SUMMARY
The jasmonates (JAs) comprise a family of plant hormones that regulate several developmental processes and mediate responses to various abiotic and biotic stresses, including pathogens. JA signalling is manipulated by several strains of the bacterial pathogen Pseudomonas syringae, including P. syringae strain DC3000, using the virulence factor coronatine (COR) as a mimic of jasmonyl‐l‐isoleucine (JA‐Ile). To better understand the JA‐Ile‐mediated processes contributing to P. syringae disease susceptibility, it is important to investigate the regulation of JA signalling during infection. In Arabidopsis thaliana, JASMONATE ZIM‐DOMAIN (JAZ) proteins are negative regulators of JA signalling. The transcription factor JASMONATE INSENSITIVE1 (JIN1/ATMYC2) has been implicated in the regulation of JAZ gene expression. To investigate the regulation of JAZ genes during P. syringae pathogenesis, we examined JAZ gene expression during infection of Arabidopsis by DC3000. We found that eight of the 12 JAZ genes are induced during infection in a COR‐dependent manner. Unexpectedly, the induction of the majority of JAZ genes during infection was not dependent on JIN1, indicating that JIN1 is not the only transcription factor regulating JAZ genes. A T‐DNA insertion mutant and an RNA interference line disrupted for the expression of JAZ10, one of the few JAZ genes regulated by JIN1 during infection, exhibited enhanced JA sensitivity and increased susceptibility to DC3000, with the primary effect being increased disease symptom severity. Thus, JAZ10 is a negative regulator of both JA signalling and disease symptom development.
INTRODUCTION
Plants use hormones to regulate development, growth and responses to external stimuli. Oxylipin compounds, collectively referred to as jasmonates (JAs), are an important family of plant hormones. JAs regulate aspects of development, such as root growth, stamen and pollen development, and senescence, as well as responses to environmental stresses, such as wounding, insect attack and pathogen infection (Browse, 2009; McConn and Browse, 1996; McConn et al., 1997; Staswick et al., 1998; Vijayan et al., 1998). Many plant pathogens have evolved virulence strategies to modulate hormone signalling in their hosts to facilitate infection and disease production (Grant and Jones, 2009). For example, several strains of the bacterial pathogen Pseudomonas syringae, including P. syringae strain DC3000, manipulate the JA signalling pathway by producing the virulence factor coronatine (COR) (Brooks et al., 2005; Kunkel and Chen, 2006). COR is a molecular mimic of jasmonyl‐l‐isoleucine (JA‐Ile), the endogenous active form of JA (Fonseca et al., 2009; Staswick and Tiryaki, 2004), and is required for the colonization of host tissue, suppression of salicylic acid (SA)‐mediated host defences and production of disease symptoms (Bender et al., 1998; 2004, 2005; Melotto et al., 2006).
In Arabidopsis, JA signalling is initiated when JA‐Ile binds to a receptor complex formed by CORONATINE INSENSITIVE1 (COI1) and a JASMONATE ZIM‐DOMAIN (JAZ) protein (2008a, 2008b; Sheard et al., 2010; Yan et al., 2009). COI1, initially identified in a screen for mutants exhibiting insensitivity to COR (Feys et al., 1994), is the F‐box protein in the SCFCOI1 ubiquitin E3 ligase complex that targets repressors of JA signalling, such as JAZ proteins, for degradation (Devoto et al., 2002; Katsir et al., 2008a; Xie et al., 1998). In the presence of JA‐Ile, JAZ proteins interact with COI1 and are degraded, leading to the activation of JA‐responsive gene expression (Chini et al., 2007; Thines et al., 2007). Although these findings uncover a mechanism whereby plants sense and respond to JA, it is still unclear how multiple JA‐regulated cues are translated into highly specific responses.
The regulation of JA responses by JAZ proteins provides one possible mechanism for the fine‐tuning of responses. There are 12 JAZ genes in Arabidopsis (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). JAZ proteins share two highly conserved motifs: the TIFY/ZIM domain important for JAZ homo‐ and heterodimerization, and the Jas motif which is required for JAZ–COI1 interactions and degradation in response to JA‐Ile (Chini et al., 2009; Chung and Howe, 2009; Melotto et al., 2008; Thines et al., 2007). As JAZ proteins do not have a DNA‐binding domain, it is likely that they repress JA responses indirectly, possibly by interacting with one or more transcription factors (Chini et al., 2007; Thines et al., 2007). Several dominant negative mutants encoding truncated JAZ proteins missing the Jas motif (JAZΔJas) have been characterized (Chini et al., 2007; Chung et al., 2010; Chung and Howe, 2009; Thines et al., 2007; Yan et al., 2007). JAZΔJas proteins are resistant to COI1‐mediated degradation and most confer JA insensitivity, demonstrating the role of JAZ proteins as repressors of JA signalling. Loss‐of‐function jaz mutants, however, are predicted to exhibit enhanced responses to JA. RNA interference (RNAi) lines have been generated for JAZ1 and JAZ10; the plants are hypersensitive to JA, consistent with a role for these two proteins as repressors (Grunewald et al., 2009; Yan et al., 2007). A mutant line carrying a T‐DNA insertion in the JAZ1 regulatory region has recently been reported to be hypersensitive to JA (Grunewald et al., 2009). Preliminary examination of several other JAZ insertion mutants has not identified any with JA‐related phenotypes (Chini et al., 2007; Thines et al., 2007); however, a systematic examination of mutants in the gene family or of higher order mutants has not been reported.
Differential regulation of JAZ gene expression is another possible mechanism to provide specificity to JA responses. JAZ genes are rapidly induced by JA, suggesting a negative feedback loop to replenish the JAZ protein pool and to dampen the response to JA (Chini et al., 2007; Thines et al., 2007). Evidence points to a role for the transcription factor JASMONATE INSENSITIVE1 (JIN1/AtMYC2) in the regulation of JAZ genes. The expression of several JAZ genes is reduced in jin1 mutants treated with JA, and JIN1 binds directly to the promoter region of JAZ3 (Chini et al., 2007). JIN1 has also been shown to interact with most JAZ proteins in vitro, leading to the hypothesis that the binding of JAZs to JIN1 in the absence of JA prevents JIN1 from transcribing JA‐responsive genes, including the JAZ genes themselves (2007, 2009; Melotto et al., 2008). The role of JIN1 in JA signalling is well established. JIN1 positively regulates wound‐responsive genes and negatively regulates several pathogen‐responsive genes (Lorenzo et al., 2004; Nickstadt et al., 2004). Thus, given that jin1 mutants are still partially JA responsive, there are likely to be additional transcription factors regulating JA responses and, possibly, JAZ gene expression as well.
Recently, transcriptional analysis of JAZ genes in response to herbivory and wounding has provided evidence of specificity in JAZ gene induction in response to these two stimuli (Chung et al., 2008). Specifically, JAZ7 and JAZ8 were induced much more robustly after mechanical wounding compared with after feeding by Spodoptera exigua larvae. These observations suggest that JAZ genes are regulated at the transcriptional level and that specificity in the JA response may be the result of their differential regulation. This finding also raises the possibility that the manipulation of JAZ gene expression to alter JA signalling could be a virulence strategy.
Pseudomonas syringae strain DC3000 colonizes the apoplast of tomato and Arabidopsis. Stimulation of JA‐mediated responses by COR is an important component of DC3000 pathogenesis and promotes both bacterial growth and disease symptom development. DC3000 COR‐deficient mutants are severely compromised for virulence (2004, 2005). In addition, Arabidopsis mutants impaired in JA signalling, such as jin1, coi1 and several JAZΔJas mutants, have reduced sensitivity to COR and are less susceptible to DC3000 (Kloek et al., 2001; Laurie‐Berry et al., 2006; Thines et al., 2007).
Defence responses to P. syringae are mediated by SA, and Arabidopsis mutants impaired in SA biosynthesis, such as those disrupting SA INDUCTION DEFICIENT 2/ISOCHORISMATE SYNTHASE 1 (SID2/ICS1), are highly susceptible to P. syringae (Dewdney et al., 2000; Wildermuth et al., 2001). Regulatory crosstalk between the JA and SA signalling pathways is well established and the stimulation of JA signalling can result in the downregulation of SA‐mediated defences (Brooks et al., 2005; Kunkel and Brooks, 2002; Spoel and Dong, 2008). jin1 and coi1 mutants exhibit enhanced expression of SA‐mediated defences during infection (Kloek et al., 2001; Laurie‐Berry et al., 2006; Nickstadt et al., 2004), indicating that one role of COR is to stimulate JA signalling, thereby suppressing SA‐mediated defences and promoting bacterial growth. Interestingly, although bacterial growth is restored to wild‐type levels in jin1 sid2 double mutants, these plants develop only mild disease symptoms (Laurie‐Berry et al., 2006). These results indicate that there are two branches of the JA signalling pathway manipulated by DC3000, one to suppress SA defences and a distinct pathway to enhance symptom production.
To better understand the JA‐Ile‐mediated processes that contribute to P. syringae disease susceptibility, it is important to investigate the regulation of JA signalling during infection. In this study, we investigated the regulation of JAZ genes during DC3000 pathogenesis and found that eight of the 12 JAZ genes are induced after infection and that this induction is dependent on COR. JAZ gene induction was independent of JIN1 for most JAZ genes, indicating that JIN1 is not the sole transcription factor involved in JAZ regulation during infection. In addition, we showed that JAZ10 is a negative regulator of the branch of the JA pathway required for DC3000 symptom development.
RESULTS
Most JAZ genes are induced during DC3000 infection
Whole‐genome transcriptional analysis of Arabidopsis, 24 h after infiltration with DC3000, revealed that a subset of JAZ genes, JAZ2, JAZ5, JAZ6, JAZ7, JAZ9 and JAZ10, is induced by DC3000 (Thilmony et al., 2006). JAZ4 and JAZ11 are not represented on Affymetrix ATH1 arrays, and so their expression during DC3000 infection is unknown. To more thoroughly investigate JAZ gene expression during DC3000 infection, we examined JAZ transcript levels at several time points after dip infection or mock infection (Fig. 1). Most JAZ genes were induced by mock treatment at 2–6 h, but induction subsided to basal levels by 24 h. Only JAZ11 and JAZ12 were unaffected by mock treatment. JAZ4 was expressed at very low levels, consistent with previous observations (Chung et al., 2008). During DC3000 infection, JAZ genes were induced similarly to mock treatment at early time points. However, for most genes, expression remained high 12–48 h after infection (Fig. 1). All JAZ genes, except JAZ3, JAZ4, JAZ11 and JAZ12, were induced by DC3000 infection. Overall, our data suggest that the expression of JAZ genes at 24 and 48 h after infection is a specific response to DC3000 and not a result of wounding or other abiotic responses related to the treatment.
Figure 1.
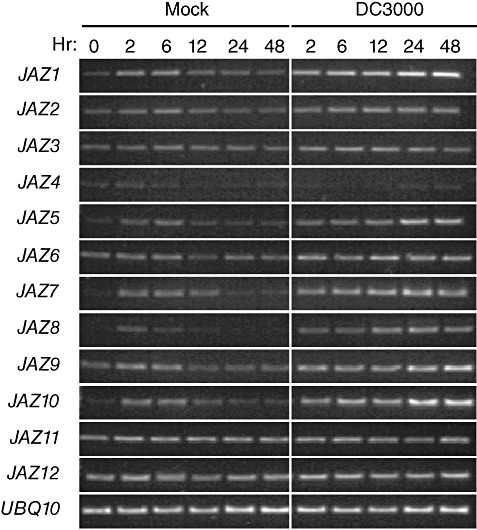
JASMONATE ZIM‐DOMAIN (JAZ) gene expression during DC3000 infection. Arabidopsis (Col‐0) plants were dip inoculated in DC3000 or MgCl2 for mock inoculation. Untreated plants were used for the 0‐h time point. Transcript levels were monitored by reverse transcription‐polymerase chain reaction (RT‐PCR) in RNA samples isolated from tissue harvested at the indicated times (hours post‐infection) after inoculation. UBQ10 (At4g05320) is shown as an internal control.
Induction of JAZ genes during infection is dependent on COR
It is likely that the induction of JAZ genes during infection is caused, at least in part, by the activity of COR, as it is a mimic of JA‐Ile. However, it is possible that other virulence factors could contribute to the induction of JAZ genes. To determine the extent to which the induction of JAZ genes during DC3000 infection is dependent on COR, we examined the expression of JAZ genes after inoculation with wild‐type DC3000 or a DC3000 COR biosynthetic mutant (COR–) (Brooks et al., 2004). We used real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) to discern small changes in transcript levels, focusing on the 24‐ and 48‐h time points, as responses to mock treatment have diminished by this time. All JAZ genes induced during DC3000 infection in the previous experiment (Fig. 1) were also induced in these experiments (Fig. 2). JAZ4 and JAZ11 were not induced significantly, whereas JAZ3 and JAZ12 were minimally induced in some, but not all, experiments (for example, see Fig. 3); therefore, it is unlikely that the induction of these genes is solely a result of DC3000 infection.
Figure 2.
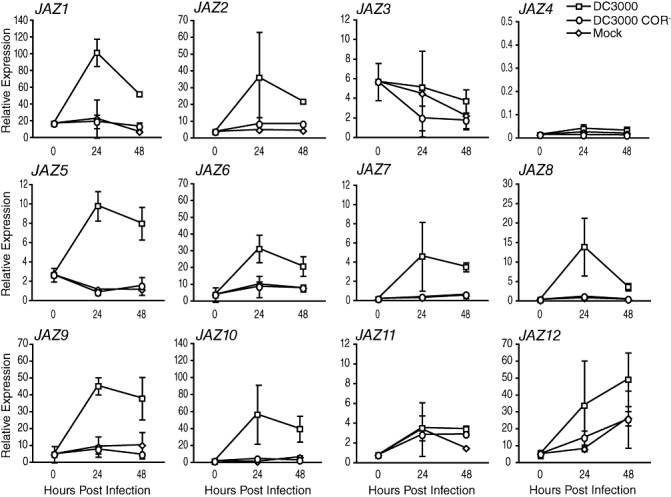
Induction of JASMONATE ZIM‐DOMAIN (JAZ) genes during infection is dependent on coronatine (COR). Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of JAZ gene expression in wild‐type plants after dip infection with DC3000 (squares), the DC3000 COR– mutant DB29 (circles) or MgCl2 for mock infection (diamonds). Untreated plants were used for the 0‐h time point. Gene expression at each time‐point is represented relative to the internal control PP2AA3 (At1g13320; note different scales). Error bars represent ±SD of two biological replicates. Bacterial growth over the course of infection for this experiment is shown in Fig. S1a. Similar results were obtained in a second independent experiment.
Figure 3.
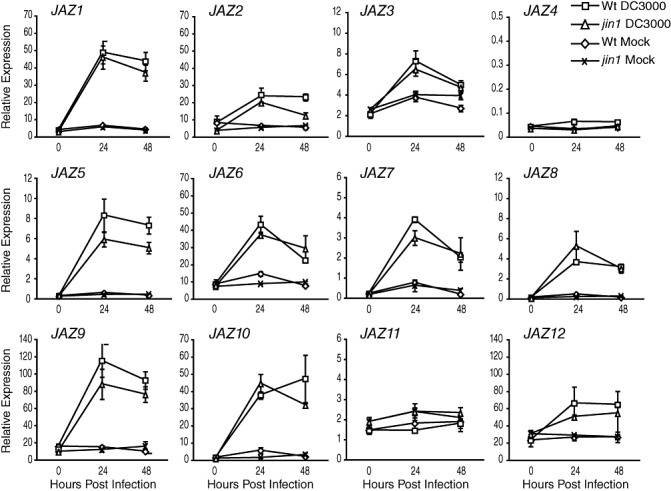
JASMONATE ZIM‐DOMAIN (JAZ) gene expression in jin1 mutant plants during DC3000 infection. Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of JAZ gene expression during DC3000 infection in wild‐type (squares) and jin1‐1 (triangles) plants, and mock (MgCl2) inoculation of wild‐type (diamonds) and jin1‐1 (×) plants. Experiments were performed and analysed as described for Fig. 2. Error bars represent ±SE of three biological replicates. Bacterial growth over the course of infection for this experiment is shown in Fig. S1b. Similar results were obtained in three additional independent experiments, one of which is shown in Fig. S2.
The induction of JAZ genes during infection is strongly dependent on COR. During infection with the DC3000 COR– strain, the level of JAZ gene transcripts remained similar to mock treatment for all JAZ genes (Fig. 2). We also examined the expression of several JAZ genes 72 h after infection, but did not observe any additional increase in expression in plants infected with the COR– strain, whereas the expression in wild‐type infections remained elevated (data not shown). The DC3000 COR– strain grows to essentially wild‐type levels early during infection (Fig. S1; (Brooks et al., 2004), and so it is unlikely that the lack of JAZ gene induction by the COR– strain is a result of differences in bacterial growth. These results demonstrate that COR is the main virulence factor affecting JA signalling during DC3000 infection.
Induction of JAZ genes during infection is not dependent on JIN1
To determine whether the induction of JAZ genes during DC3000 infection is dependent on JIN1, we examined JAZ transcript levels by real‐time RT‐PCR, 24 and 48 h after DC3000 treatment, in wild‐type and jin1‐1 mutant plants (Fig. 3, Fig. S2). Surprisingly, unlike during infection with DC3000 COR–, JAZ genes were induced in jin1 plants. We found that no JAZ gene was entirely dependent on JIN1. Indeed, only JAZ5 reproducibly exhibited reduced expression at both time points during infection in jin1 mutants. A second gene, JAZ10, exhibited partial JIN1 dependence in three of four experiments. For example, in the experiment shown in Fig. 3, we observed JIN1‐independent expression of JAZ10 but, in other experiments (Fig. S2), JAZ10 expression was partially dependent on JIN1. The remainder of the genes that were induced in wild‐type plants during infection were similarly induced in jin1 plants, although there was some variability from experiment to experiment.
The variability observed in JAZ gene expression in our experiments is probably a result of responses to multiple environmental stimuli that varied in our experimental conditions, and also possibly caused by differences in bacterial growth. The fact that the expression level of several JAZ genes was reduced at 48 h in jin1 mutants compared with the wild‐type could be caused, in part, by lower levels of bacterial growth in jin1 mutants at this time point (Fig. S1; Laurie‐Berry et al., 2006; Nickstadt et al., 2004). At the same time, it is important to note that, despite the reduced bacterial growth observed in jin1 mutants, JAZ gene induction was not impaired significantly.
As jin1‐1 is probably a null allele (Lorenzo et al., 2004), our results indicate that the induction of JAZ genes is not dependent on JIN1, and thus one or more additional unknown transcription factors regulate JAZ gene expression during infection. We also observed that, in untreated plants, the basal expression of several JAZ genes was reduced in jin1 mutants compared with the wild‐type (data not shown), suggesting that JIN1 plays a role in the basal expression of JAZ genes. Our results also do not rule out a primary role for JIN1 in the regulation of JAZ genes in other JA‐mediated responses.
Identification and characterization of T‐DNA insertional mutants in JAZ1, JAZ5 and JAZ10
The induction of many JAZ genes during DC3000 infection suggests that one or more are involved in the regulation of responses to P. syringae. To investigate the roles of JAZ genes in disease susceptibility, we assayed the responses of mutants in several JAZ genes hypothesized to be likely to be involved in pathogenesis based on our expression data. In particular, we were interested in two genes, JAZ5 and JAZ10, as these genes exhibited partial JIN1 dependence in all or some experiments (Figs 3 and S2, and data not shown). For comparison, we also included JAZ1, a highly induced gene whose expression is independent of JIN1.
We obtained a T‐DNA insertional mutant of JAZ10 from the SAIL collection (Sessions et al., 2002), a T‐DNA insertional mutant of JAZ1 from the SALK collection (Alonso et al., 2003) (Fig. 4a) and a previously described mutation in JAZ5, jaz5‐1 (Thines et al., 2007). The mutants in JAZ1 and JAZ10, designated jaz1‐1 and jaz10‐1, accumulated no transcript, and thus are likely to be null mutants (Fig. 4b). However, we detected a transcript in jaz5‐1, indicating that it may not be a loss‐of‐function mutant (Fig. 4b).
Figure 4.
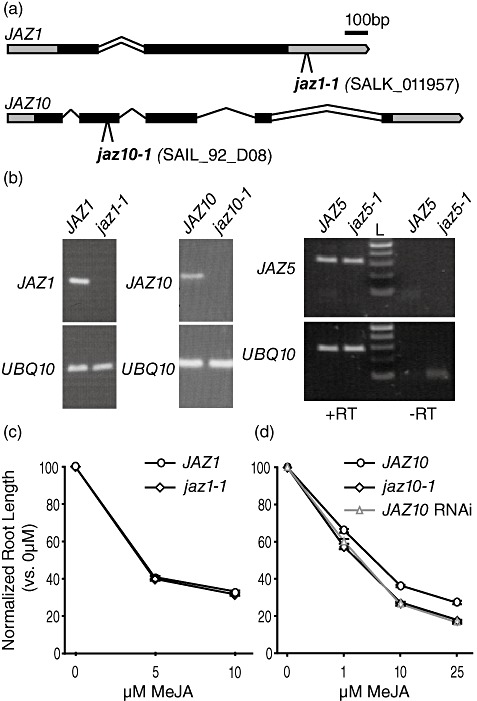
Characterization of jaz1, jaz5 and jaz10 mutants. (a) Schematic diagrams of the JAZ1 (At1g19180) and JAZ10 (At5g13220) loci. Bars represent exons with untranslated regions in grey. Lines represent introns, with double lines indicating introns that are retained in some splice variants. The T‐DNA insertion in jaz1‐1 is approximately 80 bp downstream of the end of the translated region of the gene. The T‐DNA insertion in jaz10‐1 is located 20 bp from the 3′ end of the second exon. (b) Reverse transcription‐polymerase chain reaction (RT‐PCR) of the indicated transcripts in wild‐type and jaz mutant plants. Transcript levels were monitored in seedlings after 10 days of growth on 10 µm methyl jasmonate (MeJA). UBQ10 (At4g05320) was used as an internal control. For JAZ5, a minus reverse transcriptase (–RT) control was included to demonstrate that the PCR product corresponds to JAZ5 transcript, not genomic DNA. (c) Jasmonate (JA) root growth inhibition assay of wild‐type (circles) and jaz1‐1 (diamonds) seedlings using 0, 5 and 10 µm MeJA. The normalized root length of at least 34 plants per treatment was calculated as a percentage of the average root length of plants grown without MeJA. Error bars represent ±SE. Experiments were performed three times with similar results. (d) JA root growth inhibition assay of wild‐type (circles), jaz10‐1 (diamonds) and JAZ10 RNAi (triangles) seedlings at increasing concentrations of MeJA. The normalized root length of at least 28 plants was calculated as described in (c).
RNAi has been used previously to generate plants with reduced JAZ1 or JAZ10 levels, and these plants exhibited enhanced sensitivity to methyl jasmonate (MeJA) (Grunewald et al., 2009; Yan et al., 2007). We assayed jaz1‐1 and jaz10‐1 for altered JA sensitivity in seedling root inhibition assays (Fig. 4c,d). We found that jaz1‐1 plants were essentially identical to the wild‐type at all MeJA concentrations tested, including 0 µm (Fig. 4c). In previous experiments, two lines with reduced JAZ1 expression were reported to be hypersensitive to MeJA. However, in these experiments, the mutant plants also appeared to have shorter roots when grown on medium lacking MeJA (Grunewald et al., 2009); thus, it is not clear whether these plants are actually hypersensitive to MeJA.
In contrast with jaz1‐1, we found that jaz10‐1 was hypersensitive to JA (Fig. 4d) as, at every concentration tested, root growth in jaz10‐1 plants was inhibited to a greater extent than that of wild‐type plants. jaz10‐1 behaved virtually identically in this assay to the JAZ10 RNAi line, confirming that the RNAi line disrupts JAZ10 function. Furthermore, this confirms that JAZ10 is a negative regulator of JA signalling (Chung and Howe, 2009; Yan et al., 2007). We did not observe any alteration in seedling root growth in the presence of MeJA in jaz5‐1 mutants (data not shown).
jaz10 mutants exhibit enhanced susceptibility to DC3000
Arabidopsis mutants with reduced sensitivity to JA and COR exhibit reduced susceptibility to P. syringae (Feys et al., 1994; Kloek et al., 2001; Laurie‐Berry et al., 2006). Thus, we hypothesized that the JA‐hypersensitive jaz10‐1 mutant would exhibit enhanced susceptibility to DC3000 infection. To test this, we used syringe infiltration with a low level of DC3000 to infect the wild‐type, a JAZ10 RNAi line and jaz10‐1 mutant plants. We included sid2‐2 mutant plants as a control for enhanced susceptibility (Wildermuth et al., 2001). Four days after infection, many leaves of wild‐type plants had no disease symptoms, and those that did develop symptoms exhibited mild to moderate chlorosis over the infiltrated area (Fig. 5a). In contrast, JAZ10 RNAi, jaz10‐1 and sid2 mutants exhibited significantly more severe disease symptoms than the wild‐type, although jaz10 mutant symptoms were not as severe as sid2 (Fig. 5a). We quantified disease symptoms in each genotype by examining each infiltrated leaf to assess the percentage of the infiltrated area showing chlorosis (Table 1). Although over 50% of wild‐type leaves showed no symptoms or less than 10% chlorosis over the infiltrated area, very few (approximately 12%) jaz10‐1 or JAZ10 RNAi leaves exhibited these mild symptoms. Leaves of both jaz10 lines generally developed more extensive chlorosis than did wild‐type leaves. As expected, leaves of sid2 plants showed the most severe symptoms, with the majority of infected leaves having over 50% of the infiltrated area showing chlorosis.
Figure 5.
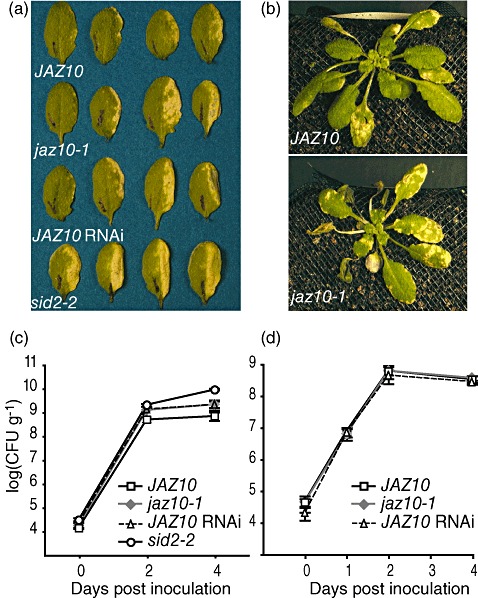
jaz10‐1 is more susceptible to Pseudomonas syringae infection. (a) Disease symptoms of wild‐type (JAZ10), jaz10 lines and sid2 leaves, 4 days after hand infiltration. The right halves (to mid‐vein) of the leaves of each genotype were infiltrated with DC3000 at 5 × 105 colony‐forming units (CFU)/mL. Leaves shown represent the range of symptoms exhibited by each genotype, from the mildest on the left to the most severe on the right. The black marks on the leaves are from a pen used to mark treated leaves at the time of infection. (b) Representative wild‐type and jaz10‐1 mutant plants, 4 days after dip inoculation with DC3000. (c) Growth of DC3000 in wild‐type (squares), jaz10‐1 (grey diamonds), JAZ10 RNAi‐9 (triangles) and sid2‐2 (circles) plants after hand infiltration. Bacteria were isolated from leaves at the indicated times after inoculation. Data points represent the average of four samples ± SE. Similar results were obtained in two additional independent experiments. (d) Growth of DC3000 in wild‐type (squares), jaz10‐1 (grey diamonds) and JAZ10 RNAi‐9 (triangles) after dip inoculation. Data were collected and analysed as in (c). Similar results were obtained in two additional independent experiments.
Table 1.
Quantification of disease symptoms 4 days after hand inoculation.
| Symptom severity | Arabidopsis genotype, number of leaves (% of total) | |||
|---|---|---|---|---|
| Wild‐type | jaz10‐1 | JAZ10 RNAi‐9 | sid2‐2 | |
| No symptoms | 14 (11.1) | 2 (1.7) | 2 (1.9) | 0 (0) |
| <10% chlorosis | 51 (40.5) | 12 (10.3) | 12 (11.3) | 0 (0) |
| 10%–50% chlorosis | 59 (46.8) | 71 (60.7) | 60 (56.6) | 0 (0) |
| 50%–100% chlorosis | 2 (1.6) | 31 (26.5) | 30 (28.3) | 65 (64.4) |
| 100% chlorosis + collapse | 0 (0) | 1 (0.9) | 2 (1.9) | 44 (43.6) |
| Total leaves examined | 126 | 117 | 106 | 101 |
Symptoms were quantified 4 days after infiltration as described for Fig. 5. Leaves with no symptoms were indistinguishable from uninfected leaves. Leaves with disease symptoms were grouped according to the amount of infiltrated area showing chlorosis. Similar results were seen in an independent experiment.
We also observed that jaz10‐1 mutants exhibited enhanced symptom production after dip infection (Fig. 5b). Four days after infection, jaz10‐1 plants developed chlorosis and disease lesions on more leaves than wild‐type plants. In addition, in jaz10‐1 plants, older leaves occasionally underwent complete collapse, a response rarely observed in wild‐type plants.
We measured bacterial growth over the course of infection in both the infiltration (Fig. 5c) and dip inoculation (Fig. 5d) experiments, and found that bacterial levels in the wild‐type, jaz10‐1 and JAZ10 RNAi line were not statistically significantly different at any time point (Fig. 5c,d). The sid2 mutant supported higher bacterial growth than both jaz10 lines and the wild‐type plants (Fig. 5c). Our results show that JAZ10 is involved in a branch of the JA signalling pathway targeted by DC3000 to enhance susceptibility. Interestingly, defects in JAZ10 have a more pronounced effect on symptom production than on bacterial growth.
We also used hand inoculation to assay jaz1‐1 and jaz5‐1 mutant lines for altered disease susceptibility. Bacterial growth and symptom production were similar to the wild‐type in both mutants (data not shown). However, as jaz5‐1 may not be a loss‐of‐function mutant (Fig. 4b), we cannot conclude that JAZ5 is not required for wild‐type levels of susceptibility.
DISCUSSION
JAZ genes are differentially regulated in response to different stimuli
Our results, together with those of several other studies, suggest that JA‐mediated responses to different environmental stimuli involve the differential regulation of JAZ genes. The expression of JAZ genes has been examined in only a few conditions to date. Microarray experiments examining expression at a single time point have shown that the 10 JAZ genes represented on the Affymetrix full genome chip are induced by MeJA treatment (Chini et al., 2007; Dombrecht et al., 2007; Thines et al., 2007) and six are induced by 24 h post‐infection with DC3000 (Thilmony et al., 2006). An additional study, examining responses to wounding and herbivory by S. exigua, examined JAZ expression at multiple time points, and found that 11 JAZ genes are induced by mechanical wounding and herbivory (Chung et al., 2008). We examined JAZ gene expression at several time points during infection with DC3000 and found that eight of 12 JAZ genes are reproducibly induced by infection. Six of these have been observed previously to be induced by DC3000 (Thilmony et al., 2006). Differences in inoculation techniques (dip infection vs. infiltration) may account for the observed induction of the additional JAZ genes in our study. Our experiments also show that JAZ induction in response to DC3000 is distinct from the response to mock treatment. All JAZ genes, except JAZ11 and JAZ12, were induced quickly after mock treatment and returned to untreated levels by 24 h (Fig. 1). This induction is probably a result of wounding and other stresses that may occur in the inoculation process, and is consistent with a previous study showing that wounding rapidly induces all JAZ genes, except JAZ11 (Chung et al., 2008). It is likely that endogenous JA signalling accounts for this early induction. During DC3000 infection, JAZ expression remained induced 24 and 48 h after infection. As a result of the rapid initial stress response, it is difficult to determine exactly when pathogen‐induced expression begins, but it is likely that pathogen‐derived signals, such as COR, continue to stimulate JA signalling after the initial stress signal subsides.
Qualitatively, the pattern of JAZ gene expression in response to DC000 infection is similar to that observed in response to herbivory (Chung et al., 2008). However, there are also some notable differences in the level of gene induction for a few specific genes. For example, JAZ7 and JAZ8 were robustly induced in response to DC3000 (1, 2, 3), whereas these genes were only weakly induced after S. exigua feeding (Chung et al., 2008). In addition, although JAZ10 was only moderately induced after herbivory, JAZ10 was one of the most highly induced genes in our experiments. Thus, although both responses are mediated via JA signalling, there are also unique regulatory events governing the expression of specific JAZ genes in response to different stimuli. All of these results highlight the need for further studies of JAZ gene expression in response to different stimuli and for the identification of the signalling components responsible for the differential regulation of JAZ genes.
COR is the primary virulence factor responsible for the induction of JAZ genes during DC3000 infection
To test whether COR is responsible for the induction of JAZ genes, we examined JAZ gene expression during infection with a DC3000 COR– mutant. We found that JAZ gene induction was essentially eliminated on infection with DC3000 COR– (Fig. 2), indicating that COR is the primary virulence factor stimulating JA signalling during DC3000 infection. However, there is evidence from other studies that additional virulence factors also modulate JA signalling. For example, AvrB, a type III secreted effector protein, induced a COI1‐dependent gene, RAP2.6, in the absence of COR (He et al., 2004). It is possible that other P. syringae virulence factors have an impact on JA signalling without affecting the expression of JAZ genes.
JIN1 is not the only transcription factor regulating JAZ gene expression
Currently, JIN1 is the only transcription factor implicated in the JA‐mediated induction of JAZ gene expression (2007, 2009; Chung and Howe, 2009). We investigated the contribution of JIN1 to the expression of JAZ genes in response to DC3000 infection. Surprisingly, we found that, for most DC3000‐responsive JAZ genes, induction during infection was independent of JIN1 (Fig. 3 and S2).
Previous studies implicating JIN1 in JAZ induction examined expression in seedlings in response to JA, and found that only JAZ6 and JAZ8 appeared to be induced by JA independently of JIN1 (Chini et al., 2007). Expression of the other JAZ genes examined was reduced in jin1 plants compared with the wild‐type. As the previous experiment only examined expression at one time point after JA treatment, it is impossible to obtain any information about the kinetics of JAZ gene expression. Thus, it is possible that other JAZ genes were induced in jin1 mutants, but that their expression reached wild‐type levels at a time point not included in the study. Alternatively, this could provide additional evidence of the differential regulation of JAZ gene expression in response to different stimuli. JAZ gene induction during infection could be mediated by other transcription factors, or even other signalling pathways, than those responsible for the regulation of JAZ gene expression in seedlings treated with MeJA. In support of this, ARF6 and ARF8 have been implicated recently in the mediation of JAZ1 induction by auxin (Grunewald et al., 2009). Thus, the expression of JAZ genes in response to specific stimuli is not regulated solely by JIN1. Rather, it is likely that JAZ genes are regulated by a combination of transcription factors, and that JIN1 is required for JAZ expression in certain conditions, but other transcription factors are important in others. Based on our observations, it is likely that any role played by JIN1 in the regulation of JAZ genes during P. syringae infection is in association with, or secondary to, one or more additional factors.
Transcription factors from several gene families have been implicated in JA signalling (Wasternack, 2007), and it is likely that these also regulate JAZ gene expression and may be targets of repression by JAZ proteins. Currently, JAZ proteins are believed to repress transcription factor activity via protein–protein interactions (Chico et al., 2008). The majority of JAZ proteins interact with JIN1 in vitro (2007, 2009; Melotto et al., 2008) but, as this has not been confirmed in planta, the role of these interactions in regulating JA‐mediated responses is unclear, and it is probable that JAZ proteins also interact with other transcription factors. As JIN1 has been shown to act with the AtMYB2 transcription factor to regulate abscisic acid (ABA)‐responsive gene expression (Abe et al., 2003), it is tempting to speculate that JIN1 may act with MYB proteins to regulate responses to JA. In addition, processes that are dependent on COI1, but independent of JIN1, such as pollen development and wounding‐induced trichome development (Mandaokar and Browse, 2009; Yoshida et al., 2009), may involve additional transcription factors that also regulate JAZ genes. It will be important to identify the additional transcription factors regulating JAZ gene expression to fully understand the mechanism by which JAZ proteins act as repressors of JA signalling.
JAZ10 is a negative regulator of JA signalling and disease
JIN1 is required for full susceptibility to P. syringae (Laurie‐Berry et al., 2006; Nickstadt et al., 2004). In examining JAZ gene expression during DC3000 infection in jin1 mutants, we observed that JAZ5 and JAZ10 transcript levels were reduced compared with those in wild‐type plants, implicating these genes in the regulation of JA‐mediated responses to DC3000. No phenotype was observed in jaz1‐1 and jaz5‐1 mutant plants, which do not exhibit altered JA sensitivity in seedlings (Fig. 4), although a role for JAZ5 in P. syringae pathogenesis cannot be ruled out as the allele examined is not a null. However, the jaz10‐1 T‐DNA mutant identified in this study, like JAZ10 RNAi lines (Yan et al., 2007), is both hypersensitive to MeJA in root growth inhibition assays and develops more severe disease symptoms on infection with DC3000 (Fig. 5). Thus, JAZ10 is a negative regulator of JA signalling in seedlings and of disease susceptibility to DC3000. Interestingly, although symptom production was clearly enhanced in the JAZ10 RNAi and jaz10‐1 mutants, bacterial growth was not affected significantly. There is evidence that the branch of the JA signalling pathway regulating symptom production is distinct from the branch regulating bacterial growth (Laurie‐Berry et al., 2006); therefore, the jaz10 phenotype could be a result of the hyperactivation of JA responses that contribute specifically to symptom production. The identification of the genes that are upregulated in this mutant during infection may provide clues to the mechanisms responsible for the production of disease symptoms.
The enhanced symptom production in JAZ10 RNAi and jaz10‐1 plants could reflect an overall enhancement of JA signalling in these lines. Several transcripts can be produced from JAZ10, including JAZ10.4, a splice variant missing the important Jas motif (Chung et al., 2010; Chung and Howe, 2009). This truncated JAZ10 protein does not interact with COI1, is resistant to degradation in the presence of JA, but can still interact with JIN1 and several JAZ proteins. It has been proposed that, as JAZ10.4 is not degraded in the presence of JA, it can accumulate in stimulated cells, and dampen JA responses after JA signalling has been initiated (Chung and Howe, 2009). This model provides an explanation for the hypersensitive phenotype that appears to be specific to jaz10‐1 mutants (Thines et al., 2007; Yan et al., 2007). Plants overexpressing a genomic clone of JAZ10 exhibited reduced sensitivity to MeJA, suggesting an important regulatory role for JAZ10.4 (Chung et al., 2010). It is still unclear whether JAZ10.4 acts systemically or is tissue specific. It is also possible that JAZ10.4 is only produced during a subset of JA‐mediated responses.
Future prospects
The results of our study, taken in the context of recent studies on wounding and herbivory, illustrate that JAZ genes are differentially regulated in response to various environmental stresses. Likewise, certain JAZ genes may be developmentally regulated or show tissue‐specific expression. The examination of JAZ gene expression in these contexts, as well as in response to additional stresses, such as ozone or fungal infection, may reveal additional layers of regulation. It is still unclear how differences in JAZ gene expression patterns relate to JAZ protein function. Current models of JA signalling hypothesize that JAZ genes are induced in response to JA in order to replenish the pool of JAZ proteins degraded by SCFCOI1 and thus to attenuate signalling (Chico et al., 2008). A reasonable hypothesis based on this model is that the induction of a particular JAZ gene reflects the degradation of that particular protein. This has not been demonstrated experimentally but, if this is the case, the induction of a particular JAZ gene in response to a specific stimulus implicates that protein in the regulation of responses to that stimulus. In all studies to date, overlapping sets of JAZ genes were induced in response to various conditions. This suggests that individual JAZ proteins are involved in multiple signalling events, and that the various JA‐mediated responses probably result from the combined activity of several JAZ proteins and other transcriptional regulators.
Loss‐of‐function mutants can be used to address important questions regarding the role of JAZ proteins in JA signalling. Mutants in several JAZ genes have been examined but, to date, JA‐related phenotypes have been reported only for jaz1 and jaz10 mutants (Chini et al., 2007; Grunewald et al., 2009; Thines et al., 2007). Several JAZ genes are the result of genomic duplication events during the evolution of Arabidopsis (Vanholme et al., 2007). This fact and the extensive co‐regulation of JAZ genes suggest that there is redundancy in this family (Chico et al., 2008; Thines et al., 2007). It is worthwhile to examine double mutants, and perhaps higher order mutants, of various JAZ genes to identify roles of specific JAZ genes in JA signalling. The regulation of responses to JA is likely to be complex and may include interactions among JAZ proteins, interactions between JAZ proteins and other transcription factors, and the regulation of JAZ gene expression by several transcription factors. In addition, the possibility of multiple active JA family members has not yet been excluded as a means of response modulation (Ribot et al., 2008; Stintzi et al., 2001; Wasternack, 2007). Multiple avenues of research will be necessary to unravel this web of regulatory interactions and functional redundancy.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
All Arabidopsis lines used in this study were in the Columbia (Col‐0) background. The jin1‐1 mutant (Berger et al., 1996) was chosen because no full‐length transcript is present in this line (Lorenzo et al., 2004). jaz5‐1 (SALK_053775) (Thines et al., 2007) was obtained from John Browse (Washington State University, Pullman, WA, USA). SAIL_92_D08, a T‐DNA insertional mutant in JAZ10 (At5g13220) (Sessions et al., 2002), and SALK_011957, a T‐DNA insertional mutant in JAZ1 (At1g19180) (Alonso et al., 2003), designated jaz10‐1 and jaz1‐1, respectively, were obtained from the Arabidopsis Biological Resource Center (ABRC) (http://abrc.osu.edu). jaz1‐1 was backcrossed to wild‐type Col‐0 once, jaz5‐1 was backcrossed to the wild‐type twice and jaz10‐1 was backcrossed three times, and a homozygous line for each mutant was chosen for further characterization. For all experiments involving these mutants, a wild‐type sibling identified from the same F2 population was used as the wild‐type control. The JAZ10 RNAi‐9 line (Yan et al., 2007) was obtained from Edward Farmer (Univeristé de Lausanne, Lausanne, Switzerland). sid2‐2 (eds16) (Dewdney et al., 2000; Wildermuth et al., 2001) was obtained from Mary Wildermuth (University of California at Berkeley, Berkeley, CA, USA). Plants used for infection assays were grown in soil in growth chambers with an 8‐h photoperiod at 22 °C and 75% humidity. Plants used for root inhibition assays were grown on one‐half strength Murashige and Skoog (0.5 × MS) (Murashige and Skoog, 1962) plates with a 16‐h photoperiod.
Bacterial strains and infection experiments
Pseudomonas syringae pv. tomato strain DC3000 (Cuppels, 1986) and DC3000 DB29 cmaA cfa6 double mutant (COR–) strain (Brooks et al., 2004) were used in the infection experiments. Bacteria were grown on King's B medium (King et al., 1954) or nutrient–yeast extract–glucose (NYG) (Daniels et al., 1988) containing rifampicin at 100 µg/mL at 28 °C. For dip inoculation, whole rosette leaves of 4‐week‐old plants were immersed in 10 mm MgCl2 with suspensions of approximately 4 × 108 colony‐forming units (CFU)/mL bacteria and 0.02% (v/v) Silwet L‐77 (OSi Specialties Inc., Danbury, CT, USA). Mock inoculations used 10 mm MgCl2 and 0.02% Silwet L‐77. Plants were kept in growth chambers under clear plastic domes, except untreated samples, for the first 24 h after treatment. For hand inoculations, bacterial suspensions of 5 × 105 CFU/mL or 1 × 106 CFU/mL (as indicated) were made in 10 mm MgCl2 and syringe infiltrated into one half of fully expanded leaves of 4‐week‐old plants. Bacterial growth in plant tissue after dip infection or infiltration was monitored as described previously (Laurie‐Berry et al., 2006).
MeJA root inhibition assays
Sterilized seeds were germinated and grown vertically in square plates with 0.5 × MS, pH 6.0, 1% (w/v) agar, 1% (w/v) sucrose and various concentrations (0, 1, 5, 10 and 25 µM) of MeJA (Sigma Aldrich, St. Louis, MO, USA), as described previously (Laurie‐Berry et al., 2006). After 10 days of growth, root length was measured using NIH ImageJ (Abramoff et al., 2004).
Identification of jaz1‐1 and jaz10‐1
To identify plants containing a T‐DNA insertion in JAZ1, seedlings of the SALK_011957 line were genotyped by PCR using the LBa1 (5′‐TGGTTCACGTAGTGGGCCATCG‐3′) (Alonso et al., 2003) primer and gene‐specific primers (Table S1). Individual plants homozygous for the insertion were chosen for further characterization. To identify plants containing a T‐DNA insertion in JAZ10, seedlings of the SAIL_92_D08 line were analysed by PCR for the presence of the T‐DNA using the LB1 (5′‐GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC‐3′) (Sessions et al., 2002) primer and gene‐specific primers (Table S1). In lines containing the T‐DNA insertion in JAZ10, gene‐specific primers generated a slightly larger PCR product than the wild‐type (Fig. S3). Plants containing both the larger PCR product and the T‐DNA‐specific product were crossed to the wild‐type three successive times and the final F2 population was plated on 0.5 × MS plates with 10 µm MeJA. Several plants with either wild‐type root length or short roots were allowed to self‐pollinate, and self‐seed was plated on 0.5 × MS plates with 10 µm MeJA to identify homozygous T‐DNA insertion or wild‐type siblings. These plants were examined for the presence or absence of the JAZ10 transcript (Table S1). Only plants that had short roots on MeJA, the larger gene‐specific PCR product and a T‐DNA‐specific product lacked JAZ10 transcript. The larger gene‐specific band was sequenced, and the presence of a short, approximately 90‐bp, insertion in the second exon of JAZ10 was confirmed.
RNA extraction and gene expression
To extract RNA, all above‐ground tissue from two to six adult plants was frozen in liquid nitrogen, ground to a powder and mixed with extraction buffer [250 mm Tris/HCl, pH 8.5, 375 mm NaCl, 25 mm ethylenediaminetetraacetic acid (EDTA), 1% sodium dodecylsulphate (SDS), 1% β‐mercaptoethanol]. The homogenate was extracted twice with phenol–chloroform. RNA was precipitated with 4 m LiCl and resuspended in diethylpyrocarbonate‐treated sterile water. To extract RNA from seedlings, 30 seedlings were ground in liquid nitrogen, and TriZol (Invitrogen, Carlsbad, CA, USA) was used according to the manufacturer's instructions. cDNA was generated using 1 µg of DNaseI (New England Biolabs, Ipswich, MA, USA)‐treated RNA with an Oligo‐dT primer and SuperscriptII (Invitrogen), following the manufacturer's instructions. RT‐PCR was performed with the following cycling conditions: 95 °C for 30 s, 60 °C for 30 s and 68 °C for 40 s. The number of cycles varied from 26 to 32 depending on the gene. Real‐time RT‐PCRs were run using an Applied Biosystems (Carlsbad, CA, USA) model 7500 thermocycler with SYBR Green JumpStart Taq ReadyMix (Invitrogen). Results were analysed using a sigmoidal model for relative expression (Chervoneva et al., 2007; Rutledge and Stewart, 2008) and normalized to an internal standard, PP2AA3 (At1g13320) (Czechowski et al., 2005). Primer pairs were designed to amplify all known splice variants of JAZ genes and are listed in Table S1. The A. thaliana genome sequence and annotation information used to design all the primers for this study were obtained from The Arabidopsis Information Resource TAIR (Swarbreck et al., 2008).
Supporting information
Fig. S1 Bacterial growth curves from experiments used for data collected in Figs 2 and 3. (a) Growth of Pseudomonas syringae strain DC3000 (diamond) and the DC3000 COR– mutant DB29 (square) after dip infection of wild‐type plants. Bacteria were isolated from leaves at the indicated times after inoculation. The zero time point represents bacterial levels approximately 1 h after inoculation. For the first time point, the leaves were surface sterilized with 15% H2O2 prior to grinding. Data points represent the average of four samples ± SE. (b) Growth of DC3000 after dip infection of wild‐type (diamond) or jin1‐1 (triangle) plants. Data were collected and analysed as in (a).
Fig. S2. Additional independent experiment showing JAZ gene expression in jin1 mutant plants during DC3000 infection. Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of JAZ gene expression during DC3000 infection in wild‐type (squares) and jin1‐1 (triangles) plants, and mock (MgCl2) inoculation of wild‐type (diamonds) plants. Tissue from uninfected plants was used for the 0‐h time point. mRNA levels were monitored by real‐time RT‐PCR with PP2AA3 (At1g13320) as an internal control. The expression of each gene at each time point is represented relative to PP2AA3 (note different scales). Error bars represent ±SE of three technical replicates.
Fig. S3. Genotyping of jaz10‐1. Agarose gel (2.5%) showing polymerase chain reaction (PCR) amplification products of genomic DNA from wild‐type and jaz10‐1 plants using gene‐specific primers (a) or a gene‐specific primer and LB1, a T‐DNA specific primer (b). Gene‐specific primers that span the T‐DNA insertion site amplify a slightly larger PCR product in jaz10‐1 plants than in wild‐type plants.
Table S1 List of primers used for reverse transcription‐polymerase chain reaction (RT‐PCR), real‐time RT‐PCR and genotyping.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank Edward Farmer for providing the JAZ10 RNAi line and for helpful comments on the manuscript. We are also grateful to Travis Dittmer, Andrzej Wierzbicki and Robert Kranz for critical discussions and technical advice, and to Andrew Mutka and Surobhi Lahiri for helpful discussions and comments. This study was supported by National Science Foundation grants IOB‐0620469 and IOS‐0818793.
REFERENCES
- Abe, H. , Urao, T. , Ito, T. , Seki, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell, 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramoff, M.D. , Magelhaes, P.J. and Ram, S.J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42. [Google Scholar]
- Alonso, J.M. , Stepanova, A.N. , Leisse, T.J. , Kim, C.J. , Chen, H. , Shinn, P. , Stevenson, D.K. , Zimmerman, J. , Barajas, P. , Cheuk, R. , Gadrinab, C. , Heller, C. , Jeske, A. , Koesema, E. , Meyers, C.C. , Parker, H. , Prednis, L. , Ansari, Y. , Choy, N. , Deen, H. , Geralt, M. , Hazari, N. , Hom, E. , Karnes, M. , Mulholland, C. , Ndubaku, R. , Schmidt, I. , Guzman, P. , Aguilar‐Henonin, L. , Schmid, M. , Weigel, D. , Carter, D.E. , Marchand, T. , Risseeuw, E. , Brogden, D. , Zeko, A. , Crosby, W.L. , Berry, C.C. and Ecker, J.R. (2003) Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science, 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Bender, C.L. , Palmer, D.A. , Penaloza‐Vazquez, A. , Rangaswamy, V. and Ullrich, M. (1998) Biosynthesis and regulation of coronatine, a non‐host‐specific phytotoxin produced by Pseudomonas syringae . Subcell Biochem. 29, 321–341. [DOI] [PubMed] [Google Scholar]
- Berger, S. , Bell, E. and Mullet, J.E. (1996) Two methyl jasmonate‐insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, D.M. , Hernandez‐Guzman, G. , Kloek, A.P. , Alarcon‐Chaidez, F. , Sreedharan, A. , Rangaswamy, V. , Penaloza‐Vazquez, A. , Bender, C.L. and Kunkel, B.N. (2004) Identification and characterization of a well‐defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant–Microbe Interact. 17, 162–174. [DOI] [PubMed] [Google Scholar]
- Brooks, D.M. , Bender, C.L. and Kunkel, B.N. (2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid‐dependent defences in Arabidopsis thaliana . Mol. Plant Pathol. 6, 629–639. [DOI] [PubMed] [Google Scholar]
- Browse, J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205. [DOI] [PubMed] [Google Scholar]
- Chervoneva, I. , Li, Y. , Iglewicz, B. , Waldman, S. and Hyslop, T. (2007) Relative quantification based on logistic models for individual polymerase chain reactions. Stat. Med. 26, 5596–5611. [DOI] [PubMed] [Google Scholar]
- Chico, J.M. , Chini, A. , Fonseca, S. and Solano, R. (2008) JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 11, 486–494. [DOI] [PubMed] [Google Scholar]
- Chini, A. , Fonseca, S. , Fernandez, G. , Adie, B. , Chico, J.M. , Lorenzo, O. , Garcia‐Casado, G. , Lopez‐Vidriero, I. , Lozano, F.M. , Ponce, M.R. , Micol, J.L. and Solano, R. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature, 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Chini, A. , Fonseca, S. , Chico, J.M. , Fernandez‐Calvo, P. and Solano, R. (2009) The ZIM domain mediates homo‐ and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 9, 77–87. [DOI] [PubMed] [Google Scholar]
- Chung, H.S. and Howe, G.A. (2009) A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM‐domain protein JAZ10 in Arabidopsis. Plant Cell, 21, 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, H.S. , Koo, A.J. , Gao, X. , Jayanty, S. , Thines, B. , Jones, A.D. and Howe, G.A. (2008) Regulation and function of Arabidopsis JASMONATE ZIM‐domain genes in response to wounding and herbivory. Plant Physiol. 146, 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, H.S. , Cooke, T.F. , DePew, C.L. , Patel, L.C. , Ogawa, N. , Kobayashi, Y. and Howe, G.A. (2010) Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63, 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppels, D.A. (1986) Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51, 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T. , Stitt, M. , Altmann, T. , Udvardi, M.K. and Scheible, W.R. (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, J.M. , Dow, J.M. and Osbourn, A.E. (1988) Molecular genetics of pathogenicity in phytopathogenic bacteria. Annu. Rev. Phytopathol. 26, 285–312. [Google Scholar]
- Devoto, A. , Nieto‐Rostro, M. , Xie, D. , Ellis, C. , Harmston, R. , Patrick, E. , Davis, J. , Sherratt, L. , Coleman, M. and Turner, J. (2002) COI1 links jasmonate signalling and fertility to the SCF ubiquitin–ligase complex in Arabidopsis. Plant J. 32, 457–466. [DOI] [PubMed] [Google Scholar]
- Dewdney, J. , Reuber, T.L. , Wildermuth, M.C. , Devoto, A. , Cui, J. , Stutius, L.M. , Drummond, E.P. and Ausubel, F.M. (2000) Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24, 205–218. [DOI] [PubMed] [Google Scholar]
- Dombrecht, B. , Xue, G.P. , Sprague, S.J. , Kirkegaard, J.A. , Ross, J.J. , Reid, J.B. , Fitt, G.P. , Sewelam, N. , Schenk, P.M. , Manners, J.M. and Kazan, K. (2007) MYC2 differentially modulates diverse jasmonate‐dependent functions in Arabidopsis. Plant Cell, 19, 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B. , Benedetti, C.E. , Penfold, C.N. and Turner, J.G. (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell, 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, S. , Chini, A. , Hamberg, M. , Adie, B. , Porzel, A. , Kramell, R. , Miersch, O. , Wasternack, C. and Solano, R. (2009) (+)‐7‐iso‐Jasmonoyl‐L‐isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. [DOI] [PubMed] [Google Scholar]
- Grant, M.R. and Jones, J.D. (2009) Hormone (dis)harmony moulds plant health and disease. Science, 324, 750–752. [DOI] [PubMed] [Google Scholar]
- Grunewald, W. , Vanholme, B. , Pauwels, L. , Plovie, E. , Inze, D. , Gheysen, G. and Goossens, A. (2009) Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 10, 923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, P. , Chintamanani, S. , Chen, Z. , Zhu, L. , Kunkel, B.N. , Alfano, J.R. , Tang, X. and Zhou, J.M. (2004) Activation of a COI1‐dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 37, 589–602. [DOI] [PubMed] [Google Scholar]
- Katsir, L. , Chung, H.S. , Koo, A.J. and Howe, G.A. (2008a) Jasmonate signaling: a conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11, 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir, L. , Schilmiller, A.L. , Staswick, P.E. , He, S.Y. and Howe, G.A. (2008b) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA, 105, 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, E.O. , Ward, M.K. and Raney, D.E. (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307. [PubMed] [Google Scholar]
- Kloek, A.P. , Verbsky, M.L. , Sharma, S.B. , Schoelz, J.E. , Vogel, J. , Klessig, D.F. and Kunkel, B.N. (2001) Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine‐insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26, 509–522. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N. and Brooks, D.M. (2002) Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N. and Chen, Z. (2006) Virulence strategies of plant pathogenic bacteria In: The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Scheifer K.‐H. and Stackebrandt E., eds), pp. 421–440. New York: Springer. [Google Scholar]
- Laurie‐Berry, N. , Joardar, V. , Street, I.H. and Kunkel, B.N. (2006) The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid‐dependent defenses during infection by Pseudomonas syringae . Mol. Plant–Microbe Interact. 19, 789–800. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O. , Chico, J.M. , Sanchez‐Serrano, J.J. and Solano, R. (2004) JASMONATE‐INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate‐regulated defense responses in Arabidopsis. Plant Cell, 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar, A. and Browse, J. (2009) MYB108 acts together with MYB24 to regulate jasmonate‐mediated stamen maturation in Arabidopsis. Plant Physiol. 149, 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M. and Browse, J. (1996) The critical requirement for linolenic acid in pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell, 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M. , Creelman, R.A. , Bell, E. , Mullet, J.E. and Browse, J. (1997) Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. USA, 94, 5473–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto, M. , Underwood, W. , Koczan, J. , Nomura, K. and He, S.Y. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell, 126, 969–980. [DOI] [PubMed] [Google Scholar]
- Melotto, M. , Mecey, C. , Niu, Y. , Chung, H.S. , Katsir, L. , Yao, J. , Zeng, W. , Thines, B. , Staswick, P. , Browse, J. , Howe, G.A. and He, S.Y. (2008) A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine‐ and jasmonoyl isoleucine‐dependent interactions with the COI1 F‐box protein. Plant J. 55, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T. and Skoog, F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nickstadt, A. , Thomma, B.P.H.J. , Feussner, I. , Kangasjarvi, J. , Zeier, J. , Loeffler, C. , Scheel, D. and Berger, S. (2004) The jasmonate‐insensitive mutant jin1 shows increased resistance to biotrophic as well as necrotrophic pathogens. Mol. Plant Pathol. 5, 425–434. [DOI] [PubMed] [Google Scholar]
- Ribot, C. , Zimmerli, C. , Farmer, E.E. , Reymond, P. and Poirier, Y. (2008) Induction of the Arabidopsis PHO1;H10 gene by 12‐oxo‐phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1‐dependent pathway. Plant Physiol. 147, 696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge, R.G. and Stewart, D. (2008) A kinetic‐based sigmoidal model for the polymerase chain reaction and its application to high‐capacity absolute quantitative real‐time PCR. BMC Biotechnol. 8, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A. , Burke, E. , Presting, G. , Aux, G. , McElver, J. , Patton, D. , Dietrich, B. , Ho, P. , Bacwaden, J. , Ko, C. , Clarke, J.D. , Cotton, D. , Bullis, D. , Snell, J. , Miguel, T. , Hutchison, D. , Kimmerly, B. , Mitzel, T. , Katagiri, F. , Glazebrook, J. , Law, M. and Goff, S.A. (2002) A high‐throughput Arabidopsis reverse genetics system. Plant Cell, 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard, L.B. , Tan, X. , Mao, H. , Withers, J. , Ben‐Nissan, G. , Hinds, T.R. , Kobayashi, Y. , Hsu, F.‐F. , Sharon, M. , Browse, J. , He, S.Y. , Rizo, J. , Howe, G.A. and Zheng, N. (2010) Jasmonate perception by inositol‐phosphate‐potentiated COI1‐JAZ co‐receptor. Nature, 468, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel, S.H. and Dong, X. (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe, 3, 348–351. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E. and Tiryaki, I. (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell, 16, 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E. , Yuen, G.Y. and Lehman, C.C. (1998) Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare . Plant J. 15, 747–754. [DOI] [PubMed] [Google Scholar]
- Stintzi, A. , Weber, H. , Reymond, P. , Browse, J. and Farmer, E.E. (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc. Natl. Acad. Sci. USA, 98, 12 837–12 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck, D. , Wilks, C. , Lamesch, P. , Berardini, T.Z. , Garcia‐Hernandez, M. , Foerster, H. , Li, D. , Meyer, T. , Muller, R. , Ploetz, L. , Radenbaugh, A. , Singh, S. , Swing, V. , Tissier, C. , Zhang, P. and Huala, E. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 36, D1009–D1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilmony, R. , Underwood, W. and He, S.Y. (2006) Genome‐wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 46, 34–53. [DOI] [PubMed] [Google Scholar]
- Thines, B. , Katsir, L. , Melotto, M. , Niu, Y. , Mandaokar, A. , Liu, G. , Nomura, K. , He, S.Y. , Howe, G.A. and Browse, J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature, 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Vanholme, B. , Grunewald, W. , Bateman, A. , Kohchi, T. and Gheysen, G. (2007) The tify family previously known as ZIM. Trends Plant Sci. 12, 239–244. [DOI] [PubMed] [Google Scholar]
- Vijayan, P. , Shockey, J. , Levesque, C.A. , Cook, R.J. and Browse, J. (1998) A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA, 95, 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100, 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Xie, D.X. , Feys, B.F. , James, S. , Nieto‐Rostro, M. and Turner, J.G. (1998) COI1: an Arabidopsis gene required for jasmonate‐regulated defense and fertility. Science, 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yan, J. , Zhang, C. , Gu, M. , Bai, Z. , Zhang, W. , Qi, T. , Cheng, Z. , Peng, W. , Luo, H. , Nan, F. , Wang, Z. and Xie, D. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell, 21, 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y. , Stolz, S. , Chetelat, A. , Reymond, P. , Pagni, M. , Dubugnon, L. and Farmer, E.E. (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell, 19, 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, Y. , Sano, R. , Wada, T. , Takabayashi, J. and Okada, K. (2009) Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development, 136, 1039–1048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Bacterial growth curves from experiments used for data collected in Figs 2 and 3. (a) Growth of Pseudomonas syringae strain DC3000 (diamond) and the DC3000 COR– mutant DB29 (square) after dip infection of wild‐type plants. Bacteria were isolated from leaves at the indicated times after inoculation. The zero time point represents bacterial levels approximately 1 h after inoculation. For the first time point, the leaves were surface sterilized with 15% H2O2 prior to grinding. Data points represent the average of four samples ± SE. (b) Growth of DC3000 after dip infection of wild‐type (diamond) or jin1‐1 (triangle) plants. Data were collected and analysed as in (a).
Fig. S2. Additional independent experiment showing JAZ gene expression in jin1 mutant plants during DC3000 infection. Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of JAZ gene expression during DC3000 infection in wild‐type (squares) and jin1‐1 (triangles) plants, and mock (MgCl2) inoculation of wild‐type (diamonds) plants. Tissue from uninfected plants was used for the 0‐h time point. mRNA levels were monitored by real‐time RT‐PCR with PP2AA3 (At1g13320) as an internal control. The expression of each gene at each time point is represented relative to PP2AA3 (note different scales). Error bars represent ±SE of three technical replicates.
Fig. S3. Genotyping of jaz10‐1. Agarose gel (2.5%) showing polymerase chain reaction (PCR) amplification products of genomic DNA from wild‐type and jaz10‐1 plants using gene‐specific primers (a) or a gene‐specific primer and LB1, a T‐DNA specific primer (b). Gene‐specific primers that span the T‐DNA insertion site amplify a slightly larger PCR product in jaz10‐1 plants than in wild‐type plants.
Table S1 List of primers used for reverse transcription‐polymerase chain reaction (RT‐PCR), real‐time RT‐PCR and genotyping.
Supporting info item
Supporting info item
Supporting info item
Supporting info item


