Summary
Fimbrin is an actin‐bundling protein found in intestinal microvilli, hair cell stereocilia and fibroblast filopodia. Its homologue Sac6p has been shown to play a critical role in endocytosis and diverse cellular processes in Saccharomyces cerevisiae. FgFim from the wheat scab pathogenic fungus Fusarium graminearum strain Y2021A, which is highly resistant to the fungicide JS399‐19, was identified by screening a mutant library generated by HPH‐HSV‐tk cassette‐mediated integration. The functions of FgFim were evaluated by constructing a deletion mutant of FgFim, designated ΔFgFim ‐15. The deletion mutant exhibited a reduced rate of mycelial growth, reduced conidiation, delayed conidium germination, irregularly shaped hyphae, a lack of sexual reproduction on autoclaved wheat kernels and a dramatic decrease in resistance to JS399‐19. ΔFgFim ‐15 also exhibited increased sensitivity to diverse metal cations, to agents that induce osmotic stress and oxidative stress, and to agents that damage the cell membrane and cell wall. Pathogenicity assays showed that the virulence of the FgFim deletion mutant on flowering wheat heads was impaired, which was consistent with its reduced production of the toxin deoxynivalenol in host tissue. All of these defects were restored by genetic complementation of the mutant with the parental FgFim gene. Quantitative real‐time polymerase chain reaction (PCR) assays showed that the basal expression of three Cyp51 genes, which encode sterol 14α‐demethylase, was significantly lower in the mutant than in the parental strain. The results of this study indicate that FgFim plays a critical role in the regulation of resistance to JS399‐19 and in various cellular processes in F. graminearum.
Keywords: FgFim, Fungicide, Fusarium graminearum, JS399‐19 resistance
Introduction
Fusarium graminearum (teleomorph Gibberella zeae) is a causal agent of Fusarium head blight (FHB) or scab of wheat and barley, a disease that causes severe yield and economic losses in these crops worldwide (Bai and Shaner, 2004; Goswami and Kistler, 2004). From 1991 to 2001 across nine states in the USA, for example, the total economic impact from FHB was estimated to be $7.7 billion (Nganje et al., 2004). In addition to reducing grain yield and quality, the fungus produces harmful mycotoxins in infected grain; these mycotoxins, which include deoxynivalenol (DON), nivalenol (NIV) and zearalenones (ZEA), are a threat to human and animal health (Desjardins, 2006; Sutton, 1982). Because of the lack of type I resistance genes and the complexity of resistance identified in germplasm (Mesterhazy, 1995; Rudd et al., 2001), the most efficient strategy for the control of FHB is through the application of fungicides during wheat anthesis. Only a few fungicides (including benzimidazoles, triazoles and carboximides) have been registered for the control of FHB, and they typically reduce the FHB index by only ∼50% and the DON level by only ∼40% (Blandino et al., 2006). A novel cyanoacrylate fungicide, JS399‐19 (experimental code; a.i. 2‐cyano‐3‐amino‐3‐phenylancryic acetate), which was discovered and patented by the Jiangsu Branch of the National Pesticide Research and Development South Center of China, reduced both the FHB index and DON level by 80% (Chen and Zhou, 2009; Li et al., 2008; Zhang et al., 2010).
Although JS399‐19 strongly inhibited the mycelial growth of F. graminearum and provided excellent control of FHB in a field in which benzimidazole carbamate (MBC) was ineffective (Li et al., 2008), F. graminearum frequently developed resistance to JS399‐19 under selection pressure from the fungicide in laboratory trials (Chen et al., 2008; Chen and Zhou, 2009). It is important to clarify the resistance mechanism of F. graminearum to JS399‐19 so as to avoid unexpected control failures and to sustain the usefulness of the new product.
This article investigates the role of fimbrin in the resistance of F. graminearum to JS399‐19 and in other aspects of F. graminearum biology. Fimbrin is a cytoskeletal protein associated with microfilament core bundles in microvilli, microspikes, stereocilia, membrane ruffles and cell–substratum attachment sites. Fimbrin, which has been purified from intestinal epithelial cell brush borders, is a monomeric protein with a molecular mass of 68 000 (Bretscher, 1981; Glenney et al., 1981), and is conserved from yeast to humans (Adams et al., 1991). Fimbrin binds to F‐actin, and the binding is mediated by two pairs of calponin homology (CH) domains (Klein et al., 2004; Nakano et al., 2001). Saccharomyces cerevisiae fimbrin was isolated in a screen for suppressors of a temperature‐sensitive mutation in the actin gene, and named SAC6 (Adams and Botstein, 1989; Adams et al., 1989). Sac6p localizes to cortical actin patches and actin cables, and is important for actin organization, endocytosis and cell polarity in vivo (Adams et al., 1991; Drubin et al., 1988; Kubler and Riezman, 1993). In vitro studies suggest that fimbrin not only cross‐links F‐actin, but also provides rigidity to the actin bundle (Bretscher, 1981; Cheng et al., 1999). Similarly, in Schizosaccharomyces pombe, Fim1 co‐localizes to actin patches and the actin ring, but not within actin cables (Nakano et al., 2001; Wu et al., 2001). Fim1 deletion mutants exhibit partial loss of polarization, but also disorganized and mislocalized actin patches at restrictive temperature (Wu et al., 2001). In Aspergillus nidulans, FimA‐green fluorescent protein (GFP) localizes in a patch‐like pattern at the cell cortex; deletion of fimA resulted in polarity defects, particularly during spore germination, as well as in endocytosis defects (Upadhyay and Shaw, 2008). In Ashbya gossypii, AgSAC6 is important for polarized hyphal growth and endocytosis, but is dispensable for septation or vacuolar organelle movement (Jorde et al., 2011). Human T‐ and L‐plastin are not only able to complement the temperature‐sensitive growth defect in S. cerevisiae, but are also able to restore cell morphology, actin cytoskeleton organization and sporulation defects of the SAC6 null mutant (Adams et al., 1995).
Here, we identified F. graminearum fimbrin FgFim by screening a mutant library generated by HPH‐HSV‐tk cassette‐mediated integration in F. graminearum strain Y2021A on potato sucrose agar (PSA) amended with 15 or 75 μg/mL of JS399‐19 (Chen et al., 2008). Our results suggest that FgFim plays an important role in the resistance of F. graminearum to JS399‐19. We also show that FgFim is involved in asexual and sexual development, stress responses and virulence in F. graminearum.
Results
Identification of an F. graminearum insertion mutant with significantly reduced resistance to JS399‐19 and characterization of the FgFim gene
We generated a library of HHtCMI mutants from F. graminearum strain Y2021A; strain Y2021A is highly resistant to JS399‐19 and was generated by treating the wild‐type strain 2021 with JS399‐19 (Chen et al., 2007). We screened 4336 transformants and identified mutant S4‐13 for further analysis because of its significantly reduced resistance to JS399‐19.
Southern blot analysis and thermal asymmetric interlaced‐polymerase chain reaction (TAIL‐PCR) revealed that S4‐13 contained a single‐copy insertion of the HPH‐HSV‐tk cassette in locus FGSG_09862.3. Based on data from the Broad Institute (http://www.broadinstitute.org/annotation/genome/fusarium_group/MultiHome.html), this locus comprises five extrons and six introns spanning 2507 bp and encodes fimbrin. The encoded fimbrin contains 649 amino acids and four CH domains. Sequencing of the 1950‐bp cDNA of FgFim verified the existence of the six introns.
Phylogenetic analysis using the neighbour‐joining method (Kumar et al., 2008) showed that the amino acid sequence of FgFim is highly homologous to that from yeasts and other filamentous fungi. The deduced amino acid sequence of FgFim from F. graminearum shares 81% identity with FimA of A. nidulans (CBF70817.1) and 64% identity with Sac6p of S. cerevisiae (CAA88210.1) (Fig. S1A, see Supporting Information). The alignment of the amino acid sequences of FgFim with those from A. nidulans, A. gossypii and S. cerevisiae showed that the CH domains and actin‐binding domains of fimbrin are highly conserved from yeast to F. graminearum (Fig. S1B).
Deletion and complementation of FgFim in F. graminearum
To investigate the functions of FgFim, we generated targeted deletion mutants by transformation of the gene replacement cassette HPH‐HSV‐tk in the F. graminearum strains (Table 1). To describe the method, we use the parental strain Y2021A ΔFgFim as an example. Following purification by single‐spore isolation, 45 FgFim transformants were obtained, and six (3, 5, 9, 14, 15 and 18) were randomly selected for further validation. Putative deletion strains were examined by PCR amplification using different primer combinations to detect the integration of the left and right portion of the deletion cassette and the coding sequence replacement by the HPH‐HSV‐tk cassette (Fig. 1B). Reverse transcription‐polymerase chain reaction (RT‐PCR) analyses of RNA extracted from the mutants showed that they had completely lost the target transcript (Fig. 1C). Southern blot analysis using genomic DNA of the parental strain and mutants showed a profile consistent with the insertion of the HPH‐HSV‐tk cassette in the target locus by a double recombination event (Fig. 1D). One of the ΔFgFim mutants (ΔFgFim‐15) was complemented with the parental gene FgFim. The putative complementations were examined by RT‐PCR and Southern blot analysis (Fig. 1C,D).
Table 1.
Fusarium graminearum strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| 2021 | Wild‐type | Chen et al. (2009a) |
| ΔFgFim‐2021D23 | FgFim deletion mutant in 2021 genetic background | This study |
| ΔFgβ2‐tub‐2021D59 | β2 tub deletion mutant in 2021 genetic background | This study |
| Y2021A | Isolate resistant to JS399‐19 was generated from the wild‐type strain 2021 by fungicide treatment | Chen et al. (2007) |
| ΔFgFim‐15 | FgFim deletion mutant in Y2021A genetic background | This study |
| ΔFgβ2‐tub‐22 | β2 tub deletion mutant in Y2021A genetic background | This study |
| ΔFgβ2 tubΔFgFim‐39 | β2 tub and FgFim deletion mutant in Y2021A genetic background | This study |
| ΔFgβ2 tubΔFgFim‐46 | β2 tub and FgFim deletion mutant in Y2021A genetic background | This study |
| ΔFgFim‐15C | FgFim complement mutant in Y2021A genetic background | This study |
Figure 1.
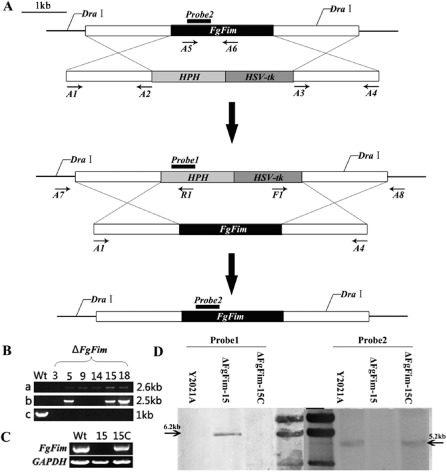
Generation and identification of Fusarium graminearum FgFim gene deletion mutants. (A) Gene replacement and complementation strategy for the FgFim gene. The gene replacement cassette HPH‐HSV‐tk contains the hygromycin resistance gene and the herpes simplex virus thymidine kinase gene. Primer binding sites are indicated by arrows (see Table S1 for the primer sequences). (B) Polymerase chain reaction (PCR) strategy to screen ΔFgFim transformants: a, PCR performed with primer pair A7/R1; a 2.6‐kb amplified fragment indicates ΔFgFim integration at the left junction; b, PCR performed with primer pair F1/A8; a 2.5‐kb amplified fragment indicates ΔFgFim integration at the right junction; c, PCR performed with primer pair A5/A6; a 1.0‐kb amplified fragment indicates a parental gene locus. (C) Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis for the expression of the FgFim gene in the parental isolate (Y2021A), the gene disruption mutant ΔFgFim ‐15 and the complemented transformant ΔFgFim ‐15 C using cDNA as template. GAPDH, glyceraldehyde‐3‐phosphate dehydrogenase. (D) Southern blot hybridization analysis of Y2021A, ΔFgFim ‐15 and ΔFgFim ‐15C using a 485‐bp hygromycin‐resistant gene (hph) fragment as probe1, a 460‐bp FgFim fragment as probe2, and genomic DNA digested with DraI.
Involvement of FgFim in hyphal growth and asexual development of F. graminearum
The FgFim deletion mutant grew significantly more slowly than the parental strain Y2021A (Table 2) and had a distinctive colony morphology on PSA plates (Fig. 2A). To determine whether the growth defects were medium dependent, we grew the FgFim deletion mutant on complete medium (CM) and minimal medium (MM). The colony defects observed on PSA plates were also observed on MM and CM plates (Fig. 2A). In addition, ΔFgFim‐15 exhibited reduced aerial hyphal growth on MM plates (Fig. 2A). Microscopic examination revealed that the hyphae of ΔFgFim‐15 were swollen and irregularly shaped when growing in yeast extract–peptone–dextrose (YEPD) medium (Fig. 2B). In mung bean liquid (MBL) medium, ΔFgFim‐15 produced significantly fewer conidia than the parental or the complemented strain (Table 2). In YEPD medium, only ∼20% conidia of ΔFgFim‐15 germinated within 6 h, but almost all conidia of the parental strain and of the complemented strain germinated under the same conditions (Table S2, see Supporting Information). When the incubation time was extended to 8 and 12 h, most of the conidia of ΔFgFim‐15 had germinated, but had formed short and unbranched germ tubes (Fig. 3), indicating that deletion of FgFim led to a delay in conidial germination and germ tube elongation. These results indicate that FgFim plays a role in hyphal growth and conidial differentiation and germination in F. graminearum.
Table 2.
Phenotypes of the FgFim deletion mutant (ΔFgFim ‐15), the parental strain (Y2021A) and the complemented strain (ΔFgFim ‐15 C) in terms of growth, conidiation and virulence*
| Strain | Growth rate on three media (mm/day)† | Conidia produced (×105/mL) | Percentage of diseased spikelets‡ | ||
|---|---|---|---|---|---|
| PSA | CM | MM | |||
| Y2021A | 25.3 ± 0.2a | 23.1 ± 0.3a | 22.4 ± 0.3a | 3.53 ± 0.38a | 26.46 ± 5.41a |
| ΔFgFim‐15 | 13.6 ± 0.3b | 11.4 ± 0.1b | 8.2 ± 0.3b | 0.63 ± 0.15b | 3.61 ± 1.40b |
| ΔFgFim‐15C | 25.7 ± 0.4a | 22.9 ± 0.4a | 22.0 ± 0.2a | 3.41 ± 0.24a | 27.23 ± 3.28a |
CM, complete medium; MM, minimal medium; PSA, potato sucrose agar.
*Values are means and standard deviations. Means in a column followed by the same letter are not significantly different (P > 0.05).
†Growth rate and conidiation were measured after incubation of three replicates for 3 and 5 days, respectively.
‡Percentage of diseased spikelets per spike, 15 days after inoculation. Thirty spikes were inoculated for each strain.
Figure 2.
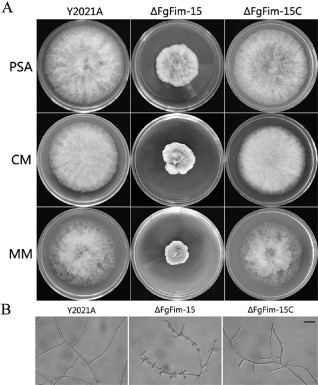
Effect of FgFim on Fusarium graminearum colony and hyphal morphology. (A) The parental strain Y2021A, deletion mutant ΔFgFim ‐15 and complemented strain ΔFgFim ‐15 C were grown on solid media [potato sucrose agar (PSA), complete medium (CM) and minimal medium (MM)] for 3 days at 25 °C. (B) Hyphal morphology of Y2021A, ΔFgFim ‐15 and ΔFgFim ‐15 C after 36 h in liquid yeast extract–peptone–dextrose (YEPD) medium. Bar, 40 μm.
Figure 3.
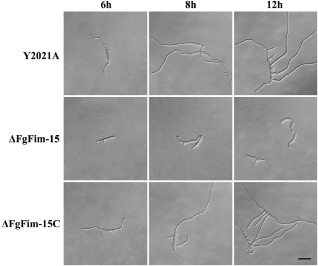
Conidia germination and germ tube elongation of the parental strain Y2021A, deletion mutant ΔFgFim ‐15 and complemented strain ΔFgFim ‐15 C after 6, 8 and 12 h in liquid yeast extract–peptone–dextrose (YEPD) medium. Bar, 40 μm.
FgFim affects resistance or sensitivity to JS399‐19 in F. graminearum
To confirm that the reduced resistance to JS399‐19 of mutant S4‐13 resulted from the disruption of FgFim, we performed fungicide resistance and sensitivity tests. As shown in Table 3, knockout of FgFim in strain Y2021A resulted in significantly reduced 50% effective concentration (EC50) and minimal inhibitory concentration (MIC) values for strain ΔFgFim‐15. Similarly, knockout of FgFim in strain 2021 resulted in significantly reduced EC50 and MIC values for strain ΔFgFim‐2021D23. In other words, knockout of FgFim reduced the resistant strain's resistance to JS399‐19 and increased the wild‐type strain's sensitivity to JS399‐19. The resistance of ΔFgFim‐15 was restored by complementation (Table 3). To exclude effects of growth defects, we disrupted the FgFim gene in strain ΔFgβ2‐tub‐22, which significantly reduced the growth rate, but did not significantly affect the resistant strain's resistance to JS399‐19. As indicated by growth on PSA amended with 50 or 100 μg/mL of JS399‐19, the resistance of ΔFgβ2‐tubΔFgFim‐39 and ΔFgβ2‐tubΔFgFim‐46 was significantly less than that of ΔFgβ2‐tub‐22 (Fig. 4). These results indicate that FgFim is involved in resistance or sensitivity to JS399‐19 in F. graminearum.
Table 3.
Resistance of F usarium graminearum strains to JS399‐19*
| Strain | Regression equation† | EC50 (μg/mL)‡ | MIC (μg/mL)§ |
|---|---|---|---|
| 2021 | Y = 1.779x + 5.992 | 0.277 | <4 |
| ΔFgFim‐2021D23 | Y = 3.781x + 8.716 | 0.104 | ≤0.4 |
| ΔFgβ2‐tub‐2021D59 | Y = 3.872x + 7.427 | 0.236 | <4 |
| Y2021A | Y = 1.677x + 1.119 | 206.122 | >1000 |
| ΔFgFim‐15 | Y = 1.676x + 2.128 | 51.724 | <100 |
| ΔFgFim‐15C | Y = 1.689x + 1.075 | 210.830 | >1000 |
| ΔFgβ2‐tub‐22 | Y = 2.644x – 1.201 | 221.409 | >1000 |
*Results are means of three experiments [differences among the experiments were not significant, i.e. P > 0.05, Fisher's least significant difference (LSD) test].
†Y refers to the percentage of inhibition and x refers to the logarithm of concentration (μg/mL) of JS399‐19.
‡Fungicide concentration that resulted in 50% mycelial growth inhibition.
§Minimal inhibitory concentration of JS399‐19 to Fusarium graminearum.
Figure 4.
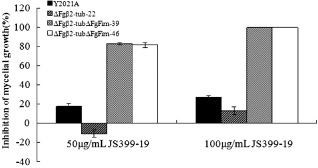
JS399‐19 inhibition of mycelial growth of the parental strain Y2021A, the β2 microtubule deletion mutant ΔFgβ2‐tub‐22 and the Fgβ2‐tub/FgFim double deletion mutants ΔFgβ2tubΔFgFim ‐39 and ΔFgβ2tubΔFgFim ‐46 on potato sucrose agar (PSA) medium. The PSA medium contained 0 (control), 50 or 100 μg/mL JS399‐19, and inhibition is expressed relative to the control. Values are means ± standard error (SE) of three repeated experiments.
Sensitivity of the FgFim deletion mutant to metal cations, to agents that damage the cell wall and cell membrane, and to agents that induce osmotic and oxidative stress
Because fimbrin is a Ca2+‐binding protein (Glenney et al., 1981) and the Sac6 deletion mutant decreases the resistance of Saccharomyces to diverse metals according to the Genome Database (http://www.yeastgenome.org/), we investigated the sensitivity of the FgFim deletion mutant to several metal cations, including calcium, lithium and zinc. As shown in Fig. 5 and Fig. S3 ΔFgFim‐15 exhibited a significantly increased sensitivity to LiCl, CaCl2 and ZnCl2. Ruotolo et al. (2008) suggested that membrane transporters and protein traffic networks could affect multi‐metal tolerance in yeast, and that changes in membrane permeability and fluidity or cell wall integrity might interfere with these membrane transporters and thereby reduce metal tolerance. To investigate the role of the cell membrane and cell wall in the increased sensitivity to metals in ΔFgFim‐15, we determined the sensitivity of ΔFgFim‐15 to the cell membrane‐damaging agent sodium dodecylsulphate (SDS) and to the cell wall‐damaging agents Congo red and caffeine. Compared with the parental strain and the complemented strain, ΔFgFim‐15 displayed increased sensitivity to these compounds (Fig. 6A, Fig. S2). To further confirm the involvement of FgFim in membrane permeability or cell wall integrity, we quantified the expression of the following six genes in F. graminearum: FgMkk1 (FGSG_07295), FgSlt2 (FGSG_10313), FgGls2 (FGSG_07946), Cyp51A, Cyp51B and Cyp51C. FgMkk1 and FgSlt2 are homologous to the S. cerevisiae cell wall integrity core element genes, Mkk1 and Slt2, respectively. FgGls2 encodes 1,3‐β‐glucan synthase. Cyp51A, Cyp51B and Cyp51C are involved in the biosynthesis of ergosterol, which regulates cell membrane fluidity and the permeability of fungal cells, and is essential for cell survival (Rodriguez et al., 1985). As shown in Fig. 6B, the expression levels of the six genes were significantly down‐regulated in ΔFgFim‐15 relative to the parental strain. These results further indicate that FgFim is associated with membrane permeability and cell wall integrity.
Figure 5.
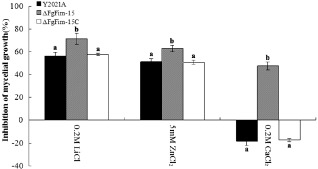
Metal cation inhibition of the parental strain Y2021A, deletion mutant ΔFgFim ‐15 and complemented strain ΔFgFim ‐15 C on potato sucrose agar (PSA) medium. PSA was not amended with metal cations (control) or was amended with the concentration indicated. Inhibition is expressed relative to the control. The different lowercase letters above the bars indicate a significant difference (P > 0.05). Values are means ± standard error (SE) of three repeated experiments.
Figure 6.
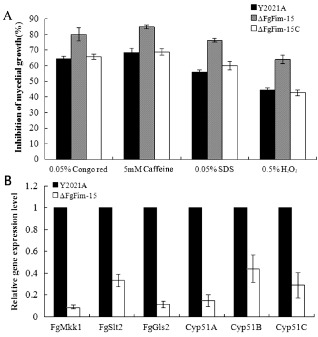
Effects of FgFim on the sensitivity of Fusarium graminearum strains to agents that generate oxidative stress or that damage cell membranes and cell walls, and on the expression of genes. (A) Sensitivity of the parental strain Y2021A, deletion mutant ΔFgFim ‐15 and complemented strain ΔFgFim ‐15 C to cell wall‐damaging agents (Congo red and caffeine), a cell membrane‐damaging agent (sodium dodecylsulphate, SDS) and an oxidative stressor (H2O2). (B) Expression levels of genes associated with cell membrane permeability or cell wall integrity (FgSlt1, FgMkk1, FgGls2, Cyp51A, Cyp51B and Cyp51C) in the mutant ΔFgFim ‐15 relative to that in the parental strain Y2021A. Values are the means ± standard error (SE) of three repeated experiments.
To investigate the role of FgFim in osmoregulation and oxidative stresses, we incubated ΔFgFim‐15, Y2021A and ΔFgFim‐15C on PSA amended with 1.2 m NaCl or KCl to generate osmotic stress and with 0.5% H2O2 to generate oxidative stress. As shown in Fig. 7A and Fig. S3, ΔFgFim‐15 exhibited increased sensitivity to 1.2 m NaCl, but reduced sensitivity to 1.2 m KCl. Previous studies have shown that intercellular glycerol plays an important role in the response of fungi to osmotic stress (Gustin et al., 1998; Wojda et al., 2003). Therefore, we quantified intercellular glycerol in mycelia of the deletion mutant (ΔFgFim‐15), the parental strain (Y2021A) and the complementary strain (ΔFgFim‐15C) after 2 h of treatment with NaCl and KCl. Glycerol levels were higher with 1.2 m NaCl than with 1.2 m KCl for all three strains, but the difference was statistically significant only for ΔFgFim‐15 (Fig. 7B). The sensitivity to oxidative stress generated by 0.5% H2O2 in PSA medium was greater in ΔFgFim‐15 than in the parental strain and complementary strain (Fig. 6A, Fig. S2).
Figure 7.
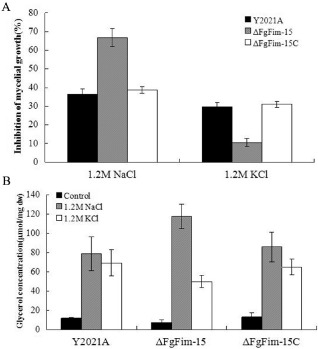
Effects of FgFim on the sensitivity to osmotic stresses. (A) Inhibition of mycelial growth of the parental strain Y2021A, deletion mutant ΔFgFim ‐15 and complemented strain ΔFgFim ‐15 C by the addition of 1.2 m NaCl or 1.2 m KCl to potato sucrose agar (PSA); inhibition is expressed relative to growth on PSA without added NaCl or KCl. (B) Intracellular glycerol concentration (μmol/mg of dried mycelia) in mycelia of the parental strain Y2021A, deletion mutant ΔFgFim ‐15 and complemented strain ΔFgFim ‐15 C after treatment with 1.2 m NaCl or 1.2 m KCl for 2 h. The untreated mycelia were used as controls. Values are the means ± standard error (SE) of three repeated experiments.
FgFim is required for sexual development and full virulence of F. graminearum
The production of perithecia on autoclaved wheat kernels was determined for ΔFgFim‐15, Y2021A and ΔFgFim‐15C. As a homothallic fungus, both the parental strain Y2021A and the complementary strain ΔFgFim‐15C alone produced abundant perithecia after 15 days. However, the FgFim mutant ΔFgFim‐15 failed to produce any perithecia under selfing conditions (Fig. 8). The absence of perithecia indicated that autonomous sexual reproduction was abolished in the FgFim deletion mutant.
Figure 8.
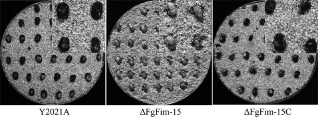
Formation of perithecia by strains Y2021A, ΔFgFim ‐15 and ΔFgFim ‐15 C on autoclaved wheat kernels. After the fungal strains had been incubated on autoclaved wheat kernels for 10 days, the kernels were transferred to sterile wet sand and incubated in a humid room [relative humidity (RH), 80%] at 25 °C and with a 12 h : 12 h light : dark photoperiod for 15 days to induce the formation of perithecia.
The virulence of the deletion mutant ΔFgFim‐15 was compared with that of the parental strain (Y2021A) and the complemented strain (ΔFgFim‐15C) by inoculating spikelets of the wheat cultivar Zhenmai 5 with macroconidia. The parental strain and complemented strain induced typical symptoms by 3 days post‐inoculation (dpi), and the disease had spread to the adjacent spikelets by 15 dpi (Fig. 9). The deletion mutant, in contrast, induced disease symptoms only at the point of inoculation and had not spread by 15 dpi (Fig. 9). At 15 dpi, the percentage of spikelets with symptoms was significantly lower with ΔFgFim‐15 than with the parental strain or the complemented strain (Table 2). These results indicate that the disruption of FgFim reduces the virulence of F. graminearum on wheat.
Figure 9.

Virulence of the parental strain Y2021A, deletion mutant ΔFgFim ‐15 and complemented strain ΔFgFim ‐15C on wheat heads. Each floret of wheat cultivar Zhenmai 5 was injected with 10 μL of a conidial suspension (5 × 105/mL) and maintained in a humid glasshouse. Wheat heads were photographed after incubation for 15 days.
FgFim affects TRI5 and TRI6 expression and DON biosynthesis
Trichothecene mycotoxins are sesquiterpenes and secondary metabolites of F. graminearum strains, and the trichothecene mycotoxin DON is an important virulence factor in F. graminearum (Desjardins et al., 1996; Proctor et al., 1995; Seong et al., 2009). Because deletion of FgFim reduced the virulence of F. graminearum on wheat, we determined how deletion of FgFim affected DON production. Compared with the parental strain and the complemented strain, ΔFgFim‐15 produced significantly less DON (Fig. 10A). To expand on these results, we analysed the expression levels of two trichothecene biosynthesis genes, TRI5 and TRI6, by quantitative real‐time PCR using RNA samples isolated from mycelia grown in glucose–yeast extract–peptone (GYEP) medium. The expression levels of TRI5 and TRI6 were substantially lower in ΔFgFim‐15 than in the parental strain (Fig. 10B). These results indicate that FgFim may indirectly affect DON biosynthesis in F. graminearum.
Figure 10.
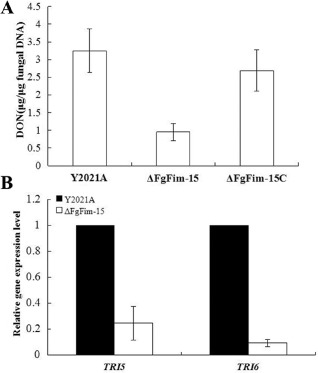
Deoxynivalenol (DON) production by the parental strain Y2021A, deletion mutant ΔFgFim ‐15 and complemented strain ΔFgFim ‐15C, and TRI5 and TRI6 expression by the parental strain and deletion mutant. (A) DON production (μg/μg of fungal DNA) by the parental strain, deletion mutant and complemented strain in infected wheat kernels. (B) Expression level of TIR5 and TRI6 in ΔFgFim ‐15 relative to that in Y2021A. Values are the means ± standard error (SE) of three repeated experiments.
Discussion
JS399‐19 is a Fusarium‐specific fungicide that controls FHB. According to the results of previous studies, resistance to JS399‐19 in F. graminearum is governed by a single major gene, and there is no cross‐resistance between JS399‐19 and well‐known fungicides belonging to other chemical classes, such as ergosterol biosynthesis inhibitors (tebuconazole and prochloraz) and strobilurins (azoxystrobin), suggesting that the mode of action and resistance mechanisms of JS399‐19 are different from those of other fungicides (Chen et al., 2008, 2009b; Li et al., 2008). In this article, disruption of FgFim, which was identified by screening a library of HHtCMI mutants, severely decreased the resistance to JS399‐19 (Table 3). However, we did not find any mutation in the FgFim gene of strain Y2021A relative to the FgFim gene of the wild‐type strain 2021 (data not shown), indicating that the high level of resistance to JS399‐19 in strain Y2021A could not be explained by a mutation in the FgFim gene. However, the expression level of FgFim in strain Y2021A relative to that in wild‐type 2021 differs over time. Although the expression level of another conserved gene β 2‐tub shows the same trend (Fig. S4, see Supporting Information), knockout of β 2‐tub did not significantly reduce the resistant strain's resistance to JS399‐19 and increased the wild‐type strain's sensitivity to JS399‐19 (Table 3). Previous studies have shown that deletion of SCP1 in yeast lacking the actin‐bundling protein, fimbrin (Sac6p), enhances sensitivity to the actin‐depolymerizing drug latrunculin‐A (Winder et al., 2003). In addition, Deoara et al. (2008), who studied the oomycete Aphanomyces cochlioides, reported that treatment with latrunculin‐B caused abnormal morphology and swelling of germ tubes from encysted zoospores, which correlated with defects in actin organization. Our previous study has shown that mycelium treated with JS399‐19 is distorted and swollen (Hou et al., 2013). Therefore, we suspect that JS399‐19 might work by disrupting actin organization in F. graminearum. To further determine the relationship between FgFim and JS399‐19, we studied the functions of FgFim in F. graminearum.
The results of this study demonstrate a critical role for FgFim in hyphal growth and conidiation. Phenotypes of ΔFgFim‐15 include a reduced mycelial growth rate, reduced conidiation, delayed conidium germination and irregularly shaped hyphae with an abnormal branching pattern. In addition, ΔFgFim‐15 exhibited no sexual reproduction on autoclaved wheat kernels. That disruption of the FgFim gene interfered with both asexual growth and sexual reproduction in F. graminearum indicates the important role of FgFim in proliferation and cytokinesis in yeast and other fungi (Adams et al., 1991; Jorde et al., 2011; Skau et al., 2011; Skau and Kovar, 2010; Upadhyay and Shaw, 2008). Furthermore, ascospores from perithecia of F. graminearum are responsible for the primary infection of wheat spikes during wheat flowering, i.e. infection by ascospores results in FHB in China (Lu et al., 2001). Based on its function and importance, FgFim is a good target for fungicides against FHB.
Fimbrin is an actin‐bundling protein, and fimbrin defects in S. cerevisiae alter actin cable assembly and stability (Moseley and Goode, 2006). In fungi, the actin cytoskeleton is involved in numerous cellular processes, including cell polarity, cellular signalling, intracellular trafficking, cytokinesis, endocytosis, exocytosis, bud site selection, cell wall remodelling and cell shape determination (Drubin et al., 1988; Harris, 2006; Karpova et al., 1998; Kubler and Riezman, 1993; Pruyne and Bretscher, 2000; Torralba et al., 1998). In this study, the FgFim deletion mutant displayed increased sensitivity to cell membrane and cell wall‐damaging agents, which is in agreement with the down‐regulation of genes related to cell wall integrity, cell wall remodelling and cell membrane fluidity and permeability (Fig. 6A,B). In addition, because the mutant showed a reduction in cell membrane stability, JS399‐19 and H2O2 may penetrate the plasma membrane of the mutant more easily than that of the parental strain, resulting in increased sensitivity of the mutant to these stress agents.
In the osmoregulation mitogen‐activated protein kinase (MAPK) pathway, the osmotica NaCl and KCl always trigger similar adaptive responses in eukaryotic cells (Duan et al., 2013; Jiang et al., 2011a, b). In this study, however, ΔFgFim‐15 exhibited increased sensitivity to 1.2 m NaCl, but reduced sensitivity to 1.2 m KCl, which was consistent with the level of glycerol accumulated in response to NaCl and KCl. K+ is not only an important osmoticum, but also promotes the activities of diverse endoenzymes. Working with yeast, Pastor et al. (2009) tested various mutants in mitochondrial function that cause strong hypersensitivity to osmotic stress; they observed that β‐galactosidase was induced more slowly by Na+ than K+. In addition, Cabrera et al. (2012) showed that calcineurin is directly involved in regulating HAK‐type K+ transporters. Because FgFim contains four CH domains, the disruption of FgFim may affect the K+ transporters. Moreover, because the adaptive responses orchestrated by the high‐osmolarity glycerol pathway in response to osmostress are complex and involve the direct regulation of plasma membrane cation transporters (Proft and Struhl, 2004), it is reasonable that ΔFgFim‐15 displayed increased sensitivity to metal cations.
Pathogenicity tests demonstrated that the ΔFgFim mutant could successfully infect wheat heads, but had lost its aggressiveness (Fig. 9). The inability to spread in planta suggests a reduced virulence of the ΔFgFim mutant. Our results further showed that the cell wall might be disorganized in the ΔFgFim mutant. This may contribute to reduced virulence because the fungal cell wall directly contacts plant cells and protects the pathogen from stress factors, including reactive oxygen species (ROS) and pathogenesis‐related proteins, produced by the host (Bowman and Free, 2006). Under pathogen attack, plants use the oxidative burst as an early defence reaction. The FgFim mutant showed increased sensitivity to H2O2, which may be related to the reduced virulence of the FgFim mutant in plant tissues. Previous studies have shown that the trichothecene toxin DON, although not required for infection, is an important virulence factor that is required for F. graminearum spread within the rachis tissue (Bai et al., 2002; Desjardins et al., 1996; Jansen et al., 2005; Maier et al., 2006; Proctor et al., 1995; Seong et al., 2009). In this study, ΔFgFim‐15 produced significantly less DON than the parental strain. This result is consistent with the observation that ΔFgFim‐15 causes scab symptoms in inoculated or nearby spikelets, but cannot spread within the rachis.
In conclusion, we have compared the F. graminearum ΔFgFim mutant with the parental strain, and have shown that the FgFim protein plays a significant role in growth, virulence, sexual and asexual development, and responses to various environmental stresses. Although the FgFim gene is not the target of fungicide JS399‐19, the deletion of FgFim may increase cell membrane permeability and thereby indirectly reduce resistance to JS399‐19. Our results also provide the basis for further investigation of the mechanisms controlling cytokinesis and endocytosis in F. graminearum, and may facilitate the development of fungicides that target FgFim for the control of FHB and the reduction of mycotoxin levels in staple cereal crops. The mutation that results in resistance to JS399‐19 in strain Y2021A remains to be identified.
Experimental Procedures
Strains, plasmids and culture conditions
The F. graminearum strains used in this study are listed in Table 1. For mycelial growth assays, the strains were grown at 25 °C on PSA (200 g of potato, 20 g of sucrose, 15 g of agar and 1 L of water), MM or CM (Klittich and Leslie, 1987; Marui et al., 2012). For sporulation assays, the strains were grown at 25 °C in MBL medium (30 g of mung beans boiled in 1 L of water for 20 min and then filtered through cheesecloth) (Bai and Shaner, 1996). Escherichia coli strain DH5α was used for general cloning and was cultured in Luria–Bertani broth at 37 °C.
TAIL‐PCR and identification of the FgFim gene
TAIL‐PCR amplification was performed as described previously (Liu and Chen, 2007) with six specific primers that were designed in this study and an arbitrary degenerate (AD) primer (Table S1, see Supporting Information). Among the six specific primers, TAILP1, TAILP2 and TAILP3 were used for left border flanking sequence amplification, and TAILP4, TAILP5 and TAILP6 were used for the right border flanking sequences. The target fragments were cloned into the pMD18‐T vector, sequenced and finally identified as FgFim in the F. graminearum genome database (MIPS: Munich Information Centre for Protein Sequences) by alignment using blastx (available at http://www.broadinstitute.org/annotation/genome/fusarium_group/MultiHome.html).
To verify the existence of FgFim and the size of the introns in F. graminearum strain Y2021A, RNA was extracted from the mycelia of the parental strain Y2021A with the RNeasy kit (Tiangen, Beijing, China) and was used for reverse transcription with the PrimeScript® RT reagent kit (TaKaRa, Dalian, China). RT‐PCR amplification of the cDNA was conducted with the primers FimF/FimR. The resulting PCR product was purified, cloned and sequenced; the sequence was used to construct the phylogenetic tree with homologous proteins from other fungi.
Construction of vectors for the deletion and complementation of FgFim using the double‐joint PCR technique
To investigate the functions of FgFim in F. graminearum, we generated an FgFim deletion mutant. A gene replacement cassette ΔFgFim carrying the hygromycin resistance gene and the herpes simplex virus thymidine kinase gene, flanked by DNA sequences corresponding to those located at the 5′ (left junction) and 3′ (right junction) ends of the FgFim gene, was constructed by double‐joint PCR as described previously (Yu et al., 2004). In the first PCR round, fragments 1.6 kb upstream and 1.5 kb downstream of FgFim were amplified from the genomic DNA of strain Y2021A using the primer pairs A1/A2 and A3/A4, respectively. The primers HTF/HTR were used to amplify a 3.5‐kb fragment encoding the HPH‐HSV‐tk cassette containing the hygromycin resistance gene, the herpes simplex virus thymidine kinase gene and the Aspergillus nidulans trpC promoter. This cassette was initially amplified from the PtrpChptA‐PItk plasmid (data not shown). The three amplicons (left junction, HPH‐HSV‐tk cassette and right junction) were gel purified using the OMEGA BIO‐TEK (Shanghai, China) gel purification kit, mixed at a 1:3:1 molar ratio and used as a template for the fusion round, which was performed using La Taq Polymerase (TaKaRa) without primers. In the third PCR round, 1 μL of product from the second PCR round was used as DNA template to amplify a 6.6‐kb DNA fragment using the primers A1/A4. The DNA fragment generated from the third PCR round, which carried the HPH‐HSV‐tk cassette fused to the FgFim flanking regions, was gel purified and used to transform protoplasts of F. graminearum strains (Table 1).
One of the FgFim deletion mutants (ΔFgFim‐15) was complemented with the full‐length FgFim gene to confirm that the phenotypic changes of the FgFim deletion mutant were caused by the disruption of the gene. The vector for the complementation of FgFim was amplified from the genomic DNA of strain Y2021A using primers A1/A4. Before this vector was transformed into strain Y2021A, FgFim in the vector was sequenced to ensure the flawlessness of the sequence. The complemented strain was designated ΔFgFim‐15C.
To construct Fgβ2‐tub and FgFim double mutants, a NEO cassette (geneticin resistance) was amplified from plasmid pII99‐Pro(DOHH) GFP (Ge et al., 2013) with the primer pair neo‐F and neo‐R (Table S1). Then, the PCR products containing the FgFim‐upstream‐NEO‐downstream cassette were transformed into protoplasts of the Fgβ2‐tub deletion mutant, ΔFgβ2‐tub‐22. Geneticin (100 μg/mL) was used for the selection of transformants.
Protoplast preparation and transformation of F. graminearum
For the preparation of protoplasts, five mycelial plugs (5 mm in diameter) taken from the margin of a 3‐day‐old colony of strain Y2021A were added to YEPD liquid medium (w/v, 1% peptone, 0.3% yeast extract, 2% glucose). After 36 h at 25 °C, the young mycelium in the YEPD liquid medium was filtered, washed with 0.7 m NaCl and treated with lysing enzyme (5 mg/mL of 0.7 m NaCl; Sigma, St. Louis, MO, USA), driselase (5 mg/mL of 0.7 m NaCl; Sigma) and snailase (10 mg/mL of 0.7 m NaCl; Kayon, Shanghai, China). After 2 h at 30 °C, the enzyme solution was filtered through three layers of lens paper to eliminate mycelial residues. The protoplasts in the filtrate were then washed with 0.7 m NaCl and with STC (0.8 m sorbitol, 0.05 m Tris, pH 8.0, 50 mm CaCl2) and resuspended in SPTC (STC with 40%, w/v, PEG6000) buffer (STC : SPTC = 4:1).
For the transformation, 107 protoplasts in 200 μL of SPTC buffer and 40 μL (100 μg/μL) of target DNA in 10 μL of heparin sodium were mixed and incubated on ice for 30 min; a 1‐mL volume of SPTC was mixed with the suspension, which was incubated at room temperature for 20 min. Protoplasts were mixed into 200 mL of regeneration medium (0.1% yeast extract, 0.1% casein hydrolysate, 1.0 m sucrose, 1.6% granulated agar) at 43 °C, which was poured into 9‐cm‐diameter Petri plates (20 mL per plate) and incubated at 25 °C. After 12–24 h, the plates were overlaid with 10 mL of selective agar (1.2% granulated agar in water containing 100 μg/mL of hygromycin B) and incubated further. Transformants were obtained 4 days post‐transformation. They were transferred to fresh PSA with 100 μg/mL of hygromycin B (which supports the growth of the transformants but not the growth of the complementations) and 0.2 μm floxuridine (which supports the growth of the complementations but not the growth of transformants). The putative transformants were purified by single‐spore isolation. Transformation of ΔFgFim‐15 with the full‐length FgFim gene was conducted as described above, except that floxuridine was used as a selection agent.
Test for mycelial growth, conidiation, perithecial development and pathogenicity
The parental strain Y2021A, deletion mutant ΔFgFim‐15 and complemented strain ΔFgFim‐15C were routinely cultured on PSA, CM and MM plates at 25 °C for 3 days. To test the sensitivities to various stresses, mycelial growth was assayed after incubation at 25 °C for 3–5 days in the dark on PSA plates with 1.2 m NaCl or KCl (osmotic stress agents), 0.05% (w/v) Congo red or 5 mm caffeine (cell wall‐damaging agents), 0.05% SDS (w/v, a cell membrane‐damaging agent), 0.5% H2O2 (v/v, an oxidative stress agent) or metal cation stressors (0.2 m LiCl, 0.2 m CaCl2 and 5 mm ZnCl2). Each plate was inoculated with a 5‐mm‐diameter mycelial plug taken from the margin of a 3‐day‐old colony. There were three replicate plates for each treatment, and the colony diameter in each plate was measured; the diameter of the original mycelial plug (5 mm) was subtracted from each measurement. The percentage of mycelial growth inhibition (RGI) was calculated using the formula RGI = [(A – B)/(A – 5)] × 100, where A is the colony diameter of the control and B is the colony diameter of a treatment. Each experiment was independently repeated three times.
For conidiation assays, five mycelial plugs (5 mm in diameter) taken from the margin of a 3‐day‐old colony of each strain were added to a 50‐mL flask containing 30 mL of MBL medium. Each strain was represented by three flasks. The flasks were incubated at 25 °C for 5 days with shaking (185 rpm). The number of conidia in the MBL broth in each flask was determined with a haemacytometer and microscope. The experiment was repeated three times. Perithecial development was examined on autoclaved wheat kernels, as described previously (Liang and Wang, 1981; Qiu et al., 2011; Xu et al., 2010).
For pathogenicity assays, conidia harvested from 7‐day‐old MBL cultures of the parental strain Y2021A, the FgFim deletion mutant (ΔFgFim‐15) and the complemented strain (ΔFgFim‐15C) were harvested and suspended in sterile distilled water at 5 × 105 condia/mL. Wheat heads of cultivar Zhenmai 5 (growing in pots) were inoculated with 10 μL of the conidial suspensions, as described by Gale et al. (2002). There were 30 replicates for each strain. After inoculation, each wheat head was placed in a plastic bag for 48 h to maintain moisture, and the wheat plants were then maintained in a glasshouse. Disease severity was calculated as the percentage of blighted spikelets in each head, 15 days after inoculation. The pathogenicity among the parental strain and the mutants was compared using Fisher's least‐significant difference (LSD) test.
Microscopic examination of mycelia and germ tubes
For the investigation of conidium germination and mycelial morphology, freshly harvested conidia and mycelial plugs of the parental strain (Y2021A) and the FgFim deletion mutant (ΔFgFim‐15) were cultured in liquid YEPD medium for 12 h and 36 h, respectively. Microscopic examination was carried out using germ tubes germinated from spores for 6, 8 and 12 h and young mycelium for 36 h in YEPD. Sections were prepared and visualized using an Olympus IX‐71 microscope (Tokyo, Japan).
Quantitative RT‐PCR
RNA samples were isolated with the RNeasy kit (Tiangen) from 2‐day‐old mycelia grown in potato sucrose broth (PSB) or GYEP liquid medium (5% glucose, 0.1% yeast extract and 0.1% peptone). First‐strand cDNA was synthesized with the PrimeScript® RT reagent kit (TaKaRa). All quantitative RT‐PCRs were performed with an ABI 7500 real‐time detection system (Applied Biosystems, Foster City, CA, USA). The primers used for quantitative RT‐PCR analysis are listed in Table S1. The expression of the measured genes in each sample was normalized to glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) gene expression, and relative changes in gene expression levels were analysed with ABI 7500 SDS software (Applied Biosystems, Foster City, CA, USA), which automatically set the baseline. Data from three biological replicates were used to calculate the mean and standard deviation.
Determination of the sensitivity or resistance to the fungicide JS399‐19
The F. graminearum parental strain Y2021A and the FgFim deletion mutant (ΔFgFim‐15) differed in their resistance to the novel cyanoacrylate fungicide JS399‐19, which was kindly provided by the Institute for the Control of Agrochemicals, Ministry of Agriculture (ICAMA), Hangzhou, China. To determine the sensitivity or resistance of each strain in Table 1 to JS399‐19, a mycelial plug (5 mm in diameter) taken from the margin of a 3‐day‐old colony was placed on the centre of a PSA plate amended with JS399‐19 at: 0, 0.025, 0.05, 0.075, 0.1, 0.2 or 0.4 μg/mL; 0, 10, 20, 30, 40, 50 or 75 μg/mL; or 50, 100, 200, 300, 400 or 500 μg/mL. Three replicates for each concentration were used for each strain. After incubation at 25 °C for 3–5 days, the colony diameter in each plate was measured in two perpendicular directions; the diameter (5 mm) of the original mycelial plug was subtracted from each measurement. For each plate, the average of the colony diameters was used to calculate the fungicide concentration that resulted in 50% mycelial growth inhibition (observed EC50). The observed EC50 values were calculated with the Data Processing System (DPS) computer program (Hangzhou Reifeng Information Technology Ltd, Hangzhou, China). The experiment was performed twice.
Determination of intracellular glycerol accumulation and DON production
For intracellular glycerol assays, each strain was grown in PSB for 2 days at 25 °C on a shaker. After treatment with 1.2 m NaCl or KCl for 2 h, mycelia of each strain were harvested and ground in liquid nitrogen. Then, 100 mg of the mycelial powders was transferred to a 2‐mL microcentrifuge tube containing 1 mL of Millipore water. After the tubes had been vortex mixed three times for 30 s each time, the tubes were centrifuged at 5000 g for 20 min. The supernatant of each sample was transferred to a new tube, and a 5‐μL aliquot of each supernatant was mixed with 195 μL of glycerol detection buffer (Applygen Technologies Inc., Beijing, China). After the mixture had been incubated at 37 °C for 15 min, the glycerol concentration was determined with a SpectraMax M5 microplate reader (Molecular Devices Inc., Sunnyvale, CA, USA) at 550 nm. The experiment was independently repeated three times.
Diseased wheat kernels were harvested from inoculated spikelets at 30 dpi for the determination of DON production using a protocol described previously (Bluhm et al., 2007; Goswami and Kistler, 2005; Mirocha et al., 1998). The amount of F. graminearum DNA in each sample was determined using a quantitative real‐time PCR method (Yin et al., 2009). The amount of DON (per milligram of fungal DNA) in each sample was detected by gas chromatography with an electron capture detector (GC‐ECD) (Agilent Technologies Inc., Wilmington, DE, USA). Three biological replicates were tested for each strain.
Supporting information
Fig. S1 Alignment of FgFim from Fusarium graminearum with those from Saccharomyces cerevisiae and other fungi. (A) Phylogenetic tree generated by the neighbour‐joining method with Mega 4.1 software on the basis of the deduced amino acid sequences of FgFim from Fusarium graminearum isolate Y2021A and those from Ashbya gossypii (AgSAC6, GenBank accession no. AAS54558.2), Aspergillus nidulans (FimA, CBF70817.1), Aspergillus oryzae (fimbrin, XP_001826508.1), Aspergillus fumigatus (Sac6, XP_755078.1), Ajellomyces dermatitidis (Sac6, EEQ83496.1), Colletotrichum orbiculare (fimbrin, ENH84123.1), Fusarium oxysporum (fimbrin, ENH62771.1), Fusarium sambucinum (fimbrin, CAA10667.1), Magnaporthe oryzae (fimbrin, XP_003710980.1), Mycosphaerella populorum (Sac6, EMF12866.1), Paracoccidioides brasiliensis (plastin‐3, EEH22505.1), Neurospora crassa (fimbrin, XP_956577.2), Verticillium dahliae (fimbrin, EGY15515.1), Schizosaccharomyces pombe (Fim1, CAB39801.1) and Saccharomyces cerevisiae (Sac6p, CAA88210.1). The bootstrap values are indicated on the phylogenetic tree. (B) The alignment of the amino acid sequences of FgFim with those from Ashbya gossypii, A. nidulans and S. cerevisiae. 1, 2, 3 and 4 represent calponin homology (CH) domains.
Fig. S2 Assays for sensitivity to agents that damage cell walls (Congo red and caffeine) and cell membranes (sodium dodecylsulphate, SDS) and induce oxidative stress (H2O2). Cultures of the parental strain Y2021A, deletion mutant ΔFgFim‐15 and complemented strain ΔFgFim‐15C were incubated on potato sucrose agar (PSA) with/without 0.05% Congo red, 5 mm caffeine, 0.05% SDS and 0.5% H2O2 at 25 °C. Cultures were photographed at 3–5 days post‐inoculation (dpi).
Fig. S3 Assays for sensitivity to agents that induce osmotic stress and to metal cations. Cultures of the parental strain Y2021A and the deletion mutant ΔFgFim‐15 were incubated on potato sucrose agar (PSA) with/without 1.2 m NaCl or KCl, 0.2 m LiCl, 0.2 m CaCl2 or 5 mm ZnCl2 at 25 °C. Cultures were photographed at 3–5 days post‐inoculation (dpi).
Fig. S4 Expression level of FgFim (A) and Fgβ 2 ‐tub (B) in strain Y2021A relative to that in wild‐type 2021 during germ tube elongation after 24, 48 and 72 h in liquid yeast extract–peptone–dextrose (YEPD) medium. Values are the means ± standard error of three repeated experiments.
Table S1 Oligonucleotide primers used in this study.
Table S2 Percentages of Y2021A, ΔFgFim‐15 and ΔFgFim‐15C conidia that germinated after incubation in yeast extract–peptone–dextrose (YEPD) liquid medium at 6 or 12 h. The experiment was performed three times. Values are means ± standard error.
Acknowledgements
This work was supported by the Chinese 973 Program (2012CB114000), the National Science Foundation of China (31201543) and The Ph.D. Programs Foundation of the Ministry of Education of China (No. 20120097120009).
References
- Adams, A.E.M. and Botstein, D. (1989) Dominant suppressors of yeast actin mutations that are reciprocally suppressed. Genetics, 121, 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, A.E.M. , Botstein, D. and Drubin, D.G. (1989) A yeast actin‐binding protein is encoded by SAC6, a gene found by suppression of an actin mutation. Science, 243, 231–233. [DOI] [PubMed] [Google Scholar]
- Adams, A.E.M. , Botstein, D. and Drubin, D.G. (1991) Requirement of yeast fimbrin for actin organization and morphogenesis in vivo . Nature, 354, 404–408. [DOI] [PubMed] [Google Scholar]
- Adams, A.E.M. , Shen, W.Y. , Lin, C.S. , Leavitt, J. and Matsudaira, P. (1995) Isoform‐specific complementation of the yeast Sac6 null mutation by human fimbrin. Mol. Cell Biol. 15, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, G.H. and Shaner, G. (1996) Variation in Fusarium graminearum and cultivar resistance to wheat scab. Plant Dis. 80, 975–979. [Google Scholar]
- Bai, G.H. and Shaner, G. (2004) Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. [DOI] [PubMed] [Google Scholar]
- Bai, G.H. , Desjardins, A.E. and Plattner, R.D. (2002) Deoxynivalenol nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia, 153, 91–98. [DOI] [PubMed] [Google Scholar]
- Blandino, M. , Minelli, L. and Reyneri, A. (2006) Strategies for the chemical control of Fusarium head blight: effect on yield, alveographic parameters and deoxynivalenol contamination in winter wheat grain. Eur. J. Agron. 25, 193–201. [Google Scholar]
- Bluhm, B.H. , Zhao, X. , Flaherty, J.E. , Xu, J.R. and Dunkle, L.D. (2007) RAS2 regulates growth and pathogenesis in Fusarium graminearum . Mol. Plant–Microbe Interact. 20, 627–636. [DOI] [PubMed] [Google Scholar]
- Bowman, S.M. and Free, S.J. (2006) The structure and synthesis of the fungal cell wall. Bioessays, 28, 799–808. [DOI] [PubMed] [Google Scholar]
- Bretscher, A. (1981) Fimbrin is a cytoskeletal protein that crosslinks F‐actin in vitro. Proc. Natl. Acad. Sci. USA, 78, 6849–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera, E. , Alvarez, M.C. , Martin, Y. , Siverio, J.M. and Ramos, J. (2012) K+ uptake systems in the yeast Hansenula polymorpha. Transcriptional and post‐translational mechanisms involved in high‐affinity K+ transporter regulation. Fungal Genet. Biol. 49, 755–763. [DOI] [PubMed] [Google Scholar]
- Chen, C.J. , Yu, J.J. , Bi, C.W. , Zhang, Y.N. , Xu, J.Q. and Wang, J.X. (2009a) Mutations in a β‐tubulin confer resistance of Gibberella zeae to benzimidazole fungicides. Phytopathology, 99, 1403–1411. [DOI] [PubMed] [Google Scholar]
- Chen, Y. and Zhou, M.G. (2009) Characterization of Fusarium graminearum isolates resistant to both carbendazim and a new fungicide JS399‐19. Phytopathology, 99, 441–446. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Chen, C. , Wang, J. , Jin, L. and Zhou, M. (2007) Genetic study on JS399‐19 resistance in hyphal fusion of Fusarium graminearum by using nitrate nonutilizing mutants as genetic markers. J. Genet. Genomics, 34, 469–476. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Li, H.K. , Chen, C.J. and Zhou, M.G. (2008) Sensitivity of Fusarium graminearum to fungicide JS399‐19: in vitro determination of baseline sensitivity and the risk of developing fungicide resistance. Phytoparasitica, 36, 326–337. [Google Scholar]
- Chen, Y. , Chen, C. , Zhou, M. , Wang, J. and Zhang, W. (2009b) Monogenic resistance to a new fungicide, JS399‐19, in Gibberella zeae . Plant Pathol. 58, 565–570. [Google Scholar]
- Cheng, D.M. , Marner, J. and Rubenstein, P.A. (1999) Interaction in vivo and in vitro between the yeast fimbrin, SAC6P, and a polymerization‐defective yeast actin (V266G and L267G). J. Biol. Chem. 274, 35 873–35 880. [DOI] [PubMed] [Google Scholar]
- Deoara, A. , Hashidoko, Y. and Tahara, S. (2008) Actin filaments predominate in morphogenic cell stages, whereas plaques predominate in non‐morphogenic cell stages in Peronosporomycetes. Mycol. Res. 112, 868–882. [DOI] [PubMed] [Google Scholar]
- Desjardins, A.E. (2006) Fusarium Mycotoxins: Chemistry, Genetics, and Biology. St. Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Desjardins, A.E. , Proctor, R.H. , Bai, G. , McCormick, S.P. , Shaner, G. and Buechley, G. (1996) Reduced virulence of trichothecene‐nonproducing mutants of Gibberella zeae in wheat field tests. Mol. Plant–Microbe Interact. 9, 775–781. [Google Scholar]
- Drubin, D.G. , Miller, K.G. and Botstein, D. (1988) Yeast actin‐binding proteins: evidence for a role in morphogenesis. J. Cell Biol. 107, 2551–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Y. , Ge, C. , Liu, S. , Wang, J. and Zhou, M. (2013) A two‐component histidine kinase Shk1 controls stress response, sclerotial formation and fungicide resistance in Sclerotinia sclerotiorum . Mol. Plant Pathol. 14, 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, L.R. , Chen, L.F. , Hernick, C.A. , Takamura, K. and Kistler, H.C. (2002) Population analysis of Fusarium graminearum from wheat fields in eastern China. Phytopathology, 92, 1315–1322. [DOI] [PubMed] [Google Scholar]
- Ge, C.Y. , Duan, Y.B. , Zhou, M.G. and Chen, C.J. (2013) A protoplast transformation system for gene deletion and complementation in Sclerotinia sclerotiorum . J. Phytopathol. 161, 800–806. [Google Scholar]
- Glenney, J. , Kaulfus, P. , Matsudaira, P. and Weber, K. (1981) F‐actin binding and bundling properties of fimbrin, a major cytoskeletal protein of microvillus core filaments. J. Biol. Chem. 256, 9283–9288. [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2005) Pathogenicity and in planta mycotoxin accumulation among members of the Fusarium graminearum species complex on wheat and rice. Phytopathology, 95, 1397–1404. [DOI] [PubMed] [Google Scholar]
- Gustin, M.C. , Albertyn, J. , Alexander, M. and Davenport, K. (1998) MAP kinase pathways in the yeast Saccharomyces cerevisiae . Microbiol. Mol. Biol. Rev. 62, 1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S.D. (2006) Cell polarity in filamentous fungi: shaping the mold. Int. Rev. Cytol. 251, 41–77. [DOI] [PubMed] [Google Scholar]
- Hou, Y. , Zheng, Z. , Xu, S. , Chen, C. and Zhou, M. (2013) Proteomic analysis of Fusarium graminearum treated by the fungicide JS399‐19. Pestic. Biochem. Physiol. 107, 86–92. [DOI] [PubMed] [Google Scholar]
- Jansen, C. , Wettstein, D.V. , Schäfer, W. , Kogel, K.H. , Felk, A. and Maier, F.J. (2005) Infection patterns in barley and wheat spikes inoculated with wild‐type and trichodiene synthase gene disrupted Fusarium graminearum . Proc. Natl. Acad. Sci. USA, 102, 16 892–16 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J. , Liu, X. , Yin, Y. and Ma, Z. (2011a) Involvement of a velvet protein FgVeA in the regulation of asexual development, lipid and secondary metabolisms and virulence in Fusarium graminearum . PloS ONE, 6, e28291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J.H. , Yun, Y.Z. , Fu, J. , Shim, W.B. and Ma, Z.H. (2011b) Involvement of a putative response regulator FgRrg‐1 in osmotic stress response, fungicide resistance and virulence in Fusarium graminearum . Mol. Plant Pathol. 12, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde, S. , Walther, A. and Wendland, J. (2011) The Ashbya gossypii fimbrin SAC6 is required for fast polarized hyphal tip growth and endocytosis. Microbiol. Res. 166, 137–145. [DOI] [PubMed] [Google Scholar]
- Karpova, T.S. , Moltz, S.L. , Riles, L.E. , Guldener, U. , Hegemann, J.H. and Veronneau, S. (1998) Depolarization of the actin cytoskeleton is a specific phenotype in Saccharomyces cerevisiae . J. Cell Sci. 111, 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, M.G. , Shi, W. , Ramagopal, U. , Tseng, Y. , Wirtz, D. and Kovar, D.R. (2004) Structure of the actin crosslinking core of fimbrin. Structure, 12, 999–1013. [DOI] [PubMed] [Google Scholar]
- Klittich, C. and Leslie, J. (1987) Nitrate nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology, 77, 1640–1646. [Google Scholar]
- Kubler, E. and Riezman, H. (1993) Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 12, 2855–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Nei, M. , Dudley, J. and Tamura, K. (2008) MEGA: a biologist‐centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.K. , Diao, Y.M. , Wang, H.X. , Chen, C.J. , Ni, J.P. and Zhou, M.G. (2008) JS399‐19, a new fungicide against wheat scab. Crop Prot. 27, 90–95. [Google Scholar]
- Liang, X.Y. and Wang, Z.C. (1981) On developing a method for rapid formation of perithecia of Gibberella zeae (SCHW.) petch and its test in diagnosing the species. Acta Phytopathol. Sin. 11, 1–6. [Google Scholar]
- Liu, Y.G. and Chen, Y. (2007) High‐efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques, 43, 649–656. [DOI] [PubMed] [Google Scholar]
- Lu, W. , Chen, S. and Wang, Y. (2001) Research on Wheat Scab. Beijing: Science Publication House, 14–23. [Google Scholar]
- Maier, F.J. , Miedaner, T. , Hadeler, B. , Felk, A. , Salomon, S. and Lemmens, M. (2006) Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 7, 449–461. [DOI] [PubMed] [Google Scholar]
- Marui, J. , Matsushita, M.M. , Tada, S. , Hattori, R. , Suzuki, S. and Amano, H. (2012) Comparison of expression and enzymatic properties of Aspergillus oryzae lysine aminopeptidases ApsA and ApsB. World J. Microbiol. Biotechnol. 28, 2643–2650. [DOI] [PubMed] [Google Scholar]
- Mesterhazy, A. (1995) Types and components of resistance to Fusarium head blight of wheat. Plant Breed. 114, 377–386. [Google Scholar]
- Mirocha, C.J. , Kolaczkowski, E. , Xie, W.P. , Yu, H. and Jelen, H. (1998) Analysis of deoxynivalenol and its derivatives (batch and single kernel) using gas chromatography mass spectrometry. J. Agric. Food Chem. 46, 1414–1418. [Google Scholar]
- Moseley, J.B. and Goode, B.L. (2006) The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev. 70, 605–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, K. , Satoh, K. , Morimatsu, A. , Ohnuma, M. and Mabuchi, I. (2001) Interactions among a fimbrin, a capping protein, and an actin‐depolymerizing factor in organization of the fission yeast actin cytoskeleton. Mol. Biol. Cell, 12, 3515–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nganje, W.E. , Johnson, D.D. , Wilson, W.W. , Leistritz, F.L. , Bangsund, D.A. and Tiapo, N.M. (2004) Economic Impacts of Fusarium Head Blight in Wheat and Barley: 1993–2001. Fargo, ND: Department of Agribusiness and Applied Economics, Agricultural Experiment Station, North Dakota State ; University. [Google Scholar]
- Pastor, M.M. , Proft, M. and Pascual‐Ahuir, A. (2009) Mitochondrial function is an inducible determinant of osmotic stress adaptation in yeast. J. Biol. Chem. 284, 30 307–30 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor, R.H. , Hohn, T.M. and McCormick, S.P. (1995) Reduced virulence of Gibberella zeae caused by disruption of a trichthecine toxin biosynthetic gene. Mol. Plant–Microbe Interact. 8, 593–601. [DOI] [PubMed] [Google Scholar]
- Proft, M. and Struhl, K. (2004) MAP kinase‐mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell, 118, 351–361. [DOI] [PubMed] [Google Scholar]
- Pruyne, D. and Bretscher, A. (2000) Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J. Cell Sci. 113, 571–586. [DOI] [PubMed] [Google Scholar]
- Qiu, J. , Xu, J. , Yu, J. , Bi, C. , Chen, C. and Zhou, M. (2011) Localisation of the benzimidazole fungicide binding site of Gibberella zeae beta2‐tubulin studied by site‐directed mutagenesis. Pest Manag. Sci. 67, 191–198. [DOI] [PubMed] [Google Scholar]
- Rodriguez, R.J. , Low, C. , Bottema, C.D.K. and Parks, L.W. (1985) Multiple functions for sterols in Saccharomyces‐cerevisiae . Biochim. Biophys. Acta, 837, 336–343. [DOI] [PubMed] [Google Scholar]
- Rudd, J.C. , Horsley, R.D. , McKendry, A.L. and Elias, E.M. (2001) Host plant resistance genes for fusarium head blight: sources, mechanisms, and utility in conventional breeding systems. Crop Sci. 41, 620–627. [Google Scholar]
- Ruotolo, R. , Marchini, G. and Ottonello, S. (2008) Membrane transporters and protein traffic networks differentially affecting metal tolerance: a genomic phenotyping study in yeast. Genome Biol. 9, R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong, K.Y. , Pasquali, M. , Zhou, X. , Song, J. , Hilburn, K. and McCormick, S. (2009) Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 72, 354–367. [DOI] [PubMed] [Google Scholar]
- Skau, C.T. and Kovar, D.R. (2010) Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr. Biol. 20, 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau, C.T. , Courson, D.S. , Bestul, A.J. , Winkelman, J.D. , Rock, R.S. and Sirotkin, V. (2011) Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J. Biol. Chem. 286, 26 964–26 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, J.C. (1982) Epidemiology of wheat head blight and maize ear rot caused by Fusarium graminearum . Can. J. Plant Pathol. 4, 195–209. [Google Scholar]
- Torralba, S. , Raudaskoski, M. , Pedregosa, A.M. and Laborda, F. (1998) Effect of cytochalasin A on apical growth, actin cytoskeleton organization and enzyme secretion in Aspergillus nidulans . Microbiology, 144, 45–53. [DOI] [PubMed] [Google Scholar]
- Upadhyay, S. and Shaw, B.D. (2008) The role of actin, fimbrin and endocytosis in growth of hyphae in Aspergillus nidulans . Mol. Microbiol. 68, 690–705. [DOI] [PubMed] [Google Scholar]
- Winder, S. , Jess, T. and Ayscough, K. (2003) SCP1 encodes an actin‐bundling protein in yeast. Biochemistry, 375, 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojda, I. , Alonso‐Monge, R. , Bebelman, J.P. , Mager, W.H. and Siderius, M. (2003) Response to high osmotic conditions and elevated temperature in Saccharomyces cerevisiae is controlled by intracellular glycerol and involves coordinate activity of MAP kinase pathways. Microbiol‐Sgm. 149, 1193–1204. [DOI] [PubMed] [Google Scholar]
- Wu, J.Q. , Bahler, J. and Pringle, J.R. (2001) Roles of a fimbrin and an alpha‐actinin‐like protein in fission yeast cell polarization and cytokinesis. Mol. Biol. Cell, 12, 1061–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.B. , Li, H.P. , Zhang, J.B. , Song, B. , Chen, F.F. and Duan, X.J. (2010) Disruption of the chitin synthase gene CHS1 from Fusarium asiaticum results in an altered structure of cell walls and reduced virulence. Fungal Genet. Biol. 47, 205–215. [DOI] [PubMed] [Google Scholar]
- Yin, Y. , Liu, X. and Ma, Z. (2009) Simultaneous detection of Fusarium asiaticum and Fusarium graminearum in wheat seeds using a real‐time PCR method. Lett. Appl. Microbiol. 48, 680–686. [DOI] [PubMed] [Google Scholar]
- Yu, J.H. , Hamari, Z. , Han, K.H. , Seo, J.A. , Reyes‐Dominguez, Y. and Scazzocchio, C. (2004) Double‐joint PCR: a PCR‐based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.J. , Zhang, X. , Chen, C.J. , Zhou, M.G. and Wang, H.C. (2010) Effects of fungicides JS399‐19, azoxystrobin, tebuconazloe, and carbendazim on the physiological and biochemical indices and grain yield of winter wheat. Pestic. Biochem. Phys. 98, 151–157. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Alignment of FgFim from Fusarium graminearum with those from Saccharomyces cerevisiae and other fungi. (A) Phylogenetic tree generated by the neighbour‐joining method with Mega 4.1 software on the basis of the deduced amino acid sequences of FgFim from Fusarium graminearum isolate Y2021A and those from Ashbya gossypii (AgSAC6, GenBank accession no. AAS54558.2), Aspergillus nidulans (FimA, CBF70817.1), Aspergillus oryzae (fimbrin, XP_001826508.1), Aspergillus fumigatus (Sac6, XP_755078.1), Ajellomyces dermatitidis (Sac6, EEQ83496.1), Colletotrichum orbiculare (fimbrin, ENH84123.1), Fusarium oxysporum (fimbrin, ENH62771.1), Fusarium sambucinum (fimbrin, CAA10667.1), Magnaporthe oryzae (fimbrin, XP_003710980.1), Mycosphaerella populorum (Sac6, EMF12866.1), Paracoccidioides brasiliensis (plastin‐3, EEH22505.1), Neurospora crassa (fimbrin, XP_956577.2), Verticillium dahliae (fimbrin, EGY15515.1), Schizosaccharomyces pombe (Fim1, CAB39801.1) and Saccharomyces cerevisiae (Sac6p, CAA88210.1). The bootstrap values are indicated on the phylogenetic tree. (B) The alignment of the amino acid sequences of FgFim with those from Ashbya gossypii, A. nidulans and S. cerevisiae. 1, 2, 3 and 4 represent calponin homology (CH) domains.
Fig. S2 Assays for sensitivity to agents that damage cell walls (Congo red and caffeine) and cell membranes (sodium dodecylsulphate, SDS) and induce oxidative stress (H2O2). Cultures of the parental strain Y2021A, deletion mutant ΔFgFim‐15 and complemented strain ΔFgFim‐15C were incubated on potato sucrose agar (PSA) with/without 0.05% Congo red, 5 mm caffeine, 0.05% SDS and 0.5% H2O2 at 25 °C. Cultures were photographed at 3–5 days post‐inoculation (dpi).
Fig. S3 Assays for sensitivity to agents that induce osmotic stress and to metal cations. Cultures of the parental strain Y2021A and the deletion mutant ΔFgFim‐15 were incubated on potato sucrose agar (PSA) with/without 1.2 m NaCl or KCl, 0.2 m LiCl, 0.2 m CaCl2 or 5 mm ZnCl2 at 25 °C. Cultures were photographed at 3–5 days post‐inoculation (dpi).
Fig. S4 Expression level of FgFim (A) and Fgβ 2 ‐tub (B) in strain Y2021A relative to that in wild‐type 2021 during germ tube elongation after 24, 48 and 72 h in liquid yeast extract–peptone–dextrose (YEPD) medium. Values are the means ± standard error of three repeated experiments.
Table S1 Oligonucleotide primers used in this study.
Table S2 Percentages of Y2021A, ΔFgFim‐15 and ΔFgFim‐15C conidia that germinated after incubation in yeast extract–peptone–dextrose (YEPD) liquid medium at 6 or 12 h. The experiment was performed three times. Values are means ± standard error.


