Summary
Mitogen‐activated protein kinase (MAPK) cascades are involved in plant development, stress responses and hormonal signal transduction. MAPK kinases (MAPKKs), as the key nodes in these cascades, link MAPKs and MAPKK kinases (MAPKKKs). In this study, GhMKK4, a novel group C MAPKK gene from cotton (Gossypium hirsutum), was isolated and identified. Its expression can be induced by various stresses and signalling molecules. The overexpression of GhMKK4 in Nicotiana benthamiana enhanced its susceptibility to bacterial and fungal pathogens, but had no significant effects on salt or drought tolerance. Notably, the overexpressing plants showed increased sensitivity to abscisic acid (ABA) and gibberellin A3 (GA3), and ABA and gibberellin (GA) signalling were affected on infection with Ralstonia solanacearum bacteria. Furthermore, the overexpressing plants showed more reactive oxygen species (ROS) accumulation and stronger inhibition of catalase (CAT), a ROS‐scavenging enzyme, than control plants after salicylic acid (SA) treatment. Interestingly, two genes encoding ornithine decarboxylase (ODC) and S‐adenosylmethionine decarboxylase (SAMDC), the key enzymes in polyamine synthesis, exhibited reduced R. solanacearum‐induced expression in overexpressing plants. These findings broaden our knowledge about the functions of MAPKKs in diverse signalling pathways and the negative regulation of disease resistance in the cotton crop.
Introduction
The adaptive responses of plants to environmental changes are primarily conducted through the perception of external signals. These perceived signals are amplified through multiple signal transduction pathways (Shinozaki et al., 2003). Mitogen‐activated protein kinase (MAPK) cascades have emerged as common signal transduction pathways for the translation of external stimuli into cellular responses (MAPK Group, 2002).
A typical MAPK cascade consists of a MAPK kinase kinase (MAPKKK), a MAPK kinase (MAPKK) and a MAPK. Activation of MAPKKKs by upstream signals results in the sequential phosphorylation of their downstream MAPKKs and MAPKs (Kong et al., 2012; Rodriguez et al., 2010). MAPK cascades are highly conserved and have been regarded as the integrative points of multiple pathways (Jonak et al., 2002). In Arabidopsis, there are approximately 60 MAPKKKs, 10 MAPKKs and 20 MAPKs (Rodriguez et al., 2010). There are fewer MAPKKs than MAPKKKs and MAPKs, which suggests that individual MAPKKs may function as bifurcation points and are likely to be involved in multiple MAPK cascades to perform diverse biological functions (Andreasson and Ellis, 2010; Heinrich et al., 2011; Xu et al., 2008). Therefore, the functions of plant MAPKKs have been a major focus of ongoing research. At present, several MAPKKs have been identified in different plants, including Arabidopsis AtMKK1‐10; alfalfa MsSIMKK and MsPRKK; tomato LeMEK1; tobacco NtMEK1‐2 and NtSIPKK; rice OsMKK1, 3, 4, 5, 6 and 10; maize ZmMEK1 and ZmMAPKK1; and cotton GhMKK5 (Cardinale et al., 2002; Doczi et al., 2012; Kong et al., 2011; Zhang et al., 2012). MAPKKs are divided into four groups (A, B, C and D) based on the phylogenetic analyses of their amino acid sequences and phosphorylation motifs (Hamel et al., 2006; MAPK Group, 2002). Arabidopsis AtMKK4 and AtMKK5 are two widely investigated group C MAPKKs that can activate the MAPKs AtMPK3 and AtMPK6, which participate in development, abiotic stress, flagellin perception and innate immunity, as well as in the regulation of the biosynthesis of camalexin (Asai et al., 2002; Ren et al., 2008). These reports suggest that group C MAPKKs have pivotal roles in plants. Although several group C MAPKKs in other plant species have been isolated recently (Kishi‐Kaboshi et al., 2010; Kong et al., 2011; Zhang et al., 2012), the functions of group C MAPKKs have not been investigated fully.
Emerging evidence has revealed the importance of MAPKKs in plant abiotic and biotic stress responses and development. In Arabidopsis, constitutively activated AtMKK9 can increase the sensitivity of transgenic plants to salt tolerance (Xu et al., 2008). However, AtMKK4 mediates osmotic stress tolerance (Kim et al., 2011). The overexpression of AtMKK3 enhances the resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 (Doczi et al., 2007). Similarly, the ectopic expression of AtMKK7 induces systemic resistance to Pseudomonas syringae pv. maculicola (Psm) ES4326 (Zhang et al., 2007). However, some MAPKKs have negative effects on pathogen resistance. Arabidopsis AtMKK1/MKK2 can activate AtMPK4 to negatively regulate immunity (Kong et al., 2012). AtMKK2‐overexpressing transgenic plants exhibit a markedly enhanced susceptibility to the fungal necrotroph Alternaria brassicicola, even though these plants show some resistance to Pst DC3000 and Erwinia carotovora ssp. carotovora (Brader et al., 2007). Similarly, our previous study has indicated that the overexpression of GhMKK5 in N. benthamiana enhances the resistance of the plants to the bacterial pathogen Ralstonia solanacearum, but increases their susceptibility to the oomycete pathogen Phytophthora parasitica var. nicotianae Tucker (Zhang et al., 2012). During development, Arabidopsis AtMKK7 negatively regulates polar auxin transport (Dai et al., 2006), and the YODA–MKK4/MKK5–MPK3/MPK6 cascade is involved in stomatal development (Wang et al., 2007). In tobacco, NtMEK2 can regulate osmotic stress signals and pollen germination (Mikolajczyk et al., 2000). Although these results have broadened our knowledge about plant MAPKKs, the functions of MAPKKs are diverse, and further study of their roles is required.
As the nodal point in signal transduction, MAPKKs are involved in the modulation of the levels of many plant hormones. In apple, the MAPK signalling cascade MdMKK1–MdMPK1 functions in abscisic acid (ABA) signalling by regulating ABI5 or ABI5‐like transcription factors (Wang et al., 2010). The Arabidopsis AtMKK9–MPK3/MPK6 cascade can promote ethylene (ET)‐insensitive 3 (EIN3)‐mediated transcription in ET signalling, and the mkk9 mutant exhibits a broad spectrum of moderate ET‐insensitive phenotypes (Yoo et al., 2008). Furthermore, AtMPK3/MPK6 can also be activated by AtMKK5, resulting in an increase in ET level (Liu et al., 2008). The constitutively active form of AtMKK3 has been reported to participate in jasmonic acid (JA) signalling by regulating the expression of JA‐related genes (Takahashi et al., 2007a). On infection, AtMKK2‐overexpressing plants show a weaker increase in the production of JA and salicylic acid (SA), suggesting that AtMKK2 functions in the modulation of hormone levels in response to pathogen infection (Brader et al., 2007). Previous reports have focused mainly on the identification of the stressors and signals to which the MAPK cascades respond, especially in model plants. Data describing the functions of MAPKKs in the regulation of hormone levels and the cross‐talk among diverse signals during stress responses are limited.
At present, the investigation of plant MAPKKs has focused mainly on model plants and food crops. Cotton (Gossypium hirsutum) is one of the oldest and most important fibre and oil crops. Research on the cotton MAPKKs has far‐reaching significance in improving the growth and yield under various stress conditions. In this study, a novel group C MAPKK gene from cotton, GhMKK4, was isolated and identified. The expression of GhMKK4 could be induced by biotic and abiotic stresses and various signalling molecules. The ectopic expression of GhMKK4 in N. benthamiana enhanced its susceptibility to bacterial and fungal pathogens, but did not affect significantly its salt or drought tolerance. The GhMKK4‐overexpressing plants showed elevated ABA levels, but not GA levels, under normal conditions, and increased reactive oxygen species (ROS) accumulation after SA treatment. These results suggest that GhMKK4 plays a pivotal role in pathogen responses and multiple signal transduction pathways.
Results
Sequence analysis of GhMKK4
The full‐length cDNA of GhMKK4 (FJ966886) consisted of 1338 nucleotides, containing a 1056‐bp open reading frame (ORF), a 26‐bp 5′‐untranslated region (5′‐UTR) and a 256‐bp 3′‐UTR. GhMKK4 was predicted to encode a protein of 351 amino acid residues with a putative molecular weight of 39.07 kDa and an isoelectric point of 9.55. The GhMKK4 protein exhibited the same family signature as other plant MAPKKs, including 11 conserved subdomains, a conserved S/TXXXXXS/T motif, an activation loop and a docking site, as demonstrated by multiple sequence alignments. Furthermore, GhMKK4 showed 66.93% homology to AtMKK4 and 65.58% to AtMKK5 from Arabidopsis thaliana, 64.38% to NtMEK2 from Nicotiana tabacum, 64.19% to LeMKK2 from Lycopersicon esculentum and 77.11% to GhMKK5 from Gossypium hirsutum (Fig. 1A). As shown in Fig. 1B, GhMKK4 was highly similar to group C MAPKKs, such as AtMKK4, AtMKK5, NtMEK2 and GhMKK5. These results suggest that GhMKK4 is a group C MAPKK.
Figure 1.
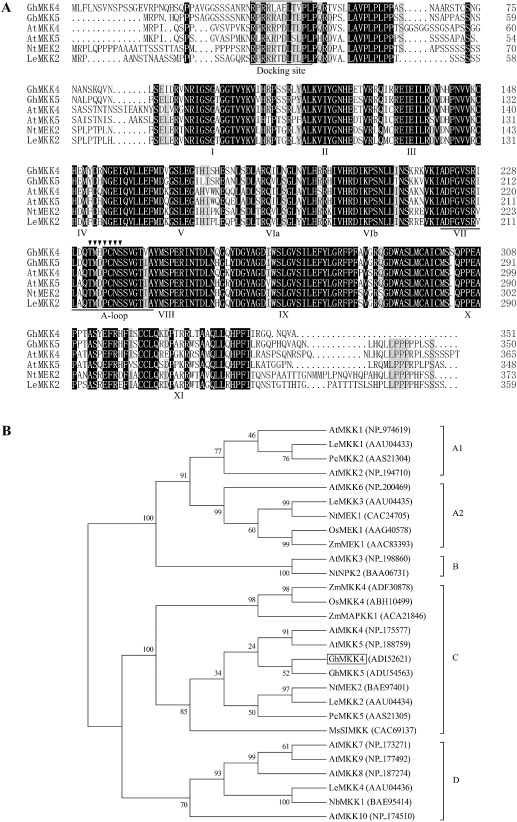
Comparison of the deduced amino acid sequences of GhMKK4 and closely related plant mitogen‐activated protein kinase (MAPK) kinases (MAPKKs). (A) The amino acid sequence alignment of GhMKK4 (ADI52621), GhMKK5 (ADU54563), AtMKK4 (NP_175577), AtMKK5 (NP_188759), NtMEK2 (BAE97401) and LeMKK2 (AAU04434). Identical amino acids are shaded in black. The protein kinase subdomains are shown with Roman numerals (I–XI) at the bottom of the sequences, and the activation loop (A‐loop) is underlined. The serine (Ser) and/or threonine (Thr) residues in the conserved S/TXXXXXS/T consensus motif between MAPKK subdomains VII and VIII are marked with arrowheads (▼). The docking site is boxed. (B) The phylogenetic relationships between GhMKK4 and other plant MAPKK proteins. The neighbour‐joining phylogenetic tree was created with ClustalW in MEGA 4.1. The numbers above and below the branches indicate the bootstrap values (>50%) from 500 replicates. The gene name is followed by the protein ID. The species of origin of the MAPKKs is indicated by the abbreviation before the gene names: At, Arabidopsis thaliana; Gh, Gossypium hirsutum; Le, Lycopersicon esculentum; Ms, Medicago sativa; Nb, Nicotiana benthamiana; Nt, Nicotiana tabacum; Os, Oryza sativa; Pc, Petroselinum crispum; Zm, Zea mays.
The genome sequence of GhMKK4 was also analysed. Sequence comparisons revealed that GhMKK4 had no intron structure, similar to other group C and D MAPKKs (Fig. 2). This result further indicates that GhMKK4 is a member of group C MAPKKs.
Figure 2.
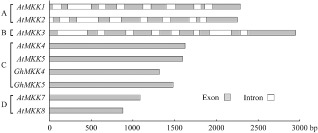
Comparison of the genomic DNA sequences of GhMKK4 and several mitogen‐activated protein kinase (MAPK) kinase (MAPKK) genes of Arabidopsis available in GenBank. The white boxes indicate the introns, and the grey boxes represent exons. The scale indicates the length of the sequence. A, B, C and D indicate the MAPKK groups.
Expression patterns of GhMKK4 mRNA
Reverse transcription‐polymerase chain reaction (RT‐PCR) was conducted to investigate the expression patterns of GhMKK4 in 7‐day‐old cotton seedlings. The relative expression of GhMKK4 is shown in Fig. 3. GhMKK4 was expressed much more strongly in the leaves and roots than in the stems (Fig. 3A). This tissue‐specific expression suggests that GhMKK4 may serve specific functions in different tissues.
Figure 3.
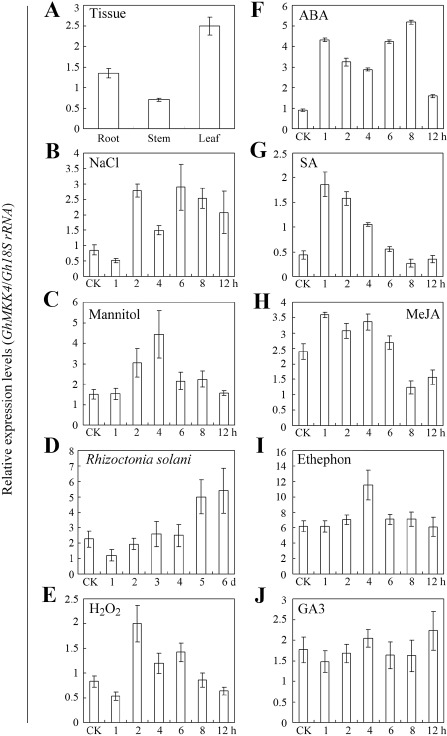
Relative expression of GhMKK4 in different tissues and in response to different stress factors. These results are the quantification of the GhMKK4 band intensity over the expression of Gh18S rRNA genes by Quantity One software. For the tissue‐specific expression of GhMKK4, the roots, stems and cotyledon leaves of 7‐day‐old cotton seedlings were used. For different treatments, the seedlings were treated with 200 mm NaCl (B), 200 mm mannitol (C), Rhizoctonia solani (D), 100 μm H2O2 (E), 100 μm abscisic acid (ABA) (F), 10 mm salicylic acid (SA) (G), 100 μm methyl jasmonate (MeJA) (H), 5 mm ethylene (ET) released from ethephon (I) or 10 μm gibberellins A3 (GA3) (J).
The environmental stress response assays showed that the expression of GhMKK4 was induced significantly by NaCl and mannitol, and that NaCl had a relatively long‐lasting effect (Fig. 3B,C). Treatment with Rhizoctonia solani significantly induced the expression of GhMKK4, peaking at day 5 or 6 during the indicated time (Fig. 3D). To explore the mechanism through which GhMKK4 is involved in signal transduction, the responsiveness of GhMKK4 to diverse signalling molecules was also examined. As shown in Fig. 3E, H2O2 markedly induced the accumulation of GhMKK4 at 2 h, with GhMKK4 expression then decreasing gradually to the level of the control. Notably, GhMKK4 expression was rapidly and strongly induced by ABA, SA and methyl jasmonate (MeJA) (Fig. 3F–H). Another signalling molecule, ET, which is released from ethephon, also induced GhMKK4 accumulation (Fig. 3I). However, 10 μm GA3 had a negligible effect on GhMKK4 expression (Fig. 3J). These results indicate that GhMKK4 is responsive to environmental stresses and may be involved in multiple signal transduction pathways in stress responses.
Enhanced susceptibility of GhMKK4‐overexpressing plants to bacterial pathogens
Transgenic N. benthamiana plants overexpressing GhMKK4 were produced to further evaluate the role of GhMKK4. The GhMKK4 expression levels in 12 randomly selected overexpressing (OE) lines of T1 progeny plants were detected using RT‐PCR (Fig. 4A). Three representative lines (1#, 2# and 4#) were selected, and their T3 progeny were used for functional analyses. Interestingly, the OE lines, especially lines 2# and 4#, grew relatively more slowly than the vector control (Vec) plants at the early vegetative stage (Fig. 4B). However, the differences in plant size diminished gradually by the late vegetative stage (data not shown).
Figure 4.
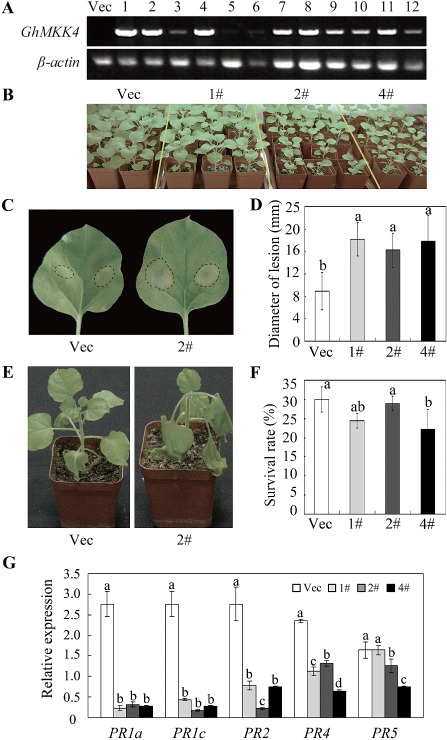
Identification of transgenic plants and subsequent bacterial pathogen resistance analysis. (A) The evaluation of GhMKK4 expression in the T1 progeny of transgenic plants. (B) The developmental phenotype of the transgenic plants. The photograph was taken approximately 6 weeks after the seedlings had been transplanted into the tubs. (C) The symptoms of detached leaves from 2‐month‐old T3 generation transgenic plants at 4 days after Ralstonia solanacearum injection. The lesion diameters are indicated in (D). Different letters above the columns indicate significant differences (P < 0.05) according to Duncan's multiple range test. (E) The phenotypes of the transgenic plants after inoculation with R. solanacearum using the trickle irrigation method. The survival rates of these plants are shown in (F). (G) The expression of the pathogenesis‐related (PR) genes in transgenic plants as analysed by quantitative real‐time polymerase chain reaction (qPCR). Different letters above the columns indicate significant differences (P < 0.05) according to Duncan's multiple range test. Each experiment was repeated at least three times. Vec, vector control; 1#, 2#, 4#, overexpressing lines.
When the detached leaves were injected with R. solanacearum over 4 days, the leaves of the OE plants showed notable signs of chlorosis and relatively larger lesion diameters than those of the Vec plants (Fig. 4C,D). The trickle irrigation method was also used to examine the response to the pathogen. The OE plants displayed marked wilting, whereas the Vec plants showed a relatively higher survival rate than the OE plants (Fig. 4E,F). These results indicate that the overexpression of GhMKK4 enhances the susceptibility to the bacterial pathogen in the transgenic plants.
To elucidate the reasons underlying the enhanced susceptibility, the relative expression levels of pathogenesis‐related protein (PR) genes were examined using quantitative real‐time PCR (qPCR). The PR genes, including PR1a, PR1c, PR2, PR4 and PR5, were expressed at much lower levels in the OE plants than in the Vec plants (Fig. 4G). This reduced expression of PR genes may be the reason for the enhanced susceptibility to the bacterial pathogen in the transgenic plants.
Enhanced susceptibility of GhMKK4‐overexpressing plants to a fungal pathogen
To test the response to a fungal pathogen, the detached leaves from 2‐month‐old T3 generation transgenic plants were incubated with R. solani. After incubation for 6 days, marked lesions were observed in the leaves of both Vec and OE plants. However, the lesions on the OE leaves were much larger than those on the Vec leaves, and many more hyphae and spores were present in the OE leaves than in the Vec leaves (Fig. 5A,B). When the plants were infected by the trickle irrigation method for approximately 10 days, the OE plants displayed significant wilting, with more serious caudex rot and a much lower survival rate than the Vec plants (Fig. 5C,D). These results indicate that the transgenic plants overexpressing GhMKK4 show an enhanced susceptibility to the fungal pathogen.
Figure 5.
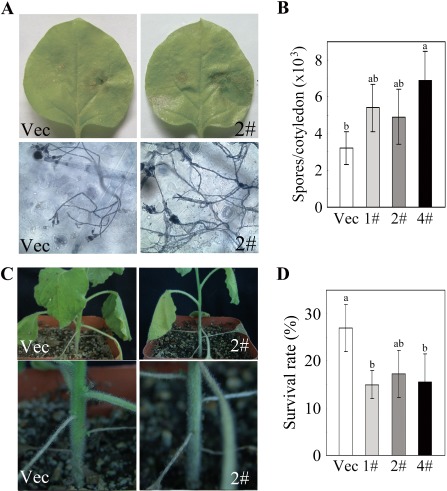
GhMKK4‐overexpressing plants exhibit enhanced susceptibility to Rhizoctonia solani. (A) Signs of disease on the detached leaves of transgenic plants 6 days after inoculation. The hyphae were visualized by trypan blue staining. (B) The number of spores per cotyledon at 6 days after R. solani infection. The data shown represent the means ± standard errors of three independent experiments. Different letters above the columns indicate significant differences (P < 0.05) according to Duncan's multiple range test. (C) The symptoms of the transgenic plants inoculated with R. solani using the trickle irrigation method. (D) The survival rates of the transgenic plants during R. solani infection using the trickle irrigation method. The data shown indicate the means ± standard errors of three independent experiments (n ≥ 30). Different letters above the columns indicate significant differences (P < 0.05) according to Duncan's multiple range test. Vec, vector control; 1#, 2#, 4#, overexpressing lines.
GhMKK4‐overexpressing plants do not show significant salt or drought stress tolerance
To test the function of GhMKK4 in response to abiotic stresses, transgenic plants were subjected to NaCl and drought stress. There was no significant difference in the growth and germination rates of the transgenic seeds during germination on water‐moistened filter paper. Under the 50 mm NaCl condition, the germination of both OE and Vec seeds was inhibited slightly. Although a larger number of OE seeds germinated slightly earlier at the very beginning of the experimental period, there was no significant difference between the overall germination rates of the transgenic seeds during germination (Fig. 6A,B). Similar results were observed under the 200 mm NaCl condition (data not shown). When germinated on filter paper moistened with 100 mm mannitol, OE seeds showed a relatively higher germination rate than Vec seeds, but this difference was not significant. A similar result was observed under the 200 mm mannitol condition (data not shown). Furthermore, the withholding of water for 12 days during the vegetable growth stage (drought stress) caused both OE and Vec plants to wilt. No noticeable differences were found between these plants during the drought or recovery periods, and the plant survival rate was similar in both groups (Fig. 6C,D). In addition, there was no significant difference in the rate of water loss during the drought treatment between OE and Vec plants (Fig. 6E). These results indicate that GhMKK4‐overexpressing plants do not show significant abiotic stress tolerance.
Figure 6.
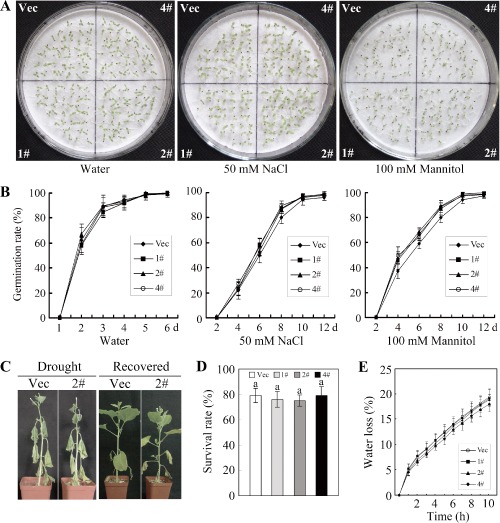
Tolerance analysis of transgenic plants to salt and drought stresses. (A) The seed germination phenotype. The seeds were germinated on filter paper soaked with 50 mm NaCl or 100 mm mannitol. The photographs shown were taken approximately 14 days after the seeds had been sown. (B) The germination rates of the seeds under normal and stress conditions. Germination was scored daily, and the results of germination on filter paper soaked with 50 mm NaCl or 100 mm mannitol are presented. (C) The phenotypes of plants subjected to drought stress at the late vegetable stage. Water was withheld from transgenic plants for 12 days, and the plants were then watered for 2 days to allow recovery. (D) The survival rates of the transgenic plants under drought conditions. The data shown represent the means ± standard errors of three independent experiments (n ≥ 30). (E) The water loss of detached leaves from the OE and Vec plants. The rates of water loss were calculated by the loss in fresh weight of the samples. The data shown represent the means ± standard errors of three independent experiments (n = 6). OE, overexpressing; Vec, vector control; 1#, 2#, 4#, overexpressing lines.
Overexpression of GhMKK4 affects ABA and GA signalling in transgenic plants
To identify the signal transduction mechanisms in which GhMKK4 might be involved, we evaluated the germination of transgenic seeds treated with signalling molecules. When germinated on filter paper moistened with 5 μm ABA, the green cotyledon rate of OE seeds was more than 10% lower than that of Vec seeds (Fig. 7A). To confirm this change in response to ABA, the post‐germination growth of seedlings was measured on filter paper moistened with 0.1 or 0.3 μm ABA. There were no significant differences in the root length or fresh weight at 0.1 μm ABA between OE and Vec seedlings. However, when treated with 0.3 μm ABA, OE seedlings showed a noticeably shorter root length and lower fresh weight than Vec seedlings (Fig. 7B). On GA3 treatment, although there was no noticeable difference in the seed germination rate (data not shown), the OE seedlings exhibited a shorter root length and lower fresh weight post‐germination (Fig. 7C).
Figure 7.
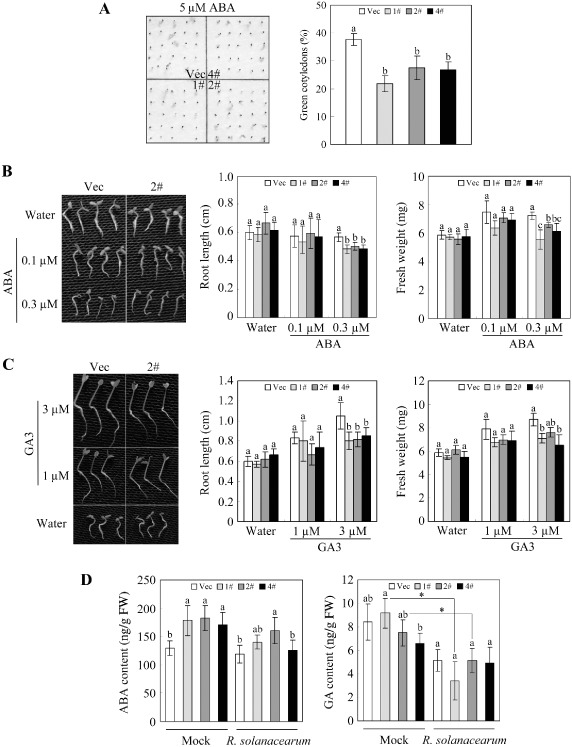
The influence of GhMKK4 overexpression on abscisic acid (ABA) and gibberellin (GA) signalling. (A) Seed germination under the ABA condition. The seeds germinated and developed green cotyledons on filter paper soaked with 5 μm ABA. (B) The post‐germination growth of seedlings under the 0.1 or 0.3 μm ABA condition. The root length and fresh weight (weight of 10 seedlings) of the seedlings were recorded 10 days after sowing. (C) The post‐germination growth of seedlings under the 1 or 3 μm GA3 condition. The root length and fresh weight (weight of 10 seedlings) of seedlings were recorded 7 days after sowing. (D) The ABA and GA levels in the transgenic plants determined by enzyme‐linked immunosorbent analysis (ELISA) under normal and Ralstonia solanacearum‐induced conditions. Leaves of 2‐month‐old plants were used for hormone measurement. The data shown indicate the means ± standard errors of three independent experiments. Different letters above the columns indicate significant differences (P < 0.05) according to Duncan's multiple range test. FW, fresh weight; Vec, vector control; 1#, 2#, 4#, overexpressing lines.
Previous experiments have demonstrated that OE plants show an enhanced susceptibility to R. solanacearum. Here, we measured the ABA and GA levels in 2‐month‐old transgenic plants to better understand the basis of the increased susceptibility. Notably, the ABA levels in OE plants were higher than those in Vec plants (Fig. 7D). In contrast, the GA levels in OE plants were lower than those in Vec plants (except for line 1#), which was consistent with the developmental phenotype described previously (Fig. 4B). When infected with R. solanacearum, although the difference was not significant, the ABA levels in the OE plants were depressed slightly, and the ABA levels in the Vec plants were barely affected. Interestingly, the GA levels were significantly depressed in all lines except for line 4# after R. solanacearum treatment. However, there was no significant difference in the GA levels between the OE and Vec plants after treatment. Together, these results indicate that overexpression of GhMKK4 affects ABA and GA signalling in transgenic plants.
Overexpression of GhMKK4 increases SA‐induced ROS accumulation in transgenic plants
SA is essential for the establishment of systemic acquired resistance (SAR) (Loake and Grant, 2007). In this study, we observed that SA affected H2O2 accumulation in transgenic plants, consistent with previous reports. Without any treatment, there was no significant difference in H2O2 accumulation in the plants (Fig. 8A). After SA treatment, both the Vec and OE plants exhibited a marked increase in H2O2 levels. However, H2O2 accumulation in OE plants was much higher than that in Vec plants. Moreover, the accumulation of superoxide anion (O2 −), another primary component of ROS, showed a similar pattern to that of H2O2 ( Fig. S1, see Supporting information). Too much ROS accumulation can result in cell death. After SA treatment, the OE plants showed a more serious cell death phenotype than Vec plants, as visualized by trypan blue staining (Fig. 8B). To investigate the possible mechanisms underlying the increase in SA‐induced ROS accumulation in OE plants, we evaluated the relative expression of genes that encode ROS‐scavenging enzymes, such as superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX) and the antioxidative enzyme glutathione‐S‐transferase (GST), as well as the ROS‐producing enzymes, respiratory burst oxidase homologue (RbohA and RbohB), using qPCR (Fig. S2, see Supporting information). In the absence of SA treatment, the expression of SOD, CAT, APX and GST showed different levels of decrease in OE plants relative to Vec plants. After SA treatment, the expression of SOD in Vec plants and of CAT in both Vec and OE plants was decreased, but the difference in the expression of SOD and CAT between OE and Vec plants was not significant. The expression of APX, especially for lines 1# and 4#, showed a significantly low level in OE plants. By contrast, the expression of GST, RbohA and RbohB was relatively higher in OE plants than in Vec plants. To confirm these results, the activities of SOD, peroxidase (POD) and CAT were measured. Prior to SA treatment, there was no marked difference in the activities of these enzymes between OE and Vec plants. After SA treatment, SOD activity increased slightly, and POD activity was significantly induced (Fig. 8C,D). Interestingly, the activity of CAT was more significantly repressed in OE lines after SA treatment (Fig. 8E). These results indicate that GhMKK4 overexpression confers increased SA‐induced ROS accumulation in transgenic plants, probably by the inhibition of CAT activity.
Figure 8.
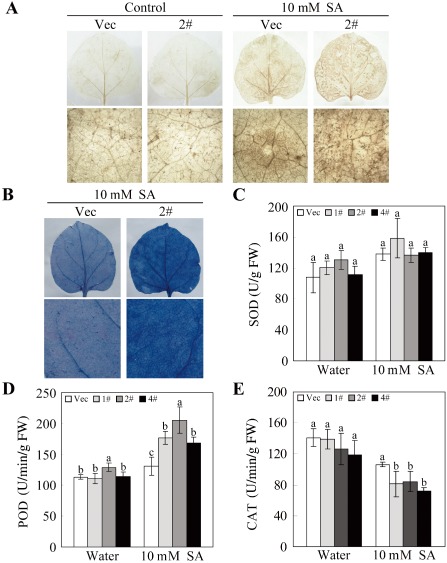
The increased accumulation of H2O2 induced by salicylic acid (SA) in the overexpressing plants. (A) The accumulation of H2O2 in transgenic plants, visualized by 3,3‐diaminobenzidine (DAB) staining under normal and SA‐induced conditions. (B) The trypan blue staining of the leaves after SA treatment. (C–E) The activities of the reactive oxygen species (ROS)‐scavenging enzymes under normal and SA‐induced conditions. Leaves were treated with 10 mm SA for 2 h, and three leaves at least of each line were used. The data shown indicate the means ± standard errors of three independent experiments. Different letters above the columns indicate significant differences (P < 0.05) according to Duncan's multiple range test. FW, fresh weight; Vec, vector control; 1#, 2#, 4#, overexpressing lines.
Expression profiling of signalling pathway‐related genes in transgenic plants
To test other possible signalling pathways during defence responses that involve GhMKK4, certain signalling pathway‐related genes were analysed using qPCR after R. solanacearum treatment. As shown in Fig. 9, the expression of 1‐aminocyclopropane‐1‐carboxylate oxidase (ACO) (except line 4#) and nitric oxide‐associated 1 (NOA1), which are involved in ET and nitric oxide (NO) signalling, respectively, was lower in OE plants than in Vec plants. The expression of NONEXPRESSER OF PR GENES1 (NPR1), which is essential for SA‐mediated signal transduction, did not show a significant difference between OE and Vec plants (except for line 1#). In addition, the polyamine synthesis‐related genes arginine decarboxylase (ADC), ornithine decarboxylase (ODC) and S‐adenosylmethionine decarboxylase (SAMDC) were tested. There was a slightly lower expression of ADC in OE plants than in Vec plants after treatment (except line 4#), whereas the transcriptional levels of ODC and SAMDC were much lower in OE plants than in Vec plants. These results suggest that GhMKK4 may have negative effects on pathogen‐induced polyamine synthesis.
Figure 9.
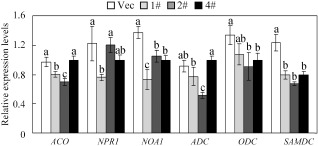
Quantitative real‐time polymerase chain reaction (qPCR) analysis of the expression of signalling‐related genes in overexpressing (OE) and vector control (Vec) plants under Ralstonia solanacearum‐inoculated conditions. Different letters above the columns indicate significant differences (P < 0.05) according to Duncan's multiple range test. ACO, 1‐aminocyclopropane‐1‐carboxylate oxidase; ADC, arginine decarboxylase; NOA1, nitric oxide‐associated 1; NPR1, NONEXPRESSER OF PR GENES1; ODC, ornithine decarboxylase; SAMDC, S‐adenosylmethionine decarboxylase; 1#, 2#, 4#, overexpressing lines.
Discussion
As the key node of MAPK cascades, MAPKKs may integrate multiple signal transduction pathways and may be involved in various biological processes. In this study, we identified a novel group C MAPKK gene, GhMKK4, from cotton. As a result of the difficulty of genetic transformation in cotton, we performed the initial functional analyses of the gene by ectopic expression in N. benthamiana.
Most of the plant MAPKKs reported recently have shown positive roles in pathogen resistance, whereas negative effects on resistance are rare. For example, AtMKK1 can be activated by pathogen elicitors and confers plant resistance to both virulent and avirulent bacterial pathogens (Meszaros et al., 2006; Teige et al., 2004). The overexpression of Arabidopsis AtMKK7 contributes to the activation of plant basal resistance and SAR in plants (Zhang et al., 2007). Furthermore, N. benthamiana NbMKK1 mediates the nonhost resistance to Pseudomonas cichorii (Takahashi et al., 2007b). As a rarely negative event, AtMKK1 and AtMKK2 can interact with MPK4 to negatively regulate the innate immune response in plants (Gao et al., 2008). In particular, the group C MAPKKs reported to date, including AtMKK4/MKK5 and rice OsMKK4, confer pathogen‐associated molecular pattern (PAMP)‐triggered defence responses (Asai et al., 2002; Kishi‐Kaboshi et al., 2010). Our previous study has revealed that GhMKK5, belonging to group C MAPKKs, confers resistance to the bacterial pathogen R. solanacearum in transgenic plants, but increases the susceptibility to P. parasitica var. nicotianae Tucker (Zhang et al., 2012). In this study, GhMKK4‐overexpressing plants were more susceptible to both bacterial and fungal pathogens than control (Vec) plants (Figs 4 and 5). These results provide further evidence of the negative roles of MAPKKs in disease resistance.
Emerging evidence has revealed that ABA plays a negative role in disease resistance. The resistance to the biotrophic pathogens Hyaloperonospora parasitica and Blumeria graminis was enhanced in the ABA biosynthesis mutants (Jensen et al., 2008; Mohr and Cahill, 2003). Similarly, ABA suppresses the resistance to necrotrophic pathogens, including Botrytis cinerea, Fusarium oxysporum, Plectosphaerella cucumerina and Erwinia chrysanthemi (Anderson et al., 2004; Asselbergh et al., 2008; Audenaert et al., 2002; Hernandez‐Blanco et al., 2007). The gain‐of‐function mutation cds2‐1D leads to the constitutive accumulation of high levels of endogenous ABA, which enhances the susceptibility to Pseudomonas syringae (Fan et al., 2009). However, some reports have also revealed that ABA may positively regulate the resistance to some pathogens (Adie et al., 2007; Kaliff et al., 2007; Ton and Mauch‐Mani, 2004; Wawrzynska et al., 2008). In this study, OE plants showed higher ABA levels than Vec plants. Therefore, we infer that the relatively higher ABA levels may be involved in the regulation of the enhanced pathogen susceptibility of OE plants. Furthermore, OE plants exhibited relatively lower pathogen‐induced expression of ODC and SAMDC, which are rate‐limiting enzymes in polyamine synthesis, suggesting that there is a lower pathogen‐induced polyamine level in OE plants. Polyamines have been suggested to play important roles in pathogen resistance (Walters, 2003). Together, these mechanisms may explain the decreased disease resistance in OE plants. The resistance mechanisms of plants to pathogens are very sophisticated. The molecular mechanisms through which GhMKK4 is involved in the negative regulation of the pathogen response have still not been elucidated fully and further investigation is ongoing.
A large body of evidence has shown that exogenous SA treatment can induce the accumulation of H2O2 in plant tissues (Agarwal et al., 2005; Chao et al., 2010; Harfouche et al., 2008; Rao et al., 1997). In this study, OE plants exhibited greater H2O2 accumulation after SA treatment. One possible explanation is that SA inhibits the H2O2‐degrading activity of CAT in OE plants. However, the activity of POD, the other major enzyme involved in the scavenging of H2O2, was elevated in OE plants after SA treatment. The elevated activity of POD may be a result of induction by increased H2O2. These results are consistent with those of previous studies (Fang et al., 2009). SA treatment can initiate lipid peroxidation, cell death and the activation of several other defence mechanisms through H2O2 (Rao et al., 1997). In this study, SA treatment induced greater accumulation of H2O2 and more serious cell death in OE plants than in Vec plants. Thus, GhMKK4 may be involved in an H2O2‐mediated defence mechanism, which may compensate for the enhanced susceptibility to the pathogens in OE plants.
It is noteworthy that GhMKK4 has high homology (77.11%) to GhMKK5, which has been identified previously. However, their functions are somewhat different. According to the amino acid alignment, although most subdomains are the same, some amino acids in subdomains between GhMKK4 and GhMKK5 are different, including subdomains I, VIa, VII and XI (Fig. 1A). In particular, several amino acids in the docking sites and one amino acid in the S/TXXXXXS/T motif of GhMKK4 and GhMKK5 are different. This information suggests that GhMKK4 and GhMKK5 may have different interactional substrates, and may be activated by different upstream MAPKKKs. In addition, several amino acids in the docking sites and A‐loops of GhMKK4 and GhMKK5 are different from those of AtMKK4 and AtMKK5. This suggests that GhMKK4 and GhMKK5 may have somewhat different roles from AtMKK4 and AtMKK5. Although there is high homology of GhMKK4 and GhMKK5 with AtMKK4 and AtMKK5, their genetic backgrounds are quite different. Furthermore, their overexpression in plants may result in various biochemical or physiological changes. MAPKKs classified into the same subgroups based on sequence similarity may not necessarily have identical functions (Lee et al., 2004). This may also explain the contrary role of the other group C member NtMEK2 from GhMKK4 in polyamine synthesis during pathogen resistance (Jang et al., 2009). Although the cascades in which GhMKK4 is involved are currently unknown, this should be the subject of future studies.
MAPKKs are the central components in plant MAPK cascades and play important roles in abiotic stresses, such as salt, drought and low temperatures. In this study, OE plants did not show significant tolerance to salt or drought stress. Under NaCl treatment, although there was no significant difference between the overall germination rates of the transgenic seeds during germination, a greater number of OE seeds germinated slightly earlier at the very beginning of treatment. Under mannitol treatment, although the difference was not significant, OE seeds showed a relatively higher germination rate than Vec seeds. Therefore, we cannot conclude that there is no response to salt or drought stress in OE plants. We assume that the potentially enhanced salt or drought stress tolerance may be impaired by a negative regulatory mechanism existing in the response to biotic stress. Furthermore, previous studies have reported that the overexpression of heterologous SAMDC in plants generally results in an improvement in the tolerance to abiotic stress, including salt (Roy and Wu, 2002) and drought (Waie and Rajam, 2003). Here, OE plants showed decreased expression of SAMDC and ODC, which may be another explanation for the lack of significant salt or drought stress tolerance. In addition, OE plants showed relatively lower GA levels than Vec plants, which means that OE seeds should show stunted germination or seedling growth relative to Vec seeds. However, there were no significant differences in germination or seedling growth between OE and Vec plants under stress conditions. Therefore, GhMKK4 may have potential effects on tolerance to salt or drought stress.
As a result of the different genetic background between N. benthamiana and cotton, altered expression of GhMKK4 in cotton itself may result in different phenotypes. In this study, overexpression of GhMKK4 did not enhance the tolerance to stresses, which probably limits its application in genetic improvement. However, these results may provide good reference, and the potential defence mechanism of GhMKK4 may provide clues on cotton genetic improvement.
Experimental Procedures
Plant material, growth conditions and treatments
Cotton (G. hirsutum L. cv. lumian 22) seeds were germinated in a wet cloth, and the seedlings were maintained in hydroponic culture for growth under glasshouse conditions at 26 ± 1 °C with a 16‐h light/8‐h dark cycle (relative humidity of 60%–75%). Nicotiana benthamiana seeds were placed on moist filter paper under glasshouse conditions for germination. Three‐ or four‐leaf‐stage N. benthamiana seedlings were transplanted into pots with soil and maintained under glasshouse conditions. For NaCl or mannitol treatments, uniformly developed cotton seedlings were sprayed with and dipped in these two solutions. For H2O2, ABA, SA, MeJA, ethephon and GA3 treatment, the uniformly developed cotton seedlings were sprayed with each chemical and then transferred to a box to maintain humidity. For pathogen treatment, 7‐day‐old cotton seedlings were inoculated with conidial suspensions of R. solani (105 conidia/mL) using the root dip method. The treated cotyledons were collected for RNA extraction. Each treatment was repeated at least twice.
Gene isolation, vector construction and genetic transformation
The GhMKK4 gene was isolated as described previously (Shi et al., 2011). The primer sequences used are provided in Table S1 (see Supporting Information). The vector construction and genetic transformation were performed as described by Zhang et al. (2012). Plants transformed with vector pBI121 were used as controls. The T3 progeny of OE and Vec plants were used for further experiments.
Semi‐quantitative RT‐PCR and qPCR analyses
Total RNA was isolated with Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol, and used for first‐strand cDNA synthesis with a reverse transcriptase system (TransGen Biotech, Beijing, China). Semi‐quantitative RT‐PCR was used to detect the expression levels of GhMKK4 under different treatments. The expression of the cotton 18S rRNA gene was used as a control. qPCR was performed using the SYBR PrimeScriptTM RT‐PCR Kit (TaKaRa, Dalian, China) in a 25‐μL volume on a CFX96TM Real‐time System (Bio‐Rad, Hercules, CA, USA), as described previously (Yu et al., 2012). The house‐keeping gene N. benthamiana β‐actin was used as control gene for qPCR analysis. The relative expression levels of the stress‐related genes were determined using the 2−ΔΔCT method.
Pathogen infection assay
The bacterial pathogen R. solanacearum was cultured overnight at 37 °C in Luria–Bertani (LB) broth, harvested by centrifugation and resuspended in sterile tap water. The fungal pathogen R. solani was cultured on potato dextrose agar (PDA) medium at 28 °C for 2 weeks, and the spores were then suspended in 1% glucose. For the infection assay, detached uniform leaves from 2‐month‐old T3 generation OE and Vec plants were inoculated with suspensions of R. solanacearum bacteria [optical density at 600 nm (OD600) = 0.6] or R. solani spores (105 spores/mL). At least three leaves from each line were detached for the infection assay. The infected leaves were kept in a transparent box under glasshouse conditions. Furthermore, the infection was confirmed by inoculation with the above suspensions of R. solanacearum bacteria or R. solani spores using the trickle irrigation method. The inoculated plants were kept in a moist chamber. Bacterial growth was monitored by performing serial dilutions onto King's B agar medium. The disease resistance analysis was repeated three times.
3,3‐Diaminobenzidine (DAB), nitroblue tetrazolium (NBT) and trypan blue staining assays
The DAB, NBT and trypan blue staining assays were performed as described previously (Zhang et al., 2011, 2012).
Salt and drought stress analyses
For the germination analysis, T3 generation OE and Vec seeds were placed on filter papers moistened with NaCl (50 and 200 mm) or mannitol (100 and 200 mm) solutions. The germination percentage was measured daily. For salt treatment, 2‐month‐old OE and Vec plants were irrigated with NaCl solutions (50 and 200 mm) for over 2 weeks, and the survival rate (the number of surviving plants relative to the total number of treated plants) was recorded. For the drought treatment, water was withheld completely from the plants for 12 days, and the plants were then watered regularly for 2 days to allow them to recover. For the water loss measurements, detached leaves from OE and Vec plants were incubated at 37 °C and weighed on an electronic balance at the indicated times. The rate of water loss was calculated as the percentage of the initial fresh weight at each time point. The salt and drought stress analyses were repeated at least three times.
Seed germination and growth in response to exogenous ABA and GA
For the germination analysis, the seeds were placed on filter papers moistened with ABA (5 μm) or GA3 (5 μm) solutions. The germination percentage was measured daily. To determine the seedling growth rate, the seeds were placed on filter papers moistened with ABA (0.1 and 0.3 μm) or GA3 (1 and 3 μm) solutions for germination and growth. The plates were maintained under glasshouse conditions for more than 10 days, and the root length and fresh weight (weight of 10 seedlings) were recorded.
Quantification of endogenous ABA and GA by enzyme‐linked immunosorbent assay (ELISA)
The samples were homogenized in liquid nitrogen and extracted in ice‐cold phosphate‐buffered saline (PBS, pH 7.4). After centrifugation at 4000 g (4 °C) for 20 min, the supernatant was stored at −20 °C for ELISA. ELISA was performed on a 96‐well microtitration plate, as described previously (Yang et al., 2001).
Antioxidative enzyme activity assays
For the enzyme activity assays, 0.5 g of leaf tissue was homogenized in 5 mL of ice‐cold extraction buffer (50 mm PBS, pH 7.8) using a prechilled mortar and pestle on ice. The homogenate was centrifuged at 12 000 g for 20 min at 4 °C. The resulting supernatant was collected, and the enzymatic activities of SOD, POD and CAT were measured, as described previously (Kong et al., 2011; Yang et al., 2008).
Statistical analysis
The results are expressed as the mean ± standard error (SE) of triplicate experiments. Statistical significance between measurements for different treatments or times was subjected to Duncan's multiple range test with an analysis of variance (ANOVA) using Statistical Analysis System (SAS) version 9.1 software (SAS Institute, Cary, NC, USA).
Supporting information
Fig. S1 The accumulation of superoxide anion (O2 −) induced by salicylic acid (SA) in the transgenic plants. The content of superoxide anion was visualized by nitroblue tetrazolium (NBT) staining under normal and SA‐induced conditions. Leaves were treated with 10 mm SA for 2 h, and three leaves at least of each line were used. Vec, vector control; 2#, overexpressing lines.
Fig. S2 The expression patterns of genes related to reactive oxygen species (ROS) production and scavenging in transgenic plants under normal and salicylic acid (SA)‐induced conditions analysed by quantitative real‐time polymerase chain reaction (qPCR). Leaves were treated with 10 mm SA for 2 h, and three leaves at least of each line were used. Vec, vector control; 1#, 2#, 4#, overexpressing lines.
Table S1 The primers used in this study.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31171837; 30970225) and the Genetically Modified Organisms Breeding Major Projects of China (2009ZX08009‐092B). We are grateful to Dr Xiangpei Kong (Shandong Agricultural University, Taian, China) for critical reading of the manuscript.
References
- Adie, B.A.T. , Perez‐Perez, J. , Perez‐Perez, M.M. , Godoy, M. , Sanchez‐Serrano, J.J. , Schmelz, E.A. and Solano, R. (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis . Plant Cell, 19, 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal, S. , Sairam, R.K. , Srivastava, G.C. , Tyagi, A. and Meena, R.C. (2005) Role of ABA, salicylic acid, calcium and hydrogen peroxide on antioxidant enzymes induction in wheat seedlings. Plant Sci. 169, 559–570. [Google Scholar]
- Anderson, J.P. , Badruzsaufari, E. , Schenk, P.M. , Manners, J.M. , Desmond, O.J. , Ehlert, C. , Maclean, D.J. , Ebert, P.R. and Kazan, K. (2004) Antagonistic interaction between abscisic acid and jasmonate‐ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis . Plant Cell, 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson, E. and Ellis, B. (2010) Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 15, 106–113. [DOI] [PubMed] [Google Scholar]
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.L. , Gomez‐Gomez, L. , Boller, T. , Ausubel, F.M. and Sheen, J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Asselbergh, B. , Achuo, A.E. , Hofte, M. and Van Gijsegem, F. (2008) Abscisic acid deficiency leads to rapid activation of tomato defence responses upon infection with Erwinia chrysanthemi . Mol. Plant Pathol. 9, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert, K. , De Meyer, G.B. and Hofte, M.M. (2002) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid‐dependent signaling mechanisms. Plant Physiol. 128, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader, G. , Djamei, A. , Teige, M. , Palva, E.T. and Hirt, H. (2007) The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis . Mol. Plant–Microbe Interact. 20, 589–596. [DOI] [PubMed] [Google Scholar]
- Cardinale, F. , Meskiene, I. , Ouaked, F. and Hirt, H. (2002) Convergence and divergence of stress‐induced mitogen‐activated protein kinase signaling pathways at the level of two distinct mitogen‐activated protein kinase kinases. Plant Cell, 14, 703–711. [PMC free article] [PubMed] [Google Scholar]
- Chao, Y.Y. , Chen, C.Y. , Huang, W.D. and Kao, C.H. (2010) Salicylic acid‐mediated hydrogen peroxide accumulation and protection against Cd toxicity in rice leaves. Plant Soil, 329, 327–337. [Google Scholar]
- Dai, Y. , Wang, H. , Li, B. , Huang, J. , Liu, X. , Zhou, Y. , Mou, Z. and Li, J. (2006) Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis . Plant Cell, 18, 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doczi, R. , Brader, G. , Pettko‐Szandtner, A. , Rajh, I. , Djamei, A. , Pitzschke, A. , Teige, M. and Hirt, H. (2007) The Arabidopsis mitogen‐activated protein kinase kinase MKK3 is upstream of group C mitogen‐activated protein kinases and participates in pathogen signaling. Plant Cell, 19, 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doczi, R. , Okresz, L. , Romero, A.E. , Paccanaro, A. and Bogre, L. (2012) Exploring the evolutionary path of plant MAPK networks. Trends Plant Sci. 17, 518–525. [DOI] [PubMed] [Google Scholar]
- Fan, J. , Hill, L. , Crooks, C. , Doerner, P. and Lamb, C. (2009) Abscisic acid has a key role in modulating diverse plant–pathogen interactions. Plant Physiol. 150, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, C.X. , Xiong, J. , Qiu, L. , Wang, H.B. , Song, B.Q. , He, H.B. , Lin, R.Y. and Lin, W.X. (2009) Analysis of gene expressions associated with increased allelopathy in rice (Oryza sativa L.) induced by exogenous salicylic acid. Plant Growth Regul. 57, 163–172. [Google Scholar]
- Gao, M. , Liu, J. , Bi, D. , Zhang, Z. , Cheng, F. , Chen, S. and Zhang, Y. (2008) MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen‐activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18, 1190–1198. [DOI] [PubMed] [Google Scholar]
- Hamel, L.P. , Nicole, M.C. , Sritubtim, S. , Morency, M.J. , Ellis, M. , Ehlting, J. , Beaudoin, N. , Barbazuk, B. , Klessig, D. , Lee, J. , Martin, G. , Mundy, J. , Ohashi, Y. , Scheel, D. , Sheen, J. , Xing, T. , Zhang, S. , Seguin, A. and Ellis, B.E. (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11, 192–198. [DOI] [PubMed] [Google Scholar]
- Harfouche, A.L. , Rugini, E. , Mencarelli, F. , Botondi, R. and Muleo, R. (2008) Salicylic acid induces H2O2 production and endochitinase gene expression but not ethylene biosynthesis in Castanea sativa in vitro model system. J. Plant Physiol. 165, 734–744. [DOI] [PubMed] [Google Scholar]
- Heinrich, M. , Baldwin, I.T. and Wu, J. (2011) Two mitogen‐activated protein kinase kinases, MKK1 and MEK2, are involved in wounding‐ and specialist lepidopteran herbivore Manduca sexta‐induced responses in Nicotiana attenuata . J. Exp. Bot. 62, 4355–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez‐Blanco, C. , Feng, D.X. , Hu, J. , Sanchez‐Vallet, A. , Deslandes, L. , Llorente, F. , Berrocal‐Lobo, M. , Keller, H. , Barlet, X. , Sanchez‐Rodriguez, C. , Anderson, L.K. , Somerville, S. , Marco, Y. and Molina, A. (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell, 19, 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, E.K. , Min, K.H. , Kim, S.H. , Nam, S.H. , Zhang, S. , Kim, Y.C. , Cho, B.H. and Yang, K.Y. (2009) Mitogen‐activated protein kinase cascade in the signaling for polyamine biosynthesis in tobacco. Plant Cell Physiol. 50, 658–664. [DOI] [PubMed] [Google Scholar]
- Jensen, M.K. , Hagedorn, P.H. , de Torres‐Zabala, M. , Grant, M.R. , Rung, J.H. , Collinge, D.B. and Lyngkjaer, M.F. (2008) Transcriptional regulation by an NAC (NAM‐ATAF1,2‐CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp hordei in Arabidopsis . Plant J. 56, 867–880. [DOI] [PubMed] [Google Scholar]
- Jonak, C. , Okresz, L. , Bogre, L. and Hirt, H. (2002) Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 5, 415–424. [DOI] [PubMed] [Google Scholar]
- Kaliff, M. , Staal, J. , Myrenas, M. and Dixelius, C. (2007) ABA is required for Leptosphaeria maculans resistance via ABI1‐ and ABI4‐dependent signaling. Mol. Plant–Microbe Interact. 20, 335–345. [DOI] [PubMed] [Google Scholar]
- Kim, S.H. , Woo, D.H. , Kim, J.M. , Lee, S.Y. , Chung, W.S. and Moon, Y.H. (2011) Arabidopsis MKK4 mediates osmotic‐stress response via its regulation of MPK3 activity. Biochem. Biophys. Res. Commun. 412, 150–154. [DOI] [PubMed] [Google Scholar]
- Kishi‐Kaboshi, M. , Okada, K. , Kurimoto, L. , Murakami, S. , Umezawa, T. , Shibuya, N. , Yamane, H. , Miyao, A. , Takatsuji, H. , Takahashi, A. and Hirochika, H. (2010) A rice fungal MAMP‐responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 63, 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, Q. , Qu, N. , Gao, M. , Zhang, Z. , Ding, X. , Yang, F. , Li, Y. , Dong, O.X. , Chen, S. , Li, X. and Zhang, Y. (2012) The MEKK1‐MKK1/MKK2‐MPK4 kinase cascade negatively regulates immunity mediated by a mitogen‐activated protein kinase kinase kinase in Arabidopsis . Plant Cell, 24, 2225–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, X. , Pan, J. , Zhang, M. , Xing, X. , Zhou, Y. , Liu, Y. and Li, D. (2011) ZmMKK4, a novel group C mitogen‐activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis . Plant Cell Environ. 34, 1291–1303. [DOI] [PubMed] [Google Scholar]
- Lee, J. , Rudd, J.J. , Macioszek, V.K. and Scheel, D. (2004) Dynamic changes in the localization of MAPK cascade components controlling pathogenesis‐related (PR) gene expression during innate immunity in parsley. J. Biol. Chem. 279, 22 440–22 448. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Wang, Y. , Xu, J. , Su, T. , Liu, G. and Ren, D. (2008) Ethylene signaling is required for the acceleration of cell death induced by the activation of AtMEK5 in Arabidopsis . Cell Res. 18, 422–432. [DOI] [PubMed] [Google Scholar]
- Loake, G. and Grant, M. (2007) Salicylic acid in plant defence—the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472. [DOI] [PubMed] [Google Scholar]
- MAPK Group (2002) Mitogen‐activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7, 301–308. [DOI] [PubMed] [Google Scholar]
- Meszaros, T. , Helfer, A. , Hatzimasoura, E. , Magyar, Z. , Serazetdinova, L. , Rios, G. , Bardoczy, V. , Teige, M. , Koncz, C. , Peck, S. and Bogre, L. (2006) The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 48, 485–498. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk, M. , Awotunde, O.S. , Muszynska, G. , Klessig, D.F. and Dobrowolska, G. (2000) Osmotic stress induces rapid activation of a salicylic acid‐induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell, 12, 165–178. [PMC free article] [PubMed] [Google Scholar]
- Mohr, P.G. and Cahill, D.M. (2003) Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica . Funct. Plant Biol. 30, 461–469. [DOI] [PubMed] [Google Scholar]
- Rao, M.V. , Paliyath, C. , Ormrod, D.P. , Murr, D.P. and Watkins, C.B. (1997) Influence of salicylic acid on H2O2 production, oxidative stress, and H2O2‐metabolizing enzymes—salicylic acid‐mediated oxidative damage requires H2O2 . Plant Physiol. 115, 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, D. , Liu, Y. , Yang, K.Y. , Han, L. , Mao, G. , Glazebrook, J. and Zhang, S. (2008) A fungal‐responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis . Proc. Natl. Acad. Sci. USA, 105, 5638–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, M.C. , Petersen, M. and Mundy, J. (2010) Mitogen‐activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649. [DOI] [PubMed] [Google Scholar]
- Roy, M. and Wu, R. (2002) Overexpression of S‐adenosylmethionine decarboxylase gene in rice increases polyamine level and enhances sodium chloride‐stress tolerance. Plant Sci. 163, 987–992. [Google Scholar]
- Shi, J. , Zhang, L. , An, H. , Wu, C. and Guo, X. (2011) GhMPK16, a novel stress‐responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol. Biol. 12, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K. , Yamaguchi‐Shinozaki, K. and Seki, M. (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. [DOI] [PubMed] [Google Scholar]
- Takahashi, F. , Yoshida, R. , Ichimura, K. , Mizoguchi, T. , Seo, S. , Yonezawa, M. , Maruyama, K. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2007a) The mitogen‐activated protein kinase cascade MKK3–MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis . Plant Cell, 19, 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y. , Nasir, K.H. , Ito, A. , Kanzaki, H. , Matsumura, H. , Saitoh, H. , Fujisawa, S. , Kamoun, S. and Terauchi, R. (2007b) A high‐throughput screen of cell‐death‐inducing factors in Nicotiana benthamiana identifies a novel MAPKK that mediates INF1‐induced cell death signaling and non‐host resistance to Pseudomonas cichorii . Plant J. 49, 1030–1040. [DOI] [PubMed] [Google Scholar]
- Teige, M. , Scheikl, E. , Eulgem, T. , Doczi, R. , Ichimura, K. , Shinozaki, K. , Dangl, J.L. and Hirt, H. (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis . Mol. Cell, 15, 141–152. [DOI] [PubMed] [Google Scholar]
- Ton, J. and Mauch‐Mani, B. (2004) β‐Amino‐butyric acid‐induced resistance against necrotrophic pathogens is based on ABA‐dependent priming for callose. Plant J. 38, 119–130. [DOI] [PubMed] [Google Scholar]
- Waie, B. and Rajam, M.V. (2003) Effect of increased polyamine biosynthesis on stress responses in transgenic tobacco by introduction of human S‐adenosylmethionine gene. Plant Sci. 164, 727–734. [Google Scholar]
- Walters, D. (2003) Resistance to plant pathogens: possible roles for free polyamines and polyamine catabolism. New Phytol. 159, 109–115. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Ngwenyama, N. , Liu, Y. , Walker, J.C. and Zhang, S. (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen‐activated protein kinases in Arabidopsis . Plant Cell, 19, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.J. , Zhu, S.Y. , Lu, Y.F. , Zhao, R. , Xin, Q. , Wang, X.F. and Zhang, D.P. (2010) Two coupled components of the mitogen‐activated protein kinase cascade MdMPK1 and MdMKK1 from apple function in ABA signal transduction. Plant Cell Physiol. 51, 754–766. [DOI] [PubMed] [Google Scholar]
- Wawrzynska, A. , Christiansen, K.M. , Lan, Y. , Rodibaugh, N.L. and Innes, R.W. (2008) Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol. 148, 1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Li, Y. , Wang, Y. , Liu, H. , Lei, L. , Yang, H. , Liu, G. and Ren, D. (2008) Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis . J. Biol. Chem. 283, 26 996–27 006. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Zhang, J. , Wang, Z. , Zhu, Q. and Wang, W. (2001) Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 127, 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Tang, R. , Zhu, J. , Liu, H. , Mueller‐Roeber, B. , Xia, H. and Zhang, H. (2008) Enhancement of stress tolerance in transgenic tobacco plants constitutively expressing AtIpk2β, an inositol polyphosphate 6‐/3‐kinase from Arabidopsis thaliana . Plant Mol. Biol. 66, 329–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. , Tena, G. , Xiong, Y. and Sheen, J. (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature, 451, 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. , Huaxia, Y. , Lu, W. , Wu, C. , Cao, X. and Guo, X. (2012) GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol. 12, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Xi, D. , Luo, L. , Meng, F. , Li, Y. , Wu, C.A. and Guo, X. (2011) Cotton GhMPK2 is involved in multiple signaling pathways and mediates defense responses to pathogen infection and oxidative stress. FEBS J. 278, 1367–1378. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Li, Y. , Lu, W. , Meng, F. , Wu, C.A. and Guo, X. (2012) Cotton GhMKK5 affects disease resistance, induces HR‐like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana . J. Exp. Bot. 63, 3935–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Dai, Y. , Xiong, Y. , DeFraia, C. , Li, J. , Dong, X. and Mou, Z. (2007) Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. Plant J. 52, 1066–1079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The accumulation of superoxide anion (O2 −) induced by salicylic acid (SA) in the transgenic plants. The content of superoxide anion was visualized by nitroblue tetrazolium (NBT) staining under normal and SA‐induced conditions. Leaves were treated with 10 mm SA for 2 h, and three leaves at least of each line were used. Vec, vector control; 2#, overexpressing lines.
Fig. S2 The expression patterns of genes related to reactive oxygen species (ROS) production and scavenging in transgenic plants under normal and salicylic acid (SA)‐induced conditions analysed by quantitative real‐time polymerase chain reaction (qPCR). Leaves were treated with 10 mm SA for 2 h, and three leaves at least of each line were used. Vec, vector control; 1#, 2#, 4#, overexpressing lines.
Table S1 The primers used in this study.


