SUMMARY
The elucidation of the structures of viroids, noncoding infectious RNA species, is paramount to obtain an understanding of the various aspects of their life cycles (including replication, transport and pathogenesis). In general, the secondary structures of viroids have been predicted using computer software programs which have been shown to possess several important limitations. Clearly, the predicted structure of a viroid needs to receive physical support prior to its use in the accurate interpretation of any mechanistic studies. Here, SHAPE probing coupled to computer‐assisted structure prediction using the RNAstructure software program was employed to determine the structures of five viroids. These species belong to four genera of the Pospiviroidae family, and none have had their structure characterized in solution. In addition, several interesting questions were addressed by either studying various sequence variants or varying the SHAPE conditions. More importantly, this approach is novel in the study of viroids, and should be of significant aid in the determination of the structures of other RNA species.
INTRODUCTION
Viroids are covalently closed, single‐stranded, noncoding RNAs, and are the smallest infectious agents (245–401 nucleotides) replicating in plants. They occur worldwide, cause economic losses and affect crop quality. According to their molecular and biological properties, they have been classified into either the Pospiviroidae family, which includes five genera (Pospiviroid, Hostuviroid, Cocadviroid, Coleviroid and Apscaviroid), or the Avsunviroidae family, which includes two genera (Avsunviroid and Pelamoviroid) (Flores et al., 2005). The Pospiviroidae family, whose type species is the Potato spindle tuber viroid (PSTVd), includes more than 30 species and is characterized by a rod‐like secondary structure with five structural/functional domains [Terminal Left (TL), Pathogenicity (P), Central (C), which includes a central conserved region (CCR), Variable (V) and Terminal Right (TR)]. The members of this family replicate and accumulate in the nucleus via an asymmetric rolling‐circle mechanism. The Avsunviroidae family, whose type species is the Avocado sunblotch viroid (ASBVd), includes ASBVd and the Eggplant latent viroid (ELVd), which both adopt a rod‐like secondary structure, as well as the Peach latent mosaic viroid (PLMVd) and the Chrysanthemum chlorotic mottle viroid (CChMVd), which fold into branched secondary structures. These last four viroids replicate and accumulate in chloroplasts through a symmetric rolling‐circle mechanism.
Viroids are noncoding infectious RNA species; therefore, the elucidation of their adopted structures is paramount to obtain an understanding of the various aspects of their life cycles, including replication, transport and pathogenesis. To date, the secondary structures of only three viroids [PSTVd, PLMVd and the Citrus exocortis viroid (CEVd)] have been determined in solution (Bussière et al., 2000; Domdey et al., 1978; Dubéet al., 2010; Gast et al., 1996; 1978, 1982). The reason for the lack of information on viroid structures in solution is that their determination remains laborious. In general, the secondary structures of viroids have been predicted using computer software programs (Bussière et al., 1996; Fadda et al., 2003; Navarro and Flores, 1997; Symons, 1981). The prediction of secondary structures, which is based on thermodynamic approaches using dynamic programming, such as RNAstructure (Reuter and Mathews, 2010) and Mfold (Zuker, 2003), has met with some success in the case of small RNAs (<200 nucleotides) (Masquida et al., 2010). However, the main drawback of these software programs is that the folding of any single RNA sequence into its energetically most favoured structure may not produce the biologically active RNA structure. For longer RNAs, computer predictions are less helpful as they tend to yield ‘closed’ structures, whereas comparative approaches exploiting both sequence and structure conservation information afford more reliable results. Clearly, any predicted secondary structure needs to receive physical support in vitro or in vivo prior to being used to accurately interpret any mechanistic studies.
A new technique, RNA‐selective 2′‐hydroxyl acylation analysed by primer extension (SHAPE), has been shown to be useful for the characterization of both RNA structure and dynamics at a single‐nucleotide resolution (Steen et al., 2010). The principle of the method is that benzoyl cyanide (BzCN) reacts with the 2′‐hydroxyl groups of flexible single‐stranded nucleotides, but not with those of base‐paired or constrained nucleotides (Mortimer and Weeks, 2008). As a result, during primer extension, the reaction stops one nucleotide before either one of these situations, and the flexible nucleotide is thereby identified. SHAPE chemistry rarely needs significant optimization, and requires only 2 days to complete for an RNA of 100–200 nucleotides in length (Wilkinson et al., 2006). Recently, the accuracy of SHAPE probing was validated by characterizing the structure of PLMVd as a model viroid (Dubéet al., 2011). For the same PLMVd sequence variant, a structure was obtained that was in agreement with that previously reported based on a combination of enzymatic mapping and oligonucleotide binding shift assays (Bussière et al., 2000). Moreover, the probing of several sequence variants, as well as of derived RNA molecules, led to the identification of new structural motifs. In addition, these results have shown that the heterogeneity of a viroid is not limited to its nucleotide sequence, but may also occur at the structural level.
A combination of computer‐assisted structure prediction and SHAPE probing has recently been used, and has been shown to substantially improve the structures of several RNAs deduced by software alone (Deigan et al., 2009; Kladwang et al., 2011). The aim of this study was to develop a fast, reliable and easily applicable standard procedure, based on a combination of probing in solution and computer prediction, for the determination of a viroid's secondary structure. This report presents the elucidation of the secondary structures of five viroid species belonging to four genera for which almost no structure had previously been characterized in solution. In addition, several interesting questions were addressed either by the study of various sequence variants or by varying the SHAPE conditions.
RESULTS AND DISCUSSION
Selection of viroids for probing
With the aim of developing a simple systematic procedure for the determination of a viroid's structure, viroids infecting citrus plants were defined as constituting a suitable set of viroids with which to work. Apart from CEVd (Gross et al., 1982), to our knowledge, no report has been published describing the structural features of these viroids. The structural information currently available comes primarily from computer‐assisted structure predictions (Steger et al., 1984; Szychowski et al., 2005; Visvader and Symons, 1985, 1986). Specifically, this set of viroids includes the Citrus bent leaf viroid (CBLVd, 315–329 nucleotides in size), the Hop stunt viroid (HSVd, 295–303 nucleotides), the Citrus dwarfing viroid[CDVd, 291–297 nucleotides; also known as Citrus viroid III (CVd‐III)], the Citrus bark cracking viroid[CBCVd, 284–286 nucleotides; also known as Citrus viroid IV (CVd‐IV)], CEVd (370–375 nucleotides), the Citrus viroid V (CVd‐V, 290–294 nucleotides) and the Citrus viroid VI [329–331 nucleotides; also known as Citrus viroid OS (CVd‐OS)] (Table 1). Figure 1 illustrates the phylogenetic relationships between these viroid species that belong to four different genera of the Pospiviroidae family. At least one species of each genera was retained for structural analysis (underlined). Moreover, because CEVd strains are classified into severe and mild strains (classes A and B, respectively; Chaffai et al., 2007; Visvader and Symons, 1985, 1986), representative sequences of both clusters were selected.
Table 1.
Characterized viroids and their respective National Center for Biotechnology Information (NCBI) accession numbers, together with the primers used to produce the DNA templates and in the primer extension reactions during the SHAPE experiments.
| Viroid | NCBI accession number | Size (nucleotides) | Primer name* | Sequence (5′→3′)† |
|---|---|---|---|---|
| CEVd.188 | HQ284011 | 370 | ||
| CEVd.189 | HQ284014 | 372 | ||
| CEVd.190 | HQ284018 | 372 | ||
| S95–117 | TAATACGACTCACTATAGGGGAAACCTGGAGGAAGTCGAG | |||
| S203–225 | TAATACGACTCACTATAGGCTCCACATCCGATCGTCGCTG | |||
| AS77–94 | GGGGATCCCTGAAGGACT | |||
| AS343–359 | GCTAGGGTTCCGAGGGCT | |||
| AS258–276 | TCCACCGGGTAGTAGCCAG | |||
| AS185–202 | AGCAGCGAAAGGAAGGAG | |||
| AS157–174 | GTTTCTCCGCTGGACGCC | |||
| CBLVd.032 | JF742600 | 328 | ||
| S2–25 | TAATACGACTCACTATAGGAGACTTCTTGTGGTTCCTGTGG | |||
| S104–122 | TAATACGACTCACTATAGGCTCGTCAGCTGCGGAGG | |||
| AS84–103 | TTCGTCGACGACGACCAGTC | |||
| AS200–217 | GCAGCGCTCGGGTGAAGG | |||
| AS311–329 | GAGGAGCCCTCAGGGGTTC | |||
| HSVd.cit106 | JF742601 | 296 | ||
| S82–104 | TAATACGACTCACTATAGGCAACTCTTCTCAGAATCCAGC | |||
| S195–214 | TAATACGACTCACTATAGGCTCTTCCGATGAGACGCG | |||
| AS174–194 | AGAAGGAGGTAAAGAAGAAGG | |||
| AS262–280 | ATCCGCGGCAGAGGCTCAG | |||
| AS58–81 | CCGGGGCTCCTTTCTCAGGTAAGT | |||
| CVd‐III.072 | JF742602 | 295 | ||
| S130–156 | TAATACGACTCACTATAGGTAGAGTCTCCGCTAGTCGGAAAG | |||
| S202–221 | TAATACGACTCACTATAGGACAACTGAGTGAGTTGTC | |||
| AS182–201 | GCTCGACTAGCGGGAGCTAG | |||
| AS9–27 | CCACAGGAACCACACGGAG | |||
| AS113–130 | GTCTTTACTCCACTCGGCG | |||
| CVd IV | — | 285 | ||
| S1–19 | TAATACGACTCACTATAGGGGAAGTTTCTCTGCGGG | |||
| S99–116 | TAATACGACTCACTATAGGGCGCCGCGGATCACTG | |||
| AS268–285 | AGAGGGGTCTCTAACGCG | |||
| AS174–192 | GAAGGAAGAAGCGACGATC | |||
| AS79–98 | TCCCCTCGACGAGTCTGAAG |
AS, antisense primer; S, sense primer. The numbers after the S or AS indicate the nucleotide positions bound by the primer.
The italic sequence is the T7 promoter sequence.
Figure 1.
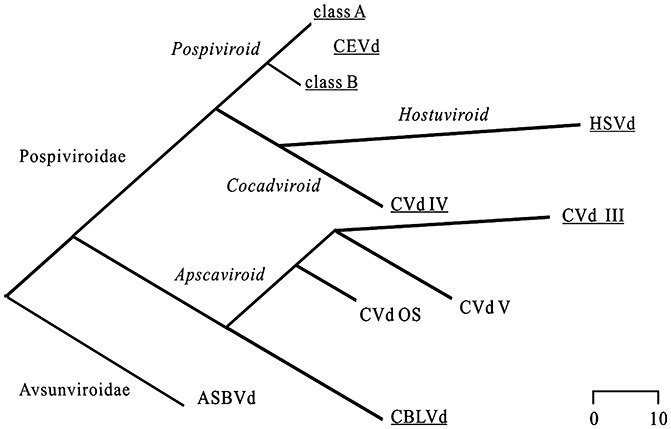
Phylogenetic relationship of the viroids probed in this study. The phylogenetic tree of the citrus viroids was generated using the MegAlign program with the ClustalV method in the DNASTAR software package (Madison, WI, USA). The clustering groups fit those reported previously (Elena et al., 2001). The viroids probed in this study are underlined. The scale bar represents the number of nucleotide substitutions. ASBVd, Avocado sunblotch viroid; CBLVd, Citrus bent leaf viroid; CEVd, Citrus exocortis viroid; CVd III/IV, Citrus viroid III/IV; CVdOS, Citrus viroid OS; HSVd, Hop stunt viroid.
Elucidation of the secondary structures of three CEVd variants
Three CEVd variants were chosen for structural analysis, and were named according to the Subviral Database (Rocheleau and Pelchat, 2006) which contains the sequences of all viroids and related RNA species: CEVd.188 [National Center for Biotechnology Information (NCBI) accession number HQ284011], CEVd.189 (NCBI accession number HQ284014) and CEVd.190 (NCBI accession number HQ284018). CEVd.188 is a class A viroid, whereas both CEVd.189 and CEVd.190 belong to class B (Visvader and Symons, 1985). Comparison of the three genomes revealed that CEVd.189 and CEVd.190 differ by 33 and 35 nucleotides, respectively, from CEVd.188, and that these two variants differ by only five nucleotides between them. More importantly, the use of software structure prediction tools, such as Mfold and RNAstructure, provided a good range of motifs that are potentially adopted by the TL region (i.e. rod‐like, Y shape or cruciform motifs) in the most stable structures (Fig. S1, see Supporting Information). As a result, the secondary structures of these three CEVd variants were probed by the SHAPE method.
The DNA for the in vitro transcription reactions was prepared by a circular permuted reminiscent strategy (Beaudry et al., 1995). Briefly, it involved a polymerase chain reaction (PCR) amplification of a unit‐length CEVd genome from a tandemly duplicated gene template (i.e. head‐to‐tail copies), and was used to determine the 5′‐end of the subsequent RNA molecule. The amplification was performed using a forward primer that contained the T7 RNA polymerase promoter sequence at its 5′‐end, followed by two or three guanosine residues for more efficient transcriptional initiation, and ended with the 5′‐terminal sequence desired for the subsequent RNA molecule. The 3′‐terminus of this RNA was similarly determined by the reverse primer used. By tandemly moving both primers along the template, it is possible to position the starting site at any desired nucleotide. Thus, this permits the probing of the entire RNA molecule, as the 3′‐end region cannot be probed because it is bound by the primer during the primer extension step. In the present case, two DNA templates were synthesized in order to produce transcripts that started at either position 94 or position 202 (i.e. near the CCR on either the upper or lower strands) for each of the CEVd variants. Subsequently, in vitro run‐off transcriptions were performed and the resulting RNA molecules were purified by denaturing 5% polyacrylamide gel electrophoresis (PAGE). Only the full‐length transcripts were recovered. After purification, the transcripts were dissolved in 0.5 × TE buffer (see Experimental procedures), heat denatured at 65 °C for 2 min and snap‐cooled on ice for 5 min. Folding buffer was then added and the samples were incubated at 37 °C for 5 min. Subsequently, MgCl2 (or water) was added, followed by an incubation of 10 min at 37 °C. Finally, the BzCN reagent [or dimethylsulphoxide (DMSO) for the negative control] was added. The reactions occurred almost instantaneously with the 2′‐hydroxyl groups of the ribose residues, and were performed either in the absence or presence of 10 mM MgCl2 in order to reveal the existence of certain tertiary interactions, such as pseudoknots, that could require metal ions for their formation. As most of the remaining BzCN was inactivated by hydrolysis within a few seconds, forming a nonreactive product (Mortimer and Weeks, 2008), there was no need to stop the reactions. After ethanol precipitation, the RNA pellets were dissolved and the primer extension reactions were performed using a 32P‐radiolabelled primer complementary to the 3′ end of the analysed transcript. The reverse transcriptase stops one nucleotide prior to that bearing the adduct formed with the BzCN reagent; hence, electrophoresis of the primer extension products on sequencing gels leads to the identification of the flexible residues. After electrophoresis, the gels were exposed to phosphor screens, which were then revealed using a Typhoon apparatus.
A typical gel for the analysis of the primer extension reactions, performed with a CEVd.188 transcript starting at position 94 in the presence of an oligonucleotide complementary to positions 276 to 258 of the lower region, is illustrated in Fig. 2A. The intensity of each band was subsequently analysed using SAFA software (Laederach et al., 2008), and is shown in histogram form in Fig. 2B. In the present case, there was no significant difference observed when the reactions were performed in the absence or presence of MgCl2 (compare the top and bottom graphs in Fig. 2B). The flexible nucleotides (i.e. those located mainly in single‐stranded regions) were those showing the higher intensities, and vice versa (Fig. 2C).
Figure 2.
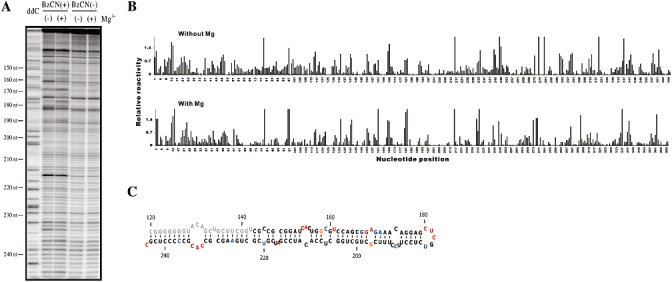
Typical example of the SHAPE probing of the Citrus exocortis viroid clone CEVd.188. (A) Typical SHAPE experiment gel obtained using the primer CEVd AS 258–276 in the primer extension reaction. The lane ddC represents the ladder produced during primer extension using dideoxycytosine. The viroid was probed in either the presence or absence of benzoyl cyanide (BzCN+/–), and either without or with 10 mm Mg2+(–/+). The nucleotide positions on the viroid's structure are indicated on the left of the gel. (B) Histograms generated following analysis of the raw data using the SAFA software after background subtraction and normalization in the absence (top graph) and presence (bottom graph) of magnesium ions. (C) Secondary structure of the region probed in A (from nucleotides 144 to 244) according to SHAPE data incorporated into the RNAstructure software. The region in grey was impossible to analyse as it was located in a poorly resolved region of the gel. The colours are attributed according to the relative SHAPE intensities (black ≤ 0.3, 0.3 < blue < 0.5, 0.5 ≤ orange < 0.7, 0.7 ≤ red).
According to the method developed by Deigan et al. (2009), SHAPE data can be incorporated into RNAstructure software (Reuter and Mathews, 2010). In this case, SHAPE data are converted into a pseudo‐free energy change and are used in conjunction with conventional nearest‐neighbour parameters to predict RNA secondary structure. In other words, the structural study is not solely based on a thermodynamic approach, but also includes biochemical data, which significantly increases the accuracy of the results and provides an experimental correction to the conventional nearest‐neighbour parameters. In the present case, all flexible residues were found to be located in single‐stranded regions, although they did not all react efficiently with the BzCN reagent. Moreover, a certain number of single‐stranded nucleotides were observed not to react with the chemical reagent.
Each primer extension reaction provided information on a region spanning approximately 100–125 nucleotides. Therefore, using several primers complementary to distinct regions located around the viroid, and by generating overlapping data to facilitate the integration of the reads, a total of three or four primers was required in order to completely cover a viroid's genome. More specifically, in the case of CEVd, the data from four primers were compiled and analysed together. All gels were analysed using the SAFA software (Laederach et al., 2008). The signals obtained from different primers for each variant were normalized to a uniform scale as reported by Low and Weeks (2010). Briefly, the 2% most reactive signals of all intensities taken together were removed from the pool, and the intensities of the next 8% most reactive were averaged. The intensity obtained for each nucleotide was then divided by this average value. This normalization step was performed manually using the Excel software program.
According to the approach described above, the CEVd.188 viroid (class A) has a long rod‐like secondary structure (Fig. 3A). The structure obtained differs slightly in the TL region from CEVd‐t, a class A viroid (Szychowski et al., 2005). In the latter variant, a Y‐shaped structure was proposed for this region; however, under the conditions used here, this structure was not detected. The TL region of this variant is more rod‐like than Y‐shaped. Even so, the two structures possess 79.2% structural identity when the percentage identity is evaluated by determining the number of mismatched nucleotides of the total number of nucleotides when the structure determined here is compared with those predicted previously. However, when the structure reported here is compared with that of CEV‐A, a previously described class A CEVd variant (Visvader and Symons, 1985, 1986), the two structures are quite similar, with 91.6% of the secondary structure being analogous. Exceptions are found only in the TL and P regions. The two other CEVd sequence variants (i.e. CEVd.189 and CEVd.190; described as being class B) also appear to fold into long rod‐like structures (Fig. 3B, C). The incorporation of SHAPE data into the software prediction did not reveal the presence of any complex secondary structures, such as either cruciform or Y‐shaped structures. Indeed, when the structures determined in this study were compared with that of CEV‐DE26, which has been classified as class B (Visvader and Symons, 1985, 1986), only 77.3% of the nucleotides were found to have the same structure. The rearrangements are found in every region, except for the C region, whose structure seems to be well conserved (data not shown).
Figure 3.

Secondary structures of Citrus exocortis viroid (CEVd) clones CEVd.188 (A), CEVd.189 (B) and CEVd.190 (C) as determined by integration of the SHAPE data in the presence of 10 mm MgCl2 into the RNAstructure 5.1 software program. The colours are attributed according to the relative SHAPE intensities (black ≤ 0.3, 0.3 < blue < 0.5, 0.5 ≤ orange < 0.7, 0.7 ≤ red). TL (Terminal Left), P (Pathogenicity), C (Central), V (Variable) and TR (Terminal Right) refer to the structural domains defined elsewhere (Chaffai et al., 2007; Keese and Symons, 1985). The A motif on each structure is indicated by a box.
The only structural differences between the class A and B CEVd viroids probed in this study were found to be located mainly in the PL motif (the pathogenicity‐modulating domain; Visvader and Symons, 1986). In actuality, these regions are highly variable in terms of sequence across all CEVd variants. Specifically, CEVd.189 and CEVd.190 (class B) demonstrated a more open structure in this motif than did CEVd.188 (class A). In addition, the PL motif folds differently in these two class B viroids, a difference that may be explained by a U318 to C318 mutation in CEVd.190 that disables the base pairing with A57.
When the SHAPE reactions were repeated in the absence of MgCl2, only minor differences were observed between CEVd.188 and CEVd.189 (compare Fig. 3A with Fig. S2A and Fig. 3B with Fig. S2B, see Supporting Information). Specifically, no stretch of three to five nucleotides was found that showed an increased flexibility in the absence of magnesium. Had such a stretch been found, it would have been indicative of a possible tertiary interaction, such as the formation of a pseudoknot. Usually, it was mainly the nucleotides located at the ends of paired regions that appeared to be less flexible, indicating that the metal ion contributes to the closing of the helical region. In the case of CEVd.190, a major local rearrangement of the structure in the P region results in the formation of a cruciform structure. This results in four nucleotides that are significantly more flexible (A50, A52, A53 and A54) in the absence of MgCl2 (Figs 3C and S2C). Indeed, A53 and A54 base pair with U314 and U313, respectively, in the presence of magnesium ions, but are not base paired at all in their absence.
Concurrently, RNase mapping was performed in the presence of magnesium ions on the CEVd.188 variant in order to obtain additional physical support for the SHAPE data. Enzymatic probing using RNase T1 (which cleaves after G residues located in single‐stranded regions), RNase A (which cleaves after U and C residues located in single‐stranded regions) and RNase V1 (a nonspecific RNase which cleaves residues located in double‐stranded regions) also supports the folding into a long rod‐like structure that excludes the formation of a Y motif in the TL region (Fig. S3, see Supporting Information).
Finally, CEVd.188 RNA was also subjected to SHAPE probing at temperatures ranging from 5 °C to 45 °C (i.e. 5, 15, 25, 35 and 45 °C). Previous work has shown that CEVd infection can vary from a nonsymptomatic condition to a severe reaction when grown at 40 °C (Skoric et al., 2001). We were interested to investigate whether the temperature to which a viroid is subjected can affect the structure. The transcripts were preincubated at the same temperature at which the reaction occurred. Surprisingly, almost identical SHAPE banding patterns were obtained regardless of the incubation temperature (data not shown). This result indicates that the viroid secondary structure is not significantly affected by the incubation temperature, even at temperatures as high as 45 °C. This is a clear demonstration of the stability of the viroid secondary structure. Moreover, it suggests that the normal environment temperature should not alter the structure of a viroid species.
Probing the secondary structure of four viroid species
Subsequently, this new protocol was used to probe the structure of four other viroid species that infect citrus trees. Specifically, the strategy based on the SHAPE probing of two transcripts possessing distinct 5′‐end positions, coupled to bioinformatic secondary structure prediction using RNAstructure, was employed. Details on the sequence variants of each viroid are given in Table 1, and the resulting secondary structures are depicted in Fig. 4. Except for CVd‐III.072, in which a Y‐shaped structure was detected in the TL domain, all the viroids folded into rod‐like structures. In all cases, several differences were observed when these structures were compared with those obtained by computer prediction based solely on thermodynamic data (Figs S4 and S5, see Supporting Information). For example, in the case of CVd‐III.072, which belongs to the Apscaviroid genus, the folding obtained using the SHAPE data yields two different structures possessing the same ΔG value (Fig. 4A). Specifically, the TL region can form either a Y‐shaped structure or one that appears to be rod‐like. All of the remaining regions of the viroid are identical. However, when the structures predicted with (Fig. 4A) and without (Fig. S4B) SHAPE data were compared, the main differences were found to be located in the TL and TR regions. Indeed, the TL region predicted by RNAstructure alone is clearly rod‐like. Moreover, the right terminal loop of the structure was formed by nucleotides 145–149, instead of nucleotides 137–144 as in the previously predicted structure. Apart from the alternative Y‐shaped motif of the TL region, the general structure of this viroid is very similar to that obtained in the absence of magnesium ions (Fig. S6A, see Supporting Information).
Figure 4.

Secondary structures of Citrus viroid III clone CVd‐III.072 (A), Citrus bent leaf viroid clone CBLVd.032 (B), Hop stunt viroid clone HSVd.cit106 (C) and Citrus viroid IV clone CVd‐IV (D) as determined by integration of the SHAPE data into the RNAstructure software. The colours are attributed according to the relative SHAPE intensities (black ≤ 0.3, 0.3 < blue < 0.5, 0.5 ≤ orange < 0.7, 0.7 ≤ red). TL (Terminal Left), P (Pathogenicity), C (Central), V (Variable) and TR (Terminal Right) refer to the structural domains defined elsewhere (Ashulin et al., 1991; Francis et al., 1995; Stasys et al., 1995), except for CVd‐IV, which was defined in this study by comparing its structure with those of other members of the Pospiviroidae family. The A motif on each structure is indicated by a box.
In the case of CBLVd.032 (Apscaviroid), the inclusion of SHAPE data revealed the presence of a large bulge in the TR domain between nucleotides 169 and 186 (Fig. 4B), which is not present in the prediction based on thermodynamic data alone (Fig. S4D). This bulge was not detected in the other viroids probed in this study and its presence remains to be confirmed. The remaining part of the molecule is similar, even when the Mfold prediction is considered (Fig. S4C). Without MgCl2, the structure produced is very similar, and no pseudoknots were detected under the experimental conditions used (Fig. S6B).
The SHAPE probing highlights two major loops in HSVd.cit106 (Hostuviroid) that are not predicted by the software alone (Figs 4C and S5A, B). First, the loop A motif appears to be larger when the SHAPE data are considered in the folding of the molecule by RNAstructure. Second, the nucleotides spanning the regions 90–99 and 197–206 are highly sensitive to BzCN, generating a large loop in the C region. The absence of magnesium ions seems to close this large loop, within which some nucleotides are base paired together (Fig. S6C)
Finally, the structures predicted by the different software programs are in agreement with that incorporating the SHAPE data in the case of CVd‐IV (which belongs to the Cocadviroid genus) (Figs 4D and S5C, D), as well as with the structure probed in the absence of magnesium (Fig. S6D).
Unlike the case of CVd‐IV, the loop A motif in all of the other viroids always appears to be more single stranded in the structure based on the SHAPE data than in that based on the thermodynamic data (Figs 4, S4 and S5). This is in good agreement with a previous report showing the existence of a relatively large single‐stranded region forming the A motif (also named pre‐melt loop) in PSTVd (Gast et al., 1996). This structure is also detected in CEVd, being quite accessible in CEVd.190 and less so in CEVd.188 and CEVd.189 (Fig. 3).
Reproducibility of the probings
During their replication cycle, viroids undergo a ligation step of their linear monomeric species. The latter species are considered to be the genomic version of the viroids. In this study, linear molecules were probed because they are easier to produce in vitro. Consequently, one can ask whether or not the conclusions drawn are applicable to natural circular conformers. Recently, the circular species of PLMVd was probed, and the results obtained were similar to those for the corresponding linear molecule (data not shown). This most probably results from the high stability of the secondary structure of the viroids. Importantly, the latter result supports the idea that the probing of a linear transcript is relevant to the determination of the structure of a circular viroid. It is important to note that, in this study, the structure of each viroid was determined by probing two different constructs that overlapped. The results of the overlapping regions of both transcripts for each viroid were compared, and the level of reproducibility was almost perfect in each case.
CONCLUDING REMARKS
In this report, the structures of seven RNA molecules corresponding to five distinct viroids that infect citrus trees were probed. This work, which is based on biochemical data, confirmed that viroids belonging to the Pospiviroidae family fold into long rod‐like structures regardless of their genus. Previously, the assumption of the adoption of this structure was based solely on computer modelling, with the exceptions of the viroids CEVd, PSTVd and PLMVd, which were probed in solution (Bussière et al., 2000; Domdey et al., 1978; Dubéet al., 2010; Gast et al., 1996; 1978, 1982). Moreover, the A‐rich motif was detected in all of the viroids probed during the course of this study, suggesting that it is conserved in all Pospiviroidae species, and that it is most probably important in their biology. Previously, this loop was designated as the left‐most pre‐melt loop located in the P region of PSTVd (Gast et al., 1996). It corresponds to loops 8 and 9 of PSTVd, which have been suggested to impair trafficking when mutated following the infection of Nicotiana benthamiana plants (Zhong et al., 2008). Other reports have claimed that the mutation of this region affects the infection mechanisms of both PSTVd (Wiesyk et al., 2011) and CEVd (Murcia et al., 2011; Visvader and Symons, 1986).
The analysis of RNA species for which crystal structures have been reported previously led to an estimation that the SHAPE‐supported structure prediction has an overall false negative rate (FNR) of 17% and a false discovery rate (FDR) of 21% (Kladwang et al., 2011). The use of SHAPE in combination with software prediction increases the accuracy as, in the absence of in‐solution data, FNR and FDR are 38% and 45%, respectively, for these RNA species. In the current study, comparison of the variant structure predicted by the RNAstructure software alone, with that obtained using the SHAPE data as input, revealed that, on average, 19.9% of nucleotides have a changed state, i.e. single‐stranded nucleotides appear to be double stranded or vice versa, or are base paired with a different nucleotide. Moreover, the SHAPE data help to reposition accessible nucleotides within single‐stranded regions that the software alone predicted to be double stranded. In addition, prediction in the absence of in‐solution data led to structures displaying, on average, 59.3% of accessible nucleotides, i.e. the nucleotides which appear in red in 3, 4, located in single‐stranded regions. Considering SHAPE data, this number increases to a mean value of 72.1%. A variation of only 0.9% was observed for the ‘non accessible’ nucleotides—the black nucleotides in 3, 4. The latter result is probably associated with the fact that all of these viroids are relatively GC rich, i.e. between 54% and 60%. It would be interesting to elucidate other viroid structures, including that of ASBVd, which has a relatively low GC content (∼38%). Importantly, combining the power of SHAPE with computer‐assisted structure prediction obtained using the RNAstructure software, which is specifically designed to easily integrate probing data, offers an interesting alternative method for the elucidation of RNA structure.
Despite the fact that only minor changes are observed when the structures obtained using the RNAstructure software with and without the SHAPE data are compared, the impact on the host can be dramatic. Recent studies on PTSVd have shown that the closing of loop 6, after a substitution of only three nucleotides, abolishes systemic trafficking (Takeda et al., 2011). Thus, an accurate knowledge of a viroid's structure is a key aspect in the elucidation of its pathogenicity. Clearly, SHAPE probing coupled to structure prediction is a fast, reliable and easily applicable standard procedure for the characterization of viroid secondary structures. We sincerely believe that this procedure should be of great assistance for the determination of the structures of many other RNA species.
EXPERIMENTAL PROCEDURES
Viroid sources
CBLVd.032 (NCBI accession number JF742600), CVd‐III.072 (NCBI accession number JF742602), CEVd.188 (NCBI accession number HQ284011), CEVd.189 (NCBI accession number HQ284014) and CEVd.190 (NCBI accession number HQ284011) were isolated from an 861‐S1 ‘Etrog’ citron seedling grafted on to a trifoliate orange rootstock showing both serious bark cracking and bent leaf symptoms. The rootstock originated from a diseased Citrus sinensis Osbeck cv. Robertson citrus tree from Hubei Province (China) that showed both bark cracking and symptoms of decline. HSVd.cit106 (NCBI accession number JF742601) was isolated from a diseased Citrus unshiu Marcov citrus tree from Jiangxi Province (China) that showed bent leaves. Each viroid sequence was dimerized and cloned into pGEM‐T vector (Promega Corp., Madison, WI, USA), and its identity was confirmed by DNA sequencing in both directions. CVd‐IV CA clone was synthesized as a dimer by the Geneart Company (Life Technologies c/o GENEART Inc., Burlington, ON, Canada) according to the reported sequence (Francis et al., 1995). All of these clones were clustered into five genera according to the phylogenetic tree and classification scheme of viroids defined previously (Elena et al., 2001; Flores et al., 2005) (Fig. 1).
In vitro transcription and purification of RNAs
Each clone was amplified using purified Pfu DNA polymerase from one of the plasmids described above, employing a pair of oligonucleotides in which the forward primer contained a T7 promoter and which produced a full‐length, double‐stranded DNA monomer. The entire collection of oligonucleotides used in this study is given in Table 1. For CEVd, PCR amplification was performed using 35 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s, which were preceded by a 3‐min denaturation at 94 °C, and were followed by a final extension of 7 min at 72 °C. All steps were performed using an Eppendorf Mastercycler Gradient (Mississauga, ON, Canada). PCR amplifications for all other viroids were performed using five cycles of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 30 s, followed by 30 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 30 s, and with the same denaturation and final extension steps. PCR products were electrophoresed in 1% agarose gels in order to confirm the synthesis of the expected DNA fragment. The full‐sized DNA of each sample was then gel purified. Transcriptions were performed as described previously using the purified DNA as template and purified T7 RNA polymerase (Dubéet al., 2010). All transcriptions reactions were fractionated by 5% denaturing (8 m urea) PAGE. The target RNA products were excised under UV shadowing and eluted overnight in elution buffer [500 mm NH4OAc, 1 mm ethylenediaminetetraacetic acid (EDTA) and 0.1% sodium dodecylsulphate (SDS)]. The RNA products were then ethanol precipitated, washed with 70% ethanol and dissolved in water before quantification by UV spectrophotometry.
5′‐end labelling of primers
Oligonucleotides (8 pmol) were 5′‐end labelled in the presence of 3.4 pmol [γ32P]‐ATP (6000 Ci/mmol; PerkinElmer NEN Radiochemicals, Boston, MA, USA) and 3 U of T4 polynucleotide kinase (USB Corporation, Cleveland, OH, USA). The reactions were performed at 37 °C for 60 min. The labelled primers were then purified using ProbeQuant™G‐50 Microcolumns according to the manufacturer's recommended protocol (GE Healthcare, Baie d'Urfe, QC, Canada).
SHAPE
SHAPE was performed on RNA aliquots (5 pmol) diluted in 8 µL of 0.5 × TE buffer [1 × stock: 10 mm Tris‐HCl (pH 7.5), 1 mm EDTA]. The reactions were incubated at 95 °C for 3 min, followed by 5 min on ice, prior to the addition of 1 µL of folding solution [500 mm Tris‐HCl (pH 7.5), 500 mm NaCl]. The mixtures were then incubated at 37 °C for 5 min prior to the addition of 1 µL of either 100 mm MgCl2 or water and incubation at 37 °C for 10 min. Finally, 1 µL of either 600 mm BzCN diluted in DMSO or DMSO was added. The latter was used as a negative control. There was no need to stop the reaction as it is instantaneous, and any remaining reagent is hydrolysed with a half‐life of 0.5 s at 37 °C. All reactions were then ethanol precipitated in the presence of glycogen and NaOAc, and the resulting pellets were washed with 70% ethanol and dried.
Primer extensions were performed using SuperScript III reverse transcriptase (Invitrogen, Burlington, ON, Canada) and radiolabelled oligonucleotides (see Table 1 for the complete set of oligonucleotides used to probe each viroid) complementary to the RNA. Initially, all of the pellets from the above reactions were dissolved in 12 µL of water, and 1 µL (1 pmol) of labelled oligonucleotide was added. The resulting mixtures were then incubated for 5 min at 65 °C, followed by 5 min at 37 °C and 1 min at 4 °C. A mixture of 4 µL of 5 × first strand buffer, 1 µL of 0.1 m dithiothreitol (DTT), 1 µL of 10 mm deoxynucleoside triphosphates (dNTPs) and 2 µL of DMSO was then added to each sample and the reactions were incubated at 50–61 °C for 1 min depending on the viroid species. The temperatures of the primer extension reactions varied from one viroid to another as not all of the reactions were productive at the same temperature. The control reactions generating RNA ladders also contained either 1 µL of 10 mm 2′,3′‐dideoxycytosine‐5′‐triphosphate (ddCTP) or 2 µL of 10 mm 2′,3′‐dideoxyguanosine‐5′‐triphosphate (ddGTP). Finally, 1 µL of Superscript III reverse transcriptase was added and extension reactions were incubated at 50–61 °C for 20 min prior to the addition of 2 µL of 2 m NaOH. The reactions were then incubated at 95 °C for 5 min and ethanol precipitated in the presence of glycogen and NaOAc. After washing with 70% ethanol, the precipitates were dissolved in 20 µL of 95% formamide, 10 mm EDTA solution, and the radioactivity was evaluated by Cerenkov counting. Equivalent amounts of radioactivity were fractionated on 8% denaturing PAGE gels. The gels were subsequently dried and visualized by exposure to phosphor imaging screens. All gels were analysed using the SAFA software (http://safa.stanford.edu) (Laederach et al., 2008). Each experiment was repeated twice, and the results are presented as an average of the two experiments. The signals obtained from different primers for each variant were normalized to a uniform scale as reported by Low and Weeks (2010). Briefly, the 2% most reactive signals of all intensities were removed from the pool, and the intensities of the next 8% most reactive were averaged. The intensity obtained for each nucleotide was then divided by this averaged value. This normalization step was performed manually using the Excel software program.
RNAstructure
The SHAPE constraints were incorporated into RNAstructure using the .txt files generated following SAFA analysis, and the normalization was performed using the Excel software program. The file comprises two columns: the first is the nucleotide number, and the second is the normalized SHAPE reactivity. The structure was then folded by choosing the menu option ‘Force ‐ > Read SHAPE Reactivity—Pseudo‐Energy Constraints’. In this case, the SHAPE intensities are converted into a pseudo‐free energy change term in the RNAstructure program during the course of the folding (Deigan et al., 2009).
RNAse probing
All RNA transcripts that were probed were synthesized as described above. Specifically, instead of adding BzCN to the reaction, the RNAs were cleaved by adding RNase V1, T1 or A in solution with MgCl2. The mixtures were incubated for 1 min at 37 °C in the presence of RNase V1 (Pierce Molecular Biology, Rockford, IL, USA) at a final concentration of 0.002 U/µL, RNase T1 (Roche, Laval, QC, Canada) at a final concentration of 0.2 U/µL or RNase A (USB Corporation) at a final concentration of 1 pg/µL. The mixtures were quenched by the addition of 90 µL of water and the products were purified by phenol–chlorofom extraction and ethanol precipitation. The reactions were then reverse transcribed and treated as described for SHAPE probing. Once again, SAFA software analysis and signal normalization were performed.
Supporting information
Fig. S1 Predicted secondary structures of the three Citrus exocortis viroid (CEVd) variants used in this study. The most stable structures of CEVd.188, as predicted by Mfold (A) and RNAstructure 5.1 (B), of CEVd.189, as predicted by Mfold (C) and RNAstructure 5.1 (D), and of CEVd.190, as predicted by Mfold (E) and RNAstructure 5.1 (F), are shown.
Fig. S2 Secondary structures of Citrus exocortis viroid (CEVd) clones CEVd.188 (A), CEVd.189 (B) and CEVd.190 (C) as determined by integration of the SHAPE data without Mg ions into the RNAstructure 5.1 software program. TL (Terminal Left), P (Pathogenicity), C (Central), V (Variable) and TR (Terminal Right) refer to the structural domains defined in Fig. 3.
Fig. S3 RNase probing results performed on Citrus exocortis viroid clone CEVd.188. The nucleotides are highlighted according to the RNase mapping data; the green nucleotides are cleaved by RNase V1, the red ones by RNase T1 and the blue ones by RNase A. TL (Terminal Left), P (Pathogenicity), C (Central), V (Variable) and TR (Terminal Right) refer to the structural domains defined in Fig. 3.
Fig. S4 Predicted secondary structures of the Citrus viroid III variant CVd‐III.072 and Citrus bent leaf viroid variant CBLVd.032. The most stable structures of CVd‐III.072, as predicted by Mfold (A) and RNAstructure 5.1 (B), and of CBLVd.032, as predicted by Mfold (C) and RNAstructure 5.1 (D), are shown.
Fig. S5 Predicted secondary structures of the Hop stunt viroid variant HSVd.cit106 and Citrus viroid IV (CVd‐IV) variant. The most stable structures of HSVd.cit106, as predicted by Mfold (A) and RNAstructure 5.1 (B), and of CVd‐IV, as predicted by Mfold (C) and RNAstructure 5.1 (D), are shown.
Fig. S6 Secondary structures of the Citrus viroid III variant CVd‐III.072 (A), Citrus bent leaf viroid variant CBLVd.032 (B), Hop stunt viroid variant HSVd.cit106 (C) and Citrus viroid IV (CVd‐IV) variant (D), as determined by the integration of the SHAPE data without Mg ions into the RNAstructure 5.1 software program. TL (Terminal Left), P (Pathogenicity), C (Central), V (Variable) and TR (Terminal Right) refer to the structural domains defined in Fig. 4.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
This work was supported by a grant from the Natural Sciences and Engineering Research Council (NSERC Canada, grant number 155219‐07) to Jean‐Pierre Perreault, and a grant from the National Science Foundation of China (No. 31071662) and funding from Hubei Province (No. 2010CBB 02001) to Xu Wenxing. The RNA group is supported by grants from the Université de Sherbrooke and the Canadian Institutes of Health Research (CIHR, grant number PRG‐80169). JPP holds the Canada Research Chair in Genomics and Catalytic RNA and is a member of the Centre de Recherche Clinique Étienne‐Lebel.
REFERENCES
- Ashulin, L. , Lachman, O. , Hadas, R. and Bar‐Joseph, M. (1991) Nucleotide sequence of a new viroid species, citrus bent leaf viroid (CBLVd) isolated from grapefruit in Israel. Nucleic Acids Res. 19, 4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudry, D. , Bussière, F. , Lareau, F. , Lessard, C. and Perreault, J.P. (1995) The RNA of both polarities of the peach latent mosaic viroid self‐cleaves in vitro solely by single hammerhead structures. Nucleic Acids Res. 23, 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussière, F. , Lafontaine, D. and Perreault, J.P. (1996) Compilation and analysis of viroid and viroid‐like RNA sequences. Nucleic Acids Res. 24, 1793–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussière, F. , Ouellet, J. , Côté, F. , Lévesque, D. and Perreault, J.P. (2000) Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J. Virol. 74, 2647–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffai, M. , Serra, P. , Gandia, M. , Hernandez, C. and Duran‐Vila, N. (2007) Molecular characterization of CEVd strains that induce different phenotypes in Gynura aurantiaca: structure–pathogenicity relationships. Arch. Virol. 152, 1283–1294. [DOI] [PubMed] [Google Scholar]
- Deigan, K.E. , Li, T.W. , Mathews, D.H. and Weeks, K.M. (2009) Accurate SHAPE‐directed RNA structure determination. Proc. Natl. Acad. Sci. USA, 106, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domdey, H. , Jank, P. , Sanger, L. and Gross, H.J. (1978) Studies on the primary and secondary structure of potato spindle tuber viroid: products of digestion with ribonuclease A and ribonuclease T1, and modification with bisulfite. Nucleic Acids Res. 5, 1221–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé, A. , Baumstark, T. , Bisaillon, M. and Perreault, J.P. (2010) The RNA strands of the plus and minus polarities of peach latent mosaic viroid fold into different structures. RNA, 16, 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé, A. , Bolduc, F. , Bisaillon, M. and Perreault, J.P. (2011) Mapping studies of the Peach latent mosaic viroid reveal novel structural features. Mol. Plant. Pathol. 12, 688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena, S.F. , Dopazo, J. , de la Pena, M. , Flores, R. , Diener, T.O. and Moya, A. (2001) Phylogenetic analysis of viroid and viroid‐like satellite RNAs from plants: a reassessment. J. Mol. Evol. 53, 155–159. [DOI] [PubMed] [Google Scholar]
- Fadda, Z. , Daros, J.A. , Fagoaga, C. , Flores, R. and Duran‐Vila, N. (2003) Eggplant latent viroid, the candidate type species for a new genus within the family Avsunviroidae (hammerhead viroids). J. Virol. 77, 6528–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, R. , Randles, J.W. , Owens, R.A. , Bar‐Joseph, M. and Diener, T.O. (2005) Viroids In: Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses (Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U. and Ball A.L., eds), pp. 1145–1159. San Diego: CA: Elsevier Academic Press. [Google Scholar]
- Francis, M.I. , Szychowski, J.A. and Semancik, J.S. (1995) Structural sites specific to citrus viroid groups. J. Gen. Virol. 76, 1081–1089. [DOI] [PubMed] [Google Scholar]
- Gast, F.U. , Kempe, D. , Spieker, R.L. and Sanger, H.L. (1996) Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicons provides evidence for central non‐canonical base‐pairs, large A‐rich loops, and a terminal branch. J. Mol. Biol. 262, 652–670. [DOI] [PubMed] [Google Scholar]
- Gross, H.J. , Domdey, H. , Lossow, C. , Jank, P. , Raba, M. , Alberty, H. and Sanger, H.L. (1978) Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature, 273, 203–208. [DOI] [PubMed] [Google Scholar]
- Gross, H.J. , Krupp, G. , Domdey, H. , Raba, M. , Jank, P. , Lossow, C. , Alberty, H. , Ramm, K. and Sanger, H.L. (1982) Nucleotide sequence and secondary structure of citrus exocortis and chrysanthemum stunt viroid. Eur. J. Biochem. 121, 249–257. [DOI] [PubMed] [Google Scholar]
- Keese, P. and Symons, R.H. (1985) Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. USA, 82, 4582–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang, W. , VanLang, C.C. , Cordero, P. and Das, R. (2011) Understanding the errors of SHAPE‐directed RNA structure modeling. Biochemistry, 50, 8049–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laederach, A. , Das, R. , Vicens, Q. , Pearlman, S.M. , Brenowitz, M. , Herschlag, D. and Altman, R.B. (2008) Semiautomated and rapid quantification of nucleic acid footprinting and structure mapping experiments. Nat. Protoc. 3, 1395–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, J.T. and Weeks, K.M. (2010) SHAPE‐directed RNA secondary structure prediction. Methods, 52, 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquida, B. , Beckert, B. and Jossinet, F. (2010) Exploring RNA structure by integrative molecular modelling. New Biotechnol. 27, 170–183. [DOI] [PubMed] [Google Scholar]
- Mortimer, S.A. and Weeks, K.M. (2008) Time‐resolved RNA SHAPE chemistry. J. Am. Chem. Soc. 130, 16178–16180. [DOI] [PubMed] [Google Scholar]
- Murcia, N. , Bernad, L. , Duran‐Vila, N. and Serra, P. (2011) Two nucleotide positions in the Citrus exocortis viroid RNA associated with symptom expression in Etrog citron but not in experimental herbaceous hosts. Mol. Plant. Pathol. 12, 203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, B. and Flores, R. (1997) Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of self‐cleaving viroids with hammerhead ribozymes. Proc. Natl. Acad. Sci. USA, 94, 11262–11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, J.S. and Mathews, D.H. (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics, 11, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau, L. and Pelchat, M. (2006) The Subviral RNA Database: a toolbox for viroids, the hepatitis delta virus and satellite RNAs research. BMC Microbiol. 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoric, D. , Conerly, M. , Szychowski, J.A. and Semancik, J.S. (2001) CEVd‐induced symptom modification as a response to a host‐specific temperature‐sensitive reaction. Virology, 280, 115–123. [DOI] [PubMed] [Google Scholar]
- Stasys, R.A. , Dry, I.B. and Rezaian, M.A. (1995) The termini of a new citrus viroid contain duplications of the central conserved regions from two viroid groups. FEBS Lett. 358, 182–184. [DOI] [PubMed] [Google Scholar]
- Steen, K.A. , Malhotra, A. and Weeks, K.M. (2010) Selective 2′‐hydroxyl acylation analyzed by protection from exoribonuclease. J. Am. Chem. Soc. 132, 9940–9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger, G. , Hofmann, H. , Fortsch, J. , Gross, H.J. , Randles, J.W. , Sanger, H.L. and Riesner, D. (1984) Conformational transitions in viroids and virusoids: comparison of results from energy minimization algorithm and from experimental data. J. Biomol. Struct. Dyn. 2, 543–571. [DOI] [PubMed] [Google Scholar]
- Symons, R.H. (1981) Avocado sunblotch viroid: primary sequence and proposed secondary structure. Nucleic Acids Res. 9, 6527–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szychowski, J.A. , Vidalakis, G. and Semancik, J.S. (2005) Host‐directed processing of Citrus exocortis viroid. J. Gen. Virol. 86, 473–477. [DOI] [PubMed] [Google Scholar]
- Takeda, R. , Petrov, A.I. , Leontis, N.B. and Ding, B. (2011) A three‐dimensional RNA motif in Potato spindle tuber viroid mediates trafficking from palisade mesophyll to spongy mesophyll in Nicotiana benthamiana . Plant Cell, 23, 258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader, J.E. and Symons, R.H. (1985) Eleven new sequence variants of citrus exocortis viroid and the correlation of sequence with pathogenicity. Nucleic Acids Res. 13, 2907–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader, J.E. and Symons, R.H. (1986) Replication of in vitro constructed viroid mutants: location of the pathogenicity‐modulating domain of citrus exocortis viroid. EMBO J. 5, 2051–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesyk, A. , Candresse, T. , Zagorski, W. and Gora‐Sochacka, A. (2011) Use of randomly mutagenized genomic cDNA banks of potato spindle tuber viroid to screen for viable versions of the viroid genome. J. Gen. Virol. 92, 457–466. [DOI] [PubMed] [Google Scholar]
- Wilkinson, K.A. , Merino, E.J. and Weeks, K.M. (2006) Selective 2′‐hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616. [DOI] [PubMed] [Google Scholar]
- Zhong, X. , Archual, A.J. , Amin, A.A. and Ding, B. (2008) A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell, 20, 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Predicted secondary structures of the three Citrus exocortis viroid (CEVd) variants used in this study. The most stable structures of CEVd.188, as predicted by Mfold (A) and RNAstructure 5.1 (B), of CEVd.189, as predicted by Mfold (C) and RNAstructure 5.1 (D), and of CEVd.190, as predicted by Mfold (E) and RNAstructure 5.1 (F), are shown.
Fig. S2 Secondary structures of Citrus exocortis viroid (CEVd) clones CEVd.188 (A), CEVd.189 (B) and CEVd.190 (C) as determined by integration of the SHAPE data without Mg ions into the RNAstructure 5.1 software program. TL (Terminal Left), P (Pathogenicity), C (Central), V (Variable) and TR (Terminal Right) refer to the structural domains defined in Fig. 3.
Fig. S3 RNase probing results performed on Citrus exocortis viroid clone CEVd.188. The nucleotides are highlighted according to the RNase mapping data; the green nucleotides are cleaved by RNase V1, the red ones by RNase T1 and the blue ones by RNase A. TL (Terminal Left), P (Pathogenicity), C (Central), V (Variable) and TR (Terminal Right) refer to the structural domains defined in Fig. 3.
Fig. S4 Predicted secondary structures of the Citrus viroid III variant CVd‐III.072 and Citrus bent leaf viroid variant CBLVd.032. The most stable structures of CVd‐III.072, as predicted by Mfold (A) and RNAstructure 5.1 (B), and of CBLVd.032, as predicted by Mfold (C) and RNAstructure 5.1 (D), are shown.
Fig. S5 Predicted secondary structures of the Hop stunt viroid variant HSVd.cit106 and Citrus viroid IV (CVd‐IV) variant. The most stable structures of HSVd.cit106, as predicted by Mfold (A) and RNAstructure 5.1 (B), and of CVd‐IV, as predicted by Mfold (C) and RNAstructure 5.1 (D), are shown.
Fig. S6 Secondary structures of the Citrus viroid III variant CVd‐III.072 (A), Citrus bent leaf viroid variant CBLVd.032 (B), Hop stunt viroid variant HSVd.cit106 (C) and Citrus viroid IV (CVd‐IV) variant (D), as determined by the integration of the SHAPE data without Mg ions into the RNAstructure 5.1 software program. TL (Terminal Left), P (Pathogenicity), C (Central), V (Variable) and TR (Terminal Right) refer to the structural domains defined in Fig. 4.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


