SUMMARY
In Arabidopsis, gene expression studies and analysis of knock‐out (KO) mutants have been instrumental in building an integrated view of disease resistance pathways. Such an integrated view is missing in rice where shared tools, including genes and mutants, must be assembled. This work provides a tool kit consisting of informative genes for the molecular characterization of the interaction of rice with the major fungal pathogen Magnaporthe oryzae. It also provides for a set of eight KO mutants, all in the same genotypic background, in genes involved in key steps of the rice disease resistance pathway. This study demonstrates the involvement of three genes, OsWRKY28, rTGA2.1 and NH1, in the establishment of full basal resistance to rice blast. The transcription factor OsWRKY28 acts as a negative regulator of basal resistance, like the orthologous barley gene. Finally, the up‐regulation of the negative regulator OsWRKY28 and the down‐regulation of PR gene expression early during M. oryzae infection suggest that the fungus possesses infection mechanisms that enable it to block host defences.
INTRODUCTION
In order to face attack by pathogens, plants have evolved sophisticated defence pathways. The current model states that several layers of defence exist (Chisholm et al., 2006). The first layer consists of preformed barriers that block or inhibit pathogen growth. They involve the cuticle (Skamnioti and Gurr, 2007), cell wall strengthening (Juge, 2006) and the constitutive expression of defence‐related genes (Vergne et al., 2010), and represent mostly broad‐spectrum pathogen resistance. The second layer responds to so‐called pathogen‐associated molecular patterns (PAMPs) from pathogens that are detected by pattern recognition receptors (PRRs). This layer is often neutralized by the pathogen through the secretion of effector molecules. A third layer of defence classically involves nucleotide‐binding site‐leucine‐rich repeat (NBS‐LRR) proteins from the plant that recognize effectors.
Our current view is that inducible lines of defence comprise four major steps: recognition, signal transduction, transcription activation and defence gene expression. Pathogen recognition involves PRRs, such as the receptor‐like kinases FLS2 (Takai et al., 2008; Zipfel, 2009) and CERK1 (Miya et al., 2007; Shimizu et al., 2010) in Arabidopsis and the chitin‐binding protein CEBiP in rice (Kaku et al., 2006; Kishimoto et al., 2010). Some proteins, such as RAR1, are also required at this stage to maintain appropriate NBS‐LRR protein steady‐state levels (da Silva Correia et al., 2007). Subsequent signal transduction involves mitogen‐activated protein (kinase kinase) [MAP(KK)] kinases (e.g. OsMAPK5a, OsEDR1; for a review, see Pitzschke et al., 2009) and other regulatory proteins (Park et al., 2008). Several other genes are required but their function is less well characterized (Fujiwara et al., 2010). Transcription activation occurs through the action of several classes of protein (Eulgem, 2005), including WRKY and TGA proteins. The regulation of WRKY transcription factors is complex (Rushton et al., 2010) and the regulation of TGAs occurs through the action of the central regulator NPR1 (Fobert and Després, 2005). Following these signal transduction events, defences are activated (Hammond‐Kosack and Jones, 1996). They include cell wall strengthening and the production of antimicrobial phytoalexins and a wide array of pathogenesis‐related (PR) proteins.
Genetic and genomic analyses performed over several decades in the model plant Arabidopsis have yielded important insights into how these pathways are controlled (Nishimura and Dangl, 2010). In rice, 45 genes are now known to be required for disease resistance [Delteil et al., 2010; see Table S1 (Supporting Information) for an updated version]. These genes are called ‘disease regulators’ hereafter. In Arabidopsis, the generalized use of knock‐out (KO) mutants in one major background (Col‐0 for most cases) greatly simplified the analysis of disease resistance pathways. Indeed, plant–pathogen interactions are very sensitive to genetic background. As a result, complex integrated pathways could be built (Hammond‐Kosack and Parker, 2003). This is not the case in rice where more than 20 genetic backgrounds from indica and japonica subspecies have been used to study mutants in 45 rice disease regulator genes involved in the disease resistance pathway. In the majority of cases (26 of 45), overexpressors alone were studied, raising the possibility that artefacts may be responsible for the phenotype. Silencing was used in the completely sequenced genotype Nipponbare in only four of 12 cases, shedding some doubt on the specificity of the RNAi construct in other genetic backgrounds. The genes listed in Table 1 provide examples of the limitations of such studies. First, it can be seen that data on Magnaporthe oryzae resistance are often missing (15 of 45 genes; Table S1), despite the importance of this fungal pathogen, the causal agent of rice blast. Quite strikingly, information on cell culture is often available, but not on whole‐plant KO (e.g. OsBWMK1; Koo et al., 2009b). Thus, the assembled puzzle in rice suffers from several weaknesses.
Table 1.
Available and missing data for the nine genes selected for this study.
| Gene name | Plant | Type | Altered resistance | Altered defence expression | Reference | ||
|---|---|---|---|---|---|---|---|
| M. oryzae | X. oryzae pv. oryzae | Before infection | After infection | ||||
| NH1 | Rice plant (TP309) | Constitutive overexpression | Yes* | Yes | + | nd | Yuan et al. (2007); Chern et al. (2005) |
| BWMK1 | Tobacco | Constitutive overexpression | nd | na | + | na | Cheong et al. (2003) |
| OsWRKY28 | Rice plant (Kitaake) | Multiple overexpression of WRKYs | nd | Yes | + | nd | Peng et al. (2010) |
| OsEDS5 | na | — | nd | nd | na | na | Vergne et al. (2007) |
| rTGA2.1 | Rice plants (Liao Geng) | Constitutive RNAi | nd | Yes | + | No change | Fitzgerald et al. (2005) |
| CEBiP | Rice plant | Constitutive RNAi | Yes | na | nd | na | Kishimoto et al. (2010) |
| Pi21 | Rice plant (Aichiasahi) | Constitutive RNAi | Yes | No effect | No change | Enhanced | Fukuoka et al. (2009) |
| SPL7 | Rice plant (Norin 8) | Point mutation | Yes | nd | nd | nd | Yamanouchi et al. (2002); Yin et al. (2000) |
NH1 required for benzothiadiazole‐induced resistance.
na, not applicable; nd, not determined.
Little comprehensive information is provided on disease regulators and PR gene expression in rice mutants, as well as during infection. Although some studies have reported on PR gene expression before infection (25 of 45 genes), few have reported on PR gene expression after infection (nine of 45). Finally, with the exception of a few microarray experiments (e.g. OsWRKY13; Qiu et al., 2008), there are few reports on the expression of disease regulator genes in mutants. This should provide interesting information for the building of regulatory networks. Overall, we lack an integrated view of these 45 rice regulators and many gaps must be filled.
In order to start bridging these gaps, we initiated a rice resource consisting of nine disease regulator mutants (all in one genetic background—Nipponbare) and 20 genes representative of the disease resistance pathway. They were used for expression studies aimed at building an integrated view of the molecular events taking place during rice blast infection.
RESULTS
Early and strong transcriptional regulation of disease regulators
In order to compare fully susceptible plants with fully resistant plants, Nipponbare plants were inoculated with M. oryzae isolates FR13 and CL3.6.7, respectively. Typical symptoms (Fig. S1A, see Supporting Information) were observed 4 days post‐inoculation, as compared with intermediate symptoms produced by the GY11 isolate (partial resistance in this case). The early cytological events associated with these interactions were very similar in the early phases and up to 48 h post‐inoculation (hpi) (Fig. S1B).
The expression of 20 genes (Fig. 1 and Table 2) was measured by reverse transcription‐quantitative polymerase chain reaction (RT‐QPCR) as early as 1 h after inoculation and compared with that of mock‐treated plants (see Experimental procedures) in order to eliminate the effect of the inoculation procedure on gene expression. Overall, the responses to virulent and avirulent strains of M. oryzae were qualitatively similar, although expression levels were often different. With a few exceptions (OsBIRH1 and OsEDS5), the expression patterns observed late after infection were similar to those published previously. For OsBIRH1, Li et al. (2008) reported an induction, whereas we observed a clear repression (Fig. 1). For OsEDS5, we did not observe a change in expression (Fig. 1), whereas this gene was reported to be induced by Vergne et al. (2007). These discrepancies may be a result of the fact that different genetic backgrounds were used (indica in other studies, japonica in this work).
Figure 1.
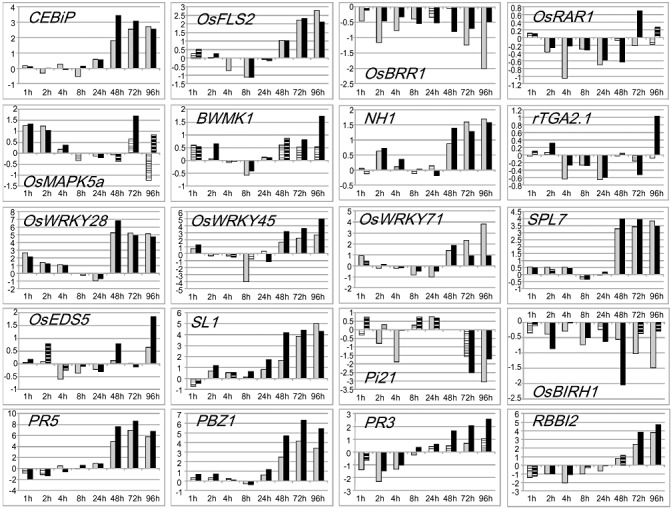
Early and late regulation of defence‐related genes during rice blast infection. Gene expression was measured by reverse transcription‐quantitative polymerase chain reaction in plants inoculated with gelatin only (mock treatment) or with Magnaporthe oryzae (virulent isolate FR13, grey bars; avirulent isolate CL3.6.7, black bars) at different time points after treatment. Gene expression was normalized using actin. The results are the log2 values of the ratio of the mean transcript levels for inoculated vs. mock‐treated plants from four independent biological replicates. A t‐test was performed to establish whether the expression of one given gene inoculated with the virulent or avirulent isolate was different from its corresponding expression in the mock‐treated plants (plain black or grey bars, P < 0.05; no statistically significant difference, hatched bars).
Table 2.
Primers used for reverse transcription‐quantitative polymerase chain reaction gene expression studies.
| Name | Gene | Forward | Reverse |
|---|---|---|---|
| Actin | Os03g50885 | GCGTGGACAAAGTTTTCAACCG | TCTGGTACCCTCATCAGGCATC |
| CEBiP | Os03g04110 | CACTTGTACGGCTGCTTGAA | GGAAGGTGGGAAGTCCATTC |
| OsFLS2 | Os04g52780 | TGGGTTACATGGCTCCAGAGTTCG | AAGCTGAACACGTCCACCTTCGTC |
| OsBRR1 | Os03g12730 | TGTACGTCTTGGCGTACTCCTG | GAGATTTGCCTCTACTTGGACTCG |
| OsRAR1 | Os02g33180 | TTTACCGTGCTCTGTGTGACTG | GCCAACGAACGAGACCGAAG |
| OsMAPK5a | Os03g17700 | CCGCTGCAGAGAATCACAGTTG | ATCGGCGATGTCGTGCAATCTC |
| BWMK1 | Os06g49430 | CGTCGAGAGCCACAAGAAGAAC | TGCCAATGACTTCCTGGATCTGG |
| NH1 | Os01g09800 | CCTGATGGTTGCCTTCTGTC | ATTCAAGCACTTGTATTACACCTC |
| rTGA2.1 | Os07g48820 | CCATGCCATGAGTGGAAATGGG | TGCATGAGCATTCACTGCTGTC |
| OsWRKY28 | Os06g44010 | CGCCGATGAACTTTGCTC | CCACCTTGGCACGTGTAGA |
| OsWRKY45 | Os05g25770 | ACGACGAGGTTGTCTTCGATCTG | GCCCGTGTCCATCCATGATTCTTC |
| OsWRKY71 | Os02g08440 | CCGAGCAGATGGCGATGAC | AGGCAGAGACAGGAGAGGATG |
| SPL7 | Os05g45410 | CGGATTAGAGGCTTGCGTGTTAC | GCACAGTAGTCAGCGGATAGAAC |
| OsEDS5 | Os02g02980 | CACGGCTAGGTTCAGTTCCAATG | CCAATCCATCAGCAAGAAGAGACG |
| SL1 | Os12g16720 | TGTGACTAAGCAGAGAAGCAAG | AAGAGAAATACGCCACTTATTGAC |
| Pi21 | Os04g32850 | GGTCATCTTGGTGGACCTGCAATG | CGATGCAGTACTCCTCTTCAAGGC |
| OsBIRH1 | Os03g01830 | GAGGGTGATGGAGAGGAACA | GACTAGACGCATGGCACATC |
| PR5 | Os12g43430 | CGCTGCCCCGACGCTTAC | ACGACTTGGTAGTTGCTGTTGC |
| PBZ1 | Os12g36880 | AGGCATCAGTGGTCAGTAGAG | CGGGTCTTGTATGTGCTTCC |
| PR3 | Os04g41620 | CGTGTCTGTGGAGAGCGTGGTC | TCGTCGTTGGTGCGGTCATTGG |
| RBBI2 | Os01g03390 | ATCTGTGTCCGTCAATAAAACTCG | TTGCTCTTGGTCACTGGCTAG |
The OsWRKY45 gene is the gene described in Shimono et al. (2007).
As expected, all PR genes tested were induced at 24–48 hpi to higher levels in resistant plants than in susceptible plants (Fig. 1). This is a typical feature of the interaction (Ribot et al., 2008). With the exception of two genes (OsRAR1 and OsEDS5), the other 18 of the 20 genes tested were, to a variable extent, differentially expressed on infection, particularly at late time points (after 24 hpi). This confirms previous findings that there is a strong transcriptional control of disease regulators in rice (Vergne et al., 2008). Although the majority of the regulatory genes were up‐regulated, three (OsBRR1, Pi21 and OsBIRH1; Fig. 1) were down‐regulated.
Several disease regulators were differentially expressed during the very early steps of infection, before penetration of the fungus into the first infected cells (Fig. S1B). Four genes (OsMAPK5a, SL1, OsWRKY28 and OsWRKY45) were up‐regulated two‐ to three‐fold and as early as 1 hpi (OsMAPK5a and OsWRKY28; Fig. 1), irrespective of the type of interaction. Quite surprisingly, three of the four PR genes tested were down‐regulated (from two‐ to four‐fold) in the early time points after inoculation (Fig. 1). To our knowledge, these are the earliest transcriptional responses ever reported during rice blast infection.
Phenotyping of rice mutants for resistance
Insertion mutants and corresponding null‐segregant plants (wild‐type, WT) were identified for eight genes (see Experimental procedures for more details) in the OryzaTagLine mutant collection (Larmande et al., 2008; Sallaud et al., 2004). For each insertion line, PCR was used to select null and homozygous mutant plants in a segregating T2 family. The primers used are given in Table S2 (see Supporting Information) These plants were allowed to self and the genotypes were confirmed in the T3 generation. Transcript levels for the mutated gene (Fig. 2) were strongly reduced in four lines (cebip, pi21, oswrky28 and rtga2.1), slightly reduced in three cases (nh1, bwmk1 and spl7) and not affected in the oseds5 line.
Figure 2.
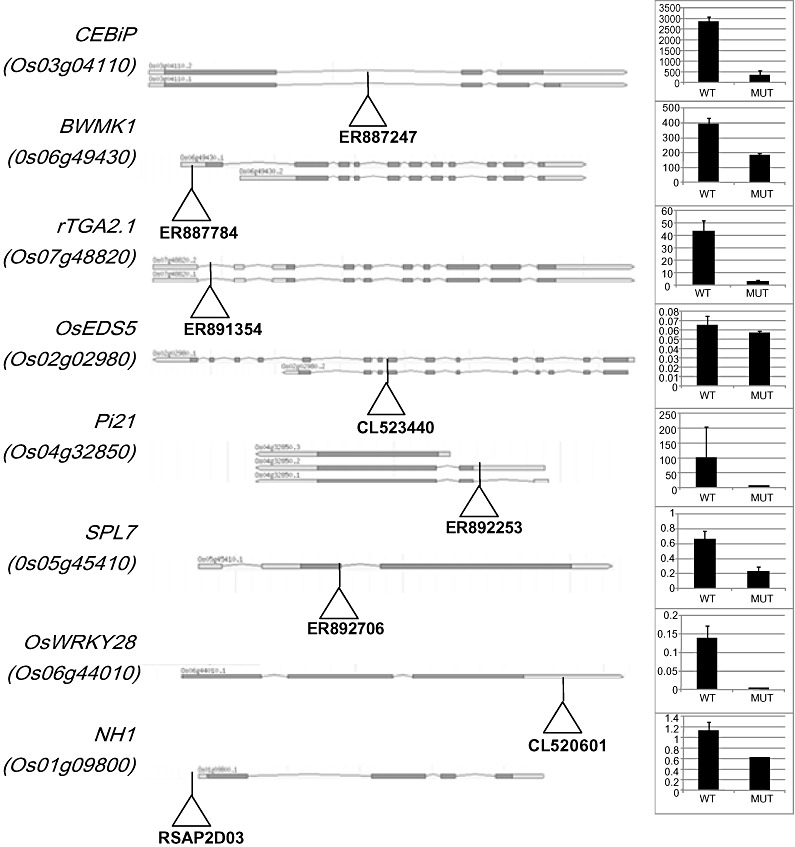
Genomic structure and transcript levels in the insertion mutants for eight disease regulators. Left: the different splice forms for each gene, together with the position of the T‐DNA insertion site. The primers used to genotype the plants are shown in Table S2. Right: transcript levels for the corresponding gene, as measured by reverse transcription‐quantitative polymerase chain reaction, in mutant plants (MUT) compared with the corresponding null segregant plants (WT), 2 days after infection. The mean and standard deviation of four independent biological replicates, expressed in arbitrary units, are shown.
The homozygous mutant lines and corresponding null segregant lines were inoculated three times with the moderately virulent M. oryzae GY11 isolate in order to measure blast disease resistance. Typical symptoms are shown in Fig. 3. Three mutant lines (nh1, rtga2.1 and cebip) reproducibly showed more symptoms (Fig. 3) and a significantly increased lesion number (Fig. 4) compared with control null segregant lines. In contrast, the lines mutated in the OsWRKY28 and SPL7 genes showed fewer symptoms (Fig. 3) and a significantly lower lesion number when compared with their respective WT plants (Fig. 4). The increased resistance conferred by the spl7 mutation before spontaneous necrosis developed (Fig. 4) was even stronger in adult plants, when necrosis had developed (Fig. S2, see Supporting Information). The pi21, bwmk1 and oseds5 lines displayed WT responses to infection.
Figure 3.
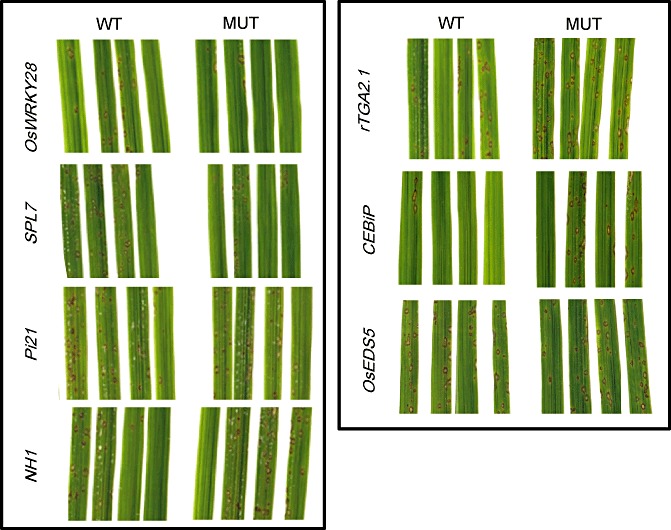
Symptoms of the mutant lines on Magnaporthe oryzae infection. Plants were inoculated with the virulent isolate GY11 of M. oryzae. Photographs were taken at 5 days post‐inoculation. For each line, a representative sample of leaves from mutant (MUT) and corresponding null segregant (WT) plants is shown. The photographs for cebip were taken in a separate experiment.
Figure 4.
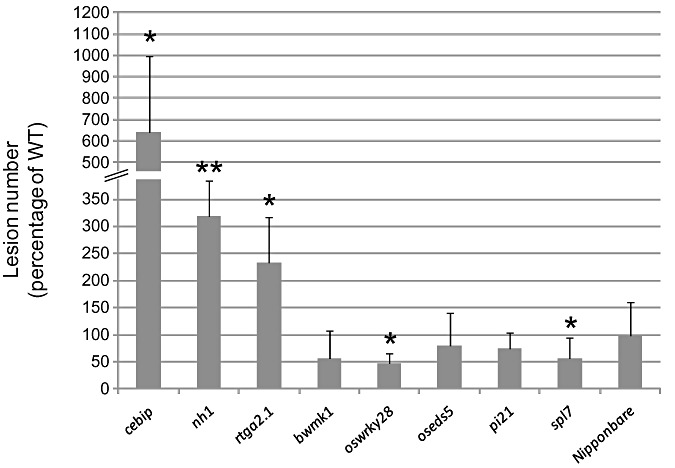
Quantitative effect on blast resistance of the mutations in disease regulators. The total number of lesions was counted for each leaf after infection by the virulent isolate GY11 of Magnaporthe oryzae. For each mutant line, the average number of lesions over more than eight plants was calculated for the corresponding null segregant line. This value was used to calculate the percentage of lesions per individual mutant plant relative to the mean of the null segregant plants (WT). The mean and standard deviation were then calculated. This experiment was repeated three times and one representative experiment is shown. A t‐test was performed to establish whether one given mutant line was different from its corresponding null segregant line (*P < 0.05; **P < 0.001).
Expression of defence‐related genes in rice mutants
Next, we evaluated the expression of 13 selected genes, representative of the expression patterns observed (Fig. 1 and Table 2), to test whether the genetic defects in the mutants could alter defence expression in the absence or presence of the fungus at 48 hpi. The 48‐hpi time point was selected on the basis of the observation that most genes tested were induced at that time (Fig. 1). Moreover, this set of genes included the eight genes for which mutants were available in our rice mutant resource. The majority of the mutants did not show a significant modification of gene expression in the absence or presence of the fungus at 48 hpi (data not shown). However, three mutants (cebip, nh1 and oswrky28) showed strong modifications of gene expression when compared with their respective WT plants (Fig. 5). Five genes (OsMAPK5a, SPL7, OsWRKY45, OsWRKY71 and PR5) were significantly less induced in the cebip mutant than in the corresponding WT plants at 48 hpi. The up‐regulation of five genes, including CEBiP, SL1, SPL7, OsWRKY28 and rTGA2.1, was reduced in the nh1 background after infection (see Fig. 5). In the oswrky28 mutant, three PR genes (PBZ1, PR5 and RBBI2) were more strongly induced after infection and RBBI2 transcript levels were higher before infection.
Figure 5.
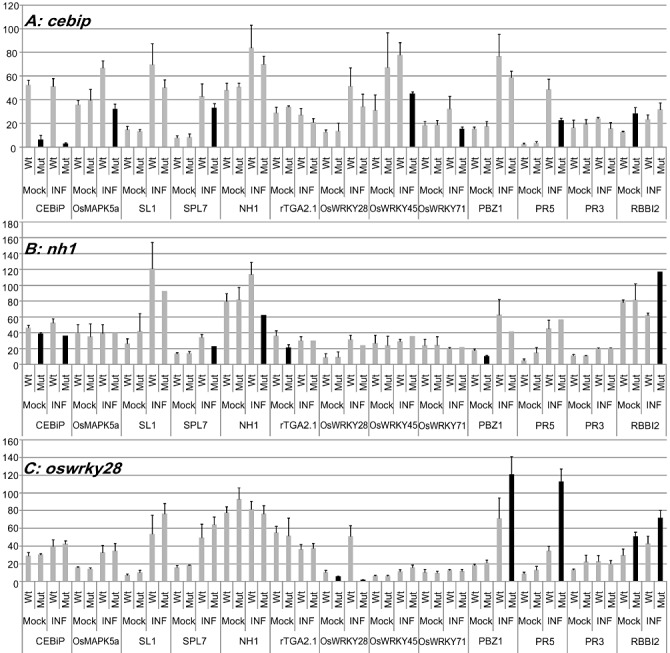
Mis‐regulation of defence‐related genes in cebip, nh1 and oswrky28 mutants. Mutant lines (Mut) and their corresponding null segregant lines (Wt) were treated with gelatin only (Mock) or inoculated with the virulent GY11 isolate of Magnaporthe oryzae. Three mutant lines are shown: cebip (A), nh1 (B) and oswrky28 (C). RNAs were then extracted 2 days after infection, at a time at which most defence genes show differential expression (see Fig. 1). The expression of 13 genes was measured by reverse transcription‐quantitative polymerase chain reaction. The transcript levels in mutant plants for which there is a significant difference between Wt and Mut are highlighted in black. The values represent the mean and standard deviation of four independent biological replicates. A t‐test was performed to establish whether the level of gene expression in the mutant line was different from that of the null segregant line under the same treatment (black bars, P < 0.05).
Thus, the cebip, nh1 and oswrky28 mutants show very specific and contrasted molecular phenotypes when measuring defence gene expression.
DISCUSSION
The NH1, rTGA2.1 and OsWRKY28 genes are required for full blast resistance
The OsWRKY28 gene has been shown recently to be important for blight resistance (Peng et al., 2010). This was demonstrated by collectively silencing three WRKY genes and the individual role of each WRKY gene could not be established. Moreover, the individual role in blast resistance of the OsWRKY28 gene was not demonstrated. Here, we show that constitutive knocking down of OsWRKY28 leads to a two‐fold increase in resistance to blast when compared with WT (Fig. 4). This is consistent with the observation that the oswrky28 mutant overexpresses several PR genes (Fig. 5). The OsWRKY28 gene is a putative orthologue of the barley HvWRKY1/2 genes (data not shown). The HvWRKY1/2 genes are negative regulators of basal defence in barley (Shen et al., 2007). The results obtained with OsWRKY28 are consistent with this finding. Thus, our results suggest that OsWRKY28 has a conserved function in barley and rice.
Despite its importance in many plant–pathogen interactions (Dong, 2004), the NH1 gene has never been shown to be required for blast resistance. However, this gene has been shown to be required for the chemical induction of blast resistance (Sugano et al., 2010). The T‐DNA in the nh1 mutant is inserted in the promoter region (Fig. 2). Inducible but not constitutive expression of this mutant allele is affected by the insertion (Fig. 5B). Thus, the inducible expression of NH1 could require some elements in the promoter region that are mutated by the insertion element. Here, we show that rice mutants for the NH1gene are affected in blast resistance (Fig. 4), although to a lesser extent than the cebip mutants (see below). Thus, NH1 is a key regulator of blast disease resistance.
The rTGA2.1 gene has been shown previously to play a negative role in rice basal defence responses to bacterial pathogens (Fitzgerald et al., 2005). Moreover, the rtga2.1 mutant plants produced using RNAi were stunted and possibly affected in closely related TGA genes. Here, we show that rtga2.1 mutants that exhibit no obvious developmental defect (data not shown) are more susceptible to blast fungus than their respective WT plants (3, 4). This is consistent with the published role of the orthologous gene in Arabidopsis (Kesarwani et al., 2007; Zhang et al., 2003).
The data presented here thus demonstrate the involvement of three genes, rTGA2.1, NH1 and OsWRKY28, in the establishment of full basal resistance to rice blast.
Confirmation of the role of CEBiP and SPL7 in blast resistance
The CEBiP protein has been shown to bind chitin in vitro and in cell cultures (Kaku et al., 2006). The CEBiP gene is also required for the massive transcriptional response to chitin in cell cultures. For example, the OsMAPK5a gene is induced 15‐fold by chitin in WT cells, but only seven‐fold in cells silenced for CEBiP. Similarly, the PR5 gene is induced 17‐fold by chitin, but only seven‐fold in cells silenced for CEBiP (Kaku et al., 2006). Recently, Kishimoto et al. (2010) have demonstrated that KO of CEBiP expression reduces blast resistance. We also show that the CEBiP gene is required for blast resistance (Fig. 4), as mutants for this gene exhibit six‐fold more lesions than the corresponding WT plants. This confirms that, together with the OsCERK1 gene, the CEBiP gene is a key element regulating PAMP‐triggered immunity in rice plants.
As predicted from the literature (Yamanouchi et al., 2002), the KO mutant for the SPL7 gene shows spontaneous lesions (Fig. S2), as well as a slight increase in resistance (Fig. 4).
Mutants not displaying altered responses to blast infection
The insertion line for the Pi21 gene was not affected significantly for resistance, although slightly fewer lesions were observed (3, 4). This may be a result of the fact that Nipponbare already contains the nonfunctional allele of Pi21 (Fukuoka et al., 2009). Thus, the removal of further activity of Pi21 in Nipponbare does not have a strong effect on disease resistance.
In the case of OsEDS5 and BWMK1, we did not observe an altered response to blast infection in the corresponding insertion mutants (3, 4). This may be explained by the fact that the expression of the corresponding genes is not affected strongly in these mutants (Fig. 2). In addition, for BWMK1, at least two alternative splice forms exist and only one is affected by the insertion (Fig. 2). The splice form that is disrupted may not be involved in resistance, consistent with the observation that they are differentially regulated (Koo et al., 2009a; Koo et al., 2007). Alternatively, the BWMK1 and OsEDS5 genes may not be required for blast resistance.
Repression of defence in the very early phases of infection
Most published studies on gene expression during blast infection have been performed at time points after 24 hpi (Vergne et al., 2008) and information on the early time points is missing. We decided to explore earlier time points, before cell penetration (Fig. S1B), in order to identify early transcriptional changes that may occur before the fungus completely penetrates the first host cells. Of the 20 genes tested, five genes (Table S1), that are either known positive (SL1, OsWRKY45) or negative (OsMAPK5a, Pi21 and OsWRKY28) disease resistance regulators, are differentially expressed very early during infection (Fig. 1). Consistent with the onset of resistance, the positive regulators SL1 and OsWRKY45 are induced, whereas the negative regulator Pi21 is repressed.
Strikingly, the activation of two negative regulators (OsMAPK5a and OsWRKY28) and the repression of positive regulators of defence, such as three PR genes (PR3, PR5 and RBBI2), were also observed (Fig. 1). These observations hold true for both virulent and avirulent isolates, suggesting that this is a common response of the plant to infection by the fungus. Inhibition of basal defence by molecules produced by pathogens is now well established (Alfano, 2009). The current view is that PAMPs, such as flagellin for bacteria and chitin for fungi, can trigger basal defence after recognition by PRRs. In rice, the CEBiP and OsCERK1 proteins are such receptors for chitin (Kishimoto et al., 2010; Shimizu et al., 2010). Pathogens have evolved effectors to inhibit basal defence. These effectors control host cell functions, such as defence gene expression, through various biochemical activities, including protein modification and hormone mimicry (Block et al., 2008). There are also cases in which transcriptional control is involved, as in the case of the Xanthomonas transcription activator‐like (TAL) effectors (Boch and Bonas, 2010). In the case of rice blast, there are only indirect indications that the inhibition of basal defence occurs. For example, the MIG1/RLM1 gene of M. oryzae is required to overcome rice defence (Mehrabi et al., 2008). Similarly, rice plants inoculated with the M. oryzae des1 mutant exhibit strong defence responses, including PR gene induction in neighbouring tissues (Chi et al., 2009). Thus, our results are an indication that basal defences might be inhibited at the transcriptional level during rice blast infection. We hypothesize that the blast fungus represses the transcription of key regulators in order to lower basal defence.
Here, we show that, in the oswrky28 mutant, PR gene expression is up‐regulated (Fig. 5). Thus, the OsWRKY28 gene is a negative regulator of PR expression. Similarly, PR gene expression has been shown to be higher in plants silenced for the OsMAPK5a gene (Xiong and Yang, 2003). Thus, the up‐regulation of OsWRKY28 and/or OsMAPK5a in the early phases of infection could explain the observed down‐regulation of the PR genes. Whether the fungus is directly manipulating the expression of OsWRKY28 and/or OsMAPK5a remains to be demonstrated.
Concluding remarks
Little is known about the regulation of defence mechanisms in rice. In this study, we have addressed this issue by examining the effect of mutations on rice genes orthologous to known defence regulators in Arabidopsis and barley.
The available information on disease regulators in rice is largely incomplete (Delteil et al., 2010 and Table S1 for an updated list of the genes involved in disease resistance). For example, many regulators are known to be involved in resistance to bacterial blight caused by Xanthomonas oryzae pv. oryzae, but the corresponding data for rice blast are still missing. In order to build an integrated view of these regulators, we initiated a mutant collection targeting disease resistance regulators. This work provides a tool kit, including genes and KO mutants, available to the community to study rice disease resistance. Although insertional mutations do not always lead to complete KO of gene expression, they remain a very valuable resource for establishing regulation networks, in particular through expression studies. Insertion lines for 16 other regulatory genes that were not tested in this study are available in the Nipponbare background (Table S1). The sharing and studying of these mutants should help us put together the regulatory network leading to disease resistance in rice.
EXPERIMENTAL PROCEDURES
Infection assays
Fungi were grown as described in Berruyer et al. (2003). Rice plants were grown as described in Faivre‐Rampant et al. (2008). One rice cultivar, Nipponbare (Oryza sativa L.), and three isolates, FR13, GY11 and CL3.6.7, of the blast fungus (M. oryzae) were used. The isolate CL3.6.7 is incompatible and isolates FR13 and GY11 are compatible with Nipponbare (Fig. S1). It is a common observation that gene expression can be altered by the inoculation procedure (e.g. Vergne et al., 2007). For this reason, a mock treatment (gelatin only) was included to normalize gene expression.
For mutant phenotyping, inoculation was carried out by spraying 2.5 × 104 conidia/mL of GY11 isolate (a compatible strain which leads to partial resistance), whereas, for expression analyses, we used 2 × 105 conidia/mL of GY11 conidial suspension, on 4‐week‐old plants. All treated seedlings were transferred to 100% relative humidity for 24 h. For mutant phenotyping, the fourth leaves were harvested and scanned at 5 days after infection for lesion observations and quantifications, whereas, for expression analyses, the fourth leaves were collected at 48 h after infection for RNA extractions.
Building a rice mutant resource
Insertion mutants in the Nipponbare background are currently available for 21 of the 43 rice disease resistance regulator genes. Ten of these insertion lines were available through the OryzaTagLine (Larmande et al., 2008; Sallaud et al., 2004) collection, and we selected CEBiP, rTGA2.1, NH1, Pi21, SPL7 and OsWRKY28 (Table 1) as representative of the disease resistance pathway. The CEBiP protein was shown in cell culture to be a chitin receptor (Kaku et al., 2006), and it has been shown recently (Kishimoto et al., 2010) that KO of CEBiP expression leads to increased fungal growth. rTGA2.1 is a negative regulator of defence to bacterial pathogens in rice (Fitzgerald et al., 2005). NH1 is an orthologue of NPR1, a central regulator in many plant species, and is known to be required for bacterial resistance in rice (Dong, 2004). The Pi21 gene has been identified recently as the first gene underlying a rice blast quantitative trait locus that confers a durable and broad‐spectrum resistance (Fukuoka et al., 2009). The SPL7 gene encodes a heat‐shock transcription factor (Yamanouchi et al., 2002) and spl7 mutants are known to display spontaneous lesions resembling hypersensitive response lesions, as well as increased levels of resistance to rice blast (Yin et al., 2000). The OsWRKY28 gene is probably the orthologous copy of either the HvWRKY1 or HvWRKY2 gene, both of which have been shown to be required for basal immunity in barley (Shen et al., 2007).
We also selected the BWMK1gene as representative of the signal transduction events. The function of this gene has only been demonstrated in cell cultures of heterologous species (Koo et al., 2009b). Finally, we selected the rice putative orthologue of the Arabidopsis EDS5 gene (Nawrath et al., 2002), which has been shown previously to be differentially expressed on infection in the indica cv. IR64, but not in Nipponbare (Vergne et al., 2007). Thus, a role in rice blast disease resistance is not established for four of the eight genes selected (Table 1).
Expression analysis
For RT‐QPCR applications, frozen tissues were ground in liquid nitrogen. Approximately 500 µL of powder was then treated with 1 mL of TRIZOL (Invitrogen, Carlsbad, CA, USA) as recommended. RNA samples (5 µg) were denatured for 5 min at 65 °C with oligo(dT)18 (3.5 µm) and deoxynucleoside triphosphate (dNTP) (1.5 µm). They were then subjected to reverse transcription for 60 min at 37 °C with 200 U of reverse transcriptase M‐MLV (Promega, Madison, WI, USA) in the appropriate buffer. Two microlitres of cDNA (dilution 1:10) were then used for RT‐QPCR. RT‐QPCR mixtures contained PCR buffer, dNTP (0.25 mm), MgCl2 (2.5 mm), forward and reverse primers (final concentration of 150, 300 or 600 nm), 1 U of HotGoldStar polymerase and SYBR Green PCR mix as per the manufacturer's recommendations (Eurogentec, Seraing, Belgium). Amplification was performed as follows: 95 °C for 10 min; 40 cycles of 95 °C for 15 s, 62 °C for 1 min and 72 °C for 30 s; finally, 95 °C for 1 min and 55 °C for 30 s. The RT‐QPCRs were performed using an MX3000P machine (Stratagene, La Jolla, CA, USA) and data were extracted using MX3000P software. The amount of plant RNA in each sample was normalized using actin (Os03g50890) as an internal control. The calculation of gene expression was performed using the measured efficiency for each primer pair as described in Vergne et al. (2007). The list of primers used is provided in Table 2.
Supporting information
Fig. S1 Macroscopic symptoms and microscopic events during Magnaporthe oryzae infection. (A) Characteristic symptoms on Nipponbare leaves inoculated with different M. oryzae isolates. (B) Magnaporthe oryzae infection process in rice at the cellular and subcellular levels.
Fig. S2 The spl7 mutant shows massive spontaneous lesions and enhanced resistance in adult plants.
Table S1 Updated list of disease regulators.
Table S2 Polymerase chain reaction (PCR) primers used for genotyping insertion lines.
Supporting info item
ACKNOWLEDGEMENTS
AD was funded by a joint grant between centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD) and the Languedoc‐Rousillon region. Most of this work was supported by the Génoplante Programme [project Interaction Rice MAgnaporthe (IRMA)], and the work relative to OsWRKY28 was supported by the CerealImmunity Project (Generation Challenge Programme). OFR was funded by a post‐doctoral grant from the Cassa di Risparmio delle Provincie Lombarde (CARIPLO) Foundation. We thank Loïc Fontaine for technical assistance in the glasshouse and Delphine Mieulet for providing rice mutant seeds.
REFERENCES
- Alfano, J.R. (2009) Roadmap for future research on plant pathogen effectors. Mol. Plant Pathol. 10, 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruyer, R. , Adreit, H. , Milazzo, J. , Gaillard, S. , Berger, A. , Dioh, W. , Lebrun, M.H. and Tharreau, D. (2003) Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor. Appl. Genet. 107, 1139–1147. [DOI] [PubMed] [Google Scholar]
- Block, A. , Li, G. , Fu, Z.Q. and Alfano, J.R. (2008) Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 11, 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch, J. and Bonas, U. (2010) Xanthomonas AvrBs3 family‐type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. [DOI] [PubMed] [Google Scholar]
- Cheong, Y.H. , Moon, B.C. , Kim, J.K. , Kim, C.Y. , Kim, M.C. , Kim, I.H. , Park, C.Y. , Kim, J.C. , Park, B.O. , Koo, S.C. , Yoon, H.W. , Chung, W.S. , Lim, C.O. , Lee, S.Y. and Cho, M.J. (2003) BWMK1, a rice mitogen‐activated protein kinase, locates in the nucleus and mediates pathogenesis‐related gene expression by activation of a transcription factor. Plant Physiol. 132, 1961–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern, M. , Fitzgerald, H.A. , Canlas, P.E. , Navarre, D.A. and Ronald, P.C. (2005) Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant–Microbe Interact. 18, 511–520. [DOI] [PubMed] [Google Scholar]
- Chi, M.‐H. , Park, S.‐Y. , Kim, S. and Lee, Y.‐H. (2009) A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. Plos Pathog. 5, e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Delteil, A. , Zhang, J. , Lessard, P. and Morel, J.‐B. (2010) Potential candidate genes for improving rice disease resistance. Rice, 3, 56–71. [Google Scholar]
- Dong, X. (2004) NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10, 71–78. [DOI] [PubMed] [Google Scholar]
- Faivre‐Rampant, O. , Thomas, J. , Allègre, M. , Morel, J.‐B. , Tharreau, D. , Nottéghem, J.L. , Lebrun, M.H. , Schaffrath, U. and Piffanelli, P. (2008) Characterization of the model system rice–Magnaporthe for the study of nonhost resistance in cereals. New Phytol. 180, 899–910. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, H.A. , Canlas, P.E. , Chern, M.S. and Ronald, P.C. (2005) Alteration of TGA factor activity in rice results in enhanced tolerance to Xanthomonas oryzae pv. oryzae . Plant J. 43, 335–347. [DOI] [PubMed] [Google Scholar]
- Fobert, P.R. and Després, C. (2005) Redox control of systemic acquired resistance. Curr. Opin. Plant Biol. 8, 378–382. [DOI] [PubMed] [Google Scholar]
- Fujiwara, T. , Maisonneuve, S. , Isshiki, M. , Mizutani, M. , Chen, L. , Wong, H.L. , Kawasaki, T. and Shimamoto, K. (2010) Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J. Biol. Chem. 285, 11 308–11 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka, S. , Saka, N. , Koga, H. , Ono, K. , Shimizu, T. , Ebana, K. , Hayashi, N. , Takahashi, A. , Hirochika, H. , Okuno, K. and Yano, M. (2009) Loss of function of a proline‐containing protein confers durable disease resistance in rice. Science, 325, 998–1001. [DOI] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. and Jones, J. (1996) Resistance gene‐dependent plant defense responses. Plant Cell, 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. and Parker, J.E. (2003) Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14, 177–193. [DOI] [PubMed] [Google Scholar]
- Juge, N. (2006) Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 11, 359–367. [DOI] [PubMed] [Google Scholar]
- Kaku, H. , Nishizawa, Y. , Ishii‐Minami, N. , Akimoto‐Tomiyama, C. , Dohmae, N. , Takio, K. , Minami, E. and Shibuya, N. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA, 103, 11 086–11 091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani, M. , Yoo, J. and Dong, X. (2007) Genetic interactions of TGA transcription factors in the regulation of pathogenesis‐related genes and disease resistance in Arabidopsis. Plant Physiol. 144, 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto, K. , Kouzai, Y. , Kaku, H. , Shibuya, N. , Minami, E. and Nishizawa, Y. (2010) Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J. 64, 343–354. [DOI] [PubMed] [Google Scholar]
- Koo, S. , Choi, M. , Chun, H. , Park, H. , Kang, C. , Shim, S. , Chung, J. , Cheong, Y. , Lee, S. , Yun, D.‐J. , Chung, W. , Cho, M. and Kim, M. (2009a) Identification and characterization of alternative promoters of the rice MAP kinase gene OsBWMK1. Mol. Cells, 27, 467–473. [DOI] [PubMed] [Google Scholar]
- Koo, S.C. , Yoon, H.W. , Kim, C.Y. , Moon, B.C. , Cheong, Y.H. , Han, H.J. , Lee, S.M. , Kang, K.Y. , Kim, M.C. , Lee, S.Y. , Chung, W.S. and Cho, M.J. (2007) Alternative splicing of the OsBWMK1 gene generates three transcript variants showing differential subcellular localizations. Biochem. Biophys. Res. Commun. 360, 188–193. [DOI] [PubMed] [Google Scholar]
- Koo, S.C. , Moon, B.C. , Kim, J.K. , Kim, C.Y. , Sung, S.J. , Kim, M.C. , Cho, M.J. and Cheong, Y.H. (2009b) OsBWMK1 mediates SA‐dependent defense responses by activating the transcription factor OsWRKY33. Biochem. Biophys. Res. Commun. 387, 365–370. [DOI] [PubMed] [Google Scholar]
- Larmande, P. , Gay, C. , Lorieux, M. , Perin, C. , Bouniol, M. , Droc, G. , Sallaud, C. , Perez, P. , Barnola, I. , Biderre‐Petit, C. , Martin, J. , Morel, J.B. , Johnson, A.A.T. , Bourgis, F. , Ghesquiere, A. , Ruiz, M. , Courtois, B. and Guiderdoni, E. (2008) Oryza Tag Line, a phenotypic mutant database for the Genoplante rice insertion line library. Nucleic Acids Res. 36, D1022–D1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D.Y. , Liu, H.Z. , Zhang, H.J. , Wang, X.E. and Song, F.M. (2008) OsBIRH1, a DEAD‐box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J. Exp. Bot. 59, 2133–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi, R. , Ding, S. and Xu, J.‐R. (2008) MADS‐box transcription factor Mig1 is required for infectious growth in Magnaporthe grisea . Eukaryot. Cell, 7, 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya, A. , Albert, P. , Shinya, T. , Desaki, Y. , Ichimura, K. , Shirasu, K. , Narusaka, Y. , Kawakami, N. , Kaku, H. and Shibuya, N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA, 104, 19 613–19 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C. , Heck, S. , Parinthawong, N. and Metraux, J.‐P. (2002) EDS5, an essential component of salicylic acid‐dependent signaling for disease resistance in arabidopsis, is a member of the mate transporter family. Plant Cell, 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M.T. and Dangl, J.L. (2010) Arabidopsis and the plant immune system. Plant J. 61, 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.‐J. , Peng, Y. , Chen, X. , Dardick, C. , Ruan, D. , Bart, R. , Canlas, P.E. and Ronald, P.C. (2008) Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21‐mediated innate immunity. PLoS Biol. 6, e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. , Bartley, L. , Canlas, P. and Ronald, P. (2010) OsWRKY IIa transcription factors modulate rice innate immunity. Rice, 3, 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke, A. , Schikora, A. and Hirt, H. (2009) MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12, 421–426. [DOI] [PubMed] [Google Scholar]
- Qiu, D. , Xiao, J. , Xie, W. , Liu, H. , Li, X. , Xiong, L. and Wang, S. (2008) Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol. Plant, 1, 538–551. [DOI] [PubMed] [Google Scholar]
- Ribot, C. , Hirsch, J. , Batzergue, S. , Tharreau, D. , Notteghem, J.L. , Lebrun, M.H. and Morel, J.B. (2008) Susceptibility of rice to the blast fungus, Magnaporthe grisea . J. Plant Physiol. 165, 114–124. [DOI] [PubMed] [Google Scholar]
- Rushton, P.J. , Somssich, I.E. , Ringler, P. and Shen, Q.J. (2010) WRKY transcription factors. Trends Plant Sci. 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Sallaud, C. , Gay, C. , Larmande, P. , Bès, M. , Piffanelli, P. , Piégu, B. , Droc, G. , Regad, F. , Bourgeois, E. , Meynard, D. , Périn, C. , Sabau, X. , Ghesquière, A. , Glaszmann, J.C. , Delseny, M. and Guiderdoni, E. (2004) High throughput T‐DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J. 39, 450–464. [DOI] [PubMed] [Google Scholar]
- Shen, Q.‐H. , Saijo, Y. , Mauch, S. , Biskup, C. , Bieri, S. , Keller, B. , Seki, H. , Ulker, B. , Somssich, I.E. and Schulze‐Lefert, P. (2007) Nuclear activity of MLA immune receptors links isolate‐specific and basal disease‐resistance responses. Science, 315, 1098–1103. [DOI] [PubMed] [Google Scholar]
- Shimizu, T. , Nakano, T. , Takamizawa, D. , Desaki, Y. , Ishii‐Minami, N. , Nishizawa, Y. , Minami, E. , Okada, K. , Yamane, H. , Kaku, H. and Shibuya, N. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono, M. , Sugano, S. , Nakayama, A. , Jiang, C.J. , Ono, K. , Toki, S. and Takatsuji, H. (2007) Rice WRKY45 plays a crucial role in benzothiadiazole‐inducible blast resistance. Plant Cell, 19, 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Correia, J. , Miranda, Y. , Leonard, N. and Ulevitch, R. (2007) SGT1 is essential for Nod1 activation. Proc. Natl. Acad. Sci. USA, 104, 6764–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skamnioti, P. and Gurr, S.J. (2007) Magnaporthe grisea Cutinase2 mediates appressorium differentiation and host penetration and is required for full virulence. Plant Cell, 19, 2674–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano, S. , Jiang, C.‐J. , Miyazawa, S.‐I. , Masumoto, C. , Yazawa, K. , Hayashi, N. , Shimono, M. , Nakayama, A. , Miyao, M. and Takatsuji, H. (2010) Role of OsNPR1 in rice defense program as revealed by genome‐wide expression analysis. Plant Mol. Biol. 74, 549–562. [DOI] [PubMed] [Google Scholar]
- Takai, R. , Isogai, A. , Takayama, S. and Che, F.S. (2008) Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol. Plant–Microbe Interact. 21, 1635–1642. [DOI] [PubMed] [Google Scholar]
- Vergne, E. , Ballini, E. , Marques, S. , Mammar, B.S. , Droc, G. , Gaillard, S. , Bourot, S. , DeRose, R. , Tharreau, D. , Notteghem, J.L. , Lebrun, M.H. and Morel, J.B. (2007) Early and specific gene expression triggered by rice resistance gene Pi33 in response to infection by ACE1 avirulent blast fungus. New Phytol. 174, 159–171. [DOI] [PubMed] [Google Scholar]
- Vergne, E. , Ballini, E. , Droc, G. , Tharreau, D. , Notteghem, J.‐L. and Morel, J.‐B. (2008) ARCHIPELAGO: a dedicated resource for exploiting past, present, and future genomic data on disease resistance regulation in rice. Mol. Plant–Microbe Interact. 21, 869–878. [DOI] [PubMed] [Google Scholar]
- Vergne, E. , Grand, X. , Ballini, E. , Chalvon, V. , Saindrenan, P. , Tharreau, D. , Nottéghem, J.L. and Morel, J.B. (2010) Preformed expression of defense is a hallmark of partial resistance to rice blast fungal pathogen Magnaporthe oryzae . BMC Plant Biol. 10, 206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L.Z. and Yang, Y.N. (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid‐inducible mitogen‐activated protein kinase. Plant Cell, 15, 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi, U. , Yano, M. , Lin, H. , Ashikari, M. and Yamada, K. (2002) A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. USA, 99, 7530–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Z. , Chen, J. , Zeng, L. , Goh, M. , Leung, H. , Khush, G.S. and Wang, G.‐L. (2000) Characterizing rice lesion mimic mutants and identifying a mutant with broad‐spectrum resistance to rice blast and bacterial blight. Mol. Plant–Microbe Interact. 13, 869–876. [DOI] [PubMed] [Google Scholar]
- Yuan, Y.X. , Zhong, S.H. , Li, Q. , Zhu, Z.R. , Lou, Y.G. , Wang, L.Y. , Wang, J.J. , Wang, M.Y. , Li, Q.L. , Yang, D.L. and He, Z.H. (2007) Functional analysis of rice NPR1‐like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5, 313–324. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Tessaro, M.J. , Lassner, M. and Li, X. (2003) Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell, 15, 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. (2009) Early molecular events in PAMP‐triggered immunity. Curr. Opin. Plant Biol. 12, 414–420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Macroscopic symptoms and microscopic events during Magnaporthe oryzae infection. (A) Characteristic symptoms on Nipponbare leaves inoculated with different M. oryzae isolates. (B) Magnaporthe oryzae infection process in rice at the cellular and subcellular levels.
Fig. S2 The spl7 mutant shows massive spontaneous lesions and enhanced resistance in adult plants.
Table S1 Updated list of disease regulators.
Table S2 Polymerase chain reaction (PCR) primers used for genotyping insertion lines.
Supporting info item


