Summary
Viruses are likely to be the most dangerous parasites of living organisms because of their widespread occurrence, possible deleterious effects on their hosts and high rates of evolution. Virus host‐to‐host transmission is a critical step in the virus life cycle, because it enables survival in a given environment and efficient dissemination. As hosts of plant viruses are not mobile, these pathogens have adopted diverse transmission strategies involving various vector organisms, mainly arthropods, nematodes, fungi and protists. In nature, plants are often infected with more than one virus at a time, thereby creating potential sources for vectors to acquire and transmit simultaneously two or more viruses. Simultaneous transmission can result in multiple infections of new host plants, which become subsequent potential sources of the viruses, thus enhancing the spread of the diseases caused by these pathogens. Moreover, it can contribute to the maintenance of viral genetic diversity in the host communities. However, despite its possible significance, the problem of the simultaneous transmission of plant viruses by vectors has not been investigated in detail. In this review, the current knowledge on multiple viral transmissions by aphids, whiteflies, leafhoppers, planthoppers, nematodes and fungi is outlined.
Introduction
Viruses are considered as the most threatening human, animal and plant parasites because of their widespread occurrence, high rates of evolution and often highly destructive effects on their hosts (Duffy et al., 2008; Elena and Sanjuán, 2005; Froissart et al., 2010; Woolhouse et al., 2005). Viruses affecting plants can cause substantial yield losses to the cereal, vegetable, fruit and floral industries, and greatly decrease the quality of crop products.
To survive in nature, viruses must be efficiently spread to infect more hosts. Peculiar to the biological cycles of plant viruses is that their hosts are not mobile and direct contact between them is rare or nonexistent. For transmission from one host plant to another (horizontal transmission), most plant viruses depend on vectors, which include arthropods, nematodes, fungi and protists (Andret‐Link and Fuchs, 2005; Blanc, 2007; Campbell, 1996; Hogenhout et al., 2008; Nault, 1997; Sylvester, 1985). The principal vectors are arthropods, among which a significant role is played by aphids, whiteflies, leafhoppers and delphacid planthoppers (Blanc et al., 2011; Hogenhout et al., 2008; Nault, 1997; Spence, 2001). Aphids are the most common and versatile vectors (Ng and Perry, 2004), responsible for dissemination of 28% of the nearly 700 vector‐transmitted virus species recorded so far (Hogenhout et al., 2008). The remaining 72% are transmitted by whiteflies (18%), leafhoppers and planthoppers (7%), beetles (7%), nematodes (7%), fungi (4%), other hemiptera, thrips, mites and unidentified vectors (29%).
The diversity of strategies used by vector‐borne plant viruses is much higher than that in animal viruses (Blanc et al., 2011). Vector transmission is a complex and variable process, involving highly specific interactions between viral, host and vector determinants (Andret‐Link and Fuchs, 2005; Martinière et al., 2013; Purcell and Almeida, 2005). These multi‐component interactions are certainly even more complex if the host plant is infected by more than one virus at a time (Salvaudon et al., 2013). A mixed infection may result from multiple transmission events that involve numerous vectors, each potentially carrying a different virus or viral isolate. In addition, it may be the effect of a single transmission performed by an individual vector carrying two viruses simultaneously. Plant co‐infection may affect individual viral properties in terms of accumulation, virulence and transmission. If the viruses co‐infecting the plant share the same vector(s), each has a chance to be acquired and transmitted to another host plant. As multiple viral infections are frequent among crop plants and arable weeds (Syller, 2012), the questions arise as to whether simultaneous vector transmission of two or more viruses occurs frequently, and what outcomes of this phenomenon can be expected. The latter are rather easy to predict. Simultaneous transmission can contribute to the maintenance of viral genetic diversity in the host communities. Moreover, it can result in multiple infections of new host plants, thereby creating potential sources of the viruses for subsequent vector transmissions, and thus increasing the spread of the diseases caused by the viruses involved. Simultaneous transmission can thus influence the epidemiology of individual viruses, and the severity and rate of expansion of a given outbreak (Srinivasan and Alvarez, 2007). Therefore, increased attention should be given to the co‐transmission of viruses, for which in‐depth characterization is greatly needed.
However, despite its importance, the ability of natural vectors of plant viruses to transmit more than one virus at a time has been only fragmentarily recognized and characterized. The literature on this subject is comparatively richer, although still poor, for aphid‐transmitted viruses than for viruses transmitted by other types of vector. In this article, the current knowledge on multiple transmissions of plant viruses by aphids, whiteflies, leafhoppers, planthoppers, nematodes and fungi is outlined. No case reports from field or glasshouse trials, or from laboratory work, are available in relation to other types of vector, such as beetles, thrips or mites. The review focuses essentially on viruses that are naturally transmitted by vectors as independent entities. A list of viruses addressed in this review is shown in Table 1.
Table 1.
List of plant viruses addressed in this article, grouped according to the type of vector
| Acronym | Name of species | Genus | Family |
|---|---|---|---|
| Aphids | |||
| AMV | Alfalfa mosaic virus | Alfamovirus | Bromoviridae |
| BBWV | Broad bean wilt virus | Fabavirus | Comoviridae |
| BYDV‐PAS | Barley yellow dwarf virus‐PAS | Luteovirus | Luteoviridae |
| BYDV‐PAV | Barley yellow dwarf virus‐PAV | Luteovirus | Luteoviridae |
| ClYVV | Clover yellow vein virus | Potyvirus | Potyviridae |
| CMV | Cucumber mosaic virus | Cucumovirus | Bromoviridae |
| PAMV | Potato aucuba mosaic virus | Potexvirus | Alphaflexiviridae |
| PeMV | Peanut mottle virus | Potyvirus | Potyviridae |
| PPV | Plum pox virus | Potyvirus | Potyviridae |
| PRSV‐W | Papaya ringspot virus‐type W | Potyvirus | Potyviridae |
| PStV | Peanut stripe virus | Potyvirus | Potyviridae |
| PSV | Pea streak virus | Carlavirus | Betaflexiviridae |
| PVY | Potato virus Y | Potyvirus | Potyviridae |
| SPFMV | Sweet potato feathery mottle virus | Potyvirus | Potyviridae |
| SPVG | Sweet potato virus G | Potyvirus | Potyviridae |
| TVMV | Tobacco vein mottling virus | Potyvirus | Potyviridae |
| WMV | Watermelon mosaic virus | Potyvirus | Potyviridae |
| ZYMV | Zucchini yellow mosaic virus | Potyvirus | Potyviridae |
| Whiteflies | |||
| CpMMV | Cowpea mild mottle virus | Carlavirus | Betaflexiviridae |
| MYaV | Melon yellowing‐associated virus | Carlavirus | Betaflexiviridae |
| OYVMV | Okra yellow vein mosaic virus | Begomovirus | Geminiviridae |
| PepGMV | Pepper golden mosaic virus | Begomovirus | Geminiviridae |
| PHYVV | Pepper huasteco yellow vein virus | Begomovirus | Geminiviridae |
| TICV | Tomato infectious chlorosis virus | Crinivirus | Closteroviridae |
| TLCV | Tobacco leaf curl virus | Begomovirus | Geminiviridae |
| ToCV | Tomato chlorosis virus | Crinivirus | Closteroviridae |
| Leafhoppers/planthoppers | |||
| RBSDV | Rice black‐streaked dwarf virus | Fijivirus | Reoviridae |
| RSV | Rice stripe virus | Tenuivirus | Unassigned |
| Nematodes | |||
| ArMV | Arabis mosaic virus | Nepovirus | Secoviridae |
| GFLV | Grapevine fanleaf virus | Nepovirus | Secoviridae |
| PEBV | Pea early browning virus | Tobravirus | Virgaviridae |
| SLRSV | Strawberry latent ringspot virus | Nepovirus | Secoviridae |
| TRV | Tobacco rattle virus | Tobravirus | Virgaviridae |
| Fungi and plasmodiophorids | |||
| BNYVV | Beet necrotic yellow vein virus | Benyvirus | Benyviridae?* |
| BSMV | Beet soil‐borne mosaic virus | Benyvirus | Benyviridae?* |
| LBVaV | Lettuce big‐vein associated virus | Varicosavirus | Unassigned |
| TNV | Tobacco necrosis virus | Necrovirus | Tombusviridae |
| TStV | Tobacco stunt virus | Varicosavirus | Unassigned |
*New family has been proposed and is under consideration at the International Committee on Taxonomy of Viruses (ICTV).
Simultaneous Virus Transmission by Aphids
Aphids (order Hemiptera, family Aphididae) are ubiquitous and highly versatile plant viral vectors, transmitting viruses in a nonpersistent (noncirculative), semipersistent (noncirculative) or persistent (circulative) manner. As these strategies of virus transmission have been described in numerous comprehensive reviews (see, for example, Andret‐Link and Fuchs, 2005; Brault et al., 2010; Gray and Banerjee, 1999; Hogenhout et al., 2008; Ng and Falk, 2006; Ng and Perry, 2004; Syller, 2006), only concise characteristics are given here.
Nonpersistent (noncirculative) transmission is the most common manner of virus dissemination by aphids. In this way, the causative agents of many economically important viral diseases are transmitted. The virus is acquired by an aphid during brief probing made by the insect in the search for a host to feed. As the virus does not require a latent period, i.e. the time between the acquisition access period (AAP) and inoculation access period (IAP), it can be transmitted immediately to another plant during subsequent probing of the insect. To be transmitted, the virions attach to the epicuticle which lines the stylets (mouthparts) of the aphid. It is noteworthy that the same virus can be transmitted by more than one aphid species, and one aphid species can readily transmit several viruses.
The viruses vectored in a persistent (circulative) manner are acquired by aphids during longer feeds, lasting from several hours to several days. After the AAP, a latent period is required for the virus to be transmitted to another plant. The ingested virions are internalized in the aphid's body, in which the virus passes through the gut wall to the haemolymph and then to the salivary glands, being actively transported across several cell membranes. During the passage, the viral nucleic acids must be packaged in capsid proteins (CPs). For successful virus transmission, mutual recognition between specific sites on the CP and on the membranes of the aphid's gut and salivary glands is mandatory. Even a minor change in the coat protein or the relevant vector's membrane alters the permeability of the membranes to virions, which makes it impossible for the virus to ultimately pass into the salivary glands to be introduced into subsequent plant(s) in the vector's saliva excreted when the insect feeds again.
A semipersistent (noncirculative) manner of transmission is an intermediate category, which has been separated for viruses which have longer AAPs and IAPs than nonpersistent viruses, but shorter than persistent viruses. Like the former and unlike the latter, the viruses vectored in a semipersistent way do not circulate within their vectors.
Mixed viral infections of plants occur commonly in nature, thereby creating potential sources for aphid vectors to acquire and transmit two or more viruses simultaneously (Rochow, 1972; Syller, 2012). It is therefore surprising to note that multiple viral transmission events have been documented so far in few reports, in most cases relating to nonpersistent viruses. Hampton and Sylvester (1969) reported the simultaneous transmission of Alfalfa mosaic virus (AMV, Alfamovirus) and Pea streak virus (PSV; Carlavirus) by the pea aphid Acyrthosiphon pisum Harris from doubly infected hosts. Interestingly, transmission of AMV was increased by co‐infection with PSV, whereas transmission of PSV was decreased by co‐infection with AMV, compared with those from singly infected plants. Another member of the family Bromoviridae, Cucumber mosaic virus (CMV), and Broad bean wilt virus (BBWV) have been proven to be transmitted simultaneously from co‐infected Nicotiana tabacum to healthy N. glutinosa plants by single aphids of Myzus persicae Sulz. (Makram et al., 1976). The transmission efficiency was low, but did not differ from that from singly infected plants. In addition, in studies on Peanut mottle virus (PeMV) and Peanut stripe virus (PStV), both potyviruses, simultaneous transmission of the two viruses by single aphids from double‐infected to healthy assay plants was very poor with M. persicae, and absent with Aphis craccivora Koch (Sreenivasulu and Demski, 1988). In the same study, a low rate (M. persicae) or failure (A. craccivora) of transmission was also recorded when individual aphids acquired PeMV and PStV during sequential feeding on singly infected peanut plants. However, unlike BBWV and CMV (Makram et al., 1976), PeMV and, particularly, PStV were transmitted with significantly higher efficiencies, reaching up to nearly 30% in the PStV–M. persicae combination, from singly infected than from double‐infected source plants (Sreenivasulu and Demski, 1988). Similar results were obtained with CMV, which has been reported to be transmitted simultaneously, but at a rate not exceeding 8%, with the potyviruses Papaya ringspot virus ‐ type W (PRSV‐W) and Zucchini yellow mosaic virus (ZYMV) from doubly infected to healthy zucchini squash plants by M. persicae and Aphis gossypii Glov. (Pinto et al., 2008). As in the case of PeMV and PStV (Sreenivasulu and Demski, 1988), the transmission of CMV, PRSV‐W and ZYMV was more efficient in single infections than in double infections. Interestingly, contrasting results have been obtained recently with another potyvirus, Sweet potato feathery mottle virus (SPFMV), which was transmitted by A. gossypii at a greater rate from plants co‐infected with Sweet potato virus G (SPVG; also potyvirus) than from singly infected plants (Wosula et al., 2012). However, the results of the most recent study (Salvaudon et al., 2013) show that, even within the same viral genus (Potyvirus), diverse outcomes of within‐plant interactions between viruses can be observed, depending on the species of the competing viruses. Isolates of closely related Watermelon mosaic virus (WMV) and ZYMV were transmitted by A. gossypii at generally greater rates from singly infected plants (80% and 40%, respectively) than from co‐infected plants (20%, as a total for co‐infections). In the source plants, ZYMV isolates reached similar concentrations (measured as the number of RNA copies in plant tissues) in single and mixed infections, whereas WMV isolates accumulated to significantly lower levels in the presence of ZYMV. It may be concluded that, despite being the weaker competitor in the plants co‐infected with ZYMV, WMV was still quite efficiently acquired and transmitted by A. gossypii (Salvaudon et al., 2013).
The reasons for the diverse effects of mixed infections on the rates of transmission of particular viruses by aphids are unclear and seem to be complex. The AMV/PSV, BBWV/CMV, PRSV‐W/CMV and ZYMV/CMV pairs are combinations of viruses representing different families, whereas the PeMV/PStV, SPFMV/SPVG and WMV/ZYMV combinations contain viruses belonging to the same genus. In mixed infections, viruses interact with each other in synergistic (facilitative), competitive (antagonistic) or neutral ways (reviewed by Syller, 2012). Depending on the type of interaction, the viral titre can be enhanced or decreased, compared with that in singly infected plants, or remain unchanged. Taking into account that vector transmission has, in general, been considered to be positively correlated with virus accumulation in the host plant (discussed by Froissart et al., 2010), the effect of one virus on the titre of a second virus might be expected to have considerable implications for the rates of acquisition and transmission of the latter by its vector. Mutual relationships of closely related viruses are predominantly competitive. Although classified in the same genus, PeMV and PStV are serologically unrelated viruses, and probably do not cross‐protect against each other (Sreenivasulu and Demski, 1988). It has been proven, however, that competitive virus–virus interactions can also occur between distinct viral species representing the same genus, as found for certain whitefly‐transmitted criniviruses (Wintermantel et al., 2008) (see the relevant section below). Hence, it seems possible that, in a doubly infected host plant, PeMV and PStV interact with each other in a competitive way. This type of mutual relationship is likely to have a great impact on the distribution and localization in the host of the competing viruses, which tend to separate from one another in host cells/tissues. Such a phenomenon, termed spatial separation, has been demonstrated by Dietrich and Maiss (2003) using differentially labelled cDNA clones of the potyviruses Plum pox virus (PPV), Tobacco vein mottling virus (TVMV) and Clover yellow vein virus (ClYVV). Particles of spatially separated viruses can mix only in a few cells at the border of viral subpopulations, which consequently would markedly decrease the chance of the simultaneous acquisition and transmission of particles of both viruses by individual aphids, as illustrated in Fig. 1. Therefore, it may be further speculated that the failure of aphids to transmit simultaneously PeMV and PStV from mixed infections (Sreenivasulu and Demski, 1988) could result from the within‐host spatial separation of these viruses. Another hypothesis to explain the reduced rates of simultaneous transmission of the cucumovirus and potyviruses by aphid vectors from mixed infections has been offered by Pinto et al. (2008). As proposed, this might be the effect of competition between virions of the two viruses for access to binding sites on the aphid's stylets, which would reduce the number of available infective particles of each virus for further transmission. Both explanations given above can be combined, thus proposing that the low rates of simultaneous transmission of two viruses from mixed infections are the effect of strong competition for binding sites on the aphid's stylets between virions of both viruses mixed within a thin border zone separating viral subpopulations in the host tissue (Fig. 1).
Figure 1.
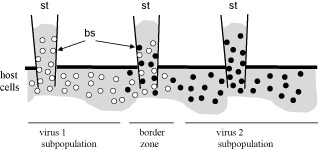
Acquisition by aphids of particles of virus 1 (○) and virus 2 (●), interacting in a competitive way and manifesting spatial separation in the co‐infected host plant. Virions of spatially separated viruses 1 and 2 mix only within a thin border zone separating viral subpopulations in the host tissue. There they compete with one another for access to binding sites (bs) on the aphid's stylets (st). Consequently, spatial separation decreases the probability of the simultaneous acquisition and transmission of both viruses by individual aphids probing the plant tissue at the border zone.
The assumed within‐host competitiveness among closely related viruses, with isolates/strains of the same virus expressing the highest levels of relatedness, has recently received strong support from a study showing the effects of co‐infection by different isolates of Potato virus Y (PVY) on the titres of particular isolates in potato and tobacco plants (Syller and Grupa, 2013). Single aphids were found to be able to transmit simultaneously two PVY isolates, but the frequency of transmission was quite low. The relevant isolates were detected in aphid‐inoculated assay plants by enzyme‐linked immunosorbent analysis (ELISA), reverse transcription‐polymerase chain reaction (RT‐PCR) and mechanical back‐inoculations to healthy tobacco plants. The generally low rates of transmission by M. persicae of two PVY isolates from mixed infections, reported by Gibson et al. (1988), Srinivasan et al. (2012) and Syller and Grupa (2013), can be explained as proposed above, and illustrated in Fig. 1. Surprisingly, M. persicae successfully acquired and transmitted both isolates in certain combinations of PVY isolates, despite the fact that the concentration of one of them in the source plant dropped below the limit of detection by ELISA, most probably as a result of the suppression of activity by the competitor isolate (Syller and Grupa, 2013). In this respect, this finding is consistent with that reported by Salvaudon et al. (2013), at the same time giving no support to the common opinion on the positive correlation between virus accumulation in the plant and the rate of its transmission by aphids.
When assuming that two related viruses are spatially separated in the epidermal and mesophyll tissues of the host plant, it also seems possible that their simultaneous transmission by an individual aphid may result from sequential acquisition of the viruses by the aphid wandering around on a leaf surface and making short shallow exploratory probes on leaf areas occupied by single viruses. Low rates of simultaneous transmission of viruses taken up by an aphid during consecutive probes (see Syller and Grupa, 2013) may be because particles of a nonpersistent virus have a very short retention time (minutes) in the aphid vector and can be easily lost during subsequent probing (Hooks et al., 2007; Ng and Falk, 2006).
It is worth noting that many of the viruses transmitted by aphids in a non‐ and semipersistent manner, namely potyviruses and caulimoviruses, use a helper strategy for transmission, based on the encoding of a helper component (HC) (reviewed by Syller, 2006). The HC, termed the helper component‐proteinase (HC‐Pro) for potyviruses, is a nonstructural protein encoded by the virus during plant infection. It acts as a reversible ‘bridge’ (‘bridge’ hypothesis) in attaching the virion to the cuticle of the maxillary food canal of an aphid vector. The virion is released into a plant when the aphid feeds again. Experiments in which aphids were fed on purified HC‐Pro/virion preparations through parafilm membranes showed that, to mediate virus transmission, HC‐Pro must be delivered to vectors either before or together with virions (Pirone and Blanc, 1996). In mixed infections, a biologically active HC‐Pro of one virus can assist aphid transmission of the second virus. A potyviral HC‐Pro has been proven to assist the transmission of virions located in the same cell, in other cells or in other host plants in which the vector makes subsequent probes, a phenomenon termed HC‐transcomplementation (Froissart et al., 2002). Consequently, the HC‐Pro produced in the host plant by a certain potyvirus can facilitate transmission of HC‐Pro‐deficient and thus nontransmissible potyviral isolates, as well as of unrelated viruses, the most well‐known experimental evidence being obtained for Potato aucuba mosaic virus (PAMV). As pointed out by Froissart et al. (2002), in planta or natural HC‐transcomplementation can only be detected when the phenomenon occurs between different virus species (or strains) that are easily detectable. Therefore, it cannot be excluded that parallel transmission of some viruses using the HC‐transcomplementation strategy may not have been detected or investigated. More recently, few virologists have paid attention to these research problems.
It may seem questionable whether the helper strategy lies within the scope of this review. After all, in a ‘helper‐dependent’ virus complex, there is a complete unilateral assistance of the dependent virus by the helper (Zhang et al., 2000). In such a complex, the helper virus can be transmitted independently by the vector, whereas the dependent virus relies entirely on the helper for transmission by the vector. However, as mentioned above, the dependent virus might be an HC‐Pro‐deficient (presumably as a result of a minor amino acid mutation) and thus nontransmissible isolate of the virus (e.g. PVY) that is normally readily transmitted by the aphid as an independent entity. Therefore, simultaneous aphid transmission of both the helper virus and the dependent viral isolate lies within the scope of this article. More obviously, beyond the scope of this review is another helper dependence strategy, in which the species belonging to the genus Umbravirus, grouping viruses that do not encode for a CP, and thereby cannot be transmitted by aphids, become aphid transmissible if they use a CP of a suitable luteovirid, co‐infecting the host plant (Syller, 2003). In addition, we do not discuss a parallel aphid transmission of a virus (e.g. luteovirid) and a viroid encapsidated by the viral CP whilst sharing the host (Syller et al., 1997).
Multiple transmissions of viruses vectored by aphids in a persistent (circulative) manner have received less attention from virologists or are more difficult to study, as suggested by the scarcity of the available literature. Noteworthy observations have recently been made on the transmission of PAV and PAS species of Barley yellow dwarf virus (BYDV) by Rhopalosiphum padi L. from single and multiple infections (Hall and Little, 2013). Multiple infections with different BYDV species are quite common in grasses. However, as pointed out by the authors, no studies have been performed previously on within‐host interactions between PAV and PAS, or have been able to relate virus population size in BYDV mixed infections to the probability of aphid transmission. PAV and PAS are indistinguishable by ELISA with polyclonal antibodies, but can be distinguished using RT‐PCR followed by restriction enzyme analysis. The results obtained indicate that single aphids of R. padi are able to simultaneously transmit PAV and PAS from co‐infected plants (Hall and Little, 2013). It was also found that, from mixed infections, PAV was more readily transmitted than PAS. It has been concluded that within‐host interactions between PAV and PAS create conditions that promote both the competitive exclusion of PAS and co‐existence of these species, and thus the maintenance of genetic diversity in the host community (Hall and Little, 2013).
Simultaneous Virus Transmission by Whiteflies
Whiteflies (order Hemiptera, family Aleyrodidae) are mainly responsible for the transmission of begomoviruses (family Geminiviridae), but these insects are also vectors of criniviruses (Closteroviridae), ipomoviruses (Potyviridae), torradoviruses (Secoviridae) and two species of carlaviruses (Betaflexiviridae): Cowpea mild mottle virus (CpMMV) and Melon yellowing‐associated virus (MYaV) (Navas‐Castillo et al., 2011). Begomoviruses have quite recently been reported to be transmitted by whiteflies in a persistent circulative way, and criniviruses and ipomoviruses in a semipersistent manner; the manner of transmission of torradoviruses is still unknown (Navas‐Castillo et al., 2011). With regard to carlaviruses, there are some discrepancies concerning the manner of whitefly transmission of CpMMV. Because of some differences in the AAP and IAP and the virus retention time in whitefly vectors, the virus has been reported to be transmitted by Bemisia tabaci Genn. in either a semipersistent (Iwaki et al., 1982) or nonpersistent (Muniyappa and Reddy, 1983) manner. More recently, both studies have been referred to by Menzel et al. (2010), who categorized CpMMV as transmitted by B. tabaci in a nonpersistent manner, thus making no distinction between the two modes of virus transmission. In contrast, Navas‐Castillo et al. (2011) described the virus as vectored by B. tabaci in a semipersistent manner. As can be seen, there is no conclusive evidence for a mode of CpMMV transmission by whiteflies.
There have been very few reports published on the simultaneous transmission of plant viruses by whiteflies. The geminiviruses Tobacco leaf curl virus (TLCV) and Okra yellow vein mosaic virus (OYVMV) were found to be transmitted in parallel by B. tabaci following their sequential acquisition from TLCV‐infected tobacco and OYVMV‐infected okra plants, regardless of the chronological order of the virus acquisition (Tsering and Patel, 1990). More recently, another two geminiviruses, Pepper huasteco yellow vein virus (PHYVV) and Pepper golden mosaic virus (PepGMV), have been reported to be simultaneously acquired and transmitted by B. tabaci to pepper (Capsicum annuum cv. Sonora Anaheim) in Mexico (Medina‐Ramos et al., 2008). In mixed infection, the viruses induce ‘rizado amarillo del chile’, a disease devastating crops of pepper, which is one of the most important horticultural plants in Mexico. In the co‐infected host, PHYVV and PepGMV displayed a synergistic interaction, which was associated with an increase in the number of infected cells and in the concentrations of both viruses, but had no noticeable influence on the localization of either virus in plant tissue (Rentería‐Canett et al., 2011). The results obtained by Medina‐Ramos et al. (2008) indicated that, from single infections, PepGMV was transmitted by B. tabaci less readily than PHYVV, and co‐infection with PHYVV could facilitate its acquisition and transmission by whiteflies, thereby enhancing its dispersion in pepper crops.
In addition, two criniviruses, Tomato chlorosis virus (ToCV) and Tomato infectious chlorosis virus (TICV), have been reported to be simultaneously transmitted by whiteflies (Wintermantel et al., 2008). ToCV and TICV induce similar yellowing diseases in tomato (Lycopersicon esculentum L.) crops worldwide (Anfoka and Abhary, 2007; Dalmon et al., 2009; Wintermantel et al., 2008; and references therein). According to current knowledge, ToCV is transmitted by the glasshouse whitefly Trialeurodes vaporariorum West., the banded‐wing whitefly T. abutilonea Hald. and B. tabaci biotypes A and B, whereas TICV is only transmitted by T. vaporariorum (Navas‐Castillo et al., 2011). Both viruses were simultaneously transmitted by T. vaporariorum from doubly infected Physalis wrightii Gray and Nicotiana benthamiana Domin plants to assay plants of the two species (Wintermantel et al., 2008). Interestingly, TICV was occasionally transmitted to both hosts by T. abutilonea, known as a nonvector of this virus, from plants co‐infected with ToCV. Although T. abutilonea entirely failed to transmit TICV from double infections with ToCV in additional experiments, the results suggest that, in mixed infections, one virus can facilitate the transmission of another virus by a whitefly that normally is not a vector of the latter (Wintermantel et al., 2008); this phenomenon has been demonstrated for other virus vector systems and is called transcomplementation (Latham and Wilson, 2008).
However, no functional transcomplementation of TICV by ToCV for transmission by B. tabaci, another nonvector of ToCV, was found in other studies, employing a large number of whiteflies per test plant to increase the probability of occurrence of this event (Dalmon et al., 2009). The result suggests that, if this phenomenon occurs in nature, its frequency is very low.
Simultaneous Virus Transmission by Leafhoppers and Planthoppers
These insects belong to the order Hemiptera, leafhoppers representing the family Cicadellidae in the suborder Clypeorrhyncha (Cicadomorpha), and planthoppers representing the family Delphacidae in the suborder Archaeorrhyncha (Fulgoromorpha) (Hogenhout et al., 2008). So far, leafhoppers have been shown to transmit 27 plant viruses, 13 in a persistent circulative nonpropagative manner, 10 in a persistent propagative way and four in a semipersistent manner (Hogenhout et al., 2008). According to the same article, delphacid planthoppers transmit 18 viruses in a persistent propagative way.
One of the most important delphacid planthoppers is the small brown planthopper (Laodelphax striatellus Fall.), which occurs worldwide, mainly in temperate regions, and plays a key role in the spread of two economically important rice viruses: Rice black‐streaked dwarf virus (RBSDV) and Rice stripe virus (RSV) (Li et al., 2013). After the RSV‐infected planthoppers L. striatellus had been released under laboratory conditions to feed for 2 days on RBSDV‐infected rice plants, the planthoppers became infected simultaneously with both viruses (Li et al., 2013). Double infection increased the accumulation of particular RBSDV RNA segments and the abundance of RBSDV‐derived small interfering RNAs (siRNAs), but had no effect on the abundance of RSV siRNAs (Li et al., 2013). The enhanced accumulation of specific RBSDV genome segments in L. striatellus during mixed infection with RSV may be seen as an effect of a synergistic within‐host interaction between the viruses. It is worth emphasizing that synergistic interactions are quite common in mixed infections of plant viruses, the best characterized being double infections involving potyviruses (reviewed by Syller, 2012). The facilitative effect of a potyvirus on the accumulation of its unrelated counterpart results from the suppression of antiviral RNA silencing by the HC‐Pro encoded by potyviruses (Syller, 2006, 2012). As pointed out by Li et al. (2013), there are still important questions regarding mixed infections with RBSDV and RSV in L. striatellus, which await clarification. First, it seems worthy of elucidation whether the insect infection with RBSDV prior to RSV enhances the accumulation of the latter (or of some segments of the RSV genome). Second, it is not known whether double infection of L. striatellus by RBSDV and RSV, and, in consequence, simultaneous transmission of the two viruses to new host plants, occurs in the field. If so, it is not known what are the implications of the phenomenon for the epidemiology and pathogenesis of the diseases caused by both pathogens.
Simultaneous Virus Transmission by Nematodes
Nematode species capable of transmitting plant viruses belong to two families: Longidoridae (Dorylaimida; genera Longidorus and Xiphinema) and Trichodoridae (Triplonchida; genera Trichodorus and Paratrichodorus) (Bileva et al., 2009; Hull, 2009). Twenty‐four species of Longidoridae transmit 12 viruses of the genus Nepovirus and one of Sadwavirus, whereas 13 species of Trichodoridae transmit all three members of the genus Tobravirus.
Serious obstacles have been encountered in investigations on virus transmission by nematodes, mainly because these organisms live in the soil, feeding on plant roots, have specific requirements with respect to soil moisture content and cannot be maintained in pure culture (Hull, 2009; MacFarlane, 2003). Nevertheless, recent progress in the development of molecular techniques has made it possible to obtain a deeper insight into the molecular mechanism governing the specific binding of a plant virus to its nematode vector (MacFarlane, 2003; Schellenberger et al., 2011). In this process, the C‐terminal domain of the viral CP plays a key role (MacFarlane et al., 1996; Vassilakos et al., 2001). Successful transmission of PaY4 and PpK20 isolates of Tobacco rattle virus (TRV) by their vector Paratrichodorus pachydermus Seinh. was found to require a specific interaction between this domain and the nonstructural viral protein 2b (Vassilakos et al., 2001; Vellios et al., 2002; Visser and Bol, 1999). It has been proposed that the tobravirus 2b protein acts as a bridge to link the virus particle with cuticle lining the feeding apparatus of the nematode vector (Visser and Bol, 1999), a model resembling that developed to explain the function of HC‐Pro in aphid transmission of potyviruses (Pirone and Blanc, 1996). It seems to be possible that a compatible binding interaction between the tobravirus CP and 2b proteins is mandatory to prevent rapid degradation of the 2b protein (Vellios et al., 2002). Although there is interaction between the TRV PpK20 CP and the PpK20 2b protein, and between the TRV PaY4 CP and the PaY4 2b protein, there is no compatibility between PpK20 CP and PaY4 2b, and probably vice versa. In these combinations, the 2b protein is rapidly degraded, which precludes nematode transmission of the recombinant virus. Thus, the 2b protein appears to be a significant factor in determining the specificity of transmission of different viruses or viral isolates by nematode vectors (Vellios et al., 2002).
It must be emphasized that high specificity is a characteristic feature of virus–nematode relationships (Hull, 2009; MacFarlane, 2003; Visser and Bol, 1999). It has been fairly well recognized with respect to two distantly related nepoviruses, Arabis mosaic virus (ArMV) and Grapevine fanleaf virus (GFLV), specifically transmitted by different Xiphinema nematode species, X. diversicaudatum Micol. and X. index Thorne and Allen, respectively (MacFarlane, 2003; Marmonier et al., 2010). The virus–vector specificity is even more pronounced in exclusive associations between virus isolates and nematode species, as shown, for example, for the TpA56 isolate of Pea early browning virus (PEBV) and PpK20 and PaY4 isolates of TRV (Vassilakos et al., 2001; Vellios et al., 2002).
Perhaps, high specificity in virus–nematode interactions is a major constraint in the simultaneous transmission by nematodes of two viruses/isolates from mixed infections. Indeed, no such transmission has been documented so far, but some findings seem to be worth mentioning as they throw some light on the problem. When X. diversicaudatum nematodes, which had fed on Petunia plants infected with both ArMV and Strawberry latent ringspot virus (SLRSV), were transferred singly to healthy Petunia seedlings, 20 of 706 were found to be infected with ArMV, one with SLRSV and none of the plants contained both viruses (Lister, 1964). More recently, to evaluate the outcomes of plant co‐infection by different viral subpopulations, N. benthamiana plants were inoculated with a mixture of TRV PpK20 labelled with green (TRV‐GFP) and red (TRV‐RFP) fluorescent protein (Vassilakos et al., 2001). The study revealed that the viruses were able to simultaneously infect the same lateral roots, but did not infect the same root cells. The molecular mechanism of this interference is not known, but the phenomenon may have implications for the nematode transmission of viruses. During the initial stage of feeding, trichodorid nematodes inject secretions of pharyngeal glands into the root cell to aggregate the cytoplasm at the feeding site and liquefy the cell contents. In most instances, the feeding process causes individual cell death, and can cause physiological, nonlethal changes in other cells. These changes may lead to the leakage of cell contents and, consequently, to the mixing of viruses inhabiting the cells. Afterwards, the viral mixture may be acquired by the feeding nematode. An alternative explanation has also been proposed, in which the complementation may occur inside the nematode oesophagus, with active 2b protein being retained during a first feed, and the virus particle interacting with this protein during subsequent feeding (Vassilakos et al., 2001). As concluded, the process of transmission of tobraviruses by nematodes resembles that of HC‐dependent virus transmission by aphids in a parafilm membrane feeding assay, where HC is physically mixed with viral particles.
Simultaneous Virus Transmission by Fungal Vectors
Although quite a number of soil‐borne plant viruses are transmitted by several species of fungal and plasmodiophorid vectors (Adams et al., 2001; Campbell, 1996; Rochon et al., 2004), the literature on the simultaneous transmission of two viral agents by these vectors is scarce. The fungus‐ and plasmodiophorid‐transmitted viruses include about 20 rod‐shaped species (mainly belonging to the furo‐ and bymoviruses), which are acquired in an in vivo manner and survive in the resting spores of their vectors: the fungus Olpidium brassicae (Wor.) Dangeard and the plasmodiophorid protists Polymyxa graminis L., P. betae Kesk. or Spongospora subterranea (Wallr.) Lagerheim (Campbell, 1996; Rochon et al., 2004). Among the fungal vectors of viruses, O. brassicae has long been known as a vector of three important viruses, Tobacco necrosis virus (TNV), Tobacco stunt virus (TStV) and Lettuce big‐vein associated virus (LBVaV), the latter formerly known as Lettuce big‐vein virus (LBVV) (Hayes et al., 2006; Hiruki, 1994). These three viruses in various combinations were vectored by zoospores of O. brassicae (Hiruki, 1994). Zoospores, obtained by immersing roots of tobacco plants infected with TStV‐Olpidium (tobacco strain) or roots of lettuce plants infected with LBVV‐Olpidium (lettuce strain) in the respective buffer, transmitted TNV that had been added to the zoospores before dip inoculation of mung bean assay plants. TNV was also transmitted by zoospores obtained from virus‐free tobacco or lettuce strains of the fungus. No significant differences in TNV acquisition and transmission between virus‐free Olpidium and the fungus from TStV‐ or LBVV‐infected source plants were found. When inocula, prepared by crushing the roots of mung bean plants inoculated with TNV + TStV‐Olpidium zoospores or TNV + LBVV‐Olpidium zoospores, were poured into soil around tobacco or lettuce plants, TNV proved to be dominant over each of the two other viruses (Hiruki, 1994). The above findings are believed to be the first evidence for multiple transmission of plant viruses using different strains of a fungal vector.
In the field, mixed infections of plants by fungus‐ or plasmodiophorid‐transmitted viruses are not rare. For example, Beet necrotic yellow vein virus (BNYVV) and Beet soil‐borne mosaic virus (BSMV), members of the genus Benyvirus, are widespread and frequently infect the same beet plants (Rush, 2003). As pointed out by the author, the implications of this close vicinity, with regard to disease incidence and severity, and for recombination, are uncertain. It may be added that such close proximity of BNYVV and BSMV may also have significant implications for the transmission of both viruses by their plasmodiophorid vector, P. betae, including the possibility of simultaneous transmission. However, no information on the occurrence of such events to be included in this review has been found.
Final Remarks
As pointed out elsewhere (Syller, 2012), more attention in virological research has long been paid to properties of individual virus species than to intra‐host interactions among viruses in mixed infections. In addition, control strategies have mostly aimed to eliminate vectors and sources of infection rather than to target the interactions between pathosystem components (Killiny et al., 2012). Consequently, many crucial pathogen–vector and pathogen–pathogen relationships remain insufficiently recognized, which may hinder our understanding of the epidemiological aspects of the diseases caused by these pathogens.
The rate of simultaneous vector transmission of two plant viruses, demonstrated in the studies reviewed, was generally low. However, it must be emphasized that simultaneous vector transmission was assessed in experimental conditions. Glasshouse and/or laboratory conditions may not reflect natural conditions, undoubtedly ensuring a greater variety of both virus–vector and virus–virus combinations. It may be assumed that multiple viral transmissions contribute more strongly to the dissemination of these pathogens than is presently thought. Moreover, as mentioned, they may play an important role in the maintenance of genetic diversity in viral populations. However, the above outcomes of the simultaneous transmission of vector‐borne viruses from mixed infections have probably never been evaluated under natural conditions.
It is not rare under natural conditions that two, or more, viruses occur as closely associated species, sharing both hosts and vectors. ToCV and TICV have been found together in tomato, which indicates that infection with one crinivirus does not protect against infection with a second (Wintermantel et al., 2008). The transmission efficiency of these two viruses by the glasshouse whitefly, T. vaporariorum, corresponded with virus titres in both singly and doubly infected hosts. Intriguingly, aphids were found to readily transmit isolates of WMV (Salvaudon et al., 2013) or PVY (Syller and Grupa, 2013), despite the lower (or even undetectable using an immunoenzymatic assay) virus concentration in mixed infected plants. As pointed out by Salvaudon et al. (2013), such findings may be relevant in understanding how two viruses coexist whilst inhabiting very similar ecological niches (i.e. overlapping host and vector ranges). It has been suggested that the fitter of the two viruses competing within the host for suitable replication conditions and vector transmission can even completely eliminate the less fit or unfit competitor (Lecoq et al., 2011; Power, 1996). However, the spatial separation of related viruses in the co‐infected plant generates a specific bottleneck, preventing multiple infection of plant cells by several viral genomes, as discussed in earlier articles (Gutiérrez et al., 2012; Syller, 2012). The evaluation of the kinetics and progress of multiple infections is performed using the multiplicity of infection (MOI) parameter, which determines the number of viral genomes that enter and effectively replicate in a cell. Relevant for viral populations may be the fact that spatial separation reduces the opportunities for competition between different viral genetic variants, and thereby restricts the possibilities to displace unfit variants, which consequently leads to decreasing fitness and competitiveness of the entire population (Elena et al., 2011).
In the present article, it has been attempted to show that multiple transmission of plant viruses by diverse vector organisms is not merely an accidental effect of feeding of the vector in a cell/tissue accidentally containing virions of two different viral species or strains, but, behind this phenomenon, are complex within‐host interactions between the viruses involved, as well as mutual relationships between each of the viruses and the vector. There is increasing evidence that plant viruses may interact directly or indirectly with their insect vectors, modifying their behaviour and/or preferences to enhance their own spread (e.g. Ingwell et al., 2012; Moreno‐Delafuente et al., 2013; Salvaudon et al., 2013; Srinivasan and Alvarez, 2007; Stafford et al., 2011). Hence, the complexity of the pathosystem involving the viral pathogen(s), host(s) and vector(s) makes many of the biological and molecular events in multiple vector transmission difficult to explain on the basis of our current knowledge of mutual relationships between these components, also influenced by numerous biotic and abiotic factors. Therefore, further investigations are needed to obtain a deeper insight into the molecular mechanisms behind these relationships.
Acknowledgements
The author thanks anonymous reviewers for valuable remarks and suggestions to improve the manuscript.
References
- Adams, M.J. , Antoniw, J.F. and Mullins, J.G.L. (2001) Plant virus transmission by plasmodiophorid fungi is associated with distinctive transmembrane regions of virus‐encoded proteins. Arch. Virol. 146, 1139–1153. [DOI] [PubMed] [Google Scholar]
- Andret‐Link, P. and Fuchs, M. (2005) Transmission specificity of plant viruses by vectors. J. Plant Pathol. 87, 153–165. [Google Scholar]
- Anfoka, G.H. and Abhary, M.K. (2007) Occurrence of Tomato infectious chlorosis virus (TICV) in Jordan. Bull. OEPP, 37, 186–190. [Google Scholar]
- Bileva, T. , Choleva, B. , Hockland, S. and Ciancio, A. (2009) Management of virus‐transmitting nematodes with special emphasis on South‐East Europe In: Integrated Management of Fruit Crops and Forest Nematodes (Ciancio A. and Mukerji K.G., eds), pp. 215–242. Dordrecht: Springer Science+Business Media ; B.V. [Google Scholar]
- Blanc, S. (2007) Virus transmission—getting out and in In: Viral Transport in Plants (Waigman E. and Heinlein M., eds), pp. 1–28. Berlin, Heidelberg: Springer. [Google Scholar]
- Blanc, S. , Uzest, M. and Drucker, M. (2011) New research horizons in vector‐transmission of plant viruses. Curr. Opin. Microbiol. 14, 483–491. [DOI] [PubMed] [Google Scholar]
- Brault, V. , Uzest, M. , Monsion, B. , Jacquot, E. and Blanc, S. (2010) Aphids as transport devices for plant viruses. C. R. Biol. 333, 524–538. [DOI] [PubMed] [Google Scholar]
- Campbell, R.N. (1996) Fungal transmission of plant viruses. Annu. Rev. Phytopathol. 34, 87–108. [DOI] [PubMed] [Google Scholar]
- Dalmon, A. , Fabre, F. , Guilbaud, L. , Lecoq, H. and Jacquemond, M. (2009) Comparative whitefly transmission of Tomato chlorosis virus and Tomato infectious chlorosis virus from single or mixed infections. Plant Pathol. 58, 221–227. [Google Scholar]
- Dietrich, C. and Maiss, E. (2003) Fluorescent labelling reveals spatial separation of potyvirus populations in mixed infected Nicotiana benthamiana plants. J. Gen. Virol. 84, 2871–2876. [DOI] [PubMed] [Google Scholar]
- Duffy, S. , Shackelton, L.A. and Holmes, E.C. (2008) Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9, 267–276. [DOI] [PubMed] [Google Scholar]
- Elena, S.F. and Sanjuán, R. (2005) Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 79, 11 555–11 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena, S.F. , Bedhomme, S. , Carrasco, P. , Cuevas, J.M. , de la Iglesia, F. , Lafforgue, G. , Lalić, J. , Pròsper, À. , Tromas, N. and Zwart, M.P. (2011) The evolutionary genetics of emerging plant RNA viruses. Mol. Plant–Microbe Interact. 24, 287–293. [DOI] [PubMed] [Google Scholar]
- Froissart, R. , Michalakis, Y. and Blanc, S. (2002) Helper component‐transcomplementation in the vector transmission of plant viruses. Phytopathology, 92, 576–579. [DOI] [PubMed] [Google Scholar]
- Froissart, R. , Doumayrou, J. , Vuillaume, F. , Alizon, S. and Michalakis, Y. (2010) The virulence–transmission trade‐off in vector‐borne plant viruses : a review of (non‐) existing studies. Philos. Trans. R. Soc. London, B: Biol. Sci. 365, 1907–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, R.W. , Payne, R.W. and Katis, N. (1988) The transmission of potato virus Y by aphids of different vectoring abilities. Ann. Appl. Biol. 113, 35–43. [Google Scholar]
- Gray, S.M. and Banerjee, N. (1999) Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 63, 128–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, S. , Michalakis, Y. and Blanc, S. (2012) Virus population bottlenecks during within‐host progression and host‐to‐host transmission. Curr. Opin. Virol. 2, 546–555. [DOI] [PubMed] [Google Scholar]
- Hall, G.S. and Little, D.P. (2013) Within‐host competition between barley yellow dwarf‐PAV and ‐PAS. Virus Res. 174, 148–151. [DOI] [PubMed] [Google Scholar]
- Hampton, R.O. and Sylvester, E.S. (1969) Simultaneous transmission of two pea viruses by Acyrthosiphon pisum quantified on sweetpea as diagnostic local lesions. Phytopathology, 59, 1663–1667. [PubMed] [Google Scholar]
- Hayes, R.J. , Wintermantel, W.M. , Nicely, P.A. and Ryder, E.J. (2006) Host resistance to Mirafiori lettuce big‐vein virus and Lettuce big‐vein associated virus and virus sequence diversity and frequency in California. Plant Dis. 90, 233–239. [DOI] [PubMed] [Google Scholar]
- Hiruki, C. (1994) Multiple transmission of plant viruses by Olpidium brassicae . Can. J. Plant Pathol. 16, 261–265. [Google Scholar]
- Hogenhout, S.A. , Ammar, E.‐D. , Whitfield, A.E. and Redinbaugh, M.G. (2008) Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46, 327–359. [DOI] [PubMed] [Google Scholar]
- Hooks, C.R. , Fereres, A. and Wang, K.‐H. (2007) Using protector plants to guard crops from aphid‐borne non‐persistent viruses. Soil Crop Manage. 18, 1–7. [Google Scholar]
- Hull, R. (2009) Plant‐to‐plant movement In: Comparative Plant Virology, 2nd edn. (Hull R., ed.), pp. 223–243. Amsterdam: Elsevier Academic ; Press. [Google Scholar]
- Ingwell, L.L. , Eigenbrode, S.D. and Bosque‐Pérez, N.A. (2012) Plant viruses alter insect behavior to enhance their spread. Sci. Rep. 2, 578. doi: 10.1038/srep00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki, M. , Thongmeearkom, P. , Prommin, M. , Honda, Y. and Hibi, T. (1982) Whitefly transmission and some properties of cowpea mild mottle virus on soybean in Thailand. Plant Dis. 66, 365–368. [Google Scholar]
- Killiny, N. , Rashed, A. and Almeida, R.P.P. (2012) Disrupting the transmission of a vector‐borne plant pathogen. Appl. Environ. Microbiol. 78, 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham, J.R. and Wilson, A.K. (2008) Transcomplementation and synergism in plants: implications for viral transgenes? Mol. Plant Pathol. 9, 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq, H. , Fabre, F. , Joannon, B. , Wipf‐Scheibel, C. , Chandeysson, C. , Schoeny, A. and Desbiez, C. (2011) Search for factors involved in the rapid shift in Watermelon mosaic virus (WMV) populations in south‐eastern France. Virus Res. 159, 115–123. [DOI] [PubMed] [Google Scholar]
- Li, J. , Andika, I.B. , Shen, J. , Lv, Y. , Ji, Y. , Sun, L. and Chen, J. (2013) Characterization of Rice black‐streaked dwarf virus‐ and Rice stripe virus‐derived siRNAs in singly and doubly infected insect vector Laodelphax striatellus . PLoS ONE 8, e66007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, R.M. (1964) Strawberry latent ringspot: a new nematode‐borne virus. Ann. Appl. Biol. 54, 167–176. [Google Scholar]
- MacFarlane, S.A. (2003) Molecular determinants of the transmission of plant viruses by nematodes. Mol. Plant Pathol. 4, 211–215. [DOI] [PubMed] [Google Scholar]
- MacFarlane, S.A. , Wallis, C.V. and Brown, D.J.F. (1996) Multiple genes involved in the nematode transmission of pea early browning virus. Virology, 219, 417–422. [DOI] [PubMed] [Google Scholar]
- Makram, M.W. , Schmelzer, K. and Karl, E. (1976) Simultaneous transmission of broad bean wilt and cucumber mosaic viruses by single Myzus persicae (Sulz.). Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. 131, 120–121. [DOI] [PubMed] [Google Scholar]
- Marmonier, A. , Schellenberger, P. , Esmenjaud, D. , Schmitt‐Keichinger, C. , Ritzenthaler, C. , Andret‐Link, P. , Lemaire, O. , Fuchs, M. and Demangeat, G. (2010) The coat protein determines the specificity of virus transmission by Xiphinema diversicaudatum . J. Plant Pathol. 92, 275–279. [Google Scholar]
- Martinière, A. , Bak, A. , Macia, J.‐L. , Lautredou, N. , Gargani, D. , Doumayrou, J. , Garzo, E. , Moreno, A. , Fereres, A. , Blanc, S. and Drucker, M. (2013) A virus responds instantly to the presence of the vector on the host and forms transmission morphs. Elife 2, e00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina‐Ramos, G. , De La Torre‐Almaráz, R. , Bujanos‐Muñiz, R. , Guevara‐González, R.G. , Tierranegra‐García, N. , Guevara‐Olvera, L. , González‐Chavira, M.M. and Torres‐Pacheco, I. (2008) Co‐transmission of Pepper huasteco yellow vein virus and Pepper golden mosaic virus in chilli pepper by Bemisia tabaci (Genn.). J. Entomol. 5, 176–184. [Google Scholar]
- Menzel, W. , Winter, S. and Vetten, H.J. (2010) Complete nucleotide sequence of the type isolate of Cowpea mild mottle virus from Ghana. Arch. Virol. 155, 2069–2073. [DOI] [PubMed] [Google Scholar]
- Moreno‐Delafuente, A. , Garzo, E. , Moreno, A. and Fereres, A. (2013) A plant virus manipulates the behavior of its whitefly vector to enhance its transmission efficiency and spread. PLoS ONE 8, e61543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniyappa, V. and Reddy, D.V.R. (1983) Transmission of cowpea mild mottle virus by Bemisia tabaci in a nonpersistent manner. Plant Dis. 67, 391–393. [Google Scholar]
- Nault, L.R. (1997) Arthropod transmission of plant viruses: a new synthesis. Ann. Entomol. Soc. Am. 90, 521–541. [Google Scholar]
- Navas‐Castillo, J. , Fiallo‐Olivé, E. and Sánchez‐Campos, S. (2011) Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. [DOI] [PubMed] [Google Scholar]
- Ng, J.C. and Falk, B.W. (2006) Virus–vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu. Rev. Phytopathol. 44, 183–212. [DOI] [PubMed] [Google Scholar]
- Ng, J.C.K. and Perry, K.L. (2004) Transmission of plant viruses by aphid vectors. Mol. Plant Pathol. 5, 505–511. [DOI] [PubMed] [Google Scholar]
- Pinto, Z.V. , Rezende, J.A.M. , Yuki, V.A. and de Stefano Piedade, S.M. (2008) Ability of Aphis gossypii and Myzus persicae to transmit Cucumber mosaic virus in single and mixed infection with two potyviruses to zucchini squash. Summa Phytopathol. 34, 183–185. [Google Scholar]
- Pirone, T.P. and Blanc, S. (1996) Helper‐dependent vector transmission of plant viruses. Annu. Rev. Phytopathol. 34, 227–247. [DOI] [PubMed] [Google Scholar]
- Power, A.G. (1996) Competition between viruses in a complex plant–pathogen system. Ecology, 77, 1004–1010. [Google Scholar]
- Purcell, A.H. and Almeida, R.P.P. (2005) Insects as vectors of disease agents In: Encyclopedia of Plant and Crop Science (Goodman R.M., ed.), p. 5 London: Taylor and Francis. [Google Scholar]
- Rentería‐Canett, I. , Xoconostle‐Cázares, B. , Ruiz‐Medrano, R. and Rivera‐Bustamante, R.F. (2011) Geminivirus mixed infection on pepper plants: synergistic interaction between PHYVV and PepGMV. Virol. J. 8, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochon, D. , Kakani, K. , Robbins, M. and Reade, R. (2004) Molecular aspects of plant virus transmission by Olpidium and plasmodiophorid vectors. Annu. Rev. Phytopathol. 42, 211–241. [DOI] [PubMed] [Google Scholar]
- Rochow, W.F. (1972) The role of mixed infections in the transmission of plant viruses by aphids. Annu. Rev. Phytopathol. 10, 101–124. [Google Scholar]
- Rush, C.M. (2003) Ecology and epidemiology of Benyviruses and plasmodiophorid vectors. Annu. Rev. Phytopathol. 41, 567–592. [DOI] [PubMed] [Google Scholar]
- Salvaudon, L. , De Moraes, C.M. and Mescher, M.C. (2013) Outcomes of co‐infection by two potyviruses: implications for the evolution of manipulative strategies. Proc. R. Soc. B 280, 20122959 Available at 10.1098/rspb.2012.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger, P. , Sauter, C. , Lorber, B. , Bron, P. , Trapani, S. , Bergdoll, M. , Marmonier, A. , Schmitt‐Keichinger, C. , Lemaire, O. , Demangeat, G. and Ritzenthaler, C. (2011) Structural insights into viral determinants of nematode mediated Grapevine fanleaf virus transmission. PLoS Pathog. 7, e1002034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence, N.J. (2001) Virus–vector interactions in plant virus disease transmission and epidemiology In: Biotic Interactions in Plant–Pathogen Associations (Jeger M.J. and Spence N.J., eds), pp. 15–26. New York: Oxford University Press. [Google Scholar]
- Sreenivasulu, P. and Demski, J.W. (1988) Transmission of peanut mottle and peanut stripe viruses by Apis craccivora and Myzus persicae . Plant Dis. 72, 722–723. [Google Scholar]
- Srinivasan, R. and Alvarez, J.M. (2007) Effect of mixed viral infections (Potato virus Y–Potato leafroll virus) on biology and preference of vectors Myzus persicae and Macrosiphum euphorbiae (Hemiptera: Aphididae). J. Econ. Entomol. 100, 646–655. [DOI] [PubMed] [Google Scholar]
- Srinivasan, R. , Hall, D.G. , Cervantes, F.A. , Alvarez, J.M. and Whitworth, J.L. (2012) Strain specificity and simultaneous transmission of closely related strains of a Potyvirus by Myzus persicae . J. Econ. Entomol. 105, 783–791. [DOI] [PubMed] [Google Scholar]
- Stafford, C.A. , Walker, G.P. and Ullman, D.E. (2011) Infection with a plant virus modifies feeding behavior. Proc. Natl. Acad. Sci. USA, 108, 9350–9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syller, J. (2003) Molecular and biological features of umbraviruses, the unusual plant viruses lacking genetic information for capsid protein. Physiol. Mol. Plant Pathol. 63, 35–46. [Google Scholar]
- Syller, J. (2006) The roles and mechanisms of helper component proteins encoded by potyviruses and caulimoviruses. Physiol. Mol. Plant Pathol. 67, 119–130. [Google Scholar]
- Syller, J. (2012) Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 13, 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syller, J. and Grupa, A. (2013) The effects of co‐infection by different Potato virus Y (PVY) isolates on virus concentration in solanaceous hosts and efficiency of transmission. Plant Pathol. doi: 10.1111/ppa.12095. [DOI] [Google Scholar]
- Syller, J. , Marczewski, W. and Pawłowicz, J. (1997) Transmission by aphids of potato spindle tuber viroid encapsidated by potato leafroll luteovirus particles. Eur. J. Plant Pathol. 103, 285–289. [Google Scholar]
- Sylvester, E.S. (1985) Multiple acquisition of viruses and vector‐dependent prokaryotes: consequences on transmission. Annu. Rev. Entomol. 30, 71–88. [Google Scholar]
- Tsering, K. and Patel, B.N. (1990) Simultaneous transmission of tobacco leaf curl virus and yellow‐vein mosaic virus of Abelmoschus esculentus (L.) Moench by Bemisia tabaci Genn. Tob. Res. 16, 127–128. [Google Scholar]
- Vassilakos, N. , Vellios, E.K. , Brown, E.C. , Brown, D.J.F. and MacFarlane, S.A. (2001) Tobravirus 2b protein acts in trans to facilitate transmission by nematodes. Virology, 279, 478–487. [DOI] [PubMed] [Google Scholar]
- Vellios, E. , Duncan, G. , Brown, D. and MacFarlane, S. (2002) Immunogold localization of tobravirus 2b nematode transmission helper protein associated with virus particles. Virology, 300, 118–124. [DOI] [PubMed] [Google Scholar]
- Visser, P.B. and Bol, J.F. (1999) Nonstructural proteins of Tobacco rattle virus which have a role in nematode‐transmission: expression pattern and interaction with viral coat protein. J. Gen. Virol. 80, 3272–3280. [DOI] [PubMed] [Google Scholar]
- Wintermantel, W.M. , Cortez, A.A. , Anchieta, A.G. , Gulati‐Sakhuja, A. and Hladky, L.L. (2008) Co‐infection by two criniviruses alters accumulation of each virus in a host‐specific manner and influences efficiency of virus transmission. Phytopathology, 98, 1340–1345. [DOI] [PubMed] [Google Scholar]
- Woolhouse, M.E.J. , Haydon, D.T. and Antia, R. (2005) Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20, 238–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosula, E.N. , Clark, C.A. and Davis, J.A. (2012) Effect of host plant, aphid species, and virus infection status on transmission of Sweetpotato feathery mottle virus . Plant Dis. 96, 1331–1336. [DOI] [PubMed] [Google Scholar]
- Zhang, X.‐S. , Holt, J. and Colvin, J. (2000) Mathematical models of host plant infection by helper‐dependent virus complexes: why are helper viruses always avirulent? Phytopathology, 90, 85–93. [DOI] [PubMed] [Google Scholar]


