SUMMARY
Recent discoveries regarding small RNAs and the mechanisms of gene silencing are providing new opportunities to explore fungal pathogen–host interactions and potential strategies for novel disease control. Plant pathogenic fungi are a constant and major threat to global food security; they represent the largest group of disease‐causing agents on crop plants on the planet. An initial understanding of RNA silencing mechanisms and small RNAs was derived from model fungi. Now, new knowledge with practical implications for RNA silencing is beginning to emerge from the study of plant–fungus interactions. Recent studies have shown that the expression of silencing constructs in plants designed on fungal genes can specifically silence their targets in invading pathogenic fungi, such as Fusarium verticillioides, Blumeria graminis and Puccinia striiformis f.sp. tritici. Here, we highlight the important general aspects of RNA silencing mechanisms and emphasize recent findings from plant pathogenic fungi. Strategies to employ RNA silencing to investigate the basis of fungal pathogenesis are discussed. Finally, we address important aspects for the development of fungal‐derived resistance through the expression of silencing constructs in host plants as a powerful strategy to control fungal disease.
INTRODUCTION
More than 25 years ago, Sanford and Johnston (1985) proposed the concept of parasite‐derived resistance (PDR) as a radical strategy to control plant disease. In PDR, the expression of a parasite gene in the plant host interferes with the parasite's ability to grow and develop, resulting in disease resistance. Perhaps the most successful application of PDR is the use of transgenic papaya plants expressing Papaya ringspot virus (PSRV) coat protein to protect against virus infection (Gonsalves, 1998). With the exception of viruses, it was generally believed that the lack of ready macromolecule exchange between host and parasites would be a major impediment to more widespread development of PDR.
Over the past decade or so, the discovery of small RNAs and the elucidation of RNA silencing mechanisms have rekindled an interest in PDR. Today, it is strongly suggested, on the basis of reporter gene silencing, that RNA molecules traffic between host and parasites (Tinoco et al., 2010; Tomilov et al., 2008; Westwood et al., 2009). Growing evidence suggests that RNA silencing constitutes a crucial component of many plant–parasite interactions in addition to viruses, including responses to insects, bacteria, nematodes and fungal infections (Huang et al., 2006; Katiyar‐Agarwal and Jin, 2010; Mao et al., 2007; Ruiz‐Ferrer and Voinnet, 2009; Turner et al., 2006). Several very recent published reports on the use of host‐induced gene silencing (HIGS) to control invading fungi are probable forerunners of many more applications to come.
In this article, we describe: (i) the central components of RNA silencing, emphasizing recent discoveries in plant pathogenic fungi (PPF); (ii) the fundamental principles of the use of RNA silencing to study PPF; (iii) applications in HIGS using fungus‐derived genes to confer disease resistance; and (iv) future perspectives of HIGS for the control of fungal disease.
GENERAL MECHANISM OF EUKARYOTIC RNA SILENCING
Approximately 20 years ago, anomalies observed during efforts to functionally characterize genes led to the discovery of post‐transcriptional gene silencing (PTGS). This phenomenon is known as co‐suppression in plants, quelling in Neurospora crassa and RNA interference (RNAi) in Caenorhabditis elegans and other animals (Lee et al., 1993; Napoli et al., 1990; Romano and Macino, 1992). Subsequent further independent investigations revealed that much of the underlying machinery and mechanisms is conserved across organisms.
PTGS can be generalized as a sequence‐specific regulatory mechanism for the silencing of gene expression triggered by long double‐stranded (dsRNA) or hairpin‐structured RNAs. These molecules are processed into 21–35‐nucleotide‐long single‐stranded RNAs that associate with and direct the RNA‐induced silencing complex (RISC) to the target transcript. This results in mRNA degradation or translational repression (Fig. 1) (Esquela‐Kerscher and Slack, 2006). There are distinct classes of small RNA molecules, such as short interfering RNAs (siRNAs), microRNAs (miRNAs), tRNA‐derived RNA fragments (tRFs) and Piwi‐interacting small RNAs (piRNAs). Currently, siRNAs and miRNAs are the best elucidated classes of small RNAs in plants and animals, and are discussed in more detail. Although it is clear that siRNA and miRNA processing pathways are distinct, in plants they typically begin in the nucleus, where Dicer and members of the ribonuclease III (RNase III) family bind to long dsRNA or hairpin RNAs and process them into siRNAs or miRNAs, respectively. In animals, Drosha is responsible for the slicing off of the long stem‐loop dsRNA structure, which is then processed into siRNAs or miRNAs by Dicer (for more details, see elsewhere: Baulcombe, 2004; Carthew and Sontheimer, 2009; Miyoshi et al., 2010; Sato et al., 2011). To our knowledge, it is unclear how siRNA is exported from the nucleus. In contrast, endogenously derived short hairpin RNA molecules (miRNA) are transported to the cytoplasm by a nuclear pore protein named Exportin‐5 (Katahira and Yoneda, 2011; Kim, 2004; Lund et al., 2004; Yi et al., 2003). In other instances, short dsRNA molecules may be derived from exogenous sources, such as viruses replicating in plant cells. Once in the cytoplasm, endogenously or exogenously derived short dsRNA molecules are processed into small dsRNA, and loaded into RISC. Here the ‘passage’ strand is eliminated, and the ‘guide’ RNA strand directs Argonaute to specifically degrade the target RNA transcript through the slicing ribonuclease activity of Argonaute or to interfere with translation (Esquela‐Kerscher and Slack, 2006). These small RNAs can also be used as primers by RNA dependent‐RNA polymerase (RdRp) to resynthesize dsRNAs (Melnyk et al., 2011). This amplification step ensures that several rounds of RNA silencing are triggered. Several members of the Argonaute family have been described. For example, AGO associates with 21–25‐nucleotide‐long small RNAs (siRNAs and miRNAs), whereas PIWI binds to 23–30‐nucleotide‐long RNA (piRNAs) molecules to silence repetitive element transcripts (Brennecke et al., 2007; Hartig et al., 2007). Here, we refer to the principle RNA silencing proteins as Dicer, Argonaute and RdRp.
Figure 1.
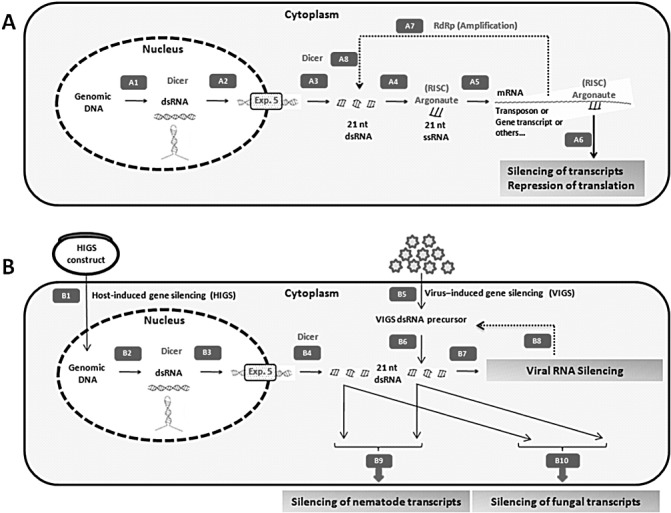
General mechanism and potential application of RNA silencing for the development of plant disease control strategies. In plant cells (A), endogenous RNA silencing is triggered by transcription of double‐stranded RNA (dsRNA) (or hairpin) molecules in the nucleus (A1). These precursor molecules are processed by ribonuclease III (RNase III) enzymes, such as Dicer (or Drosha in animals), producing RNA molecules, which are loaded and transported from the nucleus by Exportin‐5. In the cytoplasm, 21‐nucleotide dsRNAs are produced by Dicer activity (A3). On elimination of the passage strand, the guide single‐stranded RNA (ssRNA) associates with Argonaute (A4) and directs the RNA‐induced silencing complex (RISC) to bind to target endogenous transcripts (A5), resulting in the degradation of transcripts (A6). Unbound siRNA binds to mRNAs (A7) through the action of RNA‐dependent RNA polymerase (RdRp) and resynthesizes dsRNA which is cut into 21‐nt dsRNA molecules by Dicer (A8), leading to RNA silencing amplification. In pathogen‐infected plant cells (B), integration (B1) and transcription (B2) of the host‐induced gene silencing (HIGS) construct results in HIGS dsRNA molecules (or hairpins) that are processed into 21‐nt dsRNA molecules (B3 and B4; as explained in A). Alternatively, virus‐induced gene silencing (VIGS) (B5) is used to produce VIGS dsRNA precursors that are processed into 21‐nt dsRNA molecules by Dicer (B6). VIGS dsRNA molecules lead directly to the silencing of viral target transcripts in the plant cytoplasm (B7). The action of plant RdRp also leads to viral RNA silencing amplification (B8). HIGS dsRNA molecules are exported from plant cells and lead to the silencing of nematode targets on nematode feeding (B9), or the silencing of fungal transcripts on fungal infection (B10).
In fungi, a diverse number of small RNA classes have been described over the last few years. Recently, microRNA‐like RNAs (milRNAs) have been discovered in N. crassa (Lee et al., 2010). The molecular machinery involved in the biogenesis of milRNAs appears to be significantly different from that reported for miRNAs in plants and animals. Thus, it is still a matter of debate whether genuine miRNAs are present in filamentous fungi. Other small RNAs in fungi include quelling‐deficient 2 (QDE‐2)‐interacting small RNAs (qiRNAs) and Dicer‐independent small interfering RNAs (disiRNAs) in N. crassa (Lee et al., 2009, 2010). Endogenous short RNAs (esRNAs) have been discovered in Mucor circinelloides, and LTR retrotransposon‐siRNAs (LTR‐siRNAs) and tRFs in Magnaporthe oryzae and Aspergillus fumigatus (Jöchl et al., 2008; Nicolas et al., 2010; Nunes et al., 2011).
FUNDAMENTAL PRINCIPLES OF RNA SILENCING IN PPF
A knowledge of RNA silencing protein machinery in fungi was initially derived from studies of N. crassa and referred to as ‘quelling’ (Catalanotto et al., 2002, 2004; Cogoni and Macino, 1997; Fulci and Macino, 2007; Lee et al., 2009; Li et al., 2010b; Maiti et al., 2007; Nakayashiki, 2005; Nolan et al., 2008; Romano and Macino, 1992; Schumann et al., 2010; Xiao et al., 2010). Although quelling was first described in 1992, it was several years later that Cogoni and Macino discovered three distinct loci responsible for PTGS through the characterization of quelling‐deficient (qde) mutants (Cogoni and Macino, 1997). The qde‐1, qde‐2 and qde‐3 loci encode proteins that correspond to RdRp, Argonaute and RecQ DNA helicase, respectively. Small RNAs produced by Dicer associate with qde‐2 and require qde‐1 and qde‐3 for biogenesis (Catalanotto et al., 2002, 2004). Over the past few years, there have been several examples of RNA silencing in PPF, including members of the Ascomycota, Basidiomycota and Zygomycota, as well as in Phytophthora spp. (fungus‐like organisms; Oomycetes within the Kingdom Stramenopila) (Table 1).
Table 1.
Summary of RNA silencing in plant pathogenic fungi.
| Phylum | Species | Targeted gene | Promoter used | RNA precursor | Comments | Reference |
|---|---|---|---|---|---|---|
| Ascomycota | Alternaria alternata | Putative hydrolase (ACTT2), a host‐selective ACT‐toxin | trpC * | Hairpin† | Silenced transformants were nonpathogenic | Miyamoto et al. (2008) |
| Green fluorescent protein (GFP) | trpC * | Hairpin | Silencing of GFP suggests RNA silencing functionality | Miyamoto et al. (2008) | ||
| Enoyl‐reductase (ACTTS2), a host‐selective ACT‐toxin | trpC * | Hairpin | Silencing of ACCTS2 transcripts resulted in lack of ACT‐toxin production and complete loss of pathogenicity | Ajiro et al. (2010) | ||
| Aspergillus flavus and A. parasiticus | Transcription factor (aflR)‡ | gpd * | Hairpin | The atoxigenic silenced strain was stable in corn and wheat | McDonald et al. (2005) | |
| Bipolaris oryzae | Polyketide synthase gene (PKS1) | trpC * | Hairpin | The silencing construct was stable during asexual propagation | Moriwaki et al. (2007) | |
| Botrytis cinerea | Superoxide dismutase (BCSOD1) | oliC * | Sense and antisense constructs | Both constructs reduced transcript levels but had no effect on virulence | Patel et al. (2008) | |
| Cladosporium fulvum | Hydrophobin gene (HCf‐1) | n/a | Antisense RNA | Silenced transformants reverted to the wild‐type | Hamada and Spanu (1998) | |
| First exons of six hydrophobin coding genes | gpd * | Chimeric hairpin | This work highlights that changes in gene expression of nontargeted genes pose a challenge for the silencing of multiple genes | Lacroix and Spanu (2009) | ||
| Cochliobolus sativus | GFP, a host‐selective toxin (ToxA) and a polyketide synthase (CsPKS1) | Hairpin | Development of pSGate1 silencing vector | Leng et al. (2011) | ||
| CsPKS1‐silenced strains resulted in albino phenotypes | ||||||
| Colletotrichum gloeosporioides | Transcription factor (PAC1) | trpC * | Hairpin | pTroya, a Gateway RNA silencing vector, was developed from pSilent‐1 backbone for high‐throughput application in fungi | Shafran et al. (2008) | |
| Colletotrichum lagenarium | eGFP | trpC * | Hairpin | Development and application of pSilent‐1 to silence fungal transcripts | Nakayashiki et al. (2005) | |
| Fusarium graminearum | Transcription factor (Tri6) | gpd * | Hairpin | Silencing transformants were less virulent on wheat | McDonald et al. (2005) | |
| Fusarium solani f.sp. pisi | β(1,3)‐d‐glucan synthase (FsFKS1) | alcA | Hairpin | Imperfect cell wall formation led to reduced spore viability and lysis of spore and mycelia in silenced transformants | Ha et al. (2006) | |
| Fusarium solani | Chitosanase (CSN1) | gpd * | Hairpin | Silenced transformants were more virulent than wild‐type | Liu et al. (2010) | |
| Magnaporthe oryzae | eGFP | trpC * | Sense, antisense and hairpin RNA constructs | Hairpin construct more efficiently triggered eGFP silencing | Kadotani et al. (2003) | |
| MPG1 | trpC * | Hairpin | Development of pSilent‐1 vector as tool for gene silencing for Ascomycete fungi | Nakayashiki et al. (2005) | ||
| PKS‐like gene | trpC * | Hairpin | Extends application of the pSilent‐1 vector | Nakayashiki et al. (2005) | ||
| eGFP | trpC * | dsRNA | Development of a vector with convergent opposing promoters vector [pSilent‐Dual1 (pSD1)]; | Nguyen et al. (2008) | ||
| gpd * | ||||||
| pSD1 triggers RNA silencing slightly less efficiently than pSilent‐1 | ||||||
| 37 genes involved in calcium signalling | trpC * | Chimeric dsRNA | This co‐silencing approach facilitates screening of transformants based on GFP fluorescence | Nguyen et al. (2008) | ||
| gpd * | ||||||
| Work identifies many calcium signalling proteins involved in hyphal growth, sporulation and pathogenicity | ||||||
| Ophiostoma novo‐ulmi | Endopolygalacturonase (Epg1) | gdp | Hairpin | A longer stem‐loop RNA fragment (409 bp) resulted in greater reduction of mRNA transcripts compared with a 200‐bp fragment | Carneiro et al. (2010) | |
| Sclerotinia sclerotiorum | B regulatory subunit (rgb1) of 2A phosphoprotein phosphatase (PP2A) | trpC * | Hairpin | pSilent‐1 vector triggered high levels of silencing in two transformants which exhibited reduced hyphal radial growth, impaired sclerotial maturation and were unable to cause disease on tomato and Arabidopsis thaliana | Erental et al. (2007) | |
| Verticillium longisporum | Chorismate synthase (Vlaro2) | trpC * | Hairpin | pSilent‐1 vector was used and silenced 80% of Vlaro2 transcripts | Singh et al. (2010) | |
| Silenced transformants were able to infect Arabidopsis thaliana and Brassica napus | ||||||
| Venturia inaequalis | Trihydroxynaphthalene reductase (THN) and GFP | gpdA § | Chimeric hairpin | Silenced transformants had a light brown phenotype and were able to infect apple (Royal Gala) | Fitzgerald et al. (2004) | |
| Basidiomycota | ||||||
| Melampsora lini | Effector protein (AvrL567) | AvrL567 native promoter | Hairpin | Silenced rust lines were virulent on flax plants carrying L6 immune receptor | Lawrence et al. (2010) | |
| Moniliophthora perniciosa | GFP, hydrophobin (MpHYD3) and 1‐cys peroxiredoxin (MpPRX1) | n/a | In vitro synthesized dsRNA | dsRNA shown to be feasible for transforming this fungus | Caribé dos Santos et al. (2009) | |
| Ustilago hordei | GUS and mating‐type gene (bW) | Hsp70 native promoter | Hairpin | U. hordei transformants (MAT1) mated with MAT‐2 exhibited reduced production of mating hyphae | Laurie et al. (2008) | |
| Zygomycota | ||||||
| Mucor circinelloides ¶ | Carotenogenic gene (carB) | Native promoter | 3% of silenced transformants showed albino phenotype | Nicolas et al. (2003) | ||
| Fungi‐like organism | ||||||
| Phytophthora infestans | G‐protein β‐subunit encoding gene (Pigpb1) | Pigpb1 native promoter | Pigpb1 sense strand | Silenced transformants failed to sporulate | Latijnhouwers and Govers (2003) | |
| Cdc 14 coding gene (PiCdc14) | HAM34 | Sense orientation | Transformants showed reduced sporulation | Ah Fong and Judelson (2003) | ||
| G‐protein α‐subunit gene (Pigpa1) | Pigpa1 native promoter | Pigpa1 sense orientation | Transformants exhibited reduced zoospore production and infection on potato leaves | Latijnhouwers et al. (2004) | ||
| GFP inf1 and cdc14 | n/a | In vitro synthesized dsRNA | GFP, inf1 and cdc14 expression levels were reduced after exposure to dsRNA | Whisson et al. (2005) | ||
| bZIP transcription factor (Pibzp1) | HAM34 | Sense and antisense orientations | Silenced transformants showed abnormal zoospore movement, failed to develop appressoria and were incapable of infecting tomato leaflets | Blanco and Judelson (2005) | ||
| Nuclear LIM interactor‐interacting factors (NIFC1 and NIFC2) | HAM34 | Hairpin | Zoospore cyst germination was impaired by 60% in silenced NIFC transformants | Judelson and Tani (2007) | ||
| Silencing occurred at the transcription level | ||||||
| Inf1 | Sense, antisense and hairpin RNA constructs | Hairpin most efficient method of silencing | Ah‐Fong et al. (2008) | |||
| Putative glycosylated protein (Pihmp1) | n/a | In vitro synthesized dsRNA | Silenced lines showed loss of pathogenicity | Avrova et al. (2008) | ||
| Putative ATP‐dependent DEAD‐box RNA‐helicase gene (Pi‐RNH1) | n/a | In vitro synthesized dsRNA | Pi‐RNH1‐silenced lines formed large aberrant zoospores that had multiple flagella and underwent partial cleavage | Walker et al. (2008) | ||
| Four members of the CesA encoding for cellulose synthase genes | n/a | In vitro synthesized CesA1‐, CesA2‐, CesA3‐ and CesA4‐dsRNA | Silenced strains contained disrupted cell wall surrounding appressoria | Grenville‐Briggs et al. (2008) | ||
| Cellulose content of the silenced strains was >50% lower than that of nonsilenced strains | ||||||
| Effector protein (PiAVR3a) | HAM34 | Hairpin | A PiAVR3a‐silenced line (CS12) was significantly reduced in pathogenicity on Solanum tuberosum cv Bintje (susceptible) and on Nicotiana benthamiana | Bos et al. (2010) | ||
| Dicer‐like (Pidcl1), Argonaute (Piago1/2) and Histone deacetylase (Pihda1) | n/a | In vitro synthesized dsRNA | Stable Ns (nonsporulating, Picdc14‐silenced) transformant protoplasts were treated with dsRNA homologous to Pidcl1, Piago1/2 and Pihda1 | Vetukuri et al. (2011) | ||
| Regenerated lines of Pidcl1, Piago1/2 and Pihda1 showed an obvious sporulation | ||||||
| P. parasitica var. nicotianae | A coding gene considered to be involved in cellulose‐binding (CB), elicitor (E) of defence in plants and lectin (L)‐like activities (CBEL) | Bremica lactucae HSP70 | Antisense or sense constructs | Silenced transformants showed reduced attachment to cellulosic surfaces and cell wall thickening | Gaulin et al. (2002) | |
| P. sojae | Heterotrimeric G‐protein α subunit (PsGPA1) | HAM34 | Antisense construct | Silenced PsGPA1 lines had abnormal zoospore chemotaxis, encystment and germination | Hua et al. (2008) | |
| Silenced transformants were unable to infect soybean | ||||||
| C2H2 zinc finger transcription factor (PsCZF1) | HAM34 and native PsCZF1 | Antisense construct | Hyphal growth rate of silenced transformants was reduced about 50% and oospore production, zoospore and cyst germination were impaired | Wang et al. (2009) | ||
| Silenced strains lost virulence in soybean | ||||||
| MAP kinase encoding gene (PsSAK1) | HAM34 | Antisense construct | Silenced transformants showed faster encystment, reduced germination rate and longer germ tubes compared with wild‐type | Li et al. (2010a) | ||
| Transformants were unable to colonize wounded and unwounded soybean leaves | ||||||
| Putative seven‐transmembrane G‐protein‐coupled receptor (GPR11) | HAM34 | Antisense construct | Zoospore release from sporangia was drastically impaired as well as zoospore encystment and germination | Wang et al. (2010) | ||
| Silenced transformants lost pathogenicity to soybean | ||||||
| PsYKT6, a conserved member gene of the soluble N‐ethylmaleimide‐sensitive factor attachment protein receptors (SNAREs) | HAM34 | Antisense construct | Silencing of PsYKT6 revealed involvement of this gene in asexual development, sexual reproduction and pathogenesis on soybean cultivars | Zhao et al. (2011) | ||
| Antisense constructs were stable in all three transformants | ||||||
| Crinkling‐ and necrosis‐inducing proteins (CRN) (PsCRN63 and PsCRN115) | HAM34 | Sense construct | Silenced transformants were unable to suppress host cell death | Liu et al. (2011) | ||
Aspergillus nidulans promoters.
Hairpin = inverted repeats.
Delivery of synthetic siRNAs into Aspergillus flavus protoplasts resulted in significant reduction in aflD transcripts (Abdel‐Hadi et al., 2011).
Glomerella cingulata.
Nonpathogenic fungus, but it is a relevant organism within Zygomycota.
n/a, not applicable.
As shown in Table 1, a variety of target genes have been used to test whether silencing is functional in any given PPF. Transgenes, such as β‐glucuronidase (GUS) and green fluorescent protein (GFP) (reporter genes), or endogenous fungal genes with an easily screened phenotype have proven to be useful as markers for PTGS (Caribé dos Santos et al., 2009; Fitzgerald et al., 2004; Kadotani et al., 2003; Laurie et al., 2008; Miyamoto et al., 2008; Nguyen et al., 2008; Whisson et al., 2005). Further, reporters, such as a GFP, can be fused to a target gene as an endogenous positive control to provide visible evidence that silencing is functional in a particular GFP‐expressing fungal transformant. This approach is particularly useful for high‐throughput screens. Exploring this strategy, Nguyen et al. (2008) developed a pSilent‐Dual1 (pSD1) vector to study 37 genes involved in calcium signalling in M. oryzae. Although this is a powerful approach, it requires the ability to create a GFP‐tagged strain. Alternatively, endogenous genes leading to easily scorable traits, such as melanin biosynthesis genes, are ideal and have been used in Bipolaris oryzae, Cochliobolus sativus, M. oryzae and Venturia inaequalis (Fitzgerald et al., 2004; Leng et al., 2011; Moriwaki et al., 2007; Nakayashiki et al., 2005). Other suitable target genes include those coding for hydrophobins as a result of the easily wettable phenotype of silenced transformants (Nakayashiki et al., 2005). Screens for silenced hydrophobin transcripts have been used in Cladosporium fulvum, M. oryzae and Moniliophthora perniciosa (Caribé dos Santos et al., 2009; Hamada and Spanu, 1998; Lacroix and Spanu, 2009; Nakayashiki et al., 2005). Other endogenous genes for screens include enzymes whose activities can be measured directly, such as polygalacturonase in Ophiostoma novo‐ulmi (Carneiro et al., 2010).
RNA silencing has been explored in a number of PPF and Phytophthora spp. (Table 1) amenable to genetic transformation. In general, constitutive promoters are effective for the expression of silencing constructs. The Aspergillus nidulans trpC and gpd are the promoters of choice in PPF within the Ascomycota. In addition, the oliC (A. nidulans) and gpdA (Glomerella cingulata) promoters have been used effectively in Botrytis cinerea and V. inaequalis, respectively (Fitzgerald et al., 2004; Patel et al., 2008). For PPF within the Basidiomycota, the Ustilago maydis HSP70 promoter has been used successfully in Moniliophthora perniciosa (Caribé dos Santos et al., 2009). Native and species‐specific promoters have also been employed, such as AvrL567 in Melampsora lini and an Ustilago‐specific HSP70 in Ustilago hordei (Laurie et al., 2008; Lawrence et al., 2010). Alternatively, in vitro‐synthesized double‐stranded RNA (dsRNA) molecules have been used successfully for RNA silencing. In this approach, sense and antisense strands of the target gene are synthesized by in vitro transcription and then mixed to form dsRNA molecules. This eliminates many cloning steps to construct the silencing vector prior to fungal transformation (Grenville‐Briggs et al., 2008; Whisson et al., 2005). In the Basidiomycota, Moniliophthora perniciosa, hydrophobin and peroxiredoxin transcripts were successfully silenced using in vitro‐synthesized dsRNA (Caribé dos Santos et al., 2009). In Phytophthora infestans, synthesized dsRNA was used to silence exogenous and endogenous genes, such as GFP, inf1 and cdc14 (Whisson et al., 2005). As Phytophthora spp. are diploid and homologous recombination has not been demonstrated conclusively, RNA silencing is currently the only option for functional gene analysis in Oomycetes. As listed in Table 1, the HAM34 promoter has been widely used in Phytophthora spp., as well as native promoters, such as Pigpa1 and Pigpb1. Although silencing mediated by synthesized dsRNA is typically transient, this does not appear to be the case in P. infestans, where silencing may persist throughout the pathogen's entire life cycle. The use of synthesized dsRNA is likely to be valuable for high‐throughput functional characterization of genes and/or silencing of gene families, such as those for the cellulose synthase gene family (CesA) in P. infestans (Grenville‐Briggs et al., 2008). Nevertheless, the main drawback of the use of synthesized dsRNA is the constant need to generate new silenced lines for long‐term studies.
As listed in Table 1, the source of the dsRNA precursor molecule is also critical. This precursor molecule can be transcribed in trans independently in sense or antisense orientations (Kadotani et al., 2003; Patel et al., 2008). Alternatively, a short spacer may be used between cis‐located sense and antisense sequences corresponding to the target to allow the self‐formation of a hairpin RNA molecule (Kadotani et al., 2003; Miyamoto et al., 2008; Nakayashiki et al., 2005). The hairpin structure has been used extensively with PPF (Table 1). Kadotani et al. (2003) showed that a hairpin‐generating construct more effectively silenced GFP in M. oryzae than did sense and antisense constructs. The principle constraint to the use of a hairpin‐like structure is that it requires the cloning of inverted fragments, which can be time consuming. To simplify cloning, Nguyen et al. (2008) developed the pSD1 vector, which contains opposing A. nidulans trpC and gpd promoters separated by a multiple cloning site. pSD1 offers the great advantage of a single‐step, nonoriented cloning procedure.
Although gene silencing has a number of advantages compared with gene knockout, studies to date have produced variable results in terms of efficiency. For example, in Moniliophthora perniciosa, the silencing efficiency of hydrophobin and peroxiredoxin transcripts varied from 18% to 97% and 23% to 87%, respectively (Caribé dos Santos et al., 2009). Results using RNA hairpins are also highly variable. For example, a RNA hairpin precursor used to transform the Ascomycota Ophiostoma novo‐ulmi resulted in three knockdown transformants having 6%, 22% and 31% expression relative to the wild‐type (Carneiro et al., 2010). A number of factors may explain the inconsistent efficiency of silencing. For instance, the genomic context of the inserted RNA silencing vector and/or number of integration events may influence the transcriptional activity of the precursor RNA molecule. An important drawback regarding the application of gene silencing to simultaneously silence multiple genes is that it may lead to the silencing of nontarget transcripts. As an illustration, the analysis of the expression levels of C. fulvum transformants carrying various hydrophobin silencing constructs resulted in a reduction in the expression levels of several off‐target transcripts (Lacroix and Spanu, 2009).
RNA silencing has been applied successfully to discover and characterize a number of fungal pathogenicity genes. In Alternaria alternata, the silencing of two host‐selective ACT‐toxins resulted in the complete loss of pathogenicity (Ajiro et al., 2010; Miyamoto et al., 2008). In M. oryzae, high‐throughput silencing of 37 genes involved in calcium signalling revealed that many genes affected hyphal growth, sporulation and pathogenicity (Nguyen et al., 2008). The silencing of mycotoxin‐specific genes in Fusarium graminearum resulted in reduced virulence on wheat (McDonald et al., 2005). In Melampsora lini, transformants silenced for the AvrL567 avirulence effector were able to cause disease on flax plants carrying the corresponding L6 immune receptor (Lawrence et al., 2010).
APPLICATIONS IN PDR USING HIGS
Plants naturally utilize the RNA silencing machinery to defend against invading viruses (Csorba et al., 2009; Harvey et al., 2011; Hu et al., 2011). HIGS has also been shown to be effective against other plant pathogens (Huang et al., 2006). In contrast with plant viruses, most of the nematode body resides outside of the plant cell; nevertheless, RNA silencing has been shown to target nematode parasitism genes (Urwin et al., 2002). Arabidopsis thaliana expressing a dsRNA targeting the root‐knot nematode parasitism gene 16D10 led to effective disease resistance against four major Meloidogyne spp. (Huang et al., 2006). The expression of MjTis11 (Meloidogyne javanica putative transcription factor) in tobacco resulted in the silencing of the target gene in feeding root‐knot nematodes (Fairbairn et al., 2007). Alternatively, dsRNA delivered by the viral infection of host plants has the potential to induce gene silencing in nematodes (Dubreuil et al., 2009).
RNA silencing has been implicated in resistance to bacterial pathogens, including crown gall disease caused by Agrobacterium tumefaciens (Escobar and Dandekar, 2003; Escobar et al., 2001, 2002; Viss et al., 2003). To induce gall formation, iaaM (tryptophan monooxygenase) and ipt (AMP isopentenyl transferase) genes are horizontally transferred from the bacterium to the host genome. Escobar et al. (2001) showed that the silencing of iaaM and ipt oncogenes in Arabidopsis thaliana and Lycopersicon esculentum resulted in high levels of resistance to crown gall. The same approach was also effective in producing resistance to crown gall in walnut (Julglans regia L.) and apple (Escobar et al., 2002; Viss et al., 2003). Other work has suggested that a number of small RNAs participate in the bacterium–plant interaction. miRNAs (miR393, miR160, miR167 and mR825) were upregulated in plants after challenge with bacterial pathogens (Katiyar‐Agarwal and Jin, 2010). In insects, a reduction in damage caused by the coleopteran western corn rootworm was obtained by expression of dsRNA V‐ATPase A in transgenic corn plants (Baum et al., 2007). For other examples, see Mao et al., 2007; Turner et al., 2006.
Examples of the application of RNA silencing in fungus–plant interactions are now beginning to emerge. Recently, Tinoco et al. (2010) showed that tobacco transformed to express a GUS hairpin‐structured dsRNA specifically silenced GUS transcripts in a GUS‐expressing strain of Fusarium verticillioides during infection. Other recent publications have shown that the in planta expression of fungal dsRNA results in HIGS in the biotrophic pathogens Blumeria graminis and Puccinia striiformis f.sp. tritici. In the barley–B. graminis pathosystem, 76 fungal genes found to be expressed during the interaction were subjected to RNA silencing by HIGS; 21% (16 of 76) of plants transiently transformed with each of the fungal RNA silencing constructs exhibited a reduction in the number of B. graminis spores able to develop haustoria (Nowara et al., 2010). Two genes coding for 1,3‐β‐glucanosyl‐transferase (GTF1 and GTF2) were further investigated. Virus‐induced gene silencing (VIGS), using Barley stripe mosaic virus (BSMV) to produce antisense GTF1 and GTF2 transcripts, led to a significant reduction in haustorium formation and the rate of secondary hyphae elongation, respectively. The analysis of independent T1 transgenic plants carrying inverted repeats (hairpin) targeting GTF1 showed that the level of dsRNA‐derived hairpin transcript decreased rapidly in noninoculated plants. However, some transgenic plants inoculated with B. graminis f.sp. hordei exhibited reduced symptoms compared with control plants. The same study also used HIGS to silence Avra10 transcripts produced by B. graminis f.sp. hordei. In susceptible barley hosts, Avra10 acts as a virulence effector, whereas, in plants containing the R gene Mla10, it is additionally recognized as an avirulence factor. As predicted, the expression of the silencing construct targeting the Avra10 transcript in susceptible plants resulted in a reduction in pathogen development (haustorium formation) (Nowara et al., 2010). Furthermore, the abundance of the Avra10 transcript was reduced in plants co‐bombarded with the cognate resistance gene Mla10 and the Avra10 silencing construct following fungal infection. The authors concluded that these data are consistent with gene silencing occurring within young haustoria. Also using VIGS mediated by BSMV, Yin et al. (2010) demonstrated HIGS in wheat targeting Puccinia striiformis f.sp. tritici (PST) genes. These authors initially showed that a highly abundant haustorial PST transcript (PSTha12J12) was silenced by the expression of dsRNA in VIGS‐mediated BSMV–wheat transformants. Further examination of other PST genes revealed that only genes highly expressed in haustoria were effectively silenced. Three predicted genes coding for secreted proteins and a fourth gene predicted to code for a chitinase with expression patterns similar to PSTha12J12 were silenced to a high level. Similarly two genes, one with unknown function and a Uromyces fabae hexose transporter homologue having high expression levels in purified haustoria cells and relatively high levels in urediniospores, were silenced. Other genes that were expressed constitutively in a variety of fungal cell types, such as an elongation factor‐1 homologue, predicted β‐tubulin, glyceraldehyde 3‐phosphate dehydrogenase and actin coding genes, were not silenced (Yin et al., 2010). Although VIGS mediated by BSMV in wheat appears to be restricted to the silencing of only pathogen genes highly expressed in haustoria, these recent reports using different pathosystems strongly suggest that RNA molecules are able to move from plants into fungal cells and effectively silence their target genes. We expect more examples to be forthcoming in the near future.
FUTURE PERSPECTIVES OF HIGS IN FUNGUS–PLANT INTERACTIONS
The application of PDR to engineer transgenic plants has led to valuable outcomes. The development of transgenic virus‐resistant plants represents the foremost example, and the deployment of such plants has been recognized as saving an entire commercial agricultural industry (Gonsalves, 1998). More recently, the coupling of PDR to HIGS has emerged as a feasible strategy to develop plants resistant to other important pathogens, including nematodes and fungi.
Although PDR was discovered more than 25 years ago, it is only now that promising applications of fungus‐derived resistance mediated by HIGS are ready for take off. In this concept, the source of resistance is derived from the parasite's own genome. The public availability of more than 100 fungal genome sequences and many plant genome sequences, combined with transcriptome data, now offers the scientific community an arsenal of candidate genes for testing and for the design of silencing constructs to avoid off‐target transcripts. Possible targets may include virulence determinants, genes essential for fungal infection development and known fungicide targets. The availability of host genome sequence data helps to ensure that HIGS constructs do not target and negatively affect the host.
The ability to identify fungal targets, as well as the means to develop stable and environmentally safe transgenic crop plants, holds great promise for the fulfilment of future food needs. The concept and use of transgenic pathogen‐resistant crops are well established (Collinge et al., 2010), and the recent growing acceptance of genetically modified plants around the world, including the European Union and China, is likely to attract the attention of government (funds) and researchers (scientific knowledge) to further develop PDR. Although the application of the concept of fungus‐derived resistance mediated by HIGS appears to be very encouraging, a series of challenges needs to be carefully addressed before application of this strategy can be fully implemented. We conclude by highlighting a few fundamental questions.
How do RNA molecules traffic from the plant into fungal cells?
A knowledge of the mechanisms involved in the movement of protein effector molecules from pathogens into host cells is now beginning to emerge (Dean, 2011; Rooney et al., 2005; Schornack et al., 2009; Tyler, 2009; White et al., 2009). In Arabidopsis, the intercellular and systemic movement of small RNAs is mediated through the plasmodesmata and phloem, respectively (Melnyk et al., 2011). Although virus particles are also capable of moving through plasmodesmata and in the phloem, the mechanism by which naked RNA moves between two organisms is unknown. It remains to be determined how RNA is delivered from the plant nucleus, across the plasma membrane and plant cell wall into fungal cells. Is it a passive or active process? If active, what host and pathogen genes are involved and how is the process regulated?
Can HIGS be effective for other types of fungal pathogen?
HIGS has been shown to be effective in silencing the transcripts of obligate biotrophic invading fungi, at least for B. graminis and Puccinia striiformis f.sp. tritici. Perhaps the intimate interaction of this group of pathogens allows sufficient opportunity for the trafficking of RNA molecules. Considering hemibiotrophic and necrotrophic fungi, is there adequate time for RNA movement from host into fungal cells before host cells die?
What are the most promising fungal targets?
In the gene‐for‐gene concept, pathogen avirulence (Avr) genes are constantly evolving to escape recognition by the corresponding host resistance (R) gene product (Stukenbrock and McDonald, 2009). To date, the number of successful HIGS candidates leading to reduced fungal growth development remains limited and includes effectors, a cell wall elongation gene, a chitinase, a hexose transporter and a gene with unknown function. Much work remains to be done to identify suitable fungal candidate genes. Fortunately, as we have described, opportunities exist to establish high‐throughput screening pipelines to identify strong candidates.
What are the best strategies for applying HIGS to control fungal disease?
A number of possible strategies are feasible to exploit HIGS for durable resistance. The use of (i) a silencing construct that targets entire gene families, (ii) multiple lines deployed in rotation to minimize the selection pressure on pathogens, (iii) a ‘polygenic’ HIGS line conferring resistance to single or multiple fungal pathogens or (iv) a combination of both classical R genes and HIGS cassettes in the same host, which may synergistically boost resistance, are all strategies worthy of investigation.
ACKNOWLEDGEMENTS
This work is part of the doctorial dissertation of Cristiano C. Nunes and was supported by the CAPES Foundation (Brazilian Educational Agency), Fulbright Program and the Graduate School at North Carolina State University. Thanks are due to Dr Gary Payne and members of the Center for Integrated Fungal Research for valuable discussions and critical reading of the manuscript.
REFERENCES
- Abdel‐Hadi, A.M. , Caley, D.P. , Carter, D.R.F. and Magan, N. (2011) Control of aflatoxin production of Aspergillus flavus and Aspergillus parasiticus using RNA silencing technology by targeting aflD (nor‐1) gene. Toxins, 3, 647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ah Fong, A. and Judelson, H.S. (2003) Cell cycle regulator Cdc14 is expressed during sporulation but not hyphal growth in the fungus‐like oomycete Phytophthora infestans . Mol. Microbiol. 50, 487–494. [DOI] [PubMed] [Google Scholar]
- Ah‐Fong, A.M. , Bormann‐Chung, C.A. and Judelson, H.S. (2008) Optimization of transgene‐mediated silencing in Phytophthora infestans and its association with small‐interfering RNAs. Fungal Genet. Biol. 45, 1197–1205. [DOI] [PubMed] [Google Scholar]
- Ajiro, N. , Miyamoto, Y. , Masunaka, A. , Tsuge, T. , Yamamoto, M. , Ohtani, K. , Fukumoto, T. , Gomi, K. , Peever, T.L. , Izumi, Y. , Tada, Y. and Akimitsu, K. (2010) Role of the host‐selective ACT‐toxin synthesis gene ACTTS2 encoding an enoyl‐reductase in pathogenicity of the tangerine pathotype of Alternaria alternata . Phytopathology, 100, 120–126. [DOI] [PubMed] [Google Scholar]
- Avrova, A.O. , Boevink, P.C. , Young, V. , Grenville‐Briggs, L.J. , van West, P. , Birch, P.R.J. and Whisson, S.C. (2008) A novel Phytophthora infestans haustorium‐specific membrane protein is required for infection of potato. Cell. Microbiol. 10, 2271–2284. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Baum, J.A. , Bogaert, T. , Clinton, W. , Heck, G.R. , Feldmann, P. , Ilagan, O. , Johnson, S. , Plaetinck, G. , Munyikwa, T. , Pleau, M. , Vaughn, T. and Roberts, J. (2007) Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. [DOI] [PubMed] [Google Scholar]
- Blanco, F.A. and Judelson, H.S. (2005) A bZIP transcription factor from Phytophthora interacts with a protein kinase and is required for zoospore motility and plant infection. Mol. Microbiol. 56, 638–648. [DOI] [PubMed] [Google Scholar]
- Bos, J.I. , Armstrong, M.R. , Gilroy, E.M. , Boevink, P.C. , Hein, I. , Taylor, R.M. , Zhendong, T. , Engelhardt, S. , Vetukuri, R.R. , Harrower, B. , Dixelius, C. , Bryan, G. , Sadanandom, A. , Whisson, S.C. , Kamoun, S. and Birch, P. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA, 107, 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J. , Aravin, A.A. , Stark, A. , Dus, M. , Kellis, M. , Sachidanandam, R. and Hannon, G.J. (2007) Discrete small RNA‐generating loci as master regulators of transposon activity in Drosophila . Cell, 128, 1089–1103. [DOI] [PubMed] [Google Scholar]
- Caribé dos Santos, A.C. , Sena, J.A.L. , Santos, S.C. , Dias, C.V. , Pirovani, C.P. , Pungartnik, C. , Valle, R.R. , Cascardo, J.C.M. and Vincentz, M. (2009) dsRNA‐induced gene silencing in Moniliophthora perniciosa, the causal agent of witches' broom disease of cacao. Fungal Genet. Biol. 46, 825–836. [DOI] [PubMed] [Google Scholar]
- Carneiro, J.S. , Bastide, P.Y. , Chabot, M. , Lerch, L. and Hintz, W.E. (2010) Suppression of polygalacturonase gene expression in the phytopathogenic fungus Ophiostoma novo‐ulmi by RNA interference. Fungal Genet. Biol. 47, 399–405. [DOI] [PubMed] [Google Scholar]
- Carthew, R.W. and Sontheimer, E.J. (2009) Origins and mechanisms of miRNAs and siRNAs. Cell, 136, 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto, C. , Azzalin, G. , Macino, G. and Cogoni, C. (2002) Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora . Genes Dev. 16, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto, C. , Pallotta, M. , ReFalo, P. , Sachs, M.S. , Vayssie, L. , Macino, G. and Cogoni, C. (2004) Redundancy of the two dicer genes in transgene‐induced posttranscriptional gene silencing in Neurospora crassa . Mol. Cell. Biol. 24, 2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C. and Macino, G. (1997) Isolation of quelling‐defective (qde) mutants impaired in posttranscriptional transgene‐induced gene silencing in Neurospora crassa . Proc. Natl. Acad. Sci. USA, 94, 10 233–10 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge, D.B. , Jorgensen, H.J. , Lund, O.S. and Lyngkjaer, M.F. (2010) Engineering pathogen resistance in crop plants: current trends and future prospects. Annu. Rev. Phytopathol. 48, 269–291. [DOI] [PubMed] [Google Scholar]
- Csorba, T. , Pantaleo, V. and Burgyán, J. (2009) RNA silencing: an antiviral mechanism In: Advances in Virus Research, Vol. 75 (Loebenstein G. and Carr J.P., eds), pp. 35–71, 230. New York, NY: Academic Press. [DOI] [PubMed] [Google Scholar]
- Dean, P. (2011) Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 35, 1100–1125. [DOI] [PubMed] [Google Scholar]
- Dubreuil, G. , Magliano, M. , Dubrana, M.P. , Lozano, J. , Lecomte, P. , Favery, B. , Abad, P. and Rosso, M.N. (2009) Tobacco rattle virus mediates gene silencing in a plant parasitic root‐knot nematode. J. Exp. Bot. 60, 4041–4050. [DOI] [PubMed] [Google Scholar]
- Erental, A. , Harel, A. and Yarden, O. (2007) Type 2A phosphoprotein phosphatase is required for asexual development and pathogenesis of Sclerotinia sclerotiorum . Mol. Plant–Microbe Interact. 20, 944–954. [DOI] [PubMed] [Google Scholar]
- Escobar, M.A. and Dandekar, A.M. (2003) Agrobacterium tumefaciens as an agent of disease. Trends Plant Sci. 8, 380–386. [DOI] [PubMed] [Google Scholar]
- Escobar, M.A. , Civerolo, E.L. , Summerfelt, K.R. and Dandekar, A.M. (2001) RNAi‐mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc. Natl. Acad. Sci. USA, 98, 13 437–13 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar, M.A. , Leslie, C.A. , McGranahan, G.H. and Dandekar, A.M. (2002) Silencing crown gall disease in walnut (Juglans regia L.). Plant Sci. 163, 591–597. [Google Scholar]
- Esquela‐Kerscher, A. and Slack, F.J. (2006) Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer, 6, 259–269. [DOI] [PubMed] [Google Scholar]
- Fairbairn, D.J. , Cavallaro, A.S. , Bernard, M. , Mahalinga‐Iyer, J. , Graham, M.W. and Botella, J.R. (2007) Host‐delivered RNAi: an effective strategy to silence genes in plant parasitic nematodes. Planta, 226, 1525–1533. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, A. , Van Kan, J.A. and Plummer, K.M. (2004) Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet. Biol. 41, 963–971. [DOI] [PubMed] [Google Scholar]
- Fulci, V. and Macino, G. (2007) Quelling: post‐transcriptional gene silencing guided by small RNAs in Neurospora crassa . Curr. Opin. Microbiol. 10, 199–203. [DOI] [PubMed] [Google Scholar]
- Gaulin, E. , Jauneau, A. , Villalba, F. , Rickauer, M. , Esquerre‐Tugaye, M.T. and Bottin, A. (2002) The CBEL glycoprotein of Phytophthora parasitica var‐nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J. Cell Sci. 115, 4565–4575. [DOI] [PubMed] [Google Scholar]
- Gonsalves, D. (1998) Control of papaya ringspot virus in papaya: a case study. Annu. Rev. Phytopathol. 36, 415–437. [DOI] [PubMed] [Google Scholar]
- Grenville‐Briggs, L.J. , Anderson, V.L. , Fugelstad, J. , Avrova, A.O. , Bouzenzana, J. , Williams, A. , Wawra, S. , Whisson, S.C. , Birch, P.R. , Bulone, V. and van West, P. (2008) Cellulose synthesis in Phytophthora infestans is required for normal appressorium formation and successful infection of potato. Plant Cell, 20, 720–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, Y. , Covert, S.F. and Momany, M. (2006) FsFKS1, the 1,3‐B‐glucan synthase from the caspofungin‐resistant fungus Fusarium solani . Eukaryot. Cell, 5, 1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, W. and Spanu, P.D. (1998) Co‐suppression of the hydrophobin gene HCf‐1 is correlated with antisense RNA biosynthesis in Cladosporium fulvum . Mol. Gen. Genet. 259, 630–638. [DOI] [PubMed] [Google Scholar]
- Hartig, J.V. , Tomari, Y. and Forstemann, K. (2007) piRNAs—the ancient hunters of genome invaders. Genes Dev. 21, 1707–1713. [DOI] [PubMed] [Google Scholar]
- Harvey, J.J. , Lewsey, M.G. , Patel, K. , Westwood, J. , Heimstadt, S. , Carr, J.P. and Baulcombe, D.C. (2011) An antiviral defense role of AGO2 in plants. PLoS ONE, 6, e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Q. , Niu, Y. , Zhang, K. , Liu, Y. and Zhou, X. (2011) Virus‐derived transgenes expressing hairpin RNA give immunity to Tobacco mosaic virus and Cucumber mosaic virus . Virol. J. 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, C. , Wang, Y. , Zheng, X. , Dou, D. , Zhang, Z. , Govers, F. and Wang, Y. (2008) A Phytophthora sojae G‐protein alpha subunit is involved in chemotaxis to soybean isoflavones. Eukaryot. Cell, 7, 2133–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, G. , Allen, R. , Davis, E.L. , Baum, T.J. and Hussey, R.S. (2006) Engineering broad root‐knot resistance in transgenic plants by RNAi silencing of a conserved and essential root‐knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA, 103, 14 302–14 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jöchl, C. , Rederstorff, M. , Hertel, J. , Stadler, P.F. , Hofacker, I.L. , Schrettl, M. , Haas, H. and Huttenhofer, A. (2008) Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 36, 2677–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judelson, H.S. and Tani, S. (2007) Transgene‐induced silencing of the zoosporogenesis‐specific NIFC gene cluster of Phytophthora infestans involves chromatin alterations. Eukaryot. Cell, 6, 1200–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadotani, N. , Nakayashiki, H. , Tosa, Y. and Mayama, S. (2003) RNA silencing in the phytopathogenic fungus Magnaporthe oryzae . Mol. Plant–Microbe Interact. 16, 769–776. [DOI] [PubMed] [Google Scholar]
- Katahira, J. and Yoneda, Y. (2011) Nucleocytoplasmic transport of microRNAs and related small RNAs. Traffic, 12, 1468–1474. [DOI] [PubMed] [Google Scholar]
- Katiyar‐Agarwal, S. and Jin, H. (2010) Role of small RNAs in host–microbe interactions. Annu. Rev. Phytopathol. 48, 225–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, V.N. (2004) MicroRNA precursors in motion: exportin‐5 mediates their nuclear export. Trends Cell Biol. 14, 156–159. [DOI] [PubMed] [Google Scholar]
- Lacroix, H. and Spanu, P.D. (2009) Silencing of six hydrophobins in Cladosporium fulvum: complexities of simultaneously targeting multiple genes. Appl. Environ. Microbiol. 75, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers, M. and Govers, F. (2003) A Phytophthora infestans G‐protein beta subunit is involved in sporangium formation. Eukaryot. Cell, 2, 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers, M. , Ligterink, W. , Vleeshouwers, V.G.A. , Van West, P. and Govers, F. (2004) A G‐alpha subunit controls zoospore motility and virulence in the potato late blight pathogen Phytophthora infestans . Mol. Microbiol. 51, 925–936. [DOI] [PubMed] [Google Scholar]
- Laurie, J.D. , Linning, R. and Bakkeren, G. (2008) Hallmarks of RNA silencing are found in the smut fungus Ustilago hordei but not in its close relative Ustilago maydis . Curr. Genet. 53, 49–58. [DOI] [PubMed] [Google Scholar]
- Lawrence, G.J. , Dodds, P.N. and Ellis, J.G. (2010) Transformation of the flax rust fungus, Melampsora lini: selection via silencing of an avirulence gene. Plant J. 61, 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.C. , Chang, S.S. , Choudhary, S. , Aalto, A.P. , Maiti, M. , Bamford, D.H. and Liu, Y. (2009) qiRNA is a new type of small interfering RNA induced by DNA damage. Nature, 459, 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.C. , Li, L. , Gu, W. , Xue, Z. , Crosthwaite, S.K. , Pertsemlidis, A. , Lewis, Z.A. , Freitag, M. , Selker, E.U. , Mello, C.C. and Liu, Y. (2010) Diverse pathways generate microRNA‐like RNAs and dicer‐independent small interfering RNAs in fungi. Mol. Cells, 38, 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R.C. , Feinbaum, R.L. and Ambros, V. (1993) The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14 . Cell, 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Leng, Y. , Wu, C. , Liu, Z. , Friesen, T.L. , Rasmussen, J.B. and Zhong, S. (2011) RNA‐mediated gene silencing in the cereal fungal pathogen Cochliobolus sativus . Mol. Plant Pathol. 12, 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, A. , Wang, Y. , Tao, K. , Dong, S. , Huang, Q. , Dai, T. , Zheng, X. and Wang, Y. (2010a) PsSAK1, a stress‐activated MAP kinase of Phytophthora sojae, is required for zoospore viability and infection of soybean. Mol. Plant–Microbe Interact. 23, 1022–1031. [DOI] [PubMed] [Google Scholar]
- Li, L. , Chang, S.S. and Liu, Y. (2010b) RNA interference pathways in filamentous fungi. Cell. Mol. Life Sci. 67, 3849–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Zhang, B. , Li, C. and Bao, X. (2010) Knock down of chitosanase expression in phytopathogenic fungus Fusarium solani and its effect on pathogenicity. Curr. Genet. 56, 275–281. [DOI] [PubMed] [Google Scholar]
- Liu, T. , Ye, W. , Ru, Y. , Yang, X. , Gu, B. , Tao, K. , Lu, S. , Dong, S. , Zheng, X. , Shan, W. , Wang, Y. and Dou, D. (2011) Two host cytoplasmic effectors are required for pathogenesis of Phytophthora sojae by suppression of host defenses. Plant Physiol. 155, 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, E. , Guttinger, S. , Calado, A. , Dahlberg, J.E. and Kutay, U. (2004) Nuclear export of microRNA precursors. Science, 303, 95–98. [DOI] [PubMed] [Google Scholar]
- Maiti, M. , Lee, H. and Liu, Y. (2007) QIP, a putative exonuclease, interacts with the Neurospora argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 21, 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y.B. , Cai, W.J. , Wang, J.W. , Hong, G.J. , Tao, X.Y. , Wang, L.J. , Huang, Y.P. and Chen, X.Y. (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant‐mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. [DOI] [PubMed] [Google Scholar]
- McDonald, T. , Brown, D. , Keller, N.P. and Hammond, T.M. (2005) RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol. Plant–Microbe Interact. 18, 539–545. [DOI] [PubMed] [Google Scholar]
- Melnyk, C.W. , Molnar, A. and Baulcombe, D.C. (2011) Intercellular and systemic movement of RNA silencing signals. EMBO J. 30, 3553–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, Y. , Masunaka, A. , Tsuge, T. , Yamamoto, M. , Ohtani, K. , Fukumoto, T. , Gomi, K. , Peever, T.L. and Akimitsu, K. (2008) Functional analysis of a multicopy host‐selective ACT‐toxin biosynthesis gene in the tangerine pathotype of Alternaria alternata using RNA silencing. Mol. Plant–Microbe Interact. 21, 1591–1599. [DOI] [PubMed] [Google Scholar]
- Miyoshi, K. , Miyoshi, T. and Siomi, H. (2010) Many ways to generate microRNA‐like small RNAs: non‐canonical pathways for microRNA production. Mol. Genet. Genomics, 284, 95–103. [DOI] [PubMed] [Google Scholar]
- Moriwaki, A. , Ueno, M. , Arase, S. and Kihara, J. (2007) RNA‐mediated gene silencing in the phytopathogenic fungus Bipolaris oryzae . FEMS Microbiol. Lett. 269, 85–89. [DOI] [PubMed] [Google Scholar]
- Nakayashiki, H. (2005) RNA silencing in fungi: mechanisms and applications. FEBS Lett. 579, 5950–5957. [DOI] [PubMed] [Google Scholar]
- Nakayashiki, H. , Hanada, S. , Quoc, N.B. , Kadotani, N. , Tosa, Y. and Mayama, S. (2005) RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol. 42, 275–283. [DOI] [PubMed] [Google Scholar]
- Napoli, C. , Lemieux, C. and Jorgensen, R. (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co‐suppression of homologous genes in trans. Plant Cell, 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, Q.B. , Kadotani, N. , Kasahara, S. , Tosa, Y. , Mayama, S. and Nakayashiki, H. (2008) Systematic functional analysis of calcium‐signalling proteins in the genome of the rice‐blast fungus, Magnaporthe oryzae, using a high‐throughput RNA‐silencing system. Mol. Microbiol. 68, 1348–1365. [DOI] [PubMed] [Google Scholar]
- Nicolas, F.E. , Torres‐Martinez, S. and Ruiz‐Vazquez, R.M. (2003) Two classes of small antisense RNAs in fungal RNA silencing triggered by non‐integrative transgenes. EMBO J. 22, 3983–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, F.E. , Moxon, S. , de Haro, J.P. , Calo, S. , Grigoriev, I.V. , Torres‐Martinez, S. , Moulton, V. , Ruiz‐Vazquez, R.M. and Dalmay, T. (2010) Endogenous short RNAs generated by dicer 2 and RNA‐dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides . Nucleic Acids Res. 38, 5535–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan, T. , Cecere, G. , Mancone, C. , Alonzi, T. , Tripodi, M. , Catalanotto, C. and Cogoni, C. (2008) The RNA‐dependent RNA polymerase essential for post‐transcriptional gene silencing in Neurospora crassa interacts with replication protein A. Nucleic Acids Res. 36, 532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowara, D. , Gay, A. , Lacomme, C. , Shaw, J. , Ridout, C. , Douchkov, D. , Hensel, G. , Kumlehn, J. and Schweizer, P. (2010) HIGS: host‐induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis . Plant Cell, 22, 3130–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes, C.C. , Gowda, M. , Sailsbery, J. , Xue, M. , Chen, F. , Brown, D. , Oh, Y. , Mitchell, T.M. and Dean, R.A. (2011) Diverse and tissue‐enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae . BMC Genomics, 12, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, R.M. , van Kan, J.A. , Bailey, A.M. and Foster, G.D. (2008) RNA‐mediated gene silencing of superoxide dismutase (bcsod1) in Botrytis cinerea . Phytopathology, 98, 1334–1339. [DOI] [PubMed] [Google Scholar]
- Romano, N. and Macino, G. (1992) Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6, 3343–3353. [DOI] [PubMed] [Google Scholar]
- Rooney, H.C. , Van't Klooster, J.W. , van der Hoorn, R.A. , Joosten, M.H. , Jones, J.D. and de Wit, P.J. (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for cf‐2‐dependent disease resistance. Science, 308, 1783–1786. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Ferrer, V. and Voinnet, O. (2009) Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60, 485–510. [DOI] [PubMed] [Google Scholar]
- Sanford, J.C. and Johnston, S.A. (1985) The concept of parasite‐derived resistance—deriving resistance genes from the parasite's own genome. J. Theor. Biol. 113, 395–405. [Google Scholar]
- Sato, F. , Tsuchiya, S. , Meltzer, S.J. and Shimizu, K. (2011) MicroRNAs and epigenetics. FEBS J. 278, 1598–1609. [DOI] [PubMed] [Google Scholar]
- Schornack, S. , Huitema, E. , Cano, L.M. , Bozkurt, T.O. , Oliva, R. , Van Damme, M. , Schwizer, S. , Raffaele, S. , Chaparro‐Garcia, A. , Farrer, R. , Segretin, M.E. , Bos, J. , Haas, B.J. , Zody, M.C. , Nusbaum, C. , Win, J. , Thines, M. and Kamoun, S. (2009) Ten things to know about oomycete effectors. Mol. Plant. Pathol. 10, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann, U. , Ayliffe, M. , Kazan, K. and Wang, M. (2010) RNA silencing in fungi. Front. Biol. 5, 478–494. [Google Scholar]
- Shafran, H. , Miyara, I. , Eshed, R. , Prusky, D. and Sherman, A. (2008) Development of new tools for studying gene function in fungi based on the gateway system. Fungal Genet. Biol. 45, 1147–1154. [DOI] [PubMed] [Google Scholar]
- Singh, S. , Braus‐Stromeyer, S.A. , Timpner, C. , Tran, V.T. , Lohaus, G. , Reusche, M. , Knufer, J. , Teichmann, T. , von Tiedemann, A. and Braus, G.H. (2010) Silencing of Vlaro2 for chorismate synthase revealed that the phytopathogen Verticillium longisporum induces the cross‐pathway control in the xylem. Appl. Microbiol. Biotechnol. 85, 1961–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock, E.H. and McDonald, B.A. (2009) Population genetics of fungal and oomycete effectors involved in gene‐for‐gene interactions. Mol. Plant–Microbe Interact. 22, 371–380. [DOI] [PubMed] [Google Scholar]
- Tinoco, M.L. , Dias, B.B. , Dall'Astta, R.C. , Pamphile, J.A. and Aragao, F.J. (2010) In vivo trans‐specific gene silencing in fungal cells by in planta expression of a double‐stranded RNA. BMC Biol. 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomilov, A.A. , Tomilova, N.B. , Wroblewski, T. , Michelmore, R. and Yoder, J.I. (2008) Trans‐specific gene silencing between host and parasitic plants. Plant J. 56, 389–397. [DOI] [PubMed] [Google Scholar]
- Turner, C.T. , Davy, M.W. , MacDiarmid, R.M. , Plummer, K.M. , Birch, N.P. and Newcomb, R.D. (2006) RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double‐stranded RNA feeding. Insect Mol. Biol. 15, 383–391. [DOI] [PubMed] [Google Scholar]
- Tyler, B.M. (2009) Entering and breaking: virulence effector proteins of oomycete plant pathogens. Cell. Microbiol. 11, 13–20. [DOI] [PubMed] [Google Scholar]
- Urwin, P.E. , Lilley, C.J. and Atkinson, H.J. (2002) Ingestion of double‐stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant–Microbe Interact. 15, 747–752. [DOI] [PubMed] [Google Scholar]
- Vetukuri, R.R. , Avrova, A.O. , Grenville‐Briggs, L.J. , van West, P. , Soderbom, F. , Savenkov, E.I. , Whisson, S.C. and Dixelius, C. (2011) Evidence for involvement of Dicer‐like, Argonaute and histone deacetylase proteins in gene silencing in Phytophthora infestans . Mol. Plant Pathol. 12, 772–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viss, W.J. , Pitrak, J. , Humann, J. , Cook, M. , Driver, J. and Ream, W. (2003) Crown‐gall‐resistant transgenic apple trees that silence Agrobacterium tumefaciens oncogenes. Mol. Breed. 12, 283–295. [Google Scholar]
- Walker, C.A. , Koppe, M. , Grenville‐Briggs, L.J. , Avrova, A.O. , Horner, N.R. , McKinnon, A.D. , Whisson, S.C. , Birch, P.R. and van West, P. (2008) A putative DEAD‐box RNA‐helicase is required for normal zoospore development in the late blight pathogen Phytophthora infestans . Fungal Genet. Biol. 45, 954–962. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Dou, D. , Wang, X. , Li, A. , Sheng, Y. , Hua, C. , Cheng, B. , Chen, X. , Zheng, X. and Wang, Y. (2009) The PsCZF1 gene encoding a C2H2 zinc finger protein is required for growth, development and pathogenesis in Phytophthora sojae . Microb. Pathog. 47, 78–86. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Li, A. , Wang, X. , Zhang, X. , Zhao, W. , Dou, D. , Zheng, X. and Wang, Y. (2010) GPR11, a putative seven‐transmembrane G protein‐coupled receptor, controls zoospore development and virulence of Phytophthora sojae . Eukaryot. Cell, 9, 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood, J.H. , Roney, J.K. , Khatibi, P.A. and Stromberg, V.K. (2009) RNA translocation between parasitic plants and their hosts. Pest Manag. Sci. 65, 533–539. [DOI] [PubMed] [Google Scholar]
- Whisson, S.C. , Avrova, A.O. , Van West, P. and Jones, J.T. (2005) A method for double‐stranded RNA‐mediated transient gene silencing in Phytophthora infestans . Mol. Plant Pathol. 6, 153–163. [DOI] [PubMed] [Google Scholar]
- White, F.F. , Potnis, N. , Jones, J.B. and Koebnik, R. (2009) The type III effectors of Xanthomonas . Mol. Plant Pathol. 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, H. , Alexander, W.G. , Hammond, T.M. , Boone, E.C. , Perdue, T.D. , Pukkila, P.J. and Shiu, P.K. (2010) QIP, a protein that converts duplex siRNA into single strands, is required for meiotic silencing by unpaired DNA. Genetics, 186, 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, R. , Qin, Y. , Macara, I.G. and Cullen, B.R. (2003) Exportin‐5 mediates the nuclear export of pre‐microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, C. , Jurgenson, J. and Hulbert, S. (2010) Development of a host‐induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici . Mol. Plant–Microbe Interact. 24, 554–561. [DOI] [PubMed] [Google Scholar]
- Zhao, W. , Dong, S. , Ye, W. , Hua, C. , Meijer, H.J.G. , Dou, X. , Govers, F. and Wang, Y. (2011) Genome‐wide identification of Phytophthora sojae SNARE genes and functional characterization of the conserved SNARE PsYKT6. Fungal Genet. Biol. 48, 241–251. [DOI] [PubMed] [Google Scholar]


