Summary
Recently, Pepino mosaic virus (PepMV) infections causing severe yellowing symptoms in tomato plants have been reported in glasshouse tomato crops. When studying this phenomenon in commercial glasshouses, two different types of yellowing symptoms, occurring in adjacent plants, were distinguished: interveinal leaf yellowing and yellow mosaics. After several weeks, the interveinal leaf yellowing symptoms gradually disappeared and the plant heads became green again, with yellow mosaic patterns on the leaves as an intermediate stage. The sequencing of multiple isolates causing interveinal leaf yellowing identified two point mutations, occurring in positions 155 and 166 of the coat protein (CP), as unique to the yellowing pathotype. Site‐directed mutagenesis of infectious clones confirmed that both CP mutations are determinants of the interveinal leaf yellowing symptoms. Sequencing of CP clones from plants or plant parts with the yellow mosaic symptoms resulted in a mixture of wild‐type and mutated sequences, whereas sequencing of CP clones from the green heads of recovered plants resulted in only wild‐type sequences. Yellow mosaic symptoms could be reproduced by inoculation of an artificial 1:1 mixture of RNA transcripts from the wild‐type and mutated infectious clones. These results show that the ratio of mutated versus wild‐type sequences can determine the nature and severity of symptom development. The gradual recovery of the plants, which coincides with the disappearance of the yellowing mutations, suggests that selection pressure acts to the advantage of the wild‐type virus. Experiments with wild‐type and mutated infectious clones showed that reverse mutation events from mutant to wild‐type occur and that the wild‐type virus does not have a replicative advantage over the mutant. These results suggest that reverse mutation events occur, with subsequent selection pressure acting in favour of the wild‐type virus in the growing plant parts, possibly related to a lower long‐distance movement efficiency of the mutant.
Introduction
Pepino mosaic virus (PepMV) is a Potexvirus (family Alphaflexiviridae) which infects tomato (Solanum lycopersicum) crops worldwide. The virus is very efficiently transmitted mechanically and was first detected in Peru in pepino plants (Solanum muricatum) displaying yellow leaf mosaics (Jones et al., 1980). In 1999, PepMV was observed for the first time in glasshouse tomato plants in the Netherlands (van der Vlugt et al., 2000) and, since then, it has spread rapidly to all economically important glasshouse tomato‐growing areas worldwide (Hanssen and Thomma, 2010; Werkman and Sansford, 2010). The host range is mainly restricted to plant species from the Solanaceae family, with tomato as the only crop for which economic damage has been reported. Eggplant (S. melongena) and potato (S. tuberosum) can be infected experimentally without displaying symptoms (Martin and Mousserion, 2002; Werkman and Sansford, 2010). The PepMV genome consists of a single RNA strand of approximately 6.4 kb, containing five open reading frames with a replicase gene comprising methyltransferase, nucleoside triphosphate (NTP)‐binding and polymerase motifs, a triple gene block (TGB) gene encoding three proteins, TGBp1, TGBp2 and TGBp3, involved in viral movement, and a coat protein (CP) gene.
PepMV symptomatology in glasshouse tomato crops is highly variable and has therefore been studied extensively. The virus affects the ripening process of tomatoes, leading to fruit discoloration symptoms, such as marbling, blotchy ripening and flaming (Hanssen et al., 2011). Symptoms on the vegetative plant parts comprise nettleheads, leaf bubbling, mild chlorosis, small yellow spots on the leaves and, in some cases, even leaf mosaics and leaf or stem necrosis (Hanssen et al., 2009; Hanssen and Thomma, 2010; Hasiów‐Jaroszewska et al., 2009b, 2011; Roggero et al., 2001; van der Vlugt et al., 2000).
Four different genotypes or strains of PepMV can be distinguished on the basis of sequence comparisons: (i) the Peruvian genotype, first isolated from S. muricatum and generally referred to as ‘LP’ because the reference sequence LP2001 was isolated from Lycopersicum peruvianum (now called S. peruvianum); (ii) the European (tomato) genotype ‘EU’, which was the first PepMV strain detected in glasshouse tomato crops in Europe; (iii) the Chilean strain ‘CH2’, which was first identified by Ling (2007) on a seed lot from Chile and is now dominating the PepMV population in Europe; and (iv) the American strain US1, which was isolated from a seed lot by Maroon‐Lango et al. (2005) and, since then, has only been reported in a tomato crop in the Canary Islands (Alfaro‐Fernández et al., 2008) and in mixed infection in the USA (Ling, 2008).
Apart from the fact that the LP strain is asymptomatic in tomato, no clear correlation could be found between symptom severity (aggressiveness) and PepMV strain. However, mixed infections of two strains, more specifically EU and CH2, or LP and CH2, have been shown to cause more damage to the crop (Hanssen et al., 2008, 2009; Hanssen and Thomma, 2010; Pagan et al., 2006). The high variability in symptom severity in PepMV‐infected tomato crops can be attributed in part to factors such as plant stage at the time of infection, light intensity, temperature fluctuations, nutrition and the tomato cultivar. In addition, minor genetic differences between isolates can result in large differences in isolate aggressiveness (severity of symptoms) and symptomatology (nature of symptoms) (Hanssen et al., 2009; Hasiów‐Jaroszewska et al., 2009b). It has been shown recently that one point mutation in the viral gene encoding TGBp3 converts a mild isolate to a necrotic one (Hasiów‐Jaroszewska et al., 2011).
Recently, new virus‐like symptoms, consisting of severe interveinal leaf yellowing or bright yellow leaf mosaics, often covering the entire foliage and coinciding with serious fruit deformations, have been noticed in commercial tomato production in several European countries (Poland, Belgium, France and the Netherlands). In this work, PepMV (CH2 strain) was identified as the causal agent of these symptoms and the isolates were characterized by genotyping, phenotyping, site‐directed mutagenesis on infectious clones and epidemiological studies. Our results show that the yellowing pathotype is caused by point mutations in the CP gene. The ratio of mutated versus wild‐type PepMV sequences within the plant was shown to determine the severity and nature of the yellowing symptoms. In addition, the yellowing mutations appear to be unstable, as back‐mutation towards the wild‐type form was found to occur in both natural infections and in experiments with infectious clones.
Results
Occurrence, description and association of PepMV with the yellowing phenotypes
In 2010–2012, several PepMV infections in glasshouse tomato crops in Poland, Belgium, the Netherlands and France were reported to coincide with severe yellowing symptoms. Two different phenotypes, often coinciding in the same plant row, were distinguished: (i) interveinal leaf yellowing with dark‐green vein‐banding symptoms covering the entire foliage (hereafter referred to as interveinal leaf yellowing or IY; Fig. 1A); and (ii) yellow mosaic patterns consisting of irregular, bright yellow areas on the leaves (hereafter referred to as yellow mosaic or YM; Fig. 1B). This YM pattern is clearly distinct from the typical small yellow spots because of the much larger size of the yellow areas and the general occurrence all over the plant. Fruits produced from the IY plants were nonmarketable because of severe deformation and discoloration. The distribution pattern of the yellowing symptoms in the commercial tomato glasshouses suggested mechanical transmission of the virus through crop handling practices, as affected plants mainly occurred in the same plant rows, where YM and IY plants were adjacent, and, in addition, plants with only mild nettlehead (NH) symptoms occurred in the same rows. A Belgian glasshouse (code 22) with one row of adjacent YM, IY and NH plants was used as a case study. Leaf samples were collected from plants with these three different phenotypes (sample codes B‐2212‐IY, B‐2212‐YM and B‐2212‐NH) (Fig. 2A,B). In the same period, samples of plants with very similar yellowing symptoms (IY type) were collected in Poland, and designated P‐1‐IY, P‐3‐IY and P‐5‐IY. The inoculation of these different samples on tomato test plants in climate chambers showed that all three symptom types were distinct. Only filamentous particles about 500–530 nm in length, typical for potexviruses, were observed by electron microscopy. Reverse transcription‐polymerase chain reaction (RT‐PCR) analyses for pospiviroids were negative. PepMV analyses using a strain‐specific TaqMan RT‐quantitative (q)PCR assay confirmed the presence of PepMV in all samples. A high concentration of the CH2 strain was found, whereas specific quantitative RT‐PCR tests for the EU, LP and US1 strains were negative. Therefore, the symptoms were assumed to be caused by a PepMV mutant. The Polish isolates P‐1‐IY, P‐3‐IY and P‐5‐IY, together with the Belgian isolate B‐2212‐IY, were used as reference isolates to study the yellowing pathotypes. Subsequently, seven additional yellowing isolates collected from Belgian (isolates B‐0111‐IY; B‐1812‐IY; B‐1012‐IY), French (isolate FR‐1911‐IY) and Dutch (isolates NL‐NS12‐IY; NL‐2411‐IY; NL‐9412‐IY) tomato glasshouses were included in the study to verify the results obtained with the four reference isolates. The Polish PepMV CH2 isolate P‐22, which is asymptomatic in tomato, was used as a reference. All the isolates used in this study are listed in Table 1.
Figure 1.

Yellowing symptoms (IY, interveinal leaf yellowing; YM, yellow mosaics) observed in glasshouse conditions. (A) IY symptoms, variety Admiro; (B) YM symptoms, variety Admiro.
Figure 2.

Yellowing symptoms. (A) Interveinal leaf yellowing (IY) symptoms after inoculation of isolate B‐2212‐IY on tomato plants (variety Tricia). (B) Yellow mosaic (YM) symptoms after inoculation of isolate B‐2212‐YM on tomato plants (variety Tricia). (C) IY symptoms after inoculation of original isolate P‐5‐IY on tomato plants (variety Moneymaker). (D) IY symptoms after inoculation of RNA transcripts of infectious clone P‐5‐IY flc (variety Moneymaker). (E) IY symptoms after inoculation of RNA transcripts of mutated infectious clone P‐22flc‐CPE155K (variety Moneymaker). (F) YM symptoms after inoculation of a 1:1 ratio of RNA transcripts from P‐22flc and P‐5‐IY flc infectious clones (variety Moneymaker).
Table 1.
List of Pepino mosaic virus (PepMV) isolates used in this study
| Sample code* | Country of origin | Year of origin | Phenotype | Codon 155 | AA | Codon 166 | AA |
|---|---|---|---|---|---|---|---|
| P‐1‐IY | Poland | 2010 | IY | AAA | K | GAT | D |
| P‐3‐IY | Poland | 2010 | IY | AAA | K | GAT | D |
| P‐5‐IY | Poland | 2010 | IY | AAA | K | GAT | D |
| P‐22 | Poland | 2005 | No symptoms | GAA | E | GAT | D |
| B‐2212‐IY | Belgium | 2012 | IY | AAA | K | GAT | D |
| B‐2212‐YM | Belgium | 2012 | YM |
AAA GAA |
K E |
GAT | D |
| B‐2212‐NH | Belgium | 2012 | Mild NH | GAA | E | GAT | D |
| B‐2212‐IY‐GH | Belgium | 2012 | GH from recovered IY plant | GAA | E | GAT | D |
| B‐2212‐IY‐YM | Belgium | 2012 | Middle YM from ‘recovered’ IY plant |
AAA GAA |
K E |
GAT | D |
| B‐0111‐IY | Belgium | 2011 | IY | AAA | K | GAT | D |
| B‐1812‐IY | Belgium | 2012 | IY | AAA | K | GAT | D |
| B‐1012‐IY | Belgium | 2012 | IY | GAA | E | GGT | G |
| NL‐NS12‐IY | Netherlands | 2012 | IY | GAA | E | GGT | G |
| NL‐NS12‐IY‐GH | Netherlands | 2012 | GH from recovered IY plant | GAA | E | GAT | D |
| NL‐NS12‐IY‐YM | Netherlands | 2012 | Middle YM leaves from recovered IY plant | GAA | E |
GAT GGT |
D G |
| NL‐NS12‐IY‐IY | Netherlands | 2012 | Older, IY leaves from recovered IY plant | GAA | E | GGT | G |
| NL‐2411‐IY | Netherlands | 2011 | IY | GAA | E | GGT | G |
| NL‐9412‐IY | Netherlands | 2012 | IY | GAA | E | GGT | G |
| FR‐1911‐IY | France | 2011 | IY | GAA | E | GGT | G |
*Sample codes consist of: (i) a letter code which refers to the country of origin (B, Belgium; FR, France; NL, the Netherlands); (ii) a unique number code for the glasshouse; (iii) the year of isolation (not included for the Polish isolates); and (iv) a symptom code: IY, interveinal leaf yellowing; YM, severe yellow mosaics; NH, nettlehead; GH, green head.
AA, amino acids; D, aspartic acid; E, glutamic acid; G, glycine; K, lysine.
Identification of unique CP mutation that correlates with the yellowing phenotype
Full‐length clones from Polish yellowing isolates P‐1‐IY, P‐3‐IY and P‐5‐IY were constructed and the complete genomes were sequenced. Sequence comparison of these three clones with other nonyellowing PepMV reference identified several point mutations between the yellowing and the reference isolates. However, only one point mutation, located at position 155 of CP, was shared by all three yellowing isolates and not by the other isolates. This codon consists of GAA in the non yellowing isolates, but AAA in the three yellowing clones, resulting in an amino acid substitution from glutamic acid to the highly basic lysine. Subsequently, nine CP clones of the Belgian reference yellowing isolate B‐2212‐IY were sequenced and the same mutation was found in all clones. Comparison with all currently available PepMV sequences in GenBank revealed that this mutation has never been reported previously, and thus is unique to the yellowing phenotype. This mutation is further referred to as the CP155 mutation.
Site‐directed mutagenesis confirms that the CP155 mutation causes IY symptoms
To confirm the hypothesis that the CP155 mutation is responsible for the IY phenotype, site‐directed mutagenesis was performed on full‐length infectious clones of P‐5‐IY and P‐22. The infectivity of the full‐length clone (flc) P‐22flc (asymptomatic isolate) has been documented in previous studies (Hasiów‐Jaroszewska et al., 2011). The full‐length clone P‐5‐IYflc was shown to be infectious on Nicotiana benthamiana plants, whereas clones P‐1‐IYflc and P‐3‐IYflc were not. Therefore, only P‐22flc (asymptomatic) and P‐5‐IYflc (yellowing) were used in further studies. No differences in host range or symptomatology were observed between the IY isolate P‐5‐IY and the full‐length clone P‐5‐IYflc derived therefrom (Fig. 2C,D). The CP155 mutation was introduced into P‐22flc, in which GAA in codon 155 was replaced by AAA, resulting in an amino acid substitution from glutamic acid to lysine (mutant clone designated P‐22flc‐CPE155K). In addition, the reverse mutant P‐5flc‐CPK155E was constructed starting from P‐5‐IYflc. Here, AAA in codon 155 was replaced by GAA, resulting in an amino acid substitution from lysine to glutamic acid. To verify whether other amino acid substitutions at position 155, with similar or different chemical properties (alkaline–acid–neutral), would have an impact on the observed symptoms, mutants of P‐5‐IYflc and P‐22flc encoding arginine, aspartic acid and alanine were also constructed. All the constructed mutant clones are listed in Table 2.
Table 2.
Site‐directed mutagenesis: overview of the constructed and inoculated mutants
| Mutant name | Primers 5′–3′ | Codon From–to | AA change | Symptoms on tomato* |
|---|---|---|---|---|
| P‐22flc‐CPE155K | TTGGGCTATCAGAAAGATACCAAG | 155 | E–K | IY |
| TTTTGCCCAGTTGGCAGGTGGCAC | GAA–AAA | |||
| P‐22flc‐CPE155R | TTGGGCTATCAGAGAGATACCAAG | 155 | E–R | Yellow dots |
| TTTTGCCCAGTTGGCAGGTGGCAC | GAA–AGA | |||
| P‐22flc‐CPE155A | TTGGGCTATCAGGCAGATACCAAGT | 155 | E–A | None |
| TTTTGCCCAGTTGGCAGGTGGCAC | GAA–GCA | |||
| P‐22flc‐CPE155D | GGGCTATCAGGATGATACCAAGT | 155 | E–D | None |
| AATTTTGCCCAGTTGGCAGGTGGC | GAA–GAT | |||
| P‐22flcCPD166G | TGACTTCTTTGGTGGAGTCACAAA | 166 | D–G | IY |
| AGAGCAGCAAACTTGGTATCTTCC | GAT–GGT | |||
| P‐5flc‐CPK155E | TTGGGCTATCAGGAAGATACCAAG | 155 | K–E | None |
| TTTTGCCCAGTTGGCAGGTGGCAC | AAA–GAA | |||
| P‐5flc‐CPK155R | TTGGGCTATCAGAGAGATACCAAG | 155 | K–R | Yellow dots |
| TTTTGCCCAGTTGGCAGGTGGCAC | AAA–AGA | |||
| P‐5flc‐CPK155A | TTGGGCTATCAGGCAGATACCAAGT | 155 | K–A | None |
| TTTTGCCCAGTTGGCAGGTGGCAC | AAA–GCA | |||
| P‐22flc‐CPS8A | ACCTACAGCTGCTAACCCCATC | 8 | S–A | None |
| TGGTTTTCCATAGTTGAGTTA | TCT–GCT | |||
| P‐22flc‐CPV43I | GATTTGAAGAAGGTCAAATACGTG | 43 | I–V | None |
| ACTTAGGGAAGGAGCTGTATTAG | ATC–GTC | |||
| P‐5flc‐CPS9N | CTACAGCTTCTAACCCATCAGATGC | 9 | S–N | None |
| GTTGGTTTTCCATAGTTGAGTTAA | AGC–AAC |
The italic letters indicate the codons that were introduced.
AA, amino acid.
*IY, interveinal leaf yellowing.
All mutant clones obtained by site‐directed mutagenesis were infectious as confirmed by quantitative RT‐PCR in the inoculated plants. As expected, the substitution from glutamic acid to lysine at position 155 of CP (P‐22flc‐CPE155K) resulted in the development of IY symptoms on tomato varieties Moneymaker (Table 2; Fig. 2E), Beta Lux and Malinowy Ożarowski (photographs not shown). Similarly, the reverse mutation in P‐5flc‐CPK155E resulted in a change from the yellowing pathotype to a mild one. The substitution from glutamic acid to arginine, an amino acid from the same group as lysine, at position 155 of CP resulted in yellow spots on the leaves. The substitution from glutamic acid to aspartic acid at position 155 of CP (P‐22flc‐CPE155D) resulted in mild symptoms without yellowing. These results show that the point mutation from glutamic acid to lysine in codon 155 of CP is a determinant for the IY phenotype.
CP sequence of other yellowing isolates confirms the relevance of the CP155 mutation and identifies a second yellowing mutation at position 166
To verify whether the point mutation CP155 is the only determinant of the PepMV IY phenotype, CP genes of the seven additional yellowing isolates from Belgium, the Netherlands and France were cloned and sequenced.
Unexpectedly, from these seven additional IY isolates, only two contained the identified CP155 mutation, whereas the other five shared another mutation with codon 166 consisting of GGT rather than GAT in all other PepMV isolates [GenBank and PepMV collections at Scientia Terrae Research Institute (STRI), Sint‐Katelijne‐Waver, Belgium and Institute of Plant Protection‐NRI (IPP‐NRI), Poznan, Poland] (Table 1). This point mutation results in an amino acid substitution from aspartic acid in the nonyellowing reference isolates to glycine in the five yellowing isolates. These results suggest that there are two different point mutations in CP that can lead to the yellowing phenotype. To verify the role of the CP166 mutation, the mutation was introduced into P‐22flc, resulting in the mutant clone P‐22flc‐CPD166G (Table 2). Similar to P‐22flc‐CPE155K, inoculation with RNA transcripts derived from P‐22flc‐CPD166G resulted in clear IY symptoms in tomato plants.
Based on alignments of yellowing with reference isolates, three other point mutations (in codons 8, 9 and 43 of the CP gene), which did not show consistent correlation with the yellowing pathotypes, were chosen for site‐directed mutagenesis to verify whether other substitutions in CP may similarly affect symptomatology. All these mutant clones were infectious, but the mutations did not influence symptomatology (Table 2).
Mixtures of mutated and wild‐type virus variants modulate PepMV symptomatology
In the Belgian case study glasshouse, samples from the two distinct but related yellowing phenotypes IY and YM, occurring adjacently in the same plant row, were collected. Cloning and sequencing of the CP gene revealed that the yellowing mutation CP155 was present in all nine clones obtained from sample B‐2212‐IY. However, in the sample B‐2212‐YM, taken from plants with YM but no IY symptoms, only two of six CP clones contained this mutation. In addition, the mutation was absent in all three CP clones obtained from sample B‐2212‐NH (plant in the same row, with only mild nettlehead symptoms but no yellowing). These results suggest that the YM phenotype is an intermediate phenotype caused by a mixture of wild‐type and mutated viruses within the viral population within one plant. To further confirm this hypothesis, clone libraries were constructed from these three samples. From each sample, 30 CP clones were obtained and sequenced. As expected, all 30 clones obtained from sample B‐2212‐IY and none of the 30 clones obtained from sample B‐2212‐NH contained the CP155 mutation. However, 14 of the 30 clones obtained from the YM sample B‐2212‐YM contained the CP155 mutation, whereas the other 16 clones showed the wild‐type sequence. These results suggest that the ratio of mutated versus wild‐type CP PepMV variants modulates symptomatology. To further confirm this hypothesis, an artificial mixture of mutated and nonmutated RNA transcripts was inoculated onto test plants. RNA was transcribed from P‐5‐IYflc, P‐22flc, P‐22flc‐CPE155K and P5flc‐CPK155E. The RNA transcripts were mixed as follows and used for the inoculation of tomato test plants: P‐5‐IYflc/P‐22flc, P‐22flc/P‐22flcCPE155K and P‐5‐IYflc/P5flc‐CPK155E in a 1:1 ratio. As expected, all the inoculated plants displayed YM but no IY symptoms after 2 weeks (Fig. 2F).
Reduced fitness of yellowing mutants?
Observations in commercial glasshouses are indicative of a loss of fitness associated with the yellowing mutation, as the yellowing symptoms gradually disappear. After several weeks, the heads of the plants turn green again, usually with the YM phenotype as an intermediate. As we have shown that the YM phenotype is caused by a mixture of wild‐type and mutated sequences, the intermediate stage suggests a selection pressure to the advantage of the wild‐type virus in the plants affected by the yellowing mutant, or a higher fitness of the wild‐type virus relative to the yellowing mutant. Alternatively, the disappearance of the symptoms might be related to the recovery of the plant rather than to a change in the viral population. To further study this phenomenon, affected plants were sampled during the recovery process. A recovered plant with IY symptoms on the lower leaves, YM symptoms on the middle leaves and normal green leaves in the head was sampled in the Belgian case study glasshouse (code 22). CP clones were prepared from a sample from the green head (B‐2212‐IY‐GH) and from the intermediate leaves displaying YM symptoms (B‐2212‐IY‐YM). None of the three CP clones sequenced from B‐2212‐IY‐GH contained the CP155 mutation, whereas two of four CP clones obtained from B‐2212‐IY‐YM contained the CP155 mutation. Similar results were obtained for the CP166 mutant, as observations in the Dutch glasshouse with code NS (sample NL‐NS12‐IY) also revealed recovery of the affected plants. Here, three different samples were taken from the different parts of a recovered IY plant: (i) the yellow, older leaves (NL‐NS12‐IY‐IY); (ii) the middle leaves with YM symptoms (NL‐NS12‐IY‐YM); and (iii) the young, green leaves from the head of the plant (NL‐NS12‐IY‐GH). Four CP clones were sequenced from each sample. All sequences obtained from the lower leaf sample NL‐NS12‐IY‐IY contained the CP166 mutation, two of four sequences from the middle leaf sample NL‐NS12‐IY‐YM contained the mutated sequence, and none of the four sequences from the green head NL‐NS12‐IY‐GH contained the CP166 mutation. These results were further confirmed by an inoculation experiment with mixtures of mutated and nonmutated RNA transcripts. The plants inoculated with 1:1 mixtures of P‐5‐IYflc/P‐22flc, P‐22flc/P‐22flcCPE155K and P‐5‐IYflc/P5flc‐CPK155E, which initially displayed YM symptoms, turned green again in the head after 5–6 weeks. The absence of the CP155 mutant in the recovered plant heads was confirmed by cloning and sequencing of the CP gene (10 clones from each treatment, 30 clones in total). A TaqMan RT‐qPCR experiment was set up to check whether the yellowing mutation results in a reduced replicative fitness. Therefore, RNA transcripts from infectious clones differing only in the CP155 mutation, more specifically P‐5‐IYflc, P‐5flc‐CPK155E, P‐22flc and P‐22flc‐CPE155K, were inoculated onto N. benthamiana plants. Leaves of these N. benthamiana plants were then used to produce equally concentrated inocula to infect tomato plants. Viral titres in the young leaves of these tomato plants were followed by sampling and TaqMan RT‐qPCR analyses at 3, 7, 14, 22 and 28 days post‐inoculation (dpi). The Ct values obtained for the four isolates were very similar at all time points (Fig. 3A), with the lowest values of around 7–8 at 7 dpi and a gradual increase in Ct values later on, to around 15 at 28 dpi. No significant difference in accumulation speed or level could be measured between the different infectious clones. These results are in contrast with the hypothesis of a replicative advantage for the wild‐type virus. Remarkably, although tomato plants inoculated with the yellowing clones P‐5‐IYflc and P‐22flc‐CPE155K initially displayed clear IY symptoms, at around 40 dpi the heads of these plants started to turn green again (Fig. 3B,C). Based on the results presented in the former paragraphs and the fact that infectious clones containing the CP155 mutation were used in this experiment, these findings suggest the occurrence of reverse mutation events towards the wild‐type, and subsequent selection pressure to the advantage of the wild‐type. A confirmation experiment to verify the occurrence of back‐mutation events was performed in tomato plants inoculated with the P‐5flc clone containing the CP155 mutation. After inoculation, the plants were grown for a 2‐month period and sampled at 6 and 8 weeks. Initially, clear IY symptoms developed, but, after 5–6 weeks, recovery was seen in the heads of the plants. In total, 40 CP clones derived from the green heads were sequenced and analysed for the presence of the CP155 mutation. As expected, the CP155 mutation was absent in all clones.
Figure 3.
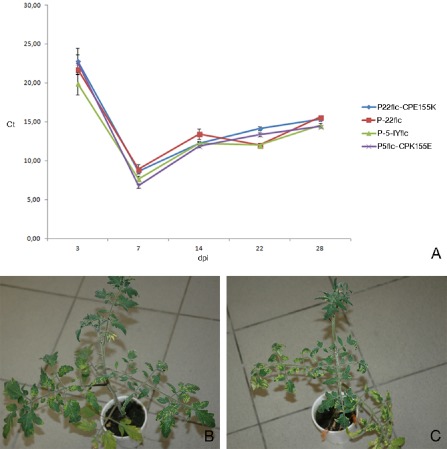
Study of replicative fitness. (A) Graph showing the Ct values (y axis) obtained using the CH2‐specific TaqMan reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) assay in tomato plants inoculated with P‐5‐IY flc P‐5flc‐CPK155E, P‐22flc and P‐22flc‐CPE155K and sampled at 3, 7, 14, 22 and 28 days post‐inoculation (dpi; x axis). Each data point represents a mean value of 10 samples from 10 different plants, with error bars representing the standard errors. (B) Tomato plant (Tricia) inoculated with P‐5‐IY flc, displaying yellowing symptoms on the lower leaves and recovery in the head at 40 dpi. (C) Tomato plant (Tricia) inoculated with P‐22flc‐CPE155K, displaying yellowing symptoms on the lower leaves and recovery in the head at 40 dpi.
Yellowing mutations change CP surface properties
To study the impact of the CP155 and CP166 mutations on the CP structure, several protein modelling methods (de novo and homology modelling) were tested and evaluated on the sequence of the Polish necrotic isolate P‐19. The models obtained were evaluated and ranked as ‘fairly and very good models’ by the ProQ method (LGscore for P‐19‐2,25, P‐22‐2,34 and P‐5‐IY‐2,45). MetaMQAPII assessment of the generated models is shown in Fig. 4. The purpose of the modelling procedure was to evaluate the effect of CPE155K and CPD166G mutations on the tertiary CP structure or surface properties. The analyses revealed that amino acids 155 and 166 are located on the surface of the protein and in flexible loop regions (regions with no secondary structural elements) (Fig. 4). Both mutations lead to changes in acidity and electric charge, which may affect the physical–chemical properties and bonds between amino acids or other molecules. It is thus probable that the region of the CP containing amino acids 155 and 166 plays an important role in the interaction between the virus and certain host factors, and that the identified amino acid substitutions alter the defence responses in the host plant, resulting in yellowing symptoms.
Figure 4.
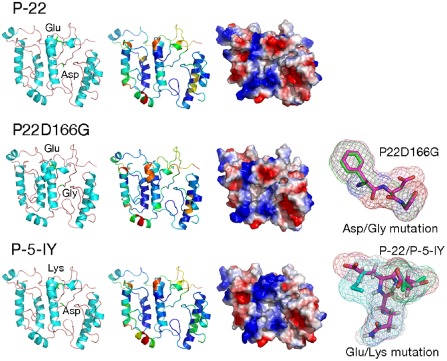
Three‐dimensional models of three isolates of CP protein—P‐5‐IY, P‐22 and P‐22‐D166G. All representations of the given proteins are shown in the same orientation and scale. All panels: the first column presents models in the ribbon representation, showing secondary structure elements with side chains of mutated amino acids; the second column shows models in the ribbon representation, presenting model assessment, coloured according to the predicted local deviation from the real structure (e.g. the predicted error of the model), as calculated by the MetaMQAP method (blue indicates low predicted deviation of Cα atoms down to 0 Å, red indicates unreliable regions with deviation >5 Å, green to orange indicates intermediate values); the third column shows models in the surface representation, coloured according to the distribution of the electrostatic surface potential calculated with ABPS (PyMol) (blue indicates positively charged regions, red indicates negatively charged regions). In addition, the last column presents superimposed three‐amino‐acid fragments with point mutations (superimposed fragments of FDG and FGG, QED and QKD amino acids).
Discussion
Successive epidemics of PepMV seem to be associated with the appearance of viral variants with different biological and genetic properties. During early PepMV epidemics in Europe, the PepMV population was shown to be rather homogeneous. The high level of genetic identity of isolates from Spain, England, France and the Netherlands suggested their common origin (Cotillon et al., 2002; Mumford and Metcalfe, 2001; van der Vlugt et al., 2000; Verhoeven et al., 2003). Since 2004, the situation has changed and, in particular, the CH2 strain has been emerging. The CH2 strain has now become dominant in Spain (Gómez et al., 2009), Belgium (Hanssen et al., 2009) and Poland (Hasiów et al., 2008), and is also emerging in North America (Ling, 2008).
It has been shown that the CH2 strain accumulates more rapidly than the EU strain in single infections (Gómez et al., 2009). The level of genetic identity between isolates representing the CH2 strain is very high, but small genetic differences account for large differences in symptom severity on tomato and other test plants of the Solanaceae family—from latent or symptomless infections, through mild nettlehead symptoms to severe necrosis. Minor nucleotide sequence differences between isolates from the same genotype have been shown to have an impact on symptom severity (Hanssen and Thomma, 2010; Hasiów‐Jaroszewska et al., 2011). The establishment of a correlation between virus genotype and disease phenotype seems to be particularly complex for PepMV, probably reflecting the numerous factors that influence the expression of symptoms. It has been shown recently that the rate of PepMV molecular evolution is, on average, 5.570 × 10−3 substitutions/site/year (Gómez et al., 2012). This value is somewhat higher than the rates reported recently for other plant RNA viruses, which may suggest that PepMV evolves more rapidly than other plant viruses.
In this work, PepMV isolates causing severe yellowing symptoms, which were recently isolated from tomato crops in several European countries, were studied. Cloning and subsequent sequence analyses resulted in the identification of two point mutations in positions 155 and 166 of the CP gene that could both, independently, be determinants of the yellowing phenotype. Both mutations result in an amino acid substitution changing the chemical properties of the protein. At position 155, a change of codon from GAA to AAA was associated with the yellowing symptoms, resulting in an amino acid substitution from glutamic acid to the highly basic lysine. At position 166, a change of codon from GAT to GGT, resulting in an amino acid substitution from aspartic acid to glycine, could be linked to the yellowing symptoms. The role of these CP mutations in IY was confirmed using infectious clones and site‐directed mutagenesis. The role of point mutations in the CP gene in the appearance of yellowing symptoms has also been demonstrated previously for Tobacco mosaic virus (Banerjee et al., 1995; Lindbeck et al., 1992) and Tomato mosaic virus (Ohnishi et al., 2009).
Modelling of the tridimensional structure of the yellowing mutants revealed that positions 155 and 166 are localized on the surface of the protein. This suggests that these positions or this region of the protein plays a role in virus infection by interaction with one or more host factors. A change in physical and chemical properties of this protein region is anticipated when glutamic acid is replaced by lysine, or when aspartic acid is replaced by glycine. These changes can affect protein–protein interactions and thus alter the chain of plant response events that take place during viral infection. CP has been shown to interact with tomato heat shock protein cognate 70 during viral infection (Matthaios et al., 2012). Previously, it has been shown that a single nucleotide substitution in TGBp3, changing K to E at position 67 of the protein, results in the development of necrotic symptoms on tomato leaves (Hasiów‐Jaroszewska and Borodynko, 2012; Hasiów‐Jaroszewska et al., 2011). Amino acid 67 is also located on the surface of TGBp3.
Two distinct yellowing phenotypes, often occurring adjacently in the same plant row in the same crop, could be discriminated. IY symptoms, which cover the entire plant, coincide with stunting and fruit deformation, whereas YM symptoms do not influence plant vigour or fruit formation. Further study of the YM phenotype revealed that the YM symptoms are caused by a mixture of mutated and nonmutated variants. In glasshouse tomato plants displaying YM symptoms, the co‐existence of viral variants with and without the CP155 mutation, or with and without the CP166 mutation, was shown. Artificial mixtures of RNA transcripts from P‐22flc/P‐5‐IYflc and P‐22flc/P‐22flc‐CPE155K also resulted in the development of YM symptoms. Interestingly, after several weeks, the plants seemed to have recovered, and no IY or YM symptoms were visible any longer in the upper (youngest) parts of the plants. The same phenomenon was observed in commercial glasshouses, as the IY and YM symptoms gradually disappeared, with the plant heads turning green again. Indeed, sequencing of multiple clones derived from the green heads of these previously yellow plants showed that the mutants had disappeared in the plants growing in commercial glasshouses, as well as in the plants inoculated with the transcript mixtures. In an additional experiment using RNA transcripts of P‐5‐IYflc as inoculum, recovery was also observed after the initial IY and subsequent YM symptoms, and absence of the CP155 mutation was demonstrated in the recovered green plant heads. These results show that reverse mutation events towards the wild‐type virus had occurred. Back‐mutation has also been reported for the type species of the Potexvirus genus. In the Potato virus X genome, a mutation converting glycine to alanine in the GKS motif of the RNA‐dependent RNA polymerase reverted to the wild‐type after passage on N. clevelandii plants (Davenport and Baulcombe, 1997). To test whether the wild‐type virus has a replicative advantage over the mutant, a TaqMan RT‐qPCR experiment using infectious clones only differing in the CP155 mutation was performed. At all four time points, equal titres were obtained from plants inoculated with the different clones, and the hypothesis of a replicative advantage of the wild‐type virus over the mutants could be eliminated. These results suggest that the recovery process is initiated by the occurrence of back‐mutation events from mutant to wild‐type, with subsequent selection pressure acting to the advantage of the wild‐type virus in the growing parts of the plants. Possibly, the mutant virus has a lower movement efficiency relative to the wild‐type, resulting in a gradual re‐installation of the wild‐type virus in the younger plant plants. The role of CP in long‐distance movement through the phloem has been studied and confirmed for many plant viruses (Hull, 2002).
The impact of CP point mutations and mutant mixtures on replication capacities and biological properties has been studied for both plant and animal viruses (Lindbeck et al., 1992; Mo et al., 2004). These and our results demonstrate how one single mutation in one viral gene, as well as the composition of mixtures of mutant and wild‐type virus variants, can greatly modify symptomatology. In this work, we have shown that the composition of PepMV mutant mixtures and the forces of selection pressure acting on them modulate symptom expression and thus play an important role in PepMV symptomatology and genetic variability.
Experimental Procedures
Identification of PepMV in field samples
Three Polish PepMV isolates causing severe IY symptoms in tomato, designated P‐1‐IY, P‐3‐IY and P‐5‐IY, were collected in commercial tomato crops by IPP‐NRI in Poznan, Poland in 2010. The code IY for yellowing was included in the names to discriminate these isolates from other PepMV isolates that do not cause yellowing. In 2011–2012, eight other PepMV isolates causing similar IY symptoms were collected from commercial tomato glasshouses in Belgium, France and the Netherlands by STRI (Table 1). The samples were analysed for other viruses by electron microscopy (National Institute of Biology, Ljubljana, Slovenia) and for viroids by a pospiviroid‐specific RT‐PCR analysis using the primers Vir1/Vir2 (Mumford et al., 2000). Inoculations on test plants were performed to verify mechanical transmission of the yellowing symptoms, using N. benthamiana and tomato plants (variety SG 46‐649, Syngenta Seeds, Enkhuizen, the Netherlands; variety Tricia, Monsanto Vegetable Seeds, Bergschenhoek, the Netherlands; Moneymaker, Beta Lux and Malinowy Ożarowski varieties, PNOS, Ożarów Mazowiecki, Poland). Inoculation was performed on seedlings after dusting the leaves with carborundum powder by rubbing a 1:4 dilution of ground leaf material in phosphate buffer. All experiments were performed in three replicates. Plants were maintained in climate chamber conditions at 23–24 °C with a 16‐h photoperiod. Symptoms were monitored until 24 dpi. Leaves with IY symptoms were collected and total RNA was isolated using an RNeasy Plant Mini Kit (Qiagen, Hilde, Germany) or Spectrum Plant Total RNA Kit (Sigma‐Aldrich, Seelze, Germany) following the manufacturers' instructions. The RNA concentrations were measured using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA). PepMV detection and strain identification were performed by RT‐PCR following Ling et al. (2007) and by TaqMan RT‐qPCR assay following Gutiérrez‐Aguirre et al. (2009).
Construction of infectious clones
The construction of infectious clones was performed following Hasiów‐Jaroszewska et al. (2009a), with some modifications. Complete viral genomes of the Polish isolates P‐1‐IY, P‐3‐IY and P‐5‐IY were amplified by PCR after reverse transcription. cDNA template was generated from 1 μL of total RNA (adjusted to 1 μg/μL) using a Transcriptor High Fidelity cDNA Synthesis Kit (Roche, Mannheim, Germany) and antisense primer polyTTTNotI 5′‐GCGGCCGCT25ATTTAGTAGATTTAGATACTAA‐3′ with a NotI restriction site (in italic), according to the manufacturer's recommendations, in a 20‐μL reaction volume. The cDNA of about 6.4 kb was amplified using Expand Long Range dNTPack (Roche) starting from 1 μL cDNA in a 50‐μL reaction volume according to the manufacturer's instructions, with the primers and reaction conditions as described previously (Hasiów‐Jaroszewska et al., 2009a). Full‐length clones were produced by ligating the PCR product into the pCR‐TOPO‐XL vector (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. For each isolate, two plasmids were sequenced. The 5′ and 3′ ends of genomic RNA were verified by sequencing with M13 F and M13 R primers. BioEdit software (Hall, 1999) was used to perform sequence editing and compilation. Sequences were aligned with reference PepMV sequences downloaded from GenBank using the ClustalW (ver. 1.4) algorithm embedded in BioEdit software. The nucleotide sequence of the full‐length clone of P‐5‐IY has been submitted to GenBank under accession number JX417070.
To verify the infectivity of the full‐length clones, the plasmids were cut with NotI (Thermo Scientific) and purified using JETQUICK PCR (Genomed, Warsaw, Poland). The purified, linear plasmid was subsequently used for transcription. Transcription was performed using 1 μg of linear plasmid as a template and mMessage mMachine T7 (Applied Biosystems Foster City, CA, USA), according to the manufacturer's instructions. The RNA obtained after transcription was measured at least twice using a NanoDrop spectrophotometer (Thermo Scientific) and inoculated mechanically onto N. benthamiana and subsequently onto tomato plants. Rub inoculation was performed with 40 μL of RNA transcripts (about 6–8 μg of RNA) after dusting the leaf surfaces with carborundum powder. The experiments were repeated twice, each time using five N. benthamiana plants and five tomato plants. Mock inoculation was performed by rubbing plants with phosphate buffer. Inoculated plants were maintained in glasshouses under the same conditions as described above. Plants were monitored for the development of symptoms up to 24 dpi. Young leaves of inoculated plants were sampled at 7 and 12 dpi, and analysed for the presence of PepMV by enzyme‐linked immunosorbent assay (ELISA) using a commercial antiserum (DSMZ, Braunschweig, Germany) and RT‐PCR (Ling et al., 2007).
Cloning and sequencing of the CP genes
To determine the CP sequences, cDNA transcripts were amplified by PCR using the PepMV‐specific primers Ker 1 nt 5379–5400 (5′‐CACCAATAAATTTAGTTTTAGC‐3′) and PepMV‐FL‐R nt 6359–6376 (5′‐TAGAAAACCCCACTCTGA‐3′) at an annealing temperature of 61 °C. PCR products were loaded onto an agarose gel, bands of the correct size (998 bp) were excised and DNA was purified using the Qiaquick Gel Extraction Kit (Qiagen) following the manufacturer's instructions. Cloning was performed using the TA cloning kit (Invitrogen) following the user's manual. For each isolate, between two and 30 clones, depending on the specific purpose of the sequence determination, were sequenced (Macrogen Inc., Seoul, South Korea). Sequences were aligned using MEGA 5.05 software (Tamura et al., 2011).
Site‐directed mutagenesis
The P‐5‐IY full‐length clone (P‐5‐IYflc) and the previously obtained full‐length clone of the asymptomatic Polish PepMV isolate P‐22 (P‐22flc) (Hasiów‐Jaroszewska et al., 2011) served as templates for site‐directed mutagenesis. An overview of the constructed mutants is provided in Table 2. To introduce point mutations into P‐22flc and P‐5‐IYflc, PCR‐based mutagenesis was carried out using the Phusion® Site‐Directed Mutagenesis Kit (Finnzymes, Wilmington, DE, USA) and the primers specified in Table 2. The reaction mixtures contained 5′ phosphorylated primers and 2–10 ng of each plasmid as a template. PCR was performed with an initial denaturation step at 98 °C for 30 s, followed by 25 cycles of: (i) denaturation at 98 °C for 10 s; (ii) annealing at 60 °C for 30 s; and (iii) extension at 68 °C for 4 min. The amplified, linear PCR product was circularized in a 5‐min ligation reaction with Quick T4 DNA Ligase (Finnzymes) and used for the transformation of One Shot TOP10 Chemically Competent Cells (Invitrogen). The plasmid DNA was isolated from individual colonies using the Pure Yield Plasmid Miniprep System (Promega, Madison, WI, USA). The resulting clones were sequenced. In vitro transcription and inoculation onto N. benthamiana and tomato plants were performed as described above. Wild‐type PepMV isolates and mock‐inoculated plants served as controls. In vitro transcription, followed by plant inoculation, was repeated at least three times.
Study of viral replicative fitness
A TaqMan RT‐qPCR experiment was set up to verify the hypothesis of the mutants having a reduced replicative fitness. Equal amounts of RNA transcripts from infectious clones differing only in the CP155 mutation, more specifically P‐5‐IYflc, P‐5flc‐CPK155E, P‐22flc and P‐22flcCPE155K, were inoculated onto N. benthamiana plants. At 14 dpi, N. benthamiana leaves from the same stage were sampled for the different inoculations and the viral concentration was measured by TaqMan RT‐qPCR. Inocula with similar Ct values (around 11) were prepared for all four isolates, using dilutions if needed. Each inoculum was used to inoculate 10 tomato plants (Tricia, Monsanto Vegetable Seeds, Bergschenhoek, the Netherlands). At 3, 7, 14, 22 and 28 dpi, the plants were sampled (always the youngest fully developed leaf) and the viral concentrations were measured with TaqMan RT‐qPCR (Gutiérrez‐Aguirre et al., 2009).
Confirmation of back‐mutation
An experiment was performed to verify the occurrence of back‐mutation from the yellowing mutants towards the wild‐type sequence. Inoculation with the yellowing infectious clone P‐5‐IYflc containing CP155 was performed in two different tomato varieties: Moneymaker and Beta Lux. P‐5‐IYflc transcripts were used to inoculate N. benthamiana plants and, subsequently, tomato plants, as described above. Three plants from each variety were inoculated and grown for a 2‐month period in quarantine conditions in a glasshouse. At 40 and 60 days after inoculation, one mixed sample per variety was taken, from which 10 CP clones were produced and sequenced. In total, 40 clones were sequenced and studied for the retention of the CP155 mutation.
Modelling of the CP tertiary structure
The CP secondary structure prediction and tertiary fold recognition (FR) were carried out via the GeneSilico metaserver gateway (http://genesilico.pl/meta2/) (Kurowski and Bujnicki, 2003). For proteins for which FR methods did not report any good template for the homology modelling procedure, de novo methods for structural modelling were applied. To model the CP tertiary structure, we used the ROSETTA algorithm for de novo modelling (Simons et al., 1997). Hundreds of thousands of decoys were generated and clustered to identify the best low‐energy conformations. The selection of models based on the average energy clusters, size, density and visual inspection of final structures was performed. Final models were evaluated using MetaMQAP II (Pawlowski et al., 2008) and ProQ server (Siew et al., 2000; Wallner and Elofsson, 2003). Quality was assessed using the ProQ method: Lgscore >1.5, fairly good model; Lgscore >2.5, very good model; Lgscore >4, extremely good model; MaxSub >0.1, fairly good model; MaxSub >0.5, very good model; MaxSub >0.8, extremely good model. Mapping of the electrostatic potential on protein surfaces was calculated with APBS (Adaptive Poisson–Boltzmann Solver) (Baker et al., 2001). Homology modelling of the proteins was performed using the ‘FRankenstein's monster’ approach (Kosinski et al., 2003). First preliminary models were built with MODELLER (Sali and Blundell, 1993) based on the sequence alignment between target and template (models obtained from de novo modelling). Models were scored by MetaMQAP and ProQ to predict their accuracy at the level of individual residues. Missing parts of proteins or those poorly scored were refined with the REFINER method (Boniecki et al., 2003).
Acknowledgements
The Polish research (IPP‐NRI, Poznan) was supported by project Iuventus IP2011 017171 (2012–2014) from the Ministry of Science and Higher Education and 2011/01/D/NZ9/00279 (2011–2014) from the National Science Centre in Poland. The Belgian researchers (STRI, Sint‐Katelijne‐Waver) are supported by IWT Vlaanderen (IWT 080501; Brussels, Belgium). We would like to thank Lieve Wittemans from ‘Proefstation voor de Groenteteelt’ and Rob Moerkens from ‘Proefcentrum Hoogstraten’ for the follow up of the yellowing symptoms in commercial glasshouses.
References
- Alfaro‐Fernández, A. , Cebrián, M.C. , Córdoba‐Sellés, C. , Herrera‐ Vásquez, J.A. and Jordá, C. (2008) First report of the US1 strain of Pepino mosaic virus in tomato in the Canary Islands, Spain. Plant Dis. 92, 11. [DOI] [PubMed] [Google Scholar]
- Baker, N.A. , Sept, D. , Joseph, S. , Holst, M.J. and McCammon, J.A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA, 98, 10 037–10 041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, N. , Wang, J.Y. and Zaitlin, M. (1995) A single nucleotide change in the coat protein gene of tobacco mosaic virus is involved in the induction of severe chlorosis. Virology, 207, 234–239. [DOI] [PubMed] [Google Scholar]
- Boniecki, M. , Rotkiewicz, P. , Skolnick, J. and Kolinski, A. (2003) Protein fragment reconstruction using various modeling techniques. J. Comput. Aided Mol. Des. 17, 725–738. [DOI] [PubMed] [Google Scholar]
- Cotillon, A.C. , Girard, M. and Ducouret, S. (2002) Complete nucleotide sequence of the genomic RNA of a French isolate of Pepino mosaic virus (PepMV). Arch. Virol. 147, 2231–2238. [DOI] [PubMed] [Google Scholar]
- Davenport, G.F. and Baulcombe, D.C. (1997) Mutation of the GKS motif of the RNA dependent RNA polymerase from potato virus X disables or eliminates virus replication. J. Gen. Virol. 78, 1247–1251. [DOI] [PubMed] [Google Scholar]
- Gómez, P. , Sempere, R.N. , Elena, S.F. and Aranda, M.A. (2009) Mixed infections of Pepino mosaic virus strains modulate the evolutionary dynamics of this emergent virus. J. Virol. 83, 12 378–12 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, P. , Sempere, R.N. , Aranda, M.A. and Elena, S.F. (2012) Phylodynamics of Pepino mosaic virus in Spain. Eur. J. Plant Pathol. 3, 445–449. [Google Scholar]
- Gutiérrez‐Aguirre, I. , Mehle, N. , Delic, D. , Gruden, K. , Mumford, R. and Ravnikar, M. (2009) Real‐time quantitative PCR based sensitive detection and genotype discrimination of Pepino mosaic virus . J. Virol. Methods, 162, 46–55. [DOI] [PubMed] [Google Scholar]
- Hall, T.A. (1999) BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Hanssen, I.M. and Thomma, B. (2010) Pepino mosaic virus: a successful pathogen that rapidly evolved from emerging to endemic in tomato crops. Mol. Plant Pathol. 11, 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen, I.M. , Paeleman, A. , Wittemans, L. , Goen, K. , Lievens, B. , Bragard, C. , Vanachter, A.C.R.C. and Thomma, B.P.H.J. (2008) Genetic characterization of Pepino mosaic virus isolates from Belgian greenhouse tomatoes reveals genetic recombination. Eur. J. Plant Pathol. 121, 131–146. [Google Scholar]
- Hanssen, I.M. , Paeleman, A. , Vandewoestijne, E. , Van Bergen, L. , Bragard, C. , Lievens, B. , Vanachter, A.C.R.C. and Thomma, B.P.H.J. (2009) Pepino mosaic virus isolates and differential symptomatology in tomato. Plant Pathol. 58, 450–460. [Google Scholar]
- Hanssen, I.M. , van Esse, H.P. , Ballester, A.‐R. , Hogewoning, S.W. , Parra, N.O. , Paeleman, A. , Lievens, B. , Bovy, A.G. and Thomma, B.P.H. (2011) Differential tomato transcriptomic responses induced by Pepino mosaic virus isolates with differential aggressiveness. Plant Physiol. 156, 301–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasiów, B. , Borodynko, N. and Pospieszny, H. (2008) Complete genomic RNA sequence of the Polish Pepino mosaic virus isolate belonging to the US2 strain. Virus Genes, 36, 209–214. [DOI] [PubMed] [Google Scholar]
- Hasiów‐Jaroszewska, B. and Borodynko, N. (2012) Characterization of the necrosis determinants of the European genotype of pepino mosaic virus by site specific mutagenesis of an infectious cDNA clone. Arch. Virol. 157, 337–341. [DOI] [PubMed] [Google Scholar]
- Hasiów‐Jaroszewska, B. , Borodynko, N. and Pospieszny, H. (2009a) Infectious RNA transcripts derived from cloned cDNA of a pepino mosaic virus isolate. Arch. Virol. 154, 853–856. [DOI] [PubMed] [Google Scholar]
- Hasiów‐Jaroszewska, B. , Pospieszny, H. and Borodynko, N. (2009b) New necrotic isolates of Pepino mosaic virus representing the CH2 genotype. J. Phytopathol. 157, 494–496. [Google Scholar]
- Hasiów‐Jaroszewska, B. , Borodynko, N. , Jackowiak, P. , Figlerowicz, M. and Pospieszny, H. (2011) Single mutation converts mild pathotype of the Pepino mosaic virus into necrotic one. Virus Res. 159, 57–61. [DOI] [PubMed] [Google Scholar]
- Hull, R. (2002) Matthews' Plant Virology, 4th edn Oxford: Elsevier Academic Press. [Google Scholar]
- Jones, R.A.C. , Koenig, R. and Lesemann, D.E. (1980) Pepino mosaic virus, a new Potexvirus from pepino (Solanum muricatum). Ann. Appl. Biol. 94, 61–68. [Google Scholar]
- Kosinski, J. , Cymerman, I. , Feder, M. , Kurowski, M.A. , Sasin, J.M. and Bujnicki, J.M. (2003) A ‘FRankenstein's monster’ approach to comparative modeling: merging the finest fragments of Fold‐Recognition models and iterative model refinement aided by 3D structure evaluation. Proteins, 53, 369–379. [DOI] [PubMed] [Google Scholar]
- Kurowski, M.A. and Bujnicki, J.M. (2003) GeneSilico protein structure prediction meta‐server. Nucleic Acids Res. 31, 3305–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbeck, A.G.C. , Lewandowski, D.J. , Culver, J.N. , Thomson, W.W. and Dawson, W.O. (1992) Mutant coat protein of tobacco mosaic virus induces acute chlorosis in expanded and developing tobacco leaves. Mol. Plant–Microbe Interact. 5, 235–241. [Google Scholar]
- Ling, K.S. (2007) Molecular characterization of two Pepino mosaic virus variants from imported tomato seed reveals high levels of sequence identity between Chilean and US isolates. Virus Genes, 34, 1–8. [DOI] [PubMed] [Google Scholar]
- Ling, K.S. (2008) Genetic composition of Pepino mosaic virus population in North American greenhouse tomatoes. Plant Dis. 92, 1683–1688. [DOI] [PubMed] [Google Scholar]
- Ling, K.S. , Wechter, W.P. and Jordan, R. (2007) Development of a one‐step immunocapture real‐time TaqMan RT‐PCR assay for the broad spectrum detection of Pepino mosaic virus . J. Virol. Methods, 144, 65–72. [DOI] [PubMed] [Google Scholar]
- Maroon‐Lango, C.J. , Guaragna, M.A. , Jordan, R.L. , Hammond, J. , Bandla, M. and Marquardt, S.K. (2005) Two unique US isolates of Pepino mosaic virus from a limited source of pooled tomato tissue are distinct from a third (European‐like) US isolate. Arch. Virol. 150, 1187–1201. [DOI] [PubMed] [Google Scholar]
- Martin, J. and Mousserion, C. (2002) Potato varieties which are sensitive to the tomato strains of Pepino mosaic virus (PepMV). Phytoma, 552, 26–28. [Google Scholar]
- Matthaios, M. , Veiga, R. , Ghita, M. , Tsikou, D. , Medina, V. , Canto, T. , Makris, A.M. and Livieratos, I.C. (2012) Pepino mosaic virus capsid protein interacts with a tomato heat shock protein cognate 70. Virus Res. 163, 28–39. [DOI] [PubMed] [Google Scholar]
- Mo, H. , Lu, L. , Pithawalla, R. , Kempf, D.J. and Molla, A. (2004) Complementation in cells cotransfected with a mixture of wild‐type and mutant human immunodeficiency virus (HIV) influences the replication capacities and phenotypes of mutant variants in a single‐cycle HIV resistance assay. J. Clin. Microbiol. 42, 4169–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumford, R.A. and Metcalfe, E.J. (2001) The partial sequencing of the genomic RNA of a UK isolate of Pepino mosaic virus and the comparison of the coat protein sequence with other isolates from Europe and Peru. Arch. Virol. 146, 2455–2460. [DOI] [PubMed] [Google Scholar]
- Mumford, R.A. , Walsh, K. and Boonham, N. (2000) A comparison of molecular methods for the routine detection of viroids. Bull. EPPO 30, 431–436. [Google Scholar]
- Ohnishi, J. , Hirai, K. , Kanda, A. , Usugi, T. , Meshi, T. and Tsuda, S. (2009) The coat protein of Tomato mosaic virus L11Y is associated with virus‐induced chlorosis on infected tobacco plants. J. Gen. Plant Pathol. 75, 297–306. [Google Scholar]
- Pagan, I. , Córdoba‐Selles, M. , Martinez‐Priego, L. , Fraile, A. , Malpica, J. , Jorda, C. and Garcia‐Arenal, F. (2006) Genetic structure of the population of Pepino mosaic virus infecting tomato crops in Spain. Phytopathology, 96, 274–279. [DOI] [PubMed] [Google Scholar]
- Pawlowski, M. , Gajda, M.J. , Matlak, R. and Bujnicki, J.M. (2008) MetaMQAP: a meta‐server for the quality assessment of protein models. BMC Bioinformatics, 9, 403. doi: 10.1186/1471-2105-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggero, P. , Masenga, V. , Lenzi, R. , Coghe, F. , Ena, S. and Winter, S. (2001) First report of Pepino mosaic virus in tomato in Italy. Plant Pathol. 50, 798. [Google Scholar]
- Sali, A. and Blundell, T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Siew, N. , Elofsson, A. , Rychlewski, L. and Fischer, D. (2000) MaxSub: an automated measure for the assessment of protein structure prediction quality. Bioinformatics, 16, 776–785. [DOI] [PubMed] [Google Scholar]
- Simons, K.T. , Kooperberg, C. , Huang, E. and Baker, D. (1997) Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and bayesian scoring functions. J. Mol. Biol. 268, 209–225. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven, J.T.J. , van der Vlugt, R.A.A. and Roenhorst, J.W. (2003) High similarity between isolates of pepino mosaic virus suggests a common origin. Eur. J. Plant Pathol. 109, 419–425. [Google Scholar]
- van der Vlugt, R.A. , Stijger, C.M. , Verhoeven, J.T.J. and Lesemann, D.E. (2000) First report of Pepino mosaic virus on tomato. Plant Dis. 84, 103. [DOI] [PubMed] [Google Scholar]
- Wallner, B. and Elofsson, A. (2003) Can correct protein models be identified. Protein Sci. 12, 1073–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werkman, A.W. and Sansford, C.E. (2010) Pest Risk Analysis for Pepino mosaic virus for the EU. Deliverable Report 4.3. EU Sixth Framework Project. PEPEIRA. Available at http://www.pepeira.com [accessed on November 6, 2010].


