Summary
Although the Sw‐5 gene cluster has been cloned, and Sw‐5b has been identified as the functional gene copy that confers resistance to Tomato spotted wilt virus (TSWV), its avirulence (Avr) determinant has not been identified to date. Nicotiana tabacum ‘SR1‘ plants transformed with a copy of the Sw‐5b gene are immune without producing a clear visual response on challenge with TSWV, whereas it is shown here that N. benthamiana transformed with Sw‐5b gives a rapid and conspicuous hypersensitive response (HR). Using these plants, from all structural and non‐structural TSWV proteins tested, the TSWV cell‐to‐cell movement protein (NSM) was confirmed as the Avr determinant using a Potato virus X (PVX) replicon or a non‐replicative pEAQ‐HT expression vector system. HR was induced in Sw‐5b‐transgenic N. benthamiana as well as in resistant near‐isogenic tomato lines after agroinfiltration with a functional cell‐to‐cell movement protein (NSM) from a resistance‐inducing (RI) TSWV strain (BR‐01), but not with NSM from a Sw‐5 resistance‐breaking (RB) strain (GRAU). This is the first biological demonstration that Sw‐5‐mediated resistance is triggered by the TSWV NSM cell‐to‐cell movement protein.
Keywords: avirulence, cell‐to‐cell movement, NSM, resistance gene, Sw‐5, tospovirus, TSWV
Introduction
Tospoviruses are amongst the most destructive pathogens known for tomatoes and are a limiting factor in vegetable production worldwide (Aramburu and Marti, 2003; Boiteux and Giordano, 1993; Finetti Sialer et al., 2002; Pappu et al., 2009; Williams et al., 2001). Tomato spotted wilt virus (TSWV) is the best studied member of the genus Tospovirus and represents a great burden to many economically important agricultural and ornamental crops in (sub)tropical and temperate climatic regions. The virus is transmitted in a propagative manner by various species of thrip (Thripidae, main vector—Frankliniella occidentalis), and currently ranks second on the list of the most important plant viruses in the world (Scholthof et al., 2011).
Since TSWV was identified and classified as the first phytopathogenic virus in the Bunyaviridae, a large family of mostly arthropod‐borne animal‐infecting RNA viruses, more than 20 new tospovirus species have been identified and distinguished on the basis of the nucleoprotein (N) gene sequence and vector specificity (King et al., 2011). Based on phylogenetic analysis of the N gene, all tospovirus species are grouped into two major clusters that correlate with their geographical distribution, i.e. the New World and Old World tospoviruses. Like all members of the Bunyaviridae, tospoviruses consist of enveloped virus particles (Ø 80–120 nm) and contain three RNA segments which, according to their size, are denoted S (small), M (medium) and L (large) RNA. The S RNA encodes the N protein and the RNA silencing suppressor protein NSS (Bucher et al., 2003; Kormelink et al., 1991; Takeda et al., 2002). The M RNA encodes the glycoprotein precursor (GP) to the glycoproteins GN and GC (N and C refer to the amino‐ and carboxy‐terminal positions in the precursor) and the cell‐to‐cell movement protein NSM (Kormelink et al., 1992, 1994; Storms et al., 1995). The L RNA encodes the viral polymerase (de Haan et al., 1991).
Several strategies for the chemical and cultural control of tospoviruses and their vectors have been shown to be ineffective. The most successful, however, is the cultivation of resistant cultivars, which are usually developed by the introgression of resistance (R) genes into commercial genotypes. Some tomato accessions are sources of natural resistance to TSWV, and are being used in breeding programmes throughout the world (Finlay, 1953; Price et al., 2007; Saidi and Warade, 2008). In recent decades, two single dominant R genes have received most attention because of their applicability for commercial resistance breeding against tospoviruses, i.e. Sw‐5 and Tsw. The first, Sw‐5 (Aramburu et al., 2011; Cho et al., 1995; Stevens et al., 1991), is the most interesting as it confers a broad tospovirus resistance against TSWV, Chrysanthemum stem necrosis virus (CSNV), Tomato chlorotic spot virus (TCSV) and Groundnut ringspot virus (GRSV) (Boiteux and Giordano, 1993; Stevens et al., 1991, 1994, 1995). The resistance derives from a tomato cultivar (Stevens), which was obtained in South Africa by a cross between Solanum peruvianum and S. lycopersicum (Van Zijl et al., 1985), and protects against virus invasion by the induction of a hypersensitive response (HR). Little is yet known about the mechanism of defence driven by this R gene. The second, single dominant, R gene is Tsw (Boiteux, 1995). This gene originates from distinct Capsicum chinense accessions and is highly specific as it only confers resistance against TSWV isolates (Boiteux, 1995).
Single dominant R genes generally make up the second line of defence of the plant immune system against pathogens, a battle that is generally illustrated with the zig–zag model (Jones and Dangl, 2006) and involves RNA silencing as one of the first lines of defence against plant viruses. The first lines of defence are triggered by the so‐called microbial‐ or pathogen‐associated molecular patterns (MAMPs or PAMPs) (Nürnberger and Brunner, 2002), which are perceived by pattern recognition receptors (PRRs) (Postel and Kemmerling, 2009) and lead to the onset of PAMP‐triggered immunity (PTI) (Chisholm et al., 2006). Viruses encode virulence factors (effectors) that counteract PTI, and thereby enable them to achieve a successful infection. In the next phase, these same effectors are specifically recognized directly or indirectly by protein products from R genes, referred to as effector‐triggered immunity (ETI). This recognition generally leads to a rapid HR, and involves programmed cell death (PCD) at the site of infection. Recently, the RNA silencing suppressor NSS has been identified as the effector triggering Tsw‐mediated resistance (Ronde et al., 2013), but the effector that triggers Sw‐5b‐governed resistance has remained unknown to date.
Proteins encoded by single dominant R genes typically contain a nucleotide‐binding leucine‐rich repeat (NB‐LRR) domain (Dangl and Jones, 2001). In plants, the NB‐LRR‐type R genes are further divided into two groups based on the structure of the conserved N‐terminal domain: TIR‐NB‐LRRs, which share homology to Toll/interleukin receptors, and the coiled coil (CC)‐NB‐LRRs (Pan et al., 2000). Sw‐5 belongs to the CC‐NB‐LRR group and its locus contains at least six paralogues, denoted Sw‐5a to Sw‐5f (Folkertsma et al., 1999; Rehman et al., 2009). The Sw‐5b gene represents the functional resistance gene copy and is able to provide broad tospovirus resistance, as demonstrated in transgenic tobacco plants expressing Sw‐5b (Spassova et al., 2001).
The possible occurrence of genome rearrangements and the ongoing evolution of new TSWV variants by mutations pose a constant threat to the broad resistance against tospoviruses conferred by the Sw‐5 gene. Some natural field isolates of TSWV able to breakdown Sw‐5 resistance have already been reported in tomato crops in Hawaii, Australia, South Africa (JF1 isolate), Spain (GRAU isolate) and Italy (Aramburu and Marti, 2003; Cho et al., 1995; Ciuffo et al., 2005; Latham and Jones, 1998; Thompson and Van Zijl, 1995). Studies on TSWV reassortants have indicated that the genetic determinant involved in overcoming the resistance is associated with the M RNA (Hoffmann et al., 2001). More recently, López et al. (2011) proposed, by in silico analysis, that two amino acid substitutions in the NSM protein of TSWV could be responsible for overcoming the resistance of Sw‐5, but no experimental evidence was provided to support this assumption.
In the present study, the TSWV cell‐to‐cell movement protein (NSM) has been identified as the Avr determinant for the Sw‐5 resistance gene in Nicotiana benthamiana transformed with the functional Sw‐5b copy and in resistant near‐isogenic tomato lines. In addition, a tospovirus challenge assay has shown that Sw‐5‐mediated resistance also protects against the more distinct Impatiens necrotic spot virus (INSV), demonstrating its broad resistance profile against several tospovirus species.
Results
Nicotiana benthamiana transformed with Sw‐5b shows HR on challenge with a TSWV resistance‐inducing (RI) isolate
Previously, the Sw‐5b resistance gene copy was identified as the functional resistance gene, as Nicotiana tabacum ‘SR1’ transformed with this copy conferred resistance against TSWV (Spassova et al., 2001). These transgenic lines, however, did not reveal a clear HR and appeared totally immune, which hampered the identification of the TSWV Avr determinant of Sw‐5. To determine whether transformation of another host with Sw‐5b would provide similar results or, in contrast, would reveal a visual HR and thereby allow the identification of the TSWV Avr determinant, N. benthamiana was transformed and subsequently analysed for an HR. Although TSWV BR‐01 (RI) and GRAU (RB, resistance breaking) both produced a systemic infection on untransformed N. benthamiana (Fig. 1A,B), isolate BR‐01 (RI) triggered an HR within 3–4 days, as visualized by necrotic lesions only on inoculated leaves of transformed N. benthamiana [Fig. 1, photographs in C and D were taken at 5–7 days post‐inoculation (dpi) and in E at 15 dpi], indicative of an Sw‐5b‐induced resistance response. Challenge with TSWV‐RB isolate GRAU did not trigger an HR‐like response, but, instead, resulted in a systemic infection, as in untransformed N. benthamiana (Fig. 1F,G). Transformed N. benthamiana was additionally challenged with INSV, belonging to the same phylogenetic clade of American tospoviruses, and Melon yellow spot virus [MYSV, Physalis severe mottle virus (PSMV) isolate], another more distinct tospovirus from the Eur‐Asian clade. Although MYSV systemically infected Sw‐5b‐transformed N. benthamiana, INSV surprisingly triggered an HR, as observed with TSWV‐RI, suggesting that the spectrum of Sw‐5b‐mediated resistance covers a large range of members from the ‘American’ tospovirus clade (Fig. S1, see Supporting Information). Although no systemic symptoms were observed in Sw‐5b‐transformed N. benthamiana on challenge with TSWV‐RI and INSV, the absence of virus from the upper (non‐inoculated) leaves was verified by reverse transcription‐polymerase chain reaction (RT‐PCR) at 10 dpi using systemically infected wild‐type N. benthamiana as a positive control. As expected, viral RNA was detected in wild‐type plants and in Sw‐5b‐transformed N. benthamiana inoculated with TSWV‐RB or MYSV, but not on challenge with RI tospoviruses TSWV and INSV (Fig. 1H).
Figure 1.
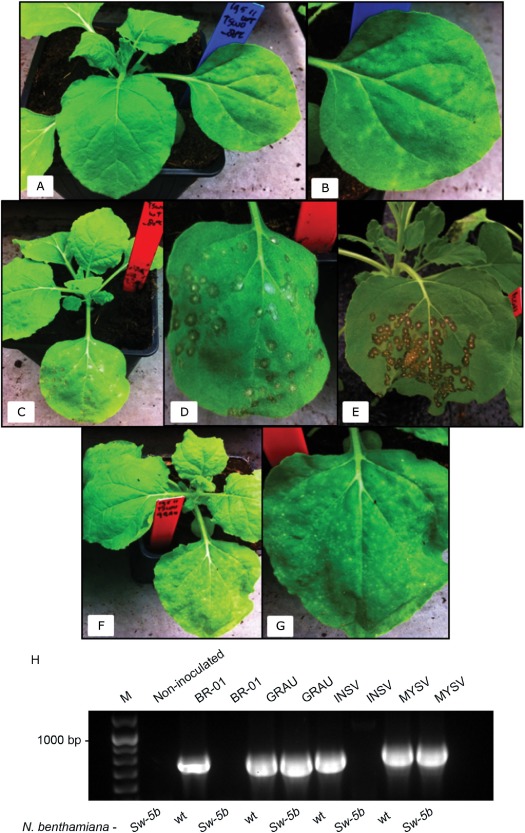
Challenge of untransformed and Sw‐5b‐transformed Nicotiana benthamiana with resistance‐inducing Tomato spotted wilt virus (TSWV) BR‐01 and resistance‐breaking TSWV GRAU isolates. Photographs from all leaves were taken at 5–7 days post‐inoculation (dpi), with the exception of those shown in (E). (A) Untransformed N. benthamiana plant challenged with TSWV BR‐01 and showing typical chlorotic lesions on locally infected leaves. (B) Enlarged view of a locally infected leaf from (A). (C) Nicotiana benthamiana Sw‐5b challenged with TSWV BR‐01. (D) Enlarged view of the virus‐challenged leaf from (C). (E) Nicotiana benthamiana Sw‐5b challenged with TSWV BR‐01 showing the challenged leaf at 15 dpi. (F) Nicotiana benthamiana Sw‐5b challenged with TSWV GRAU. (G) Enlarged view of the virus‐challenged leaf from (F). (H) Reverse transcription‐polymerase chain reaction (RT‐PCR) for monitoring systemic infection in wild‐type (wt) and Sw‐5b N. benthamiana plants inoculated with TSWV (BR‐01 and GRAU), Impatiens necrotic spot virus (INSV) and Melon yellow spot virus (MYSV). Total RNA of non‐inoculated upper leaves was used as a template for N (nucleocapsid) gene amplification at 10 dpi. M, GeneRuler 100‐bp Plus DNA ladder.
Transient expression of TSWV genes from a non‐replicative pEAQ‐HT and from a Potato virus X (PVX) replicon identifies NSM as the TSWV Avr determinant of Sw‐5b
Earlier studies to identify the TSWV Avr determinant that triggers Tsw‐governed resistance resulted in different and conflicting reports (Lovato et al., 2008; Margaria et al., 2007). Recent analysis unambiguously identified the TSWV RNA silencing suppressor NSS as the Avr determinant (Ronde et al., 2013), but this was only successful with the use of a non‐replicative transient pEAQ‐HT expression vector (Sainsbury et al., 2009), but not when using PVX. To identify the Avr determinant, genes coded from TSWV M RNA (NSM and GP) and possible candidates for the Avr‐determinant of Sw‐5 were cloned into both PVX (pGR107) and pEAQ‐HT (Fig. 2) for agroinfiltration into Sw‐5b‐transformed N. benthamiana. Cloning of the GP into pGR107 failed, probably because of size constraints and stability. As controls, constructs for the S RNA‐encoded N and NSS genes were included (Fig. 2; Ronde et al., 2013). All gene constructs were verified for their translatability from pEAQ‐HT or PVX during agroinfiltration of the constructs into untransformed N. benthamiana and subsequent Western immunoblot analysis. Although all NSM (Fig. 3C), NSS and N gene constructs (Ronde et al., 2013) were well expressed, the viral glycoproteins GN and GC processed from GP could not be detected (data not shown), probably because of the low transient expression levels, as witnessed earlier (D. Ribeiro, personal communication).
Figure 2.
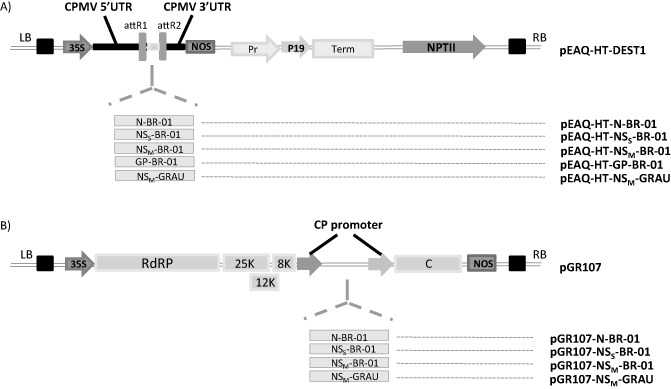
Schematic representation of the expression vector constructs used to challenge Nicotiana benthamiana transformants containing Sw‐5b. (A) Tomato spotted wilt virus (TSWV) genes (NSM of BR‐01 and GRAU isolates; N, NSS and the glycoprotein precursor (GP) from BR‐01) cloned individually into the pEAQ‐HT vector. AttR1/R2, Gateway recombination sites. (B) Tomato spotted wilt virus (TSWV) genes (NSM of BR‐01 and GRAU isolate, N, NSS) cloned individually in the appropriated restriction sites (ClaI and SalI) of the pGR107 vector (Lovato et al., 2008). LB, left border; RB, right border; 35S, Cauliflower mosaic virus promoter region; RdRP, Potato virus X (PVX) RNA‐dependent RNA polymerase; 25K, 12K and 8K, PVX movement proteins (triple gene block); CP promoter, PVX coat protein subgenomic RNA promoter; CP, PVX coat protein; NOS, nopaline synthase terminator.
Figure 3.
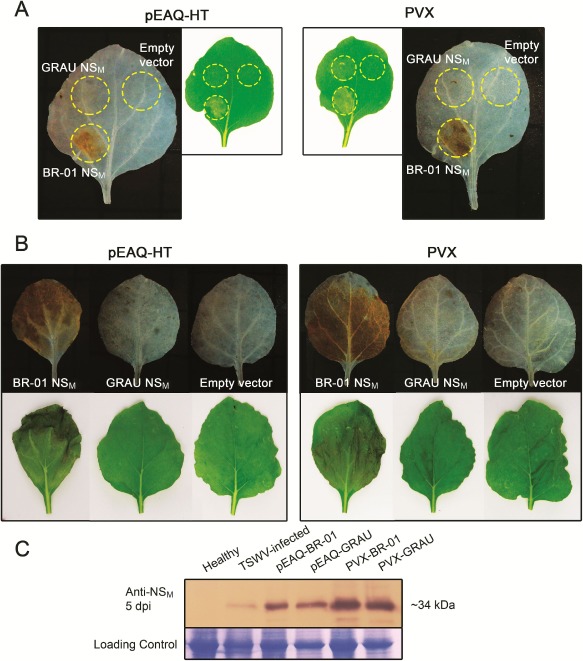
Response of transgenic Nicotiana benthamiana (Sw‐5b) leaves infiltrated with pEAQ‐HT and Potato virus X (PVX) constructs expressing NSM from isolates BR‐01 and GRAU, respectively. Photographs of leaves shown in (A) and (B) were taken 5–6 days post‐inoculation (dpi). In both panels, leaves were submitted to treatment with ethanol (destaining) for removal of chlorophyll to facilitate hypersensitive response (HR) visualization. (A) Individual leaf simultaneously infiltrated with three constructs pEAQ‐HT‐NSM ‐ BR ‐01, pEAQ‐HT‐NSM ‐ GRAU and pEAQ‐HT empty vector (left side) and with Potato virus X (PVX)‐Gw‐NSM ‐ BR ‐01, PVX‐Gw‐NSM ‐ GRAU and PVX‐Gw empty vector (right side), showing the visual response on infiltration. (B) Whole leaves infiltrated with pEAQ‐HT‐NSM ‐ BR ‐01, pEAQ‐HT‐NSM ‐ GRAU and pEAQ‐HT empty vector (left side) and with PVX‐Gw‐NSM ‐ BR ‐01, PVX‐Gw‐NSM ‐ GRAU and PVX‐Gw empty vector (right side), showing the visual response on infiltration. (C) Western immunoblot analysis with specific polyclonal antiserum against NSM protein (34 kDa) of Tomato spotted wilt virus (TWSV). 34 kDa, molecular weight marker protein. Samples were taken from N. benthamiana (Sw‐5b) plants inoculated with the wild‐type TSWV virus and infiltrated with the constructs pEAQ‐HT and PVX‐Gw containing NSM BR‐01 and NSM GRAU. Extract from healthy plants was used as a negative control.
Although, as expected, a visual HR was observed on challenge of Sw‐5b‐transformed N. benthamiana with TSWV BR‐01 (RI), and a systemic infection with TSWV GRAU (RB) (Fig. 1), repeated analysis consistently revealed a dark brownish necrosis, typical of an (upcoming) HR‐like response, only in leaves infiltrated with pEAQ‐HT‐NSM‐BR‐01 or pGR107+NSM‐BR‐01 (Fig. 3). Although photographs were taken at 5 and 6 dpi for better digital images, the HR‐like response was already observed at 3 dpi. This necrosis became more distinct later, but was not observed with pEAQ‐HT‐NSM‐GRAU or pGR107+NSM‐GRAU (Fig. 3A,B). The last two constructs revealed only a weak chlorosis after agroinfiltration of Sw‐5b‐transgenic N. benthamiana, similar to that caused by pEAQ‐HT or pGR107 without any insert (Fig. 3A,B), or containing GP, N or NSS from the RI BR‐01 strain, or just blank Agrobacterium tumefaciens. On chlorophyll removal of agroinfiltrated leaves, the HR‐like response after infiltration with NSM,BR‐01 was even more distinct (Fig. 3A,B). Infiltration of NSM‐BR‐01 and NSM‐GRAU constructs on leaves from untransformed N. benthamiana only revealed a weak interveinal chlorosis (Fig. S2, see Supporting Information).
Induction of HR in Sw‐5 near‐isogenic tomato lines
To further substantiate the observation that NSM represented the Avr determinant for Sw5‐b‐mediated resistance in a natural and commercial host fashion, a tomato breeding isoline (CNPH ‘LAM 147’) harbouring the Sw‐5 gene was agroinfiltrated with pEAQ‐HT constructs of NSM RI and NSM RB. At 4 dpi, leaves infiltrated with NSM RI showed a clear HR, whereas those infiltrated with NSM RB did not, which was more evident after chlorophyll removal from the leaves (Fig. 4).
Figure 4.
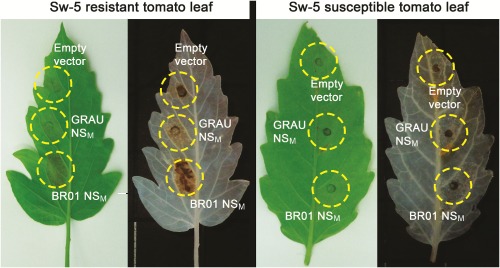
Response of susceptible and resistant (harbouring the Sw‐5 gene) tomato near‐isogenic line leaves infiltrated with individual pEAQ‐HT constructs expressing NSM from isolate BR‐01 or GRAU. Photographs of leaves shown were taken 4 days post‐inoculation (dpi). In both cases, leaves were submitted to treatment with ethanol (destaining) for removal of chlorophyll to facilitate hypersensitive response (HR) visualization. Individual leaves from the resistant line (left side) and susceptible isoline (right side) were simultaneously infected with three constructs pEAQ‐HTNSM ‐ BR ‐01, pEAQ‐HT‐NSM ‐ GRAU and pEAQ‐HT empty vector. Leaves of the resistant line showed the visual necrotic response (HR lesions) on infiltration.
Discussion
During earlier studies in which N. tabacum ‘SR1’ was transformed with Sw‐5 resistance gene candidates, Sw‐5b was identified as the functional resistance gene copy (Spassova et al., 2001). However, in these transgenic lines, no clear HR was observed on challenge with TSWV, and transgenic plants were totally immune. Here, we have successfully generated transgenic N. benthamiana containing Sw‐5b that responded with an HR on challenge with the RI TSWV BR‐01 (wild‐type reference strain), but not with the RB TSWV GRAU isolate (Aramburu and Marti, 2003). Although initially not reported for transgenic N. tabacum (Spassova et al., 2001), challenge of the Sw‐5b‐transgenic N. benthamiana also yielded resistance to GRSV and TCSV (data not shown), two tospovirus species phylogenetically close to TSWV and already reported to induce Sw‐5 resistance (Boiteux and Giordano, 1993). Surprisingly, protection was also observed against INSV, but not against MYSV, which belongs to a distant phylogenetic lineage of the Eur‐Asian tospoviruses (de Oliveira et al., 2012), indicating that Sw‐5b resistance acts against a broad spectrum of related tospoviruses belonging to the American tospovirus clade.
Using the Sw‐5b‐transformed N. benthamiana, and confirmed with a near‐isogenic tomato line, it was shown that Sw‐5b‐governed resistance is triggered by the NSM cell‐to‐cell movement protein and not by the (precursor to the) glycoproteins, both candidates for the avirulence determinant (Hoffmann et al., 2001), as visualized by the induction of HR after expression of NSM from the RI TSWV BR‐01 isolate, but not from the RB TSWV GRAU isolate. Neither gene triggered a response when agroinfiltrated into untransformed N. benthamiana (Fig. S1). In contrast with earlier work on the identification of the NSS RNA silencing suppressor as the Avr determinant of the single dominant Tsw resistance gene (Ronde et al., 2013), NSM was able to induce Sw‐5b‐mediated HR when expressed not only from the non‐replicative pEAQ‐HT vector system (Sainsbury et al., 2009), but also from a PVX replicon (pGR107).
An HR response is not restricted to R‐gene‐mediated defence, as it can also result from basal defence and non‐host resistance (Somssich and Hahlbrock, 1998), and can be uncoupled from the resistance response conferred by the R gene (Bendahmane et al., 1999); however, in general, it is activated after R genes have been triggered and is thereby used as an indirect indicator for R‐gene activation. The latter is assumed to involve direct or indirect interactions between the plant resistance gene product (R) and the pathogen avirulence factor (Avr) (Morel and Dangl, 1997). So far, only a small number of single dominant resistance genes against plant viruses have been cloned, and for these, as well as others for which the R genes have not yet been cloned, the polymerase protein (Kim and Palukaitis, 1997; Les Erickson et al., 1999), movement protein (Weber et al., 1993; Yoshikawa et al., 2006), coat protein (Asurmendi et al., 2004; Bendahmane et al., 2002) and RNA silencing suppressor (Li et al., 1999; Malcuit et al., 1999; Oh, 1995; Román et al., 2011; Ronde et al., 2013) have been identified as the Avr determinant.
Nowadays, RNA silencing is generally accepted as a virus‐triggered immunity mechanism in plants and is suppressed by viral RNA silencing suppressor proteins, also referred to as effectors. R‐gene‐mediated immunity is a second line of defence that is triggered by effectors. The resulting arms race, nicely illustrated by the zig–zag model (Dangl and Jones, 2001), thereby implies a link between RNAi and R‐gene‐mediated immunity for viral pathogens with a key role for viral RNA silencing suppressor proteins as effectors. Having identified the NSM movement protein as the Avr determinant for Sw‐5b raises the question as to how this protein should be projected as an effector in this same zig–zag model. Only a few cases have been reported in which a plant viral cell‐to‐cell movement protein has been identified as an effector and has been simultaneously shown to modulate the antiviral RNAi response (Lanfermeijer et al., 2003; Vogler et al., 2008). Whether TSWV NSM also contains such modulating activities remains to be investigated. Previously, TSWV NSM has been reported to provoke the deposition of 1,3‐β‐d‐glucan (GLU) or callose in mesophyll plasmodesmata (Pd), resembling the GLU depositions observed during an HR on viral infections (Rinne et al., 2005).
A recent bioinformatics analysis (López et al., 2011) on NSM sequences available from databases indicated that a few amino acid substitutions are responsible for the breakdown of resistance in tomato lines carrying the Sw‐5 gene. Although no experimental data were provided in support, the authors suggested that the ability of TSWV to infect resistant plants was related to the replacement of a cysteine (C) with tyrosine (Y) at position 118 (C118Y) or a threonine (T) with asparagine (N) at position 120 (T120N). The RB isolate TSWV GRAU used in this study contained C and N at positions 118 and 120, respectively, whereas the RI isolate TSWV BR‐01 contained C and T at these positions. Although TCSV and GRSV are classified as distinct tospovirus species, they are closely related to TSWV and are also able to induce the Sw‐5 resistance gene. Their NSM amino acid sequences share ∼85% similarity with TSWV NSM from the RI isolate, and contain C and N residues at similar positions. Whether a change in these residues is indeed responsible (only) for overcoming the resistance conferred by the dominant Sw‐5 gene remains to be investigated. In the light of this, it is interesting to note that INSV also triggers Sw‐5 resistance and its NSM (∼68% identity to TSWV NSM) contains residues H and T at amino acid positions 118 and 120, whereas MYSV does not trigger Sw‐5 resistance (sharing ∼40% identity) and contains residues I and T at these positions (Fig. S3, see Supporting Information).
Although the Sw‐5b gene was cloned a decade ago (Folkertsma et al., 1999; Spassova et al., 2001) and the Avr determinant of Sw‐5b‐governed resistance was mapped to the M RNA (Hoffmann et al., 2001), we have provided evidence that identifies NSM as the trigger for Sw‐5 resistance. Unravelling the mechanism by which Sw‐5b is triggered will now become a challenge and will not only contribute to provide an insight into dominant resistance genes, but may also help us to understand the broad‐spectrum resistance of Sw‐5b (against TSWV, TCSV, GRSV and INSV), a feature that is quite uncommon for a single dominant resistance gene, and which will be essential for the development of broad‐spectrum resistance strategies.
Experimental Procedures
Virus and plant material
Two TSWV isolates were used in this study, i.e. the Sw‐5b RI reference strain BR‐01 (de Ávila et al., 1993) and the RB isolate GRAU (Aramburu and Marti, 2003). Isolates were maintained by mechanical inoculations on N. benthamiana and grown under glasshouse conditions (24 °C with a 16‐h light/8‐h dark regime).
Nicotiana benthamiana transformation (Sw‐5b gene)
The construct pBIN+Sw‐5b containing the functional resistance gene copy Sw‐5b was introduced into A. tumefaciens strain LBA4404 and used to generate N. benthamiana transformants, in a similar manner to the procedure described previously to generate transgenic N. tabacum SR1 containing various Sw‐5 gene copies (Spassova et al., 2001).
Development of near‐isogenic tomato lines harbouring the Sw‐5 gene
The S. lycopersicum cultivar ‘Santa Clara’ (highly susceptible to Tospovirus species) was crossed (as female parent) with the resistant cultivar ‘Viradoro’, a germplasm source of the Sw‐5 locus derived from the tomato cultivar ‘Stevens’ (Dianese et al., 2011). After seven generations of backcross breeding (using ‘Santa Clara’ as a recurrent parent), a Tospovirus‐resistant near‐isogenic line was selected from a segregating F8 population. This inbred line (named ‘CNPH LAM 147’) was phenotypically very similar to ‘Santa Clara’, differing, however, in its resistance response to Tospovirus, because of the presence of the genomic segment encompassing the locus with the Sw‐5 gene cluster (Dianese et al., 2010). Progeny tests were carried out with ‘Santa Clara’ and ‘CNPH LAM 147’ in order to confirm the homozygous condition of both inbred lines. Seeds of these two genotypes were sown in 5‐L pots filled with sterile soil and maintained in a glasshouse. Mechanical inoculation with TSWV (BR‐01 isolate) was performed in 20 plants of ‘Santa Clara’ and 20 plants of the ‘CNPH LAM 147’ line, following standard procedures (Boiteux and Giordano, 1993). A co‐dominant molecular marker system (Dianese et al., 2010), able to discriminate between heterozygous resistant (Sw‐5/sw‐5) and homozygous resistant (Sw‐5/Sw‐5) plants, was employed in conjunction with the inoculation assays, aiming to obtain two pure and contrasting near‐isogenic lines. Total genomic DNA was extracted from asymptomatic ‘CNPH LAM 147’ and also from the mock‐inoculated ‘Santa Clara’ plants. A single dominant homozygous resistant (Sw‐5/Sw‐5) plant of the line ‘CNPH LAM 147’ and a single recessive homozygous susceptible (sw‐5/sw‐5) ‘Santa Clara’ plant were selected. Seeds of these two contrasting genotypes were multiplied under glasshouse conditions and used in all subsequent assays.
Cloning of N, NSM, NSS and the GP genes into binary vectors
The TSWV BR‐01 GP gene was excised with BamHI from pBIN19‐GP (Bucher et al., 2003) and, after agarose gel electrophoresis, purified and treated with T4 DNA polymerase. The pENTR11 entry vector was digested with EcoRI and, after agarose gel electrophoresis, was purified and religated to remove the ccdB region. The resulting pENTR11 vector, lacking the ccdB region, was digested with EcoRV and, after dephosphorylation, was used for cloning of the blunt‐ended GP gene, yielding the vector pENTR11+GPBR‐01. The TSWV BR‐01 NSM gene was amplified by PCR from pGR107+NSM‐BR‐01 (Lovato et al., 2008), and the GRAU NSM gene was amplified by RT‐PCR from total RNA of TSWV GRAU‐infected N. benthamiana extracted with Trizol (Invitrogen, Carlsbad, CA, USA). For the cDNA synthesis, M‐MLV reverse transcriptase was used (Promega, Madison, WI, USA). The PCRs were performed with primers (listed below) containing attB sites (Gateway Technology, Invitrogen), and the resulting PCR products were recombined into pDONR207 entry vector using BP Clonase (Invitrogen), yielding pDONR207+NSM‐BR‐01 and pDONR207+NSM‐GRAU. Primers for BR‐01 and GRAU NSM amplification: NSM‐BR01‐pD1 (GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGTTGACTCTTTTCGGTAACAA), NSM‐BR01‐pD2 (GGGGACCACTTTGTACAAGAAAGCTGGGTCCTATATTTCATCAAAGGATAACTG), NSM‐GRAU‐pD1 (GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGTTGACTTTTTTCAGCAATAAG) and NSM‐GRAU‐pD2 (GGGGACCACTTTGTACAAGAAAGCTGGGTCCTATATTTCATCAAAAGATA ACTG).
The entry vectors pENTR11+GPBR‐01, pDONR207+NSM‐BR‐01 and pDONR207+NSM‐GRAU were recombined into Gateway‐compatible pEAQ‐HT and pGR107 (Lacorte et al., 2010) by LR Clonase (Invitrogen), yielding pEAQ‐HT‐GPBR‐01, pEAQ‐HT‐NSM‐BR01, pEAQ‐HT‐NSM‐GRAU, pGR107‐NSM‐BR01 and pGR107‐NSM‐GRAU. The binary vectors pEAQ‐HT harbouring TSWV BR‐01 N and NSS genes were previously built and used as described (Ronde et al., 2013). All constructs were transformed into A. tumefaciens strain Cor308 or LBA4044. All standard procedures were performed following the manufacturers’ recommendations and Green and Sambrook (2012).
Agroinfiltration and monitoring of HR
Agrobacterium infiltration assays were performed according to the protocol of Bucher et al. (2003), with slight modifications. To this end, A. tumefaciens Cor308 and LBA4044 Ooms et al. (1982), harbouring single constructs, were grown overnight at 28 °C in LB3 medium containing 2 μg/mL tetracycline and 100 μg/mL kanamycin selection pressure. From this culture, 600 μL were freshly inoculated into 3 mL of induction medium and grown overnight. Leaves were agroinfiltrated with a suspension containing a final optical density at 600 nm (OD600 nm) of 1.0 per construct. The development of HR was monitored daily up to 12 days after agroinfiltration. For ease of monitoring and confirmation of HR, chlorophyll was removed from infiltrated leaves with ethanol and acetic acid (3:1, v/v).
RNA extraction and RT‐PCR
RNA extraction from wild‐type and Sw‐5b‐transformed N. benthamiana leaves was carried out using Trizol reagent (Invitrogen). Amplification of N genes via RT‐PCR was performed using M‐MLV reverse transcriptase (Promega) and Go Taq DNA polymerase (Promega) with the specific primers: TSWV‐N‐F (ATGTCTAAGGTTAAGCTCACTA), TSWV‐N‐R (TCAAGCAAGTTCTGCGAGTTTT), INSV‐N‐F (ATGAACAAAGCAAAGATTACCA), INSV‐N‐R (TTAAATAGAATCATTTTTCCCA), MYSV‐N‐F (ATGTCTACCGTTGCTAAGCTGA) and MYSV‐N‐R (TTAAACTTCAATGGACTT AGAT). All procedures followed the manufacturers’ instructions.
Western immunoblot detection of viral proteins
For Western immunoblot detection of TSWV proteins, four leaf discs from N. benthamiana leaves were macerated in 400 μL of phosphate buffer (0.01 m, pH 7.0). The supernatant was collected and supplemented with 2 × Laemmli buffer (Sigma‐Aldrich, St. Louis, MO, USA). Denatured proteins were resolved on 12.5% sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and, after electrophoresis, were transferred to Immobilon‐P membrane (Millipore, Billerica, MA, USA). Blotted membranes were blocked with PBS (50 mm sodium phosphate buffer; 10 mm NaCl) containing 2% non‐fat milk; the blots were then incubated using polyclonal antibodies (1 μg/mL) specific for TSWV N, NSS, NSM and GP proteins (de Avila et al., 1990; Kikkert et al., 1999; Kormelink et al., 1991, 1994).
Supporting information
Fig. S1 Response of Sw‐5b transgenic Nicotiana benthamiana on challenge with Tomato spotted wilt virus (TSWV), Impatiens necrotic spot virus (INSV) and Melon yellow spot virus (MYSV). Although TSWV and INSV induce a clear local hypersensitive response (HR), MYSV inoculation leads to a local and systemic infection.
Fig. S2 Response of untransformed, wild‐type Nicotiana benthamiana leaves infiltrated with pEAQ‐HT and Potato virus X (PVX) constructs expressing cell‐to‐cell movement protein (NSM) from isolates BR‐01 and GRAU.
Fig. S3 Alignment of the cell‐to‐cell movement proteins (NSM) discussed in this study. Identical amino acid residues are shaded in grey. The arrow indicates position 120 for Tomato spotted wilt virus (TSWV) NSM. GenBank accession numbers of the sequences: S58512 (TSWV BR‐01), FM163370 (TSWV GRAU), AF513220 (Groundnut ringspot virus, GRSV), AF213674 (Tomato chlorotic spot virus, TCSV), M74904 (Impatiens necrotic spot virus, INSV) and AB061773 (Melon yellow spot virus, MYSV). The alignment was performed via Vector NTI (Invitrogen).
Acknowledgements
This research was financially supported in part by the Dutch Technology Foundation STW (Stichting voor de Technische Wetenschappen), Applied Science Division of NWO (Nederlandse Organisatie voor Wetenschappelijk Onderzoek), and by CAPES (Coordenação de Aperfeiçoamento de Pssoal de Nível Superior) and WUR (Wageningen University) fellowships to MH, ASdO and EdCD. We would like to thank Dr George Lomonossoff for providing the pEAQ‐HT expression vector. The authors declare no conflicts of interest.
References
- Aramburu, J. and Marti, M. (2003) The occurrence in north‐east Spain of a variant of Tomato spotted wilt virus (TSWV) that breaks resistance in tomato (Lycopersicon esculentum) containing the Sw‐5 gene. Plant Pathol. 52, 407. [Google Scholar]
- Aramburu, J. , Galipienso, L. , Soler, S. and López, C. (2011) Characterization of Tomato spotted wilt virus isolates that overcome the Sw‐5 resistance gene in tomato and fitness assays. Phytopathol. Mediterr. 49, 342–351. [Google Scholar]
- de Ávila, A.C. , De Haan, P. , Smeets, M.L.L. , de Resende, R.O. , Kormelink, R. , Kitajima, E.W. , Goldbach, R.W. and Peters, D. (1993) Distinct levels of relationships between tospovirus isolates. Archives of Virology, 128, 211–227. [DOI] [PubMed] [Google Scholar]
- de Avila, A.C. , Huguenot, C. , Resende, R.O. , Kitajima, E.W. , Goldbach, R.W. and Peters, D. (1990) Serological differentiation of 20 isolates of Tomato spotted wilt virus. Journal of General Virology, 71: 2801–2807. [DOI] [PubMed] [Google Scholar]
- Asurmendi, S. , Berg, R. , Koo, J. and Beachy, R. (2004) Coat protein regulates formation of replication complexes during tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA, 101, 1415–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane, A. , Kanyuka, K. and Baulcombe, D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell Online, 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane, M. , Szécsi, J. , Chen, I. , Berg, R.H. and Beachy, R.N. (2002) Characterization of mutant tobacco mosaic virus coat protein that interferes with virus cell‐to‐cell movement. Proc. Natl. Acad. Sci. USA, 99, 3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux, L. and Giordano, L.d.B. (1993) Genetic basis of resistance against two Tospovirus species in tomato (Lycopersicon esculentum). Euphytica, 71, 151–154. [Google Scholar]
- Boiteux, L.S. (1995) Allelic relationships between genes for resistance to tomato spotted wilt tospovirus in Capsicum chinense . Theor. Appl. Genet. 90, 146–149. [DOI] [PubMed] [Google Scholar]
- Bucher, E. , Sijen, T. , de Haan, P. , Goldbach, R. and Prins, M. (2003) Negative‐strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J. Virol. 77, 1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Cho, J. , Custer, D. , Brommonschenkel, S. and Tanksley, S. (1995) Conventional breeding: host‐plant resistance and the use of molecular markers to develop resistance to tomato spot wilt virus in vegetables. Acta Hort. 431, 367–378. [Google Scholar]
- Ciuffo, M. , Finetti‐Sialer, M. , Gallitelli, D. and Turina, M. (2005) First report in Italy of a resistance‐breaking strain of Tomato spotted wilt virus infecting tomato cultivars carrying the Sw5 resistance gene. Plant Pathol. 54, 564. [Google Scholar]
- Dangl, J.L. and Jones, J.D. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dianese, E.C. , Fonseca, M.E.N. , Goldbach, R. , Kormelink, R. , Inoue‐Nagata, A.K. , Resende, R.O. and Boiteux, L.S. (2010) Development of a locus‐specific, co‐dominant SCAR marker for assisted‐selection of the Sw‐5 (Tospovirus resistance) gene cluster in a wide range of tomato accessions. Molecular Breeding, 25, 133–142. [Google Scholar]
- Dianese, E.C. , Fonseca, M.E.N. , Inoue‐Nagata, A.K. , Resende, R.O. and Boiteux, L.S. (2011) Search in Solanum (section Lycopersicon) germplasm for sources of broad‐spectrum resistance to four Tospovirus species. Euphytica, 180, 307–319. [Google Scholar]
- Finetti Sialer, M. , Lanave, C. , Padula, M. , Vovlas, C. and Gallitelli, D. (2002) Occurrence of two distinct Tomato spotted wilt virus subgroups in southern Italy. J. Plant Pathol. 84, 145–152. [Google Scholar]
- Finlay, K. (1953) Inheritance of spotted wilt resistance in the tomato II. Five genes controlling spotted wilt resistance in four tomato types. Aust. J. Biol. Sci. 6, 153–163. [PubMed] [Google Scholar]
- Folkertsma, R.T. , Spassova, M.I. , Prins, M. , Stevens, M.R. , Hille, J. and Goldbach, R.W. (1999) Construction of a bacterial artificial chromosome (BAC) library of Lycopersicon esculentum cv. Stevens and its application to physically map the Sw‐5 locus. Mol. Breed. 5, 197–207. [Google Scholar]
- Green, M.R. and Sambrook, J. (2012) Molecular Cloning: Laboratory Manual, 4th Edition Cold Spring Harbor: Laboratory Press. [Google Scholar]
- de Haan, P. , Kormelink, R., de , Oliveira Resende, R. , van Poelwijk, F. , Peters, D. and Goldbach, R. (1991) Tomato spotted wilt virus L RNA encodes a putative RNA polymerase. J. Gen. Virol. 72, 2207–2216. [DOI] [PubMed] [Google Scholar]
- Hoffmann, K. , Qiu, W. and Moyer, J. (2001) Overcoming host‐ and pathogen‐mediated resistance in tomato and tobacco maps to the M RNA of Tomato spotted wilt virus. Mol. Plant–Microbe Interact. 14, 242–249. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kikkert, M. , van Len, J. , Storms, M. , Bodegom, P. , Kormelink, R. and Goldbach, R. (1999) Tomato spotted wild virus particle morphogenesis in plant cells. Journal of Virology, 73, 2288–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.‐H. and Palukaitis, P. (1997) The plant defense response to cucumber mosaic virus in cowpea is elicited by the viral polymerase gene and affects virus accumulation in single cells. EMBO J. 16, 4060–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, A.M. , Adams, M.J. and Lefkowitz, E. (2011) Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Amsterdam: Elsevier. [Google Scholar]
- Kormelink, R. , Kitajima, E.W. , Haan, P.D. , Zuidema, D. , Peters, D. and Goldbach, R. (1991) The nonstructural protein (NSS) encoded by the ambisense S RNA segment of tomato spotted wilt virus is associated with fibrous structures in infected plant cells. Virology, 181, 459–468. [DOI] [PubMed] [Google Scholar]
- Kormelink, R. , de Haan, P. , Meurs, C. , Peters, D. and Goldbach, R. (1992) The nucleotide sequence of the M RNA segment of tomato spotted wilt virus, a bunyavirus with two ambisense RNA segments. J. Gen. Virol. 73, 2795–2804. [DOI] [PubMed] [Google Scholar]
- Kormelink, R. , Storms, M. , Van Lent, J. , Peters, D. and Goldbach, R. (1994) Expression and subcellular location of the NSM protein of Tomato Spotted Wilt Virus (TSWV), a putative viral movement protein. Virology, 200, 56–65. [DOI] [PubMed] [Google Scholar]
- Lacorte, C. , Ribeiro, S.G. , Lohuis, D. , Goldbach, R. and Prins, M. (2010) Potato virus X and Tobacco mosaic virus‐based vectors compatible with the Gateway™ cloning system. J. Virol. Meth. 164, 7–13. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer, F.C. , Dijkhuis, J. , Sturre, M.J. , de Haan, P. and Hille, J. (2003) Cloning and characterization of the durable tomato mosaic virus resistance gene Tm‐2(2) from Lycopersicon esculentum . Plant Mol. Biol. 52, 1037–1049. [DOI] [PubMed] [Google Scholar]
- Latham, L. and Jones, R. (1998) Selection of resistance breaking strains of tomato spotted wilt tospovirus. Ann. Appl. Biol. 133, 385–402. [Google Scholar]
- Les Erickson, F. , Holzberg, S. , Calderon‐Urrea, A. , Handley, V. , Axtell, M. , Corr, C. and Baker, B. (1999) The helicase domain of the TMV replicase proteins induces the N‐mediated defence response in tobacco. Plant J. 18, 67–75. [DOI] [PubMed] [Google Scholar]
- Li, H.‐W. , Lucy, A.P. , Guo, H.‐S. , Li, W.‐X. , Ji, L.‐H. , Wong, S.‐M. et al (1999) Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 18, 2683–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López, C. , Aramburu, J. , Galipienso, L. , Soler, S. , Nuez, F. and Rubio, L. (2011) Evolutionary analysis of tomato Sw‐5 resistance‐breaking isolates of Tomato spotted wilt virus. J. Gen. Virol. 92, 210–215. [DOI] [PubMed] [Google Scholar]
- Lovato, F.A. , Inoue‐Nagata, A.K. , Nagata, T. , de Ávila, A.C. , Pereira, L.A.R. and Resende, R.O. (2008) The N protein of Tomato spotted wilt virus (TSWV) is associated with the induction of programmed cell death (PCD) in Capsicum chinense plants, a hypersensitive host to TSWV infection. Virus Res. 137, 245–252. [DOI] [PubMed] [Google Scholar]
- Malcuit, I. , Marano, M.R. , Kavanagh, T.A. , De Jong, W. , Forsyth, A. and Baulcombe, D.C. (1999) The 25‐kDa movement protein of PVX elicits Nb‐mediated hypersensitive cell death in potato. Mol. Plant–Microbe Interact. 12, 536–543. [Google Scholar]
- Margaria, P. , Ciuffo, M. , Pacifico, D. and Turina, M. (2007) Evidence that the nonstructural protein of Tomato spotted wilt virus is the avirulence determinant in the interaction with resistant pepper carrying the Tsw gene. Mol. Plant–Microbe Interact. 20, 547–558. [DOI] [PubMed] [Google Scholar]
- Morel, J.‐B. and Dangl, J.L. (1997) The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T. and Brunner, F. (2002) Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen‐associated molecular patterns. Curr. Opin. Plant Biol. 5, 318–324. [DOI] [PubMed] [Google Scholar]
- Oh, J.‐W. (1995) Open reading frames of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. Mol. Plant–Microbe Interact. 8, 979–987. [DOI] [PubMed] [Google Scholar]
- de Oliveira, A.S. , Melo, F.L. , Inoue‐Nagata, A.K. , Nagata, T. , Kitajima, E.W. and Resende, R.O. (2012) Characterization of bean necrotic mosaic virus: a member of a novel evolutionary lineage within the Genus Tospovirus. PLoS ONE, 7, e38634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms, G. , Hooykaas, P.J.J. , Van Veen, R.J.M. , Van Beelen, P. , Regensburg‐Tuïnk, T.J.G. and Schilperoort, R.A. (1982) Octopine Ti‐plasmid deletion mutants of agrobacterium tumefaciens with emphasis on the right side of the T‐region. Plasmid, 7, 15–29. [DOI] [PubMed] [Google Scholar]
- Pan, Q. , Wendel, J. and Fluhr, R. (2000) Divergent evolution of plant NBS‐LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol. 50, 203–213. [DOI] [PubMed] [Google Scholar]
- Pappu, H. , Jones, R. and Jain, R. (2009) Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 141, 219–236. [DOI] [PubMed] [Google Scholar]
- Postel, S. and Kemmerling, B. (2009) Plant systems for recognition of pathogen‐associated molecular patterns. Seminars in Cell & Developmental Biology, 20, 1025–1031. [DOI] [PubMed] [Google Scholar]
- Price, D.L. , Memmott, F.D. , Scott, J.W. , Olson, S.M. and Stevens, M.R. (2007) Identification of molecular markers linked to a new Tomato spotted wilt virus resistance source in tomato. Tomato Genet. Coop. 57, 35–36. [Google Scholar]
- Rehman, S. , Postma, W. , Tytgat, T. , Prins, P. , Qin, L. , Overmars, H. et al (2009) A secreted SPRY domain‐containing protein (SPRYSEC) from the plant‐parasitic nematode Globodera rostochiensis interacts with a CC‐NB‐LRR protein from a susceptible tomato. Mol. Plant–Microbe Interact. 22, 330–340. [DOI] [PubMed] [Google Scholar]
- Rinne, P.L. , van den Boogaard, R. , Mensink, M.G. , Kopperud, C. , Kormelink, R. , Goldbach, R. et al (2005) Tobacco plants respond to the constitutive expression of the tospovirus movement protein NS(M) with a heat‐reversible sealing of plasmodesmata that impairs development. Plant J. 43, 688–707. [DOI] [PubMed] [Google Scholar]
- Román, A.C. , González‐Rico, F.J. , Moltó, E. , Hernando, H. , Neto, A. , Vicente‐Garcia, C. , Ballestar E., Gómez‐Skarmeta J.L., Vavrova‐Anderson J., White R.J., Montoliu L. and Fernández‐Salguero, P.M. (2011) Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 21, 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronde, D. , Butterbach, P. , Lohuis, D. , Hedil, M. , Lent, J.W. and Kormelink, R. (2013) Tsw gene‐based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus. Mol. Plant Pathol. 14, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi, M. and Warade, S. (2008) Tomato breeding for resistance to Tomato spotted wilt virus (TSWV): an overview of conventional and molecular approaches. Czech J. Genet. Plant Breed. 44, 83–92. [Google Scholar]
- Sainsbury, F. , Thuenemann, E.C. and Lomonossoff, G.P. (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693. [DOI] [PubMed] [Google Scholar]
- Scholthof, K.B.G. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. et al (2011) Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich, I.E. and Hahlbrock, K. (1998) Pathogen defence in plants—a paradigm of biological complexity. Trends Plant Sci. 3, 86–90. [Google Scholar]
- Spassova, M.I. , Prins, T.W. , Folkertsma, R.T. , Klein‐Lankhorst, R.M. , Hille, J. , Goldbach, R.W. et al (2001) The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine‐rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol. Breed. 7, 151–161. [Google Scholar]
- Stevens, M. , Scott, S. and Gergerich, R. (1991) Inheritance of a gene for resistance to tomato spotted wilt virus (TSWV) from Lycopersicon peruvianum Mill. Euphytica, 59, 9–17. [Google Scholar]
- Stevens, M. , Scott, S. and Gergerich, R. (1994) Evaluation of seven Lycopersicon species for resistance to tomato spotted wilt virus (TSWV). Euphytica, 80, 79–84. [Google Scholar]
- Stevens, M. , Lamb, E. and Rhoads, D. (1995) Mapping the Sw‐5 locus for tomato spotted wilt virus resistance in tomatoes using RAPD and RFLP analyses. Theor. Appl. Genet. 90, 451–456. [DOI] [PubMed] [Google Scholar]
- Storms, M.M.H. , Kormelink, R. , Peters, D. , Van Lent, J.W.M. and Goldbach, R.W. (1995) The nonstructural NSm protein of tomato spotted wilt virus induces tubular structures in plant and insect cells. Virology, 214, 485–493. [DOI] [PubMed] [Google Scholar]
- Takeda, A. , Sugiyama, K. , Nagano, H. , Mori, M. , Kaido, M. , Mise, K. , Tsuda, S. and Okuno, T. (2002) Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus . FEBS Lett. 532, 75–79. [DOI] [PubMed] [Google Scholar]
- Thompson, G. and Van Zijl, J. (1995) Control of tomato spotted wilt virus in tomatoes in South Africa. Tospoviruses and Thrips of Floral and Vegetable Crops 431, 379–384. [Google Scholar]
- Van Zijl, J. , Bosch, S. and Coetzee, C. (1985) Breeding tomatoes for processing in South Africa International Symposium on Fruit & Vegetables for Processing 194 (Strydom D.K., ed), pp. 69–76. Cape Town: Acta Horticulturae. [Google Scholar]
- Vogler, H. , Kwon, M.O. , Dang, V. , Sambade, A. , Fasler, M. , Ashby, J. and Heinlein, M. (2008) Tobacco mosaic virus movement protein enhances the spread of RNA silencing. Plos Pathog. 4, e1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, H. , Schultze, S. and Pfitzner, A. (1993) Two amino acid substitutions in the tomato mosaic virus 30‐kilodalton movement protein confer the ability to overcome the Tm‐2 (2) resistance gene in the tomato. J. Virol. 67, 6432–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, L. , Lambertini, P.L. , Shohara, K. and Biderbost, E. (2001) Occurrence and geographical distribution of tospovirus species infecting tomato crops in Argentina. Plant Dis. 85, 1227–1229. [DOI] [PubMed] [Google Scholar]
- Yoshikawa, N. , Okada, K. , Asamuma, K. , Watanabe, K. , Igarasi, A. , Li, C. and Isogai, M. (2006) A movement protein and three capsid proteins are all necessary for the cell‐to‐cell movement of apple latent spherical cheravirus. Arch. Virol. 151, 837–848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Response of Sw‐5b transgenic Nicotiana benthamiana on challenge with Tomato spotted wilt virus (TSWV), Impatiens necrotic spot virus (INSV) and Melon yellow spot virus (MYSV). Although TSWV and INSV induce a clear local hypersensitive response (HR), MYSV inoculation leads to a local and systemic infection.
Fig. S2 Response of untransformed, wild‐type Nicotiana benthamiana leaves infiltrated with pEAQ‐HT and Potato virus X (PVX) constructs expressing cell‐to‐cell movement protein (NSM) from isolates BR‐01 and GRAU.
Fig. S3 Alignment of the cell‐to‐cell movement proteins (NSM) discussed in this study. Identical amino acid residues are shaded in grey. The arrow indicates position 120 for Tomato spotted wilt virus (TSWV) NSM. GenBank accession numbers of the sequences: S58512 (TSWV BR‐01), FM163370 (TSWV GRAU), AF513220 (Groundnut ringspot virus, GRSV), AF213674 (Tomato chlorotic spot virus, TCSV), M74904 (Impatiens necrotic spot virus, INSV) and AB061773 (Melon yellow spot virus, MYSV). The alignment was performed via Vector NTI (Invitrogen).


