Summary
Sclerotinia sclerotiorum is a serious pathogen of numerous crops around the world. The major virulence factor of this pathogen is oxalic acid (OA). Mutants that cannot produce OA do not cause disease, and plants that express enzymes that degrade OA, such as oxalate oxidase (OxO), are very resistant to S. sclerotiorum. To examine the effect of OA on plants, we infiltrated soybean leaves with 5 mm OA and examined the gene expression changes at 2 h post‐infiltration. By comparing the gene expression levels between leaves of a transgenic soybean carrying an OxO gene (OxO) and its parent AC Colibri (AC) infiltrated with OA (pH 2.4) or water (pH 2.4 or 5.5), we were able to compare the effects of OA dependent or independent of its pH. Gene expression by microarray analysis identified 2390 genes that showed changes in expression, as determined using an overall F‐test P‐value cut‐off of 0.001. The additional requirement that at least one pairwise t‐test false discovery rate (FDR)‐corrected P value should be less than 0.001 reduced the list of the most highly significant differentially expressed genes to 1054. Independent of pH, OA altered the expression levels of 78 genes, with ferritin showing the strongest induction by OA. The combination of OA plus its low pH caused 1045 genes (99% of all significant genes) to be differentially expressed, with many of the up‐regulated genes being related to basal defence, such as genes of the phenylpropanoid pathway and various cytochrome P450s. RNA‐seq was also conducted on four samples: OxO and AC genotypes infiltrated with either OA pH 2.4 or water pH 2.4. The RNA‐seq analysis also identified ferritin paralogues as being strongly induced by OA. As the expression of ferritin, a gene that encodes for an iron storage protein, is induced by free iron, these results suggest that S. sclerotiorum benefits from the ability of OA to free iron from plant proteins, as this induces host cell death, and also allows the uptake and assimilation of the iron for its own metabolic needs.
Keywords: iron, leaf, oxalic acid, pathology, redox, Sclerotinia
Introduction
Oxalic acid (OA) is deemed to be the main virulence factor of the plant pathogen Sclerotinia sclerotiorum (Marciano et al., 1983). Mutants of S. sclerotiorum deficient in the production of OA were first identified in screens of mycelia grown from ascospores after UV irradiation (Godoy et al., 1990). These mutants are unable to invade plant tissue, although they produce and secrete a full range of degradative enzymes. The exact mutation of these strains has never been identified and it is unclear whether other genes involved in virulence are also affected because of the random nature of the mutagenesis method. More recently, another mutant was obtained by site‐directed mutagenesis of a single S. sclerotiorum gene, oxalate acetylhydrolase (Oah), a gene that catalyses the hydrolytic cleavage of oxaloacetate to acetate and OA (J. Rollins, University of Florida, Gainesville, FL, USA, personal communication). Like the UV‐induced OA‐deficient mutants, the Oah mutant is unable to either colonize healthy plant tissue or produce sclerotial bodies when growing on rich medium. The observation that Oah mutants can only infect a host via a wound, and that the spread of the mutant does not extend beyond the infection point, further confirms that OA should be classified as a virulence factor rather than a pathogenicity factor.
Although OA is the main virulence factor of S. sclerotiorum, its exact mechanism of action is not fully understood. Different studies have suggested that OA plays several roles, ranging from very obvious effects, such as acidification of the medium, to numerous subtle effects that aid the pathogen during infection and invasion. Marciano et al. (1983) proposed that OA chelates Ca2+ ions from pectin in the plant middle lamellae to facilitate the degradation of the plant cell wall by fungal pectin‐degrading enzymes. OA also creates an acidic environment favourable to activation and increased expression of various exo‐ and endopolygalacturonases (Favaron et al., 2004; Riou et al., 1991). Guimaraes and Stotz (2004) demonstrated that OA can act at the cellular level to regulate the opening of stomata by disrupting the abscisic acid (ABA)‐dependent stomatal closure, making the tissue more easily penetrable by fungal mycelia. Studies in soybean and Nicotiana tabacum have shown that OA can act independently of calcium to inhibit the defence‐associated oxidative burst, presum ably by restraining the production of hydrogen peroxide (Cessna et al., 2000).
Programmed cell death (PCD) can be a very effective plant defence mechanism against biotrophic pathogens that are generally race specific and able to elicit effector‐triggered immunity (ETI) (Jones and Dangl, 2006). In contrast, S. sclerotiorum is a non‐race‐specific necrotroph that would possibly benefit from PCD. Kim et al. (2008) provided evidence supporting the hypothesis that OA induces PCD, allowing S. sclerotiorum to use the dead and dying cells to facilitate infection of the tissue. More recently, the same group used a green fluorescent protein (GFP)‐tagged reporter construct (ro‐GFP) capable of changing its excitation state according to the oxidation state of the cell environment. The ro‐GFP reporter enabled a comparison of the oxidative states between leaves inoculated with either wild‐type S. sclerotiorum or an OA‐deficient mutant (Williams et al., 2011). The authors concluded that OA creates an initial reduced environment as early as 3 h after inoculation, which suppresses the oxidative burst, thus supporting the theory advocated by Cessna et al. (2000). Therefore, according to these studies, OA initially reduces the cell redox environment and suppresses the oxidative burst and reactive oxygen species (ROS) production, but later OA is involved in inducing ROS production and eliciting PCD. This scenario seems complex and it remains unclear as to the exact role of OA in these observed redox changes.
In the work presented here, we used transcriptome profiling with oligo microarrays and RNA‐seq to analyse the changes in mRNA levels in leaves following infiltration of an OA solution. Infiltrations were carried out on both a transgenic soybean line expressing oxalate oxidase (OxO) and its parental line AC Colibri (AC), in an attempt to distinguish the effects of OA from other S. sclerotiorum virulence factors. Our findings confirm that OA is capable of modulating the oxidative stress response of the plant, and that this modulation appears to be fostered by the release of iron ions, as reflected by the high induction of ferritin and iron‐associated genes.
Results
Microarrays
OxO transgenic soybean plants and parental AC plants were infiltrated with 5 mm OA (pH 2.4), acidified water (pH 2.4) or water more reflective of plant physiological acidity (pH 5.5), and sampled 2 h following infiltration to examine differential gene expression using soybean oligo‐based microarrays (Gonzalez and Vodkin, 2007). Pairwise hybridizations followed a loop design (Fig. 1). Analysis of the microarray data revealed a total of 2390 sequences which changed significantly in abundance between the transgenic and parent plants, or between treatments, at overall F‐test P values of <0.001 for genotype, treatment or genotype‐by‐treatment (Table S1, see Supporting Information). In addition to the F‐tests, 13 pairwise t‐tests between the least‐square means estimated from analysis of variance (ANOVA) were run to estimate the effect of the combination of both OA and its acidic pH, OA independent of its pH, transgene and acidity (summarized in Table 1, detailed in Table S1). The list of significantly differentially expressed genes, as determined by overall F‐test P values of <0.001, was further narrowed by selecting only genes that also had at least one specific pairwise t‐test false discovery rate (FDR)‐corrected P value of <0.001, giving a list of 1054 genes deemed to be the most highly significant (Table S2, see Supporting Information), and which serve as the focus of discussion for this article.
Figure 1.
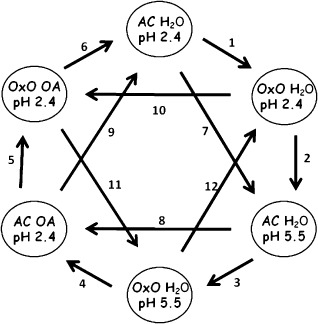
Microarray slide hybridization layout showing the design used in the oxalic acid infiltration experiment. Arrows represent oligo microarray slides with two hybridized samples, arrowheads indicate cyanine 5 (Cy5)‐labelled samples and the tails represent Cy3‐labelled samples; circles show the treated samples. AC, AC Colibri parental soybean line; OA, oxalic acid; OxO, transgenic soybean line expressing oxalate oxidase.
Table 1.
Pairwise t‐tests performed between the microarray experiment treatment factors. Each of the 13 tests aids in the understanding of one of the four possible effects. Detailed results are shown in Table S1
| Effect | Factor 1 | Factor 2 | (Factor 1/Factor 2) estimated value is: |
|---|---|---|---|
| Transgene | OxO | AC | Overall effect of the transgene |
| OxO_H2O_pH 5.5 | AC_H2O_pH 5.5 | Effect of the transgene on water at physiological pH | |
| OxO_H2O_pH 2.4 | AC_H2O_pH 2.4 | Effect of the transgene on infiltration of acidic water | |
| OxO_OA_pH 2.4 | AC_OA_pH 2.4 | Effect of the transgene on infiltration of OA | |
| pH | H2O_pH 2.4 | H2O_pH 5.5 | Overall effect of the pH change 2.4 vs. 5.5 |
| AC_H2O_pH 2.4 | AC_H2O_pH 5.5 | Effect of pH 2.4 vs. pH 5.5 on AC plants | |
| OxO_H2O_pH 2.4 | OxO_H2O_pH 5.5 | Effect of pH 2.4 vs. pH 5.5 on OxO plants | |
| OA independent of its pH | OA_pH 2.4 | H2O_pH 2.4 | Overall effect of OA infiltration independent of its pH |
| AC_OA_pH 2.4 | AC_H2O_pH 2.4 | Effect of OA infiltration in AC independent of OA pH | |
| OxO_OA_pH 2.4 | OxO_H2O_pH 2.4 | Effect of OA infiltration in OxO independent of OA pH | |
| OA and its pH | OA_pH 2.4 | H2O_pH 5.5 | Overall effect of OA infiltration |
| AC_OA_pH 2.4 | AC_H2O_pH 5.5 | Effect of OA infiltration in AC plants | |
| OxO_OA_pH 2.4 | OxO_H2O_pH 5.5 | Effect of OA infiltration in OxO plants |
AC, AC Colibri parental soybean line; OA, oxalic acid; OxO, transgenic soybean line expressing oxalate oxidase.
Effects of OA and its pH
Of the 1054 genes selected as being the most significant for this study, nearly all (1045) showed a change in expression as a result of a combination of OA and its low pH, as seen when comparing leaves infiltrated with OA pH 2.4 versus water pH 5.5 (Table S3, see Supporting Information). The vast majority of these changing genes (95.3%) showed the same direction of induction or repression in both plant genotypes, but AC showed stronger responses in general, reflective of a gradual OA removal by OxO enzyme activity in the transgenic line. The strongest effects of OA and its low pH were seen in AC; there were 46 oligos with at least three‐fold increased expression, which can be summarized as being ferritins, members of the phenylpropanoid pathway, cytochrome P450s, glutathione S‐transferases, peroxidases or pathogenesis‐related (PR) proteins. The oligos that decreased by one‐third or more in AC plants as a result of OA at pH 2.4 were related mainly to primary metabolism and redox, including several iron‐associated, redox‐related proteins, such as ferric reductase, iron superoxide dismutase and ferredoxin. Comparing the AC and OxO genotypes, of the 10 genes most reduced in AC versus OxO, six were unknowns or no hits, one was an aminotransferase, one was a ferric reductase and two were ferredoxins. The 10 oligos most induced in AC versus OxO reflected the activation of defences: three peroxidases, three no hits or unknowns, two to the same PR protein, one MLO10 and one β‐glucosidase. The observed results point to a defensive response being elicited by the combined effect of OA and its pH, in addition to alterations in plant iron homeostasis, as indicated by the strong induction of ferritin.
Effects of OA, independent of pH
By screening the fold‐change values in the comparisons between infiltrations with OA versus infiltrations with water at pH 2.4, we were able to identify genes that changed in expression as a result of OA independent of its acidic pH. Of the 1054 differentially regulated oligo spots selected for discussion, 78 showed a significant differential expression comparing OA infiltration versus water infiltration at pH 2.4 (Table 2). The direction of expression (induced or repressed) was the same for all 78 genes for both AC and OxO, but the strength of the differential expression tended to be stronger in AC, again presumably because of the removal of OA in the OxO‐expressing plants. Only three genes showed a stronger change in OxO than AC in response to OA versus water at pH 2.4: Glyma01g26840.2, an unknown (2.13‐ versus 1.08‐fold); Glyma10g32820.1, a protease inhibitor (2.60‐ versus 1.82‐fold); and Glyma10g44170.2, a homogentisate phytylprenyltransferase (1.95‐ versus 1.70‐fold). Notably, among the highest OA‐induced microarray spots were eight genes coding for ferritin or ferritin subunits. These eight oligo spots correspond to seven predicted coding sequences, i.e. Glyma models (the computationally predicted set of coding sequences in the assembled soybean genome available at http://www.phytozome.net/soybean.php; Schmutz et al., 2010). These data show that ferritin is induced by the infiltration of OA, independent of the pH change. Many of the other genes induced were consistent with a defensive/stress response, such as genes of the phenylpropanoid pathway, a protease inhibitor, peroxidases, ethylene related and cytochrome P450s. The genes showing the greatest down‐regulation in response to OA relative to water at pH 2.4 tended to be poorly characterized, but probably within chloroplast or mitochondria.
Table 2.
Differentially regulated genes as a result of oxalic acid (OA) infiltration independent of its pH. Comparisons between infiltrations of OA in leaves of OxO (the transgenic soybean expressing oxalate oxidase) and its parent AC (AC Colibri)
| Oligo array ID | Log2 ratio | Best match to Glyma models | Short annotation | ||
|---|---|---|---|---|---|
| OA/H2O pH 2.4 | |||||
| Overall | OxO | AC | |||
| GM18751P049N07 | 4.23 | 3.39 | 4.04 | Glyma18g43650.1 | Ferritin light chain |
| GM18681P049K09 | 4.00 | 2.98 | 4.20 | Glyma18g43650.1 | Ferritin |
| GM14987P040A11 | 3.49 | 2.72 | 3.62 | Glyma03g06420.2 | Ferritin 2 |
| GM05373P014P21 | 2.41 | 1.71 | 2.40 | Glyma11g35610.1 | Ferritin 2 |
| GM12232P032N16 | 2.32 | 1.74 | 2.28 | Glyma01g31300.1 | Ferritin 3 |
| GM18577P049G01 | 1.90 | 1.18 | 2.01 | Glyma14g06160.1 | Ferritin 4 |
| GM04737P013F09 | 1.79 | 1.42 | 1.91 | Glyma02g43040.2 | Ferritin 4 |
| GM14944P039O16 | 1.72 | 1.13 | 1.52 | Glyma18g02800.2 | Ferritin 3 |
| GM03715P010K19 | 1.40 | 1.60 | 1.65 | Glyma09g25470.4 | Putative acyl‐CoA synthetase |
| GM09947P026O11 | 1.35 | 1.38 | 0.87 | Glyma10g32820.1 | Protease inhibitor 2 |
| GM10142P027G14 | 1.15 | 0.75 | 1.13 | Glyma07g02180.2 | Long‐chain fatty acid CoA ligase |
| GM04776P013G24 | 1.06 | 0.69 | 1.24 | Glyma07g33780.2 | Caffeoyl‐CoA O‐methyltransferase |
| GM07411P020E19 | 1.01 | 0.96 | 1.29 | Glyma09g39390.1 | No hits |
| GM04726P013E22 | 0.97 | 0.82 | 1.39 | Glyma13g01080.2 | 4‐Coumarate:coenzyme A ligase |
| GM10325P027O05 | 0.90 | 0.89 | 1.06 | Glyma08g23560.2 | Hydroxycinnamoyl transferase |
| GM18849P050B09 | 0.89 | 0.69 | 0.98 | No match | Caffeic acid O‐methyltransferase |
| GM12430P033F22 | 0.87 | 1.09 | 0.11 | Glyma01g26840.2 | Unknown |
| GM38358P100O06 | 0.87 | 0.98 | 1.17 | No match | Phenylalanine ammonia‐lyase |
| GM14959P039P07 | 0.87 | 0.86 | 1.08 | Glyma10g06600.1 | Phenylalanine ammonia‐lyase |
| GM18945P050F09 | 0.86 | 0.68 | 0.83 | No match | Caffeic acid O‐methyltransferase |
| GM19041P050J09 | 0.86 | 0.71 | 0.95 | No match | Caffeic acid O‐methyltransferase |
| GM13565P036F05 | 0.85 | 0.83 | 1.19 | Glyma13g20800.1 | Phenylalanine ammonia‐lyase |
| GM35930P094J02 | 0.85 | 0.55 | 0.92 | Glyma13g06550.1 | Quercetin 3‐O‐glucoside‐6″‐O‐malonyltransferase |
| GM04128P011L24 | 0.84 | 0.69 | 0.97 | Glyma14g38580.1 | trans‐Cinnamate 4‐monooxygenase |
| GM01662P005F06 | 0.83 | 0.97 | 0.76 | Glyma10g44170.2 | Homogentisate phytylprenyltransferase |
| GM15500P041F20 | 0.83 | 1.08 | 1.27 | Glyma01g33150.1 | Putative cytochrome P450 |
| GM31871P083P23 | 0.81 | 0.65 | 1.50 | Glyma10g40750.1 | Unknown |
| GM04366P012F22 | 0.81 | 0.61 | 1.22 | Glyma11g15680.5 | Ascorbate peroxidase |
| GM06501P017O21 | 0.78 | 0.71 | 0.86 | Glyma13g20800.1 | Phenylalanine ammonia‐lyase |
| GM36437P095O05 | 0.78 | 0.10 | 1.26 | Glyma14g05350.1 | 1‐Aminocyclopropane‐1‐carboxylate oxidase |
| GM18406P048O22 | 0.77 | 0.48 | 1.24 | Glyma12g07780.3 | Ascorbate peroxidase 2 |
| GM19137P050N09 | 0.76 | 0.63 | 0.87 | No match | Caffeic acid O‐methyltransferase |
| GM14124P037M12 | 0.75 | 0.69 | 1.31 | Glyma01g32400.1 | Protein kinase |
| GM38070P100C06 | 0.75 | 0.82 | 1.14 | No match | Phenylalanine ammonia‐lyase |
| GM12369P033D09 | 0.75 | 0.85 | 1.47 | Glyma19g01470.1 | α‐Dioxygenase |
| GM38262P100K06 | 0.75 | 0.94 | 1.12 | No match | Phenylalanine ammonia‐lyase |
| GM19158P050O06 | 0.75 | 0.69 | 1.05 | No match | Phenylalanine ammonia‐lyase |
| GM07122P019I18 | 0.74 | 0.97 | 1.08 | Glyma10g06600.1 | Phenylalanine ammonia‐lyase |
| GM18660P049J12 | 0.73 | 0.84 | 1.01 | Glyma18g45260.1 | 2′‐Hydroxydihydrodaidzein reductase |
| GM38166P100G06 | 0.69 | 0.73 | 1.12 | No match | Phenylalanine ammonia‐lyase |
| GM18870P050C06 | 0.66 | 0.72 | 0.77 | No match | Phenylalanine ammonia‐lyase |
| GM19150P050N22 | 0.66 | 0.77 | 0.91 | No match | Cinnamoyl CoA dehydrogenase |
| GM19062P050K06 | 0.65 | 0.61 | 0.80 | No match | Phenylalanine ammonia‐lyase |
| GM18966P050G06 | 0.65 | 0.59 | 0.84 | No match | Phenylalanine ammonia‐lyase |
| GM04388P012G20 | 0.65 | 0.63 | 0.97 | Glyma10g40870.3 | Cinnamyl‐alcohol dehydrogenase |
| GM18958P050F22 | 0.63 | 0.76 | 0.81 | No match | Cinnamoyl CoA dehydrogenase |
| GM12707P034B11 | 0.60 | 0.57 | 0.87 | Glyma0169s00210.1 | Monodehydroascorbate reductase |
| GM03619P010G19 | 0.59 | 0.52 | 0.78 | Glyma01g41450.1 | Lipase class 3 |
| GM18862P050B22 | 0.59 | 0.62 | 0.84 | No match | Cinnamoyl CoA dehydrogenase |
| GM12288P032P24 | 0.57 | 0.71 | 0.93 | Glyma13g36110.1 | Putative cytochrome P450 |
| GM35875P094G19 | 0.57 | 0.59 | 0.99 | Glyma02g40550.1 | 2‐Hydroxyphytanoyl‐CoA lyase |
| GM31372P082L04 | 0.55 | 0.73 | 1.04 | Glyma12g01780.1 | Alcohol dehydrogenase |
| GM15220P040K04 | 0.48 | 0.36 | 0.58 | Glyma01g36890.1 | Dehydroquinate synthase |
| GM38242P100J10 | 0.47 | 0.40 | 0.84 | No match | Cinnamoyl CoA reductase |
| GM10980P029J12 | 0.44 | 0.72 | 0.87 | Glyma02g04120.2 | ATP citrate lyase b‐subunit |
| GM13944P037E24 | 0.44 | 0.52 | 0.85 | Glyma15g15020.1 | ATP citrate lyase a‐subunit |
| GM19054P050J22 | 0.43 | 0.55 | 0.72 | No match | Cinnamoyl CoA dehydrogenase |
| GM34640P091D08 | 0.39 | 0.41 | 1.04 | Glyma16g01710.1 | Zinc finger |
| GM19138P050N10 | 0.36 | 0.33 | 0.54 | No match | Cinnamoyl CoA reductase |
| GM05086P014D22 | 0.33 | 0.37 | 0.76 | Glyma02g11740.1 | No hits |
| GM04356P012F12 | 0.32 | 0.43 | 0.72 | Glyma06g19820.3 | Aldehyde dehydrogenase |
| GM03791P010N23 | 0.30 | 0.32 | 0.56 | Glyma18g50940.1 | frnE protein‐like |
| GM12611P033N11 | −0.16 | −0.46 | −0.73 | Glyma01g42360.1 | Unknown |
| GM07176P019K24 | −0.28 | −0.40 | −0.65 | Glyma05g02250.1 | Inositol 1,3,4‐trisphosphate 5/6‐kinase |
| GM07686P021A06 | −0.31 | −0.50 | −0.97 | Glyma04g01750.2 | Glyceraldehyde‐3‐phosphate dehydrogenase |
| GM11660P031F20 | −0.32 | −0.22 | −0.41 | Glyma07g13210.1 | No hits |
| GM12224P032N08 | −0.32 | −0.49 | −1.12 | Glyma03g42240.3 | Unknown |
| GM01627P005D19 | −0.35 | −0.27 | −0.81 | Glyma12g06920.2 | PPF‐1 protein |
| GM02011P006D19 | −0.38 | −0.62 | −1.00 | Glyma04g02100.1 | FtsH protease |
| GM07366P020C22 | −0.38 | −0.60 | −1.00 | Glyma20g12740.1 | Phosphate translocator |
| GM22715P060C11 | −0.42 | −0.44 | −1.12 | Glyma04g24430.1 | Unknown |
| GM17124P045J12 | −0.43 | −0.51 | −0.76 | Glyma18g06510.1 | Cinnamoyl CoA reductase |
| GM12235P032N19 | −0.47 | −0.46 | −1.47 | Glyma09g34410.1 | Protein kinase |
| GM07160P019K08 | −0.69 | −0.80 | −0.92 | Glyma12g06760.2 | Unknown |
| GM14246P038B14 | −1.12 | −0.66 | −0.69 | Glyma10g06620.1 | RNA‐binding protein |
| GM17482P046I10 | −1.12 | −0.86 | −1.26 | Glyma11g11350.3 | Unknown |
| GM27219P071O03 | −1.32 | −1.15 | −1.49 | Glyma11g11350.3 | Nodulin |
| GM25844P068E20 | −1.46 | −1.24 | −1.79 | Glyma12g03520.2 | Unknown |
Effects of the OxO transgene
The overall effect of the OxO transgene across the three treatments (OA, pH 2.4 and pH 5.5) was statistically low, and there were no genes that met the strict selection criteria of an overall F‐test P value of <0.001, as well as a t‐test FDR‐corrected P value of <0.001. Therefore, to identify the genes with the highest probability of fluctuating differently between OxO and AC in response to OA, we selected genes that had an F‐test P value of 0.001 for genotype‐by‐treatment effect, as well as a t‐test FDR‐corrected P‐value cut‐off of <0.05 (Table 3). It should be noted that, of the 12 genes identified using these criteria, all but two showed higher expression in AC than OxO, and eight genes showed at least a 1.5‐fold (log2 > 0.56) increase when AC was infiltrated with OA versus water at pH 2.4. None of these genes fluctuated in OxO. One gene, chloroplast ferredoxin, was expressed more strongly in OxO versus AC, but that was caused by the reduced expression (log2 = −1.096) in AC on OA treatment, with little effect (log2 = –0.244) on this gene in OxO. Genes induced in AC included (in addition to a few unknowns and no hits): a PR protein, regulator of gene silencing, NADPH isocitrate dehydrogenase, gibberellin 2‐oxidase and an R2R3 Myb transcription factor (Table 3).
Table 3.
Differentially regulated genes in OxO versus AC, determined by selecting oligo spots with an overall F‐test P value of <0.001 for genotype effect
| Oligo array ID | Genotype‐by‐treatment P valuea | OxO/AC OA_pH 2.4 | Best match to Glyma models | Short annotation | |
|---|---|---|---|---|---|
| FDR‐corrected P valueb | Log2 ratio | ||||
| GM09502P025L22 | 6.919E‐07 | 0.0133 | −1.40 | Glyma06g12890.3 | Pathogenesis‐related protein |
| GM04200P011O24 | 2.101E‐06 | 0.0126 | 1.16 | Glyma08g13360.1 | Chloroplast ferredoxin |
| GM36357P095K21 | 4.480E‐06 | 0.0050 | −0.78 | Glyma04g37040.1 | Calmodulin‐related protein |
| GM04742P013F14 | 1.011E‐05 | 0.0289 | −0.73 | Glyma02g40820.4 | NADPH‐specific isocitrate dehydrogenase |
| GM12430P033F22 | 4.652E‐05 | 0.0218 | 0.76 | Glyma01g26840.2 | Unknown, PAR domain |
| GM28544P075F08 | 5.330E‐05 | 0.0245 | −0.89 | Glyma03g02600.1 | No hits |
| GM06066P016M18 | 1.296E‐04 | 0.0033 | −0.42 | Glyma20g02040.1 | Serine carboxypeptidase S10 family |
| GM30894P081H06 | 1.429E‐04 | 0.0362 | −0.58 | Glyma17g11610.1 | No hits |
| GM11391P030K15 | 1.882E‐04 | 0.0075 | −1.01 | Glyma15g39750.1 | Gibberellin 2‐oxidase |
| GM01974P006C06 | 5.961E‐04 | 0.0075 | −1.10 | Glyma01g41290.2 | No hits |
| GM26086P068O22 | 6.299E‐04 | 0.0339 | −0.59 | Glyma07g06990.2 | Unknown, PAR domain |
| GM24080P063L08 | 9.244E‐04 | 0.0362 | −0.64 | Glyma12g32540.1 | Transcription factor Myb protein |
AC, AC Colibri parental soybean line; FDR, false discovery rate; OA, oxalic acid; OxO, transgenic soybean line expressing oxalate oxidase.
Overall genotype‐by‐treatment F‐test P value.
Pairwise t‐test FDR‐corrected P value.
Effects of pH on the plant response
Relative expression changes between the treatments involving water at pH 2.4 (pH of the OA solution) versus water at pH 5.5 (pH of the apoplast) can aid in the understanding of how plants respond to pH changes, specifically the acidification of the apoplast, as well as the role of the acidic nature of OA on pathogenesis. Seven genes made our stringent significance cut‐off (Table 4) and, in all cases, the OxO and AC lines showed the same direction of differential expression; AC showed a stronger expression level modulation for all seven genes, whether increasing or decreasing, than OxO. The most significant up‐regulated genes were two unknowns, a cytochrome P450 (possibly an isoflavone synthase) and a protein disulphide isomerase. Three of the genes significantly affected by pH were down‐regulated, and included a light‐responsive gene, a proline‐rich protein and a solanesyl diphosphate synthase.
Table 4.
Differentially regulated genes as a result of pH change. Comparisons between infiltrations with water at pH 2.4 and water at pH 5.5 in OxO (the transgenic soybean expressing oxalate oxidase) and its parent AC (AC Colibri)
| Oligo array ID | Ratio H2O_pH 2.4/H2O_pH 5.5 | Best match to Glyma models | Short annotation | ||
|---|---|---|---|---|---|
| Overall | OxO | AC | |||
| GM04281P012C09 | 1.05 | 0.72 | 1.11 | Glyma02g15430.1 | Unknown |
| GM04288P012C16 | 0.88 | −0.49 | 1.52 | Glyma10g36650.1 | Light stress responsive |
| GM06874P018O10 | 0.77 | 0.42 | 1.12 | Glyma06g46570.1 | Proline‐rich family |
| GM19147P050N19 | 0.58 | 0.52 | 1.08 | Glyma15g39070.1 | Solanesyl diphosphate synthase 1 |
| GM16856P044O08 | −0.61 | −0.17 | −0.91 | Glyma06g12090.3 | Protein disulphide isomerase |
| GM09369P025G09 | −0.83 | −0.15 | −1.02 | Glyma17g14620.1 | Unknown |
| GM07293P019P21 | −1.08 | −0.36 | −1.23 | No match | Cytochrome P450 |
Differentially expressed genes detected by RNA‐seq
To increase gene coverage and to verify and validate the microarray data, four of the same RNA samples used in the microarray study were deep sequenced for RNA species (RNA‐seq) via Solexa/Illumina technology. The four samples sequenced were individual replicates of OxO infiltrated with water at pH 2.4, OxO infiltrated with OA at pH 2.4, AC infiltrated with water at pH 2.4 and AC infiltrated with OA at pH 2.4. Each of the four libraries yielded between 30 and 42 million 100‐bp reads (Table 5). After quality checking and trimming, reads were aligned to the publicly available soybean genome. About 88% of the reads from each library were successfully aligned; 82% aligned uniquely on the genome and about 78% aligned uniquely to a predicted coding region (Table 5). A total of 39 885 coding sequences, representing 71% of the predicted genes in the soybean genome, were matched by at least one sequence read in at least one of the four libraries. All further discussion of these RNA‐seq data focuses on the set of reads uniquely aligned to predicted coding genes.
Table 5.
Summary of reads obtained by RNA‐seq analysis. Sequenced RNA from OxO (the transgenic soybean expressing oxalate oxidase) and its parent AC (AC Colibri) after vacuum infiltration treatments with water, pH adjusted to 2.4 or oxalic acid (OA) (pH 2.4)
| Library name | Soybean line | Total reads sequenced | Total number of uniquely aligned reads* |
|---|---|---|---|
| AC_H2O | AC‐Colibri parent line | 34 633 488 | 26 882 986 (77.6%) |
| AC_OA | AC‐Colibri parent line | 41 734 185 | 32 735 158 (78.4%) |
| OxO_H2O | 80(30)‐1 transgenic | 30 066 023 | 23 311 016 (77.5%) |
| OxO_OA | 80(30)‐1 transgenic | 31 037 297 | 24 306 468 (78.3%) |
*Reads were quality checked and trimmed; trimmed reads of less than 30 bp were removed and the remaining reads were aligned to the soybean genome. Only reads that uniquely mapped to coding sequences with less than two mismatches every 36 bp are reported. See Experimental procedures for more details.
Four comparisons were performed by calculating the fold change (log2 ratios of reads per kilobase per million, RPKM) between the different libraries for each of the reads, namely: OxO_H2O_pH 2.4 vs. AC_H2O_pH 2.4, OxO_OA vs. AC_OA, AC_OA vs. AC_H2O_pH 2.4, and OxO_OA vs. OxO_H2O_pH 2.4. The data were then filtered by selecting those reads having fold‐change differences greater or equal to eight, or smaller or equal to 1/8 (log2 of 3 or –3). Reads having less than 10 raw counts in all four libraries were removed from the analysis. A total of 936 predicted genes were selected as having the greatest changes in abundance between the plant lines or between the treatments (Table S4, see Supporting Information), and were classified into functional categories. Genes that changed in abundance the most were related directly to nucleic acids (DNA/RNA); of the 111 genes falling into this category, 96 were putative transcription factors, constituting 10% of all differentially regulated genes. The second largest category was signalling genes. Genes related to primary metabolism comprised 9.7% of the changing genes, whereas membrane and signalling‐related genes each accounted for 9% of the total. Many of these genes were related to upstream physiological events, suggesting that we were observing early responses to OA and acidity. Of the 936 differentially expressed genes selected, 630 showed up‐regulation in both AC and OxO lines infiltrated with OA, relative to the samples infiltrated with water, at pH 2.4. Once again, ferritin was one of the strongest genes induced by OA. Also, among this group of induced genes were 13 auxin‐related genes, 20 genes involved in secondary metabolism, including seven genes involved in terpenoid biosynthesis, and six genes in the lignin biosynthesis pathway. Eight peroxidases and six heavy metal transport genes were also induced in both samples. Only 49 genes were down‐regulated in both lines as a result of OA infiltration; among these repressed genes were several membrane transporters and disease resistance genes.
Cross‐platform comparison: microarray versus RNA‐seq
The RNA‐seq results confirmed and validated the differential expression of genes between the OxO transgenic line and its parent identified by the oligo microarray assays. Of the 39 885 reads from RNA‐seq that were successfully aligned to a predicted soybean coding sequence (Glyma model), 2158 matched one of the 2390 significantly differentially regulated sequences from the oligo‐based microarray study which assayed the same RNA samples (Table S1).
To assess the degree of correlation between the two platforms, the Pearson correlation coefficient (ρ) was calculated for each of the 2158 genes on corresponding comparisons between the log2 intensity ratios of the microarrays and the log2 RPKM ratios of the RNA‐seq data. A highly significant positive linear correlation (P < 0.001) was found in the four comparisons tested (Fig. 2). To verify whether the RNA‐seq method was introducing biases caused by transcript length in the set of differentially regulated genes identified in the microarrays, the differential regulation of the 2158 coding sequences was plotted as a function of their lengths. For this experiment, the RNA‐seq method tended to detect higher modulation than the microarrays for most of the differentially regulated transcripts, which fall in the range of 200–3000 bases in length. In turn, expression values for longer transcripts of 4000 or more bases showed little or no differences between the two platforms (Fig. S1, see Supporting Information).
Figure 2.
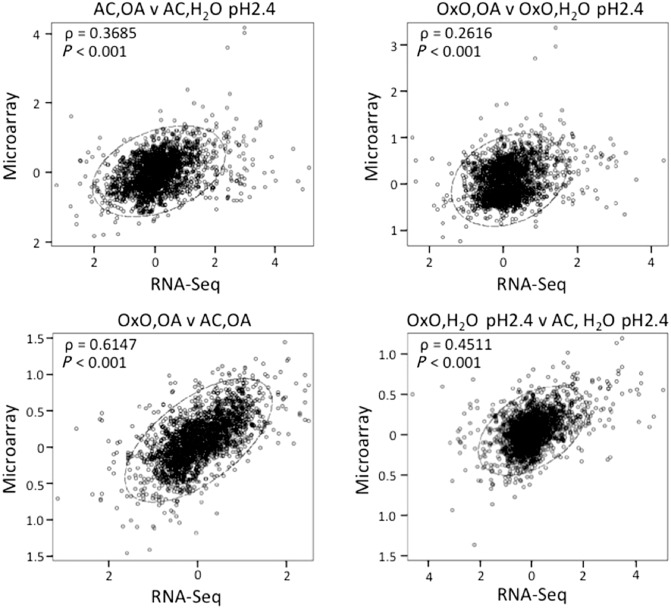
Correlation between microarray and RNA‐Seq data. y axes, microarray data as ratio of treatment over control; x axes, RNA‐seq data as ratio of treatment over control. The Pearson correlation coefficient (ρ) and its P value are given within each graph. AC, AC Colibri parental soybean line; OA, oxalic acid; OxO, transgenic soybean line expressing oxalate oxidase.
The 31 spots showing the strongest differential expression in the microarray experiment, selected as having a log2 fold change greater than 2 or lower than −2 (Table 6), were compared side by side with their best Glyma model match in the RNA‐seq data. Three of the microarray spots did not match with a Glyma model from the RNA‐seq data and, of the 28 remaining sequences, 21 showed the same direction of expression (Fig. 3). In addition, although the differential expression values of the microarray were lower in all cases, they were proportional in value to their RNA‐seq counterparts. Discrepancies between RNA‐seq and microarrays were found in seven sequences. For one of these sequences, the difference was consistent across the four treatments. For this case, the oligo sequence probably represented a different gene than that as the best match in the Glyma models. The other six discrepancies were non‐consistent across the treatments, and had extremely low differential expression values in the treatments disagreeing in the direction of expression (less than 0.5 up or down in either the microarray or RNA‐seq), perhaps making them more susceptible to analysis artefacts, such as microarray data normalization. In general, a good agreement was shown between microarray and RNA‐seq (especially for the stronger differentially expressed genes), and the data obtained by RNA‐seq served to validate and expand the microarray results.
Table 6.
Top 31 genes modulated in expression in OxO (the transgenic soybean expressing oxalate oxidase) and its parent AC (AC Colibri) after infiltration with oxalic acid, water at pH 5.5 or water at pH 2.4, based on the microarray results. These data were used in the comparison with the RNA‐seq results in Fig. 3
| Oligo array ID | Best match to Glyma models | Short annotation | DR | Comparison |
|---|---|---|---|---|
| GM18681P049K09 | Glyma18g43650.1 | Ferritin | UP | OA vs. H2O at pH 2.4 in OxO |
| GM18751P049N07 | Glyma18g43650.1 | Ferritin | UP | OA vs. H2O at pH 2.4 |
| GM14987P040A11 | Glyma03g06420.2 | Ferritin 2 | UP | OA vs. H2O at pH 2.4 in OxO |
| GM05373P014P21 | Glyma11g35610.1 | Ferritin 3 | UP | OA vs. H2O at pH 2.4 |
| GM12232P032N16 | Glyma01g31300.1 | Ferritin 2 | UP | OA vs. H2O at pH 2.4 |
| GM18577P049G01 | Glyma14g06160.1 | Ferritin 4 | UP | OA vs. H2O at pH 2.4 in OxO |
| GM37303P098C07 | Glyma01g36190.2 | Auxin related | UP | OA vs. H2O at pH 5.5 in AC |
| GM32514P085K18 | Glyma01g02580.1 | CAD | UP | OA vs. H2O at pH 5.5 in AC |
| GM35484P093G12 | Glyma02g43990.2 | Glucosidase | UP | OA vs. H2O at pH 5.5 in AC |
| GM37068P097I12 | Glyma08g18220.1 | No hits | UP | OA vs. H2O at pH 5.5 in AC |
| GM03538P010D10 | Glyma13g36110.1 | No hits | UP | OA vs. H2O at pH 5.5 in AC |
| GM18756P049N12 | Glyma11g01350.2 | CHS | UP | OA vs. H2O at pH 5.5 in AC |
| GM00967P003I07 | Glyma01g43880.1 | CHS | UP | OA vs. H2O at pH 5.5 |
| GM09502P025L22 | Glyma06g12890.3 | PR protein | UP | OA vs. H2O at pH 5.5 in AC |
| GM36533P096C05 | Glyma09g03100.1 | Electron carrier | UP | OA vs. H2O at pH 5.5 |
| GM37533P098L21 | Glyma18g44310.1 | Cationic peroxidase | UP | OA vs. H2O at pH 5.5 in AC |
| GM34930P091P10 | Glyma09g41450.1 | Peroxidase | UP | OA vs. H2O at pH 5.5 in AC |
| GM36297P095I09 | Glyma03g37390.1 | Pectinesterase | UP | OA vs. H2O at pH 5.5 |
| GM18021P047O21 | Glyma06g26610.1 | Blue copper‐binding protein | UP | OA vs. H2O at pH 5.5 |
| GM32575P085N07 | Glyma08g02160.1 | No hits | UP | OA vs. H2O at pH 5.5 |
| GM12095P032H23 | Glyma19g45030.1 | No hits | DOWN | OA vs. H2O at pH 5.5 in AC |
| GM34828P091L04 | Glyma08g28370.1 | CONSTANS‐like | DOWN | OA vs. H2O at pH 5.5 in AC |
| GM18471P049B15 | Glyma15g06270.1 | Unknown | DOWN | OA vs. H2O at pH 5.5 in AC |
| GM08939P024E11 | Glyma10g12370.1 | No hits | DOWN | OA vs. H2O at pH 5.5 in AC |
| GM22661P060A05 | Glyma07g08150.1 | Unknown | DOWN | OA vs. H2O at pH 5.5 in AC |
| GM18501P049C21 | Glyma16g10880.3 | Terpenoids | DOWN | OA vs. H2O at pH 5.5 in AC |
| GM17072P045H08 | Glyma08g41460.2 | Aminotransferase | DOWN | OA vs. H2O at pH 5.5 in AC |
| GM24040P063J16 | Glyma17g04650.1 | No hits | DOWN | OA vs. H2O at pH 5.5 in AC |
| GM07660P020P04 | Glyma14g03580.1 | No hits | DOWN | OA vs. H2O at pH 5.5 in AC |
| GM11550P031B06 | Glyma02g45170.1 | No hits | DOWN | OA vs. H2O at pH 5.5 in AC |
| GM23696P062L08 | Glyma11g21400.1 | No hits | DOWN | H2O at pH 2.4 vs. H2O at pH 5.5 |
CAD, cinnamyl‐alcohol dehydrogenase; CHS, chalcone synthase; DR, direction of differential expression; OA, oxalic acid; PR, pathogenesis related. Italic oligo ID did not have a match in the RNA‐seq experiment.
Figure 3.
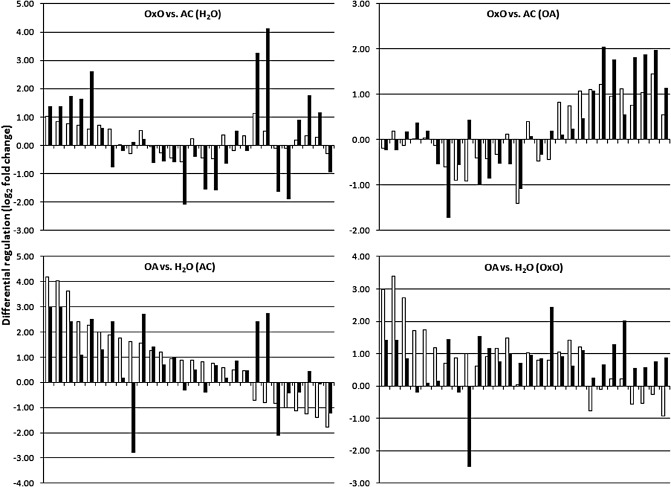
Comparison between values of differential expression detected by microarrays (open bars) and RNA‐seq (filled bars) for the top 28 differentially regulated oligo‐spots detected in the microarray experiment, maintained in the same order along the x axis across the four images. AC, AC Colibri parental soybean line; OA, oxalic acid; OxO, transgenic soybean line expressing oxalate oxidase.
Ferritin and iron homeostasis changes induced by OA as detected by RNA‐seq
The RNA‐seq data provided an increased coverage of the transcriptome, enabling further investigation of changes in iron homeostasis. Genes related to iron metabolism were selected from the RNA‐seq data using keyword searches, and a total of 103 sequences was found. Sequences with less than 10 counts in all the libraries were removed, leaving a total of 82 genes (Table S5, see Supporting Information). These iron‐related genes were classified according to their annotation in Phytozome v8.0. Nine groups were identified: vacuolar iron transporters (VIT), stabilizers of iron transporter (SufD), iron‐dependent oxygenases, iron–sulphur cluster proteins, iron‐regulated transporters, ferritins, iron reduction‐oxidases, iron superoxide dismutases and cytochrome b561. In addition to the ferritin genes that showed the highest expression changes, a cytochrome b561 showed very high induction in the OA‐infiltrated samples. Other iron‐related genes that were highly induced were AtNRAMP1 in AC treated with water and OxO treated with OA. Genes coding for iron–sulphur cluster‐containing proteins showed a general induction with some degree of difference between the four samples. Both OxO samples consistently showed more induction of ferric reductases/oxidases compared with the AC samples. An iron superoxide dismutase (FeSOD) was highly induced when acidic water was infiltrated into both genotypes.
Effect of pretreatment with iron on the infection ability of the OA‐deficient mutant
The gene expression results suggest that iron homeostasis is disturbed in the leaf by infiltration with OA. To test whether one possible benefit to S. sclerotiorum from freeing iron from the leaf would be to make this essential element available to itself, we infiltrated Arabidopsis thaliana leaves with solutions of either FeSO4 or FeCl3 (ferrous and ferric iron, respectively), and inoculated the leaves with an OA‐deficient S. sclerotiorum, an Oah– knockout mutant. All leaves infiltrated with 1% and 0.1% FeCl3 and/or with 1% FeSO4 turned necrotic and died within 3 h. Leaves infiltrated with 0.01% FeSO4 did not show any visible response. Leaves infiltrated with 0.01% FeCl3 or 0.1% FeSO4 showed similar responses: Oah–‐inoculated leaves showed agar plug attachment after 24 h, indicating tissue penetration by mycelia. The inoculum did not show any plug attachment on leaves without iron pretreatment (including on leaves infiltrated with water), indicating no penetration by mycelia. Curiously, for treatments that showed plug attachment (Oah– inoculation on 0.01% FeCl3‐ or 0.1% FeSO4‐infiltrated leaves), Oah–‐induced disease did not progress normally. Rather, a general necrosis spread through the leaf and, after 20 days, the leaves were dead. No further growth of mycelia was observed in the necrotized tissue, indicating that, although iron infiltration might have initially helped the pathogen to penetrate and infect the host, the host tissue managed to defend the plant by limiting infection (Fig. S2, see Supporting Information). Non‐inoculated control plants infiltrated with 0.01% FeCl3 or 0.1% FeSO4 solution only showed slight variations in chlorosis and no necrosis.
Discussion
Soybean oligo microarrays and RNA‐seq were utilized to analyse soybean leaf response to OA, the main virulence factor of S. sclerotiorum, a serious pathogen of many crops world‐wide. The microarrays surveyed the expression of over 38 000 genes, and identified 2390 spots as being significant at F‐test cut‐offs of 0.001. To focus only on the genes that were statistically the most robust, this gene list was narrowed to 1054 genes that also showed an FDR‐corrected P value of <0.001 in a treatment pairwise t‐test. The RNA‐seq analysis identified 936 genes that changed at least eight‐fold, and recovered many of the same genes as being differentially expressed, showing that the expression data are sound. However, some discrepancies were identified between the two methods used to study RNA levels. Unknown cross‐hybridization can be expected to contribute to error in the microarray analysis, but RNA‐seq procedures and data analysis are also not free of introduced biases. It has been suggested that the length of the transcripts may be a source of bias in RNA‐seq experiments, as the density of reads would not be uniform along the transcript; small sequencing errors would have more of an effect in shorter transcripts than in long ones, affecting multigene families (Fu et al., 2009; Marioni et al., 2008; Mortazavi et al., 2008). Oshlack and Wakefield (2009) hypothesized that the ability of RNA‐seq to detect a differentially regulated transcript depends on the length of the transcript, because this method, which uses RNA fragmentation, will give rise to more reads mapping to a longer transcript than to a shorter one. Microarrays, although not perfect, do not present this type of bias. In addition to a small bias against short‐length transcripts, RNA‐seq did not have the benefit of replication in this study, whereas our microarray experiment (Fig. 1) consisted of three biological replications and, each with four technical replications, enhancing the recovery of highly significant data.
Physiological studies have been reported on plant responses to OA (Cessna et al., 2000; Kim et al., 2008; Williams et al., 2011), but there have not been any reports on gene expression studies. The gene expression data collected here complement the physiological experiments, and suggest certain underlying mechanisms to plausibly explain some of the observed effects of OA. The expression data point to a disturbance of iron homeostasis. OA is a well‐known chelator of iron, and has even been implicated in robbing iron from our diet, if the food we eat is also high in OA (Bataille and Fournier, 2001).
Ferritin is a ubiquitous iron storage protein that uses multiple subunits to surround an estimated 4500 atoms of iron per ferritin shell (Harrison and Arosio, 1996). A biochemical study of the ability of various organic acids to remove iron from ferritin has been conducted (Macur et al., 1991), and has found that OA is very effective, being better than citric, tartaric, succinic and fumaric acids at releasing iron from purified ferritin. Considering its high affinity for iron, we hypothesize that OA from S. sclerotiorum causes the release of iron from cellular storage compartments, including redox proteins and enzymes, and that this benefits the pathogen by inducing cell death and making iron readily available for sequestration by this necrotrophic pathogen. That iron is being released from host storage by OA is further supported by the findings that the main inducer of ferritin transcription in plants is iron (Briat et al., 1999; Torti and Torti, 2002).
Iron that has been released from ferritin and/or other cellular components may exist as Fe(II) or Fe(III) ions, which are both very reactive, and thus able to generate ROS, which can damage nucleic acids, proteins and lipids. Therefore, it is essential that iron regulation is tightly controlled to minimize this toxic condition; ferritin is a key component limiting the extent and character of the oxidative stress that can occur in cells. Ferritin has enzymatic activity via ferroxidase, which converts ferrous iron [Fe(II)] to ferric iron [Fe(III)] to allow internalization and sequestration within the ferritin core. Ferritin also serves as an antioxidant by capturing ROS‐inducing iron, which protects cells from damage caused by excess iron (Lawen and Lane, 2013; Torti and Torti, 2002).
It has been shown previously, and our results support this finding, that, although OA exerts most of its function in a manner independent of its pH and its ability to acidify the environment (Coumo et al., 2005; Kim et al., 2008; Williams et al., 2011), low acidity favours the release of iron from ferritin (Laulhere and Briat, 1993; Macur et al., 1991). We noted what could be a synergistic effect of an acidic milieu and OA, as many more genes were significantly differentially expressed when comparing OA with water at pH 5.5 than with water at pH 2.4. RNA‐seq results showed that ferritin was induced in plants infiltrated with water at pH 2.4.
In addition to ferritin, other proteins in the cells require iron to function. Cytochromes are haem‐containing proteins which mostly use iron to perform their redox chemistry and electron transfer, and include enzymes such as cytochrome P450s, as well as components of chloroplast and mitochondrial electron transfer chains (Ross and Jakubowski, 2011). Restricting available iron from cytochromes would rapidly kill cells and greatly benefit this necrotrophic fungus.
The oxidative states of tomato leaves inoculated with either wild‐type S. sclerotiorum or an OA‐deficient mutant were investigated using the ro‐GFP reporter (Williams et al., 2011). The authors concluded that OA creates an initial reduced environment as early as 3 h after inoculation, which suppresses the oxidative burst. This result is also consistent with the theory advocated by Cessna et al. (2000). Therefore, according to these studies, OA initially reduces the cell redox environment and suppresses the oxidative burst and ROS production, but later OA is involved in inducing ROS production and eliciting PCD. It appears that, if OA removes and chelates iron from iron‐containing cellular proteins, this would lead to a reduced cellular environment. Higher ROS accumulation later could be the result of the absorption of light energy into the photosystem centres that have lost the capacity to pass that energy through the electron transfer chain (Allen et al., 1999) because of the release of iron.
Interestingly, although ferritin was strongly induced by OA infiltration, no ferritin genes were significantly induced in microarray experiments involving S. sclerotiorum‐infected plants (Calla et al., 2009, 2014), indicating that, if ferritin transcripts increase in abundance as a result of increased iron levels elicited by OA, in the presence of the fungus these levels are not increased within the cell to a sufficient extent to induce detectable changes in ferritin expression levels. This observation suggests that S. sclerotiorum is using OA to both release iron from the host and for rapid uptake by the pathogen. The effect of removing excess iron from infected cells would also reduce their toxicity to the pathogen, as it would minimize the induction of ROS, as free iron can react with H2O2 and produce the highly reactive and damaging hydroxyl radical (OH–) (Briat et al., 1999; Ravet et al., 2009). It would be very beneficial to the pathogen to use OA to induce cell death by the removal of iron from cytochromes, including the electron transfer chains, stopping/reducing electron flow through chloroplast and mitochondria, and then absorbing the OA‐chelated iron for its own metabolic needs during infection of the dead/dying cells. Our gene expression studies (this article and Calla et al., 2009, 2014) support this scenario.
The iron infiltration experiment showed that the provision of excess iron may complement the Oah– pathogen which lacks OA, as this mutant gains the ability to attach to the leaves, supporting the suggestion of a role for OA in assisting the acquisition of iron for the pathogen. That the fungus was unable to advance very far into the tissue further confirms that the plant cells need to be dead or dying prior to infection, and that, in the absence of OA, S. sclerotiorum is unable to complete this task. However, the available data are insufficient to prove this theory as the effect was minor, and plug attachment, indicative of initial tissue penetration by the pathogen, may be a result of unmeasured effects induced in the plant by iron. However, the unexpected necrosis observed on leaves that were both infiltrated and inoculated cannot be attributed to iron or pathogen alone. Leaves that were iron infiltrated but not inoculated, would also have died if iron toxicity were responsible for necrosis. However, the leaves did not die, indicating that the necrosis was in response to the pathogen, and that iron and pathogen together were needed to induce necrosis. How this occurs is a subject for future investigations.
Experimental Procedures
Plant growth and infiltration with OA
Seeds of the OxO transgenic soybean line 80(30)‐1 (Donaldson et al., 2001) and its parental line AC were placed in trays of damp vermiculite (Holiday, Toronto, ON, Canada) to germinate for 7 days in a growth chamber with the following conditions: 14‐h photoperiod, 26/24 °C day/night temperatures and 50% humidity. Illumination was provided by white fluorescent and incandescent bulbs at a light intensity of 300–400 μmol/m2/s and maintained about 30 cm from the top of the growing area. Seedlings were transplanted to 10.5‐cm‐diameter plastic pots (Kord Products, Brampton, ON, Canada) containing a blend of sterilized topsoil, sand and ProMix 3:2:1 (Ritchie Seed and Feed, Ottawa, ON, Canada) for an additional 8 days. Fertilizer (20.20.20 N.P.K., Plant Products Co. Ltd, Brampton, ON, Canada) was applied once a week after emergence of the first trifoliate. Prior to infiltration, the tops of the pots were covered with plastic wrap (Saran) and secured with elastic bands to prevent soil from dropping into the infiltration solutions. Plants with fully opened V2 trifoliates were subjected to vacuum infiltration by drawing a vacuum and then releasing after 2 min whilst the leaves were submerged in a 500‐mL solution of one of the following: 5 mm OA pH 2.4, H2O pH 5.5 or H2O pH 2.4 (pH adjusted with 1 m HCl). All solutions contained 0.005% Silwet L‐77 (Vac‐in‐stuff; Lehle Seeds, Round Rock, TX, USA) as a surfactant to enhance infiltration. One leaflet from each of trifoliates 1 and 2 was collected from the plants 2 h post‐infiltration, flash frozen in liquid nitrogen and stored at −80 °C. Samples from 12 AC or 80(30)‐1 OxO plants were bulked for each sample and RNA was isolated at a later date. The experiment was repeated three times to generate three biological/experimental replications.
RNA extraction, purification and quantification
Total RNA was isolated using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA). Tubes containing Phase Lock Gel™ (Brinkmann Instruments, Inc., Westbury, NY, USA) were used to improve the separation of the organic and inorganic phases. After extraction, RNA concentrations were estimated using a NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). For microarray experiments, RNA was further purified through Qiagen RNeasy columns (Qiagen, Valencia, CA, USA), and final quality was determined by a combination of spectrophotometry (NanoDrop ND‐1000) and gel electrophoresis (BioAnalyzer 2100, Agilent Technologies, Palo Alto, CA, USA).
Microarrays
Microarray labelling and hybridization procedures followed closely those described by Zou et al. (2005). RNA from three independent experiments was reverse transcribed into cDNA using SuperScript III Reverse Transcriptase (Invitrogen) in the presence of aminoallyl‐dUTP. Labelled cDNA was purified (Qiagen) and quantified (NanoDrop ND‐1000) to assess the adequate incorporation of the dye necessary for microarray analysis. Samples were mixed in pairs according to an interconnecting loop design (Fig. 1), suspended in hybridization buffer and applied onto the corresponding microarray slides. The oligo (70‐mer) slides representing approximately 38 000 soybean transcripts (Gonzalez and Vodkin, 2007) were incubated within a sealed hybridization chamber submerged in a water bath in the dark at 42 °C for 48 h. After incubation, a series of washes was performed on the slides to remove non‐binding probe (Zou et al., 2005), and median intensity values were determined by GenePix 5.0 (Axon, Milpitis, CA, USA) and normalized using maanova/R. Analysis of variance (ANOVA) was run on the normalized intensity values to assess the significance of the calculated differential expression, and the statistical model included the array as a random effect, and genotype and treatment as fixed effects (Calla et al., 2009).
RNA‐seq
Total RNA from the same samples as used in the OA infiltration microarray study was digested with DNAse I (Ambion, Austin, TX, USA) and purified with Qiagen RNeasy columns as described above. RNA samples were quantified and assessed for quality using a BioAnalyzer 2100 (Agilent Technologies). One microgram of each sample (i.e. OA pH 2.4 infiltration of OxO, OA pH 2.4 infiltration of AC, H2O pH 2.4 infiltration of OxO and H2O pH 2.4 infiltration of AC) was deep sequenced, using single end reads with the ‘sequencing‐by‐synthesis’ (SBS) technology at the W. M. Keck Center for Comparative and Functional Genomics, University of Illinois at Urbana‐Champaign, IL, USA. At the sequencing facility, cDNA libraries from each of the samples were constructed, quantified by quantitative polymerase chain reaction (qPCR), pooled in equimolar concentrations in pools of two samples, sequenced on eight lanes (two samples per lane) for 100 cycles on a HiSeq2000 using a TruSeq SBS sequencing kit version 2, and analysed with the Illumina Pipeline version 1.8 (Illumina, San Diego, CA, USA). An error rate using the phiX DNA was calculated as ∼1%. Raw reads of 100 bases were then processed by trimming off the adapters and aligning to the soybean genome using commercial services provided by Data2Bio (http://www.Data2Bio.com). In this pipeline, nucleotides of each raw read were scanned for low‐quality bases with a PHRED quality value of <15 (out of 40) and then aligned in the soybean genome (Schmutz et al., 2010) release 1.0 available in Phytozome v 7.0 (http://www.phytozome.net. Reads were normalized by calculating RPKM, which accounts for gene and library size. In addition, log2 ratios between reads on different libraries were calculated for the relative assessment of changes in abundance. Furthermore, the data were filtered for coding sequences having at least four counts in at least one of the four samples sequenced. Counts of value zero were changed to unity to allow for the calculation of approximate ratios.
Arabidopsis iron infiltration
Four‐week‐old Arabidopsis thaliana ecotype Columbia plants, grown in Sunshine Mix LC1 (SunGro, Vancouver, BC, Canada) with a 13‐h photoperiod and at 22/20 °C day/night temperatures, were hand infiltrated with 1%, 0.1% or 0.01% (w/v) solution of FeSO4 or FeCl3 on two individual leaves using a 1‐mL syringe without a needle. One of the infiltrated leaves was immediately inoculated with a 5‐mm‐diameter potato dextrose agar (PDA) plug containing an overnight, actively growing culture of the S. sclerotiorum Oah– mutant (kindly provided by J. Rollins). One non‐infiltrated leaf was also inoculated with the same mutant. In this manner, three treatments were applied to the same plant: infiltrated, infiltrated and inoculated, and inoculated only (without infiltration). Each of the six different iron treatments was repeated twice, for a total of 12 plants in the whole experiment. Immediately after inoculation, the plants were returned to the growth chamber where the humidity was raised to 100%.
Disclaimer
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Supporting information
Fig. S1 Effects of transcript length on the estimated differential expression by RNA‐seq compared with microarray. Longer transcripts showed higher agreement between the two platforms. AC, AC Colibri parental soybean line; OA, oxalic acid; OxO, transgenic soybean line expressing oxalate oxidase.
Fig. S2 Response of Arabidopsis thaliana to infiltration with 0.1% FeSO4 and/or inoculation with actively growing mycelia of Sclerotinia sclerotiorum Oah– mutant. Photographed 25 days after treatment immediately after leaves were detached. Three independent replications (plants) are shown.
Table S1 List of 2390 sequences significantly changing in abundance between the transgenic and parent plants, or between treatments, at overall F‐test P values of <0.001 for genotype, treatment or genotype‐by‐treatment.
Table S2 List of 1054 significantly differentially expressed genes as determined by overall F‐test P values of <0.001, and that also had at least one specific pairwise t‐test false discovery rate (FDR)‐corrected P value of <0.001
Table S3 List of 1045 genes that showed a change in expression as a result of a combination of oxalic acid (OA) and its low pH, as seen when comparing leaves infiltrated with OA pH 2.4 versus water at pH 5.5.
Table S4 List of the 936 predicted genes selected by RNA‐seq analysis as having the greatest changes in abundance between plant lines or between treatments by comparisons of fold change of reads per kilobase per million (RPKM). Data were filtered by selecting those reads having differences greater or equal to eight‐fold, or smaller or equal to 1/8‐fold (log2 of 3 or −3); reads having less than 10 raw counts in all four libraries were removed from the analysis.
Table S5 List of 103 genes related to iron metabolism selected from the RNA‐seq data using keyword searches.
Acknowledgements
We are grateful to Dr J. Rollins (University of Florida, Gainesville, FL, USA) for providing the Sclerotinia sclerotiorum Oah– mutant and freely sharing his helpful insights. The authors express gratitude for generous funding through the United States Department of Agriculture (USDA)‐Agricultural Research Services, USDA National Sclerotinia Initiative and Agriculture and Agri‐Food Canada.
Reproduced with the permission of the Minister of the Department of Agriculture and Agri‐Food, Government of Canada.
This article has been contributed to by U.S. Government employees and their work is in the public domain in the USA.
References
- Allen, L.J. , MacGregor, K.B. , Koop, R.S. , Bruce, D.H. , Karner, J. and Brown, A.W. (1999) The relationship between photosynthesis and a mastoparan‐induced hypersensitive response in isolated mesophyll cells. Plant Physiol. 119, 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille, P. and Fournier, A. (2001) Calcium supply in calcium lithiasis. Med. Nutr. 37, 9–12. [Google Scholar]
- Briat, J.F. , Lobréaux, S. , Grignon, N. and Vansuyt, G. (1999) Regulation of plant ferritin synthesis: how and why. Cell. Mol. Life Sci. 56, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calla, B. , Vuong, T. , Radwan, O. , Hartman, G.L. and Clough, S.J. (2009) Gene expression profiling soybean stem tissue early response to Sclerotinia sclerotiorum and in silico mapping in relation to resistance markers. Plant Genome, 2, 149–166. [Google Scholar]
- Calla, B. , Blahut‐Beatty, L. , Koziol, L. , Zhang, Y. , Neece, D.J. , Carbajulca, D. , Garcia, A. , Simmonds, D.H. and Clough, S.J. (2014) Genomic evaluation of oxalate‐degrading transgenic soybean in response to Sclerotinia sclerotiorum infection. Mol. Plant Pathol. in press. Available at 10.1111/mpp.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cessna, S.G. , Sears, V.E. , Dickman, M.B. and Low, P.S. (2000) Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell, 12, 2191–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumo, C. , Dickman, M.B. , Kohn, L. and Rolling, J. (2005) Sclerotinia Sclerotiorum Sequencing Project. Cambridge, MA: Broad Institute of Harvard and ; MIT. [Google Scholar]
- Donaldson, P.A. , Anderson, T. , Lane, B.G. , Davidson, A.L. and Simmonds, D.H. (2001) Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf‐2.8 (germin) gene are resistant to the oxalate‐secreting pathogen Sclerotinia sclerotiorum . Physiol. Mol. Plant Pathol. 59, 297–307. [Google Scholar]
- Favaron, F. , Sella, L. and D'Ovidio, R. (2004) Relationships among endo‐polygalacturonase, oxalate, pH, and plant polygalacturonase‐inhibiting protein (PGIP), in the interaction between Sclerotinia sclerotiorum and soybean. Mol. Plant–Microbe Interact. 17, 1402–1409. [DOI] [PubMed] [Google Scholar]
- Fu, X. , Fu, N. , Guo, S. , Yan, Z. , Xu, Y. , Hu, H. , Menzel, C. , Chen, W. , Li, Y. , Zeng, R. and Khaitovic, P. (2009) Estimating accuracy of RNA‐Seq and microarrays with proteomics. BMC Genomics, 10, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy, G. , Steadman, J.R. , Dickman, M.B. and Dam, R. (1990) Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris . Physiol. Mol. Plant Pathol. 37, 179–191. [Google Scholar]
- Gonzalez, D. and Vodkin, L. (2007) Specific elements of the glyoxylate pathway play a significant role in the functional transition of the soybean cotyledon during seedling development. BMC Genomics, 8, 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes, R.L. and Stotz, H.U. (2004) Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol. 136, 3703–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, P.M. and Arosio, P. (1996) The ferritins: molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta, 1275, 161–203. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kim, K.S. , Min, J.‐Y. and Dickman, M.B. (2008) Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant–Microbe Interact. 21, 605–612. [DOI] [PubMed] [Google Scholar]
- Laulhere, J.P. and Briat, J.F. (1993) Iron release and uptake by plant ferritin: effects of pH, reduction and chelation. Biochem. J. 290, 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawen, A. and Lane, D.J.R. (2013) Mammalian iron homeostasis in health and disease: uptake, storage, transport, and molecular mechanisms of action. Antioxid. Redox Signal. 18, 2473–2507. [DOI] [PubMed] [Google Scholar]
- Macur, R.E. , Olsen, R.A. and Inskeep, W.P. (1991) Photochemical mobilization of ferritin iron. Plant Soil, 130, 69–74. [Google Scholar]
- Marciano, P. , Di Lenna, P. and Magro, P. (1983) Oxalic acid, cell wall‐degrading enzymes and pH in pathogenesis and their significance in the virulence of two Sclerotinia sclerotiorum isolates on sunflower. Physiol. Plant Pathol. 22, 339–345. [Google Scholar]
- Marioni, J.C. , Mason, C.E. , Mane, S.M. , Stephens, M. and Gilad, Y. (2008) RNA‐seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18, 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi, A. , Williams, B.A. , McCue, K. , Schaeffer, L. and Wold, B. (2008) Mapping and quantifying mammalian transcriptomes by RNA‐Seq. Nat. Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Oshlack, A. and Wakefield, M. (2009) Transcript length bias in RNA‐seq data confounds systems biology. Biol. Direct, 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet, K. , Touraine, B. , Boucherez, J. , Briat, J.‐F. , Gaymard, F. and Cellier, F. (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J. 57, 400–412. [DOI] [PubMed] [Google Scholar]
- Riou, C. , Freyssinet, G. and Fevre, M. (1991) Production of cell wall‐degrading enzymes by the phytopathogenic fungus Sclerotinia sclerotiorum . Appl. Environ. Microbiol. 57, 1478–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, P.H. and Jakubowski, N. (2011) Cytochromes—fascinating molecular machines. Metallomics, 3, 316–318. [DOI] [PubMed] [Google Scholar]
- Schmutz, J. , Cannon, S.B. , Schlueter, J. , Ma, J. , Mitros, T. , Nelson, W. et al (2010) Genome sequence of the palaeopolyploid soybean. Nature, 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Torti, F.M. and Torti, S.V. (2002) Regulation of ferritin genes and protein. Blood, 99, 3505–3516. [DOI] [PubMed] [Google Scholar]
- Williams, B. , Kabbage, M. , Kim, H.‐J. , Britt, R. and Dickman, M.B. (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7, e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J. , Rodriguez‐Zas, S. , Aldea, M. , Li, M. , Zhu, J. , Gonzalez, D. et al (2005) Expression profiling soybean response to Pseudomonas syringae reveals new defense‐related genes and rapid HR‐specific downregulation of photosynthesis. Mol. Plant–Microbe Interact. 18, 1161–1174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Effects of transcript length on the estimated differential expression by RNA‐seq compared with microarray. Longer transcripts showed higher agreement between the two platforms. AC, AC Colibri parental soybean line; OA, oxalic acid; OxO, transgenic soybean line expressing oxalate oxidase.
Fig. S2 Response of Arabidopsis thaliana to infiltration with 0.1% FeSO4 and/or inoculation with actively growing mycelia of Sclerotinia sclerotiorum Oah– mutant. Photographed 25 days after treatment immediately after leaves were detached. Three independent replications (plants) are shown.
Table S1 List of 2390 sequences significantly changing in abundance between the transgenic and parent plants, or between treatments, at overall F‐test P values of <0.001 for genotype, treatment or genotype‐by‐treatment.
Table S2 List of 1054 significantly differentially expressed genes as determined by overall F‐test P values of <0.001, and that also had at least one specific pairwise t‐test false discovery rate (FDR)‐corrected P value of <0.001
Table S3 List of 1045 genes that showed a change in expression as a result of a combination of oxalic acid (OA) and its low pH, as seen when comparing leaves infiltrated with OA pH 2.4 versus water at pH 5.5.
Table S4 List of the 936 predicted genes selected by RNA‐seq analysis as having the greatest changes in abundance between plant lines or between treatments by comparisons of fold change of reads per kilobase per million (RPKM). Data were filtered by selecting those reads having differences greater or equal to eight‐fold, or smaller or equal to 1/8‐fold (log2 of 3 or −3); reads having less than 10 raw counts in all four libraries were removed from the analysis.
Table S5 List of 103 genes related to iron metabolism selected from the RNA‐seq data using keyword searches.


