Summary
A dimeric PR‐1‐type pathogenesis‐related protein (PR‐1‐5), recently identified in wheat, was found to interact with Stagonospora nodorum ToxA in both yeast two‐hybrid and co‐immunoprecipitation assays. Site‐specific mutational analyses revealed that the RGD motif of ToxA is not targeted by PR‐1‐5, whereas two surface‐exposed asparagine residues are essential for the interaction: the N102 residue of the turning loop between β2 and β3 in ToxA and the N141 residue of the turning loop between βC and βD in PR‐1‐5. Recombinant PR‐1‐5 and ToxA mutant proteins carrying alanine substitutions at the interacting sites were expressed in Pichia pastoris, together with the wild‐type proteins. Native polyacrylamide gel electrophoresis (PAGE) confirmed that the PR‐1‐5‐N141A mutant retains the ability to form dimers. Plant assays indicated that the ToxA‐N102A mutant fails to induce necrosis, whereas the PR‐1‐5‐N141A mutant is impaired in the ‘necrosis‐promoting’ activity shown by the wild‐type PR‐1‐5 when co‐infiltrated with ToxA in sensitive wheat. Reverse transcriptase‐polymerase chain reaction and Western blot analyses revealed that the native PR‐1‐5 protein is differentially expressed between ToxA‐sensitive and ToxA‐insensitive wheat lines in response to ToxA treatment. These results suggest that PR‐1‐5 is a potential target of ToxA and the site‐specific interaction between PR‐1‐5 and ToxA may mediate ToxA‐induced necrosis in sensitive wheat.
Keywords: host‐selective toxin, inducible defence‐related protein, protein dimerization, protein–protein interaction
Introduction
PR‐1‐type pathogenesis‐related (PR) proteins are best known among the plant PR families for being hallmarks of the activation of hypersensitive response/defence pathways and the development of salicylic acid‐dependent systemic acquired resistance (Buchel and Linthorst, 1999; van Loon et al., 2006; van Loon and van Strien, 1999). The accumulation of PR‐1 proteins in a wide range of plant species has been associated with both incompatible and compatible pathogen–host interactions culminating in cell death (Gruner et al., 2003; Hong et al., 2005; Lazniewska and Macioszek, 2010; Leon‐Reyes et al., 2009; Lu et al., 2011; Santamaria et al., 2001). However, the biochemical function and biological activity of PR‐1 proteins are still obscure, although antifungal activities have been described for a few proteins purified from dicotyledonous plants (Kiba et al., 2007; Niderman et al., 1995; Rauscher et al., 1999), and enhanced disease resistance has been observed in transgenic tobacco plants over‐expressing certain PR‐1 genes (Alexander et al., 1993; Li et al., 2011). Direct roles in the mediation of plant cell death associated with host–pathogen interactions have not been demonstrated for any members of the plant PR‐1 family.
Homologues of PR‐1 proteins are widely spread among other kingdoms, including Animalia, Fungi and Bacteria (Cantacessi et al., 2012; Gibbs et al., 2008; Henriksen et al., 2001; Schuren et al., 1993). Among these PR‐1‐like proteins, the best known is the human glioma pathogenesis‐related (GliPR) protein implicated in cancer development in brain and prostate cells (Bonafe et al., 2010; Szyperski et al., 1998). Early studies on the tomato PR‐1 protein P14a (Fernandez et al., 1997) and GliPR (Szyperski et al., 1998) revealed a conserved ‘PR‐1 fold’ (also called SCP‐like extracellular protein domain, pfam00188) that features four α‐helices and one four‐strand β‐sheet, with histidine (His)‐72, glutamic acid (Glu)‐77, Glu‐98 and His‐117 (in the P14a numeration) identified to be potential active sites. Homology‐based modelling for the cone snail (Conus textile) Tex31 (Milne et al., 2003) and human GAPR‐1 (Serrano et al., 2004) proteins suggested a ‘catalytic triad’ consisting of serine (Ser)‐73 with Glu‐98/His‐117 (or Glu‐77/His‐72) within the largest cavity in the PR‐1 fold, which is reminiscent of that of serine proteases, but none of these PR‐1‐like proteins has been confirmed to possess protease activity. Recent studies in human/animal systems have suggested ‘novel’ functions for PR‐1‐like proteins based on several newly identified properties, including coordination with divalent metal ions, such as Zn2+ (Asojo et al., 2011; Wang et al., 2010), and binding to negatively charged membrane‐associated lipids (van Galen et al., 2012) or certain fatty acid‐derived lipid signalling molecules, such as leukotrienes (Xu et al., 2012). The importance of these unusual features in specific physiological processes has not been well established.
ToxA is the first discovered fungal proteinaceous host‐selective toxin originally identified from the tan spot fungus Pyrenophora tritici‐repentis (Ptr) (Ballance et al., 1989, 1996; Ciuffetti et al., 1997; Tomas et al., 1990) and, more recently, from the leaf/glume blotch fungus Stagonospora nodorum (Sn) (Friesen et al., 2006). SnToxA is nearly identical (>99% similarity) to PtrToxA and is thought to have been acquired by Ptr from Sn through a recent horizontal gene transfer event (Friesen et al., 2006). How ToxA induces cell death in sensitive wheat is still under investigation. A current model suggests that ToxA binds to a membrane‐located receptor through its RGD motif and is internalized through a yet‐to‐be‐identified mechanism; within the plant cells, ToxA targets ToxABP1 (Manning et al., 2007), a chloroplastic protein homologous to the Arabidopsis plastid membrane protein THYLAKOID FORMATION 1 (THF1) (Wang et al., 2004), essential for photosynthesis, thus leading to cell death (reviewed by Ciuffetti et al., 2010). It has also been reported that ToxA interacts with a second chloroplast‐associated protein, plastocyanin (Tai et al., 2007). Sensitivity to ToxA and susceptibility to the ToxA‐producing fungus are controlled by a single dominant gene Tsn1 (Faris et al., 1996), which encodes a protein belonging to the nucleotide‐binding site‐leucine‐rich repeat (NBS‐LRR) family of plant disease resistance proteins, but featuring an unusual N‐terminal fusion with a serine/threonine protein kinase (S/TPK) domain (Faris et al., 2010). Tsn1 is unlikely to be the long‐assumed ToxA receptor as it lacks a transmembrane domain and does not interact directly (at least in yeast) with ToxA (Faris et al., 2010). ToxA may act as both an elicitor and a virulence factor because a large number of defence‐related genes, including those for PR proteins, are up‐regulated in ToxA‐treated sensitive wheat, like those observed in classical gene‐for‐gene interactions (Adhikari et al., 2009; Pandelova et al., 2009, 2012). It has been proposed that ToxA, and perhaps other fungal effectors, may exploit plant defence mechanisms to induce cell death for survival of the necrotrophic pathogen (Ciuffetti et al., 2010; Friesen et al., 2008; Mengiste, 2012; Oliver et al., 2012).
Previously, we have obtained two partial cDNA clones that encode PR‐1‐like proteins potentially interacting with SnToxA from a yeast two‐hybrid (Y2H) library screening experiment (Lu et al., 2009). Subsequent genome‐wide analysis of the wheat PR‐1 family identified the corresponding full‐length genes, named TaPr‐1‐1 and TaPr‐1‐5 (Lu et al., 2011). Recent characterization of the recombinant PR‐1‐1 and PR‐1‐5 proteins revealed that the former exists as monomers, whereas the latter forms dimers contributing to protease resistance (Lu et al., 2013). Here, we report that the dimeric PR‐1‐5 physically interacts with SnToxA in a sequence‐specific manner, and that the interacting sites on both proteins are important for their respective biological activities. The data suggest that PR‐1‐5 is a potential target of ToxA and that the site‐specific interaction between PR‐1‐5 and ToxA may mediate ToxA‐induced necrosis governed by the cognate Tsn1 gene in sensitive wheat.
Results
PR‐1‐5 physically interacts with ToxA
Our initial Y2H library screening (Lu et al., 2009) identified two partial cDNA clones encoding PR‐1‐like proteins (named PR‐1‐1 and PR‐1‐5) potentially interacting with SnToxA (hereafter ToxA unless otherwise noted). To confirm the PR‐1–ToxA interaction, we performed additional Y2H assays using the full‐length cDNA sequences encoding the mature protein of ToxA and PR‐1‐1 or PR‐1‐5. The ToxA protein was N‐terminally fused to the GAL4‐DNA‐binding domain (BD) and the two PR‐1 proteins were fused to the GAL4‐activation domain (AD). ‘Positive’ interactions were determined by blue coloration derived from α‐galactosidase activity on selective agar plates, and the strength of the interaction was simultaneously ‘quantified’ using serially diluted cell suspensions of the transformant co‐expressing the bait/prey proteins. In all tests, the yeast transformant co‐expressing the full‐length mature ToxA and PR‐1‐5 proteins produced blue coloration, with the intensity observed in undiluted cells comparable with that of 10−2‐diluted cells in a standard positive control (Fig. 1A, compare rows 1 and 5). In contrast, the transformant co‐expressing the full‐length mature ToxA and PR‐1‐1 proteins did not produce any blue coloration (row 2). Replacement of either the PR‐1‐5 prey or the ToxA bait with the yeast proteins RecT or p53 caused a loss of blue coloration (rows 3 and 4). A bait–prey swap assay was pursued, but was found not to be feasible because the GAL4‐AD–ToxA fusion appeared to be ‘toxic’ when expressed in yeast (data not shown). Nevertheless, the Y2H assay results were sufficient to show that the full‐length mature PR‐1‐5 protein, but not PR‐1‐1 protein, interacted with ToxA, although the interaction appeared to be weak (about two orders of magnitude lower than the standard control).
Figure 1.
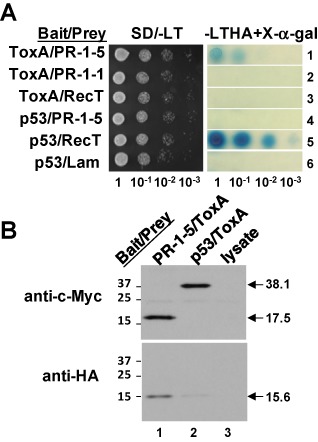
Characterization of the interaction between PR‐1‐5 and ToxA. (A) Yeast two‐hybrid assay. Serially diluted cell suspensions from a single colony of the transformant co‐expressing the bait/prey proteins were inoculated onto selective agar plates and incubated at 30 °C for 36 h. p53/RecT and p53/Lam are standard ‘positive’ and ‘negative’ controls, respectively. Blue coloration on the X‐α‐gal plates reflects the strength of protein–protein interactions. (B) Co‐immunoprecipitation assay. Epitope‐tagged bait and prey proteins were separated by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred onto poly(vinylidene difluoride) (PVDF) membrane; the blot was probed first with anti‐c‐Myc antibody (top panel) and then with anti‐haemagglunin (HA) antibody (bottom panel) after stripping. Lane 3 was loaded with a ‘blank’ control. Arrows indicate the respective bait and prey proteins (molecular masses in kDa). The faint band found in the bottom panel (lane 2) was caused by non‐specific binding of the HA tag to the protein G beads. Numbers on the left indicate protein size markers.
The interaction was further validated by co‐immunoprecipitation assays in which PR‐1‐5 and ToxA were N‐terminally labelled with c‐Myc and haemagglutinin (HA) epitope tags, respectively, and translated in vitro, and the protein complex was detected using anti‐c‐Myc and anti‐HA antibodies. As shown in Fig. 1B, the 17.5‐kDa c‐Myc‐tagged PR‐1‐5 protein (15.1‐kDa PR‐1‐5 plus 2.4‐kDa c‐Myc) was detected using anti‐c‐Myc antibody, whereas the 15.6‐kDa HA‐tagged ToxA protein (13.2‐kDa ToxA plus 2.4‐kDa HA) was detected in the same protein sample using anti‐HA antibody (lane 1). In the negative control, the 38.1‐kDa c‐Myc‐tagged p53 protein (35.7‐kDa p53 plus 2.4‐kDa c‐Myc) was detected with a signal intensity equal to the 17.5‐kDa c‐Myc‐tagged PR‐1‐5 protein, and a faint signal was seen in the vicinity of the 15.6‐kDa HA‐tagged ToxA protein (lane 2). The latter was a result of the non‐specific binding of the HA tag to protein G beads, as confirmed in a separate experiment (data not shown). Nothing was detected in the background control (lane 3), which only contained the lysate used for in vitro translation. These experiments demonstrate that PR‐1‐5 physically interacts with ToxA both in vivo (at least in yeast) and in vitro. Attempts to pull down the native PR‐1‐5 protein in leaf tissue extracts isolated from ToxA‐treated sensitive wheat using the 15.6‐kDa HA‐tagged ToxA protein were unsuccessful. This may have been a result of the fact that the concentration of the native PR‐1‐5 protein in the wheat leaf extracts was relatively lower than that of the in vitro‐translated c‐Myc‐tagged PR‐1‐5 protein. However, it was more likely a result of competitive binding from other ToxA‐interacting proteins, e.g. ToxABP1 (Manning et al., 2007) and plastocyanin (Tai et al., 2007), both of which are constitutively expressed at high levels (see the final section of the Results).
Two surface‐exposed asparagine residues are essential for the interaction between PR‐1‐5 and ToxA
A series of deletion constructs for both PR‐1‐5 and ToxA proteins was tested by Y2H assays to locate regions containing the interacting sites. In PR‐1‐5, the deletion at the N‐terminal region [up to 62 amino acids (aa)] did not affect the interaction (Fig. 2A, row 2, right), whereas the two deletions (13 and 36 aa, respectively) at the C‐terminal region greatly reduced or abolished the interaction (rows 3 and 4). These results delineate the ToxA‐interacting region to positions 128–151, which include the two β strands (βC and βD) and the turning loop in the PR‐1 fold. In ToxA, the 39‐aa deletion at the C‐terminus, which includes three β strands (β6, β7and β8) and the entire RGD motif, did not affect the interaction (Fig. 2B, row 2), whereas the two deletions (31 and 44 aa) at the N‐terminal region apparently reduced or abolished the interaction (rows 3 and 4). These results indicate that the ToxA RGD motif, which is essential for ToxA activity (Manning et al., 2004, 2008; Sarma et al., 2005; Tuori et al., 2000), is not involved in the PR‐1‐5–ToxA interaction. Instead, the two β strands (β2 and β3) and the turning loop (positions 92–105) contain the interacting sites.
Figure 2.
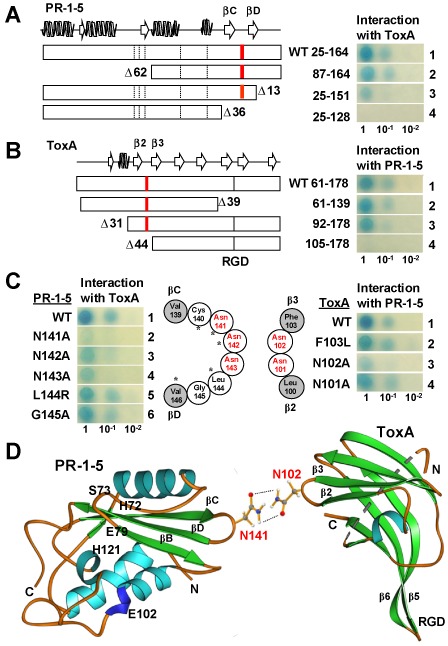
Identification of residues essential for the PR‐1‐5–ToxA interaction. (A, B) Deletion analyses. Open boxes indicate the mature proteins with secondary structures drawn at the top following P14a (Fernandez et al., 1997) and Ptr ToxA (Sarma et al., 2005) for PR‐1‐5 and SnToxA, respectively. Broken and full bars indicate the positions of the five putative active sites in the PR‐1 domain and the ToxA RGD motif, respectively. Red bars indicate the positions of the asparagine‐rich loops. Residue numbering is based on the full‐length PR‐1‐5 protein (GenBank accession number HQ541965) and SnToxA (DQ423483) proteins with ‘Δ’ indicating the deleted residues. (C) Site‐specific mutagenesis. Open and filled circles indicate residues consisting of the loops and the connecting β‐strands, respectively. Asparagine residues are highlighted in red. Asterisks indicate residues conserved in P14a. (D) Ribbon diagrams showing the predicted site‐specific interaction between PR‐1‐5 and ToxA. α helices, β strands and loops are in light blue, green and orange, respectively. The side‐chains of the two interacting asparagine residues are shown in a ‘stick–ball’ scheme with red, grey, blue and white balls for oxygen, carbon, nitrogen and hydrogen atoms, respectively. Broken lines indicate potential hydrogen bonds.
In order to identify critical residues in the targeted interacting regions, we performed site‐specific mutagenesis, with a focus on the residues within the turning loops, because mutations in these amino acids are less likely to be structurally disruptive. In PR‐1‐5, the turning loop between βC and βD contains six residues (Fig. 2C, left). We mutated all of these residues individually, except for cysteine (Cys)‐140, which is predicted to be part of a disulphide bridge. The N141A mutation affected the interaction most significantly, as indicated by the complete loss of blue coloration in the 10−1‐diluted cells and the faint coloration in the undiluted cells (Fig. 2C, left, row 2). The N142A mutation reduced the interaction slightly, as indicated by the weakened coloration in the 10−1‐diluted cells (Fig. 2C, left, row 3). The N143A mutation also caused the loss of blue coloration in the 10−1‐diluted cells, but only reduced the coloration slightly in the undiluted cells (Fig. 2C, left, row 4). In contrast, the L144R and G145A mutations did not change the blue coloration patterns (Fig. 2C, left, rows 5 and 6). These results suggest that L144 and G145 residues are not essential for interaction with ToxA. They also indicate that a single‐residue mutation within the turning loop is less likely to affect the expression level/stability of the PR‐1‐5 protein. In ToxA, the turning loop between β2 and β3 contains two asparagine residues only (Fig. 2C, right). We mutated both asparagine residues individually, and also substituted the adjacent phenylalanine (F103) residue with leucine (L) to generate a site‐related control with a conservative substitution (F and L are both hydrophobic). The N101A and F103L mutations did not affect the interaction (Fig. 2C, right, rows 2 and 4). In contrast, the N102A mutation greatly reduced the interaction, as indicated by the complete loss of blue coloration in the 10−1‐diluted cells and the weak coloration in the undiluted cells (Fig. 2C, right, row 3). Thus, the N141 residue in PR‐1‐5 and the N102 residue in ToxA were identified as the primary interacting sites. Interestingly, mutations at the five putative active sites in the PR‐1‐5 protein did not affect the interaction significantly (data not shown). The N141A mutation did not affect dimerization (see next section) or the protease resistance characteristics of the wild‐type PR‐1‐5 protein (Lu et al., 2013).
To determine the surface accessibility of the two asparagine residues, we examined the structural data published for P14a [Protein Data Bank (PDB) ID: 1CFE; Fernandez et al., 1997] and PtrToxA (PDB ID: 1ZLE; Sarma et al., 2005) using the Jmol (Hanson, 2010) and Ribbon (Carson, 1997) programs. The ‘PR‐1 fold’ is highly conserved among PR‐1‐like proteins, as demonstrated for the human GliPR protein, which has only 35% identity to P14a, but nevertheless adopts the same three‐dimensional structure (Szyperski et al., 1998). The sequences that comprise the βC, βD strands and the turning loop between P14a (RLGCGRAR CNNGWWFISCNYD) and PR‐1‐5 (SIGCARVVCNNNLGVFITCNYE) (italics indicate similar residues) have 67% similarity and two conserved asparagine residues at the targeted sites (N137/N138 in P14a aligned to N141/N142 in PR‐1‐5). PtrToxA and SnToxA are nearly identical (Friesen et al., 2006; Sarma et al., 2005) and have 100% similarity within the β2, β3 strands and the turning loop. Both Jmol and Ribbon programs placed the side chains of the N102 residue in ToxA and the N137 residue in P14a at a position protruding from the surface of the folded protein when the corresponding PDB data were examined. We also confirmed that the asparagine residues at the PR‐1‐5–ToxA interacting sites were not likely to be glycosylated, as predicted by several web servers specializing in the prediction of N‐linked glycosylation sites in eukaryotic proteins (e.g. GlycoEP; Chauhan et al., 2013). Thus, we predicted that the interaction between PR‐1‐5 and ToxA is probably established through hydrogen bonding between the side chains of the N102 and N141 residues (Fig. 2D).
The recombinant ToxA‐N102A mutant protein fails to induce necrosis in ToxA‐sensitive wheat
For the functional characterization of the PR‐1‐interacting site in ToxA, we expressed the wild‐type ToxA and the N102A mutant proteins in Pichia pastoris. Very low yields were obtained in initial trials with the constructs for the 118‐aa mature ToxA proteins [not detectable by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE)]. We then attempted to express the 17‐kDa proToxA with the N‐domain (Tuori et al., 2000). To our surprise, a single major species of 13 kDa was detected for both proteins (Fig. 3A, lanes 1 and 2). Matrix‐assisted laser desorption ionization‐time of flight/time of flight (MALDI‐TOF/TOF) analysis confirmed the sequence identity of the 13‐kDa bands in which the N‐domain was apparently removed, suggesting that ToxA was ‘correctly’ processed in Pichia, probably through a mechanism common in ascomycete fungi. No significant background proteins were detected in the mock strain transformed with the expression vector only (lane 3). The amount of recombinant ToxA proteins was estimated at ∼5 ng/μL. Examination of the tandem mass spectrometry (MS/MS) data confirmed the presence of the 11‐aa protein fragments containing the interacting site (LNNFITIGLNR, positions 100–110 in the full‐length ToxA protein) in the wild‐type ToxA and the alanine substitution (LNAFITIGLNR) in the mutant protein (Fig. 3B, MS data are shown for convenience; detailed MS/MS data are available on request).
Figure 3.

Characterization of the recombinant ToxA proteins. (A) Sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel showing ToxA and the ToxA‐N102A mutant proteins expressed in Pichia pastoris. Each lane was loaded with 20 μL of cell‐free culture supernatant. The control sample was from a ‘mock’ strain transformed with the expression vector only. Numbers on the left indicate the molecular masses (in kDa) of protein markers. (B) Matrix‐assisted laser desorption ionization‐time of flight/time of flight mass spectrometry (MALDI‐TOF/TOF MS) data showing the 11‐amino‐acid fragments containing the wild‐type sequence (top, peak = 1274.71) and the alanine substitution (highlighted in red) in the mutant protein (bottom, peak = 1231.75). (C) Plant assays showing the activity of the recombinant ToxA proteins. ToxA‐sensitive wheat (Grandin) plants were infiltrated with ToxA at the estimated final concentrations (ng/μL) indicated on the right. Control plants were treated with the protein sample of the ‘mock’ strain shown in (A) (lane 3). Marks delimit the infiltrated area. Photographs were taken at 72 h post‐infiltration.
The ToxA‐sensitive wheats Grandin (hexaploid) and Langdon (tetraploid) and ToxA‐insensitive wheat BR34 (hexaploid) were infiltrated with varying amounts of recombinant ToxA proteins (ranging from 0.1 to 1 ng/μL) to determine their activity and specificity. Typical necrosis symptoms were observed in Grandin plants treated with 0.25–1 ng/μL of ToxA at 72 h post‐infiltration (Fig. 3C, top); this activity was comparable with that of purified PtrToxA, which induces the same necrosis symptoms when applied at 0.1–0.5 μm (roughly 1–5 ng/μL) (Manning et al., 2008). Necrosis symptoms were also observed in ToxA‐treated plants of Langdon, which appeared to be less sensitive than Grandin to ToxA, as indicated by the weaker necrosis at lower concentrations (e.g. 0.25 ng/μL) (Fig. S1, see Supporting Information). The ToxA‐insensitive BR34 plants did not show any necrosis in the same treatments (data not shown). These observations indicate that the recombinant ToxA protein is active with the same specificity as the native toxin. Under the same conditions, the ToxA‐N102A mutant failed to induce necrosis in sensitive wheat; some weak chlorosis‐like symptoms were observed only in plants treated with the highest concentration (1 ng/μL) (Fig. 3C, middle). These results confirm that the surface‐exposed, PR‐1‐5‐interacting N102 residue is essential for ToxA activity. The control plants treated with the cell‐free culture supernatant from a yeast strain transformed with the ‘empty’ expression vector did not show any symptoms (Fig. 3C, bottom), indicating that the ToxA activity was solely responsible for the observed necrosis symptoms in sensitive wheat.
The recombinant PR‐1‐5‐N141A mutant protein shows impaired necrosis‐promoting activity compared with the wild‐type
For the functional characterization of the ToxA‐interacting site in PR‐1‐5, we expressed the PR‐1‐5‐N141A mutant protein in P. pastoris and obtained its purified form, together with the previously expressed wild‐type PR‐1‐5 and the dimerization‐defective E102A mutant proteins (Lu et al., 2013). The three purified PR‐1‐5 proteins were all found to have the expected molecular mass of 15 kDa as determined by SDS‐PAGE analysis (Fig. 4A). Low‐temperature (LT) SDS‐PAGE analysis indicated that the PR‐1‐5‐N141A mutant protein retained the ability to form dimmers, in contrast with the E102A mutant, which was found only as a monomer (Fig. 4B). Native PAGE analysis confirmed that the PR‐1‐5 dimers show the expected molecular masses of ∼30 kDa as compared with the native proteinase K (EC 3.4.21.14; molecular mass, 28.9 kDa; isoelectric point, 8.9), which migrated to a position very close to the PR‐1‐5 dimers, but distant to the E102A monomers (Fig. 4C). The monomeric bands detected in LT SDS‐PAGE for the two dimeric PR‐1‐5 proteins were absent in native PAGE (compare arrows with ‘m’ in B and C), suggesting that the PR‐1‐5 protein may exist exclusively as a dimer in its native configuration. Proteolytic digestion tests indicated that the PR‐1‐5‐N141A mutant protein remained resistant to both serine and cysteine proteases, as did the wild‐type (data not shown). To ensure sequence identity, all three purified PR‐1‐5 proteins were re‐validated by size exclusion chromatography and/or MALDI‐TOF/TOF analysis, and the MS/MS data confirmed the presence of the 18‐aa protein fragments containing the mutated interacting site (VVCANNLGVFITCNYEPR) in the N141A mutant protein (Fig. S2, see Supporting Information).
Figure 4.
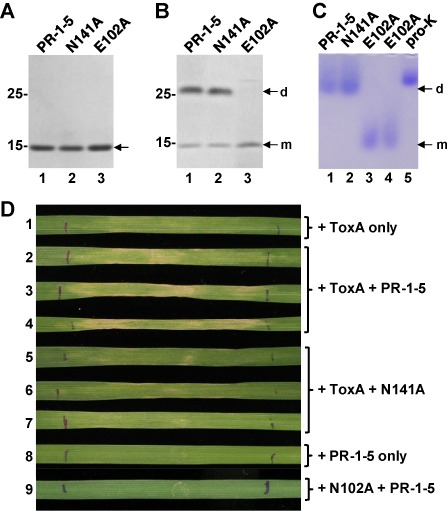
Characterization of the recombinant PR‐1‐5 proteins. (A) Sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel showing the purified PR‐1‐5 proteins (arrow). E102A is a dimerization‐defective mutant (Lu et al., 2013) included as a control. (B) Low‐temperature SDS‐PAGE gel showing the dimers (d) and monomers (m) of the PR‐1‐5 proteins under non‐reducing conditions. (C) Native PAGE gel showing the ∼30‐kDa PR‐1‐5 dimers (d) in comparison with the 15‐kDa E102A monomers (m) (loaded twice in lanes 3 and 4). Lane 5 was loaded with proteinase K (pro‐K, MW = 28.9 kDa; isoelectric point, 8.9). Numbers on the left in (A) and (B) indicate the molecular masses (in kDa) of the protein markers. (D) Plant assays showing the necrosis‐promoting activity of the PR‐1‐5 protein. ToxA‐sensitive wheat (Langdon) plants were infiltrated with the recombinant ToxA and/or PR‐1‐5 proteins as indicated on the right. Photographs were taken at 5 days post‐infiltration (one or three representative leaves of each treatment are shown).
The biological activity of the PR‐1‐5 proteins was tested by co‐infiltration assays in different wheat lines. In attempts to mimic a natural situation, we adjusted the ToxA concentration to 0.05 ng/μL at which ToxA alone does not induce apparent necrosis at 48 h (or even longer) post‐infiltration; high concentrations (>0.1 ng/μL) of ToxA were not suitable for these assays because they cause rapid cell death, making it difficult to detect the activity of the recombinant PR‐1‐5 proteins. The final concentrations of the PR‐1‐5 proteins were adjusted to 0.25–0.5 ng/μL, which is close to the concentration of the native PR‐1‐5 protein in ToxA‐treated sensitive wheat (based on a comparison with serially diluted recombinant PR‐1‐5 protein in Western blots, data not shown). The best results were obtained when the co‐infiltration assays were performed using tetraploid Langdon wheat (which is less sensitive to ToxA, as mentioned above) with an extended incubation time (i.e. 5 days) as used in previous studies (e.g. Manning et al., 2008). At the defined concentrations, the Langdon plants treated with ToxA alone did not show apparent necrosis symptoms at 5 days post‐infiltration (Fig. 4D, leaf 1). In contrast, those co‐infiltrated with ToxA and the wild‐type PR‐1‐5 protein all developed necrotic symptoms with tissue collapse seen across the infiltration area (Fig. 4D, leaves 2–4). Under the same conditions, plants co‐infiltrated with the PR‐1‐5‐N141A mutant protein all looked similar to those treated with ToxA alone, although some minor necrosis was seen near the infiltration site (Fig. 4D, leaves 5–7), indicating that the mutant protein is impaired in its ability to promote the development of necrosis. Infiltration with PR‐1‐5 or the PR‐1‐5‐N141A mutant protein alone did not induce necrosis (Fig. 4D, leaf 8), and the same was true for co‐infiltration with the ToxA‐N102A mutant and the PR‐1‐5 protein (Fig. 4D, leaf 9). When tested with hexaploid wheat lines, PR‐1‐5‐associated necrosis‐promoting activity was observed in ToxA‐sensitive Grandin, but not in ToxA‐insensitive BR34, wheat plants (Fig. S3, see Supporting Information). No necrosis was observed when several ToxA‐insensitive tsn1 mutant wheat lines (Faris et al., 2010) were tested under the same co‐infiltration conditions (S. Lu and J. D. Faris, unpublished data). These results demonstrate that the expression of the PR‐1‐5‐related necrosis‐promoting activity depends on the co‐existence of ToxA and its cognate sensitivity gene Tsn1 (Faris et al., 2010), and such ‘death‐promoting’ activity requires a physical interaction between PR‐1‐5 and ToxA.
In addition, preliminary experiments suggest that the monomeric E102A mutant protein is less active in promoting necrosis, but the results are not conclusive because the tested protein sample contains a second minor species detectable under non‐reducing conditions (Fig. 4B, lane 3). We will try to further purify this mutant protein, together with the monomeric PR‐1‐1 protein (Lu et al., 2013), to address whether the dimerization of PR‐1‐5 is essential for the observed ‘death‐promoting’ activity.
The native PR‐1‐5 protein is differentially expressed between ToxA‐sensitive and ToxA‐insensitive wheat lines
The PR‐1‐5 gene is present in the genomes of both ToxA‐sensitive and ToxA‐insensitive wheat lines, but has been shown to be differentially expressed in response to infection by a ToxA‐producing isolate of Sn (Lu et al., 2009, 2011). To determine whether the native PR‐1‐5 protein is expressed in response to ToxA in the absence of the pathogen, we analysed mRNA and protein samples extracted from ToxA‐treated plants of different wheat lines. Reverse transcriptase‐polymerase chain reaction (RT‐PCR) revealed that PR‐1‐5 transcripts were absent in mock‐infiltrated ToxA‐insensitive wheat BR34 (Fig. 5, lanes 1–4) and detectable only at low levels (24 and 48 h post‐infiltration) in mock‐infiltrated ToxA‐sensitive wheat Grandin (Fig. 5, lanes 11 and 12). When BR34 plants were treated with ToxA, only low levels of PR‐1‐5 transcripts became detectable at 48 h (Fig. 5, lane 8). In contrast, the transcripts were up‐regulated sharply in Grandin, as indicated by the strong band seen as early as 12 h, when necrosis had just became visible (Fig. 5, lane 14), and by the greatly intensified band seen at 48 h, at which time the entire infiltrated area had collapsed (Fig. 5, lane 16). In Western blot analysis, the anti‐PR‐1‐5 antibody (Lu et al., 2013) hybridized to a 15‐kDa band in ToxA‐treated Grandin plants at 12, 24 and 48 h, with the signal intensity correlating with the abundance of the PR‐1‐5 transcripts shown in the RT‐PCR results (Fig. 5, lanes 14–16, compare white and red asterisks). These data confirm that the native PR‐1‐5 protein is expressed and accumulates coincidentally with necrosis development in ToxA‐sensitive wheat. In contrast, under the same conditions, the transcripts encoding three other proteins were found to be either up‐regulated in ToxA‐treated plants (as for the PR‐1‐1 protein) or constitutively expressed at high levels in all plants [as for the two ToxA‐interacting chloroplast proteins ToxABP1 (Manning et al., 2007) and plastocyanin (Tai et al., 2007)] of both sensitive and insensitive wheat lines (Fig. S4, see Supporting Information). The up‐regulation of PR‐1‐1 in both wheat lines indicates that ToxA may induce the expression of multiple PR‐1 genes through Tsn‐1‐independent pathways not necessarily associated with the development of necrosis. Functional tests of the recombinant PR‐1‐1 protein, as mentioned above, would help to determine whether the native PR‐1‐1 protein also contributes to ToxA‐induced necrosis in sensitive wheat.
Figure 5.
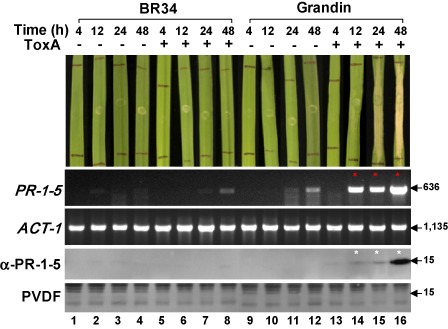
Differential expression of the native PR‐1‐5 protein between ToxA‐insensitive (BR34) and ToxA‐sensitive (Grandin) wheat lines in response to ToxA treatment. Plant assays (top panel): second leaves of 2‐week‐old plants were ‘mock’ infiltrated (−) or infiltrated with ToxA protein (+) at a final concentration of 1.0 ng/μL. Photographs were taken at four time points post‐infiltration as indicated at the top. A representative leaf is shown for each treatment. Reverse transcriptase‐polymerase chain reaction (RT‐PCR) (second and third panels): PCR products were separated on ethidium bromide‐stained 1% agarose gels. Numbers on the right indicate size (in base pairs) of the amplicons. Transcripts up‐regulated relative to those of controls are indicated by red asterisks. ACT‐1, wheat actin gene (internal control). The oligonucleotide sequences of the primers used for RT‐PCR are given in Table S1. Western blot analysis (bottom two panels): protein samples isolated from the plants shown at the top were separated by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE), transferred onto poly(vinylidene difluoride) (PVDF) membrane and probed with the anti‐PR‐1‐5 antibody (α‐PR‐1‐5) (Lu et al., 2013). Signals were detected by exposure to X‐ray films for 30 s. Membranes were stained with Coomassie blue after signal detection (bottom). Numbers on the right indicate the positions of size markers (molecular masses in kDa).
Discussion
Our initial Y2H library screening experiments identified PR‐1 proteins as potential targets of ToxA (Lu et al., 2009). In the same experiments, we also recovered several cDNA clones encoding plastocyanin identical to that reported by Tai et al. (2007), but were unable to identify ToxABP1 (Manning et al., 2007). We noticed that the ToxA bait used by Manning et al. (2007) included residues 23–178, with the first 22 amino acids removed based on the original signal peptide analysis (Ciuffetti et al., 1997). In our experiments, we used residues 17–178 with the first 16 amino acids removed based on the prediction from the SignalP server (Petersen et al., 2011). In addition, we used mRNA for cDNA synthesis instead of total RNA as used by Manning et al. (2007). These technical differences may have affected the chance to pull out ToxABP1 from a particular cDNA library, and may also explain why no PR‐1‐like clones were recovered from previous studies using the same Y2H library screening systems (Manning et al., 2007; Tai et al., 2007).
In this study, we confirmed the physical interaction between PR‐1‐5 and ToxA through Y2H and in vitro co‐immunoprecipitation assays, and determined that the interaction is mediated by two surface‐exposed asparagine residues. The polar side‐chain of asparagine can act as both a hydrogen bond donor and an acceptor, thus playing an important role in intermolecular interactions (Carpena et al., 2009; Shimoni and Glusker, 1995). The binding of PR‐1‐5 to ToxA may rely on hydrogen bonds formed between the oxygen (or hydrogen) atom in the side‐chain of the N141 residue in PR‐1‐5 and the hydrogen (or oxygen) atom in the side‐chain of the N102 residue in ToxA (Fig. 2D). Amino acid sequence alignments revealed that the N141 residue in the PR‐1‐5 protein is actually a part of a unique motif‐like sequence (NNNL) found only in PR‐1‐5 and PR‐1‐4 proteins in the wheat PR‐1 family; extensive database searches failed to identify a homologue having this ‘NNNL’ motif in plant PR‐1 proteins (Fig. S5, see Supporting Information). However, the N102 residue in ToxA is located within a region whose counterpart is completely missing in the mammalian FnIII proteins topologically most closely related to ToxA (Sarma et al., 2005). Thus, the specificity of the interaction between PR‐1‐5 and ToxA may have stemmed from a beneficial ‘matching’ of two unusual structures, each acquired by an unrelated protein during natural selection. Like the RGD motif, the N101–N102 loop is conserved; no substitutions were found at either asparagine residue in the natural ToxA isoforms investigated in a previous study (Tan et al., 2012). Further mutational analysis is needed to determine the functional importance of the two asparagine‐rich motifs in the PR‐1‐5 and ToxA interaction. It remains to be determined whether PR‐1‐5 interacts directly with ToxA in planta. The HA‐tagged ToxA protein failed to pull down the native PR‐1‐5 protein from the plant extracts used for co‐immunoprecipitation assays, probably as a result of potential competitive binding from ToxABP1 (Manning et al., 2007) and plastocyanin (Tai et al., 2007), as mentioned in the previous section; however, the negative results may also suggest that PR‐1‐5 does not interact directly with ToxA in planta or the interaction is too weak to be detected using the pull‐down method. Bimolecular fluorescence complementation assays (reviewed by Kodama and Hu, 2012) will be pursued using an Agrobacterium‐based transient expression system to further evaluate the PR‐1‐5–ToxA interaction in planta.
Previous studies on ToxA have been largely focused on the RGD motif (Manning et al., 2004, 2008; Meinhardt et al., 2002; Sarma et al., 2005; Tuori et al., 1995, 2000; Zhang et al., 1997) because of its similarity to the classical mammalian RGD‐containing domains that bind to membrane‐associated integrin receptors. More than 20 single‐residue mutations have been made to ToxA (Manning et al., 2004, 2008; Meinhardt et al., 2002); none of them targeted the N102 residue or immediately adjacent amino acids. Despite the demonstrated importance of the RGD motif, the fact that plants lack any bona fide integrins makes it difficult to draw parallels with the mammalian systems. Some integrin‐like proteins have been identified in Arabidopsis (Gouget et al., 2006; Knepper et al., 2011), but their homologues in wheat have not been reported. The RGD‐independent interaction between PR‐1‐5 and ToxA identified in this study opens a door to explore alternative receptors directly linked to plant pathways. The successful expression of ToxA in Pichia will facilitate functional studies. Unlike the Escherichia coli‐expressed PtrToxA (Manning et al., 2008) and SnToxA (Tan et al., 2012), which are His‐tagged and less active, the Pichia‐expressed SnToxA obtained in this study is tag free and appears to be as active as the native PtrToxA, although further purification is needed to precisely quantify the activity.
By using LT SDS‐PAGE and native PAGE analyses, we confirmed that PR‐1‐5 represents the first dimeric member in the plant PR‐1 family, and may exist exclusively as a dimer, at least under the defined native PAGE conditions (Fig. 4). The fact that the PR‐1‐5‐N141A mutant protein retains the ability to form dimers (Fig. 4) indicates that the ToxA‐binding site is not essential for dimerization in PR‐1‐5. This is in contrast with the E102A mutant, which fails to form dimers because of the mutation at one of the active sites known to be involved in dimerization (Lu et al., 2013; Serrano et al., 2004). In ToxA, the RGD motif is not targeted by PR‐1‐5 (Fig. 2). Together, these results suggest that the binding of ToxA to PR‐1‐5 (or vice versa) would not disturb the active sites in either protein; thus, the resulting protein complex would be dually functional in downstream operations. Recent studies have shown that the dimerization of the GAPR‐1 protein is required for its function involving membrane‐binding activity (Eberle et al., 2002; Serrano et al., 2004; van Galen et al., 2012). It will be important to determine whether the PR‐1‐5 dimer is the functional subunit in the PR‐1‐5–ToxA protein complex and how dimerization affects the necrosis‐promoting activity.
In planta characterization of PR‐1‐5 function is a challenging task because of the genomic redundancy of the PR‐1 family. There are an estimated total of >60 PR‐1‐like genes in the hexaploid wheat genome, with several having >80% nucleotide identity to PR‐1‐5 (Lu et al., 2011). Attempts to specifically knock down the PR‐1‐5 gene by virus‐induced gene silencing (Holzberg et al., 2002) have been unsuccessful in our laboratory. The co‐infiltration assays presented in this study provide an alternative way to test the biological activity of the PR‐1 proteins in their native host. The concentration of ToxA is important for co‐infiltration assays. It is known that ToxA produced by Ptr isolates in liquid culture under laboratory conditions can reach a final concentration of 1–2 ng/μL (Tomas et al., 1990). Purified native or recombinant ToxA proteins are routinely used for infiltration assays with a final concentration of 1–25 ng/μL; the minimal concentration required for the induction of necrosis has been reported to be 0.7–1.3 ng/μL (Manning et al., 2004; Meinhardt et al., 2002; Tomas et al., 1990). Realistically, the apoplastic concentrations of ToxA secreted by invading mycelia of the fungus in a susceptible host may never reach such minimal levels. Thus, we believe that the use of subminimal concentrations (below 0.7 ng/μL) for co‐infiltration assays is a legitimate choice to mimic a natural situation. Similarly, the amount of the PR‐1 protein must be taken into account. It is essential to use a concentration close to its native status, because the impact of forcefully delivered excess ‘exogenous’ proteins on biological processes associated with plant apoplastic spaces, cell walls or cell wall–plasma membrane interfaces cannot be predicted.
How the PR‐1‐5–ToxA interaction mediates ToxA‐induced necrosis in wheat remains to be investigated. One possibility is that the PR‐1‐5–ToxA interaction may be involved in the activation of cell death pathway(s) in sensitive wheat. The native PR‐1‐5 protein is also expressed in mock‐treated sensitive wheat and is easily detectable at 24 and 48 h post‐infiltration (Fig. 5, lanes 11 and 12). We speculate that, in sensitive wheat, the PR‐1‐5 protein can be induced by certain stress conditions, such as cell wall damage associated with artificial infiltration or fungal penetration in a disease‐favourable natural environment. ToxA can target these ‘stress‐induced’ PR‐1‐5 proteins (although with low abundance) to establish an initial interaction, which is sufficient for the activation of Tsn1‐controlled cell death pathway(s). Once the pathway(s) is initiated and local cell death starts, there could be a feedback control loop that up‐regulates the PR‐1‐5 protein whilst necrosis advances (Fig. 5, lanes 14–16). We noticed that the sensitive wheat plants treated with ToxA alone in the co‐infiltration assays (Figs 4D and S3A, leaf 1) eventually developed necrosis symptoms, similar to plants co‐infiltrated with ToxA and PR‐1‐5, if incubated for a longer period (4–5 days instead of 48 h post‐infiltration in the case of Grandin). Thus, the exogenous application of PR‐1‐5 through co‐infiltration might mimic an ‘ahead of time’ expression that allows necrosis to develop earlier than in the case of infiltration with ToxA alone, in which the expression of necrosis may depend on the progressive up‐regulation of the native PR‐1‐5 protein.
It is known that, in the presence of specific death stimuli, plant programmed cell death pathways can be activated by extracellular enzymes, such as serine protease‐related saspases/phytaspases having caspase‐like activities essential for apoptosis in human/animal systems (Bonneau et al., 2008; Chichkova et al., 2010; Vartapetian et al., 2011). Although lacking similarity to saspases/phytaspases, PR‐1‐5 shares certain commonalities with caspases, including resistance to serine/cysteine proteases, formation of dimers and conserved motif‐like sequences (Lu et al., 2013). The necrosis‐promoting activity observed in this study further supports the notion that PR‐1‐5 may have caspase‐like activities. It is possible that the binding of ToxA to PR‐1‐5 causes a specific conformational change in the PR‐1‐5 dimer, leading to the activation of its substrate‐specific protease activity, which would turn on (probably through interaction with other death signalling‐related enzymes/proteins) Tsn1‐controlled defence‐related pathways, such as those in classical gene‐for‐gene interactions, eventually resulting in cell death. The identification of the potential substrates of PR‐1‐5 activity through protease profiling may provide useful information with regard to this hypothesis. Interestingly, a recent study (Zhang et al., 2012) has reported that the powdery mildew effector CSEP0055 targets three barley PR proteins, including PR17c, PR1a and PR1b, as revealed by Y2H library screening, and the CSEP0055–PR17c interaction may contribute to disease resistance, although the functional significance of the CSEP0055–PR‐1a/PR1b interaction has not been tested. Nevertheless, these findings suggest further that plant PR‐1 proteins can be potential targets of both necrotrophic and biotrophic effectors.
In summary, the PR‐1‐5–ToxA interaction identified in this study suggests that specific recognition between a host‐selective toxin and its target (or cofactor) may be achieved through affinity binding mediated by unique surface‐exposed sites on the two interacting partners. The ToxA‐binding asparagine‐rich loop identified in the dimeric PR‐1‐5 protein may represent an effector‐binding domain in the PR‐1 family. Thus, PR‐1, and perhaps also other extracellular defence proteins, may be directly targeted by specific proteinaceous toxins produced by necrotrophic pathogens, such as Ptr and Sn. Further characterization of the PR‐1‐5–ToxA interaction will help us to understand how necrotrophic pathogens exploit plant defence mechanisms to induce diseases in the host plants.
Experimental Procedures
Y2H assays
The targeted coding regions in the mature proteins of SnToxA (bait) and PR‐1‐1/PR‐1‐5 (prey) were PCR amplified from laboratory cDNA clones of SnToxA (GenBank accession number DQ423483; Friesen et al., 2006) and PR‐1‐1 (HQ541961)/PR‐1‐5 (HQ541965) (Lu et al., 2011), respectively. Mutations were generated by PCR fusion using the primers listed in Table S1 (see Supporting Information). PCR products were subcloned into the pGBK‐T7 (bait) or the pGAD‐T7 (prey) vector and transformed into yeast strain AH109, as described previously (Lu, 2012). For α‐galactosidase activity assays, yeast cells were picked up from a single colony of the transformant grown on agar plates containing synthetic defined medium lacking leucine and tryptophan (SD/–LT, selecting for the bait and prey proteins) and suspended in 50 μL of sterile water, followed by three 1 : 10 dilutions in series. Five microlitres of each cell suspension in the dilution sets were inoculated onto agar plates of SD/–LT and SD/–LTHA + X‐α‐gal (lacking leucine, tryptophan, histidine and adenine, and supplemented with the chromogenic substrate 5‐bromo‐4‐chloro‐3‐indolyl‐α‐d‐galactopyranoside) at the same time to select for reporter genes. Plates were incubated at 30 °C for 36–48 h until blue coloration became visible in the 10−3‐diluted cells of the standard positive control (p53/RecT) before photographing. All α‐galactosidase activity assays were duplicated and repeated at least once.
Co‐immunoprecipitation assays
The cMyc‐PR‐1‐5 (bait) and HA‐SnToxA (prey) epitope‐tagged proteins (constructed in the pGBK‐T7/pGAD‐T7 plasmids with the T7 promoter) were generated by in vitro translation using the TNT® Coupled Reticulocyte Lysate System (Promega, Madison, WI, USA), and co‐immunoprecipitation assays were performed using the Dynabeads Protein G package (Life Technologies, Grand Island, NY, USA) following the manufacturer's instructions. The protein complex was finally eluted from the magnetic beads in 20 μL of elution buffer with 10 μL of reducing agent provided by the manufacturer. The co‐precipitated bait and prey proteins were detected by Western blot analysis using horseradish peroxidase (HRP)‐conjugated anti‐c‐Myc (Sigma‐Aldrich, St. Louis, MO, USA) and HRP‐conjugated anti‐HA (Roche Diagnostics, Indianapolis, IN, USA) antibodies.
Expression of the recombinant SnToxA and PR‐1‐5 proteins in yeast
The coding region corresponding to the 17‐kDa proToxA (Tuori et al., 2000) was PCR amplified from the SnToxA cDNA clone. ToxA‐N102A and PR‐1‐5‐N141A mutant constructs were generated by PCR fusion. PCR primers are listed in Table S2 (see Supporting Information). Yeast transformation and protein expression/isolation were performed as described previously (Lu et al., 2013). The concentration of the target protein was estimated by SDS‐PAGE in comparison with a series of dilutions of bovine serum albumin (BSA) (New England Biolabs, Ipswich, MA, USA) included in the same gel. Large‐scale expression (from the same yeast strain) and subsequent purification of the PR‐1‐5‐N141A mutant protein were performed through a commercial service (NeoBioLab, Cambridge, MA, USA), together with the wild‐type PR‐1‐5 and the E102A mutant proteins (Lu et al., 2013). The purified PR‐1‐5 proteins were finally provided in phosphate‐buffered saline with a concentration of 0.2–0.27 mg/mL. The sequence identity of the recombinant SnToxA and PR‐1‐5 proteins was confirmed by MALDI‐TOF/TOF analysis.
Protein gel electrophoresis and MS analysis
SDS‐PAGE was performed following standard protocols (Green and Sambrook, 2012). Low‐temperature SDS‐PAGE was performed as described by Lu et al. (2013). Size exclusion chromatography and MS analyses were performed at the Proteomics and Mass Spectrometry Facility, Cornell University Biotechnology Resource Center, Ithaca, NY, USA. Native PAGE was performed using procedures adapted from a Bio‐Rad (Hercules, CA, USA) protocol. Purified proteins (1 μg per lane) were separated on SDS‐free 8% straight polyacrylamide gels without stacking gel in SDS‐free Tris‐glycine running buffer at 4 °C, followed by Coomassie blue staining. The native proteinase K (EC 3.4.21.14) (New England Biolabs, Ipswich, MA, USA) was included as a reference protein because it has a molecular mass of 28.9 kDa, close to that of the PR‐1‐5 dimers (∼30 kDa), and is a basic protein [isoelectric point (pI), 8.9], like the PR‐1‐5 protein (pI, 8.3).
Plant assays
ToxA‐sensitive hexaploid (cultivar Grandin) and tetraploid (cultivar Langdon) wheat and ToxA‐insensitive hexaploid wheat (cultivar BR34) were used for infiltration/co‐infiltration assays. Plants were grown in 6‐in (15.24‐cm) pots (10–16 plants per pot) containing SB100 professional growing mix (Sungrow Horticulture, Dellevue, WA, USA) in a growth chamber at an average temperature of 21 °C with a 16‐h photoperiod for 2 weeks. For ToxA activity assays, the second leaf of each plant was infiltrated with ∼20 μL of cell‐free culture supernatant (diluted in series with water) containing the recombinant ToxA protein (Fig. 3A) at the desired concentrations using a 1‐mL syringe without the needle (Delasco, Council Bluffs, IA, USA). Control plants were infiltrated with the culture supernatant from a yeast strain transformed with the expression vector only (Fig. 3A). Treated plants were kept in a growth chamber and symptoms were examined 2–5 days after infiltration. Each treatment included at least five plants. For co‐infiltration assays, ToxA and PR‐1‐5 proteins were diluted with water and mixed together, with the final concentrations adjusted to 0.05 ng/μL for ToxA and 0.25 ng/μL for PR‐1‐5. The protein solution was incubated at room temperature for 10–60 min before use. Plants were treated in the same way as mentioned above, except that both the first and second leaves of the plants were infiltrated. Each treatment included 5–10 plants and was repeated at least twice.
RT‐PCR and Western blot analysis
mRNA samples were isolated from leaf tissues of 2‐week‐old ToxA‐treated and control plants of cultivars BR34 and Grandin that were collected at different time points (4, 12, 24 and 48 h post‐infiltration). RT‐PCR was performed as described previously (Lu et al., 2011) using the following primers: 5′‐AATACGCCGCAGGACTACGTT‐3′ and 5′‐AAGACTGCCATAGAGAAGCTCA‐3′ for PR‐1‐5 and 5′‐ ATGGCTGACGGTGAGGACAT‐3′ and 5′‐GATCAGAAGCACTTCCTGTGGA‐3′ for ACT‐1 (internal control) (Lu et al., 2011). Protein extraction and Western blot analysis using the HRP‐conjugated anti‐PR‐1‐5 polyclonal antibody were performed as described by Lu et al. (2013).
Sequence analysis
Amino acid sequences of plant PR‐1 proteins from wheat and other species were retrieved from GenBank databases available online from the National Center for Biotechnology Information. Alignments were generated using the MegAlign programs from Lasergene 8.1 software (DNASTAR Inc., Madison, WI, USA). Structural data for P14a (Fernandez et al., 1997) and PtrToxA (Sarma et al., 2005) were retrieved from PDB available online from the European Bioinformatics Institute; the corresponding PDB files were examined using the Jmol (Hanson, 2010) and Ribbon (Carson, 1997) programs.
Supporting information
Fig. S1 Plant assays showing activity of the recombinant ToxA proteins in the ToxA‐sensitive tetraploid Langdon wheat. Second leaves of 2‐week‐old plants were infiltrated with ToxA at the estimated final concentrations (ng/μL) indicated on the right (highest concentration shown for the N102A mutant). Control plants were treated with the protein sample of the ‘mock’ strain shown in Fig. 3A (lane 3). Marks delimit the infiltrated area. Photographs were taken at 72 h post‐infiltration (one representative leaf of each treatment is shown).
Fig. S2 Characterization of the purified recombinant PR‐1‐5 proteins. (A) Size exclusion chromatography analysis of the recombinant PR‐1‐5 (left) and PR‐1‐5‐N141A (right) proteins. A single peak was detected for both proteins (samples were separated using Phenomenex Yarra SEC‐2000 columns; Torrance, CA, USA). (B) Mass spectrum showing the tryptic fragment (peak, 41.85) containing the single‐residue mutation site in the recombinant PR‐1‐5‐N141A mutant protein. The amino acid (aa) sequence of the 18‐aa fragment (corresponding positions of 138–155 in the PR‐1‐5 protein) is shown with the mutated residue highlighted in red [detailed tandem mass spectrometry (MS/MS) data are available on request].
Fig. S3 Plant assays showing the necrosis‐promoting activity of the PR‐1‐5 protein in hexaploid wheat cultivars. ToxA‐sensitive (Grandin, A) and insensitive (BR34, B) wheat plants were infiltrated with the recombinant ToxA and/or PR‐1‐5 proteins as indicated on the right. Photographs were taken at 48 h post‐infiltration (one or three representative leaves of each treatment are shown).
Fig. S4 Expression patterns of PR‐1‐1, ToxABP1 and PCN in comparison with that of PR‐1‐5 (Fig. 5) in response to ToxA treatment. mRNA samples were collected from the second leaves of ToxA‐insensitive (BR34) and ToxA‐sensitive (Grandin) wheat plants that were ‘mock'‐infiltrated (–) or infiltrated with ToxA protein (+) at a final concentration of 1.0 ng/μL. Polymerase chain reaction (PCR) products were separated on ethidium bromide‐stained 1% agarose gels. Numbers on the right indicate size (in base pairs) of the amplicons. Oligonucleotide sequences of the primers used for reverse transcriptase‐polymerase chain reaction (RT‐PCR): 5′‐CCTTTTAGACACCGAACCAGGAAGT‐3′ (forward) and 5′‐ATGGCGGCCATATCGTCGCTTCCT‐3′ (forward) and 5′‐TTAATGCTTCAAGGGATAAGCGCT‐3′ (reverse) for TOXABP1 (Manning et al., 2007), and 5′‐ATGGCCGCCCTCTCCTCTGCA‐3′ (forward) and 5′‐TTAGTTGACGGTGACCTTGCCGA‐3′ (reverse) for PCN (Tai et al., 2011).
Table S1 Polymerase chain reaction (PCR) primers used for the generation of yeast two‐hybrid constructs.
Table S2 Polymerase chain reaction (PCR) primers used for the generation of Pichia expression constructs.
Fig. S5 Amino acid sequence alignment showing the residues consisting of the turning loop between βC and βD in the PR‐1 domains of plant PR‐1 proteins. Residues shown correspond to positions 135–150 of the PR‐1‐5 protein (GenBank accession number HQ541965). Arrows indicate residues aligned to the N141 site of PR‐1‐5 and its ‘sister’ protein PR‐1‐4 (underlined), both of which have a unique ‘NNNL’ motif (highlighted in yellow). Numbers on the right indicate the positions in the full‐length proteins. Alignments were generated using the MegAlign program (DNASTAR). Species abbreviations: At, Arabidopsis thaliana; Bn, Brassica napus; Cc, Capsicum chinense; Ca, C. annuum; Ew, Eutrema wasabi; Gm, Glycine max; Hv, Hordeum vulgare; Le, Lycopersicon esculentum; Md, Malus × domestica; Nt, Nicotiana tabacum; Os, Oryza sativa; Sb, Sorghum bicolor; St, Solanum tuberosum; Ta, Triticum aestivum; Tm, T. monococcum; Zm, Zea mays. The 23 wheat (Ta) PR‐1‐like proteins (PR‐1‐1 to PR‐1‐23) have been reported previously by Lu et al. (2011).
Acknowledgements
We thank Kelsey Dunnell and Tyler Lewandowski for technical assistance. This study was supported by USDA‐ARS CRIS project 5442‐21000‐037‐00D.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture (USDA). USDA is an equal opportunity provider and employer.
References
- Adhikari, T.B. , Bai, J. , Meinhardt, S.W. , Gurung, S. , Myrfield, M. , Patel, J. , Ali, S. , Gudmestad, N.C. and Rasmussen, J.B. (2009) Tsn1‐mediated host responses to ToxA from Pyrenophora tritici‐repentis. Mol Plant Microbe Interact, 22, 1056–1068. [DOI] [PubMed] [Google Scholar]
- Alexander, D. , Goodman, R.M. , Gut‐Rella, M. , Glascock, C. , Weymann, K. , Friedrich, L. , Maddox, D. , Ahl‐Goy, P. , Luntz, T. and Ward, E. (1993) Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis‐related protein 1a. Proc. Natl. Acad. Sci. USA, 90, 7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asojo, O.A. , Koski, R.A. and Bonafe, N. (2011) Structural studies of human glioma pathogenesis‐related protein 1. Acta Crystallogr. D: Biol. Crystallogr. 67, 847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballance, G.M. , Lamari, L. and Bernier, C.C. (1989) Purification and characterization of a host‐selective necrosis toxin from Pyrenophora tritici‐repentis . Physiol. Mol. Plant Pathol. 35, 203–213. [Google Scholar]
- Ballance, G.M. , Lamari, L. , Kowatsch, R. and Bernier, C.C. (1996) Cloning, expression and occurrence of a gene encoding the Ptr necrosis toxin from Pyrenophora tritici‐repentis Mol. Plant Pathol. On‐Line (http://www.bspp.org.uk/mppol/1996/1209ballance).
- Bonafe, N. , Zhan, B. , Bottazzi, M.E. , Perez, O.A. , Koski, R.A. and Asojo, O.A. (2010) Expression, purification, crystallization and preliminary X‐ray analysis of a truncated soluble domain of human glioma pathogenesis‐related protein 1. Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 66, 1487–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau, L. , Ge, Y. , Drury, G.E. and Gallois, P. (2008) What happened to plant caspases? J. Exp. Bot. 59, 491–499. [DOI] [PubMed] [Google Scholar]
- Buchel, A.S. and Linthorst, H.J.M. (1999) PR‐1: a group of plant proteins induced upon pathogen infection In: Pathogenesis‐Related Proteins in Plants (Datta S.K. and Muthukrishnan S., eds), pp. 21–47. Boca Raton, FL: CRC Press. [Google Scholar]
- Cantacessi, C. , Hofmann, A. , Young, N.D. , Broder, U. , Hall, R.S. , Loukas, A. et al (2012) Insights into SCP/TAPS proteins of liver flukes based on large‐scale bioinformatic analyses of sequence datasets. PLoS ONE, 7, e31164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpena, X. , Vidossich, P. , Schroettner, K. , Calisto, B.M. , Banerjee, S. , Stampler, J. et al (2009) Essential role of proximal histidine–asparagine interaction in mammalian peroxidases. J. Biol. Chem. 284, 25 929–25 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson, M. (1997) Ribbons. Methods Enzymol. 277, 493–505. [PubMed] [Google Scholar]
- Chauhan, J.S. , Rao, A. and Raghava, G.P. (2013) In silico platform for prediction of N‐, O‐ and C‐glycosites in eukaryotic protein sequences. PLoS ONE, 8, e67008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichkova, N.V. , Shaw, J. , Galiullina, R.A. , Drury, G.E. , Tuzhikov, A.I. , Kim, S.H. et al (2010) Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO J. 29, 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffetti, L.M. , Tuori, R.P. and Gaventa, J.M. (1997) A single gene encodes a selective toxin causal to the development of tan spot of wheat. Plant Cell, 9, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffetti, L.M. , Manning, V.A. , Pandelova, I. , Betts, M.F. and Martinez, J.P. (2010) Host selective toxins, Ptr ToxA and Ptr ToxB, as necrotrophic effectors in the Pyrenophora tritici‐repentis–wheat interaction. New Phytol. 187, 911–919. [DOI] [PubMed] [Google Scholar]
- Eberle, H.B. , Serrano, R.L. , Fullekrug, J. , Schlosser, A. , Lehmann, W.D. , Lottspeich, F. , Kaloyanova, D. , Wieland, F.T. and Helms, J.B. (2002) Identification and characterization of a novel human plant pathogenesis related protein that localizes to lipid‐enriched microdomains in the Golgi complex. J. Cell Sci. 115, 827–838. [DOI] [PubMed] [Google Scholar]
- Faris, J.D. , Anderson, J.A. , Francl, L.J. and Jordahl, J.G. (1996) Chromosomal location of a gene conditioning insensitivity in wheat to a necrosis‐inducing culture filtrate from Pyrenophora tritici‐repentis . Phytopathology, 86, 459–463. [Google Scholar]
- Faris, J.D. , Zhang, Z. , Lu, H. , Lu, S. , Reddy, L. , Cloutier, S. , Fellers, J.P. , Meinhardt, S.W. , Rasmussen, J.B. , Xu, S.S. , Oliver, R.P. , Simons, K.J. and Friesen, T.L. (2010) A unique wheat disease resistance‐like gene governs effector‐triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA, 107, 13 544–13 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, C. , Szyperski, T. , Bruyere, T. , Ramage, P. , Mosinger, E. and Wuthrich, K. (1997) NMR solution structure of the pathogenesis‐related protein P14a. J. Mol. Biol. 266, 576–593. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Stukenbrock, E.H. , Liu, Z. , Meinhardt, S. , Ling, H. , Faris, J.D. , Rasmussen, J.B. , Solomon, P.S. , McDonald, B.A. and Oliver, R.P. (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Faris, J.D. , Solomon, P.S. and Oliver, R.P. (2008) Host‐specific toxins: effectors of necrotrophic pathogenicity. Cell. Microbiol. 10, 1421–1428. [DOI] [PubMed] [Google Scholar]
- van Galen, J. , Olrichs, N.K. , Schouten, A. , Serrano, R.L. , Nolte‐'t Hoen, E.N. , Eerland, R. et al (2012) Interaction of GAPR‐1 with lipid bilayers is regulated by alternative homodimerization. Biochim. Biophys. Acta, 1818, 2175–2183. [DOI] [PubMed] [Google Scholar]
- Gibbs, G.M. , Roelants, K. and O'Bryan, M.K. (2008) The CAP superfamily: cysteine rich secretory proteins, antigen 5, and pathogenesis‐related 1 proteins – roles in reproduction, cancer, and immune defense. Endocr. Rev. 29, 865–897. [DOI] [PubMed] [Google Scholar]
- Gouget, A. , Senchou, V. , Govers, F. , Sanson, A. , Barre, A. , Rouge, P. , Pont‐Lezica, R. and Canut, H. (2006) Lectin receptor kinases participate in protein–protein interactions to mediate plasma membrane cell wall adhesions in Arabidopsis. Plant Physiol. 140, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, M.R. and Sambrook, J. (2012) Molecular Cloning: A Laboratory Manual, 4th edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Gruner, R. , Strompen, G. , Pfitzner, A.J. and Pfitzner, U.M. (2003) Salicylic acid and the hypersensitive response initiate distinct signal transduction pathways in tobacco that converge on the AS‐1‐like element of the PR‐1a promoter. Eur. J. Biochem. 270, 4876–4886. [DOI] [PubMed] [Google Scholar]
- Hanson, R.M. (2010) Jmol – a paradigm shift in crystallographic visualization. J. Appl. Crystallogr. 43, 1250–1260. [Google Scholar]
- Henriksen, A. , King, T.P. , Mirza, O. , Monsalve, R.I. , Meno, K. , Ipsen, H. , Larsen, J.N. , Gajhede, M. and Spangfort, M.D. (2001) Major venom allergen of yellow jackets, Ves v 5: structural characterization of a pathogenesis‐related protein superfamily. Proteins, 45, 438–448. [DOI] [PubMed] [Google Scholar]
- Holzberg, S. , Brosio, P. , Gross, C. and Pogue, G.P. (2002) Barley stripe mosaic virus induced gene silencing in a monocot plant. Plant J. 30, 315–327. [DOI] [PubMed] [Google Scholar]
- Hong, J.K. , Lee, S.C. and Hwang, B.K. (2005) Activation of pepper basic PR‐1 gene promoter during defense signaling to pathogen, abiotic and environmental stresses. Gene, 356, 169–180. [DOI] [PubMed] [Google Scholar]
- Kiba, A. , Nishihara, M. , Nakatsuka, T. and Yamamura, S. (2007) Pathogenesis related protein 1 homologue is an antifungal protein in Wasabia japonica leaves and confers resistance to Botrytis cinerea in transgenic tobacco. Plant Biotechnol. 24, 247–253. [Google Scholar]
- Knepper, C. , Savory, E.A. and Day, B. (2011) Arabidopsis NDR1 is an integrin‐like protein with a role in fluid loss and plasma membrane‐cell wall adhesion. Plant Physiol. 156, 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama, Y. and Hu, C.D. (2012) Bimolecular fluorescence complementation (BiFC): a 5‐year update and future perspectives. Biotechniques, 53, 285–298. [DOI] [PubMed] [Google Scholar]
- Lazniewska, J. and Macioszek, V.K. (2010) Fight to the death: Arabidopsis thaliana defense response to fungal necrotrophic pathogens. Acta Physiol. Plant. 32, 1–10. [Google Scholar]
- Leon‐Reyes, A. , Spoel, S.H. , De Lange, E.S. , Abe, H. , Kobayashi, M. , Tsuda, S. et al (2009) Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 149, 1797–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z.T. , Dhekney, S.A. and Gray, D.J. (2011) PR‐1 gene family of grapevine: a uniquely duplicated PR‐1 gene from a Vitis interspecific hybrid confers high level resistance to bacterial disease in transgenic tobacco. Plant Cell Rep. 30, 1–11. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C. and van Strien, E.A. (1999) The families of pathogenesis‐related proteins, their activities, and comparative analysis of PR‐1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M.J. (2006) Significance of inducible defense related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Lu, S. (2012) Use of the yeast two‐hybrid system to identify targets of fungal effectors. Methods Mol. Biol. 835, 165–189. [DOI] [PubMed] [Google Scholar]
- Lu, S. , Friesen, T.L. and Faris, J.D. (2009) Identification of ToxA‐interacting proteins suggests a possible role for pathogenesis‐related protein 1 (PR‐1) in mediating Stagonospora nodorum–wheat interactions. In: XIV International Congress on Molecular Plant–Microbe Interactions, Quebec City, Canada, IS‐MPMI 2009 XIV Congress 2097 .
- Lu, S. , Friesen, T.L. and Faris, J.D. (2011) Molecular characterization and genomic mapping of the pathogenesis‐related protein 1 (PR‐1) gene family in hexaploid wheat (Triticum aestivum L.). Mol. Genet. Genomics, 285, 485–503. [DOI] [PubMed] [Google Scholar]
- Lu, S. , Faris, J.D. , Sherwood, R. and Edwards, M.C. (2013) Dimerization and protease resistance: new insight into the function of PR‐1. J. Plant Physiol. 170, 105–110. [DOI] [PubMed] [Google Scholar]
- Manning, V.A. , Andrie, R.M. , Trippe, A.F. and Ciuffetti, L.M. (2004) Ptr ToxA requires multiple motifs for complete activity. Mol Plant Microbe Interact, 17, 491–501. [DOI] [PubMed] [Google Scholar]
- Manning, V.A. , Hardison, L.K. and Ciuffetti, L.M. (2007) Ptr ToxA interacts with a chloroplast‐localized protein. Mol. Plant–Microbe Interact. 20, 168–177. [DOI] [PubMed] [Google Scholar]
- Manning, V.A. , Hamilton, S.M. , Karplus, P.A. and Ciuffetti, L.M. (2008) The Arg‐Gly‐Asp‐containing, solvent‐exposed loop of Ptr ToxA is required for internalization. Mol. Plant–Microbe Interact. 21, 315–325. [DOI] [PubMed] [Google Scholar]
- Meinhardt, S.W. , Cheng, W. , Kwon, C.Y. , Donohue, C.M. and Rasmussen, J.B. (2002) Role of the arginyl‐glycyl‐aspartic motif in the action of Ptr ToxA produced by Pyrenophora tritici‐repentis . Plant Physiol. 130, 1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste, T. (2012) Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294. [DOI] [PubMed] [Google Scholar]
- Milne, T.J. , Abbenante, G. , Tyndall, J.D. , Halliday, J. and Lewis, R.J. (2003) Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis‐related protein superfamily. J. Biol. Chem. 278, 31 105–31 110. [DOI] [PubMed] [Google Scholar]
- Niderman, T. , Genetet, I. , Bruyere, T. , Gees, R. , Stintzi, A. , Legrand, M. , Fritig, B. and Mosinger, E. (1995) Pathogenesis‐related PR‐1 proteins are antifungal. Isolation and characterization of three 14‐kilodalton proteins of tomato and of a basic PR‐1 of tobacco with inhibitory activity against Phytophthora infestans . Plant Physiol. 108, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, R.P. , Friesen, T.L. , Faris, J.D. and Solomon, P.S. (2012) Stagonospora nodorum: from pathology to genomics and host resistance. Annu. Rev. Phytopathol. 50, 23–43. [DOI] [PubMed] [Google Scholar]
- Pandelova, I. , Betts, M.F. , Manning, V.A. , Wilhelm, L.J. , Mockler, T.C. and Ciuffetti, L.M. (2009) Analysis of transcriptome changes induced by Ptr ToxA in wheat provides insights into the mechanisms of plant susceptibility. Mol. Plant, 2, 1067–1083. [DOI] [PubMed] [Google Scholar]
- Pandelova, I. , Figueroa, M. , Wilhelm, L.J. , Manning, V.A. , Mankaney, A.N. , Mockler, T.C. and Ciuffetti, L.M. (2012) Host‐selective toxins of Pyrenophora tritici‐repentis induce common responses associated with host susceptibility. PLoS ONE, 7, e40240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, T.N. , Brunak, S. , von Heijne, G. and Nielsen, H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods, 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Rauscher, M. , Adam, A.L. , Wirtz, S. , Guggenheim, R. , Mendgen, K. and Deising, H.B. (1999) PR‐1 protein inhibits the differentiation of rust infection hyphae in leaves of acquired resistant broad bean. Plant J. 19, 625–633. [DOI] [PubMed] [Google Scholar]
- Santamaria, M. , Thomson, C.J. , Read, N.D. and Loake, G.J. (2001) The promoter of a basic PR1‐like gene, AtPRB1, from Arabidopsis establishes an organ‐specific expression pattern and responsiveness to ethylene and methyl jasmonate. Plant Mol. Biol. 47, 641–652. [DOI] [PubMed] [Google Scholar]
- Sarma, G.N. , Manning, V.A. , Ciuffetti, L.M. and Karplus, P.A. (2005) Structure of Ptr ToxA: an RGD‐containing host‐selective toxin from Pyrenophora tritici‐repentis . Plant Cell, 17, 3190–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuren, F.H. , Asgeirsdottir, S.A. , Kothe, E.M. , Scheer, J.M. and Wessels, J.G. (1993) The Sc7/Sc14 gene family of Schizophyllum commune codes for extracellular proteins specifically expressed during fruit‐body formation. J. Gen. Microbiol. 139, 2083–2090. [DOI] [PubMed] [Google Scholar]
- Serrano, R.L. , Kuhn, A. , Hendricks, A. , Helms, J.B. , Sinning, I. and Groves, M.R. (2004) Structural analysis of the human Golgi‐associated plant pathogenesis related protein GAPR‐1 implicates dimerization as a regulatory mechanism. J. Mol. Biol. 339, 173–183. [DOI] [PubMed] [Google Scholar]
- Shimoni, L. and Glusker, J.P. (1995) Hydrogen bonding motifs of protein side chains: descriptions of binding of arginine and amide groups. Protein Sci. 4, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyperski, T. , Fernandez, C. , Mumenthaler, C. and Wuthrich, K. (1998) Structure comparison of human glioma pathogenesis‐related protein GliPR and the plant pathogenesis‐related protein P14a indicates a functional link between the human immune system and a plant defense system. Proc. Natl. Acad. Sci. USA, 95, 2262–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, Y.‐S. , Bragg, J. and Meinhardt, S.W. (2007) Functional characterization of ToxA and molecular identification of its intracellular targeting protein in wheat. Am. J. Plant Physiol. 2, 76–89. [Google Scholar]
- Tan, K.C. , Ferguson‐Hunt, M. , Rybak, K. , Waters, O.D. , Stanley, W.A. , Bond, C.S. et al (2012) Quantitative variation in effector activity of ToxA isoforms from Stagonospora nodorum and Pyrenophora tritici‐repentis . Mol. Plant–Microbe Interact. 25, 515–522. [DOI] [PubMed] [Google Scholar]
- Tomas, A. , Feng, G.H. , Reeck, G.R. , Bockus, W.W. and Leach, J.E. (1990) Purification of a cultivar‐specific toxin from Pyrenophora tritici‐repentis, causal agent of tan spot of wheat. Mol. Plant–Microbe Interact. 3, 221–224. [Google Scholar]
- Tuori, R.P. , Wolpert, T.J. and Ciuffetti, L.M. (1995) Purification and immunological characterization of toxic components from cultures of Pyrenophora tritici‐repentis . Mol Plant–Microbe Interact. 8, 41–48. [DOI] [PubMed] [Google Scholar]
- Tuori, R.P. , Wolpert, T.J. and Ciuffetti, L.M. (2000) Heterologous expression of functional Ptr ToxA. Mol. Plant–Microbe Interact. 13, 456–464. [DOI] [PubMed] [Google Scholar]
- Vartapetian, A.B. , Tuzhikov, A.I. , Chichkova, N.V. , Taliansky, M. and Wolpert, T.J. (2011) A plant alternative to animal caspases: subtilisin‐like proteases. Cell Death Differ. 18, 1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Sullivan, R.W. , Kight, A. , Henry, R.L. , Huang, J. , Jones, A.M. and Korth, K.L. (2004) Deletion of the chloroplast‐localized thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 136, 3594–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.L. , Kuo, J.H. , Lee, S.C. , Liu, J.S. , Hsieh, Y.C. , Shih, Y.T. , Chen, C.J. , Chiu, J.J. and Wu, W.G. (2010) Cobra CRISP functions as an inflammatory modulator via a novel Zn2+‐ and heparan sulfate dependent transcriptional regulation of endothelial cell adhesion molecules. J. Biol. Chem. 285, 37 872–37 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Francischetti, I.M. , Lai, R. , Ribeiro, J.M. and Andersen, J.F. (2012) Structure of protein having inhibitory disintegrin and leukotriene scavenging functions contained in single domain. J. Biol. Chem. 287, 10 967–10 976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H.F. , Francl, L.J. , Jordahl, J.G. and Meinhardt, S.W. (1997) Structural and physical properties of a necrosis‐inducing toxin from Pyrenophora tritici‐repentis . Phytopathology, 87, 154–160. [DOI] [PubMed] [Google Scholar]
- Zhang, W.J. , Pedersen, C. , Kwaaitaal, M. , Gregersen, P.L. , Morch, S.M. , Hanisch, S. , Kristensen, A. , Fuglsang, A.T. , Collinge, D.B. and Thordal‐Christensen, H. (2012) Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Mol. Plant Pathol. 13, 1110–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Plant assays showing activity of the recombinant ToxA proteins in the ToxA‐sensitive tetraploid Langdon wheat. Second leaves of 2‐week‐old plants were infiltrated with ToxA at the estimated final concentrations (ng/μL) indicated on the right (highest concentration shown for the N102A mutant). Control plants were treated with the protein sample of the ‘mock’ strain shown in Fig. 3A (lane 3). Marks delimit the infiltrated area. Photographs were taken at 72 h post‐infiltration (one representative leaf of each treatment is shown).
Fig. S2 Characterization of the purified recombinant PR‐1‐5 proteins. (A) Size exclusion chromatography analysis of the recombinant PR‐1‐5 (left) and PR‐1‐5‐N141A (right) proteins. A single peak was detected for both proteins (samples were separated using Phenomenex Yarra SEC‐2000 columns; Torrance, CA, USA). (B) Mass spectrum showing the tryptic fragment (peak, 41.85) containing the single‐residue mutation site in the recombinant PR‐1‐5‐N141A mutant protein. The amino acid (aa) sequence of the 18‐aa fragment (corresponding positions of 138–155 in the PR‐1‐5 protein) is shown with the mutated residue highlighted in red [detailed tandem mass spectrometry (MS/MS) data are available on request].
Fig. S3 Plant assays showing the necrosis‐promoting activity of the PR‐1‐5 protein in hexaploid wheat cultivars. ToxA‐sensitive (Grandin, A) and insensitive (BR34, B) wheat plants were infiltrated with the recombinant ToxA and/or PR‐1‐5 proteins as indicated on the right. Photographs were taken at 48 h post‐infiltration (one or three representative leaves of each treatment are shown).
Fig. S4 Expression patterns of PR‐1‐1, ToxABP1 and PCN in comparison with that of PR‐1‐5 (Fig. 5) in response to ToxA treatment. mRNA samples were collected from the second leaves of ToxA‐insensitive (BR34) and ToxA‐sensitive (Grandin) wheat plants that were ‘mock'‐infiltrated (–) or infiltrated with ToxA protein (+) at a final concentration of 1.0 ng/μL. Polymerase chain reaction (PCR) products were separated on ethidium bromide‐stained 1% agarose gels. Numbers on the right indicate size (in base pairs) of the amplicons. Oligonucleotide sequences of the primers used for reverse transcriptase‐polymerase chain reaction (RT‐PCR): 5′‐CCTTTTAGACACCGAACCAGGAAGT‐3′ (forward) and 5′‐ATGGCGGCCATATCGTCGCTTCCT‐3′ (forward) and 5′‐TTAATGCTTCAAGGGATAAGCGCT‐3′ (reverse) for TOXABP1 (Manning et al., 2007), and 5′‐ATGGCCGCCCTCTCCTCTGCA‐3′ (forward) and 5′‐TTAGTTGACGGTGACCTTGCCGA‐3′ (reverse) for PCN (Tai et al., 2011).
Table S1 Polymerase chain reaction (PCR) primers used for the generation of yeast two‐hybrid constructs.
Table S2 Polymerase chain reaction (PCR) primers used for the generation of Pichia expression constructs.
Fig. S5 Amino acid sequence alignment showing the residues consisting of the turning loop between βC and βD in the PR‐1 domains of plant PR‐1 proteins. Residues shown correspond to positions 135–150 of the PR‐1‐5 protein (GenBank accession number HQ541965). Arrows indicate residues aligned to the N141 site of PR‐1‐5 and its ‘sister’ protein PR‐1‐4 (underlined), both of which have a unique ‘NNNL’ motif (highlighted in yellow). Numbers on the right indicate the positions in the full‐length proteins. Alignments were generated using the MegAlign program (DNASTAR). Species abbreviations: At, Arabidopsis thaliana; Bn, Brassica napus; Cc, Capsicum chinense; Ca, C. annuum; Ew, Eutrema wasabi; Gm, Glycine max; Hv, Hordeum vulgare; Le, Lycopersicon esculentum; Md, Malus × domestica; Nt, Nicotiana tabacum; Os, Oryza sativa; Sb, Sorghum bicolor; St, Solanum tuberosum; Ta, Triticum aestivum; Tm, T. monococcum; Zm, Zea mays. The 23 wheat (Ta) PR‐1‐like proteins (PR‐1‐1 to PR‐1‐23) have been reported previously by Lu et al. (2011).


