SUMMARY
Type IV pilin (PilA) is a major constituent of pilus and is required for bacterial biofilm formation, surface motility and virulence. It is known that mature PilA is produced by cleavage of the short leader sequence of the pilin precursor, followed by methylation of N‐terminal phenylalanine. The molecular mass of the PilA mature protein from the tobacco bacterial pathogen Pseudomonas syringae pv. tabaci 6605 (Pta 6605) has been predicted to be 12 329 Da from its deduced amino acid sequence. Previously, we have detected PilA as an approximately 13‐kDa protein by immunoblot analysis with anti‐PilA‐specific antibody. In addition, we found the putative oligosaccharide‐transferase gene tfpO downstream of pilA. These findings suggest that PilA in Pta 6605 is glycosylated. The defective mutant of tfpO (ΔtfpO) shows reductions in pilin molecular mass, surface motility and virulence towards host tobacco plants. Thus, pilin glycan plays important roles in bacterial motility and virulence. The genetic region around pilA was compared among P. syringae pathovars. The tfpO gene exists in some strains of pathovars tabaci, syringae, lachrymans, mori, actinidiae, maculicola and P. savastanoi pv. savastanoi. However, some strains of pathovars tabaci, syringae, glycinea, tomato, aesculi and oryzae do not possess tfpO, and the existence of tfpO is independent of the classification of pathovars/strains in P. syringae. Interestingly, the PilA amino acid sequences in tfpO‐possessing strains show higher homology with each other than with tfpO‐nonpossessing strains. These results suggest that tfpO and pilA might co‐evolve in certain specific bacterial strains.
INTRODUCTION
Type IV pili (T4P) are filamentous polymers of pilin protein subunits (PilA) that extend from many Gram‐negative bacterial surfaces (reviewed in Mattick, 2002; Nudleman and Kaiser, 2004; Strom and Lory, 1993). These pili contribute to various bacterial behaviours, including adhesion to biotic and abiotic surfaces, surface twitching and swarming motilities, biofilm formation and bacteriophage adsorption. Thus, they are thought to be involved in bacterial pathogenicity by initiating the colonization on host tissue through adhesion to and motility across the surface (Burdman et al., 2011; Mattick, 2002). In phytopathogenic bacteria, such as Pseudomonas syringae pv. tabaci 6605 (Pta 6605), Acidovorax avenae ssp. citrulli and P. syringae pv. tomato DC3000, T4P promote virulence towards their respective host plants (Bahar et al., 2009; Burdman et al., 2011; Roine et al., 1998; Taguchi and Ichinose, 2011). Furthermore, T4P in Pta 6605 are known to be involved in the hypersensitive reaction‐inducing activity in nonhost Arabidopsis thaliana leaves, as the pilA‐defective mutant (ΔpilA) of this pathogen shows a remarkably reduced expression of hypersensitive reaction and pathogenicity (hrp)‐related genes (Taguchi and Ichinose, 2011).
Recently, protein glycosylation has been observed not only in eukaryotes, but also in prokaryotes, in both Gram‐negative and Gram‐positive bacteria (Benz and Schmidt, 2002; Ichinose et al., 2011; Logan, 2006; Power and Jennings, 2003). Glycans are often used to decorate proteins at the bacterial surface or components of appendages, such as flagella and pili. Pilin glycosylation has been reported in a modest number of animal pathogenic bacteria. However, pilin glycosylation is not species specific, but strain specific. Among the five distinct phylogenetic groups (I–V) based on the PilA amino acid sequence and the presence of specific accessory genes in P. aeruginosa (Pa), pilins of bacteria from groups I (e.g. Pa 1244) and IV (e.g. Pa 5196) were found to be glycosylated (Castric, 1995; 2004, 2008). Similar strain‐specific pilin glycosylation was also found in Neisseria meningitidis and N. gonorrhoeae (Aas et al., 2007; Virji et al., 1993). Pilins of both N. meningitidis and N. gonorrhoeae are glycosylated at serine 63 (Ser63) by the attachment of a short oligosaccharide of up to three sugar residues in length by the glycosyltransferases PglL and PglO, respectively (Parge et al., 1995; Power et al., 2006; Stimson et al., 1995). In the case of P. aeruginosa, Pa 1244 requires TfpO (originally called PilO) in order to transfer a single O‐antigen repeating unit of lipopolysaccharide (LPS) to the C‐terminal Ser residue of PilA (Castric, 1995; Castric et al., 2001; Comer et al., 2002), and Pa 5196 pilins are modified at multiple sites with trisaccharides of α‐1,5‐d‐Araf by arabinosyltransferase TfpW (Kus et al., 2008; Voisin et al., 2007). Both tfpO in Pa 1244 and tfpW in Pa 5196, encoding pilin glycosyltransferases TfpO and TfpW, respectively, localize immediately downstream of pilA. The Pa 1244 ΔtfpO mutants produced functional and nonglycosylated pili with reduced twitching motility, increased surface hydrophobicity and less virulence relative to the wild‐type (WT) strain (Smedley et al., 2005). Similarly, the defective mutant strains for tfpW and the glycan biosynthesis‐related genes in Pa 5196 showed reductions in motility, molecular mass of the pilin protein and pilus assembly relative to the WT strain (Harvey et al., 2011; Kus et al., 2008). These reports suggest that the glycan may contribute to T4P adhesiveness, thereby enhancing colonization and virulence.
Pseudomonas syringae, an opportunistic plant pathogenic bacterial species, can be classified into pathovars according to their host ranges and disease symptoms. Pta 6605 is a bacterium causing wildfire disease in tobacco plants (Ichinose et al., 2003). Previously, another filamentous fibre, flagellin of Pta 6605, was revealed to be glycosylated by three glycosyltransferases: Fgt1, Fgt2 and VioT (Ichinose et al., 2011; Nguyen et al., 2009; Taguchi et al., 2006). The fgt1 and fgt2 genes are located just upstream of the flagellin gene fliC, whereas vioT is located in the gene cluster to synthesize the substrate of VioT, dTDP‐N‐(3‐hydroxy‐1‐oxobutyl)‐2‐O‐methylviosamine. Glycosylation of the flagellin protein was found to be essential for surface motility, the stability of flagellar filaments and total virulence (Ichinose et al., 2011; 2008, 2009). Recently, we have found that this bacterial strain possesses T4P composed of major PilA subunits and several minor pilin subunits (Taguchi and Ichinose, 2011). Notably, the ΔpilA mutant exhibits the absence of T4P, followed by the loss of surface swarming motility and reduced biofilm formation and virulence. This indicates that T4P are required for virulence in this pathogen. The amino acid sequence of PilA in Pta 6605 was deduced from the pilA nucleotide sequence (Taguchi and Ichinose, 2011). Interestingly, the C‐terminal residue of PilA in Pta 6605 was Ser, similar to that of Pa 1244 pilin. It is known that the C‐terminal Ser residue is a unique glycosylation site in Pa 1244 pilin (Comer et al., 2002). However, pilin glycosylation has not been reported in any phytopathogenic bacteria, including P. syringae. In the present study, we found a putative pilin glycosyltransferase gene, tfpO, immediately downstream of pilA in Pta 6605. We report here on pilin glycosylation in Pta 6605 and the roles of pilin glycan in surface swarming motility and virulence.
RESULTS
tfpO gene in Pta 6605
Based on the genomic information at the Pseudomonas Genome Database V2 website (http://v2.pseudomonas.com), we attempted to isolate a DNA fragment containing pilA and its downstream region in Pta 6605. A pair of polymerase chain reaction (PCR) primers (PilA‐F and PilA‐R) was designed based on the registered sequence (accession number CP000058) of pilB (PSPPH0820) and the open reading frame 1 (orf1, PSPPH0825) in P. syringae pv. phaseolicola (Pph) 1448A. A 5533‐bp DNA fragment including pilA was amplified by PCR and the sequence was analysed. There are two open reading frames (orf) downstream of pilA (Fig. 1). The putative function and amino acid identity of each protein product are shown in Table 1. The first ORF (immediately downstream of pilA) has 86% amino acid identity to a putative glycosyltransferase of P. savastanoi pv. savastanoi 3335 (Psav 3335) and 37% identity to the pilin glycosyltransferase TfpO of Pa 1244 (Castric, 1995). Therefore, we refer to this gene as tfpO hereafter. Furthermore, TfpO in Pta 6605 has 19% identity over 225 residues and 17% identity over 388 residues with oligosaccharyltransferases TfpW of Pa 5196 (Kus et al., 2004) and PglL of N. meningitidis (Power et al., 2006), both glycosylates of the respective pilin proteins. These findings led us to hypothesize that this gene is involved in pilin glycosylation. The second ORF showed significant homology with the gene encoding the polysaccharide biosynthesis protein in Xanthomonas axonopodis pv. citri strain 306, with 41% amino acid identity, and Acidovorax avenae ssp. avenae ATCC 19860, with 34% identity, and was named psb.
Figure 1.
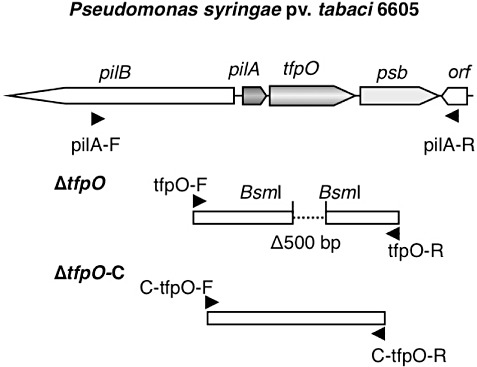
Schematic organization of pilA and its surrounding region in Pseudomonas syringae pv. tabaci 6605. The construction of the tfpO mutant and its complemented strain with a DNA fragment containing pilA and tfpO is shown. Sets of polymerase chain reaction (PCR) primers were used for PCR, as indicated by the arrows. To generate the internal deletion, digestion of the PCR product by primers (tfpO‐F and tfpO‐R) was performed with BsmI for ΔtfpO. For complementation of the ΔtfpO mutant, a DNA fragment containing pilA, tfpO and a putative promoter sequence was amplified with C‐tfpO‐F and C‐tfpO‐R.
Table 1.
Identities and putative functions of open reading frames (ORFs) in the pilA region of Pseudomonas syringae pv. tabaci 6605.
| ORF | Size (amino acids) | Putative function of homologues | Organisms | NCBI protein accession number | Amino acids (%) |
|---|---|---|---|---|---|
| pilA | 131 | Type IV pilin | Psav 3335 | ZP_07003592.1 | 97 |
| Pa 1244 | CAE18458 | 28 | |||
| Pa 5196 | AAM52059 | 29 | |||
| tfpO | 476 | Glycosyltransferase | Psav 3335 | ZP_07003593 | 86 |
| Pa 1244 | CAA58769 | 37 | |||
| Pa 5196 | AAM52060 | 19 | |||
| Nm MC58 | AAF41024 | 17 | |||
| Psb | 428 | Polysaccharide biosynthesis protein | Xac 306 | NP_640412.1 | 41 |
| Aaa | YP_004233424.1 | 34 |
Abbreviations for organisms: Aaa, Acidovorax avenae ssp. avenae ATCC19860; Nm, Neisseria meningitidis; Pa, Pseudomonas aeruginosa; Psav, Pseudomonas savastanoi pv. savastanoi; Xac, Xanthomonas axonopodis pv. citri. NCBI, National Center for Biotechnology Information.
Influence of tfpO mutation on pilin structure
The amino acid sequence of PilA Pta 6605 has been reported previously (Taguchi and Ichinose, 2011). According to Strom et al. (1993), a short leader sequence of a pilin precursor was cleaved, and the mature PilA was produced by the methylation of N‐terminal phenylalanine by a bifunctional PilD protein with prepilin peptidase and N‐methyltransferase. The molecular mass of the PilA mature protein was predicted to be 12 329 Da. We detected PilA as an approximately 13‐kDa protein in the whole‐cell lysate of Pta 6605 by immunoblot analysis with anti‐PilA‐specific antibody (Fig. 2A). A glycostaining experiment of partially purified pilin protein from the Pta 6605 WT strain showed a positive signal, indicating that PilA is glycosylated (Fig. 2B).
Figure 2.
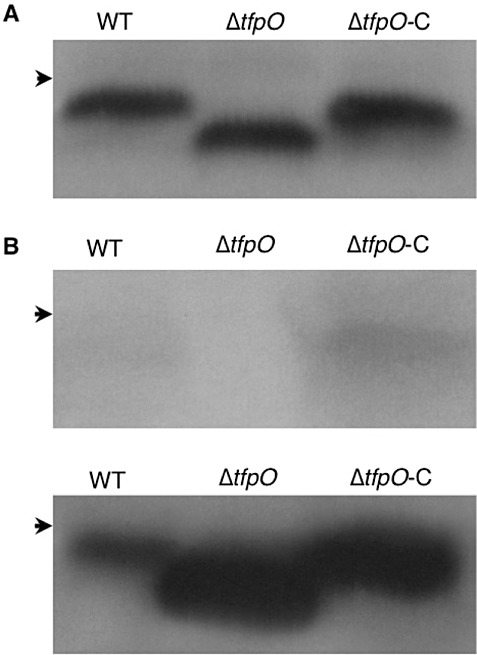
Detection of the type IV pilin (PilA) protein from the wild‐type (WT), its ΔtfpO mutant and the complemented strain of Pseudomonas syringae pv. tabaci 6605. (A) Immunoblot analysis of the PilA protein in whole‐cell lysates with anti‐PilA‐specific antibody. (B) Staining of glycoproteins (top panel) and immunoblot analysis (bottom panel) of 1 µg of partially purified PilA protein from bacterial strains. Arrowheads indicate the positions at 15 kDa.
To examine the potential role of TfpO in pilin glycosylation, the tfpO‐defective mutant and its complemented strain in Pta 6605 were generated, and designated as ΔtfpO and ΔtfpO‐C, respectively. As expected, the ΔtfpO mutant pilin migrated more rapidly than that of the WT strain on sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE), whereas that of the ΔtfpO‐C strain showed similar mobility to the WT pilin (Fig. 2A). In addition, the glycostaining experiment indicated that PilA prepared from the ΔtfpO mutant was not stained as a glycoprotein, whereas a positive band was again detected in PilA of the ΔtfpO‐C strain (Fig. 2B). Therefore, it is evident that pilin is glycosylated and that tfpO is required for pilin glycosylation in Pta 6605.
Involvement of flagellin glycosylation‐related genes in pilin glycosylation
Flagellin and T4P are two well‐known examples of bacterial glycoproteins, and we have identified three genes for flagellin glycosyltransferases (fgt1, fgt2 and vioT) and several genes, including vioA and vioB, that are required for the biosynthesis of the precursor of the flagellin glycan, the D‐Quip4N(3‐hydroxy‐1‐oxobutyl)2Me residue, in Pta 6605 (Ichinose et al., 2011; Nguyen et al., 2009; Taguchi et al., 2006; Yamamoto et al., 2011). To investigate the potential involvement of these genes in pilin glycosylation, we carried out SDS‐PAGE and Western blot analyses using the whole‐cell lysates from WT, ΔtfpO, Δfgt1, Δfgt2, ΔvioT, ΔvioA and ΔvioB strains in Pta 6605 (Fig. S1, see Supporting Information). All T4P, except that from ΔtfpO, showed the same mobility in SDS‐PAGE analysis, indicating that fgt1, fgt2, vioT, vioA and vioB are not involved in pilin glycosylation.
Surface motility and adhesion ability of the ΔtfpO mutant
To determine the role of pilin glycan on bacterial behaviour, the surface swimming and swarming motilities of the mutant were examined and compared with those of the WT and nonpiliated ΔpilA‐defective mutant strains (Fig. 3). Although no surface swimming motility was observed at 2 days after inoculation in the nonpiliated ΔpilA and piliated ΔtfpO mutant strains, that of each mutant strain was partially recovered at 5 days after inoculation. However, the surface swarming motility of ΔpilA and also the ΔtfpO mutant strain was lost even at 5 days after inoculation. Meanwhile, both surface swimming and swarming were partially restored in the ΔtfpO‐C strain after 5 days of incubation. We have observed that the ΔpilA mutant retains the WT level of swimming motility in liquid culture medium (Taguchi and Ichinose, 2011). Under microscopic observation, the ΔtfpO mutant still exhibited swimming ability in liquid medium, similar to that of the WT strain (data not shown). These results suggest that tfpO is required for the surface motility of Pta 6605.
Figure 3.
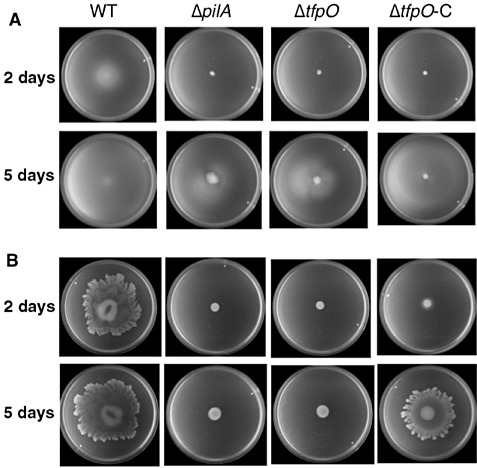
Motility test. Surface swimming motility [MMMF (see Experimental procedures) with 0.25% agar] at 25 °C (A) and surface swarming motility [SWM (see Experimental procedures) with 0.45% agar] at 27 °C (B) of the wild‐type (WT), ΔpilA and ΔtfpO mutants and ΔtfpO complemented strain. The photographs show representative results obtained from three independent experiments after incubation for 2 and 5 days.
We also analysed the ability of each strain to form a biofilm. As observed previously, the ΔpilA mutant showed a reduction in biofilm formation (Taguchi and Ichinose, 2011). However, both the ΔtfpO and ΔtfpO‐C strains showed similar levels of biofilm formation to the WT strain (Fig. 4), and pilin glycan did not seem to be required for biofilm formation.
Figure 4.
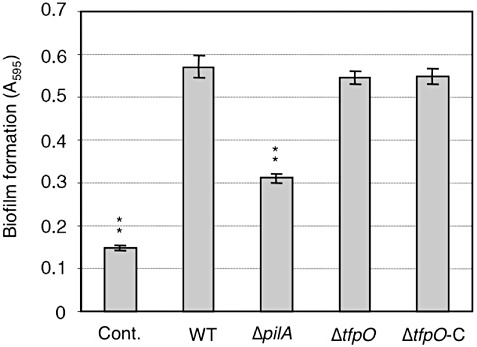
Biofilm formation by the wild‐type (WT), ΔpilA and ΔtfpO mutants and the ΔtfpO complemented strain. Each bacterium was grown in Luria–Bertani (LB) broth medium with 10 mm MgCl2, transferred to fresh MMMF medium (see Experimental procedures) and incubated for 48 h at 25 °C without agitation. Averages obtained from three independent experiments are shown. Asterisks indicate a significant difference from the WT in a t‐test (**P < 0.005).
Virulence of the ΔtfpO mutant
To investigate the effects of ΔtfpO mutation on the virulence towards host tobacco leaves, dip and infiltration inoculations were carried out. Although there were no significant differences in disease symptoms in plants inoculated with the WT, ΔpilA, ΔtfpO and ΔtfpO‐C strains by the infiltration inoculation method (Fig. 5A), the disease symptoms of ΔpilA‐ and ΔtfpO‐inoculated tobacco leaves by the dip inoculation method were reduced significantly, whereas those of the complemented strain were partially restored to the WT level (Fig. 5B). The populations of the WT strain, the ΔpilA and ΔtfpO mutant strains and the ΔtfpO complemented strain were examined at 1, 3 and 6 days after dip inoculation. As shown in Fig. 5C, the ΔpilA and ΔtfpO mutant strains and the ΔtfpO‐C strain grew less than the WT strain. In the case of the ΔtfpO‐C strain, the population was increased and the difference between the WT and ΔtfpO‐C strain was reduced by 6 days after inoculation. This observation is consistent with the late restoration of the ΔtfpO‐C strain in the swarming motility test. The results obtained from both experiments suggest that pilin glycan is important for surface motility and virulence.
Figure 5.
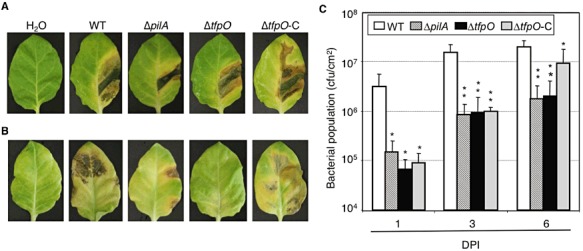
Virulence assay on host tobacco plants. Tobacco leaves were inoculated with each bacterium by the infiltration method at 2 × 105 colony‐forming units (cfu)/mL (A) and by the dip inoculation method at 2 × 108 cfu/mL (B), and were photographed at 9 and 12 days post‐inoculation (DPI), respectively. (C) Bacterial populations were measured at 1, 3 and 6 DPI by the dip inoculation method using three tobacco leaf discs (8 mm in diameter). The bars represent standard deviations for at least three independent experiments. Asterisks indicate a significant difference from the wild‐type (WT) in a t‐test (*P < 0.05, **P < 0.005).
The genetic organization of pilA and its surrounding region in P. syringae
The genomic regions around pilA in P. syringae pathovars were compared. The DNA sequence, location and direction of pilB (e.g. PSPPH0820 in Pph 1448A) and orf (e.g. PSPPH0825 in Pph 1448A) are well conserved in P. syringae pathovars. However, the genomic regions between pilB and orf were quite variable (Fig. 6). The tfpO orthologues in Pta 6605 were also found in P. syringae pv. mori str. 301020, pv. actinidiae str. M302091, pv. lachrymans str. M301315, pv. syringae 642, pv. maculicola str. ES4326 and Psav NCPPB 3335. Their deduced amino acid sequences showed homology to TfpOPta6605 with 99%, 78%, 79%, 88%, 84% and 86% identities, respectively. Pph 1448A possesses only the C‐terminal half as an incomplete fusion gene with the ISPsy18 transposase gene. The tfpO gene was always located upstream from the putative polysaccharide biosynthetic gene, psb. However, many other strains, such as pv. syringae B728a, pv. syringae FF5, pv. glycinea (Pgl) race 4, pv. tabaci ATCC 11528, pv. tomato DC3000 and T1, and pv. oryzae 1_6, do not possess the tfpO or psb genes. Pathovar syringae B728a and pv. aesculi str. NCPPB3681 only possess the pilA gene between pilB and orf1 (orthologue of PSPPH0825), and other tfpO‐nonpossessing strains possess ORFs for glutathione S‐transferase, transcription factor and unknown proteins. Each pathovar in P. syringae has been classified into five major clades by comprehensive genome sequencing analyses (Studholme, 2011). However, the distribution of the tfpO and psb genes is independent of clades and pathovars (Fig. 6).
Figure 6.
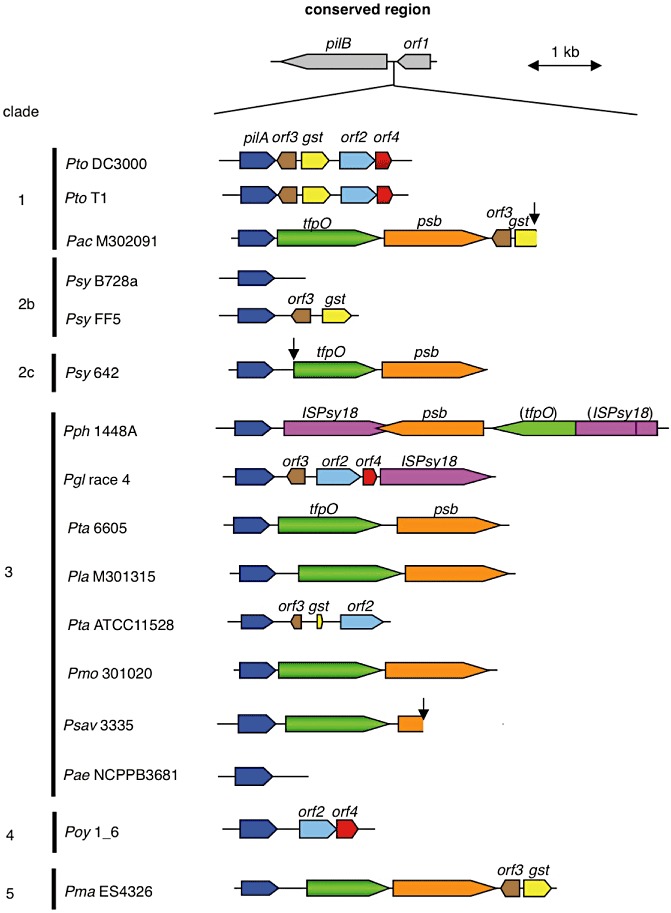
Schematic organization of pilA and its surrounding region in Pseudomonas syringae pathovars. The pilA clusters are flanked by conserved genes encoding for type IV pilus biogenesis protein pilB and orf1. The genes having the same hypothetical function are shown in matching colours: pilA (blue), tfpO (green), psb (orange), gst (for glutathione S‐transferase, yellow), orf2 (light blue), orf3 (brown), orf4 (red) and ISPsy18 (for ISPsy18 transposase, violet). The clusters are compared among P. syringae pathovars/strains whose genome sequences have been analysed so far. The abbreviations of each bacterial name and those of each Entrez Genome Project number (or RefSeq accession number where there is no Genome Project entry) are as follows: P. syringae pv. tomato DC3000 (Pto DC3000, AE016853), pv. tomato T1 (Pto T1, RefSeq accession NZ_ABSM00000000), pv. actinidiae M302091 (Pac M302091, AEAL00000000), pv. syringae B728a (Psy B728a, CP000075), pv. syringae FF5 (Psy FF5, ACXZ00000000), pv. syringae 642 (Psy 642, ADGB00000000), pv. phaseolicola 1448A (Pph 1448A, CP000058), pv. glycinea race 4 (Pgl race4, AB683862), pv. tabaci 6605 (Pta 6605, AB571112), pv. lachrymans M301315 (Pla M301315, AEAF00000000), pv. tabaci ATCC 11528 (Pta ATCC11528, ACHU00000000), pv. mori 301020 (Pmo 301020, AEAG00000000), P. savastanoi pv. savastanoi NCPPB 3335 (Psav 3335, ADMI00000000), pv. aesculi NCPPB3681 (Pae NCPPB3681, RefSeq accession NZ_ACXS01000159), pv. oryzae 1_6 (Poy 1_6, ABZR00000000) and pv. maculicola ES4326 (Pma ES4326, AEAK00000000). The clades of P. syringae species were classified by the high‐throughput genome sequencing approach as reported by Studholme (2011). The arrows indicate the truncated sequence contigs.
We also analysed the DNA sequences of this pilA‐containing region in several isolates of pv. tabaci, and found that isolates 7375 and MD340a possess tfpO and show the same genomic organization as Pta 6605, whereas isolates 6823 and 113R do not possess tfpO and show the same genomic organization as Pta ATCC11528 (data not shown). The deduced amino acid sequences of TfpO from pv. tabaci isolates 7375 and MD340a are 100% identical to that from the isolate 6605. Interestingly, the deduced amino acid sequences of PilA protein from tfpO‐possessing strains in pv. tabaci are identical to each other, and those from tfpO‐nonpossessing strains are also identical. However, PilA from tfpO‐possessing strains in pv. tabaci showed only 41% identity to those from tfpO‐nonpossessing strains. Thus, as expected, only PilA proteins from tfpO‐possessing strains were detected with the same mobility by anti‐Pta 6605 pilin‐specific antibody (Taguchi and Ichinose, 2011), but those from tfpO‐nonpossessing strains were not (Fig. S2, see Supporting Information). This result also suggests that the pilin proteins from strains 7375 and MD340a are glycosylated. The amino acid sequences of PilA from all P. syringae pathovars/strains investigated were aligned, and a phylogenetic tree was generated using the neighbour‐joining (NJ) method (Saitou and Nei, 1987) (Fig. 7). We classified all PilA proteins into groups I, II, IIIa and IIIb. All PilA proteins in groups I, II and IIIa belong to tfpO‐possessing strains, whereas all other PilA proteins in group IIIb belong to tfpO‐nonpossessing strains, indicating that PilA proteins are at least partially divided into tfpO‐possessing and tfpO‐nonpossessing strains.
Figure 7.
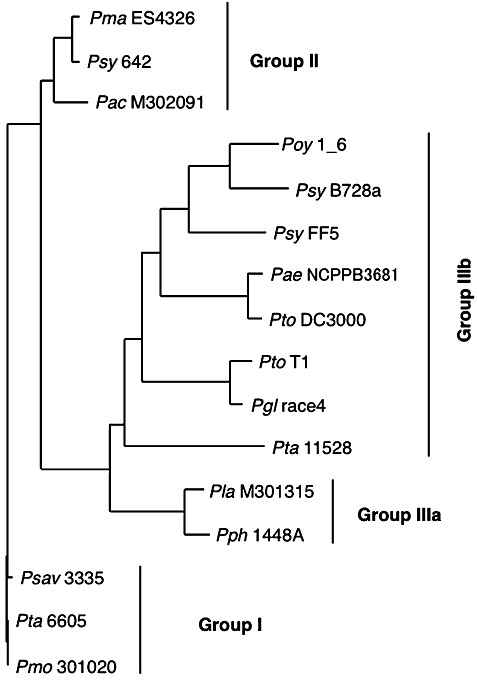
Phylogenetic tree comparing the type IV pilin (PilA) protein sequences of Pseudomonas syringae pathovars. A phylogenetic tree was constructed from a multiple sequence alignment with the neighbour‐joining method using GENETYX Ver. 15 software. The abbreviations of each bacterial strain are indicated in the legend of Fig. 6.
DISCUSSION
Function of TfpO, a pilin glycosyltransferase
In this study, we introduced a mutation into tfpO, a gene encoding the putative pilin glycosyltransferase in Pta 6605. Because the PilA protein from the WT strain, but not that from the ΔtfpO mutant, was positively detected by glycostaining, and because the molecular mass of the PilA protein from the ΔtfpO mutant was lower than that from the WT strain, TfpO was thought to be a pilin glycosyltransferase. It is known that the pilin of the Pa 1244 strain is glycosylated with an oligosaccharide that is structurally identical to the O‐antigen repeating unit in LPS in this bacterium (Castric et al., 2001). It is also known that TfpOPa1244 catalyses the attachment of an O‐antigen repeating unit to the β‐carbon of the pilin C‐terminal residue, Ser (Comer et al., 2002). Thus, the glycan in the pilin protein originates in the same metabolic pathway as O‐antigen biosynthesis, because the mutation of the genes involved in the initial steps of O‐antigen biosynthesis (either wbpM or wbpL) abolishes pilin glycosylation (DiGiandomenico et al., 2002). Because LPS is normally synthesized in the Pta 6605 ΔtfpO mutant strain (data not shown), TfpO is not involved in O‐antigen biosynthesis in Pta 6605. However, the substrate specificity of TfpOPa1244 is extremely low (DiGiandomenico et al., 2002). The crucial requirement for TfpOPa1244 activity is the presence of a Ser residue at the C‐terminus in pilin as a glycosylated site, and the pilin common sequence is not required for glycosylation (Qutyan et al., 2010). In contrast with TfpOPa1244, both TfpW from Pa 5196 and PglO/PglL from Neisseria species are able to glycosylate the internal Ser/threonine of the pilin protein (Kus et al., 2008; Parge et al., 1995; Stimson et al., 1995). Furthermore, the group IV PilA protein of Pa 5196 is more similar to the PilE pilin protein of Neisseria species than to those of other P. aeruginosa groups (Kus et al., 2004). TfpOPta6605 showed greater identity to TfpOPa1244 (37% over 441 residues) than TfpWPa5196 (19% over 225 residues) or PglL/pglO (17% over 388 residues). Therefore, the function of TfpOPta6605 may be more similar to that of TfpOPa1244 than to that of TfpWPa5196 or PglO/PglL.
Position of the glycosylation site and structure of pilin glycan
We revealed that the mutation of tfpO resulted in a reduction in the molecular mass of PilA protein by Western blot analysis in Pta 6605. Liquid chromatography‐electrospray ionization mass analysis revealed that the molecular masses of four trypsin‐digested peptide fragments in pilin protein (F1‐R30, A31‐K44, L45‐R81 and T82‐R89; the numbers indicate the positions from the N‐terminus of the putative mature PilA protein) had the same values as those calculated from amino acid sequences (data not shown; F1‐R30 is calculated as the peptide with a methyl group at the N‐terminal phenylalanine), suggesting that these four peptides do not possess glycan. However, the remaining peptide L90‐S125 was not found in the analysis. The glycosylated amino acid residue(s) should be clarified by mass spectrometric analysis of dissected PilA peptides and of the mutated PilA protein with Ser/alanine substitution.
Although the structure of the pilin glycan has not been elucidated as yet, the molecular mass of PilA was not affected in the Δfgt1, Δfgt2, ΔvioT, ΔvioA and ΔvioB mutant strains (Fig. S1). Because fgt1, fgt2, vioT, vioA and vioB are required for flagellin glycosylation by rhamnose and viosamine‐related sugars in Pta 6605 (Ichinose et al., 2011; Nguyen et al., 2009; Taguchi et al., 2006), pilin glycan may not contain these sugars, and the pilin glycosylation system may be quite different from the flagellin glycosylation system.
Furthermore, Pa 1244 is known to modify its pilin with N‐hydroxylbutyryl‐N‐formyl‐Pse‐xylose‐N‐acetylfucosamine, which has antigenic similarity to LPS (Castric et al., 2001). In a related study, the O‐antigen of several P. syringae pv. tabaci distinct strains was found to be composed of three rhamnose residues and one N‐acetylglucosamine/fucosamine residue (Zdorovenko et al., 1997). It should be elucidated whether or not the pilin glycan of Pta 6605 has the same structure as the O‐antigen repeating unit.
Function of pilin glycan
Although we have clarified previously, by dip inoculation assay, that T4P of Pta 6605 is required for surface motility, biofilm formation and virulence on its host tobacco plant (Taguchi and Ichinose, 2011), the role of pilin glycosylation is still unclear. The glycosylation of Neisseria pilins at Ser63 facilitates the solubilization of pilin monomer, but does not play a role in pilus‐mediated adhesion (Marceau et al., 1998; Marceau and Nassif, 1999). In addition, analysis of the pilin allele distribution among isolates of P. aeruginosa suggests that group I pilin, which can be post‐translationally glycosylated via the action of TfpO, may confer a colonization or persistence advantage in a cystic fibrosis host (Kus et al., 2004). The ΔtfpO mutant in Pa 1244, a representative strain of group I, has also been reported to reduce T4P‐mediated twitching motility relative to the WT strain, but forms a normal biofilm (Smedley et al., 2005). In our study, the ΔtfpO mutant in Pta 6605 also lost all of its T4P‐mediated surface swarming motility and part of its virulence, but did not lose biofilm formation ability. Although the T4P‐mediated surface motility is different in Pa 1244 and Pta 6605, pilin glycan is required for these surface motilities, and its function may be similar in both bacteria.
T4P is required for twitching motility and biofilm formation in P. aeruginosa (Chiang and Burrows, 2003), and T4P is required for swarming motility and biofilm formation in Pta 6605 (Taguchi and Ichinose, 2011). Bacterial twitching motility occurs by extension, tethering and retraction of polar T4P (Mattick, 2002). Although swarming motility is different from twitching motility, the former in Pta 6605 also seems to require all the extension, tethering and retraction activities. Because the ΔtfpO mutant strain retains its biofilm formation activity, it may also retain the activities for extension and tethering and lose its retraction activity.
In P. aeruginosa, the T4P group I strains in which pilin is glycosylated are most prevalent in patients with cystic fibrosis (Kus et al., 2004), and the pilin glycan in the Pa 1244 strain may promote lung colonization (Smedley et al., 2005). Similarly, a lack of surface motility results in a reduction in virulence of the ΔtfpO mutant of Pta 6605. Overall, these data suggest that the pilus of Pta 6605 aids bacterial adherence to host tissue and that pilin glycan contributes to bacterial virulence through surface motility.
Co‐evolution of pilA and tfpO
Previously, P. aeruginosa pilin was divided into five distinct phylogenetic groups (I–V) based on its amino acid sequence and the existence of unique accessory genes immediately downstream of pilA (Kus et al., 2004). Thus, it was reported that the P. aeruginosa pilin allele is strongly associated with cystic fibrosis isolates (Kus et al., 2004). However, the relationship between the intraspecies classification of P. aeruginosa based on the pilin allele and housekeeping genes is not known. In P. syringae pathovars, intrapathovar variation has been investigated by the high‐throughput genome sequencing approach, and the pathovars of P. syringae can be classified into five major clades (Studholme, 2011). However, the genomic organization between pilB and orf1 is independent of pathovars and clades, and more variable when compared with that in P. aeruginosa. Among 16 strains in 12 pathovars, the genomic organization around pilA and its surrounding region can be divided into at least nine types (three tfpO‐possessing types and six ftpO‐nonpossessing types), and there are many short orfs here (Fig. 6). Some orfs may encode hypothetical proteins, and some seem to be part of the disrupted gene. The existence of tfpO/psb genes is also independent of pathovars and clades, and tfpO/psb genes are distributed among pathovars/strains of P. syringae. For example, there are both tfpO‐possessing and tfpO‐nonpossessing strains in the same pathovars of syringae and tabaci. In Pph 1448A, the truncated tfpO gene and psb gene are placed between two transposons, both of which are ISPsy18 genes, indicating that tfpO and psb were horizontally inserted by transposition. Pgl race 4 also retains IsPsy18 in this region, indicating a horizontally active site.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 2. Each strain of P. syringae pv. tabaci was maintained in King's B (KB) medium at 27 °C, and Escherichia coli strains were grown at 37 °C in Luria–Bertani (LB) medium.
Table 2.
Bacterial strains and plasmids used in this study.
| Bacterial strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Escherichia coli strain | ||
| DH5a | F–λ–ø80dLacZΔM15 Δ(lacZYA‐argF)U169 recA1 endA1 hsdR17 (rK–mK+) supE44 thi‐1gyrA relA1 | Takara, Kyoto, Japan |
| S17‐1 | thi pro hsdR–hsdM + recA [chr::RP4‐2‐Tc::Mu‐Km::Tn7] | Schäfer et al. (1994) |
| S17‐1 λ‐pir | λ pir lysogen of S17 (Tpr Smr thi pro hsdR‐M+recA RP::2‐Tc::Mu‐Km::Tn7 | Simon et al. (1983) |
| Pseudomonas syringae pv. tabaci | ||
| Isolate 6605 | Wild‐type, Nalr | Taguchi et al. (2001) |
| 6605‐ΔpilA | Isolate 6605 ΔpilA, Nalr | Taguchi and Ichinose (2011) |
| 6605‐ΔtfpO | Isolate 6605 ΔtfpO, Nalr | This study |
| 6605‐ΔtfpO‐C | Isolate 6605 ΔtfpO complemented, Nalr (pBSLtfpO) | This study |
| Isolate 6823 | Wild‐type | Taguchi et al. (2001) |
| Isolate 7375 | Wild‐type | Taguchi et al. (2001) |
| Isolate MD340a | Wild‐type | Gift from Dr J. Lydon |
| Isolate 113R | Wild‐type | Gift from Dr J. Lydon |
| Isolate ATCC 11528 | Wild‐type | Rico and Preston (2008) |
| Plasmid | ||
| pCR‐Blunt II‐TOPO | Cloning vector, Kmr | Invitrogen |
| pTOPO‐pilA | 5533‐bp insert containing pilA surrounding, Kmr | This study |
| pGEM‐T easy | Cloning vector, Ampr | Promega, Madison, WI, USA |
| pGEM‐tfpO | 3704‐bp insert containing pilA and tfpO, Ampr | This study |
| pGEM‐ΔtfpO | 3204‐bp insert containing pilA and 500‐bp interval deleted tfpO, Ampr | This study |
| pGEM‐tfpO‐C | 2339‐bp insert containing pilA and tfpO, Ampr | This study |
| pK18mobsacB | Small mobilizable vector, Kmr, sucrose‐sensitive (sacB) | Schäfer et al. (1994) |
| pK18‐ΔtfpO | 3204‐bp insert through EcoRI site containing pilA and 500‐bp interval deleted tfpO, Kmr | This study |
| pBSL118 | Mini‐Tn5 derived plasmid vector for insertion mutagenesis, Ampr, Kmr | Alexeyev et al. (1995) |
| pBSLtfpO‐C | 2339‐bp insert containing pilA and tfpO, Ampr, Kmr | This study |
Ampr, ampicillin resistance; Kmr, kanamycin resistance; Nalr, nalidixic acid resistance.
Plant materials and inoculation experiments
Tobacco plants (Nicotiana tabacum L. cv. Xanthi NC) were grown at 26 °C with a 16‐h photoperiod, and leaves of 2–3‐month‐old plants were used for inoculation experiments. For the dip inoculation experiments, bacterial strains cultured overnight in LB broth medium supplemented with 10 mm MgCl2 were harvested after centrifugation, and then suspended in 10 mm MgSO4 and 0.02% Silwet L77 (OSI Specialties, Danbury, CT, USA) at a density of 2 × 108 colony‐forming units (cfu)/mL. The leaves were detached at the petiole (three leaves for each strain) and dipped in the bacterial suspension for 20 min. The leaves were then incubated in 85% humidity in a growth cabinet at 23 °C with an 18‐h photoperiod and water supply. To examine the bacterial growth, five leaf discs (8 mm in diameter) were dip inoculated in the same bacterial suspension for 20 min. For the infiltration experiment, leaves (three leaves for each strain) were infiltrated with bacterial suspension in 10 mm MgSO4 without Silwet at a density of 2 × 105 cfu/mL. The leaves were incubated in the same way as in the dip inoculation. We then examined the virulence in terms of symptom severity and bacterial growth. To measure the bacterial populations, dip‐inoculated leaf discs were soaked in 15% H2O2 for 1 min to sterilize the leaf surfaces, and then washed with sterile distilled water. Then, three dip‐inoculated leaf discs were ground with a mortar and pestle. The homogenates in 10 mm MgSO4 were plated on KB plates after serial dilutions. After 48 h of incubation at 27 °C, the colonies were counted and the bacterial populations were calculated.
DNA cloning and the generation of mutant and complemented strains
To investigate the genomic information on the pilA region in Pta strains, two primers, pilA‐F (5′‐GTTGACTTCTTCAAGGCTGG‐3′) and pilA‐R (5′‐CCCGTGGTACTTCAAATTCG‐3′), were designed on the basis of two conserved genes, pilB (PSPPH_0820) and orf1 (PSPPH_0825), in the registered sequences of Pph 1448A (accession number CP000058). The resulting PCR products were cloned into pCR‐Blunt II‐TOPO plasmid, and DNA sequencing was performed using an ABI PRISM 3130xl (Applied Biosystems, Chiba, Japan) and a BigDye Terminator Cycle Sequencing Kit.
To generate the ΔtfpO mutant strain, a pair of primers (5′‐GTCAGCGTCACTCTGAGTAG‐3′ for tfpO‐F and 5′‐AACGCACCAATGGCAAAAGC‐3′ for tfpO‐R) was used to amplify the DNA region containing tfpO with PCR, and the DNA product was subsequently isolated by the pGEM‐T‐Easy vector cloning system. Digestion was performed with BsmI to delete an internal 500‐bp region for the generation of the mutant plasmid. The internally deleted DNA fragment was excised with EcoRI digestion and introduced into an EcoRI site of the pK18mobsacB plasmid (Schäfer et al., 1994). Deletion mutant strains were obtained by conjugation and homologous recombination according to the methods reported previously (Taguchi et al., 2006).
For complementation of the ΔtfpO mutant, the primers 5′‐CAGACATATAACCACCAGAC‐3′ for C‐tfpO‐F and 5′‐TGTAACCAGCGTAATTGCGC‐3′ for C‐tfpO‐R were used to amplify the predicted promoter region and the entire pilA and tfpO ORFs. The amplified DNA fragment was inserted into the EcoRI sites of pBSL118, a transposon vector (Alexeyev et al., 1995), to generate pBSLtfpO‐C, and introduced into the ΔtfpO mutant by conjugation using the E. coli S17‐l λpir strain to generate ΔtfpO‐C. All sequences of recombinant DNA were confirmed by an ABI PRISM 3130xl using a BigDye Terminator Cycle Sequencing Kit.
The nucleotide sequences of the DNA fragments for the region from pilB to orf1 for Pta 6605, MD340a, 7375, 113R, 6823 and Pgl race 4 have been deposited under the respective accession numbers AB571112, AB677536, AB677537, AB677538, AB677539 and AB683862 in the DDBJ, EMBL and GenBank nucleotide sequence databases.
Motility assay
Bacteria cultured overnight in LB medium containing 10 mm MgCl2 at 25 °C were resuspended in 10 mm MgSO4 and adjusted to an optical density at 600 nm (OD600) of 0.1. For the assessment of surface motilities on semi‐solid agar medium, 3‐µL aliquots were inoculated in the centre of 0.25% agar MMMF plates [50 mm potassium phosphate buffer, 7.6 mm (NH4)2SO4, 1.7 mm MgCl2 and 1.7 mm NaCl, pH 5.7, supplemented with 10 mm each of mannitol and fructose] for the swimming assay and of 0.45% agar SWM plates (0.5% peptone, 0.3% yeast extract) for the swarming assay, as described in Taguchi et al. (2006). The motility was examined using three plates for each strain, and was observed 2 and 5 days after incubation at 25 °C for the swimming assay and at 27 °C for the swarming assay. The swimming motility in liquid MMMF medium was observed using a phase‐contrast microscope.
Biofilm formation
Each bacterial strain was grown in LB broth medium containing 10 mm MgCl2 overnight, and the concentration was adjusted to OD600= 0.1 with 3 mL of fresh MMMF medium in a polystyrene tube (three tubes for each strain). After 48 h of incubation at 25 °C without agitation, adherent bacteria were stained with 0.5% crystal violet for 1 h, and loosely bound bacteria were removed by washing with distilled H2O three times. For quantitative analysis of the biofilms, crystal violet was extracted from stained cells by 3 mL of 95% ethanol, and OD595 values were measured (Taguchi et al., 2006).
Preparation of the pilus‐containing surface fraction
Pili were partially purified as described previously (Taguchi and Ichinose, 2011) with a minor modification. Each bacterium was grown on 1.5% agar KB plates for 48 h at 27 °C. Cells from 20 plates were gently scraped off the agar surface and resuspended in 200 mL of phosphate buffer (50 mm sodium phosphate, pH 6.8). Pili were sheared off from the cells by stirring for 30 min, and cells were removed by centrifugation at 10 000 g for 20 min. After filtration of the supernatant through a 0.45‐µm pore filter, 30% polyethylene glycol (PEG8000) and 5 m NaCl were added to final concentrations of 3% and 0.5 m, respectively. The mixture was incubated at 4 °C overnight to allow the precipitation of proteins. Pilus‐containing fractions were obtained as pellets by centrifugation at 100 000 g for 30 min, and were resolved in water. The dissolved pilin fraction was loaded through Detoxi‐Gel Endotoxin Removing Gel (Thermo Scientific, Yokohama, Japan) to avoid the contamination of LPS and exopolysaccharide, and was then re‐extracted after the appropriate band of pilin had been cut from 15% SDS‐PAGE.
Western blot analysis and detection of glycoproteins
Bacterial whole‐cell lysate was prepared from bacterial cells harvested from an overnight culture in KB broth medium with shaking at 27 °C. PilA proteins in bacterial whole‐cell lysates or pilus‐containing surface fractions were separated on a 15% SDS‐PAGE gel. After transfer to a poly(vinylidene difluoride) (PVDF) membrane (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, UK), the proteins were subjected to Western blot analysis using a rabbit polyclonal anti‐PilAPta6605‐specific peptide antibody with a goat anti‐rabbit secondary antibody conjugated with alkaline phosphatase (Bio‐Rad Laboratories, Hercules, CA, USA) and an alkaline phosphatase‐based chemiluminescent detection system (CDP‐Star Reagent, New England Biolabs, Ipswich, MA, USA), as described previously (Taguchi and Ichinose, 2011). The anti‐PilAPta6605‐specific antibody was generated with a synthetic peptide (VAENYSNGSAQADVC).
The partially purified pilin proteins from Pta 6605 were separated by SDS‐PAGE, and then subjected to staining with a GelCode® Glycoprotein Staining Kit (Pierce, Rockford, IL, USA), which specifically stains glycoproteins as magenta bands on SDS‐PAGE gel.
Statistical analysis
The results of the biofilm formation assay and the bacterial populations in host tobacco leaves are expressed as means with standard deviations (SDs). The two‐tailed t‐test was performed for comparisons between the quantitative measurement of the WT, each mutant and the complemented strains. Values of P < 0.05 were considered to be statistically significant.
Supporting information
Fig. S1 Detection of type IV pilin (PilA) protein. Anti‐pilAPta6605‐specific antibody was used to detect PilA protein in the whole‐cell lysates from wild‐type (WT), Δfgt1, Δfgt2, ΔvioT, ΔvioA and ΔvioB strains in Pseudomonas syringae pv. tabaci. Arrowheads indicate the positions at about 15 kDa.
Fig. S2 Detection of type IV pilin (PilA) protein. Anti‐pilAPta6605‐specific antibody was used to detect PilA protein in the whole‐cell lysates from several isolates of Pseudomonas syringae pv. tabaci. Arrowheads indicate the positions at about 15 kDa.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We would like to thank the Leaf Tobacco Research Laboratory of Japan Tobacco Inc.; Dr John Lydon (Sustainable Agricultural Systems Laboratory, US Department of Agriculture); and Dr Gail Preston (Department of Plant Sciences, University of Oxford, UK) for providing Pta strains 6605, 6823 and 7375; 113R and MD340a; and ATCC11528, respectively. This work was supported in part by the Program for Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN).
REFERENCES
- Aas, F.E. , Vik, A. , Vedde, J. , Koomey, M. and Egge‐Jacobsen, W. (2007) Neisseria gonorrhoeae O‐linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol. Microbiol. 65, 607–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyev, M.F. , Shokolenko, I.N. and Croughan, T.P. (1995) New mini‐Tn5 derivatives for insertion mutagenesis and genetic engineering in gram‐negative bacteria. Can. J. Microbiol. 41, 1053–1055. [DOI] [PubMed] [Google Scholar]
- Bahar, O. , Goffer, T. and Burdman, S. (2009) Type IV pili are required for virulence, twitching motility, and biofilm formation of Acidovorax avenae subsp. citrulli . Mol. Plant–Microbe Interact. 22, 909–920. [DOI] [PubMed] [Google Scholar]
- Benz, I. and Schmidt, M.A. (2002) Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45, 267–276. [DOI] [PubMed] [Google Scholar]
- Burdman, S. , Bahar, O. , Parker, J.K. and Fuente, L. (2011) Involvement of type IV pili in pathogenicity of plant pathogenic bacteria. Genes 2, 706–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric, P. (1995) pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology, 141, 1247–1254. [DOI] [PubMed] [Google Scholar]
- Castric, P. , Cassels, F.J. and Carlson, R.W. (2001) Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 276, 26 479–26 485; correction: 38, 36058. [DOI] [PubMed] [Google Scholar]
- Chiang, P. and Burrows, L.L. (2003) Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa . J. Bacteriol. 185, 2374–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer, J.E. , Marshall, M.A. , Blanch, V.J. , Deal, C.D. and Castric, P. (2002) Identification of the Pseudomonas aeruginosa 1244 pilin glycosylation site. Infect. Immun. 70, 2837–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiandomenico, A. , Matewish, M.J. , Bisaillon, A. , Stehle, J.R. , Lam, J.S. and Castric, P. (2002) Glycosylation of Pseudomonas aeruginosa 1244 pilin: specificity of glycan substrate. Mol. Microbiol. 46, 519–530. [DOI] [PubMed] [Google Scholar]
- Harvey, H. , Kus, J.V. , Tessier, L. , Kelly, J. and Burrows, L.L. (2011) Pseudomonas aeruginosad‐arabinofuranose biosynthetic pathway and its role in type IV pilus assembly. J. Biol. Chem. 286, 28 128–28 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose, Y. , Shimizu, R. , Ikeda, Y. , Taguchi, F. , Marutani, M. , Mukaihara, T. , Inagaki, Y. , Toyoda, K. and Shiraishi, T. (2003) Need for flagella for complete virulence of Pseudomonas syringae pv. tabaci: genetic analysis with flagella‐defective mutants ΔfliC and ΔfliD in host tobacco plants. J. Gen. Plant Pathol. 69, 244–249. [Google Scholar]
- Ichinose, Y. , Taguchi, F. , Nguyen, L.C. , Naito, K. , Suzuki, T. , Inagaki, Y. , Toyoda, K. and Shiraishi, T. (2011) Glycosylation of bacterial flagellins and its role in motility and virulence In: Genome‐Enabled Analysis of Plant–Pathogen Interactions (Wolpert T., Shiraishi T., Collmer A., Glazebrook J. and Akimitsu K., eds), pp. 215–224. St. Paul, MN: APS Press. [Google Scholar]
- Kus, J.V. , Tullis, E. , Cvitkovitch, D.G. and Burrows, L.L. (2004) Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non‐CF patients. Microbiology, 150, 1315–1326. [DOI] [PubMed] [Google Scholar]
- Kus, J.V. , Kelly, J. , Tessier, L. , Harvey, H. , Cvitkovitch, D.G. and Burrows, L.L. (2008) Modification of Pseudomonas aeruginosa Pa5196 type IV pilins at multiple sites with D‐Araf by a novel GT‐C family arabinosyltransferase, TfpW. J. Bacteriol. 190, 7464–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan, S.M. (2006) Flagellar glycosylation—a new component of the motility repertoire? Microbiology, 152, 1249–1262. [DOI] [PubMed] [Google Scholar]
- Marceau, M. and Nassif, X. (1999) Role of glycosylation at Ser63 in production of soluble pilin in pathogenic Neisseria . J. Bacteriol. 181, 656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau, M. , Forest, K. , Beretti, J.L. , Tainer, J. and Nassif, X. (1998) Consequences of the loss of O‐linked glycosylation of meningococcal type IV pilin on piliation and pilus‐mediated adhesion. Mol. Microbiol. 27, 705–715. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. (2002) Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314. [DOI] [PubMed] [Google Scholar]
- Nguyen, L.C. , Yamamoto, M. , Ohnishi‐Kameyama, M. , Andi, S. , Taguchi, F. , Iwaki, M. , Yoshida, M. , Ishii, T. , Konishi, T. , Tsunemi, K. and Ichinose, Y. (2009) Genetic analysis of the genes involved in the synthesis of modified 4‐amino‐4,6‐dideoxyglucose in flagellin of Pseudomonas syringae pv. tabaci . Mol. Genet. Genomics, 282, 595–605. [DOI] [PubMed] [Google Scholar]
- Nudleman, E. and Kaiser, D. (2004) Pulling together with type IV pili. J. Mol. Microbiol. Biotechnol. 7, 52–62. [DOI] [PubMed] [Google Scholar]
- Parge, H.E. , Forest, K.T. , Hickey, M.J. , Christensen, D.E. , Getzoff, E.D. and Tainer, J.A. (1995) Structure of the fibre‐forming protein pilin at 2.6 A resolution. Nature, 378, 32–38. [DOI] [PubMed] [Google Scholar]
- Power, P.M. and Jennings, M.P. (2003) The genetics of glycosylation in Gram‐negative bacteria. FEMS Microbiol. Lett. 218, 211–222. [DOI] [PubMed] [Google Scholar]
- Power, P.M. , Seib, K.L. and Jennings, M.P. (2006) Pilin glycosylation in Neisseria meningitidis occurs by a similar pathway to wzy‐dependent O‐antigen biosynthesis in Escherichia coli . Biochem. Biophys. Res. Commun. 347, 904–908. [DOI] [PubMed] [Google Scholar]
- Qutyan, M. , Henkel, M. , Horzempa, J. , Quinn, M. and Castric, P. (2010) Glycosylation of pilin and nonpilin protein constructs by Pseudomonas aeruginosa 1244. J. Bacteriol. 192, 5972–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico, A. and Preston, G.M. (2008) Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast‐induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol. Plant–Microbe Interact. 21, 269–282. [DOI] [PubMed] [Google Scholar]
- Roine, E. , Raineri, D.M. , Romantschuk, M. , Wilson, M. and Nunn, D.N. (1998) Characterization of type IV pilus genes in Pseudomonas syringae pv. tomato DC3000. Mol. Plant–Microbe Interact. 11, 1048–1056. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. (1987) The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Schäfer, A. , Tauch, A. , Jager, W. , Kalinowski, J. , Thierbach, G. and Puhler, A. (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene, 145, 69–73. [DOI] [PubMed] [Google Scholar]
- Simon, R. , Priefer, U. and Pühler, A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram‐negative bacteria. Biotechnology, 1, 784–791. [Google Scholar]
- Smedley, J.G. III , Jewell, E. , Roguskie, J. , Horzempa, J. , Seyboldt, A. , Stolz, D.B. and Castric, P. (2005) Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. J. Bacteriol. 73, 7922–7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimson, E. , Virji, M. , Makepeace, K. , Dell, A. , Morris, H.R. , Payne, G. , Saunders, J.R. , Jennings, M.P. , Barker, S. and Panico, M. (1995) Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4‐diacetamido‐2,4,6‐trideoxyhexose. Mol. Microbiol. 17, 1201–1214. [DOI] [PubMed] [Google Scholar]
- Strom, M.S. and Lory, S. (1993) Structure–function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47, 565–596. [DOI] [PubMed] [Google Scholar]
- Strom, M.S. , Nunn, D.N. and Lory, S. (1993) A single bifunctional enzyme, PilD, catalyzes cleavage and N‐methylation of proteins belonging to the type IV pilin family. Proc. Natl. Acad. Sci. USA, 90, 2404–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme, D.J. (2011) Application of high‐throughput genome sequencing to intrapathovar variation in Pseudomonas syringae . Mol. Plant Pathol. 12, 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi, F. and Ichinose, Y. (2011) Role of type IV pili in virulence of Pseudomonas syringae pv. tabaci 6605: correlation of motility, multidrug resistance, and HR‐inducing activity on a nonhost plant. Mol. Plant–Microbe Interact. 24, 1001–1011. [DOI] [PubMed] [Google Scholar]
- Taguchi, F. , Tanaka, R. , Kinoshita, S. , Ichinose, Y. , Imura, Y. , Andi, S. , Toyoda, K. , Shiraishi, T. and Yamada, T. (2001) HarpinPsta from Pseudomonas syringae pv. tabaci is defective and deficient in its expression and HR‐inducing activity. J. Gen. Plant Pathol. 67, 116–123. [Google Scholar]
- Taguchi, F. , Takeuchi, K. , Katoh, E. , Murata, K. , Suzuki, T. , Marutani, M. , Kawasaki, T. , Eguchi, M. , Katoh, S. , Kaku, H. , Yasuda, C. , Inagaki, Y. , Toyoda, K. , Shiraishi, T. and Ichinose, Y. (2006) Identification of glycosylation genes and glycosylated amino acids of flagellin in Pseudomonas syringae pv. tabaci . Cell. Microbiol. 8, 923–938. [DOI] [PubMed] [Google Scholar]
- Taguchi, F. , Shibata, S. , Suzuki, T. , Ogawa, Y. , Aizawa, S. , Takeuchi, K. and Ichinose, Y. (2008) Effects of glycosylation on swimming ability and flagella polymorphic transformation of Pseudomonas syringae pv. tabaci 6605. J. Bacteriol. 190, 764–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi, F. , Takeuchi, K. , Inagaki, Y. , Toyoda, K. , Shiraishi, T. and Ichinose, Y. (2009) Glycosylation of flagellin from Pseudomonas syringae pv. tabaci 6605 contributes to evasion of host tobacco plant surveillance system. Physiol. Mol. Plant Pathol. 74, 11–17. [Google Scholar]
- Virji, M. , Saunders, J.R. , Sims, G. , Makepeace, K. , Maskell, D. and Ferguson, D.J.P. (1993) Pilus‐facilitated adherence of Neisseria meningitidis to human epithelial and endothelial cells: modulation of adherence phenotype occurs concurrently with changes in primary amino acid sequence and the glycosylation status of pilin. Mol. Microbiol. 10, 1013–1028. [DOI] [PubMed] [Google Scholar]
- Voisin, S. , Kus, J.V. , Houliston, S. , St‐Michael, F. , Watson, D. , Cvitkovitch, D.G. , Kelly, J. , Brisson, J.R. and Burrows, L.L. (2007) Glycosylation of Pseudomonas aeruginosa strain Pa5196 type IV pilins with mycobacterium‐like α‐1,5‐linked D‐Araf oligosaccharides. J. Bacteriol. 189, 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M. , Ohnishi‐Kameyama, M. , Nguyen, L.C. , Taguchi, F. , Chiku, K. , Ishii, T. , Ono, H. , Yoshida, M. and Ichinose, Y. (2011) Identification of genes involved in the glycosylation of modified viosamine of flagellins in Pseudomonas syringae by mass spectrometry. Genes, 2, 788–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdorovenko, G.M. , Solyanik, L.P. , Yahovleva, L.M. and Paramonov, N.A. (1997) Characterization of O‐antigens from different strains of Pseudomonas syringae pv. tabaci . Biochemistry (Moscow), 62, 28–37. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Detection of type IV pilin (PilA) protein. Anti‐pilAPta6605‐specific antibody was used to detect PilA protein in the whole‐cell lysates from wild‐type (WT), Δfgt1, Δfgt2, ΔvioT, ΔvioA and ΔvioB strains in Pseudomonas syringae pv. tabaci. Arrowheads indicate the positions at about 15 kDa.
Fig. S2 Detection of type IV pilin (PilA) protein. Anti‐pilAPta6605‐specific antibody was used to detect PilA protein in the whole‐cell lysates from several isolates of Pseudomonas syringae pv. tabaci. Arrowheads indicate the positions at about 15 kDa.
Supporting info item
Supporting info item


