Summary
The avirulence determinant triggering the resistance conferred by the tomato gene Sw‐5 against Tomato spotted wilt virus (TSWV) is still unresolved. Sequence comparison showed two substitutions (C118Y and T120N) in the movement protein NSm present only in TSWV resistance‐breaking (RB) isolates. In this work, transient expression of NSm of three TSWV isolates [RB1 (T120N), RB2 (C118Y) and non‐resistance‐breaking (NRB)] in Nicotiana benthamiana expressing Sw‐5 showed a hypersensitive response (HR) only with NRB. Exchange of the movement protein of Alfalfa mosaic virus (AMV) with NSm supported cell‐to‐cell and systemic transport of the chimeric AMV RNAs into N. tabacum with or without Sw‐5, except for the constructs with NBR when Sw‐5 was expressed, although RB2 showed reduced cell‐to‐cell transport. Mutational analysis revealed that N120 was sufficient to avoid the HR, but the substitution V130I was required for systemic transport. Finally, co‐inoculation of RB and NRB AMV chimeric constructs showed different prevalence of RB or NBR depending on the presence or absence of Sw‐5. These results indicate that NSm is the avirulence determinant for Sw‐5 resistance, and mutations C118Y and T120N are responsible for resistance breakdown and have a fitness penalty in the context of the heterologous AMV system.
Keywords: Avr, competition assays, movement protein NSm, Tospovirus, transient expression
Introduction
Tomato spotted wilt virus (TSWV) is the type member of the plant‐infecting Tospovirus genus in the family Bunyaviridae (Milne and Francki, 1984). The viral genome organization consists of three single‐stranded RNAs: the large (L) negative‐sense RNA and the middle (M) and small (S) ambisense RNAs. Segment L (8.9 kb) encodes an RNA‐dependent RNA polymerase (RdRp) (de Haan et al., 1991); segment M (4.8 kb) expresses from viral‐sense RNA the NSm which operates as a movement protein (MP) (Lewandowski and Adkins, 2005; Li et al., 2009; Storms et al., 1995), and from viral‐complementary sense RNA the precursor of surface glycoproteins GN/GC containing determinants for thrip transmission (Sin et al., 2005); and segment S (2.9 kb) encodes a silencing suppressor NSS (Takeda et al., 2002) in the viral sense and the nucleocapsid protein (N) from the viral‐complementary sense, used for the encapsidation of viral RNA and, according to recent studies, facilitating long‐distance movement (de Haan et al., 1990; Feng et al., 2013).
The management of the disease caused by TSWV has been extremely difficult because of its broad host range and the resistance of the thrip vectors to insecticides (Boiteux and Giordano, 1993). The highest level of resistance to TSWV was obtained by the introgression of the dominant single resistance genes Tsw in pepper and Sw‐5 in tomato. These genes were derived from Capsicum chinense and Solanum peruvianum, respectively (Boiteux, 1995; Moury et al., 1998; Stevens et al., 1991). The resistance mediated by Sw‐5 follows the gene‐for‐gene relationship (Staskawicz et al., 1995) by triggering the typical hypersensitive response (HR) around the TSWV infection foci, limiting virus spread to distal parts of the plant. The avirulence (Avr) protein targeted by the resistance Sw‐5 gene is unknown to date. Previous work has revealed that the Sw‐5 locus contains at least five paralogues (denoted Sw‐5a to Sw‐5e), but only the Sw‐5b gene is necessary and sufficient to confer resistance against TSWV (Spassova et al., 2001). The Sw‐5b gene encodes a protein of 1246 amino acids and is classified as a member of the coiled‐coil, nucleotide‐binding adaptor shared by APAF‐1, certain R gene products and CED‐4 (NB‐ARC) and leucine‐rich repeat group of resistance gene candidates (Meyers et al., 1999).
Control strategies based on the Sw‐5 gene are affected by the emergence of TSWV resistance‐breaking (RB) isolates able to overcome the resistance, which have been reported in South Africa (Thompson and vanZijl, 1995), Hawaii (Canady et al., 2001; Gordillo et al., 2008), Australia (Latham and Jones, 1998), Spain (Aramburu and Marti, 2003) and Italy (Ciuffo et al., 2005; Zaccardelli et al., 2008). The lack of a TSWV infectious clone has hampered the study of the molecular mechanisms associated with Sw‐5 RB isolates. Previous analysis based on a complete set of reassortants generated from an infectious mixture of two isolates of TSWV showed that the M segment has a major role in overcoming Sw‐5 resistance (Hoffmann et al., 2001). Moreover, the comparative analysis of nucleotide and amino acid sequences of RNA M from RB and non‐resistance‐breaking (NRB) isolates has revealed that the capacity to overcome Sw‐5 resistance is associated with the presence of a tyrosine or asparagine at positions 118 (Y118) or 120 (N120) of the NSm protein, respectively (López et al., 2011).
In this work, we analysed the role of the NSm protein in the resistance mediated by the Sw‐5 gene by: (i) transient expression of the protein in Sw‐5‐resistant plants (tomato, Nicotiana tabacum and N. benthamiana) in the absence of other TSWV components; and (ii) using the heterologous viral system based on Alfalfa mosaic virus (AMV), which allows the functional exchangeability of viral MPs assigned to the ‘30 K family’ (Fajardo et al., 2013; Melcher, 2000; Sánchez‐Navarro et al., 2006). The results indicate that the NSm is the Avr factor of the Sw‐5b gene, in which the Y118 and N120 residues are crucial to overcome the HR.
Results
Transient expression of the TSWV NSm protein in Sw5‐b transgenic N. benthamiana plants
To assess the direct role of the NSm protein of TSWV in the resistance mediated by the Sw‐5 gene, in the absence of other viral components, we performed a transient expression of the NSm protein in resistant Sw5‐b transgenic N. benthamiana and/or N. tabacum lines. Both transgenic lines contain the same expression cassette, allowing the constitutive expression of the Sw5‐b protein (Spassova et al., 2001; kindly provided by Dr M. Prins, Keygene N.V./Amsterdam University). For this purpose, three NSm genes derived from two Sw5‐RB (GRAU and Llo2TL3) and one Sw5‐NRB (Gr1NL2) TSWV isolates (López et al., 2011) were used in this study. Each NSm of the RB isolates is representative of one of the two amino acids proposed by López et al. (2011) to be associated with overcoming Sw‐5 resistance. Thus, although the NRB Gr1NL2 NSm (hereafter referred to as NRB) contains a cysteine and a threonine at positions 118 (118C) and 120 (120T), respectively, the NSm proteins of the RB Llo2TL3 (hereafter referred to as RB2) and GRAU (hereafter referred to as RB1) isolates contain a tyrosine at position 118 (118Y) or an asparagine at position 120 (120N), respectively (Fig. S1, see Supporting Information). In a preliminary study we observed that the TSWV isolates GRAU and Gr1NL2 reproduced the expected phenotypes in Sw5‐b N. benthamiana plants (Table S1, see Supporting Information). In the case of the RB TSWV Llo2TL3 isolate and as a result of the lack of infectious tissue, we used an AMV hybrid containing the NSm RB2 gene (see below). This hybrid virus infected locally and systemically the Sw5‐b N. benthamiana plants without inducing any necrotic response. Later, the NSm genes were cloned into a binary plasmid, being fused to the haemagglutinin (HA) epitope at its C‐terminus, and were transiently expressed by A. tumefaciens in wild‐type N. benthamiana (Nb/wt) or N. tabacum (Nt/wt) plants. Western blot analysis revealed that the three NSm proteins accumulated in agroinfiltrated leaves when transiently expressed in either Nb/wt or Nt/wt leaf with an electrophoretic mobility of the expected 35 kDa (Fig. 1A). However, protein accumulation in N. tabacum plants was considerably lower (5–10 times) when compared with N. benthamiana plants. When transient expression of these three NSm proteins was assayed in susceptible and resistant tomato cultivars carrying the Sw‐5 gene, no expression at all was detected for any of the three NSm proteins or for the control construct that carries the green fluorescent protein (GFP) (data not shown). Therefore, to overcome this problem, the different constructs were transiently expressed in transgenic N. benthamiana or N. tabacum plants constitutively expressing the Sw‐5 gene (Nb/Sw5‐b; Nt/Sw5‐b). The clearest results were observed in N. benthamiana plants. As shown in Fig. 1B, 6 days post‐agroinfiltration, only the construct that contains the NRB gene triggered the hypersensitive‐like response on the Nb/Sw5‐b leaf (Fig. 1B, panel 3). These results clearly identify the NSm gene as the only TSWV component required to trigger the HR mediated by the Sw‐5 gene.
Figure 1.
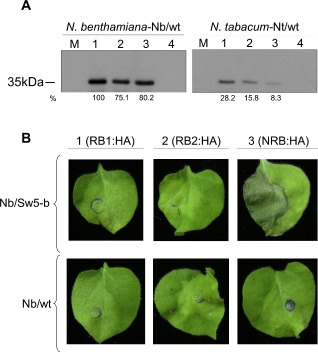
Transient expression of the Tomato spotted wilt virus (TSWV) NSm movement protein (MP) in wild‐type Nicotiana benthamiana (Nb/wt) or N. tabacum (Nt/wt) and transgenic N. benthamiana plants carrying the resistance gene Sw5‐b (Nb/Sw5‐b). (A) Western blot analysis of the Nb/wt and Nt/wt infiltrated leaves at 3 days post‐inoculation expressing RB1:HA (lane 1), RB2:HA (lane 2) and NRB:HA (lane 3). Lanes M and 4 correspond to non‐agroinfiltrated leaves and leaves infiltrated with cultures carrying the empty binary plasmid, respectively. The numbers below the panel represent the relative percentages of the intensity of each band with respect to the more intense band in lane 1. (B) Photographs of Nb/Sw5‐b (top) and Nb/wt (bottom) leaves expressing RB1:HA (1), RB2:HA (2) and NRB:HA (3) at 6 days post‐agroinfiltration. HA, haemagglutinin; NRB, non‐resistance‐breaking; RB, resistance‐breaking.
Cell‐to‐cell and systemic movement of the chimeric AMV constructs containing TSWV NSm in P12 N. tabacum plants
We analysed the role of the NSm gene in the resistance mediated by the Sw‐5 gene, but in a viral context. For this purpose and because of the lack of an infectious TSWV clone, we used the heterologous AMV model system, which has been demonstrated to allow the functional exchangeability for the local (Sánchez‐Navarro et al., 2006) and systemic (Fajardo et al., 2013) transport of MPs assigned to the 30K family. First, we analysed the capacity of the three NSm proteins (NRB, RB1 and RB2) to support the local and systemic transport of chimeric AMV. To do this, the NSm gene was exchanged with the corresponding AMV MP gene in the AMV RNA 3 wt (pAL3NcoP3) (van der Vossen et al., 1993) or in an RNA 3 derivative that expresses the GFP (pGFP/A255/CP) (Sánchez‐Navarro et al., 2001). In the chimeric constructs, the heterologous NSm proteins were extended with the C‐terminal 44 residues (A44) of the AMV MP, to allow a compatible interaction with the AMV coat protein (CP) (Sánchez‐Navarro et al., 2006).
Cell‐to‐cell movement of the AMV RNA 3 hybrids was studied by inoculation of T7 transcripts generated from the pGFP/NRB:A44/CP, pGFP/RB1:A44/CP and pGFP/RB2:A44/CP plasmids into transgenic N. tabacum plants that express constitutively the P1 and P2 polymerase proteins of AMV (P12) (Fig. 2A). All constructs resulted in clear fluorescent infection foci at 2 days post‐inoculation (dpi) (Fig. 2A), indicating that the three NSm proteins were competent to support the local transport of the hybrid AMV RNA 3. However, the analysis of the area of 50 independent foci at 2 and 3 dpi revealed that the foci derived from the pGFP/RB2:A44/CP construct were significantly smaller than those generated by pGFP/NRB:A44/CP and pGFP/RB1:A44/CP constructs (Student's t‐test, P < 0.05) (Fig. 2B). Analysis of the replication of the constructs on P12 protoplasts (Fig. 2C) did not suggest significant RNA accumulation level differences that could account for the differences observed in the cell‐to‐cell movement.
Figure 2.
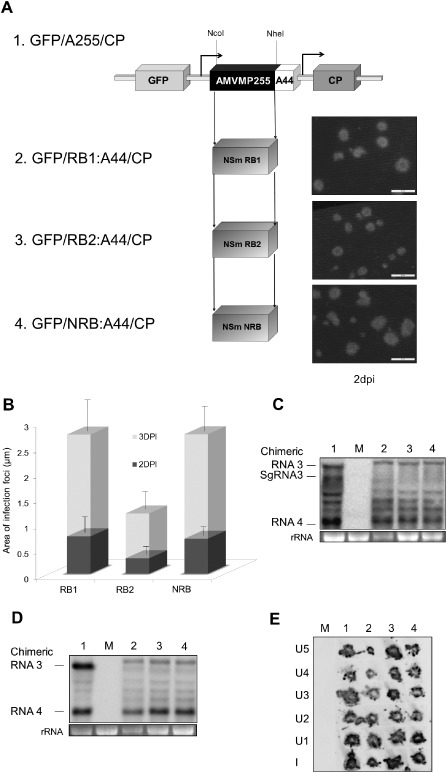
Analysis of the accumulation, cell‐to‐cell and systemic transport of the Alfalfa mosaic virus (AMV) chimeric RNAs carrying the movement protein (MP) of Tomato spotted wilt virus (TSWV) isolates. (A) Infection foci observed in P12 plants inoculated with RNA 3 transcripts from pGFP/A255/CP derivatives, which contain the TSWV NSm RB1 (2), RB2 (3) and NRB (4) genes. The schematic representation shows the GFP/A255/CP AMV RNA 3 (1), in which the open reading frames corresponding to the green fluorescent protein (GFP), the movement protein (MP) and the coat protein (CP) are represented by large boxes. The number shown in the MP box represents the total amino acid residues of the AMV MP (255) exchanged for the TSWV NSm, represented by single boxes below. The NcoI and NheI restriction sites used to exchange the MP genes are indicated. The arrows indicate the subgenomic promoters. The C‐terminal 44 amino acids of the AMV MP are indicated as A44. Images correspond to representative photographs of the infection foci observed at 2 days post‐inoculation (dpi) using a Leica stereoscopic microscope. The scale bar corresponds to 2 mm. (B) Histograms representing the average area of 50 independent infection foci at 2 and 3 dpi developed in P12 plants inoculated with transcripts derived from the AMV RNA 3 variants shown in (A). Error bars indicate the standard deviation. (C) Northern blot analysis of the accumulation of the chimeric AMV RNAs in P12 protoplasts inoculated with RNA transcribed from the constructs shown in (A). (D) Northern blot analysis of the accumulation of the chimeric AMV RNAs lacking the 5′ proximal GFP gene in P12 protoplasts. P12 protoplasts were inoculated with RNA transcribed from plasmid pAL3NcoP3 derivatives, expressing the AMV MP (lane 1) or the NSm RB1 (lane 2; plasmid pRB1:A44/CP), RB2 (lane 3; plasmid pRB2:A44/CP) or NRB (lane 4; plasmid pNRB:A44/CP). The positions of the chimeric RNAs 3 and 4 and additional subgenomic RNA (sgRNA) are indicated in the left margin. (E) Tissue printing analysis of P12 plants inoculated with the AMV RNA 3 derivatives used in (D). Plants were analysed at 14 dpi by printing the transverse section of the corresponding petiole from inoculated (I) and upper (U) leaves. The position of each leaf is indicated by numbers which correspond to the position of the leaves in the plant from the lower to the upper part, in which U1 corresponds to the closest leaf to that inoculated. rRNA indicates 23S RNA loading control. M refers to mock inoculated plant. NRB, non‐resistance‐breaking; RB, resistance‐breaking.
The capacity of the different TSWV MPs to support the systemic transport of the AMV RNA 3 was also analysed. For this purpose, we used the wt AMV RNA 3 constructs, as the RNA 3 derivatives carrying the GFP reporter gene do not support systemic movement in P12 tobacco plants (Sánchez‐Navarro et al., 2001). First, we observed that the different AMV RNA 3 hybrids accumulated comparable levels of RNAs 3 and 4 in P12 protoplasts (Fig. 2D). The accumulation and distribution of the chimeric RNA 3s were then analysed in inoculated and upper non‐inoculated leaves of P12 plants by tissue printing of petiole cross‐sections, in which a positive hybridization signal always correlated with the presence of the virus in the corresponding leaf, as described previously (Fajardo et al., 2013; Mas and Pallás, 1995; Sánchez‐Navarro et al., 2010). The results showed that, despite the differences observed in local movement, all AMV RNA 3 constructs were able to support systemic movement, infecting all upper leaves of P12 plants (Fig. 2E).
Analysis of the capability of the different AMV derivatives to overcome the resistance conferred by Sw‐5 in tomato and transgenic N. tabacum plants
In the next step, we analysed the capacity of the hybrid AMV to infect Sw‐5‐resistant (Cultivar ‘Verdi’; lane 1 in Fig. 3) and TSWV‐susceptible (‘Marmande’; lane 2 in Fig. 3) tomato cultivars. Therefore, the tomato plants were inoculated with wt AMV RNA 1 and RNA 2, purified CP and wt or chimeric RNA 3 constructs. Northern blot analysis of the inoculated tomato leaves in Fig. 3 shows the accumulation of the RNA 4, derived from the corresponding viral RNA 3. Similar accumulation levels were observed in the resistant and susceptible tomato cultivars tested when the plants were inoculated with the AMV wt (Fig. 3, AMV RNA 4 band intensities: 40.7% vs. 35.3%, respectively), indicating that the genetic differences between the two tomato cultivars do not affect significantly virus accumulation. A high accumulation level was observed in the susceptible tomato cultivar (Fig. 3, lane 2) when inoculated with the chimeric AMV RNA 3 expressing the NRB protein (100%), followed by the chimeric AMV RNA 3 expressing the RB1 (56.6%) and RB2 (21.0%) proteins. These results indicate that, in the tomato lacking the Sw5‐b resistance gene, the NRB NSm protein provides an advantage when compared with the RB NSm proteins or even with the wt AMV MP. In the same tomato cultivar, the low accumulation level observed with the hybrid RNA 3 expressing the RB2 protein, whose sequence differs only with regard to two or three residues relative to the RB1 or NRB protein, respectively, is remarkable (see Fig. S1). However, in the Sw‐5‐resistant tomato cultivar (Fig. 3, lane 1), the presence of the NRB gene resulted in a significantly reduced (93%) accumulation (6.2% vs. 100%), whereas such a reduction was only 5% (51.6% vs. 56.5%) or 9% (10.6% vs. 21.0%) for the AMV RNA 3 variant carrying the RB1 or RB2 gene, respectively. These results confirm that the presence of the NRB NSm protein negatively affects AMV accumulation in the Sw‐5‐resistant tomato cultivar.
Figure 3.
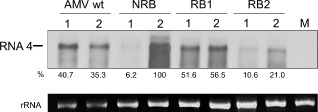
Northern blot analysis of the Alfalfa mosaic virus (AMV) chimeric RNA accumulation in the inoculated leaves of Sw‐5‐resistant ‘Verdi’ (lane 1) and Tomato spotted wilt virus (TSWV)‐susceptible ‘Marmande’ (lane 2) tomato cultivars. The tomato plants were inoculated with the corresponding RNA 3 transcript expressing the AMV movement protein (MP) (AMV wt) or the NSm of the TSWV isolates Gr1NL2 (NRB), GRAU (RB1) and Llo2TL3 (RB2). Mock (M) represents total RNA extraction of healthy tissue. The position of the RNA 4 is indicated in the left margin of the photograph. rRNA indicates 23S RNA loading control. The numbers below the panel represent the relative percentages of the intensity of each band with respect to the more intense band (lane 2/NRB). NRB, non‐resistance‐breaking; RB, resistance‐breaking.
The presence of viral RNAs in the upper non‐inoculated leaves of the tomato cultivars was analysed at 14 and 21 dpi by tissue printing analysis. No hybridization signal was detected in any of the plants analysed, including those inoculated with the wt AMV, indicating that the AMV variant used to perform the analysis is unable to move systemically in tomato. To circumvent this limitation, we used N. tabacum plants, which supported local and systemic AMV accumulation (see above). We then tested the chimeric AMV constructs in both transgenic N. tabacum plants that express constitutively the Sw5‐b gene (Nt/Sw5‐b) (Spassova et al., 2001) and wt N. tabacum plants (Nt/wt). Nt/wt and Nt/Sw5‐b plants were inoculated as described above. The accumulation of the viral RNA on inoculated leaves was analysed by Northern blot at 7 dpi (Fig. 4). All AMV RNA 3 derivatives supported comparable levels of viral RNA 3 and 4 accumulation in Nt/wt and Nt/Sw5‐b plants, except for the construct containing the NRB gene in Nt/Sw5‐b plants, which accumulated 65% less efficiently (Fig. 4, lane 3). These results were equivalent to those obtained in resistant tomato plants (see above). We also analysed the capacity of the heterologous MPs (NSm) to support the systemic transport of AMV RNA 3 by tissue printing (Sánchez‐Navarro et al., 2010). The analysis of all upper non‐inoculated leaves of the Nt/wt and Nt/Sw5‐b plants at 14 dpi revealed that all constructs rendered a positive hybridization signal in both hosts, except for the NRB:A44/CP RNA 3 hybrid, which was exclusively detected in the susceptible Nt/wt plants (data not shown). To further confirm the accumulation of viral RNAs in the upper leaves, we performed a Northern blot analysis of total RNA extracted from a mixture of the upper leaves U1, U2 and U3 (Fig. 4). The results showed that the three AMV RNA 3 chimeric variants, NRB:A44/CP, RB1:A44/CP and RB2:A44/CP, accumulated comparable levels of RNAs 3 and 4 in the upper leaves of Nt/wt plants (Fig. 4, wt/systemic), indicating that the three NSm proteins are competent to support the systemic transport of viral RNAs. However, only the two RNA 3 constructs, RB1:A44/CP and RB2:A44/CP, were detected in the upper leaves of the resistant Nt/Sw5‐b plants (Fig. 4, Sw5‐b/systemic). This indicates that the NSm is the Avr determinant responsible for overcoming the resistance mediated by the Sw5‐b gene in the AMV viral context.
Figure 4.
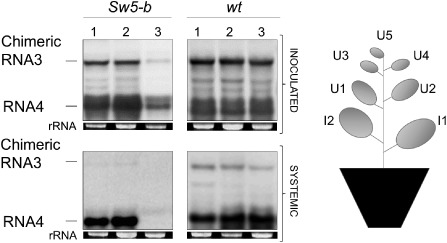
Northern blot analysis of Alfalfa mosaic virus (AMV) chimeric RNA accumulation in transgenic Nicotiana tabacum plants that express constitutively the Sw5‐b gene (Nt/Sw5‐b). Nicotiana tabacum wild‐type (Nt/wt) and Nt/Sw5‐b plants were inoculated as described in Fig. 3, in which the RNA 3 transcript expresses the NSm RB1(lane 1), RB2 (lane 2) and NRB (lane 3). The analysed RNAs from inoculated leaves corresponded to a mixture of total RNA extracted from the two inoculated leaves (I1 and I2) at 7 days post‐inoculation (dpi), whereas the analysed RNA from systemic leaves corresponded to a mixture of the total RNAs extracted from the upper (U) leaves U1, U2 and U3 at 14 dpi. The positions of the chimeric RNA 3 and RNA 4 are indicated in the left margin of the photographs. rRNA indicates 23S RNA loading control. NRB, non‐resistance‐breaking; RB, resistance‐breaking.
Mutational analysis of the RB and NRB NSm proteins
The amino acid alignments among RB and NRB NSm proteins indicated that the capability of TSWV to overcome the resistance mediated by Sw‐5 might be exclusively a result of single changes present at residues 118 (Y) and 120 (N) of the NSm protein (López et al., 2011), which are representative of the RB2 and RB1 NSm isolates analysed herein. We cannot exclude, however, that other residues might also contribute. Therefore, to analyse this aspect, we performed a mutational analysis using the RB1 and NRB NSm proteins, which differ only in two residues (RB2 and NRB differ in three residues) at positions 120 (N in RB1 and T in NRB) and 130 (I in RB1 and V in NRB). By directed mutagenesis, we synthesized two variants of RNA 3 for the heterologous AMV model system shown above, pGFP/RB1:A44/CP and pRB1:A44/CP constructs, in which the asparagine at position 120 was changed to a threonine (pGFP/RB1‐T120:A44/CP and pRB1‐T120:A44/CP) or the isoleucine at position 130 was changed to a valine (pGFP/RB1‐V130:A44/CP and pRB1‐V130:A44/CP). The analysis of the cell‐to‐cell movement of the chimeric mutants expressing the GFP in N. tabacum P12 plants revealed that, at 3 dpi, the presence of a T at position 120 in the RB1 protein (RB1‐T120) increased significantly the area of the foci; meanwhile, the presence of a V at position 130 (RB1‐V130) resulted in the opposite effect (Student's t‐test, P < 0.05) (Fig. 5A). These differences were not caused by changes in the replication capability, as all constructs accumulated comparable levels of viral RNAs on P12 protoplasts (Fig. 5B). The capacity of both RB1 mutants to overcome the resistance mediated by the Sw5‐b gene was analysed by the inoculation of N. tabacum Nt/wt and Nt/Sw5‐b plants with transcripts derived from the two pRB1‐T120:A44/CP and pRB1‐V130:A44/CP mutant constructs, using the pRB1:A44/CP construct as control (Fig. 5C). Northern blot analysis of the inoculated and upper non‐inoculated leaves of Nt/wt plants revealed that the three constructs accumulated comparable levels of viral RNAs 3 and 4, indicating that neither of the two changes introduced in the RB1 gene affected the capacity of the NSm protein to support the local and/or systemic transport of viral progeny. However, when the same analysis was performed with Nt/Sw5‐b plants, we observed that the three constructs were competent to infect the inoculated leaves, as shown by the accumulation of the viral RNAs 3 and 4, but only the construct containing the RB1 gene was detected in the upper non‐inoculated leaves. In addition, we observed differences in the symptomatology on the inoculated leaves of the resistant Nt/Sw5‐b plants. Thus, whereas RB1 and RB1‐V130 resulted in similar chlorotic spots, the construct carrying RB1‐T120 reproduced the typical necrotic lesions observed for the construct that expressed the NRB protein. Similar results were observed when the three NSm proteins (RB1, RB1‐T120 and RB1‐V130) were transiently expressed in Nb/Sw‐5b plants, in which only RB1‐T120 triggered the hypersensitive‐like response (Fig. 5E, panel 3). Together, these results demonstrate that mutations at position 120 are responsible for evading the HR mediated by Sw‐5, but also that, in the context of the AMV system, other changes are required to compensate for the putative fitness cost associated with the incorporation of the critical residue.
Figure 5.
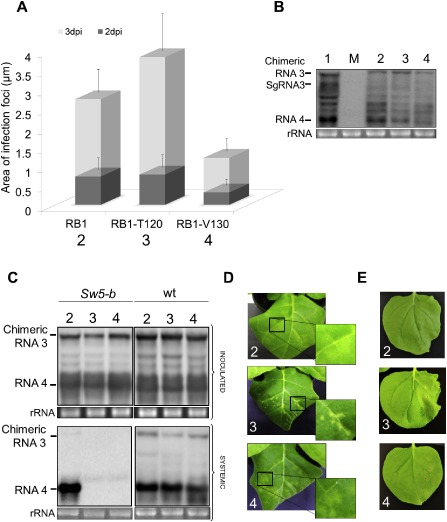
Functional characterization of NSm RB1 single mutants. (A) Histograms represent the average of the area of 50 independent infection foci at 2 and 3 days post‐inoculation (dpi) observed in Nicotiana tabacum P12 plants inoculated with transcripts from Alfalfa mosaic virus (AMV) RNA 3 pGFP/A255/CP derivatives pGFP/RB1:A44/CP (lane 2), pGFP/RB1‐T120:A44/CP (lane 3) and pGFP/RB1‐V130:A44/CP (lane 4). The fluorescent infection foci were visualized using a Leica stereoscopic microscope. Error bars indicate the standard deviation. (B) Northern blot analysis of the accumulation of chimeric AMV RNAs in P12 protoplasts inoculated with RNA transcripts derived from the constructs used in (A) plus the plasmid pGFP/A255/CP (lane 1). (C) Northern blot analysis of the accumulation of the chimeric AMV RNAs in N. tabacum plants that express constitutively the Sw5‐b gene (Nt/Sw5‐b) or N. tabacum wild‐type (Nt/wt) plants. All plants were inoculated as described in Fig. 3 in which the chimeric RNA 3 corresponds to the transcripts derived from the constructs pRB1:A44/CP (lane 2), pRB1‐T120:A44/CP (lane 3) and pRB1‐V130:A44/CP (lane 4). Total RNA was extracted from inoculated and upper leaves as described in Fig. 4. The positions of the chimeric RNA 3 and RNA 4 are indicated in the left margin of the photographs. (D) Symptomatology observed in Nt/Sw5‐b plants inoculated with chimeric AMV derivatives used in (C) at 6 dpi. (E) Photographs of Nb/Sw5‐b leaves expressing TSWV NSm RB1:HA (2), RB1‐T120:HA (3) and RB1‐V130:HA (4) at 6 days post‐agroinfiltration. Mock (M) represents total RNA extraction of healthy tissue. rRNA indicates 23S RNA loading control. HA, haemagglutinin; NRB, non‐resistance‐breaking; RB, resistance‐breaking.
Competition assays
The presence of the TSWV RB isolates was associated mainly with Sw‐5‐resistant tomato crops, with scarce or null presence of these isolates in susceptible crops. This observation could suggest a fitness cost for RB TSWV isolates. The results obtained with the AMV model system and the different NSm proteins could suggest a possible fitness cost associated with RB1 and RB2 (e.g. the reduced RNA accumulation in tomato or cell‐to‐cell transport in N. tabacum). Although we cannot rule out that these effects may be caused by the heterologous AMV system or, perhaps, that other TSWV components may compensate for the putative fitness cost effects (see below), we analysed the relative fitness of chimeric AMV constructs carrying RB and NRB NSm genes to determine whether the pressure of the Sw‐5 gene could be sufficient to select the RB NSm proteins. To do this, a competition assay between RB1, RB2 and NRB NSm chimeric constructs was conducted by co‐inoculation of N. tabacum P12 and Sw5‐b‐expressing (Nt/Sw5‐b) plants with an infectious mixture containing equivalent transcript amounts. After two serial passages at 7‐day intervals using extracts of the inoculated leaves as inoculum, the prevalent isolate present in the inoculated infected tissue was determined by direct sequencing of the reverse transcription‐polymerase chain reaction (RT‐PCR) amplicons encompassing the full‐length NSm gene. The results obtained in three independent experiments revealed that, in P12 plants, all sequenced NSm amplicons corresponded to the NRB isolate, whereas, in Nt/Sw5‐b plants, all the sequences corresponded to the RB1 isolate. These results suggest a fitness cost for RB strains in the absence of Sw‐5 gene pressure, whereas, in Sw‐5‐resistant genotypes, the AMV hybrid carrying the RB1 gene prevailed. In addition, it should be noted that, in the latter case, only the hybrid RNA 3 containing the RB1 gene was detected, thus suggesting a better fitness provided by this NSm under Sw‐5 pressure.
Discussion
The present analysis was addressed to experimentally confirm previous data suggesting that the NSm protein is the Avr determinant of TSWV in the resistance mediated by the Sw‐5 gene (López et al., 2011). The initial results obtained by transient expression of RB and NRB NSm proteins in transgenic N. benthamiana cultivars carrying the Sw5‐b gene (Nb/Sw5‐b) revealed a hypersensitive‐like response only with the NRB NSm protein, thus indicating unequivocally that NSm is the Avr determinant for the resistance provided by the Sw‐5 gene. However, we were unable to reproduce the typical necrotic reaction to TSWV infection associated with the resistance mediated by the Sw‐5 gene, indicating that other factors could be modulating such a phenotypic response, e.g. a putative high protein accumulation in the infected cells or an enhanced effect caused by other cell responses associated with the viral infection. The observation that the HR‐like response was clearly developed in N. benthamiana (Nb/Sw5‐b) plants, but not in N. tabacum (Nt/Sw5‐b) plants, a host that accumulates a 5–10 times lower protein titre in transient expression, could support the concept of a minimal protein accumulation threshold required to trigger the typical HR.
Furthermore, the differences between the NRB and RB1 NSm proteins are exclusively located at positions 120 (T or N) and 130 (V or I), but only the former has been suggested previously by López et al. (2011) as being responsible for overcoming Sw‐5 resistance, and is necessary and sufficient to trigger the necrotic response (see below). Here, we demonstrated that two (RB1) or three (RB2) residues confer the capacity to overcome the Sw‐5 resistance. Based on the gene‐for‐gene model of disease resistance described by Flor (1971), the few amino acid changes observed in the RB NSm proteins will maintain the pathogenic function, but not the participation in the recognition event with the host resistance factor (Fraser, 1990). In agreement with this, we demonstrated that the two RB1 and RB2 proteins are still competent for local and systemic viral transport in the AMV heterologous system. The observation that few changes are associated with the capacity of an Avr gene to overcome host resistance is a common property for different viral proteins, such as the MP (Calder and Palukaitis, 1992; Meshi et al., 1989), RNA polymerase (Meshi et al., 1988; Padgett and Beachy, 1993) and CP (Dawson et al., 1988; Saito et al., 1987) of tobamoviruses, and the NSs protein of TSWV (de Ronde et al., 2013; Margaria et al., 2007).
Another aspect was to determine how the critical residues required to overcome the Sw‐5 resistance affect the functionality of the NSm proteins. This aspect was studied using the AMV model system. The absence of other TSWV components in the AMV system allowed us to correlate any effect on the viral transport with the different residues present in the NSm protein, although we cannot discount that the observed effect could be specific to the heterologous AMV system. Taking this into consideration, we observed that the three NSm proteins used in the analysis were competent to support local and systemic transport of AMV into N. tabacum plants. However, we observed that the cell‐to‐cell transport of the chimeric AMV RNA 3 expressing the RB2 protein was significantly affected, showing infection foci with a reduced area. The differences in the amino acid NSm sequences observed among the RB2, RB1 and NRB proteins analysed are located at positions 118 (Y), 130 (I) and 188 (T). Y118 and I130 are present in NSm proteins of other TSWV isolates, but T188 is exclusive of RB2 and the P321 isolates (GenBank accession number 307572726). This observation opens up the possibility that T188 may affect the transport capacity of the NSm protein. Further research is needed to confirm this hypothesis.
The AMV hybrids carrying the NSm genes were used to inoculate different plant species containing the Sw‐5 gene. Thus, we observed that the presence of the NRB NSm gene was always correlated with a significant reduction in the accumulation of viral RNAs in the inoculated leaves of tomato or Nicotiana species tested, carrying the resistance gene Sw‐5. This phenotype was also correlated with the absence of systemic virus infection. This result reproduces the same phenotype as observed for the TSWV wt in these resistant plants, in which the NRB isolates are able to infect the inoculated leaves, but have lost the capacity to move to the upper part of the plant. Together, the results obtained in the present work indicate that the NSm protein is the Avr determinant for the resistance mediated by the dominant gene Sw‐5.
Here, we also analysed whether the critical Y118 and N120 residues, proposed by López et al. (2011) to be responsible for overcoming Sw‐5 resistance, are sufficient to trigger this phenotype. To answer this question, we performed a mutational analysis using the RB1 protein that differs only in two residues (N120 and I130) from those of the NRB protein used herein. The analysis revealed that N120 was required to avoid the HR associated with Sw‐5‐resistant plants, but also that this residue negatively affected the cell‐to‐cell transport in the AMV heterologous system. The conservation of this amino acid in all members of the genus Tospovirus, except in the TSWV RB isolates (López et al., 2011), supports the functional importance (strong negative selection) of this amino acid residue. However, I130 significantly increased cell‐to‐cell transport, but triggered the HR in infected Nt/Sw5‐b or transiently expressed Nb/Sw5‐b plants. Interestingly, neither of the two single mutants was able to infect systemically the Nt/Sw5‐b plants. These results suggest that the change T120N, present in RB1, induces a fitness cost in the local movement of the chimeric construct, which was confirmed by competition experiments. However, with the AMV experimental system used, we cannot rule out the possibility that this fitness cost could be specific to the heterologous system or perhaps be overcome through secondary mutations (Sanjuán et al., 2004, 2005) located outside the NSm protein. In addition, the change V130I, present in NSm of most of the TSWV isolates available in databases (503 of 504 sequences), seems to be a positively selected residue for efficient cell‐to‐cell viral movement. Our results suggest that the RB isolates appear only in an I130 background. The fitness penalty is a prerequisite for both the resistance genes (R) and Avr genes in the different models proposed for the co‐evolution of the host–parasite in a gene‐for‐gene system (Bergelson et al., 2001; Burdon and Thrall, 2003; Sasaki, 2000; Segarra, 2005). This assumption is also supported by the small size of virus genomes, in which any modification of the few encoded multifunctional proteins could result in a fitness cost (Fraile and Garcia‐Arenal, 2010; Sacristán and Garcia‐Arenal, 2008). It was suggested that even a limited number of nucleotide changes in the virus genome may have strong pleiotropic effects. Mutations responsible for gains of virulence frequently induce fitness costs to the virus in plants which are devoid of the corresponding resistance. This has been shown in several instances (Agudelo‐Romero et al., 2008; Ayme et al., 2006; Desbiez et al., 2003; Goulden et al., 1993; Jenner et al., 2002; Lanfermeijer et al., 2003), although it cannot be generalized because there are examples of virulent strains that are at least as fit as the avirulent ones (Chain et al., 2007; Sorho et al., 2005). High fitness penalties associated with increased pathogenicity have been inferred for different plant viruses from direct (Fraile et al., 2011) and indirect (Culver et al., 1994; Hanada and Harrison, 1977; Mestre et al., 2003; Murant et al., 1968) evidence. The results presented herein support a high fitness penalty associated with the RB NSm gene, at least in the AMV system. This was confirmed experimentally by competition experiments in which the chimeric NRB RNA 3 outcompeted the RB1 and RB2 constructs in the absence of the Sw‐5 resistance gene, whereas the RB1 variant was prevalent in the Sw‐5‐resistant background, even outcompeting RB2. This latter result also suggests that the RB1 NSm isolate has less fitness penalty than RB2, at least in the resistant genotype, an effect that could be the consequence of a more permissive amino acid change or a more competitive evolved NSm gene. It is remarkable that most codons of NSm were found to be under neutral or purifying selection, and a positive selection was only detected at codon 118 as a result of the adaptation to overcome the resistance conferred by the Sw‐5 gene (López et al., 2011). The same observation was suggested for the substitution T120N, although the small number of isolates showing this change might have precluded its detection by the statistical methods used (López et al., 2011). The results presented herein support a positive selection for N120 under the selection pressure of the resistance gene Sw‐5. In addition, the observation of different fitness penalties between the two RB NSm forms may indicate that both genes are evolving to compensate for the fitness loss associated with these amino acid changes (Y118 and N120). If this is the correct scenario, it can be questioned how long it will take for other mutations to appear in RB NSm able to compete (with similar or higher fitness) with the NRB NSm in a context in which absence of the resistance gene Sw‐5 occurs. Further research is needed to study this aspect and to confirm whether the results obtained with the AMV system could be applied to TSWV.
Experimental Procedures
Recombinant plasmids for the introduction of the NSm genes in the AMV RNA 3 and for transient expression
A modified infectious AMV cDNA 3 clone, which expresses GFP (pGFP/A255/CP) (Sánchez‐Navarro and Bol, 2001), was used to exchange the N‐terminal 255 amino acids of the AMV MP gene with the corresponding MP gene (NSm) of TSWV. Three TSWV isolates derived from natural infections of tomato, two Sw‐5‐RB [named GRAU (GenBank FM163370) and Llo2TL3 (GenBank HM015518)] and one Sw5‐NRB [Gr1NL2 (GenBank HM015513)] were used as templates to amplify the MP gene (López et al., 2011) employing specific primers. The MP genes are referred to as RB1 (GRAU isolate), RB2 (Llo2TL3 isolate) and NRB (Gr1NL2 isolate). The digested fragments were used to replace the NcoI‐NheI fragment of pGFP/A255/CP, corresponding to the N‐terminal 255 amino acids of the AMV MP, to generate the constructs pGFP/RB1:A44/CP, pGFP/RB2:A44/CP and pGFP/NRB:A44/CP, respectively.
In addition, the TSWV MP genes were introduced into an infectious cDNA 3 clone of AMV wt (pAL3NcoP3) (van der Vossen et al., 1993) by exchanging the NcoI‐PstI fragment between the pAL3NcoP3 plasmid and the pGFP/A255/CP derivatives, described above. The resultant chimeric plasmids were referred as pRB1:A44/CP, pRB2:A44/CP and pNRB:A44/CP.
The pGFP/RB1:A44/CP and pRB1:A44/CP plasmids were used as templates to introduce, by directed mutagenesis, the substitutions T120 (substitution N for T at position 120) and V130 (substitution I for V at position 130) of the MP, resulting in the mutant constructs pGFP/RB1‐T120:A44/CP or pGFP/RB1‐V130:A44/CP and pRB1‐T120:A44/CP or pRB1‐V130:A44/CP, respectively.
For the transient expression of the different TSWV MPs, the previously amplified MP genes were introduced into the expression cassette of the plasmid pSK+ 35S‐MPPNRSV:HA‐PoPit (Martinez‐Gil et al., 2009) by exchanging the Prunus necrotic ringspot virus (PNRSV) MP gene. The resulting cassettes contain the corresponding TSWV MP fused to the HA epitope at its C‐terminus. Each cassette was introduced into the pMOG800 binary vector using a unique XhoI restriction site.
Inoculation of N. tabacum plants and tomato cultivars
pAL3NcoP3, pGFP/A255/CP and the corresponding NSm derivatives were linearized with PstI and transcribed with T7 RNA polymerase. The transcripts were inoculated onto transgenic N. tabacum plants that express constitutively the P1 and P2 polymerase proteins of AMV (P12), as described previously (Taschner et al., 1991). The fluorescence derived from the chimeric AMV RNA 3, carrying the GFP, was monitored using a Leica stereoscopic microscope (Wetzlar, Germany). The area of the infection foci was measured at 2 and 3 dpi using Image J software (Wayne Rasband, National Institutes of Health, Bethesda, MD, USA; http://rsbweb.nih.gov/ij).
Nicotiana tabacum wt plants (Nt/wt) and N. tabacum plants expressing constitutively the resistance gene Sw5‐b (Nt/Sw5‐b) (Spassova et al., 2001), and the tomato cultivars ‘Verdi’ (heterozygous for the Sw‐5 resistance gene) and ‘Marmande’ (which does not carry Sw‐5) (provided by Semillas Fitó, Barcelona, Spain), were inoculated with a mixture of capped transcripts corresponding to AMV RNAs 1, 2, the wt or chimeric RNA 3 plus few micrograms of purified AMV CP, as described previously (Neeleman and Bol, 1999).
For the competition assays, the inoculum contained a mixture of AMV RNAs 1 and 2 plus the three RNA 3 transcripts, at the same concentration, derived from the pRB1:A44/CP, pRB2:A44/CP and pNRB:A44/CP plasmids. P12 and Nt/Sw5‐b plants were inoculated as described above and two serial passages at 7 dpi were performed using an extract of the inoculated leaves as inoculum.
Northern blot and tissue printing assays
Tissue printing analyses were performed using a transverse section of the corresponding petiole, as described previously (Fajardo et al., 2013). Total RNA was extracted from inoculated (I) and upper (U) non‐inoculated leaves at 7 and 14 dpi, as described previously (Sánchez‐Navarro et al., 1997). In the case of the upper leaves, the RNA extraction was performed using a mixture of U3, U4 and U5 leaves, in which U1 corresponds to that closest to the inoculated leaf. Hybridization and detection were conducted as described previously (Pallás et al., 1998) using a dig‐riboprobe (Roche Mannheim, Germany) complementary to the AMV 3′‐untranslated region (3′‐UTR). The intensity of the bands was quantified using Image J 1.48c software (http://imagej.nih.gov/ij).
Transient expression of the TSWV MPs in planta and Western blot assay
Agrobacterium tumefaciens, strain C58, transformed with the corresponding binary pMOG 800 plasmid, was grown overnight in a shaking incubator at 28 °C in Luria–Bertani (LB) medium supplemented with the appropriate antibiotic. Cultures were collected by centrifugation and adjusted to an optical density at 600 nm (OD600) of 0.5 with 10 mm MgCl2, 10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES), pH 5.6, and 150 μm acetosyringone. These suspensions were used to infiltrate the different plants, as described previously (Herranz et al., 2005). The expression of the different viral MPs was analysed by Western blot assay, as described previously (Martinez‐Gil et al., 2009). Blots were developed using an ECL+ Plus Western Blotting Detection System (Amersham; Little Chalfont, Buckinghamshire, UK) and the LAS‐3000 digital imaging system (FujiFilm; Tokyo, Japan). The intensity of the bands was quantified using ImageGauge 4.0 software (FujiFilm).
Supporting information
Fig. S1 Sequence alignment of the three NSm proteins derived from the two Tomato spotted wilt virus (TSWV) resistance‐ breaking (RB) isolates GRAU (RB1; GenBank FM163370) and Llo2TL3 (RB2; GenBank HM015518) and the non‐resistance‐breaking (NRB) isolate Gr1NL2 (GenBank HM015513). The amino acids of the RB isolates that differ from those of the NRB variant are indicated in red.
Table S1 Symptoms observed in Nicotiana benthamiana (Nb) and N. tabacum (Nt) plants, wild‐type (wt) or carrying the Sw5‐b gene (Sw5‐b), inoculated with Tomato spotted wilt virus (TSWV) or chimeric Alfalfa mosaic virus (AMV) constructs.
Acknowledgements
A.P. was a recipient of a JAE‐Pre contract from the Consejo Superior de Investigaciones Científicas (CSIC), and M.C.C was a recipient of an I3P contract from CSIC (co‐financed by Fondo Social Europeo, FSE). We thank L. Corachán for excellent technical assistance and Dr Marcel Prins for providing the Nt/Sw5‐b and Nb/Sw5‐b seeds. This work was supported by grant BIO2011‐25018 from the Spanish granting agency DGICYT, grant PAID05‐11/2888 from the Universidad Politécnica de Valencia and by grant RTA2008‐00010‐C03 from the Instituto Nacional de Investigaciones Agrarias (INIA). All authors have no conflicts of interest to declare.
References
- Agudelo‐Romero, P. , de la Iglesia, F. and Elena, S.F. (2008) The pleiotropic cost of host‐specialization in Tobacco etch potyvirus. Infect. Genet. Evol. 8, 806–814. [DOI] [PubMed] [Google Scholar]
- Aramburu, J. and Marti, M. (2003) The occurrence in north‐east Spain of a variant of Tomato spotted wilt virus (TSWV) that breaks resistance in tomato (Lycopersicon esculentum) containing the Sw‐5 gene. Plant Pathol. 52, 407. [Google Scholar]
- Ayme, V. , Souche, S. , Caranta, C. , Jacquemond, M. , Chadoeuf, J. , Palloix, A. and Moury, B. (2006) Different mutations in the genome‐linked protein VPg of potato virus Y confer virulence on the pvr2(3) resistance in pepper. Mol. Plant–Microbe Interact. 19, 557–563. [DOI] [PubMed] [Google Scholar]
- Bergelson, J. , Dwyer, G. and Emerson, J.J. (2001) Models and data on plant–enemy coevolution. Annu. Rev. Genet. 35, 469–499. [DOI] [PubMed] [Google Scholar]
- Boiteux, L.S. (1995) Allelic relationships between genes for resistance to tomato spotted wilt tospovirus in Capsicum chinense . Theor. Appl. Genet. 90, 146–149. [DOI] [PubMed] [Google Scholar]
- Boiteux, L.S. and Giordano, L.D. (1993) Genetic‐basis of resistance against 2 tospovirus species in tomato (Lycopersicon‐esculentum). Euphytica, 71, 151–154. [Google Scholar]
- Burdon, J.J. and Thrall, P.H. (2003) The fitness costs to plants of resistance to pathogens. Genome Biol. 4, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder, V.L. and Palukaitis, P. (1992) Nucleotide‐sequence analysis of the movement genes of resistance breaking strains of tomato mosaic‐virus. J. Gen. Virol. 73, 165–168. [DOI] [PubMed] [Google Scholar]
- Canady, M.A. , Stevens, M.R. , Barineau, M.S. and Scott, J.W. (2001) Tomato spotted wilt virus (TSWV) resistance in tomato derived from Lycopersicon chilense Dun. LA 1938. Euphytica, 117, 19–25. [Google Scholar]
- Chain, F. , Riault, G. , Trottet, M. and Jacquot, E. (2007) Evaluation of the durability of the Barley yellow dwarf virus‐resistant Zhong ZH and TC14 wheat lines. Eur. J. Plant Pathol. 117, 35–43. [Google Scholar]
- Ciuffo, M. , Finetti‐Sialer, M.M. , Gallitelli, D. and Turina, M. (2005) First report in Italy of a resistance‐breaking strain of Tomato spotted wilt virus infecting tomato cultivars carrying the Sw5 resistance gene. Plant Pathol. 54, 564. [Google Scholar]
- Culver, J.N. , Stubbs, G. and Dawson, W.O. (1994) Structure–function relationship between tobacco mosaic virus coat protein and hypersensitivity in Nicotiana sylvestris . J. Mol. Biol. 242, 130–138. [DOI] [PubMed] [Google Scholar]
- Dawson, W.O. , Bubrick, P. and Grantham, G.L. (1988) Modifications of the tobacco mosaic virus coat protein gene affecting replication, movement, and symptomatology. Phytopathology, 78, 783–789. [Google Scholar]
- Desbiez, C. , Gal‐On, A. , Girard, M. , Wipf‐Scheibel, C. and Lecoq, H. (2003) Increase in Zucchini yellow mosaic virus symptom severity in tolerant zucchini cultivars is related to a point mutation in P3 protein and is associated with a loss of relative fitness on susceptible plants. Phytopathology, 93, 1478–1484. [DOI] [PubMed] [Google Scholar]
- Fajardo, T.V. , Peiro, A. , Pallás, V. and Sánchez‐Navarro, J. (2013) Systemic transport of Alfalfa mosaic virus can be mediated by the movement proteins of several viruses assigned to five genera of the 30K family. J. Gen. Virol. 94, 677–681. [DOI] [PubMed] [Google Scholar]
- Feng, Z. , Chen, X. , Bao, Y. , Dong, J. , Zhang, Z. and Tao, X. (2013) Nucleocapsid of Tomato spotted wilt tospovirus forms mobile particles that traffic on an actin/endoplasmic reticulum network driven by myosin XI‐K. New Phytol. 200, 1212–1224. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971) Current status of gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Fraile, A. and Garcia‐Arenal, F. (2010) The coevolution of plants and viruses: resistance and pathogenicity In: Natural and Engineered Resistance to Plant Viruses, Pt II (Carr J.P. and Loebenstein G., eds), pp. 1–32. London: Academic Press. [DOI] [PubMed] [Google Scholar]
- Fraile, A. , Pagan, I. , Anastasio, G. , Saez, E. and Garcia‐Arenal, F. (2011) Rapid genetic diversification and high fitness penalties associated with pathogenicity evolution in a plant virus. Mol. Biol. Evol. 28, 1425–1437. [DOI] [PubMed] [Google Scholar]
- Fraser, R.S.S. (1990) The genetics of resistance to plant‐viruses. Annu. Rev. Phytopathol. 28, 179–200. [Google Scholar]
- Gordillo, L.F. , Stevens, M.R. , Millard, M.A. and Geary, B. (2008) Screening two Lycopersicon peruvianum collections for resistance to Tomato spotted wilt virus. Plant Dis. 92, 694–704. [DOI] [PubMed] [Google Scholar]
- Goulden, M.G. , Kohm, B.A. , Cruz, S.S. , Kavanagh, T.A. and Baulcombe, D.C. (1993) A feature of the coat protein of potato virus‐x affects both induced virus‐resistance in potato and viral fitness. Virology, 197, 293–302. [DOI] [PubMed] [Google Scholar]
- de Haan, P. , Wagemakers, L. , Peters, D. and Goldbach, R. (1990) The S RNA segment of tomato spotted wilt virus has an ambisense character. J. Gen. Virol. 71, 1001–1007. [DOI] [PubMed] [Google Scholar]
- de Haan, P. , Kormelink, R. , de Oliveira Resende, R. , van Poelwijk, F. , Peters, D. and Goldbach, R. (1991) Tomato spotted wilt virus L RNA encodes a putative RNA polymerase. J. Gen. Virol. 72, 2207–2216. [DOI] [PubMed] [Google Scholar]
- Hanada, K. and Harrison, B.D. (1977) Effects of virus genotype and temperature on seed transmission of nepo‐viruses. Ann. Appl. Biol. 85, 79–92. [Google Scholar]
- Herranz, M.C. , Sánchez‐Navarro, J.A. , Sauri, A. , Mingarro, I. and Pallás, V. (2005) Mutational analysis of the RNA‐binding domain of the Prunus necrotic ringspot virus(PNRSV) movement protein reveals its requirement for cell‐to‐cell movement. Virology, 339, 31–41. [DOI] [PubMed] [Google Scholar]
- Hoffmann, K. , Qiu, W.P. and Moyer, J.W. (2001) Overcoming host‐ and pathogen‐mediated resistance in tomato and tobacco maps to the M RNA of Tomato spotted wilt virus. Mol. Plant–Microbe Interact. 14, 242–249. [DOI] [PubMed] [Google Scholar]
- Jenner, C.E. , Wang, X. , Ponz, F. and Walsh, J.A. (2002) A fitness cost for Turnip mosaic virus to overcome host resistance. Virus Res. 86, 1–6. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer, F.C. , Dijkhuis, J. , Sturre, M.J.G. , de Haan, P. and Hille, J. (2003) Cloning and characterization of the durable tomato mosaic virus resistance gene Tm‐2(2) from Lycopersicon esculentum . Plant Mol. Biol. 52, 1037–1049. [DOI] [PubMed] [Google Scholar]
- Latham, L.J. and Jones, R.A.C. (1998) Selection of resistance breaking strains of tomato spotted wilt tospovirus. Ann. Appl. Biol. 133, 385–402. [Google Scholar]
- Lewandowski, D.J. and Adkins, S. (2005) The tubule‐forming NSm protein from Tomato spotted wilt virus complements cell‐to‐cell and long‐distance movement of Tobacco mosaic virus hybrids. Virology, 342, 26–37. [DOI] [PubMed] [Google Scholar]
- Li, W. , Lewandowski, D.J. , Hilf, M.E. and Adkins, S. (2009) Identification of domains of the Tomato spotted wilt virus NSm protein involved in tubule formation, movement and symptomatology. Virology, 390, 110–121. [DOI] [PubMed] [Google Scholar]
- López, C. , Aramburu, J. , Galipienso, L. , Soler, S. , Nuez, F. and Rubio, L. (2011) Evolutionary analysis of tomato Sw‐5 resistance‐breaking isolates of Tomato spotted wilt virus. J. Gen. Virol. 92, 210–215. [DOI] [PubMed] [Google Scholar]
- Margaria, P. , Ciuffo, M. , Pacifico, D. and Turina, M. (2007) Evidence that the nonstructural protein of Tomato spotted wilt virus is the avirulence determinant in the interaction with resistant pepper carrying the TSW gene. Mol. Plant–Microbe Interact. 20, 547–558. [DOI] [PubMed] [Google Scholar]
- Martinez‐Gil, L. , Sánchez‐Navarro, J.A. , Cruz, A. , Pallás, V. , Perez‐Gil, J. and Mingarro, I. (2009) Plant virus cell‐to‐cell movement is not dependent on the transmembrane disposition of its movement protein. J. Virol. 83, 5535–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas, P. and Pallás, V. (1995) Non‐isotopic tissue‐printing hybridization: a new technique to study long‐distance plant virus movement. J. Virol. Methods, 52, 317–326. [DOI] [PubMed] [Google Scholar]
- Melcher, U. (2000) The ‘30K’ superfamily of viral movement proteins. J. Gen. Virol. 81, 257–266. [DOI] [PubMed] [Google Scholar]
- Meshi, T. , Motoyoshi, F. , Adachi, A. , Watanabe, Y. , Takamatsu, N. and Okada, Y. (1988) 2 concomitant base substitutions in the putative replicase genes of tobacco mosaic‐virus confer the ability to overcome the effects of a tomato resistance gene, tm‐1. EMBO J. 7, 1575–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshi, T. , Motoyoshi, F. , Maeda, T. , Yoshiwoka, S. , Watanabe, H. and Okada, Y. (1989) Mutations in the tobacco mosaic‐virus 30‐kD protein gene overcome tm‐2 resistance in tomato. Plant Cell, 1, 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre, P. , Brigneti, G. , Durrant, M.C. and Baulcombe, D.C. (2003) Potato virus Y NIa protease activity is not sufficient for elicitation of Ry‐mediated disease resistance in potato. Plant J. 36, 755–761. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C. , Dickerman, A.W. , Michelmore, R.W. , Sivaramakrishnan, S. , Sobral, B.W. and Young, N.D. (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide‐binding superfamily. Plant J. 20, 317–332. [DOI] [PubMed] [Google Scholar]
- Milne, R.G. and Francki, R.I. (1984) Should tomato spotted wilt virus be considered as a possible member of the family Bunyaviridae? Intervirology, 22, 72–76. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Selassie, K.G. , Marchoux, G. , Daubeze, A.M. and Palloix, A. (1998) High temperature effects on hypersensitive resistance to Tomato Spotted wilt Tospovirus (TSWV) in pepper (Capsicum chinense Jacq.). Eur. J. Plant Pathol. 104, 489–498. [Google Scholar]
- Murant, A.F. , Taylor, C.E. and Chambers, J. (1968) Properties, relationships and transmission of a strain of raspberry ringspot virus infecting raspberry cultivars immune to common Scottish strain. Ann. Appl. Biol. 61, 175–186. [Google Scholar]
- Neeleman, L. and Bol, J.F. (1999) Cis‐acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA. Virology, 254, 324–333. [DOI] [PubMed] [Google Scholar]
- Padgett, H.S. and Beachy, R.N. (1993) Analysis of a tobacco mosaic virus strain capable of overcoming N gene‐mediated resistance. Plant Cell, 5, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallás, V. , Mas, P. and Sánchez‐Navarro, J.A. (1998) Detection of plant RNA viruses by nonisotopic dot‐blot hybridization. Methods Mol. Biol. 81, 461–468. [DOI] [PubMed] [Google Scholar]
- de Ronde, D. , Butterbach, P. , Lohuis, D. , Hedil, M. , van Lent, J.W.M. and Kormelink, R. (2013) Tsw gene‐based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus. Mol. Plant Pathol. 14, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacristán, S. and Garcia‐Arenal, F. (2008) The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9, 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, T. , Meshi, T. , Takamatsu, N. and Okada, Y. (1987) Coat protein gene sequence of tobacco mosaic‐virus encodes a host response determinant. Proc. Natl. Acad. Sci. USA, 84, 6074–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Navarro, J.A. and Bol, J.F. (2001) Role of the Alfalfa mosaic virus movement protein and coat protein in virus transport. Mol. Plant–Microbe Interact. 14, 1051–1062. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Navarro, J.A. , Reusken, C.B.E. , Bol, J.F. and Pallás, V. (1997) Replication of alfalfa mosaic virus RNA 3 with movement and coat protein genes replaced by corresponding genes of Prunus necrotic ringspot ilarvirus. J. Gen. Virol. 78, 3171–3176. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Navarro, J.A. , Miglino, R. , Ragozzino, A. and Bol, J.F. (2001) Engineering of alfalfa mosaic virus RNA 3 into an expression vector. Arch. Virol. 146, 923–939. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Navarro, J.A. , Herranz, M.C. and Pallás, V. (2006) Cell‐to‐cell movement of Alfalfa mosaic virus can be mediated by the movement proteins of Ilar‐, bromo‐, cucumo‐, tobamo‐ and comoviruses and does not require virion formation. Virology, 346, 66–73. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Navarro, J.A. , Fajardo, T. , Zicca, S. , Pallás, V. and Stavolone, L. (2010) Caulimoviridae tubule‐guided transport is dictated by movement protein properties. J. Virol. 84, 4109–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán, R. , Moya, A. and Elena, S.F. (2004) The contribution of epistasis to the architecture of fitness in an RNA virus. Proc. Natl. Acad. Sci. USA, 101, 15 376–15 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán, R. , Cuevas, J.M. , Moya, A. and Elena, S.F. (2005) Epistasis and the adaptability of an RNA virus. Genetics, 170, 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A. (2000) Host–parasite coevolution in a multilocus gene‐for‐gene system. Proc. R. Soc. B: Biol. Sci. 267, 2183–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra, J. (2005) Stable polymorphisms in a two‐locus gene‐for‐gene system. Phytopathology, 95, 728–736. [DOI] [PubMed] [Google Scholar]
- Sin, S.H. , McNulty, B.C. , Kennedy, G.G. and Moyer, J.W. (2005) Viral genetic determinants for thrips transmission of Tomato spotted wilt virus. Proc. Natl. Acad. Sci. USA, 102, 5168–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorho, F. , Pinel, A. , Traoré, O. , Bersoult, A. , Ghesquière, A. , Hébrard, E. , Konaté, G. , Séré, Y. and Fargette, D. (2005) Durability of natural and transgenic resistances in rice to Rice yellow mottle virus. Eur. J. Plant Pathol. 112, 349–359. [Google Scholar]
- Spassova, M.I. , Prins, T.W. , Folkertsma, R.T. , Klein‐Lankhorst, R.M. , Hille, J. , Goldbach, R.W. and Prins, M. (2001) The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine‐rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol. Breed. 7, 151–161. [Google Scholar]
- Staskawicz, B.J. , Ausubel, F.M. , Baker, B.J. , Ellis, J.G. and Jones, J.D.G. (1995) Molecular‐genetics of plant‐disease resistance. Science, 268, 661–667. [DOI] [PubMed] [Google Scholar]
- Stevens, M.R. , Scott, S.J. and Gergerich, R.C. (1991) Inheritance of a gene for resistance to tomato spotted wilt virus (TSWV) from Lycopersicon peruvianum Mill. Euphytica, 59, 9–17. [Google Scholar]
- Storms, M.M. , Kormelink, R. , Peters, D. , van Lent, J.W. and Goldbach, R.W. (1995) The nonstructural NSm protein of tomato spotted wilt virus induces tubular structures in plant and insect cells. Virology, 214, 485–493. [DOI] [PubMed] [Google Scholar]
- Takeda, A. , Sugiyama, K. , Nagano, H. , Mori, M. , Kaido, M. , Mise, K. , Tsuda, S. and Okuno, T. (2002) Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett. 532, 75–79. [DOI] [PubMed] [Google Scholar]
- Taschner, P.E. , Van der Kuyl, A.C. , Neeleman, L. and Bol, J.F. (1991) Replication of an incomplete alfalfa mosaic virus genome in plants transformed with viral replicase genes. Virology, 181, 445–450. [DOI] [PubMed] [Google Scholar]
- Thompson, G.J. and vanZijl, J.J.B. (1995) Control of tomato spotted wilt virus in tomatoes in South Africa. Acta Hortic. 431, 379–384. [Google Scholar]
- van der Vossen, E.A. , Neeleman, L. and Bol, J.F. (1993) Role of the 5′ leader sequence of alfalfa mosaic virus RNA 3 in replication and translation of the viral RNA. Nucleic Acids Res. 21, 1361–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccardelli, M. , Perrone, D. , Del Galdo, A. , Campanile, F. , Parrella, G. and Giordano, I. (2008) Tomato genotypes resistant to tomato spotted wilt virus evaluated in open field crops in southern Italy. Acta Hortic. 789, 147–149. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Sequence alignment of the three NSm proteins derived from the two Tomato spotted wilt virus (TSWV) resistance‐ breaking (RB) isolates GRAU (RB1; GenBank FM163370) and Llo2TL3 (RB2; GenBank HM015518) and the non‐resistance‐breaking (NRB) isolate Gr1NL2 (GenBank HM015513). The amino acids of the RB isolates that differ from those of the NRB variant are indicated in red.
Table S1 Symptoms observed in Nicotiana benthamiana (Nb) and N. tabacum (Nt) plants, wild‐type (wt) or carrying the Sw5‐b gene (Sw5‐b), inoculated with Tomato spotted wilt virus (TSWV) or chimeric Alfalfa mosaic virus (AMV) constructs.


