Summary
Virus‐induced gene silencing (VIGS) is a powerful reverse genetics tool in plant science. In this study, we investigated the temporal and spatial silencing patterns achieved by Bean pod mottle virus (BPMV)‐based VIGS in soybean using virus constructs targeting green fluorescence protein (GFP). Silencing GFP enabled an in‐depth analysis of silencing in soybean tissues over time in a transgenic line constitutively expressing GFP. We discovered evidence for variable GFP silencing based on insert orientation and targeted region in the coding sequence. A 3′ sequence in reverse orientation produced the strongest silencing phenotypes. Furthermore, we documented that BPMV VIGS can achieve widespread silencing in a broad range of tissues, including leaves, stems, flowers and roots. Near‐complete silencing was attained in leaves and flowers. Although weaker than in shoots, the observed gene silencing in soybean roots will also allow reverse genetics studies in this tissue. When GFP fluorescence was assayed in cross‐sections of stems and leaf petioles, near‐complete and uniform silencing was observed in all cell types. Silencing was observed from as early as 2 weeks post‐virus inoculation in leaves to 7 weeks post‐virus inoculation in flowers, suggesting that this system can induce and maintain silencing for significant durations.
Continued technical advances are making large sequencing projects significantly easier. With the help of such technologies, scientists have successfully sequenced the genomes of crop plants including soybean (Schmutz et al., 2010). Subsequent data analysis has resulted in the identification of large numbers of genes. Although the availability of these data offers an unprecedented opportunity for scientists to conduct in‐depth studies, assigning functions to the genes identified remains problematic.
Traditionally, forward and reverse genetics are employed to study gene function. Although forward genetics has been extremely powerful, it is time consuming in crop plants, as it requires the generation of mutant populations, screening for mutants with desired phenotypes, and mapping and cloning the genes responsible for the phenotypes (Alonso and Ecker, 2006). The relatively more rapid reverse genetics approaches are becoming increasingly important. In the past decade, reverse genetics strategies, such as T‐DNA insertion mutagenesis (Azpiroz‐Leehan and Feldmann, 1997), TILLING (targeting induced local lesions in genomes) (Henikoff et al., 2004) and RNA interference (McGinnis et al., 2005), have been developed. Unfortunately, reverse genetics tools are not uniformly available for economically important crop species, such as soybean. The recalcitrant nature of soybean for genetic transformation hampers the application of conventional reverse genetics approaches. Thus, to perform large‐scale, high‐throughput genomic studies in soybean, an economically feasible, rapid system that bypasses transformation is required.
Several plant virus‐induced gene silencing (VIGS) systems have been designed to address reverse genetics needs in various plants including soybean (Burch‐Smith et al., 2004; Constantin et al., 2004; Ding et al., 2006; Grønlund et al., 2008; Igarashi et al., 2009; Meng et al., 2009; Nagamatsu et al., 2007; Pogue et al., 2002; Zhang and Ghabrial, 2006; Zhang et al., 2009, 2010). VIGS relies on the sequence‐specific degradation of endogenous mRNA. Generally, a partial sequence of a target gene is inserted into the viral genome and the resultant virus is used to infect a host plant. Virus replication triggers plant defence responses that identify and cleave double‐stranded RNA (dsRNA), an intermediate product of viral replication. dsRNA cleavage produces short interfering RNA (siRNA) which, in association with the RNA‐induced silencing complex (RISC), targets homologous RNA for degradation and silences the targeted endogenous gene (Burch‐Smith et al., 2004; Purkayastha and Dasgupta, 2009).
VIGS is used to silence genes in various plant tissues, including leaves (Liu et al, 2002), roots (Ryu et al., 2004), tubers (Faivre‐Rampant et al., 2004), flowers (Chen et al., 2004; Liu et al., 2004) and seedlings, and can sometimes persist to progeny (Kanazawa et al., 2011a, b; Senthil‐Kumar and Mysore, 2011; Yamagishi and Yoshikawa, 2009). To use VIGS to its full potential, an in‐depth understanding of temporal and spatial silencing patterns is necessary. Here, we report a systematic analysis of VIGS in soybean using a Bean pod mottle virus (BPMV) vector (Zhang et al., 2010) over time in leaves, flowers, stems and roots.
Hernandez‐Garcia et al. (2009) have developed a transgenic soybean Jack cultivar that expresses the green fluorescent protein gene (GFP) under the control of the G. max ubiquitin promoter. Silencing of this ubiquitously expressed transgene can easily be visualized and measured in various organs and tissues, and it has provided an ideal experimental system to generate a silencing atlas over time at the whole‐plant level. In addition to being a visible marker, silencing of GFP has minimal effects on cellular processes. Here, we report the use of this transgenic line to temporally follow BPMV VIGS in soybean tissues over time. We demonstrate the feasibility of using a BPMV‐based VIGS approach for reverse genetics screens in various soybean tissues, and we establish the time parameters for this VIGS system to silence genes in various shoot tissues, as well as in roots. The use of a GFP reporter gene allowed the assessment of temporal and spatial silencing patterns, as well as the approximate degree of silencing achieved by BPMV. The silencing of GFP in transgenic soybean plants that express GFP under the control of the Glycine max ubiquitin promoter facilitated the localization and quantification of the silencing pattern in various organs and tissues over time.
First, we identified the most effective GFP silencing construct to use in these experiments, because insert size, location of the target region and cloning orientation can affect the silencing potential of VIGS vectors (Bennypaul et al., 2011; Liu and Page, 2008; Yuan et al., 2011; Zhang et al., 2010). We made four constructs that targeted the 5′ or 3′ halves of the GFP coding sequence in the sense and antisense orientations using the pBPMV‐IA‐V1 vector (Zhang et al., 2010) (Fig. S1A, see Supporting Information). At 21 days post‐inoculation (dpi), GFP fluorescence was reduced in plants inoculated with all four GFP silencing constructs. However, plants inoculated with the 3′‐end antisense construct (1102‐A) showed the most consistent and dramatic reduction of GFP fluorescence (Fig. S1B) and GFP mRNA expression (Fig. S1C).
To examine the temporal and spatial GFP silencing patterns induced by BPMV in different soybean tissues, we used construct 1102‐A, because it was the most effective. A significant reduction in GFP fluorescence was observed in the leaflets of the first trifoliate as early as 14 dpi (Fig. 1A). Examination of the second and fourth trifoliates at 21 and 35 dpi, respectively, showed significant silencing up to 35 dpi when compared with controls (Fig. 1B,C), documenting a long‐lasting VIGS effect. However, the relative levels of GFP mRNA demonstrated that the magnitude of GFP silencing at 35 dpi was not as strong as that at 14 and 21 dpi (Fig. 1D) (Appendix S1, see Supporting Information).
Figure 1.
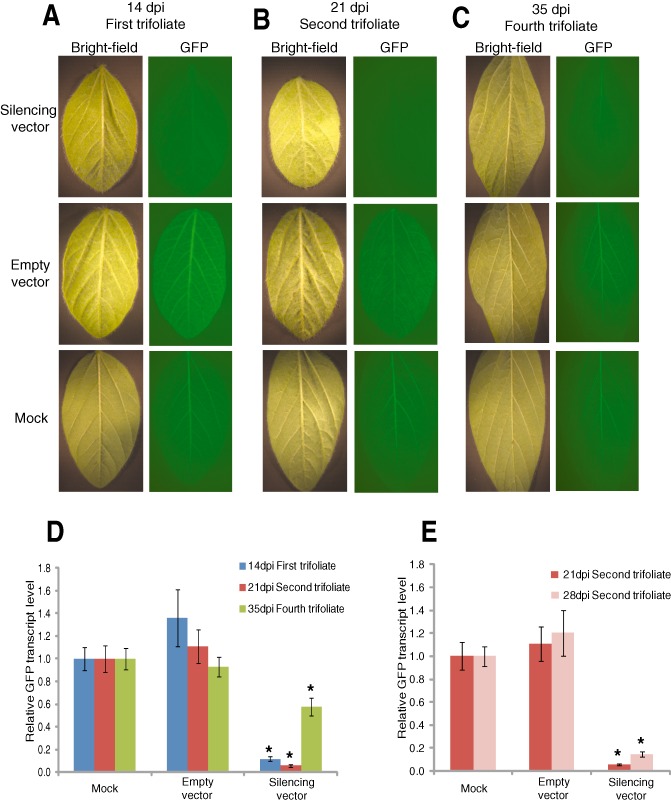
Bean pod mottle virus (BPMV)‐induced gene silencing of the green fluorescent protein transgene (GFP) in leaves. (A–C) Silencing of GFP in the first trifoliate (A), second trifoliate (B) and fourth trifoliate (C) at 14, 21 and 35 days post‐inoculation (dpi), respectively. Leaves inoculated with silencing vector (top panel), BPMV empty vector (middle panel) or mock (bottom panel) were visualized under white light (left panel) and UV light (right panel) and photographed (3‐s exposure on a Zeiss Stemi SV11 stereoscope). Similar results were obtained from two independent experiments. (D) Quantification of GFP silencing in the first, second and fourth trifoliates at 14, 21 and 35 dpi, respectively. Quantitative polymerase chain reaction (qPCR) was used to quantify the silencing of GFP in RNA isolated from the first, second and fourth trifoliates treated with silencing vector, empty vector or mock. The expression levels were calculated using the 2–ΔΔCT method and represent changes in mRNA abundance in BPMV‐inoculated plants relative to the mock‐treated control. Data are the average of three biological samples, each with three technical replicates. Mean values significantly different from the mock are indicated by an asterisk as determined by paired t‐test (P < 0.01). (E) Quantification of GFP silencing in the second trifoliates at 21 and 28 dpi using qPCR. RNA samples were isolated from the second trifoliates treated with the silencing vector, empty vector or mock at the 21‐ and 28‐dpi time points. The expression levels of GFP were calculated using the 2–ΔΔCT method and represent changes in mRNA abundance in BPMV‐inoculated plants relative to the mock‐treated control. Data are the average of three biological samples, each with three technical replicates. Mean values significantly different from the mock are indicated by an asterisk as determined by paired t‐test (P < 0.01).
In order to examine the duration of silencing at a particular location, we monitored the GFP fluorescence in one leaflet from the first trifoliate at 14 dpi and in a second leaflet from the same trifoliate at 21 dpi. Similarly, GFP fluorescence was monitored in two leaflets of the second trifoliate at 21 and 28 dpi and in the third trifoliate at 28 and 35 dpi. Although significant silencing was observed in the inoculated plants up to 35 dpi when compared with the controls, no significant differences occurred within the leaflets of the same trifoliate (Fig. S2, see Supporting Information), showing a sustained level of VIGS during the observation period. The GFP observations were supported by quantitative polymerase chain reaction (qPCR) analysis of GFP transcript levels in leaflets of the second trifoliate collected at 21 and 28 dpi (Fig. 1E).
To test silencing in flowers, GFP fluorescence in floral organs from plants inoculated with the silencing vector was compared with that of control treatments. Under our experimental conditions, plants produced their first flowers at 7 weeks post‐virus inoculation. At this time, flowers were assayed for GFP fluorescence. After this 7‐week period, high levels of silencing were observed in all floral parts, including petals, sepals and reproductive whorls (Fig. 2A,B). The 95% reduction in GFP mRNA levels in the most silenced flowers confirms the silencing efficiency in floral tissues (Fig. 2C). When GFP fluorescence was compared among all flowers collected from each individual plant in the experiment, silencing was observed in all flowers, indicating that silencing in floral tissue was independent of the flower's developmental stage and/or its location on the plant (Fig. S3, see Supporting Information). The fact that BPMV‐based VIGS attained high levels of silencing and sustained the silencing for significant durations makes it possible to employ this system to dissect the molecular mechanisms underlying numerous biological phenomena in leaves and flowers.
Figure 2.
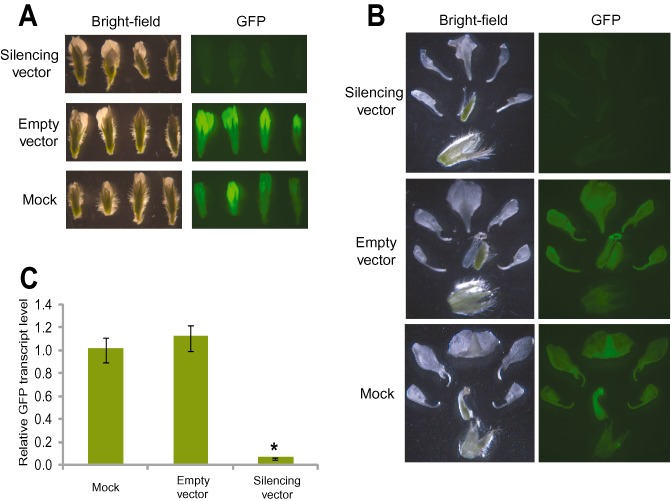
Bean pod mottle virus (BPMV)‐induced gene silencing of the green fluorescent protein transgene (GFP) in floral tissue. (A) GFP silencing in flowers. Flowers at different developmental stages were collected from transgenic GFP soybean plants treated with silencing vector (top panel), BPMV empty vector (middle panel) or mock (bottom panel) and visualized under white light (left panel) and UV light (right panel) and photographed (1‐s exposure on a Zeiss Stemi SV11 stereoscope). Similar results were obtained from three independent experiments. (B) GFP silencing in floral whorls. Hand‐dissected flowers were collected from transgenic GFP soybean plants inoculated with silencing vector (top panel), BPMV empty vector (middle panel) or mock (bottom panel) at 7 weeks post‐virus inoculation and visualized under white light (left panel) and UV light (right panel) and photographed (0.5‐s exposure on a Zeiss Stemi SV11 stereoscope). Note the reduced GFP fluorescence in the flowers and floral whorls collected from the transgenic plants inoculated with silencing construct compared with those obtained from the control treatments. (C) Quantification of gene silencing induced by BPMV in floral tissues using quantitative polymerase chain reaction (qPCR). The mRNA expression levels of GFP were measured in RNA samples isolated from floral tissues of the transgenic GFP plants at 7 weeks post‐virus inoculation and compared with those treated with empty vector or mock. The expression level was calculated using the 2–ΔΔCT method. Data are the average of three biological samples, each with three technical replicates. Mean values significantly different from the mock are indicated by an asterisk as determined by paired t‐test (P < 0.01).
The duration of silencing in leaves and flowers is dependent in part on the stability of the target gene sequence cloned into the vector. Insertion of GFP into the BPMV RNA2 polyprotein between the movement protein and large subunit of the coat protein has been shown to be stable after four passages (Diaz‐Camino et al., 2011). However, the stability of inserts cloned into the 3′ end of BPMV after the stop codon has not been investigated systematically. We performed reverse transcription‐polymerase chain reaction (RT‐PCR) on RNA samples from leaves at 14, 21, 28 and 35 dpi and flowers collected at 49 dpi using two sets of primers. The first primer set included a reverse primer in GFP and a forward primer in the viral genome, and the second primer set included forward and reverse primers in the BPMV genome that flanked the insertion site. The expected PCR products were obtained using these primers in all leaf and flower samples (Fig. S4, see Supporting Information). These data demonstrated that the GFP insert was stable in the BPMV‐IA‐V1 vector for the tissues and time courses used in the presented studies.
Mosaic patterns of infection are prominent features of virus infection in plants. In addition, viruses may not infect all cell types equally, which may result in sectored silencing or silencing confined to certain regions. For example, a Tobacco rattle virus (TRV) vector could not invade meristematic tissue and was excluded from lateral roots (Valentine et al., 2002). Our previous results using a BPMV construct tagged with GFP that also silenced the phytoene desaturase gene (PDS) indicated that the most extensive silencing occurred in tissues that contained GFP fluorescence and, presumably, BPMV (Zhang et al., 2010). This observation was consistent with a mosaic pattern of silencing. However, in the present study, we observed that the silencing of GFP by BPMV VIGS was surprisingly uniform. To verify the uniformity of silencing triggered by the BPMV system, we examined fluorescence in cross‐sections of stem regions between the second and third trifoliates and in petioles of the second trifoliates at 21 dpi. Although cross‐sections of the mock and control plants showed bright GFP fluorescence in various cell types and tissues, such as vascular tissue, parenchyma, chlorenchyma, pith as well as cortical cells, the cross‐sections from plants inoculated with silencing constructs showed almost complete, and, most importantly, uniform and section‐wide silencing of GFP (Fig. 3A,B). One potential explanation for the uniformity of GFP silencing is that in tissues that are not directly infected by BPMV, the transgene may be more effectively silenced than endogenous genes.
Figure 3.
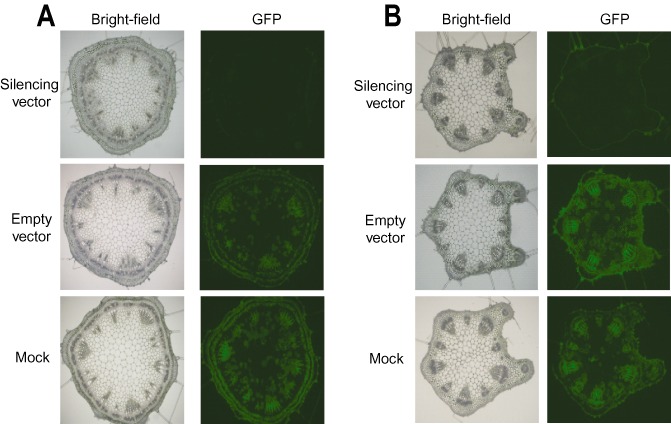
Bean pod mottle virus (BPMV)‐induced gene silencing of the green fluorescent protein transgene (GFP) in internal tissues. (A, B) GFP silencing in the stem (A) and leaf petiole (B). Cross‐sections (thickness, 80 μm) prepared from transgenic GFP soybean plants inoculated with the silencing vector (top panel), BPMV empty vector (middle panel) or mock (bottom panel) were visualized under white light (left panel) and UV light (right panel) and photographed (500‐ms exposure on a Zeiss Axiovert 100 microscope). Similar results were obtained from two independent experiments.
In general, most applications of VIGS systems, including BPMV, have been directed towards traits associated with the shoots, such as leaves, flowers and fruits. Some vectors, such as Pea early browning virus (PEBV), have been shown to be effective at silencing genes in the roots, as well as aerial plant tissues. PEBV VIGS can be used to dissect genetic requirements for symbiotic relationships with Rhizobium species and vesicular–arbuscular mycorrhizae (Constantin et al., 2008; Grønlund et al., 2010). In contrast, studies with other viruses have suggested that VIGS might not be as effective in roots (Dalmay et al., 2000; Kaloshian, 2007; Palauqui et al., 1997; Saedler and Baldwin, 2004; Sonoda and Nishiguchi, 2000; Valentine et al., 2002). Because we have an interest in developing BPMV as a tool to investigate interactions between soybean and soybean cyst nematode, we chose to investigate silencing in the roots. Previous silencing experiments with essential genes, such as actin and ribosomal proteins, have suggested that root growth is inhibited and the accumulation of transcripts is reduced (Zhang et al., 2009). However, the dramatic effect of the silencing of these genes on the shoot could have affected the expression of these genes in the root. The use of the GFP transgene in our present study overcame these complications, because there was no apparent effect on the shoot from the silencing of GFP. Thus, the silencing responses in the root were not expected to be coupled necessarily to pleiotropic effects in the shoot. As maximum silencing in foliar tissues was observed between 14 and 28 dpi, root assays were conducted at 21 dpi. When compared with the control, distinct GFP silencing was observed in roots from plants inoculated with the silencing construct. The qPCR analysis confirmed that GFP mRNA was reduced by over 65% (Fig. S5, see Supporting Information).
Closer observation of the GFP fluorescence revealed greater silencing in the upper, more mature, root tissue than in the lower, newly growing, tissues (Fig. 4A). To provide additional evidence for this observation, we cut the root system of treatment and control plants approximately at the midpoint and processed the upper and lower halves separately for RNA extraction and qPCR analysis. Data obtained from three biological replicates showed more extensive gene silencing in the upper root tissues relative to the lower region. More specifically, although GFP silencing in the upper root regions was about 80% of the control treatments, in the lower root region, GFP silencing was calculated to be 50% (Fig. 4B). Significant GFP silencing induced by BPMV in soybean roots demonstrates the potential of this system to accelerate functional and genomics studies of root biology. However, our results from experiments involving BPMV VIGS in soybean roots also show that silencing is not uniform throughout the root system.
Figure 4.
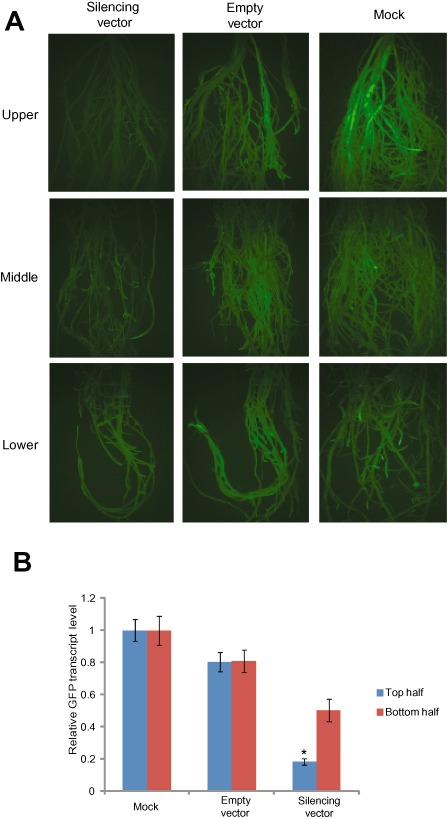
Bean pod mottle virus (BPMV)‐induced gene silencing of the green fluorescent protein transgene (GFP) in roots. (A) BPMV triggered higher silencing in the upper part of the roots than in the lower root tissues. Root tissues were collected from transgenic GFP soybean plants treated with silencing vector (left panel), BPMV empty vector (middle panel) or mock (right panel) and GFP fluorescence was visualized in the upper, middle and lower parts of the roots under UV light. Note the gradual decrease in GFP fluorescence from the upper to lower region of the root system of the silenced plants (left panel) compared with control treatments (middle and right panels).Similar results were obtained from two independent experiments. (B) Comparison of silencing efficiency induced by BPMV in the upper and lower halves of the roots. Quantitative polymerase chain reaction (qPCR) was used to measure the abundance of GFP transcript in RNA samples isolated from the upper and lower halves of the root system. The root tissues were collected from transgenic GFP soybean plants inoculated with BPMV vector, BPMV empty vector or mock at 21 days post‐inoculation (dpi). The expression levels were calculated using the 2–ΔΔCT method and represent changes in mRNA abundance in BPMV‐inoculated plants relative to the mock‐treated control. Data are the average of three biological samples, each with three technical replicates. Mean values significantly different from the mock are indicated by an asterisk as determined by paired t‐test (P < 0.01).
Another key question that we were interested in was the timing and duration of BPMV VIGS. Silencing of target genes using VIGS systems can be detected in some cases as early as 3 dpi (Hein et al., 2005; Scofield et al., 2005). However, between 1 and 2 weeks is generally the time span required to observe efficient silencing using the majority of VIGS systems. In a few cases, 4 weeks post‐virus inoculation is required to see efficient silencing (Bennypaul et al., 2011; Turnage et al., 2002). Similar to the majority of VIGS systems, we were able to detect significant silencing caused by BPMV VIGS within 2 weeks post‐inoculation. Over time the silencing was reduced, but was still significant. An interesting question that remains is whether BPMV VIGS can extend to the seed or, possibly, to the next generation, as has been observed for TRV in Nicotiana benthamiana and tomato and Cucumber mosaic virus (CMV) in petunia, N. benthamiana and tomato (Kanazawa et al., 2011b; Senthil‐Kumar and Mysore, 2011). Nagamatsu et al. (2007) have shown that CMV can induce VIGS of the chalcone synthase gene (CHS) in the soybean seed coat, demonstrating that VIGS in soybean seeds is a possibility.
In conclusion, our report details the characteristics of BPMV VIGS in soybean and establishes an atlas of VIGS over a 5‐week time course. We have demonstrated that BPMV VIGS is effective in achieving long‐lasting, uniform and consistent gene silencing in various tissues, including leaves, flowers and roots. These results can be used to guide the design of experiments that utilize BPMV VIGS in a variety of soybean tissues. This information is critical for extending and maximizing the utility of BPMV VIGS for large‐scale genomic studies in soybean and other legumes, such as common bean (Diaz‐Camino et al., 2011; Zhang et al., 2010).
Supporting information
Appendix S1. Experimental procedures
Fig. S1 Sequence location and cloning orientation influence silencing potential triggered by Bean pod mottle virus (BPMV). (A) Schematic representation of partial green fluorescent protein (GFP) sequences used for transgene silencing. Two GFP fragments targeting the 3′ or 5′ half were cloned in sense or antisense orientation in the BPMV vector and the silencing responses were quantified using quantitative polymerase chain reaction (qPCR). The silencing efficiencies are indicated. Arrows indicate the orientation of the insert. The expression levels were calculated using the 2–ΔΔCT method and represent changes in mRNA abundance in BPMV‐inoculated plants relative to the mock‐treated control. Data are the average of three biological samples, each with three technical replicates. Mean values significantly different from the mock are indicated by an asterisk as determined by paired t‐test (P < 0.01). (B) Quantification of gene silencing induced by BPMV in leaves using qPCR. The expression levels of GFP mRNA were measured in RNA samples isolated from the second trifoliate of the transgenic GFP plants at 21 days post‐inoculation (dpi) and compared with those receiving empty vector or mock. The expression levels were calculated using the 2–ΔΔCT method and represent changes in mRNA abundance in BPMV‐inoculated plants relative to the mock‐treated control. Data are the average of three biological samples, each with three technical replicates. Mean values significantly different from the mock are indicated by an asterisk as determined by paired t‐test (P < 0.01). (C) Silencing of GFP in the second trifoliate at 21 dpi by the indicated silencing constructs. Mock leaves, leaves inoculated with empty vector and silencing vectors 1101‐A, 1101‐C, 1102‐A and 1102‐D are shown. The leaves were visualized under UV light and photographed (3‐s exposure on a Zeiss Stemi SV11 stereoscope; Carl Zeiss, Thornwood, NY, USA). Note the strongest reduction in GFP fluorescence in the trifoliates from the plants inoculated with silencing construct 1102‐A compared with the other treatments.
Fig. S2 Sustained Bean pod mottle virus (BPMV)‐induced silencing in foliar tissue. (A, B) Silencing of green fluorescent protein (GFP) in the first trifoliate at 14 days post‐inoculation (dpi) (A) and 21 dpi (B). (C, D) Silencing of GFP in the second trifoliate at 21 dpi (C) and 28 dpi (D). (E, F) Silencing of GFP in the third trifoliate at 28 dpi (E) and 35 dpi (F). In each group, the top panel shows a single leaflet from an individual mock plant and the bottom panel shows a single leaflet from an individual plant inoculated with the silencing construct. Each leaflet was visualized with UV light and photographed (3‐s exposure on a Zeiss Stemi SV11 stereoscope). Note the persistent silencing in the leaflets from all three spatial locations for the duration of 7 days.
Fig. S3 Bean pod mottle virus (BPMV)‐induced silencing of the transgene in flowers. Flowers at different developmental stages from individual transgenic green fluorescent protein (GFP) soybean plants treated with silencing vector (top three panels), BPMV empty vector (middle three panels) or mock (bottom three panels) visualized under white light (left panels) and UV light (right panels) and photographed (1‐s exposure on a Zeiss Stemi SV11 stereoscope) at 7 weeks post‐inoculation. Each panel shows flowers from a single plant. Similar results were obtained from three independent experiments.
Fig. S4 Long‐lasting insert stability of Bean pod mottle virus (BPMV) virus‐induced gene silencing (VIGS) in soybean. Panels show amplified BPMV RNA2 products. Each bracket denotes three individual polymerase chain reactions (PCRs) conducted on cDNA from a transgenic soybean plant inoculated with a virus harbouring either a silencing construct or an empty vector, or a mock‐inoculated plant, respectively. The vector harbouring the 1102‐A silencing construct was used as a positive control. Top panel: PCR products were amplified using a vector‐specific forward primer (BP‐R2‐3195F) and an insert‐specific reverse primer sGFP‐349F (Table S1). Bottom panel: PCR products were amplified using vector‐specific primers (BP‐R2‐3195F and BP‐R2‐3603R) that flank the insert cloning site.
Fig. S5 Bean pod mottle virus (BPMV)‐induced silencing of the transgene in roots. Silencing efficiency induced by BPMV in roots. Quantitative polymerase chain reaction (qPCR) was used to measure the abundance of green fluorescent protein (GFP) transcript in RNA samples isolated from root systems. The root tissues were collected from transgenic GFP soybean plants inoculated with BPMV vector with silencing construct and BPMV empty vector at 21 days post‐inoculation (dpi). The expression levels were calculated using the 2–ΔΔCT method and represent changes in mRNA abundance in silenced plants relative to the empty vector inoculated control. Data are the average of three biological samples, each with three technical replicates. Mean values significantly different from the mock are indicated by an asterisk as determined by paired t‐test (P < 0.01).
Table S1 Primer sequences used in this study.
Acknowledgements
This work was supported by the NSF Plant Genome Research Program (award number 0820642), Iowa Soybean Association, United Soybean Board and North Central Soybean Research Program. This journal paper of the Iowa Agriculture and Home Economics Experiment Station, Ames, IA, project no. 3608, was supported, in part, by Hatch Act and State of Iowa Funds. We thank Dr John Finer for providing seeds of the GFP soybean line.
References
- Alonso, J.M. and Ecker, J.R. (2006) Moving forward in reverse: genetic technologies to enable genome‐wide phenomic screens in Arabidopsis. Nat. Rev. Genet. 7, 524–536. [DOI] [PubMed] [Google Scholar]
- Azpiroz‐Leehan, R. and Feldmann, K.A. (1997) T‐DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet. 13, 152–156. [DOI] [PubMed] [Google Scholar]
- Bennypaul, H.S. , Mutti, J.S. , Rustgi, S. , Kumar, N. , Okubara, P.A. and Gill, K.S. (2011) Virus‐induced gene silencing (VIGS) of genes expressed in root, leaf, and meiotic tissues of wheat. Funct. Integr. Genomics, 12, 143–156. [DOI] [PubMed] [Google Scholar]
- Burch‐Smith, T.M. , Anderson, J.C. , Martin, G.B. and Dinesh‐Kumar, S.P. (2004) Applications and advantages of virus‐induced gene silencing for gene function studies in plants. Plant J. 39, 734 – 746. [DOI] [PubMed] [Google Scholar]
- Chen, J.C. , Jiang, C.Z. , Gookin, T.E. , Hunter, D.A. , Clark, D.G. and Reid, M.S. (2004) Chalcone synthase as a reporter in virus‐induced gene silencing studies of flower senescence. Plant Mol. Biol. 55, 521–530. [DOI] [PubMed] [Google Scholar]
- Constantin, G.D. , Krath, B.N. , MacFarlane, S.A. , Nicolaisen, M. , Johansen, I.E. and Lund, O.S. (2004) Virus‐induced gene silencing as a tool for functional genomics in a legume species. Plant J. 40, 622–631. [DOI] [PubMed] [Google Scholar]
- Constantin, G.D. , Grønlund, M. , Johansen, I.E. , Stougaard, J. and Lund, O.S. (2008) Virus‐induced gene silencing (VIGS) as a reverse genetic tool to study development of symbiotic root nodules. Mol. Plant–Microbe Interact. 21, 720–727. [DOI] [PubMed] [Google Scholar]
- Dalmay, T. , Hamilton, A. , Rudd, S. , Angell, S. and Baulcombe, D.C. (2000) An RNA‐dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Diaz‐Camino, C. , Annamalai, P. , Sanchez, F. , Kachroo, A. and Ghabrial, S.A. (2011) An effective virus‐based gene silencing method for functional genomics studies in common bean. Plant Methods, 7, 16–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, X.S. , Schneider, W.L. , Chaluvadi, S.R. , Rouf, M.A. and Nelson, R.A. (2006) Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Mol. Plant–Microbe Interact. 19, 1229–1239. [DOI] [PubMed] [Google Scholar]
- Faivre‐Rampant, O. , Gilroy, E.M. , Hrubikova, K. , Hein, I. , Millam, S. , Loake, G.J. , Birch, P. , Taylor, M. and Lacomme, C. (2004) Potato virus X‐induced gene silencing in leaves and tubers of potato. Plant Physiol. 134, 1308–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grønlund, M. , Constantin, G. , Piednoir, E. , Kovacev, J. , Johansen, I.E. and Lund, O.S. (2008) Virus‐induced gene silencing in Medicago truncatula and Lathyrus odorata . Virus Res. 135, 345–349. [DOI] [PubMed] [Google Scholar]
- Grønlund, M. , Olsen, A. , Johansen, E.I. and Jakobsen, I. (2010) Protocol: using virus‐induced gene silencing to study the arbuscular mycorrhizal symbiosis in Pisum sativum . Plant Methods, 6, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein, I. , Barciszewska‐Pacak, M. , Hrubikova, K. , Williamson, S. , Dinesen, M. , Soenderby, I.E. , Sundar, S. , Jarmolowski, A. , Shirasu, K. and Lacomme, C. (2005) Virus‐induced gene silencing‐based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol. 138, 2155–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S. , Bradley, J. , Till, B.J. and Comai, L. (2004) TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135, 630–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez‐Garcia, C.M. , Martinelli, A.P. , Bouchard, R.A. and Finer, J.J. (2009) A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep. 28, 837–849. [DOI] [PubMed] [Google Scholar]
- Igarashi, A. , Yamagata, K. , Sugai, T. , Takahashi, Y. , Sugawara, E. , Tamura, A. , Yaegashi, H. , Yamagishi, N. , Takahashi, T. and Isogai, M. (2009) Apple latent spherical virus vectors for reliable and effective virus‐induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology, 386, 407–416. [DOI] [PubMed] [Google Scholar]
- Kaloshian, I. (2007) Virus‐induced gene silencing in plant roots. Methods Mol. Biol. 354, 173–181. [DOI] [PubMed] [Google Scholar]
- Kanazawa, A. , Inaba, J. , Shimura, H. , Otagaki, S. , Tsukahara, S. , Matsuzawa, A. , Kim, B.M. , Goto, K. and Masuta, C. (2011a) Virus‐mediated efficient induction of epigenetic modifications of endogenous genes with phenotypic changes in plants. Plant J. 65, 156–168. [DOI] [PubMed] [Google Scholar]
- Kanazawa, A. , Inaba, J. , Kasai, M. , Shimura, H. and Masuta, C. (2011b) RNA‐mediated epigenetic modifications of an endogenous gene targeted by a viral vector: a potent gene silencing system to produce a plant that does not carry a transgene but has altered traits. Plant Signal Behave. 6, 1090–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, E. and Page, J.E. (2008) Optimized cDNA libraries for virus‐induced gene silencing (VIGS) using tobacco rattle virus. Plant Methods, 22, 4–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. and Dinesh‐Kumar, S.P. (2002) Virus‐induced gene silencing in tomato. Plant J., 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Nakayama, N. , Schiff, M. , Litt, A. , Irish, V.F. and Dinesh‐Kumar, S.P. (2004) Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana . Plant Mol. Biol. 54, 701–711. [DOI] [PubMed] [Google Scholar]
- McGinnis, K. , Chandler, V. , Cone, K. , Kaeppler, H. , Kaeppler, S. , Kerschen, A. , Pikaard, C. , Richards, E. , Sidorenko, L. , Smith, T. , Springer, N. and Wulan, T. (2005) Transgene‐induced RNA interference as a tool for plant functional genomics. Methods Enzymol. 392, 1–24. [DOI] [PubMed] [Google Scholar]
- Meng, Y. , Moscou, M.J. and Wise, R.P. (2009) Blufensin1 negatively impacts basal defense in response to barley powdery mildew. Plant Physiol. 149, 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamatsu, A. , Matsuta, A. , Senda, M. , Matsuura, H. , Kasai, A. , Hong, J.S. , Kitanura, K. , Abe, J. and Kanazawa, A. (2007) Functional analysis of soybean genes involved in flavonoid biosynthesis by virus‐induced gene silencing. Plant Biotechnol. J. 5, 778–790. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.C. , Elmayan, T. , Pollien, J.M. and Vaucheret, H. (1997) Systemic acquired silencing: transgene‐specific post‐transcriptional silencing is transmitted by grafting from silenced stocks to non‐silenced scions. EMBO J. 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue, G.P. , Lindbo, J.A. , Garger, S.J. and Fitzmaurice, W.P. (2002) Making an ally from an enemy: plant virology and the new agriculture. Annu. Rev. Phytopathol. 40, 45–74. [DOI] [PubMed] [Google Scholar]
- Purkayastha, A. and Dasgupta, I. (2009) Virus‐induced gene silencing: a versatile tool for discovery of gene functions in plants. Plant Physiol. Biochem. 47, 967–976. [DOI] [PubMed] [Google Scholar]
- Ryu, C.M. , Anand, A. , Kang, L. and Mysore, K.S. (2004) Agrodrench: a novel and effective agroinoculation method for virus‐induced gene silencing in roots and diverse solanaceous species. Plant J. 40, 322–331. [DOI] [PubMed] [Google Scholar]
- Saedler, R. and Baldwin, I.T. (2004) Virus‐induced gene silencing of jasmonate‐induced direct defences, nicotine and trypsin proteinase‐inhibitors in Nicotiana attenuata . J. Exp. Bot. 55, 151–157. [DOI] [PubMed] [Google Scholar]
- Schmutz, J. , Cannon, S.B. , Schlueter, J. , Ma, J. , Mitros, T. , Nelson, W. , Hyten, D.L. , Song, Q. , Thelen, J.J. , Cheng, J. , Xu, D. , Hellsten, U. , May, G.D. , Yu, Y. , Sakurai, T. , Umezawa, T. , Bhattacharyya, M.K. , Sandhu, D. , Valliyodan, B. , Lindquist, E. , Peto, M. , Grant, D. , Shu, S. , Goodstein, D. , Barry, K. , Futrell‐Griggs, M. , Abernathy, B. , Du, J. , Tian, Z. , Zhu, L. , Gill, N. , Joshi, T. , Libault, M. , Sethuraman, A. , Zhang, X.C. , Shinozaki, K. , Nguyen, H.T. , Wing, R.A. , Cregan, P. , Specht, J. , Grimwood, J. , Rokhsar, D. , Stacey, G. , Shoemaker, R.C. and Jackson, S.A. (2010) Genome sequence of the palaeopolyploid soybean. Nature, 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Scofield, S.R. , Huang, L. , Brandt, A.S. and Gill, B.S. (2005) Development of a virus‐induced gene‐silencing system for hexaploid wheat and its use in functional analysis of the Lr21‐mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. and Mysore, K.S. (2011) Virus‐induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato. Plant Biotechnol. J. 9, 797–806. [DOI] [PubMed] [Google Scholar]
- Sonoda, S. and Nishiguchi, M. (2000) Graft transmission of post‐transcriptional gene silencing: target specificity for RNA degradation is transmissible between silenced and non‐silenced plants, but not between silenced plants. Plant J. 21, 1–8. [DOI] [PubMed] [Google Scholar]
- Turnage, M.A. , Muangsan, N. , Peele, C.G. and Robertson, D. (2002) Geminivirus‐based vectors for gene silencing in Arabidopsis. Plant J. 30, 107–117. [DOI] [PubMed] [Google Scholar]
- Valentine, T.A. , Roberts, I.M. and Oparka, K.J. (2002) Inhibition of tobacco mosaic virus replication in lateral roots is dependent on an activated meristem‐derived signal. Protoplasma, 219, 184–196. [DOI] [PubMed] [Google Scholar]
- Yamagishi, N. and Yoshikawa, N. (2009) Virus induced gene silencing in seeds and emergence stage of soybean plants with Apple latent spherical virus vectors. Plant Mol. Biol. 71, 15–24. [DOI] [PubMed] [Google Scholar]
- Yuan, C. , Li, C. , Yan, L. , Jackson, A.O. , Liu, Z. , Han, C. , Yu, J. and Li, D. (2011) A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. Plos ONE 6, e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. and Ghabrial, S.A. (2006) Development of Bean pod mottle virus‐based vectors for stable protein expression and sequence‐specific virus‐induced gene silencing in soybean. Virology, 344, 401–411. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Yang, C. , Whitham, S.A. and Hill, J.H. (2009) Development and use of an efficient DNA‐based viral gene‐silencing vector for soybean. Mol. Plant–Microbe Interact. 22, 123–131. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Bradshaw, J.D. , Whitham, S.A. and Hill, J.H. (2010) The development of an efficient multipurpose Bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol. 153, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Experimental procedures
Fig. S1 Sequence location and cloning orientation influence silencing potential triggered by Bean pod mottle virus (BPMV). (A) Schematic representation of partial green fluorescent protein (GFP) sequences used for transgene silencing. Two GFP fragments targeting the 3′ or 5′ half were cloned in sense or antisense orientation in the BPMV vector and the silencing responses were quantified using quantitative polymerase chain reaction (qPCR). The silencing efficiencies are indicated. Arrows indicate the orientation of the insert. The expression levels were calculated using the 2–ΔΔCT method and represent changes in mRNA abundance in BPMV‐inoculated plants relative to the mock‐treated control. Data are the average of three biological samples, each with three technical replicates. Mean values significantly different from the mock are indicated by an asterisk as determined by paired t‐test (P < 0.01). (B) Quantification of gene silencing induced by BPMV in leaves using qPCR. The expression levels of GFP mRNA were measured in RNA samples isolated from the second trifoliate of the transgenic GFP plants at 21 days post‐inoculation (dpi) and compared with those receiving empty vector or mock. The expression levels were calculated using the 2–ΔΔCT method and represent changes in mRNA abundance in BPMV‐inoculated plants relative to the mock‐treated control. Data are the average of three biological samples, each with three technical replicates. Mean values significantly different from the mock are indicated by an asterisk as determined by paired t‐test (P < 0.01). (C) Silencing of GFP in the second trifoliate at 21 dpi by the indicated silencing constructs. Mock leaves, leaves inoculated with empty vector and silencing vectors 1101‐A, 1101‐C, 1102‐A and 1102‐D are shown. The leaves were visualized under UV light and photographed (3‐s exposure on a Zeiss Stemi SV11 stereoscope; Carl Zeiss, Thornwood, NY, USA). Note the strongest reduction in GFP fluorescence in the trifoliates from the plants inoculated with silencing construct 1102‐A compared with the other treatments.
Fig. S2 Sustained Bean pod mottle virus (BPMV)‐induced silencing in foliar tissue. (A, B) Silencing of green fluorescent protein (GFP) in the first trifoliate at 14 days post‐inoculation (dpi) (A) and 21 dpi (B). (C, D) Silencing of GFP in the second trifoliate at 21 dpi (C) and 28 dpi (D). (E, F) Silencing of GFP in the third trifoliate at 28 dpi (E) and 35 dpi (F). In each group, the top panel shows a single leaflet from an individual mock plant and the bottom panel shows a single leaflet from an individual plant inoculated with the silencing construct. Each leaflet was visualized with UV light and photographed (3‐s exposure on a Zeiss Stemi SV11 stereoscope). Note the persistent silencing in the leaflets from all three spatial locations for the duration of 7 days.
Fig. S3 Bean pod mottle virus (BPMV)‐induced silencing of the transgene in flowers. Flowers at different developmental stages from individual transgenic green fluorescent protein (GFP) soybean plants treated with silencing vector (top three panels), BPMV empty vector (middle three panels) or mock (bottom three panels) visualized under white light (left panels) and UV light (right panels) and photographed (1‐s exposure on a Zeiss Stemi SV11 stereoscope) at 7 weeks post‐inoculation. Each panel shows flowers from a single plant. Similar results were obtained from three independent experiments.
Fig. S4 Long‐lasting insert stability of Bean pod mottle virus (BPMV) virus‐induced gene silencing (VIGS) in soybean. Panels show amplified BPMV RNA2 products. Each bracket denotes three individual polymerase chain reactions (PCRs) conducted on cDNA from a transgenic soybean plant inoculated with a virus harbouring either a silencing construct or an empty vector, or a mock‐inoculated plant, respectively. The vector harbouring the 1102‐A silencing construct was used as a positive control. Top panel: PCR products were amplified using a vector‐specific forward primer (BP‐R2‐3195F) and an insert‐specific reverse primer sGFP‐349F (Table S1). Bottom panel: PCR products were amplified using vector‐specific primers (BP‐R2‐3195F and BP‐R2‐3603R) that flank the insert cloning site.
Fig. S5 Bean pod mottle virus (BPMV)‐induced silencing of the transgene in roots. Silencing efficiency induced by BPMV in roots. Quantitative polymerase chain reaction (qPCR) was used to measure the abundance of green fluorescent protein (GFP) transcript in RNA samples isolated from root systems. The root tissues were collected from transgenic GFP soybean plants inoculated with BPMV vector with silencing construct and BPMV empty vector at 21 days post‐inoculation (dpi). The expression levels were calculated using the 2–ΔΔCT method and represent changes in mRNA abundance in silenced plants relative to the empty vector inoculated control. Data are the average of three biological samples, each with three technical replicates. Mean values significantly different from the mock are indicated by an asterisk as determined by paired t‐test (P < 0.01).
Table S1 Primer sequences used in this study.


