Summary
Genetic resistance provides efficient control of crop diseases, but is limited by pathogen evolution capacities which often result in resistance breakdown. It has been demonstrated recently, in three different pathosystems, that polygenic resistances combining a major‐effect gene and quantitative resistance controlled by the genetic background are more durable than monogenic resistances (with the same major gene in a susceptible genetic background), but the underlying mechanisms are unknown. Using the pepper–Potato virus Y system, we examined three mechanisms that could account for the greater durability of the polygenic resistances: (i) the additional quantitative resistance conferred by the genetic background; (ii) the increase in the number of mutations required for resistance breakdown; and (iii) the slower selection of adapted resistance‐breaking mutants within the viral population. The three mechanisms were experimentally validated. The first explained a large part of the variation in resistance breakdown frequency and is therefore expected to be a major determinant of resistance durability. Quantitative resistance factors also had an influence on the second mechanism by modifying the virus mutational pathways towards resistance breakdown and could also have an influence on the third mechanism by increasing genetic drift effects on the viral population. The relevance of these results for other plant–pathogen systems and their importance in plant breeding are discussed.
Introduction
Plant breeding for disease resistance has abundantly and successfully exploited single resistance genes with major phenotypic effects. These resistances usually confer efficient protection with low environmental impacts and low production costs. However, the major limit of genetic control is resistance breakdown (RB) resulting from adaptation of the target pathogen. RB usually involves three steps (Moury et al., 2011). The first is the appearance of an adapted mutant in the pathogen population. This mutant further competes with the other genotypes of the pathogen population and may accumulate substantially in primary infected plants depending on its competitive ability (step 2). The more competitive the mutant, the higher its probability of transmission to other plants (step 3). RB occurs, sometimes very rapidly after resistance deployment (García‐Arenal and McDonald, 2003), when these three steps take place sufficiently frequently and efficiently in pathogen populations. Thus, as major resistance genes have a restricted availability among genetic resources, they should be preserved and used judiciously.
Strategies to increase the durability of genetic resistances have been discussed in several articles (Johnson, 1981; Lindhout, 2002; Parlevliet, 2002; Pink, 2002; Wolfe, 1985) and one of the most interesting is the combination of a major resistance gene with a partially resistant genetic background (quantitative resistance). For a long time hypothesized, the efficiency of this strategy has been validated recently for three highly different plant–pathogen systems. Palloix et al. (2009) showed, in the pepper (Capsicum annuum)–Potato virus Y (PVY) system, that a major resistance gene (pvr23), which proved to be very poorly durable when introgressed in a susceptible cultivar, could become highly durable in the laboratory, as well as in field conditions, when introgressed into a partially resistant genetic background. The higher durability of a polygenic resistance, composed of one major gene combined with partial resistance quantitative trait loci (QTLs) in the genetic background, relative to a monogenic resistance, with the same major gene in a susceptible background, was confirmed for a plant–fungus interaction with the oilseed rape (Brassica napus)–Leptosphaeria maculans system (Brun et al., 2010). Moreover, Fournet et al. (2012) showed, for the potato cyst nematode Globodera pallida, that the genetic background in which a major resistance factor is introgressed plays an important role in the durability of the resistance. These results are very encouraging for plant breeders, and their extrapolation to other pathosystems will gain from exploring the mechanisms responsible for this enhancement in durability.
Three mechanisms could explain how a quantitative resistance can increase the durability of a major resistance gene.
The additional quantitative resistance level conferred by the genetic background could reduce the multiplication of the pathogen and its probability to accumulate mutations responsible for RB (Brun et al., 2010). This ‘resistance efficiency’ mechanism would act on the first step of RB.
The number and nature of mutations required by the pathogen for RB also condition the probability of appearance of adapted variants. Indeed, Harrison (2002) proposed that the combination of several genes was more durable because the pathogen requires more mutations to break down the resistance. Moreover, the nature (transition or transversion) and the fitness cost of these mutations are also involved in the capacity of RB pathogens to appear and to accumulate (Ayme et al., 2007; Fabre et al., 2009; Janzac et al., 2010). These mechanisms would act on the first and second steps of RB.
Finally, the selection pressure exerted by a quantitative resistance on pathogen populations could be lower than that exerted by a major resistance gene, slowing down the emergence of adapted variants, as hypothesized by several authors (Keller et al., 2000; Kliebenstein and Rowe, 2009; Knott, 1988; Palloix et al., 2009). This mechanism would act on the second and third steps of RB.
In order to explain how the plant genetic background protects a major resistance gene from breakdown, we tested these three mechanisms using the C. annuum–PVY system.
Results
Relationship between the breakdown of the pvr23 resistance gene and PVY accumulation
To measure the effect of the first mechanism (‘resistance efficiency’), Perennial and a set of 15 doubled haploid (DH) lines carrying the pvr23 resistance allele, but segregating for the genetic background, were tested for relationships between the rate of pvr23 RB and virus accumulation in plants. The breakdown frequency of pvr23 was estimated after inoculation with a recombinant PVY clone, the ‘CI chimera’ (Montarry et al., 2011). The ‘CI chimera’ is not infectious per se in plants carrying the pvr23 gene, i.e. in these plants, mutants of the ‘CI chimera’ possessing single nonsynonymous substitutions in the viral protein genome‐linked (VPg) cistron were detected, but not the ‘CI chimera’ itself. In addition, these substitutions were shown to determine the breakdown of pvr23 (Ayme et al., 2006). The ‘CI chimera’ showed a particularly high capacity of RB (Montarry et al., 2011). Indeed, 5 weeks after inoculation, the plants in which PVY was detected at the systemic level correspond to RBs (Montarry et al., 2011). Three independent replicates of the test were performed and a high correlation (0.88 < ρ Spearman < 0.96; P < 0.001) was observed between them. Among the 16 lines, pvr23 breakdown frequency varied from 0% to 100%, with highly significant differences between lines (Fig. 1, Table S2, see Supporting Information). The pvr23 breakdown frequency observed with another PVY clone (SON41p) in the 16 lines was highly correlated with that observed with the ‘CI chimera’ (ρ Spearman = 0.91; P < 0.001), but eight of the lines showed a very low (<10%) breakdown frequency (data not shown), which decreased the accuracy of correlation analysis between RB and PVY accumulation.
Figure 1.
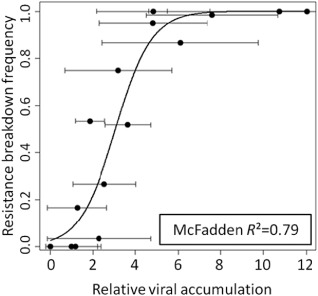
Relationship between resistance breakdown frequency and viral accumulation among Perennial and 14 doubled haploid (DH) lines issued from the F1 hybrid between Capsicum annuum genotypes Perennial and Yolo Wonder and carrying the pvr2 3 major resistance gene. The evaluation of resistance breakdown frequency was performed with 60 plants inoculated with Potato virus Y (PVY) ‘CI chimera’, and the evaluation of viral accumulation was performed by double antibody sandwich enzyme‐linked immunosorbent assay (DAS‐ELISA) on 10 plants inoculated with a variant of ‘CI chimera’ breaking down pvr2 3 (mutant ‘CI chimera VPg‐N’). Black dots correspond to experimental values and bars correspond to the standard deviation for each value. The full line corresponds to the logistic regression model fitted to the data.
A single mutant of the ‘CI chimera’, named ‘CI chimera VPg‐N’, was used to measure the virus accumulation at the systemic level in the lines by quantitative double antibody sandwich enzyme‐linked immunosorbent assay (DAS‐ELISA). The ‘CI chimera VPg‐N’ differs from the ‘CI chimera’ by a single nucleotide (changing an aspartic acid to an asparagine at position 119 of the VPg), allowing the breakdown of pvr23. This variant permits the elimination of the effect of the pvr23 resistance in order to reveal the quantitative resistance caused by the genetic background. For all DH lines, 100% infection was observed after inoculation with ‘CI chimera VPg‐N’, but the virus could not be detected in Perennial by DAS‐ELISA. The ‘CI chimera’ and ‘CI chimera VPg‐N’ are much more aggressive than SON41p. For DH285, all plants died rapidly and no virus accumulation value could be obtained for this plant genotype. For the 14 other lines, significant differences in virus accumulation (Kruskal–Wallis test; χ 2 = 87.3; P < 0.001; Table S2), with means in the range 1–12, were observed between DH lines. Two independent replicates of the virus accumulation test were performed and a high correlation was observed between them (ρ Spearman = 0.77; P = 0.0019).
The effect of the factor ‘viral accumulation’ on the binomial response variable ‘number of plants for which pvr23 was broken down’ was tested with a one‐way generalized linear model using a logit link function. It was highly significant (P < 10−5) and the model fit was highly satisfactory (McFadden R 2 = 0.79; Fig. 1) (McFadden, 1973). Overall, this analysis reveals a strong correlation between viral accumulation and RB frequency across pepper genotypes.
Selection of PVY variants breaking down the polygenic resistance
To evaluate the effect of the second and third mechanisms potentially involved in RB (‘number of mutations’ and ‘selection pressure’ mechanisms), we compared two pepper lines: DH285, carrying pvr23 and a susceptible genetic background, further named the ‘monogenic resistance’, and Perennial, carrying pvr23 and a partially resistant genetic background, further named the ‘polygenic resistance’.
Because direct mechanical inoculation of Perennial (polygenic resistance) with the PVY SON41p clone did not succeed in RB (Palloix et al., 2009), we performed graft inoculations that increase the inoculum pressure, as a permanent inoculum is multiplied in the susceptible Yolo Wonder (YW) rootstock and loaded into the scions of DH285 or Perennial. Six weeks after inoculation, all YW rootstocks showed severe mosaic symptoms and a large proportion of DH285 (monogenic resistance) scions showed a combination of mosaic and necrotic symptoms. By contrast, all Perennial scions were symptom free. DAS‐ELISA tests revealed that 72% (23/32) of DH285 scions and 0% (0/91) of Perennial scions were infected. The VPg cistron of PVY populations present in infected DH285 scions was sequenced and each PVY variant carried one nonsynonymous nucleotide substitution in the VPg cistron compared with the initial SON41p inoculum. Four different variants were found and named G, K, N and C according to the amino acid substitutions carried in the VPg: serine to glycine substitution at amino acid position 101, threonine to lysine substitution at position 115, aspartic acid to asparagine substitution at position 119 and serine to cysteine substitution at position 120, respectively. Ayme et al. (2006) demonstrated that each of these four substitutions was sufficient for the breakdown of pvr23.
Three months after inoculation, two scions of Perennial showed a single chlorotic lesion on a single leaf. Leaf tissues surrounding the chlorotic lesions in these two symptomatic leaves were used as inoculum for direct mechanical back‐inoculation to 20 Perennial plants each, which were demonstrated to be 100% infected at 14 days post‐inoculation (dpi). The VPg cistron of PVY populations present in these Perennial plants was sequenced and each PVY variant was shown to carry one distinct pair of nonsynonymous nucleotide substitutions in the VPg cistron compared with the initial SON41p inoculum. One mutant carried the same substitutions as those present in mutants G and K, and the other mutant carried the same substitutions as those present in mutants K and N. In summary, all of the 23 plants carrying the monogenic resistance were broken down by single VPg mutants, whereas two of two plants carrying the polygenic resistance were broken down by double VPg mutants, a very significant difference (Fisher's exact test, P = 0.003). These two pairs of nucleotide substitutions were introduced into the SON41p cDNA clone by directed mutagenesis, producing the two double mutants GK and KN.
Capacity of double and single VPg mutants of PVY to infect plants with the polygenic resistance
In order to explain why only double VPg mutants were successful in breaking down the polygenic resistance in the graft inoculation experiment, we tested the ability of the two double mutants (GK and KN) and of the corresponding single mutants G, K and N to infect, at the systemic level, Perennial plants carrying the polygenic resistance after direct mechanical inoculation with calibrated inocula (Ayme et al., 2006). The capacity to infect Perennial plants varied largely according to the mutant considered. The single mutant K and the two double mutants GK and KN had similar infectivities, with 46% (21/46), 41% (19/46) and 43% (13/30) of systemic infection, respectively. The single mutants G and N were significantly less infectious (P < 0.001, Fisher's exact test), with 9% (4/46) and 3% (1/29) of infected plants, respectively. By contrast, all five mutants infected 100% (25 of 25) of DH285 plants carrying the monogenic resistance.
Competitiveness of PVY variants in susceptible and monogenically or polygenically resistant plants
The competitiveness of the two double mutants KN and GK against each of the corresponding single mutants (G, K and N) was evaluated after co‐inoculation to susceptible plants (YW), to plants carrying the monogenic resistance (DH285) and to plants carrying the polygenic resistance (Perennial). To compare the competitiveness of the co‐inoculated mutants, the ratio of the number of plants in which each mutant was detected singly by derived cleaved amplified polymorphic sequence (dCAPS) analysis was compared with the result expected if the two viruses had equal fitnesses, i.e. a 1:1 ratio identical to the inoculum composition.
In the susceptible YW plants, there was no difference in competitiveness between mutants KN and K, or between KN and N, as the number of plants infected by each mutant was not significantly different from an expected 1:1 ratio (Fig. 2). For competition GK vs. K, the single mutant was significantly more competitive than the double mutant. In contrast, for competition GK vs. G, the double mutant showed a higher competitiveness than the single mutant. In the monogenic resistance (DH285), there was no difference in competitiveness between mutants KN and K. For the other three competition experiments, the double mutants showed a higher competitiveness than the corresponding single mutants (Fig. 2). In the polygenic resistance (Perennial, Per), there was no significant difference in competitiveness between mutants KN and K (as observed in the other two plant genotypes) or between mutants GK and K. For competitions GK vs. G and KN vs. N, the double mutants were significantly more competitive than the single mutants (Fig. 2). Similar statistical results were obtained when taking into account only single‐infected plants and when half of the mixed‐infected plants were grouped with plants infected by each PVY variant.
Figure 2.
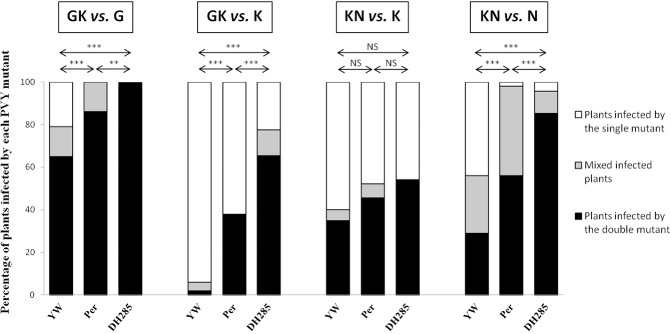
Competitiveness of double viral protein genome‐linked (VPg) mutants of Potato virus Y (PVY) SON41p against the two corresponding single mutants in the susceptible pepper genotype (Yolo Wonder, YW) and in pepper genotypes carrying a polygenic (Perennial, Per) or monogenic (DH285) resistance. For each competition, 46–50 plants were inoculated with an inoculum composed of a 1:1 ratio between the two PVY mutants. The percentages of plants infected by single mutant, double mutant or both were assessed by derived cleaved amplified polymorphic sequence (dCAPS) analysis (Table S1, see Supporting Information). Symbols ** and *** within the histogram bars correspond to significantly different distributions between virus competitors (single versus double mutant) within each plant genotype, compared with the inoculum ratio (1:1) at the 1% and 0.1% type I error thresholds (Fisher's exact test). Mixed‐infected plants (in grey) were excluded from this analysis. Symbols **, *** presented above the histogram bars correspond to significant differences between plant genotypes for the distributions of plants infected by single mutant, double mutant or mutant mixture at the 1% and 0.1% type I error thresholds (Fisher's exact test). NS, no significant differences (P > 5%).
The outcome of the competitions was also compared between the monogenic and polygenic resistances. In competition KN vs. K, there was no difference in competitiveness between mutants, either in the polygenic or monogenic resistance. For the other three competitions (i.e. GK vs. G, GK vs. N and KN vs. N), one of the mutants was more competitive than the other in at least one of the two resistant plant genotypes, and we analysed the virus frequencies in each plant population in order to compare the speed of selection of the fittest mutant between the two resistant genotypes. For this, we distinguished between plants in which the most competitive mutant (the double mutant in all cases) only was detected by dCAPS and the other plants (i.e. those in which both mutants or the single mutant only was detected). Indeed, in mixed‐infected plants, selection in favour of one or the other variant had not yet occurred and, for plants in which only the less competitive single mutant was detected, the effect of genetic drift exceeded that of selection. In contrast, for plants in which only the more competitive double mutant was detected, we expected that selection was the major evolutionary force that had acted on the virus population. For all three competitions, these comparisons revealed a significantly higher frequency of the fittest variant among monogenically resistant plants than among polygenically resistant plants (Fig. 2). Similar results were obtained when half of the mixed‐infected plants were grouped with plants infected by each variant. Among the susceptible YW plants, the double mutant showed a lower frequency than among the two resistant genotypes, for these three competitions.
Discussion
The durability of major resistance genes has been shown to be highly dependent on the genetic background of the host plant in virus–plant, fungus–plant and nematode–plant interactions (Brun et al., 2010; Fournet et al., 2012; Palloix et al., 2009). The present work aimed to highlight how the plant genetic background protects a major resistance gene from breakdown. We examined three different mechanisms that could be involved: (i) the effect of the additional quantitative resistance level conferred by the genetic background; (ii) the alteration in the number and/or nature of mutations required for RB; and (iii) the speed of selection of adapted RB mutants within the virus population. Here, we were interested mostly in the first two steps of RB (i.e. the infection of resistant plants by a virus variant), and plant to plant dispersion of virus variants was not studied. Nevertheless, the appearance and selection of RB variants are two crucial stages without which the third (dispersion of variants) does not arise.
The strength of the resistance explains most of the variations in pvr23 RB
In order to test the effect of the quantitative resistance (conferred by the plant genetic background) on the capacity of PVY to overcome the pvr23 gene, we tested Perennial and 15 DH lines carrying pvr23, but segregating for the genetic background. These lines were tested for two different traits: the pvr23 RB frequency (with PVY ‘CI chimera’) and the virus accumulation (with PVY ‘CI chimera VPg‐N’). In all DH lines carrying pvr23, the observed differences resulted from the effect of genetic factors present in the genetic background. The use of a pvr23‐breaking PVY mutant (PVY ‘CI chimera VPg‐N’) to measure virus accumulation enabled us to free ourselves from the effect of pvr23. Indeed, the single amino acid change in ‘CI chimera VPg‐N’ (compared with ‘CI chimera’) restored the interaction between the VPg protein of the virus and the eIF4E protein of the plant (coded by pvr23), resulting in pvr23 breakdown (Charron et al., 2008).
We observed significant differences in viral accumulation and RB frequency between the 15 lines (accumulation data were not available for DH285, Table S2). In addition, the RB frequency was highly correlated with the accumulation of the pvr23‐breaking PVY mutant, which depends on the quantitative resistance level caused by the plant genetic background (Fig. 1). A decrease in virus accumulation corresponds to a decrease in virus replication in the plant and, consequently, to a decrease in the probability of appearance of mutations in the virus genome, including the RB mutations. In turn, a lower probability of appearance of RB mutations would decrease the frequency of pvr23 breakdown. This mechanism could explain the higher or lower RB frequency between DH lines if pvr23 does not confer an extreme resistance and if the RB mutants appear in these lines but not in the inoculum used to inoculate these plants. Accordingly, Montarry et al. (2011) provided evidence that a PVY population which does not carry any RB mutation was able to accumulate slightly, even at the systemic level, in plants carrying pvr23 resistance. Moreover, the data of Montarry et al. (2011) also suggested that the RB mutants appeared in the pvr23 plants and not in the initial inoculum. In polygenically resistant plants (pvr23 in a partially resistant genetic background), the quantitative resistance controlled by the genetic background added to the incomplete effect of pvr23, and can be expected to decrease the RB frequency and thus to enhance the durability of resistance.
A more complex mutational pathway is required to break down the pvr23 resistance in a polygenic background
Seven single‐nucleotide nonsynonymous substitutions in the VPg cistron of SON41p (including those present in mutants G, K and N) allow the breakdown of the monogenic resistance (Ayme et al., 2006; Montarry et al., 2011). In contrast, only two mutants, showing combinations of two of the previous VPg mutations, infected plants carrying the polygenic resistance after graft inoculation with SON41p. We hypothesized that two VPg mutations would need to be combined in SON41p for breakdown of the polygenic resistance. To test this hypothesis further, we measured the capacity of the two double mutants (GK and KN) and the three corresponding single mutants (G, K and N) to infect plants with the monogenic or polygenic resistance. Although mutants K, KN and GK, sharing the mutation corresponding to mutant K, were relatively well adapted to the polygenic resistance with more than 40% of infection, the other two mutants (G and N) had a much lower infectivity (less than 10% of infection). However, these five mutants infected 100% of plants with the monogenic resistance. This suggests that the fitness threshold required by the virus to infect the polygenic resistance is higher than that required to infect the monogenic resistance, which consequently reduces the possibilities of breakdown of the polygenic resistance, as observed for mutants G and N, which were only able to infect efficiently the monogenic resistance.
There were no significant infectivity differences between the K single mutant and the two double mutants GK or KN, questioning why mutant K was not observed in plants carrying the polygenic resistance. The nature of the nucleotide substitution corresponding to these PVY mutants and the relative competitiveness of the double and single mutants provide a plausible explanation. Indeed, mutants G and N required a transition to appear, whereas mutant K required a transversion and, in PVY, transitions are on average five to eight times more frequent than transversions (Ayme et al., 2006). As a consequence, the emergence of mutant N occurred in 70% of plants carrying the monogenic resistance (Montarry et al., 2011), even though it has a lower competitiveness than mutant K. In plants with the polygenic resistance, virus multiplication is reduced and the occurrence of mutations in the virus population is decreased accordingly (i.e. first mechanism). Consequently, compared with the monogenic resistance, a smaller number of RB mutational events are expected, especially for those involving a transversion, such as mutant K. Then, for the two double mutants GK and KN, the transitions corresponding to mutants G or N probably occurred as a first step. These first mutations conferred a low level of adaptation to the polygenic resistance, which subsequently favoured the appearance of the second, less likely, transversion (that present in mutant K), which conferred a higher level of adaptation to the polygenic resistance and a higher competitiveness (Fig. 2).
To summarize, we have shown that only the fittest RB mutants can infect the polygenic resistance, whereas all RB mutants can infect the monogenic resistance. Thus, in the polygenic resistance, the set of resistance‐breaking mutations is reduced. The only way to reconcile all our experimental data and to explain why the K mutant was not observed in the polygenic resistance is to take into account the frequency of appearance of each mutant, which depends on both the nature of the RB mutation (transition or transversion) and the level of host resistance (by reducing virus replication and the mutation probability) (Fig. 3). Taken together, these results indicate that the larger number of mutations observed in the polygenic resistance is the consequence of the genetic determinism of RB, including the effect of RB mutations on virus fitness. Consequently, this mechanism is clearly specific to the plant genotype–virus genotype interaction considered, and is barely predictable.
Figure 3.

Effect of the quantitative resistance conferred by the genetic background on the durability of a major resistance gene. The scheme represents the three mechanisms shown to be involved in the higher durability of polygenic relative to monogenic resistances and the hypothetical relationships between these mechanisms. N e, effective population size; RB, resistance breakdown or resistance breaking; Δs, difference in selection coefficients between pathogen variants.
In our work, only the mutations in the VPg cistron were considered, but hypothetical mutations in other regions of the viral genome may also affect the adaptation to the polygenic resistance. Such additional mutation(s) were suspected by Palloix et al. (2009) in experimental evolution in field conditions, but no evidence of such events occurred in serial inoculations in the laboratory. The occurrence of such additional mutations would also argue in favour of a higher mutational distance to breakdown in a polygenic resistance context.
Selection of adapted RB mutants by the polygenic resistance is slower than by the monogenic resistance
To compare the selection pressure exerted by the monogenic and polygenic resistances, we initially planned to co‐inoculate plants with mixtures of PVY variants carrying or not carrying the RB mutations and to further check the composition of the PVY populations. This strategy proved to be inefficient because the PVY variant (SON41p) which did not carry RB mutations was rapidly counter‐selected and could not be retrieved from the viral populations in both types of plants (Ayme et al., 2006 and data not shown). Consequently, we used the double and single VPg mutants which showed smaller differences in competitiveness as tools to quantify the selection pressure exerted by both monogenically and polygenically resistant genotypes. Competition results showed that the fixation of the fittest variant was less frequent in the polygenic than in the monogenic resistance for the three competitions in which the double mutant was more competitive than the single mutant in the monogenic resistance. In the fourth competition (KN vs. K), in which there was no difference in competitiveness between the mutants, we did not observe differences between the host genotypes for the selection of viral mutants (Fig. 2).
These differences in competitiveness between PVY variants in the different plant genotypes could result from differences in infectivity and/or in the following steps of plant colonization by the viruses (virus multiplication, cell‐to‐cell or systemic movements).
The speed of fixation of the fittest mutants depends on the difference in their selection coefficient from that of other mutants (Δs) and on the action of genetic drift, which is influenced by the effective population size N e. The effect of genetic drift on the selection of RB mutants could be all the more significant as several articles have shown that N e can be very small for plant RNA viruses (usually < 10) and that genetic drift could be important in virus evolution (French and Stenger, 2003; Sacristán et al., 2003; Zwart et al., 2011). This is also the case for the infection of susceptible YW pepper plants with PVY (Fabre et al., 2012), and N e can be expected to be even smaller in more resistant pepper genotypes. The value of the product N e × Δs determines whether the viral population will be mainly under the influence of selection or of genetic drift (Charlesworth, 2009; Crow and Kimura, 1970; Fraser, 1972), i.e. the greater N e × Δs, the greater the effects of selection. Both N e and Δs are likely to vary simultaneously between the monogenic and polygenic resistances. The quantitative resistance caused by the plant genetic background reduces PVY accumulation and N e, potentially increasing genetic drift and therefore weakening the effectiveness of selection. However, it is not presently possible to disentangle the effects of reduced N e or reduced Δs on the slower fixation of the double mutants in the polygenic resistance. Our results show that a quantitative resistance can reduce the selection pressure exerted by a major resistance gene.
Relationships and hierarchy between the three mechanisms involved in the higher durability of the polygenic resistance
Altogether, these results allow us to estimate the relative contribution of the three mechanisms involved in the higher durability of the polygenic resistance and to suggest a model of the links between them (Fig. 3). We have shown that a large part of the observed variation in breakdown frequency can be explained by the factor ‘viral accumulation’, which depends on the quantitative resistance level caused by the genetic background (first mechanism). We assume that this mechanism has a direct effect on the risk of appearance of RB variants, but also on the two other mechanisms involved in resistance durability. Indeed, the combined effect of resistance efficiency and of the number and nature of RB mutations complicates the mutational pathways responsible for RB in the polygenic resistance. Moreover, the selection of adapted RB mutants observed in the polygenic resistance could also be partly compromised by a lower effective population size (N e) and thus enhanced genetic drift.
The possibility of generalizing the latter mechanism depends on the relative contributions of the genetic drift (N e) and differential selection coefficient (Δs). Indeed, if Δs is the most important factor, it is certainly specific to the plant–pathogen interaction and cannot be general. In contrast, if N e is the prime factor, it is a direct consequence of the resistance efficiency conferred by the genetic background (Fig. 3), and is consequently as general as the first mechanism. It will consequently be important to estimate the relative contributions of the genetic drift (N e) and differential selection coefficient (Δs) to the decrease in selection of adapted PVY variants.
The two effects of the host genetic background on the probability of appearance of RB mutants and on the speed of fixation of these mutants can be expected to apply in many plant–pathogen systems. Indeed, most major resistance genes do not completely abolish the multiplication of pathogens and may be reinforced by genetic factors, further reducing pathogen multiplication (Kang et al., 2005). In contrast, the RB mutation pathways remain specific to molecular interactions between plant resistance factors and pathogen pathogenicity factors.
Quantitative resistance factors are widespread among genetic resources and their introgression into cultivars are easier and easier. Consequently, it seems judicious for breeders to associate major resistance genes with QTLs that reduce pathogen multiplication and decrease the risks of appearance of pathogen RB variants.
Experimental Procedures
Plant and virus material
All pepper genotypes were C. annuum inbred lines. YW (allele pvr2 +) is susceptible to PVY, whereas Perennial carries the pvr23 major resistance allele plus three QTLs conferring partial resistance to PVY (Caranta et al., 1997). The three QTLs of Perennial are involved in symptom expression (area under the disease progress curve, AUDPC) and explain approximately 30% of the phenotypic variation.
Fifteen DH lines issued from the F1 hybrid (Perennial × YW) were used. All DH lines were confirmed to be homozygous for the pvr23 allele by genotyping with a tetraprimer amplification refractory mutation system‐polymerase chain reaction (ARMS‐PCR) marker which targets single‐nucleotide polymorphism (SNP) signatures differentiating pvr2 + from pvr23 (Rubio et al., 2008). These 15 DH lines were chosen on the basis of their variable ability to select RB PVY variants obtained from a preliminary phenotypic test, as indicated below. The DH line DH285 was chosen to represent the monogenic resistance as it carries the pvr23 allele in a susceptible genetic background (susceptibility alleles at the three known QTLs) (Palloix et al., 2009).
The PVY infectious clones and mutants were derived from isolates SON41p and LYE84.2 (Montarry et al., 2011). SON41p infects YW, but is not infectious per se in Perennial and DH lines which carry pvr23 (Ayme et al., 2006). Five SON41p mutants, named G, K, N, GK and KN according to the amino acid substitutions carried in the VPg cistron, were used in this study. Mutants G, K and N carry single‐nucleotide substitutions corresponding to the serine to glycine substitution at amino acid position 101 of VPg, the threonine to lysine substitution at amino acid position 115 of VPg and the aspartic acid to asparagine substitution at amino acid position 119 of VPg, respectively. Each of these three substitutions was shown to be sufficient for the breakdown of pvr23 (Ayme et al., 2006; Palloix et al., 2009). Mutants GK and KN carry simultaneously the VPg mutations present in mutants G and K or K and N, respectively. For the evaluation of RB frequencies in Perennial and the 15 DH lines, we used the ‘CI chimera’, which was obtained by homologous recombination in Saccharomyces cerevisiae between PVY clones SON41p and LYE84.2, and was chosen for its greater ability to break down the pvr23 resistance relative to the parental clone SON41p (Montarry et al., 2011), allowing a more precise comparison with PVY accumulation data. The ‘CI chimera’ is identical to SON41p, except that its CI cistron was substituted by that of LYE84.2. Both PVY clones carry identical VPg cistrons, the virus genome region directly involved in pvr23 RB. They were not infectious per se in plants carrying pvr23, and they showed the same distributions of RB mutations when inoculated to HD285 plants carrying pvr23 (Montarry et al., 2011).
To evaluate the level of quantitative resistance controlled by the plant genetic background, we used a single‐nucleotide mutant of the ‘CI chimera’ carrying the same VPg mutation as that of mutant N (named ‘CI chimera VPg‐N’), which conferred the breakdown capacity towards the pvr23 resistance gene.
Measurement of RB frequency
The breakdown frequency of pvr23 was tested in a climate‐controlled room after mechanical inoculation of Perennial and the 15 DH lines with the ‘CI chimera’. Inoculum was obtained after virus propagation in Nicotiana tabacum cv. Xanthi plants. Five grams of Xanthi leaves showing PVY symptoms were crushed in 25 mL of 0.03 m phosphate buffer (pH 7.0) supplemented with 2% (w/v) diethyldithiocarbamate, 400 mg of active charcoal and 400 mg of carborundum. Sixty pepper seedlings with two expanded cotyledons (2–3 weeks after sowing) per genotype were inoculated mechanically on their cotyledons. At 38 dpi, all plants with visible symptoms were considered to be infected by PVY. Plants without symptoms were submitted to virus detection by DAS‐ELISA, as described by Legnani et al. (1995). In these conditions, every systemic plant infection has been shown previously to result from the occurrence of one RB mutation in the VPg cistron (Montarry et al., 2011). Thus, for each pepper line, the RB frequency was calculated from the ratio of the number of systemically infected plants to the total number of inoculated plants (n = 60). This test was repeated twice.
In addition, 32 DH285 plants and 91 Perennial plants were grafted onto YW susceptible rootstocks and, 1 week later, the rootstocks were inoculated with SON41p. PVY detection in the resistant scions was performed from 6 weeks to 3 months after inoculation by DAS‐ELISA, as described by Legnani et al. (1995). The sequence of the VPg cistron of PVY populations in ELISA‐positive plants was determined as described in Moury et al. (2004).
Estimation of viral accumulation
To evaluate the level of quantitative resistance caused by the genetic background, we inoculated mechanically Perennial and the 15 DH lines with PVY ‘CI chimera VPg‐N’, which is infectious towards pvr23 resistant plants and therefore reveals the quantitative resistance controlled by the genetic background. The virus accumulation was evaluated at 36 dpi by DAS‐ELISA using a dilution range of PVY‐infected plant extracts as described in Ayme et al. (2006), i.e. relative to a common reference sample incorporated in each ELISA plate. This test was repeated once.
Measurement of virus competitiveness
The relative competitiveness of single and double VPg mutants of SON41p was measured in the monogenically resistant genotype DH285 and in the polygenically resistant genotype Perennial. Four different competition experiments were performed between pairs of viruses including a double mutant and one or the other corresponding single mutant (GK vs. G or K and KN vs. K or N). Inocula were obtained by independent propagation of each virus in N. tabacum cv. Xanthi. Crude extracts from infected Xanthi leaves were calibrated by DAS‐ELISA (Ayme et al., 2006) and mixed in a 1:1 ratio to mechanically inoculate 50 seedlings per genotype and per competition. Plants were kept in a climatic room and, at 34 dpi, total RNAs were extracted from pools of three leaves from each inoculated plant with the Tri‐Reagent kit (Molecular Research Center Inc., Cincinnati, OH, USA). These RNAs were used as template to amplify the central part of the VPg cistron (from position 5930 to position 6168), and the competitiveness of the PVY variants was analysed using dCAPS analysis, as described in Neff et al. (1998). Oligonucleotide primers and restriction endonucleases were used according to the genomic sequences of the PVY mutants to be detected (Table S1, see Supporting Information). Two dCAPS markers were used for each competition experiment, one corresponding to the single mutant and the other to the double mutant. After 2.5% agarose gel electrophoresis, plants were classified into three categories depending on the observation of cleaved or uncleaved reverse transcription‐polymerase chain reaction (RT‐PCR) products (presence of one or the other PVY mutant), or both (mixture of the two mutants).
We measured virus competitiveness as the capacity of the virus to become predominant, according to the dCAPS results, in a population of inoculated plants. Therefore, this reflects the result of competitions occurring at all steps of plant infection, from the inoculation process to plant colonization by the virus. Accordingly, virus competitiveness between pairs of variants was tested by comparing the relative proportion of single and double mutants in each plant genotype with that of the initial inoculum ratio (1:1) (Fisher's exact test) omitting mixed‐infected plants. Similar Fisher's exact tests were performed after grouping half of the mixed‐infected plants with plants infected by each PVY variant.
We also investigated the dynamics of the competition process between plant genotypes by comparing, using Fisher's exact tests, the proportion of plants infected by the fittest mutant with the proportion of the two other types of plant (mixed‐infected and infected by the less competitive mutant). In other words, we compared the proportion of plants in which the selection had acted on the virus population (plants infected by the fittest variant) with the proportion of plants in which selection had not yet occurred (mixed‐infected plants) or in which genetic drift effects exceeded selection effects (plants infected by the less competitive variant). All the statistical analyses were performed with R software (http://www.r‐project.org/).
Supporting information
Table S1 Oligonucleotide primers and endonucleases used for derived cleaved amplified polymorphic sequence (dCAPS) detection of Potato virus Y (PVY) viral protein genome‐linked (VPg) mutants.
Table S2 Frequency of resistance breakdown (RB) by Potato virus Y (PVY) ‘CI chimera’ and concentration of PVY ‘CI chimera VPg‐N’ at 36 days post‐inoculation (dpi) for Perennial and 15 doubled haploid (DH) lines issued from the F1 hybrid (Perennial × Yolo Wonder) and carrying the pvr2 3 resistance gene.
Acknowledgements
We thank V. Simon, G. Nemouchi, B. Savio and P. Mistral for technical assistance. We acknowledge Dr Frédéric Fabre for useful discussions about the present work and for help with statistical analyses. This work was financially supported by the Comité Technique Permanent de la Sélection (CTPS, French Ministry of Agriculture and Fisheries), the Agence Nationale de la Recherche (ANR) , the Pôle Européen d'Innovation Fruits et Légumes (PEIFL) and the Région Provence Alpes Côte d'Azur (PACA), with the contribution of the seed companies Gautier Semences, Clause Vegetable Seeds, Vilmorin SA, Rijk Zwaan and Sakata Vegetables Europe.
References
- Ayme, V. , Souche, S. , Caranta, C. , Jacquemond, M. , Chadoeuf, J. , Palloix, A. and Moury, B. (2006) Different mutations in the genome‐linked protein VPg of Potato virus Y confer virulence on the pvr23 resistance in pepper. Mol. Plant Microbe Interact. 19, 557–563. [DOI] [PubMed] [Google Scholar]
- Ayme, V. , Petit‐Pierre, J. , Souche, S. , Palloix, A. and Moury, B. (2007) Molecular dissection of the Potato virus Y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. Gen. Virol. 88, 1594–1601. [DOI] [PubMed] [Google Scholar]
- Brun, H. , Chèvre, A.‐M. , Fitt, B.D. , Powers, S. , Besnard, A.‐L. , Ermel, M. , Huteau, V. , Marquer, B. , Eber, F. , Renard, M. and Andrivon, D. (2010) Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus . New Phytol. 185, 285–299. [DOI] [PubMed] [Google Scholar]
- Caranta, C. , Palloix, A. , Lefebvre, V. and Daubeze, A.M. (1997) QTLs for a component of partial resistance to cucumber mosaic virus in pepper: restriction of virus installation in host‐cells. Theor. Appl. Genet. 94, 431–438. [Google Scholar]
- Charlesworth, B. (2009) Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 10, 195–205. [DOI] [PubMed] [Google Scholar]
- Charron, C. , Nicolai, M. , Gallois, J.L. , Robaglia, C. , Moury, B. , Palloix, A. and Caranta, C. (2008) Natural variation and functional analyses provide evidence for coevolution between plant eIF4E and potyviral VPg. Plant J. 54, 56–68. [DOI] [PubMed] [Google Scholar]
- Crow, J.F. and Kimura, M. (1970) An Introduction to Population Genetics Theory. New York: Harper and Row. [Google Scholar]
- Fabre, F. , Bruchou, C. , Palloix, A. and Moury, B. (2009) Key determinants of resistance durability to plant viruses: insights from a model linking within‐ and between‐host dynamics. Virus Res. 141, 140–149. [DOI] [PubMed] [Google Scholar]
- Fabre, F. , Montarry, J. , Coville, J. , Senoussi, R. , Simon, V. and Moury, B. (2012) Modelling the evolutionary dynamics of viruses within their hosts: a case study using high‐throughput sequencing. PLoS Pathog. 8, e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournet, S. , Kerlan, M.C. , Renault, L. , Dantec, J.P. , Rouaux, C. and Montarry, J. (2012) Selection of nematodes by resistant plants has implications for local adaptation and cross‐virulence. Plant Pathol. in press. doi: 10.1111/j.1365-3059.2012.02617.x. [DOI] [Google Scholar]
- Fraser, A.S. (1972) An introduction to population genetic theory In: Teratology, Vol. 5 (Crow J.F. and Kimura M., eds), pp. 386–387. New York: Harper and Row. [Google Scholar]
- French, R. and Stenger, D.C. (2003) Evolution of wheat streak mosaic virus: dynamics of population growth within plants may explain limited variation. Annu. Rev. Phytopathol. 41, 199–214. [DOI] [PubMed] [Google Scholar]
- García‐Arenal, F. and McDonald, B.A. (2003) An analysis of the durability of resistance to plant viruses. Phytopathology, 93, 941–952. [DOI] [PubMed] [Google Scholar]
- Harrison, B.D. (2002) Virus variation in relation to resistance‐breaking in plants. Euphytica, 124, 181–192. [Google Scholar]
- Janzac, B. , Montarry, J. , Palloix, A. , Navaud, O. and Moury, B. (2010) A point mutation in the polymerase of Potato virus Y confers virulence toward the Pvr4 resistance of pepper and a high competitiveness cost in susceptible cultivar. Mol. Plant Microbe Interact. 23, 823–830. [DOI] [PubMed] [Google Scholar]
- Johnson, R. (1981) Durable resistance: definition of, genetic control, and attainment in plant breeding. Phytopathology, 71, 567–568. [Google Scholar]
- Kang, B.‐C. , Yeam, I. and Jahn, M. (2005) Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. [DOI] [PubMed] [Google Scholar]
- Keller, B. , Feuillet, C. and Messmer, M. (2000) Genetics of disease resistance – basic concepts and its application in resistance breeding In: Mechanisms of Resistance to Plant Diseases (Slusarenko A., Fraser R. and van Loon K., eds), pp. 101–160. London: Kluwer Academic Publishers. [Google Scholar]
- Kliebenstein, D.J. and Rowe, H.C. (2009) Anti‐rust antitrust. Science, 323, 1301–1302. [DOI] [PubMed] [Google Scholar]
- Knott, D.R. (1988) Strategies for the utilization of partial resistance for the control of cereal rust In: Breeding Strategies for Resistance to the Rusts of Wheat (Simmonds N.W. and Rajaram S., eds), pp. 48–62. Mexico City: International Maize and Wheat Improvement Center (CIMMYT). [Google Scholar]
- Legnani, R. , Gerbe‐Selassie, K. , Nono Womdim, R. , Gognalons, P. , Moretti, A. , Laterrot, H. and Marchoux, G. (1995) Evaluation and inheritance of the Lycopersicon hirsutum resistance against Potato virus Y . Euphytica, 86, 219–226. [Google Scholar]
- Lindhout, P. (2002) The perspectives of polygenic resistance in breeding for durable disease resistance. Euphytica, 124, 217–226. [Google Scholar]
- McFadden, D. (1973) Conditional logit analysis of qualitative choice behavior In: Frontiers in Econometrics (Zarembka P., ed.), pp. 105–142. New York: Academic Press. [Google Scholar]
- Montarry, J. , Doumayrou, J. , Simon, V. and Moury, B. (2011) Genetic background matters: a plant–virus gene for gene interaction is strongly influenced by genetic contexts. Mol. Plant Pathol. 12, 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury, B. , Morel, C. , Johansen, E. , Guilbaud, L. , Souche, S. , Ayme, V. , Caranta, C. , Palloix, A. and Jacquemond, M. (2004) Mutations in Potato virus Y genome‐linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum . Mol. Plant Microbe Interact. 17, 322–329. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Fereres, A. , García‐Arenal, F. and Lecoq, H. (2011) Sustainable management of plant resistance to viruses. In: Recent Advances in Plant Virology (Caranta C., Aranda M.A., Tepfer M. and Lopez‐Moya J.J., eds.), pp. 219–336. Norfolk: Caister Academic Press. [Google Scholar]
- Neff, M. , Neff, J. , Chory, J. and Pepper, A. (1998) dCAPS a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14, 387–392. [DOI] [PubMed] [Google Scholar]
- Palloix, A. , Ayme, V. and Moury, B. (2009) Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol. 183, 190–199. [DOI] [PubMed] [Google Scholar]
- Parlevliet, J.E. (2002) Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica, 124, 147–156. [Google Scholar]
- Pink, D. (2002) Strategies using genes for non‐durable disease resistance. Euphytica, 124, 227–236. [Google Scholar]
- Rubio, M. , Caranta, C. and Palloix, A. (2008) Functional markers for selection of potyvirus resistance alleles at the pvr2‐eIF4E locus in pepper using tetra‐primer ARMS‐PCR. Genome, 51, 767–771. [DOI] [PubMed] [Google Scholar]
- Sacristán, S. , Malpica, J.M. , Fraile, A. and García‐Arenal, F. (2003) Estimation of population bottlenecks during systemic movement of tobacco mosaic virus in tobacco plants. J. Virol. 77, 9906–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, M. (1985) The current status and prospects of multiline cultivars and variety mixtures for disease resistance. Annu. Rev. Phytopathol. 23, 251–273. [Google Scholar]
- Zwart, M.P. , Daròs, J.‐A. and Elena, S.F. (2011) One is enough: in vivo effective population size is dose‐dependent for a plant RNA virus. PLoS Pathog. 7, e1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Oligonucleotide primers and endonucleases used for derived cleaved amplified polymorphic sequence (dCAPS) detection of Potato virus Y (PVY) viral protein genome‐linked (VPg) mutants.
Table S2 Frequency of resistance breakdown (RB) by Potato virus Y (PVY) ‘CI chimera’ and concentration of PVY ‘CI chimera VPg‐N’ at 36 days post‐inoculation (dpi) for Perennial and 15 doubled haploid (DH) lines issued from the F1 hybrid (Perennial × Yolo Wonder) and carrying the pvr2 3 resistance gene.


