SUMMARY
Two tobacco vein necrosis (TVN) determinants, the residues K400 and E419, have been identified previously in the helper component‐protease (HC‐Pro) protein sequence of Potato virus Y (PVY). However, since their description, non‐necrotic PVY isolates with both K400 and E419 necrotic determinants have been reported in the literature. This suggests the presence in the viral genome of other, as yet uncharacterized, TVN determinant(s). The identification of PVYN pathogenicity determinants was approached through the replacement of genomic regions of the necrotic PVYN‐605 infectious clone by corresponding sequences from the non‐necrotic PVYO‐139 isolate. Series of PVYN/O chimeras and site‐directed PVY mutants were constructed to test the involvement of different parts of the PVY genome (from nucleotide 421 to nucleotide 9629) in the induction of TVN symptoms. The analysis of both the genomic characteristics and biological properties of these mutants made it possible to highlight the involvement, in addition to residues K400 and E419, of the residue N339 of the HC‐Pro protein and two regions in the cytoplasmic inclusion (CI) protein to nuclear inclusion protein a‐protease (NIa‐Pro) sequence (nucleotides 5496–5932 and 6233–6444) in the induction of vein necrosis in tobacco infected by PVY isolates.
INTRODUCTION
Potato virus Y (PVY), one of the most important plant viruses (Scholthof et al., 2011), is the type member of the genus Potyvirus (family Potyviridae). The PVY genome, a single‐stranded positive sense RNA of approximately 10 kb, encodes a polyprotein that is cleaved by three virus‐encoded proteases into 10 products (Dougherty and Carrington, 1988) corresponding, from the N‐terminus to the C‐terminus of the polyprotein, to P1, helper component‐protease (HC‐Pro), P3, 6K1, cytoplasmic inclusion (CI) protein, 6K2, genome‐linked viral protein (VPg), nuclear inclusion protein a‐protease (NIa‐Pro), nuclear inclusion protein b (NIb) and coat protein (CP). A short overlapping gene (PIPO), embedded within the previously described large open reading frame (ORF), has been proposed recently for some potyviruses, including PVY (Chung et al., 2008). PVY is an economically important plant virus and a damaging virus affecting a wide host range, including Solanaceae family members, such as potato and tobacco (Valkonen, 2007). Biological, serological and molecular properties of PVY isolates have been used to create a complex PVY classification (Fauquet et al., 2005), which is still being discussed by international experts working on this virus (Singh et al., 2008). Thus, PVY is subdivided into strains (according to the host from which isolates were originally collected), groups (based mainly on symptoms induced in indicator hosts and on abilities to overcome selected resistance sources) and putative subgroups (containing isolates with particular properties). PVY isolates collected from potato plants have been classified into five groups, including the two main PVYN and PVYO groups, in which isolates that are either able to induce (PVYN) or not (PVYO) veinal necrosis symptoms on Nicotiana tabacum cv. Xanthi leaves are classified. Necrotic symptoms induced by PVY infection result in yield and quality reduction. In potato crops, PVY isolates cause major yield losses of up to 80% (Bokx and Hunttinga, 1981; Van der Zaag, 1987). In addition to the yield reduction, PVY can seriously affect the quality of the harvested tubers as a result of necrotic ringspot disease (Kerlan, 2006). In tobacco crops, infection by PVY causes height reduction, induces veinal necrosis symptoms and modifies the chemical composition of cured leaves, especially the nicotine content (Latorre et al., 1984). Consequently, tobacco yield losses resulting from PVY infections can reach 100%. Other major crop species affected by PVY include pepper and tomato, where emerging strains of PVY cause serious damage to yields and fruit quality (Kerlan and Moury, 2008). As a result of the agronomical impacts of the necrotic properties of PVY, the identification of the molecular determinants involved in the pathogenicity of this viral species has always been an important scientific challenge.
Comparisons between the biological properties and molecular characteristics of PVYN and PVYO isolates have suggested the involvement of the region from the 3′ end of the P1 gene to the 5′ end of the P3 gene in the necrotic capacity of PVY isolates (Glais et al., 2002). Moreover, a reverse genetics approach has demonstrated the role of amino acids K400 and E419 of the C‐terminal part of the HC‐Pro protein (Tribodet et al., 2005) in the induction of tobacco vein necrosis (TVN) symptoms. However, in this work, Tribodet et al. (2005) restricted their genome scanning procedure for the presence of the molecular determinants involved in TVN to the 2086–2763 nucleotide region of the PVY genome. Consequently, they did not rule out the possible presence of other TVN determinants in other parts of the viral sequence. Some PVY isolates, e.g. L26 (Hu et al., 2009), SASA‐61 (Barker et al., 2009; Schubert et al., 2007) and LW (Schubert et al., 2007), code for an HC‐Pro protein with both K400 and E419 residues, but do not induce veinal necrosis symptoms on infected tobacco plants. This clearly indicates that other, as yet unidentified, molecular determinants are involved in addition to, or as an alternative to, the K400/E419 residues in the necrotic ability of PVY isolates. In the case of the L26 isolate, sequence alignment performed using genomic data from both necrotic and non‐necrotic isolates suggested that the replacement of an aspartic acid by a glycine at position 205 (D205 to G205) in the HC‐Pro protein sequence was linked to the non‐necrotic property of the L26 isolate (Hu et al., 2009). This suggests an important role for the HC‐Pro residue D205 in the induction of TVN symptoms in tobacco plants infected by PVY isolates. However, all reported partial or full‐length sequenced genomes of PVY isolates, except L26, encode a D205 residue that is not correlated with the necrotic/non‐necrotic ability, reducing the possible impact of this residue in the biological properties of natural PVY isolates. Thus, further analyses need to be carried out for the accurate identification of the molecular determinants involved in the necrotic property of PVY isolates.
In this study, approaches were used to identify new viral molecular determinants involved in the expression of symptoms in PVY‐infected tobacco plants. First, chimeras resulting from the introduction of non‐necrotic PVYO‐139 sequences in the necrotic PVYN‐605 infectious clone (Jakab et al., 1997) were constructed and tested for their biological properties on tobacco plants. Thus, regions located in the HC‐Pro, CI, VPg and NIa‐Pro proteins were tested for their involvement in the PVY necrosis capacity. Then, using a series of point‐mutated versions of either the PVYN‐605 infectious clone or a non‐necrotic PVYN/O chimera, a residue located at the C‐terminal part of the HC‐Pro protein and two domains located at the CI–VPg–NIa‐Pro region were identified as new molecular determinants crucial for the TVN property of PVY isolates. Finally, alignments of 85 sequences from necrotic/non‐necrotic PVY isolates were used to analyse the link between the polymorphism of HC‐Pro residues, known for their role in the necrotic property of PVY, and the biological properties of isolates on tobacco plants.
RESULTS
Genomic regions involved in the induction of TVN in N. tabacum cv. Xanthi
The identification of PVYN pathogenicity determinants was approached through a strategy based on the construction of PVYN/O chimeras resulting from genomic exchanges between the infectious clone PVYN‐605 and the reference PVYO‐139 isolate. Five different regions of the 5′ half of the PVYN‐605 genomic sequence (nucleotides 421–4278) and four different regions of the 3′ half of the PVYN‐605 genomic sequence (nucleotides 4278–9629) were replaced by the corresponding regions of the PVYO‐139 genome. To extend the procedure to the complete PVY genome (9701 nucleotides), nucleotides 1–420 and 9629–9701 need to be tested. However, modification of the 5′ end (nucleotides 1–420) of the genome in the PVY infectious clone was not possible. Indeed, attempts to modify this region using standard molecular biology procedures resulted in unexpected modifications of the genomic organization of the viral sequence present in the recombinant plasmid. The 9629–9701 nucleotide region corresponds to the 3′ untranslated region of the PVY genome and contains only seven PVYN‐605/PVYO‐139 polymorphic nucleotides. Thus, genomic exchange for this region was not included in this work. Consequently, the presented procedure makes it possible to test the involvement of 94.9% of the viral genome and 97.5% of the coding sequence in the necrotic properties of PVYN‐605. Chimeric PVYN/O full‐length clones were created from a ligation of one genetically modified subclone (either modified N‐605 5′ half or modified N‐605 3′ half subclone) and the other wild‐type subclone (either N‐605 3′ half or N‐605 5′ half subclone, respectively). These viral constructs (Fig. 1) were inoculated using a previously published biolistically based procedure (see Experimental procedures and Tribodet et al., 2005) onto either N. clevelandii or N. benthamiana plants. The infection efficiency of the wild‐type PVYN‐605 infectious clone was, on average, 27% for five independent inoculation experiments [infection efficiencies ranged from 0% (0/15) to 54% (8/15)]. The variation of the infection efficiency obtained for the wild‐type infectious PVYN‐605 clone highlights the lack of repeatability of the biolistically based inoculation procedure used under our experimental conditions. Thus, the percentage of infected plants obtained for a single inoculation experiment performed with a clone should not be used to determine the level of infectivity. The detection of virus in the inoculated plants was performed on non‐inoculated leaves at 3 weeks post‐inoculation using enzyme‐linked immunosorbent analysis (ELISA). The chimeric PVYN/O clones tested were all infectious, as denoted by the production of at least one infected plant for each construct (Fig. 1). The ELISA results [optical density at 405 nm (OD405) above 2.0] associated with the non‐inoculated leaves from infected plants indicated that viral progenies present at 21 days post‐inoculation of the hosts had efficiently spread from inoculated tissue to the whole plant. In addition to the previously tested PVYN/ONrBg clone (Tribodet et al., 2005), eight PVYN/O clones (PVYN/OBsAg, PVYN/OAgNr, PVYN/OFusion, PVYN/ODrBs, PVYN/OBsSw, PVYN/OSwNc, PVYN/ONcBg and PVYN/OBgTt) were used to localize, in the 421–9629 nucleotide region of the PVY genome, molecular determinants involved in the PVY necrosis property. To determine the capacity of PVYN/O clones to induce TVN, the leaf symptoms of infected N. tabacum cv. Xanthi were monitored (1, 2). As expected, N. tabacum plants infected by the PVYN/ONrBg clone expressed mosaic symptoms. However, mosaic was also observed on PVYN/OAgNr‐ and PVYN/OSwNc‐infected plants. The size of the viral progeny present in N. tabacum cv. Xanthi‐infected plants (data resulting from five to seven plants for each viral construct) at 21 days post‐inoculation was calculated for the non‐necrotic PVYN/ONrBg (on average 5.18 × 1010 copies/plant), PVYN/OAgNr (on average 6.94 × 1010 copies/plant) and PVYN/OSwNc (on average 4.26 × 1010 copies/plant) chimeric mutants and compared with the size of the progeny obtained under similar conditions for the necrotic PVYN/OBsAg (on average 5.52 × 1010 copies/plant), PVYN/OFusion (on average 2.21 × 1010 copies/plant), PVYN/ODrBs (on average 7 × 1010 copies/plant) and PVYN/OBsSw (on average 5.24 × 1010 copies/plant). No obvious differences were noted between these viral quantities, indicating that necrotic and non‐necrotic PVYs accumulate similarly in infected plants. These results highlight the role of the PVYN‐605 regions between AgeI (nucleotide 1356) and NruI (nucleotide 2086) restriction sites and between SwaI (nucleotide 5496) and NcoI (nucleotide 6444) restriction sites in the induction of TVN. For each chimeric construct, the sequence of the viral progeny produced in N. tabacum cv. Xanthi was compared with the genomic sequence of the inoculated PVYN/O infectious clone. No genetic difference was denoted by these sequence analyses (data not shown).
Figure 1.
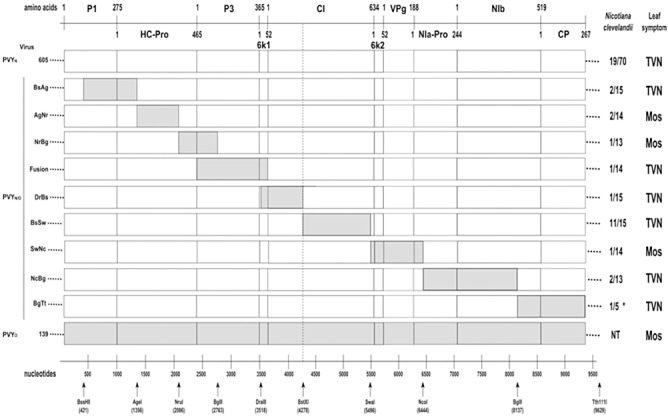
Schematic representation of the Potato virus Y (PVY) genome (PVYN‐605, PVYO‐139 and PVYN/O chimeric constructs) used to identify regions involved in the induction of tobacco vein necrosis symptoms. White and grey boxes correspond to PVYN‐605 and PVYO‐139 sequences, respectively. Amino acids and nucleotide scales are presented according to Jakab et al. (1997). Results of biolistic inoculations onto Nicotiana clevelandii are presented as ‘number of infected plants/number of inoculated plants’. TVN and Mos leaf symptoms denote the ability of the PVY isolates to induce tobacco vein necrosis or mosaic symptoms, respectively, on N. tabacum cv. Xanthi leaves. NT, not tested. *PVYN/O‐BgTt was inoculated onto N. benthamiana instead of N. clevelandii.
Figure 2.
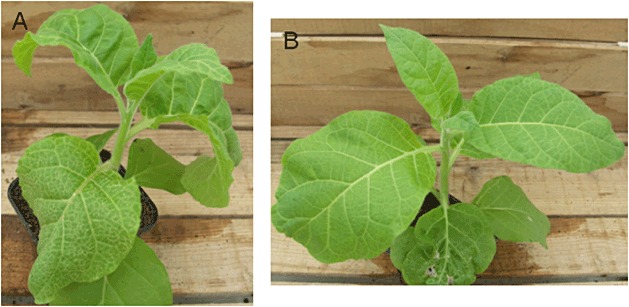
Symptoms observed on Potato virus Y (PVY)‐infected Nicotiana tabacum cv. Xanthi. Tobacco vein necrosis (A) and mosaic (B) symptoms observed 14 days after mechanical inoculations performed with PVYN‐605 and PVYO‐139, respectively.
Molecular determinants of the AgeI‐NruI (AgNr) region
In order to determine the identity of molecular determinants of the AgNr region involved in the induction of TVN in PVY‐infected N. tabacum cv. Xanthi plants, the PVYN/OO3N and PVYN/ON3O chimeric clones were constructed (Fig. 3A). These two PVYN/O chimeras made it possible to test the involvement of the 1356–1900 and 1901–2086 nucleotide regions in the necrotic property of PVY. The results of the biological characterization showed that the infection of plants by PVYN/ON3O was associated with the expression of mosaic symptoms, whereas the infection of N. tabacum by PVYN/OO3N resulted in the necrosis of infected leaves. Alignment and comparison of the amino acid sequences of the HC‐Pro protein domain overlapping the 1901–2086 nucleotide region revealed, between the PVYN‐605 and PVYO‐139 isolates, six polymorphic residues located at positions 310, 332, 339, 347, 350 and 355 of the HC‐Pro protein (Fig. 3B). Thus, six point‐mutated versions of the PVYN‐605 infectious clone, each with a single substitution at one of the candidate positions listed above, were created and used to test the link between these residues and the ability of the PVY sequence to induce TVN in N. tabacum cv. Xanthi. PVYN‐605 mutants with a PVYO‐type residue at position 310, 332, 347, 350 or 355 were able to induce necrotic symptoms on infected plants, similar to the parental PVYN‐605 infectious clone. However, the replacement of the PVYN‐type amino acid located at position 339 by the residue present at the same position in the PVYO‐139 sequence resulted in the modification of the PVY biological property (from necrotic to mosaic) on infected N. tabacum cv. Xanthi (Fig. 3A, PVYN/Oaa339‘O’). Thus, the asparagine at position 339 (N339) in the C‐terminal part of the HC‐Pro protein seems to be crucial in the process that leads to the induction of TVN in PVY‐infected N. tabacum cv. Xanthi plants.
Figure 3.
Genomic organization and biological properties of PVYN/O chimeric constructs (A), and amino acid alignment of the region N3O of PVYN‐605 and PVYO‐139 (B). White and grey boxes correspond to PVYN‐605 and PVYO‐139 sequences, respectively. Amino acids and nucleotide scales are presented according to Jakab et al. (1997). Grey stars denote PVYN to PVYO point mutations. White stars denote N/D339, K/R400 and E/D419 point mutations. The results of biolistic inoculations onto Nicotiana clevelandii are presented as ‘number of infected plants/number of inoculated plants’. TVN and Mos leaf symptoms denote the ability of the PVY isolates to induce tobacco vein necrosis or mosaic symptoms, respectively, on N. tabacum cv. Xanthi leaves. Black boxes: consensus sequence. Sequence highlighted in the grey box corresponds to the HC‐Pro PVYO sequence on the PVYN/ON3O clone. *Data resulting from two independent inoculation experiments.
Necrotic property of HC‐Pro protein requires residues N339, K400 and E419
The PVYN/OHC‐Pro‘O’ clone possesses a PVYN‐605 genetic background and a type‐O HC‐Pro coding sequence. Nicotiana tabacum cv. Xanthi plants infected by this viral chimera expressed mosaic symptoms (Fig. 3A). This is in accordance with the results associated with the previously reported data, as the PVYN/OHC‐Pro‘O’ clone encodes a type‐O HC‐Pro protein with D339, R400 and D419 residues. However, to test directly the impact of residues N339, K400 and E419 on the necrotic property of PVY, type‐N point mutations at positions corresponding to residues 339, 400 and 419 were introduced in the PVYN/OHC‐Pro‘O’ clone. The resulting PVYN/OHC‐Pro‘O’/aa‘N’ clone was inoculated onto N. tabacum cv. Xanthi plants. As infected plants expressed TVN symptoms (Fig. 3A), the presence of N339, K400 and E419 in a type‐O HC‐Pro protein is sufficient to induce TVN symptoms. To extend this result to a larger number of isolates, HC‐Pro sequence data from 85 PVY isolates known for their biological properties were retrieved from GenBank or collected from the wide sequencing programme carried out by the PVYwide Organization (http://www.inra.fr/pvy_organization). This analysis showed that 46 of the 47 isolates able to induce TVN encode a HC‐Pro protein with the ‘NKE’ triplet. As already reported by Tribodet et al. (2005), the necrotic N‐Sc isolate encodes a G419 instead of E419. Most (26/38) of the non‐necrotic PVY isolates encode the non‐necrotic ‘DRD’ triplet instead of ‘NKE’ (Table 1). Some (7/38) non‐necrotic isolates have a single variation in the necrotic triplet that obviously alters their necrotic abilities. The non‐necrotic status of the L26 isolate, which encodes the necrotic ‘NKE’ triplet, is supported by the residue G205, as proposed by Hu et al. (2009). Finally, four isolates (SASA‐61, LW, 26 and PVY‐12) of the tested viral collection induced mosaic symptoms on N. tabacum cv. Xanthi, but encoded a HC‐Pro protein with the necrotic ‘NKE’ triplet.
Table 1.
Characteristics (strain and biological property on Nicotiana tabacum cv. Xanthi) of Potato virus Y (PVY) isolates and identity of helper component‐protease (HC‐Pro) residues 205, 339, 400 and 419.
Two subregions of the SwaI‐NcoI (SwNc) region are linked to the necrotic property of PVY isolates
To determine which part of the SwNc region is involved in the induction of TVN, the sequence located between nucleotides 5496 and 6444 [positions according to Jakab et al. (1997)] was divided into three regions: R1 (nucleotides 5496–5932), R2 (nucleotides 5932–6233) and R3 (nucleotides 6233–6444). Each of these type‐N regions was replaced (alone or in combination) in the PVYN‐605 infectious clone by the corresponding type‐O sequence to produce six different mutants (SwNc_R1, SwNc_R2, SwNc_R3, SwNC_R1/R3, SwNc_R2/R3 and SwNc_R1/R2; Fig. 4). All constructs were checked to be error free. Five independent inoculations (using 15 test plants each) were performed with the PVYN/OSwNc_R1/R2 mutant. None produced infected N. clevelandii, whereas a single inoculation procedure performed with the other constructs (chimera or point‐mutated versions of the wild‐type PVYN‐605 infectious clone) produced at least one infected plant. Thus, it seems that the PVYN/OSwNc_R1/R2 construct made using the genetic background of the PVYN‐605 infectious clone and R1/R2 sequence (nucleotides 5496–6233) from the PVYO‐139 isolate is not infectious. The noninfectious status of PVYN/OSwNc_R1/R2 was unexpected. Indeed, to our knowledge, none of the PVYN/PVYO chimeric and none of the type‐O point‐mutated versions of the PVYN‐605 sequence tested so far (Bukovinszki et al., 2007; Moury et al., 2011; Rolland et al., 2009; Tribodet et al., 2005; E. Jacquot, unpublished data) has been described as a noninfectious construction when inoculated onto Nicotiana hosts. Thus, it seems that the genomic organization of the CI–6K2–NIa region of the PVYN/OSwNc_R1/R2 genome alters at least one of the critical steps of the viral cycle, resulting in a lack of production of viral progeny from the full‐length sequence inoculated onto test plants. Consequently, it was not possible to test the corresponding genomic organization (i.e. SwNc_R1/R2) for its necrosis capacity. Results of the biological characterization carried out with the other chimeras showed that the infection of plants by PVYN/OSwNc_R1, PVYN/OSwNc_R2, PVYN/OSwNc_R3 and PVYN/OSwNc_R2/R3 clones was associated with the expression of necrotic symptoms, whereas the infection of tobacco by the PVYN/OSwNc_R1/R3 mutant resulted in mosaic symptoms (Fig. 4). Alignment and comparison of the amino acid sequences corresponding to the R1/R3 region revealed, between the PVYN‐605 and PVYO‐139 isolates, 16 polymorphic residues. These polymorphic residues correspond to N/D613 and M/I622 of the CI protein, T/A2, V/T14, V/A21, Q/K22 and L/I25 of the 6K2 protein, and R/K59 and V/I61 of the VPg protein, and to I/V173, D/N176 and K/A182 of the VPg protein, and V/L21, V/A43, K/R49 and F/Y51 of the NIa‐Pro protein, for R1 and R3 regions, respectively.
Figure 4.
Genomic organization and biological properties of PVYN/O chimeric constructs. White and grey boxes correspond to PVYN‐605 and PVYO‐139 sequences, respectively. Amino acids and nucleotide scales are presented according to Jakab et al. (1997). The results of biolistic inoculations onto Nicotiana clevelandii are presented as ‘number of infected plants/number of inoculated plants’. TVN and Mos leaf symptoms denote the ability of the PVY isolates to induce tobacco vein necrosis or mosaic symptoms, respectively, on N. tabacum cv. Xanthi leaves. *Data resulting from five independent inoculation experiments.
DISCUSSION
The identification of the molecular determinants involved in pathogenesis is important to better understand plant–pathogen interactions and to answer different fundamental and applied questions linked to plant protection. Necrotic symptoms induced by PVY isolates cause major yield losses in both potato and tobacco crops (Latorre et al., 1984; Le Romancer et al., 1994). The optimization of control methods against PVY needs to be at least partly based on processes that target the necrotic capacity of PVY isolates. In a previous study, two TVN determinants, K400 and E419, were localized in the C‐terminal part of the HC‐Pro protein encoded by necrotic PVY isolates (Tribodet et al., 2005). However, this study did not rule out the presence of other TVN determinant(s) in the PVY genomic sequence, as only a short part (23.27%) of the candidate region P1/HC‐Pro/P3 (nucleotides 421–2591; positions according to Jakab et al., 1997) was tested. Recently, Hu et al. (2009) have suggested the involvement of the residue D205 of the HC‐Pro protein in the induction of TVN. These authors based their approach on comparisons between the molecular and biological properties of a series of necrotic and non‐necrotic PVY isolates. The resulting analysis showed that all except the non‐necrotic L26 isolate encoded a D205 residue. Consequently, the authors concluded that D205 is a molecular determinant involved in the necrotic properties of PVY isolates. However, even though D205 obviously plays an important role in the ability of PVY isolates to induce necrotic symptoms, the absence of polymorphism for this residue in natural PVY populations (including PVYN‐605 and PVYO‐139) does not allow the identification of this residue as a molecular marker for this biological property through genomic exchange between PVY isolates. Moreover, these data suggest that this residue does not play an important role in the necrotic vs. non‐necrotic properties for natural PVY populations, and that other molecular determinants directly involved in the differentiation of necrotic and non‐necrotic isolates should exist in the PVY genomic sequence. Thus, in order to localize and identify TVN determinant(s) in the 421–9629 nucleotide region of the PVY genome, chimeras and site‐directed mutants were constructed and tested for their biological properties on N. tabacum cv. Xanthi. The procedure used made it possible to map, in the HC‐Pro protein, the position of a new TVN determinant: the residue N339. The results demonstrated that the triple substitution of N339, K400 and E419 for D339, R400 and D419 in a type‐O HC‐Pro sequence is sufficient to modify the phenotype of the corresponding PVY isolate from mosaic to TVN. Moreover, the modification of any single residue of the ‘NKE’ triplet in the viral sequence of the necrotic isolate leads to the expression of mosaic symptoms instead of necrosis of tobacco leaves, as demonstrated by: (i) the non‐necrotic PVYKR and PVYED (Tribodet et al., 2005) and PVYN/O aa339‘O’ (this work) point‐mutated versions of the necrotic infectious PVYN‐605 clone; and (ii) the non‐necrotic ‘N:Ominus’ isolates (e.g. 11, 25, 31 and 34, Table 1) without necrotic‐type residues at position 339 or 400. Surprisingly, our PVY library does not include a wild‐type non‐necrotic isolate with N339, K400 and not E419. We cannot propose an explanation for this observation, but, based on our library, it seems that the polymorphism of residue 339 is more frequent in natural PVY populations than polymorphism at positions 400 and 419.
The K/R400 and E/D419 modifications of the HC‐Pro sequence could be considered as slight modifications of the protein characteristics. Indeed, the differences between residues for these pairs of amino acids correspond to a single carbon in their lateral arms. A reduction in enzyme activity caused by substitution from lysine (K) to arginine (R) has been reported in the literature (Liu and Roy, 2001). The N/D339 polymorphism resulted in a change from a polar (N) to a negatively charged (D) amino acid. However, the protein structure prediction proposed by the Phyre server (Kelley and Sternberg, 2009) does not seem to be influenced by the modification of the ‘NKE’/‘DRD’ triplet in the HC‐Pro protein sequence (data not shown). Moreover, all tested point‐mutated versions of the PVYN‐605 genetic background with single [PVYKR and PVYED (Rolland et al., 2009) and PVYN/Oaa339‘O’] and multiple [PVYN/O‐KE/RD (Tribodet et al., 2005) and PVYN/OHC‐Pro‘O’/aa‘N’] modifications of the ‘NKE’ triplet are replication competent, suggesting that polymorphisms of residues N/D339, K/R400 and/or E/D419 in the HC‐Pro sequence do not drastically influence the multiple biological functions of the HC‐Pro protein (for a review, see Urcuqui‐Inchima et al., 2001). The HC‐Pro protein has long been identified as a major symptom determinant (Atreya et al., 1992; Gal‐On, 2000; Klein et al., 1994; Tribodet et al., 2005). Moreover, HC‐Pro can interact with the NtMind protein of N. tabacum cv. Xanthi NN, which plays an important role in chloroplast division (Jin et al., 2007) and, consequently, in the physiology of tobacco plants. The C‐terminal part of HC‐Pro is involved in the movement of virus in infected plants (Maia et al., 1996), in RNA silencing suppression (RSS) activity (Plisson et al., 2003; Varrelmann et al., 2007), in the cleavage (protease activity) of the viral polyprotein in functional proteins (Maia et al., 1996) and in the induction of local lesions on leaves of potato genotypes carrying the Ncspl resistance genes (Moury et al., 2011). Moreover, a recent study has proposed HC‐Pro as an interaction partner of the translation initiation factor with a 4E‐binding site located at the C‐terminal part of the potyviral protein (Ala‐Poikela et al., 2011). Thus, the identification in the C‐terminal part of the HC‐Pro protein of molecular markers linked to the TVN property is both coherent with current knowledge linked to this multifunctional protein and strengthens the complex and important role of HC‐Pro in the multiple steps of the viral infection cycle.
In different pathogen–plant interactions, necrosis of infected tissues is likely to prevent movement of the pathogen in the host (Eggenberger et al., 2008; Lorrain et al., 2004; Yambao et al., 2008). It would be interesting to investigate whether the modification of residues of the ‘NKE’ triplet influence, in addition to the phenotype of the infected plant, the qualitative and/or quantitative characteristics of the viral progeny produced in the infected host. As the HC‐Pro protein is a suppressor of post‐transcriptional gene silencing (PTGS), this protein has a direct impact on the limitation of viral RNAs in the infected plant (Fukuzawa et al., 2010; Urcuqui‐Inchima et al., 2001). Indeed, different levels of HC‐Pro‐dependent suppression (hypo‐ or hyper‐suppressor) were observed for variants of Tobacco etch potyvirus (Torres‐Barcelo et al., 2008). Studies have shown that virus‐induced gene silencing delays cell death (Garcia‐Marcos et al., 2009), decreases symptoms and reduces the accumulation level of viral progeny (Yambao et al., 2008). However, some mutations in RSS motifs that result in an attenuation of symptoms without affecting virus accumulation have also been reported (Desbiez et al., 2010; Saenz et al., 2001; Torres‐Barcelo et al., 2008). The change in ‘NKE’ amino acids could modify qualitative and/or quantitative parameters of the viral cycle. Indeed, it has been demonstrated that the amino acids K/R400 and E/D419 have an impact on the accumulation of PVY in Nicotiana hosts (Rolland et al., 2009). Moreover, the fitness [i.e. the viral load in infected plants estimated by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) assays] of non‐necrotic mutants resulting from the introduction of point mutation(s) in the PVYN‐605 infectious clone at residue(s) 400 and/or 419 of the HC‐Pro sequence (i.e. PVYKR, PVYED and PVYN/O‐KR/ED) is lower than the fitness of non‐necrotic PVYO‐139, but higher than the fitness of necrotic PVYN‐605 (Rolland et al., 2009). Thus, the modification of PVYN‐605 necrotic properties through point mutation(s) seems to have a positive impact on the replication, movement and/or accumulation of the virus in its host. However, the quantitative data, collected with certain constructs used during this work employing real‐time RT‐PCR, did not show variations between isolates/mutants for their ability to systematically infect and accumulate in host plants. Thus, the PVYN/PVYO genetic exchanges of the genomic background tested in this study modify the symptoms observed on infected leaves, but do not have an impact on the capacity of PVY to systemically infect and accumulate in its host. Consequently, in addition to its role in the induction of necrotic symptoms, impacts of N/D339 mutation on the viral cycle need to be investigated to complete the data previously collected on both K/R400 and E/D419 (2009, 2010) and to improve our knowledge on the biological properties of PVY populations.
The molecular identity of the ‘K400E419’/‘R400D419’ residues in the HC‐Pro sequence allows the description of the necrotic ability of 89.4% (76/85) of the isolates present in our international PVY library. The use of the N/D339 determinant as a third marker for the TVN property raised the percentage of accurate assignment of isolates to their appropriate necrotic/non‐necrotic group to 95.3% (81/85). According to the knowledge on the characteristics of the L26 isolate (Hu et al., 2009), only isolates PVY‐26, PVY‐12, SASA‐61 and LW, i.e. 4.7% of the tested isolates (4/85), are not accurately characterized by the described TVN markers for their biological behaviour on N. tabacum cv. Xanthi. Recently published studies have described non‐necrotic PVYE isolates with the ‘NKE’ triplet in their HC‐Pro sequence (Galvino‐Costa et al., 2012) and rare necrotic PVY isolates with the non‐necrotic ‘DRD’ triplet (Tian et al., 2011). These nonconventional isolates suggest the existence, in addition to or as an alternative to the ‘NKE’ triplet, of viral genetic determinant(s) of vein necrosis in tobacco that could correspond to either PVYN‐605/PVYO‐139 conserved HC‐Pro residues not tested in Tribodet et al. (2005) or in this study, or to PVY genetic information located in another part of the viral genome. Indeed, in addition to its capacity to interact with different host proteins, HC‐pro is known to form multimers (Guo et al., 1999) and to interact with several other potyviral proteins, including P1 (Merits et al., 1999), CI (Guo et al., 2001), VPg (Roudet‐Tavert et al., 2007), NIa (Guo et al., 2001) and CP (Roudet‐Tavert et al., 2002). Moreover, a recent study has demonstrated that HC‐Pro, linked to its host partner 4E, is found in infected cells in association with the viral 6K2‐induced vesicles, suggesting an interaction between 4E–HC‐Pro and 6K2 proteins (Ala‐Poikela et al., 2011). However, it is important to keep in mind that other parameters [e.g. RNA conformation (Krause‐Sakate et al., 2005) or viral quasispecies (Sanz‐Ramos et al., 2008)] could also influence the symptomatology of viral infections.
The involvement of multiple proteins of potyviruses in overcoming resistance responses has been reported. One determinant in the HC‐Pro region and two determinants in the P3 region have been described for their involvement in the virulence of SMV‐G7 on Rsv1‐genotype soybean (Eggenberger et al., 2008; Hajimorad et al., 2008). Turnip mosaic virus (TuMV) requires mutations in both P3 and CI regions to overcome the resistance response, but these determinants are attributed to the existence of two independent R genes against the virus in the host (Jenner et al., 2002). In the present study, R1 (nucleotides 5496–5932, corresponding to the C‐terminal part of the CI protein, 6K2 and the N‐terminal part of the VPg protein) and R3 (nucleotides 5933–6444, corresponding to the C‐terminal part of the VPg protein and the N‐terminal part of the NIa‐Pro protein) domains, containing polymorphic residues in the C‐terminus of CI (2/16), in the 6K2 sequence (5/16), in both the N‐ and C‐termini of the VPg protein (2/16 and 3/16, respectively) and in the N‐terminus of the NIa‐Pro protein (4/16), were described for their involvement in the induction of TVN symptoms. According to our results and to current knowledge on protein–protein interactions involving potyviral products, the 16 polymorphic residues described in regions R1 and R3 are found in proteins known to interact with HC‐Pro. All these residues/proteins constitute good candidates for the further characterization of PVY–tobacco interactions that lead to the expression of leaf necrosis symptoms. Thus, the investigation of the role of each polymorphic residue in the R1/R3 regions constitutes one of the next steps in the identification of the determinants involved in the pathogenicity of PVY isolates.
EXPERIMENTAL PROCEDURES
Plants, viruses and PVYN‐605 infectious clone
Nicotiana clevelandii or N. benthamiana, which are not able to respond with TVN symptoms to infection by PVYN group members, were used to initiate infection with wild‐type and mutated versions of the PVYN‐605 infectious clone. Nicotiana tabacum cv. Xanthi was used as indicator host to monitor for TVN symptoms induced by necrotic PVY isolates. Healthy and infected plants were grown in separate regulated insect‐proof glasshouses at 20 °C. The PVYN‐605 (GenBank accession no. X97895; Jakab et al., 1997) and PVYO‐139 (GenBank accession no. U09509; Singh and Singh, 1996) isolates were used as references for PVYN and PVYO groups, respectively. PVYO‐139 and PVYN‐605 isolates were maintained on N. tabacum cv. Xanthi by mechanical inoculation. The infectious PVYN‐605 clone (Jakab et al., 1997), used to construct PVY chimeras and mutants, is a bipartite system constituted by the N‐605p5′ and N‐605p3′ subclones corresponding to the 5′ (nucleotides 1–4278) and 3′ (nucleotides 4278–9701) halves of the viral genome, respectively. Prior to being inoculated, the infectious full‐length clone requires a reconstruction step as described previously (Tribodet et al., 2005). Briefly, 100 µg of the two purified subclones were digested using BstXI (300 U) and KpnI (300 U). N‐605p5′ was digested in the presence of Calf Intestinal alkaline Phosphatase (CIP) (10 U). The digested plasmids were purified using the phenol–chloroform extraction procedure. Then, DNA fragments were mixed in the presence of T4 DNA ligase (150 U) for 16 h at 16 °C. Ligated DNAs were extracted by the phenol–chloroform procedure, resuspended in 50 µL of nuclease‐free water and stored at −20 °C until use for DNA‐coated gold particle preparation.
PVY genomic sequences overlapping the HC‐Pro coding region were retrieved from the GenBank database or kindly provided by the PVYwide Organization (http://www.inra.fr/pvy_organization). Information associated with the PVY sequences used in this study is listed in Table 1.
Cloning of PVYN/O chimeras and mutants
Selected unique restriction sites (illustrated in Fig. 1), present in the N‐605p5′ or N‐605p3′ subclone, were used to create PVYN/OBsAg, PVYN/OAgNr, PVYN/ONrBg, PVYN/ODrBs, PVYN/OBsSw, PVYN/OSwNc, PVYN/ONcBg and PVYN/OBgTt clones (Fig. 1 and Table S1, see Supporting Information). Each of these PVYN/O‐605 subclones was produced in two successive cloning steps as described previously (Tribodet et al., 2005). Briefly, the first step of the cloning procedure consisted of the amplification of the PVYO targeted region (e.g. nucleotides 421–1356, i.e. between the BssHII and AgeI restriction sites) by RT‐PCR using an appropriate primer pair containing restriction sites which frame the targeted sequence (e.g. BssHII and AgeI restriction sites) and viral RNA extracted from PVYO‐139‐infected N. tabacum. The second step of the cloning procedure involved the insertion of the RT‐PCR fragment in a modified version of the appropriate N‐605p5′ or N‐605p3′ subclone, where the corresponding PVYN sequence (e.g. the region between BssHII and AgeI restriction sites) had been previously deleted. The PVYN/OFusion, PVYN/OHC‐Pro‘O’, PVYN/OO3N, PVYN/ON3O, PVYN/OSwNc_R1, PVYN/OSwNc_R2, PVYN/OSwNc_R3, PVYN/OSwNc_R1/R2, PVYN/OSwNc_R2/R3 and PVYN/OSwNc_R1/R3 subclones (Table S2, see Supporting Information) were created using the fusion‐PCR method described by Catlett et al. (2003). For these constructs, a PCR‐fusion step was performed to link two or more PCR‐amplified DNA fragments [e.g. (i) PVYN P1 fragment with a BssHII restriction site; (ii) a PVYO HC‐Pro fragment framed at 5′ and 3′ ends by short overlap PVYN P1 and P3 sequences, respectively; and (iii) a PVYN P3 fragment with a Bstz17I restriction site). The resulting DNA fragment was subsequently cloned (e.g. using the BssHII and Bstz17I restriction sites) into a modified version of the appropriate N‐605p5′ or N‐605p3′ subclone in which the corresponding PVYN sequence (e.g. the region between BssHII and Bstz17I restriction sites) had been previously deleted.
Six point‐mutated versions of the PVYN‐605 infectious clone (PVYNaa310O, PVYNaa332O, PVYNaa339O, PVYNaa347O, PVYNaa350O and PVYNaa355O; Table S1), each with a single nucleotide substitution, were constructed using a megaprimer approach (Tyagi et al., 2004) to introduce a mutation at nucleotide positions 1942, 2009, 2029, 2054, 2063 and 2077 in the wild‐type N‐605p5′ subclone. Finally, PVYN/O‘O’/aa‘N’ was created by a megaprimer approach to introduce type‐N point mutations at nucleotide positions 2029, 2213 and 2271 in the PVYN/OHC‐Pro‘O’ construct (Table S1). Each of the mutated PCR fragments produced in the megaprimer procedure was cloned into a pSC‐A‐amp/kan vector (Agilent Technologies, La Jolla, CA, USA). The resulting pSC‐A‐amp/kan recombinant clones were digested by BstEII, BmgBI and HindIII to make possible the construction of a pSC‐A‐amp/kan vector containing a type‐O HC‐Pro coding sequence with the three type‐N point mutations. Finally, the HC‐Pro‘O’/aa‘N’ fragment from the pSC‐A‐amp/kan recombinant plasmid was inserted into PVYN/OHC‐Pro‘O’ by a restriction–ligation procedure using BstEII and Bstz17I restriction sites.
The nucleotide sequences (produced by Eurofins MWG Operon, Ebersberg, Germany) of the chimeras and mutated versions of the PVYN‐605 infectious clone were checked to be error free prior to being used in the biolistic‐mediated inoculation process (sequence of the viral subclones).
PVY inoculation, virus detection in plants and molecular analysis of the produced viral progenies
The different constructions were introduced into N. clevelandii or N. benthamiana by bombardment as described by Tribodet et al. (2005). Briefly, 25‐mg gold particles (1 µm in diameter) were mixed with 100 µL of spermidine (50 mm), sonicated for 4 s and added to 100 µg of ligated DNA. Then, cold CaCl2 (100 µL) was added slowly. The mixture was kept at room temperature for 10 min. After a centrifugation step at 10 000 g for 15 s, DNA‐coated gold particles were washed three times with 1 mL of cold absolute ethanol and transferred to 3 mL of ethanol containing 0.05 mg/mL PVP 360K. At this step, the DNA‐coated particles were transferred into a 63.5‐cm polypropylene tube, dried and cut into cartridges (each containing ± 2 µg DNA). Bombardments were performed at 200 psi with a distance of 3 cm between the gun (Helios Gene gun, Bio‐Rad, Hercules, CA, USA) and the targeted leaf. Each plant was inoculated with three cartridges on three separate leaves. After the bombardment, plants were kept under glasshouse conditions for 3 weeks. Viral progenies were detected and/or quantified in inoculated plants at 3 weeks post‐inoculation using a serological approach (ELISA) and PVY polyclonal antibodies, kindly provided by Maryse Guillet (INRA‐FNPPPT, Rennes, France), as described previously (Jacquot et al., 2005), and/or a molecular approach (real‐time RT‐PCR) according to the procedure described by Balme‐Sinibaldi et al. (2006). Viral populations present in infected N. clevelandii or N. benthamiana plants were transferred onto N. tabacum cv. Xanthi by mechanical inoculation. The sanitary status of inoculated N. tabacum cv. Xanthi plants was monitored by symptom observations (mosaic vs. vein necrosis) for 3 weeks after the inoculation step and confirmed by ELISA and/or real‐time RT‐PCR at the end of the monitored period (i.e. 21 days after inoculation). Then, the viral progenies present in infected plants were amplified by immunocapture RT‐PCR (IC‐RT‐PCR) as described previously (Glais et al., 1998) using the 3′NTR‐reverse primer (5′‐GTCTCCTGATTGAAGTTTAC‐3′) for the production of cDNA and appropriate primer pairs. The IC‐RT‐PCR procedure was repeated at least twice for each viral progeny, and each PCR product was sent to Eurofins MWG Operon (Germany). The sequences of the viral progenies present in infected N. tabacum cv. Xanthi were then compared with the genomic sequences of the corresponding wild‐type, chimera and mutated versions of the infectious PVYN‐605 clones.
Alignment of PVY sequences
The PVY sequences collected from the different databases were analysed using Geneious Pro 4.7.6 software (Biomatters Ltd., Auckland, New Zealand) (Drummond et al., 2009), and amino acids 339, 400 and 419 located at the C‐terminal part of the HC‐Pro protein were identified using the translation tool of the software. During the molecular analysis of the data, PVYN‐605 and PVYO‐139 sequences were used as references.
Supporting information
Table S1 Primers for the construction of Potato virus Y (PVY) infectious clones.
Table S2 Primers for the construction of Potato virus Y (PVY) infectious clones.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We are grateful to Dr Stewart Gray (Cornell University, Ithaca, NY, USA) from the PVYwide Organization who kindly provided some PVY isolates. The presented work was supported by the Institut National de la Recherche Agronomique (France) and the Fédération Nationale des Producteurs de Plants de Pommes de Terre (France).
REFERENCES
- Ala‐Poikela, M. , Goytia, E. , Haikonen, T. , Rajamäki, M.‐L. and Valkonen, J.P.T. (2011) Helper component proteinase of the genus potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif. J. Virol. 85, 6784–6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya, C.D. , Atreya, P.L. , Thornbury, D.W. and Pirone, T.P. (1992) Site‐directed mutations in the potyvirus HC‐Pro gene affect helper component activity, virus accumulation, and symptom expression in infected tobacco plants. Virology, 191, 106–111. [DOI] [PubMed] [Google Scholar]
- Balme‐Sinibaldi, V. , Tribodet, M. , Croizat, F. , Lefeuvre, P. , Kerlan, C. and Jacquot, E. (2006) Improvement of Potato virus Y (PVY) detection and quantitation using PVYN‐ and PVYO‐specific real‐time RT‐PCR assays. J. Virol. Methods, 134, 261–266. [DOI] [PubMed] [Google Scholar]
- Barker, H. , McGeachy, K.D. , Toplak, N. , Gruden, K. , Žel, J. and Browning, I. (2009) Comparison of genome sequence of PVY isolates with biological properties. Am. J. Pot. Res. 86, 227–238. [Google Scholar]
- Bokx, J.A. and Hunttinga, H. (1981) Potato virus Y. AAB/CMI Description of Plant Viruses, 242.
- Bukovinszki, Á. , Götz, R. , Johansen, E. , Maiss, E. and Balázs, E. (2007) The role of the coat protein region in symptom formation on Physalis floridana varies between PVY strains. Virus Res. 127, 122–125. [DOI] [PubMed] [Google Scholar]
- Catlett, N.L. , Lee, B.‐N. , Yoder, O.C. and Turgeon, B.G. (2003) Split‐marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Newsl. 50, 9–11. [Google Scholar]
- Chung, B.Y.W. , Miller, W.A. , Atkins, J.F. and Firth, A.E. (2008) An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA, 105, 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbiez, C. , Girard, M. and Lecoq, H. (2010) A novel natural mutation in HC‐Pro responsible for mild symptomatology of Zucchini yellow mosaic virus (ZYMV, Potyvirus) in cucurbits. Arch. Virol. 155, 397–401. [DOI] [PubMed] [Google Scholar]
- Dougherty, W.G. and Carrington, J.C. (1988) Expression and function of potyviral gene products. Annu. Rev. Phytopathol. 26, 123–143. [Google Scholar]
- Drummond, A. , Ashton, B. , Cheung, M. , Heled, J. , Kearse, M. , Moir, R. , Stones‐Havas, S. , Thierer, T. and Wilson, A. (2009) Geneious v4.7. Available at http://www.geneious.com/[accessed on April 4, 2012].
- Eggenberger, A.L. , Hajimorad, M.R. and Hill, J.H. (2008) Gain of virulence on Rsv1‐genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC‐Pro. Mol. Plant–Microbe Interact. 21, 931–936. [DOI] [PubMed] [Google Scholar]
- Fauquet, C.M. , Mayo, M.A. , Maniloff, J. , Desselberger, U. and Ball, L.A. (2005) Virus taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses. Academic Press edn (June 28, 2005), pp. 1162.
- Fukuzawa, N. , Itchoda, N. , Ishihara, T. , Goto, K. , Masuta, C. and Matsumura, T. (2010) HC‐Pro, a potyvirus RNA silencing suppressor, cancels cycling of Cucumber mosaic virus in Nicotiana benthamiana plants. Virus Genes, 40, 440–446. [DOI] [PubMed] [Google Scholar]
- Gal‐On, A. (2000) A point mutation in the FRNK motif of the potyvirus helper component‐protease gene alters symptom expression in cucurbits and elicits protection against the severe homologous virus. Phytopathology, 90, 467–473. [DOI] [PubMed] [Google Scholar]
- Galvino‐Costa, S.B.F. , dos Reis Figueira, A. , Camargos, V.V. , Geraldino, P.S. , Hu, X.‐J. , Nikolaeva, O.V. , Kerlan, C. and Karasev, A.V. (2012) A novel type of Potato virus Y recombinant genome, determined for the genetic strain PVYE. Plant Pathol. 61, 388–398. [Google Scholar]
- Garcia‐Marcos, A. , Pacheco, R. , Martianez, J. , Gonzalez‐Jara, P. , Diaz‐Ruiz, J.R. and Tenllado, F. (2009) Transcriptional changes and oxidative stress associated with the synergistic interaction between Potato virus X and Potato virus Y and their relationship with symptom expression. Mol. Plant–Microbe Interact. 22, 1431–1444. [DOI] [PubMed] [Google Scholar]
- Glais, L. , Tribodet, M. , Gauthier, J.P. , Astier‐Manifacier, S. , Robaglia, C. and Kerlan, C. (1998) RFLP mapping of the whole genome of ten viral isolates representative of different biological groups of potato virus Y. Arch. Virol. 143, 2077–2091. [DOI] [PubMed] [Google Scholar]
- Glais, L. , Tribodet, M. and Kerlan, C. (2002) Genomic variability in Potato potyvirus Y (PVY): evidence that PVY NW and PVY NTN variants are single to multiple recombinants between PVY O and PVY N isolates. Arch. Virol. 147, 363–378. [DOI] [PubMed] [Google Scholar]
- Guo, D. , Merits, A. and Saarma, M. (1999) Self‐association and mapping of interaction domains of helper component‐proteinase of potato A potyvirus. J. Gen. Virol. 80, 1127–1131. [DOI] [PubMed] [Google Scholar]
- Guo, D.Y. , Rajamäki, M.L. , Saarma, M. and Valkonen, J.P.T. (2001) Towards a protein interaction map of potyviruses: protein interaction matrixes of two potyviruses based on the yeast two‐hybrid system. J. Gen. Virol. 82, 935–939. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2008) Adaptation of Soybean mosaic virus avirulent chimeras containing P3 sequences from virulent strains to Rsv1‐genotype soybeans is mediated by mutations in HC‐Pro. Mol. Plant–Microbe Interact. 21, 937–946. [DOI] [PubMed] [Google Scholar]
- Hu, X. , Meacham, T. , Ewing, L. , Gray, S.M. and Karasev, A.V. (2009) A novel recombinant strain of Potato virus Y suggests a new viral genetic determinant of vein necrosis in tobacco. Virus Res. 143, 68–76. [DOI] [PubMed] [Google Scholar]
- Jacquot, E. , Tribodet, M. , Croizat, F. , Balme‐Sinibaldi, V. and Kerlan, C. (2005) A single nucleotide polymorphism‐based technique for specific characterization of YO and YN isolates of Potato virus Y (PVY). J. Virol. Methods, 125, 83–93. [DOI] [PubMed] [Google Scholar]
- Jakab, G. , Droz, E. , Brigneti, G. , Baulcombe, D. and Malnoe, P. (1997) Infectious in vivo and in vitro transcripts from a full‐length cDNA clone of PVY‐N605, a Swiss necrotic isolate of potato virus Y. J. Gen. Virol. 78, 3141–3145. [DOI] [PubMed] [Google Scholar]
- Jenner, C.E. , Tomimura, K. , Ohshima, K. , Hughes, S.A. and Wash, J.A. (2002) Mutations in Turnip mosaic virus P3 and cylindrical inclusion protein are separately required to overcome two Brassica napus resistance genes. Virology, 300, 50–59. [DOI] [PubMed] [Google Scholar]
- Jin, Y. , Ma, D. , Dong, J. , Li, D. , Deng, C. , Jin, J. and Wang, T. (2007) The HC‐pro protein of potato virus Y interacts with NtMinD of tobacco. Mol. Plant–Microbe Interact. 20, 1505–1511. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A. and Sternberg, M.J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. [DOI] [PubMed] [Google Scholar]
- Kerlan, C. (2006) Potato virus Y. AAB/CMI Description of Plant Viruses, 414.
- Kerlan, C. and Moury, B. (2008) Potato virus Y In: Encyclopedia of Virology, 3rd edn. (Mahy B.W.J. and Regenmorte M.I., eds), pp. 287–296. Tennessee: Academic Press. [Google Scholar]
- Klein, P.G. , Klein, R.R. , Rodriguez‐Cerezo, E. , Hunt, A.G. and Shaw, J.G. (1994) Mutational analysis of the tobacco vein mottling virus genome. Virology, 204, 759–769. [DOI] [PubMed] [Google Scholar]
- Krause‐Sakate, R. , Redondo, E. , Richard‐Forget, F. , Jadao, A.S. , Houvenaghel, M.C. , German‐Retana, S. , Pavan, M.A. , Candresse, T. , Zerbini, F.M. and Le Gall, O. (2005) Molecular mapping of the viral determinants of systemic wilting induced by a Lettuce mosaic virus (LMV) isolate in some lettuce cultivars. Virus Res. 109, 175–180. [DOI] [PubMed] [Google Scholar]
- Latorre, B.A. , Flores, V. and Marholz, G. (1984) Effect of potato virus Y on growth, yield, and chemical composition of flue‐cured tobacco in Chile. Plant Dis. 68, 884–886. [Google Scholar]
- Le Romancer, M. , Kerlan, C. and Nedellec, M. (1994) Biological characterisation of various geographical isolates of potato virus Y inducing superficial necrosis on potato tubers. Plant Pathol. 43, 138–144. [Google Scholar]
- Liu, X. and Roy, R. (2001) Mutation at active site lysine 212 to arginine uncouples the glycosylase activity from the lyase activity of human endonuclease III. Biochemistry, 40, 13 617–13 622. [DOI] [PubMed] [Google Scholar]
- Lorrain, S. , Lin, B. , Auriac, M.C. , Kroj, T. , Saindrenan, P. , Nicole, M. , Balagué, C. and Roby, D. (2004) Vascular associated death1, a novel GRAM domain‐containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell, 16, 2217–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia, I.G. , Haenni, A. and Bernardi, F. (1996) Potyviral HC‐Pro: a multifunctional protein. J. Gen. Virol. 77, 1335–1341. [DOI] [PubMed] [Google Scholar]
- Merits, A. , Guo, D. , Järvekülg, L. and Saarma, M. (1999) Biochemical and genetic evidence for interactions between Potato A potyvirus‐encoded proteins P1 and P3 and proteins of the putative replication complex. Virology, 263, 15–22. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Caromel, B. , Johansen, E. , Simon, V. , Chauvin, L. , Jacquot, E. , Kerlan, C. and Lefebvre, V. (2011) The helper component proteinase cistron of potato virus Y induces hypersensitivity and resistance in potato genotypes carrying dominant resistance genes on chromosome IV. Mol. Plant–Microbe Interact. 24, 787–797. [DOI] [PubMed] [Google Scholar]
- Plisson, C. , Drucker, M. , Blanc, S. , German‐Retana, S. , Le Gall, O. , Thomas, D. and Bron, P. (2003) Structural characterization of HC‐Pro, a plant virus multifunctional protein. J. Biol. Chem. 278, 23 753–23 761. [DOI] [PubMed] [Google Scholar]
- Rolland, M. , Kerlan, C. and Jacquot, E. (2009) The acquisition of molecular determinants involved in potato virus Y necrosis capacity leads to fitness reduction in tobacco plants. J. Gen. Virol. 90, 244–252. [DOI] [PubMed] [Google Scholar]
- Rolland, M. , Delaunay, A. , Baldwin, T.K. , Kerlan, C. and Jacquot, E. (2010) Complementation and exclusions between mutated versions of a Potato Virus Y genotype during mixed infections of Nicotiana hosts. Virus Res. 153, 197–204. [DOI] [PubMed] [Google Scholar]
- Roudet‐Tavert, G. , German‐Retana, S. , Delaunay, T. , Delécolle, V. , Candresse, T. and Le Gall, O. (2002) Interaction between potyvirus helper component‐proteinase and capsid protein in infected plants. J. Gen. Virol. 83, 1765–1770. [DOI] [PubMed] [Google Scholar]
- Roudet‐Tavert, G. , Michon, T. , Walter, J. , Delaunay, T. , Redondo, E. and Le Gall, O. (2007) Central domain of a potyvirus VPg is involved in the interaction with the host translation initiation factor eIF4E and the viral protein HCPro. J. Gen. Virol. 88, 1029–1033. [DOI] [PubMed] [Google Scholar]
- Saenz, P. , Quiot, L. , Quiot, J.B. , Candresse, T. and Garcia, J.A. (2001) Pathogenicity determinants in the complex virus population of a Plum pox virus isolate. Mol. Plant–Microbe Interact. 14, 278–287. [DOI] [PubMed] [Google Scholar]
- Sanz‐Ramos, M. , Diaz‐San Segundo, F. , Escarmis, C. , Domingo, E. and Sevilla, N. (2008) Hidden virulence determinants in a viral quasispecies in vivo. J. Virol. 82, 10 465–10 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof, K.‐B.G. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. , Hohn, B. , Saundners, K. , Candresse, T. , Ahlquist, P. , Hemenway, C. and Foster, G.D. (2011) Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, J. , Fomitcheva, V. and Sztangret‐Wisniewska, J. (2007) Differentiation of Potato virus Y strains using improved sets of diagnostic PCR‐primers. J. Virol. Methods, 140, 66–74. [DOI] [PubMed] [Google Scholar]
- Singh, M. and Singh, R.P. (1996) Nucleotide sequence and genome organization of a Canadian isolate of the common strain of potato virus Y (PVYO). Can. J. Plant Pathol. 18, 209–224. [Google Scholar]
- Singh, R.P. , Valkonen, J.P.T. , Gray, S.M. , Boonham, N. , Jones, R.A.C. , Kerlan, C. and Schubert, J. (2008) Discussion paper: the naming of Potato virus Y strains infecting potato. Arch. Virol. 153, 1–13. [DOI] [PubMed] [Google Scholar]
- Tian, Y.P. , Liu, J.L. , Zhang, C.L. , Liu, Y.Y. , Wang, B. , Li, X.D. , Guo, Z.K. and Valkonen, J.P.T. (2011) Genetic diversity of Potato virus Y infecting tobacco crops in China. Phytopathology, 101, 377–387. [DOI] [PubMed] [Google Scholar]
- Torres‐Barcelo, C. , Martin, S. , Daros, J.A. and Elena, S.F. (2008) From hypo‐ to hypersuppression: effect of amino acid substitutions on the RNA‐silencing suppressor activity of the Tobacco etch potyvirus HC‐Pro. Genetics, 180, 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribodet, M. , Glais, L. , Kerlan, C. and Jacquot, E. (2005) Characterization of Potato virus Y (PVY) molecular determinants involved in the vein necrosis symptom induced by PVYN isolates in infected Nicotiana tabacum cv. Xanthi. J. Gen. Virol. 86, 2101–2105. [DOI] [PubMed] [Google Scholar]
- Tyagi, R. , Lai, R. and Duggleby, R.G. (2004) A new approach to ‘megaprimer’ polymerase chain reaction mutagenesis without an intermediate gel purification step. BMC Biotechnol. 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urcuqui‐Inchima, S. , Haenni, A.L. and Bernardi, F. (2001) Potyvirus proteins: a wealth of functions. Virus Res. 74, 157–175. [DOI] [PubMed] [Google Scholar]
- Valkonen, J. (2007) Viruses: economical losses and biotechnological potential In: Potato Biology and Biotechnology: Advances and Perspectives (Vreugdenhil D., Bradshaw J., Gebhardt C., Govers F., MacKerron D., Taylor M. and Ross H., eds), pp. 619–641. Dundee: Elsevier. [Google Scholar]
- Van der Zaag, D.E. (1987) Yield reduction in relation to virus infection In: Viruses of Potatoes and Seed‐Potato Production (De Bokx J.A. and Van der Want J.P.H., eds), pp. 149–150. Wageningen: Pudoc. [Google Scholar]
- Varrelmann, M. , Maiss, E. , Pilot, R. and Palkovics, L. (2007) Use of pentapeptide‐insertion scanning mutagenesis for functional mapping of the plum pox virus helper component proteinase suppressor of gene silencing. J. Gen. Virol. 88, 1005–1015. [DOI] [PubMed] [Google Scholar]
- Yambao, M.L. , Yagihashi, H. , Sekiguchi, H. , Sekiguchi, T. , Sasaki, T. , Sato, M. , Atsumi, G. , Tacahashi, Y. , Nakahara, K.S. and Uyeda, I. (2008) Point mutations in helper component protease of clover yellow vein virus are associated with the attenuation of RNA‐silencing suppression activity and symptom expression in broad bean. Arch. Virol. 153, 105–115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primers for the construction of Potato virus Y (PVY) infectious clones.
Table S2 Primers for the construction of Potato virus Y (PVY) infectious clones.
Supporting info item
Supporting info item


