SUMMARY
Pectobacterium carotovorum (formerly Erwinia carotovora ssp. carotovora) is a phytopathogenic bacterium that causes soft rot disease, characterized by water‐soaked soft decay, resulting from the action of cell wall‐degrading exoenzymes secreted by the pathogen. Virulence in soft rot bacteria is regulated by environmental factors, host and bacterial chemical signals, and a network of global and gene‐specific bacterial regulators. We isolated a mini‐Tn5 mutant of P. carotovorum that is reduced in the production of extracellular pectate lyase, protease, polygalacturonase and cellulase. The mutant is also decreased in virulence as it macerates less host tissues than its parent and is severely impaired in multiplication in planta. The inactivated gene responsible for the reduced virulent phenotype was identified as corA. CorA, a magnesium/nickel/cobalt membrane transporter, is the primary magnesium transporter for many bacteria. Compared with the parent, the CorA‐ mutant is cobalt resistant. The mutant phenotype was confirmed in parental strain P. carotovorum by marker exchange inactivation of corA. A functional corA + DNA from P. carotovorum restored exoenzyme production and pathogenicity to the mutants. The P. carotovorum corA + clone also restored motility and cobalt sensitivity to a CorA‐ mutant of Salmonella enterica. These data indicate that CorA is required for exoenzyme production and virulence in P. carotovorum.
INTRODUCTION
Pectobacterium carotovorum (formerly Erwinia carotovora ssp. carotovora) is a Gram‐negative, plant pathogenic bacterium in the family Enterobacteriaceae. Together with Dickeya species, Pectobacterium species are commonly referred to as soft rot Erwinia because they cause the soft rot and blackleg disease of mainly soft and succulent fruits and vegetables. This disease involves rotting and maceration in a wide variety of important horticultural plant hosts, such as celery, potato, carrot and tomato (Barras et al., 1994; Ian et al., 2003). Indeed, more than 80 plant species have been documented to be hosts to the soft rot pathogens in the natural environment and this number could possibly increase in the future (Ma et al., 2007). Soft rot bacteria have been described as ‘brute force’ pathogens (Toth and Birch, 2005) because they employ an array of pathogenicity factors to inflict damage on their host. In particular, the production and secretion of copious amounts of several plant cell wall‐degrading exoenzymes, such as pectate lyase (Pel), polygalacturonase (Peh), cellulase (Cel) and protease (Prt), ultimately lead to the maceration of plant tissues (for reviews on the role of enzymes in pathogenesis, see Charkowski, 2006 and Robert‐Baudouy et al., 2000). However, recent findings on the regulation of pathogenicity in these bacteria have revealed a more complex pathogenicity system that is more than simple brute force.
The coordinate production of extracellular enzymes and pathogenicity in soft rot Erwinia are the result of a complex system of positive and negative regulatory factors interacting with environmental and chemical signals from within and outside the bacterium. Many regulatory proteins belonging to different families of bacterial regulators have been identified and characterized in these bacteria to control exoenzyme production and virulence (for reviews on the major regulators controlling virulence in soft rot bacteria, see Charkowski, 2006, Chatterjee et al., 2000 and Thompson, 1999).
Environmental factors, such as temperature, also control the production of Pel, Peh, Cel and Prt as these exoenzymes are reduced in P. atrosepticum and in most P. carotovorum strains at elevated temperatures because of the stimulated expression of RsmA (Hasegawa et al., 2005). Oxygen levels also influence the synthesis of exoenzymes in Dickeya dadantii (formerly Erwinia chrysanthemi), with anaerobic conditions resulting in an increased overall expression of Pels (Hugouvieux‐Cotte‐Pattat et al., 1992). Nitrogen availability, iron levels and osmolarity are other environmental factors that have also been reported to influence exoenzyme production in soft rot Erwinia (Hugouvieux‐Cotte‐Pattat et al., 1992; Sauvage and Expert, 1994).
Soft rot Erwinia also use chemical signals to regulate virulence and exoenzyme production. Notable among these is the bacterially produced quorum‐sensing signal in which N‐acyl homoserine lactone (AHL) molecules are used by the bacterium to ‘measure’ cell density during pathogenesis and are involved in exoenzyme synthesis through their interaction with ExpR1 and ExpR2, the LuxR‐type receptors (2005, 2006; Jones et al., 1993; Nasser et al., 1998). In addition, the pectate catabolite intermediates [5‐keto‐4‐deoxyuronate (DKI), 2,5‐diketo‐3‐deoxygluconate (DKII) and 2‐keto‐3‐deoxygluconate (KDG)] interact with the global transcriptional repressor KdgR and allow gene expression by dissociating KdgR protein from regulatory regions of exoenzyme target genes (Liu et al., 1999; Nasser et al., 1992). Components of host extracts also induce exoenzyme production, but the identification of the inducing molecules and the mechanism by which the inducer acts remain unknown.
Divalent cations have also been reported to affect virulence in pathogenic bacteria. For example, calcium influences exoenzyme production in P. carotovorum for which elevated extracellular calcium levels repress pehA gene expression, resulting in reduced virulence (Flego et al., 1997) In D. dadantii, the response to host extract with increased production of Pel is also dependent on Mg2+ concentration (Haque and Tsuyumu, 2005). In prokaryotes, Mg2+ transport is achieved by three classes of transporters: CorA, MgtA/MgtB and MgtE (Kehres and Maguire, 2002). CorA is a ubiquitous inner membrane protein belonging to the family of divalent metal ion transporters. In Escherichia coli and Salmonella enterica, in which this protein has been well characterized, CorA is the major Mg2+ transporter, and also transports Ni2+ and Co2+. The role of CorA in pathogenesis has been studied in a number of organisms, including S. enterica serovar Typhimurium, in which it is required for full virulence on a mouse model in spite of the presence of additional Mg2+ transport systems (Papp‐Wallace et al., 2008). In the human gastric pathogen Helicobacter pylori, CorA is required for viability under limiting Mg2+ conditions (Pfeiffer et al., 2002). A role has also been established for CorA homologues in eukaryotic pathogens, in which a pair of CorA‐like magnesium transporters, MGT1 and MGT2, plays a critical role in the growth, development and virulence of the sandfly‐transmitted parasite Leishmania major, the causal agent of leishmaniasis (Zhu et al., 2009).
Although Mg2+ is necessary for bacterial growth, is a cofactor in ATP‐requiring enzymatic reactions (Reinhart, 1988) and is directly involved in membrane stability (Nikaido and Vaara, 1985), its role and essentiality during the pathogenesis of P. carotovorum have not been reported. We report here the isolation and characterization of a P. carotovorum mutant which is reduced in exoenzyme production and virulence, and further show that the mutant gene is corA, a magnesium/nickel/cobalt transporter, and that this membrane protein is required for full virulence in P. carotovorum.
RESULTS
Isolation and characterization of the CorA mutant
We isolated a P. carotovorum mutant with altered extracellular Prt production on nutrient gelatin (NG) agar medium using mini‐Tn5 lacZ1 (de Lorenzo et al., 1990). The mutant, designated KD101, exhibited complete absence of Prt activity on NG agar medium (Fig. 1a) and showed reduced, but not total lack of, activity of Pel, Peh and Cel enzymes on agar medium (data not shown).
Figure 1.
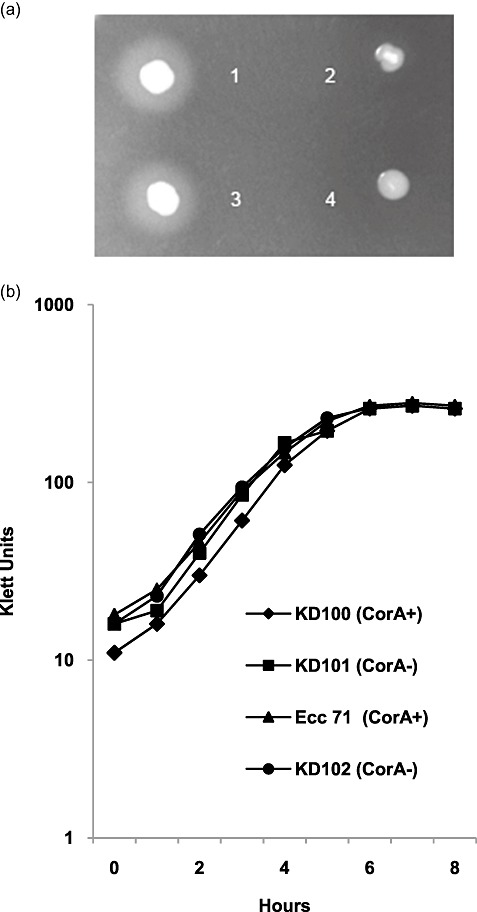
Growth curve and extracellular protease (Prt) activity of Pectobacterium carotovorum strains. (a) Prt activity produced by P. carotovorum strains on nutrient gelatin (NG) agar medium. Bacteria were patched on NG agar plates and incubated at 28 °C overnight. Prt activity was determined visually by the production of a halo around the colony: 1, parent KD100 (top left); 2, CorA‐ mutant KD101 (top right); 3, parent Ecc71 (bottom left); 4, CorA‐ mutant KD102 (bottom right). (b) Growth curves of parents KD100 and Ecc71 and CorA‐ mutants KD101 and KD102. Bacteria were grown in minimal salts (MM) medium at 28 °C and growth was monitored hourly by Klett meter.
As KD101 totally lacked Prt activity on NG agar medium, we tested whether the mutation might have adversely affected other physiological processes, such as growth, by measuring the growth rate of mutant and parent in minimal salts (MM) medium. As shown in Fig. 1b, there were no observable differences in growth between the mutant and its parental strain. Therefore, despite the reduced enzyme phenotype, the growth of mutant KD101 was not altered by the mutation. The mutant KD101 and parent KD100 were grown in MM, MM with celery extract (MM + CE) and MM with potato extract (MM + PE), and the culture supernatants were assayed quantitatively for Pel and Prt and semiquantitatively for Peh and Cel activities. In both parent and mutant, enzymatic activities of Pel, Prt (Table 1), Cel and Peh (data not shown) were induced in host extract‐supplemented media (MM + CE and MM + PE) relative to MM or MM + gelatin, indicating that the mutant, like the parent, is still inducible by the host signal. However, although exoenzyme levels were induced in extract‐supplemented media, a reduction in enzymatic activity of mutant KD101 relative to the parent was still observed regardless of the growth medium. Compared with the parental strain, the Tn5 mutant KD101 showed approximately 3.2‐, 2.0‐ and 1.3‐fold reductions in Pel when grown in MM, MM + CE and MM + PE media, respectively (Table 1). Similarly, Prt activities in these media were reduced by 2.0‐, 2.0‐ and 1.2‐fold, respectively (Table 1). The enzymatic activities of Peh and Cel, measured on semiquantitative agar media using culture supernatant, were also reduced for the mutant KD101 relative to the parent KD100 (data not shown).
Table 1.
The levels of pectate lyase and protease in corA mutants and parents.
| Bacterial strain | Relevant phenotype | Pectate lyase | Protease | ||||
|---|---|---|---|---|---|---|---|
| MM† | MM + CE‡ | MM + PE§ | MM¶ | MM + CE | MM + PE | ||
| KD100 | Wild‐type | 0.13 | 7.11 | 2.58 | 0.37 | 0.23 | 0.23 |
| KD101 | CorA‐ | 0.04* | 3.18* | 1.95* | 0.18* | 0.11* | 0.19 |
| Ecc71 | Wild‐type | 0.14 | 1.79 | 3.77 | 0.09 | 0.17 | 0.30 |
| KD102 | CorA‐ | 0.07* | 0.72* | 2.24* | 0.06 | 0.07* | 0.18* |
Student's t‐tests were performed comparing pectate lyase and protease activities of the CorA+ parent and CorA‐ strains in each medium.
Activities are expressed as units/mL/A 600.
Significantly different from the parent with P < 0.05.
MM, minimal salts medium.
CE, celery extract.
PE, potato extract.
MM, minimal salts medium supplemented with 1% gelatin.
We wanted to determine whether the reduction in the levels of extracellular Pel, Peh, Cel and Prt in mutant KD101 was also reflected at the transcript level. Using reverse transcriptase‐quantitative polymerase chain reaction (RT‐qPCR), we measured the levels of pel‐1, peh‐1, celV and prtW transcripts in the total RNA of both parent and mutant. Each of these represents a gene for one of the isozymes for extracellular Pel, Peh, Cel and Prt, respectively, described in different strains of Pectobaterium species (Chatterjee et al., 1995b; Cooper and Salmond, 1993; Liu et al., 1994; Marits et al., 1999). These were chosen as representative genes because enzymatic activities were similarly reduced for all of the exoenzymes and, with a few exceptions, the expression of these enzyme genes is generally coordinately regulated. As with enzymatic activities, Table 2 shows that the levels of the four transcripts in the mutant KD101 were reduced by a factor of approximately two‐fold in celV to six‐fold in prtW relative to parent KD100. Thus, the reduction in exoenzyme activity observed in KD101 also occurs at the transcript level. These data suggest that the isozymes of each enzyme might be similarly affected because of the reasons mentioned above.
Table 2.
Reduction in the expression of exoenzyme genes in the corA ‐ mutant.
Measured by reverse transcriptase‐quantitative polymerase chain reaction (RT‐qPCR).
Expressed as the ratio of gene expression in parent KD100 compared with that of the corA ‐ mutant KD101 normalized to the level of expression of the recA gene using the method of Schmittgen and Livak (2008). Values are the mean ± standard deviation.
Identification and characterization of corA 71
We cloned the transposon with the flanking genomic DNA sequence of KD101 into the cosmid vector pLAFR5, and used the resulting Kmr and Tcr cosmid, pCKD120, to isolate marker exchanged mutant KD102 from Ecc71. We again verified that the inactivated gene in KD102 did not affect the growth of the marker exchanged mutant (Fig. 1b). Moreover, Table 1 shows that enzyme production in KD102 was also induced in the presence of host extracts as in KD101. Pel, Prt, Cel and Peh enzyme levels of KD102 were lower in all media relative to the levels of these enzymes in its parent (data for Peh and Cel not shown). These phenotypes of marker exchanged KD102 demonstrate that Tn5 insertion is responsible for the altered phenotypes seen in KD101, as these phenotypes were transferred into the wild‐type.
Sequencing across the transposon junction revealed that the truncated gene is corA, homologue of PC1_3969 from P. carotovorum PC1 and ECA_4177 from P. atrosepticum SCRI1043. The fully functional Ecc71 corA gene (corA71) was PCR amplified and unidirectionally cloned into the low copy vector pTH19cr to generate pCKD121. The PCR‐amplified insert in pCKD121 was sequenced to generate the sequence of the full length corA gene from P. carotovorum Ecc71. corA 71 has an open reading frame of 948 base pairs (bp) and is 92% identical to the corA gene of P. carotovorum PC1. The alignments of the deduced amino acid sequences of CorA homologues from Ecc71, P. carotovorum PC1, P. atrosepticum SCRI1043, D. dadantii ECH5886 and two other bacteria from the family Enterobacteriaceae (E. coli K‐12 substrain MG1655 and S. enterica serovar Typhimurium strain LT2) are shown in Fig. 2. corA71 encodes a protein of 316 amino acids that has 98% identity and 99.6% similarity with the amino acid sequence of the P. carotovorum PC1 homologue. Five of the six differences in the amino acid residues are conservative substitutions (Fig. 2). The deduced molecular mass of Ecc71 CorA is 36.57 kDa and is almost identical to the mass of CorA reported in S. typhimurium (Smith et al., 1993). Although the CorA amino acid sequences seen in Fig. 2 are very similar, the identity is lowest at the N‐terminus and increases towards the C‐terminus. This alignment follows the trend in sequence homology between the CorA family of proteins reported previously (Niegowski and Eshaghi, 2007). In addition, the most characteristic signatures reported in the CorA proteins, the MPEL and YGMNF motifs, are present in each of the sequences (Fig. 2).
Figure 2.
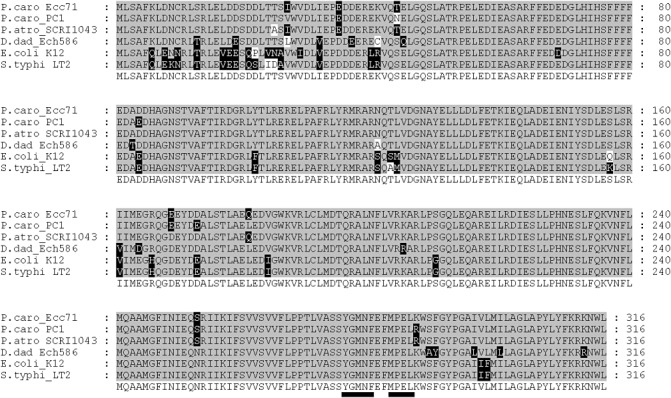
Alignment of deduced amino acid sequences of Pectobacterium carotovorum Ecc71 CorA with homologues from other bacteria. The consensus amino acid sequence from these strains is shown at the bottom of the alignment. Residues that are identical to the consensus sequence are shaded in grey, conservative substitutions are shaded in black and nonconservative substitutions are shaded in white. The signature sequences, YGMNF and MPEL, are located at the C‐terminus and are underlined in the consensus sequence. The multiple sequence alignment was generated using T‐COFEE (European Bioinformatics Institute) and the alignment output was produced using GeneDoc (Nicholas et al., 1997). Sequence IDs are abbreviated as follows: Ecc71, ‘P.caro_Ecc71’; P. carotovorum PC1(PC1 _3969), ‘P.caro_PC1’; P. atrosepticum SCRI 1043 (ECA_4177), ‘P.atro_SCRI1043’; Dicteya dadantii (Dd586_3867), ‘D.dad_Ech586’; Escherichia coli K‐12 substr. MG1655(AAA67612), ‘E.coli_K12’; Salmonella typhimurium LT2 (STM3952), ‘S.typhi_LT2’.
CorA is a membrane protein whose primary function is the transport of magnesium, but can also transport nickel and cobalt (Moncrief and Maguire, 1999). Mutants in corA first described in E. coli (Nelson and Kennedy, 1971, 1972) were resistant to lethal concentrations of Co2+ as the transport of this toxic metal into the cell was prevented, resulting in the ability of the corA mutant to grow in Co2+ concentrations lethal to the wild‐type. We verified that the corA mutant of Ecc71 also possessed the Co2+ resistance phenotype reported in E. coli and S. enterica CorA‐ mutants. As shown in Fig. 3a, mutants KD101 and KD102 grew in Co2+ concentrations of up to 200 µm, whereas the growth of parental strains KD100 and Ecc71 was inhibited at 150 µm. The cobalt‐resistant phenotype of KD101 and KD102 provides further proof that these strains are CorA‐ mutants.
Figure 3.
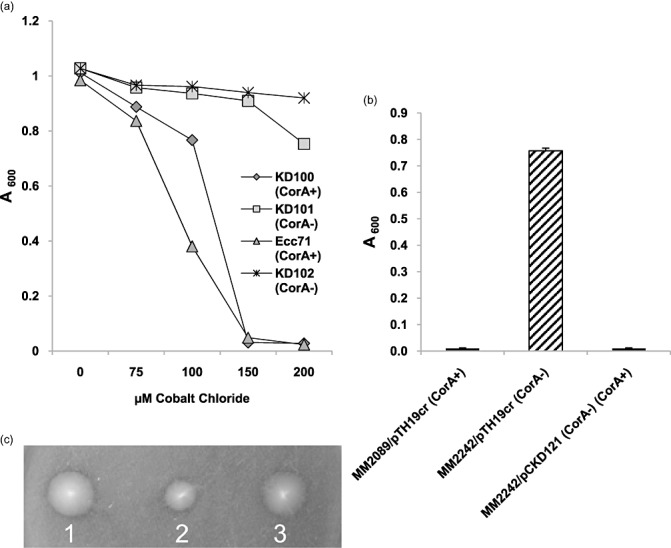
Cobalt‐resistant phenotype of CorA‐ mutants in Pectobacterium carotovorum strains and complementation of cobalt sensitivity and motility in a Salmonella enterica serovar Typhimurium corA mutant with corA 71 from Ecc71. (a) Growth of parent and CorA‐ mutant strains of Ecc71 in minimal salts (MM) medium supplemented with cobalt chloride. The parents, KD100 and Ecc71 and their respective CorA‐ mutants KD101 and KD102 were grown in MM supplemented with 0, 50, 100 and 200 µm CoCl2 at 28 °C for approximately 16 h. The absorbance at 600 nm was measured to determine growth. (b) Complementation of cobalt sensitivity in S. enterica serovar Typhimurium CorA‐ mutant with Ecc71 corA + clone pCKD121. Parent S. enterica serovar Typhimurium (MM2089) carrying pTH19cr, CorA‐ mutant strain S. enterica serovar Typhimurium (MM2242) carrying pTH19cr and CorA‐ mutant strain S. enterica serovar Typhimurium carrying pCKD121 were grown at 28 °C in Medium N supplemented with 150 µm CoCl2. The absorbance at 600 nm was measured to determine growth. (c) Complementation of motility in CorA‐ mutant S. enterica serovar Typhimurium with corA + clone of Ecc71, pCKD121: 1, MM2089/pTH19cr (CorA+); 2, MM2242/pTH19cr (CorA‐); 3, MM2242/pCKD121 (CorA‐) (CorA+). A single colony from each culture was stab inoculated onto a Luria–Bertani (LB) swimming agar plate and grown at 37 °C for 7 h.
Complementation of the mutant phenotype by extrachromosomal copies of corA
Pectobacterium carotovorum strains KD100 and Ecc71, both corA +, and strains KD101 and KD102, both corA ‐, were each transformed with corA + pCKD121 or the low copy vector pTH19cr. Quantitative and semiquantitative exoenzyme assays were performed on culture supernatants from constructs grown in MM, MM + CE and MM + PE media. The complementary effect of pCKD121 on mutants KD101 and KD102 was observed in all three media. The presence of pCKD121 restored Pel and Prt (Table 3), as well as Peh and Cel (data not shown), activities to levels approaching those of the parent, as the repressed phenotype was almost, if not completely, restored after complementation (Table 3). However, the presence of multiple copies of corA on pCKD121 in parental strains did not affect the levels of Pel, Prt, Cel and Peh (data not shown). This suggests that, among other possibilities, there could be a threshold number of copies of CorA required by the cell.
Table 3.
Complementation of exoenzyme production in corA mutants by extrachromosomal copies of corA.
| Construct | Relevant phenotype | Pectate lyase | Protease | ||||
|---|---|---|---|---|---|---|---|
| MM* | MM + CE† | MM + PE‡ | MM§ | MM + CE | MM + PE | ||
| KD100/pTH19cr | Wild‐type | 0.036 | 5.77 | 1.96 | 0.19 | 0.18 | 0.18 |
| KD101/pTH19cr | CorA‐ | 0.016 | 1.49 | 0.85 | 0.03 | 0.04 | 0.10 |
| KD101/pCKD121 | CorA‐ (CorA+) | 0.038¶ | 5.87¶ | 1.72 | 0.15 | 0.17¶ | 0.17¶ |
| Ecc71/pTH19cr | Wild‐type | 0.006 | 2.31 | 2.81 | 0.05 | 0.14 | 0.25 |
| KD102/pTH19cr | CorA‐ | 0.003 | 0.70 | 1.27 | 0.01 | 0.05 | 0.14 |
| KD102/pCKD121 | CorA‐ (CorA+) | 0.009 | 1.72 | 2.09 | 0.07 | 0.16¶ | 0.21¶ |
Student's t‐tests were performed comparing pectate lyase and protease activities of the CorA+ parent carrying pTH19cr with complemented CorA‐ (CorA+) mutants in each medium.
Activities are expressed as units/mL/A 600.
MM, minimal salts medium.
CE, celery extract.
PE, potato extract.
MM, minimal salts medium supplemented with 1% gelatin.
Not significantly different from the parent with P > 0.05.
We tested whether corA 71 from Ecc71 would complement the cobalt‐resistant phenotype of the CorA‐ strain of S. enterica serovar Typhimurium. Salmonella enterica serovar Typhimurium parent strain (MM2089) and the CorA‐ mutant strain (MM2242), each carrying vector pTH19cr or pCKD121, were grown in Medium N supplemented with 150 µm CoCl2. As shown in Fig. 3b, corA mutant strain MM2242 was resistant to the toxic effects of CoCl2 but when complemented with pCKD121, growth was completely inhibited like that of parent MM2089. Another altered phenotype reported in CorA‐ MM2242 is reduced motility (Papp‐Wallace et al., 2008). As in many pathogenic bacteria, motility is an important virulence determinant in P. carotovorum (Hossain et al., 2005). However, strain Ecc71 and its derivatives are not motile, and so we tested the effect of CorA71 on motility in Salmonella. The corA71+ clone, pCKD121, restored the swimming ability to S. enterica serovar Typhimurium corA mutant MM2242 to approximately the same level as its parent (Fig. 3c).
Pathogenicity assays and bacterial population in planta
The disruption of corA in P. carotovorum reduces the production of exoenzymes (Table 1) which play a key role in the pathogenicity of soft rot Erwinia. We therefore tested whether this decrease in enzyme production would lead to less tissue maceration. We compared the tissue‐macerating ability of CorA‐ mutants in celery petioles and carrot root discs with that of their parent strains. Figure 4a shows that, after 24 h, celery petioles were visibly less macerated by the CorA‐ mutants KD101 and KD102 relative to their parent strains. Complementation of corA mutants KD101 and KD102 with pCKD121 restored the maceration levels in celery petioles to those seen in the parent strains (Fig. 4). We used carrot disc assays to quantify the differences seen in virulence between CorA‐ mutants and their parents. The amounts of macerated tissue in carrot discs inoculated with KD100 and Ecc71 were 21.5% and 26.7%, respectively, of the original weight of the disc. By contrast, their respective CorA‐ mutants, KD101 and KD102, macerated only 0.7% and 1.1%, respectively (Fig. 5). In carrot discs, as in celery petioles, KD101 and KD102 complemented with corA 71 clone pCKD121 also resulted in these strains macerating carrot tissue to levels similar to those of their parents (Fig. 5). These observations suggest possible ways in which CorA might affect virulence, including impaired enzyme production, multiplication or survival in the host. We therefore performed an experiment to determine how well corA strains multiply in host tissues by counting the colony‐forming units (CFU) of bacteria in the macerated tissues. Figure 4b shows that the CorA‐ mutant exhibited much lower CFU in macerated tissues relative to the CorA+ parent after starting with approximately the same levels of inocula. These results demonstrate that a mutation in corA reduces exoenzyme production and virulence in P. carotovorum, and also possibly impairs bacterial multiplication or survival in host tissues.
Figure 4.
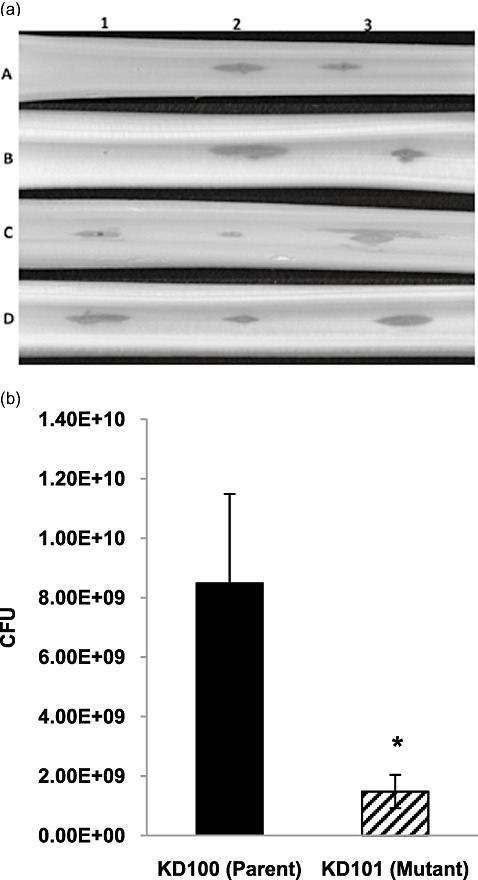
Celery tissue maceration and bacterial population in planta of Pectobacterium carotovorum strains. (a) Ten microlitres of a bacterial suspension of approximately 3 × 108 colony‐forming units (CFU)/mL was inoculated onto each site of celery petioles. Strains that carried plasmids had chloramphenicol (Cm) added to the suspension. Inoculated celery petioles were placed in a moist container and incubated at 28 °C for approximately 24 h. 1: A, H2O; B, KD100 (CorA+); C, KD101 (CorA‐). 2: A, H2O; B, Ecc71 (CorA+); C, KD102 (CorA‐). 3: A, KD100/pTH19cr (CorA+); B, KD101/pTH19cr (CorA‐); C, KD101/pCKD121 (CorA‐) (CorA+). 4: A, Ecc71/pTH19cr (CorA+); B, KD102/pTH19cr (CorA‐); C, KD102/pCKD121 (CorA‐) (CorA+). (b) Bacterial suspension (10 µL) of approximately 3.5 × 107 CFU/mL was inoculated onto each site of celery petioles. After 20 h of incubation, each inoculated petiole segment of equal size was ground in 25 mm phosphate buffer, diluted and plated. Each bar represents the mean total CFU plus the standard deviation of five replicates. Asterisk indicates significance at P < 0.05.
Figure 5.
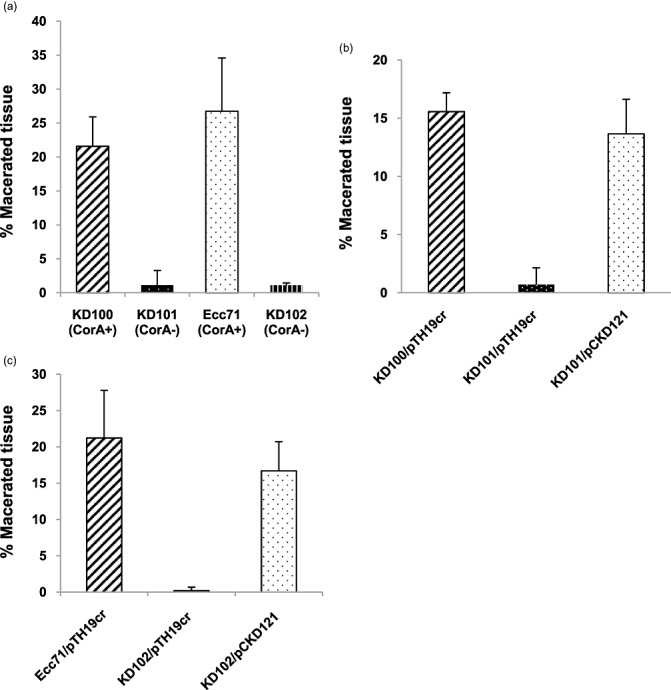
Carrot tissue maceration by Pectobacterium carotovorum strains. A small well was bored on one side of the carrot disc and inoculated with 50 µL of a bacterial suspension of approximately 7.5 × 107 colony‐forming units (CFU)/mL. Strains that carried plasmids had chloramphenicol (Cm) added to the suspension. Control carrot discs were inoculated with H2O or H2O + Cm. Carrot discs (four replicates each) were placed in a moist container at 28 °C and incubated for 72 h. After 72 h, the amount of tissue maceration was determined (see Experimental procedures). (a) KD100 (CorA+), KD101 (CorA‐), Ecc71 (CorA+) and KD102 (CorA‐); (b) KD100/pTH19cr (CorA+), KD101/pTH19cr (CorA‐) and KD101/pCKD121 (CorA‐) (CorA+); (c) Ecc71/pTH19cr (CorA+), KD102/pTH19cr (CorA‐) and KD102/pCKD121 (CorA‐) (CorA+). The error bars represent the standard deviations.
DISCUSSION
We have shown that a corA mutant of P. carotovorum Ecc71 is reduced in both the production of exoenzymes which are essential for virulence and in the multiplication or survival in host tissues. The levels of the major exoenzymes, including Pel, Peh, Cel and Prt, produced by the CorA‐ mutant were all equally affected. This suggests that CorA has a generalized regulatory effect, rather than a specific effect on any particular exoenzyme, as CorA appears to be required for the normal production of all the major exoenzymes. Although we did not test the mutants for other enzymes which make up the enzymatic artillery of soft rot Erwinia, the fact that all the major enzymes are affected suggests that the role of CorA is global. As we noted that the transcript levels of pel‐1, peh‐1, celV and prtW were dependent on the presence CorA, it is evident that, whatever the role of CorA in enzyme production, the effect is at least partly at the transcript level, where CorA may affect the synthesis of the transcripts, their stability or both. The effects of corA deficiency and the associated reduction in plant cell wall‐degrading enzymes were also manifested clearly in vivo, as CorA‐ mutants were unable to cause tissue rotting to the level of their parents in both celery petioles and carrot root discs. That exoenzyme activity was induced in the presence of celery or potato extract in both parent and CorA‐ mutant, when compared with MM medium alone, also suggests that the induction of exoenzymes by both celery and potato extract is not dependent on CorA.
The mutants were more severely affected in their maceration of carrot discs than of celery petioles. This follows another observation that the mutants are less severely affected in enzyme production in the extract of one host (potato) than another (celery) (Table 1). We speculate that these observations are reflective of the differences in the chemical constituents between the different hosts and/or their extracts. These differences could be in the form of different chemical molecules or different concentrations of the same or related molecules. Extracts from many host species of soft rot bacteria, including potato, celery and carrots, all contain an as yet uncharacterized chemical constituent(s) which serves as the inducer of exoenzyme production and pathogen virulence (Bourson et al., 1993; Murata et al., 1994; Nomura et al., 1998). How this molecule(s) is linked to the transport of Mg2+, Ni2+ or Co2+ in the cell is not clear at this point.
CorA from P. carotovorum complemented a CorA‐ mutant of S. enterica serovar Typhimurium in both a functional and regulatory manner. Functionally, when complemented with CorA from P. carotovorum, the transport function of CorA was restored, and the previous Co2+ resistance phenotype seen in corA ‐ S. enterica serovar Typhimurium was reversed, resulting in growth inhibition at lethal Co2+ levels. From a regulatory viewpoint, CorA from P. carotovorum restored motility to parental levels in the CorA‐ mutant of Salmonella. A phylogenetically related CorA has been suggested to be necessary for complementation of a corA mutant of S. enterica serovar Typhimurium, as complementation was seen with CorA from E. coli, but not that of the archaeal organism Methanococcus jannaschii (Papp‐Wallace and Maguire, 2008). As CorA from P. carotovorum is able to complement closely related CorA‐ S. enterica serovar Typhimurium, our data support this suggestion.
Possibly owing to the central role of Mg2+ in the biology of bacterial pathogens, many enterobacterial pathogens, including P. carotovorum, S. enterica serovar Typhimurium and E. coli, have multiple magnesium transporter proteins, including MgtA and MgtB, which together form an alternative Mg2+ transport system (Moncrief and Maguire, 1999). Despite this redundancy, CorA is required for full virulence in S. enterica serovar Typhimurium, as a corA mutant strain was attenuated in mice and defective in invasion and replication in epithelial cells (Papp‐Wallace et al., 2008). Interestingly, the altered phenotypes of a CorA‐ mutant of S. enterica did not appear to be a result of Mg2+ deficiency, as intracellular Mg2+ levels in the mutant remained comparable with the levels in the parent strain. The MgtA/MgtB system is normally tightly regulated by the periplasmic Mg2+ concentration, which represses these proteins under normal conditions (Snavely et al., 1991). Under Mg2+ deprivation, their expression is induced through the PhoP–PhoQ two‐component Mg2+ sensing regulatory system (Tao et al., 1998), and the induced MgtA/MtgB alternative transporters then assume Mg2+ transport into the cell. Because alternative transporters MgtA, MgtB and MgtE are present in P. carotovorum, further investigation is needed to determine whether, like S. enterica serovar Typhimurium, the altered phenotypes seen in a CorA‐ mutant of P. carotovorum are not the result of magnesium deficiency.
In the related soft rot bacterium D. dadantii (formally Erwinia chrysanthemi), hyperinduction of Pel production by host extract is mediated by the IclR‐type regulator, Pir, and controlled by the PhoP–PhoQ system, pH and Mg2+ concentration. Mutants of phoP and phoQ genes were not hyperinduced at low Mg2+ and neutral pH (Haque and Tsuyumu, 2005; Nomura et al., 1998). There are, however, some differences between the response of P. carotovorum to host extract and hyperinduction in D. dadantii. First, the genomes of many sequenced Pectobacterium species lack a genetic homologue of the Pir regulator. Second, host extract induces all the major enzymes in Pectobacterium and the PehR–PehS (PhoP–PhoQ homologue in P. carotovorum) system regulates only Peh production in P. carotovorum. Lastly, the PehR–PehS system regulates gene expression and virulence through extracellular calcium rather than magnesium (Flego et al., 2000). In spite of the differences between these two soft rot species, the ability of P. carotovorum corA strains to respond to host extract in enzyme production and the control of at least Peh production by the PhoP–PhoQ homologue could mean that the induction of enzymes in P. carotovorum is also under the control of the same or similar factors as pH and Mg2+.
One obvious question arising from the data presented here is whether the effect of the corA mutation on virulence in P. carotovorum can be explained by the modest reduction in the levels of exoenzymes alone. In culture, CorA71 had a generalized effect on exoenzyme production and virulence in P. carotovorum and motility in S. enterica serovar Typhimurium. Although we do not know whether CorA interacts with any component of the gene expression apparatus, there is precedence for a generalized regulatory role for CorA in S. enterica serovar Typhimurium. Mutation of corA in S. enterica serovar Typhimurium produced a global effect on gene expression, resulting in an increased expression of some genes and a reduction in others (Papp‐Wallace et al., 2008). More than 100 genes with diverse functions ranging from pathogenesis to transcriptional regulation were differentially regulated in corA strains of this organism. In planta, we noted a significant reduction in the CFU of the CorA‐ mutant recovered from infected tissues relative to the CorA+ parent. Therefore, it is possible that the phenotype of corA strains of P. carotovorum is the result of additive effects of this mutation on many virulence‐associated factors, including exoenzymes, and a reduction in survival or multiplication in planta.
EXPERIMENTAL PROCEDURES
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are shown in Table 4. The strains carrying drug markers were maintained on media supplemented with appropriate antibiotics. Strains of P. carotovorum were maintained on Luria–Bertani (LB) or MM medium at 28 °C and E. coli on LB at 37 °C.
Table 4.
Bacterial strains and plasmids.
| Bacterial strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Bacterial strain | ||
| Pectobacterium carotovorum | ||
| Ecc71 | Wild‐type | Zink et al. (1984) |
| AC5006 | Lac‐ mutant of Ecc71 | Murata et al. (1990) |
| KD100 | Nalr derivative of AC5006 | This study |
| KD101 | CorA ‐ Kmr derivative of KD100 by mini‐Tn5‐Km lacZ1 mutagenesis | This study |
| KD102 | CorA ‐ Kmr derivative of Ecc71 by marker exchange with pCKD120 | This study |
| Escherichia coli | ||
| DH5α | Φ80lacZΔM15 Δ(lacZYA‐argF)U169 hsdR17 recA1 endA1 thi‐1 | Invitrogen |
| HB101 | proA1 lacY hsdS20 (rB‐ mB‐) recA56 rpsL20 | Boyer and Roulland‐Dussoix (1969) |
| S17‐1 | F2 pro recA1 rB‐ mB+ | Simon et al. (1983) |
| RP4‐2 integrated (Tc::Mu) | ||
| (Km::Tn7[Smr Tpr]) | ||
| LE392 | F ‐ e14 ‐ (McrA ‐ ) hsdR514 (rk ‐ ,mk + ) supE44 supF58 lacY1 or Δ(lacIZY)6 galK2 galT22 metB1 trpR55 | Promega Corp. |
| Salmonella enterica serovar Typhimurium | ||
| MM2089 | SL1344 hisG rpsL xyl (parent) | Papp‐Wallace et al. (2008) |
| MM2242 | SL1344 corA52::Tn10Δ16Δ17 (corA mutant) | Papp‐Wallace et al. (2008) |
| Plasmid | ||
| pUT mini‐Tn5 lacZ1 | A lambda‐pir vector containing mini‐Tn5‐Km lacZ1 transposon | de Lorenzo et al. (1990) |
| pRK2013 | IncP Kmr TraRk2+ΔrepRK2 repE1 | Figurski and Helinski (1979) |
| pLAFR5 | Tcr, cosmid cloning vector | Keen et al. (1988) |
| pCKD120 | Kmr, Tcr corA ‐::Tn5 lacZ1 in pLAFR5 (cosmid vector) | This study |
| pTH19cr | Cmr, low copy cloning vector, pSC101 replicon | Hashimoto‐Gotoh et al. (2000) |
| pCKD121 | Cmr, corA + DNA in pTH19cr | This study |
Media
The compositions of LB, NG agar, Cel detection agar, polygalacturonate yeast extract agar (PYA) and Medium N have been described previously (1995a, 1985; Gibson et al., 1991). MM medium contained 400 µm of MgSO4, 7.5 mm of (NH4)2SO4, 20 mm of K2HPO4, 15 mm of KH2PO4 and 0.2% (w/v) sucrose. MM + CE and MM + PE were supplemented at a concentration of 30% extract (v/v). For cobalt sensitivity tests, CoCl2 was supplemented into MM medium at the desired concentration and the test was performed as described previously (Gibson et al., 1991). Celery and potato extracts were prepared by grating celery petioles or potato tubers obtained from a local supermarket. After the grated potato had been allowed time to sediment, the liquid layer was poured off. For celery, the pulp was squeezed and the liquid was collected. The liquid extracts were then autoclaved at 120 °C for 15 min and allowed to cool before centrifugation (5400 g for 15 min). The supernatant was kept as extract. When required, antibiotics and drugs were supplemented (µg/mL) as follows: chloramphenicol (Cm), 15; kanamycin (Km), 50; nalidixic acid (Nal), 50; tetracycline (Tc), 10. Media were solidified by the addition of 1.5% (w/v) agar.
Enzyme assays
Quantitative assays for Pel have been described previously (Chatterjee et al., 1985; Starr et al., 1977; Zink et al., 1985), but were modified for use with a 96‐well quartz microtitre plate in a plate reader. In brief, supernatants from bacterial cultures grown in MM, MM + CE or MM + PE were harvested at approximately 250, 350 and 400 Klett units, respectively. For each well, 50 µL of sample + H2O were added to 250 µL of mastermix made up of 130 µL Pel buffer (0.23 m Tris/HCl and 0.78 mm CaCl2 at pH 8.5) and 120 µL Pel substrate made up of 0.575% (w/v) polygalacturonic acid (PGA), pH 5.5. The microtitre plate was placed in a Spectra Max M5 microplate reader (Molecular Devices, Sunnyvale, CA, USA) and the enzyme was assayed using a kinetic protocol. The kinetics for substrate degradation, measured at A 235, were determined for 10 min at 40‐s intervals. The activity of Pel was determined by dividing V max (units per minute) by A 600 of the culture and sample volume in millilitres.
Assays for Prt were performed as described previously (Braun and Schmitz, 1980). Prt activity was characteristically low in MM, so that cultures were grown in MM supplemented with 1.0% (w/v) gelatin for detectable Prt activity in MM. The enzyme sample (0.8 mL) was added to 1 m Tris/HCl, pH 8.0 (0.2 mL) and 1.0% (w/v) azocasein substrate (1.0 mL) and incubated at 30 °C. Reaction samples (0.5 mL) were taken at appropriate time intervals (e.g. 0, 30, 60 and 120 min) and the reaction was stopped by the addition of 14% (v/v) perchloric acid (0.25 mL). The mixture was centrifuged at 11 000 g for 1 min and the supernatant (0.5 mL) was added to 10 m NaOH (0.05 mL). Aliquots of the developed reaction mixture were placed in a microtitre plate and A 436 was measured.
Molecular techniques
DNA manipulations, including genomic DNA isolation, DNA precipitation, transformation of bacteria by electroporation, restriction digest, DNA ligation and gel electrophoresis, were performed using standard methods (Sambrook and Russell, 2006). Plasmid isolation was performed using a QIAprep Spin MiniPrep Kit (Qiagen, Valencia, CA, USA) and by the alkaline lysis method. All nucleic acid quantification was performed on a NanoDrop ND‐100 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Mutant isolation and cosmid cloning
Biparental mating was used to generate Tn5 lacZ1 mutants, essentially as described previously (Chatterjee et al., 1995a). KD100 was mutagenized with mini‐Tn5 lacZ1 using E. coli S17‐1(λ pir)/pUT::mini‐Tn5 lacZ1 as the donor strain. Transconjugants were plated on NG agar media containing Km and Nal. Mutants were selected for their altered production of Prt.
To clone the transposon flanking sequence in the mutant, genomic DNA was digested with Sau3A. Fragments of an estimated size of 20–30 kb were ligated into BamHI‐ and SacI‐digested pLAFR5, and the ligation product was packaged into λ phage, with a packaging extract obtained from Promega (Madison, WI, USA), and transduced into E. coli LE392. Transductants were selected on LB agar supplemented with Km. A cosmid DNA clone, pCDK120, was isolated from a selected transconjugant.
Linkage of the altered phenotype to transposon insertion
The cosmid pCKD120, carrying the inactivated gene sequence of corA, was marker exchanged into Ecc71 by triparental mating. Escherichia coli DH5α was transformed with the cosmid containing the inactivated corA genomic sequence (pCKD120). The cosmid clone pCKD120 was mobilized into Ecc71 using E. coli HB101/pRK2013 as the helper. Transconjugants were selected on MM + Km plates and double patched on Tc plates. Marker exchange of the truncated gene was confirmed in a selected Kmr transconjugant (KD102) by PCR, as the same size of PCR product was amplified from KD102, KD101 and cosmid clone pCKD120 using primers specific to Tn5 lacZ1 and corA. The phenotypes of KD102 were also compared with KD101 to confirm marker exchange.
corA cloning and sequencing
Oligonucleotide primers were obtained from MWG Operon Biotechnologies (Madison, AL, USA). The primer Tn‐LacZ P6 (Table 5) was used to sequence across the transposon junction into the flanking genomic sequence of cosmid clone pCKD120, and the generated DNA sequence was used to search against genomic databases of Pectobacterium and Dickeya spp. The search revealed a 92% identity (E‐value of 0.0) with segments of the P. carotovora ssp. carotovorum PC1 (PC1_3969) and E. carotovora ssp. atroseptica SCRI1043 (ECA_4177) homologous gene corA. Thus, the truncated gene in KD101 was designated corA71 in keeping with the nomenclature used in E. coli and S. enterica serovar Typhimurium.
Table 5.
Primers used in this study.
| Primer | Sequence (5′→ 3′)* | Location |
|---|---|---|
| BamHI‐ECA‐CorA‐P3 | AATGGATCCGTCTACCGGCGCTTATATTCAC | Upstream of corA start facing downstream |
| PstI‐ECA‐CorA‐P1 | TACCTGCAGAATGAGACCGATAGCAGGATTC | Downstream of corA stop facing upstream |
| TnlacZ‐P9 | ATGAGTCAGCAACACCTTCT | Downstream of Tn5‐LacZ1 facing downstream |
| TnlacZ‐P6 | TCGGTTGTACAAAAACTTTC | Upstream of Tn5‐LacZ1 facing upstream |
| qPCRcorA‐P4 | CGGCAACAGAGATTCGATA | Internal downstream corA primer facing upstream |
| corA‐qPCR‐P2 | CTGAGCGCATTTAAACTGGA | Internal upstream corA primer facing downstream |
| Pel1‐qRTPCR‐P1 | CCAACTTCGGTGTCTGGATT | Internal upstream pel1 primer facing downstream |
| Pel1‐qRTPCR‐P2 | CGGGGAGTTATCGATACGAA | Internal downstream pel1 primer facing upstream |
| Peh1‐qRTPCR‐P1 | GGATAAAAAGGTGAGCTGGTG | Internal downstream peh‐1 primer facing upstream |
| Peh1‐qRTPCR‐P2 | CGGAGAATTAATGAGAGAGACG | Internal upstream peh‐1 primer facing downstream |
| PrtW‐qRTPCR‐P1 | GATGAGCTACTGGAGTGAGCAG | Internal upstream prtW primer facing downstream |
| PrtW‐qRTPCR‐P2 | ATACCGTATCACCGGTACGC | Internal downstream prtW primer facing upstream |
| CELVP1 | CAGCATTATCCGCCACGCCAGTA | Internal upstream celV primer facing downstream |
| CELVP2 | CATGGCGACGCGGAATACGTTA | Internal downstream celV primer facing upstream |
| Pc‐RecAP1 | GGTGAGCTGGTTGATCTGGG | Internal upstream recA primer facing downstream |
| Pc‐RecAP2 | GCATTTGCTTTGCCCTGACC | Internal downstream recA primer facing upstream |
Restriction enzyme sites engineered into the primers are shown in italic.
Using Pectobacterium genome sequences, primers were designed and used to verify by PCR that the transposon insertion into the mutants was in the corA71 gene. Using cosmid clone pCKD120, mutant KD101 and marker exchanged mutant KD102 as template, a set of PCRs was performed using a combination of transposon and corA‐specific primers (Table 5). The upstream transposon primer Tn‐LacZ P6, facing upstream, was paired with the upstream corA primer BamHI_ECA_CorA P3, facing downstream, and resulted in the amplification of a 1‐kb fragment in KD101, KD102 and pCKD120. Similarly, another combination pairing the primer located on the downstream end of the transposon facing downstream, Tn‐LacZ P9, and downstream corA primer facing upstream, PstI_ECA_CorA P1, resulted in the amplification of a 1.2‐kb fragment from KD101, KD102 and pCKD120. Using the same combination of primers mentioned above, no amplified products were observed when parental genomic DNAs of KD100 and Ecc71 were used as templates.
The PCR products amplified from KD101 genomic DNA using the primer sets Tn‐LacZ P6 +BamHI_ECA_CorA P3 and Tn‐lacZ P9 +PstI_ECA_CorA P1 (Table 5) were sequenced, and the sequences generated were compared with genomic databases to determine the exact location of the Tn5 lacZ1 insertion into the corA71 gene. Based on the genomic sequence of P. carotovorum PC1 (NC_012917.1), the transposon was inserted 481 bp from the putative start codon with a 9‐bp repeat of nucleotides 4 479 643 and 4 479 651 to either side of the transposon.
To facilitate the cloning of corA from Ecc71, restriction enzyme sites were engineered into the 5′ end of primers of the amplified sequences. The primers BamHI_ECA_CorA P3 and PstI_ECA_CorA P1, located approximately 500 bp adjacent to the start and stop codon of corA 71, respectively, were used to PCR amplify the corA gene from Ecc71 genomic DNA. An approximately 2.0‐kb product was amplified and subsequently double digested with BamHI and PstI. This fragment was unidirectionally cloned into pTH19cr, generating pCKD121. Insertion of the corA gene in pCKD121 was verified by double digestion with BamHI and PstI and by PCR using corA primers. DNA sequencing was performed on an ABI Prism 3100 Avant Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) using a Big Dye Terminator V.3.1 Cycle Sequencing Kit (Applied Biosystems). The sequence of corA71 and the deduced amino acid sequence of the product have been deposited in GenBank at the National Center for Biotechnology Information (NCBI) under the accession number HM852064.
RT‐qPCR
Parent KD100 and corA mutant KD101 were grown in MM, harvested at 150 Klett units and total RNA was isolated using the hot phenol–chloroform extraction method, as described previously (Liu et al., 1994) A modification consisted of the addition of a 1/10th volume of precooled (−80 °C) stop solution [10% (v/v) phenol, pH 7.4, in 95% (v/v) ethanol] to the culture samples immediately before initial centrifugation. Total RNA was quantified and DNase digested using a TURBO DNA‐free™ Kit (Applied Biosystems), and cDNA was prepared using random hexamers (New England Biolabs, Ipswich, MA, USA). A 20‐µL total cDNA reaction was prepared in two PCR steps. Step 1: RNA primer annealing: 70 °C for 1 min, followed by 5 min each at 60, 50, 40, 30, 20 and 15 °C. Step 2: the cDNA synthesis reaction from step 1 was added to 1 µL M‐MuLV RT (New England Biolabs), 1 µm deoxynucleoside triphosphates (dNTPs) mix (20 mm) and 1 µL RNase inhibitor, and set at 50 °C for 1 h. Real‐time PCR was performed in an ABI Prism 7000 Sequence Detection System (Applied Biosystems) using a RealMasterMix Kit (Eppendorf, Dordrecht, the Netherlands) with the primers of pel‐1, peh‐1, prtW, celV and recA (Table 5). The PCR amplification conditions used were as follows: initial denaturation for 5 min at 94 °C, followed by 40 cycles of 94 °C for 45 s, 50 °C for 1 min and 68 °C for 2 min. The comparative C T method described by Schmittgen and Livak (2008) was used for quantitative analysis of the calculated C T values using the following formula:
where
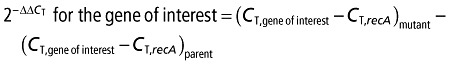 |
Transcript levels were normalized with the housekeeping gene recA.
Motility
The motility test was performed as described previously with modifications (Papp‐Wallace et al., 2008). LB swimming agar medium containing Cm was stab inoculated with sterile toothpicks from a single colony of overnight culture. Plates were placed at 37 °C and incubated for approximately 7 h.
Pathogenesis
Assays were performed essentially as described previously (Murata et al., 1991). Celery petioles were washed thoroughly with distilled water before inoculation. Inocula of the parents and their CorA‐ mutants, with and without plasmids, were prepared by suspending cells from a culture originally from a single colony into sterile water. Strains carrying plasmids had Cm added to the sterile water. Celery petioles were inoculated with 10 µL of a bacterial suspension containing approximately 3 × 108 CFU/mL. Inoculated petioles were placed in a moist container and incubated at 28 °C for 24–48 h. The extent of maceration was measured visually.
Pathogenicity tests using carrots were performed essentially as described previously (Farrar et al., 2000). Carrots were washed thoroughly with distilled water and chopped into discs approximately 5 mm thick. To minimize internal variation, all comparisons of maceration came from disks from the same carrot stick. A small well was bored in the centre of each disc using a number 1 cork borer, and 50 µL of bacterial suspension containing approximately 7.5 × 107 CFU/mL + Cm were added. Carrot discs were placed in a moist plastic chamber and incubated at 28 °C for 72 h. Maceration was measured by weighing the disc after 72 h, dislodging macerated tissue with a stream of water and reweighing the disc again. The difference between the initial and final weights calculated from the carrot disc was taken as the weight of the macerated tissue. Four replicates of the carrot disc were used for each inoculation.
In planta bacterial populations of KD100 and KD101 were determined by inoculating celery petioles with 10 µL of bacterial suspensions containing about 3.5 × 107 CFU/mL in five replicates. Petioles were placed in a moist container and incubated at 28 °C. After 20 h, equal‐sized petioles containing macerated tissue were ground in 25 mm phosphate buffer, and serial dilutions were plated on LB agar containing Nal. The plates were incubated at 28 °C and the number of colonies on appropriate dilutions were counted and used to determine the population counts. The means were compared using Student's t‐test.
ACKNOWLEDGEMENTS
This research was supported by US Department of Agriculture Evans‐Allen Funds. We thank A. Chatterjee, M. McGuire, T. Hashimoto‐Gotoh and V. de Lorenzo for bacterial strains and plasmids. We also thank A. Chatterjee and R. Olatinwo for critical review of the manuscript.
REFERENCES
- Barras, F. , van Gijsegem, F. and Chatterjee, A.K. (1994) Extracellular enzymes and pathogenesis of soft‐rot Erwinia . Annu. Rev. Phytopathol. 32, 201–234. [Google Scholar]
- Bourson, C. , Favey, S. , Reverchon, S. and Robert‐Baudouy, J. (1993) Regulation of the expression of a pelA:uidA fusion in Erwinia chrysanthemi and demonstration of the synergistic action of plant extract with polygalacturonate on pectate lyase synthesis. J. Gen. Microbiol. 139, 1–9. [DOI] [PubMed] [Google Scholar]
- Boyer, H.W. and Roulland‐dussoix, D. (1969) A complementation analysis of the restriction and modification of DNA in Escherichia coli . J. Mol. Biol. 41, 459–472. [DOI] [PubMed] [Google Scholar]
- Braun, V. and Schmitz, G. (1980) Excretion of a protease by Serratia marcescens . Arch. Microbiol. 124, 55–61. [DOI] [PubMed] [Google Scholar]
- Charkowski, A.O. (2006) The soft rot Erwinia In: Plant‐Associated Bacteria (Gnanamanickam S., ed.), pp. 423–505. Dordrecht, the Netherlands: Springer. [Google Scholar]
- Chatterjee, A. , Cui, Y. , Liu, Y. , Dumenyo, C.K. and Chatterjee, A.K. (1995a) Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density‐sensing signal, N‐(3‐oxohexanoyl)‐L‐homoserine lactone. Appl. Environ. Microbiol. 61, 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, A. , Liu, Y. and Chatterjee, A.K. (1995b) Nucleotide sequence of a pectate lyase structural gene, pel1, of Erwinia carotovora subsp. carotovora strain 71 and structural relationship of pel1 with other pel genes of Erwinia species. Mol. Plant–Microbe Interact. 8, 92–95. [DOI] [PubMed] [Google Scholar]
- Chatterjee, A.K. , Ross, L.M. , McEvoy, J.L. and Thurn, K.K. (1985) pULB113, an RP4:mini‐Mu plasmid, mediates chromosomal mobilization and R‐prime formation in Erwinia amylovora, Erwinia chrysanthemi, and subspecies of Erwinia carotovora . Appl. Environ. Microbiol. 50, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, A.K. , Dumenyo, C.K. , Liu, Y. and Chatterjee, A. (2000) Erwinia: genetics of pathogenicity factors In: Encylopedia of Microbiology (Lederberg J., ed.), pp. 236–260. San Diego: Academic Press. [Google Scholar]
- Cooper, V.J. and Salmond, G.P. (1993) Molecular analysis of the major cellulase (CelV) of Erwinia carotovora: evidence for an evolutionary ‘mix‐and‐match’ of enzyme domains. Mol. Gen. Genet. 241, 341–350. [DOI] [PubMed] [Google Scholar]
- Cui, Y. , Chatterjee, A. , Hasegawa, H. , Dixit, V. , Leigh, N. and Chatterjee, A.K. (2005) ExpR, a LuxR homolog of Erwinia carotovora subsp. carotovora, activates transcription of rsmA, which specifies a global regulatory RNA‐binding protein. J. Bacteriol. 187, 4792–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y. , Chatterjee, A. , Hasegawa, H. and Chatterjee, A.K. (2006) Erwinia carotovora subspecies produce duplicate variants of ExpR, LuxR homologs that activate rsmA transcription but differ in their interactions with N‐acylhomoserine lactone signals. J. Bacteriol. 188, 4715–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar, J.J. , Nunez, J.J. and Davis, R.M. (2000) Influence of soil saturation and temperature on Erwinia chrysanthemi soft rot of carrot. Plant Dis. 84, 665–668. [DOI] [PubMed] [Google Scholar]
- Figurski, D.H. and Helinski, D.R. (1979) Replication of an origin‐containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA, 76, 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flego, D. , Pirhonen, M. , Saarilahti, H. , Palva, T.K. and Palva, E.T. (1997) Control of virulence gene expression by plant calcium in the phytopathogen Erwinia carotovora . Mol. Microbiol. 25, 831–838. [DOI] [PubMed] [Google Scholar]
- Flego, D. , Marits, R. , Eriksson, A.R.B. , Kõiv, V. , Karlsson, M.‐B. , Heikinheimo, R. and Palva, E.T. (2000) A two‐component regulatory system, pehR–pehS, controls endopolygalacturonase production and virulence in the plant pathogen Erwinia carotovora subsp. carotovora . Mol. Plant–Microbe Interact. 13, 447–455. [DOI] [PubMed] [Google Scholar]
- Gibson, M.M. , Bagga, D.A. , Miller, C.G. and Maguire, M.E. (1991) Magnesium transport in Salmonella typhimurium: the influence of new mutations conferring Co2+ resistance on the CorA Mg2+ transport system. Mol. Microbiol. 5, 2753–2762. [DOI] [PubMed] [Google Scholar]
- Haque, M.M. and Tsuyumu, S. (2005) Virulence, resistance to magainin II, and expression of pectate lyase are controlled by the PhoP–PhoQ two‐component regulatory system responding to pH and magnesium in Erwinia chrysanthemi 3937. J. Gen. Plant Pathol. 71, 47–53. [Google Scholar]
- Hasegawa, H. , Chatterjee, A. , Cui, Y. and Chatterjee, A.K. (2005) Elevated temperature enhances virulence of Erwinia carotovora subsp. carotovora strain EC153 to plants and stimulates production of the quorum sensing signal, N‐acyl homoserine lactone, and extracellular proteins. Appl. Environ. Microbiol. 71, 4655–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto‐Gotoh, T. , Yamaguchi, M. , Yasojima, K. , Tsujimura, A. , Wakabayashi, Y. and Watanabe, Y. (2000) A set of temperature sensitive‐replication/‐segregation and temperature resistant plasmid vectors with different copy numbers and in an isogenic background (chloramphenicol, kanamycin, lacZ, repA, par, polA). Gene, 241, 185–191. [DOI] [PubMed] [Google Scholar]
- Hossain, M.M. , Shibata, S. , Aizawa, S.‐I. and Tsuyumu, S. (2005) Motility is an important determinant for pathogenesis of Erwinia carotovora subsp. carotovora . Physiol. Mol. Plant Pathol. 66, 134–143. [Google Scholar]
- Hugouvieux‐Cotte‐Pattat, N. , Dominguez, H. and Robert‐Baudouy, J. (1992) Environmental conditions affect transcription of the pectinase genes of Erwinia chrysanthemi 3937. J. Bacteriol. 174, 7807–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ian, K.T. , Kenneth, S.B. , Maria, C.H. and Paul, R.J.B. (2003) Soft rot erwiniae: from genes to genomes. Mol. Plant Pathol. 4, 17–30. [DOI] [PubMed] [Google Scholar]
- Jones, S. , Yu, B. , Bainton, N.J. , Birdsall, M. , Bycroft, B.W. , Chhabra, S.R. , Cox, A.J. , Golby, P. , Reeves, P.J. , Stephens, S. , Winson, M.K. , Salmond, G.P.C. , Stewart, G.S.A.B. and Williams, P. (1993) The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa . EMBO J. 12, 2477–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen, N.T. , Tamaki, S. , Kobayashi, D. and Trollinger, D. (1988) Improved broad‐host‐range plasmids for DNA cloning in gram‐negative bacteria. Gene, 70, 191–197. [DOI] [PubMed] [Google Scholar]
- Kehres, D.G. and Maguire, M.E. (2002) Structure, properties and regulation of magnesium transport proteins. Biometals, 15, 261–270. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Chatterjee, A. and Chatterjee, A.K. (1994) Nucleotide sequence and expression of a novel pectate lyase gene (pel‐3) and a closely linked endopolygalacturonase gene (peh‐1) of Erwinia carotovora subsp. carotovora 71. Appl. Environ. Microbiol. 60, 2545–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Jiang, G. , Cui, Y. , Mukherjee, A. , Ma, W.L. and Chatterjee, A.K. (1999) kdgREcc negatively regulates genes for pectinases, cellulase, protease, HarpinEcc, and a global RNA regulator in Erwinia carotovora subsp. carotovora . J. Bacteriol. 181, 2411–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo, V. , Herrero, M. , Jakubzik, U. and Timmis, K.N. (1990) Mini‐Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram‐negative eubacteria. J. Bacteriol. 172, 6568–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, B. , Hibbing, M.E. , Kim, H.‐S. , Reedy, R.M. , Yedidia, I. , Breuer, J. , Breuer, J. , Glasner, J.D. , Perna, N.T. , Kelman, A. and Charkowski, A.O. (2007) Host range and molecular phylogenies of the soft rot Enterobacterial genera Pectobacterium and Dickeya . Phytopathology, 97, 1150–1163. [DOI] [PubMed] [Google Scholar]
- Marits, R. , Koiv, V. , Laasik, E. and Mae, A. (1999) Isolation of an extracellular protease gene of Erwinia carotovora subsp. carotovora strain SCC3193 by transposon mutagenesis and the role of protease in phytopathogenicity. Microbiology, 145, 1959–1966. [DOI] [PubMed] [Google Scholar]
- Moncrief, M.B. and Maguire, M.E. (1999) Magnesium transport in prokaryotes. J. Biol. Inorg. Chem. 4, 523–527. [DOI] [PubMed] [Google Scholar]
- Murata, H. , Fons, M. , Chatterjee, A. , Collmer, A. and Chatterjee, A.K. (1990) Characterization of transposon insertion Out‐ mutants of Erwinia carotovora subsp. carotovora defective in enzyme export and of a DNA segment that complements out mutations in E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, and Erwinia chrysanthemi . J. Bacteriol. 172, 2970–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, H. , McEvoy, J.L. , Chatterjee, A. , Collmer, A. and Chatterjee, A.K. (1991) Molecular cloning of an aepA gene that activates production of extracellular pectolytic, cellulolytic, and proteolytic enzymes in Erwinia carotovora subsp. carotovora . Mol. Plant–Microbe Interact. 4, 239–246. [Google Scholar]
- Murata, H. , Chatterjee, A. , Liu, Y. and Chatterjee, A.K. (1994) Regulation of the production of extracellular pectinase, cellulase, and protease in the soft rot bacterium Erwinia carotovora subsp. carotovora: evidence that aepH of E. carotovora subsp. carotovora 71 activates gene expression in E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, and Escherichia coli . Appl. Environ. Microbiol. 60, 3150–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser, W. , Reverchon, S. and Robert‐Baudouy, J. (1992) Purification and functional characterization of the KdgR protein, a major repressor of pectinolysis genes of Erwinia chrysanthemi . Mol. Microbiol. 6, 257–265. [DOI] [PubMed] [Google Scholar]
- Nasser, W. , Bouillant, M.L. , Salmond, G. and Reverchon, S. (1998) Characterization of the Erwinia chrysanthemi expI‐expR locus directing the synthesis of two N‐acyl‐homoserine lactone signal molecules. Mol. Microbiol. 29, 1391–1405. [DOI] [PubMed] [Google Scholar]
- Nelson, D.L. and Kennedy, E.P. (1971) Magnesium transport in Escherichia coli . J. Biol. Chem. 246, 3042–3049. [PubMed] [Google Scholar]
- Nelson, D.L. and Kennedy, E.P. (1972) Transport of magnesium by a repressible and a nonrepressible system in Escherichia coli . Proc. Natl. Acad. Sci. USA, 69, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas, K.B. , Nicholas, H.B. and Deerfield, D.W. (1997) GeneDoc: analysis and visualization of genetic variation. EMBNEW NEWS 4, 1–4. [Google Scholar]
- Niegowski, D. and Eshaghi, S. (2007) The CorA family: structure and function revisited. Cell. Mol. Life Sci. 64, 2564–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido, H. and Vaara, M. (1985) Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, K. , Nasser, W. , Kawagishi, H. and Tsuyumu, S. (1998) The pir gene of Erwinia chrysanthemi EC16 regulates hyperinduction of pectate lyase virulence genes in response to plant signals. Proc. Natl. Acad. Sci. USA, 95, 14 034–14 039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp‐Wallace, K.M. and Maguire, M.E. (2008) Regulation of CorA Mg2+ channel function affects the virulence of Salmonella enterica Serovar Typhimurium. J. Bacteriol. 190, 6509–6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp‐Wallace, K.M. , Nartea, M. , Kehres, D.G. , Porwollik, S. , McClelland, M. , Libby, S.J. , Fang, F.C. and Maguire, M.E. (2008) The CorA Mg2+ channel is required for the virulence of Salmonella enterica Serovar Typhimurium. J. Bacteriol. 190, 6517–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer, J. , Guhl, J. , Waidner, B. , Kist, M. and Bereswill, S. (2002) Magnesium uptake by CorA is essential for viability of the gastric pathogen Helicobacter pylori . Infect. Immun. 70, 3930–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, R.A. (1988) Magnesium metabolism: a review with special reference to the relationship between intracellular content and serum levels. Arch. Intern. Med. 148, 2415–2420. [DOI] [PubMed] [Google Scholar]
- Robert‐Baudouy, J. , Nasser, J.W. , Condemine, G. , Reverchon, S. , Shevchik, V.E. and Hugovieux‐Cotte‐Pattat, N. (2000) Pectic enzymes of Erwinia chrysanthemi regulation and role in pathogenesis In: Plant–Microbe Interact (Stacey G. and Keen N.T., eds), pp. 221–368. St. Paul, MN: APS Press. [Google Scholar]
- Sambrook, J. and Russell, D.W. (2006) The Condensed Protocols from Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sauvage, C. and Expert, D. (1994) Differential regulation by iron of Erwinia chrysanthemi pectate lyases: pathogenicity of iron transport regulatory (cbr) mutants. Anglais, 7, 71–77. [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Simon, R. , Priefer, U. and Puhler, A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1, 784–791. [Google Scholar]
- Smith, R.L. , Banks, J.L. , Snavely, M.D. and Maguire, M.E. (1993) Sequence and topology of the CorA magnesium transport systems of Salmonella typhimurium and Escherichia coli. Identification of a new class of transport protein. J. Biol. Chem. 268, 14 071–14 080. [PubMed] [Google Scholar]
- Snavely, M.D. , Gravina, S.A. , Cheung, T.T. , Miller, C.G. and Maguire, M.E. (1991) Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression. J. Biol. Chem. 266, 824–829. [PubMed] [Google Scholar]
- Starr, M.P. , Chatterjee, A.K. , Starr, P.B. and Buchanan, G.E. (1977) Enzymatic degradation of polygalacturonic acid by Yersinia and Klebsiella species in relation to clinical laboratory procedures. J. Clin. Microbiol. 6, 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, T. , Grulich, P.F. , Kucharski, L.M. , Smith, R.L. and Maguire, M.E. (1998) Magnesium transport in Salmonella typhimurium: biphasic magnesium and time dependence of the transcription of the mgtA and mgtCB loci. Microbiology, 144, 655–664. [DOI] [PubMed] [Google Scholar]
- Thompson, N.R. (1999) Virulence determinants in the bacterial phytopathogen Erwinia . Methods Microbiol. 29, 347–426. [Google Scholar]
- Toth, I.K. and Birch, P.R.J. (2005) Rotting softly and stealthily. Curr. Opin. Plant Biol. 8, 424–429. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Davis, A. , Smith, B.J. , Curtis, J. and Handman, E. (2009) Leishmania major CorA‐like magnesium transporters play a critical role in parasite development and virulence. Int. J. Parasitol. 39, 713–723. [DOI] [PubMed] [Google Scholar]
- Zink, R.T. , Kemble, R.J. and Chatterjee, A.K. (1984) Transposon Tn5 mutagenesis in Erwinia carotovora subsp. carotovora and E. carotovora subsp. atroseptica . J. Bacteriol. 157, 809–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink, R.T. , Engwall, J.K. , McEvoy, J.L. and Chatterjee, A.K. (1985) recA is required in the induction of pectin lyase and carotovoricin in Erwinia carotovora subsp. carotovora . J. Bacteriol. 164, 390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]


