Summary
Taxonomy
Potato virus Y (PVY) is the type member of the genus Potyvirus in the family Potyviridae.
Virion and genome properties
PVY virions have a filamentous, flexuous form, with a length of 730 nm and a diameter of 12 nm. The genomic RNA is single stranded, messenger sense, with a length of 9.7 kb, covalently linked to a viral‐encoded protein (VPg) at the 5’ end and to a 3’ polyadenylated tail. The genome is expressed as a polyprotein of approximately 3062 amino acid residues, processed by three virus‐specific proteases into 11 mature proteins.
Hosts
PVY is distributed worldwide and has a broad host range, consisting of cultivated solanaceous species and many solanaceous and nonsolanaceous weeds. It is one of the most economically important plant pathogens and causes severe diseases in cultivated hosts, such as potato, tobacco, tomato and pepper, as well as in ornamental plants.
Transmission
PVY is transmitted from plant to plant by more than 40 aphid species in a nonpersistent manner and, in potato, by planting contaminated seed tubers.
Diversity
Five major clades, named C1, C2, Chile, N and O, have been described within the PVY species. In recent decades, a strong increase in prevalence of N × O recombinant isolates has been observed worldwide. A correlation has been observed between PVY phylogeny and certain pathogenicity traits.
Genetic control of PVY
Resistance genes against PVY have been used widely in breeding programmes and deployed in the field. These resistance genes show a large diversity of spectrum of action, durability and genetic determinism. Notably, recessive and dominant major resistance genes show highly contrasting patterns of interaction with PVY populations, displaying rapid co‐evolution or stable relationships, respectively.
Potato Virus Y , The Type Member of the Genus Potyvirus
General description
Potato virus Y (PVY) is the type member of the genus Potyvirus, which includes some of the most destructive plant viruses (Kerlan, 2006; Scholthof et al., 2011), and of the family Potyviridae, a group of single‐stranded RNA viruses. PVY was first associated with a disease causing potato degeneration in the early 1930s (Smith, 1931) and has a rather large natural host range, comprising nine botanical families (Kerlan, 2006). The most economically important hosts are potato (Solanum tuberosum), tobacco (Nicotiana spp.), tomato (S. lycopersicum) and pepper (Capsicum spp.), but PVY also infects other cultivated plants, such as ornamentals (dahlia, petunia), and numerous weeds.
The principal modes of PVY transmission are by the vegetative propagation of infected material, aphid transmission and, to a lesser extent, contact. PVY is transmitted by more than 40 aphid species (Edwardson and Christie, 1997; Kerlan, 2006) (family Aphididae) in a nonpersistent manner, i.e. a few minutes are sufficient for PVY acquisition or inoculation during aphid probing or feeding, and there is no latent period required by the virus to be transmissible after acquisition. Myzus persicae is the vector with the highest transmission efficiency and, as a result of its large geographical distribution, it is responsible for a large part of the natural spread of PVY, although many other species, such as cereal aphids, are also involved in epidemics.
PVY is the major viral threat to potato cultivation, affecting both yield and tuber quality, and results in yield losses of up to 80% (De Bokx and Huttinga, 1981; Van der Zaag, 1987). In addition, PVY control in potato production requires costly certification programmes. Symptoms caused by PVY infection on potato depend on the virus isolate, host cultivar, environmental conditions and whether they are produced by aphid‐mediated horizontal transmission or vertical transmission through infected tubers (Draper et al., 2002). Symptoms on the aerial part of the plant include leaf mosaic, mottle and crinkling, vein necrosis (Fig. 1a) and necrotic spots, stem and petiole necrosis, leaf drop and stunting of the plants in the case of tuber transmission. Some PVY variants also induce potato tuber necrotic ringspot disease, characterized by superficial annular or arch‐shaped necroses, protruding from the tuber skin (Beczner et al., 1984; Kerlan, 2006) (Fig. 1b).
Figure 1.
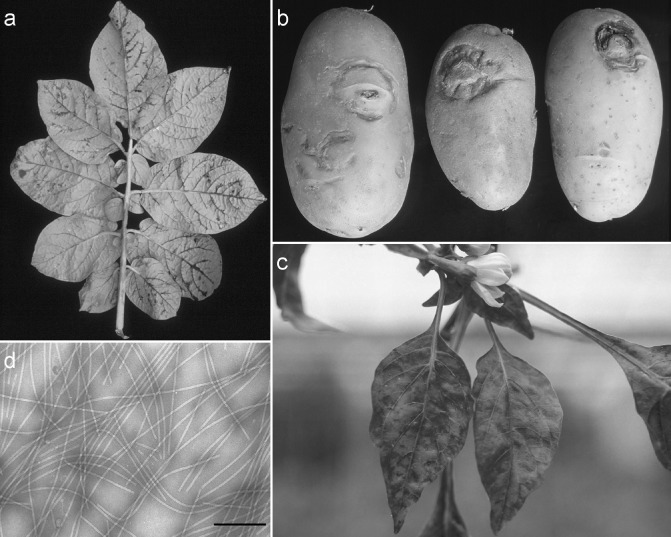
Symptoms induced by Potato virus Y (PVY). (a) Vein necrosis in potato leaf (courtesy of Nikon Vassilakos). (b) Necrotic rings on potato tubers (courtesy of the Laboratory of Virology, Benaki Phytopathological Institute). (c) Leaf mosaic on pepper [courtesy of Alain Palloix, INRA, Provence Alpes Côte d'Azur, Génétique et Amélioration des Fruits et Légumes (GAFL)]. (d) Electron micrograph of PVY virions [courtesy of Isabelle Bornard, INRA, Provence Alpes Côte d'Azur (PACA), Pathologie Végétale]. The scale bar corresponds to 200 nm.
PVY symptoms in tobacco, tomato and pepper often consist of mild mottling or mosaic (Fig. 1c), although stunting and necrosis may be observed. In addition, in tobacco, PVY modifies the chemical composition of cured leaves, especially the nicotine content (Latorre et al., 1984), resulting in high economic losses. In recent decades, tomato and pepper PVY isolates inducing leaf and stem necrosis have been reported in the Mediterranean region (d'Aquino et al., 1995; Fanigliulo et al., 2005; Gebre Selassie et al., 1985; Mascia et al., 2010). Mixed infection of PVY and Cucumber mosaic virus (CMV) also results in severe diseases in tomato crops.
Control strategies are based on breeding for resistance, the use of certified potato seeds and quarantine regulations to prevent the spread of nonindigenous isolates. The control of aphid vectors is usually of limited effectiveness, because of the nonpersistent mode of transmission. Genetically engineered resistance has been studied extensively, usually by the incorporation of partial or entire sequences of the coat protein (CP), P1 and NIb cistrons into potato or tobacco plants. PVY‐resistant genetically engineered cultivars have been registered in the USA (Solomon‐Blackburn and Barker, 2001). Most recently, potato plants transformed with a chimeric transgene containing PVY CP sequences have been evaluated in the field for a period of 6 years in Argentina. The plants showed null or negligible infection and preserved their agronomical traits and biochemical characteristics (Bravo‐Almonacid et al., 2012).
Genome organization and protein functions
PVY virions have a filamentous and flexuous form (Fig. 1d) with a helical symmetry, a length of 730–740 nm and a diameter of 12 nm. The genome consists of a messenger sense, single‐stranded RNA of approximately 9.7 kb. It is covalently linked to a single molecule of genome‐linked viral protein (VPg) at the 5’ end, and contains a polyadenosine (polyA) tail at the 3’ end. The genome contains a single open reading frame (ORF) flanked by untranslated regions, and is expressed as a polyprotein of approximately 3062 amino acid residues, which is cleaved into 10 mature proteins by three viral proteases (Carrington and Freed, 1990) (Fig. 2). An additional protein is produced from an overlapping ORF after +2 frame shifting of the P3 cistron, as fusion to the amino (N)‐terminal part of P3 (P3N‐PIPO) (Chung et al., 2008).
Figure 2.
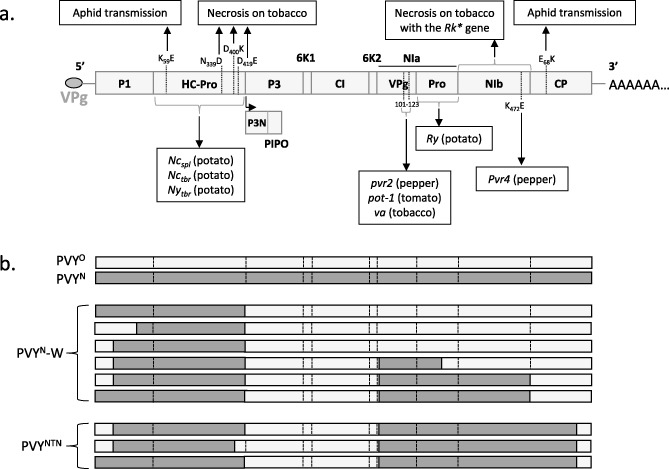
Determinants of pathogenicity mapped in the Potato virus Y (PVY) genome and recombination pattern of PVYNTN and PVYN‐W isolates. (a) PVY factors and/or mutations involved in resistance breakdown are indicated below the genome scheme. The dominant genes are specified with a capital letter and the host is indicated n parentheses. Above the genome scheme, PVY determinants of virulence or aphid transmission are noted. (b) Schematic representation of the different recombination patterns between PVYO (light grey) and PVYN (dark grey) clades (modified from Hu et al., 2009). *The tobacco Rk gene confers a nematode resistance and it (or a very tightly linked gene) induces leaf necrosis on PVY inoculation.
As a result of the high genome similarities within the genus Potyvirus, the functions of PVY proteins have been inferred largely by analogy with other members of the group. Most of them are multifunctional. The first protein coded by the 5’ region of the PVY genome is P1, the most variable protein among potyviruses. It has a protease domain in the carboxy (C)‐terminal region and cleaves itself from the adjacent helper‐component protease (HC‐Pro) (Verchot et al., 1991). It has been shown to bind single‐ and double‐stranded RNA (Brantley and Hunt, 1993; Soumounou and Laliberté, 1994), to be involved in genome amplification (Verchot and Carrington, 1995) and to enhance the suppression of plant defences based on RNA silencing (Brigneti et al., 1998; Pruss et al., 1997).
The next protein, HC‐Pro, is involved in aphid transmission (Govier et al., 1977), virus multiplication, cell‐to‐cell (Rojas et al., 1997) and systemic (Sáenz et al., 2002) movements and symptom intensity (Redondo et al., 2001; Sáenz et al., 2002; Shiboleth et al., 2007; Torres‐Barcelo et al., 2008; Yambao et al., 2008). Two conserved motifs of HC‐Pro, the N‐terminal ‘KITC’ and the C‐terminal ‘PTK’ motifs, are required for aphid transmission, with the first being involved in the interaction with the aphid stylet (Blanc et al., 1998) and the latter being a possible binding site for virions (Peng et al., 1998). The lysine to glutamic acid (K59E within the KITC motif) mutation observed in some PVY isolates renders HC‐Pro unable to interact with aphid stylets and abolishes PVY aphid transmissibility (Blanc et al., 1998). It has been suggested that HC‐Pro acts as a bridge between the virion CP and an unknown receptor in the aphid stylet (Govier and Kassanis, 1974; Pirone and Blanc, 1996). The C‐terminal part of HC‐Pro is involved in the suppression of plant defences based on the RNA silencing machinery, by binding to small interfering RNAs (siRNAs) (Brigneti et al., 1998; Lakatos et al., 2006; Llave et al., 2000; Shiboleth et al., 2007; Varrelmann et al., 2007). This property could account for the general involvement of potyvirus HC‐Pro in symptomatology and synergy with co‐infecting viruses for symptom severity (Pruss et al., 1997), but also for its roles in virus multiplication and systemic movement (Kasschau and Carrington, 2001). Additional plant defence inhibition properties are also suggested by the fact that PVY HC‐Pro interacts with subunits of the 20S proteasome, which is related to the antiviral response (Jin et al., 2007). A recent study has also shown an interaction between PVY HC‐Pro and the eukaryotic initiation factor 4E (eIF4E) and its isoform eIFiso4E of pepper and tobacco, making HC‐Pro a putative partner of the translation initiation complex (Ala‐Poikela et al., 2011).
The protein P3, together with 6K2, are the two potyviral membrane proteins (Eiamtanasate et al., 2007; Restrepo‐Hartwig and Carrington, 1994). P3 contains two hydrophobic domains located at the N‐ and C‐termini. Several studies have suggested its involvement in virus replication, systemic infection, pathogenicity and movement (Chu et al., 1997; Cui et al., 2010; Johansen et al., 2001; Merits et al., 1999). A translational frameshift on the P3 cistron leads to the production of P3N‐PIPO (Chung et al., 2008), which has been shown to be located to plasmodesmata and to participate in the viral cell‐to‐cell movement in conjunction with the CI (cylindrical or cytoplasmic inclusion) protein (Wei et al., 2010b). The function of the following 6K1 protein is unknown.
The CI protein forms the laminate cytoplasmic inclusion bodies (pinwheel‐shaped) (Edwardson, 1992), typical of potyviral infections. Its involvement in cell‐to‐cell virus movement is well established (Carrington et al., 1998; de Cedrón et al., 2006; Roberts et al., 1998; Wei et al., 2010b), whereas the RNA‐binding, RNA helicase and ATPase activities strongly suggest an important role in virus replication (Fernández et al., 1995, 1997; Merits et al., 1998). The C‐terminal part of the CI protein has been shown to interact with the plant eIF4E (Tavert‐Roudet et al., 2012), and the virus VPg and CI could therefore be recruited into the translation initiation complex, which is essential for virus multiplication and/or cell‐to‐cell movement (Abdul‐Razzak et al., 2009).
The 6K2 protein is membrane bound (Restrepo‐Hartwig and Carrington, 1994) and has been shown to play an essential role in virus replication (Wei and Wang, 2008; Wei et al., 2010a) by anchoring the viral replication complex to the endoplasmic reticulum (Schaad et al., 1997).
The NIa (first nuclear inclusion) protein possesses two domains: VPg and a protease domain which cleaves most proteins of the precursor polyprotein (Carrington and Dougherty, 1987). VPg is an intrinsically disordered protein and its structural flexibility has been proposed to be the basis of its capacity to interact physically with many viral and plant proteins (Elena and Rodrigo, 2013 and of its functional diversity (Rantalainen et al., 2011). Binding of PVY VPg with the host eIF4E is a key component of virus multiplication (translation and/or replication) (Deom et al., 1997; Robaglia and Caranta, 2006). VPg is localized in different cellular compartments depending on its combination with the NIa protease and/or the 6K2 protein. The VPg–NIa protease combination is located exclusively in the nucleolus, whereas the 6K2–VPg–NIa protease combination is found within vesicular structures derived from the endoplasmic reticulum (Jiang and Laliberte, 2011). Finally, VPg alone is linked covalently to the 5’ extremity of the viral RNA via a tyrosine residue (Murphy et al., 1990, 1991).
The NIb (second nuclear inclusion) protein is the RNA‐dependent RNA polymerase (RdRp) involved in the replication of the viral RNA (Hong and Hunt, 1996). Finally, CP is required for virion assembly, cell‐to‐cell (Rojas et al., 1997) and systemic (Andersen and Johansen, 1998; Dolja et al., 1995) movements, and aphid transmission (Atreya et al., 1990; Blanc et al., 1997). CP is a three‐domain protein with variable N‐ and C‐terminal domains exposed on the virion surface and a core region that binds RNA. The highly conserved ‘DAG’ motif at the N‐terminus of CP is essential for aphid transmissibility (Atreya et al., 1995). The context of the DAG motif can modulate the efficiency of potyvirus aphid transmission (López‐Moya et al., 1999). Remarkably, amino acid positions 9, 10 and 11 of the PVY CP, immediately adjacent to the DAG triplet (located at positions 6–8), has been shown to be subjected to positive selection (Moury and Simon, 2011). Hence, variations at these positions could modulate PVY transmission efficiency, as demonstrated for position 9 of Tobacco etch virus (TEV) CP (López‐Moya et al., 1999). In addition, the internal amino acid position 68 of PVY CP has been shown to modify quantitatively its aphid transmission efficiency by Myzus persicae or Aphis gossypii (Moury and Simon, 2011). The core region of CP has an essential role in virus assembly and cell‐to‐cell movement, suggesting the involvement of virions in cell‐to‐cell traffic (Rojas et al., 1997). Both the N‐ and C‐terminal regions are not required for assembly, but are involved in systemic movement.
What is Structuring PVY Diversity?
Where, how and when did PVY emerge?
The species PVY belongs to a large clade of 19 potyvirus species, sometimes referred to as the ‘PVY group’ or ‘PVY clade’ (Gibbs and Ohshima, 2010; Li et al., 2012; Moury and Verdin, 2012). It represents one of the four potyvirus groups that infect solanaceous crops, together with the TEV/Potato virus A clade, the Pepper veinal mottle virus (PepVMV)/Chilli veinal mottle virus clade and the Colombian datura virus (synonymous to Petunia flower mottle virus), which does not show any close relationship to other potyviruses. Viruses from the PVY clade are mostly present in America and, more particularly, in South America. Among the 19 species of the clade, 16 are present in America, with 11 being present exclusively on this continent. By contrast, only eight species have been described outside the Americas, indicating a significantly higher richness of viruses belonging to this clade in the American continents (P = 0.017; Fisher's exact test). Three species (Amazon lily mosaic virus, Alstroemeria mosaic virus and Amaranthus leaf mottle virus) have been observed only in the Old World, but only sporadically. Moreover, the natural hosts of these three species originate from South America. It is therefore likely that they are (or were) also present in America, but have not been detected because of limited sampling.
There is a significant association between the potyvirus species of the PVY clade that are found outside the American continents and their occurrence in crops propagated by vegetative propagation (Table 1; P = 0.018, Fisher's exact test). These data suggest that America (and, more specifically, South America) could be the centre of origin and diversification of the PVY clade, and that a worldwide dispersal of some of its members has involved mostly human activities (trade of potato tubers and bulbs or rhizomes of ornamental plants). The same trend is observed for the PVY species. Among the five major clades within PVY species, four (clade C, which includes the two subclades C1 and C2, and clades N and O; see below) are infectious in potato and show a worldwide distribution. The fifth (Chilean) clade does not seem to infect potato and has been described only in Chile (Moury, 2010; Sudarsono et al., 1993). These data are also in accordance with the fact that Central and South America are centres of origin and diversification for most of the hosts of viruses in the PVY clade, including PVY itself, such as species of the families Solanaceae, Asteraceae (Barreda et al., 2010) and Amaranthaceae (Segundo et al., 2007). These three botanical families also include large numbers of PVY experimental hosts (Edwardson and Christie, 1997; Kerlan, 2006), which could be a ‘genetic remnant’ of host range and infectivity properties derived from the common ancestor of the PVY clade. Reciprocally, viral species of the PVY clade that are adapted to the Asteraceae are also able to infect species in the family Solanaceae (Dujovny et al., 1998; Bidens mottle virus; GenBank accession number EF467235).
Table 1.
A test of association between geographical distribution and host characteristics among the 19 species of the Potato virus Y (PVY) group (Gibbs and Ohshima, 2010). Viruses that infect commercialized plants which multiply vegetatively have a greater chance of being present outside the American continents which contain the putative centre of diversification of the group (P = 0.018 or P = 0.0013 when considering separately the five major PVY clades; Fisher's exact tests). The five PVY clades are shown in bold type
| Present outside Americas | Present only in Americas | |
|---|---|---|
| Infectious in commercialized plants with vegetative propagation (tubers, bulbs or rhizomes) |
Alstroemeria mosaic virus
Amazon lily mosaic virus Potato virus V Potato virus Y (PVY): PVY‐N PVY‐O PVY‐C1 PVY‐C2 |
None reported |
| Not infectious in commercialized plants with vegetative propagation |
Amaranthus leaf mottle virus
Bidens mottle virus Pepper mottle virus Sunflower chlorotic mottle virus |
Alternanthera mild mosaic virus
Bidens mosaic virus Brugmansia suaveolens mottle virus Ecuadorian rocoto virus Pepper severe mosaic virus Pepper yellow mosaic virus Peru tomato mosaic virus Pfaffia mosaic virus Tomato necrotic stunt virus Verbena virus Y Wild potato mosaic virus PVY‐Chile |
Recent attempts to place a timeframe on the diversification of PVY have been unsuccessful (Cuevas et al., 2012) because of a lack of temporal structure in the PVY sequence dataset available, precluding the estimation of divergence times within the phylogenetic tree. As the initial radiation of the whole Potyvirus genus has been estimated to have occurred about 6600 years ago (Gibbs et al., 2008), a period which coincided with the development of agriculture, PVY diversification can be considered as very recent and may have been influenced strongly by human activities.
Host and geography as drivers of PVY diversity
PVY has been the focus of interest for many agronomists and virologists, and its biological and genetic diversities have been explored extensively, allowing a rather exhaustive image of its diversity to be drawn. Genome analyses have revealed five major clades within the PVY species (Fig. 3). The most widespread clades are the C1, N and O groups, whereas the C2 and Chilean groups are more restricted, either because of their narrower host range or their limited geographical distribution. Recombinant isolates with different genome parts clustering with distinct clades are also widespread.
Figure 3.
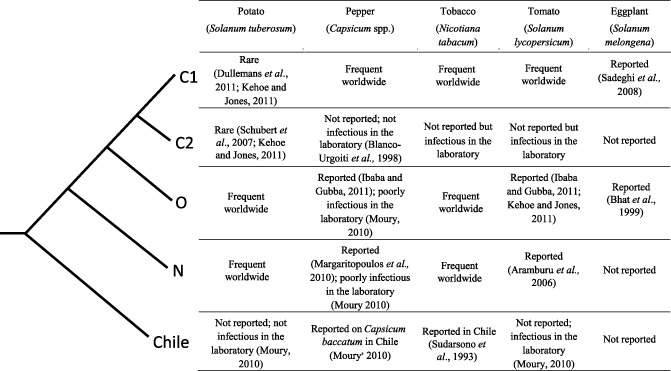
Simplified Potato virus Y (PVY) phylogeny (excluding the recombinant isolates) and infectivity of PVY isolates from the five major clades in crop plants of the family Solanaceae.
The first descriptions of PVY infections date back to the 1930s (Cockerham, 1943; Salaman and Le Pelley, 1930; Smith, 1931). PVY clades O and C (without distinguishing subclades C1 and C2) were predominant until the 1950s (Rolland et al., 2008). PVY C isolates can be distinguished from O isolates by the hypersensitive reactions which they induce in potato cultivars carrying the Nc resistance gene. PVY O isolates are a major problem in potato production and induce severe leaf mosaic symptoms and yield losses in potato cultivars lacking major resistance genes. PVY C isolates cause less severe disease in potato, with systemic mosaic or stipple streak symptoms. However, they are still predominant in other crops, such as pepper (mainly the C1 subgroup), where they induce severe mosaic and/or necrotic symptoms and high yield reduction (Gebre Selassie et al., 1985).
PVY N isolates induce severe veinal necrosis symptoms in most tobacco cultivars, whereas PVY O and C isolates usually induce mild mosaic symptoms. PVY N isolates have been reported since the 1940s in potato plants in Peru and Bolivia (Nobrega and Silberschmidt, 1944). Such isolates have been described in Europe and the USA since the 1950s (Kahn and Monroe, 1963). Compared with PVY O isolates, they induce less severe symptoms on potato plants, mostly mild mosaic or mottle in leaves. Finally, PVY isolates from the Chilean group were described initially in tobacco plants, where they induced veinal necrosis similar to that induced by N isolates (Sudarsono et al., 1993), and later in pepper (Capsicum baccatum) (Moury, 2010). Until now, this clade contains only isolates from Chile. According to phylogeny, the Chilean clade was the first to diverge during PVY evolution (Fig. 3).
Before the development of molecular biology, PVY classification was based on serology and on infectivity and symptomatology on different cultivated hosts. A remarkable property of PVY diversity is the correlation between the phylogenetic classification and some host range properties (Fig. 3). Examples include the infectivity in pepper and potato (isolates of clades C2, N and O are not or are poorly infectious in pepper, whereas isolates of clades C1 and Chile are not or are poorly infectious in potato; Fereres et al., 1993; Gebre Selassie et al., 1985; Moury, 2010; Romero et al., 2001) and the interaction with potato resistance genes Nc (specific for PVY C isolates) and Ny (specific for PVY O isolates). This suggests that relatively few host jumps and few changes in pathogenicity have occurred during PVY history. Moreover, it indicates that only few genetic exchanges have occurred between these major clades, as they would have obscured these correlations. Some of the pathogenicity traits, however, do not match with the phylogenetic clustering of PVY isolates, such as infectivity towards pepper or tobacco genotypes carrying recessive resistance, interaction with the resistance of the potato cultivar Maris Bard and symptomatology in tobacco varieties carrying the Nk nematode resistance gene.
Phylogenetic analysis has also revealed an effect of the continental origin on the genetic structuring of PVY populations, mainly for Europe, Japan, South Africa and North America (Cuevas et al., 2012). More particularly, the PVY N group splits into European and North American subclades and, although few isolates have been characterized, the Chilean group seems to be restricted to South America. For potato PVY isolates, the geographical structuring is probably hindered by the international trade of seed tubers and repeated introductions of PVY‐infected material (for example, at least five independent introductions have been postulated in Australia; Kehoe and Jones, 2011). Evidence of geographical structure could therefore reflect a smaller amount of potato material exchange or more efficient quarantine procedure or sanitary control policies.
The emergence of PVY recombinants
Since the end of the 1970s, PVY isolates inducing necrotic ringspot in potato tubers have been described and, later, were shown to be recombinants, with parts of their genome clustering with PVY O isolates and others with PVY N isolates (Le Romancer et al., 1994). Isolates showing this recombination pattern (grouped as PVYNTN; Fig. 2b) showed a dramatic increase in frequency in European potato crops in the 1990s and 2000s, representing more than 50% of isolates in France and Germany (Rolland et al., 2008). Another recombinant group (PVYN‐W; Fig. 2b) also showed an increase in prevalence in Europe and northern America during the early 2000s (Rolland et al., 2008). By contrast, PVY C isolates were no longer detected in European or American potato crops, which could be a result of the wide distribution of the Nc resistance gene in potato cultivars (Cockerham, 1943). The reasons for these spectacular increases in frequency are still unknown. These recombinant isolates possess a clear fitness advantage over nonrecombinants, as these increases in prevalence occurred independently in different geographical regions worldwide. They have a clear advantage over PVY O isolates, because they can infect potato cultivars carrying the Ny gene, which is present in many cultivars. However, why they outcompete isolates of the N clade remains unexplained. It has been shown that the molecular determinants of veinal necrosis in tobacco (cultivar Xanthi) present among PVY N, but not O, isolates confer a fitness cost in terms of the accumulation and competitiveness in Xanthi (Rolland et al., 2009). The recombination events that lead to the PVYNTN group might compensate for this fitness penalty. There is probably no direct link between the recombinant nature of PVYNTN isolates and the tuber necrosis that they induce. Indeed, similar symptoms can be induced by nonrecombinant isolates of the N clade (Nie and Singh, 2003; Singh et al., 2008).
Other recombinant PVY isolates, mostly issued from recombination between PVY O and N isolates, have been described, but show more limited geographical ranges. Although many different recombination patterns have been observed among PVY isolates, they seem to involve a limited number of independent recombination events. For example, PVYNTN and PVYN‐W isolates share three recombination breakpoints and may therefore have common origins (Schubert et al., 2007; Fig. 2).
PVY and Its Hosts: Models of Pathogenicity Evolution in Plant Viruses
eIF4E‐mediated resistance and PVY: evidence of recent and rapid co‐evolution
The first cloned natural recessive gene conferring pathogen resistance in plants was the pvr2 gene of pepper (Capsicum sp.) which provides PVY resistance (Ruffel et al., 2002). It was also the first cloned PVY resistance gene (a list of the identified PVY resistance genes is provided in Table 2). pvr2 encodes an eIF4E, which is involved in the initiation of translation of mRNAs in eukaryotes. eIF4E binds the cap attached to the 5’ extremity of mRNAs and initiates the recruitment of a scaffold of plant proteins, ending up with the translation process (Robaglia and Caranta, 2006). In plants, eIF4E belongs to a small gene family (Patrick and Browning, 2012). There are three copies of eIF4E and one isoform (eIFiso4E) in the genome of Arabidopsis thaliana. Pepper and tomato possess one copy of eIFiso4E and at least two copies of eIF4E (named eIF4E1 and eIF4E2). It has been suggested that, among these copies, one is constitutively expressed and possesses ubiquitous translation functions, whereas others are more specialized and may also possess functions unrelated to translation (Hernández and Vazquez‐Pianzola, 2005). Potyviruses have no caps at the 5’ end of their genomic RNAs, but a viral protein: VPg. Potyvirus VPgs are able to interact physically with one or other eIF4E isoform, which suggests that they have hijacked the translation machinery of eukaryotic cells for their own multiplication. Indeed, it has been shown that virus VPg and plant mRNAs compete for binding to eIF4E forms (Léonard et al., 2000). In addition, direct binding between eIF4E and potyvirus VPg has been demonstrated with yeast two‐hybrid experiments (Wittmann et al., 1997), and has been shown to be essential for virus infectivity. Indeed, plants with eIF4E or eIFiso4E knockout mutations or encoding eIF4E containing amino acid substitutions that abolish interaction with VPg are resistant to potyvirus infection (Duprat et al., 2002; Lellis et al., 2002; Ruffel et al., 2002). In turn, viruses are able to counter‐adapt through VPg mutations that restore interaction with the mutated eIF4E (Charron et al., 2008) or, in some cases, possibly through the use of an eIF4E‐independent pathway for their infectivity (Gallois et al., 2010). The PVY VPg has been shown to be the resistance‐breaking (RB) factor towards recessive resistance in pepper (Moury et al., 2004), the tomato relative Solanum habrochaites (Moury et al., 2004) and tobacco (Lacroix et al., 2011; Masuta et al., 1999). These recessive resistance traits are conferred by resistance alleles at the pvr2 locus, the pot‐1 orthologue of pvr2 (Ruffel et al., 2005) and alleles at the va locus, respectively.
Table 2.
Genes conferring resistance to Potato virus Y (PVY)
| Plant species | Resistance gene | Chromosome location | Nature of resistance | Spectrum of resistance | PVY pathogenicity factor | RB mutation and cost associated | References |
|---|---|---|---|---|---|---|---|
| Pepper | pvr21 | P4 | Recessive | Medium; some RB isolates | VPg | N121H, no cost | Charron et al. (2008); Fabre et al. (2012); Kyle and Palloix (1997); Montarry et al. (2011); Moury et al (2004); Ruffel et al. (2002) |
| pvr22 | P4 | Recessive | Large; very few RB isolates | VPg | N119D + N121H, costly | ||
| pvr23 | P4 | Recessive | Narrow; many RB isolates | VPg | Many single‐nucleotide mutations, no cost | ||
| pvr24 to pvr29 | P4 | Recessive | Not reported | Not reported | Not reported | ||
| Pvr4 | P10 | Dominant (HR/ER) | All clades | NIb | K472E, costly | Dogimont et al. (1996); Janzac et al. (2010) | |
| QTLs | P1, P6, P9, P10 | Polygenic | All clades | Not reported | Not reported | Caranta et al., 1997 | |
| Potato | Ncspl | IV | Dominant (HR) | Clade C | HC‐Pro | Not reported | Moury et al. (2011) |
| Nctbr | Not reported | Dominant (HR) | Clade C | HC‐Pro | Not reported | Cockerham (1970); Jones (1990); Moury et al. (2011) | |
| Nytbr | IV | Dominant (HR) | Clade O | HC‐Pro | Not reported | Celebi‐Toprak et al. (2002); Cockerham (1970); Jones (1990); Moury et al. (2011) | |
| Ryadg | XI | Dominant (ER) | All clades | Not reported | Not reported | Hämäläinen et al. (1997); Ross (1986) | |
| Rysto | XII | Dominant (ER) | All clades | NIa‐Pro | Not reported | Cockerham (1943); Mestre et al. (2000); Song et al. (2005) | |
| Tobacco | va0 to va2 | E | Recessive | All clades | VPg | Not reported | Masuta et al. (1999); Lacroix et al. (2011) |
| va2 | Not reported | Recessive | Not reported | Not reported | Not reported | Acosta‐Leal and Xiong (2008) | |
| Tomato | pot‐1 | T3 | Recessive | All clades | VPg | (R119H) | Moury et al. (2004); Parrella et al. (2002) |
ER, extreme resistance; HC‐Pro, helper‐component proteinase; HR, hypersensitive response; NIa, first nuclear inclusion; NIb, second nuclear inclusion; QTL, quantitative trait locus; RB, resistance breaking; VPg, genome‐linked viral protein.
Among solanaceous plants, most eIF4E‐encoding resistance genes have been identified in pepper and some in tomato relatives; in tobacco, the va resistance can be related to eIF4E, given the breakdown mechanisms, but this has not yet been demonstrated. No eIF4E‐mediated resistance has been identified in potato (S. tuberosum), which is probably a result of the tetraploid nature of this plant species, or in potato relatives. In pepper, 10 different pvr2 alleles have been described from a collection of 25 inbred lines (Charron et al., 2008), with the most widespread corresponding to the susceptibility allele pvr2 +. In spite of this relatively high allelic richness, overall, there is very limited variability in the pvr2 sequences. The other nine alleles differ from pvr2 + by only one to four amino acid substitutions in the encoded eIF4Es. Almost only nonsynonymous substitutions have been observed in the sequence of pvr2 and most polymorphic positions show signatures of positive selection (Cavatorta et al., 2008). Polymorphism is concentrated in three eIF4E domains, two coinciding with the catalytic site (mRNA cap‐binding site). Remarkably, all pepper genotypes carrying alleles that differ from pvr2 + show resistance to PVY. In spite of their high sequence similarity, these resistance alleles show a contrasting resistance spectrum and durability. Among the nine pvr2 alleles conferring PVY resistance, three (pvr2 1, pvr2 2 and pvr2 7) also confer resistance to PepVMV, when combined with the pvr6‐eIFiso4E gene (Rubio et al., 2009), and one (pvr2 2) confers resistance to TEV (Charron et al., 2008). These three species belong to three distinct clades in the genus Potyvirus, showing a large spectrum of action of eIF4E‐mediated resistance and that the infection strategies of many potyviruses rely on similar patterns of interaction with eIF4E. The pvr2 alleles also differ largely in durability. pvr2 1 and pvr2 2 have been deployed in pepper crops worldwide for more than 50 years. Almost no natural PVY isolates able to infect plants with pvr2 2 have been described so far (Moury and Verdin, 2012). pvr2 1‐breaking isolates are more frequent in natural conditions, but tend to be less prevalent than wild‐type (i.e. non‐RB) isolates (Luis‐Arteaga and Gil‐Ortega, 1986). The pvr2 3 allele is not largely deployed, except in traditional cultivars. However, pvr2 3‐breaking isolates are highly prevalent in pepper crops (Ben Khalifa et al., 2012) and can be selected easily in laboratory conditions (Ayme et al., 2006; Montarry et al., 2011). pvr2 3 is therefore expected to have a very low durability potential. The differences in durability between pvr2 1, pvr2 2 and pvr2 3 have been linked to the number and complexity of mutational pathways in the VPg coding region conferring RB capacity to PVY (Moury and Verdin, 2012) and to the pleiotropic fitness costs caused by these mutations (Ben Khalifa et al., 2009; Fabre et al., 2012).
The pattern of variation of VPg of pepper PVY isolates shows striking similarities to that of its pvr2‐encoded eIF4E ligand among pepper cultivars. Most of VPg is highly constrained, shows very limited amino acid variation and evolves under significant negative selection (B. Moury, unpublished data). By contrast, a small central region (amino acid positions 101–123) shows extensive amino acid variation, associated with significant positive selection (Moury et al., 2004). The functional relevance of these amino acid substitutions is further highlighted by the fact that almost all substitutions involved in the breakdown of the pvr2 alleles of pepper during laboratory experimental evolution coincide with these positively selected positions (Ayme et al., 2006). This correlation also implies that the RB events observed in the laboratory are representative of those which occur in natural epidemiological conditions, and that experimental evolution can be used, to a certain extent, to evaluate the durability potential of resistance. The eIF4E‐VPg pattern of diversity has been suggested to be emblematic of the matching allele model of plant–pathogen interactions (by contrast with the gene‐for‐gene model of interaction; Ben Khalifa et al., 2012; Sacristán and García‐Arenal, 2008), where each pathogen genotype is adapted to only one (or a very limited number of) resistance allele(s) in the host population, and where the RB capacity incurs no fitness cost to the pathogen.
Taken together, (i) the high variability and positive selection observed at a very small number of amino acid positions of VPg and eIF4E; the facts that (ii) the remainder of these two proteins are highly conserved and/or constrained and that (iii) almost all amino acid variability is linked to gains of function (resistance of the plant or pathogenicity of the virus); and (iv) the recent emergence of potyviruses provide rare evidence of a recent and rapid co‐evolution in plant‐pathogen interactions.
Steady relationships between PVY and dominant resistance
Dominant monogenic resistance to PVY has been characterized in potato and pepper. The pattern of interaction between these resistance genes and PVY isolates shows a remarkable stability, which contrasts sharply with the rapid diversification observed for recessive resistance alleles and PVY VPg ligands. Two kinds of phenotypic reaction have been distinguished among dominant resistance: extreme resistance (ER) and hypersensitive response (HR). The former includes the Ry sto and Ry adg genes from the wild potato relatives S. stoloniferum and S. tuberosum ssp. andigena, respectively, and the Pvr4 gene from pepper. No PVY accumulation can be detected in inoculated organs of plants carrying these ER genes (Janzac et al., 2009; Jones, 1990) and no necrotic lesions typical of a HR can be observed. The ability of Ry sto to reduce PVY accumulation at the within‐cell level has been demonstrated by protoplast experiments (Barker and Harrison, 1984). Several lines of experimental evidence support links between ER and HR. For the Pvr4 resistance in pepper, some PVY isolates are able to induce HR‐like necrotic lesions in inoculated cotyledons, and PVY can be detected in these organs (Janzac et al., 2009). However, none of the isolates was able to induce necrotic lesions or to be detected by enzyme‐linked immunosorbent assay (ELISA) or reverse transcription‐polymerase chain reaction (RT‐PCR) in Pvr4 pepper leaves. HR‐like reactions also occur in Pvr4 pepper under high inoculation pressures, such as in graft inoculations, in which the infected rootstock continuously provides virus to the scion (Janzac et al., 2009), and in Ry sto potato plants by Agrobacterium‐mediated expression of the resistance elicitor (Mestre et al., 2000). Therefore, ER and HR seem to be manifestations of the same resistance mechanisms, ER possibly being an HR which is expressed more rapidly and/or with a higher intensity by the plant, which can circumvent more efficiently the virus infection (Bendahmane et al., 1999). Remarkably, Pvr4 and Ry show a large spectrum of action: Ry genes are efficient against all PVY isolates and Pvr4 against all PVY isolates and five additional potyvirus species. Both are also highly durable as no RB by PVY has been observed in field conditions (for more than 20 years for Pvr4). Graft inoculation of Pvr4 scions enabled the selection of RB PVY isolates that contained a single‐nucleotide RB substitution in the NIb coding region. This nonsynonymous mutation changes a lysine residue at position 472 in NIb to glutamic acid and induces a high competitiveness cost to PVY in a susceptible pepper genotype. Although this mutation did not seem to modify the three‐dimensional structure of the protein, the high cost it induced to the virus could be linked to the overall high evolutionary constraint that is exerted on NIb (Janzac et al., 2010).
As no RB isolate was available, the viral factor corresponding to Ry sto was identified through transient expression of the PVY proteins in potato leaves. Only the NIa protease was able to induce necrotic HR‐like reactions. It is also not excluded that the elicitor of Ry sto is a host factor derived from the protease activity of NIa, and not the NIa protease directly. These reactions were specific of plants carrying Ry sto in a segregating population of potato plants (Mestre et al., 2000). The capacity of the NIa protease to induce the HR‐like response was further shown to overlap largely with the amino acid positions involved in the protease activity to the protein (Mestre et al., 2003). This suggests that putative RB mutations in PVY NIa protease would abolish or decrease the protease activity, would alter the maturation of most viral proteins and hence would be extremely costly for the virus. Overall, the high durability of the Ry and Pvr4 ER is most probably a result of the fitness cost, rather than the mutational pathways associated with RB.
Dominant PVY resistance genes conferring HR have been described only in potato or potato relatives in the genus Solanum. The Nc tbr and Nc spl genes, from S. tuberosum and S. sparsipilum, respectively, confer HR to PVY C (C1 and C2) isolates only, and the Ny tbr gene from S. tuberosum confers HR resistance to PVY O isolates only. Nc spl and Ny tbr map to the same region of chromosome IV in the potato genome and could therefore be allelic or belong to the same gene cluster (Moury et al., 2011). PVY HC‐Pro is the viral factor that corresponds to Nc tbr, Nc spl and Ny tbr (Moury et al., 2011), and the HR‐eliciting regions of Nc spl and Ny tbr seem to be contiguous in the C‐terminal end of HC‐Pro (Moury et al., 2011; Tian and Valkonen, 2013). The fact that the specificity of the HR of potato correlates largely with the clustering of PVY clades suggests few, if any, resistance breakdowns by the accumulation of point mutations along PVY evolution. Instead, breakdown of Ny tbr by recombination was suggested for PVYNTN and PVYN‐W (Moury et al., 2011).
Consequently, in contrast with recessive resistance genes, dominant resistance genes show rather stable patterns of interaction with PVY diversity, as they reveal: (i) no (or very little) evidence of breakdown; (ii) little allelic variability; and (iii) no evidence of positive selection in the corresponding viral factors (HC‐Pro, NIa protease and NIb) (Moury et al., 2002, 2006).
Insights into the durability of oligo‐ and polygenic resistance
Combining different resistance genes against the same pathogen in the same plant genotype (gene pyramiding strategy) has been proposed to improve resistance durability. PVY has provided experimental evidence of this concept and insights about the mechanisms involved in this enhanced durability. Acosta‐Leal and Xiong (2008) studied the tobacco cultivars ‘VAM’ and ‘NC745’, both carrying the va recessive resistance gene, and observed a rapid selection of RB variants in NC745, but not in VAM, for which no RB variants were obtained. The va gene confers resistance to PVY cell‐to‐cell movement. In VAM, a second unlinked recessive gene, named va2, also conferred resistance to within‐cell PVY accumulation, as observed in transfected protoplasts. Therefore, the higher durability of VAM resistance was caused by a combination of resistance genes that acted at two different steps of the virus infection cycle.
A similar situation was observed in pepper, where the durability of the pvr23 gene could be enhanced strongly by the combination with quantitative trait loci (QTLs) that increased the resistance efficiency (Palloix et al., 2009). Several mechanisms seemed to contribute to the higher durability of the polygenic resistance (pvr23 + QTLs), including higher efficiency, more complex mutational pathways leading to RB and slower selection of RB variants (Quenouille et al., in press). Interestingly, by themselves, the QTLs had only a small resistance effect, decreasing PVY accumulation slightly at the systemic level and delaying symptom onset and severity. Moreover, the QTLs were not durable by themselves, as eight serial passages in a pepper genotype carrying these QTLs showed a drastic increase in virus accumulation (Montarry et al., 2012). These results suggest that the increase in durability of the polygenic resistance composed of pvr23 and QTLs was caused by more‐than‐additive epistatic effects of the genetic components.
PVY infectivity, virulence and fitness: correlated evolution of pathogenicity traits
Beyond their interaction with particular resistance genes, PVY isolates more generally show large differences in virulence, such as the quantitative level of damage in infected hosts, and in infectivity in different host species. Variation of these traits has, for a long time, been the basis of PVY classification (Singh et al., 2008).
With regard to virulence, the most obvious differences reside in the capacity of some PVY isolates to induce necrotic (versus mosaic) reactions in their hosts, mainly in tobacco, pepper and potato. Usually, genetic variation for the necrotic phenotype also exists in the host. As mentioned above, PVY N isolates induce veinal necrosis in most tobacco cultivars, such as Xanthi. The necrotic phenotype is mostly a result of the combination of three amino acid substitutions in HC‐Pro (at positions 339, 400 and 419) (Faurez et al., 2012; Tribodet et al., 2005). The same three substitutions are present in isolates from the Chilean group, which also induce veinal necrosis in tobacco (Moury, 2010). As the Chilean group was the first to diverge in PVY history, this suggests that tobacco necrosis is an ancestral PVY trait. There is also genetic variation among potato and PVY genotypes for the induction of tuber necrosis, but the genetic bases of this phenotype have not been determined, in either PVY or the potato host.
In addition, major differences in host species adaptation are observed for potato and pepper, whereas few differences in infectivity are observed in tobacco and tomato (both susceptible to most PVY isolates) or S. melongena eggplant (resistant to most PVY isolates) (Fig. 3).
Remarkably, several pathogenicity traits are not independently distributed among PVY isolates, but are strongly associated, suggesting both common evolutionary history and genetic correlation (by linkage or pleiotropy). For example, among the five major PVY clades, a negative correlation exists between the capacities to infect pepper and potato plants. This suggests very limited numbers of host jumps during PVY evolution, followed by secondary specialization onto the new host (Moury, 2010). Similarly, all PVY isolates able to induce potato tuber necrosis belong to group N or N × O recombinant groups that are also able to induce veinal necrosis in Xanthi tobacco. Because the genetic determinism of most of these traits in PVY is still unknown, we do not know whether these correlations are caused by linkage or by pleiotropic effects of mutations. By contrast, pleiotropic effects of PVY mutations involved in fitness and/or pathogenicity traits have already been evidenced. Mutations involved in tobacco necrosis at positions 400 and 419 of HC‐Pro confer simultaneously a fitness cost in terms of competitiveness and accumulation in Xanthi (Rolland et al., 2009). Similarly, in the CP coding region of PVY N isolates, positive selection has been detected at amino acid position 25, indicating that variation at this position might have played an important role in PVY adaptation. Confirming this assumption, the mutation of this position has been shown to increase significantly PVY competitiveness in tobacco, but to decrease competitiveness in potato (Moury and Simon, 2011).
To conclude, the fact that distinct clades within PVY species show limited genetic exchanges and are correlated with adaptation to different host species and/or genotypes are indicative of a speciation in progress. The genetic differentiation into separate clades could be reinforced by host barriers, similar to those observed between pepper and potato, so that PVY could eventually evolve to form distinct viral species.
Visser et al. (2012) (PLoS ONE 7:e50631) recently obtained a slightly different phylogeny for the major PVY groups, where clades ‘Chile’ and ‘N’ were inverted compared to Fig. 3. They obtained an oldest divergence time corresponding to 970 years before present for the PVY species.
Acknowledgements
We thank Professor Dawn Arnold for her suggestion that we should write this Pathogen Profile and Isabelle Bornard for preparing the picture plate. Julie Quenouille is supported by INRA and the Région Provence Alpes Côte d'Azur (PACA). We acknowledge the financial support of the Comité Technique Permanent de la Sélection (CTPS, French Ministry of Agriculture and Fisheries), the Agence Nationale de la Recherche (ANR), the Pôle Européen d'Innovation Fruits et Légumes (PEIFL) and the seed companies Gautier Semences, Clause Vegetable Seeds, Vilmorin SA, Rijk Zwaan and Sakata Vegetables Europe.
References
- Abdul‐Razzak, A. , Guiraud, T. , Peypelut, M. , Walter, J. , Houvenaghel, C. , Candresse, T. , Le Gall, O. and German‐Retana, S. (2009) Involvement of the cylindrical inclusion (CI) protein in the overcoming of an eIF4E‐mediated resistance against Lettuce mosaic potyvirus. Mol. Plant Pathol. 10, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta‐Leal, R. and Xiong, Z. (2008) Complementary functions of two recessive R‐genes determine resistance durability of tobacco Virgin A Mutant (VAM) to Potato virus Y . Virology, 379, 275–283. [DOI] [PubMed] [Google Scholar]
- Ala‐Poikela, M. , Goytia, E. , Haikonen, T. , Rajamäki, M.L. and Valkonen, J.P.T. (2011) Helper component proteinase of the genus potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif. J. Virol. 85, 6784–6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, K. and Johansen, I. (1998) A single conserved amino acid in the coat protein gene of pea seed‐borne mosaic potyvirus modulates the ability of the virus to move systemically in Chenopodium quinoa . Virology, 241, 304–311. [DOI] [PubMed] [Google Scholar]
- d'Aquino, L. , Dalmay, T. , Burgyán, J. , Ragozzino, A. and Scala, F. (1995) Host range and sequence analysis of an isolate of Potato virus Y inducing veinal necrosis in pepper. Plant Dis. 79, 1046–1050. [Google Scholar]
- Aramburu, J. , Galipienso, L. and Matas, M. (2006) Characterization of Potato virus Y isolates from tomato crops in northeast Spain. Eur. J. Plant Pathol. 115, 247–258. [Google Scholar]
- Atreya, C. , Raccah, B. and Pirone, T. (1990) A point mutation in the coat protein abolishes aphid transmissibility of a potyvirus. Virology, 178, 161–165. [DOI] [PubMed] [Google Scholar]
- Atreya, P.L. , Lopez‐Moya, J.J. , Chu, M. , Atreya, C.D. and Pirone, T.P. (1995) Mutational analysis of the coat protein N‐terminal amino acids involved in potyvirus transmission by aphids. J. Gen. Virol. 76, 265–270. [DOI] [PubMed] [Google Scholar]
- Ayme, V. , Souche, S. , Caranta, C. , Jacquemond, M. , Chadœuf, J. , Palloix, A. and Moury, B. (2006) Different mutations in the genome‐linked protein VPg of Potato virus Y confer virulence on the pvr23 resistance in pepper. Mol. Plant–Microbe Interact. 19, 557–563. [DOI] [PubMed] [Google Scholar]
- Barker, H. and Harrison, B.D. (1984) Expression of genes for resistance to Potato virus Y in potato plants and protoplasts. Ann. Appl. Biol. 105, 539–545. [Google Scholar]
- Barreda, V.D. , Palazzesi, L. , Tellería, M.C. , Katinas, L. , Crisci, J.V. , Bremer, K. , Passalia, M.G. , Corsolini, R. , Rodríguez Brizuela, R. and Bechis, F. (2010) Eocene Patagonia fossils of the daisy family. Science, 329, 1621. [DOI] [PubMed] [Google Scholar]
- Beczner, L. , Horváth, J. , Romhányi, I. and Förster, H. (1984) Studies on the etiology of tuber necrotic ringspot disease in potato. Potato Res. 27, 339–352. [Google Scholar]
- Ben Khalifa, M. , Simon, V. , Marrakchi, M. , Fakhfakh, H. and Moury, B. (2009) Contribution of host plant resistance and geographic distance to the structure of Potato virus Y (PVY) populations in pepper in northern Tunisia. Plant Pathol. 58, 763–772. [Google Scholar]
- Bendahmane, A. , Kanyuka, K. and Baulcombe, D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell, 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Khalifa, M. , Simon, V. , Fakhfakh, H. and Moury, B. (2012) Tunisian Potato virus Y isolates with unnecessary pathogenicity towards pepper: support for the matching allele model in eIF4E resistance–potyvirus interactions. Plant Pathol. 61, 441–447. [Google Scholar]
- Bhat, A.I. , Varma, A. , Pappu, H.R. , Rajamannar, M. , Jain, R.K. and Praveen, S. (1999) Characterization of a potyvirus from eggplant (Solanum melongena) as a strain of potato virus Y by N‐terminal serology and sequence relationships. Plant Pathol. 48, 648–654. [Google Scholar]
- Blanc, S. , López‐Moya, J.J. , Wang, R. , García‐Lampasona, S. , Thornbury, D.W. and Pirone, T.P. (1997) A specific interaction between coat protein and helper component correlates with aphid transmission of a potyvirus. Virology, 231, 141–147. [DOI] [PubMed] [Google Scholar]
- Blanc, S. , Ammar, E.D. , Garcia‐Lampasona, S. , Dolja, V.V. , Llave, C. , Baker, J. and Pirone, T.P. (1998) Mutations in the potyvirus helper component protein: effects on interactions with virions and aphid stylets. J. Gen. Virol. 79, 3119–3122. [DOI] [PubMed] [Google Scholar]
- Blanco‐Urgoiti, B. , Sanchez, F. , Perez de san Roman, C. , Dopazo, J. and Ponz, F. (1998) PVY‐C isolates are a homogeneous pathotype but two different genetic strains. J. Gen. Virol. 79, 2037–2042. [DOI] [PubMed] [Google Scholar]
- Brantley, J.D. and Hunt, A.G. (1993) The N‐terminal protein of the polyprotein encoded by the potyvirus tobacco vein mottle virus is an RNA‐binding protein. J. Gen. Virol. 74, 1157–1162. [DOI] [PubMed] [Google Scholar]
- Bravo‐Almonacid, F. , Rudoy, V. , Welin, B. , Segretin, M.E. , Bedogni, M.C. , Stolowicz, F. , Criscuolo, M. , Foti, M. , Gomez, M. , López, M. , Serino, G. , Cabral, S. , Dos Santos, C. , Huarte, M. and Mentaberry, A. (2012) Field testing, gene flow assessment and pre‐commercial studies on transgenic Solanum tuberosum spp. tuberosum (cv. Spunta) selected for PVY resistance in Argentina. Transgenic Res. 21, 967–982. [DOI] [PubMed] [Google Scholar]
- Brigneti, G. , Voinnet, O. , Li, W.X. , Ji, L.H. , Ding, S.W. and Baulcombe, D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana . EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Caranta, C. , Lefebvre, V. and Palloix, A. (1997) Polygenic resistance of pepper to potyviruses consists of a combination of isolate‐specific and broad‐spectrum quantitative trait loci. Mol. Plant‐Microbe Interact. 10, 872–878. [Google Scholar]
- Carrington, J.C. and Dougherty, W.G. (1987) Small nuclear inclusion protein encoded by plant potyvirus genome is a protease. J. Virol. 61, 2540–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C. and Freed, D.D. (1990) Cap‐independent enhancement of translation by a plant potyvirus 5’ nontranslated region. J. Virol. 64, 1590–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C. , Jensen, P.E. and Schaad, M.C. (1998) Genetic evidence for an essential role for potyvirus CI protein in cell‐to‐cell movement. Plant J. 14, 393–400. [DOI] [PubMed] [Google Scholar]
- Cavatorta, J.R. , Savage, A.E. , Yeam, I. , Gray, S.M. and Jahn, M.M. (2008) Positive Darwinian selection at single amino acid sites conferring plant virus resistance. J. Mol. Evol. 67, 551–559. [DOI] [PubMed] [Google Scholar]
- de Cedrón, M.G. , Osaba, L. , Lopez, L.L. and García, A.J. (2006) Genetic analysis of the function of the plum pox virus CI RNA helicase in virus movement. Virus Res. 116, 136–145. [DOI] [PubMed] [Google Scholar]
- Celebi‐Toprak, F. , Slack, S.A. and Jahn, M.M. (2002) A new gene, Ny tbr, for hypersensitivity to Potato virus Y from Solanum tuberosum maps to chromosome IV. Theor. Appl. Genet. 104, 669–674. [DOI] [PubMed] [Google Scholar]
- Charron, C. , Nicolaï, M. , Gallois, J. , Robaglia, C. , Moury, B. , Palloix, A. and Caranta, C. (2008) Natural variation and functional analyses provide evidence for co‐evolution between plant eIF4E and potyviral VPg. Plant J. 54, 56–68. [DOI] [PubMed] [Google Scholar]
- Chu, M. , Lopez‐Moya, J.J. , Llave‐Correas, C. and Pirone, T.P. (1997) Two separate regions in the genome of the Tobacco etch virus contain determinants of the wilting response of Tabasco pepper. Mol. Plant–Microbe Interact. 10, 472–480. [DOI] [PubMed] [Google Scholar]
- Chung, B.Y.W. , Miller, W.A. , Atkins, J.F. and Firth, A.E. (2008) An overlapping essential gene in the Potyviridae . Proc. Natl. Acad. Sci. USA, 105, 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham, G. (1943) The reactions of potato varieties to viruses X, A, B and C. Ann. Appl. Biol. 30, 338–344. [Google Scholar]
- Cockerham, G. (1970) Genetical studies on resistance to potato viruses X and Y. Heredity, 25, 309–348. [Google Scholar]
- Cuevas, J.M. , Delaunay, A. , Visser, J.C. , Bellstedt, D.U. , Jacquot, E. and Elena, S.F. (2012) Phylogeography and molecular evolution of Potato virus Y . PLoS ONE, 7, e37853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, X. , Wei, T. , Chowda‐Reddy, R.V. , Sun, G. and Wang, A. (2010) The Tobacco etch virus P3 protein forms mobile inclusions via the early secretory pathway and traffics along actin microfilaments. Virology, 397, 56–63. [DOI] [PubMed] [Google Scholar]
- De Bokx, J.A. and Huttinga, H. (1981) Potato virus Y . CMI/AAB Descriptions of Plant Viruses , 242 Available at http://www.dpvweb.net/dpv/showdpv.php?dpvno=242.
- Deom, C.M. , Murphy, J.F. and Paguio, O.R. (1997) Resistance to tobacco etch virus in Capsicum annuum: inhibition of virus RNA accumulation. Mol. Plant–Microbe Interact. 7, 917–921. [Google Scholar]
- Dogimont, C. , Palloix, A. , Daubèze, A.M. , Marchoux, G. , Gebre Selassie, K. and Pochard, E. (1996) Genetic analysis of broad spectrum resistance to potyviruses using doubled haploid lines of pepper (Capsicum annuum L.). Euphytica, 88, 231–239. [Google Scholar]
- Dolja, V.V. , Haldeman‐Cahill, R. , Montgomery, A.E. , Vandenbosch, K.A. and Carrington, J.C. (1995) Capsid protein determinants involved in cell‐to‐cell and long distance movement of Tobacco etch potyvirus . Virology, 206, 1007–1016. [DOI] [PubMed] [Google Scholar]
- Draper, M.D. , Pasche, J.S. and Gudmestad, N.C. (2002) Factors influencing PVY development in three potato cultivars. Am. J. Potato Res. 79, 155–165. [Google Scholar]
- Dujovny, G. , Usugi, T. , Shohara, K. and Lenardon, S.L. (1998) Characterization of a new Potyvirus infecting sunflower in Argentina. Plant Dis. 82, 470–474. [DOI] [PubMed] [Google Scholar]
- Dullemans, A.M. , Cuperus, C. , van der Verbeek, M. and Vlugt, R.A.A. (2011) Complete nucleotide sequence of a potato isolate of strain group C of Potato virus Y from 1938. Arch. Virol. 156, 473–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprat, A. , Caranta, C. , Revers, F. , Menand, B. , Browning, K.S. and Robaglia, C. (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32, 927–934. [DOI] [PubMed] [Google Scholar]
- Edwardson, J.R. (1992) Inclusion bodies. Arch. Virol. Suppl. 5, 25–30. [DOI] [PubMed] [Google Scholar]
- Edwardson, J.R. and Christie, R.G. (1997) Potyviruses In: Florida Agricultural Experiment Station Monograph Series 18‐II – Viruses Infecting Pepper and Other Solanaceous Crops (University of Florida , ed.), pp. 424–524. Gainesville, FL: University of ; Florida. [Google Scholar]
- Eiamtanasate, S. , Juricek, M. and Yap, Y.K. (2007) C‐terminal hydrophobic region leads PRSV P3 protein to endoplasmic reticulum. Virus Genes, 35, 611–617. [DOI] [PubMed] [Google Scholar]
- Elena, S.F. and Rodrigo, G. (2013) Towards an integrated molecular model of plant–virus interactions. Curr. Opin. Virol. 2, 719–724. [DOI] [PubMed] [Google Scholar]
- Fabre, F. , Montarry, J. , Coville, J. , Senoussi, R. , Simon, V. and Moury, B. (2012) Modelling the evolutionary dynamics of viruses within their hosts: a case study using high‐throughput sequencing. PLoS Pathog. 8, e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanigliulo, A. , Comes, S. , Pacella, R. , Harrach, B. , Martin, D.P. and Crescenzi, A. (2005) Characterisation of Potato virus Y nnp strain inducing veinal necrosis in pepper: a naturally occurring recombinant strain of PVY. Arch. Virol. 150, 709–720. [DOI] [PubMed] [Google Scholar]
- Faurez, F. , Baldwin, T. , Tribodet, M. and Jacquot, E. (2012) Identification of new Potato virus Y (PVY) molecular determinants for the induction of vein necrosis in tobacco. Mol. Plant Pathol. 13, 948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fereres, A. , Perez, P. , Gemeno, C. and Ponz, F. (1993) Transmission of Spanish pepper‐PVY and potato‐PVY isolates by aphid (Homoptera, Aphididae) vectors—epidemiological implications. Environ. Entomol. 22, 1260–1265. [Google Scholar]
- Fernández, A. , Laın, S. and García, J.A. (1995) RNA helicase activity of the plum pox potyvirus CI protein expressed in Escherichia coli. Mapping of an RNA binding domain. Nucleic Acids Res. 23, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, A. , Guo, H.S. , Sáenz, P. , de Simón‐Buela, L., Cedrón, M.G. and García, J.A. (1997) The motif V of plum pox potyvirus CI RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. Nucleic Acids Res. 25, 4474–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois, J.‐L. , Charron, C. , Sánchez, F. , Pagny, G. , Houvenaghel, M.‐C. , Moretti, A. , Ponz, F. , Revers, F. , Caranta, C. and German‐Retana, S. (2010) Single amino acid changes in the turnip mosaic virus viral genome‐linked protein (VPg) confer virulence towards Arabidopsis thaliana mutants knocked out for eukaryotic initiation factors eIF(iso)4E and eIF(iso)4G. J. Gen. Virol. 91, 288–293. [DOI] [PubMed] [Google Scholar]
- Gebre Selassie, K. , Marchoux, G. , Delecolle, B. and Pochard, E. (1985) Variabilité naturelle des souches du virus Y de la pomme de terre dans les cultures de piment du sud‐est de la France. Caractérisation et classification en pathotypes. Agronomie, 5, 621–630. [Google Scholar]
- Gibbs, A. and Ohshima, K. (2010) Potyviruses and the digital revolution. Ann. Rev. Phytopathol. 48, 205–223. [DOI] [PubMed] [Google Scholar]
- Gibbs, A.J. , Ohshima, K. , Phillips, M.J. and Gibbs, M.J. (2008) The prehistory of potyviruses: their initial radiation was during the dawn of agriculture. PLoS ONE, 3, e2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govier, D.A. and Kassanis, B. (1974) A virus‐induced component of plant sap needed when aphids acquire potato virus Y from purified preparations. Virology, 61, 420–426. [DOI] [PubMed] [Google Scholar]
- Govier, D.A. , Kassanis, B. and Pirone, T.P. (1977) Partial purification and characterization of Potato virus Y helper component. Virology, 78, 306–314. [DOI] [PubMed] [Google Scholar]
- Hämäläinen, J.H. , Watanabe, K.N. , Valkonen, J.P.T. , Arihara, A. , Plaisted, R.L. , Pehu, E. , Miller, L. and Slack, S.A. (1997) Mapping and marker‐assisted selection for a gene for extreme resistance to potato virus Y. Theor. Appl. Genet. 94, 192–197. [Google Scholar]
- Hernández, G. and Vazquez‐Pianzola, P. (2005) Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech. Dev. 122, 865–876. [DOI] [PubMed] [Google Scholar]
- Hong, Y. and Hunt, A.G. (1996) RNA polymerase activity catalyzed by potyvirus‐encoded RNA‐dependent RNA polymerase. Virology, 226, 146–151. [DOI] [PubMed] [Google Scholar]
- Hu, X. , Karasev, A.V. , Brown, C.J. and Lorenzen, J.H. (2009) Sequence characteristics of Potato virus Y recombinants. J. Gen. Virol. 90, 3033–3041. [DOI] [PubMed] [Google Scholar]
- Ibaba, J.D. and Gubba, A. (2011) Diversity of Potato virus Y isolates infecting solanaceous vegetables in the province of KwaZulu‐Natal in the Republic of South Africa. Crop Prot. 30, 1404–1408. [Google Scholar]
- Janzac, B. , Fabre, M.F. , Palloix, A. and Moury, B. (2009) Phenotype and spectrum of action of the Pvr4 resistance in pepper against potyviruses, and selection for virulent variants. Plant Pathol. 58, 443–449. [Google Scholar]
- Janzac, B. , Montarry, J. , Palloix, A. , Navaud, O. and Moury, B. (2010) A point mutation in the polymerase of Potato virus Y confers virulence toward the Pvr4 resistance of pepper and a high competitiveness cost in susceptible cultivar. Mol. Plant–Microbe Interact. 23, 823–830. [DOI] [PubMed] [Google Scholar]
- Jiang, J. and Laliberte, J.‐F. (2011) The genome‐linked protein VPg of plant viruses—a protein with many partners. Curr. Opin. Virol. 1, 347–354. [DOI] [PubMed] [Google Scholar]
- Jin, Y. , Ma, D. , Dong, J. , Jin, J. , Li, D. , Deng, C. and Wang, T. (2007) HC‐Pro protein of Potato Virus Y can interact with three Arabidopsis 20S proteasome subunits in planta . J. Virol. 81, 12 881–12 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, I.E. , Lund, O.S. , Hjulsager, C.K. and Laursen, J. (2001) Recessive resistance in Pisum sativum and potyvirus pathotype resolved in a gene‐for‐cistron correspondence between host and virus. J. Virol. 75, 6609–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R.A.C. (1990) Strain group specific and virus specific hypersensitive reactions to infection with potyviruses in potato cultivars. Ann. Appl. Biol. 117, 93–105. [Google Scholar]
- Kahn, R.P. and Monroe, R.L. (1963) Detection of the tobacco veinal necrosis strain of potato virus Y in Solanum cardenasii and S. andigenum introduced into the United States. Phytopathology, 53, 1356–1359. [Google Scholar]
- Kasschau, K.D. and Carrington, J.C. (2001) Long‐distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC‐Pro. Virology, 285, 71–81. [DOI] [PubMed] [Google Scholar]
- Kehoe, M.A. and Jones, R.A.C. (2011) A proposal to help resolve the disagreement between naming of Potato virus Y strain groups defined by resistance phenotypes and those defined by sequencing. Arch. Virol. 156, 2273–2278. [DOI] [PubMed] [Google Scholar]
- Kerlan, C. (2006) Potato virus Y . AAB/CMI Descriptions of Plant Viruses 414 Available at http://www.dpvweb.net/dpv/showdpv.php?dpvno=414.
- Kyle, M.M. and Palloix, A. (1997) Proposed revision of nomenclature for potyvirus resistance genes in Capsicum . Euphytica, 97, 183–188. [Google Scholar]
- Lacroix, C. , Glais, L. , Verrier, J.‐L. and Jacquot, E. (2011) Effect of passage of a Potato virus Y isolate on a line of tobacco containing the recessive resistance gene va2 on the development of isolates capable of overcoming alleles 0 and 2. Eur. J. Plant Pathol. 130, 259–269. [Google Scholar]
- Lakatos, L. , Csorba, T. , Pantaleo, V. , Chapman, E.J. , Carrington, J.C. , Liu, Y. , P., Dolja, V.V. , Calvino, L.F. , Lopez‐Moya, J.J. and Burgyan, J. (2006) Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25, 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre, B.A. , Flores, V. and Marholz, G. (1984) Effect of potato virus Y on growth, yield, and chemical composition of flue‐cured tobacco in Chile. Plant Dis. 68, 884–886. [Google Scholar]
- Le Romancer, M. , Kerlan, C. and Nedellec, M. (1994) Biological characterization of various geographical isolates of Potato virus Y inducing superficial necrosis on potato tubers. Plant Pathol. 43, 138–144. [Google Scholar]
- Lellis, A.D. , Kasschau, K.D. , Whitham, S.A. and Carrington, J.C. (2002) Loss‐of‐susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12, 1046–1051. [DOI] [PubMed] [Google Scholar]
- Léonard, S. , Plante, D. , Wittmann, S. , Daigneault, N. , Fortin, M.G. and Laliberte, J.‐F. (2000) Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 74, 7730–7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R. , Gao, S. , Hernandez, A.G. , Wechter, W.P. , Fei, Z. and Ling, K.S. (2012) Deep sequencing of small RNAs in tomato for virus and viroid identification and strain differentiation. PLoS ONE, 7, e37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C. , Kasschau, K.D. and Carrington, J.C. (2000) Virus‐encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA, 97, 13 401–13 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Moya, J.J. , Wang, R.Y. and Pirone, T.P. (1999) Context of the coat protein DAG motif affects potyvirus transmissibility by aphids. J. Gen. Virol. 80, 3281–3288. [DOI] [PubMed] [Google Scholar]
- Luis‐Arteaga, M. and Gil‐Ortega, R. (1986) Biological characterization of PVY as isolated from pepper in Spain. In: VI Meeting on Capsicum and Eggplant, Zaragoza, Spain, October 21–24 , 183–188.
- Margaritopoulos, J.T. , Dovas, C.I. , Gounaris, J. , Skouras, P.J. , Kanavaki, O.M. , Katis, N.I. and Tsitsipis, J.A. (2010) Molecular analysis of the coat protein of Potato virus Y isolates in Greece suggests multiple introduction from different genetic pools. J. Phytopathol. 158, 73–80. [Google Scholar]
- Mascia, T. , Finetti‐Sialer, M.M. , Cillo, F. and Gallitelli, D. (2010) Biological and molecular characterization of a recombinant isolate of Potato virus Y associated with a tomato necrotic disease occurring in Italy. J. Plant Pathol. 92, 131–138. [Google Scholar]
- Masuta, C. , Nishimura, M. , Morishita, H. and Hataya, T. (1999) A single amino acid change in viral genome‐associated protein of Potato virus Y correlates with resistance breaking in ‘Virgin A Mutant’ tobacco. Phytopathology, 89, 118–123. [DOI] [PubMed] [Google Scholar]
- Merits, A. , Guo, D.Y. and Saarma, M. (1998) VPg, coat protein and five non‐structural proteins of potato A potyvirus bind RNA in a sequence‐unspecific manner. J. Gen. Virol. 79, 3123–3127. [DOI] [PubMed] [Google Scholar]
- Merits, A. , Guo, D. , Jarvekulg, L. and Saarma, M. (1999) Biochemical and genetic evidence for interactions between potato A potyvirus‐encoded proteins P1 and P3 and proteins of the putative replicative complex. Virology, 263, 15–22. [DOI] [PubMed] [Google Scholar]
- Mestre, P. , Brigneti, G. and Baulcombe, D.C. (2000) An Ry‐mediated resistance response in potato requires the intact active site of the NIa proteinase from potato virus Y. Plant J. 23, 653–661. [DOI] [PubMed] [Google Scholar]
- Mestre, P. , Brigneti, G. , Durrant, M.C. and Baulcombe, D.C. (2003) Potato virus Y NIa protease activity is not sufficient for elicitation of Ry‐mediated disease resistance in potato. Plant J. 36, 755–761. [DOI] [PubMed] [Google Scholar]
- Montarry, J. , Doumayrou, J. , Simon, V. and Moury, B. (2011) Genetic background matters: a plant–virus gene‐for‐gene interaction is strongly influenced by genetic contexts. Mol. Plant Pathol. 12, 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarry, J. , Cartier, E. , Jacquemond, M. , Palloix, A. and Moury, B. (2012) Virus adaptation to quantitative plant resistance: erosion or breakdown? J. Evol. Biol. 25, 2242–2252. [DOI] [PubMed] [Google Scholar]
- Moury, B. (2010) A new lineage sheds light on the evolutionary history of Potato virus Y . Mol. Plant Pathol. 11, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury, B. and Simon, V. (2011) dN/dS‐based methods detect positive selection linked to trade‐offs between different fitness traits in the coat protein of Potato virus Y . Mol. Biol. Evol. 28, 2707–2717. [DOI] [PubMed] [Google Scholar]
- Moury, B. and Verdin, E. (2012) Viruses of pepper crops in the Mediterranean basin: a remarkable stasis. Adv. Virus Res. 84, 127–162. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Morel, C. , Johansen, E. and Jacquemond, M. (2002) Evidence for diversifying selection in Potato virus Y and in the coat protein of other potyviruses. J. Gen. Virol. 83, 2563–2573. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Morel, C. , Johansen, E. , Guilbaud, L. , Souche, S. , Ayme, V. , Caranta, C. , Palloix, A. and Jacquemond, M. (2004) Mutations in Potato virus Y genome‐linked protein determine virulence towards recessive resistances in Capsicum annuum and Lycopersicon hirsutum . Mol. Plant–Microbe Interact. 17, 322–329. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Desbiez, C. , Jacquemond, M. and Lecoq, H. (2006) Genetic diversity of plant virus populations: towards hypothesis testing in molecular epidemiology. Adv. Virus Res. 67, 49–87. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Caromel, B. , Johansen, E. , Simon, V. , Chauvin, L. , Jacquot, E. , Kerlan, C. and Lefebvre, V. (2011) The helper component proteinase cistron of Potato virus Y induces hypersensitivity and resistance in potato genotypes carrying dominant resistance genes on chromosome IV. Mol. Plant–Microbe Interact. 24, 787–797. [DOI] [PubMed] [Google Scholar]
- Murphy, J.F. , Rhoads, R.E. , Hunt, A.G. and Shaw, J.G. (1990) The VPg of tobacco etch virus RNA is the 49‐kDa proteinase or the N‐terminal 24‐kDa part of the proteinase. Virology, 178, 285–288. [DOI] [PubMed] [Google Scholar]
- Murphy, J.F. , Rychlik, W. , Rhoads, R.E. , Hunt, A.G. and Shaw, J.G. (1991) A tyrosine residue in the small nuclear inclusion protein of Tobacco vein mottling virus links the VPg to the viral RNA. J. Virol. 65, 511–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, X.Z. and Singh, R.P. (2003) Evolution of North American PVYNTN strain Tu 660 from local PVYN by mutation rather than recombination. Vir. Genes, 26, 39–47. [DOI] [PubMed] [Google Scholar]
- Nobrega, N.R. and Silberschmidt, K. (1944) Sobre una provavel variante do virus ‘Y’ da batatinha que tem a peculiaridade de provocar necroses em plantas de fumo. Arquiv. Inst. Biol. São Paulo 15, 307–330. [Google Scholar]
- Palloix, A. , Ayme, V. and Moury, B. (2009) Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol. 183, 190–199. [DOI] [PubMed] [Google Scholar]
- Parrella, G. , Ruffel, S. , Moretti, A. , Morel, C. , Palloix, A. and Caranta, C. (2002) Recessive resistance genes against potyviruses are localized in colinear genomic regions of the tomato (Lycopersicon spp.) and pepper (Capsicum spp.) genomes. Theor. Appl. Genet. 105, 855–861. [DOI] [PubMed] [Google Scholar]
- Patrick, R.M. and Browning, K.S. (2012) The eIF4F and eIFiso4F complexes of plants: an evolutionary perspective. Comp. Funct. Genomics. ID287814, 12 pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y.‐H. , Kadoury, D. , Gal‐On, A. , Huet, H. , Wang, Y. and Raccah, B. (1998) Mutations in the HC‐Pro gene of zucchini yellow mosaic potyvirus: effects on aphid transmission and binding to purified virions. J. Gen. Virol. 79, 897–904. [DOI] [PubMed] [Google Scholar]
- Pirone, T.P. and Blanc, S. (1996) Helper‐dependent vector transmission of plant viruses. Annu. Rev. Phytopathol. 34, 227–247. [DOI] [PubMed] [Google Scholar]
- Pruss, G. , Ge, X. , Shi, X.M. , Carrington, J.C. and Vance, V.B. (1997) Plant viral synergism: the potyviral genome encodes a broad‐range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell, 9, 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenouille, J. , Montarry, J. , Palloix, A. and Moury, B. (in press) Farther, slower, stronger: how the plant genetic background protects a major resistance gene from breakdown. Mol. Plant Pathol. 14, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantalainen, K.I. , Eskelin, K. , Tompa, P. and Makinen, K. (2011) Structural flexibility allows the functional diversity of potyvirus genome‐linked protein VPg. J. Virol. 85, 2449–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo, E. , Krause‐Sakate, R. , Yang, S.J. , Lot, H. , Le Gall, O. and Candresse, T. (2001) Lettuce mosaic virus pathogenicity determinants in susceptible and tolerant lettuce cultivars map to different regions of the viral genome. Mol. Plant–Microbe Interact. 14, 804–810. [DOI] [PubMed] [Google Scholar]
- Restrepo‐Hartwig, M.A. and Carrington, J.C. (1994) The tobacco etch potyvirus 6‐kilodalton protein is membrane associated and involved in viral replication. J. Virol. 68, 2388–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia, C. and Caranta, C. (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 11, 40–45. [DOI] [PubMed] [Google Scholar]
- Roberts, I.M. , Wang, D. , Findlay, K. and Maule, A.J. (1998) Ultrastructural and temporal observations of the potyvirus cylindrical inclusions (CIs) show that the CI protein acts transiently in aiding virus movement. Virology, 245, 173–181. [DOI] [PubMed] [Google Scholar]
- Rojas, M.R. , Zerbini, F.M. , Allison, R.F. , Gilbertson, R.L. and Lucas, W.J. (1997) Capsid protein and helper component‐proteinase function as potyvirus cell‐to‐cell movement proteins. Virology, 237, 283–295. [DOI] [PubMed] [Google Scholar]
- Rolland, M. , Lacroix, C. , Blanchard, A. , Baldwin, T. , Kerlan, C. and Jacquot, E. (2008) Potato virus Y (PVY): from its discovery to the latest outbreaks. Virologie, 12, 261–273. [DOI] [PubMed] [Google Scholar]
- Rolland, M. , Kerlan, C. and Jacquot, E. (2009) The acquisition of molecular determinants involved in Potato virus Y necrosis capacity leads to fitness reduction in tobacco plants. J. Gen. Virol. 90, 244–252. [DOI] [PubMed] [Google Scholar]
- Romero, A. , Blanco‐Urgoiti, B. , Soto, M. , Fereres, A. and Ponz, F. (2001) Characterization of typical pepper‐isolates of PVY reveals multiple pathotypes within a single genetic strain. Virus Res. 79, 71–80. [DOI] [PubMed] [Google Scholar]
- Ross, H. (1986) Potato breeding—problems and perspectives. J. Plant Breed, Suppl. 13, 132 pp. [Google Scholar]
- Rubio, M. , Nicolai, M. , Caranta, C. and Palloix, A. (2009) Allele mining in the pepper gene pool provided new complementation effects between pvr2‐eIF4E and pvr6‐eIF(iso)4E alleles for resistance to pepper veinal mottle virus. J. Gen. Virol. 90, 2808–2814. [DOI] [PubMed] [Google Scholar]
- Ruffel, S. , Dussault, M.H. , Palloix, A. , Moury, B. , Bendahmane, A. , Robaglia, C. and Caranta, C. (2002) A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32, 1067–1075. [DOI] [PubMed] [Google Scholar]
- Ruffel, S. , Gallois, J.L. , Lesage, M.L. and Caranta, C. (2005) The recessive potyvirus resistance gene pot‐1 is the tomato orthologue of the pepper pvr2‐eIF4E gene. Mol. Gen. Genomics, 274, 346–353. [DOI] [PubMed] [Google Scholar]
- Sacristán, S. and García‐Arenal, F. (2008) The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9, 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi, M.S. , Behjatnia, S.A.A. , Masumi, M. and Izadpanah, K. (2008) Characterisation of a strain of Potato virus Y causing eggplant mosaic in southern Iran. Australas. Plant Pathol. 37, 79–86. [Google Scholar]
- Sáenz, P. , Salvador, B. , Simon‐Mateo, C. , Kasschau, K.D. , Carrington, J.C. and García, J.A. (2002) Host‐specific involvement of the HC protein in the long‐distance movement of potyviruses. J. Virol. 76, 1922–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaman, R.N. and Le Pelley, R.H. (1930) Paracrinkle: a potato disease of the virus group. Proc. R. Soc. London, 106, 140–175. [Google Scholar]
- Schaad, M.C. , Jensen, P.E. and Carrington, J.C. (1997) Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum‐targeted viral protein. EMBO J. 16, 4049–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof, K.B. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. , Hohn, B. , Saunders, K. , Candresse, T. , Ahlquist, P. , Hemenway, C. and Foster, G. (2011) Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, J. , Fomitcheva, V. and Sztangret‐Wisniewska, J. (2007) Differentiation of Potato virus Y strains using improved sets of diagnostic PCR‐primers. J. Virol. Methods, 140, 66–74. [DOI] [PubMed] [Google Scholar]
- Segundo, E. , Lesemann, D.E. , Martín, G. , Carmona, M. , Ruiz, L. , Cuadrado, I.M. , Velasco, L. and Janssen, D. (2007) Amaranthus leaf mottle virus: 3'‐end RNA sequence proves classification as distinct virus and reveals affinities within the genus Potyvirus. Eur. J. Plant Pathol. 117, 81–87. [Google Scholar]
- Shiboleth, Y.M. , Haronsky, E. , Leibman, D. , Arazi, T. , Wassenegger, M. , Whitham, S.A. , Gaba, V. and Gal‐On, A. (2007) The conserved FRNK box in HC‐Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J. Virol. 81, 13 135–13 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R.P. , Valkonen, J.P.T. , Gray, S.M. , Boonham, N. , Jones, R.A.C. , Kerlan, C. and Schubert, J. (2008) Discussion paper: the naming of Potato virus Y strains infecting potato. Arch. Virol. 153, 1–13. [DOI] [PubMed] [Google Scholar]
- Smith, K.M. (1931) Composite nature of certain potato viruses of the mosaic group as revealed by the use of plant indicator. Proc. R. Soc. London, Ser. B: Biol. Sci. 109, 251–267. [Google Scholar]
- Solomon‐Blackburn, R.M. and Barker, H. (2001) A review of host major‐gene resistance to potato viruses X, Y, A and V in potato: genes, genetics and mapped locations. Heredity, 86, 8–16. [DOI] [PubMed] [Google Scholar]
- Song, Y.S. , Hepting, L. , Schweizer, G. , Hartl, L. , Wenzel, G. and Schwarzfischer, A. (2005) Mapping of extreme resistance to PVY (Ry sto) on chromosome XII using anther‐culture‐derived primary dihaploid potato lines. Theor. Appl. Genet. 111, 879–887. [DOI] [PubMed] [Google Scholar]
- Soumounou, Y. and Laliberté, J.‐F. (1994) Nucleic‐acid binding properties of the P1 protein of turnip mosaic potyvirus produced in Escherichia coli . J. Gen. Virol. 75, 2567–2573. [DOI] [PubMed] [Google Scholar]
- Sudarsono, Woloshuk, S.L. , Xiong, Z. , Hellmann, G.M. , Wernsman, E.A. , Weissinger, A.K. and Lommel, S.A. (1993) Nucleotide sequence of the capsid protein cistrons from six Potato virus Y (PVY) isolates infecting tobacco. Arch. Virol. 132, 161–170. [DOI] [PubMed] [Google Scholar]
- Tavert‐Roudet, G. , Abdul‐Razzak, A. , Doublet, B. , Walter, J. , Delaunay, T. , German‐Retana, S. , Michon, T. , Le Gall, O. and Candresse, T. (2012) The C terminus of lettuce mosaic potyvirus cylindrical inclusion helicase interacts with the viral VPg and with lettuce translation eukaryotic initiation factor 4E. J. Gen. Virol. 93, 184–193. [DOI] [PubMed] [Google Scholar]
- Tian, Y.‐P. and Valkonen, J.P.T. (2013) Genetic determinants of Potato virus Y required to overcome or trigger hypersensitive resistance to PVY strain group O controlled by the gene Ny in potato. Mol. Plant–Microbe Interact. 26, 297–205. [DOI] [PubMed] [Google Scholar]
- Torres‐Barcelo, C. , Martin, S. , Daros, J.‐A. and Elena, S.F. (2008) From hypo‐ to hypersuppression: effect of amino acid substitutions on the RNA‐silencing suppressor activity of the tobacco etch potyvirus HC‐Pro. Genetics, 180, 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribodet, M. , Glais, L. , Kerlan, C. and Jacquot, E. (2005) Characterization of Potato virus Y (PVY) molecular determinants involved in the vein necrosis symptom induced by PVYN isolates in infected Nicotiana tabacum cv. Xanthi. J. Gen. Virol. 86, 2101–2105. [DOI] [PubMed] [Google Scholar]
- Van der Zaag, D.E. (1987) Yield reduction in relation to virus infection In: Viruses of Potatoes and Seed‐Potato Production (de Bokx J.A. and van der Want J.P.H., eds), pp. 146–150. Wageningen: Pudoc. [Google Scholar]
- Varrelmann, M. , Maiss, E. , Pilot, R. and Palkovics, L. (2007) Use of pentapeptide‐insertion scanning mutagenesis for functional mapping of the plum pox virus helper component proteinase suppressor of gene silencing. J. Gen. Virol. 88, 1005–1015. [DOI] [PubMed] [Google Scholar]
- Verchot, J. and Carrington, J.C. (1995) Evidence that the potyvirus P1 proteinase functions in trans as an accessory factor for genome amplification. J. Virol. 69, 3668–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot, J. , Koonin, E.V. and Carrington, J.C. (1991) The 35‐kDa protein from the N‐terminus of the potyviral polyprotein functions as a 3rd virus‐encoded proteinase. Virology, 185, 527–535. [DOI] [PubMed] [Google Scholar]
- Wei, T. and Wang, A. (2008) Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI‐ and COPII‐dependent manner. J. Virol. 82, 12 252–12 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. , Huang, T.‐S. , McNeil, J. , Laliberté, J.‐F. , Hong, J. , Nelson, R.S. and Wang, A. (2010a) Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 84, 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. , Zhang, C. , Hong, J. , Xiong, R. , Kasschau, K.D. , Zhou, X. , Carrington, J.C. and Wang, A. (2010b) Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N‐PIPO. PLoS Pathog. 6, e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, S.S. , Chatel, H. , Fortin, M.G. and Laliberté, J.‐F. (1997) Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso)4E of Arabidopsis thaliana using the yeast two‐hybrid system. Virology, 234, 84–92. [DOI] [PubMed] [Google Scholar]
- Yambao, M.L.M. , Yagihashi, H. , Sekiguchi, H. , Sekiguchi, T. , Sasaki, T. , Sato, M. , Atsumi, G. , Takahashi, Y. , Nakahara, K.S. and Uyeda, I. (2008) Point mutations in helper component protease of clover yellow vein virus are associated with the attenuation of RNA‐silencing suppression activity and symptom expression in broad bean. Arch. Virol. 153, 105–115. [DOI] [PubMed] [Google Scholar]


