Summary
Boosted responsiveness of plant cells to stress at the onset of pathogen‐ or chemically induced resistance is called priming. The chemical β‐aminobutyric acid (BABA) enhances Arabidopsis thaliana resistance to hemibiotrophic bacteria through the priming of the salicylic acid (SA) defence response. Whether BABA increases Arabidopsis resistance to the necrotrophic bacterium Pectobacterium carotovorum ssp. carotovorum (Pcc) is not clear. In this work, we show that treatment with BABA protects Arabidopsis against the soft‐rot pathogen Pcc. BABA did not prime the expression of the jasmonate/ethylene‐responsive gene PLANT DEFENSIN 1.2 (PDF1.2), the up‐regulation of which is usually associated with resistance to necrotrophic pathogens. Expression of the SA marker gene PATHOGENESIS RELATED 1 (PR1) on Pcc infection was primed by BABA treatment, but SA‐defective mutants demonstrated a wild‐type level of BABA‐induced resistance against Pcc. BABA primed the expression of the pattern‐triggered immunity (PTI)‐responsive genes FLG22‐INDUCED RECEPTOR‐LIKE KINASE 1 (FRK1), ARABIDOPSIS NON‐RACE SPECIFIC DISEASE RESISTANCE GENE (NDR1)/HAIRPIN‐INDUCED GENE (HIN1)‐LIKE 10 (NHL10) and CYTOCHROME P450, FAMILY 81 (CYP81F2) after inoculation with Pcc or after treatment with purified bacterial microbe‐associated molecular patterns, such as flg22 or elf26. PTI‐mediated callose deposition was also potentiated in BABA‐treated Arabidopsis, and BABA boosted Arabidopsis stomatal immunity to Pcc. BABA treatment primed the PTI response in the SA‐defective mutants SA induction deficient 2‐1 (sid2‐1) and phytoalexin deficient 4‐1 (pad4‐1). In addition, BABA priming was associated with open chromatin configurations in the promoter region of PTI marker genes. Our data indicate that BABA primes the PTI response upon necrotrophic bacterial infection and suggest a role for the PTI response in BABA‐induced resistance.
Introduction
Plants are equipped to sense evolutionarily conserved microbial molecular signatures, collectively called microbe‐associated molecular patterns (MAMPs). Recognition of MAMPs activates immune responses (Ausubel, 2005; Boller and Felix, 2009). This general, early defence response is called pattern‐triggered immunity (PTI) (Zhang and Zhou, 2010; Zipfel, 2009), and is characterized by the accumulation of reactive oxygen species, callose deposition and the expression of defence‐related genes (Boudsocq et al., 2010; Gómez‐Gómez et al., 1999; Zhang and Zhou, 2010). Bacterial‐induced stomatal closure is also characteristic of the PTI response (Desclos‐Theveniau et al., 2012; Melotto et al., 2006; Singh et al., 2012; Zeng et al., 2010). The virulent Pseudomonas syringae pv. tomato (Pst) DC3000 reopens stomata in a coronatine (COR)‐dependent manner (Melotto et al., 2006; Zeng et al., 2010).
In Arabidopsis, salicylic acid (SA)‐dependent defence responses are considered to be effective mainly against biotrophic pathogens, such as the oomycete Hyaloperonospora arabidopsidis, the fungus Erysiphe orontii and the hemibiotrophic bacterium P. syringae (Glazebrook, 2005). Accordingly, impaired SA production leads to increased susceptibility to these pathogens. For example, reduction of SA production in SA induction deficient (sid) mutant plants results in increased susceptibility to both virulent and avirulent forms of P. syringae and H. arabidopsidis (Nawrath and Métraux, 1999). Activation of SA signalling can, however, also be effective against necrotrophic pathogens, such as the fungus Botrytis cinerea (Ferrari et al., 2003; Zimmerli et al., 2001). Jasmonic acid (JA)‐dependent signalling mostly mediates resistance to insects and necrotrophic microbial pathogens (Creelman and Mullet, 1997; Norman‐Setterblad et al., 2000; Overmyer et al., 2000; Reymond and Farmer, 1998). The coronatine‐insensitive 1 (coi1) mutant, which is impaired in the perception of JA, generally exhibits enhanced susceptibility to a variety of necrotrophic pathogens, including Alternaria brassicicola and B. cinerea fungi, oomycete Pythium sp. and the bacterium Pectobacterium carotovorum ssp. carotovorum (Pcc) (Norman‐Setterblad et al., 2000; Thomma et al., 1998). It is noteworthy that some coi1 alleles do not show enhanced susceptibility to Pcc (Kariola et al., 2003). The production of ethylene (ET) is one of the earliest plant responses to pathogens (Thomma et al., 1998). The necrotroph Pcc activates the ET‐dependent defence response and the Arabidopsis ethylene‐insensitive 2 (ein2) mutant displays enhanced susceptibility to Pcc (Kazan and Manners, 2008; Norman‐Setterblad et al., 2000). Defence pathways influence each other through a network of regulatory interactions, and thus plant responses to various stress stimuli are a result of this complex interplay (Bostock, 2005; Kunkel and Brooks, 2002). Several studies have described crosstalk among SA, JA and ET signalling pathways (Kunkel and Brooks, 2002; Pieterse et al., 2009). SA and JA signalling pathways interact on many levels and, in most cases, this relationship seems to be mutually antagonistic (Kunkel and Brooks, 2002).
On infection by pathogens or after treatment with various chemicals, plants can establish a unique physiological condition, called the ‘primed’ state (Conrath et al., 2002; Prime‐A‐Plant Group et al., 2006; Van der Ent et al., 2009). Priming is the phenomenon that enables cells to respond to much lower levels of a stimulus in a more rapid and robust manner than do nonprimed cells (Conrath et al., 2002; Conrath, 2011). Plants primed by treatments that induce resistance show a faster and/or stronger activation of defence responses when subsequently challenged by pathogens or abiotic stresses (Beckers et al., 2009; Conrath, 2011; Prime‐A‐Plant Group et al., 2006). Priming is observed in plants and also in animals (Beckers et al., 2009; Jung et al., 2009; Pham et al., 2007). Priming provides low‐cost protection in relatively high disease pressure conditions (van Hulten et al., 2006). Many different organic and inorganic compounds have been shown to induce resistance in plants (Kuć, 2001). Among these, the nonprotein amino acid β‐aminobutyric acid (BABA) has been shown to be a potent inducer of resistance against abiotic stress (Jakab et al., 2005; Zimmerli et al., 2008), nematodes (Oka et al., 1999), insects (Hodge et al., 2005) and microbial pathogens (Cohen, 2002; Jakab et al., 2001). BABA protects plants by priming pathogen‐specific defence responses, notably BABA induces priming of PATHOGENESIS RELATED 1 (PR1) expression on Arabidopsis infection by Pst DC3000 in an SA‐dependent manner (Ton et al., 2005; Zimmerli et al., 2000).
BABA‐induced resistance is efficient against plant microbial pathogens (Cohen, 2002; Jakab et al., 2001; Prime‐A‐Plant Group et al., 2006; Zimmerli et al., 2001), but whether BABA protects Arabidopsis plants against necrotrophic bacteria is not known. We report here that BABA provides Arabidopsis resistance against necrotrophic Pcc bacteria and that resistance is correlated with a primed Arabidopsis PTI response. In addition, we show that BABA treatment induces an open chromatin configuration at the promoter region of primed genes.
Results
BABA‐induced resistance to Pcc SCC1
The priming agent BABA protects Arabidopsis against hemibiotrophic bacteria (Ton et al., 2005; Tsai et al., 2011; Zimmerli et al., 2000), but the effect of this chemical in Arabidopsis against necrotrophic bacteria, such as Pcc, is not known. To further evaluate the spectrum of action of BABA, the resistance of BABA‐ and water‐treated Arabidopsis to virulent Pcc SCC1 infections was compared. Pretreatment with BABA enhanced Arabidopsis resistance to Pcc SCC1 at both symptom and titre levels at 1 day post‐inoculation (dpi) (Fig. 1A,B). BABA‐treated Arabidopsis indeed demonstrated less Pcc SCC1‐mediated water‐soaked lesions (Fig. 1A) and Pcc SCC1 titres per leaf area were about 50 times lower in BABA‐treated Arabidopsis when compared with water control plants (Fig. 1B). The protective effect of BABA lasted for at least 2 days (Fig. S1, see Supporting Information). These data indicate that BABA pretreatment protects Arabidopsis plants against the necrotrophic bacterium Pcc.
Figure 1.
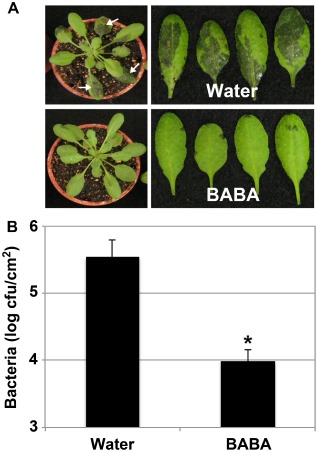
Enhanced resistance of β‐aminobutyric acid (BABA)‐treated Arabidopsis to Pectobacterium carotovorum ssp. carotovorum (Pcc) SCC1 infection. (A) Pcc SCC1‐mediated water‐soaked lesions at 1 day post‐inoculation (dpi). Five‐week‐old Arabidopsis plants were dipped in a bacterial solution of 5 × 106 colony‐forming units (cfu)/mL. White arrows indicate Pcc SCC1‐mediated water‐soaked lesions. (B) Quantification of Pcc SCC1 infection in water‐ and BABA‐treated Arabidopsis plants. Plants were dip inoculated as in (A) and bacterial titres were evaluated at 1 dpi. Data represent average values of three independent biological replicates ± standard deviation (SD) (n = 9). A statistically significant difference compared with water‐treated Col‐0 controls is indicated with an asterisk (P < 0.05) based on Student's t‐test.
Roles of JA and ET defence signalling pathways in BABA‐induced resistance to Pcc SCC1 infection
The JA and ET defence responses are activated upon Pcc infection in Arabidopsis (Kariola et al., 2003; Norman‐Setterblad et al., 2000), and defective JA/ET signalling generally leads to enhanced sensitivity to Pcc (Norman‐Setterblad et al., 2000). JA/ET defence signalling is thus critical for Arabidopsis resistance to Pcc. We therefore evaluated the expression levels of the JA/ET marker gene PLANT DEFENSIN 1.2 (PDF1.2) by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) after Pcc SCC1 infection. As expected (Kariola et al., 2003; Norman‐Setterblad et al., 2000), the up‐regulation of Arabidopsis PDF1.2 was observed at 16, 18 and 24 h post‐inoculation (hpi) with Pcc SCC1 (Fig. 2A). At these time points, water‐ and BABA‐treated Arabidopsis demonstrated similar levels of PDF1.2 mRNA accumulation, suggesting that BABA does not prime the JA/ET response upon Pcc infection. To further evaluate the role of JA/ET defence signalling cascades in BABA‐induced resistance to Pcc SCC1, we analysed the effect of BABA in JA and ET signalling mutants. Both JA‐insensitive coi1‐16 (Ellis and Turner, 2002) and ET‐insensitive ein2‐1 (Guzman and Ecker, 1990) mutants were protected by BABA application. As shown in Fig. 2B, these mutants clearly developed less Pcc SCC1‐mediated water‐soaked lesions after BABA treatment. In addition, bacterial titres were about 10 times lower in coi1‐16 and ein2‐1 mutants pretreated with BABA (Fig. 2C). Therefore, coi1‐16 and ein2‐1 mutants displayed wild‐type levels of BABA‐induced resistance against Pcc SCC1 infection. Collectively, these data suggest that JA/ET signalling is not critical for BABA‐mediated Arabidopsis resistance to Pcc.
Figure 2.

Role of jasmonic acid/ethylene (JA/ET) defence signalling cascades in β‐aminobutyric acid (BABA)‐induced resistance to Pectobacterium carotovorum ssp. carotovorum (Pcc). (A) Time course of PLANT DEFENSIN 1.2 (PDF1.2) expression in Arabidopsis on Pcc SCC1 infection. Arabidopsis plants were treated with water or BABA and challenged 48 h later with virulent Pcc SCC1 at a concentration of 5 × 106 colony‐forming units (cfu)/mL. Leaves were harvested at 16, 18 and 24 h post‐inoculation (hpi). Transcript levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to both UBQ10 and EF‐1. Relative gene expression levels were compared with water controls (defined value of one). Data represent the means and standard deviation (SD) of three independent biological replicates (n = 9). Values of BABA‐treated plants were not significantly different from those of water‐treated controls based on Student's t‐test (P < 0.01). (B) Pcc SCC1‐mediated symptoms of water‐ and BABA‐treated JA/ET signalling mutants. Five‐week‐old Arabidopsis plants were dip inoculated in 5 × 106 cfu/mL P cc SCC1 and photographs were taken 16 h (ethylene‐insensitive 2‐1, ein2‐1) or 1 day (coronatine‐insensitive 1‐16, coi1‐16) later. White arrows indicate Pcc SCC1‐mediated water‐soaked lesions. (C) Pcc SCC1 titres in JA/ET signalling mutants. Water (W)‐ and BABA (B)‐treated plants were dip inoculated as in (B) and bacterial titres were evaluated at 1 day post‐inoculation (dpi). Average values ± SD from three independent experiments are presented (n = 9). Asterisks indicate a significant difference from water controls based on Student's t‐test (P < 0.05).
Priming of the SA‐dependent defence signalling cascade by BABA upon Pcc SCC1 infection
BABA primes the expression of SA‐responsive genes upon infection with hemibiotrophic Pst DC3000 bacteria (Ton et al., 2005; Tsai et al., 2011; Zimmerli et al., 2000) and the necrotrophic fungus B. cinerea (Zimmerli et al., 2001). To determine whether BABA also potentiates the Arabidopsis SA defence signalling cascade on Pcc infection, we monitored the transcript accumulation of PR1 by qRT‐PCR at 16, 18 and 24 hpi with the wild‐type Pcc SCC1. Expression levels of the SA‐responsive PR1 were higher in Pcc‐inoculated BABA‐treated plants than in water controls, suggesting that BABA potentiates PR1 expression upon Pcc SCC1 infection (Fig. 3A). To further evaluate the role of SA signalling in BABA‐induced Arabidopsis resistance to Pcc, we tested the responses of SA biosynthesis (sid2‐1) (Nawrath and Métraux, 1999) and SA signalling (phytoalexin deficient 4‐1, pad4‐1) (Glazebrook et al., 1997) mutants to Pcc SCC1 infection. As expected, sid2‐1 and pad4‐1 did not show up‐regulation or BABA‐mediated priming of PR1 at 18 hpi with Pcc SCC1 (Fig. S2, see Supporting Information). However, both BABA‐treated SA‐defective mutants demonstrated a reduction in the formation of Pcc SCC1‐mediated water‐soaked lesions when compared with water‐treated controls (Fig. 3B). In addition, BABA treatment induced a reduction in bacterial titres in BABA‐treated sid2‐1 and pad4‐1, suggesting a functional BABA‐induced resistance against Pcc SCC1 infection in these SA‐defective mutants (Fig. 3C). Collectively, these data suggest that, although BABA primes SA‐dependent defence responses upon Pcc SCC1 infection, SA signalling is not critical for BABA‐induced resistance to Pcc SCC1.
Figure 3.

Involvement of salicylic acid (SA) signalling in β‐aminobutyric acid (BABA)‐induced resistance to Pectobacterium carotovorum ssp. carotovorum (Pcc) SCC1. (A) Expression analyses of the SA‐responsive PATHOGENESIS RELATED 1 (PR1). Arabidopsis plants were treated with water or BABA and challenged 48 h later with Pcc SCC1 at a concentration of 5 × 106 colony‐forming units (cfu)/mL. RNAs were harvested 16, 18 and 24 h post‐inoculation (hpi). Transcript levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to both UBQ10 and EF‐1. Relative gene expression levels were compared with water controls (defined value of one). Data represent the means and standard deviation (SD) of three independent biological replicates (n = 9). Asterisks indicate a significant difference from water controls based on Student's t‐test (P < 0.01). (B) Pathogen‐induced lesion formation in SA‐defective mutants. Five‐week‐old SA induction deficient 2‐1 (sid2‐1) and phytoalexin deficient 4‐1 (pad4‐1) mutant plants were dip inoculated in 5 × 106 cfu/mL P cc SCC1 and photographs were taken 1 day later. White arrows indicate Pcc SCC1‐mediated water‐soaked lesions. (C) Pcc SCC1 proliferation in Col‐0 and SA‐defective mutants. Water (W)‐ and BABA (B)‐treated plants were dip‐inoculated as in (B) and bacterial titres were evaluated at 1 day post‐inoculation (dpi). Average values ± SD from three independent experiments are presented (n = 9). Asterisks indicate a significant difference from water controls based on Student's t‐test (P < 0.05).
BABA‐induced potentiation of PTI‐responsive gene expression and callose deposition
PTI is the first active defence layer in plant immunity (Jones and Dangl, 2006), and our data suggest that JA/ET and SA defence signalling cascades are not critical for BABA‐induced Arabidopsis resistance to Pcc (Figs 2, 3). To test whether the chemical BABA primes the PTI response in Arabidopsis, expression levels of the PTI‐responsive genes FLG22‐INDUCED RECEPTOR‐LIKE KINASE 1 (FRK1), ARABIDOPSIS NON‐RACE SPECIFIC DISEASE RESISTANCE GENE (NDR1)/HAIRPIN‐INDUCED GENE (HIN1)‐LIKE 10 (NHL10) and CYTOCHROME P450, FAMILY 81 (CYP81F2) (Boudsocq et al., 2010) were analysed after inoculation with Pcc strain WPP17. This Pcc strain is deficient in several hypersensitive response and pathogenicity (hrp) and hrp conserved (hrc) genes, and thus mainly activates the host PTI response (Yap et al., 2004). Accordingly, strain Pcc WPP17 is much less infective than the wild‐type Pcc SCC1 (Fig. S3, see Supporting Information). On Pcc WPP17 infection, BABA‐treated Arabidopsis accumulated more FRK1, NHL10 and CYP81F2 mRNAs than the water controls, suggesting that BABA primes bacterial‐mediated PTI marker gene expression (Fig. 4A). Priming of the expression of PTI‐responsive genes was also observed after treatment with the peptide flg22, the biologically active epitope of the bacterial MAMP flagellin (Fig. 4B), and with the purified bacterial MAMP EF‐Tu (elf26) (Fig. 4C).
Figure 4.
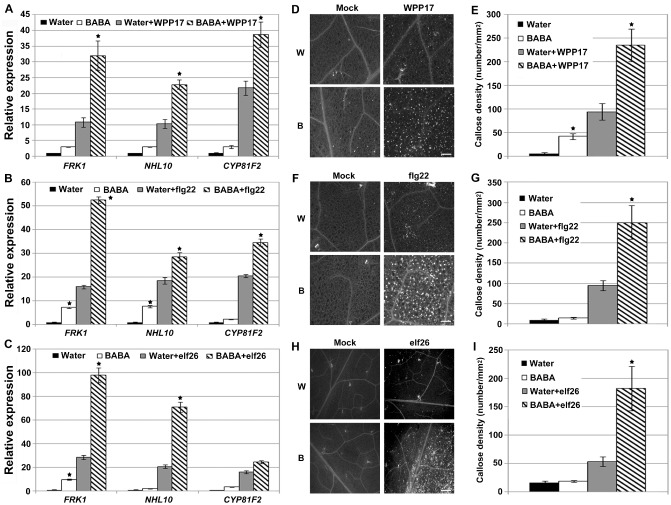
Effect of β‐aminobutyric acid (BABA) treatments on the Arabidopsis pattern‐triggered immunity (PTI) response. (A) BABA potentiates the expression of FLG22‐INDUCED RECEPTOR‐LIKE KINASE 1 (FRK1), ARABIDOPSIS NON‐RACE SPECIFIC DISEASE RESISTANCE GENE (NDR1)/HAIRPIN‐INDUCED GENE (HIN1)‐LIKE 10 (NHL10) and CYTOCHROME P450, FAMILY 81 (CYP81F2) on Pectobacterium carotovorum ssp. carotovorum (Pcc) WPP17 infection. Transcript levels were analysed at 2 h post‐inoculation (hpi) with 1 × 108 colony‐forming units (cfu)/mL Pcc WPP17. (B,C) Potentiated expression of PTI‐responsive genes 30 min after infiltration with 1 μM flg22 (B) or elf26 (C). For (A–C), transcript levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to both UBQ10 and EF‐1. Relative gene expression levels were compared with water controls (defined value of one). Data represent the means and standard deviation (SD) of three independent biological replicates (n = 9). (D–I) BABA primes callose deposition upon Pcc WPP17 infection (D,E), flg22 (F,G) and elf26 (H,I). Leaves were syringe infiltrated with Pcc WPP17 (1 × 108 cfu/mL), 1 μM flg22 or elf26, and samples were collected 6 h later for aniline blue staining. Graph represents the average number of deposits observed per square millimetre. Biological triplicates were averaged ± SD (n = 27). For A, B, C, E, G and I, asterisks indicate a significant difference from water controls based on Student's t‐test (P < 0.01). White bars, 200 μm. B, BABA treated; W, water treated.
As the accumulation of callose is a typical marker for the PTI response (Gómez‐Gómez et al., 1999), we compared callose deposition in water‐ and BABA‐treated Arabidopsis after inoculation with Pcc WPP17. Aniline blue staining and image analysis revealed a higher level of callose deposition in BABA‐treated Arabidopsis than in water controls at 6 hpi with Pcc WPP17 (Fig. 4D,E). Callose deposition was also enhanced in BABA‐treated Arabidopsis leaves after treatment with MAMPs flg22 or elf26 (Fig. 4F–I). In addition, BABA‐mediated priming of PTI was evaluated upon infection with the wild‐type Pcc SCC1. As expected, PTI‐responsive FRK1, NHL10 and CYP81F2 expression and callose deposition were primed in BABA‐treated Arabidopsis (Fig. S4A,B, see Supporting Information). Collectively, these data suggest that BABA primes the Arabidopsis PTI response.
Reopening of stomata upon Pcc infection is blocked by BABA
Plants close stomata during bacterial invasion. Stomatal closure occurs through MAMP recognition and is therefore considered as an early PTI response (Melotto et al., 2006; Zeng and He, 2010). Virulent bacteria, such as Pst DC3000, reopen stomata in a COR‐dependent manner (Melotto et al., 2006; Zeng and He, 2010). BABA blocks the COR‐dependent reopening of stomata upon Pst DC3000 infection (Tsai et al., 2011). To test whether BABA also inhibits stomatal reopening upon Pcc infection, stomatal movements were observed in epidermal peels from water‐ and BABA‐treated Arabidopsis kept in a solution containing Pcc bacteria. Similar to Pst DC3000 (Melotto et al., 2006; Zeng and He, 2010), Pcc bacteria induced stomatal closure at 1.5 hpi and stomatal reopening was observed at 5 hpi with Pcc in epidermal peels from water‐treated Arabidopsis (Fig. 5). By contrast, at 5 hpi, stomatal guard cells from BABA‐treated Arabidopsis were still closed (Fig. 5). Thus, as on Pst DC3000 inoculation (Tsai et al., 2011), BABA inhibited the reopening of stomata in contact with Pcc SCC1. These results further support the idea that BABA potentiates the PTI response in Arabidopsis.
Figure 5.
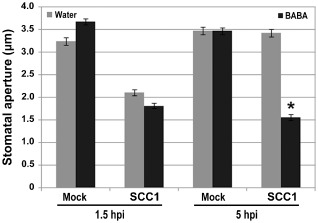
β‐Aminobutyric acid (BABA) effect on stomatal movements upon Pectobacterium carotovorum ssp. carotovorum (Pcc) SCC1 infection. Stomatal apertures in epidermal peels exposed to buffer MgSO4 (Mock) or Pcc SCC1 [1 × 106 colony‐forming units (cfu)/mL] were analysed at 1.5 and 5 h post‐inoculation (hpi). Results are shown as the mean (n > 60 stomata) ± standard error (SE). Asterisks indicate a significant difference from the water control based on Student's t‐test analysis (P < 0.001). Experiments were repeated three times with similar results.
Priming of the PTI response in SA‐defective mutants upon Pcc infection
Expression of the SA‐responsive gene PR1 is primed by BABA (Fig. 3A), but SA‐defective mutants show wild‐type levels of BABA‐mediated resistance to Pcc (Fig. 3B,C). Our data suggest that BABA primes the PTI response upon Pcc infection. Wild‐type levels of BABA‐mediated resistance in sid2‐1 and pad4‐1 could therefore be attributed to a potentiated PTI response. To test whether a primed PTI response is observed in SA‐defective mutants upon Pcc infection, expression levels of the PTI‐responsive gene FRK1 were evaluated 2 h after infiltration–inoculation with 1 × 108 colony‐forming units (cfu)/mL hrp‐ and hrc‐deficient bacterial mutant Pcc WPP17 or at 6 hpi with 5 × 106 cfu/mL of wild‐type Pcc SCC1. BABA‐treated SA‐defective mutants demonstrated a primed FRK1 expression upon bacterial infection (Figs 6A and S5A, see Supporting Information). In addition, BABA‐treated SA‐defective mutants accumulated more callose deposits than did water‐treated controls after Pcc inoculation (Figs 6B and S5B). Upon inoculation with Pcc WPP17, levels of callose deposition in SA‐defective mutants were lower than in wild‐type Col‐0 controls (Fig. 6B). Reduced callose accumulation in SA‐deficient mutants is, however, not observed upon infection with the nonhost bacterium Pseudomonas syringae pv. phaseolicola (Ham et al., 2007). To further evaluate the role of BABA on the PTI response of SA‐defective mutants, PTI‐mediated stomatal movements upon Pcc SCC1 infection were evaluated in sid2‐1 and pad4‐1. Similar to Pst DC3000 (Melotto et al., 2006), Pcc SCC1 did not induce stomatal closure in SA‐defective mutants (Fig. 6C). This observation demonstrates the importance of the SA signalling cascade in Pcc‐mediated stomatal closure. Although BABA did not induce constitutive stomatal closure in mock‐treated wild‐type controls (Figs 5,6C; Tsai et al., 2011), the stomata of BABA‐treated SA‐defective mutants were constitutively closed (Fig. 6C). Similar to what is observed in wild‐type controls (Figs 5, 6C), BABA blocked the Pcc‐mediated reopening of stomata in sid2‐1 and pad4‐1 (Fig. 6C). Taken together, these results indicate that BABA primes or reinforces the PTI response in SA‐deficient mutants.
Figure 6.
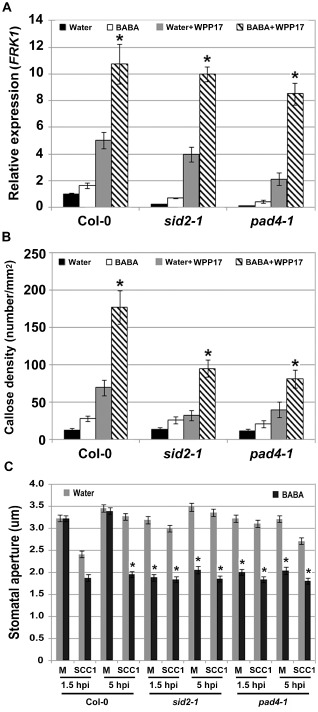
β‐Aminobutyric acid (BABA) primes the pattern‐triggered immunity (PTI) response in salicylic acid (SA)‐defective mutants. (A) BABA‐mediated priming of FLG22‐INDUCED RECEPTOR‐LIKE KINASE 1 (FRK1) expression in Col‐0, SA induction deficient 2‐1 (sid2‐1) and phytoalexin deficient 4‐1 (pad4‐1) after inoculation with Pectobacterium carotovorum ssp. carotovorum (Pcc) WPP17. Transcript levels of FRK1 were analysed 2 h post‐inoculation (hpi) with Pcc WPP17 [1 × 108 colony‐forming units (cfu)/mL]. FRK1 expression levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to both UBQ10 and EF‐1. Relative gene expression levels were compared with Col‐0 water controls (defined value of one). Data represent the means and standard deviation (SD) of three independent biological replicates (n = 9). (B) Callose deposition is primed in SA‐defective mutants. Leaves were syringe infiltrated with 1 × 108 cfu/mL P cc WPP17 and samples were collected 6 h later for aniline blue staining. Graph represents the average number of deposits observed per square millimetre. Biological triplicates were averaged ± SD (n = 27). For (A) and (B), asterisks indicate a significant difference from the water controls based on Student's t‐test (P < 0.01). (C) Role of BABA on stomatal movements in Pcc SCC1‐inoculated SA‐defective mutants. Stomatal apertures in epidermal peels exposed to buffer or Pcc SCC1 (1 × 106 cfu/mL) were analysed at 1.5 and 5 hpi in Col‐0, sid2‐1 and pad4‐1. Results are shown as mean (n > 60 stomata) ± standard error (SE). Asterisks indicate a significant difference from the water control based on Student's t‐test analysis (P < 0.001). Experiments were repeated three times with similar results.
BABA induces chromatin modifications associated with transcriptional activation
Chromatin modifications at the N‐terminal tail of histone H3 are associated with Arabidopsis pathogenesis (Jaskiewicz et al., 2011; Luna et al., 2012; Zhou et al., 2005). To address whether histone modifications occur in Arabidopsis treated with the priming agent BABA, we performed chromatin immunoprecipitation (ChIP) analyses of the promoter of PTI‐responsive FRK1, NHL10, CYP81F2 and PR1, and ACTIN 2/7 as a negative control. Within the standard histone code, acetylation of histone H3 at lysine 9 and lysine 14 (H3K9K14Ac), and dimethylation at lysine 4 (H3K4Me2), are correlated with transcriptional activation (Lusser, 2002). BABA treatment was associated with enrichment (1.5–4.0‐fold) of H3K9K14Ac and H3K4Me2 in promoter regions of PTI marker genes, whereas no enrichment was observed in the ACTIN 2/7 control (Fig. 7). BABA treatment thus induces chromatin modifications associated with increased transcriptional capacity.
Figure 7.
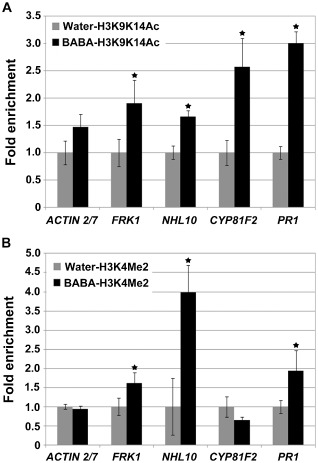
Modifications of histone H3 after β‐aminobutyric acid (BABA) treatment. Levels of H3K9K14Ac (A) and H3K4Me2 (B) at the promoter region of pattern‐triggered immunity (PTI)‐responsive FLG22‐INDUCED RECEPTOR‐LIKE KINASE 1 (FRK1), ARABIDOPSIS NON‐RACE SPECIFIC DISEASE RESISTANCE GENE (NDR1)/HAIRPIN‐INDUCED GENE (HIN1)‐LIKE 10 (NHL10), CYTOCHROME P450, FAMILY 81 (CYP81F2) and PATHOGENESIS RELATED 1 (PR1) mutants and in the nonresponsive control ACTIN 2/7. Relative enrichment levels of H3K9K14Ac and H3K4Me2 were determined in plants treated with 200 μM BABA for 2 days or in water‐treated controls. The water‐treated Col‐0 levels were set to unity after normalization by internal control TUB2. Biological duplicates were averaged and the statistical significance was determined by Student's t‐test (*P < 0.01) (comparison between BABA‐ and water‐treated plants). Error bars are the standard deviation (SD) (n = 6).
Discussion
The nonprotein amino acid BABA enhances plant resistance to numerous pathogens, notably microbial pathogens (Cohen, 2002; Jakab et al., 2001), nematodes (Oka et al., 1999) and insects such as aphids (Hodge et al., 2005). The spectrum of action of BABA against microbial pathogens is broad, with observed protection in numerous crops against very divergent groups of microbial pathogens, such as oomycetes (Cohen, 1994; Cohen et al., 2011; Hamiduzzaman et al., 2005; Papavizas, 1964; Slaughter et al., 2008), biotrophic (Shailasree et al., 2001; Worrall et al., 2012) and necrotrophic (Jakab et al., 2001) fungi and viruses (Siegrist et al., 2000). In the model plant Arabidopsis, BABA enhances resistance to necrotrophic fungi, such as B. cinerea, A. brassicicola and Plectosphaerella cucumerina (Ton and Mauch‐Mani, 2004; Zimmerli et al., 2001), oomycetes such as H. arabidopsidis and hemibiotrophic Pst bacteria (Zimmerli et al., 2000). In this study, we have demonstrated that BABA enhances Arabidopsis resistance to Pcc at both symptom and titre levels, widening the spectrum of action of BABA in Arabidopsis to necrotrophic bacteria. BABA‐induced resistance against Pcc is observed in calla lily (Luzzatto et al., 2007). Arabidopsis resistance to Pcc is usually related to concomitant activation of the JA/ET defence signalling pathways (Kariola et al., 2003; Norman‐Setterblad et al., 2000). BABA did not prime the expression of the JA/ET‐responsive PDF1.2, suggesting that the JA/ET‐dependent defence responses are not potentiated by BABA upon Pcc infection. By contrast, BABA primed the expression of the SA marker PR1 upon Pcc infection, indicating that SA signalling might be responsible for BABA‐induced Arabidopsis resistance to Pcc. Similarly, BABA primes the expression of PR1, but not PDF1.2, in Arabidopsis infected with the necrotrophic fungal pathogen B. cinerea (Zimmerli et al., 2001). Together, these observations suggest that BABA does not potentiate the JA/ET signalling that is usually associated with Arabidopsis resistance to necrotrophic microbial pathogens, such as Pcc (Kariola et al., 2003; Norman‐Setterblad et al., 2000), but rather primes the SA defence signalling that is responsive to hemibiotrophic microbial pathogens (Ton et al., 2005; Tsai et al., 2011; Zimmerli et al., 2000) and has limited effects on necrotrophs (Ferrari et al., 2003; Zimmerli et al., 2001). Mutants defective in SA‐ or JA/ET‐dependent defence signalling pathways were fully protected against Pcc by BABA. These observations further suggest that JA/ET signalling cascades are not necessary for BABA‐induced protection against Pcc and, although the SA response is boosted by BABA treatment, other layers of Arabidopsis defence may compensate and play a crucial role for full resistance in SA‐defective mutants. Such independence towards the SA and JA/ET defence signalling cascades is also observed in BABA‐treated Arabidopsis upon infection with H. arabidopsidis (Zimmerli et al., 2000) or P. cucumerina (Ton and Mauch‐Mani, 2004). In both cases, BABA pretreatments induce fast and strong callose accumulations upon pathogen attack, suggesting that BABA‐mediated cell wall reinforcement at infection sites efficiently blocks these pathogens (Ton and Mauch‐Mani, 2004; Zimmerli et al., 2000).
The PTI response is the first defence layer in plant immunity (Jones and Dangl, 2006). Similar to BABA‐induced resistance to Pcc, PTI‐induced Arabidopsis resistance mediated by flg22 is independent of SA or JA/ET defence signalling cascades (Zipfel et al., 2004). We therefore questioned whether BABA could prime the PTI response in Arabidopsis challenged with Pcc. Our data indicate that bacterial‐, flg22‐ and elf26‐induced PTIs are primed by BABA pretreatment, as illustrated by potentiated PR1, FRK1, CYP81F2 and NHL10 expression and callose deposition. BABA also primes the Arabidopsis PTI response upon infection with the hemibiotrophic bacterium Pst DC3000 (Singh et al., 2012), suggesting that BABA priming of Arabidopsis PTI is a common mechanism for the enhancement of Arabidopsis resistance to bacteria. BABA pretreatments induced an ‘open chromatin’ configuration at promoter regions of PTI‐responsive genes. Such enrichment of histone H3 activating marks may favour potentiation of PTI marker gene expressions. Chromatin structure reprogramming and priming are also correlated upon treatment with the systemic acquired resistance (SAR) chemical inducer acibenzolar S‐methyl (Jaskiewicz et al., 2011) and during transgenerational SAR (Luna et al., 2012). As suggested recently (van den Burg and Takken, 2009), modification of chromatin structures may regulate the plant defence response. Occasionally, we observed a direct BABA‐mediated up‐regulation of PTI‐responsive gene expression or callose deposition after infiltration of buffer (mock controls) (Fig. 4). As the infiltration of buffer in leaves induces wounding stress (Jaskiewicz et al., 2011), direct up‐regulation of FRK1, CYP81F2, NHL10 or callose deposition by BABA after buffer infiltration may actually reflect priming of the wounding response. Accordingly, no increase in callose deposition was observed in BABA‐treated plants that were not subjected to buffer infiltration (Fig. S6, see Supporting Information). The SA‐dependent defence response is usually associated with the Arabidopsis PTI response (Tsuda et al., 2008). Priming by BABA of the SA marker PR1 thus further suggests that BABA boosts the PTI response in Arabidopsis. The observed protective effect of BABA against Pcc in SA‐defective mutants may therefore be associated with a primed PTI response, as wild‐type levels of PTI priming were observed in sid2‐1 and pad4‐1 mutants (Fig. 6A,B). The PTI response is multilayered (Schwessinger and Zipfel, 2008) and complex, and the removal of one branch of the PTI response (i.e. SA signalling) may not be sufficient to alter BABA‐induced resistance in these SA‐defective mutants. In this context, priming of PTI‐mediated callose deposition and PTI marker gene expression, and inhibition of stomatal reopening, could be sufficient for BABA‐induced resistance to be fully functional in SA‐defective mutants.
Stomata are microscopic pores that control CO2 uptake for photosynthesis and water loss during transpiration (Acharya and Assmann, 2009). Stomatal closure upon activation of Arabidopsis PTI in response to hemibiotrophic Pst DC3000 or Pseudomonas syringae pv. syringae (Pss) infections, known as ‘stomatal immunity’, is critical for plant protection (Melotto et al., 2006, 2008; Schellenberg et al., 2010). Our data indicate that the necrotrophic bacterium Pcc, similar to hemibiotrophic bacteria, induces a rapid SA‐dependent closure of stomata. Pst DC3000 and Pss reopen Arabidopsis stomata in a COR‐ or Syringolin A‐dependent manner, respectively (Melotto et al., 2006; Schellenberg et al., 2010; Zeng et al., 2010). Although reopening mechanisms upon Pcc infection are still unknown, our data demonstrate that Arabidopsis stomata reopen at 5 hpi with Pcc (Fig. 5). BABA treatment inhibited the reopening of stomata, further suggesting that BABA potentiates or reinforces the Arabidopsis PTI response. BABA demonstrates a similar effect upon Pst DC3000 COR‐dependent reopening of stomata (Tsai et al., 2011), suggesting a common mechanism of action of BABA on both necrotrophic and hemibiotrophic bacteria, possibly through the inhibition of effector actions, as already suggested for Pst DC3000 (Tsai et al., 2011). Both the BABA‐treated SA biosynthesis mutant sid2‐1 and SA signalling mutant pad4‐1 demonstrated closed stomata even in the absence of Pcc bacteria (Fig. 6C). This observation is surprising and indicates that functional SA signalling is necessary for wild‐type levels of stomatal opening in BABA‐treated mock plants. As SA antagonizes abscisic acid (ABA) signalling (de Torres Zabala et al., 2009), BABA may activate directly ABA‐mediated stomatal closure when SA signalling is defective. Our data suggest that BABA‐mediated efficient and long‐lasting stomatal closure upon bacterial attack represents one important aspect of BABA‐induced resistance against bacterial pathogens.
In conclusion, the Arabidopsis PTI response appears to have a central role in BABA‐induced resistance to Pcc. As shown, the JA/ET defence signalling cascade, which is usually associated with Arabidopsis resistance to Pcc (Norman‐Setterblad et al., 2000), was not primed by BABA treatment. JA‐ and ET‐defective mutants, such as coi1‐16 or ein2‐1, were also fully protected by BABA, further suggesting that BABA‐induced resistance to Pcc is independent of JA/ET. BABA primed SA‐dependent defence signalling, but SA‐defective sid2‐1 and pad4‐1 mutants still demonstrated a functional BABA‐induced resistance. These SA‐defective mutants showed primed PTI‐mediated callose deposition, primed PTI‐responsive gene expression and BABA blocked stomatal reopening in SA‐defective mutants. We thus propose a model in which BABA restricts Pcc infection in Arabidopsis through priming of the PTI response (Fig. 8). Microarray analyses revealed that BABA up‐regulates directly numerous genes coding for putative kinases, notably leucine‐rich repeat protein kinases (LRR‐PKs) (Zimmerli et al., 2008). Pattern recognition receptors, such as FLS2 and EFR1, which perceive MAMPs, are LRR‐PKs (Ausubel, 2005; Boller and Felix, 2009). Direct up‐regulation by BABA of putative positive regulators of the PTI response, such as LRR‐PKs, could thus prepare Arabidopsis plants for a faster and stronger activation of the PTI response upon pathogen challenge. Prestress deposition of defence signalling components is indeed known to be critical for priming (Beckers et al., 2009; Singh et al., 2012). Alternatively, BABA‐mediated opening of chromatin at defence‐related genes may account for the primed response of PTI marker genes.
Figure 8.
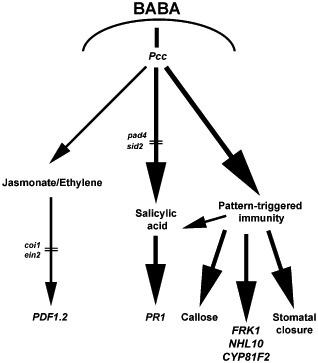
Hypothetical model illustrating β‐aminobutyric acid (BABA)‐induced resistance to Pectobacterium carotovorum ssp. carotovorum (Pcc). This model is based on the information provided in this study and references cited in the discussion. Thick bold black arrows represent priming, and thin arrows indicate no priming. Upon Pcc infection, BABA primes salicylic acid (SA)‐dependent signalling and the pattern‐triggered immunity (PTI) response. Priming leads to a potentiated expression of FLG22‐INDUCED RECEPTOR‐LIKE KINASE 1 (FRK1), ARABIDOPSIS NON‐RACE SPECIFIC DISEASE RESISTANCE GENE (NDR1)/HAIRPIN‐INDUCED GENE (HIN1)‐LIKE 10 (NHL10), CYTOCHROME P450, FAMILY 81 (CYP81F2) and PATHOGENESIS RELATED 1 (PR1), increased callose deposition and blockage of reopening of stomata. Even when the SA defence signalling cascade is disrupted in SA‐defective SA induction deficient 2‐1 (sid2‐1) and phytoalexin deficient 4‐1 (pad4‐1), priming of other PTI‐dependent defence responses may be sufficient for a functional BABA‐induced resistance. coi1, coronatine‐insensitive 1; ein2, ethylene‐insensitive 2.
Experimental Procedures
Biological materials
Arabidopsis thaliana (L. Heyhn.) ecotype Columbia (Col‐0) and derived mutant lines were grown as described previously (Tsai et al., 2011). The Col‐0 background mutant sid2‐1 was provided by C. Nawrath (University of Lausanne, Lausanne, Switzerland); pad4‐1 and ein2‐1 were obtained from the Arabidopsis Biological Resource Centre, and the Col‐6 background mutant coi1‐16 with a functional PEN2 (Westphal et al., 2008) was a gift from Dr M. R. Grant (University of Exeter, Exeter, UK). We obtained wild‐type Pcc SCC1 bacteria (Kariola et al., 2005) expressing the green fluorescent protein (GFP) (Kwon et al., 2009) from O. K. Park (Korea University, Seoul, Korea), and Pcc hrp‐ and hrc‐deficient strain WPP17 (Yap et al., 2004) was a gift from A. O. Charkowski (University of Wisconsin, Madison, WI, USA).
Bacterial inoculations
Pcc bacteria were cultivated at 28 °C, 340 rpm in Luria–Bertani (LB) medium (Bioman Scientific Co, Taipei, Taiwan) for 16–18 h without selection (Pcc WPP17) or with ampicillin (Pcc SCC1). Pcc were then collected by centrifugation, suspended in 10 mm MgSO4 at A 600 = 0.25, corresponding to a concentration of 108 cfu/mL. Five‐week‐old Arabidopsis rosettes were dipped in a bacterial solution of 5 × 106 cfu/mL (for Pcc SCC1) or 1 × 108 cfu/mL (for Pcc WPP17) in 10 mm MgSO4 containing 0.01% Silwet L‐77 for 15 min. After inoculation, plants were kept at 100% relative humidity and symptoms were evaluated 1 day later. Bacterial titres were determined by serial dilutions on LB plates, as described previously for Pst DC3000 (Zimmerli et al., 2000).
qRT‐PCR
qRT‐PCR experiments were performed as described previously (Wu et al., 2010). Briefly, leaf samples from three to five plants per treatment were harvested at the indicated time points, flash frozen in liquid N2 and kept at −80 °C. Total RNA was extracted using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) and treated with DNase I (Qiagen, Carlsbad, CA, USA) for 15 min. cDNA was synthesized from 2 μg of total RNA using oligo(dT) primers and the reverse transcriptase from the M‐MLV kit (Invitrogen; http://www.invitrogen.com/). The iCycler sequence detection system (Bio‐Rad; http://www.bio‐rad.com/) and SYBR Green PCR Master Mix (Bio‐Rad, Hercules, CA, USA) were used for real‐time PCR analysis. The thermal cycling programme was 95 °C for 3 min, followed by 40 cycles of 95 °C for 30 s and 60 °C for 30 s. Melting curve analysis was used to ensure the specificity of the product. Normalization of gene expression across different samples was performed with EF‐1a and UBQ10 as internal controls. Primer sequences were as follows: UBQ10 (At4g05320) F, 5′‐GGCCTTGTATAATCCCTGATGA‐3′/R, 5′‐AAAGAGATAACAGGAACGGAAA‐3′; EF‐1a (At5g60390) F, 5′‐TGAGCACGCTCTTCTTGCTTTC‐3′/R, 5′‐GGTGGTGGCATCCATCTTGTTA‐3′; FRK1 (AT2G19190) F, 5′‐GCCAACGGAGACATTAGAG‐3′/R, 5′‐CCATAACGACCTGACTCATC‐3′; NHL10 (AT2G35980) F, 5′‐TTCCTGTCCGTAACCCAAAC‐3′/R, 5′‐CCCTCGTAGTAGGCATGAGC‐3′; CYP81F2 (AT5G57220) F, 5′‐AAATGGAGAGAGCAACACAATG‐3′/R, 5′‐ATCGCCCATTCCAATGTTAC‐3′; PR1 (AT2G14610) F, 5′‐AAAACTTAGCCTGGGGTAGCGG‐3′/R, 5′‐CCACCATTGTTACACCTCACTTTG‐3′; PDF1.2a (AT5G44420) F, 5′‐AGAAGTTGTGCGAGAAGCCAAG‐3′/R, 5′‐GTGTGCTGGGAAGACATAGTTGC‐3′.
BABA treatment
Five‐week‐old Arabidopsis plants were soil drenched with BABA (Fluka, Steinheim, Germany) dissolved in water at a final concentration of 200 μm, or with water as control, 2 days before bacterial inoculation or MAMP treatments. Samples were collected at the indicated time points after bacterial inoculation.
MAMP treatments
The peptides representing the MAMPs flagellin (flg22) and EF‐Tu (elf26) were synthesized by Biomer Technology (Pleasanton, CA, USA); 1 μm flg22 or elf26 was dissolved in 10 mm MgSO4 and syringe infiltrated in leaves of 5‐week‐old Arabidopsis. Buffer MgSO4 only was used as a control. Leaf samples from three to five plants per treatment were harvested at the indicated time points for further gene expression or callose deposition analyses.
Callose staining
Leaves from 5‐week‐old Arabidopsis plants were syringe infiltrated with 5 × 106 cfu/mL Pcc SCC1, 1 × 108 cfu/mL Pcc WPP17, 1 μm flg22 or 1 μm elf26 in 10 mm MgSO4. Control plants were syringe infiltrated with 10 mm MgSO4 only. At the indicated time points, nine leaf discs from three different plants were collected for analyses. Harvested leaf discs were cleared overnight by soaking in 95% ethanol at room temperature and then washed three times (2 h for each washing) with sterilized water. Cleared leaves were stained with 0.05% aniline blue in 0.07 m phosphate buffer (pH 8) overnight. Callose deposits were visualized under ultraviolet illumination using an Olympus BX51 microscope digital camera and application software DP2‐BSW (Olympus, Hamburg, Germany). Callose deposits were counted using the ‘analyze particles’ function of ImageJ (http://rsb.info.nih.gov/ij/).
Stomatal assay
Plants were maintained under light (approximately 100 μmol/m2/s) for at least 3 h to allow the opening of stomata before the performance of the experiments. The epidermis of three fully expanded leaves from three plants was peeled off and placed in a plastic well containing the stomatal buffer only (10 mm MgSO4) or bacterial suspension in buffer (1 × 106 cfu/mL Pcc SCC1 in 10 mm MgSO4). At various time points, photographs were taken randomly using an Olympus BX51 microscope digital camera and application software DP2‐BSW (http://www.olympusglobal.com/). The width of the stomatal aperture was measured using the ‘measure’ function of ImageJ (http://rsb.info.nih.gov/ij/).
ChIP assay
The ChIP assay was carried out as described by Gendrel et al. (2005). Chromatin extracts were prepared from 5‐week‐old plants treated with formaldehyde. The chromatin was sheared to an average length of 500 bp by sonication and immune precipitated with specific antibodies, including anti‐acetylated histone H3K9K14 (catalogue no. 06‐599; Millipore, Temecula, CA, USA) and anti‐dimethyl histone H3K4 (catalogue no. 07‐030; Millipore). Relative enrichments in the promoter region of ACTIN 2/7, FRK1, NHL10, CYP81F2 and PR1 in Col‐0 pretreated with water or BABA were determined after normalization to TUB2. The DNA sequence from ACTIN 2/7, which is constitutively expressed in Arabidopsis, was used as a negative control (Johnson et al., 2002). Real‐time PCR was used to determine the amounts of genomic DNA immune precipitated in the ChIP experiments. The primer pairs used were as follows: TUB2 (AT5G62690) F, 5′‐ACAAACACAGAGAGGAGTGAGCA‐3′/R, 5′‐ACGCATCTTCGGTTGGATGAGTGA‐3′; ACTIN 2/7 (AT5G09810) F, 5′‐CGTTTCGCTTTCCTTAGTGTTAGCT‐3′/R, 5′‐AGCGAACGGATCTAGAGACTCACCTTG‐3′; FRK1 (AT2G19190) F, 5′‐AACCTTAGAAGATGGTTGGTTGA‐3′/R, 5′‐CAGAAGAGCAAAGCTTGTGAA‐3′; NHL10 (AT2G35980) F, 5′‐AAACTCCCGTTGGAGTTGGT‐3′/R, 5′‐GTGATGTTGCCCCAAACTTC‐3′; CYP81F2 (AT5G57220) F, 5′‐AATTTTGTATCGTGTGTAAAATTTAGCTT‐3′/R, 5′‐AAGTTAGGTTTCGTAAGCATGCC‐3′; PR1 (AT2G14610) F, 5′‐TCGGTCACCTAGAGTTTTTCAA‐3′/R, 5′‐CCGCCACATCTATGACGTAAG‐3′.
Accession numbers
FRK1 (AT2G19190); NHL10 (AT2G35980); CYP81F2 (AT5G57220); PR1 (AT2G14610); PDF1.2a (AT5G44420); UBQ10 (At4g05320); EF‐1a (At5g60390); TUB2 (AT5G62690); ACTIN 2/7 (AT5G09810).
Supporting information
Fig. S1 β‐Aminobutyric acid (BABA)‐mediated enhanced resistance at 48 h post‐inoculation (hpi) with Pectobacterium carotovorum ssp. carotovorum (Pcc) SCC1. Arabidopsis plants were treated with water (A) or BABA (B) and challenged 48 h later with Pcc SCC1 at a concentration of 5 × 106 colony‐forming units (cfu)/mL. Photographs were taken 2 days later. White arrows indicate Pcc SCC1‐mediated water‐soaked lesions.
Fig. S2 Defective PATHOGENESIS RELATED 1 (PR1) up‐regulation in salicylic acid (SA)‐defective mutants. Arabidopsis plants were treated with water or BABA and challenged 48 h later with Pectobacterium carotovorum ssp. carotovorum (Pcc) SCC1 at a concentration of 5 × 106 colony‐forming units (cfu)/mL. RNAs were harvested at 18 h post‐inoculation (hpi). Transcript levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to both UBQ10 and EF‐1. Relative gene expression levels were compared with water controls (defined value of one). Data represent the means and standard deviation (SD) of three independent biological replicates (n = 9). Asterisks indicate a significant difference from the water controls based on Student's t‐test (P < 0.01).
Fig. S3 The Pectobacterium carotovorum ssp. carotovorum (Pcc) hypersensitive response and pathogenicity (hrp)‐ and hrp conserved (hrc)‐deficient mutant strain WPP17 is less virulent than the wild‐type Pcc SCC1 strain. Five‐week‐old Col‐0 plants were dip inoculated with 5 × 106 colony‐forming units (cfu)/mL Pcc SCC1 (A) or Pcc WPP17 (B) and photographs were taken 1 day later. White arrows indicate Pcc‐mediated water‐soaked lesions.
Fig. S4 β‐Aminobutyric acid (BABA) primes the pattern‐triggered immunity (PTI) response upon infection with wild‐type Pectobacterium carotovorum ssp. carotovorum (Pcc) SCC1 bacteria. (A) BABA potentiates the expression of FLG22‐INDUCED RECEPTOR‐LIKE KINASE 1 (FRK1), ARABIDOPSIS NON‐RACE SPECIFIC DISEASE RESISTANCE GENE (NDR1)/HAIRPIN‐INDUCED GENE (HIN1)‐LIKE 10 (NHL10) and CYTOCHROME P450, FAMILY 81 (CYP81F2) upon Pcc SCC1 infection. Transcript levels were analysed at 4 h post‐inoculation (hpi) with 5 × 106 colony‐forming units (cfu)/mL Pcc SCC1. Transcript levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to both UBQ10 and EF‐1. Relative gene expression levels were compared with water controls (defined value of one). Data represent the means and standard deviation (SD) of three independent biological replicates (n = 9). (B) BABA primes callose deposition upon Pcc SCC1 infection. Leaves were syringe infiltrated with Pcc SCC1 (5 × 106 cfu/mL) and samples were collected 12 h later for aniline blue staining. The graph represents the average number of deposits observed per square millimetre. Biological triplicates were the average ± SD (n = 27). Asterisks indicate a significant difference from water controls based on Student's t‐test (P < 0.01).
Fig. S5 β‐Aminobutyric acid (BABA)‐mediated priming of pattern‐triggered immunity (PTI) upon Pectobacterium carotovorum ssp. carotovorum (Pcc) SCC1 infection in salicylic acid (SA)‐defective mutants. (A) FLG22‐INDUCED RECEPTOR‐LIKE KINASE 1 (FRK1) expression is primed in BABA‐treated Col‐0, SA induction deficient 2‐1 (sid2‐1) and phytoalexin deficient 4‐1 (pad4‐1) after inoculation with Pcc SCC1. Transcript levels of FRK1 were analysed at 6 h post‐inoculation (hpi) with 5 × 106 colony‐forming units (cfu)/mL Pcc SCC1. FRK1 expression levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to both UBQ10 and EF‐1. Relative gene expression levels were compared with Col‐0 water controls (defined value of one). Data represent the means and standard deviation (SD) of three independent biological replicates (n = 9). (B) Callose deposition is primed in SA‐defective mutants upon Pcc SCC1 infection. Leaves were syringe infiltrated with Pcc SCC1 (5 × 106 cfu/mL), and samples were collected 12 h later for aniline blue staining. Graph represents the average number of deposits observed per square millimetre. Biological triplicates were averaged ± SD (n = 27). Asterisks indicate a significant difference from water controls based on Student's t‐test (P < 0.01).
Fig. S6 Callose deposition in nonbuffer‐infiltrated Arabidopsis leaves. Arabidopsis plants were soil drenched with water or β‐aminobutyric acid (BABA), and leaves were analysed for callose deposition 2 days later. The graph represents the average number of deposits observed per square millimetre. Biological triplicates were averaged ± standard deviation (SD) (n = 27). Values of BABA‐treated plants were not significantly different from those of water‐treated controls based on Student's t‐test (P < 0.01).
Acknowledgements
We are grateful to C. Nawrath, M. R. Grant and the Arabidopsis Biological Resource Centre (ABRC) for providing seeds. We thank A. O. Charkowski and O. K. Park for the pathogens. We acknowledge the Technology Commons, College of Life Science, National Taiwan University for qRT‐PCR equipment. We also acknowledge members of L. Zimmerli's laboratory for critical comments. This work was supported by the National Science Council of Taiwan grants 96‐2628‐B‐002‐112‐MY3 and 99‐2628‐B‐002‐053‐MY3 (to L.Z.). The authors have no conflicts of interest to declare.
References
- Acharya, B. and Assmann, S. (2009) Hormone interactions in stomatal function. Plant Mol. Biol. 69, 451–462. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M. (2005) Are innate immune signalling pathways in plants and animals conserved? Nat. Immunol. 6, 973–979. [DOI] [PubMed] [Google Scholar]
- Beckers, G.M.J. , Jaskiewicz, M. , Liu, Y. , Underwood, W.R. , He, S.Y. , Zhang, S. and Conrath, U. (2009) Mitogen‐activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana . Plant Cell, 21, 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bostock, R.M. (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Ann. Rev. Phytopathol. 43, 545–580. [DOI] [PubMed] [Google Scholar]
- Boudsocq, M. , Willmann, M.R. , McCormack, M. , Lee, H. , Shan, L.B. , He, P. , Bush, J. , Cheng, S.H. and Sheen, J. (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature, 464, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg, H.A. and Takken, F. (2009) Does chromatin remodeling mark systemic acquired resistance? Trends Plant Sci. 14, 286–294. [DOI] [PubMed] [Google Scholar]
- Cohen, Y. (1994) 3‐Aminobutyric acid induces systemic resistance against Peronospora tabacina . Physiol. Mol. Plant Pathol. 44, 273–288. [Google Scholar]
- Cohen, Y. , Rubin, A.E. and Vaknin, M. (2011) Post infection application of DL‐3‐amino‐butyric acid (BABA) induces multiple forms of resistance against Bremia lactucae in lettuce. Eur. J. Plant Pathol. 130, 13–27. [Google Scholar]
- Cohen, Y.R. (2002) 3‐Aminobutyric acid‐induced resistance against plant pathogens. Plant Dis. 86, 448–457. [DOI] [PubMed] [Google Scholar]
- Conrath, U. (2011) Molecular aspects of defence priming. Trends Plant Sci. 16, 524–531. [DOI] [PubMed] [Google Scholar]
- Conrath, U. , Pieterse, C.M.J. and Mauch‐Mani, B. (2002) Priming in plant–pathogen interactions. Trends Plant Sci. 7, 210–216. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A. and Mullet, J.E. (1997) Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. [DOI] [PubMed] [Google Scholar]
- Desclos‐Theveniau, M. , Arnaud, D. , Huang, T.Y. , Lin, G.J.C. , Chen, W.Y. , Lin, Y.C. and Zimmerli, L. (2012) The Arabidopsis lectin receptor kinase LecRK‐V.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. Plos Pathog. 8, e1002513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C. and Turner, J.G. (2002) A conditionally fertile coi1 allele indicates cross‐talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta, 215, 549–556. [DOI] [PubMed] [Google Scholar]
- Ferrari, S. , Plotnikova, J.M. , De Lorenzo, G. and Ausubel, F.M. (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 35, 193–205. [DOI] [PubMed] [Google Scholar]
- Gendrel, A.V. , Lippman, Z. , Martienssen, R. and Colot, V. (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods, 2, 213–218. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defence against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. , Zook, M. , Mert, F. , Kagan, I. , Rogers, E.E. , Crute, I.R. , Holub, E.B. , Hammerschmidt, R. and Ausubel, F.M. (1997) Phytoalexin‐deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics, 146, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Gómez, L. , Felix, G. and Boller, T. (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana . Plant J. 18, 277–284. [DOI] [PubMed] [Google Scholar]
- Guzman, P. and Ecker, J.R. (1990) Exploiting the triple response of Arabidopsis to identify ethylene‐related mutants. Plant Cell, 2, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham, J.H. , Kim, M.G. , Lee, S.Y. and Mackey, D. (2007) Layered basal defenses underlie non‐host resistance of Arabidopsis to Pseudomonas syringae pv. phaseolicola . Plant J. 51, 604–616. [DOI] [PubMed] [Google Scholar]
- Hamiduzzaman, M.M. , Jakab, G. , Barnavon, L. , Neuhaus, J.M. and Mauch‐Mani, B. (2005) beta‐Aminobutyric acid‐induced resistance against downy mildew in grapevine acts through the potentiation of callose formation and jasmonic acid signalling. Mol. Plant–Microbe Interact. 18, 819–829. [DOI] [PubMed] [Google Scholar]
- Hodge, S. , Thompson, G.A. and Powell, G. (2005) Application of DL‐beta‐aminobutyric acid (BABA) as a root drench to legumes inhibits the growth and reproduction of the pea aphid Acyrthosiphon pisum (Hemiptera:Aphididae). Bull. Entomol. Res. 95, 449–455. [DOI] [PubMed] [Google Scholar]
- van Hulten, M. , Pelser, M. , van Loon, L.C. , Pieterse, C.M.J. and Ton, J. (2006) Costs and benefits of priming for defence in Arabidopsis. Proc. Natl. Acad. Sci. USA, 103, 5602–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab, G. , Cottier, V. , Toquin, V. , Rigoli, G. , Zimmerli, L. , Métraux, J.P. and Mauch‐Mani, B. (2001) Beta‐Aminobutyric acid‐induced resistance in plants. Eur. J. Plant Pathol. 107, 29–37. [Google Scholar]
- Jakab, G. , Ton, J. , Flors, V. , Zimmerli, L. , Métraux, J.P. and Mauch‐Mani, B. (2005) Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 139, 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz, M. , Conrath, U. and Peterhänsel, C. (2011) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, L. , Cao, X. and Jacobsen, S. (2002) Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12, 1360– 1367. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jung, H.W. , Tschaplinski, T.J. , Wang, L. , Glazebrook, J. and Greenberg, J.T. (2009) Priming in systemic plant immunity. Science, 324, 89–91. [DOI] [PubMed] [Google Scholar]
- Kariola, T. , Palomaki, T.A. , Brader, G. and Palva, E.T. (2003) Erwinia carotovora subsp carotovora and Erwinia‐derived elicitors HrpN and PehA trigger distinct but interacting defence responses and cell death in Arabidopsis. Mol. Plant–Microbe Interact. 16, 179–187. [DOI] [PubMed] [Google Scholar]
- Kariola, T. , Brader, G. , Li, J. and Palva, E.T. (2005) Chlorophyllase 1, a damage control enzyme, affects the balance between defence pathways in plants. Plant Cell, 17, 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K. and Manners, J.M. (2008) Jasmonate signalling: toward an Integrated view. Plant Physiol. 146, 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuć, J. (2001) Concepts and direction of induced systemic resistance in plants and its application. Eur. J. Plant Pathol. 107, 7–12. [Google Scholar]
- Kunkel, B.N. and Brooks, D.M. (2002) Cross talk between signalling pathways in pathogen defence. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Kwon, S.J. , Jin, H.C. , Lee, S. , Nam, M.H. , Chung, J.H. , Kwon, S.I. , Ryu, C.M. and Park, O.K. (2009) GDSL lipase‐like 1 regulates systemic resistance associated with ethylene signalling in Arabidopsis. Plant J. 58, 235–245. [DOI] [PubMed] [Google Scholar]
- Luna, E. , Bruce, T.J. , Roberts, M.R. , Flors, V. and Ton, J. (2012) Next‐generation systemic acquired resistance. Plant Physiol. 158, 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser, A. (2002) Acetylated, methylated, remodeled: chromatin states for gene regulation. Curr. Opin. Plant Biol. 5, 437–443. [DOI] [PubMed] [Google Scholar]
- Luzzatto, T. , Yishay, M. , Lipsky, A. , Ion, A. , Belausov, E. and Yedidia, I. (2007) Efficient, long‐lasting resistance against the soft rot bacterium Pectobacterium carotovorum in calla lily provided by the plant activator methyl jasmonate. Plant Pathol. 56, 692–701. [Google Scholar]
- Melotto, M. , Underwood, W. , Koczan, J. , Nomura, K. and He, S.Y. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell, 126, 969–980. [DOI] [PubMed] [Google Scholar]
- Melotto, M. , Underwood, W. and He, S.Y. (2008) Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 46, 101–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C. and Métraux, J.P. (1999) Salicylic acid induction‐deficient mutants of Arabidopsis express PR‐2 and PR‐5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell, 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman‐Setterblad, C. , Vidal, S. and Palva, E.T. (2000) Interacting signal pathways control defence gene expression in Arabidopsis in response to cell wall‐degrading enzymes from Erwinia carotovora . Mol. Plant–Microbe Interact. 13, 430–438. [DOI] [PubMed] [Google Scholar]
- Oka, Y. , Cohen, Y. and Spiegel, Y. (1999) Local and systemic induced resistance to the root‐knot nematode in tomato by DL‐beta‐amino‐n‐butyric acid. Phytopathology, 89, 1138–1143. [DOI] [PubMed] [Google Scholar]
- Overmyer, K. , Tuominen, H. , Kettunen, R. , Betz, C. , Langebartels, C. , Sandermann, H. and Kangasjarvi, J. (2000) Ozone‐sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signalling pathways in regulating superoxide‐dependent cell death. Plant Cell, 12, 1849–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavizas, G.C. (1964) Greenhouse control of Aphanomyces root rot of peas with aminobutyric acid and methylaspartic acid. Plant Dis. Rep. 48, 537–541. [Google Scholar]
- Pham, L.N. , Dionne, M.S. , Shirasu‐Hiza, M. and Schneider, D.S. (2007) A specific primed immune response in Drosophila is dependent on phagocytes. Plos Pathog. 3, e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Leon‐Reyes, A. , Van der Ent, S. and Van Wees, S.C.M. (2009) Networking by small‐molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Prime‐A‐Plant Group , Conrath, U. , Beckers, G.J.M. , Flors, V. , Garcia‐Agustin, P. , Jakab, G. , Mauch, F. , Newman, M.A. , Pieterse, C.M.J. , Poinssot, B. , Pozo, M.J. , Pugin, A. , Schaffrath, U. , Ton, J. , Wendehenne, D. , Zimmerli, L. and Mauch‐Mani, B. (2006) Priming: getting ready for battle. Mol. Plant–Microbe Interact. 19, 1062–1071. [DOI] [PubMed] [Google Scholar]
- Reymond, P. and Farmer, E.E. (1998) Jasmonate and salicylate as global signals for defence gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Schellenberg, B. , Ramel, C. and Dudler, R. (2010) Pseudomonas syringae virulence factor syringolin a counteracts stomatal immunity by proteasome inhibition. Mol. Plant–Microbe Interact. 23, 1287–1293. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B. and Zipfel, C. (2008) News from the frontline: recent insights into PAMP‐triggered immunity in plants. Curr. Opin. Plant Biol. 11, 389–395. [DOI] [PubMed] [Google Scholar]
- Shailasree, S. , Sarosh, B.R. , Vasanthi, N.S. and Shetty, H.S. (2001) Seed treatment with beta‐aminobutyric acid protects Pennisetum glaucum systemically from Sclerospora graminicola . Pest Manag. Sci. 57, 721–728. [DOI] [PubMed] [Google Scholar]
- Siegrist, J. , Orober, M. and Buchenauer, H. (2000) Beta‐aminobutyric acid‐mediated enhancement of resistance in tobacco to tobacco mosaic virus depends on the accumulation of salicylic acid. Physiol. Mol. Plant Pathol. 56, 95–106. [Google Scholar]
- Singh, P. , Kuo, Y.C. , Mishra, S. , Tsai, C.H. , Chien, C.C. , Chen, C.W. , Desclos‐Theveniau, M. , Chu, P.W. , Chinchilla, D. , Schulze, B. , Boller, T. and Zimmerli, L. (2012) The lectin receptor kinase‐VI.2 is required for priming and positively regulates Arabidopsis pattern‐triggered immunity. Plant Cell, 24, 1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter, A.R. , Hamiduzzaman, M.M. , Gindro, K. , Neuhaus, J.M. and Mauch‐Mani, B. (2008) β‐Aminobutyric acid induced resistance in grapevine against downy mildew: involvement of pterostilbene. Eur. J. Plant Pathol. 122, 185– 195. [Google Scholar]
- Thomma, B.P.H.J. , Eggermont, K. , Penninckx, I.A.M.A. , Mauch‐Mani, B. , Vogelsang, R. , Cammue, B.P.A. and Broekaert, W.F. (1998) Separate jasmonate‐dependent and salicylate‐dependent defence‐response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA, 95, 15 107–15 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, J. and Mauch‐Mani, B. (2004) Beta‐amino‐butyric acid‐induced resistance against necrotrophic pathogens is based on ABA‐dependent priming for callose. Plant J. 38, 119–130. [DOI] [PubMed] [Google Scholar]
- Ton, J. , Jakab, G. , Toquin, V. , Flors, V. , Lavicoli, A. , Maeder, M.N. , Métraux, J.P. and Mauch‐Mani, B. (2005) Dissecting the beta‐aminobutyric acid induced priming pathways in Arabidopsis. Plant Cell, 17, 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres Zabala, M. , Bennett, M.H. , Truman, W.H. and Grant, M.R. (2009) Antagonism between salicylic and abscisic acid reflects early host–pathogen conflict and moulds plant defence responses. Plant J. 59, 375–386. [DOI] [PubMed] [Google Scholar]
- Tsai, C.H. , Singh, P. , Chen, C.W. , Thomas, J. , Weber, J. , Mauch‐Mani, B. and Zimmerli, L. (2011) Priming for enhanced defence responses by specific inhibition of the Arabidopsis response to coronatine. Plant J. 65, 469–479. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. , Sato, M. , Glazebrook, J. , Cohen, J.D. and Katagiri, F. (2008) Interplay between MAMP‐triggered and SA‐mediated defence responses. Plant J. 53, 763–775. [DOI] [PubMed] [Google Scholar]
- Van der Ent, S. , Van Hulten, M. , Pozo, M.J. , Czechowski, T. , Udvardi, M.K. , Pieterse, C.M.J. and Ton, J. (2009) Priming of plant innate immunity by rhizobacteria and beta‐aminobutyric acid: differences and similarities in regulation. New Phytol. 183, 419–431. [DOI] [PubMed] [Google Scholar]
- Westphal, L. , Scheel, D. and Rosahl, S. (2008) The coi1‐16 mutant harbors a second site mutation rendering PEN2 nonfunctional. Plant Cell, 20, 824–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall, D. , Holroyd, G.H. , Moore, J.P. , Glowacz, M. , Croft, P. , Taylor, J.E. , Paul, N.D. and Roberts, M.R. (2012) Treating seeds with activators of plant defence generates long‐lasting priming of resistance to pests and pathogens. New Phytol. 193, 770–778. [DOI] [PubMed] [Google Scholar]
- Wu, C.C. , Singh, P. , Chen, M.C. and Zimmerli, L. (2010) L‐Glutamine inhibits beta‐aminobutyric acid‐induced stress resistance and priming in Arabidopsis. J. Exp. Bot. 61, 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap, M.N. , Barak, J.D. and Charkowski, A.O. (2004) Genomic diversity of Erwinia carotovora subsp carotovora and its correlation with virulence. Appl. Environ. Microbiol. 70, 3013–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, W. , Melotto, A.M. and He, S.Y. (2010) Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr. Opin. Biotechnol. 21, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, W.Q. and He, S.Y. (2010) A prominent role of the flagellin receptor FLAGELLIN‐SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 153, 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. and Zhou, J.M. (2010) Plant immunity triggered by microbial molecular signatures. Mol. Plant 3, 783–793. [DOI] [PubMed] [Google Scholar]
- Zhou, C. , Zhang, L. , Duan, J. , Miki, B. and Wu, K. (2005) HDA19 is involved in jasmonic acid and ethylene signalling of pathogen‐response in Arabidopsis. Plant Cell, 17, 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli, L. , Jakab, C. , Métraux, J.P. and Mauch‐Mani, B. (2000) Potentiation of pathogen‐specific defence mechanisms in Arabidopsis by beta‐aminobutyric acid. Proc. Natl. Acad. Sci. USA, 97, 12 920–12 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli, L. , Métraux, J.P. and Mauch‐Mani, B. (2001) Beta‐aminobutyric acid‐induced protection of Arabidopsis against the necrotrophic fungus Botrytis cinerea . Plant Physiol. 126, 517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli, L. , Hou, B.H. , Tsai, C.H. , Jakab, G. , Mauch‐Mani, B. and Somerville, S. (2008) The xenobiotic beta‐aminobutyric acid enhances Arabidopsis thermotolerance. Plant J. 53, 144–156. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. (2009) Early molecular events in PAMP‐triggered immunity. Curr. Opin. Plant Biol. 12, 414–420. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D.G. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 β‐Aminobutyric acid (BABA)‐mediated enhanced resistance at 48 h post‐inoculation (hpi) with Pectobacterium carotovorum ssp. carotovorum (Pcc) SCC1. Arabidopsis plants were treated with water (A) or BABA (B) and challenged 48 h later with Pcc SCC1 at a concentration of 5 × 106 colony‐forming units (cfu)/mL. Photographs were taken 2 days later. White arrows indicate Pcc SCC1‐mediated water‐soaked lesions.
Fig. S2 Defective PATHOGENESIS RELATED 1 (PR1) up‐regulation in salicylic acid (SA)‐defective mutants. Arabidopsis plants were treated with water or BABA and challenged 48 h later with Pectobacterium carotovorum ssp. carotovorum (Pcc) SCC1 at a concentration of 5 × 106 colony‐forming units (cfu)/mL. RNAs were harvested at 18 h post‐inoculation (hpi). Transcript levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to both UBQ10 and EF‐1. Relative gene expression levels were compared with water controls (defined value of one). Data represent the means and standard deviation (SD) of three independent biological replicates (n = 9). Asterisks indicate a significant difference from the water controls based on Student's t‐test (P < 0.01).
Fig. S3 The Pectobacterium carotovorum ssp. carotovorum (Pcc) hypersensitive response and pathogenicity (hrp)‐ and hrp conserved (hrc)‐deficient mutant strain WPP17 is less virulent than the wild‐type Pcc SCC1 strain. Five‐week‐old Col‐0 plants were dip inoculated with 5 × 106 colony‐forming units (cfu)/mL Pcc SCC1 (A) or Pcc WPP17 (B) and photographs were taken 1 day later. White arrows indicate Pcc‐mediated water‐soaked lesions.
Fig. S4 β‐Aminobutyric acid (BABA) primes the pattern‐triggered immunity (PTI) response upon infection with wild‐type Pectobacterium carotovorum ssp. carotovorum (Pcc) SCC1 bacteria. (A) BABA potentiates the expression of FLG22‐INDUCED RECEPTOR‐LIKE KINASE 1 (FRK1), ARABIDOPSIS NON‐RACE SPECIFIC DISEASE RESISTANCE GENE (NDR1)/HAIRPIN‐INDUCED GENE (HIN1)‐LIKE 10 (NHL10) and CYTOCHROME P450, FAMILY 81 (CYP81F2) upon Pcc SCC1 infection. Transcript levels were analysed at 4 h post‐inoculation (hpi) with 5 × 106 colony‐forming units (cfu)/mL Pcc SCC1. Transcript levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to both UBQ10 and EF‐1. Relative gene expression levels were compared with water controls (defined value of one). Data represent the means and standard deviation (SD) of three independent biological replicates (n = 9). (B) BABA primes callose deposition upon Pcc SCC1 infection. Leaves were syringe infiltrated with Pcc SCC1 (5 × 106 cfu/mL) and samples were collected 12 h later for aniline blue staining. The graph represents the average number of deposits observed per square millimetre. Biological triplicates were the average ± SD (n = 27). Asterisks indicate a significant difference from water controls based on Student's t‐test (P < 0.01).
Fig. S5 β‐Aminobutyric acid (BABA)‐mediated priming of pattern‐triggered immunity (PTI) upon Pectobacterium carotovorum ssp. carotovorum (Pcc) SCC1 infection in salicylic acid (SA)‐defective mutants. (A) FLG22‐INDUCED RECEPTOR‐LIKE KINASE 1 (FRK1) expression is primed in BABA‐treated Col‐0, SA induction deficient 2‐1 (sid2‐1) and phytoalexin deficient 4‐1 (pad4‐1) after inoculation with Pcc SCC1. Transcript levels of FRK1 were analysed at 6 h post‐inoculation (hpi) with 5 × 106 colony‐forming units (cfu)/mL Pcc SCC1. FRK1 expression levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to both UBQ10 and EF‐1. Relative gene expression levels were compared with Col‐0 water controls (defined value of one). Data represent the means and standard deviation (SD) of three independent biological replicates (n = 9). (B) Callose deposition is primed in SA‐defective mutants upon Pcc SCC1 infection. Leaves were syringe infiltrated with Pcc SCC1 (5 × 106 cfu/mL), and samples were collected 12 h later for aniline blue staining. Graph represents the average number of deposits observed per square millimetre. Biological triplicates were averaged ± SD (n = 27). Asterisks indicate a significant difference from water controls based on Student's t‐test (P < 0.01).
Fig. S6 Callose deposition in nonbuffer‐infiltrated Arabidopsis leaves. Arabidopsis plants were soil drenched with water or β‐aminobutyric acid (BABA), and leaves were analysed for callose deposition 2 days later. The graph represents the average number of deposits observed per square millimetre. Biological triplicates were averaged ± standard deviation (SD) (n = 27). Values of BABA‐treated plants were not significantly different from those of water‐treated controls based on Student's t‐test (P < 0.01).


