Summary
Bacterial blight (BB), caused by Xanthomonas oryzae pv. oryzae (Xoo), is not only a disease devastating rice production worldwide, but also an ideal model system for the study of the interaction between plants and their bacterial pathogens. The rice near‐isogenic line (NIL) CBB23, derived from a cross between a wild rice Oryza rufipogon accession (RBB16) and a susceptible indica rice variety (Jingang 30), is highly resistant to all field Xoo strains tested so far. Although the BB resistance of CBB23 has been widely used in rice breeding programmes, the mechanism of its extremely broad‐spectrum resistance remains unknown. Here, we report the molecular cloning of an avirulence gene, designated as avrXa23, from Xoo strain PXO99A. We validate that AvrXa23, a novel transcription activator‐like effector, specifically triggers the broad‐spectrum BB resistance in CBB23. The prevalence of avrXa23 in all 38 Xoo strains surveyed may explain the broad‐spectrum feature of BB resistance in CBB23. The results will significantly facilitate the molecular cloning of the corresponding resistance (R) gene in the host, and provide new insights into our understanding of the molecular mechanism for broad‐spectrum disease resistance in plants.
Keywords: avrXa23, broad‐spectrum resistance, rice, TAL effector, Xa23, Xoo
Introduction
Rice bacterial blight (BB), caused by Xanthomonas oryzae pv. oryzae (Xoo), is one of the most devastating rice diseases worldwide, and host genetic resistance is regarded as the most effective approach to the management of this disease (Mew, 1987). Over the last two decades, scientists have exerted tremendous efforts to identify and isolate BB‐resistance genes. Apart from cultivated rice, wild rice has proven to be another important and rich genetic resource for BB resistance (Khush et al., 1989) and, indeed, some elite BB‐resistance genes, such as Xa21 (Ikeda et al., 1990), Xa23 (Zhang et al., 2001) and Xa27 (Amante‐Bordeos et al., 1992; Gu et al., 2004), have been identified from wild rice Oryza longistaminata, O. rufipogon and O. minuta, respectively. Xa21 and Xa27 have been molecularly cloned and studied extensively (Chen et al., 2010; Gu et al., 2005, 2009; Song et al., 1995).
The rice near‐isogenic line (NIL) CBB23 was derived from a cross between an O. rufipogon accession (RBB16) and a susceptible indica rice variety, Jingang 30 (JG30; Zhang et al., 2001). Our previous studies have revealed that a single resistance (R) gene, designated as Xa23, in CBB23 confers completely dominant and high resistance to all the naturally occurring Xoo races tested (Wang et al., 2009; Zhang et al., 2001), and have delimited the Xa23 locus on the long arm of chromosome 11 between the molecular markers 69B and CP02662, a region with numerous repetitive sequences (Fan et al., 2006; Wang et al., 2006). Although the BB resistance of CBB23 has been widely used in rice breeding programmes in China, the mechanism for its extremely broad‐spectrum resistance has yet to be elucidated.
Transcription activator‐like (TAL) effectors are a class of proteins identified in many plant‐pathogenic Xanthomonas spp., including Xoo (Boch and Bonas, 2010; Bonas et al., 1989; Hopkins et al., 1992). The pathogens inject their TAL effectors through a type III secretion system (TTSS) into plant cells, primarily to activate the expression of host genes that contribute to disease (Yang et al., 2006). However, plants have evolved promoter elements that trap TAL effectors to trigger plant defence (Gu et al., 2005; Romer et al., 2007; Strauß et al., 2012). All TAL effectors have a conserved architecture: an N‐terminus required for type III secretion, a central region consisting of a varying number of near‐perfect 34‐amino‐acid repeats, and a C‐terminus containing nuclear localization signals (NLSs) and an acidic transcription activation domain (AD) (Boch and Bonas, 2010). TAL effectors activate host gene expression by binding to promoter sequences of target genes. The number and order of the 34‐amino‐acid repeats determine the specificity between a TAL effector and its target DNA sequence (Boch and Bonas, 2010). Amino acid polymorphism among the central repeats exists and is concentrated at residues 12 and 13 of each repeat, referred to as the repeat‐variable di‐residue (RVD) (Moscou and Bogdanove, 2009), which mediates specific DNA recognition. The code of DNA recognition between RVDs of the TAL effector and the nucleotides of its target site has been deciphered (Boch et al., 2009; Moscou and Bogdanove, 2009).
To investigate the molecular interaction between CBB23 and Xoo, we generated and identified some Tn5‐tagged PXO99A mutants that were virulent on CBB23 (Wang et al., 2009). Here, we report an extensive survey of the resistance spectrum of CBB23, the molecular cloning of the avirulence (avr) gene avrXa23 in Xoo and the molecular mechanism of the broad‐spectrum BB resistance of CBB23.
Results
CBB23 displays a broad spectrum of BB resistance
Plants of CBB23, JG30, IRBB21 and ZW1, a japonica rice line with the Xa23 locus transferred from CBB23, were inoculated by the leaf‐cutting method with 39 naturally occurring Xoo strains and three Tn5‐insertion mutants of PXO99A at the booting stage. CBB23 showed high resistance to all 39 Xoo strains, including PXO99A and nine other Xoo strains from the Philippines for which the reaction patterns have been documented previously (Zhang et al., 1998, 2001), but was susceptible to the three PXO99A mutants, P99M2, P99M4 and P99M5 (Table 1, Fig. 1). By contrast, IRBB21, a rice variety containing the broad‐spectrum resistance gene Xa21 (Ikeda et al., 1990; Khush et al., 1991), was susceptible to nine of the 39 Xoo strains (Table 1). As a control, JG30, the recurrent parent of CBB23, was highly susceptible to all the naturally occurring Xoo strains and the PXO99A Tn5‐insertion mutants (Table 1, Fig. 1). Notably, ZW1 showed the same spectrum of Xoo resistance as that of CBB23, indicating that Xa23 functions well in both indica and japonica genetic backgrounds. According to Gu et al. (2004), IRBB27 (Xa27) is susceptible to Xoo strains ZHE173, PXO71 and AXO1947 (Table 1). As Xa21 and Xa27 are widely recognized as broad‐spectrum BB‐resistance genes, it appears that Xa23 confers a broader spectrum of BB resistance than these two important genes.
Table 1.
CBB23 is highly resistant to all 39 naturally occurring Xanthomonas oryzae pv. oryzae (X oo) strains tested
| Xoo strain | Origin of Xoo | Reaction pattern of ricea | ||||
|---|---|---|---|---|---|---|
| CBB23b | JG30b | ZW1b | IRBB21b | IRBB27c | ||
| 1 HLJ‐72 | China | R | S | R | R | R |
| 2 HB17 | China | R | S | R | R | R |
| 3 NX42 | China | R | S | R | R | R |
| 4 ZHE173 | China | R | S | R | R | S |
| 5 GD1358 | China | R | S | R | R | R |
| 6 LN57 | China | R | S | R | R | R |
| 7 JS49‐6 | China | R | S | R | R | R |
| 8 HuB05‐6 | China | R | S | R | S | |
| 9 HAN06‐1 | China | R | S | R | S | |
| 10 HAN08‐2 | China | R | S | R | S | |
| 11 Yun96‐11 | China | R | S | R | R | |
| 12 Yun98‐1 | China | R | S | R | R | |
| 13 Yun98‐5 | China | R | S | R | R | |
| 14 LN44 | China | R | S | R | R | |
| 15 JL691 | China | R | S | R | R | |
| 16 PXO61 | Philippines | Rd | S | R | R | |
| 17 PXO86 | Philippines | Rd | S | R | R | R |
| 18 PXO79 | Philippines | Rd | S | R | R | R |
| 19 PXO71 | Philippines | Rd | S | R | R | MS |
| 20 PXO112 | Philippines | Rd | S | R | R | R |
| 21 PXO99 | Philippines | Rd | S | R | R | R |
| 22 PXO145 | Philippines | Rd | S | R | R | |
| 23 PXO280 | Philippines | Rd | S | R | R | |
| 24 PXO339 | Philippines | Rd | S | R | R | |
| 25 PXO341 | Philippines | Rd | S | R | S | |
| 26 T7174 | Japan | R | S | R | R | R |
| 27 T7147 | Japan | R | S | R | R | |
| 28 T7133 | Japan | R | S | R | R | |
| 29 KXO19 | Korea | R | S | R | R | |
| 30 KXO85 | Korea | R | S | R | S | |
| 31 KXO112 | Korea | R | S | R | R | |
| 32 KXO559 | Korea | R | S | R | MS | |
| 33 KXO576 | Korea | R | S | R | S | |
| 34 KXO657 | Korea | R | S | R | MS | |
| 35 KXO660 | Korea | R | S | R | R | |
| 36 PP8511 | Bangladesh | R | S | R | R | |
| 37 CN9417 | Bangladesh | R | S | R | R | |
| 38 TL9924 | Bangladesh | R | S | R | R | |
| 39 AXO1947 | Africa | R | S | R | S | S |
| 40 P99M2 | Mutant of PXO99 | S | S | S | R | |
| 41 P99M4 | Mutant of PXO99 | S | S | S | R | |
| 42 P99M5 | Mutant of PXO99 | S | S | S | R | |
MS, moderate susceptible; R, resistant; S, susceptible.
The inoculation assays were mainly performed during 2008–2010 in Beijing. For Xoo strain AXO1947, the assays were carried out in 2009 at Iowa State University.
Data of reaction patterns of IRBB27 are from Gu et al. (2004).
Figure 1.

Disease responses of CBB23 and JG30 to representative Xanthomonas oryzae pv. oryzae (Xoo) strains. Bacterial suspension was inoculated into rice leaves using the leaf clipping method. Photographs were taken at 14 days post‐inoculation.
Screening of cosmid clones containing TAL effector gene(s) in PXO99A
As no naturally occurring Xoo strain virulent to CBB23 has been identified, we generated and identified several Tn5‐tagged PXO99A mutants that were virulent in CBB23, and found that the Tn5 insertions were located in gene(s) related to TAL effector(s) in some of the virulent mutants, including P99M5 (Wang et al., 2009). Based on these findings and the fact that there is a large repertoire of TAL effector genes in PXO99A (Salzberg et al., 2008), we speculated that the avr gene corresponding to Xa23 could be a member of TAL effector‐coding genes. Thus, we performed the selection of cosmid clones containing TAL effector gene(s) from the genomic library of PXO99A by Southern blotting with the central repeat fragment of avrXa7 as a probe. A total of 34 TAL effector‐containing cosmids was selected and individually transformed into P99M5 through electroporation (Table 2).
Table 2.
Virulence assays of P99M5 transformants harbouring cosmids containing transcription activator‐like (TAL) effector gene(s) from PXO99A
| Cosmid | Transformanta | Caused BB lesion length (cm)b on | |
|---|---|---|---|
| CBB23 | JG30 | ||
| C1: pHM1/99lib‐1 | P99M5‐C1 | 10.0 ± 0.98 Aa | 11.0 ± 0.53 Aa |
| C2: pHM1/99lib‐2 | P99M5‐C2 | 11.5 ± 1.01 Aa | 11.0 ± 1.01 Aa |
| C4: pHM1/99lib‐4 | P99M5‐C4 | 10.0 ± 1.45 Aa | 11.0 ± 1.95 Aa |
| C5: pHM1/99lib‐5 | P99M5‐C5 | 13.0 ± 0.52 Aa | 13.0 ± 1.03 Aa |
| C6: pHM1/99lib‐6 | P99M5‐C6 | 13.0 ± 1.03 Aa | 14.0 ± 0.50 Aa |
| C7: pHM1/99lib‐7 | P99M5‐C7 | 9.0 ± 2.04 Aa | 9.0 ± 1.54 Ab |
| C8: pHM1/99lib‐8 | P99M5‐C8 | 10.0 ± 0.46 Aa | 9.0 ± 1.03 Ab |
| C9: pHM1/99lib‐9 | P99M5‐C9 | 11.0 ± 1.50 Aa | 10.0 ± 1.50 Aa |
| C11: pHM1/99lib‐11 | P99M5‐C11 | 12.0 ± 1.51 Aa | 10.0 ± 1.95 Aa |
| C12: pHM1/99lib‐12 | P99M5‐C12 | 11.0 ± 1.47 Aa | 13.0 ± 0.51 Aa |
| C13: pHM1/99lib‐13 | P99M5‐C13 | 10.0 ± 0.99 Aa | 12.0 ± 1.04 Aa |
| C14: pHM1/99lib‐14 | P99M5‐C14 | 12.0 ± 0.48 Aa | 11.0 ± 1.53 Aa |
| C15: pHM1/99lib‐15 | P99M5‐C15 | 1.50 ± 0.51 Cc | 12.0 ± 1.02 Aa |
| C16: pHM1/99lib‐16 | P99M5‐C16 | 14.0 ± 0.51 Aa | 14.0 ± 0.47 Aa |
| C19: pHM1/99lib‐19 | P99M5‐C19 | 11.0 ± 0.53 Aa | 12.0 ± 0.96 Aa |
| C20: pHM1/99lib‐20 | P99M5‐C20 | 10.0 ± 0.97 Aa | 10.0 ± 1.00 Aa |
| C21: pHM1/99lib‐21 | P99M5‐C21 | 13.0 ± 0.46 Aa | 12.5 ± 1.00 Aa |
| C22: pHM1/99lib‐22 | P99M5‐C22 | 12.0 ± 1.00 Aa | 10.0 ± 0.54 Aa |
| C23: pHM1/99lib‐23 | P99M5‐C23 | 13.0 ± 0.53 Aa | 12.0 ± 0.56 Aa |
| C25: pHM1/99lib‐25 | P99M5‐C25 | 9.0 ± 1.04 Ab | 11.0 ± 0.49 Aa |
| C26: pHM1/99lib‐26 | P99M5‐C26 | 12.0 ± 0.52 Aa | 13.0 ± 0.51 Aa |
| C27: pHM1/99lib‐27 | P99M5‐C27 | 11.0 ± 0.53 Aa | 12.0 ± 0.49 Aa |
| C28: pHM1/99lib‐28 | P99M5‐C28 | 10.5 ± 0.50 Aa | 12.0 ± 0.96 Aa |
| C29: pHM1/99lib‐29 | P99M5‐C29 | 11.0 ± 1.04 Aa | 12.0 ± 0.50 Aa |
| C31: pHM1/99lib‐31 | P99M5‐C31 | 9.0 ± 0.96 Ab | 10.0 ± 0.50 Aa |
| C32: pHM1/99lib‐32 | P99M5‐C32 | 13.0 ± 0.97 Aa | 12.0 ± 0.50 Aa |
| C33: pHM1/99lib‐33 | P99M5‐C33 | 11.0 ± 0.98 Aa | 12.0 ± 0.95 Aa |
| C34: pHM1/99lib‐34 | P99M5‐C34 | 10.0 ± 0.99 Aa | 10.0 ± 0.48 Aa |
| CK1 | PXO99A | 1.20 ± 0.41 Cc | 12.0 ± 1.04 Aa |
| CK2 | P99M5 | 13.0 ± 1.51 Aa | 13.5 ± 0.98 Aa |
BB, bacterial blight.
Each of the cosmids (C1–C34) was introduced into virulent mutant P99M5 and two transformants were used for leaf clipping inoculation in leaves of CBB23 and JG30. PXO99A and P99M5 were used for inoculation as controls.
Lesion length was the mean ± SD of 10 leaves, measured at 14 days post‐inoculation. The values followed by a common letter are not significantly different as determined by least‐significant difference (LSD) test at P < 0.05 (LSD0.05 = 4.49, small letter ‘a’) or P < 0.01 (LSD0.01 = 6.45, capital letter ‘A’), respectively.
The cosmid clone C15 harbours the avr gene corresponding to Xa23
To determine whether the screened PXO99A genomic cosmids harbour the avr gene corresponding to Xa23, we inoculated CBB23 leaves with the 34 individual cosmid‐derived transformants and measured the disease responses accordingly (Table 2). Inoculation assay showed that, except for P99M5‐C15, all the transformants were virulent on both CBB23 and JG30 plants; the lesion length was in the range 9–13 cm, comparable with that of the control strain P99M5 (Table 2). However, the lesion length caused by transformant P99M5‐C15 was still as long as 12 cm on JG30, but only 1.5 cm on CBB23 (Table 2). These results clearly indicated that the cosmid pHM1/99lib‐15 (C15) completely complemented the avirulence defect of P99M5 (Table 2, Fig. 2a). In other words, the cosmid C15 harbours the cognate avr gene of Xa23.
Figure 2.
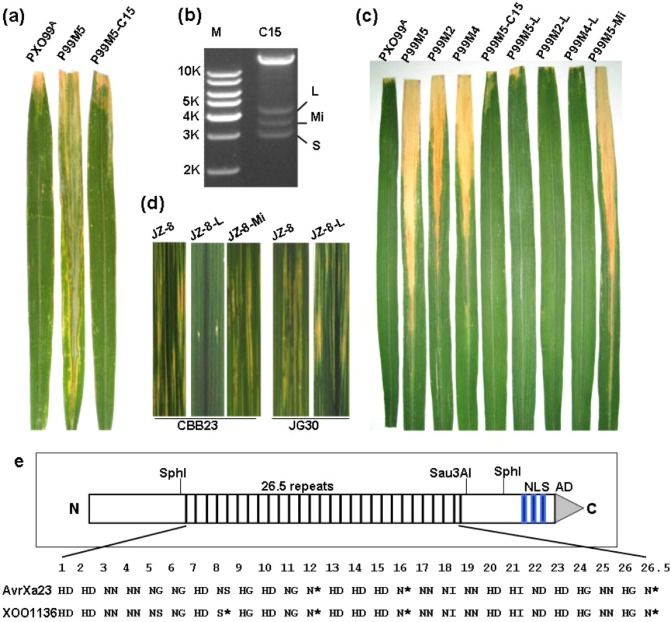
Molecular cloning of avrXa23. (a) Virulence assays of the wild‐type Xanthomonas oryzae pv. oryzae (Xoo) strain PXO99A, mutant P99M5 and the complementary strain P99M5‐C15 (P99M5 harbouring the cosmid C15) in CBB23 by artificial leaf clipping inoculation. Representative leaves are presented to show the lesions caused by the strains. (b) Cosmid C15, digested by SphI, released three SphI fragments, L (large), Mi (middle) and S (small), containing the central repeats of talC9b, talC9a and talC9c, respectively. M, molecular ladder. (c) Virulence assays of virulent mutants P99M2, P99M4, P99M5 and their transformants in CBB23 by artificial leaf clipping inoculation. The transformants harbour the cosmids pHM1‐L or pHM1‐Mi, which contain the N‐ and C‐terminal coding regions of avrXa7, but with the central repeat region being replaced with the SphI fragments of L and Mi from cosmid C15, respectively. Representative leaves are presented to show the lesions caused by the different Xoo strains. PXO99A was used as a control. (d) Virulence assays of virulent Xanthomonas oryzae pv. oryzicola (Xoc) strain JZ‐8 and its transformants JZ‐8‐L (JZ‐8 harbouring plasmid pHM1‐L) and JZ‐8‐Mi (JZ‐8 harbouring plasmid pHM1‐Mi) in CBB23 and JG30 by artificial leaf piercing inoculation. Representative leaves are presented to show the lesions caused by the Xoc strains. Photographs were taken at 4 days post‐inoculation. (e) Schematic representation of AvrXa23. Thin boxes represent the 26.5 repeats which are also represented by the di‐amino acids at the positions 12 and 13 of each repeat of AvrXa23 from PXO99A and its relative (XOO1136) from the Japanese Xoo MAFF 311018. Three blue bars indicate the nuclear localization signals (NLSs). The activation domain (AD) is represented by a grey triangle. The N‐ and C‐termini are indicated by ‘N’ and ‘C’, respectively. The SphI restriction sites and the Sau3 AI site flanking the central repeat region of avrXa23 are also indicated.
avrXa23 encodes a novel TAL effector
Subcloning and transposon‐tagging sequencing of the cosmid C15 were performed to reveal the identities of the TAL effectors. Sequencing analysis revealed that C15 had an insertion of a DNA fragment containing full‐length talC9a and talC9b, and truncated talC9c, of the TAL effector cluster 9 in the genome of PXO99A (Salzberg et al., 2008). As talC9c is avrXa27 (Gu et al., 2005), we reasoned that the avrXa23 gene could be either talC9a or talC9b. Thus, we digested plasmid DNA of C15 with SphI, and isolated the SphI fragments L and Mi (Fig. 2b), containing the central repeats of talC9b and talC9a, respectively. The SphI fragments L and Mi were then individually cloned into plasmid pSK/avrXa7ΔSphF (avrXa7 with its repeat region deleted), resulting in plasmids pSK‐L and pSK‐Mi, respectively. Next, pSK‐L and pSK‐Mi were separately linearized by HindIII digestion and ligated with HindIII‐digested pHM1 vector, resulting in plasmids pHM‐L and pHM‐Mi, respectively. Finally, pHM‐L and pHM‐Mi were separately introduced into competent cells of the Tn5‐insertion mutant P99M5, resulting in strains P99M5‐L and P99M5‐Mi, respectively. pHM‐L was also separately introduced into competent cells of the Tn5‐insertion mutants P99M2 and P99M4, resulting in strains P99M2‐L and P99M4‐L, respectively. Inoculation assay showed that CBB23 is still susceptible to P99M5‐Mi, but highly resistant to P99M5‐L, P99M2‐L and P99M4‐L (Fig. 2c), indicating that the avirulence activity of P99M5 was specifically restored by plasmid pHM‐L. As the L fragment was released from talC9b, we thought that talC9b might be the avrXa23 gene.
To further confirm the function of talC9b, we transferred pHM‐L and pHM‐Mi plasmids separately into JZ‐8, a strain of Xanthomonas oryzae pv. oryzicola (Xoc) which is highly virulent in causing bacterial leaf streak disease in CBB23 and JG30. Virulence assay clearly showed that JZ‐8‐L (JZ‐8 harbouring the pHM‐L plasmid) became avirulent on CBB23 plants, but was still virulent on JG30, whereas JZ‐8‐Mi (JZ‐8 harbouring pHM‐Mi) remained as virulent as wild‐type JZ‐8 on CBB23 plants (Fig. 2d). On the basis of these observations, we concluded that talC9b is the avrXa23 gene. Bioinformatic analysis revealed that avrXa23 (the nucleotide sequence of avrXa23 has been registered in GenBank with the accession number GU732172.1) encodes a TAL effector of 1238 amino acids, containing three C‐terminal NLSs, a transcription AD and a novel central region of 26.5 direct repeats of mostly 34 amino acids (Fig. 2e). The most likely avrXa23 in the Japanese Xoo strain MAFF 311018 is XOO1136, which also encodes 26.5 repeats with 99% identity at both the nucleotide and predicted amino acid levels to the repeat region of avrXa23 (Fig. 2e; Ochiai et al., 2005).
Mutation analysis of talC9b in Xoo mutants further confirms its avrXa23 activity
In our previous study, we could not determine, based on the flanking sequences, which member of the TAL effector gene family was disrupted by Tn5 insertion in mutants P99M2, P99M4 and P99M5, all derived from PXO99A (Wang et al., 2009). In light of the aforementioned evidence for talC9b being avrXa23, we performed a more detailed molecular analysis on the mutants. We designed polymerase chain reaction (PCR) primer P1 within the promoter region of talC9b, and performed PCRs using different primer combinations between P1 and each of the four primers locating in the Tn5‐DNA (Fig. 3a). PCR analysis and sequencing of the PCR products revealed that Tn5‐DNA indeed presented in talC9b in mutants P99M2, P99M4 and P99M5. The Tn5‐DNA inserted at nucleotide positions 282 and 2018 in P99M4 and P99M5, respectively, in sense orientation, and at 671 in P99M2, in antisense orientation (Fig. 3a). Therefore, the nested PCRs with primer combinations P1/P2 and P1/P3 amplified 1143‐ and 1979‐bp fragments, respectively, from mutant P99M2 (Fig. 3b). Similarly, nested PCRs with primer combinations P1/P4 and P1/P5 amplified 760‐ and 1229‐bp fragments from P99M4, and 2496‐ and 2965‐bp fragments from P99M5, respectively (Fig. 3b). Southern blotting of the genomic DNA digested with SphI and probed with Tn5‐DNA as probe also showed the expected 1346‐, 1883‐ and 2377‐bp hybridization bands in P99M2, P99M4 and P99M5, respectively (Fig. 3c).
Figure 3.
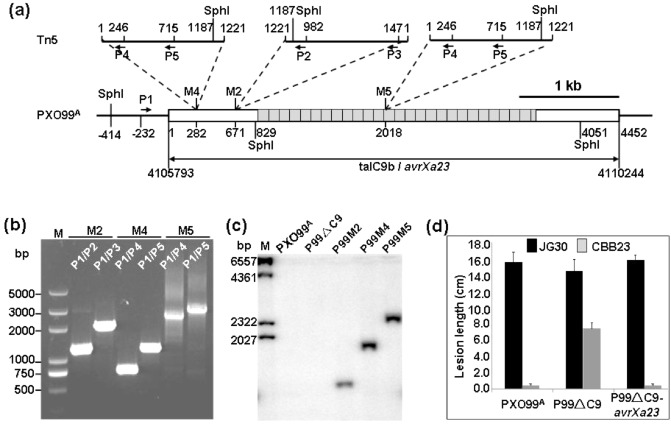
Characterization of Xanthomonas oryzae pv. oryzae (Xoo) mutants. (a) Schematic representation of insertion sites and orientations of Tn5‐DNA in mutants P99M2, P99M4 and P99M5. Black bars represent the genome of PXO99A or the Tn5‐DNA. Boxes represent talC9b located in genome region 4105793–4110244 of PXO99A (GenBank: NC_010717.1). Grey boxes represent the 26.5 repeats of talC9b. Numbers under the boxes and a nearby bar indicate the relative nucleotide positions from the first ‘A’ in the open reading frame (ORF) of talC9b. Numbers above the bars indicate the nucleotide positions of the 1221‐bp Tn5‐DNA. Broken lines indicate the Tn5 insertion sites in P99M2 (M2), P99M4 (M4) and P99M5 (M5). The nucleotide positions of the polymerase chain reaction (PCR) primers (P1–P5) and SphI restriction sites are also indicated. (b) Nested PCR analysis of mutants P99M2 (M2), P99M4 (M4) and P99M5 (M5). Primer combinations and the size (bp) of the molecular ladder (M) bands are shown. (c) Southern blotting of Xoo strains. Genomic DNA of Xoo strains were digested with SphI, separated in 1.2% (w/v) agarose gel and transferred to a nylon membrane. The 569‐bp PCR fragment amplified from Tn5‐DNA using primers P3 and P5 (a) was labelled with 32 P‐dCTP and used as the DNA probe. PXO99A and P99ΔC9 were used as negative controls. M, molecular ladder. (d) Virulence assays of the talC9b knockout mutant P99ΔC9 and its complementary strain P99ΔC9‐avrXa23 in CBB23 by artificial leaf clipping inoculation. Wild type Xoo PXO99A and susceptible rice JG30 were used as controls. The lesion length was the mean ± SD of five leaves measured at 14 days post‐inoculation.
In addition, we created a PXO99A mutant with cluster 9 of the TAL effector genes deleted (referred to as P99ΔC9). Like the Tn5‐insertion mutants, P99ΔC9 became compatible with CBB23, albeit with a shorter lesion length (c. 8 cm) than in JG30 (c. 14 cm), and reintroduction of pHM‐L completely restored the avirulence to P99ΔC9 in CBB23 (Fig. 3d). All of these observations further confirmed that talC9b is avrXa23.
Discussion
Our studies in the past decade have demonstrated that CBB23 is highly resistant to all the representative Xoo strains collected from China, the Philippines, Japan, Korea and Bangladesh, including some strains that overcame the resistance mediated by Xa21 and Xa27, the well‐known R genes with broad‐spectrum BB resistance (Table 1). Indeed, we have not identified any naturally occurring Xoo isolate that can overcome the Xa23‐dependent resistance (Wang et al., 2009). Thus, the Xa23 locus in CBB23, originally from wild rice O. rufipogon (Zhang et al., 2001), confers the broadest spectrum resistance to BB of rice.
Strong and dominant resistance to BB fits a classic gene‐for‐gene interaction between rice and Xoo. We reasoned that there would be an avr gene in Xoo corresponding to the R gene Xa23 in rice (Wang et al., 2009). In the present study, we cloned the avr gene avrXa23 from Xoo, which is talC9b that encodes a novel TAL effector with 26.5 central repeats. By introducing talC9b/avrXa23 into both CBB23‐compatible Xoo mutants and Xoc strain JZ‐8, we demonstrated that the novel TAL effector AvrXa23 specifically triggers Xa23‐dependent BB and bacterial leaf streak disease resistance (Figs 2 and 3c). The latter resistance, once Xa23 is cloned, may be engineered against Xoc if the expression of Xa23 can be activated appropriately, as demonstrated for Xa27 (Hummel et al., 2012).
As CBB23 was highly resistant to all the naturally occurring Xoo strains tested so far, and the broad‐spectrum resistance was specifically triggered by TAL effector AvrXa23, we speculated that avrXa23, or its functional equivalent, resides in all Xoo strains tested. For example, the sequenced Japanese Xoo strain (MAFF 311018) contains a TAL effector that is 99% identical to PXO99A avrXa23. The two effectors have 26.5 repeats that differ only in two repeats (Fig. 2e). Southern blotting showed that the expected 2797‐bp hybridization band (the SphI‐Sau3AI fragment containing the 26.5 central repeats of avrXa23) presented in all the 38 Xoo strains (Fig. 4), indicating that avrXa23 may be ubiquitous among Xoo strains. This might explain the extremely broad spectrum of CBB23 for BB resistance, although experimental evidence for such prevalence is needed.
Figure 4.
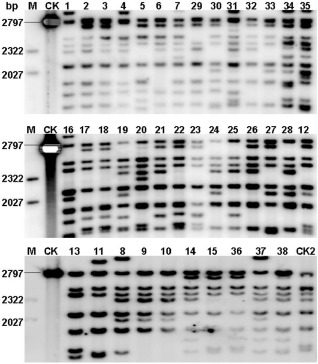
Southern blotting of 38 Xanthomonas oryzae pv. oryzae (Xoo) strains. Genomic DNA of Xoo strains was digested with SphI and Sau3 AI, separated in 1.2% (w/v) agarose gel and transferred to a nylon membrane. The SphI ‐Sau3 AI fragment containing the central repeat region of avrXa23 was labelled with 32 P‐dCTP and used as a DNA probe. M, molecular ladder; CK, the 2797‐bp SphI ‐Sau3 AI fragment of avrXa23; 1–38, numbers of the 38 Xoo strains in Table 1; CK2, P99ΔC9.
Molecular cloning of avr genes in pathogens and their cognate host R genes is necessary to fully understand the interaction between the pathogen and the host at the molecular level. So far, several avr genes from Xoo have been cloned, such as avrxa5 (Bai et al., 2000; Hopkins et al., 1992), avrXa7 (Hopkins et al., 1992; Vera Cruz et al., 2000), avrXa10 (Hopkins et al., 1992; Zhu et al., 1998) and avrXa27 (Gu et al., 2005). Primarily, Xoo strains inject their TAL effectors into rice cells to activate host susceptibility genes that contribute to disease susceptibility (Yang et al., 2006); however, rice has evolved promoter elements that trap the TAL effectors to trigger plant defence (Gu et al., 2005). In the case of avrXa27, the rice R gene Xa27 contains a TAL effector binding site in its promoter that directs transcriptional activation by the corresponding TAL effector AvrXa27, resulting in the hypersensitive response (HR), a local programmed cell death that inhibits pathogen growth within the infected site (Gu et al., 2005). The same defence mechanism has also been identified in pepper, demonstrated by the interactions between the host R gene Bs3 and the pathogen TAL effector AvrBs3 (Romer et al., 2007), and between Bs4c and AvrBs4 (Strauß et al., 2012). It seems that transcriptional activation of so‐called executor‐type R genes may be a common mechanism for plants encountering bacterial infection by exploiting the recognition of pathogen TAL effectors (Strauß et al., 2012). In this regard, it is conceivable to hypothesize that Xa23 in CBB23 is also an executor‐type R gene, as the Xa23‐dependent BB resistance is specifically triggered by the TAL effector AvrXa23. Consequently, the isolation of avrXa23 will certainly facilitate the molecular cloning of Xa23, which would further promote the investigation of the molecular interaction between Xoo and CBB23.
Because avrXa23 seems to be conserved in all Xoo isolates, we thought that AvrXa23 might contribute to the virulence of Xoo for infection or growth in host plants. However, we did not observe a significant difference between the wild‐type PXO99A and its avrXa23‐disrupted mutant (P99M5) in lesion length in the highly susceptible JG30 (Table 2). Whether AvrXa23 contributes virulence to Xoo needs to be clarified by studies using more rice varieties in strictly controlled conditions. Based on the virulence of the mutants P99M2, P99M4 and P99M5 in CBB23 (Table 2), it is possible that Xa23‐containing rice varieties might lose BB resistance if Xoo could mutate avrXa23 under the high selection pressure generated by widespread and long‐term adoption of Xa23‐containing rice. Considering the fact that the avrXa23‐disrupted mutants were still avirulent on IRBB21 (Table 2), we speculate that diversified usage of Xa23, Xa21 or other Xa genes could make Xa23‐dependent BB resistance durable.
Experimental Procedures
Plant varieties and bacterial strains
The susceptible variety JG30 and near‐isogenic resistant lines CBB23 (harbouring Xa23) and IRBB21 (harbouring Xa21, originally from the International Rice Research Institute, Los Baños, Philippines) are indica varieties of rice. ZW1 is a BC5F4 line derived by transferring the Xa23 locus from CBB23 into the japonica rice variety Wuyujing 3 through genetic crossing and backcrossings. The rice lines are stocks in Kai‐Jun Zhao's laboratory at the Chinese Academy of Agricultural Sciences. Rice plants were planted in the field or glasshouse at 28–32 °C. Xoo strain AXO1947 is a stock in Bing Yang's laboratory at Iowa State University; other Xoo strains used in this study are stocks in Kai‐Jun Zhao's laboratory (Table 1). Xoc strain JZ‐8 was kindly provided by Xiaoxiang Li, Rice Research Institute, Hunan Academy of Agricultural Sciences, Hunan, China.
Screening of cosmid clones containing TAL effector gene(s) of PXO99A
Genomic DNA of PXO99A was partially digested with Sau3AI and separated through agarose gel electrophoresis for DNA fragments larger than 20 kb. The DNA was ligated into BamHI‐digested cosmid vector pHM1 and packaged into lambda phages, and the phages were transduced into Escherichia coli XL1‐Blue MRF′ cells, resulting in a cosmid library. The library was screened for TAL effector‐containing plasmids with a 32P‐labelled repetitive DNA of avrXa7 as a probe. A collection of 34 representative cosmid clones carrying varying numbers of TAL effector genes was used in this study (data not shown).
Creation of talC9b knockout mutant of PXO99A (P99ΔC9)
Unique fragments upstream and downstream of TAL effector cluster 9 were PCR amplified with primers Tal9‐F1 (5′‐ATCAAGGCAATGGCCTTCAACGCAC‐3′) and Tal9‐R1 (5′‐AATATCCGGGTAGGCGCAATCACTTCGGCTTGCCATCGAAGTGGCAG‐3′), and Tal9‐F2 (5′‐AGCCTACACAATCGCTCAAGACGTCATCCCCGAAACAAAAACAGC‐3′) and Tal9‐R2 (5′‐AGTGGTACGCCAAGGTCAACTAC‐3′), respectively. The two fragments and a fragment of the kanamycin resistance gene cassette were overlap PCR amplified, and the replicon was cloned into the A/T cloning vector pGEM‐T, as described by Song and Yang (2009). The resulting plasmid was electroporated into PXO99A competent cells, and kanamycin‐resistant and ampicillin‐sensitive transformants were selected for further molecular and genetic confirmation, as described by Song and Yang (2009).
Transformation of X. oryzae cells with plasmid DNA
Competent cells of the Xoo mutants P99M2, P99M4, P99M5 and Xoc strain JZ‐8 were prepared as described previously (Wang et al., 2009). PPS medium (cloth‐filtered juice from cooked potato, 300 g; peptone, 5 g; sucrose, 15 g; Na2HSO4·12H2O, 2 g; Ca(NO3)2·4H2O, 0.5 g, per litre) was used to grow X. oryzae cells. An aliquot of 50 μL of bacterial competent cells was mixed with 10 ng (0.5 μL) of plasmid DNA for electroporation using a Bio‐Rad electroporation instrument (Bio‐Rad, Hercules, CA, USA). An electric field of 15 kV/cm, with a resistance of 200 Ω and a capacitance of 25 μF, was applied. After pulse delivery, cells were immediately transferred into 1 mL of SOC medium (20 g tryptone, 5 g yeast extract, 4.8 g MgSO4, 3.6 g dextrose, 0.5 g NaCl and 0.2 g KCl per liter) in a 2‐mL round‐bottomed polypropylene tube. After incubation at 28 °C with constant shaking for 1.5 h, the electroporated cells were plated onto PPS medium containing appropriate antibiotics, and incubated at 28 °C for 3 days. Non‐electroporated competent cells were used as a control. Positive clones were picked for further analysis.
Pathogenicity assessment of Xanthomonas strains or their transformants
The pathogenicity of Xoo strains or their transformants was evaluated using the leaf tip clipping method (Wang et al., 2009). Xoo strains were cultured in liquid PPS medium, with shaking at 250 rpm for 42 h at 28 °C. Bacterial inoculum was adjusted to ∼105 colony‐forming units (cfu)/mL in water and inoculated onto fully expanded rice leaves at the booting stage. Each strain was inoculated on 5–10 rice plants and three to five fully expanded leaves per plant. Disease symptoms were scored by lesion ratio against the whole leaf or lesion length (cm). Disease symptoms were scored 2 weeks after inoculation by visual assessment of the percentage of lesion to whole leaf. Leaves with a lesion area of less than 15%, 15–20% and greater than 20% were classified as resistant (R), moderate susceptible (MS) and susceptible (S), respectively, as described previously (Zhang et al., 1996).
For pathogenicity assay for Xoc strain JZ‐8 or its transformants, bacterial inoculum was prepared similarly as for Xoo. Adult rice plants were inoculated by piercing the leaves with a needle when they were submerged in the bacterial suspension in a Petri dish, as described previously (Li et al., 2011). Disease symptoms were recorded and photographs were taken at 4 days post‐inoculation.
All virulence assays were repeated at least three times.
PCR and Southern blot analysis of Xoo mutants
Genomic DNA of Xoo strains was isolated as described previously (Wang et al., 2009). PCRs were performed to check the Tn5‐insertion locations in Xoo mutants P99M2, P99M4 and P99M5, using primers P1 (5′‐AAGTGGGTTCACTCGCTGTCAGCACAG‐3′), P2 (5′‐GGCAGAGCATTACGCTGACT‐3′), P3 (5′‐ATTCAACGGGAAACGTCTTG‐3′), P4 (5′‐CTGATTGCCCGACATTATCG‐3′) and P5 (5′‐ACTGAATCCGGTGAGAATGG‐3′). The reaction (20 μL) contained 50 ng of template DNA, 1 × PCR buffer, 0.6 mmol/L deoxynucleoside triphosphates (dNTPs), 0.15 mmol/L of each primer and 0.75 U KOD Taq polymerase (TOYOBO, Osaka, Japan). PCR was initiated at 95 °C for 3 min, followed by 35 cycles of amplification at 94 °C for 40 s, 60 °C for 40 s, 72 °C for 1 min, and a final extension at 72 °C for 10 min.
For Southern blot analysis, 1 μg of bacterial genomic DNA was digested with SphI alone or together with Sau3AI, separated on a 1.2% (w/v) agarose gel and transferred to a nylon membrane (Hybond‐N+, Amersham Pharmacia Bio‐tech, Amersham, UK). A 569‐bp PCR fragment amplified from Tn5‐DNA using primers P3 and P5 (Fig. 3a), or the SphI‐Sau3AI fragment containing the central repeat region of avrXa23 (Fig. 2e), was used as DNA probe, and labelled with 32P‐dCTP by the random priming method (Sambrook et al., 1989). Hybridization was performed in a solution containing 5 × saline‐sodium citrate (SSC), 5 × Denhardt's solution, 0.6% (w/v) sodium dodecylsulphate (SDS), 10% (w/v) dextran sulphate and 20 mg/L denatured salmon sperm DNA at 65 °C for 14 h. Filters were washed twice in 2 × SSC, 0.1% SDS at 65 °C for 10 min, once in 1 × SSC, 0.1% SDS at 65 °C for 10 min, and once in 0.5 × SSC, 0.1% SDS at 65 °C for 10 min. Blots were exposed on a PhosphorImager plate and signals were detected by the Molecular imager® FX (Bio‐Rad).
Acknowledgements
We thank X. X. Li, Rice Research Institute, Hunan Academy of Agricultural Sciences, for kindly providing the Xoc strain JZ‐8. Thanks are also due to H. H. Li, Institute of Crop Science, Chinese Academy of Agricultural Sciences, for help with the statistical analysis of the data in Table 2. This work was partially supported by the National High‐Technology Research Program (The ‘863’ Program: Grant no. 2006AA10Z106) of the Ministry of Science and Technology of China, the National Natural Science Foundation of China (Grant no. 31171812) and the US National Science Foundation (Award number: 1238189 to B.Y.). No conflict of interest is declared for the other authors.
References
- Amante‐Bordeos, A. , Sitch, L.A. , Nelson, R. , Dalmacio, R.D. , Oliva, N.P. , Aswidinnoor, H. and Leung, H. (1992) Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice, Oryza sativa . Theor. Appl. Genet. 84, 345–354. [DOI] [PubMed] [Google Scholar]
- Bai, J. , Choi, S.H. and Ponciano, G. (2000) Xanthomonas oryzae pv. oryzae avirulence genes contribute differently and specifically to pathogen aggressiveness. Mol. Plant–Microbe Interact. 13, 1322–1329. [DOI] [PubMed] [Google Scholar]
- Boch, J. and Bonas, U. (2010) Xanthomonas AvrBs3 family‐type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. [DOI] [PubMed] [Google Scholar]
- Boch, J. , Scholze, H. , Schornack, S. , Landgraf, A. , Hahn, S. , Kay, S. , Lahaye, T. , Nickstadt, A. and Bonas, U. (2009) Breaking the code of DNA binding specificity of TAL‐type III effectors. Science, 326, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Bonas, U. , Stall, R.E. and Staskawicz, B. (1989) Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria . Mol. Gen. Genet. 218, 127–136. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Chern, M. , Canlas, P.E. , Ruan, D. , Jiang, C. and Ronald, P.C. (2010) The rice XB24 ATPase promotes autophosphorylation of XA21 and inhibits XA21‐mediated immunity. Proc. Natl. Acad. Sci. USA, 107, 8029–8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y.L. , Chen, X.W. , Wang, C.L. , Zhu, L.H. , Zhang, Q. and Zhao, K.J. (2006) Mapping the rice bacterial blight resistance gene Xa23 with RFLP marker and converting RFLP to STS marker. Acta Agronom. Sin. 32, 931–935 (in Chinese with English abstract). [Google Scholar]
- Gu, K. , Tian, D. , Yang, F. , Wu, L. , Sreekala, C. , Wang, D. , Wang, G.L. and Yin, Z. (2004) High‐resolution genetic mapping of Xa27(t), a new bacterial blight resistance gene in rice, Oryza sativa L. Theor. Appl. Genet. 108, 800–807. [DOI] [PubMed] [Google Scholar]
- Gu, K. , Yang, B. , Tian, D. , Wu, L. , Wang, D. , Sreekala, C. , Yang, F. , Chu, Z. , Wang, G.L. , White, F.F. and Yin, Z. (2005) R gene expression induced by a type‐III effector triggers disease resistance in rice. Nature, 435, 1122–1125. [DOI] [PubMed] [Google Scholar]
- Gu, K. , Tian, D. , Qiu, C. and Yin, Z. (2009) Transcription activator‐like type III effector AvrXa27 depends on OsTFIIAg5 for the activation of Xa27 transcription in rice that triggers disease resistance to Xanthomonas oryzae pv. oryzae . Mol. Plant Pathol. 10, 829–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, C.M. , White, F.F. , Choi, S.H. , Guo, A. and Leach, J.E. (1992) A family of avirulence genes from Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 5, 451–459. [DOI] [PubMed] [Google Scholar]
- Hummel, A.W. , Doyle, E.L. and Bogdanove, A.J. (2012) Addition of transcription activator‐like effector binding sites to a pathogen strain‐specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol. 195, 883–893. [DOI] [PubMed] [Google Scholar]
- Ikeda, R. , Khush, G.S. and Tabien, R.E. (1990) A new resistance gene to bacterial blight derived from O. longistaminata . Jpn. J. Breed. 40 (Suppl 1), 280–281. [Google Scholar]
- Khush, G.S. , Mackill, D.J. and Sidhu, G.S. (1989) Breeding rice for resistance to bacterial blight. In: Bacterial Blight of Rice , pp. 207–217. International Rice Research Institute, Manila, Philippines. [Google Scholar]
- Khush, G.S. , Bacalangco, E. and Ogawa, T. (1991) A new gene for resistance to bacterial blight from O. longistaminata . Rice Genet. Newsl. 7, 121–122. [Google Scholar]
- Li, Y.R. , Zou, H.S. , Che, Y.Z. , Cui, Y.P. , Guo, W. , Zou, L.F. , Chatterjee, S. , Biddle, E.M. , Yang, C.H. and Chen, G.Y. (2011) A novel regulatory role of HrpD6 in regulating hrp‐hrc‐hpa genes in Xanthomonas oryzae pv. oryzicola . Mol. Plant–Microbe Interact. 24, 1086–1101. [DOI] [PubMed] [Google Scholar]
- Mew, T.W. (1987) Current status and future prospects of research on bacterial blight of rice. Annu. Rev. Phytopathol. 25, 359–382. [Google Scholar]
- Moscou, M.J. and Bogdanove, A.J. (2009) A simple cipher governs DNA recognition by TAL effectors. Science, 326, 1501. [DOI] [PubMed] [Google Scholar]
- Ochiai, H. , Inoue, Y. , Takeya, M. , Sakaki, A. and Kaku, A. (2005) Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large number of effector genes and insertion sequences to its race diversity. Jpn. Agric. Res. Q. 39, 275–287. [Google Scholar]
- Romer, P. , Hahn, S. , Jordan, T. , Strauß, T. , Bonas, U. and Lahaye, T. (2007) Plant–pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science, 318, 645–648. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L. , Sommer, D.D. , Schatz, M.C. , Phillippy, A.M. , Rabinowicz, P.D. , Tsuge, S. , Furutani, A. , Ochiai, H. , Delcher, A.L. , Kelley, D. , Madupu, R. , Puiu, D. , Radune, D. , Shumway, M. , Trapnell, C. , Aparna, G. , Jha, G. , Pandey, A. , Patil, P.B. , Ishihara, H. , Meyer, D.F. , Szurek, B. , Verdier, V. , Koebnik, R. , Dow, J.M. , Ryan, R.P. , Hirata, H. , Tsuyumu, S. , Won Lee, S. , Seo, Y.S. , Sriariyanum, M. , Ronald, P.C. , Sonti, R.V. , Van Sluys, M.A. , Leach, J.E. , White, F.F. and Bogdanove, A.J. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A . BMC Genomics, 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Song, C. and Yang, B. (2009) Mutagenesis of 18 type III effectors reveals virulence function of XopZPXO99 in Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 23, 893–902. [DOI] [PubMed] [Google Scholar]
- Song, W.Y. , Wang, G.L. , Chen, L.L. , Kim, H.S. , Pi, I.Y. , Holsten, T. , Garduer, J.B. , Wang, B. , Zhai, W.X. , Zhu, L.H. , Fanquet, C. and Ronald, P. (1995) A receptor kinase‐like protein encoded by the rice disease resistance gene, Xa21 . Science, 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Strauß, T. , Van Poeckeb, R.M.P. , Strauß, A. , Römer, P. , Minsavage, G.V. , Singh, S. , Wolf, C. , Strauß, A. , Kim, S. , Lee, H.A. , Yeom, S.I. , Parniske, M. , Stall, R.E. , Jones, J.B. , Choi, D. , Prins, M. and Lahayea, T. (2012) RNA‐seq pinpoints a Xanthomonas TAL effector activated resistance gene in a large‐crop genome. Proc. Natl. Acad. Sci. USA, 109, 19 480–19 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera Cruz, C.M. , Bai, J. , Ona, I. , Leung, H. , Nelson, R.J. , Mew, T.W. and Leach, J.E. (2000) Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc. Natl. Acad. Sci. USA, 97, 13 500–13 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.L. , Chen, L.T. , Zeng, C.Z. , Zang, Q.Y. , Liu, P.Q. , Liu, Y.G. , Fan, Y.L. , Zang, Q. and Zhao, K.J. (2006) Chromosome walking for fine mapping of Xa23 gene locus by using genomic libraries. Chin. J. Rice Sci. 20, 355–360 (in Chinese with English abstract). [Google Scholar]
- Wang, C.L. , Xu, A.B. , Gao, Y. , Fan, Y.L. , Liang, Y.T. , Zheng, C.K. , Sun, L.Q. , Wang, W.Q. and Zhao, K.J. (2009) Generation and characterization of Tn5‐tagged Xanthomonas oryzae pv. oryzae mutants that overcome Xa23‐mediated resistance to bacterial blight of rice. Eur. J. Plant Pathol. 123, 343–351. [Google Scholar]
- Yang, B. , Sugio, A. and White, F.F. (2006) Os8N3 is a host disease‐susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA, 103, 10 503–10 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Shi, A.N. , Yang, W.C. and Wang, C.L. (1996) Breeding of three near‐isogenic japonica rice lines with different major genes for resistance to bacterial blight. Acta Agronom. Sin. 22, 135–141 (in Chinese with English abstract). [Google Scholar]
- Zhang, Q. , Lin, S.C. , Zhao, B.Y. , Wang, C.L. , Yang, W.C. , Zhou, Y.L. , Li, D.Y. , Chen, C.B. and Zhu, L.H. (1998) Identifying of a new gene for resistance to bacterial blight from O. rufipogon . Rice Genet. Newsl. 15, 138–142. [Google Scholar]
- Zhang, Q. , Wang, C.L. , Zhao, K.J. , Zhao, Y.L. , Caslana, V.C. , Zhu, X.D. , Li, D.Y. and Jiang, Q.X. (2001) The effectiveness of advanced rice lines with new resistance gene Xa23 to rice bacterial blight. Rice Genet. Newsl. 18, 71–72. [Google Scholar]
- Zhu, W.G. , Yang, B. , Chittoor, J.M. , Johnson, L.B. and White, F.F. (1998) AvrXa10 contains an acidic transcriptional activation domain in the functionally conserved C‐terminus. Mol. Plant–Microbe Interact. 11, 824–832. [DOI] [PubMed] [Google Scholar]


