Summary
The multigenic Rsv1 locus in the soybean plant introduction (PI) ‘PI96983’ confers extreme resistance against the majority of Soybean mosaic virus (SMV) strains, including SMV‐N, but not SMV‐G7 and SMV‐G7d. In contrast, in susceptible soybean cultivars lacking a functional Rsv1 locus, such as ‘Williams82’ (rsv1), SMV‐N induces severe disease symptoms and accumulates to a high level, whereas both SMV‐G7 and SMV‐G7d induce mild symptoms and accumulate to a significantly lower level. Gain of virulence by SMV‐N on Rsv1‐genotype soybean requires concurrent mutations in both the helper‐component proteinase (HC‐Pro) and P3 cistrons. This is because of the presence of at least two resistance (R) genes, probably belonging to the nucleotide‐binding leucine‐rich repeat (NB‐LRR) class, within the Rsv1 locus, independently mediating the recognition of HC‐Pro or P3. In this study, we show that the majority of experimentally evolved mutational pathways that disrupt the avirulence functions of SMV‐N on Rsv1‐genotype soybean also result in mild symptoms and reduced accumulation, relative to parental SMV‐N, in Williams82 (rsv1). Furthermore, the evaluation of SMV‐N‐derived HC‐Pro and P3 chimeras, containing homologous sequences from virulent SMV‐G7 or SMV‐G7d strains, as well as SMV‐N‐derived variants containing HC‐Pro or P3 point mutation(s) associated with gain of virulence, reveals a direct correlation between the perturbation of HC‐Pro and a fitness penalty in Williams82 (rsv1). Collectively, these data demonstrate that gain of virulence by SMV on Rsv1‐genotype soybean results in fitness loss in a previously susceptible soybean genotype, this being a consequence of mutations in HC‐Pro, but not in P3.
Introduction
Plants have evolved diverse mechanisms, such as dominant resistance (R) proteins encoded by R genes, to recognize invading pathogens, including viruses, and to launch a counter‐attack (Ding and Voinnet, 2007; Palukaitis and Carr, 2008). R gene‐mediated recognition is highly specific and typically operates in a manner described by the ‘gene‐for‐gene’ hypothesis, where a specific R protein recognizes, directly or indirectly, a specific pathogen protein, termed an ‘avirulence factor’ (Bonas and Lahaye, 2002; Collier and Moffett, 2009; Flor, 1971; Van der Hoorn and Kamoun, 2008). Phenotypically, the recognition of plant viruses by R proteins results in the expression of extreme resistance (ER) or a hypersensitive response (HR) (Hull, 2002), both of which are considered to be the consequence of the same recognition event (Bendahmane et al., 1999). Pathogens, such as bacteria and fungi, have evolved various strategies to evade recognition, including the loss or alteration of avirulence factors or the suppression of resistance responses (Abramovitch et al., 2003; Jackson et al., 1999; Kvitko et al., 2009; Leach et al., 2001; Luderer et al., 2002; Schürch et al., 2004; Stahl and Bishop, 2000; Tsiamis et al., 2000). However, as a result of the multifunctionality of viral gene products, minor alteration of the avirulence determinants is the only affordable mechanism available to plant viruses to evade R gene‐mediated recognition (Hull, 2002; Kang et al., 2005).
‘Gene‐for‐gene’ theory predicts that the alteration of a pathogen avirulence protein for gain of virulence on an R‐genotype plant is associated with a fitness cost (Flor, 1971). This is mainly based on the assumption that plant immune systems have evolved to target proteins with important roles in the pathogen life cycle, and that avirulence proteins are often pathogenicity factors that facilitate infection in the absence of a corresponding R gene (i.e. pathogenicity effectors) (Dodds and Rathjen, 2010). Furthermore, the ‘guard hypothesis’ proposes that at least some R proteins detect the activity of these pathogenicity factors in the host cell, rather than recognize the pathogen proteins themselves (Jones and Dangl, 2006). However, experimental studies in support of these predictions are mostly focused on nonviral pathogens that possess specialized pathogenicity and virulence genes (Kjemtrup et al., 2000; Leach et al., 2001; Sacristan and Garcia‐Arenal, 2008; Van't Slot and Knogge, 2002; Vera Cruz et al., 2000). In the case of plant viruses, limited empirical evidence has been reported, in particular when progeny viruses derived from infectious cDNA clones, rather than wild‐type populations, served as the inoculum source (Goulden et al., 1993; Janzac et al., 2010; Jenner et al., 2002a, b; Kobayashi and Hohn, 2004; Rolland et al., 2009). The utilization of progeny viruses derived from cDNA clones is advantageous over wild‐type viruses because it minimizes the chance of second site mutations being included in such analyses (Sacristan and Garcia‐Arenal, 2008). Interestingly, based on theoretical considerations, a higher cost on pathogenicity in susceptible hosts has been predicted for gain of virulence on R‐genotype plants for viruses relative to cellular pathogens (Sacristan and Garcia‐Arenal, 2008). In this study, we have taken advantage of the Soybean mosaic virus (SMV)‐Rsv1 pathosystem to investigate whether gain of virulence on soybean genotypes containing the Rsv1 locus, or Rsv1‐associated R gene(s) (Hayes et al., 2004; Wen et al., 2013), is associated with fitness loss in SMV‐susceptible soybean cultivars.
SMV, a single‐stranded, positive‐sense RNA virus, is a species within the genus Potyvirus belonging to the Potyviridae family. Its genome, approximately 9.6 kb in length, contains a long open reading frame (ORF) and a small overlapping ORF known as pipo (Adams et al., 2005; Chung et al., 2008). On expression, the resultant single large polypeptide is processed post‐translationally by three viral‐encoded proteinases to yield a number of multifunctional proteins, including helper‐component proteinase (HC‐Pro) and P3 (Urcuqui‐Inchima et al., 2001). A small ORF embedded in the P3 cistron (i.e. pipo) has the potential to encode a protein in the +2 frame in relation to the polyprotein ORF and plays a role in virus movement (Vijayapalani et al., 2012; Wen and Hajimorad, 2010).
Rsv1, a single locus in the plant introduction (PI) ‘PI96983’, confers ER against the majority of SMV strains, including SMV‐N (Cho and Goodman, 1979; Hajimorad and Hill, 2001). However, naturally occurring SMV‐G7 and its experimentally evolved variant, SMV‐G7d, overcome Rsv1‐mediated ER (Cho and Goodman, 1979; Hajimorad et al., 2003). Phenotypic responses of PI96983 (Rsv1) to inoculation with SMV‐G7 and SMV‐G7d differ, and are expressed as lethal systemic HR and systemic mosaic, respectively (Hajimorad et al., 2003). The induction of systemic HR by SMV‐G7 probably represents a weak recognition of SMV‐G7 P3 by the Rsv1 locus; however, one cannot exclude the possibility that it is a consequence of delayed induction of the biochemical and physiological events required to arrest SMV at the site of infection (Culver et al., 1991; Dinesh‐Kumar et al., 2000; Hajimorad et al., 2005). Regardless, in interactions with Rsv1‐genotype soybeans, SMV‐N is avirulent, whereas both SMV‐G7 and SMV‐G7d are virulent and are able to spread systemically. Interestingly, in susceptible soybean cultivars lacking a functional Rsv1 locus, such as Williams82 (rsv1), SMV‐N induces severe disease symptoms, whereas both SMV‐G7 and SMV‐G7d induce only mild symptoms (Hajimorad et al., 2003; Zhang et al., 2009).
The P3 protein of SMV‐G7 has been identified as the elicitor of Rsv1‐mediated lethal systemic HR (Hajimorad et al., 2005). However, both HC‐Pro and P3 of SMV‐N are recognized by the Rsv1 locus (Eggenberger et al., 2008; Hajimorad et al., 2006, 2008, 2011). By taking advantage of two soybean recombinant inbred lines (RILs), L800 (3gG2) and L943 (–3gG2), derived from a cross between PI96983 (Rsv1) and a susceptible soybean cultivar, Lee68 (rsv1) (Hayes et al., 2004), we have shown recently that P3 from SMV‐N is recognized by RIL L800 (3gG2), whereas HC‐Pro is recognized by RIL L943 (–3gG2) (Wen et al., 2013). RIL L800 contains a single PI96983‐derived member (3gG2) of an Rsv1‐associated subfamily of nucleotide‐binding leucine‐rich repeat (NB‐LRR) genes, whereas RIL L943 lacks 3gG2, but contains a suite of five other NB‐LRR genes belonging to the same family (Hayes et al., 2004; Wen et al., 2013). Thus, the recognition of HC‐Pro and P3 from SMV‐N in PI96983 (Rsv1) is mediated by distinct R genes at the Rsv1 locus. It should be noted that the P3 cistron of SMV, as in other potyviruses, contains the pipo ORF (Chung et al., 2008; Wen and Hajimorad, 2010); however, PIPO from SMV‐N is not involved in the triggering of the Rsv1‐mediated resistance response (Wen et al., 2011).
Our main objectives in this study were as follows: (i) to test the hypothesis that gain of virulence of SMV on Rsv1‐genotype soybean is associated with fitness loss in previously susceptible soybean genotypes; and (ii) to determine whether fitness loss in soybean genotypes lacking a functional Rsv1 locus is the result of gain of virulence mutations in both HC‐Pro and P3, or in only one of the two proteins.
In this article, we have adopted the definitions of ‘virulence’ and ‘pathogenicity’ according to Shaner et al. (1992) and ‘fitness’ according to Holland et al. (1991). ‘Virulence’ is defined as the ability of an SMV strain, or a variant, to overcome Rsv1‐mediated ER in PI96983 or RIL L800 (3gG2) or to move systemically in RIL L943 (–3gG2) (Wen et al., 2013), regardless of the phenotypic expression. ‘Pathogenicity’ is defined as the capability of an SMV strain, or a variant, to induce severe disease symptoms (i.e. severe mottling, stunting and leaf distortion) in the universally susceptible soybean cultivar Williams82 (rsv1), coupled with a large accumulation of virions or viral RNA. ‘Fitness’ is defined as the ability of an SMV strain, or a variant, to replicate and propagate in Williams82 (rsv1) based on disease phenotype and the accumulation of the SMV virion or SMV RNA.
Results
Virulence of SMV‐G7 and SMV‐G7d on PI96983 (Rsv1) is associated with low pathogenicity in Williams82 (rsv1)
Both SMV‐G7 and SMV‐G7d, unlike avirulent SMV‐N, are virulent on PI96983 (Rsv1), causing lethal systemic HR and systemic mosaic, respectively (Fig. 1A). In contrast with interactions with PI96983 (Rsv1), SMV‐N is highly pathogenic in Williams82 (rsv1), causing severe mottling, moderate stunting and leaf distortion, whereas both SMV‐G7 and SMV‐G7d cause only mild mottling (Fig. 1B). These phenotypic observations are all in agreement with previous reports (Hajimorad and Hill, 2001; Hajimorad et al., 2003; Zhang et al., 2009). It should be noted that the phenotypes of SMV‐N, SMV‐G7 and SMV‐G7d on Lee68 (rsv1), another universally susceptible soybean genotype to SMV inoculation, resemble those on Williams82 (rsv1) (data not shown). The evaluation of virion and viral RNA accumulations in systemically infected leaves of Williams82 (rsv1) by antigen‐coated indirect enzyme‐linked immunosorbent assay (ELISA) and real‐time quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR), respectively, showed that SMV‐G7 and SMV‐G7d accumulated to significantly lower levels than SMV‐N (Fig. 1C,D). Taken together, these observations suggest that the virulence of SMV‐G7 and SMV‐G7d on Rsv1‐genotype soybean is associated with fitness loss in Williams82 (rsv1).
Figure 1.

Phenotypic responses of PI96983 (Rsv1) (A) and Williams82 (rsv1) (B) to mechanical inoculation with progeny viruses derived from molecularly cloned Soybean mosaic virus (SMV) strains SMV‐N (N), SMV‐G7 (G7) and SMV‐G7d (G7d), and comparison of viral accumulations in systemically infected trifoliate leaves of Williams82 (rsv1) by antigen‐coated indirect enzyme‐linked immunosorbent assay (ELISA) (C) and real‐time quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) (D). The inoculated plants were maintained in a growth chamber at 22 °C and photographed at 35 days post‐inoculation (dpi). The accumulation of viruses was evaluated in leaflets from systemically infected trifoliate leaves, but harvested at 28 dpi. Each bar represents the mean values for virions (C) or viral RNA accumulations (D) from three or two independent experiments, respectively, each with five replicate plants, with the standard errors indicated. Optical density (OD at 405 nm) data are net values after deducting the background, whereas the values for viral RNA accumulation are relative to that of SMV‐N (set to 1.0) after normalization to the quantity of soybean ubiquitin transcript. Significant differences in the mean value between the accumulation of SMV‐N and SMV‐G7, as well as between SMV‐N and SMV‐G7d, were determined using Student's t‐test (P < 0.01), and are indicated by asterisks.
The majority of mutational pathways leading to gain of virulence of SMV‐N on PI96983 (Rsv1) result in fitness loss on Williams82 (rsv1)
It has been shown previously, using a comparative genomic analysis approach involving avirulent SMV‐N and virulent SMV‐G7, that an SMV‐N‐derived mutant, SMV‐NR682M+R787I+A947T, harbouring a single mutation in HC‐Pro (R682M) combined with two mutations in P3 (R787I+A947T), is virulent on Rsv1‐genotype soybean (Eggenberger et al., 2008). However, using an experimental evolutionary approach to directly adapt avirulent SMV‐N to virulence on Rsv1‐genotype soybean, Hajimorad et al. (2011) identified different mutational pathways within HC‐Pro and P3 leading to virulence. Nevertheless, all experimentally evolved SMV‐N‐derived virulent mutants also harboured mutations in both HC‐Pro and P3 (Hajimorad et al., 2011; Fig. 2A). Interestingly, phenotypic comparison of these SMV‐N‐derived virulent mutants on Williams82 (rsv1) showed that all, except SMV‐NR682M+R787I+A947T, induced mild symptoms (Figs 2A and 3A), and accumulated to significantly lower levels when compared with the parental SMV‐N (Fig. 2B). In contrast with that seen with all other virulent SMV‐N‐derived mutants, including SMV‐NG319S+K321E+A947V and SMV‐NG319S+K321E+R682M+A947V+K952E+M1157I, the phenotype associated with SMV‐NR682M+R787I+A947T infection on Williams82 (rsv1) was severe (Figs 2A and 3A; data not shown), and its accumulation was comparable with that of parental SMV‐N (Fig. 2B). Interestingly, among the SMV‐N‐derived virulent mutants, SMV‐NR682M+R787I+A947T, unlike all the other mutants, induced lethal systemic HR in PI96983 (Rsv1) (Fig. 3B). It should be noted that SMV‐NG319S+K321E+A947V induced a similar phenotype to that of SMV‐NG319S+K321E+A947V+K952E on PI96983 (Rsv1) (Fig. 3B; data not shown), whereas SMV‐NG319S+K321E+R682M+A947V+K952E+M1157I caused mild stunting and moderate leaf mottling combined with limited systemic necrosis (Fig. S1, see Supporting Information).
Figure 2.
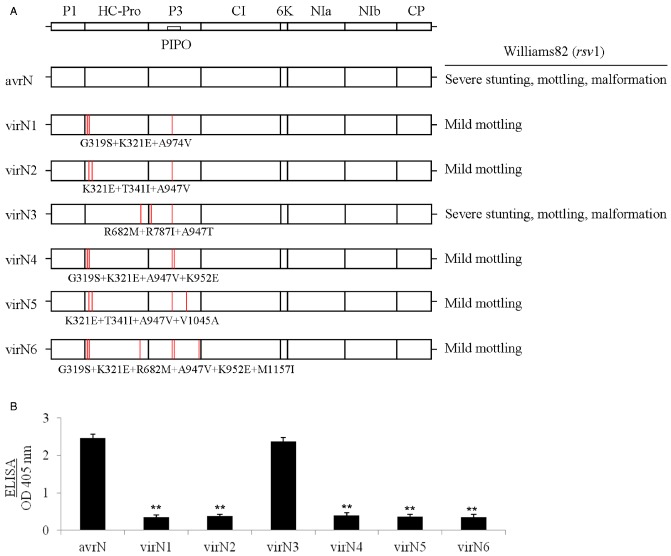
(A) Schematic representation of the genomic maps of Soybean mosaic virus (SMV), avirulent SMV‐N (avrN) and SMV‐N‐derived mutants virulent on Rsv1 genotypes (virN1–virN6) containing mutations (red lines) in both HC‐Pro and P3 (Eggenberger et al., 2008; Hajimorad et al., 2011). The disease phenotypes expressed on Williams82 (rsv1) following mechanical inoculation with progeny viruses derived from molecularly cloned genomes are shown on the right‐hand side. The inoculated plants were maintained in a growth chamber at 22 °C until evaluation for their phenotypic responses at 21 days post‐inoculation (dpi). (B) Comparison of the accumulation of virions of avrN with virulent mutants virN1–virN6 using the antigen‐coated indirect enzyme‐linked immunosorbent assay (ELISA). Systemically infected leaflets from trifoliate leaves of inoculated Williams82 (rsv1) plants were harvested for comparative analysis at 28 dpi. Optical density (OD at 405 nm) data are net values after deducting the background. Each bar represents the mean value of virion accumulation from three independent experiments, each with five replicate plants, with the standard errors indicated. Significant differences between the mean values for avrN and each of the virNs were determined using Student's t‐test (P < 0.01) and are indicated by asterisks.
Figure 3.

Phenotypic responses on Williams82 (rsv1) (A) and PI96983 (Rsv1) (B) following biolistic inoculation with molecularly cloned SMV‐NR682M +R787I+A947T (virN3), SMV‐NK321E +T341I+A947V+V1045A (virN5), SMV‐NK321E +T341I+A947V (virN2) and SMV‐NG319S +K321E+A947V+K952E (virN4). The inoculated plants were maintained in a growth chamber at 22 °C until evaluation at 28 days post‐inoculation.
Gain of virulence mutations in HC‐Pro of SMV‐N, but not those in P3, are the determinants of fitness loss in Williams82 (rsv1)
Simultaneous mutations in both HC‐Pro and P3 are essential for gain of virulence of SMV‐N on Rsv1‐genotype soybean (Eggenberger et al., 2008; Hajimorad et al., 2008, 2011; Wen et al., 2013). To determine whether gain of virulence mutations in HC‐Pro, P3, or both, are responsible for fitness loss on Williams82 (rsv1), we first evaluated the accumulation of SMV‐N‐derived variants harbouring one or two nonsynonymous mutations in HC‐Pro in comparison with parental SMV‐N (Fig. 4A). Only the accumulation of virions of SMV‐NK321E and SMV‐NR682M remained comparable with that of parental SMV‐N, whereas the accumulation of the other remaining HC‐Pro mutants, all virulent on L943 (–3gG2) (Wen et al., 2013), was reduced significantly (Fig. 4A). The phenotypes of SMV‐NK321E and SMV‐NR682M on Williams82 (rsv1) resembled that of parental SMV‐N, whereas those of the remaining mutants were expressed as mild mottling (data not shown). Interestingly, the K321E mutation in HC‐Pro, combined with R945G or A947V (both mutations in P3; Hajimorad et al., 2011), did not influence the pathogenicity or accumulation level of SMV‐NK321E+R945G or SMV‐NK321E+A947V in Williams82 (rsv1) (Fig. S2, see Supporting Information; data not shown). It should be noted that SMV‐NK321E remained avirulent on L943 (–3gG2), whereas SMV‐NR682M was virulent; however, SMV‐NR682M induced systemic necrosis (Wen et al., 2013; data not shown).
Figure 4.
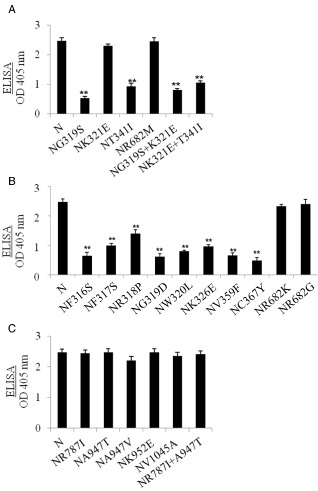
Comparison of virion accumulation in Williams82 (rsv1) of SMV‐N‐derived variants containing mutations in the HC‐Pro cistron (A, B) and P3 cistron (C) using antigen‐coated indirect enzyme‐linked immunosorbent assay (ELISA). Plants were inoculated mechanically with progeny viruses derived from molecularly cloned genomes and maintained in a growth chamber at 22 °C until trifoliate tissues were harvested at 28 days post‐inoculation. Each bar represents the mean value of virion accumulation from three independent experiments, each with five replicate plants, with the standard errors indicated. Significant differences between the mean accumulation values of SMV‐N and each of its derivative mutants were determined using Student's t‐test (P < 0.01) and are indicated by asterisks.
We also evaluated the pathogenicity on Williams82 (rsv1) of additional SMV‐N‐derived HC‐Pro mutants harbouring single nonsynonymous mutations in HC‐Pro (Fig. 4B). These HC‐Pro mutations were found to be associated with virulence on Rsv1‐genotype soybean of SMV‐N‐derived chimeras containing P3 sequences from the virulent SMV‐G7 and SMV‐G7d strains (Hajimorad et al., 2008). Except for SMV‐NR682K and SMV‐NR682G, which accumulated to levels comparable with parental SMV‐N, the other mutants, all virulent on L943 (–3gG2) (Wen et al., 2013), accumulated to significantly lower levels in Williams82 (rsv1) compared with parental SMV‐N (Fig. 4B). The pathogenicity phenotypes (i.e. the severity of disease symptoms caused) of all of these mutants on Williams82 (rsv1) showed a direct correlation with the level of each virus (data not shown).
To determine whether gain of virulence on RIL L800 (3gG2) mutations in the P3 cistron of SMV‐N, individually or in combination (Eggenberger et al., 2008; Hajimorad et al., 2011; Wen et al., 2013), affected pathogenicity on Williams82 (rsv1), we evaluated a number of SMV‐N‐derived P3 mutants. Interestingly, the disease phenotypes of all the SMV‐N‐derived P3 mutants on Williams82 (rsv1) resembled that of parental SMV‐N (data not shown), and all accumulated to a comparable level with that of SMV‐N (Fig. 4C, S2). We also evaluated the pathogenicity of two additional SMV‐N‐derived P3 mutants on Williams82 (rsv1) with gain of virulence on RIL L800 (3gG2) mutations in the P3 cistron (Hajimorad et al., 2011; Wen et al., 2013) (Fig. S2). Both SMV‐NR945G and SMV‐NP948L accumulated to comparable levels to that of parental SMV‐N, and induced similar severe phenotypes on Williams82 (rsv1) (Fig. S2; data not shown).
HC‐Pro, but not P3, of the SMV‐G7 and SMV‐G7d strains is responsible for the modulation of pathogenicity on Williams82 (rsv1)
To demonstrate that the genetic determinants of SMV‐G7 and SMV‐G7d which reduce pathogenicity on Williams82 (rsv1) also reside in HC‐Pro, but not in P3, we evaluated the pathogenicity of SMV‐G7‐ and SMV‐G7d‐derived chimeras. In these experiments, the pathogenicity of SMV‐G7‐ and SMV‐G7d‐derived chimeras containing a precise replacement of the HC‐Pro cistron with that from SMV‐N was compared with that of parental viruses (Fig. 5A). Both the SMV‐G7/NHC‐Pro and SMV‐G7d/NHC‐Pro chimeras induced severe disease phenotypes in Williams82 (rsv1) (Fig. 5A), and accumulated to significantly higher levels than parental SMV‐G7 and SMV‐G7d, irrespective of whether the accumulation of virions or viral RNA was measured (Fig. 5B,D, respectively). In the reciprocal experiments, when SMV‐N‐derived HC‐Pro chimeras containing a precise replacement of the HC‐Pro cistron with that from SMV‐G7 or SMV‐G7d were inoculated onto Williams82 (rsv1), SMV‐N/G7HC‐Pro and SMV‐N/G7dHC‐Pro both induced mild disease phenotypes (Fig. 5A). Consistent with the disease phenotypes, their respective virions and RNAs both accumulated to significantly lower levels than that of parental SMV‐N (Fig. 5B,D, respectively).
Figure 5.
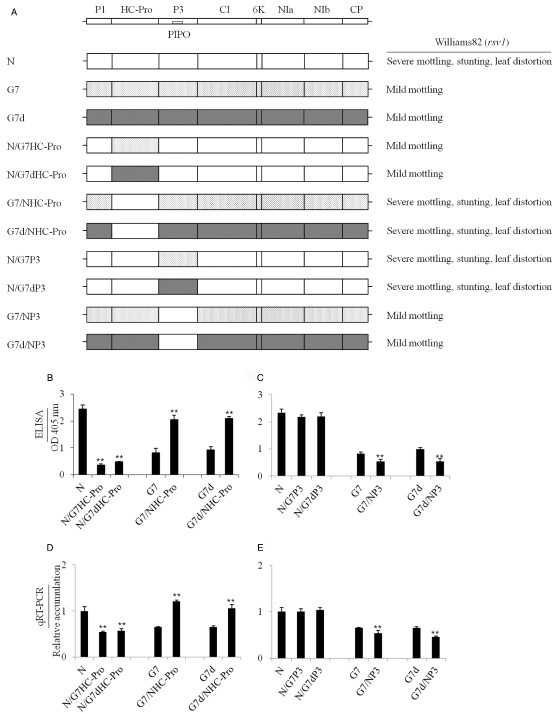
(A) Schematic representation of the genomic map of Soybean mosaic virus (SMV), parental SMV‐N (N), SMV‐G7 (G7) and SMV‐G7d (G7d), and their derivative HC‐Pro and P3 chimeras, with precise exchanges of each of the two cistrons (Hajimorad et al., 2006; Wen et al., 2013). Williams82 (rsv1) plants were inoculated mechanically with progeny viruses derived from molecularly cloned genomes, maintained in a growth chamber at 22 °C and the induced phenotypes shown on the right‐hand side were recorded at 21 days post‐inoculation (dpi). (B–E) Accumulation levels of parental viruses were compared with their derivative chimeras using antigen‐coated indirect enzyme‐linked immunosorbent assay (ELISA) (B, C) or real‐time quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) (D, E). Leaflets from systemically infected trifoliate leaves were harvested at 28 dpi for comparison. Each bar represents the mean values for virion accumulation (B, C) or viral RNA (D, E) from three and two independent experiments, respectively, each with five replicate plants, with the standard errors indicated. Optical density (OD at 405 nm) (B, C) data are net values after deducting the background, whereas those of viral RNA accumulation (D, E) are relative to SMV‐N (set to 1.0) after normalization to the quantity of soybean ubiquitin transcript. Significant differences between the mean values of the parental viruses and their derivative chimeras were determined using Student's t‐test (P < 0.01) and are indicated by asterisks.
To demonstrate that the P3 cistrons of SMV‐G7 and SMV‐G7d do not modulate pathogenicity on Williams82 (rsv1), we evaluated SMV‐G7‐ and SMV‐G7d‐derived chimeras containing a precise replacement of P3 with the corresponding region from SMV‐N (Hajimorad et al., 2006). The disease phenotypes associated with both the SMV‐G7/NP3 and SMV‐G7d/NP3 chimeras were mild and similar to that of the parental viruses (Fig. 5A); however, surprisingly, the chimeric viruses accumulated to significantly lower levels than the parents (Fig. 5C). The lower level of virion accumulation correlated with a lower level of viral RNA (Fig. 5E). In the reciprocal experiment, the disease phenotypes resulting from the inoculation of Williams82 (rsv1) with SMV‐N‐derived P3 chimeras containing a precise replacement of P3 with the corresponding region from SMV‐G7 and SMV‐G7d resembled that of parental SMV‐N (Fig. 5A). However, in this case, no significant differences in virion or viral RNA accumulation of SMV‐N/G7P3 or SMV‐N/G7dP3 chimeras were observed in comparison with that of parental SMV‐N (Fig. 5C,E). This is consistent with the unaltered pathogenicity of the SMV‐N‐derived P3 mutants on Williams82 (rsv1) (Figs 4C and S2).
Discussion
To control viral diseases of plants, the deployment of R genes is the most effective, environmentally friendly and cost‐efficient approach (Hull, 2002; Palukaitis and Carr, 2008). However, the development of elite resistant cultivars containing R genes is lengthy and costly; hence, plant breeders strive to select durable R genes. The durability of R genes against plant viruses, in particular RNA viruses, appears, at first glance, to be a challenge because of the plasticity of viruses as a direct consequence of error‐prone replication, large population size and short generation time (Domingo and Holland, 1997; Drake and Holland, 1999). Nevertheless, factors such as the number of mutations required for gain of virulence, evolutionary constraints acting against amino acid substitutions in avirulence factors and the high fitness cost associated with gain of virulence contribute to the durability of R genes (Bornemann and Varrelmann, 2013; Harrison, 2002; Janzac et al., 2009, 2010).
In proposing the ‘gene‐for‐gene’ theory, Flor (1971) predicted that R genes recognizing easily mutable genes of pathogens would be less durable than those targeting rarely mutated genes. This is probably a direct consequence of a high fitness cost to the pathogen for gain of virulence (Leach et al., 2001). Among viral pathosystems involving R‐genotype plants, the Ry gene in potato against Potato virus Y (PVY) is extremely durable because it recognizes an element that is essential for virus replication (Mestre et al., 2003). In the case of the durable potato Rx gene, which is effective against Potato virus X (PVX), only a single naturally occurring virulent strain has been isolated worldwide (Moreira et al., 1980). Here, a requirement for two simultaneous mutations in the coat protein (CP) of PVX for gain of virulence, and the associated fitness loss in susceptible hosts as a consequence (Goulden et al., 1993), possibly have limited the emergence of virulent strains. Tobacco mosaic virus (TMV) also cannot easily overcome the N′ gene of tobacco, because the alteration of the TMV CP required for the evasion of N′‐mediated recognition prevents virus assembly, and consequently inhibits systemic movement (Taraporewala and Culver, 1996).
In the SMV‐Rsv1 system, SMV‐G7 is the only naturally occurring virulent strain reported from North America to date (Cho and Goodman, 1979; Khatabi et al., 2012). Elsewhere in the world, naturally occurring SMV strains virulent on Rsv1‐genotype soybean have emerged only in South Korea, where soybean cultivars containing Rsv1 alleles were planted on a large scale (Choi et al., 2005). It should be noted that the Rsv1 locus has nine alleles with various specificities against SMV strains (Moon et al., 2009). Our main objectives in this study were to determine whether SMV gain of virulence on Rsv1‐expressing cultivars is associated with fitness loss in susceptible soybean cultivars, and to identify the SMV genetic determinant(s) responsible for loss of fitness.
Rsv3, another R gene in soybean effective against SMV, also confers ER and HR to some strains of the virus (Seo et al., 2009a; Zhang et al., 2009), but virulent strains on Rsv3, unlike Rsv1, appear to be widespread in the USA and elsewhere (Khatabi et al., 2012; Seo et al., 2009b; Viel et al., 2009). The strain‐specific cytoplasmic inclusion (CI) protein of SMV, a genetically variable cistron, can act as the avirulence factor detected by Rsv3, and even a single amino acid substitution in CI can functionally convert a strain from avirulence to virulence (Seo et al., 2009a, b; Zhang et al., 2009). Interestingly, SMV‐N, a highly pathogenic strain on susceptible soybean, is virulent on Rsv3, whereas the attenuated SMV‐G7 strain is avirulent (Khatabi et al., 2012; Zhang et al., 2009). Thus, it appears that there is no significant fitness loss to SMV‐N in susceptible soybean as a consequence of gain of virulence on Rsv3 genotypes. Probably, a lack of a fitness cost in susceptible hosts, the variability of the CI cistron and a small number of amino acid substitutions required for gain of virulence, have collectively contributed negatively to the durability of Rsv3 in soybean.
In contrast with Rsv3, gain of virulence on Rsv1‐genotype soybean requires multiple simultaneous mutations in both HC‐Pro and P3 (Eggenberger et al., 2008; Hajimorad et al., 2008, 2011; Wen et al., 2013). Both HC‐Pro and P3 are highly variable among the SMV cistrons (Seo et al., 2009b). However, as shown in this article, a significant reduction in virus accumulation and disease symptom amelioration in susceptible soybean cultivars are associated with gain of virulence on Rsv1. SMV is transmitted naturally by aphids and is also seed borne (Domier et al., 2007). In other viral pathosystems, virus concentration in the source plant has been shown to influence vector transmission efficiency (Banik and Zitter, 1990; Gray et al., 1991). Probably, as a result of the fitness penalty in susceptible soybean, strains of SMV virulent on Rsv1 do not have an advantage over avirulent isolates with respect to aphid transmission. It should be noted that HC‐Pro is involved directly in aphid transmission of potyviruses (Urcuqui‐Inchima et al., 2001), and gain of virulence mutations in HC‐Pro may also negatively impact directly its interaction with aphids. Hence, as a consequence of a requirement for concurrent multiple mutations in HC‐Pro and P3 to evade detection, loss of fitness in susceptible soybean associated with gain of virulence and, possibly, inefficient vector transmission of virulent strains, it appears that Rsv1‐mediated resistance will be relatively durable.
The Rsv1 locus is complex and contains distinct R genes which recognize independently HC‐Pro or P3 (Hayes et al., 2004; Wen et al., 2013). The P3‐specific R gene is present in RIL L800 and is associated with a member (3gG2) of a previously defined family of NB‐LRR genes (Hayes et al., 2004). Interestingly, none of the gain of virulence mutations in P3 resulted in fitness loss in Williams82 (rsv1), despite the fact that the majority of these mutations conferred virulence to SMV‐N on RIL L800 (3gG2) (Hajimorad et al., 2011; Wen et al., 2011, 2013). On Rsv1‐genotype soybean, such as PI96983 or L78‐379, some of these mutations, including R787I, A947T and A947V, were essential, and played a primary role in virulence, whereas others, such as R945G, P948L, K952E and V1045A, played a secondary role (Eggenberger et al., 2008; Hajimorad et al., 2011; Wen et al., 2013). It should be noted that all of these SMV‐N‐derived P3 mutants were virulent on RIL L800 (3gG2) and induced systemic HR (Hajimorad et al., 2011; Wen et al., 2013). One may argue that the induction of systemic HR by virulent SMV‐N‐derived P3 mutants on RIL L800 (3gG2) is an indication that the P3 structure has not been altered drastically to evade completely 3gG2‐mediated recognition; hence, the mutants remained as pathogenic on Williams82 (rsv1) as the parental SMV‐N. However, SMV‐G7d, unlike SMV‐G7, caused systemic mottling on Rsv1‐genotype soybean and RIL L800 (3gG2), indicating that SMV‐G7d evades Rsv1‐associated R‐gene recognition successfully (Hajimorad et al., 2003; Wen et al., 2011). Nevertheless, SMV‐G7d, similar to SMV‐G7, is poorly pathogenic on Williams82 (rsv1) (Fig. 1; Hajimorad et al., 2003). Interestingly, replacement of P3 in SMV‐G7 and SMV‐G7d with the same cistron from SMV‐N not only failed to enhance the pathogenicity of SMV‐G7/NP3 or SMV‐G7d/NP3 in Williams82 (rsv1), but, for an unknown reason, even had a negative impact on the accumulation of the two chimeras. However, P3 from SMV‐G7 or SMV‐G7d did not alter the pathogenicity of SMV‐N on Williams82 (rsv1). The P3 sequences of SMV‐G7 and SMV‐G7d differ from that of SMV‐N by 23 and 27 amino acids, respectively (Hajimorad et al., 2006). These observations collectively suggest that pathogenicity determinants of SMV on Williams82 (rsv1) do not reside on P3. Gain of virulence mutation in P3 of another potyvirus resulted in fitness loss on susceptible plants; however, the underlying mechanism remains unknown (Desbiez et al., 2003). P3 is known to play critical roles in potyviral replication, including movement, pathogenicity, virulence/avirulence activities and interacts physically with HC‐Pro, CI and nuclear inclusion b proteins (Cui et al., 2010; Eggenberger et al., 2008; Hajimorad et al., 2005; Jenner et al., 2003; Khatabi et al., 2012; Kim et al., 2010; Urcuqui‐Inchima et al., 2001; and references cited therein). Interestingly, all gain of virulence mutations in P3 are positioned within a short stretch of amino acids on the SMV‐N polypeptide. Among these mutants, SMV‐NA947T is the only mutant with altered P3 and PIPO proteins. However, it appears that the pipo ORF does not play a role in virulence on Rsv1‐genotype soybean (Wen et al., 2011).
In contrast with substitutions in P3, most gain of virulence mutations in HC‐Pro resulted in fitness loss on Williams 82 (rsv1). Only one of these mutations is located at the C‐terminus at polypeptide position 682, whereas all the others are clustered at the N‐terminus within a short stretch of 51 residues (polypeptide residues 316–367). HC‐Pro is 457 amino acids in length and corresponds to SMV‐N polyprotein residues 309–765 (GenBank Accession Number D00507). Eggenberger et al. (2008) reported that the replacement of lysine with methionine at position 682 of the SMV‐N polypeptide, combined with two additional mutations in P3 (R787I+A947T), conferred virulence to SMV‐N on Rsv1. However, subsequent experimental adaptation of SMV‐N to Rsv1‐genotype soybean showed that the mutation at position 682 probably plays a secondary role in virulence (Hajimorad et al., 2008, 2011). Consistent with this idea, SMV‐NR682M, SMV‐NR682G and SMV‐NR682K were all virulent on RIL L943 (–3gG2), but provoked systemic HR (data not shown). This observation suggests that substitution at position 682 does not alter HC‐Pro structure sufficiently to allow SMV‐N to completely evade recognition mediated by Rsv1‐associated R gene(s) in RIL L943 (–3gG2).
Unlike substitution at position 682, the majority of HC‐Pro mutations at the N‐terminus, except K321E, resulted in mild mottling in systemically infected RIL L943 (–3gG2) (Wen et al., 2013; data not shown). Most of the mutations at the N‐terminus played a primary role in experimental adaptations of SMV‐N or SMV‐N‐derived chimeras containing P3 sequences from SMV‐G7 or SMV‐G7d on Rsv1‐genotype soybean (Hajimorad et al., 2008, 2011). Interestingly, SMV‐NK321E was the only mutant that displayed a similar pathogenicity phenotype on Williams82 (rsv1) as that of parental SMV‐N. This is in line with its lack of virulence on RIL 943 (–3gG2) (Wen et al., 2013). Nevertheless, the K321E substitution is critical for virulence of SMV‐N on Rsv1‐genotype soybean (Hajimorad et al., 2011; Wen et al., 2013). Regardless, the functional roles of HC‐Pro mutations in virulence on Rsv1‐genotype soybean and in fitness loss on Williams82 (rsv1) remain unknown. In a number of other potyviral pathosystems, HC‐Pro also plays a role in virus accumulation and symptom expression (Atreya et al., 1992; Desbiez et al., 2010; Dolja et al., 1993; Gal‐On, 2000; Lim et al., 2007; Shiboleth et al., 2007; Stenger et al., 2006; Wu et al., 2010). Furthermore, HC‐Pro acts at other levels during potyviral infection, including avirulence activity (Faurez et al., 2012; Moury et al., 2011; Tian and Valkonen, 2013; Wen et al., 2013), genome amplification and movement (Kasschau et al., 1997; Urcuqui‐Inchima et al., 2001), suppressor of gene silencing (Ebhardt et al., 2005; Mérai et al., 2006; Moissiard et al., 2007) and disease synergism (Shi et al., 1997). In the SMV‐Rsv1 system, it appears that the virulence function of SMV HC‐Pro does not affect its synergistic activity, as transient expression of SMV‐G7 HC‐Pro via a Bean pod mottle virus‐based vector showed that it is still capable of induction of synergistic disease symptoms in soybean (Zhang and Ghabrial, 2006). This is in contrast with HC‐Pro from Plum pox virus, in which the protein's function in symptomatology and synergism has overlapping determinants (Sáenz et al., 2001). It should be noted that potyviral HC‐Pro is not the only viral suppressor of gene silencing that serves as a target for recognition by an R gene (Choi et al., 2004; Kobayashi and Hohn, 2004; Scholthof, 2006). However, a link between avirulence activity and suppression of gene silencing activity has been documented in only two pathosystems (Ishibashi et al., 2011; Ronde et al., 2013). At present, it remains unknown whether any of the gain of virulence mutations in HC‐Pro of SMV‐N impact on its gene silencing suppression function.
The Rsv1 locus represents an intriguing example of a co‐evolutionary arms race between soybean and SMV. R genes at the locus have evolved not only to target two genetically unrelated SMV cistrons (Wen et al., 2013), but also to ensure cost is associated with gain of virulence, resulting in co‐survival of both soybean and SMV. This is supported by the phenotype of SMV‐G7d on Rsv1‐genotype soybean, as well as on susceptible soybean genotypes (Hajimorad et al., 2003; Wen et al., 2013). One can speculate that the Rsv1‐associated R gene recognizing P3 (i.e. 3gG2) evolved earlier than the R gene(s) targeting HC‐Pro for recognition. This is mainly because of a lack of significant fitness loss in susceptible Williams82 (rsv1) as a result of P3 perturbation. The mechanism(s) of recognition of HC‐Pro and P3 by Rsv1‐associated R genes may also differ. However, currently, knowledge on the interactions between the Rsv1‐associated R proteins and HC‐Pro or P3 is lacking, and it is unknown whether perception of these SMV avirulence factors by the cognate R proteins is via a direct ligand receptor‐ (Flor, 1971), guard‐ (Jones and Dangl, 2006) or decoy‐ (Van der Hoorn and Kamoun, 2008) type mechanism. Scenarios can be envisioned in which any of these mechanisms could explain gain of virulence mutations that result in loss of fitness on previously susceptible soybean genotypes as SMV evolves to avoid recognition. Future research will focus on the mechanisms of recognition of HC‐Pro and P3 by the R proteins encoded by R genes at the Rsv1 locus.
Experimental Procedures
Viruses, soybean genotypes and inoculation
Molecularly cloned SMV‐N, SMV‐G7 (AY216010) and SMV‐G7d (AY216987) parental strains, and HC‐Pro and P3 chimeras with precise exchanges of each of the two cistrons among the three viruses, have been described previously (Hajimorad et al., 2003; Wang et al., 2006; Wen et al., 2013). SMV‐N‐derived HC‐Pro and P3 mutants containing one or more nonsynonymous mutation(s) in either of the cistrons, or a combination of one or more nonsynonymous mutations in both of the cistrons, have also been described previously (Eggenberger et al., 2008; Hajimorad et al., 2006, 2008, 2011). Except where stated otherwise, progeny viruses derived from molecularly cloned genomes, and passaged twice in soybean (Glycine max) cultivar Williams82 (rsv1), served as the inoculum source. To prepare a stock of inoculum, unifoliate leaves of Williams82 (rsv1) were mechanically inoculated with sap extract containing progeny viruses derived from biolistically inoculated Williams82 (rsv1). Mechanical and biolistic inoculations were performed essentially as described previously (Hajimorad and Hill, 2001; Hajimorad et al., 2003, 2008). The inoculated plants were kept in a growth chamber at 22 °C with a photoperiod of 16 h until the first and second fully developed trifoliate leaves were harvested at 14 days post‐inoculation (dpi), combined, pulverized in the presence of liquid nitrogen and stored at –80 °C. To inoculate plants for comparative analysis, sap was extracted from the frozen pulverized leaf tissues in the presence of 50 mm phosphate buffer, pH 7.0, with a ratio of 1:5 (w/v), the suspension was clarified by centrifugation at 2655 g for 2 min at 4 °C and 20 μL were rub inoculated on each of the unifoliate leaves to be inoculated. Five replicate plants were inoculated with each inoculum in each experiment. Soybean line PI96983 (Rsv1) was also used in this study (Bernard et al., 1991).
Evaluation of virion accumulation by ELISA
To quantify virion accumulation, ELISA, as described previously by Malapi‐Nelson et al. (2009), was used. Each of the viruses was mechanically inoculated onto five replicate plants, as described above, and the experiment was repeated three times. To collect tissues for analysis, the central leaflets from four fully developed trifoliate leaves positioned at the top of each of the inoculated plants were harvested at 28 dpi. The leaflets from the same plant were combined, pulverized in the presence of liquid nitrogen and stored at −80 °C. To prepare antigen, the frozen pulverized tissues were homogenized in the presence of carbonate buffer, pH 9.6, with a ratio of 1:20 (w/v), the suspension was clarified by centrifugation at 2655 g for 2 min at 4 °C, and the supernatant was used as the antigen. Polyclonal antibodies against the SMV virion (Malapi‐Nelson et al., 2009), and anti‐rabbit alkaline phosphatase conjugate (Sigma‐Aldrich, St. Louis, MO, USA), were each used at a dilution of 1:2000 in phosphate‐buffered saline containing 0.05% Tween 20 and 5% nonfat dry milk, pH 7.4. p‐Nitrophenyl phosphate served as the substrate (Sigma‐Aldrich), and the optical density (OD) at 405 nm was recorded with a Microplate Reader Model 680 (Bio‐Rad, Hercules, CA, USA). Significant differences in the accumulation of viruses were determined using Student's t‐test.
Evaluation of viral RNA accumulation by qRT‐PCR
Each of the viruses was mechanically inoculated onto five replicate plants, as described above, and the central leaflets from four fully developed trifoliate leaves positioned at the top of each plant were harvested at 28 dpi. Leaflets from each treatment were combined, pulverized in the presence of liquid nitrogen and stored at −80 °C. Each experiment was repeated twice. Total RNA was isolated from ∼100 mg of pulverized tissue using the RNeasy Plant Mini Kit (QIAGEN, Valencia, CA, USA) and quantified using a NanoDrop ND‐1000 UV/VIS spectrophotometer (NanoDrop, Wilmington, DE, USA). Total RNA (500 ng) from each treatment was reverse transcribed in 20‐μL reactions using the high‐capacity cDNA reverse transcription (RT) kit in the presence of random primers (Applied Biosystems, Foster City, CA, USA). RT reaction was incubated at 55 °C for 30 min, followed by 5 min of incubation at 85 °C. Soybean ubiquitin transcript was used as an internal control for normalization, and was quantified using the primers UBI3‐for (5′‐GTGTAATGTTGGATGTGTTCCC‐3′) and UBI3‐rev (5′‐ACACAATTGAGTTCAACACAAACCG‐3′) (Mazarei et al., 2007). The SMV primers, directed at CP nucleotide sequences conserved among SMV‐N, SMV‐G7 and SMV‐G7d, were SMV CP‐for (5′‐GCAGCTCTCTCGGGAGTTAACA‐3′) and SMV CP‐rev (5′‐CCTTGCAGTGTGCCTTTCAG‐3′). These nucleotides correspond to sequences 9192–9213 and 9281–9260 in the SMV‐N genome, respectively. Each primer set amplified a single product, as confirmed by the melting temperature of the amplicons. qRT‐PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems) according to the manufacturer's protocol. PCRs in triplicate were performed in a 96‐well reaction block in the presence of SYBR Green, essentially as described by Mazarei et al. (2003). The PCR programme consisted of 20 min at 25 °C, followed by one cycle of 10 min at 95 °C and, finally, 40 cycles of 3 s at 95 °C, 5 s at 60 °C and 18 s at 72 °C. The standard curve method was used for relative quantification, and the accumulation of viral RNAs was normalized to the quantity of soybean ubiquitin transcripts according to Pfaffl (2001). For quality assurance purposes, only qRT‐PCR assays that resulted in standard curves with the following parameters were considered: (i) linear standard curve throughout the measured area; (ii) standard curve slope between –3.5 and –3.2; and (iii) R2 value above 0.99 (Bustin, 2002). Significant differences in the accumulation of viral RNA were determined using Student's t‐test.
Supporting information
Fig. S1 The phenotype expressed on soybean plant introduction (PI) ‘PI96983 (Rsv1)’ following mechanical inoculation with progeny viruses derived from the molecularly cloned genome of SMV‐NG319S+K321E+R682M+A947V+K952E+M1157I (virN6). The inoculated plants were maintained in a growth chamber at 22 °C until being photographed at 21 days post‐inoculation.
Fig. S2 Comparison of accumulation levels of virions of SMV‐N (N) with SMV‐N‐derived P3 mutants or SMV‐N‐derived mutants with a single mutation in P3 (R945G or A947V) in combination with an amino acid substitution in HC‐Pro (K321E) in Williams82 (rsv1) using the antigen‐coated indirect enzyme‐linked immunosorbent assay (ELISA). Plants were inoculated mechanically with progeny viruses derived from molecularly cloned genomes, maintained in a growth chamber at 22 °C until trifoliate leaves were harvested at 28 days post‐inoculation. Each bar represents the mean value of virion accumulation from three independent experiments, each with five replicate plants, with the standard errors indicated. No significant differences in the accumulation level of SMV‐N relative to that of each of the mutants were detected by Student's t‐test (P < 0.01). OD, optical density.
Acknowledgements
This project was supported in part by the University of Tennessee College of Agricultural Sciences and Natural Resources, the Tennessee Agricultural Experimental Station, Knoxville, the North Central Soybean Research Program and the Tennessee Soybean Promotion Board. We are grateful to Drs T. Ashfield (Indiana University, Bloomington, IN, USA) and A. Kachroo (University of Kentucky, Lexington, KY, USA) for critical reading of the manuscript.
References
- Abramovitch, R.B. , Kim, Y.‐J. , Chen, S. , Dickman, M.B. and Martin, G.B. (2003) Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22, 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, M.J. , Antoniw, J.F. and Fauquet, C.M. (2005) Molecular criteria for genus and species discrimination within the family Potyviridae . Arch. Virol. 150, 459–479. [DOI] [PubMed] [Google Scholar]
- Atreya, C.D. , Atreya, P.L. , Thornbury, D.W. and Pirone, T.P. (1992) Site‐directed mutations in the potyvirus HC‐Pro gene affect helper component activity, virus accumulation, and symptom expression in infected tobacco plants. Virology, 191, 106–111. [DOI] [PubMed] [Google Scholar]
- Banik, M.T. and Zitter, T.A. (1990) Determination of cucumber mosaic virus titer in muskmelon by enzyme‐linked immunosorbent assay and correlation with aphid transmission. Plant Dis. 74, 857–859. [Google Scholar]
- Bendahmane, A. , Kanyuka, K. and Baulcombe, D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell, 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, R.L. , Nelson, R.L. and Cremeens, C.R. (1991) USDA soybean genetic collection: isoline collection. Soybean Genet. Newsl. 18, 27–57. [Google Scholar]
- Bonas, U. and Lahaye, T. (2002) Plant disease resistance triggered by pathogen‐derived molecules: refined models of specific recognition. Curr. Opin. Microbiol. 5, 44–50. [DOI] [PubMed] [Google Scholar]
- Bornemann, K. and Varrelmann, M. (2013) Effect of sugar beet genotype on the Beet necrotic yellow vein virus P25 pathogenicity factor and evidence for a fitness penalty in resistance‐breaking strains. Mol. Plant Pathol. 14, 356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin, S.A. (2002) Quantification of mRNA using real‐time reverse transcription PCR (RT‐PCR): trends and problems. J. Mol. Endocrinol. 29, 23–29. [DOI] [PubMed] [Google Scholar]
- Cho, E.‐K. and Goodman, R.M. (1979) Strains of soybean mosaic virus: classification based on virulence in resistant soybean cultivars. Phytopathology, 69, 467–470. [Google Scholar]
- Choi, B.K. , Koo, J.M. , Ahn, H.J. , Yum, H.J. , Choi, C.W. , Ryu, K.H. , Chen, P. and Tolin, S.A. (2005) Emergence of Rsv‐resistance breaking Soybean mosaic virus isolates from Korean soybean cultivars. Virus Res. 112, 42–51. [DOI] [PubMed] [Google Scholar]
- Choi, C.W. , Qu, F. , Ren, T. , Ye, X. and Morris, T.J. (2004) RNA silencing‐suppressor function of Turnip crinkle virus coat protein cannot be attributed to its interaction with the Arabidopsis protein TIP. J. Gen. Virol. 85, 3415–3420. [DOI] [PubMed] [Google Scholar]
- Chung, B.Y.‐W. , Miller, W.A. , Atkins, J.F. and Firth, A.E. (2008) An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA, 105, 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier, S.M. and Moffett, P. (2009) NB‐LRRs work a ‘bait and switch’ on pathogen. Trends Plant Sci. 14, 521–529. [DOI] [PubMed] [Google Scholar]
- Cui, X. , Wei, T. , Chowda‐Reddy, R.V. , Sun, G. and Wang, A. (2010) The Tobacco etch virus P3 protein forms mobile inclusions via the early secretory pathway and trafficks along actin microfilaments. Virology, 397, 56–63. [DOI] [PubMed] [Google Scholar]
- Culver, J.N. , Lindbeck, A.G.C. and Dawson, W.O. (1991) Virus–host interactions: induction of chlorotic and necrotic responses in plants by tobamoviruses. Annu. Rev. Phytopathol. 29, 193–217. [Google Scholar]
- Desbiez, C. , Gal‐On, A. , Girard, M. , Wipf‐Scheibel, C. and Lecoq, H. (2003) Increase in Zucchini yellow mosaic virus symptom severity in tolerant zucchini cultivars is related to a point mutation in P3 protein and is associated with a loss of relative fitness on susceptible plants. Phytopathology, 93, 1478–1484. [DOI] [PubMed] [Google Scholar]
- Desbiez, C. , Girard, M. and Lecoq, H. (2010) A novel natural mutation in HC‐Pro responsible for mild symptomatology of Zucchini yellow mosaic virus (ZYMV, Potyvirus) in cucurbits. Arch. Virol. 155, 397–401. [DOI] [PubMed] [Google Scholar]
- Dinesh‐Kumar, S.P. , Tham, W.‐H. and Baker, B.J. (2000) Structure–function analysis of the tobacco mosaic virus resistance gene N . Proc. Natl. Acad. Sci. USA, 97, 14 789–14 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.‐W. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dolja, V.V. , Herndon, K.L. , Pirone, T.P. and Carrington, J.C. (1993) Spontaneous mutagenesis of a plant potyvirus genome after insertion of a foreign gene. J. Virol. 67, 5968–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier, L.L. , Steinlage, T.A. , Hobbs, H.A. , Wang, Y. , Herrera‐Rodriguez, G. , Haudenshield, J.S. , McCoppin, N.K. and Hartman, G.L. (2007) Similarities in seed and aphid transmission among Soybean mosaic virus isolates. Plant Dis. 91, 546–550. [DOI] [PubMed] [Google Scholar]
- Domingo, E. and Holland, J.J. (1997) RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51, 151–178. [DOI] [PubMed] [Google Scholar]
- Drake, J.W. and Holland, J.J. (1999) Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA, 96, 13 910–13 913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt, H.A. , Thi, E.P. , Wang, M.‐B. and Unrau, P.J. (2005) Extensive 3′ modification of plant small RNAs is modulated by helper component‐proteinase expression. Proc. Natl. Acad. Sci. USA, 102, 13 398–13 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenberger, A.L. , Hajimorad, M.R. and Hill, J.H. (2008) Gain of virulence on Rsv1‐genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC‐Pro. Mol. Plant–Microbe Interact. 21, 931–936. [DOI] [PubMed] [Google Scholar]
- Faurez, F. , Baldwin, T. , Tribodet, M. and Jacquot, E. (2012) Identification of new Potato virus Y (PVY) molecular determinants for the induction of vein necrosis in tobacco. Mol. Plant Pathol. 13, 948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Gal‐On, A. (2000) A point mutation in the FRNK motif of the potyvirus helper component‐protease gene alters symptom expression in cucurbits and elicits protection against the severe homologus virus. Phytopathology, 90, 467–473. [DOI] [PubMed] [Google Scholar]
- Goulden, M.G. , Köhm, B.A. , Santa Cruz, S. , Kavanagh, T.A. and Baulcombe, D.C. (1993) A feature of the coat protein of potato virus X affects both induced virus resistance in potato and viral fitness. Virology, 197, 293–302. [DOI] [PubMed] [Google Scholar]
- Gray, S.M. , Power, A.G. , Smith, D.M. , Seaman, A.J. and Altman, N.S. (1991) Aphid transmission of barely yellow dwarf virus: acquisition access periods and virus concentration requirements. Phytopathology, 81, 539–545. [Google Scholar]
- Hajimorad, M.R. and Hill, J.H. (2001) Rsv1‐mediated resistance against Soybean mosaic virus‐N is hypersensitive response‐independent at inoculation site, but has the potential to initiate a hypersensitive response‐like mechanism. Mol. Plant–Microbe Interact. 14, 587–598. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2003) Evolution of Soybean mosaic virus‐G7 molecularly cloned genome in Rsv1‐genotype soybean results in emergence of a mutant capable of evading Rsv1‐mediated recognition. Virology, 314, 497–509. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2005) Loss and gain of elicitor function of Soybean mosaic virus G7 provoking Rsv1‐mediated lethal systemic hypersensitive response maps to P3. J. Virol. 79, 1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2006) Strain‐specific P3 of Soybean mosaic virus elicits Rsv1‐mediated extreme resistance, but absence of P3 elicitor function alone is insufficient for virulence on Rsv1‐genotype soybean. Virology, 345, 156–166. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2008) Adaptation of Soybean mosaic virus avirulent chimeras containing P3 sequences from virulent strains to Rsv1‐genotype soybeans is mediated by mutations in HC‐Pro. Mol. Plant–Microbe Interact. 21, 937–946. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Wen, R.‐H. , Eggenberger, A.L. , Hill, J.H. and Saghai Maroof, M.A. (2011) Experimental adaptation of an RNA virus mimics natural evolution. J. Virol. 85, 2557–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, B.D. (2002) Virus variation in relation to resistance‐breaking in plants. Euphytica, 124, 181–192. [Google Scholar]
- Hayes, A.J. , Jeong, S.C. , Gore, M.A. , Yu, Y.G. , Buss, G.R. , Tolin, S.A. and Saghai Maroof, M.A. (2004) Recombination within a nucleotide‐binding‐site/leucine‐rich‐repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics, 166, 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, J.J. , Torre, J.C. , Clarke, D.K. and Duarte, E. (1991) Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65, 2960–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, R. (2002) Matthew's Plant Virology. New York, NY: Academic Press. [Google Scholar]
- Ishibashi, K. , Meshi, T. and Ishikawa, M. (2011) Gaining replicability in a nonhost compromises the silencing suppression activity of Tobacco mild green mosaic virus in a host. J. Virol. 85, 1893–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.W. , Athanassopoulos, E. , Tsiamis, G. , Mansfield, J.W. , Sesma, A. , Arnold, D.L. , Gibbon, M.J. , Murillo, J. , Taylor, J.D. and Vivian, A. (1999) Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc. Natl. Acad. Sci. USA, 96, 10 875–10 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzac, B. , Fabre, F. , Palloix, A. and Moury, B. (2009) Constraints on evolution of virus avirulence factors predict the durability of corresponding plant resistances. Mol. Plant Pathol. 10, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzac, B. , Montarry, J. , Palloix, A. , Navaud, O. and Moury, B. (2010) A point mutation in the polymerase of Potato virus Y confers virulence toward the Pvr4 resistance of pepper and a high competitiveness cost in susceptible cultivar. Mol. Plant–Microbe Interact. 23, 823–830. [DOI] [PubMed] [Google Scholar]
- Jenner, C.E. , Tomimura, K. , Oshima, K. , Hughes, S.L. and Walsh, J.A. (2002a) Mutations in Turnip mosaic virus P3 and cylindrical inclusion proteins are separately required to overcome two Brassica napus resistance genes. Virology, 300, 50–59. [DOI] [PubMed] [Google Scholar]
- Jenner, C.E. , Wang, X. , Ponz, F. and Walsh, J.A. (2002b) A fitness cost for Turnip mosaic virus to overcome host resistance. Virus Res. 86, 1–6. [DOI] [PubMed] [Google Scholar]
- Jenner, C.E. , Wang, X. , Tomimura, K. , Ohshima, K. , Ponz, F. and Walsh, J.A. (2003) The dual role of the potyvirus P3 protein of Turnip mosaic virus as a symptom and avirulence determinant in Brassicas. Mol. Plant–Microbe Interact. 16, 777–784. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kang, B.‐C. , Yeam, I. and Jahn, M.M. (2005) Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D. , Cronin, S. and Carrington, J.C. (1997) Genome amplification and long‐distance functions associated with the central domain of tobacco etch potyvirus helper‐component‐proteinase. Virology, 228, 251–262. [DOI] [PubMed] [Google Scholar]
- Khatabi, B. , Fajolu, O.L. , Wen, R.‐H. and Hajimorad, M.R. (2012) Evaluation of North American isolates of Soybean mosaic virus for gain of virulence on Rsv‐genotype soybeans with special emphasis on resistance‐breaking determinants on Rsv4 . Mol. Plant Pathol. 13, 1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B.M. , Suehiro, N. , Natsuaki, T. , Inukai, T. and Masuta, C. (2010) The P3 protein of Turnip mosaic virus can alone induce hypersensitive response‐like cell death in Arabidopsis thaliana carrying TuN1 . Mol. Plant–Microbe Interact. 23, 144–152. [DOI] [PubMed] [Google Scholar]
- Kjemtrup, S. , Nimchuk, Z. and Dangl, J.L. (2000) Effector proteins of phytopathogenic bacteria: bifunctional signals in virulence and host recognition. Curr. Opin. Microbiol. 3, 73–78. [DOI] [PubMed] [Google Scholar]
- Kobayashi, K. and Hohn, T. (2004) The avirulence domain of Cauliflower mosaic virus transactivator/viroplasmin is a determinant of viral virulence in susceptible hosts. Mol. Plant–Microbe Interact. 17, 475–483. [DOI] [PubMed] [Google Scholar]
- Kvitko, B.H. , Park, D.H. , Velásquez, A.C. , Wei, C.‐F. , Russell, A.B. , Martin, G.B. , Schneider, D.J. and Collmer, A. (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5, e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, J.E. , Cruz, C.M.V. , Bai, J. and Leung, H. (2001) Pathogen fitness penalty as a predictor of durability of disease resistance genes. Annu. Rev. Phytopathol. 39, 187–224. [DOI] [PubMed] [Google Scholar]
- Lim, H.S. , Ko, T.S. , Hobbs, H.A. , Lambert, K.N. , Yu, J.M. , McCoppin, N.K. , Korban, S.S. , Hartman, G.L. and Domier, L.L. (2007) Soybean mosaic virus helper component‐protease alters leaf morphology and reduces seed production in transgenic soybean plants. Phytopathology, 97, 366–372. [DOI] [PubMed] [Google Scholar]
- Luderer, R. , Takken, F.L.W. , De Wit, P.J.G.M. and Joosten, M.A.J. (2002) Cladosporium fulvum overcomes Cf‐2‐mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 45, 875–884. [DOI] [PubMed] [Google Scholar]
- Malapi‐Nelson, M. , Wen, R.‐H. , Ownley, B.H. and Hajimorad, M.R. (2009) Co‐infection of soybean with Soybean mosaic virus and Alfalfa mosaic virus results in disease synergism and alteration in accumulation level of both viruses. Plant Dis. 93, 1259–1264. [DOI] [PubMed] [Google Scholar]
- Mazarei, M. , Lennon, K.A. , Puthoff, D.P. , Rodermel, S.R. and Baum, T.J. (2003) Expression of an Arabidopsis phosphoglycerate mutase homologue is localized to apical meristems, regulated by hormones, and induced by sedentary plant‐parasitic nematodes. Plant Mol. Biol. 53, 513–530. [DOI] [PubMed] [Google Scholar]
- Mazarei, M. , Elling, A.A. , Maier, T.R. , Puthoff, D.P. and Baum, T.J. (2007) GmEREBP1 is a transcription factor activating defense genes in soybean and Arabidopsis . Mol. Plant–Microbe Interact. 20, 107–119. [DOI] [PubMed] [Google Scholar]
- Mérai, Z. , Kérenyi, Z. , Kertész, S. , Magna, M. , Lakatos, L. and Silhavy, D. (2006) Double‐stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 80, 5747–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre, P. , Brigneti, G. , Durrant, M.C. and Baulcombe, D.C. (2003) Potato virus Y NIa protease activity is not sufficient for elicitation of Ry‐mediated disease resistance in potato. Plant J. 36, 755–761. [DOI] [PubMed] [Google Scholar]
- Moissiard, G. , Parizotto, E.A. , Himber, C. and Voinnet, O. (2007) Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer‐like 4 and Dicer‐like 2, and is compromised by viral‐encoded suppressor proteins. RNA, 13, 1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, J.‐K. , Jeong, S.‐C. , Van, K. , Saghai Maroof, M.A. and Lee, S.‐H. (2009) Marker‐assisted identification of resistance genes to soybean mosaic virus in soybean lines. Euphytica, 169, 375–385. [Google Scholar]
- Moreira, A. , Jones, R.A.C. and Fribourg, C.E. (1980) Properties of a resistance‐breaking strain of potato virus X. Ann. Appl. Biol. 95, 93–103. [Google Scholar]
- Moury, B. , Caromel, B. , Johansen, E. , Simon, V. , Chauvin, L. , Jacquot, E. , Kerlan, C. and Lefebvre, V. (2011) The helper component proteinase cistron of Potato virus Y induces hypersensitivity and resistance in potato genotypes carrying dominant resistance genes on chromosome IV. Mol. Plant–Microbe Interact. 24, 787–797. [DOI] [PubMed] [Google Scholar]
- Palukaitis, P. and Carr, J.P. (2008) Plant resistance responses to viruses. J. Plant Pathol. 90, 153–171. [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, M. , Kerlan, C. and Jacquot, E. (2009) The acquisition of molecular determinants involved in potato virus Y necrosis capacity leads to fitness reduction in tobacco plants. J. Gen. Virol. 90, 244–252. [DOI] [PubMed] [Google Scholar]
- Ronde, D.D. , Butterbach, P. , Lohuis, D. , Hedil, M. , Van Lent, J.W.M. and Kormelink, R. (2013) Tsw gene‐based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus . Mol. Plant Pathol. 14, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacristan, S. and Garcia‐Arenal, F. (2008) The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9, 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáenz, P. , Quiot, L. , Quiot, J.‐B. , Candresse, T. and Garcia, J.A. (2001) Pathogenicity determinants in the complex virus population of a Plum pox virus isolate. Mol. Plant–Microbe Interact. 14, 278–287. [DOI] [PubMed] [Google Scholar]
- Scholthof, H.B. (2006) The Tombusvirus‐encoded P19: from irrelevance to elegance. Nat. Rev. Microbiol. 4, 405–411. [DOI] [PubMed] [Google Scholar]
- Schürch, S. , Linde, C.C. , Knogge, W. , Jackson, L.F. and McDonald, B.A. (2004) Molecular population genetic analysis differentiates two virulence mechanisms of the fungal avirulence gene NIP1 . Mol. Plant–Microbe Interact. 17, 1114–1125. [DOI] [PubMed] [Google Scholar]
- Seo, J.‐K. , Lee, S.‐H. and Kim, K.‐H. (2009a) Strain‐specific cylindrical inclusion protein of Soybean mosaic virus elicits extreme resistance and a lethal systemic hypersensitive response in two resistant soybean cultivars. Mol. Plant–Microbe Interact. 22, 1151–1159. [DOI] [PubMed] [Google Scholar]
- Seo, J.‐K. , Ohshima, K. , Lee, H.‐G. , Son, M. , Choi, H.‐S. , Lee, S.‐H. , Sohn, S.‐H. and Kim, K.‐H. (2009b) Molecular variability and genetic structure of the population of Soybean mosaic virus based on the analysis of complete genome sequences. Virology, 393, 91–103. [DOI] [PubMed] [Google Scholar]
- Shaner, G. , Stromberg, E.L. , Lacy, G.H. , Barker, K.R. and Pirone, T.P. (1992) Nomenclature and concepts of pathogenicity and virulence. Annu. Rev. Phytopathol. 30, 47–66. [DOI] [PubMed] [Google Scholar]
- Shi, X.M. , Miller, H. , Verchot, J. , Carrington, J.C. and Vance, V.B. (1997) Mutations in the region encoding the central domain of helper component‐proteinase (HC‐Pro) eliminate potato virus X/potyviral synergism. Virology, 231, 35–42. [DOI] [PubMed] [Google Scholar]
- Shiboleth, Y.M. , Haronsky, E. , Leibman, D. , Arazi, T. , Wassenegger, M. , Whitham, S.A. , Gaba, V. and Gal‐On, A. (2007) The conserved FRNK box in HC‐Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J. Virol. 81, 13 135–13 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, E.A. and Bishop, J.B. (2000) Plant–pathogen arms races at the molecular level. Curr. Opin. Plant Biol. 3, 299–304. [DOI] [PubMed] [Google Scholar]
- Stenger, D.C. , Young, B.A. and French, R. (2006) Random mutagenesis of wheat streak mosaic virus HC‐Pro: non‐infectious interfering mutations in a gene dispensable for systemic infection of plants. J. Gen. Virol. 87, 2741–2747. [DOI] [PubMed] [Google Scholar]
- Taraporewala, Z.F. and Culver, J.N. (1996) Identification of an elicitor active site within the three‐dimensional structure of the tobacco mosaic tobamovirus coat protein. Plant Cell, 8, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Y.‐P. and Valkonen, J.P.T. (2013) Genetic determinants of Potato virus Y required to overcome or trigger hypersensitive resistance to PVY strain group O controlled by the gene NY in potato. Mol. Plant–Microbe Interact. 26, 297–305. [DOI] [PubMed] [Google Scholar]
- Tsiamis, G. , Mansfield, J.W. , Hockenhull, R. , Jackson, R.W. , Sesma, A. , Athanassopoulos, E. , Bennett, M.A. , Stevens, C. , Vivian, A. , Taylor, J.D. and Murillo, J. (2000) Cultivar‐specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo‐blight disease. EMBO J. 19, 3204–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urcuqui‐Inchima, S. , Haenni, A.L. and Bernardi, F. (2001) Potyvirus proteins: a wealth of functions. Virus Res. 74, 157–175. [DOI] [PubMed] [Google Scholar]
- Van Der Hoorn, R.A.L. and Kamoun, S. (2008) From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell, 20, 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van't Slot, K.A.E. and Knogge, W. (2002) A dual role for microbial pathogen‐derived effector proteins in plant disease and resistance. Crit. Rev. Plant Sci. 21, 229–271. [Google Scholar]
- Vera Cruz, C.M. , Bai, J. , Oña, I. , Leung, H. , Nelson, R.J. , Mew, T.‐W. and Leach, J.E. (2000) Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc. Natl. Acad. Sci. USA, 97, 13 500–13 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viel, C. , Ide, C. , Cui, X. , Wang, A. , Farsi, M. , Michelutti, R. and Stromvik, M. (2009) Isolation, partial sequencing, and phylogenetic analyses of Soybean mosaic virus (SMV) in Ontario and Quebec. Can. J. Plant Pathol. 31, 108–113. [Google Scholar]
- Vijayapalani, P. , Maeshima, M. , Nagasaki‐Takekuchi, N. and Miller, W.A. (2012) Interaction of the trans‐frame potyvirus protein P3N‐PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog. 8, e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Eggenberger, A. , Hill, J. and Bogdanove, A.J. (2006) Pseudomonas syringae effector avrB confers soybean cultivar‐specific avirulence on Soybean mosaic virus adapted for transgene expression but effector avrPto does not. Mol. Plant–Microbe Interact. 19, 304–312. [DOI] [PubMed] [Google Scholar]
- Wen, R.‐H. and Hajimorad, M.R. (2010) Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology, 400, 1–7. [DOI] [PubMed] [Google Scholar]
- Wen, R.‐H. , Saghai Maroof, M.A. and Hajimorad, M.R. (2011) Amino acid changes in P3, and not the overlapping pipo‐encoded protein, determine virulence of Soybean mosaic virus on functionally immune Rsv1‐genotype soybean. Mol. Plant Pathol. 12, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, R.‐H. , Khatabi, B. , Ashfield, T. , Saghai Maroof, M.A. and Hajimorad, M.R. (2013) The HC‐Pro and P3 cistrons of an avirulent Soybean mosaic virus are recognized by different resistance genes at the complex Rsv1 locus. Mol. Plant–Microbe Interact. 26, 203–215. [DOI] [PubMed] [Google Scholar]
- Wu, H.‐W. , Lin, S.‐S. , Chen, K.‐C. , Yeh, S.‐D. and Chua, N.‐H. (2010) Discriminating mutations of HC‐Pro of Zucchini yellow mosaic virus with differential effects on small RNA pathways involved in viral pathogenicity and symptom development. Mol. Plant–Microbe Interact. 23, 17–28. [DOI] [PubMed] [Google Scholar]
- Zhang, C. and Ghabrial, S.A. (2006) Development of Bean pod mottle virus‐based vectors for stable protein expression and sequence‐specific virus‐induced gene silencing in soybean. Virology, 344, 401–411. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Hajimorad, M.R. , Eggenberger, A.L. , Tsang, S. , Whitham, S.A. and Hill, J.H. (2009) Cytoplasmic inclusion of Soybean mosaic virus serves as a virulence determinant on Rsv3‐genotype soybean and a symptom determinant. Virology, 391, 240–248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The phenotype expressed on soybean plant introduction (PI) ‘PI96983 (Rsv1)’ following mechanical inoculation with progeny viruses derived from the molecularly cloned genome of SMV‐NG319S+K321E+R682M+A947V+K952E+M1157I (virN6). The inoculated plants were maintained in a growth chamber at 22 °C until being photographed at 21 days post‐inoculation.
Fig. S2 Comparison of accumulation levels of virions of SMV‐N (N) with SMV‐N‐derived P3 mutants or SMV‐N‐derived mutants with a single mutation in P3 (R945G or A947V) in combination with an amino acid substitution in HC‐Pro (K321E) in Williams82 (rsv1) using the antigen‐coated indirect enzyme‐linked immunosorbent assay (ELISA). Plants were inoculated mechanically with progeny viruses derived from molecularly cloned genomes, maintained in a growth chamber at 22 °C until trifoliate leaves were harvested at 28 days post‐inoculation. Each bar represents the mean value of virion accumulation from three independent experiments, each with five replicate plants, with the standard errors indicated. No significant differences in the accumulation level of SMV‐N relative to that of each of the mutants were detected by Student's t‐test (P < 0.01). OD, optical density.


