SUMMARY
Polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs) are the major enzymes involved in the biosynthesis of secondary metabolites, which have diverse activities, including roles as pathogenicity/virulence factors in plant pathogenic fungi. These enzymes are activated by 4′‐phosphopantetheinylation at the conserved serine residues, which is catalysed by 4′‐phosphopantetheinyl transferase (PPTase). PPTase is also required for primary metabolism (α‐aminoadipate reductase, AAR). In the genome sequence of the cereal fungal pathogen Cochliobolus sativus, we identified a gene (PPT1) orthologous to the PPTase‐encoding genes found in other filamentous ascomycetes. The deletion of PPT1 in C. sativus generated mutants (Δppt1) that were auxotrophic for lysine, unable to synthesize melanin, hypersensitive to oxidative stress and significantly reduced in virulence to barley cv. Bowman. To analyse the pleiotropic effects of PPT1, we also characterized deletion mutants for PKS1 (involved in melanin synthesis), AAR1 (for AAR) and NPS6 (involved in siderophore‐mediated iron metabolism). The melanin‐deficient strain (Δpks1) showed no differences in pathogenicity and virulence compared with the wild‐type strain. Lysine‐auxotrophic mutants (Δaar1) induced spot blotch symptoms, as produced by the wild‐type strain, when inoculated on wounded barley leaves or when lysine was supplemented. The Δnps6 strain showed a slightly reduced virulence compared with the wild‐type strain, but exhibited significantly higher virulence than the Δppt1 strain. Our results suggest that an unknown virulence factor, presumably synthesized by PKSs or NRPSs which are activated by PPTase, is directly responsible for high virulence of C. sativus on barley cv. Bowman.
INTRODUCTION
Fungi produce various secondary metabolites with diverse activities (Calvo et al., 2002; Keller et al., 2005; Stack et al., 2007). Polyketides (PKs) and nonribosomal peptides (NRPs) are among the secondary metabolites involved in pathogenicity and virulence (or avirulence) of plant pathogenic fungi, such as Cochliobolus heterostrophus (Baker et al., 2006; Lee et al., 2005; Oide et al., 2006), Cochliobolus carbonum (Walton, 2006), Alternaria alternata (Johnson et al., 2000) and Magnaporthe oryzae (Bohnert et al., 2004; Howard and Valent, 1996). Several genes for polyketide synthases (PKSs) and/or nonribosomal peptide synthetases (NRPSs) have been cloned, characterized and shown to be required for the biosynthesis of PKs and NRPs, which function as pathogenicity/virulence factors. For example, two PKS‐encoding genes, PKS1 and PKS2, are involved in the synthesis of T‐toxin, a virulence factor produced by the southern corn leaf blight pathogen C. heterostrophus (Baker et al., 2006). In M. grisea, PK‐derived melanin is required for its pathogenicity (Howard and Valent, 1996). Some mycotoxins are also synthesized by PKSs. For example, aflatoxins, mainly produced by Aspergillus flavus and A. parasiticus, are a group of PK‐derived compounds which contaminate agricultural commodities and are toxic to livestock as well as humans (Evans et al., 2011; Hoffmeister and Keller, 2007). A genome‐wide search identified 23, 15 and 25 genes for PKSs in M. grisea, Fusarium graminearum and C. heterostrophus, respectively (Cuomo et al., 2007; Kroken et al., 2003). NRPs are another class of secondary metabolites synthesized by NRPSs in bacteria and fungi (Eisfeld, 2009). Some of these NRPs play critical roles in pathogen–host interactions. For example, AM‐toxin, a cyclic peptide host‐specific toxin produced by the apple pathotype of A. alternata, is synthesized by an NRPS‐encoding gene, AMT1 (Johnson et al., 2000). Another example is HC‐toxin, a cyclic tetrapeptide produced by race 1 of the northern corn leaf blight fungus C. carbonum, whose biosynthesis requires an NRPS (Walton, 2006). Twelve NRPS‐encoding genes (NPS) have been identified in the C. heterostrophus genome, and one, NPS6, has been shown to be involved in virulence and resistance to oxidative stress (Lee et al., 2005). NPS6 was later found to be involved in iron uptake with conserved function among filamentous ascomycetes (Oide et al., 2006). In the human pathogen, A. fumigatus, at least 14 NPS genes have been found in the genome (Stack et al., 2007), and 14 and 20 NPS genes have been identified in two other important plant pathogenic fungi, M. grisea and F. graminearum, respectively (Cuomo et al., 2007). Recently, hybrid PKS–NRPSs have been discovered in several fungi, such as M. grisea, C. heterostrophus and F. graminearum (Collemare et al., 2008; Lee et al., 2005; Turgeon et al., 2008). Magnaporthe grisea seems to be rich in PKS–NRPS hybrids because 10 PKS–NRPS‐encoding genes were found in the whole genome sequence, whereas only one and two PKS–NRPS genes were identified in Neurospora crassa and F. graminearum, respectively (Collemare et al., 2008). Interestingly, ACE1 (Avirulence Conferring Enzyme1), a hybrid PKS–NRPS produced by M. grisea, is classified as an avirulence determinant (Bohnert et al., 2004).
Despite the large number and diversity of PKSs, NRPSs and hybrid PKS–NRPSs produced by fungi, all except for type III PKSs require a post‐translational modification by 4′‐phosphopantetheinylation at the conserved serine residues before they become active (Horbach et al., 2009; Marquez‐Fernandez et al., 2007; Stack et al., 2007). This enzymatic activation is controlled by members of the 4′‐phosphopantetheinyl transferase (PPTase) family (Horbach et al., 2009; Marquez‐Fernandez et al., 2007). Based on the primary sequences and substrate specificities, three groups of PPTase have been identified, including bacterial AcpS‐type PPTases, Sfp‐type PPTases and PPTases that are integral domains of type I fatty acid synthase FASII (Horbach et al., 2009; Marquez‐Fernandez et al., 2007; Mootz et al., 2002; Quadri et al., 1998). Among these three groups of PPTases, Sfp‐type PPTases are mainly associated with the biosynthesis of PKs and NRPs (Mootz et al., 2002; Quadri et al., 1998). A PPTase‐encoding gene (CfwA/NpgA) has been identified to be involved in the primary and secondary metabolism of A. nidulans (Keszenman‐Pereyra et al., 2003; Marquez‐Fernandez et al., 2007). Mutants of A. nidulans deleted in CfwA/NpgA fail to synthesize lysine and become lysine auxotrophs because the α‐aminoadipate reductase (AAR) encoded by the gene Lys2 is inactivated (Marquez‐Fernandez et al., 2007). In addition, these deletion mutants were unable to synthesize several PKs and NRPs, including sterigmatocystin, shamixantone, dehydroaustinol, pigments and siderophores (Marquez‐Fernandez et al., 2007; Oberegger et al., 2003). CfwA/NpgA is also required for asexual development, penicillin biosynthesis and the activation of all PKSs and NRPKs in A. nidulans (Calvo et al., 2002; Keszenman‐Pereyra et al., 2003; Marquez‐Fernandez et al., 2007). Recently, Horbach et al. (2009) cloned and characterized a PPTase‐encoding gene (PPT1) (orthologous to CfwA/NpgA) from the maize anthracnose fungus Colletotrichum graminicola, and showed that Δppt1 mutants were reduced in asexual sporulation and oxidative stress tolerance, failed to infect the plant host and lost the ability to synthesize lysine, melanin and siderophores. They also demonstrated that Δppt1 mutants of the rice blast fungus M. oryzae were unable to penetrate and cause disease in rice (Horbach et al., 2009). However, information about the role(s) of PPTases in the pathogenicity/virulence of other plant pathogenic fungi is still lacking.
Cochliobolus sativus (Anamorph: Bipolaris sorokiniana) is the causal agent of several important cereal diseases, including spot blotch, common root rot and black point (Kumar et al., 2002; Mathre, 1997; Wiese, 1977). Three pathotypes of C. sativus were identified by Valjavec‐Gratian and Steffenson (1997a) on the basis of their differential virulence patterns on three barley genotypes (ND5883, Bowman and B112). These three pathotypes are designated as 0, 1 and 2. Pathotype 0 isolates show low virulence on all three barley genotypes. Pathotype 1 isolates show high virulence on ND5883, but low virulence on other barley genotypes. Pathotype 2 isolates show high virulence on Bowman, but low virulence on ND5883 and B112. Genetic analysis and molecular mapping indicated that a single locus (VHv1) controls the high virulence of the pathotype 2 isolate ND90Pr on Bowman (Valjavec‐Gratian and Steffenson, 1997b; Zhong et al., 2002). However, the exact nature of this virulence factor remains unknown.
In several species of the genus Cochliobolus, host‐selective toxins (HSTs) are common and have been identified as the primary pathogenicity or virulence factors. Examples include HC‐toxin produced by C. carbonum, victorin produced by Cochliobolus victoria and T‐toxin produced by C. heterostrophus (Baker et al., 2006; Walton, 2006; Wolpert et al., 2002). Cochliobolus sativus produces typical necrosis and chlorosis on susceptible hosts, and thus HSTs are assumed to be involved in the pathogenesis. However, no HSTs were identified from the culture filtrates of isolate ND90Pr, although attempts have been made (S. Zhong et al., unpublished data). We hypothesize that isolate ND90Pr produces a virulence factor, presumably synthesized by PKSs or NRPSs during infection, which makes Bowman susceptible. To test this hypothesis, we generated and characterized knockout mutants for the PPT1 orthologue identified in the C. sativus genome.
RESULTS
Identification of an orthologue of PPT1 in C. sativus
A blast search was conducted against the C. sativus genome sequence provided by the Joint Genome Institute (JGI) with the PPT1 gene sequence of C. graminicola (Horbach et al., 2009), identifying a single copy PPT1 orthologue [deposited as HQ830035 in the National Center for Biotechnology Information (NCBI) GenBank database]. The predicted protein encoded by the C. sativus PPT1 homologue consists of 375 amino acids, which are 97%, 83%, 32%, 35%, 33% and 30% identical to the PPTases in C. heterostrophus (IPR008278, JGI), Pyrenophora tritici‐repentis (XP_001930737.1, NCBI), M. oryzae (XP_001522742.1, NCBI), Gibberella zeae (XP_388955.1, NCBI), C. graminicola (DQ028305, NCBI) and the A. fumigatus CfwA protein (EDP54396.1, NCBI), respectively.
Deletion of PPT1 in C. sativus leads to auxotrophy for lysine, nonmelanization, hypersensitivity to iron depletion and hydrogen peroxide (H2O2), and loss of virulence
To characterize the function of PPT1 in C. sativus, we replaced the coding region of PPT1 with a hygromycin B resistance gene cassette (hph) using the split marker system (Catlett et al., 2003) and generated three independent deletion mutants (Δppt1‐2, Δppt1‐3 and Δppt1‐5). These deletion mutants were confirmed by Southern hybridization analysis of EcoRV‐digested genomic DNA from the wild‐type and Δppt1 strains using a probe amplified from the 5′ end flanking sequence of C. sativus PPT1 (Fig. 1A). As indicated in Fig. 1B, the 3.5‐kb fragment in the wild‐type strain (ND90Pr) was replaced by the 6.1‐kb fragment in the Δppt1 strains (Δppt1‐2, Δppt1‐3, Δppt1‐5). To confirm gene inactivation at the transcription level, reverse transcription‐polymerase chain reaction (RT‐PCR) experiments were performed using primers (RT‐PPT1‐F/RT‐PPT1‐R) (Table 1) specific to PPT1 and primers (RT‐ACT‐F/RT‐ACT‐R) (Table 1) corresponding to the constitutively expressed β‐actin‐encoding gene (ACT) (control). Using RT‐ACT‐F/RT‐ACT‐R as primers, a 150‐bp fragment was amplified from cDNA of the wild‐type strain, as well as the various mutants analysed. However, when primers RT‐PPT1‐F/RT‐PPT1‐R were used, the 506‐bp fragment of C. sativus PPT1 was amplified from the cDNA of the wild‐type strain, but not from the cDNA of the Δppt1 strains (Fig. S1, see Supporting Information). These data indicate that PPT1 was successfully deleted in the Δppt1 strains.
Figure 1.
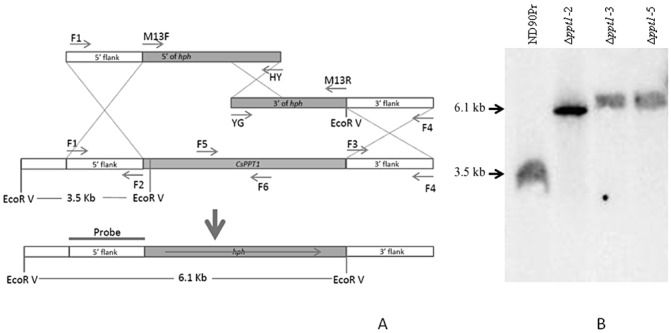
Generation of Δppt1 strains of Cochliobolus sativus. (A) Diagram showing replacement of the PPT1 gene by a 2.6‐kb fragment carrying the Escherichia coli hygromycin phosphotransferase gene (hph) using the split‐marker system (Catlett et al., 2003). (B) Southern hybridization of EcoRV‐digested genomic DNA from the wild‐type and Δppt1 strains using a probe amplified with primers CsPPT1‐F1 and CsPPT1‐F2. The 3.5‐kb fragment in the wild‐type strain (ND90Pr) was replaced by the 6.1‐kb fragment in the Δppt1 strains (Δppt1‐2, Δppt1‐3, Δppt1‐5).
Table 1.
Primers used in this study.
| Primer name | Primer sequence (5′–3′)* |
|---|---|
| M13F | GACGTTGTAAAACGACGGCCAGTG |
| M13R | CACAGGAAACAGCTATGACCATGA |
| HY | GGATGCCTCCGCTCGAAGTA |
| YG | CGTTGCAAGACCTGCCTGAA |
| CsPPT1‐F1 | AACTCCGAACATGACGCATT |
| CsPPT1‐F2 | CACTGGCCGTCGTTTTACAACGTCCCTGCTTGGTGTGAGATGTT |
| CsPPT1‐F3 | TCATGGTCATAGCTGTTTCCTGTGTCACGATGAAGGCCGATG |
| CsPPT1‐F4 | GGGGTATGAGGGAGAAGACG |
| CsPPT1‐F5 | GGCACAAGCAACTCAGCAT |
| CsPPT1‐F6 | ACCCTTGCAACGCTCATAAT |
| PPT1‐F | CCACCGATGATAGTGGTGTG |
| PPT1‐R | GGGGTATGAGGGAGAAGACG |
| RT‐PPT1‐F | CCACCGATGATAGTGGTGTG |
| RT‐PPT1‐R | GGGGTATGAGGGAGAAGACG |
| CsAAR‐F1 | GTCTCGGATTTCAGGTCAGC |
| CsAAR‐F2 | CACTGGCCGTCGTTTTACAACGTCACTGCCCCTGTACCACTTTG |
| CsAAR‐F3 | TCATGGTCATAGCTGTTTCCTGTGCGGTCTGGAGGGTATCTTGA |
| CsAAR‐F4 | TCGAGCATTTTGACTGGATG |
| CsPKS1‐F1 | AAGATGTCAACGCCTGGAAG |
| CsPKS1‐F2 | CACTGGCCGTCGTTTTACAACGTCAGAGATGTTGCCAGGAGCAG |
| CsPKS1‐F3 | TCATGGTCATAGCTGTTTCCTGTGGATATCGAGACTGCGCTTGG |
| CsPKS1‐F4 | AGTGAGGAAGGAGCCATGAA |
| CsPKS1‐F5 | TGGTATGGTCATGGGTCTCA |
| CsPKS1‐F6 | ACGCATGGAACCAAGGATAG |
| CsNPS6‐F1 | CGCTGCGACACAGTGTATTT |
| CsNPS6‐F2 | CACTGGCCGTCGTTTTACAACGTCTGCTACTGACGTCCATCTCG |
| CsNPS6‐F3 | TCATGGTCATAGCTGTTTCCTGTGCGGATAAGGTGCAGTGGTTC |
| CsNPS6‐F4 | GAGGTTACGCTGGGTGCTTA |
| CsNPS6‐F6 | GGCTGTGGGTGAGGATACAT |
| RT‐Actin‐F | GTATGGGCCAAAAGGACTCA |
| RT‐Actin‐R | CACGCAGCTCGTTGTAGAAG |
| HYG1‐F | GAATTCAGCGAGAGCCTGAC |
| HYG1‐R | GATGTTGGCGACCTCGTATT |
Italic sequences are complementary to M13F and M13R sequences, respectively.
The wild‐type strain showed normal growth on lysine‐deficient minimal medium (MM) on which Δppt1 strains were unable to grow (Fig. 2). The growth of Δppt1 strains was restored when lysine was supplemented to MM or when rich media, such as potato dextrose agar (PDA), were used (Fig. 2). In contrast with the wild‐type strain, the Δppt1 strains produced white colonies and nonmelanized conidia on all media used (Fig. 2). These results indicate that PPT1 is required for the biosynthesis of lysine and melanin.
Figure 2.
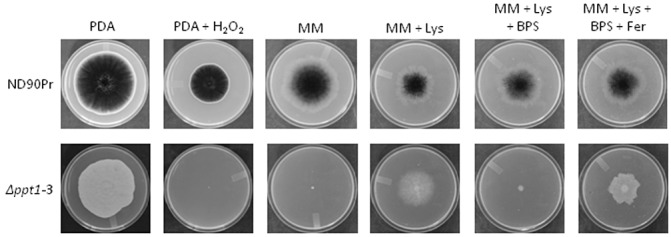
Fungal growth on different agar plates. Mycelial plugs of uniform size (2 mm × 2 mm) from 3‐day‐old fresh culture were inoculated on the centres of the plates amended with the indicated agents and grown for 6 days at 25 °C. BPS, bathophenanthroline disulphonic acid; Fer, iron‐free ferrichrome; Lys, lysine; MM, minimal medium; PDA, potato dextrose agar.
The quantification of the conidia produced by the wild‐type and Δppt1 strains on PDA plates indicated that the Δppt1 strains showed significantly reduced conidiation, as only one‐quarter as many conidia were produced compared with the wild‐type strain (Fig. 3). However, no significant differences were observed in the size and shape of the conidia produced by the Δppt1 strains and the wild‐type on PDA or MM with lysine (data not shown). In addition, the germination rates of conidia from the wild‐type and Δppt1 strains on different agar plates and leaf surfaces were similar (Fig. 4). These data indicate that PPT1 is involved in conidiation, but does not affect the size, morphology and germination of conidia in C. sativus.
Figure 3.
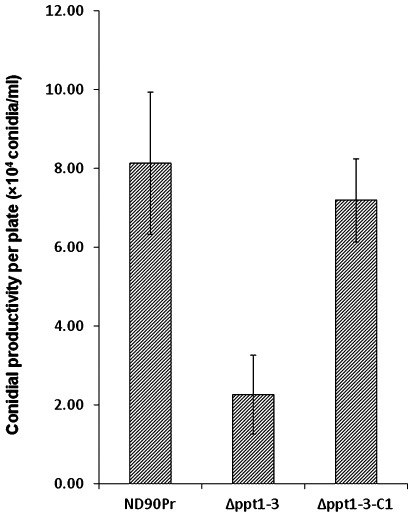
Conidial productivity of wild‐type (ND90Pr), PPT1‐deleted mutant (Δppt1‐3) and Δppt1 complementation mutant (Δppt1‐3‐C1) grown on potato dextrose agar (PDA) plates for 6 days at 25 °C in a cycle of 14 h of light and 10 h of darkness. Error bars indicate the standard deviation.
Figure 4.
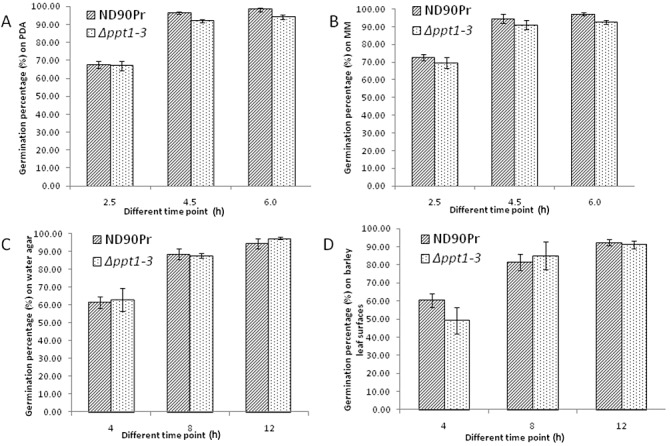
Conidial germination rates of the wild‐type strain (ND90Pr) and the PPT1‐deleted mutant (Δppt1‐3) on different agar plates and barley leaves at different time points. (A) Germination on potato dextrose agar (PDA). (B) Germination on minimal medium (MM). (C) Germination on water agar. (D) Germination on barley leaves. Error bars indicate the standard deviation.
Siderophores are NRPs that mediate iron uptake for fungal growth under iron‐limiting conditions (Haas et al., 2008) and confer tolerance to oxidative stresses (Oide et al., 2006). To test the role of PPT1 in the regulation of iron acquisition, the wild‐type and Δppt1 strains were grown on MM containing 200 µm of bathophenanthroline disulphonic acid (BPS, an iron chelator). As illustrated in Fig. 2, the wild‐type strain showed normal growth, whereas the growth of Δppt1 strains was completely inhibited. However, the growth of Δppt1 strains was restored by adding the iron‐free siderophore ferrichrome to MM with BPS (Fig. 2). This restored growth was caused by the ferrichrome outcompeting BPS for the chelation of free iron, and the ferrichrome chelate is a usable source of iron for filamentous fungi (Haas et al., 2008). In addition, the growth of Δppt1 strains was completely inhibited on PDA with H2O2 (10 mm), whereas the wild‐type strain was able to grow, but at a significantly reduced rate (Fig. 2). These results indicate that Δppt1 strains are unable to produce the siderophores that are required for iron uptake and for tolerance to oxidative stress (H2O2) during growth.
The wild‐type strain ND90Pr caused severe spot blotch disease when spray inoculated on barley cv. Bowman (Fig. 5). However, no obvious spot blotch symptoms were observed when conidia of Δppt1 strains were inoculated on seedling plants of Bowman without lysine supplementation (Fig. 5A). Based on the 1–9 disease rating scale of Fetch and Steffenson (1999), the disease levels caused by the wild‐type and Δppt1 strains were rated as 8–9 and 1–2, respectively, 5 days after inoculation (dai). In addition, the Δppt1 strains produced more infection sites on inoculated barley leaves when lysine was supplemented during and after inoculation (Fig. 5A). Nevertheless, the Δppt1 strains were unable to produce the severe spot blotch symptoms induced by the wild‐type strain (ND90Pr) regardless of lysine supplementation (Fig. 5A), indicating that the infection process was impaired. To determine whether the deletion of PPT1 affects infection structure differentiation of the fungus, we examined leaf segments of barley cv. Bowman sampled at 12, 24 and 48 h after inoculation (hai) with conidia of the wild‐type and Δppt1 strains. Microscopic observations showed that both of the strains formed appressorium‐like structures at 12 and 24 hai with lysine supplementation (Fig. 6), although the Δppt1 strain showed a reduced frequency of appressorium formation compared with the wild‐type strain (Fig. 7). At 48 hai, the wild‐type produced infection hyphae which branched and proliferated inside the plant tissue causing massive cell death (stained dark blue), whereas the Δppt1 strain formed fewer infection hyphae, which occasionally caused the death of a few plant cells (Fig. 6). These data indicate that Δppt1 strains are pathogenic (able to penetrate), but have lost the ability to continuously infect plant tissue and cause disease on barley cv. Bowman.
Figure 5.
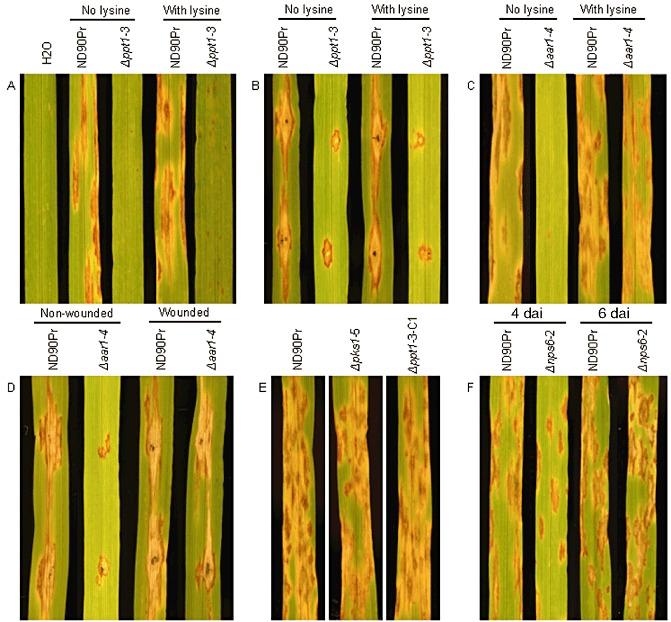
Disease symptoms of barley cv. Bowman inoculated with conidia (A, C, E and F) or small mycelial plugs (B and D) of the wild‐type (ND90Pr), knockout mutants (Δppt1‐3, Δaar1‐4, Δpks1‐5 and Δnps6‐2) and complementation strain (Δppt1‐3‐C1). All photographs were taken at 5 days after inoculation (dai), except for (F), which was taken after 4 and 6 dai. Lysine was supplemented at 50 µg/mL when needed. A syringe needle was used to make wounds on leaves for inoculation with small mycelial plugs (B and D).
Figure 6.
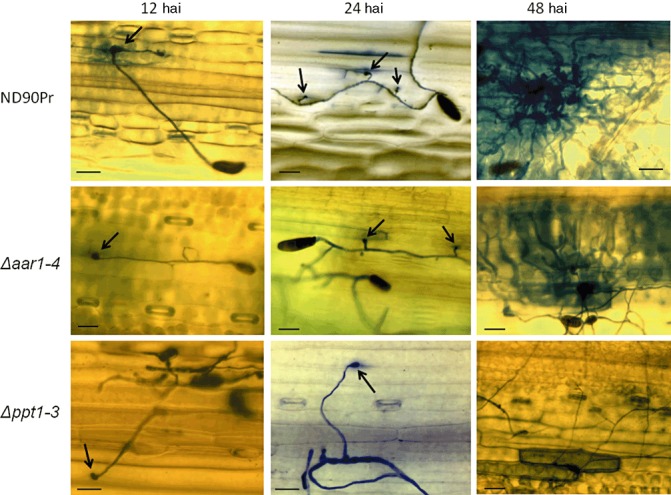
Infection structure differentiation of the wild‐type (ND90Pr), Δaar1 (Δaar1‐4) and Δppt1 (Δppt1‐3) strains at 12, 24 and 48 h after inoculation (hai) on intact barley leaves of cv. Bowman with lysine (50 µg/mL) supplementation. Fungal and dead plant cells were stained blue with trypan blue. Appressoria are indicated by black arrows. Bars, 60 µm.
Figure 7.
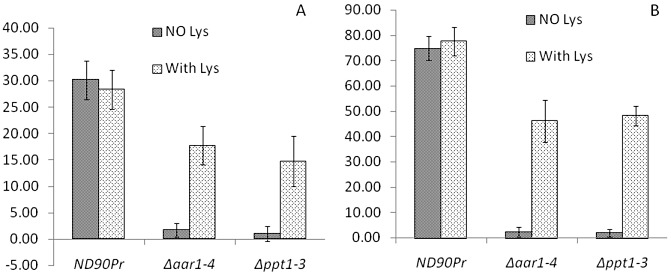
Frequency (%) of conidia with germ tubes or hyphae forming appressoria on seedling leaves of barley cv. Bowman from the wild‐type (ND90Pr), Δaar1 (Δaar1‐4) and Δppt1 (Δppt1‐3) strains at 12 h (A) and 24 h (B) after inoculation without or with lysine (Lys at 50 µg/mL) supplementation. The frequency (%) was calculated from the number of appressorium‐forming conidia relative to the total number of conidia examined. Error bars indicate the standard deviation.
When mycelial plugs of the wild‐type and Δppt1 strains were used to inoculate wounded leaves of Bowman, the wild‐type strain generated expanded lesions (approximately 2 cm in length) surrounded by chlorosis, whereas the Δppt1 strains only produced lesions restricted to the inoculation points (approximately 0.4 cm in length) without any chlorosis (Fig. 5B). Similar results were observed when lysine was supplemented during and after inoculation of the wounded barley leaves (Fig. 5B). These results further indicate that PPT1 is required for virulence in C. sativus.
We generated two complementation strains (Δppt1‐3‐C1 and Δppt1‐3‐C2) by placing the wild‐type PPT1 of C. sativus back into one of the deletion mutants, Δppt1‐3. PCR amplification analysis with primers CsPPT1‐F5 and CsPPT1‐F6, designed from the deleted region of PPT1, indicated that only the wild‐type and the complementation mutants showed the expected DNA fragment, whereas no PCR products were observed in the Δppt1 strains (data not shown). Southern hybridization analysis of EcoRV‐digested DNA from Δppt1‐3‐C1 and Δppt1‐3‐C2 also indicated that a 6.3‐kb fragment carrying PPT1 was detected in addition to the 6.1‐kb hph cassette found in the Δppt1 strains (data not shown). RT‐PCR analysis detected the expression of the PPT1 gene in the wild‐type strain and the complementation strain (Δppt1‐3‐C1), but not in the deletion mutant (Δppt1‐3) (Fig. S1), indicating that PPT1 was successfully rescued in the complementation mutants. When the complementation mutants of Δppt1 were evaluated under the conditions mentioned above, no significant phenotypic differences were observed when compared with the wild‐type strain (Fig. 5E and data not shown).
Lysine synthesis is required for fungal growth, but not for pathogenicity and virulence, in C. sativus
To determine the role of lysine biosynthesis in the pathogenicity and virulence of C. sativus, we generated deletion mutants (Δaar1) of the orthologous gene encoding for AAR (AAR1) in the isolate ND90Pr. Deletion of AAR1 in the Δaar1 strains was confirmed by Southern hybridization analysis, which indicated that a 10.2‐kb fragment containing AAR1 in the wild‐type strain was replaced by an 8.8‐kb fragment carrying hph in the three Δaar1 strains (Fig. S2, see Supporting Information). The Δaar1 strains failed to grow on MM without lysine, but were able to grow on PDA (Fig. S3, see Supporting Information) and MM with lysine (data not shown); they also produced melanized conidia with a size and morphology similar to those of the wild‐type (data not shown).
When conidia of Δaar1 strains were spray inoculated on seedling plants of barley cv. Bowman without lysine supplementation, few infection sites were induced and no obvious spot blotch lesions were observed (Fig. 5C). However, when lysine was supplemented in the conidial suspension for inoculation and lysine solution was sprayed onto the inoculated barley plants twice a day during disease development, the Δaar1 strains induced typical spot blotch symptoms with a disease severity similar to that of the wild‐type (Fig. 5C). Point inoculation of Δaar1 strains on intact barley leaves generated lesions restricted to the inoculated spots, whereas the wild‐type induced expanded lesions which eventually merged together (Fig. 5D). However, both wild‐type and Δaar1 strains produced expanded lesions and no significant differences in virulence were observed between them when inoculated on wounded barley leaves (Fig. 5D). To determine the impact of AAR1 deletion on infection structure differentiation, we inoculated 2‐week‐old seedlings of barley cv. Bowman with conidia of the wild‐type and Δaar1 strains, and examined their infection structures at 12, 24 and 48 hai. With lysine supplementation, the Δaar1 strain formed normal appressorium‐like structures at 12 and 24 hai (Fig. 6), although the frequency was relatively low compared with the wild‐type (Fig. 7). At 48 hai, and with lysine supplementation, the Δaar1 strain continued to form branching and proliferating infection hyphae inside the plant tissue, causing massive death of cells, similar to that observed for the wild‐type (Fig. 6). These results indicate that lysine biosynthesis is required to keep the fungus continuously growing on intact plants, but is not directly involved in the virulence of C. sativus.
Role of melanization in the pathogenicity and virulence of C. sativus
Melanization has been demonstrated to be required for the pathogenicity/virulence of several fungal plant pathogens, including C. lagenarium (Rasmussen and Hanau, 1989), C. graminicola (Horbach et al., 2009) and M. grisea (Howard and Valent, 1996). To demonstrate the role of melanization in the pathogenicity/virulence of C. sativus, we generated deletion mutants (Δpks1) of the C. sativus PKS1, a PKS gene involved in the melanin biosynthesis pathway of the fungus (Leng et al., 2011). Deletion of the PKS1 gene in the Δpks1 strains was confirmed by PCR amplification analysis, which indicated that an expected 700‐bp amplicon was only present in the wild‐type strain and ectopic transformant (ect.), but not in the Δpks1 strains (Δpks1‐1, Δpks1‐2, Δpks1‐3), when the PKS1‐specific primers (CsPKS1‐F5/CsPKS1‐F6) were used (Fig. S4B, see Supporting Information). However, the hph gene was amplified from the deletion mutants but not from the wild‐type and ect. strain (Fig. S4C). The Δpks1 strains showed normal growth on PDA and MM, except that their conidia were nonmelanized. When conidia of the Δpks1 strains were spray inoculated on seedling plants of barley cv. Bowman, severe spot blotch disease symptoms were induced and no significant differences in virulence were observed between the wild‐type and the Δpks1 strains (Fig. 5E). This result indicates that melanization is not required for the pathogenicity and virulence of C. sativus.
Role of NPS6 in the virulence of C. sativus
NPS6 is the gene encoding an NRPS, which is involved in siderophore‐mediated iron metabolism in C. heterostrophus, F. graminearum and A. brassicicola (Lee et al., 2005; Oide et al., 2006). Deletion of NPS6 in these plant pathogens led to a reduction in virulence in their respective hosts, suggesting the role of NPS6 as a virulence determinant (Oide et al., 2006). To determine whether the reduction in virulence of Δppt1 was caused by inactivation of NPS6, one of the other known virulence components affected by PPT1, we characterized the function of NPS6 in C. sativus. A gene orthologous to NPS6 of C. heterostrophus was identified from the genome sequence of the C. sativus strain ND90Pr and deletion mutants (Δnps6) were generated. PCR amplification analysis using primers (CsNPS6‐F1/CsNPS6‐F6) designed from the deleted region of NPS6 showed that no amplicons were generated from the Δnps6 strains, whereas a 956‐bp fragment was amplified from the wild‐type and ect. strain (Fig. S5, see Supporting Information). However, the hph gene was detected in all the deletion mutants and the ect. strain, but not in the wild‐type (Fig. S5). This result indicates that NPS6 is successfully deleted in the Δnps6 strains.
Infection assays revealed that both NPS6‐deficient mutants and the wild‐type strain showed typical spot blotch symptoms, but the Δnps6 strains of C. sativus were slightly reduced in virulence on barley cv. Bowman at 4 dai (Fig. 5F). However, at 6 dai, no significant differences in disease symptoms were observed between the wild‐type and NPS6‐deficient mutants (Fig. 5F). The results indicate that NPS6 is involved in the virulence of C. sativus by delaying disease development, but is not the major gene in ND90Pr that confers high virulence on barley cv. Bowman.
DISCUSSION
In this study, we characterized an orthologue (PPT1) of cfwA/npgA in C. sativus and demonstrated that it is not only required for primary metabolism, but is also involved in the regulation of virulence of the fungus on the barley host. Our results also suggest that virulence reduction in the ppt1 mutants is a result of the loss of production of a novel HST other than due to the lack of lysine biosynthesis or one of the other known virulence components (melanin and siderophore) affected by PPT1.
Horbach et al. (2009) found that Δppt1 mutants of C. graminicola were able to grow on rich medium, such as PDA, but failed to synthesize melanin, suggesting that the PPT1 gene is also required for melanin biosynthesis. In C. heterostrophus, melanin is synthesized through the fungal 1,8‐dihydroxynaphthalene (DHN)–melanin biosynthesis pathway, in which a PKS encoded by the PKS18 gene is the first enzyme to convert acetate to 1,3,6,8‐tetrahydroxynaphthalene (T4HN) (Eliahu et al., 2007). In a previous study (Leng et al., 2011), we identified a single copy of the PKS18 orthologue (CsPKS1) in the C. sativus genome. RNA‐mediated silencing of CsPKS1 led to an albino phenotype (Leng et al., 2011), indicating that this gene is required for melanin synthesis. In this study, the CsPKS1‐deleted mutants (Δpks1) of C. sativus also showed the same albino phenotype as those of the CsPKS1‐silenced strains, confirming the role of CsPKS1 in the melanin synthesis pathway. Lack of melanization in the Δppt1 mutants is presumably a result of the deficiency of PPTase encoded by the PPT1 gene, which is required for the activation of PKS1 through 4′‐phosphopantetheinylation at the conserved serine residues.
In A. nidulans, deletion of the CfwA/NpgA gene led to mutants that produced a cotton‐like ‘fluffy’ morphology and were unable to differentiate any conidiophore structure before 3–4 days (Marquez‐Fernandez et al., 2007). In addition, the ΔcfwA/npgA mutants showed significantly reduced conidial production and only produced 0.06% of the conidiospores formed by the wild‐type strain per square centimetre of colony after 5 days of growth on PDA plates (Marquez‐Fernandez et al., 2007). Horbach et al. (2009) also showed that conidia produced by the Δppt1 mutants of C. graminicola were smaller than those of the wild‐type and exhibited dramatic morphological defects. In our study, we did not observe any morphological defects in the conidia produced by the C. sativus Δppt1 mutants compared with the wild‐type, except that the conidia of the Δppt1 mutants were nonmelanized. Germination rates were also similar between the conidia produced by the Δppt1 strains and the wild‐type conidia on water agar, PDA, MM and leaf surfaces. However, we showed that the C. sativus Δppt1 strains produced fewer conidia on PDA plates compared with the wild‐type, as has been reported in A. nidulans (Marquez‐Fernandez et al., 2007) and C. graminicola (Horbach et al., 2009). The similarities and discrepancies between the results obtained for C. sativus, A. nidulans and C. graminicola suggest that the roles of PPT1 in conidiation may differ according to the fungus studied. Conidiation pathways of fungi are still not well understood and merit further investigation.
Pathogenicity tests revealed that the Δppt1 strains of C. sativus showed fewer infection sites and were significantly reduced in virulence compared with the wild‐type strain when their conidia were spray inoculated on intact barley plants (Fig. 5A). The reduction in the number of infection sites is partly a result of a lack of sufficient lysine for the germinated conidia to continue to grow and form appressoria for penetration. This is supported by the fact that the conidia of the Δppt1 strains germinated well on the barley leaf surface, similar to the wild‐type conidia (Fig. 4), but formed fewer appressoria without lysine supplementation (Fig. 7), whereas the frequency of appressorium formation and the number of infection sites were increased significantly when lysine was supplemented during and after inoculation (5, 7). However, a lack of lysine biosynthesis is not the direct cause of the loss of virulence in the Δppt1 strains. This conclusion is based on a comparison of the Δppt1 strains with the lysine‐deficient Δaar1 mutants with regard to plant infection and disease development. When inoculated on intact barley leaves without lysine supplementation, both mutants showed a very low frequency of appressoria, induced few infection sites and failed to cause typical spot blotch symptoms. However, when lysine was supplemented, the Δaar1 strains induced large lesions similar to those produced by the wild‐type strain, whereas the Δppt1 strains caused only pin‐sized lesions (Fig. 5), although they showed a similar appressorium formation frequency (Fig. 7). When inoculated on wounded barley leaves, the Δppt1 strains induced lesions restricted to the inoculation sites, whereas the Δaar1 mutants developed large expanded lesions from the inoculated and wounded sites and showed a disease severity comparable with that of the wild‐type. These results suggest that the loss of virulence in the Δppt1 strains is not a result of lysine deficiency, but of a lack of a PPTase‐activated PKS/NRPS, which is required for the synthesis of an unidentified virulence determinant in C. sativus.
Melanin has been demonstrated to be required for host penetration and disease development for several fungal pathogens. For example, in the rice blast fungus M. grisea, only melanized appressoria can penetrate the host and cause disease (Kawamura et al., 1997; Nosanchuk and Casadevall, 2003). In C. graminicola, melanin‐deficient mutants were also impaired in host cell penetration, but lesion formation was restored when wounded leaves were inoculated with conidia of these mutants (Horbach et al., 2009; Rasmussen and Hanau, 1989). Even in some human pathogens, such as Paracoccidioides brasiliensis, the causal agent of the human systemic disease paracoccidioidomycosis, melanin has been demonstrated to contribute to the pathogenicity/virulence (Taborda et al., 2008). However, our inoculation experiments showed no differences in pathogenicity and virulence between the wild‐type strain and the melanin‐deficient mutant (Δpks1) on cv. Bowman, suggesting that melanin is not required for the pathogenicity and virulence of C. sativus. Melanin was also found not to be required for pathogenicity and virulence in the sister species, C. heterostrophus (Eliahu et al., 2007).
Based on the typical spot blotch symptoms of necrosis and chlorosis, toxins were presumed to be involved in the ability of C. sativus to cause disease in its various hosts (Pringle, 1979). Several putative toxins have been isolated from the culture filtrates of the fungus (Gayed, 1961, 1962; Pringle, 1979), including helminthosporal (De Mayo et al., 1961), derived from its immediate precursor prehelminthosporal during the isolation procedure (De Mayo et al., 1965), 9‐hydroxyprehelminthosporal (Aldridge and Turner, 1970) and victoxinine (Pringle, 1979). However, none of these toxins produced the typical necrotic or chlorotic symptoms of spot blotch when infiltrated into detached barley leaves, nor were any of these toxins selectively toxic (Pringle, 1979). In the present study, we have shown that PPT1 is required for virulence in the C. sativus isolate ND90Pr, thus suggesting that PKSs or NRPSs are involved in the biosynthesis of virulence factors or determinants, which induce high susceptibility on barley cv. Bowman. The failure to isolate virulence factors (or toxins) in ND90Pr that induces spot blotch symptoms may be caused by several factors: (i) the fungal isolate produces very low (or no) concentrations of the toxin in culture; (ii) the optimal conditions for in vitro toxin production by the fungus have not been identified; or (iii) the toxin is induced only in vivo during interaction with an appropriate host genotype (cv. Bowman). The identification of DNA markers associated with the virulence locus for Bowman (Zhong and Steffenson, 2002; Zhong et al., 2002) and the recent availability of the genome sequence from the C. sativus isolate ND90Pr have allowed us to identify a candidate NPS gene, which is directly involved in the high virulence of ND90Pr on barley cv. Bowman (S. Zhong et al., unpublished data). Further characterization of the gene and gene product, as well as its involvement in pathogenesis and spot blotch symptom development, will provide a better understanding of this important pathosystem.
EXPERIMENTAL PROCEDURES
Fungal isolates and growth media
The C. sativus isolate ND90Pr (ATCC 201652) was used as the recipient for all transformation experiments. The media and conditions used for the culture of isolates of C. sativus have been described by Leng et al. (2011). PDA, MM (Tinline et al., 1960) and water agar plates with or without lysine (50 µg/mL) supplementation were also used for fungal growth as needed.
Assays of conidium productivity on agar plates
To compare the conidial productivity of different strains of C. sativus, small mycelial plugs (2 mm × 2 mm) from 3‐day‐old fungal cultures of each strain were inoculated on the centres of PDA plates and allowed to grow for 6 days in a cycle of 14 h of light and 10 h of darkness. Conidia were harvested by adding 10 mL of distilled water to the plate and scraping the agar surface with a rubber spatula, and then filtered through two layers of cheesecloth to remove mycelial fragments. A haemocytometer was used to count the conidia and the average number of conidia from each strain was calculated from three replicate plates.
Assays of sensitivity to oxidative and iron depletion stresses
To evaluate the sensitivity of the wild‐type and Δppt1 strains to H2O2, a small mycelial plug (2 mm × 2 mm) from 3‐day‐old fungal cultures of each strain was inoculated on the centres of PDA plates supplemented with H2O2 at final concentrations of 5, 10 and 15 mm. After 6 days of incubation at 25 °C in the dark, the radial diameter of the fungal colony was measured. PDA plates containing 10 mm H2O2 were used for the final tests because the growth of Δppt1 strains was completely inhibited, whereas the wild‐type strain was still able to grow under these conditions. Three replicates were used for each test per strain.
To test the sensitivity of knockout mutants to iron depletion conditions, MM plates were supplemented with lysine at 50 µg/mL and the iron chelator BPS (Sigma‐Aldrich, St. Louis, MO, USA) at 200 µM, with or without iron‐free ferrichrome (Sigma‐Aldrich) at 50 µM, and used to grow the cultures. The diameter of the fungal colony was measured on each of the plates after incubation at 25 °C for 6 days in a cycle of 14 h of light and 10 h of darkness. The use of MM plates with BPS at 200 µm for the experiment was based on initial tests, which demonstrated that the wild‐type strain was able to grow, whereas the Δppt1 strains did not show growth under these conditions.
Identification of orthologues of PPT1, AAR, PKS1 and NPS6 in C. sativus
A draft genome sequence of the C. sativus strain ND90Pr was provided by the JGI. A blast search was performed against the genome sequence to identify homologues of PPT1, AAR, NPS6 and PKS1 in C. sativus using the sequences of PPT1 and AAR of C. graminicola (Horbach et al., 2009) and NPS6 and PKS18 of C. heterostrophus (Lee et al., 2005; Oide et al., 2006) as queries, respectively.
Gene replacement and complementation
The split marker system (Catlett et al., 2003) was used for gene replacement (Fig. 1A). The 5′ and 3′ flanking sequences of the target gene were amplified from ND90Pr using the primers listed in Table 1. The two overlapping 3′ and 5′ constructs were generated by fusion PCR, separately, and were mixed, purified by ethanol precipitation and used for transformation.
To prepare complementation mutants for the C. sativus Δppt1 strain, the geneticin resistance gene cassette was released from pII99 (Namiki et al., 2001) by digestion with XbaI and BglII, and cloned into the BamHI/XbaI‐digested pBluescript (+) to form a new cloning vector pBG418. A 4.1‐kb DNA fragment containing the open reading frame (ORF) of PPT1, as well as its 2.6‐kb 5′ end and 0.4‐kb 3′ end flanking regions, was amplified from the C. sativus isolate ND90Pr by the primer pair PPT‐F/PPT‐R (Table 1) and cloned into the pGEM®‐T Easy Vector (Promega, Madison, WI, USA). The fragment was then released from the T‐Easy vector with NotI, and cloned into pBG418 to generate pBG418‐CsPPT1. This vector was verified by restriction enzyme digestion and linearized by XhoI for transformation.
Fungal transformation
Fungal transformants were obtained via polyethylene glycol (PEG)‐mediated transformation. Protoplast preparation was performed according to Zhong et al. (2002) and transformation was carried out according to the procedure of Turgeon et al. (1987). All transformants were purified by single spore isolation and stored on silica gels using the method of Windels et al. (1988).
Southern hybridization and PCR
Genomic DNA was isolated according to Zhong et al. (2002). Southern blot analysis was performed to confirm the deletions of PPT1 and AAR1 in C. sativus. Genomic DNA from the wild‐type strain and the deletion mutants (Δppt1 and Δaar1) was digested by EcoRV. The digests were fractionated on a 0.9% agrose gel in Tris‐Acetate‐EDTA (TAE) buffer and transferred to Hybond N+ (Amersham Biosciences, Piscataway, NJ, USA). The probes used to detect the deletion of the C. sativus PPT1 and AAR1 genes are indicated in Fig. 1A and Fig. S2A, respectively. The hybridization and detection procedures were performed according to the method described previously by Zhong et al. (2002). To detect the deletion of PKS1 and NPS6 in C. sativus, primer pairs CsPKS1‐F5/CsPKS1‐F6 and CsNPS6‐F1/CsNPS6‐F6 (Table 1), designed from the deleted region of the PKS1 and NPS6 genes, were used for PCR, respectively (Figs S4A and S5A). The primer pair Hph‐F/Hph‐R (Table 1) was used to amplify the hygromycin B resistance gene (hph) in each of the transformants.
RT‐PCR for the detection of PPT1 gene expression
RT‐PCR was performed to analyse transcript expression of the PPT1 gene in the wild‐type strain ND90Pr (WT), the knockout mutants of PPT1 (Δppt1‐3), PKS1 (Δpks1‐5) and AAR1 (Δaar1‐4), and the Δppt1 complementation strain (Δppt1‐3‐C1). Total RNA was extracted from mycelia of the fungal strains grown in potato dextrose broth (PDB) (BD, Franklin Lakes, NJ, USA) using the PureLink™ RNA Mini Kit (Invitrogen, Carlsbad, CA, USA) and purified by treatment with DNase I (Invitrogen). cDNA was generated from 2 µg of total RNA by reverse transcription using the SuperScript® III First‐Strand Synthesis System (Invitrogen), and then diluted 10 times and used as the template for RT‐PCR. The primer pair CsPPT1‐F5/CsPPT1‐F6 (Table 1), specific for PPT1, was used in each reaction (20 µL) containing 2 µL of the cDNA template, 5 pmol of each primer, 0.2 mm of each deoxynucleoside triphosphate (dNTP), 1 × reaction buffer and 2.5 U of Taq DNA polymerase (New England Biolabs, Ipswich, MA, USA), and PCR was performed using a Mastercycler PTC‐100 (MJ Research, Ramsey, MN, USA). The thermal cycling conditions were as follows: initial denaturation (95 °C, 2 min), followed by 25 or 30 cycles of denaturation (94 °C, 30 s), annealing (58 °C, 30 s) and extension (72 °C, 1 min), and one final cycle of extension (72 °C, 10 min). The RT‐Actin‐F and RT‐Actin‐R primers (Table 1), designed from the β‐actin‐encoding gene (ACT), were used in the control experiment.
Examination of spore germination in vitro and on the leaf surface
Asexual spore (conidium) germination of the wild‐type strain (ND90Pr) and the deletion mutant (Δppt1‐3) was examined in vitro by incubating the spores on PDA, MM and water agar plates. Spore suspensions at 5 × 103 spores/mL were prepared from each culture grown on PDA. Approximately 0.2 mL of spore suspension was spread on PDA, MM with lysine and water agar. The percentage of germinated spores on PDA and MM was recorded at 2.5, 4.5 and 6 hai at 28 °C. Spore germination rates on water agar plates were recorded at 4, 8 and 12 hai. For each strain, spore germination tests were performed three times on each of the solid agar plates.
To test spore germination on barley leaves, a spore suspension at a concentration of 5 × 103 spores/mL was prepared from the wild‐type and Δppt1 mutant (Δppt1‐3) and sprayed onto 2‐week‐old seedlings of barley cv. Bowman. The percentages of germinated spores on the second leaves of plants were recorded at 4, 8 and 12 hai in a humidity chamber (25–28 °C). Four leaves were counted at each time point and the average percentage of germination was calculated for each fungal strain.
Plant infection assays and microscopic examination
Pathogenicity tests were performed by spray inoculation with conidial suspension or point inoculation with mycelial plugs on 2‐week‐old seedlings of barley (cv. Bowman) plants. The spray inoculations were carried out according to Fetch and Steffenson (1999), except that the conidial suspension had a concentration of 2 × 103 conidia/mL. For point inoculation, fully expanded second leaves (intact or wounded) of 2‐week‐old barley (cv. Bowman) plants were inoculated with small mycelial plugs (2 mm × 2 mm). For inoculation of intact barley leaves, individual mycelial plugs were placed on the middle portion of the leaves, separated at a distance of 2 cm. For the inoculation of wounded leaves, individual mycelial plugs were placed at the wounded sites generated by piercing with a syringe needle (1 cm3). Inoculated plants were incubated in a humidity chamber for 18–24 h, and then transferred into a growth chamber (20 ± 2 °C) and incubated for 4–7 days before disease rating. The 1–9 rating scale of Fetch and Steffenson (1999) was used to rate the spot blotch disease for spray inoculation experiments. Lesion sizes were measured when point inoculation was used.
Infection assays of the deletion mutants (Δaar1 and Δppt1), auxotrophic for lysine, were also conducted with lysine added to the conidial suspensions for inoculation at a final concentration of 50 µg/mL. Lysine at 50 µg/mL was also supplemented in the water tank of a humidifier used for misting during the 24 h of incubation in the humidity chamber. The inoculated barley plants were moved into a growth chamber and were sprayed with a lysine solution (50 µg/mL) twice a day until the experiment ended. Controls without lysine supplementation were also included in the inoculation experiments. The disease severity of the inoculated plants was rated using the 1–9 rating scale of Fetch and Steffenson (1999).
To examine the infection structure differentiation of the wild‐type and deletion mutants (Δaar1 and Δppt1), 2‐week‐old seedlings of barley cv. Bowman were inoculated with a spore suspension (5 × 103 conidia/mL) prepared from each of the strains. At 12, 24 and 48 hai, leaf segments were sampled and subjected to treatments according to the method described by Koch and Slusarenko (1990). The stained fungal structures on and inside plant tissues were examined using an Olympus BX51 microscope (Olympus, Center Vally, PA, USA) and photographed by a CCD camera (Diagnostic Instruments, Inc., Sterling Heights, MI, USA). The average frequency of appressorium formation was calculated for each fungal strain based on the percentage of appressorium‐forming spores relative to the total number of spores examined on three inoculated leaves.
Accession numbers
The nucleotide and protein sequences of C. sativus PPT1, AAR1, PKS1 and NPS6 were deposited in GenBank under accession numbers HQ830035, HQ830034, HQ830033 and HQ830032, respectively.
Supporting information
Fig. S1 Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of PPT1 transcripts in the wild‐type strain (ND90Pr), the knockout mutant of PPT1 (Δppt1‐3), the Δppt1 complementation strain (Δppt1‐3‐C1) and the knockout mutants of PKS1 (Δpks1‐5) and AAR1 (Δaar1‐4). The expression of the β‐actin‐encoding gene (ACT) was used as a control.
Fig. S2 Generation of Δaar1 strains of Cochliobolus sativus. (A) Replacement of the AAR1 gene by a 2.6‐kb fragment carrying the Escherichia coli hygromycin phosphotransferase gene (hph) using the split‐marker system (Catlett et al., 2003). (B) Southern hybridization analysis of EcoRV‐digested genomic DNA from the wild‐type (ND90Pr), ectopic transformant (ect.) and Δaar1 strains (Δaar1‐2, Δaar1‐3, Δaar1‐4) using the probe indicated in Fig. S2A. The 10.2‐kb fragment in the wild‐type strain ND90Pr was replaced by the 8.8‐kb fragment in the three Δaar1 strains. The transformant with an ectopic integration of hph showed a 10.2‐kb band as observed in the wild‐type strain.
Fig. S3 Colonies of wild‐type, Δpks1, Δaar1 and Δnps6 grown on potato dextrose agar (PDA). Mycelial plugs of uniform size from 3‐day‐old cultures were inoculated onto the centres of the plates and grown for 6 days at 25 °C before photography. The wild‐type strain and Δaar1 and Δnps6 mutants showed black or dark brown pigments, whereas the Δpks1 strain showed a white colony, on PDA plates.
Fig. S4 Generation of Δpks1 strains of Cochliobolus sativus. (A) Replacement of the PKS1 gene by a 2.6‐kb fragment carrying the Escherichia coli hygromycin phosphotransferase gene (hph) using the split‐marker system (Catlett et al., 2003). (B) Polymerase chain reaction (PCR) amplification analysis of the wild‐type (ND90Pr), ectopic transformant (ect.) and Δpks1 strains (Δpks1‐1, Δpks1‐2, Δpks1‐3, Δpks1‐5, Δpks1‐9 and Δpks1‐10) using primers (CsPKS1‐F5/CsPKS1‐F6) designed from the deleted region of PKS1. Only the wild‐type and ect. strain showed a 700‐bp amplicon. (C) PCR amplification analysis of the wild‐type (ND90Pr), ectopic transformant (ect.) and Δpks1 strains (Δpks1‐1, Δpks1‐2, Δpks1‐3, Δpks1‐5, Δpks1‐9 and Δpks1‐10) using the primer pair (HYG1‐F/HYG1‐R) designed from the hygromycin B resistance gene (hph). A 453‐bp amplicon from hph was found in the ect. and Δpks1 strains, but not in the wild‐type strain.
Fig. S5 Generation of Δnps6 strains of Cochliobolus sativus. (A) Replacement of the NPS6 gene by a 2.6‐kb fragment carrying the Escherichia coli hygromycin phosphotransferase gene (hph) using the split‐marker system (Catlett et al., 2003). (B) Polymerase chain reaction (PCR) amplification analysis of the wild‐type (ND90Pr), ectopic transformant (ect.) and Δnps6 strains (Δnps6‐1, Δnps6‐2, Δnps6‐3, Δnps6‐4, Δnps6‐5, Δnps6‐6, Δnps6‐8 and Δnps6‐14) using primers (CsNPS6‐F1/CsNPS6‐F6) designed from the deleted region of NPS6. Only the wild‐type and ect. strain showed a 956‐bp amplicon. (C) PCR amplification analysis of the wild‐type (ND90Pr), ectopic transformant (ect.) and Δnps6 strains (Δnps6‐1, Δnps6‐2, Δnps6‐3, Δnps6‐4, Δnps6‐5, Δnps6‐6, Δnps6‐8 and Δnps6‐14) using the primer pair (HYG1‐F/HYG1‐R) designed from the hygromycin B resistance gene (hph). A 453‐bp amplicon from hph was shown in the ect. and Δnps6 strains, but not in the wild‐type strain.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
This study was partially supported by the Triticeae‐CAP project (2011‐68002‐30029) from the US Department of Agriculture National Institute of Food and Agriculture. The authors thank Dr Justin D. Faris and Dr Timothy F. Friesen for providing equipment and facilities for the Southern blot analysis and pathogenicity tests, and Dr Xiwen Cai for supplying microscopy for the observation of fungal structures. We also thank Dr Shaukat Ali and Rui Wang for assistance in fungal inoculation, and Lauren Sager for proofreading of the manuscript. The sequence data of the C. sativus strain ND90Pr were produced and provided by the US Department of Energy Joint Genome Institute (http://genome.jgi.doe.gov/Cocsa1/Cocsa1.home.html).
REFERENCES
- Aldridge, D.C. and Turner, W.B. (1970) 9‐Hydroxyprehelminthosporal, a metabolite of Cochliobolus (Helminthosporium) sativus . J. Chem. Soc. C, 686–688. [DOI] [PubMed] [Google Scholar]
- Baker, S.E. , Kroken, S. , Inderbitzin, P. , Asvarak, T. , Li, B.Y. , Shi, L. , Yoder, O.C. and Turgeon, B.G. (2006) Two polyketide synthase encoding genes are required for biosynthesis of the polyketide virulence factor, T‐toxin, by Cochliobolus heterostrophus . Mol. Plant–Microbe Interact. 19, 139–149. [DOI] [PubMed] [Google Scholar]
- Bohnert, H.U. , Fudal, I. , Dioh, W. , Tharreau, D. , Notteghem, J.L. and Lebrun, M.H. (2004) A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell, 16, 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo, A.M. , Wilson, R.A. , Bok, J.W. and Keller, N.P. (2002) Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66, 447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett, N. , Lee, B.N. , Yoder, O. and Turgeon, B. (2003) Split‐marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Newsl. 50, 9–11. [Google Scholar]
- Collemare, J. , Billard, A. , Bohnert, H.U. and Lebrun, M.H. (2008) Biosynthesis of secondary metabolites in the rice blast fungus Magnaporthe grisea: the role of hybrid PKS–NRPS in pathogenicity. Mycol. Res. 112, 207–215. [DOI] [PubMed] [Google Scholar]
- Cuomo, C.A. , Gueldener, U. , Xu, J.R. , Trail, F. , Turgeon, B.G. , Di Pietro, A. , Walton, J.D. , Ma, L.J. , Baker, S.E. , Rep, M. , Adam, G. , Antoniw, J. , Baldwin, T. , Calvo, S. , Chang, Y.L. , DeCaprio, D. , Gale, L.R. , Gnerre, S. , Goswami, R.S. , Hammond‐Kosack, K. , Harris, L.J. , Hilburn, K. , Kennell, J.C. , Kroken, S. , Magnuson, J.K. , Mannhaupt, G. , Mauceli, E. , Mewes, H.W. , Mitterbauer, R. , Muehlbauer, G. , Munsterkotter, M. , Nelson, D. , O'Donnell, K. , Ouellet, T. , Qi, W.H. , Quesneville, H. , Roncero, M.I.G. , Seong, K.Y. , Tetko, I.V. , Urban, M. , Waalwijk, C. , Ward, T.J. , Yao, J.Q. , Birren, B.W. and Kistler, H.C. (2007) The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science, 317:1400–1402. [DOI] [PubMed] [Google Scholar]
- De Mayo, P. , Spencer, E.Y. and White, R.W. (1961) Helminthosporal, the toxin from Helminthosporium sativum I. Isolation and characterization. Can. J. Chem. 39, 1608–1616. [Google Scholar]
- De Mayo, P. , Williams, R.E. and Spencer, E.Y. (1965) Terpenoids VIII. The immediate precursors of helminthosporal and helminthosporol. Can. J. Chem. 43, 1357–1365. [Google Scholar]
- Eisfeld, K. (2009) Non‐ribosomal peptide synthetases of fungi: physiology and genetics In: The Mycota: A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research. Vol. XV. Physiology and Genetics (Anke T., Weber D. and Esser K., eds), pp. 305–330. Berlin: Springer‐Verlag. [Google Scholar]
- Eliahu, N. , Igbaria, A. , Rose, M.S. , Horwitz, B.A. and Lev, S. (2007) Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen‐activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1 . Eukaryot. Cell, 6, 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, B.S. , Robinson, S.J. and Kelleher, N.L. (2011) Surveys of non‐ribosomal peptide and polyketide assembly lines in fungi and prospects for their analysis in vitro and in vivo . Fungal Genet. Biol. 48, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetch, T.G., Jr and Steffenson, B.J. (1999) Rating scales for assessing infection responses of barley infected with Cochliobolus sativus . Plant Dis. 83, 213–217. [DOI] [PubMed] [Google Scholar]
- Gayed, S.K. (1961) Production of symptoms of barley leaf spot disease by culture filtrates of Helminthosporium sativum . Nature, 191, 725–726. [Google Scholar]
- Gayed, S.K. (1962) The pathogenicity of six strains of Helminthosporium sativum to three cereals. Mycopathologia, 18, 271–279. [DOI] [PubMed] [Google Scholar]
- Haas, H. , Eisendle, M. and Turgeon, B.G. (2008) Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol. 46, 149–187. [DOI] [PubMed] [Google Scholar]
- Hoffmeister, D. and Keller, N.P. (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 24, 393–416. [DOI] [PubMed] [Google Scholar]
- Horbach, R. , Graf, A. , Weihmann, F. , Antelo, L. , Mathea, S. , Liermann, J.C. , Opatz, T. , Thines, E. , Aguirre, J. and Deising, H.B. (2009) Sfp‐type 4′‐phosphopantetheinyl transferase is indispensable for fungal pathogenicity. Plant Cell, 21, 3379–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, R.J. and Valent, B. (1996) Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea . Annu. Rev. Microbiol. 50, 491–512. [DOI] [PubMed] [Google Scholar]
- Johnson, R.D. , Johnson, L. , Itoh, Y. , Kodama, M. , Otani, H. and Kohmoto, K. (2000) Cloning and characterization of a cyclic peptide synthetase gene from Alternaria alternata apple pathotype whose product is involved in AM‐toxin synthesis and pathogenicity. Mol. Plant–Microbe Interact. 13, 742–753. [DOI] [PubMed] [Google Scholar]
- Kawamura, C. , Moriwaki, J. , Kimura, N. , Fujita, Y. , Fuji, S. , Hirano, T. , Koizumi, S. and Tsuge, T. (1997) The melanin biosynthesis genes of Alternaria alternata can restore pathogenicity of the melanin‐deficient mutants of Magnaporthe grisea . Mol. Plant–Microbe Interact. 10, 446–453. [DOI] [PubMed] [Google Scholar]
- Keller, N.P. , Turner, G. and Bennett, J.W. (2005) Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. [DOI] [PubMed] [Google Scholar]
- Keszenman‐Pereyra, D. , Lawrence, S. , Twfieg, M.E. , Price, J. and Turner, G. (2003) The npgA/cfwA gene encodes a putative 4′‐phosphopantetheinyl transferase which is essential for penicillin biosynthesis in Aspergillus nidulans . Curr. Genet. 43, 186–190. [DOI] [PubMed] [Google Scholar]
- Koch, E. and Slusarenko, A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell, 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroken, S. , Glass, N.L. , Taylor, J.W. , Yoder, O.C. and Turgeon, B.G. (2003) Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA, 100, 15 670–15 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, J. , Schäfer, P. , Hückelhoven, R. , Langen, G. , Baltruschat, H. , Stein, E. , Nagarajan, S. and Kogel, K.‐H. (2002) Bipolaris sorokiniana, a cereal pathogen of global concern: cytological and molecular approaches towards better control. Mol. Plant Pathol. 3, 185–195. [DOI] [PubMed] [Google Scholar]
- Lee, B.N. , Kroken, S. , Chou, D.Y. , Robbertse, B. , Yoder, O.C. and Turgeon, B.G. (2005) Functional analysis of all nonribosomal peptide synthetases in Cochliobolus heterostrophus reveals a factor, NPS6, involved in virulence and resistance to oxidative stress. Eukaryot. Cell, 4, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng, Y. , Wu, C. , Liu, Z. , Friesen, T.L. , Rasmussen, J.B. and Zhong, S. (2011) RNA‐mediated gene silencing in the cereal fungal pathogen Cochliobolus sativus . Mol. Plant Pathol. 12, 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez‐Fernandez, O. , Trigos, A. , Ramos‐Balderas, J.L. , Viniegra‐Gonzalez, G. , Deising, H.B. and Aguirre, J. (2007) Phosphopantetheinyl transferase CfwA/NpgA is required for Aspergillus nidulans secondary metabolism and asexual development. Eukaryot. Cell, 6, 710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathre, D.E. (1997) Compendium of Barley Diseases, 2nd edn. St. Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Mootz, H.D. , Schorgendorfer, K. and Marahiel, M.A. (2002) Functional characterization of 4′‐phosphopantetheinyl transferase genes of bacterial and fungal origin by complementation of Saccharomyces cerevisiae lys5 . FEMS Microbiol. Lett. 213, 51–57. [DOI] [PubMed] [Google Scholar]
- Namiki, F. , Matsunaga, M. , Okuda, M. , Inoue, I. , Nishi, K. , Fujita, Y. and Tsuge, T. (2001) Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis . Mol. Plant–Microbe Interact. 14, 580–584. [DOI] [PubMed] [Google Scholar]
- Nosanchuk, J.D. and Casadevall, A. (2003) The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 5, 203–223. [DOI] [PubMed] [Google Scholar]
- Oberegger, H. , Eisendle, M. , Schrettl, M. , Graessle, S. and Haas, H. (2003) 4‐Phosphopantetheinyl transferase‐encoding npgA is essential for siderophore biosynthesis in Aspergillus nidulans . Curr. Genet. 44, 211–215. [DOI] [PubMed] [Google Scholar]
- Oide, S. , Moeder, W. , Krasnoff, S. , Gibson, D. , Haas, H. , Yoshioka, K. and Turgeon, B.G. (2006) NPS6, encoding a nonribosomal peptide synthetase involved in siderophore‐mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell, 18, 2836–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, R.B. (1979) Role of toxins in etiology of spot blotch disease of barley. Can. Plant Dis. Surv. 59, 74–79. [Google Scholar]
- Quadri, L.E. , Weinreb, P.H. , Lei, M. , Nakano, M.M. , Zuber, P. and Walsh, C.T. (1998) Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry, 37, 1585–1595. [DOI] [PubMed] [Google Scholar]
- Rasmussen, J.B. and Hanau, R.M. (1989) Exogenous scytalone restores appressorial melanization and pathogenicity in albino mutants of Colletotrichum graminicola . Can. J. Plant Pathol. 11, 349–352. [Google Scholar]
- Stack, D. , Neville, C. and Doyle, S. (2007) Nonribosomal peptide synthesis in Aspergillus fumigatus and other fungi. Microbiology, 153, 1297–1306. [DOI] [PubMed] [Google Scholar]
- Taborda, C.P. , Da Silva, M.B. , Nosanchuk, J.D. and Travassos, L.R. (2008) Melanin as a virulence factor of Paracoccidioides brasiliensis and other dimorphic pathogenic fungi: a minireview. Mycopathologia, 165, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinline, R.D. , Strauffer, J.F. and Dickson, J.G. (1960) Cochliobolus sativus III. Effects of ultraviolet radiation. Can. J. Bot. 38, 275–282. [Google Scholar]
- Turgeon, B.G. , Garber, R.C. and Yoder, O.C. (1987) Development of a fungal transformation system based on selection of sequences with promoter activity. Mol. Cell. Biol. 7, 3297–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon, B.G. , Oide, S. and Bushley, K. (2008) Creating and screening Cochliobolus heterostrophus non‐ribosomal peptide synthetase mutants. Mycol. Res. 112, 200–206. [DOI] [PubMed] [Google Scholar]
- Valjavec‐Gratian, M. and Steffenson, B.J. (1997a) Pathotypes of Cochliobolus sativus on barley. Plant Dis. 81, 1275–1278. [DOI] [PubMed] [Google Scholar]
- Valjavec‐Gratian, M. and Steffenson, B.J. (1997b) Genetics of virulence in Cochliobolus sativus and resistance in barley. Phytopathology, 87, 1140–1143. [DOI] [PubMed] [Google Scholar]
- Walton, J.D. (2006) HC‐toxin. Phytochemistry, 67, 1406–1413. [DOI] [PubMed] [Google Scholar]
- Wiese, M.V. (1977) Compendium of Wheat Diseases. St. Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Windels, C.M. , Burnes, P.M. and Kommedahl, T. (1988) Five‐year preservation of Fusarium species on silica gel and soil. Phytopathology, 78, 107–109. [Google Scholar]
- Wolpert, T.J. , Dunkle, L.D. and Ciuffetti, L.M. (2002) Host‐selective toxins and avirulence determinants: what's in a name? Annu. Rev. Phytopathol. 40, 251–285. [DOI] [PubMed] [Google Scholar]
- Zhong, S. and Steffenson, B.J. (2002) Identification and characterization of DNA markers associated with a locus conferring virulence on barley in the plant pathogenic fungus Cochliobolus sativus . Theor. Appl. Genet. 104, 1049–1054. [DOI] [PubMed] [Google Scholar]
- Zhong, S. , Steffenson, B.J. , Martinez, J.P. and Ciuffetti, L.M. (2002) A molecular genetic map and electrophoretic karyotype of the plant pathogenic fungus Cochliobolus sativus . Mol. Plant–Microbe Interact. 15, 481–492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of PPT1 transcripts in the wild‐type strain (ND90Pr), the knockout mutant of PPT1 (Δppt1‐3), the Δppt1 complementation strain (Δppt1‐3‐C1) and the knockout mutants of PKS1 (Δpks1‐5) and AAR1 (Δaar1‐4). The expression of the β‐actin‐encoding gene (ACT) was used as a control.
Fig. S2 Generation of Δaar1 strains of Cochliobolus sativus. (A) Replacement of the AAR1 gene by a 2.6‐kb fragment carrying the Escherichia coli hygromycin phosphotransferase gene (hph) using the split‐marker system (Catlett et al., 2003). (B) Southern hybridization analysis of EcoRV‐digested genomic DNA from the wild‐type (ND90Pr), ectopic transformant (ect.) and Δaar1 strains (Δaar1‐2, Δaar1‐3, Δaar1‐4) using the probe indicated in Fig. S2A. The 10.2‐kb fragment in the wild‐type strain ND90Pr was replaced by the 8.8‐kb fragment in the three Δaar1 strains. The transformant with an ectopic integration of hph showed a 10.2‐kb band as observed in the wild‐type strain.
Fig. S3 Colonies of wild‐type, Δpks1, Δaar1 and Δnps6 grown on potato dextrose agar (PDA). Mycelial plugs of uniform size from 3‐day‐old cultures were inoculated onto the centres of the plates and grown for 6 days at 25 °C before photography. The wild‐type strain and Δaar1 and Δnps6 mutants showed black or dark brown pigments, whereas the Δpks1 strain showed a white colony, on PDA plates.
Fig. S4 Generation of Δpks1 strains of Cochliobolus sativus. (A) Replacement of the PKS1 gene by a 2.6‐kb fragment carrying the Escherichia coli hygromycin phosphotransferase gene (hph) using the split‐marker system (Catlett et al., 2003). (B) Polymerase chain reaction (PCR) amplification analysis of the wild‐type (ND90Pr), ectopic transformant (ect.) and Δpks1 strains (Δpks1‐1, Δpks1‐2, Δpks1‐3, Δpks1‐5, Δpks1‐9 and Δpks1‐10) using primers (CsPKS1‐F5/CsPKS1‐F6) designed from the deleted region of PKS1. Only the wild‐type and ect. strain showed a 700‐bp amplicon. (C) PCR amplification analysis of the wild‐type (ND90Pr), ectopic transformant (ect.) and Δpks1 strains (Δpks1‐1, Δpks1‐2, Δpks1‐3, Δpks1‐5, Δpks1‐9 and Δpks1‐10) using the primer pair (HYG1‐F/HYG1‐R) designed from the hygromycin B resistance gene (hph). A 453‐bp amplicon from hph was found in the ect. and Δpks1 strains, but not in the wild‐type strain.
Fig. S5 Generation of Δnps6 strains of Cochliobolus sativus. (A) Replacement of the NPS6 gene by a 2.6‐kb fragment carrying the Escherichia coli hygromycin phosphotransferase gene (hph) using the split‐marker system (Catlett et al., 2003). (B) Polymerase chain reaction (PCR) amplification analysis of the wild‐type (ND90Pr), ectopic transformant (ect.) and Δnps6 strains (Δnps6‐1, Δnps6‐2, Δnps6‐3, Δnps6‐4, Δnps6‐5, Δnps6‐6, Δnps6‐8 and Δnps6‐14) using primers (CsNPS6‐F1/CsNPS6‐F6) designed from the deleted region of NPS6. Only the wild‐type and ect. strain showed a 956‐bp amplicon. (C) PCR amplification analysis of the wild‐type (ND90Pr), ectopic transformant (ect.) and Δnps6 strains (Δnps6‐1, Δnps6‐2, Δnps6‐3, Δnps6‐4, Δnps6‐5, Δnps6‐6, Δnps6‐8 and Δnps6‐14) using the primer pair (HYG1‐F/HYG1‐R) designed from the hygromycin B resistance gene (hph). A 453‐bp amplicon from hph was shown in the ect. and Δnps6 strains, but not in the wild‐type strain.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


