Summary
The interactions between viral RNAs and coat proteins (CPs) are critical for the efficient completion of infection cycles of RNA viruses. However, the specificity of the interactions between CPs and genomic or subgenomic RNAs remains poorly understood. In this study, Bamboo mosaic virus (BaMV) was used to analyse such interactions. Using reversible formaldehyde cross‐linking and mass spectrometry, two regions in CP, each containing a basic amino acid (R99 and R227, respectively), were identified to bind directly to the 5′ untranslated region of BaMV genomic RNA. Analyses of the alanine mutations of R99 and R227 revealed that the secondary structures of CP were not affected significantly, whereas the accumulation of BaMV genomic, but not subgenomic, RNA was severely decreased at 24 h post‐inoculation in the inoculated protoplasts. In the absence of CP, the accumulation levels of genomic and subgenomic RNAs were decreased to 1.1%–1.5% and 33%–40% of that of the wild‐type (wt), respectively, in inoculated leaves at 5 days post‐inoculation (dpi). In contrast, in the presence of mutant CPs, the genomic RNAs remained about 1% of that of wt, whereas the subgenomic RNAs accumulated to at least 87%, suggesting that CP might increase the accumulation of subgenomic RNAs. The mutations also restricted viral movement and virion formation in Nicotiana benthamiana leaves at 5 dpi. These results demonstrate that R99 and R227 of CP play crucial roles in the accumulation, movement and virion formation of BaMV RNAs, and indicate that genomic and subgenomic RNAs interact differently with BaMV CP.
Introduction
The efficient completion of infection cycles of RNA viruses requires specific interactions between several RNA‐binding proteins and viral RNAs. Among the virus‐encoded RNA‐binding proteins, the coat proteins (CPs) are involved in many crucial biological functions of viruses (Asurmendi et al., 2004; Herranz et al., 2012; Kao et al., 2011; Martinez‐Turino and Hernandez, 2010; Qu et al., 2003; Reichert et al., 2007; Tremblay et al., 2006). The comprehension of the interactions between CPs and viral RNAs is also important for the development of virus‐based applications in biotechnology (Douglas and Young, 1998; Kao et al., 2011; Young et al., 2008). The interactions between CPs and viral genomic RNAs have been studied extensively for several viruses (Aparicio et al., 2003; Kwon et al., 2005; Reade et al., 2010; Sit et al., 1994; Yi et al., 2009; Zimmern and Butler, 1977). However, for many viruses that express their genes via subgenomic RNAs, whether the same mechanism also governs the interaction between CPs and the subgenomic RNAs remains largely unknown.
Bamboo mosaic virus (BaMV) is a member of the genus Potexvirus, family Alphaflexiviridae, harbouring a positive‐sense RNA genome (Adams et al., 2005). The BaMV virion is a flexible rod, 470–580 nm in length and 15 nm in diameter, composed of CP and the genomic RNA (Lin et al., 1977). The 6.4‐kb genomic RNA possess a 5′ cap structure and a 3′ poly(A) tail, resembling the messenger RNAs of host plants, except that it is multi‐cistronic containing five conserved open reading frames (ORFs) (Lin et al., 1994). BaMV ORF1 encodes a 155‐kDa replication protein with three functional domains, including the capping enzyme domain (Huang et al., 2004; Li et al., 2001a), the helicase‐like domain (HLD) (Li et al., 2001b) and the RNA‐dependent RNA polymerase (RdRp) domain from the N‐ to C‐terminus (Li et al., 1998). The three movement proteins (the triple gene block, TGB) and CP (25 kDa) are produced from subgenomic RNAs. The 5′ untranslated region (UTR) of BaMV genomic RNA folds into a conserved secondary structure designated the ‘apical hairpin stem loop (AHSL)’, which is important in replication (Chen HC et al., 2012; Chen SC et al., 2010).
The CP of potexviruses is crucial for virion assembly, symptom expression and viral cell‐to‐cell movement through plasmodesmata into adjacent cells (Kwon et al., 2005; Lan et al., 2010; Lucas, 2006; Verchot‐Lubicz et al., 2010). Previous studies have demonstrated that the RNA‐binding affinity of CP of Cymbidium mosaic potexvirus (CymMV) plays a critical role in CymMV movement (Lu et al., 2009). Based on the alignment of the amino acid sequences of several potexvirus CPs, it has been revealed that the central consensus region of potexvirus CPs is the putative RNA‐binding domain (Abouhaidar and Lai, 1989; Lu et al., 2009). The RNA‐binding domain of the CP of another potexvirus, Papaya mosaic virus (PapMV), has been localized to amino acids 91–169 of PapMV CP. Within the domain, two amino acids (K97 and E128) have been shown to be important for the formation and stability of the nucleocapsid‐like particles (Tremblay et al., 2006). Despite detailed studies on the interactions between CP and genomic RNAs, little is known about those between CP and subgenomic RNAs.
For BaMV, TGBp1 and CP are the major RNA‐binding proteins which also interact with each other (Wu et al., 2011; Wung et al., 1999). Recently, the interaction between BaMV CP and HLD of the replication protein has been shown to play an important role in virus movement (Lee et al., 2011), suggesting that the intercellular trafficking ribonucleoprotein (RNP) complex is composed of viral RNA, TGBp1, CP and viral replication protein. Therefore, the RNA‐binding properties of BaMV CP should also be important in virion assembly and RNP movement. However, the precise RNA‐binding site(s) of BaMV CP and its interaction with subgenomic RNAs remain to be elucidated.
In this study, reversible formaldehyde cross‐linking, RNA affinity purification and mass spectrometry were used to identify the RNA‐binding regions of CP. Mutational studies were conducted to reveal the key amino acids responsible for the interactions with viral RNAs. The effects of mutant CPs on the accumulation and movement, and genomic and subgenomic RNAs, were further analysed in protoplasts and plants. The results revealed that the key amino acids in BaMV CP exert a different influence on genomic and subgenomic RNAs, providing further insights into the interactions between CP and viral RNAs.
Results
Identification of RNA‐binding peptides in CP
Previous studies have demonstrated the binding of Potato virus X (PVX) and PapMV CP to the cognate 5′ UTR of viral RNA (Kwon et al., 2005; Sit et al., 1994). To test whether BaMV CP interacts with the 5′ UTR of BaMV genomic RNA, electrophoretic mobility shift assay (EMSA) was performed with a 32P‐labelled RNA probe corresponding to the 5′ UTR of BaMV (nucleotides 1–93) (Fig. 1) and the recombinant BaMV CP expressed and purified from Escherichia coli (Fig. 2A). The results indicated that BaMV CP could interact with the 5′ UTR of BaMV genomic RNA, as observed for the other potexviruses. To locate the essential RNA region for CP binding, 32P‐labelled RNAs corresponding to nucleotides 1–52, 27–69 and 54–93 of BaMV 5′ UTR were prepared and subjected to EMSA. The result (Fig. 1) revealed that nucleotides 1–52 of BaMV RNA are required for efficient binding by BaMV CP under our assay conditions.
Figure 1.
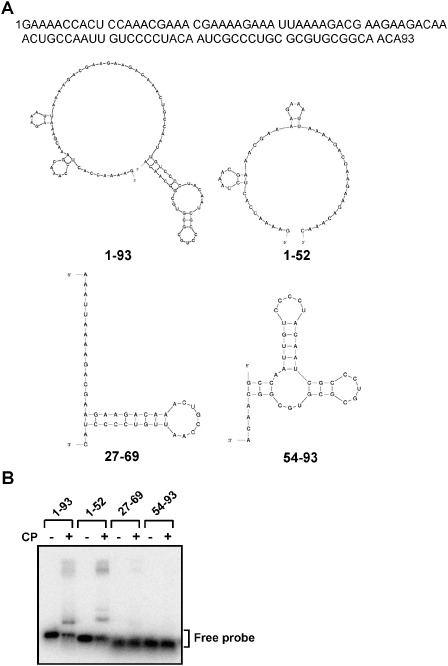
Affinities between Bamboo mosaic virus (BaMV) coat protein (CP) and various regions of BaMV RNA 5′ untranslated region (UTR). (A) The sequence and computer‐predicted (Zuker, 2003) secondary structure of radiolabelled probes of BaMV used in this figure. (B) Electrophoresis mobility shift assay (EMSA) was used to locate the RNA regions in the 5′ UTR required for interaction with BaMV CP. For each reaction, 1 fmol of radiolabelled probe, corresponding to nucleotides 1–93, 1–52, 27–69 or 54–93 in the 5′ UTR of BaMV RNA (as indicated at the top), was incubated either alone (–) or in the presence (+) of wild‐type (wt) BaMV CP (2 μm) for 10 min, followed by electrophoresis through a 1% agarose gel. The signals were analysed by the BAS‐2500 imaging system (Fujifilm). The position of the free probe is indicated on the right.
Figure 2.
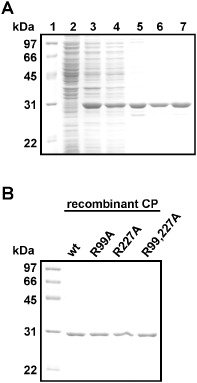
Purification of various recombinant Bamboo mosaic virus (BaMV) coat proteins (rCPs) from Escherichia coli. The proteins were purified from E. coli harbouring various constructs, including wild‐type (wt) (A), R99A, R227A and R99,227A, by chromatography using phenyl, DEAE and Superdex 200 columns sequentially. The purification products of each step were analysed by electrophoresis through a 12% polyacrylamide gel containing 1% sodium dodecylsulphate (SDS) (SDS‐PAGE) and stained with Coomassie blue. For (A): lane 1, molecular weight markers; lane 2, proteins without isopropyl‐β‐d‐thiogalactopyranoside (IPTG) induction; lane 3, IPTG‐induced proteins; lane 4, ammonium sulphate precipitation; lanes 5–7, eluted proteins from phenyl, DEAE and S200 columns, respectively. (B) Final purification products of various rCPs from E. coli. The rCPs were purified according to the same steps as in (A), and the final products were analysed by SDS‐PAGE.
The reversible formaldehyde cross‐linking method (Kim et al., 2005) was used to identify the RNA‐binding motif of BaMV CP. Biotinylated RNA of BaMV 5′ UTR 1–52 was used to map the RNA‐binding region in BaMV CP. The cross‐links between trypsin‐digested peptides and biotinylated RNA were reversed and the peptides were subjected to analysis by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS). The corresponding amino acid sequences are shown in Table 1. The experiment was repeated three times and, each time, the same results were obtained. As shown, two regions, amino acids 88–99 and 214–227, of BaMV CP were responsible for the binding to the 5′ UTR of BaMV RNA.
Table 1.
Identification of peptides cross‐linked with biotinylated RNA by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS)
| Experiment* | Amino acid range of coat protein | Observed (m/z) | Mr (expt)a | Mr (calc)b | Deltac | Sequenced,** |
|---|---|---|---|---|---|---|
| 1 | 88–99 | 666.8859 | 1331.7573 | 1331.7449 | 0.0124 | VLLGLSLEAFDR |
| 1 | 88–99 | 666.8982 | 1331.7817 | 1331.7449 | 0.0369 | VLLGLSLEAFDR |
| 1 | 214–227 | 804.4070 | 1606.7994 | 1606.7852 | 0.0142 | GFNLNYNPNVTQAR |
| 2 | 88–99 | 666.8862 | 1331.7579 | 1331.7449 | 0.0130 | VLLGLSLEAFDR |
| 2 | 88–99 | 666.8887 | 1331.7628 | 1331.7449 | 0.0179 | VLLGLSLEAFDR |
| 2 | 214–227 | 804.3960 | 1606.7775 | 1606.7852 | –0.0077 | GFNLNYNPNVTQAR |
| 3 | 88–99 | 666.8765 | 1331.7384 | 1331.7449 | –0.0065 | VLLGLSLEAFDR |
| 3 | 88–99 | 666.8838 | 1331.7530 | 1331.7449 | 0.0081 | VLLGLSLEAFDR |
| 3 | 214–227 | 804.3934 | 1606.7722 | 1606.7852 | –0.0130 | GFNLNYNPNVTQAR |
*The experiment was repeated three times and, each time, the same peptides were obtained. Two hits representing the peptide consisting of amino acid 88–99 were consistently observed.
Experimentally derived molecular mass.
Calculated molecular mass based on amino acid sequence.
Difference between experimentally derived and calculated mass.
The mutated basic residues, R99 and R227, are indicated in bold.
Formaldehyde mediates the cross‐linking between proteins and nucleic acids by the following steps (Barker et al., 2005): (i) formaldehyde reacts with the amino group of the N‐terminus or a side chain of the protein to form a Schiff base; and (ii) the Schiff base then reacts with another amino group on the base of a nucleotide to form the cross‐link. As glycine (0.2 m) was added in the cross‐linking reactions in our experiments, the amino acids that could be cross‐linked with RNAs included lysine (Lys), cysteine (Cys), tryptophan (Trp), histidine (His), arginine (Arg), asparagine (Asn), glutamine (Gln) and tyrosine (Tyr) (Metz et al., 2004, 2006; Ponicsan et al., 2013). Therefore, it is possible that the Arg residues in the peptides were available for cross‐linking with the viral RNAs. As formaldehyde has one of the shortest cross‐linking spans (about 2–3 Å) (Orlando et al., 1997), we hypothesized that the Arg residues (R99 and R227) were involved in the binding with the viral RNAs.
It is known that the cross‐links between Lys and RNA would interfere with the cleavage by trypsin (Steen and Mann, 2004). However, the cross‐links mediated by formaldehyde are between the amino groups of the side chain and the base, whereas the trypsin cleavage site is at the peptide bond C‐terminal to Lys and Arg (Olsen et al., 2004); thus, it is plausible that the cross‐linking between R99 or R227 to viral RNAs might still allow the digestion by trypsin, as shown previously by Kim et al. (2005).
Mutational analyses on the interaction between BaMV CP and viral RNAs: physical characterization of BaMV CP mutants
Each of the identified peptides contains a positively charged residue, Arg99 (R99) or Arg227 (R227), which might be crucial for RNA interaction. Therefore, alanine substitution mutants of the infectious clone pCBG, including two single mutants (R99A and R227A) and one double mutant (R99,227A), were created for functional analysis. Furthermore, the CP coding sequences of wild‐type (wt) and mutants were cloned into the pET29a expression vector to produce recombinant protein without extra amino acids and tags from the vector, and expressed in E. coli BL21(DE3) cells. These recombinant CPs were purified by sequential chromatography through phenyl‐sepharose, DEAE‐sepharose and Superdex 200 columns, as described in Experimental procedures. The quality of the CPs at the final purification step was monitored by sodium dodecylsulphate‐12% polyacrylamide gel electrophoresis (SDS‐PAGE), followed by staining with Coomassie blue (Fig. 2B). After purification, all of the recombinant CPs showed the ratio of 0.5–0.6 for the absorbance at 260 nm/280 nm, indicating that the protein preparations were free of nucleic acids (data not shown). The gel filtration results (Fig. 3A) showed that there were no significant differences among the elution profiles of the wt CP and the mutant CPs. This result suggested that the normal interactions between CP subunits were not affected significantly by the mutations at R99 and R227. To determine whether the mutations altered the secondary structures of the recombinant proteins, circular dichroism (CD) spectroscopy (Fig. 3B) was performed on the purified proteins. From the repeated CD analyses (with 10 replicates), it was shown that the main structures of the wt and mutant CPs were not significantly different. All CPs exhibited negative peaks at 208 and 222 nm and a positive peak at 193 nm, indicating that all the CPs analysed maintained primarily α‐helical structures (Holzwarth and Doty, 1965), consistent with the crystal structure of PapMV CP (Yang et al., 2012). Furthermore, the CD spectral data were analysed by the web‐based service K2D2 (Perez‐Iratxeta and Andrade‐Navarro, 2008), which predicted the same proportion of α‐helix (66.8%) and β‐strand (2.3%) in the wt and mutant CPs. Thus, the CD analysis suggested that the structures of the CPs were not affected significantly by the mutations. The differences in the intensities of the curves for different CPs might be attributed to the differences in protein concentrations (Tremblay et al., 2006).
Figure 3.
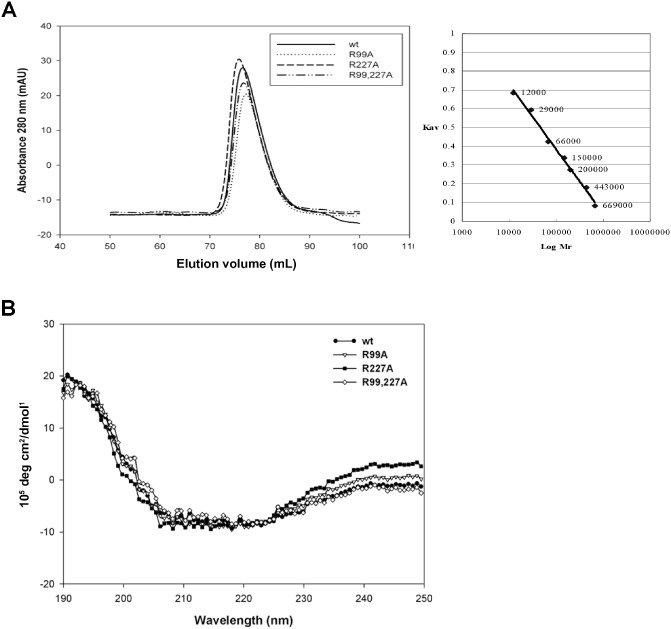
Analyses of the oligomerization and secondary structures of Bamboo mosaic virus (BaMV) coat protein (CP) wild‐type (wt) and mutants. (A) The chromatographic spectrum on a Superdex 200 column for BaMV CP wt and various mutants during purification. The calibration curve for the estimation of molecular size, inset on the right, was determined by protein standards for use in gel filtration. (B) Circular dichroism (CD) spectrum of various purified CPs (10 μg/mL) in 2 mm phosphate buffer (pH 7.2) with a 5‐mm cuvette, as determined by a Jasco J‐815 spectrometer (Jasco, Easton, MD, USA).
Mutational analyses on the interaction between BaMV CP and viral RNAs: RNA‐binding abilities of CP mutants
To investigate the effects of mutations of the putative RNA‐binding residues on RNA binding, EMSA was performed using the 32P‐labelled probe of BaMV 5′ UTR (nucleotides 1–52) and various CPs, as used in Fig. 2B, at two different concentrations. Three major shifted bands (Fig. 4A, complexes a, b and c) were detected. The appearance of multiple shifted bands suggested that the BaMV 5′ UTR might contain multiple CP‐binding sites or that the CP and BaMV 5′ UTR might form complexes with different conformations. Although the RNA‐binding capacities of CP mutants were similar to that of the wt, as estimated by the total radioactivities of the shifted bands, the abilities of the mutant CPs to form the largest complex (complex c) appeared to be disrupted. This result indicated that mutations in either of the two putative RNA‐binding residues negatively affected the interactions between the BaMV RNA and CP.
Figure 4.
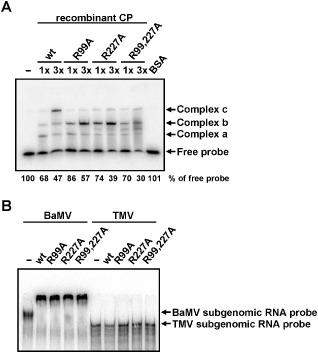
Effects of coat protein (CP) mutations on RNA binding. (A) The 32P‐labelled probe (1 fmol), representing nucleotides 1–52 of Bamboo mosaic virus (BaMV) RNA 5′ untranslated region (UTR), was incubated with 2 μm (1×) or 6 μm (3×) of various CPs as indicated at the top. Bovine serum albumin (BSA) (2 μm) was used as a negative control. The relative amounts (%) of free probe are shown at the bottom. (B) The 32P‐labelled probe (0.1 fmol) of CP subgenomic RNA of BaMV (nucleotides 5389–6366) or Tobacco mosaic virus (TMV) (nucleotides 5546–6392) was incubated with 1 μm of various CPs as indicated at the top. The samples were analysed by electrophoresis through a 5% (A) or 3.5% (B) polyacrylamide gel in 0.5 × TBE, and signals were detected by autoradiography. The positions of the free probe and shifted complexes with slower mobilities are indicated on the right. wt, wild‐type.
To further test whether BaMV CP interacts with BaMV subgenomic RNA, EMSA was performed with 32P‐labelled RNA probes representing CP subgenomic RNA of BaMV (nucleotides 5389–6366) and Tobacco mosaic virus (TMV, nucleotides 5546–6392) (Fig. 4B) and various CPs. The results showed that the wt and mutant CPs could interact with the CP subgenomic RNA of BaMV, but not with that of TMV, demonstrating the specific interaction between BaMV CP and the subgenomic RNA.
Mutational analyses on the interaction between BaMV CP and viral RNAs: the effects of CP mutations on viral accumulation in protoplasts and plants
To determine whether the mutant CPs affected the replication of BaMV, the infectious clone, pCBG, harbouring the green fluorescent protein (GFP) gene as the reporter, and the CP mutation derivatives (R99A, R227A and R99,227A) were inoculated into protoplasts of Nicotiana benthamiana. Accumulation levels of viral RNAs and CP were analysed by Northern blot and Western blot, respectively, at 16 and 24 h post‐inoculation (hpi). The accumulation levels of genomic and subgenomic RNAs were not significantly different at 16 hpi in protoplasts inoculated with wt or mutants (Fig. 5A), suggesting that the RNA replication abilities were not severely affected by different CPs. In comparison, at 24 hpi, the accumulation level of genomic RNAs was apparently decreased (to only 3.5% of wt), whereas subgenomic RNA accumulations were not severely affected (maintaining at least 59% of wt) (Fig. 5B). The wt and mutants accumulated similar levels of CPs, which were generated by translation from the 1.0‐kb subgenomic RNA, at 16 hpi (Fig. 5C). The accumulation levels of CP of the mutants were similar to those of the wt at 24 hpi (Fig. 5D). As the accumulation levels of genomic and subgenomic RNAs, and CP, were similar for wt and mutants at 16 hpi, suggesting that the replication and translation functions were not affected by the mutations, the differences in the accumulation levels of genomic RNAs of wt and mutants at 24 hpi could most probably be attributed to the reduced stability of genomic RNAs of the mutants (Fig. 5A). However, the possibility that CP might affect the replication of viral RNA could not be ruled out.
Figure 5.
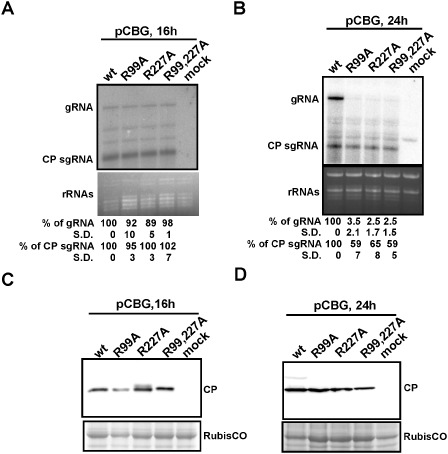
Effects of coat protein (CP) mutations on Bamboo mosaic virus (BaMV) accumulation in protoplasts. The protoplasts of Nicotiana benthamiana were inoculated with pCBG or the mutant derivatives, as indicated at the top. Total RNAs were extracted and analysed at 16 h post‐inoculation (hpi) (A) and 24 hpi (B) by Northern blot analysis using 32P‐labelled RNA probes complementary to the 3′ untranslated region (UTR) of positive‐sense BaMV RNA. Ethidium bromide‐stained rRNAs are shown as loading controls. The relative amounts (%) of viral genomic (g) and CP subgenomic (sg) RNAs, compared with that of the wild‐type (wt), are shown at the bottom. The percentage values shown represent the averages of all quantified samples. Three independent protoplast inoculation experiments were performed. The standard deviation is abbreviated as S.D. In (A), the differences between the accumulation levels of wt and each mutant were not statistically significant as analysed using Student's single‐tailed t‐test, with all P values >0.05. Total proteins were extracted from protoplasts at 16 hpi (C) and 24 hpi (D), separated by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE), transferred to a poly(vinylidene difluoride) (PVDF) membrane and probed with antiserum specific to BaMV CP. The positions of BaMV CPs are indicated on the right. The Coomassie blue‐stained gel, showing the positions around the Ribulose‐1,5‐bisphosphate carboxylase oxygenase protein (large subunit of RubisCO), was used as a loading control.
To further investigate the accumulation levels of the wt and mutants in whole plants at different time points after inoculation, the infectious clone, pKBG, and its mutant derivatives, were agroinfiltrated into N. benthamiana. Whole leaves were infiltrated with as much Agrobacterium suspension as possible to exclude the effects of mutations on cell‐to‐cell movement. The optical density at 600 nm (OD600) of all Agrobacterium suspensions was adjusted to 0.1, and the leaves were analysed at 2, 5 and 14 days post‐inoculation (dpi). Because GFP could only be expressed from the subgenomic RNAs produced during the replication of initial transcripts from pKBG and the mutant derivatives, the observation of green fluorescence may serve as an indirect indicator for the replication of BaMV genomic RNA and the transcription of subgenomic RNAs. Thus, the viral accumulations were monitored by examination of the green fluorescence levels using a Fujifilm LAS‐4000 luminescent image analyser (Fujifilm, Tokyo, Japan) and Olympus IX71 inverted microscope with an Olympus DP71 camera (Olympus, Tokyo, Japan) using GFP filters (Tseng et al., 2009) (Fig. 6A). The viral RNAs and proteins were also monitored in agroinfiltrated leaves with Northern and Western blotting at 2, 5 and 14 dpi (Fig. 6B,C). The results of Northern blot analyses revealed that only BaMV with wt CP could survive stably in agroinfiltrated leaves (Fig. 6B) and could spread to systemic leaves (data not shown) at 14 dpi. In contrast, although the accumulation of genomic RNAs for the mutants was reduced significantly (to only about 1% of that of the wt at 5 dpi), the subgenomic RNAs of the mutants accumulated to 39%–57% and 87%–95% of that of the wt at 2 and 5 dpi, respectively (Fig. 6B), consistent with the results observed in inoculated protoplasts at 24 hpi (Fig. 5B). It is conceivable that the low accumulation levels of mutant genomic RNAs might have led to the severely decreased accumulation levels of cognate subgenomic RNAs at 14 dpi (Fig. 6B). CP and GFP accumulated to similar levels in the leaves agroinfiltrated with all constructs at 5 dpi (Fig. 6C), whereas decreased amounts of CP and GFP for CP single mutants (R99A and R227A) were observed at 14 dpi (note that the protein samples analysed for wt at 14 dpi were 10‐ or two‐fold diluted) (Fig. 6C). For the double mutant (R99,227A), only trace amounts of CP and GFP were detected at 14 dpi (Fig. 6C). These results suggested that R99 and R227 of CP were both crucial for the accumulation of genomic RNAs, possibly by protecting them and increasing their stability.
Figure 6.
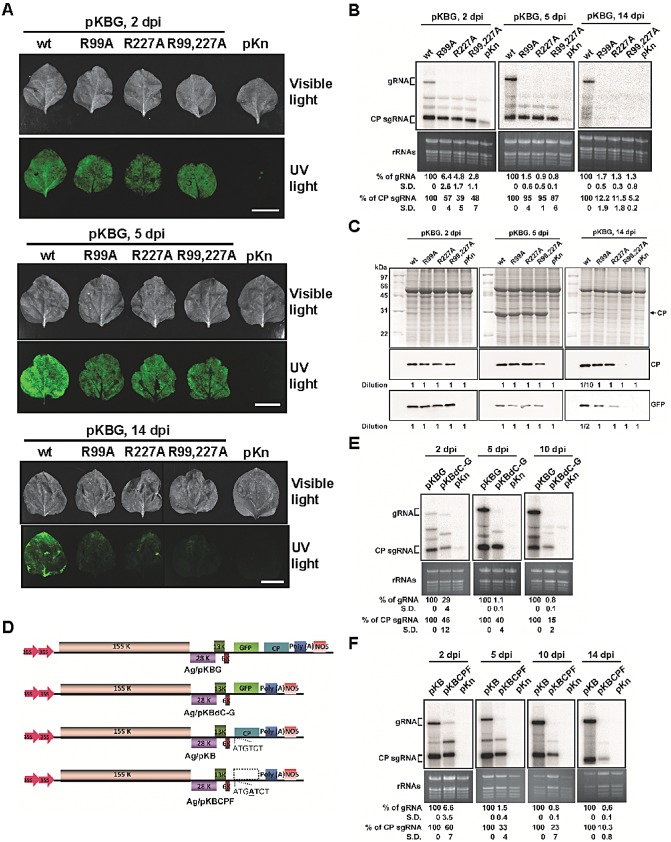
Effects of coat protein (CP) mutations on Bamboo mosaic virus (BaMV) accumulation in Nicotiana benthamiana plants. Leaves were infiltrated with Agrobacterium tumefaciens [optical density at 600 nm (OD600) of 0.1] harbouring pKBG (or pKB) or its derivatives carrying mutant CPs as indicated at the top. (A) Photographs (top panels) and fluorescence images (bottom panels) of infiltrated leaves were taken under visible light or UV illumination at 2, 5 and 14 days post‐inoculation (dpi) (scale bar, 1 cm). The vector backbone pKn was used as a negative control. wt, wild‐type. (B) Total RNAs were analysed by Northern blot hybridization to determine the accumulation of BaMV RNA in infiltrated leaves at 2, 5 and 14 dpi. The relative amounts (%) of viral genomic (g) and CP subgenomic (sg) RNAs, compared with that of the wt, are shown at the bottom. (C) Western blots were performed to detect the accumulation of various CPs and green fluorescent protein (GFP) in agroinfiltrated leaves at 2, 5 and 14 dpi using the respective antibodies as indicated on the right. As a result of the excessive difference in accumulation levels between the wt and mutants at 14 dpi, protein samples of the wt were 10‐ or two‐fold diluted, as indicated at the bottom, for Western blot analyses. (D–F) Effect of deletion of CP or its coding sequence on BaMV RNA accumulation in N. benthamiana. (D) Schematic diagrams of various BaMV‐based clones. The pKn vector was selected for use in Agrobacterium infiltration, as indicated by the ‘Ag/’ prefix. The CP deletion mutant, pKBdC‐G, was constructed by removing the CP open reading frame (ORF) in pKBG. To differentiate the effect of CP and its coding sequence, the CP ORF of pKB was disrupted by the insertion of an extra adenine to create the CP frame‐shift mutant, pKBCPF. (E) Nicotiana benthamiana leaves were infiltrated with A. tumefaciens harbouring pKBG, pKBdC‐G or pKn. Total RNAs were analysed by Northern blot hybridization to determine the accumulation of BaMV RNA in agroinfiltrated leaves at 2, 5 and 10 dpi. The relative amounts of viral RNAs are shown at the bottom. The vector backbone pKn was used as a negative control. Leaves infiltrated with pKBdC‐G became wilted at 14 dpi in all three experiments, and were not included in the Northern blot analyses. (F) Nicotiana benthamiana leaves were infiltrated with A. tumefaciens harbouring pKB, pKBCPF or pKn. Total RNAs were analysed as described above in agroinfiltrated leaves at 2, 5, 10 and 14 dpi. The relative amounts of viral RNAs are shown at the bottom. The percentages shown represent the averages of all quantified samples. Three independent experiments were performed. In each experiment, three (B, E) or four (F) 24–28‐day‐old N. benthamiana plants were used, and the third, fourth and fifth true leaves of each plant were inoculated. On days 2, 5, (10) and 14 post‐inoculation, one inoculated leaf was collected from three plants for analyses. To eliminate the differences in individual plants, the RNAs were extracted from the mixture of the three leaves and analysed by Northern blot hybridization.
The maintenance of nearly wt accumulation levels of subgenomic RNAs, but not of genomic RNAs, by the mutants at 5 dpi might have resulted from the stability of the subgenomic RNAs alone or the protection from degradation by the mutant CPs. To differentiate between the two possibilities, the CP deletion mutant of BaMV (pKBdC‐G), in which the CP ORF was replaced by that of GFP, was used to inoculate N. benthamiana via agroinfiltration as described above. The accumulation levels of viral genomic and subgenomic RNAs were monitored in agroinfiltrated leaves at 2, 5 and 10 dpi. The results revealed that, in the absence of CP, the accumulation of viral genomic and subgenomic RNAs was reduced over time, to 1.1% and 40%, respectively, of that of wt at 5 dpi (Fig. 6E). The decrease was less severe for subgenomic RNAs, suggesting that the stabilities of the subgenomic RNAs were higher than those of the genomic RNAs. To further verify whether the CP coding sequence might have contributed to the stability of the subgenomic RNAs, a CP frame‐shift mutant of BaMV (pKBCPF), in which the translation of CP was abolished but the coding sequence was retained (Fig. 6D), was constructed and assayed by agroinfiltration and Northern blot as described above. The results showed the accumulation of pKBCPF viral genomic and subgenomic RNAs to 1.5% and 33%, respectively, of that of wt at 5 dpi (Fig. 6F), similar to the values observed for pKBdC‐G. In comparison, in the presence of CPs of R99A, R227A and R99,227A, the subgenomic RNAs accumulated to 95%, 95% and 87%, respectively, of that of wt, whereas the genomic RNAs remained at about 1% (Fig. 6B). These results indicated that, although subgenomic RNAs exhibit higher stabilities without CP, the presence of CP could further increase the accumulation level of subgenomic RNAs, possibly by providing protection from degradation. In addition, the results also demonstrated that the three CP mutants (R99A, R277A and R99,227A) could help to maintain the accumulation of subgenomic RNAs, suggesting that CPs interact differently with genomic and subgenomic RNAs.
Viral movement was affected by CP mutations at the RNA‐binding motifs
The CP of potexviruses has been shown to be crucial in cell‐to‐cell movement (Lucas, 2006; Verchot‐Lubicz et al., 2010). In our unpublished work involving other CP mutants of BaMV, it was found that the mutants exhibited significantly different abilities in cell‐to‐cell movement even though the viral RNAs accumulated to a similar level (C.‐J. Hung et al., unpublished data). Hence, to investigate the effect of mutations at R99 and R227 of CP on BaMV movement, pCBG and the mutant derivatives were inoculated into the leaves of a local lesion host, Chenopodium quinoa, and the movement of the GFP‐expressing virus was examined by fluorescence microscopy. The results showed that the expression of GFP was only observed for viruses with the wt CP in inoculated leaves at 5 dpi (Fig. 7A). Furthermore, the cell‐to‐cell movement ability of the virus was monitored by an inverted microscope (Olympus IX71). The CP accumulation levels in C. quinoa leaves were also measured by Western blot analyses. It was found that the diameters of green fluorescent foci of the BaMV mutants were at least 10‐fold smaller than those of the wt in C. quinoa leaves at 5 dpi (Fig. 7B). The accumulation levels of mutant CPs were significantly lower than that of the wt, but all the mutant CPs could still be detected in inoculated leaves at 5 dpi (Fig. 7C), indicating that they were indeed translated into the infected cells, but incapable of supporting cell‐to‐cell movement. Taken together, the results suggested that R99 and R227 of CP are also required for efficient cell‐to‐cell movement of BaMV, in addition to the interaction with viral genomic RNA, although we could not rule out the possibility that the impaired cell‐to‐cell movement abilities of the mutants were the consequence of the reduced replication level of genomic RNAs. However, the results of Northern blot analysis in protoplasts (Fig. 5A) indicated that the RNA accumulation levels at 16 hpi were not significantly different among wt and mutants, suggesting that the replication abilities of the genomic RNAs were not severely affected by the mutations. Another possibility is that the stabilities of mutant genomic RNAs were reduced, which, in turn, restricted the cell‐to‐cell movement of the mutants. The stabilities of the genomic RNAs might be affected by their virion formation abilities. Thus, the effect of R99 and R227 mutations on virion formation abilities was further analysed as an indirect indication of the stabilities of viral RNAs.
Figure 7.
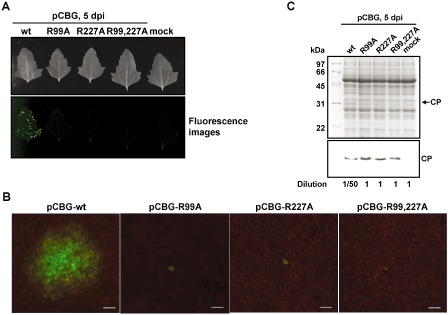
Effects of coat protein (CP) mutations on Bamboo mosaic virus (BaMV) movement in Chenopodium quinoa. (A) Photographs (top) and fluorescence images (bottom) of C. quinoa leaves inoculated with various constructs, as indicated on the top, were taken at 5 days post‐inoculation (dpi). (B) Fluorescence microscopic analysis to determine the cell‐to‐cell movement abilities of each construct. The fluorescent foci on inoculated C. quinoa leaves were visualized and measured by fluorescence microscopy. Scale bar, 200 μm. (C) Western blot analysis of BaMV CP in inoculated leaves. Total proteins extracted from C. quinoa leaves inoculated with various constructs were analysed by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE), followed by staining with Coomassie blue (top). For Western blot analysis using antiserum specific to BaMV CP, the sample of the wild‐type (wt) was diluted 50‐fold because of the excessive difference in the accumulation levels, as indicated at the bottom.
CP mutants are defective in virion formation
To determine whether R99 and R227 of CP were involved in virion assembly, the infectious clone pKBG and the three mutant derivatives (R99A, R227A and R99,227A) were inoculated via agroinfiltration into leaves of N. benthamiana, and virion formation were assayed at 5 dpi. BaMV virions were purified from agroinfiltrated leaves based on the standard protocol for BaMV virion purification (Lin and Chen, 1991). It was found that the leaf samples contained similar amounts of CP for wt and mutants, but abundant virion formation (detectable by Coomassie blue staining) was only observed in leaves inoculated with BaMV expressing the wt CP (Fig. 8A). Western blot analysis using antibodies specific to BaMV CP revealed the presence of low levels of virions of all three mutants (about 1/200 of that of the wt) (Fig. 8B), indicating that the efficiency of virion assembly was severely reduced for BaMV with the R99 and/or R227 mutations in CP. Similarly, BaMV virions were only observed in preparations from leaves inoculated with BaMV expressing the wt CP under a transmission electron microscope (Fig. 8C). These results were consistent with the notion that R99 and R227 of BaMV CP were involved in the efficient formation of viral particles. The results suggested that the reduced ability for virion assembly might be the cause of the reduced accumulation levels of the genomic RNAs.
Figure 8.
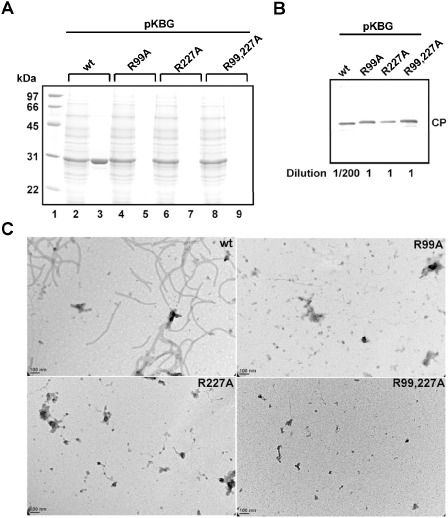
Effects of coat protein (CP) mutations on Bamboo mosaic virus (BaMV) virion formation. (A) Sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) of purified BaMV virions from leaves agroinfiltrated with various constructs, as indicated at the top, at 5 days post‐inoculation (dpi). The proteins were analysed by SDS‐PAGE, followed by Coomassie blue staining. Lane 1, protein molecular weight markers; lanes 2, 4, 6 and 8, protein samples from crude extracts following clarification with K2HPO4 and CaCl2; lanes 3, 5, 7 and 9, virion proteins from the final purification step. (B) Western blot analysis of the purified BaMV virion preparations. The blot was probed with antiserum specific to BaMV CP. The wild‐type (wt) sample was diluted 200‐fold to avoid overexposure. (C) Transmission electron micrographs of purified virions. Negative‐stained electron micrographs are shown with the same magnification (scale bar, 100 nm).
Discussion
The results presented in this study revealed the importance of the key amino acids, R99 and R229, in BaMV CP for the accumulation and cell‐to‐cell movement of viral RNAs, and formation of virions, and suggested that BaMV CPs exert different influences on the genomic and subgenomic RNAs. Recently, many RNA contact sites of different viral binding proteins have been identified by proteomics approaches and mutational analyses (Bhat and Savithri, 2011; Ji et al., 2011; Martinez‐Turino and Hernandez, 2010; Nishikiori et al., 2012; Semashko et al., 2012; Tremblay et al., 2006). The basic amino acids within the RNA‐binding motifs are frequently involved in RNA‐binding activity (Ansel‐McKinney et al., 1996; Chen and Varani, 2005; Herranz et al., 2012; Weiss and Narayana, 1998). For potexviruses, one of the two key amino acids, K97 and E128, in the RNA‐binding domain of PapMV CP is also basic (K97), which is required for nucleocapsid‐like particle formation (Tremblay et al., 2006). These previous studies highlighted the importance of the basic amino acids in the interactions between viral CP and RNAs.
However, the precise locations of the key basic amino acids within the CP of potexviruses are not conserved (Fig. S1, see Supporting Information), although amino acid sequence alignments have been applied to identify the crucial amino acids in the CPs of Clover yellow mosaic virus (Abouhaidar and Lai, 1989) and PapMV (Tremblay et al., 2006). In the current study, no RNA‐binding residue was identified in the proposed RNA‐binding motif (Tremblay et al., 2006), amino acids 119–175 of BaMV CP, based on the alignment with the known potexvirus CPs (Fig. S1). Instead, two RNA‐binding motifs of BaMV CP were identified by chemical cross‐linking (Table 1) in this study. The presence of a basic amino acid at positions relative to R227 of BaMV CP was highly conserved, whereas only Foxtail mosaic virus (FoMV) and BaMV harbour another basic amino acid (K120 and R99, respectively) within the N‐proximal RNA‐binding motif. These results suggested that, even within the genus Potexvirus, different virus species may have evolved different strategies for the encapsidation and movement of viral RNAs.
The crystal structure of PapMV has been described recently (Yang et al., 2012). The full‐length CP of PapMV is 215 amino acids in length, whereas the structure resolved by X‐ray crystallography (PDB accession number 4DOX) encompasses only the region from amino acids 11 to 174. By performing the structural modelling analysis using the Phyre2 protein fold recognition server (Kelley and Sternberg, 2009) with 4DOX as the template, a three‐dimensional model was predicted with 100% confidence, covering 66% (amino acids 43–201) of the BaMV CP sequence. The C‐terminal portion encompassing R227 was not modelled because of the lack of information in the template. However, it was shown (Fig. S2, see Supporting Information) that R99 of BaMV CP was located at the coil between helices 3 and 4, with a similar position to that of K97 of PapMV CP, which has been shown to be required for interaction with viral RNA, suggesting that R99 has the potential to interact with viral RNAs.
Apart from the motifs on viral CP, the encapsidation signals on viral RNAs are required for RNA–CP interactions and the efficient assembly of virions. The encapsidation signals have been identified for several viruses, such as the origin of assembly sequence (OAS) for TMV (Zimmern and Butler, 1977), a region encompassing 47 nucleotides at the 5′‐terminus on PapMV (Sit et al., 1994) and stem‐loop 1 structure for PVX (Kwon et al., 2005). In this study, the region in BaMV genomic RNA responsible for interacting with CP has also been narrowed down to nucleotides 1–52 of the BaMV 5′ UTR. This result is consistent with those for other potexviruses, in that the signals are located at the 5′ UTR of viral RNA.
However, our data further distinguished the functions of BaMV CP between RNA‐binding and virion formation, and demonstrated that genomic and subgenomic RNAs are affected differently by the mutations in the two key amino acids. In contrast with PapMV, in which the F13 of CP is required for the self‐assembly of CP into nucleocapsid‐like particles (Laliberte Gagne et al., 2008), R99 and R227 of BaMV CP are not required for interactions among BaMV CP subunits, as the secondary structures and the ability to oligomerize mutant CPs were not affected significantly (Fig. 3A,B), nor are they required for binding to the 5′ UTR of BaMV RNA (Fig. 4A) in the in vitro assays. The patterns of RNA–protein complexes in EMSA were affected by the mutations (Fig. 4A): the amounts of complex c were obviously reduced for the mutant CPs, which may have contributed to the severe reduction in the accumulation, cell‐to‐cell movement and encapsidation of viral genomic RNAs. In comparison, the efficiency of complex c formation by the mutant CPs had relatively little effect on the accumulation of subgenomic RNAs, in both protoplasts and plants.
Earlier studies have demonstrated that some mutations of PVX CP greatly reduce the accumulation of genomic RNA, but not subgenomic or negative‐strand RNA, in protoplasts at 16 hpi or for longer incubation times (Chapman et al., 1992). Our results (Figs 5B and 6B, E and F) provided further support for the notion that potexvirus CPs interact differently with genomic and subgenomic RNAs. In RNA viruses, deletion and replacement of the positively charged residues of CP have different effects on the recognition and packaging of different viral RNAs (Calhoun et al., 2007; Choi and Rao, 2000; Marshall and Schneemann, 2001; Venter et al., 2009). These observations indicate that different RNA‐binding sites of CP were used for the recognition and packaging of different RNAs.
In addition, it was also found that the cell‐to‐cell movement of BaMV genomic RNAs was severely hampered by the mutations of the two key amino acids (Fig. 7). The results of the electron microscopy analysis (Fig. 8) on the virion formation abilities of wt and mutants are consistent with the observations in Fig. 7 and the hypothesis that R99 and R227 of BaMV CP are important in virion formation, which, in turn, maintains the stability of viral RNAs and the cell‐to‐cell movement function of viruses. For the cell‐to‐cell movement of potexviruses, two models have been proposed: trafficking as virions or as RNP complexes (Cruz et al., 1998; Lough et al., 2000). As the mutations at R99 and/or R227 only severely abolished the assembly and accumulation of virions (Fig. 8), but not the in vitro RNA‐binding capability of CP, except for the formation of complex c (Fig. 4A), it appeared that BaMV moved between neighbouring cells in the virion form or in the RNP form as in complex c. The slower mobility of complex c suggested that more CP subunits were bound to the 32P‐labelled probes, forming a higher order structure. We have shown previously that the BaMV movement complex might be composed of viral RNA, TGBp1, CP and viral replication protein (Lee et al., 2011). Although our current data did not allow us to clearly distinguish between the two models, these results indicated that the formation of higher order complexes, such as complete virions or RNP complex c, is involved in the efficient cell‐to‐cell movement function of BaMV.
The results in this study provide further information on the differential interactions between CP and genomic or subgenomic RNAs of BaMV. The information also paves the way for the design of improved applications of BaMV as the vector or carrier for the expression of foreign proteins, as demonstrated previously (Chen TH et al., 2012; Yang et al., 2007). Studies on the mechanisms of the differential influences of BaMV CP on genomic or subgenomic RNAs are currently underway.
Experimental Procedures
Plasmid construction, in vitro transcription and recombinant protein expression
All infectious clones of BaMV were based on the backbones of pCBG (Lin et al., 2004) and pKn (Prasanth et al., 2011). Plasmid pUC119 (Vieira and Messing, 1987) was used to generate RNA transcripts in vitro. For the expression of recombinant CPs, pET29a (Novagen, Darmstadt, Germany) was used. Details on the construction of the plasmids used in this study, in vitro transcription and recombinant protein expression and purification are described in the Supporting Information Table S1.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed by the incubation of bacterium‐expressed wt and mutant CPs with in vitro transcribed radiolabelled RNA probes (1 fmol) at room temperature for 10 min in binding buffer [20 mm Tris‐HCl, pH 8.0, 3 mm MgCl2, 10 mm KCl, 2 mm dithiothreitol (DTT), 4% glycerol, 5 U RNase inhibitor]. The products were separated by electrophoresis through a 1% agarose gel or 3.5% or 5% PAGE in 0.5 × TBE (tris‐borate‐EDTA). Signals were analysed by a BAS‐2500 image analyser system (Fujifilm).
Reversible cross‐linking of CP and biotinylated RNA
The method for the cross‐linking of protein and biotinylated RNA by formaldehyde was based on a previous description (Han et al., 2009). Formaldehyde is a reversible cross‐linking agent widely used in protein–DNA, protein–RNA and protein–protein interactions (Niranjanakumari et al., 2002; Perez‐Romero and Imperiale, 2007; Sutherland et al., 2008). Briefly, 2 μm biotinylated RNA and 10 μm CP were incubated in EMSA binding buffer and 0.2% (v/v) formaldehyde at room temperature for 10 min before the addition of 0.2 m glycine to quench the reaction. The reaction was precipitated by acetone and dissolved in 50 μL of 100 mm ammonium bicarbonate (pH 8.0), followed by subsequent treatment of the cross‐linked complex with a sequencing grade trypsin (Trypsin Gold, Promega, Madison, WI, USA) at 37 °C overnight. Avidin beads (Promega) were washed with 25 mm ammonium bicarbonate (pH 7.8) and used to capture the biotinylated RNA–CP complexes. After washing 10 times with 25 mm ammonium bicarbonate (pH 7.8), the cross‐links in the samples were reversed by incubation at 70 °C for 1 h. The peptides were centrifuged at 10 000 g for 10 min and purified using a Ziptip (Millipore, Billerica, MA, USA). The identification of peptides was performed by LC‐MS/MS (Applied Biosystems Qstar‐XL Hybrid MS/MS, Foster City, CA, USA) and analysed by Mascot software (Matrix Science, Boston, MA, USA).
Inoculation of protoplasts and viral product analyses
Protocols for the preparation and inoculation of N. benthamiana protoplasts were performed as described previously (Cheng and Tsai, 1999). The protoplasts (approximately 2 × 105) were inoculated with 10 μg of plasmid DNA. Analyses of viral RNA and proteins by Northern and Western blotting in infected protoplasts were performed as described previously (Cheng and Tsai, 1999).
Agrobacterium infiltration
The pKn constructs were transformed into Agrobacterium strain pGV3850 by electroporation and grown on Luria–Bertani (LB) plates containing 50 ppm ampicillin, 10 ppm kanamycin and 10 ppm tetracycline at 28 °C (Giovannoni et al., 1989). Nicotiana benthamiana plants (24–28 days old) were grown in a glasshouse at 25 °C for leaf infiltration. The cultures of cells in LB medium with appropriate antibiotics were collected by centrifugation and resuspended in 10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES) buffer, pH 5.5, and 10 mm MgCl2 (Bendahmane et al., 2002). The bacterial suspensions were adjusted to an appropriate OD600 and infiltrated into leaves using a 1‐mL syringe without a needle.
Observation of GFP fluorescence
The expression of GFP in inoculated leaves of C. quinoa was observed with a Fujifilm LAS‐4000 and an Olympus IX71 inverted microscope with an Olympus DP71 camera (Olympus) using GFP filters (Tseng et al., 2009).
Virion purification
BaMV virions were purified as described previously (Lin and Chen, 1991), except that the final purification step was ultracentrifugation through a 20% sucrose cushion.
Transmission electron microscopy
The virions adsorbed on glow‐discharged carbon‐coated copper grids were stained with 2% uranyl acetate and examined with a transmission electron microscope (Philips CM 100 Bio, Eindhoven, The Netherlands) at 80 kV.
Supporting information
Fig. S1 Alignment of coat protein (CP) amino acid sequences of nine different potexviruses. The alignment was performed using the software ClustalW (Thompson et al., 2002) with the default parameters. The putative RNA‐binding motif (Tremblay et al., 2006) is underlined. The amino acids of Papaya mosaic virus (PapMV) (Tremblay et al., 2006) and Bamboo mosaic virus (BaMV) involved in RNA binding are indicated by open and filled stars, respectively. The amino acid sequences of the CPs of the nine potexviruses extensively characterized were selected from GenBank (Benson et al., 2013). The abbreviations and accession numbers are as follows: PAMV, Potato aucuba mosaic virus (gi|20177423); CymMV, Cymbidium mosaic virus (gi|46309858); ScaVX, Scallion virus X (gi|18652423); PVX, Potato virus X (gi|872290); HrV, Hydrangea ringspot virus (gi|62326909); CcMV, Cassava common mosaic virus (gi|9628112); PapMV, Papaya mosaic virus (gi|9629172); FoMV, Foxtail mosaic virus (gi|158268262); BaMV, Bamboo mosaic virus (gi|13182723).
Fig. S2 Comparison of the RNA‐binding sites in the Bamboo mosaic virus (BaMV) and Papaya mosaic virus (PapMV) coat protein (CP) structure. The structure of BaMV CP was modelled using the crystal structure of PapMV (PDB accession number 4DOX) as the template in the Phyre2 server (Kelley and Sternberg, 2009). The RNA‐binding residues of PapMV (K97) and BaMV (R99) CP are indicated by the arrows.
Table S1 The primers used in this study.
Acknowledgements
This research was supported by the National Science Council, Taiwan, Republic of China grants NSC‐101‐2313‐B‐005‐036‐MY3 and NSC‐97‐2752‐B‐005‐005‐PAE.
References
- Abouhaidar, M.G. and Lai, R. (1989) Nucleotide sequence of the 3′‐terminal region of clover yellow mosaic virus RNA. J. Gen. Virol. 70, 1871–1875. [DOI] [PubMed] [Google Scholar]
- Adams, M.J. , Accotto, G.P. , Agranovsky, A.A. , Bar‐Joseph, M. , Boscia, D. , Brunt, A.A. , Candresse, T. , Coutts, R.H.A. , Dolja, V. , Falk, W. , Foster, G. , Gonsalves, D. , Jelkmann, W. , Karasev, A. , Martelli, G.P. , Mawassi, M. , Milne, R.G. , Minafra, A. , Namba, S. , Rowhani, A. , Vetten, H.J. , Vishnichenko, V.K. , Wisler, G.C. , Yoshikawa, N. and Zavriev, S.K. (2005) Family Flexiviridae In: Virus Taxonomy: Classification and Nomenclature of Viruses; Eighth Report of the International Committee on Taxonomy of Viruses (Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U. and Ball L.A., eds), pp. 1089–1124. San Diego, CA: Academic Press. [Google Scholar]
- Ansel‐McKinney, P. , Scott, S.W. , Swanson, M. , Ge, X. and Gehrke, L. (1996) A plant viral coat protein RNA binding consensus sequence contains a crucial arginine. EMBO J. 15, 5077–5084. [PMC free article] [PubMed] [Google Scholar]
- Aparicio, F. , Vilar, M. , Perez‐Paya, E. and Pallas, V. (2003) The coat protein of prunus necrotic ringspot virus specifically binds to and regulates the conformation of its genomic RNA. Virology, 313, 213–223. [DOI] [PubMed] [Google Scholar]
- Asurmendi, S. , Berg, R.H. , Koo, J.C. and Beachy, R.N. (2004) Coat protein regulates formation of replication complexes during tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA, 101, 1415–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, S. , Weinfeld, M. and Murray, D. (2005) DNA–protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 589, 111–135. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Farnham, G. , Moffett, P. and Baulcombe, D.C. (2002) Constitutive gain‐of‐function mutants in a nucleotide binding site‐leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32, 195–204. [DOI] [PubMed] [Google Scholar]
- Bhat, A.S. and Savithri, H.S. (2011) Investigations on the RNA binding and phosphorylation of groundnut bud necrosis virus nucleocapsid protein. Arch. Virol. 156, 2163–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun, S.L. , Speir, J.A. and Rao, A.L. (2007) In vivo particle polymorphism results from deletion of a N‐terminal peptide molecular switch in brome mosaic virus capsid protein. Virology, 364, 407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, S. , Hills, G. , Watts, J. and Baulcombe, D. (1992) Mutational analysis of the coat protein gene of potato virus X: effects on virion morphology and viral pathogenicity. Virology, 191, 223–230. [DOI] [PubMed] [Google Scholar]
- Chen, H.C. , Kong, L.R. , Yeh, T.Y. , Cheng, C.P. , Hsu, Y.H. and Lin, N.S. (2012) The conserved 5′ apical hairpin stem loops of bamboo mosaic virus and its satellite RNA contribute to replication competence. Nucleic Acids Res. 40, 4641–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S.C. , Desprez, A. and Olsthoorn, R.C. (2010) Structural homology between bamboo mosaic virus and its satellite RNAs in the 5′ untranslated region. J. Gen. Virol. 91, 782–787. [DOI] [PubMed] [Google Scholar]
- Chen, T.H. , Hu, C.C. , Liao, J.T. , Lee, C.W. , Liao, J.W. , Lin, M.Y. , Liu, H.J. , Wang, M.Y. , Lin, N.S. and Hsu, Y.H. (2012) Induction of protective immunity in chickens immunized with plant‐made chimeric Bamboo mosaic virus particles expressing very virulent infectious bursal disease virus antigen. Virus Res. 166, 109–115. [DOI] [PubMed] [Google Scholar]
- Chen, Y. and Varani, G. (2005) Protein families and RNA recognition. FEBS J. 272, 2088–2097. [DOI] [PubMed] [Google Scholar]
- Cheng, C.P. and Tsai, C.H. (1999) Structural and functional analysis of the 3′ untranslated region of bamboo mosaic potexvirus genomic RNA. J. Mol. Biol. 288, 555–565. [DOI] [PubMed] [Google Scholar]
- Choi, Y.G. and Rao, A.L. (2000) Molecular studies on bromovirus capsid protein. VII. Selective packaging on BMV RNA4 by specific N‐terminal arginine residuals. Virology, 275, 207–217. [DOI] [PubMed] [Google Scholar]
- Cruz, S.S. , Roberts, A.G. , Prior, D.A. , Chapman, S. and Oparka, K.J. (1998) Cell‐to‐cell and phloem‐mediated transport of potato virus X. The role of virions. Plant Cell, 10, 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, T. and Young, M. (1998) Host–guest encapsulation of materials by assembled virus protein cages. Nature, 393, 152–155. [Google Scholar]
- Giovannoni, J.J. , DellaPenna, D. , Bennett, A.B. and Fischer, R.L. (1989) Expression of a chimeric polygalacturonase gene in transgenic rin (ripening inhibitor) tomato fruit results in polyuronide degradation but not fruit softening. Plant Cell, 1, 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y.T. , Hsu, Y.H. , Lo, C.W. and Meng, M. (2009) Identification and functional characterization of regions that can be crosslinked to RNA in the helicase‐like domain of BaMV replicase. Virology, 389, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz, M.C. , Pallas, V. and Aparicio, F. (2012) Multifunctional roles for the N‐terminal basic motif of Alfalfa mosaic virus coat protein: nucleolar/cytoplasmic shuttling, modulation of RNA‐binding activity, and virion formation. Mol. Plant–Microbe Interact. 25, 1093–1103. [DOI] [PubMed] [Google Scholar]
- Holzwarth, G. and Doty, P. (1965) The ultraviolet circular dichroism of polypeptides. J. Am. Chem. Soc. 87, 218–228. [DOI] [PubMed] [Google Scholar]
- Huang, Y.L. , Han, Y.T. , Chang, Y.T. , Hsu, Y.H. and Meng, M. (2004) Critical residues for GTP methylation and formation of the covalent m7GMP‐enzyme intermediate in the capping enzyme domain of bamboo mosaic virus. J. Virol. 78, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, X. , Qian, D. , Wei, C. , Ye, G. , Zhang, Z. , Wu, Z. , Xie, L. and Li, Y. (2011) Movement protein Pns6 of rice dwarf phytoreovirus has both ATPase and RNA binding activities. PLoS ONE, 6, e24986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, C.C. , Ni, P. , Hema, M. , Huang, X. and Dragnea, B. (2011) The coat protein leads the way: an update on basic and applied studies with the Brome mosaic virus coat protein. Mol. Plant Pathol. 12, 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L.A. and Sternberg, M.J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. [DOI] [PubMed] [Google Scholar]
- Kim, Y.C. , Russell, W.K. , Ranjith‐Kumar, C.T. , Thomson, M. , Russell, D.H. and Kao, C.C. (2005) Functional analysis of RNA binding by the hepatitis C virus RNA‐dependent RNA polymerase. J. Biol. Chem. 280, 38 011–38 019. [DOI] [PubMed] [Google Scholar]
- Kwon, S.J. , Park, M.R. , Kim, K.W. , Plante, C.A. , Hemenway, C.L. and Kim, K.H. (2005) cis‐Acting sequences required for coat protein binding and in vitro assembly of Potato virus X. Virology, 334, 83–97. [DOI] [PubMed] [Google Scholar]
- Laliberte Gagne, M.E. , Lecours, K. , Gagne, S. and Leclerc, D. (2008) The F13 residue is critical for interaction among the coat protein subunits of papaya mosaic virus. FEBS J. 275, 1474–1484. [DOI] [PubMed] [Google Scholar]
- Lan, P. , Yeh, W.B. , Tsai, C.W. and Lin, N.S. (2010) A unique glycine‐rich motif at the N‐terminal region of Bamboo mosaic virus coat protein is required for symptom expression. Mol. Plant–Microbe Interact. 23, 903–914. [DOI] [PubMed] [Google Scholar]
- Lee, C.C. , Ho, Y.N. , Hu, R.H. , Yen, Y.T. , Wang, Z.C. , Lee, Y.C. , Hsu, Y.H. and Meng, M. (2011) The interaction between bamboo mosaic virus replication protein and coat protein is critical for virus movement in plant hosts. J. Virol. 85, 12 022–12 031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.I. , Cheng, Y.M. , Huang, Y.L. , Tsai, C.H. , Hsu, Y.H. and Meng, M. (1998) Identification and characterization of the Escherichia coli‐expressed RNA‐dependent RNA polymerase of bamboo mosaic virus. J. Virol. 72, 10 093–10 099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.I. , Chen, Y.J. , Hsu, Y.H. and Meng, M. (2001a) Characterization of the AdoMet‐dependent guanylyltransferase activity that is associated with the N terminus of bamboo mosaic virus replicase. J. Virol. 75, 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.I. , Shih, T.W. , Hsu, Y.H. , Han, Y.T. , Huang, Y.L. and Meng, M. (2001b) The helicase‐like domain of plant potexvirus replicase participates in formation of RNA 5′ cap structure by exhibiting RNA 5′‐triphosphatase activity. J. Virol. 75, 12 114–12 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M.K. , Chang, B.Y. , Liao, J.T. , Lin, N.S. and Hsu, Y.H. (2004) Arg‐16 and Arg‐21 in the N‐terminal region of the triple‐gene‐block protein 1 of Bamboo mosaic virus are essential for virus movement. J. Gen. Virol. 85, 251–259. [DOI] [PubMed] [Google Scholar]
- Lin, M.T. , Kitajima, E.W. , Cupertino, F.P. and Costa, C.L. (1977) Partial purification and some properties of bamboo mosaic virus. Phytopathology, 67, 1439–1443. [Google Scholar]
- Lin, N.S. and Chen, C.C. (1991) Association of bamboo mosaic virus (bamv) and bamv specific electron‐dense crystalline bodies with chloroplasts. Phytopathology, 81, 1551–1555. [Google Scholar]
- Lin, N.S. , Lin, B.Y. , Lo, N.W. , Hu, C.C. , Chow, T.Y. and Hsu, Y.H. (1994) Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. J. Gen. Virol. 75, 2513–2518. [DOI] [PubMed] [Google Scholar]
- Lough, T.J. , Netzler, N.E. , Emerson, S.J. , Sutherland, P. , Carr, F. , Beck, D.L. , Lucas, W.J. and Forster, R.L. (2000) Cell‐to‐cell movement of potexviruses: evidence for a ribonucleoprotein complex involving the coat protein and first triple gene block protein. Mol. Plant–Microbe Interact. 13, 962–974. [DOI] [PubMed] [Google Scholar]
- Lu, H.C. , Chen, C.E. , Tsai, M.H. , Wang, H.I. , Su, H.J. and Yeh, H.H. (2009) Cymbidium mosaic potexvirus isolate‐dependent host movement systems reveal two movement control determinants and the coat protein is the dominant. Virology, 388, 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, W.J. (2006) Plant viral movement proteins: agents for cell‐to‐cell trafficking of viral genomes. Virology, 344, 169–184. [DOI] [PubMed] [Google Scholar]
- Marshall, D. and Schneemann, A. (2001) Specific packaging of nodaviral RNA2 requires the N‐terminus of the capsid protein. Virology, 285, 165–175. [DOI] [PubMed] [Google Scholar]
- Martinez‐Turino, S. and Hernandez, C. (2010) Identification and characterization of RNA‐binding activity in the ORF1‐encoded replicase protein of Pelargonium flower break virus. J. Gen. Virol. 91, 3075–3084. [DOI] [PubMed] [Google Scholar]
- Metz, B. , Kersten, G.F. , Hoogerhout, P. , Brugghe, H.F. , Timmermans, H.A. , de Jong, A. , Meiring, H. , ten Hove, J. , Hennink, W.E. , Crommelin, D.J. and Jiskoot, W. (2004) Identification of formaldehyde‐induced modifications in proteins: reactions with model peptides. J. Biol. Chem. 279, 6235–6243. [DOI] [PubMed] [Google Scholar]
- Metz, B. , Kersten, G.F. , Baart, G.J. , de Jong, A. , Meiring, H. , ten Hove, J. , van Steenbergen, M.J. , Hennink, W.E. , Crommelin, D.J. and Jiskoot, W. (2006) Identification of formaldehyde‐induced modifications in proteins: reactions with insulin. Bioconjug. Chem. 17, 815–822. [DOI] [PubMed] [Google Scholar]
- Niranjanakumari, S. , Lasda, E. , Brazas, R. and Garcia‐Blanco, M.A. (2002) Reversible cross‐linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods, 26, 182–190. [DOI] [PubMed] [Google Scholar]
- Nishikiori, M. , Sugiyama, S. , Xiang, H. , Niiyama, M. , Ishibashi, K. , Inoue, T. , Ishikawa, M. , Matsumura, H. and Katoh, E. (2012) Crystal structure of the superfamily 1 helicase from Tomato mosaic virus. J. Virol. 86, 7565–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, J.V. , Ong, S.E. and Mann, M. (2004) Trypsin cleaves exclusively C‐terminal to arginine and lysine residues. Mol. Cell. Proteomics, 3, 608–614. [DOI] [PubMed] [Google Scholar]
- Orlando, V. , Strutt, H. and Paro, R. (1997) Analysis of chromatin structure by in vivo formaldehyde cross‐linking. Methods, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- Perez‐Iratxeta, C. and Andrade‐Navarro, M.A. (2008) K2D2: estimation of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 8, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Romero, P. and Imperiale, M.J. (2007) Assaying protein–DNA interactions in vivo and in vitro using chromatin immunoprecipitation and electrophoretic mobility shift assays. Methods Mol. Med. 131, 123–139. [DOI] [PubMed] [Google Scholar]
- Ponicsan, S.L. , Houel, S. , Old, W.M. , Ahn, N.G. , Goodrich, J.A. and Kugel, J.F. (2013) The non‐coding B2 RNA binds to the DNA cleft and active‐site region of RNA polymerase II. J. Mol. Biol. 425, 3625–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth, K.R. , Huang, Y.W. , Liou, M.R. , Wang, R.Y. , Hu, C.C. , Tsai, C.H. , Meng, M. , Lin, N.S. and Hsu, Y.H. (2011) Glyceraldehyde 3‐phosphate dehydrogenase negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. J. Virol. 85, 8829–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F. , Ren, T. and Morris, T.J. (2003) The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77, 511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reade, R. , Kakani, K. and Rochon, D. (2010) A highly basic KGKKGK sequence in the RNA‐binding domain of the Cucumber necrosis virus coat protein is associated with encapsidation of full‐length CNV RNA during infection. Virology, 403, 181–188. [DOI] [PubMed] [Google Scholar]
- Reichert, V.L. , Choi, M. , Petrillo, J.E. and Gehrke, L. (2007) Alfalfa mosaic virus coat protein bridges RNA and RNA‐dependent RNA polymerase in vitro. Virology, 364, 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semashko, M.A. , Gonzalez, I. , Shaw, J. , Leonova, O.G. , Popenko, V.I. , Taliansky, M.E. , Canto, T. and Kalinina, N.O. (2012) The extreme N‐terminal domain of a hordeivirus TGB1 movement protein mediates its localization to the nucleolus and interaction with fibrillarin. Biochimie, 94, 1180–1188. [DOI] [PubMed] [Google Scholar]
- Sit, T.L. , Leclerc, D. and AbouHaidar, M.G. (1994) The minimal 5′ sequence for in vitro initiation of papaya mosaic potexvirus assembly. Virology, 199, 238–242. [DOI] [PubMed] [Google Scholar]
- Steen, H. and Mann, M. (2004) The ABC's (and XYZ's) of peptide sequencing. Nat. Rev. Mol. Cell Biol. 5, 699–711. [DOI] [PubMed] [Google Scholar]
- Sutherland, B.W. , Toews, J. and Kast, J. (2008) Utility of formaldehyde cross‐linking and mass spectrometry in the study of protein–protein interactions. J. Mass Spectrom. 43, 699–715. [DOI] [PubMed] [Google Scholar]
- Tremblay, M.H. , Majeau, N. , Gagne, M.E. , Lecours, K. , Morin, H. , Duvignaud, J.B. , Bolduc, M. , Chouinard, N. , Pare, C. , Gagne, S. and Leclerc, D. (2006) Effect of mutations K97A and E128A on RNA binding and self assembly of papaya mosaic potexvirus coat protein. FEBS J. 273, 14–25. [DOI] [PubMed] [Google Scholar]
- Tseng, Y.H. , Hsu, H.T. , Chou, Y.L. , Hu, C.C. , Lin, N.S. , Hsu, Y.H. and Chang, B.Y. (2009) The two conserved cysteine residues of the triple gene block protein 2 are critical for both cell‐to‐cell and systemic movement of Bamboo mosaic virus. Mol. Plant–Microbe Interact. 22, 1379–1388. [DOI] [PubMed] [Google Scholar]
- Venter, P.A. , Marshall, D. and Schneemann, A. (2009) Dual roles for an arginine‐rich motif in specific genome recognition and localization of viral coat protein to RNA replication sites in flock house virus‐infected cells. J. Virol. 83, 2872–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot‐Lubicz, J. , Torrance, L. , Solovyev, A.G. , Morozov, S.Y. , Jackson, A.O. and Gilmer, D. (2010) Varied movement strategies employed by triple gene block‐encoding viruses. Mol. Plant–Microbe Interact. 23, 1231–1247. [DOI] [PubMed] [Google Scholar]
- Vieira, J. and Messing, J. (1987) Production of single‐stranded plasmid DNA. Methods Enzymol. 153, 3–11. [DOI] [PubMed] [Google Scholar]
- Weiss, M.A. and Narayana, N. (1998) RNA recognition by arginine‐rich peptide motifs. Biopolymers, 48, 167–180. [DOI] [PubMed] [Google Scholar]
- Wu, C.H. , Lee, S.C. and Wang, C.W. (2011) Viral protein targeting to the cortical endoplasmic reticulum is required for cell–cell spreading in plants. J. Cell Biol. 193, 521–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wung, C.H. , Hsu, Y.H. , Liou, D.Y. , Huang, W.C. , Lin, N.S. and Chang, B.Y. (1999) Identification of the RNA‐binding sites of the triple gene block protein 1 of bamboo mosaic potexvirus. J. Gen. Virol. 80, 1119–1126. [DOI] [PubMed] [Google Scholar]
- Yang, C.D. , Liao, J.T. , Lai, C.Y. , Jong, M.H. , Liang, C.M. , Lin, Y.L. , Lin, N.S. , Hsu, Y.H. and Liang, S.M. (2007) Induction of protective immunity in swine by recombinant bamboo mosaic virus expressing foot‐and‐mouth disease virus epitopes. BMC Biotechnol. 7, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Wang, T. , Bohon, J. , Gagne, M.E. , Bolduc, M. , Leclerc, D. and Li, H. (2012) Crystal structure of the coat protein of the flexible filamentous papaya mosaic virus. J. Mol. Biol. 422, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, G. , Vaughan, R.C. , Yarbrough, I. , Dharmaiah, S. and Kao, C.C. (2009) RNA binding by the brome mosaic virus capsid protein and the regulation of viral RNA accumulation. J. Mol. Biol. 391, 314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, M. , Willits, D. , Uchida, M. and Douglas, T. (2008) Plant viruses as biotemplates for materials and their use in nanotechnology. Annu. Rev. Phytopathol. 46, 361–384. [DOI] [PubMed] [Google Scholar]
- Zimmern, D. and Butler, P.J. (1977) The isolation of tobacco mosaic virus RNA fragments containing the origin for viral assembly. Cell, 11, 455–462. [DOI] [PubMed] [Google Scholar]
- Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Alignment of coat protein (CP) amino acid sequences of nine different potexviruses. The alignment was performed using the software ClustalW (Thompson et al., 2002) with the default parameters. The putative RNA‐binding motif (Tremblay et al., 2006) is underlined. The amino acids of Papaya mosaic virus (PapMV) (Tremblay et al., 2006) and Bamboo mosaic virus (BaMV) involved in RNA binding are indicated by open and filled stars, respectively. The amino acid sequences of the CPs of the nine potexviruses extensively characterized were selected from GenBank (Benson et al., 2013). The abbreviations and accession numbers are as follows: PAMV, Potato aucuba mosaic virus (gi|20177423); CymMV, Cymbidium mosaic virus (gi|46309858); ScaVX, Scallion virus X (gi|18652423); PVX, Potato virus X (gi|872290); HrV, Hydrangea ringspot virus (gi|62326909); CcMV, Cassava common mosaic virus (gi|9628112); PapMV, Papaya mosaic virus (gi|9629172); FoMV, Foxtail mosaic virus (gi|158268262); BaMV, Bamboo mosaic virus (gi|13182723).
Fig. S2 Comparison of the RNA‐binding sites in the Bamboo mosaic virus (BaMV) and Papaya mosaic virus (PapMV) coat protein (CP) structure. The structure of BaMV CP was modelled using the crystal structure of PapMV (PDB accession number 4DOX) as the template in the Phyre2 server (Kelley and Sternberg, 2009). The RNA‐binding residues of PapMV (K97) and BaMV (R99) CP are indicated by the arrows.
Table S1 The primers used in this study.


