Arbuscular mycorrhizal (AM) fungi belong to the order Glomeromycota and arguably form the most successful symbiosis in plants. This is a very ancient symbiosis which originated approximately 450 million years ago and is thought to have facilitated the evolution of land plants. The ability to host AM fungi inside plant roots has since been maintained by the vast majority (>80%) of current land plant species, underlining the importance of this symbiosis (Smith and Read, 2008). AM fungi offer the plant increased access to water and scarce nutrients, especially phosphates and nitrates, in return for photosynthates. In addition, AM fungi can provide protection to pathogens. At the heart of the AM symbiosis is the formation of symbiotic host membrane compartments containing highly branched hyphae inside cortical cells of the root, termed arbuscules, where nutrients are exchanged in a controlled manner (Gutjahr and Parniske, 2013).
Strikingly, there is very little host specificity in the interaction with AM fungi, although differences in the symbiotic efficiency of different plant–fungus combinations have been observed (Smith and Read, 2008). In other words, a single/individual AM fungus has the capacity to colonize intracellularly almost all land plants. Furthermore, the same mycelium can colonize different plants at the same time (Smith and Read, 2008). This must involve an extraordinary level of compatibility and ability to adapt to different hosts by the fungus. The molecular and genetic basis for this compatibility is still largely unknown, but probably involves a complex molecular dialogue which, in part, includes the perception of (lipo‐)chitooligosaccharide signals from the fungus (Gutjahr and Parniske, 2013). Understanding the genetic make‐up of the fungus is essential to obtain an insight into this fascinating aspect of the symbiosis.
AM fungi have several unique/unusual characteristics that have severely complicated molecular genetic studies (Sanders and Croll, 2010). AM fungi are obligate biotrophs that need a plant to complete their life cycle. Their hyphal network forms a continuous (coenocytic) cytoplasmic compartment in which numerous nuclei move and migrate. Spores typically contain several hundreds of nuclei that migrate from the hypha into the spore and subsequently divide. Thus, there is never a stage during the AM life cycle in which only one nucleus initiates the next generation (Sanders and Croll, 2010). As a result, genetic transformation of an AM fungus has proven to be notoriously unstable and, despite various attempts, stable transformation of AM fungi has met with little success. Furthermore, in the >200 species of Glomeromycota that have been studied, a sexual stage has never been observed. Therefore, these fungi are considered to be ancient asexual organisms. However, hyphae of different (related) fungal individuals have been shown to fuse and exchange nuclei by anastomosis. Even more striking is the observation that a substantial level of genetic variation can be contained within a fungal individual (heterokaryosis). This has led to the highly speculative hypothesis that the genome space of a single AM fungus is, in fact, the equivalent of multiple (diverse) nuclear genomes, called nucleotypes (Sanders and Croll, 2010). However, others have argued that nuclei are identical (homokaryotic), but show increased ploidy or gene duplications and are heterozygotic.
Recently, two studies have revealed the first genome sequence of an AM fungus, Rhizophagus irregularis isolate DAOM197198, providing the first insight into the genetic make‐up of this intriguing fungus (Lin et al., 2014; Tisserant et al., 2013). Both studies revealed a strikingly low level of polymorphism and no evidence for extensive segmental duplications in the ∼150‐Mb haploid genome. Therefore, the possibility of increased ploidy or a highly duplicated genome can be rejected for this strain (Lin et al., 2014; Tisserant et al., 2013). Furthermore, the sequencing of four individual nuclei unambiguously showed that they are near identical and lack extensive genome rearrangements (Lin et al., 2014).
The new genome data strongly contradict earlier studies which suggest that nuclei of R. irregularis are highly polymorphic (Sanders and Croll, 2010). This discrepancy is, in part, explained by the markers that were used in these studies. Polymorphism studies in mycorrhizal fungi have often been conducted using the 45S rDNA marker. However, this repetitive locus is highly polymorphic within a single nucleus (Lin et al., 2014), which complicates diversity studies among nuclei from a single individual. Nevertheless, evidence remains that nuclei within a single fungus can be polymorphic, as both genome studies report slightly different intragenic levels of polymorphism, with Lin et al. (2014) showing approximately four times less [∼0.1 single nuclear polymorphism (SNP)/kb] polymorphisms than Tisserant et al. (2013) (∼0.43 SNP/kb). Comparison of the reference assemblies of both studies indicates a (low) level of approximately 0.8% polymorphisms between both assemblies. Although part of this difference might be explained by technical issues and different assembly methods, it may also be caused by the fact that the fungal material used for sequencing originated from monoaxenic Agrobacterium rhizogenes‐transformed root cultures involving different plant host species; i.e. carrot versus chicory. As both cultures were started from the same DAOM197198 strain, this suggests that the ancestral strain was heterogenic and contained genetically (slightly) different nuclei. The divergence observed between the two extant strains of these studies may be a result of selection on different host species favouring different nucleotypes and/or of genetic drift (see below). Together, these results demonstrate that strains of a mycorrhizal fungus can become basically homokaryotic on continued growth under monoaxenic conditions. However, the observed divergence between the two strains descending from a common ancestor suggests that genetically divergent nuclei can arise and coexist in a single mycelium. Indeed, the observed exchange of nuclei during anastomoses between related AM fungi makes a heterokaryotic nature of the fungus in a natural habitat very likely. Thus, despite the occurrence of genetically slightly diverse nuclei within AM individuals in nature, the near‐isogenic sequenced R. irregularis isolates support the notion that genetic homogenization occurs on continued growth in monoaxenic root organ cultures of a single host plant species.
As a result of the presence of genetically different nuclei, spores may obtain different combinations of nucleotypes from the mycelium. This segregation of genetically different nuclei was indeed suggested by the work of Angelard et al. (2010). By genotyping subcultured individual progeny spores (called single spore lines) from a parental line, it was shown that single spore lines obtained different allele frequencies for polymorphic markers present in the parental lines. This work further showed that the segregated single spore lines had different effects on plant growth and plant gene expression (Angelard et al., 2010). Some progeny lines improved significantly plant performance compared with the parental lines. This highlights the importance of understanding the maintenance and selection of genetic variation within AM fungi, and the potential to generate ‘optimized’ mycorrhizal inoculum. In contrast with the single spore lines, subculturing of many spores and hyphae together did not cause significant genetic variation in allele frequencies. To explain this, modelling approaches have confirmed that continuous exchange of nuclei through anastomosis can be sufficient to maintain the overall genetic variation within an isolate (Bever et al., 2008). However, to what extent nuclei are exchanged via anastomosis in nature is unknown. Extensive anastomosis also contrasts with the observed homokaryotic nature of the sequenced DAOM197198w strain by Lin et al. (2014). It has been reported that repeated subculturing of the same isolate negatively affects the frequency of anastomoses. Therefore, reduced anastomosis might facilitate the segregation of nuclei. At the same time, it suggests that there are effective mechanisms in the fungus to homogenize genetic differences. It also raises another intriguing question: ‘Is there selection by the host plant species on the genetic composition of the fungus?’.
A recent study by Angelard et al. (2014) suggests that a change in host plant can induce genotypic plasticity in the fungus. It was shown that the transfer of AM lines to a different host plant species, i.e. from carrot to potato, induced significant changes in polymorphic allele frequency. Furthermore, these changes appeared not to be random. Although different lines showed varying degrees of change in allele frequency, similar changes occurred among the different lines (Angelard et al., 2014). This suggests that different hosts might select for preferred allele combinations. It was therefore proposed that induced changes in nucleotype frequency might play an important role in the adaptation to a different environment as an alternative to sexual reproduction. The availability of a near‐isogenic line, strain DAOM197198w, with no to very low levels of polymorphism, as reported by Lin et al. (2014), could be used to test the relevance of genetic variability in host adaptation. If relevant, the decreased genetic variation in this line would be expected to impair the ability of the fungus to colonize a wide variety of host plants.
How genetically different nuclei can be selected is currently unknown and remains a major research objective. One possible mechanism to select for specific nucleotypes could involve the selection of nuclei at the stage of arbuscule formation. It has been shown that the plant can sanction mycorrhizal fungi in cases in which they are less cooperative, which probably occurs at the arbuscule stage (Kiers et al., 2011). Thus, when a certain nucleotype triggers (more) host sanctioning locally at the arbuscule, this could prevent the proliferation of that nucleotype and the ability to populate the hypha. Indeed, it has been shown that fungal nuclei proliferate in the arbuscule trunk as well as in the intraradical mycelium. However, it remains unclear to what extent genetically different nuclei populate the arbuscule and the associated trunk and hyphal domain. In cases in which these nuclei are clonal, a preferential proliferation of symbiotically effective nuclei in (the vicinity of) an individual arbuscule could occur. We postulate that the interplay between fungus and plant controls this process. In this way, an enrichment of a certain nucleotype in the mycelium will occur that will fill up the spores (Fig. 1).
Figure 1.
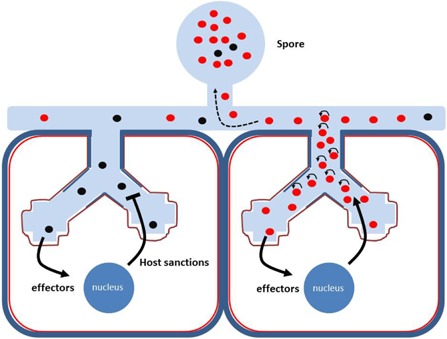
Postulated mechanistic model for host selection of preferred arbuscular mycorrhizal (AM) nucleotypes at the arbuscule. In this hypothetical example, two nucleotypes (genetically different nuclei marked in black and red) occur in the hyphal network. Arbuscule formation may involve a clonal population of nuclei originating from one of the two nucleotypes, here represented by two cells containing arbuscules. Genetically different nuclei can differ in the type/efficiency of effector proteins that are secreted and translocated across the symbiotic interface into the host cells. Such effectors can be important to suppress defence responses triggered by microbe‐associated molecular patterns (MAMPs) of the fungus and/or to stimulate the symbiotic performance of the arbuscule. Impaired suppression of defence or symbiotic performance will result in local host sanctions which impair the proliferation of the nuclei in (the vicinity of) the arbuscule. As a result, better performing nuclei (red nuclei in the right arbuscule) will proliferate more and will preferentially populate the hyphal network. As spores are filled up by the migrating nuclei, this will result in a shift in the ratio of black versus red nuclei in the progeny spores.
In this respect, it is also interesting that transcriptome and genome analyses in R. irregularis have identified a selective range of putative secreted proteins that are possibly streamlined to its obligate biotrophic lifestyle (Lin et al., 2014; Tisserant et al., 2013). For example, the observed decreased repertoire of secreted cell wall‐degrading enzymes is thought to play an important role in preventing the activation of a strong defence response during the biotrophic interaction with the plant root (Tisserant et al., 2013). Nevertheless, AM fungi contain microbe‐associated molecular patterns (MAMPs), such as chitin, which trigger the activation of defence responses in the plant. Therefore, AM fungi must have effective ways to prevent/suppress these defence responses. Several of the secreted proteins may act as effector proteins. Secreted effector proteins play a major role in biotrophic plant–pathogen interactions to suppress defence or reorganize the host cells to facilitate intracellular accommodation. Somewhat similar to arbuscules, (hemi)biotrophic pathogenic fungi and oomycetes can form feeding interfaces inside living plant cells, called haustoria, in which hyphal outgrowths are contained in a specialized host membrane through which they take up nutrients from the plant. Effectors are often secreted from these interfaces. A potential role for AM effectors is supported by the observation that numerous putative AM effectors are specifically induced during the interaction with the plant (Tisserant et al., 2013). Furthermore, the AM effector, SP7, has been identified recently and shown to be able to translocate to the nucleus of the plant cells and manipulate defence responses by interacting with an ethylene response factor (ERF) transcription factor (Kloppholz et al., 2011). We hypothesize that different effectors may have plant species‐specific effects, which could be a driving force for nucleotype selection. In other words, genetically different nuclei might express (slightly) different effectors that affect the efficiency of the symbiosis locally, i.e. at the arbuscule stage (or the associated intracellular mycelium). Local sanctions of the plant could prevent these nuclei from proliferating and thereby change the ratio of nucleotypes in the mycelium. To test this, laser microdissection could be used to isolate individual arbuscule cells to profile the fungal genotypes and (effector) transcriptomes. Application of this on various host plant species using genetically different fungal isolates could reveal whether effectors are deployed in plant species‐specific ways and whether the selection of nucleotypes occurs at the arbuscule stage. If this hypothesis is true, it would bring together two fascinating aspects of AM biology, i.e. the control of biotrophy and the selection of genomes by the plant.
References
- Angelard, C. , Colard, A. , Niculita‐Hirzel, H. , Croll, D. and Sanders, I.R. (2010) Segregation in a mycorrhizal fungus alters rice growth and symbiosis‐specific gene transcription. Curr. Biol. 20, 1216–1221. [DOI] [PubMed] [Google Scholar]
- Angelard, C. , Tanner, C.J. , Fontanillas, P. , Niculita‐Hirzel, H. , Masclaux, F. and Sanders, I.R. (2014) Rapid genotypic change and plasticity in arbuscular mycorrhizal fungi is caused by a host shift and enhanced by segregation. ISME J. 8, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever, J.D. , Kang, H.‐J. , Kaonongbua, W. and Wang, M. (2008) Genomic organization and mechanismsof inheritance in arbuscular mycorrhizal fungi: contrasting the evidence and implications of current theories In: Mycorrhiza (Varma A., ed.), pp. 135–148. Berlin, Heidelberg: Springer. [Google Scholar]
- Gutjahr, C. and Parniske, M. (2013) Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell Dev. Biol. 29, 593–617. [DOI] [PubMed] [Google Scholar]
- Kiers, E.T. , Duhamel, M. , Beesetty, Y. , Mensah, J.A. , Franken, O. , Verbruggen, E. , Fellbaum, C.R. , Kowalchuk, G.A. , Hart, M.M. , Bago, A. , Palmer, T.M. , West, S.A. , Vandenkoornhuyse, P. , Jansa, J. and Bücking, H. (2011) Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science, 333, 880–882. [DOI] [PubMed] [Google Scholar]
- Kloppholz, S. , Kuhn, H. and Requena, N. (2011) A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr. Biol. 21, 1204–1209. [DOI] [PubMed] [Google Scholar]
- Lin, K. , Limpens, E. , Zhang, Z. , Ivanov, S. , Saunders, D.G. , Mu, D. , Pang, E. , Cao, H. , Cha, H. , Lin, T. , Zhou, Q. , Shang, Y. , Li, Y. , Sharma, T. , van Velzen, R. , de Ruijter, N. , Aanen, D.K. , Win, J. , Kamoun, S. , Bisseling, T. , Geurts, R. and Huang, S. (2014) Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 10, e1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, I.R. and Croll, D. (2010) Arbuscular mycorrhiza: the challenge to understand the genetics of the fungal partner. Annu. Rev. Genet. 44, 271–292. [DOI] [PubMed] [Google Scholar]
- Smith, S.E. and Read, D. (2008) Mycorrhizal Symbiosis. San Diego, CA: Academic Press. [Google Scholar]
- Tisserant, E. , Malbreil, M. , Kuo, A. , Kohler, A. , Symeonidi, A. , Balestrini, R. , Charron, P. , Duensing, N. , Frei dit Frey, N. , Gianinazzi‐Pearson, V. , Gilbert, L.B. , Handa, Y. , Herr, J.R. , Hijri, M. , Koul, R. , Kawaguchi, M. , Krajinski, F. , Lammers, P.J. , Masclaux, F.G. , Murat, C. , Morin, E. , Ndikumana, S. , Pagni, M. , Petitpierre, D. , Requena, N. , Rosikiewicz, P. , Riley, R. , Saito, K. , San Clemente, H. , Shapiro, H. , van Tuinen, D. , Bécard, G. , Bonfante, P. , Paszkowski, U. , Shachar‐Hill, Y.Y. , Tuskan, G.A. , Young, J.P. , Sanders, I.R. , Henrissat, B. , Rensing, S.A. , Grigoriev, I.V. , Corradi, N. , Roux, C. and Martin, F. (2013) Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. USA, 110, 20 117–20 122. [DOI] [PMC free article] [PubMed] [Google Scholar]


