG‐2 in a calcium‐regulated fashion, and over‐expression of a deletion mutant of AIP1 protect HeLa and COS cells from apoptosis induced by serum starvation; thus, AIP1 might mediate, at least in part, the ALG‐2 requirement for apoptosis (Sadoul, 2006).
In the present study, it has been shown that AsALG‐2, an oat homologue to Ca2+‐binding proteins in other plant species, contains two EF‐hand Ca2+‐binding domains. To our knowledge, there is no phenotype for mutants in the orthologous gene in Arabidopsis. Victorin‐induced AsALG‐2 accumulation was strongly suppressed by the Ca2+ channel blocker nifedipine, suggesting that the accumulation of AsALG‐2 is dependent on Ca2+. DPI, an inhibitor of NADPH oxidase, showed a strong impact on the accumulation of victorin‐induced AsALG‐2 (Fig. 4). In animal cells, DPI has been shown to block thrombospondin 2 (TSP2) synthesis, which is a matricellular protein controlling the apoptosis–proliferation balance in endothelial cells. Over‐expression of a constitutively active mutant of Rac (RacV12) specifically increases TSP2 mRNA levels without affecting TSP1 in human aortic endothelial cells (HAECs). Moreover, TSP2 induction by RacV12 is dependent on ROS production, as gp91ds‐tat peptide, an inhibitor of NADPH oxidase, and the flavoprotein inhibitor DPI block TSP2 synthesis (Lopes et al., 2003). We have shown previously that calcium influx is the initial and crucial event that activates the signal transduction triggered by victorin in sensitive oat lines (Yang et al., 2004). Ca2+ is the simplest messenger, as it requires no chemical synthesis or modification, and the most versatile messenger, as it exerts its effects at numerous locations inside and outside cells. However, the understanding of intracellular Ca2+ signalling vastly eclipses what is known about extracellular Ca2+ ‘signalling’ (Hofer and Brown, 2003). In many experimental systems, it has been shown that an increased intracellular Ca2+ concentration, generated, for instance, by ionophores, induces apoptotic cell death (Nicotera et al., 1990). The removal of extracellular Ca2+ or buffering of intracellular Ca2+ can prevent DNA fragmentation and apoptotic body formation (Nicotera et al., 1990). Transiently elevated Ca2+ concentrations are essential for several pathways leading to apoptosis, such as glucocorticoid‐mediated (McConkey et al., 1989a) and T‐cell receptor‐mediated (McConkey et al., 1989b) T‐cell death. Several lines of evidence have demonstrated that alterations in intracellular calcium play important roles during apoptosis. Mostly, DNA laddering, a general hallmark of apoptosis, is a Ca2+‐dependent event that requires a Ca2+‐activated endonuclease (Cohen and Duke, 1984). It has also been demonstrated that calmodulin and its target proteins, such as calcineurin, Ca2+‐dependent kinases and the Ca2+‐dependent protease calpain, play important roles in PCD processes (Kawai et al., 1999). In animal systems, it has been shown that ALG‐2 is physiologically dispensable for apoptotic responses induced by the T‐cell receptor, Fas and glucocorticoid, and that ALG‐2 functions in regulating fundamental cellular processes in response to Ca2+ mobilization, including its involvement in signal transduction, gene expression or membrane trafficking (Jang et al., 2002; Shibata et al., 2004). Caspases are activated in ALG‐2‐depleted cells on stimulation with these reagents, suggesting that ALG‐2 exerts its function downstream or independent of protease activity in the apoptotic signalling pathway (Lacanà et al., 1997); moreover, it has another function as an alleged proapoptotic or as a player in a survival pathway (La Cour et al., 2003).
AsALG‐2 is highly associated with victorin‐induced apoptotic cell death in oats
In animals, ALG‐2 has been shown to bind to the death domain of Fas and to dissociate from Fas during Fas‐mediated apoptosis in Jurkat cells; moreover, the 22‐kDa ALG‐2 protein is cleaved and becomes a 19‐kDa protein after Fas activation (Jung et al., 2001). The authors also indicated the connection of ALG‐2 to apoptosis by its direct interaction with Fas, and enlisted ALG‐2 as a new member of the post‐translationally modified proteins during Fas‐mediated apoptosis. Interestingly, both the T‐cell receptor‐ and Fas‐stimulated lysates from ALG‐2 clones efficiently cleave human PARP from Jurkat cell lysates in vitro, suggesting that ALG‐2 exerts its function at a distal step of the apoptotic programme common to different pathways, where several death signals converge (Lacanà et al., 1997). It has been suggested that ALG‐2 might act downstream of caspase activation because ALG‐2 depletion does not affect the activation of caspases. As ALG‐2 depletion blocks apoptosis, it implies that ALG‐2 dissociates the activation of caspases and the execution of apoptosis, or is an essential molecule to complete apoptosis (Lacanà et al., 1997).
In this study, we have shown that oat's AsALG‐2 cDNA is homologous to the Ca2+‐binding proteins in A. thaliana, O. sativa and P. sativum; however, the involvement of the Ca2+‐binding protein in apoptosis in plant cells still remains to be answered. Under the conditions presented in this study, the victorin‐induced accumulation of AsALG‐2 was mostly concomitant with other macromolecular markers of apoptosis, such as nuclear DNA laddering and rRNA degradation. AsALG‐2 expression was rapidly and strongly induced by inducers of apoptotic cell death (Fig. 2). In oat–victorin interaction, the expression pattern of the AsALG‐2 gene was not similar to that of PR‐10 (Fig. 3). The up‐regulation of AsALG‐2 reached a maximum at 6 h and the down‐regulation thereafter was closely correlated with the occurrence of cell death after victorin treatment, in which almost 100% of the first layer of cells died (Hoat et al., 2006; Tada et al., 2001; Yao et al., 2002). Interestingly, AsALG‐2 mRNA was not induced by necrotic cell death inducers, such as 30 mm CuSO4 and heat shock (data not shown), or other non‐cell death triggers, including salicylic acid, chitosan and chitin (Fig. 3). These results suggest that the up‐regulation of AsALG‐2 is highly associated with PCD and could be involved in the signalling pathway of apoptotic cell death in oat cells. However, we could not detect the expression of AsALG‐2 mRNA during incompatible and compatible interactions between oat and crown rust fungi (data not shown). Previous studies have indicated that rRNA/mRNA degradation is also not detected during cell death induced by incompatible races of the crown rust fungus, whereas nuclear DNA laddering is detected (Hoat et al., 2006). As apoptosis includes a variety of different steps, the victorin‐induced PCD in Iowa × 469 cells and crown rust‐induced PCD in Shokan 1 cells may represent different ends of this spectrum (Schwartz et al., 1993). Indeed, in some cases, ALG‐2 expression is not enhanced in apoptotic cells; therefore, the activity of AsALG‐2 might be regulated by protein modification or ligand interaction (Krebs, 1998). Our results may suggest that plants show at least two types of PCD (Greenberg, 1996). It is important to investigate whether the inhibition of AsAGL2 compromises apoptotic cell death. However, to our knowledge, no gene targeting or feasible RNAi system has been reported in oat to date. A possible approach may involve the use of Barley stripe mosaic virus for virus‐induced gene silencing (VIGS) in oat. We hope that we will be able to establish a VIGS system in our laboratory and to address the above questions in a future paper.
In conclusion, our data suggest that AsALG‐2 may be involved in the signalling pathway of PCD in oat. This supports the hypothesis that animal and plant PCD may share a calcium‐mediated signalling pathway, at least in part.
Experimental Procedures
Plant materials and growth conditions
Oat (Avena sativa L.) cv. × 469, a victorin‐susceptible line, and cv. × 424, a victorin‐resistant line, were grown in a growth chamber for 7 days under a 16‐h photoperiod at 20 °C, as described previously (Mayama et al., 1986).
Elicitor and inhibitor treatment
The host‐selective toxin, victorin, kindly provided by T. J. Wolpert (Oregon State University, Corvallis, OR, USA), was used at concentrations of 0, 0.25 and 5 ng/mL. The calcium ionophore (Nacalai Tesque, Kyoto, Japan) was used at a concentration of 20 μm. The following inhibitors were used in the present study: 15 mm ZnCl2 (Nacalai Tesque) as a Ca2+ antagonist; 5 μm K252a (Sigma, St. Louis, MO, USA) as a serine/threonine kinase inhibitor; 5 mm nifedipine as a Ca2+ channel blocker; 5 μm cycloheximide (Nacalai Tesque) as a de novo protein synthesis inhibitor; 100 μg/mL SOD and 100 μM NAC as radical scavengers; 2 mm DPI (Sigma) as a NADPH oxidase–NO synthesis inhibitor; 150 μg/mL aprotinin (Nacalai Tesque) as a serine protease inhibitor; 20 μm pepstatin A (Nacalai Tesque) as an aspartic protease inhibitor, and 500 μM E‐64 (Nacalai Tesque) as a cysteine protease inhibitor. The epidermal layer was carefully peeled from the leaves, and 5‐cm segments, taken at 1–6 cm from the leaf tip, were floated on distilled water for 2 h. The peeled leaf segments were then floated with the surface in contact with the test solution or water in glass Petri dishes. Inhibitors were co‐incubated with victorin (or water for the control) at the concentrations mentioned above. After incubation for the indicated times under light conditions in the growth chamber, the treated samples were used for DNA or RNA analysis.
Preparation and analysis of DNA and RNA
Genomic DNA was extracted using the Plant Genomic DNA Mini Kit (Viogene, Sunnyvale, CA, USA). DNA was fractionated on a 1% agarose gel, stained with 0.5 μg/mL ethidium bromide and photographed with an Image Saver AE‐6905C (ATTO Corporation, Tokyo, Japan). Total RNA was extracted using the RNeasy® Plant Mini Kit (Qiagen, Gaithersburg, MD, USA) and separated on a 1.2% formaldehyde–agarose gel. Perfect RNA™ Markers, 0.2–10 kb, purchased from Novagen, Darmstadt, Germany, were used for accurate size determination. After electrophoresis, total RNA was analysed by ethidium bromide staining, and transferred to a Hybond‐N+ membrane (Amersham Biosciences UK Limited, Little Chalfont, Buckinghamshire, UK). The blot was pre‐hybridized in ULTRAhyb Hybridization Buffer (Ambion, Austin, TX, USA) for 2 h at 70 °C, followed by hybridization with a digoxigenin (DIG)‐labelled probe (Roche Diagnostics GmbH, Mannheim, Germany) for an additional 16 h at 70 °C. The blot was subsequently washed twice in 2 × saline sodium citrate (SSC) containing 0.1% SDS for 20 min at 70 °C, and then twice in 0.1 × SSC containing 0.1% SDS for 20 min at 70 °C. The hybridization signals were detected using anti‐DIG and CDP‐Star Ready to use (Roche Diagnostics GmbH), according to the manufacturer's guide.
Differential display of mRNA
Total RNA, extracted from the oat lines Iowa × 469 and Iowa × 424 treated with victorin for 4 h, was used for RT reaction. For each RNA sample, three RT reactions were set up, each containing one of the three different H‐T11M primers anchored (Table 1) with a poly(A+) tail (where H is AAGCTT; M is A, C or G). Each 20‐μL reaction mixture contained 9.4 μL of RNase‐free water, 4 μL of 5 × RT buffer, 1.6 μL of 250 μm deoxynucleoside triphosphates (dNTPs), 2 μL of 2 μm H‐T11M primer, 200 ng of total RNA and 1 μL (100 units) MMLV reverse transcriptase (GenHunter®). Reaction was performed in a programmed thermocycler at 65 °C for 5 min, 37 °C for 60 min, 75 °C for 5 min, and soaked at 4 °C. After the tubes had been incubated at 37 °C for 10 min, 1 μL of reverse transcriptase was added to each tube and quickly mixed by tapping before incubation with MMLV reverse transcriptase. The products of RT were purified by 1 volume of phenol–chloroform extraction, followed by 1 volume of chloroform extraction and ethanol precipitation.
Table 1.
Synthetic primers used for differential display polymerase chain reaction (DDPCR).
| Name | Sequence (5'–3’) |
|---|---|
| H‐AP1 | AAGCTTGATTGCC |
| H‐AP2 | AAGCTTCGACTGT |
| H‐AP3 | AAGCTTTGGTCAG |
| H‐AP4 | AAGCTTCTCAACG |
| H‐AP5 | AAGCTTAGTAGGC |
| H‐AP6 | AAGCTTGCACCAT |
| H‐AP7 | AAGCTTAACGAGG |
| H‐AP8 | AAGCTTTTACCGC |
| H‐AP9 | AAGCTTCATTCCG |
| H‐AP10 | AAGCTTCCACGTA |
| H‐AP11 | AAGCTTCGGGTAA |
| H‐AP12 | AAGCTTGAGTGCT |
| H‐AP13 | AAGCTTCGGCATA |
| H‐AP14 | AAGCTTGGAGCTT |
| H‐AP15 | AAGCTTACGCAAC |
| H‐AP16 | AAGCTTTAGAGCG |
| H‐AP25 | AAGCTTTCCTGGA |
| H‐AP26 | AAGCTTGCCATGG |
| H‐AP27 | AAGCTTCTGCTGG |
| H‐AP28 | AAGCTTACGATGC |
| H‐AP29 | AAGCTTAGCAGCA |
| H‐AP30 | AAGCTTCGTACGT |
| H‐AP31 | AAGCTTGGTGAAC |
| H‐AP32 | AAGCTTCCTGCAA |
Differential display PCR (DDPCR)
Two microlitres from the 20 μL of the first cDNA reaction were taken for DDPCR. Each PCR was carried out using the arbitrary primer H‐APi and the primer used in RT (Table 1). The PCR programme was carried out as follows: 40 cycles at 94 °C for 30 s, 40 °C for 2 min and 72 °C for 1 min. The amplified cDNA subpopulations of the 3'‐termini of mRNAs, as defined by the pair of primers, were size fractionated by a 6% DNA sequence gel. For each PCR, 3.5 μL of sample was incubated with 2 μL of loading dye [99% formamide, 1 mm ethylenediaminetetraacetic acid (EDTA), pH 8.0, 0.009% xylene cyanole FF and 0.009% bromophenol blue] at 80 °C for 2 min before loading, and then electrophoresed for 4–5 h at 1000 V.
Silver staining of gel and DNA recovery
The gel was immediately stained by a Silver Sequence DNA Sequencing System Kit (Promega, Madison, WI, USA) following the manufacturer's instructions. Bands of differentially expressed cDNA fragments were cut out with a clean razor blade and transferred to a 1.5‐mL lock‐top tube filled with 100 μL of dH2O. The gel slice was soaked for 10 min at room temperature, boiled for 30 min to elute DNA from the gel slice and collected by centrifugation. The extracts were employed as template for re‐amplification with the primer combination and PCR conditions using Pfu‐Turbo or KOD Plus (TOYOBO, Osaka, Japan). After electrophoresis, the specific bands were cut from a 1.5% LM agarose gel and purified using a MiniElute Gel Extraction Kit (Qiagen, Cat. No. 28606) following the manufacturer's instructions. The amplicon was ligated into the EcoRV site of pBluescript SKII+ using a DNA ligation Kit Ver. 2 (TaKaRa, Osaka, Japan, Cat. No. 6022).
Sequencing
Cycle sequencing reactions were performed using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Perkin‐Elmer, Waltham, MA, USA). The reaction products were purified by ethanol precipitation and analysed using the ABI ORISM 310 Genetic Analyser (Applied Biosystems, Carlsbad, CA, USA). Sequence homology was searched through the blast program.
Cloning of a full‐length AsALG‐2 cDNA clone
To clone a full‐length AsALG‐2 cDNA, we used a GeneRacer™ Kit (Invitrogen, Warrington, UK, Cat. No. L1500‐01) according to the manufacturer's instructions.
Acknowledgements
We are grateful to T. J. Wolpert (Oregon State University, Corvallis, OR, USA) for providing us with purified victorin. This work was supported in part by a Grant‐in‐Aid for Scientific Research and Special Coordination Funds for Promoting Science and Technology and by fellowships to Trinh Xuan Hoat from the Ministry of Education, Science, Sports, and Culture of Japan.
References
- Apte, S.S. , Mattei, M.G. , Seldin, M.F. and Olsen, B.R. (1995) The highly conserved defender against the death 1 (DAD1) gene maps to human chromosome 14q11‐q12 and mouse chromosome 14 and has plant and nematode homologs. FEBS Lett. 363, 304–306. [DOI] [PubMed] [Google Scholar]
- Chen, C. and Sytkowski, A.J. (2005) Apoptosis‐linked gene‐2 connects the Raf‐1 and ASK1 signalings. Biochem. Biophys. Res. Commun. 333, 51–57. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan, A.M. and Dixit, V.M. (1997) Portrait of an executioner: the molecular mechanism of Fas/APO‐1‐induced apoptosis. Semin. Immunol. 9, 69–76. [DOI] [PubMed] [Google Scholar]
- Cohen, J.J. and Duke, R.C. (1984) Glucocorticoid activation of a calcium dependent endonuclease in thymocyte nuclei leads to cell death. J. Immunol. 132, 38–42. [PubMed] [Google Scholar]
- Danon, A. , Rotari, V.I. , Gordon, A. , Mailhac, N. and Gallois, P. (2004) Ultraviolet‐C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase‐like activities and which can be suppressed by caspase inhibitors, p35 and Defender against Apoptotic Death. J. Biol. Chem. 279, 779–787. [DOI] [PubMed] [Google Scholar]
- DeLong, A. , Calderon‐Urrea, A. and Dellaporta, S.L. (1993) Sex determination gene TASSELSEED2 of maize encodes a short‐chain alcohol dehydrogenase required for stage‐specific floral organ abortion. Cell, 74, 757–768. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. (1996) Programmed cell death: a way of life for plants. Proc. Natl. Acad. Sci. USA, 93, 12 094–12 097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J.T. and Yao, N. (2004) The role and regulation of programmed cell death in plant–pathogen interactions. Cell. Microbiol. 6, 201–211. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. , Guo, A. , Klessig, D.F. and Ausubel, F.M. (1994) Programmed cell death in plants: a pathogen‐triggered response activated coordinately with multiple defense functions. Cell, 77, 551–563. [DOI] [PubMed] [Google Scholar]
- Hoat, T.X. , Nakayashiki, H. , Tosa, Y. and Mayama, S. (2006) Specific cleavage of ribosomal RNA and mRNA during victorin‐induced apoptotic cell death in oat. Plant J. 46, 922–933. [DOI] [PubMed] [Google Scholar]
- Hofer, A.M. and Brown, E.M. (2003) Extracellular calcium sensing and signaling. Nature, 4, 30–538. [DOI] [PubMed] [Google Scholar]
- Hünig, T. and Schimpl, A. (1997) Systemic autoimmune disease as a consequence of defective lymphocyte death. Curr. Opin. Immunol. 9, 826–830. [DOI] [PubMed] [Google Scholar]
- Jang, I.K. , Hu, R. , Lacana, E. , D'Adamio, L. and Gu, H. (2002) Apoptosis‐linked gene 2‐deficient mice exhibit normal T‐cell development and function. Mol. Cell. Biol. 22, 4094–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, Y.S. , Kim, K.S. , Kim, K.D. , Lim, J.S. , Kim, J.W. and Kim, E. (2001) Apoptosis‐linked gene 2 binds to the death domain of Fas and dissociates from Fas during Fas‐mediated apoptosis in Jurkat cells. Biochem. Biophys. Res. Commun. 288, 420–426. [DOI] [PubMed] [Google Scholar]
- Kawai, T. , Nomura, F. , Hoshino, K. , Copeland, N.G. , Gilbert, D.J. , Jenkins, N.A. and Akira, S. (1999) Death‐associated protein kinase 2 is a new calcium/calmodulin‐dependent protein kinase that signals apoptosis through its catalytic activity. Oncogene, 18, 3471–3480. [DOI] [PubMed] [Google Scholar]
- Kitaura, Y. , Matsumoto, S. , Satoh, H. , Hitomi, K. and Maki, M. (2001) Peflin and ALG‐2, members of the penta‐EF‐hand protein family, form a heterodimer that dissociates in a Ca2+‐dependent manner. J. Biol. Chem. 276, 14 053–14 058. [DOI] [PubMed] [Google Scholar]
- Kitaura, Y. , Watanabe, M. , Satoh, H. , Kawai, T. , Hitomi, K. and Maki, M. (1999) Peflin, a novel member of the five‐EF‐hand‐protein family, is similar to the apoptosis‐linked gene 2 (ALG‐2) protein but possesses nonapeptide repeats in the N‐terminal hydrophobic region. Biochem. Biophys. Res. Commun . 263, 68–75. [DOI] [PubMed] [Google Scholar]
- Klein, S.L. , Strausberg, R.L. , Wagner, L. , Pontius, J. , Clifton, S.W. and Richardson, P. (2002) Genetic and genomic tools for Xenopus research: the NIH Xenopus initiative. Dev. Dyn. 225, 384–391. [DOI] [PubMed] [Google Scholar]
- Krebs, J. (1998) The role of calcium in apoptosis. Biometals, 11, 375–382. [DOI] [PubMed] [Google Scholar]
- Kusaka, K. , Tada, Y. , Shigemi, T. , Sakamoto, M. , Nakayashiki, H. , Tosa, Y. and Mayama, S. (2004) Coordinate involvement of cysteine protease and nuclease in the executive phase of plant apoptosis. FEBS Lett. 578, 363–367. [DOI] [PubMed] [Google Scholar]
- La Cour, J.M. , Mollerup, J. , Winding, P. , Tarabykina, S. , Sehested, M. and Berchtold, M.W. (2003) Up‐regulation of ALG‐2 in hepatomas and lung cancer tissue. Am. J. Pathol. 163, 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacanà, E. , Ganjei, J.K. , Vito, P. and D'Adamio, L. (1997) Dissociation of apoptosis and activation of IL‐1β‐converting enzyme/Ced‐3 proteases by ALG‐2 and the truncated Alzheimer's gene ALG‐3. J. Immunol. 158, 5129–5135. [PubMed] [Google Scholar]
- Lo, K.W. , Zhang, Q. , Li, M. and Zhang, M. (1999) Apoptosis‐linked gene product ALG‐2 is a new member of the calpain small subunit subfamily of Ca2+‐binding proteins. Biochemistry, 38, 7498–7508. [DOI] [PubMed] [Google Scholar]
- Lopes, N. , Gregg, D. , Vasudevan, S. , Hassanain, H. , Goldschmidt‐Clermont, P. and Kovacic, H. (2003) Thrombospondin 2 regulates cell proliferation induced by Rac1 redox‐dependent signaling. Mol. Cell. Biol. 23, 5401–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki, M. , Narayana, S.V.L. and Hitomi, K. (1997) A growing family of the Ca2+‐binding proteins with five EF‐hand motifs. Biochem. J. 328, 718–720. [PMC free article] [PubMed] [Google Scholar]
- Mayama, S. , Tani, T. , Midland, S.L. , Sims, J.J. and Keen, N.T. (1986) The purification of victorin and its phytoalexin elicitors activity in oat leaves. Physiol. Mol. Plant Pathol. 29, 1–18. [Google Scholar]
- McConkey, D.J. , Nicotera, P. , Hartzell, P. , Bellomo, G. , Wyllie, A.H. and Orrenius, S. (1989a) Glucocorticoids activate a suicide process in thymocytes through an elevation of cytosolic Ca2+ concentration. Arch. Biochem. Biophys. 269, 365–370. [DOI] [PubMed] [Google Scholar]
- McConkey, D.J. , Hartzell, P. , Amador‐Perez, J.F. , Orrenius, S. and Jondal, M. (1989b) Calcium‐dependent killing of immature thymocytes by stimulation via the CD3/T cell receptor complex. J. Immunol. 143, 1801–1806. [PubMed] [Google Scholar]
- Miura, M. , Zhu, H. , Rotello, R. , Hartwieg, E.A. and Yuan, J. (1993) Induction of apoptosis in fibroblasts by IL‐1 beta‐converting enzyme, a mammalian homolog of the C. elegans cell death gene ced‐3. Cell, 75, 653–660. [DOI] [PubMed] [Google Scholar]
- Nicotera, P. , Bellomo, G. and Orrenius, S. (1990) The role of Ca2+ in cell killing. Chem. Res. Toxicol. 3, 484–494. [DOI] [PubMed] [Google Scholar]
- Reed, J.C. (1996) Mechanisms of Bcl‐2 family protein function and dysfunction in health and disease. Behring Inst. Mitt. 97, 72–100. [PubMed] [Google Scholar]
- Ryerson, D.E. and Heath, M.C. (1996) Cleavage of nuclear DNA into oligonucleosomal fragments during cell death induced by fungal infection or by abiotic treatments. Plant Cell, 8, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoul, R. (2006) Do Alix and ALG‐2 really control endosomes for better or for worse? Biol. Cell, 98, 69–77. [DOI] [PubMed] [Google Scholar]
- Satoh, H. , Nakano, Y. , Shibata, H. and Maki, M. (2002a) The penta‐EF‐hand domain of ALG‐2 interacts with amino‐terminal domains of both annexin VII and annexin XI in a Ca2+‐dependent manner. Biochem. Biophys. Acta, 1600, 61–67. [DOI] [PubMed] [Google Scholar]
- Satoh, H. , Shibata, H. , Nakano, Y. , Kitaura, Y. and Maki, M. (2002b) ALG‐2 interacts with the amino‐terminal domain of annexin XI in a Ca2+‐dependent manner. Biochem. Biophys. Res. Commun. 291, 1166–1172. [DOI] [PubMed] [Google Scholar]
- Schwartz, L.M. , Smitho, S. , Jones, M.E.E. and Osborne, B.A. (1993) Do all programmed cell deaths occur via apoptosis? Proc. Natl. Acad. Sci. USA, 90, 980–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, H. , Yamada, K. , Mizuno, T. , Yorikawa, C. , Takahashi, H. , Satoh, H. , Kitaura, Y. and Maki, M. (2004) The penta‐EF‐hand protein ALG‐2 interacts with a region containing PxY repeats in Alix/AIP1, which is required for the subcellular punctate distribution of the amino‐terminal truncation form of Alix/AIP1. J. Biochem. 135, 117–128. [DOI] [PubMed] [Google Scholar]
- Song, J.T. , Lu, H. and Greenberg, J.T. (2004a) Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, ABERRANT GROWTH AND DEATH2 and AGD2‐LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell, 16, 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.T. , Lu, H. , McDowell, J.N. and Greenberg, J.T. (2004b) A key role for ALD1 in activation of local and systemic defenses in Arabidopsis . Plant J. 40, 200–212. [DOI] [PubMed] [Google Scholar]
- Tada, Y. , Hata, S. , Takata, Y. , Nakayashiki, H. , Tosa, Y. and Mayama, S. (2001) Induction and signaling of an apoptotic response typified by DNA laddering in the defense response of oats to infection and elicitors. Mol. Plant–Microbe Interact. 14, 477–486. [DOI] [PubMed] [Google Scholar]
- Tanaka, K. , Kondoh, N. , Shuda, M. , Matsubara, O. , Imazeki, N. , Ryo, A. , Wakatsuki, T. , Hada, A. , Goseki, N. , Igari, T. , Hatsuse, K. , Aihara, T. , Horiuchi, S. , Yamamoto, N. and Yamamoto, M. (2001) Enhanced expression of mRNAs of antisecretory factor‐1, gp96, DAD1 and CDC34 in human hepatocellular carcinomas. Biochem. Biophys. Acta, 1536, 1–12. [DOI] [PubMed] [Google Scholar]
- Tsuda, M. , Seong, K.H. and Aigaki, T. (2006) POSH, a scaffold protein for JNK signaling, binds to ALG‐2 and ALIX in Drosophila. FEBS Lett. 580, 3296–3300. [DOI] [PubMed] [Google Scholar]
- Vito, P. , Lacana, E. and D'Adamio, L. (1996) Interfering with apoptosis: Ca2+‐binding protein ALG‐2 and Alzheimer's disease gene ALG‐3. Science, 271, 521–525. [DOI] [PubMed] [Google Scholar]
- Wiens, M. , Perović‐Ottstadt, S. , Müller, I.M. and Müller, W.E. (2004) Allograft rejection in the mixed cell reaction system of the demosponge Suberites domuncula is controlled by differential expression of apoptotic genes. Immunogenetics, 56, 597–610. [DOI] [PubMed] [Google Scholar]
- Wiens, M. , Belikov, S.I. , Kaluzhnaya, O.V. , Schroder, H.C. , Hamer, B. , Perovic‐Ottstadt, S. , Borejko, A. , Luthringer, B. , Müller, I.M. and Müller, W.E. (2006) Axial (apical‐basal) expression of pro‐apoptotic and pro‐survival genes in the Lake Baikal demosponge Lubomirskia baicalensis . DNA Cell Biol. 25, 152–164. [DOI] [PubMed] [Google Scholar]
- Yang, Q. , Hoat, T.X. , Imai, S. , Ishihara, A. , Zhang, L. , Nakayashiki, H. , Tosa, Y. and Mayama, S. (2004) Analysis of the involvement of hydroxyanthranilate hydroxycinnamoyltransferase and caffeoyl‐CoA 3‐O‐methyltransferase in phytoalexin biosynthesis in oat. Mol. Plant–Microbe Interact. 17, 81–89. [DOI] [PubMed] [Google Scholar]
- Yao, N. and Greenberg, J.T. (2006) Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell, 18, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, N. , Tada, Y. , Park, P. , Nakayashiki, H. , Tosa, Y. and Mayama, S. (2001) Novel evidence for apoptotic cell response and differential signals in chromatin condensation and DNA cleavage in victorin‐treated oats. Plant J. 28, 13–26. [DOI] [PubMed] [Google Scholar]
- Yao, N. , Tada, Y. , Sakamoto, M. , Nakayashiki, H. , Park, P. , Tosa, Y. and Mayama, S. (2002) Mitochondrial oxidative burst involved in apoptotic response in oats. Plant J. 30, 567–579. [DOI] [PubMed] [Google Scholar]


