Summary
In Erwinia amylovora, ECF (extracytoplasmic functions) alternative sigma factor HrpL regulates the transcription of hrp (hypersensitive response and pathogenicity)‐type III secretion system (T3SS) genes by binding to a consensus sequence known as the hrp box in hrp gene promoters. In turn, the expression of hrpL has been proposed to be positively controlled by alternative sigma factor 54 (σ54) (RpoN) and HrpS, a member of the σ54 enhancer‐binding proteins (EBPs). However, the function of RpoN has not been characterized genetically in E. amylovora. In this study, we investigated the role of RpoN, a nitrogen limitation sigma factor, and its modulation protein YhbH, a novel ribosome‐associated protein, in E. amylovora virulence. Our results showed that mutations in hrpS, hrpL, rpoN and yhbH, but not yfiA and rmf3, resulted in a nonpathogenic phenotype on immature pear fruits and apple shoots. Consistently, the expression of T3SS genes, including hrpL, dspE, hrpN and hrpA, was barely detected in hrpS, hrpL, rpoN and yhbH mutants. These mutants were also not capable of eliciting a hypersensitive response (HR) on tobacco; however, the overexpression of hrpL using an inducible promoter rescued the HR‐eliciting abilities of these mutants. These results suggest that a sigma factor cascade exists in the regulatory networks of E. amylovora and regulates important virulence factors. On the basis of this study and previously reported data, a model is proposed for the regulation of T3SS in E. amylovora.
Introduction
Fire blight disease of apples and pears has been considered the most destructive disease of pome fruit trees since its discovery in the 1880s (Vanneste, 2000). Its causal agent, Erwinia amylovora, is a highly virulent necrogenic vascular pathogen and is closely related to many important mammalian enterobacterial pathogens, such as Escherichia coli, Yersinia pestis and Salmonella enterica. In the past decade or so, extensive genetic and genomic studies have demonstrated that E. amylovora utilizes two essential virulence factors, the exopolysaccharide (EPS) amylovoran and a hypersensitive response and pathogenicity (hrp)‐type III secretion system (T3SS), to cause disease (Khan et al., 2012; Koczan et al., 2009; Wang et al., 2009; Zhao and Qi, 2011).
Like many other Gram‐negative plant pathogenic bacteria, E. amylovora contains an hrp‐T3SS that delivers effector proteins into host plants. The hrp‐T3SS gene cluster in E. amylovora can be divided into three subregions: the hrp/hrc region, the Hrc effector and elicitors (HEE) region and the Hrp‐associated enzymes (HAE) region (Oh and Beer, 2005; Oh et al., 2005). The hrp/hrc region contains 25 genes, including four regulatory genes (hrpL, hrpS and hrpXY) and genes encoding structural components of T3SS. So far, the T3SS of E. amylovora has been found to secrete at least 15 virulence‐associated proteins, including HrpA, HrpN, DspE, HrpW, HopC1, AvrRpt2 and Eop1 (Bogdanove et al., 1998; Nissinen et al., 2007; Zhao et al., 2005, 2006).
In E. amylovora, the transcription of hrp‐T3SS genes is activated by the master regulator HrpL, a member of the ECF (extracytoplasmic functions) subfamily of sigma factors (Wei and Beer, 1995). HrpL binds to a consensus sequence, known as the hrp box (GGAACC–N16–CCACNNA), in hrp gene promoters. Most T3SS structural (such as hrpA) and effector (such as hrpN and dspE) genes are subject to direct HrpL regulation (McNally et al., 2012; Nissinen et al., 2007). The expression of hrpL is believed to be activated by both HrpS and a two‐component regulatory system: HrpX (sensor) and HrpY (response regulator) (Wei et al., 2000). However, recent bioinformatics and genetic studies have suggested that HrpXY may not be involved in the regulation of hrpL expression as hrpXY mutants remain virulent (Zhao et al., 2009b). Furthermore, as HrpS belongs to the NtrC family of sigma 54 (σ54) enhancer‐binding proteins (EBPs) and the hrpL gene contains a σ54 promoter, it has been proposed that σ54 (RpoN) might also be involved in the regulation of hrpL gene expression (Wei et al., 2000). We have also demonstrated that rpoN expression is induced during the infection of immature pear fruit tissue (Zhao et al., 2005). However, the exact role of RpoN (and also HrpS) has not been characterized genetically in E. amylovora.
In bacteria, core RNA polymerase (RNAP) requires sigma factors for promoter recognition to initiate transcription (Ghosh et al., 2010; Österberg et al., 2011). Sigma factors can be classified into two major families, the sigma 70 (σ70) and σ54 families. Most sigma factors belong to the extensive σ70 family, including ‘housekeeping’ sigma factor σ70(RpoD) and alternative sigma factors [σ38(RpoS), σ32(RpoH) and σ24(RpoE)], and direct the binding of RNAP to the consensus −10 (TATAAT) and −35 (TTGACA) sequences for transcription initiation (Gruber and Gross, 2003; Österberg et al., 2011). In contrast, the σ54 family contains only one single member, RpoN(σ54), which is structurally and functionally distinct from all other sigma factors and directs the binding of RNAP to the conserved −12 (TGC) and −24 (GG) promoter elements (Barrios et al., 1999). The σ54–RNAP holoenzyme complex forms a closed loop, which is transcriptionally silent, and requires bacterial EBPs, such as HrpS, to open the closed DNA loop and start transcription (Bush and Dixon, 2012; Schumacher et al., 2006).
Although initially identified for its role in nitrogen assimilation, RpoN has been found to control many other physiological processes through different EBPs, especially the regulation of hrp genes and other virulence factors (Bush and Dixon, 2012; Kazmierczak et al., 2005; Shingler, 2011). In Pseudomonas syringae, the expression of hrpL was completely abolished in an rpoN mutant, which was also defective in causing disease (Hendrickson et al., 2000a, b). The expression of Pantoea stewartii hrp genes was reduced in an E. coli rpoN mutant strain (Frederick et al., 1993). In Pectobacterium carotovorum, studies have shown that the expression of hrpL and other T3SS genes is dependent on both RpoN and HrpS (Chatterjee et al., 2002). Furthermore, in P. syringae and Pseudomonas aeruginosa, the rpoN mutant has lost its ability to produce the phytotoxin coronatine and EPS alginate, respectively (Alarcón‐Chaidez et al., 2003; Hendrickson et al., 2000a, b, c; Peñaloza‐Vázquez et al., 2004). However, Vibrio cholera and Vibrio anguillarum rpoN mutants lack flagella and are nonmotile (Dong and Mekalanos, 2012; O'Toole et al., 1997).
On the basis of the genome sequence of E. amylovora strain Ea273, rpoN (Eam_3104) belongs to a large eight‐gene operon (from Eam_3098 to Eam_3105, Fig. 1A). The first six genes in the operon (from yrbG to yhbG) encode for a sodium exchanger protein, an arabinose 5‐phosphate isomerase, a phosphatase, two exported proteins and an ABC transporter, respectively (Fig. 1A). The last gene in the operon is yhbH (Eam_3105), which encodes a small protein with 95 amino acids and has been annotated as a σ54 modulation protein (Smits et al., 2010). The deduced YhbH protein shares 84% identity with that of E. coli. In E. coli, YhbH has been characterized as a ribosomal binding protein and recently renamed as hibernation promoting factor (HPF) (Ueta et al., 2005). HPF is involved in ribosome stabilization and preservation in the stationary phase by binding specifically to 90S ribosome to form 100S ribosome, which has no translational activity (Ueta et al., 2005, 2008). Other ribosome‐associated proteins involved in this process include the ribosome modulation factor (RMF) and YfiA. The binding of RMF causes the dimerization of 70S ribosomes into 90S particles, which are then stabilized as 100S dimers on HPF binding, whereas YfiA, which shares 40% homology to HPF and has the same binding sites within the ribosome as HPF, works antagonistically by preventing 90S ribosome formation (Polikanov et al., 2012; Ueta et al., 2005). In addition, YhbH has been shown to be up‐regulated by quorum sensing in E. coli and during biofilm formation in Bacillus cereus (DeLisa et al., 2001; Oosthuizen et al., 2002). However, no evidence has shown whether YhbH interacts directly with σ54 or how YhbH is involved in σ54‐mediated gene regulation.
Figure 1.
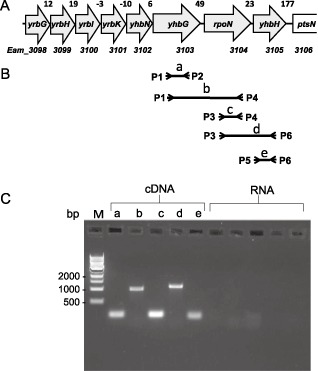
Operon determination. (A) Schematic maps of rpoN, yhbH and upstream genes (from yrbG to yhbG) in the Erwinia amylovora genome. The numbers between the arrows represent the bases separating the corresponding genes. Not drawn to scale. (B) Schematic representation of the localization of the primers used to amplify polymerase chain reaction (PCR) products from cDNAs. (C) PCR amplification of cDNAs using primer pairs P1–P2, P1–P4, P3–P4, P3–P6 and P5–P6 for a, b, c, d and e PCR products, respectively. RNA template was used as a negative control with the same primer pairs. M, molecular marker.
The purpose of this study was to determine the role of RpoN and its modulation protein YhbH in E. amylovora virulence. This is the first study involving YhbH in plant pathogenic bacteria, and the results show that YhbH, together with RpoN and HrpS, functions upstream of HrpL to regulate T3SS gene expression and plays a vital role in virulence.
Results
Mutations in rpoN, yhbH, hrpL and hrpS render E. amylovora nonpathogenic
In order to determine the role of RpoN and YhbH in E. amylovora virulence, we generated rpoN, yhbH, hrpS and hrpL, as well as yfiA (Eam_2617) and rmf3 (between Eam_1373 and Eam_1374 of Ea273; Eamy_1382 of CFBP1430), mutants by red cloning (Zhao et al., 2009a). These mutants were tested for virulence on immature pear fruits and 1‐year‐old apple shoots. In E. amylovora, the YfiA protein has 112 amino acids and was also annotated as σ54 modulation protein, whereas the small protein Rmf3 contains 55 amino acids and was not annotated in the genome of Ea273 (but is in CFBP1430). Blast search using the rmf3 (Eamy_1382) gene sequence identified the rmf3 gene locus between Eam_1373 and Eam_1374 in strain Ea273. The deduced YfiA and Rmf3 proteins are identical in 12 sequenced E. amylovora strains (Mann et al., 2013), and share 82% and 80% identity, respectively, with those of E. coli.
For the wild‐type (WT) strain, disease symptoms appeared at the inoculation site 2 days after inoculation, the necrotic lesion turned black with visible ooze formation at 4 days post‐inoculation and blackened necrotic areas covered almost the whole pear fruits at 8 days (Fig. 2). For rpoN, yhbH, hrpL and hrpS mutant strains, no symptoms were observed (Fig. 2A). However, yfiA and rmf3 mutants caused similar disease to the WT strain (Fig. 2C). Similarly, the WT strain caused typical ‘shepherd's crook’ symptoms and visible necrosis, and the length of necrosis reached 31.7 ± 3.05 cm at 7 days post‐inoculation. No symptoms were observed on apple shoots inoculated with rpoN, yhbH, hrpL or hrpS mutants (data not shown).
Figure 2.
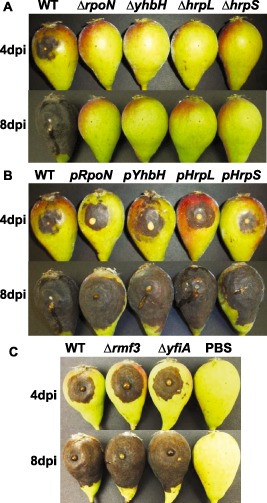
Virulence assays of Erwninia amylovora wild‐type (WT) strain, mutants and complementation strains on immature pears. (A) Symptoms caused by WT, ΔrpoN, ΔyhbH, ΔhrpL and ΔhrpS mutants on immature pears. (B) Symptoms caused by WT and complementation strains of ΔrpoN (pRpoN), ΔyhbH (pYhbH), Δ hrpL (pHrpL) and ΔhrpS (pHrpS) mutants. (C) Symptoms caused by ΔyfiA and Δrmf3. dpi, days post‐inoculation.
In order to confirm that the absence of disease symptoms on the four mutants was caused by mutations in rpoN, yhbH, hrpL and hrpS genes, we transformed the four mutants with plasmids containing rpoN, yhbH, hrpL and hrpS genes, respectively. To complement the yhbH mutant, we used the plasmid containing both rpoN and yhbH genes (Fig. 1). The four complementation strains produced comparable disease progress and symptoms as the WT strain on immature pear fruits (Fig. 2B), and the lengths of necrotic tissue on apple shoots were measured as 23 ± 4.79, 30.69 ± 4.68, 35.75 ± 5.51 and 40.75 ± 4.92 cm for yhbH, hrpL, hrpS and rpoN complementation strains, respectively. These results indicate that RpoN, YhbH, HrpL and HrpS are all required for E. amylovora to cause disease.
Expression of hrpL is under the control of RpoN , YhbH and HrpS , but expression of hrpS and rpoN is independent from each other
Although we predicted that the expression of rpoN is under the control of the promoter of an eight‐gene operon (from Eam_3098 to yhbH) using the operon prediction tool (Okuda et al., 2006), it has also been reported that rpoN contains its own promoter for independent expression in E. coli (Sperandeo et al., 2006). Based on our complementation studies described above, we confirmed that rpoN indeed contains its own promoter in E. amylovora. To further determine whether rpoN and yhbH were transcribed from both its own promoter and the operon promoter, polymerase chain reaction (PCR) amplifications were performed on cDNA derived from RNA extracted from E. amylovora WT. Products having the expected sizes for the combination of primer pairs were amplified from cDNA, but not from RNA, allowing the reconstitution of a continuous 1967‐nucleotide‐long transcript encompassing yhbG to yhbH (Fig. 1B,C).
Next, we examined how mutations of rpoN and yhbH affect hrpL and the expression of other T3SS genes using quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR). As expected, no expression of rpoN, yhbH, hrpL and hrpS genes was detected in the corresponding mutants both in vitro and in vivo (Fig. 3A,B). In hrp‐inducing medium, T3SS genes, including dspE, hrpL, hrpN and hrpA, were barely detected in all four mutants (Fig. 3A). However, the hrpS gene was expressed at a similar level in rpoN, yhbH and hrpL mutants when compared with WT. Similarly, the expression of the rpoN gene was not affected in yhbH, hrpL and hrpS mutants. Meanwhile, the expression of yhbH was about two‐fold lower in the rpoN mutant than in WT, but no change was detected in hrpL and hrpS mutants (Fig. 3A). These results indicate that RpoN, HrpS and YhbH are required for the expression of hrpL and T3SS genes, but that the expression of rpoN and hrpS is independent from each other. Furthermore, it is possible that yhbH may also contain its own weak promoter.
Figure 3.
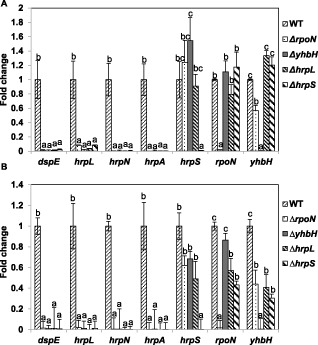
Expression of selected genes in vivo and in vitro by quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR). (A) Relative gene expression of rpoN, yhbH, hrpS and hrpL genes and three type III secretion system (T3SS) genes in ΔrpoN, ΔyhbH, ΔhrpL and ΔhrpS mutant strains compared with the wild‐type (WT) grown in hrp (hypersensitive response and pathogenicity)‐inducing minimal medium at 18 °C for 6 h. (B) Relative expression of rpoN, yhbH, hrpS and hrpL genes and three T3SS genes in ΔrpoN, ΔyhbH, ΔhrpL and ΔhrpS mutant strains compared with WT inoculated onto immature pear fruits for 6 h. The relative fold change of each gene was derived from the comparison of mutant strains versus WT control. The 16S rDNA (rrsA) gene was used as a control. The values of the relative fold change were the means of three replicates. The experiments were repeated at least twice with similar results. Error bars indicate standard deviation. One‐way analysis of variance (ANOVA) and Student–Newmans–Keuls test (P = 0.05) were used to analyse the qRT‐PCR data. For each gene tested in WT and the four mutants, fold changes marked with the same letter do not differ significantly (P < 0.05).
Under in vivo conditions, T3SS genes, including dspE, hrpL, hrpN and hrpA, were also barely detected in all four mutants (Fig. 3B). The expression of hrpS, rpoN and yhbH in the four mutants showed similar trends as described above under in vitro conditions, except that the expression of rpoN and yhbH genes in the hrpS mutant was about two‐fold lower than that in the WT strain (Fig. 3B), suggesting that HrpS may promote the expression of the rpoN/yhbH operon under in vivo conditions. Furthermore, the expression of hrpS, rpoN and yhbH was slightly lower in the hrpL mutant, suggesting a possible positive feedback loop. These data further demonstrate that RpoN, YhbH and HrpS activate hrpL expression, which then activates the expression of other T3SS genes.
Mutations in rpoN, yhbH, hrpL and hrpS abolish their abilities to elicit a hypersensitive response (HR)
Next, we tested whether RpoN and YhbH are also required for HR induction in tobacco. As expected, rpoN, yhbH, hrpL and hrpS mutants were unable to elicit HR on tobacco; meanwhile, normal tissue collapse was observed for all complementation strains, as well as WT, after 24 h (Fig. 4A). These results demonstrate that HrpL, HrpS, RpoN and YhbH are required to elicit HR on nonhost tobacco.
Figure 4.
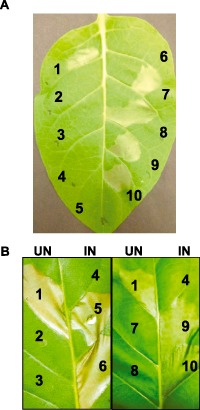
Hypersensitive response (HR) assay on tobacco leaves. (A) Eight‐week‐old tobacco leaves were infiltrated with wild‐type, mutant strains and complementation strains with cell suspensions at an optical density at 600 nm (OD 600) of 0.1:1, Ea1189 WT; 2, ΔrpoN; 3, ΔyhbH; 4, ΔhrpL; 5, ΔhrpS; 6, phosphate‐buffered saline (PBS); 7, ΔrpoN(pRpoN); 8, ΔyhbH (pYhbH); 9, Δ hrpL (pHrpL); 10, ΔhrpS (pHrpS). (B) Eight‐week‐old tobacco leaves were infiltrated with strains carrying pHrpL‐30 driven by an arabinose‐inducible promoter: 1, Ea1189 WT; 2 and 5, ΔrpoN(pHrpL‐30); 3 and 6, ΔyhbH(pHrpL‐30); 7 and 9, ΔhrpL(pHrpL‐30); 8 and 10, ΔhrpS(pHrpL‐30); 4, PBS. UN, uninduced with arabinose; IN, induced with arabinose. Photographs were taken at 24 h post‐infiltration. PBS was used as a negative control.
As suggested by the expression data (Fig. 3), it is obvious that rpoN, hrpS and yhbH regulate hrpL and the expression of other T3SS genes. We therefore cloned the hrpL gene under the control of an arabinose‐inducible promoter, and the resulting plasmid pHrpL‐30 was transformed into rpoN, yhbH, hrpL and hrpS mutants and tested for HR on tobacco under inducing conditions. As shown in Fig. 4, all four mutants containing pHrpL‐30 showed restored ability to elicit HR in tobacco under inducing conditions, but not without induction. These results demonstrate that RpoN, YhbH and HrpS function upstream of HrpL to regulate hrp gene expression.
Discussion
As a breakthrough discovery in modern molecular plant bacteriology, T3SS and effector biology represent the centre stage in the study of the pathogenesis of plant pathogenic bacteria (He et al., 2004). A full understanding of T3SS, such as the structure, gene expression, regulation, effector function, secretion and translocation, will greatly increase our knowledge in elucidating the genetic, molecular and physiological basis for pathogenesis during interactions with eukaryotic hosts (Zhao and Qi, 2011). In this study, we investigated the role of an alternative sigma factor RpoN and, for the first time, a novel ribosome‐associated protein YhbH in E. amylovora virulence. We demonstrated that RpoN and YhbH, together with HrpS, are all essential for virulence on host plants, HR induction on tobacco and the expression of hrpL and other T3SS genes. Therefore, it is apparent that a sigma factor cascade controls T3SS in E. amylovora regulatory networks, suggesting that the activation of one sigma factor by another might allow the pathogen to integrate diverse environmental signals or to withstand specific stress conditions encountered in the host environment.
Bacterial RNAP holoenzymes are composed of a core enzyme associated with one of a range of sigma factors (Ghosh et al., 2010; Österberg et al., 2011). Therefore, the central mechanism for the programming of transcription initiation in bacteria is that a pool of sigma factors competes for binding to RNAP to form holoenzymes, leading to the expression of a subset of genes controlled by the specific sigma factor (Österberg et al., 2011). It has been found that alternative sigma factor RpoN presents in a diverse phylogeny, including Pseudomonas, Listeria, Campylobacter and many enterobacteria, and controls a wide diversity of cellular processes, including nitrogen metabolism, flagellar motility and pilus‐mediated attachment (Kazmierczak et al., 2005). Most importantly, RpoN plays an important role in controlling T3SS gene expression and virulence in Pseudomonas, Pantoea and Pectobacterium (Alarcón‐Chaidez et al., 2003; Chatterjee et al., 2002; Hendrickson et al., 2000a, b, c; Peñaloza‐Vázquez et al., 2004). In this study, we have demonstrated that the alternative sigma factor RpoN is essential for the regulation of hrpL and other T3SS genes, and thus controls E. amylovora virulence.
Unlike the σ70 family, binding of σ54 to the RNAP holoenzyme confers unique features on the holoenzyme and forms a transcriptionally silent closed loop complex, which could then be activated by EBPs, such as HrpS, which provide energy through ATP hydrolysis under appropriate environmental conditions (Bush and Dixon, 2012; Shingler, 2011). EBPs, also called σ54 activators, are members of the functionally versatile AAA+ (ATPases associated with various cellular activities) family of proteins, which are highly regulated in response to environmental cues (Bush and Dixon, 2012; Schumacher et al., 2006). Therefore, the evolutionary advantages of σ54‐dependent gene regulation through the activation of promoters by EBPs are that transcription is tightly regulated and occurs rapidly and specifically (Bush and Dixon, 2012; Shingler, 2011). As a consequence, σ54‐dependent gene expression is often responsible for the creation of swift and precise responses to environmental change and regulates stress response genes, such as T3SS and nitrogen assimilation genes (Jovanovic et al., 2011; Schumacher et al., 2006).
Some EBPs contain an N‐terminal regulatory domain, such as NtrC, which is responsible for the sensing of environmental cues. However, some EBPs lack an N‐terminal regulatory domain, such as HrpRS of Pseudomonas and HrpS of Erwinia (Hutcheson et al., 2001; Schumacher et al., 2006). In P. syringae, HrpR and HrpS form heteromeric complexes that bind to the hrpL promoter and stimulate transcription by σ54–RNAP (Grimm et al., 1995; Jovanovic et al., 2011). HrpS, but not HrpR, is specifically bound by a negative regulator HrpV to prevent HrpRS from activating hrpL (Preston et al., 1998). HrpG, a chaperone‐like protein that acts as an anti‐anti‐activator, binds to HrpV, preventing its association with HrpS (Stauber et al., 2012; Wei et al., 2005). In E. amylovora, HrpS forms homohexamers to control hrpL expression. However, as both HrpR and HrpS lack the regulatory domain, it is assumed that environmental or host signals are not sensed by HrpR or HrpS. Furthermore, HrpXY, which is not present in P. syringae, does not control hrpS expression and virulence in E. amylovora (Wei et al., 2000; Zhao et al., 2009b). Therefore, how E. amylovora and P. syringae sense unknown plant or environmental signals to activate the hrp‐T3SS system is still a mystery.
In this study, we have also discovered that, for the first time, a novel ribosome‐associated protein YhbH (HPF), but not YfiA and Rmf3, is critical in mediating the σ54‐dependent regulation of hrp‐T3SS in E. amylovora. Blast search using the National Center for Biotechnology Information (NCBI) database has shown that YhbH presents in a very diverse phylogeny, including proteobacteria, firmicutes and many other bacteria. Initial analyses of sequenced genomes indicated that the RpoN‐YhbH locus is conserved among members of Enterobactericeae, such as E. coli, Salmonella, Yersinia, Shigella, Vibrio, Pectobacterium, Erwinia, Pantoea and Dickeya. Proteomic studies have shown that the YhbH protein is differentially expressed during the human macrophage response to Francisella tularensis infection (Carlson et al., 2007) and during biofilm formation in Bacillus cereus (Oosthuizen et al., 2002). However, most functional studies on YhbH so far have been limited to E. coli, and it has not been shown in any bacterium that YhbH is involved in virulence.
Bacteria usually slow down protein synthesis on entry into the stationary phase by converting ribosomes into translationally inactive 100S dimers or 70S monomers (Polikanov et al., 2012). In E. coli, 100S dimer formation is mediated by RMF and HPF, leading to ‘ribosome hibernation’ that aids cell survival, or, alternatively, another stationary phase protein YfiA promotes the formation of translationally inactive monomeric 70S ribosomes, which prevents the recycling of ribosomes for translation initiation (Ueta et al., 2005, 2008). In E. coli, all three proteins are under the control of RpoS, which is not involved in virulence in E. amylovora (Y. F. Zhao, unpublished data). It is believed that T3SS functions early in the infection cycle and that T3SS genes are expressed in actively growing bacteria, and thus it is reasonable to speculate that YhbH might have an alternative function by either interacting with the RpoN–RNAP complex as a σ54 modulation protein or promoting HrpS–RpoN interaction. However, the molecular mechanism responsible for why and how YhbH (HPF), but not YfiA and RMF, is involved in the mediation of σ54‐dependent hrp‐T3SS gene expression in E. amylovora is still unknown.
On the basis of our results and previously reported data, we propose the following model for the regulation of T3SS gene expression in E. amylovora (Fig. 5). RpoN, together with core RNAP, forms a σ54–RNAP complex and binds to the −24 (GG) and −12 (TGC) positions from the HrpL transcription start, and remains transcriptionally silent. Meanwhile, HrpS, a σ54‐activating EBP, forms a hexamer and binds to upstream DNA activator sequences (UAS) of hrpL. Hexameric HrpS contacts the σ54–RNAP–promoter complex via the consensus GAFTGA motif and the energy provided by ATP hydrolysis of the HrpS AAA+ domain triggers the opening of the σ54–RNAP–promoter complex and DNA melting. In this process, the function of YhbH might also be required to start hrpL transcription. HrpL then recognizes a conserved ‘hrp box’ at the promoter regions of HrpL‐dependent operons or genes, and regulates other T3SS gene expression. However, in this model, how E. amylovora senses host or environmental signals or whether signals are sensed through HrpX/HrpY or HrpS/RpoN remain elusive. Future studies should focus on the detection of the signals activating T3SS and how these signals are sensed by the pathogen. The determination of the molecular mechanism of how YhbH is involved in the regulation of hrpL and other T3SS genes will also be critical.
Figure 5.
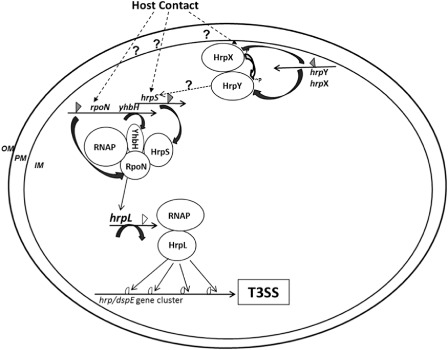
A model of type III secretion system (T3SS) regulation in Erwinia amylovora. HrpL, an ECF (extracytoplasmic functions) sigma factor and master regulator of T3SS; HrpS, a sigma factor 54 (σ54)‐dependent enhancer binding protein; HrpX/HrpY, two‐component regulatory systems; RpoN, a σ54 alternative sigma factor; RNAP, RNA polymerase; YhbH, a σ54 modulation protein (ribosome‐associated protein). OM, outer membrane; PM, plasma membrane; IM, inner membrane; P, phosphorylation; open triangle, σ54 promoter; filled triangle, σ70 promoter; circle open triangle, hrp‐box promoter. Positive regulation is indicated by an arrow; ‘?’ and broken line, unknown mechanism.
Experimental Procedures
Bacterial strain, plasmids and media
The bacterial strains and plasmids utilized in this study are listed in Table 1. Luria–Bertani (LB) broth was used for the routine growth of E. amylovora and E. coli strains. An hrp‐inducing minimal medium (HMM) [1 g (NH4)2SO4, 0.246 g MgCl2.6H2O, 0.099 g NaCl, 8.708 g K2HPO4, 6.804 g KH2PO4] containing 10 mm galactose was used to induce T3SS gene expression (Wei et al., 1992). When required, antibiotics were used at the following concentrations: 20 μg/mL kanamycin (Km), 100 μg/mL ampicillin (Ap) and 10 μg/mL chloramphenicol (Cm). The oligonucleotide primers used for mutant construction, mutant confirmation, qRT‐PCR and cloning in this study are listed in Table S1 (see Supporting Information).
Table 1.
Bacterial strains and plasmids used in this study
| Strains, plasmids | Relevant characters or sequences (5′–3′)* | Reference or source |
|---|---|---|
| Strains | ||
| Erwinia amylovora | ||
| Ea1189 | Wild‐type, isolated from apple | Burse et al. (2004) |
| ΔrpoN | rpoN::Km; KmR‐insertional mutant of rpoN of Ea1189, KmR | This study |
| ΔyhbH | yhbH::Cm; CmR‐insertional mutant of yhbH of Ea1189, CmR | This study |
| ΔhrpL | hrpL::Km; KmR‐insertional mutant of hrpL of Ea1189, KmR | This study |
| ΔhrpS | hrpS::Km; KmR‐insertional mutant of hrpS of Ea1189, KmR | This study |
| ΔyfiA (Eam_2617) | yfiA::Cm; CmR‐insertional mutant of yfiA of Ea1189, CmR | This study |
| Δrmf3 (Eam_1373‐74) | rmf3::Cm; CmR‐insertional mutant of rmf3 of Ea1189, CmR | This study |
| Escherichia coli | ||
| DH10B | F– mcrA Δ(mrr‐hsdRMS‐mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 endA1 araΔ139 Δ(ara, leu)7697 galU galK λ–rpsL (StrR) nupG | Invitrogen, Carlsbad, CA, USA |
| Plasmids | ||
| pKD46 | ApR, PBAD gam bet exo pSC101 oriTS | Datsenko and Wanner (2000) |
| pKD13 | KmR, FRT cat FRT PS1 PS2 oriR6K rgbN | Datsenko and Wanner (2000) |
| pKD3 | CmR, FRT cat FRT PS1 PS2 oriR6K rgbN | Datsenko and Wanner (2000) |
| pWSK29 | ApR; cloning vector, low copy number | Wang and Kushner (1991) |
| pBad30 | ApR, arabinose‐inducing promoter, cloning vector | Thao et al. (2010) |
| pRpoN | 1.8‐kb PCR fragment containing rpoN gene in pWSK29 | This study |
| pYhbH | 2.265‐kb PCR fragment containing both rpoN and yhbH genes in pWSK29 | This study |
| pHrpL | 1.317‐kb DNA fragment containing hrpL gene in pWSK29 vector | This study |
| pHrpS | 1.81‐kb DNA fragment containing hrpS gene in pWSK29 vector | This study |
| pHrpL‐30 | 579‐bp DNA fragment containing 30 nucleotides 5’ of the start codon of hrpL gene in pBAD30 | This study |
*KmR, CmR, ApR, kanamycin, chloramphenicol and ampicillin resistance, respectively.
DNA manipulation and the construction of plasmids
Plasmid DNA purification, PCR amplification of genes, isolation of fragments from agarose gels, cloning, restriction enzyme digestion and T4 DNA ligation were performed using standard molecular procedures (Sambrook and Russel, 2001).
Construction of mutants in E. amylovora by Lamda‐Red recombinase cloning
E. amylovora mutant strains were generated using the λ phage recombinase method, as described previously (Datsenko and Wanner, 2000; Zhao et al. 2009a, b). Briefly, overnight cultures of E. amylovora strains harbouring pKD46 were inoculated in LB broth containing 0.1% arabinose and grown to exponential phase [optical density at 600 nm (OD600) = 0.8]. Cells were harvested, made electrocompetent and stored at −80 °C. Recombination fragments consisting of a kanamycin (Kmr) or chloramphenicol (Cmr) resistance gene with its own promoter, flanked by a 50‐nucleotide homology arm, were generated by PCR from pKD13 and pKD32 plasmids, respectively. To confirm the deletion mutations, PCR amplifications from internal resistance gene primers to the external region of the target genes were performed (Table S1). For the resulting mutants, the majority of the coding region of each gene was replaced by the marker gene, except for the first and last 50 nucleotides.
Cloning of genes for complementation
For the complementation of the mutants, the regulatory and gene sequences of hrpL, hrpS, rpoN and yhbH genes were used to design primers to amplify fragments of the gene and their promoter sequences (Table S1). The PCR fragments were cloned into low‐copy‐number vector pWSK29 with KpnI–SacI restriction sites. The final plasmids were designated pHrpL, pHrpS, pRpoN and pYhbH, respectively.
The primer pair hrpL‐30F/hrpL‐30R containing EcoRI and XbaI restriction sites was used to amplify the 579‐bp promoterless in‐frame hrpL gene, which was then cloned into pBAD30 with an arabinose‐inducing promoter. Following amplification, DNA fragments and the vector were both digested by EcoRI and XbaI, and ligated together. The final plasmid was designated as pHrpL‐30. All plasmids were introduced into the E. amylovora strain by electroporation. Transformants were selected on LB plates supplemented with Km and Ap. Their genotypes were confirmed by both enzymatic digestion and sequencing.
Virulence assays on apple shoots and immature pear fruits
Virulence assays on apple shoots were performed as described previously (Wang et al., 2010). Briefly, overnight cultures of E. amylovora WT and mutant strains were harvested by centrifugation and suspended in 0.5 × phosphate‐buffered saline (PBS). Cell suspensions were adjusted to OD600 = 0.1 in PBS and inoculated onto seven actively growing ‘Gala’ apple shoots by pricking the tip with a sterile needle. Five microlitres of bacterial suspension were added to the wounding site by pipetting. Symptom development (progression of necrosis) was recorded at 7 days post‐inoculation and the length of necrotic tissue was measured. The experiment was performed at least twice.
Immature fruits of pear (Pyrus communis L. cv. Bartlett) were surface sterilized and pricked with a sterile needle (Zhao et al., 2005, 2006). Two microlitres of cell suspension (OD600 = 0.1, 100 × dilution) were added to the wounded tissue and the pears were incubated in a humidified chamber at 28 °C. Symptoms were recorded at 4 and 8 days post‐inoculation. For each strain tested, fruits were assayed in triplicate, and each experiment was performed three times.
HR assay on tobacco
Overnight cultures of E. amylovora WT and mutant strains were harvested by centrifugation and cells were resuspended in PBS to OD600 = 0.1. Bacterial suspension was infiltrated into tobacco leaves (Nicotiana tabacum) by needleless syringe. Infiltrated plants were kept in a humid growth chamber and HR symptoms were recorded at 24 h post‐infiltration. For inducible‐hrpL complementation HR assay, arabinose was added to the bacterial suspension to a final concentration of 0.1% immediately before infiltration. All experiments were repeated three times.
RNA isolation
Bacterial strains grown overnight in LB medium with appropriate antibiotics were harvested by centrifugation and washed twice with PBS before inoculating into 5 mL HMM medium at 18 °C. After 6 h in HMM medium, 2 mL of RNA protect reagent (Qiagen, Hilden, Germany) were added to 1 mL of bacterial cell cultures. For in vivo conditions, overnight bacterial cultures were harvested by centrifugation, resuspended in PBS and adjusted to an OD600 of 0.2–0.3. Immature pear fruits were cut in half and inoculated with bacterial suspension. After 6 h at 28 °C in a moist chamber, bacterial cells were collected by washing pear surfaces with RNA protect reagent (Qiagen), 2:1 with water, as described previously (Wang et al., 2012). Cells were harvested by centrifugation for 10 min at 4000 g and RNA was extracted using a Qiagen Bacterial RNA Protect Mini Kit, as recommended by the manufacturer (Qiagen). On‐column DNA digestion was performed using DNase I. RNA was quantified using a Nano‐Drop ND‐100 spectrophotometer (Nano‐Drop Technologies; Wilmington, DE, USA), and RNA quality was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
qRT‐PCR
One microgram of total RNA was reverse transcribed using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. qRT‐PCR was performed using an ABI 7300 System (Applied Biosystems, Foster City, CA, USA). Power SYBR® Green PCR master mix (Applied Biosystems) was used to detect the expression of selected genes with primers designed using Primer3 software. One microlitre of cDNA (2 ng/reaction) or water (no‐template control) was used as template for qRT‐PCRs with Fast SYBR Green PCR Master Mix (Applied Biosystems) and primers at a final concentration of 500 nm. qRT‐PCR amplifications were carried out at 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min, and a final dissociation curve analysis step from 65 to 95 °C. Technical replicate experiments were performed for each biological triplicate sample. Gene expression levels were analysed using the relative quantification (ΔΔCt) method. A 16S rDNA (rrsA) gene was used as a control. A relative quantification value was calculated for each gene with the control group as a reference. A P value was computed using a moderated t‐test to measure the significance associated with each relative quantification value. Variations were considered to be statistically significant when P < 0.05.
Supporting information
Table S1 Primers used in this study.
Acknowledgements
This project was supported by the Agriculture and Food Research Initiative Competitive Grants Program grant no. 2010‐65110‐20497 from the USDA National Institute of Food and Agriculture (YFZ). We would like to thank Dr P. Lawrence Pusey at the USDA Agricultural Research Service, Wenatchee, WA, USA for providing the immature pear fruits used in this study. The authors have no conflicts of interest to declare.
References
- Alarcón‐Chaidez, F.J. , Keith, L. , Zhao, Y.F. and Bender, C.L. (2003) RpoN (σ54) is required for plasmid‐encoded coronatine biosynthesis in Pseudomonas syringae . Plasmid. 49, 106–117. [DOI] [PubMed] [Google Scholar]
- Barrios, H. , Valderrama, B. and Morett, E. (1999) Compilation and analysis of σ54‐dependent promoter sequences. Nucleic Acids Res. 27, 4305–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Kim, J.F. , We, Z.M. , Kolchinsky, I. , Charkowski, A.O. , Conlin, A.K. , Collmer, A. and Beer, S.V. (1998) Homology and functional similarity of an hrp‐linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proc. Natl. Acad. Sci. USA. 95, 1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burse, A. , Weingart, H. and Ullrich, M.S. (2004) NorM, an Erwinia amylovora multidrug efflux pump involved in in vitro competition with other epiphytic bacteria. Appl. Environ. Microbiol. 70, 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, M. and Dixon, R. (2012) The role of bacterial enhancer binding proteins as specialized activators of σ54‐dependent transcription. Microbiol. Mol. Biol. Rev. 76, 497–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, P.E. Jr , Carroll, J.A. , O'Dee, D.M. and Nau, G.J. (2007) Modulation of virulence factors in Francisella tularensis determines human macrophage responses. Microb. Pathog. 42, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, A. , Cui, Y. and Chatterjee, A.K. (2002) Regulation of Erwinia carotovora hrpL, which encodes an extracytoplasmic function subfamily of sigma factors required for expression of the HRP regulon. Mol. Plant–Microbe Interact. 15, 971–980. [DOI] [PubMed] [Google Scholar]
- Datsenko, K.A. and Wanner, B.L. (2000) One‐step inactivation of chromosomal genes in Escherichia coli K‐12 using PCR products. Proc. Natl. Acad. Sci. USA. 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisa, M.P. , Wu, C.F. , Wang, L. , Valdes, J.J. and Bentley, W.E. (2001) DNA microarray‐based identification of genes controlled by autoinducer 2‐stimulated quorum sensing in Escherichia coli . J. Bacteriol. 183, 5239–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, T.G. and Mekalanos, J.J. (2012) Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 40, 7766–7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick, R.D. , Majerczak, D.R. and Coplin, D.L. (1993) Erwinia stewartii WtsA, a positive regulator of pathogenicity gene expression, is similar to Pseudomonas syringae pv. phaseolicola HrpS. Mol. Microbiol. 9, 477–485. [DOI] [PubMed] [Google Scholar]
- Ghosh, T. , Bose, D. and Zhang, X. (2010) Mechanisms for activating bacterial RNA polymerase. FEMS Microbiol. Rev. 34, 611–627. [DOI] [PubMed] [Google Scholar]
- Grimm, C. , Aufsatz, W. and Panopoulos, N.J. (1995) The HrpRS locus of Pseudomonas syringae pv. phaseolicola constitutes a complex regulatory unit. Mol. Microbiol. 15, 155–165. [DOI] [PubMed] [Google Scholar]
- Gruber, T.M. and Gross, C.A. (2003) Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57, 441–466. [DOI] [PubMed] [Google Scholar]
- He, S.Y. , Nomura, K. and Whittam, T.S. (2004) Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta. 1694, 181–206. [DOI] [PubMed] [Google Scholar]
- Hendrickson, E.L. , Guevera, P. and Ausubel, F.M. (2000a) The alternative sigma factor RpoN is required for hrp activity in Pseudomonas syringae pv. maculicola and acts at the level of hrpL transcription. J. Bacteriol. 182, 3508–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, E.L. , Guevera, P. , Peñaloza‐Vàzquez, A. , Shao, J. , Bender, C. and Ausubel, F.M. (2000b) Virulence of the phytopathogen Pseudomonas syringae pv. maculicola is RpoN dependent. J. Bacteriol. 182, 3498–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, E.L. , Plotnikova, J. , Mahajan‐Miklos, S. , Rahme, L.G. and Ausubel, F.M. (2000c) Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J. Bacteriol. 183, 7126–7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson, S.W. , Bretz, J. , Sussan, T. , Jin, S. and Pak, K. (2001) Enhancer‐binding proteins HrpR and HrpS interact to regulate hrp‐encoded type III protein secretion in Pseudomonas syringae strains. J. Bacteriol. 183, 5589–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic, M. , James, E.H. , Burrows, P.C. , Rego, F.G. , Buck, M. and Schumacher, J. (2011) Regulation of the co‐evolved HrpR and HrpS AAA+ proteins required for Pseudomonas syringae pathogenicity. Nat. Commun. 2, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak, M.J. , Wiedmann, M. and Boor, K.J. (2005) Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69, 527–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M.A. , Zhao, Y.F. and Korban, S.S. (2012) Molecular mechanisms of pathogenesis and resistance to the bacterial pathogen Erwinia amylovora, causal agent of fire blight disease in Rosaceae. Plant Mol. Biol. Rep. 30, 247–260. [Google Scholar]
- Koczan, J.M. , McGrath, M.J. , Zhao, Y. and Sundin, G.W. (2009) Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology. 99, 1237–1244. [DOI] [PubMed] [Google Scholar]
- Mann, R.A. , Smits, T.H.M. , Buhlmann, A. , Blom, J. , Goesmann, A. , Frey, J.E. , Plummer, K.M. , Beer, S.V. , Luck, J.E. , Duffy, B. and Rodoni, B. (2013) Comparative genomics of 12 strains of Erwinia amylovora identifies a pan‐genome with a large conserved core. PLoS ONE. 8, e55644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, R.R. , Toth, I.K. , Cock, P.J. , Pritchard, L. , Hedley, P.E. , Morris, J.A. , Zhao, Y.F. and Sundin, G.W. (2012) Genetic characterization of the HrpL regulon of the fire blight pathogen Erwinia amylovora reveals novel virulence factors. Mol. Plant Pathol. 13, 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissinen, R.M. , Ytterberg, A.J. , Bogdanove, A.J. , van Wijk, K. and Beer, S.V. (2007) Analyses of the secretomes of Erwinia amylovora and selected hrp mutants reveal novel type III secreted proteins and an effect of HrpJ on extracellular harpin levels. Mol. Plant Pathol. 8, 55–67. [DOI] [PubMed] [Google Scholar]
- O'Toole, R. , Milton, D.L. , Hörstedt, P. and Wolf‐Watz, H. (1997) RpoN of the fish pathogen Vibrio (Listonella) anguillarum is essential for flagellum production and virulence by the water‐borne but not intraperitoneal route of inoculation. Microbiology. 143, 3849–3859. [DOI] [PubMed] [Google Scholar]
- Oh, C.‐S. and Beer, S.V. (2005) Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol. Lett. 253, 185–192. [DOI] [PubMed] [Google Scholar]
- Oh, C.S. , Kim, J.F. and Beer, S.V. (2005) The Hrp pathogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infection. Mol. Plant Pathol. 6, 125–138. [DOI] [PubMed] [Google Scholar]
- Okuda, S. , Katayama, T. , Kawashima, S. , Goto, S. and Knehisa, M. (2006) ODB: a database of operons accumulating known operons across multiple genomes. Nucleic Acids Res. 34, D358–D362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosthuizen, M.C. , Steyn, B. , Theron, J. , Cosette, P. , Lindsay, D. , Holy, A. and Brözel, V.S. (2002) Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl. Environ. Microbiol. 68, 2770–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Österberg, S. , del Peso‐Santos, T. and Shingler, V. (2011) Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 65, 37–55. [DOI] [PubMed] [Google Scholar]
- Peñaloza‐Vázquez, A. , Fakhr, M.K. , Bailey, A.M. and Bender, C.L. (2004) AlgR functions in algC expression and virulence in Pseudomonas syringae pv. syringae . Microbiology. 150, 2727–2737. [DOI] [PubMed] [Google Scholar]
- Polikanov, Y.S. , Blaha, G.M. and Steitz, T.A. (2012) How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 336, 915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, G. , Deng, W.L. , Huang, H.C. and Collmer, A. (1998) Negative regulation of hrp genes in Pseudomonas syringae by HrpV. J. Bacteriol. 180, 4532–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russel, D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schumacher, J. , Joly, N. , Rappas, M. , Zhang, X. and Buck, M. (2006) Structures and organisation of AAA+ enhancer binding proteins in transcriptional activation. J. Struct. Biol. 156, 190–199. [DOI] [PubMed] [Google Scholar]
- Shingler, V. (2011) Signal sensory systems that impact σ54‐dependent transcription. FEMS Microbiol. Rev. 35, 425–440. [DOI] [PubMed] [Google Scholar]
- Smits, T.H.M. , Rezzonico, F. , Kamber, T. , Blom, J. , Goesmann, A. , Frey, J.E. and Duffy, B. (2010) Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Mol. Plant–Microbe Interact. 23, 384–393. [DOI] [PubMed] [Google Scholar]
- Sperandeo, P. , Pozzi, C. , Deho, G. and Polissi, A. (2006) Non‐essential KDO biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG‐yhbG locus. Res. Microbiol. 157, 547–558. [DOI] [PubMed] [Google Scholar]
- Stauber, J.L. , Loginicheva, E. and Schechter, L.M. (2012) Carbon source and cell density‐dependent regulation of type III secretion system gene expression in Pseudomonas syringae pathovar tomato DC3000. Res. Microbiol. 163, 531–539. [DOI] [PubMed] [Google Scholar]
- Thao, S. , Chen, C.S. , Zhu, H. and Escalante‐Semerena, J.C. (2010) Nε‐lysine acetylation of a bacterial transcription factor inhibits its DNA‐binding activity. PLoS ONE. 5, e15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta, M. , Yoshida, H. , Wada, C. , Baba, T. , Mori, H. and Wada, A. (2005) Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli . Genes Cells. 10, 1103–1112. [DOI] [PubMed] [Google Scholar]
- Ueta, M. , Ohniwa, R.L. , Yoshida, H. , Maki, Y. , Wada, C. and Wada, A. (2008) Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli . J. Biochem. 143, 425–433. [DOI] [PubMed] [Google Scholar]
- Vanneste, J. (2000) Fire Blight: The Disease and Its Causative Agent, Erwinia Amylovora. Wallingford, Oxfordshire: CABI Publishing. [Google Scholar]
- Wang, D. , Korban, S.S. and Zhao, Y.F. (2009) The Rcs phosphorelay system is essential for pathogenicity in Erwinia amylovora . Mol. Plant Pathol. 10, 277–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Korban, S.S. and Zhao, Y.F. (2010) Molecular signature of differential virulence in natural isolates of Erwinia amylovora . Phytopathology. 100, 192–198. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Qi, M. , Calla, B. , Korban, S.S. , Clough, S.J. , Cock, P.J. , Sundin, G.W. , Toth, I. and Zhao, Y. (2012) Genome‐wide identification of genes regulated by the Rcs phosphorelay system in Erwinia amylovora . Mol. Plant–Microbe Interact. 25, 6–17. [DOI] [PubMed] [Google Scholar]
- Wang, R.F. and Kushner, S.R. (1991) Construction of versatile low‐copy‐number vectors for cloning, sequencing and gene expression in Escherichia coli . Gene. 100, 195–199. [PubMed] [Google Scholar]
- Wei, C.F. , Deng, W.L. and Huang, H.C. (2005) A chaperone‐like HrpG protein acts as a suppressor of HrpV in regulation of the Pseudomonas syringae pv. syringae type III secretion system. Mol. Microbiol. 57, 520–536.15978082 [Google Scholar]
- Wei, Z. , Kim, J.F. and Beer, S.V. (2000) Regulation of hrp genes and type III protein secretion in Erwinia amylovora by HrpX/HrpY, a novel two‐component system, and HrpS. Mol. Plant–Microbe Interact. 13, 1251–1262. [DOI] [PubMed] [Google Scholar]
- Wei, Z.M. and Beer, S.V. (1995) hrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of sigma factors. J. Bacteriol. 177, 6201–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Z.M. , Sneath, B.J. and Beer, S.V. (1992) Expression of Erwinia amylovora hrp genes in response to environmental stimuli. J. Bacteriol. 174, 1875–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.F. and Qi, M.S. (2011) Comparative genomics of Erwinia amylovora and related Erwinia species – what do we learn? Genes. 2, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.F. , Blumer, S.E. and Sundin, G.W. (2005) Identification of Erwinia amylovora genes induced during infection of immature pear tissue. J. Bacteriol. 187, 8088–8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.F. , He, S.Y. and Sundin, G.W. (2006) The Erwinia amylovora avrRpt2EAgene contributes to virulence on pear and AvrRpt2EA is recognized by Arabidopsis RPS2 when expressed in Pseudomonas syringae . Mol. Plant–Microbe Interact. 19, 644–654. [DOI] [PubMed] [Google Scholar]
- Zhao, Y.F. , Sundin, G.W. and Wang, D. (2009a) Construction and analysis of pathogenicity island deletion mutants of Erwinia amylovora . Can. J. Microbiol. 55, 457–464. [DOI] [PubMed] [Google Scholar]
- Zhao, Y.F. , Wang, D. , Nakka, S. , Sundin, G.W. and Korban, S.S. (2009b) Systems level analysis of two‐component signal transduction systems in Erwinia amylovora: role in virulence, regulation of amylovoran biosynthesis and swarming motility. BMC Genomics. 10, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primers used in this study.


