Summary
Cerato‐platanin family proteins are secreted and have been found in both the fungal cell wall and the extracellular medium. They elicit defence responses in a variety of plants and have been proposed to be perceived as pathogen‐associated molecular patterns (PAMPs) by the plant immune system, although, in the case of the necrotroph Botrytis cinerea, the cerato‐platanin BcSpl1 contributes to fungal virulence instead of plant resistance. In this study, we report that BcSpl1, which was previously found in the secretome as an abundant protein, is even more abundant in the fungal cell wall. By fusion to green fluorescent protein (GFP), we also show that BcSpl1 associates with the plant plasma membrane causing rapid morphological changes at the cellular level, such as the disorganization of chloroplasts, prior to macroscopic necrosis in the treated tissue. By a combination of serial deletion studies, synthetic peptides and chimeric proteins, we mapped the eliciting activity to a two‐peptide motif in the protein surface. The expression of a chimeric protein displaying this motif in B. cinerea mutants lacking BcSpl1 undoubtedly showed that the motif is responsible for the contribution of BcSpl1 to virulence.
Introduction
Botrytis cinerea is a widely distributed and highly successful plant pathogen causing devastating losses in crops worldwide (Dean et al., 2012). Early during infection, this fungus secretes a wide array of proteins involved in a variety of different functions (Espino et al., 2010), among which is the abundant necrosis‐inducing cerato‐platanin BcSpl1. Cerato‐platanins constitute a widely distributed family of fungal secreted proteins whose founder member is cerato‐platanin from Ceratocystis platani (formerly C. fimbriata) (Pazzagli et al., 1999), a pathogen of the European plane tree (Platanus acerifolia). The three‐dimensional structure of this protein consists of a double psi‐beta‐barrel fold, similar to that of expansins and endoglucanases (Krieger et al., 2010; de Oliveira et al., 2011), a fold suggesting a role in polysaccharide metabolism, and the C. platani (de Oliveira et al., 2011) and Trichoderma atroviride (Frischmann et al., 2013) cerato‐platanins have been shown to bind to oligosaccharides, although no hydrolysis has been reported.
Cerato‐platanins have been found bound to the cell wall (Boddi et al., 2004) and in the extracellular medium (Espino et al., 2010; Frías et al., 2011), and most are able to elicit defence responses in plants, including autofluorescence (Pazzagli et al., 1999; Wilson et al., 2002), synthesis of reactive oxygen species (Djonovic et al., 2006; Frías et al., 2011), induction of defence genes (Djonovic et al., 2007; Fontana et al., 2008; Frías et al., 2011) and cell death (Frías et al., 2011; Pazzagli et al., 1999; Yang et al., 2009), with markers and symptoms of the hypersensitive response (HR). These activities suggest that a putative perception of cerato‐platanins by the plant immune system, as a pathogen‐associated molecular pattern (PAMP), could have a role in the defence against fungal plant pathogens, and indeed the elicitor activity of the cerato‐platanin BcSpl1 on Arabidopsis thaliana is partially dependent on the regulator of PAMP perception BAK1 (Frías et al., 2011). Nevertheless, BcSpl1 also contributes to the virulence of the fungus, as shown by the fact that bcspl1 mutants produce lesions that are 20%–40% smaller than those of the wild‐type (Frías et al., 2011), in agreement with the hypothesis that B. cinerea actively induces HR to generate dead tissue on which to grow (Govrin and Levine, 2000).
Here, we report that two small surface‐exposed loops of BcSpl1 are necessary and sufficient for its necrosis‐inducing ability, and also suffice for its contribution to the virulence of the fungus.
Results
BcSpl1 is partially retained in the cell wall
BcSpl1 has been found consistently in the extracellular medium in cultures of B. cinerea of different ages and growth conditions (Espino et al., 2010; Girard et al., 2012; González‐Fernández et al., 2013; Shah et al., 2009a, b), although the C. platani homologue has been found predominantly bound to the fungal cell wall (Boddi et al., 2004). In order to gather more information about the location of BcSpl1 in B. cinerea, a BcSpl1‐green fluorescent protein (GFP) fusion was generated and expressed in the wild‐type strain B05.10 under the control of the Aspergillus nidulans OliC promoter. Observation under a fluorescence microscope revealed that the B. cinerea cerato‐platanin BcSpl1 is also bound to the cell wall, especially at the septa between adjacent cells (Fig. 1A). To obtain a quantitative estimation of the proportion of BcSpl1 which is retained in the cell in comparison with the amount that ends up in the culture medium as a soluble protein, the amount of BcSpl1 in both compartments was estimated by Western blot with anti‐GFP antibodies (Fig. 1B). Surprisingly, two extra bands above and below the band of the expected size were found in the mycelium fraction, which do not seem to be the result of cross‐reaction of the anti‐GFP antibody, as no signal was observed in B. cinerea strains not expressing GFP (not shown). One possible explanation for the large band could be covalent dimerization of BcSpl1, previously reported for other fungi (Vargas et al., 2008), and, for the small band, proteolysis at the BcSpl1‐GFP linker region, given the highly abundant protease secretion in B. cinerea (ten Have et al., 2010). From the intensities of the bands obtained for four biological replicates of the same experiment, we estimated that, considering all bands for the calculation, 22.4% ± 4.2% of all BcSpl1‐GFP synthesized ends up in the culture medium. We conclude that, similar to the cerato‐platanin from C. platani (Boddi et al., 2004), BcSpl1 is basically a cell wall protein, although with the particularity that its binding to this structure is weak and about one‐fifth can be found dissolved in the extracellular medium.
Figure 1.
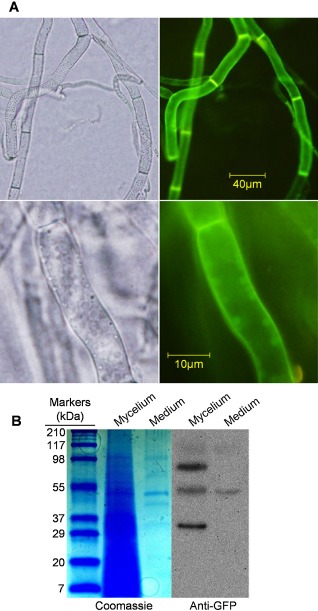
Localization of BcSpl1 in Botrytis cinerea mycelium. (A) A homokaryotic B. cinerea transformant obtained with plasmid pCRP‐GFP, and thus expressing a BcSpl1‐GFP fusion protein, was grown for 3 days in YGG medium (0.5% yeast extract, 100 mm glucose and 0.3% Gamborg's B5) and observed under a visible (left) or fluorescence (right) microscope. (B) The same transformant was grown as in (A) and the whole mycelium and an aliquot from the culture medium were boiled with sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) sample buffer; 40 μL (1/28 of total) of the former and 600 μL (1/33 of total, precipitated with methanol–chloroform) of the latter were electrophoresed and detected with Coomassie blue or anti‐green fluorescent protein (anti‐GFP) antibodies (in a Western blot) as indicated.
BcSpl1 binds to plant plasma membranes and causes cell shrinkage and chloroplast disorganization
Previous results from our group (Frías et al., 2011) have indicated that the necrosis‐inducing activity of BcSpl1 on A. thaliana requires the plant signalling protein BAK1 for full activity. BAK1 has also been shown to be required for the transduction of the signal generated by the binding of several PAMPs, such as bacterial flagellin and elongation factor Tu, to their respective membrane receptors (Segonzac and Zipfel, 2011; Zipfel and Robatzek, 2010). This led to the idea that BcSpl1 could act as a PAMP and bind to a putative receptor in the plasma membrane to initiate a cascade of events leading to HR, thus resulting in the observed necrosis. To test this hypothesis, two fusion proteins containing the whole BcSpl1 sequence plus GFP at either its N‐ or C‐terminal end were expressed in Pichia pastoris, as well as GFP alone as a control. The two fusion proteins were able to induce necrosis similarly to BcSpl1 in tomato, tobacco and Arabidopsis (Fig. S1, see Supporting Information), whereas GFP alone did not produce any effect. Incubation of tomato or tobacco protoplasts with either one of the two proteins resulted in GFP labelling of the plant plasma membrane (Fig. 2A), whereas no GFP signalling could be detected in the controls incubated with GFP alone, indicating that BcSpl1 indeed binds to plant plasma membranes. Attempts to observe, in a similar manner, the binding of BcSpl1 to A. thaliana leaf protoplasts were unsuccessful. This is probably because the leaves of this plant are much less susceptible to BcSpl1 than those of tobacco or tomato (not shown), and therefore the binding may not be sufficiently strong to be seen under the microscope. BcSpl1 binding to tomato or tobacco protoplasts also resulted in rapid and clear morphological alterations, including cell shrinkage and chloroplast disorganization. These changes were especially evident at 60 min (Fig. 2B), whereas untreated cells, or cells treated with the control proteins GFP or bovine serum albumin (BSA) (not shown), did not show any alteration.
Figure 2.
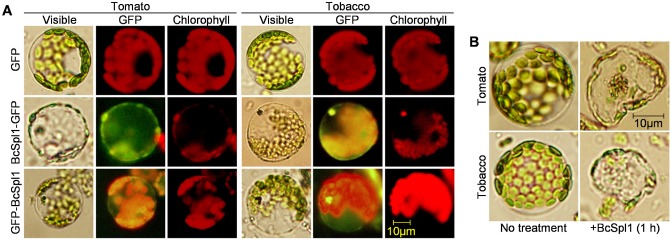
BcSpl1 binds to plant plasma membranes. (A) Tomato or tobacco protoplasts were incubated with 3.4 μm BcSpl1‐GFP, 3.4 μm GFP‐BcSpl1 or no protein in the control, and then washed and observed under a microscope with visible light or appropriate filters for the fluorescence of green fluorescent protein (GFP) and chlorophyll. (B) Effect on protoplasts of a 1‐h incubation with BcSpl1.
Several conserved regions of BcSpl1 are exposed on the protein surface and constitute potential necrosis‐inducing motifs
Protein regions recognized as PAMPs by the plant immune system are usually very well conserved in the corresponding protein families, such as in the case of bacterial flagellin (Felix et al., 1999). To search for such regions in BcSpl1, an alignment of 146 cerato‐platanin sequences was analysed in order to find parts of the sequences strongly conserved among all members of the family and among those cerato‐platanins which have been shown to elicit plant defences (Fig. 3A). Among the regions with a degree of conservation above average in the two sets (regions 1–5 in Fig. 3A), there are clearly two groups. Regions 2 and 4 (red and blue, respectively, in Fig. 3B) correspond to two loops interacting with each other, forming a small protrusion in the protein surface (de Oliveira et al., 2011), stabilized by a disulphide bond. The C‐terminal part of region 4, which is not as well conserved as the N‐terminal part (Fig. 3A), extends into the beta strand β3, according to the nomenclature of de Oliveira et al. (2011). The other three conserved regions (1, 3, 5) correspond to three beta strands (β1, β2 and β5) forming one side of the beta barrel. These highly conserved regions nicely coincide with those parts of the C. platani cerato‐platanin reported to be involved in binding to N‐acetylglucosamine oligomers (de Oliveira et al., 2011).
Figure 3.

Identification of evolutionarily conserved regions in BcSpl1. (A) Conservation indices (grey bars) were calculated with AL2CO for every BcSpl1 residue from the alignment of either 146 complete cerato‐platanin family sequences downloaded from Pfam (top panel) or the eight cerato‐platanins known (Buensanteai et al., 2010; Comparini et al., 2009; Frías et al., 2011; Pazzagli et al., 1999; Vargas et al., 2008; Wilson et al., 2002; Yang et al., 2009; Zaparoli et al., 2009) to elicit plant defences (bottom panel). Zero corresponds to the average conservation index for the whole protein. Average conservation indices were also calculated for windows of five residues for the whole protein (thin line) or after deleting the four cysteines (thick line) to avoid the effect of its absolute conservation. Broken line boxes indicate the five regions with conservation indices well above average (zero) in the two alignments. (B) Position of the five conserved regions in the structure of BcSpl1 predicted by SWISS‐MODEL in three different views.
A 40‐amino‐acid conserved BcSpl1 region induces necrosis by itself
In order to determine whether one or more of the conserved regions in the BcSpl1 sequence (Fig. 3) are necessary and sufficient for necrosis‐inducing activity, bidirectional serial deletions were generated in the bcspl1 gene, so that one or more of the five conserved regions were deleted from either the N‐ or C‐terminus (Fig. 4A). The resulting truncated bcspl1 genes were all expressed in P. pastoris as fusions to GFP, and the proteins were purified by nickel affinity chromatography. The necrosis‐inducing ability of these protein variants is presented in Fig. 4B and shows that: (i) deletion of the conserved region 1 or 5 did not produce any effect on the necrosis‐inducing activity of BcSpl1; (ii) deletion of regions 1–2 or 4–5 caused a significant reduction in the necrosis‐inducing activity; and (iii) simultaneous deletion of regions 2–4 completely abolished the necrosis‐inducing activity. These results indicate that the 40‐amino‐acid central region of the protein, comprising regions 2–4, is required for full activity. This was further confirmed by the analysis of the binding of all the truncated versions of BcSpl1 to tobacco (Fig. 4C) or tomato (Fig. S2, see Supporting Information) protoplasts. The deletion of regions 2–4 completely abolished binding to the membrane of the protoplasts of both plants, whereas regions 1 and 5 were dispensable. The deletion of region 1–2 seems to maintain sufficient binding capacity to produce green fluorescence in the membrane, and the same is true for the deletion of region 4–5. The comparison of the activity of these truncated proteins in the context of the three‐dimensional structure predicted for the protein (Fig. 3B) indicates that the two surface loops comprising the conserved motif, marked as red and blue, seem to be required for the necrosis‐inducing activity, whereas those deletions affecting the conserved regions in the beta barrel are not.
Figure 4.
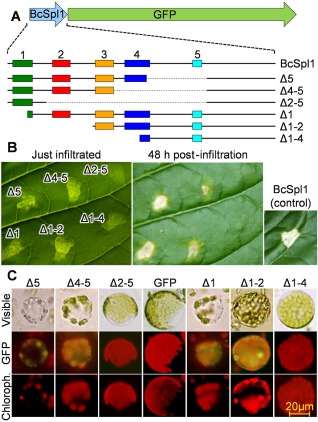
Determination of necrosis‐inducing regions in BcSpl1. (A) Scheme showing the different serial deletions that were made in the BcSpl1 part of the BcSpl1‐GFP fusion protein. The five numbers and different colours identify the five conserved regions displayed in Fig. 3. Broken lines indicate internal regions deleted from the gene. (B) Necrosis‐inducing activity of the different truncated versions of BcSpl1, at a concentration of 34 μm, on tobacco leaves. (C) Binding of the truncated proteins to tobacco protoplasts (see legend to Fig. 2 for details). GFP, green fluorescent protein.
Two short BcSpl1 fragments cooperate to induce necrosis
To analyse the hypothesis that the two conserved loops in the BcSpl1 surface may constitute the motif responsible for the necrosis‐inducing activity, two 10‐amino‐acid peptides were synthesized with the sequence of the most exposed and conserved parts of the loops: PepA (VSCSDGSNGL) and PepB (VVAGWNDANC) (Fig. 3). The sequences of the two peptides include the two cysteine residues which form a disulphide bond connecting the two loops in the predicted BcSpl1 structure (Fig. 3A). Infiltration of either one of the two peptides was able to produce necrosis in tobacco leaves, in a concentration‐dependent manner, but less efficiently than the whole BcSpl1 (Fig. 5A). Higher concentrations were needed to induce necrosis in the treated area, and even very high concentrations (136 μm) did not reproduce the effect observed with 34 μm BcSpl1. Moreover, as expected for the binding of PepA and PepB to a putative BcSpl1 receptor, the two peptides were able to act as inhibitors of the necrosis‐inducing activity of BcSpl1 (Fig. 5A), presumably by competition for the receptor. At a molar ratio of 1 : 4 between BcSpl1 and either one of the two peptides (34 μm BcSpl1 and 136 μm PepA or PepB), the necrosis produced had an appearance similar to that produced with the same concentration of the peptides alone, indicating that, at these concentrations, BcSpl1 is not contributing significantly to necrosis and is therefore inhibited, i.e. out‐competed, to a high degree.
Figure 5.

Eliciting activity of peptides PepA and PepB. (A) Necrosis‐inducing activity of different concentrations of the two peptides on tobacco leaves, either alone or mixed with BcSpl1. Controls were made with bovine serum albumin (BSA) or with BcSpl1 at a concentration of 34 μm. (B) Necrosis‐inducing activity of peptides PepA, PepB and their mixture, with or without preincubation with 1% dimethylsulphoxide (DMSO). The indicated peptides or peptide mixture (in red) were infiltrated at the indicated concentration (in black). Photographs were taken at day 3 after infiltration. (C) Ion leakage caused on tobacco leaf discs by infiltration with the indicated proteins and peptides. pA, PepA; pB, PepB. The experiment was repeated three times with similar results.
Contrary to our initial expectation, the infiltration of tobacco leaves with a mixture of PepA and PepB was not much more effective in causing necrosis than the infiltration of either peptide alone; the necrotic areas were clear only at a high concentration of the peptides in the mixture (Fig. 5B). One possible explanation for this result is that the disulphide bridge, which putatively binds the two peptides in the native BcSpl1, does not form in the peptide mixture, preventing the two peptides from reconstituting the whole motif. This possibility was explored by incubating the mixture with dimethylsulphoxide (DMSO) prior to infiltration, as this compound has been reported to encourage disulphide bond formation between peptides in aqueous solutions (Tam et al., 1991). Indeed, DMSO had a stimulating effect on the necrosis‐inducing activity of the peptide mixture (Fig. 5B), which was especially evident at 34 μm, but not of each peptide alone. DMSO alone at the same concentration, 1%, did not produce any effect on tobacco leaves or stimulate the effect of the whole BcSpl1 or of the unrelated elicitor EIX (Ron and Avni, 2004) (Fig. S3, see Supporting Information). Therefore, it seems that PepA and PepB contribute synergistically to the production of necrosis, in conditions of disulphide bond formation, in accordance with the hypothesis that the protrusion of BcSpl1, marked red and blue in the predicted structure (Fig. 3B), may constitute the necrosis‐inducing motif. A more quantitative determination of the eliciting ability of the different peptides or mixture was obtained by assaying the electrolyte leakage produced as a consequence of elicitation (Fig. 5C), and the results confirmed those of necrosis. PepA, PepB and their mixture (without DMSO) all have about the same effect, which was lower than that of BcSpl1, and the incubation of the mixture of the two peptides with DMSO had a stimulating effect on the activity.
A chimeric protein displaying the two‐peptide eliciting motif exhibits BcSpl1 necrosis‐inducing activity
To better confirm the participation of the PepA and PepB in the building of the elicitor motif, a chimeric protein was designed in which the two peptides were displayed on the surface of the small, highly stable DNA‐binding protein Sso7d (Baumann et al., 1994) from the hyperthermophilic archaeon Sulfolobus solfataricus, in an orientation designed to reproduce the putative eliciting motif (marked blue and red in Fig. 3B). For this purpose, a gene was synthesized de novo that coded for an Sso7d variant in which the N‐ and C‐terminal amino acids were substituted by PepA and PepB, respectively (Fig. 6A). The fusion was performed in such a way that the distance between the C‐terminus of PepA and the N‐terminus of PepB in the chimeric protein, assuming that it folds as the native Sso7d (protein data bank id. 1SSO), was the same as in the structure predicted for BcSpl1. The new protein (named Sso7d‐PepAB), together with a negative control without the BcSpl1 sequence (Sso7d), was expressed in P. pastoris, purified and assayed for its necrosis‐inducing activity. The necrosis observed for Sso7d‐PepAB was almost identical to that caused by the infiltration of BcSpl1, with a marginally lower necrosis‐inducing activity that was clear only at the lowest concentrations (Fig. S3). As happened with the mixture of PepA + PepB, incubation of Sso7d‐PepAB with DMSO to restore the disulphide bond had a stimulating effect, taking the activity to a level not distinguishable from that of BcSpl1 (Fig. 6B). The infiltration of the negative control (Sso7d), with (Fig. 6B) or without (not shown) DMSO, did not produce any visible effect in the leaves. In order to exclude any possible artefactual effect of the infiltration of DMSO together with the proteins, we carried out a re‐purification of the DMSO‐treated Sso7d‐PepAB protein, taking advantage of the 6 × histidine (6 × His) tag. The infiltration of the re‐purified protein, dissolved in water, also resulted in necrosis in the tobacco leaves not distinguishable from that of BcSpl1 (Fig. S3). These results were confirmed by assaying the ability of Sso7d‐PepAB to produce electrolyte leakage, which was the same as that of BcSpl1 for the DMSO‐treated protein (Fig. 6C).
Figure 6.
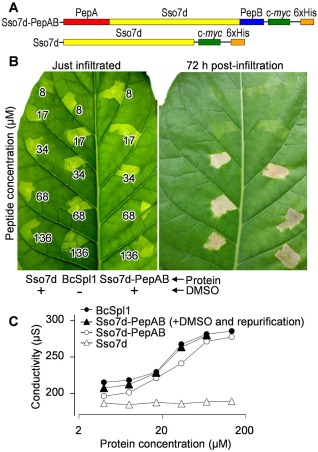
Eliciting activity of peptides PepA and PepB on a different protein backbone. (A) Scheme of the chimeric protein displaying the two peptides on the protein Sso7d from Sulfolobus solfataricus (Sso7d‐PepAB), as well as the control protein lacking the two peptides (Sso7d). (B) Necrosis‐inducing activity of the two chimeric proteins in comparison with BcSpl1. Sso7d‐PepAB and Sso7d were treated with dimethylsulphoxide (DMSO) and infiltrated without further processing. (C) Ion leakage on tobacco leaf discs caused by infiltration with the indicated proteins. Where indicated, Sso7d‐PepAB was treated with DMSO and re‐purified.
The two‐peptide eliciting motif suffices for the BcSpl1 contribution to B. cinerea virulence
As bcspl1 mutants are about 20%–40% less virulent on tomato or tobacco leaves, when compared with the wild‐type (Frías et al., 2011), we wondered whether the necrosis‐inducing activity located in the two‐peptide motif constitutes the contribution of BcSpl1 to infection. To answer this question, the chimeric protein Sso7d‐PepAB was tested for its capacity to complement the bcspl1 mutation in B. cinerea. The genes coding for the chimeric proteins Sso7d‐PepAB and Sso7d (Fig. 6A) were expressed in B. cinerea under the control of the bcspl1 promoter and signal peptide. One of the two previously characterized Δbcspl1 mutants (Frías et al., 2011) was transformed with the new genes and the secretion of the two chimeric proteins was corroborated by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and Western blot with anti c‐myc antibodies (Fig. S4, see Supporting Information). When assayed for its virulence on tobacco and tomato leaves, the strain expressing the chimeric protein with the two‐peptide eliciting motif (named Δbcspl1‐Sso7d‐PepAB) recovered the virulence of the wild‐type in both plants (Fig. 7), unequivocally showing that the two‐peptide motif accounts for the contribution of BcSpl1 to virulence. The expression of the control protein Sso7d did not increase the virulence (Fig. 7).
Figure 7.
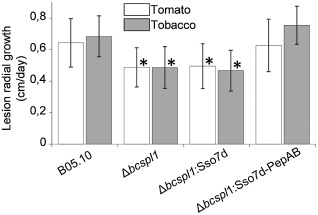
Contribution of Sso7d‐PepAB to Botrytis cinerea virulence. The strains Δbcspl1:Sso7d and Δbcspl1:Sso7d‐PepAB were generated by transformation of the bcspl1 mutant Δbcspl1.1 with the genes coding for the chimeric proteins Sso7d‐PepAB and Sso7d, respectively (see Fig. 6). Virulence for these three strains and the wild‐type (B05.10) was assayed on tomato and tobacco leaves by measuring the rate of expansion of the lesions, and is represented as mean values and standard deviation. Asterisks indicate a statistically significant difference from the wild‐type in the same plant, by Student's t‐test (n ≥ 30, P < 0.01).
Discussion
We have shown here that the cerato‐platanin BcSpl1 is able to bind to plant protoplasts, causing a rapid cell shrinking and disorganization of the chloroplast (Fig. 2), which macroscopically results in tissue necrosis. Serial deletion experiments (Fig. 4), chemically synthesized peptides (Fig. 5) and the expression of the suspected regions in a different protein backbone (Fig. 6) showed that two conserved regions (Fig. 3), interacting with each other in the protein surface, are necessary and sufficient for the phytotoxic activity of BcSpl1. Moreover, the expression of this two‐peptide motif in a Δbcspl1 mutant restores its virulence to wild‐type levels (Fig. 7), therefore accounting for the bcspl1 contribution to virulence. This is the first example, to our knowledge, of a plant eliciting motif composed of different peptides. These results corroborate previous observations obtained with two longer BcSpl1 peptides (Frías et al., 2011), which, in practice, were extended versions of PepA and PepB comprising about one‐third of the protein each. The longer peptides behaved similarly to PepA and PepB in producing necrosis when infiltrated in plant leaves, i.e. infiltration of either one of the peptides produced chlorosis in the leaves but no necrosis, whereas infiltration of the mixture produced necrosis similar to that produced by the whole BcSpl1 protein.
The three‐dimensional structure of the cerato‐platanin from C. platani (de Oliveira et al., 2011) showed a conserved region in its surface, which was proposed to have an important role in oligosaccharide binding, as shown by the fact that chemical shift perturbation of the amide proton, caused by the binding of short N‐acetylglucosamine oligomers, was high in the regions 17–29 and 51–54 of the C. platani protein (de Oliveira et al., 2011). The corresponding regions in BcSpl1 (18–30 and 53–56, respectively), according to an alignment of the whole family (de Oliveira et al., 2011), are the two regions shown here to be involved in the necrosis‐inducing activity. In addition, as shown here (Fig. 3) and by de Oliveira et al. (2011), these two regions are among the most evolutionarily conserved parts of the protein. This opens up interesting questions regarding the oligosaccharide‐binding activity and necrosis‐inducing activity of cerato‐platanins. The coincidence of both activities on the same protein region may indicate, on the one hand, that the second is just a consequence of the first. One may speculate, for example, that the binding of cerato‐platanins to a glycosylated receptor in the plant plasma membrane is the first step in the elicitation process. However, the coincidence may be just because the plant immune system has, by evolution, developed recognition of the most conserved, and thus functionally important, parts of the protein.
We have also shown here that the two‐peptide motif is responsible for the contribution of BcSpl1 to virulence. One reason why this may be so is because the necrosis‐inducing activity of BcSpl1 could generate dead tissue appropriate for the growth of B. cinerea. This necrotroph has indeed been proposed to benefit from HR (Choquer et al., 2007; Govrin and Levine, 2000; van Kan, 2006; Williamson et al., 2007), a form of programmed cell death in plants (Mur et al., 2008), and BcSpl1 has been shown previously to induce symptoms and markers of HR in plant leaves (Frías et al., 2011). An alternative, not contradictory, explanation may be that BcSpl1 could soften plant cell walls with its putative expansin activity (Baccelli et al., 2013), and therefore contribute to virulence by facilitating plant invasion by the fungus. As discussed above, the necrosis‐inducing and polysaccharide‐binding activities are located in the same protein region, making it difficult to set up experiments to discriminate between the two hypotheses. Finally, we cannot rule out possible defects in the fungal cell wall, caused by the lack of cerato‐platanin, as contributing factors in the lower virulence of bcspl1 mutants. Although no phenotypic difference, especially no alteration in the growth rate, was observed for the bcspl1 mutants in axenic culture (Frías et al., 2011), it is not possible at this point to exclude cell wall defects that become apparent only in planta.
The analysis of cerato‐platanins is further complicated because no clear biological function has yet been assigned to this protein. Its expansin‐like fold (de Oliveira et al., 2011), ability to bind polysaccharides (de Oliveira et al., 2011; Frischmann et al., 2013) and to weaken cellulosic material (Baccelli et al., 2013), and the fact that it remains mostly bound to the cell wall on secretion (Fig. 1) (Boddi et al., 2004), suggest some expansin‐like role in the fungal cell wall, in addition to its ability to induce necrosis in plants. This hypothesis also agrees with the fact that cerato‐platanins are widely distributed in fungi which are not plant pathogens, but disagrees with the fact that mutants lacking cerato‐platanin show no phenotype other than a decreased virulence (Frías et al., 2011; Frischmann et al., 2013; Jeong et al., 2007).
Experimental Procedures
Organisms, strains and culture conditions
Botrytis cinerea strain B05.10 (Büttner et al., 1994), one of the two strains with a sequenced genome (Amselem et al., 2011; Staats and van Kan, 2012), was used as wild‐type strain. bcspl1 mutants have been generated previously (Frías et al., 2011). Fungal strains were kept as conidia in 15% glycerol at −80 °C for long‐time storage, or in silica gel at 4 °C for routine use (Delcan et al., 2002). The silica stock was used to inoculate tomato‐agar plates (25% tomato fruit extract, 2% agar, pH 5.5) to obtain conidia, as described by Benito et al. (1998). The YGG medium used for B. cinerea contains 0.5% yeast extract, 100 mm glucose and 0.3% Gamborg's B5 (Duchefa Biochemie, http://www.duchefa‐biochemie.nl). Plants (Nicotiana tabacum cv. Havana, Lycopersicon esculentum cv. Moneymaker and A. thaliana Col‐0) were maintained at controlled temperature, humidity and photoperiod in a phytotron. Tobacco leaves were infiltrated in the necrotizing assays with the help of a 1‐mL syringe, without a needle, filled with the indicated proteins dissolved in water, except where otherwise indicated. Virulence assays were performed as described elsewhere (Frías et al., 2011) on tomato and tobacco leaves, and the lesion expansion rates were calculated between days 2 and 3 after inoculation, with a minimum of 30 inoculations per assay.
Standard techniques
SDS‐PAGE and Western blots were carried out according to standard procedures with Bio‐Rad (http://www.bio‐rad.com) precast gels and polyvinylidene fluoride (PVDF) membranes. Monoclonal anti‐GFP or anti‐c‐myc primary antibodies were diluted 1:1000 and detected with a 1:3000 dilution of antimouse IgG conjugated to horseradish peroxidase (HRP) (Sigma‐Aldrich, http://www.sigmaaldrich.com). The peroxidase was detected with Immobilon Western Chemiluminescent HRP Substrate (Millipore, http://www.millipore.com). Quantification of Western blot bands (Fig. 1B) was performed with the software Quantity One (Bio‐Rad) on the chemiluminescence signal recorded with a Chemidoc system (Bio‐Rad). Ion leakage was assayed as explained elsewhere (Frías et al., 2011). Protein sequence alignments were performed with Clustal Omega (Sievers et al., 2011) and processed with AL2CO (Pei and Grishin, 2001) to generate conservation indices for every amino acid, by making use of the score matrix BLOSUM62.
Plasmid constructions and the generation of new B. cinerea strains
Polymerase chain reaction (PCR) amplifications were made with Phusion polymerase (New England Biolabs, http://www.neb.com) when the DNA product was to be cloned, and Taq polymerase (GenScript, http://www.genscript.com) was used in all other cases. All oligonucleotides used (Table S1, see Supporting Information) were from Life Technologies (http://www.lifetechnologies.com). Botrytis cinerea transformations were carried out as described by Hamada et al. (1994), with the modifications introduced by van Kan et al. (1997).
To express BcSpl1‐GFP and GFP‐BcSpl1 fusion proteins in P. pastoris, two different plasmids, pCP‐GFP and pGFP‐CP, were designed as follows: the mgfp4 gene (Haseloff et al., 1997) was amplified from the nos‐GFP cassette (Hellens et al., 2000) with the primer pair GFPCP‐FW/GFPCP‐RV or CPGFP‐FW/CPGFP‐RV, and cloned at the EcoRI site at the 5′ end or the XbaI site at the 3′ end of bcspl1, respectively, in plasmid pCP (Frías et al., 2011).
Plasmid pCRP‐GFP, used to express the BcSpl1‐GFP fusion protein in B. cinerea, is derived from plasmid pOptGFP, which contains a GFP gene codon optimized for B. cinerea under the strong Aspergillus nidulans OliC promoter (Leroch et al., 2011) in a plasmid backbone previously designed for site‐directed integration in the B. cinerea genome (Noda et al., 2007). pCRP‐GFP was constructed by cloning in the SmaI site of pOptGFP the bcspl1 gene amplified by PCR from plasmid pCP (Frías et al., 2011). The primer pair used, SS‐CRP‐SmaI/CRP‐Sma1‐REV, restored the native signal sequence to the bcspl1 gene and amplified it together with the c‐myc and 6 × His epitopes originally from pPICZαA (Invitrogen, Carlsbad, CA, USA). The circular pCRP‐GFP plasmid was then transformed into B. cinerea B05.10 and the transformants were subjected to several rounds of single spore isolation. A transformant (CRP‐GFP8.1.1) producing only fluorescent conidia, and thus putatively homokaryotic, was selected.
Bidirectional deletions in the bcspl1 gene were carried out by PCR amplifying the whole pCP‐GFP plasmid described above, except the bcspl1 region to be deleted. The two primers facing outward from the intended deletion introduced KpnI sites in the borders of the amplicons, and these were then cut with KpnI and ligated with each other to generate a plasmid in which a bcspl1 region was substituted by the two amino acids coded by the KpnI site (glycine‐threonine). The resulting plasmids were then transformed into P. pastoris to express the truncated proteins.
Plasmids pSsoCP and pSso, designed to express the chimeric proteins Sso7d‐PepAB and Sso7d in yeast, were constructed by cloning the corresponding de novo‐synthesized genes (GenScript) in the P. pastoris expression vector pPICZαA. The sequences of the genes (Fig. S5, see Supporting Information) were optimized according to the codon usage in P. pastoris highly expressed genes (Bai et al., 2011) and in B. cinerea (Leroch et al., 2011), and included EcoRI and XbaI sites at the ends to clone them in the P. pastoris vector. The two chimeric proteins Sso7d‐PepAB and Sso7d were expressed in B. cinerea Δbcspl1.1 by transformation with the plasmids pNR2‐SsoCP and pNR2‐Sso. These plasmids are derived from plasmid pNR2‐CP, which contains the whole bcspl1 gene with promoter and terminator and a nourseothricin resistance cassette for selection (Frías et al., 2011). The whole pNR2‐CP plasmid, except the bcspl1 open reading frame (ORF), was amplified by inverse PCR with primers Pro‐cp‐Rv and Ter‐cp‐Fw, and ligated to the synthetic genes for Sso7d‐PepAB and Sso7d (Fig. S5) obtained with the primer pairs Ssocp‐Fw/Sso‐rv and Sso‐Fw/Sso‐rv, respectively. pNR2‐SsoCP and pNR2‐Sso were then transformed into the B. cinerea bcspl1 mutant Δbcspl1.1.
Peptides and recombinant proteins
Peptides were from GenScript. The EasySelect Pichia Expression Kit (Life Technologies) was used to express recombinant proteins in P. pastoris according to the manufacturer's instructions. Proteins were purified from the supernatants of cultures of P. pastoris induced for 2 days with 0.5% methanol in BMMH (100 mM potassium phosphate pH 6.0, 1.34% yeast nitrogen base, 4 × 10−5% biotin) medium with HisTrap FF prepacked minicolums (GE Healthcare Life Sciences, http://www.gelifesciences.com), as explained elsewhere (Frías et al., 2011). The formation of disulfide bridges in purified proteins, or in peptide mixtures, was encouraged when necessary by incubating them overnight, with slow shaking at room temperature, with 1% DMSO (Tam et al., 1991). When indicated, proteins were then re‐purified by binding to magnetic nickel beads (GenScript) for 2 h, washing four times with washing buffer and eluting with water, using the same reagents as those employed in the column purification (Frías et al., 2011).
Binding assay of GFP‐tagged proteins to plant protoplasts
Tomato, tobacco and A. thaliana protoplasts were prepared from leaf material equilibrated for 30 min in 10 mm MES–KOH, pH 5.7, 0.5 m mannitol. The protoplast solution contained 2.5 mm MES–KOH, pH 5.7, 200 mm mannitol, 5 mm KCl, 5 mm CaCl2, 2% cellulase from Trichoderma viride (Sigma‐Aldrich C1794) and 0.5% pectinase from Rhizopus sp. (Sigma‐Aldrich P2401). This solution was first infiltrated into plant tissue by applying a vacuum for 3 min, and the mixture was then incubated for 1 h at 20 °C with slow shaking. After carefully washing with 5 mm MES–KOH, pH 5.7, 400 mm mannitol, 5 mm KCl and 5 mm CaCl2, protoplasts were incubated with a 0.34 μm concentration of the appropriate protein, washed again with the same buffer and examined with an Olympus BX‐50 fluorescence microscope equipped with a U‐MWIB filter to detect GFP and a U‐MWIG filter to detect chlorophyll.
Supporting information
Fig. S1 Necrosis‐inducing activity of the two fusion proteins between BcSpl1 and green fluorescent protein (GFP) (BcSpl1‐GFP and GFP‐BcSpl1) in leaves from three different plants.
Fig. S2 Binding of the truncated BcSpl1 proteins to tomato protoplasts.
Fig. S3 Effect of dimethylsulphoxide (DMSO) on the necrosis‐inducing activities of Sso7d‐PepAB, BcSpl1 and EIX.
Fig. S4 Secretion of the chimeric proteins Sso7d and Sso7d‐PepAB in the Botrytis cinerea mutant Δbcspl1.
Fig. S5 Nucleotide sequences of the chemically synthesized genes Sso7d‐PepAB and Sso7d. The amino acid translation is shown below the DNA sequence and the relevant regions are indicated.
Table S1 Oligonucleotides used in this study.
Acknowledgements
Support for this research was provided by grants from the Ministerio de Educación y Ciencia (AGL2010‐22222) and the Agencia Canaria de Investigación, Innovación y Sociedad de la Información (ACIISI) (PI2007/009). MG and MF were supported by ACIISI. Eighty‐five per cent of funds received from ACIISI came from the European Regional Development Fund.
References
- Amselem, J. , Cuomo, C.A. , van Kan, J.A.L. , Viaud, M. , Benito, E.P. , Couloux, A. , Coutinho, P.M. , de Vries, R.P. , Dyer, P.S. , Fillinger, S. , Fournier, E. , Gout, L. , Hahn, M. , Kohn, L. , Lapalu, N. , Plummer, K.M. , Pradier, J.M. , Quévillon, E. , Sharon, A. , Simon, A. , ten Have, A. , Tudzynski, B. , Tudzynski, P. , Wincker, P. , Andrew, M. , Anthouard, V.Ã. , Beever, R.E. , Beffa, R. , Benoit, I. , Bouzid, O. , Brault, B. , Chen, Z. , Choquer, M. , Collémare, J. , Cotton, P. , Danchin, E.G. , Da Silva, C. , Gautier, A. , Giraud, C. , Giraud, T. , González, C. , Grossetete, S. , Güldener, U. , Henrissat, B. , Howlett, B.J. , Kodira, C. , Kretschmer, M. , Lappartient, A. , Leroch, M. , Levis, C. , Mauceli, E. , Neuvéglise, C. , Oeser, B. , Pearson, M. , Poulain, J. , Poussereau, N. , Quesneville, H. , Rascle, C. , Schumacher, J. , Ségurens, B. , Sexton, A. , Silva, E. , Sirven, C. , Soanes, D.M. , Talbot, N.J. , Templeton, M. , Yandava, C. , Yarden, O. , Zeng, Q. , Rollins, J.A. , Lebrun, M.H. and Dickman, M. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genet. 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccelli, I. , Luti, S. , Bernardi, R. , Scala, A. and Pazzagli, L. (2013) Cerato‐platanin shows expansin‐like activity on cellulosic materials. Appl. Microbiol. Biotechnol. in press. doi: 10.1007/s00253-013-4822-0. [DOI] [PubMed] [Google Scholar]
- Bai, J. , Swartz, D.J. , Protasevich, I.I. , Brouillette, C.G. , Harrell, P.M. , Hildebrandt, E. , Gasser, B. , Mattanovich, D. , Ward, A. , Chang, G. and Urbatsch, I.L. (2011) A gene optimization strategy that enhances production of fully functional P‐glycoprotein in Pichia pastoris . PLoS One, 6, e22577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, H. , Knapp, S. , Lundback, T. , Ladenstein, R. and Hard, T. (1994) Solution structure and DNA‐binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus . Nat. Struct. Biol. 1, 808–819. [DOI] [PubMed] [Google Scholar]
- Benito, E.P. , ten Have, A. , van't Klooster, J.W. and Van Kan, J.A.L. (1998) Fungal and plant gene expression during synchronized infection of tomato leaves by Botrytis cinerea . Eur. J. Plant Pathol. 104, 207–220. [Google Scholar]
- Boddi, S. , Comparini, C. , Calamassi, R. , Pazzagli, L. , Cappugi, G. and Scala, A. (2004) Cerato‐platanin protein is located in the cell walls of ascospores, conidia and hyphae of Ceratocystis fimbriata f. sp. platani. FEMS Microbiol. Lett. 233, 341–346. [DOI] [PubMed] [Google Scholar]
- Buensanteai, N. , Mukherjee, P.K. , Horwitz, B.A. , Cheng, C. , Dangott, L.J. and Kenerley, C.M. (2010) Expression and purification of biologically active Trichoderma virens proteinaceous elicitor Sm1 in Pichia pastoris . Protein Expr. Purif. 72, 131–138. [DOI] [PubMed] [Google Scholar]
- Büttner, P. , Koch, F. , Voigt, K. , Quidde, T. , Risch, S. , Blaich, R. , Brückner, B. and Tudzynski, P. (1994) Variations in ploidy among isolates of Botrytis cinerea: implications for genetic and molecular analyses. Curr. Genet. 25, 445–450. [DOI] [PubMed] [Google Scholar]
- Choquer, M. , Fournier, E. , Kunz, C. , Levis, C. , Pradier, J.M. , Simon, A. and Viaud, M. (2007) Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 277, 1–10. [DOI] [PubMed] [Google Scholar]
- Comparini, C. , Carresi, L. , Pagni, E. , Sbrana, F. , Sebastiani, F. , Luchi, N. , Santini, A. , Capretti, P. , Tiribilli, B. , Pazzagli, L. , Cappugi, G. and Scala, A. (2009) New proteins orthologous to cerato‐platanin in various Ceratocystis species and the purification and characterization of cerato‐populin from Ceratocystis populicola . Appl. Microbiol. Biotechnol. 84, 309–322. [DOI] [PubMed] [Google Scholar]
- Dean, R. , van Kan, J.A. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. , Dickman, M. , Kahmann, R. , Ellis, J. and Foster, G.D. (2012) The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcan, J. , Moyano, C. , Raposo, R. and Melgarejo, P. (2002) Storage of Botrytis cinerea using different methods. J. Plant Pathol. 84, 3–9. [Google Scholar]
- Djonovic, S. , Pozo, M.J. , Dagott, L. , Howell, C. and Kenerley, C. (2006) Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant–Microbe Interact. 19, 838–853. [DOI] [PubMed] [Google Scholar]
- Djonovic, S. , Vargas, W.A. , Kolomiets, M.V. , Horndeski, M. , Wiest, A. and Kenerley, C.M. (2007) A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 145, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espino, J.J. , Gutiérrez‐Sánchez, G. , Brito, N. , Shah, P. , Orlando, R. and González, C. (2010) The Botrytis cinerea early secretome. Proteomics, 10, 3020–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G. , Duran, J.D. , Volko, S. and Boller, T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Fontana, F. , Santini, A. , Salvini, M. , Pazzagli, L. , Cappugi, G. , Scala, A. , Durante, M. and Bernardi, R. (2008) Cerato‐platanin treated plane leaves restrict Ceratocystis platani growth and overexpress defence‐related genes. J. Plant Pathol. 90, 295–306. [Google Scholar]
- Frías, M. , González, C. and Brito, N. (2011) BcSpl1, a cerato‐platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192, 483–495. [DOI] [PubMed] [Google Scholar]
- Frischmann, A. , Neudl, S. , Gaderer, R. , Bonazza, K. , Zach, S. , Gruber, S. , Spadiut, O. , Friedbacher, G. , Grothe, H. and Seidl‐Seiboth, V. (2013) Self‐assembly at air/water interfaces and carbohydrate binding properties of the small secreted protein EPL1 from the fungus Trichoderma atroviride . J. Biol. Chem. 288, 4278–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard, V. , Cherrad, S. , Dieryckx, C. , Gonçalves, I. , Dupuy, J.W. , Bonneu, M. , Rascle, C. , Job, C. , Job, D. and Vacher, S. (2012) Proteomic analysis of proteins secreted by Botrytis cinerea in response to heavy metal toxicity. Metallomics, 4, 835–846. [DOI] [PubMed] [Google Scholar]
- González‐Fernández, R. , Aloria, K. , Valero‐Galván, J. , Redondo, I. , Arizmendi, J.M. and Jorrín‐Novo, J.V. (2013) Proteomic analysis of mycelium and secretome of different Botrytis cinerea wild‐type strains. J. Proteomics. in press. doi: 10.1016/j.jprot.2013.06.022. [DOI] [PubMed] [Google Scholar]
- Govrin, E.M. and Levine, A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea . Curr. Biol. 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Hamada, W. , Reignault, P. , Bompeix, G. and Boccara, M. (1994) Transformation of Botrytis cinerea with the hygromycin B resistance gene, hph . Curr. Genet. 26, 251–255. [DOI] [PubMed] [Google Scholar]
- Haseloff, J. , Siemering, K.R. , Prasher, D.C. and Hodge, S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA, 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Have, A. , Espino, J.J. , Dekkers, E. , Sluyter, S.C.V. , Brito, N. , Kay, J. , González, C. and van Kan, J.A. (2010) The Botrytis cinerea aspartic proteinase family. Fungal Genet. Biol. 47, 53–65. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P. , Edwards, E.A. , Leyland, N.R. , Bean, S. and Mullineaux, P.M. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium‐mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Jeong, J.S. , Mitchell, T.K. and Dean, R.A. (2007) The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. FEMS Microbiol. Lett. 273, 157–165. [DOI] [PubMed] [Google Scholar]
- van Kan, J.A. (2006) Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11, 247–253. [DOI] [PubMed] [Google Scholar]
- Krieger, V. , Vargas, W.A. , Kenerley, C.M. and Sacchettini, J.C. Crystal structure of Sm1, an elicitor of plant defence responses from Trichoderma virens . 2010. Protein Data Bank id.: 3M3G. Available at http://pdb.org.
- Leroch, M. , Mernke, D. , Koppenhoefer, D. , Schneider, P. , Mosbach, A. , Doehlemann, G. and Hahn, M. (2011) Living colors in the gray mold pathogen Botrytis cinerea: codon‐optimized genes encoding green fluorescent protein and mCherry, which exhibit bright fluorescence. Appl. Environ. Microbiol. 77, 2887–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur, L.A. , Kenton, P. , Lloyd, A.J. , Ougham, H. and Prats, E. (2008) The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59, 501–520. [DOI] [PubMed] [Google Scholar]
- Noda, J. , Brito, N. , Espino, J.J. and González, C. (2007) Methodological improvements in the expression of foreign genes and in gene replacement in the phytopathogenic fungus Botrytis cinerea . Mol. Plant Pathol. 8, 811–816. [DOI] [PubMed] [Google Scholar]
- de Oliveira, A.L. , Gallo, M. , Pazzagli, L. , Benedetti, C.E. , Cappugi, G. , Scala, A. , Pantera, B. , Spisni, A. , Pertinhez, T.A. and Cicero, D.O. (2011) The structure of the elicitor Cerato‐platanin (CP), the first member of the CP fungal protein family, reveals a double psibeta‐barrel fold and carbohydrate binding. J. Biol. Chem. 286, 17 560–17 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazzagli, L. , Cappugi, G. , Manao, G. , Camici, G. , Santini, A. and Scala, A. (1999) Purification, characterization, and amino acid sequence of Cerato‐platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani. J. Biol. Chem. 274, 24 959–24 964. [DOI] [PubMed] [Google Scholar]
- Pei, J. and Grishin, N.V. (2001) AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics, 17, 700–712. [DOI] [PubMed] [Google Scholar]
- Ron, M. and Avni, A. (2004) The receptor for the fungal elicitor ethylene‐inducing xylanase is a member of a resistance‐like gene family in tomato. Plant Cell, 16, 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac, C. and Zipfel, C. (2011) Activation of plant pattern‐recognition receptors by bacteria. Curr. Opin. Microbiol. 14, 54–61. [DOI] [PubMed] [Google Scholar]
- Shah, P. , Atwood, J.A. , Orlando, R. , El, M.H. , Podila, G.K. and Davis, M.R. (2009a) Comparative proteomic analysis of Botrytis cinerea secretome. J. Proteome Res. 8, 1123–1130. [DOI] [PubMed] [Google Scholar]
- Shah, P. , Gutiérrez‐Sánchez, G. , Orlando, R. and Bergmann, C. (2009b) A proteomic study of pectin‐degrading enzymes secreted by Botrytis cinerea grown in liquid culture. Proteomics, 9, 3126–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. , Gibson, T.J. , Karplus, K. , Li, W. , Lopez, R. , McWilliam, H. , Remmert, M. , Söding, J. , Thompson, J.D. and Higgins, D.G. (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats, M. and van Kan, J.A.L. (2012) Genome update of Botrytis cinerea strains B05.10 and T4. Eukaryot. Cell, 11, 1413–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, J.P. , Wu, C.R. , Liu, W. and Zhang, J.W. (1991) Disulfide bond formation in peptides by dimethyl sulfoxide. Scope and applications. J. Am. Chem. Soc. 113, 6657–6662. [Google Scholar]
- Van Kan, J.A.L. , van't Klooster, J.W. , Wagemakers, C.A.M. , Dees, D.C.T. and van der Vlugt‐Bergmans, C.J.B. (1997) Cutinase A of Botrytis cinerea is expressed, but not essential, during penetration of gerbera and tomato. Mol. Plant–Microbe Interact. 10, 30–38. [DOI] [PubMed] [Google Scholar]
- Vargas, W.A. , Djonovic, S. , Sukno, S.A. and Kenerley, C.M. (2008) Dimerization controls the activity of fungal elicitors that trigger systemic resistance in plants. J. Biol. Chem. 283, 19 804–19 815. [DOI] [PubMed] [Google Scholar]
- Williamson, B. , Tudzynski, B. , Tudzynski, P. and Van Kan, J.A.L. (2007) Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. [DOI] [PubMed] [Google Scholar]
- Wilson, L.M. , Idnurm, A. and Howlett, B.J. (2002) Characterization of a gene (sp1) encoding a secreted protein from Leptosphaeria maculans, the blackleg pathogen of Brassica napus . Mol. Plant Pathol. 3, 487–493. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Zhang, H. , Li, G. , Li, W. , Wang, X. and Song, F. (2009) Ectopic expression of MgSM1, a Cerato‐platanin family protein from Magnaporthe grisea, confers broad‐spectrum disease resistance in Arabidopsis . Plant Biotechnol. J. 7, 763–777. [DOI] [PubMed] [Google Scholar]
- Zaparoli, G. , Cabrera‐García, O. , Medrano, F.J. , Tiburcio, R. , Lacerda, G. and Guimarães‐Pereira, G. (2009) Identification of a second family of genes in Moniliophthora perniciosa, the causal agent of witches' broom disease in cacao, encoding necrosis‐inducing proteins similar to cerato‐platanins. Mycol. Res. 113, 61–72. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. and Robatzek, S. (2010) Pathogen‐associated molecular pattern‐triggered immunity: veni, vidi…? Plant Physiol. 154, 551–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Necrosis‐inducing activity of the two fusion proteins between BcSpl1 and green fluorescent protein (GFP) (BcSpl1‐GFP and GFP‐BcSpl1) in leaves from three different plants.
Fig. S2 Binding of the truncated BcSpl1 proteins to tomato protoplasts.
Fig. S3 Effect of dimethylsulphoxide (DMSO) on the necrosis‐inducing activities of Sso7d‐PepAB, BcSpl1 and EIX.
Fig. S4 Secretion of the chimeric proteins Sso7d and Sso7d‐PepAB in the Botrytis cinerea mutant Δbcspl1.
Fig. S5 Nucleotide sequences of the chemically synthesized genes Sso7d‐PepAB and Sso7d. The amino acid translation is shown below the DNA sequence and the relevant regions are indicated.
Table S1 Oligonucleotides used in this study.


