SUMMARY
Plants defend themselves against potential pathogens via the recognition of pathogen‐associated molecular patterns (PAMPs). However, the molecular mechanisms underlying this PAMP‐triggered immunity (PTI) are largely unknown. In this study, we show that tomato HP1/DDB1, coding for a key component of the CUL4‐based ubiquitin E3 ligase complex, is required for resistance to Agrobacterium tumefaciens. We found that the DDB1‐deficient mutant (high pigment‐1, hp1) is susceptible to nontumorigenic A. tumefaciens. The efficiency of callus generation from the hp1 cotyledons was extremely low as a result of the necrosis caused by Agrobacterium infection. On infiltration of nontumorigenic A. tumefaciens into leaves, the hp1 mutant moderately supported Agrobacterium growth and developed disease symptoms, but the expression of the pathogenesis‐related gene SlPR1a1 and several PTI marker genes was compromised at different levels. Moreover, exogenous application of salicylic acid (SA) triggered SlPR1a1 gene expression and enhanced resistance to A. tumefaciens in wild‐type tomato plants, whereas these SA‐regulated defence responses were abolished in hp1 mutant plants. Thus, HP1/DDB1 may function through interaction with the SA‐regulated PTI pathway in resistance against Agrobacterium infection.
INTRODUCTION
Plants are constantly exposed to potential pathogens, but are resistant to the majority of them. This is largely because plants use a complex of immune systems to defend themselves against pathogen invasion. One important component of the immune system is innate immunity, also called basal defence in many cases (Dangl and Jones, 2001). Innate immunity is elicited by the perception of pathogen‐associated molecular patterns (PAMPs), conserved among pathogens, by pattern recognition receptors (PRRs) in host plants. This leads to the activation of a series of cellular events to eventually prevent pathogen colonization, including the production of reactive oxygen species (ROS) and ethylene (ET), callose deposition at the cell wall, activation of the mitogen‐activated protein kinase (MAPK) cascade and the induction of defence‐related genes (Asai et al., 2002; Ausubel, 2005; Boller and Felix, 2009). In a given plant species, multiple PAMP‐triggered immunity (PTI) pathways may exist and are integrated into the complex of immune systems that is sufficient to prevent the colonization of plants by most pathogens.
Although a number of PAMPs and corresponding PRRs have been established (2004, 2006), the downstream signalling leading to cellular events against pathogen colonization remains to be elucidated. Recently, an emerging body of evidence has suggested that the general stress signalling molecules, including salicylic acid (SA), jasmonic acid (JA) and ET, play a role in the PTI pathway after recognition of PAMPs by PRRs (Chen et al., 2009; Halim et al., 2009; Liu et al., 2010). Despite the existing cross‐talk between signalling pathways mediated by these molecules, it is generally thought that SA‐regulated defence signalling is associated with defence responses against biotrophic pathogens, whereas JA‐ and ET‐mediated defence signalling is mainly responsible for resistance against necrotrophic pathogens, both of which are usually characterized by the induction of specific pathogenesis‐related (PR) gene expression on pathogen challenge (Durrant and Dong, 2004). For instance, induction of the PR‐1 gene has been established as a cellular marker for the activation of the SA‐regulated signalling pathway (Durrant and Dong, 2004).
Agrobacterium tumefaciens is a soil‐borne bacterium that causes crown‐gall disease in many plant species. The molecular basis of pathogenesis involves the transfer of the T‐DNA part of the tumour‐inducing (Ti) plasmid into the plant nucleus by Agrobacterium, followed by integration into the plant chromosome. The T‐DNA contains plant hormone genes that stimulate the infected plant tissue to overgrow, with the formation of tumour tissue (Gelvin, 2003). The molecular interactions between plants and Agrobacterium are very complicated. On the one hand, Agrobacterium can trigger PTI: plants perceive PAMPs from Agrobacterium and mount a defence response, including the activation of the MAPK cascade; MPK3 phosphorylates the VirE2‐interacting protein 1 (VIP1) transcription factor and promotes VIP1 shuttling into the nucleus, where it activates the transcription of PR‐1 and other stress genes (Djamei et al., 2007; Pitzschke et al., 2009). On the other, Agrobacterium can hijack this plant defence system by associating with VIP1 to move the T‐complex into the nucleus and target chromatin (Djamei et al., 2007; Pitzschke et al., 2009). In the mean time, Agrobacterium takes advantage of the host VBF ubiquitin E3 ligase, which is also activated by Agrobacterium during the early time course of defence responses, to degrade VIP1 and other Agrobacterium T‐DNA‐associated proteins to expose the T‐DNA for integration into the host chromatin (Zaltsman et al., 2010). In addition, as pathogen‐induced PR‐1 expression is mainly regulated by SA, it appears that SA also plays a significant role in the later stage of the defence response to Agrobacterium infection (Anand et al., 2008; Pruss et al., 2008). Exogenous application of SA in Nicotiana benthamiana enhanced the resistance to Agrobacterium infection, whereas decreasing the endogenous SA level by expressing the bacterial NahG (encoding salicylate hydroxylase) gene or silencing the genes involved in SA biosynthesis resulted in plants hypersusceptible to Agrobacterium infection (Anand et al., 2008).
Tomato (Solanum lycopersicum) is an economically and experimentally important crop. Several photomorphogenic mutants with exaggerated photoresponsiveness and elevated pigmentation, such as the monogenic recessive high‐pigment mutants hp1 and hp2, have been described in tomato (Kendrick et al., 1997). These mutants are characterized by higher anthocyanin levels, shorter hypocotyls and greater fruit pigmentation than their semi‐isogenic wild‐type counterparts (Mustilli et al., 1999; Yen et al., 1997). Previously, we have characterized the HP1 gene, revealing that it encodes a protein homologous to (mammalian) UV‐DAMAGED DNA‐BINDING PROTEIN‐1 (DDB1) (Lieberman et al., 2004; Liu et al., 2004; Wang et al., 2008). Conserved from fission yeast to higher eukaryotes, the CUL4–DDB1 complex has been identified as a cullin‐RING finger ubiquitin ligase that is involved in the regulation of genome stability, DNA repair, the cell cycle and histone modification, and can be subverted by pathogenic viruses to benefit viral infection (Angers et al., 2006; Braun et al., 2011; Centore et al., 2010; Jin et al., 2006; Li et al., 2006; Petroski and Deshaies, 2005).
In this study, we demonstrate that, in addition to the well‐established functions involved in photomorphogenesis and pigment development, tomato HP1/DDB1 is required for the resistance to Agrobacterium. We found that the hp1 mutant is susceptible to nontumorigenic Agrobacterium infection, as manifested by necrosis on excised cotyledons, enhanced Agrobacterium growth and the development of disease symptoms on leaves, and increased efficiency of Agrobacterium‐mediated transient expression. The expression of SlPR1a1 and several PTI marker genes was compromised in the hp1 mutant. Moreover, SlPR1a1 induction and enhanced resistance to Agrobacterium by exogenous SA were abolished in the hp1 mutant, suggesting that HP1/DDB1 may act through interaction with the SA‐mediated PTI pathway.
RESULTS
The excised cotyledons of the hp1 mutant are hypersusceptible to the nontumorigenic Agrobacterium strain
In an attempt to generate transgenic hp1 mutant plants by A. tumefaciens‐mediated transformation, we found that the excised cotyledons of the hp1 mutant were too susceptible to A. tumefaciens infection to produce callus. During the standard Agrobacterium‐mediated transformation (Fillatti et al., 1987), the excised cotyledon explants of the wild‐type (WT) AC+ and hp1 mutant were infected with nontumorigenic A. tumefaciens GV2260 containing the binary vector pBI121 (Jefferson, 1987). The pBI121 vector carried a kanamycin resistance gene for selection of positive transformation. To our surprise, under our experimental conditions (1000 lx light for 16 h, dark for 8 h), necrosis started to develop on cotyledon explants of the hp1 mutant at 10 days post‐inoculation (dpi), whereas only a few of the cotyledon explants of WT AC+ showed this cell death phenotype (Fig. 1A). At 30 dpi, most excised cotyledons of the hp1 mutant had died without generating any callus. However, the majority of cotyledon explants of WT AC+ were still alive and produced callus (Fig. 1A). The number of cotyledon explants that generated callus at this time point was also scored. As shown in Fig. 1B, only 5.8% of the hp1 cotyledon explants were able to generate callus, whereas more than 50% of the WT AC+ cotyledon explants developed callus. These results indicate that the hp1 mutant is hypersusceptible to nontumorigenic Agrobacterium, which dramatically reduces the rate of callus generation for transformation.
Figure 1.
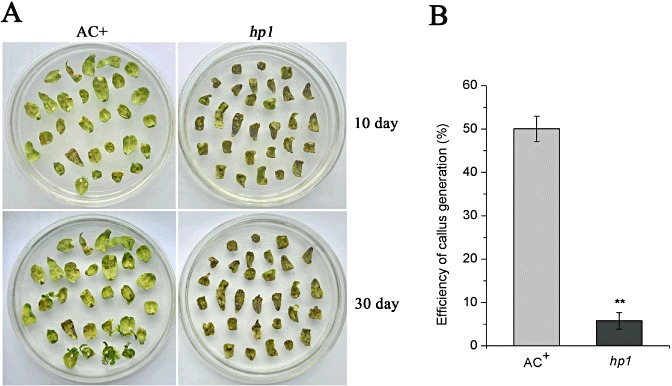
The detached cotyledons of the hp1 mutant are hypersusceptible to nontumorigenic Agrobacterium tumefaciens infection. (A) Necrosis on cotyledons caused by A. tumefaciens GV2260. Cotyledons detached from wild‐type (WT) AC+ and hp1 mutant seedlings, germinated on half‐strength Murashige and Skoog (MS) medium, were inoculated with A. tumefaciens GV2260 carrying the binary vector pBI121 (containing the kanamycin resistance gene) and incubated on regeneration medium under 1000 lx. Photographs were taken 10 and 30 days after infection. (B) Quantification of kanamycin‐resistant callus produced on the cotyledons. Each data point consists of at least three samples. Error bars indicate standard deviation. The statistical significance of the difference was confirmed by Student's t‐test (**P < 0.01). Similar results were obtained in at least two independent experiments.
The hp1 mutant supports bacterial growth and develops disease symptoms on Agrobacterium infection
In general, nontumorigenic Agrobacterium is not pathogenic to tomato plants. It does not cause disease symptoms or multiply to a large extent in leaves. The susceptibility of the hp1 cotyledon to Agrobacterium infection prompted us to examine the responses of hp1 mutant plant leaves to nontumorigenic Agrobacterium. We sought to determine whether the hp1 mutant could develop disease symptoms or support bacterial growth after Agrobacterium infection. We first assessed possible disease symptom development by inoculating WT AC+ and hp1 mutant plant leaves with A. tumefaciens GV2260 at an inoculum of 108 colony‐forming units (cfu)/mL. At 4 dpi, more than 17% of the leaves in hp1 mutant plants had wilted, whereas no WT AC+ leaves showed wilting‐like disease symptoms (Fig. 2A). In order to differentiate the subtle effect of hp1 on Agrobacterium growth in leaves, we inoculated WT AC+ and hp1 mutant plant leaves with A. tumefaciens GV2260 using a low inoculum of 104 cfu/mL. Agrobacterium populations in plant leaves were scored at 2 and 4 dpi. As expected, A. tumefaciens GV2260 showed only basal level multiplication in WT AC+ leaves. However, A. tumefaciens GV2260 multiplied five times more strongly in hp1 mutant leaves than in WT AC+ leaves (Fig. 2B). A consistently similar result was obtained when WT AC+ and hp1 mutant plant leaves were inoculated with another nontumorigenic A. tumefaciens strain EHA105 (Fig. 2C). Taken together, our results suggest that HP1/DDB1 is required for resistance to A. tumefaciens.
Figure 2.
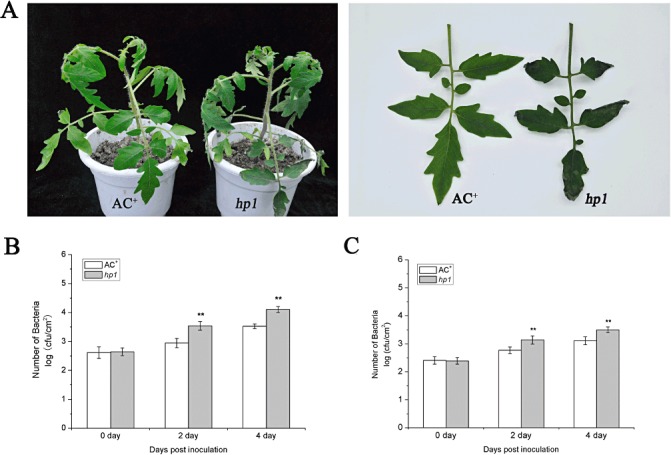
Disease symptoms and bacterial growth in leaves of wild‐type (WT) AC+ and hp1 mutant plants infected with Agrobacterium tumefaciens GV2260 or EHA105. Six‐week‐old WT AC+ and hp1 mutant plants were vacuum infiltrated with a suspension of A. tumefaciens GV2260 or EHA105, and maintained under light conditions of 200 lx 16 h/dark 8 h. (A) WT AC+ and hp1 mutant plants were vacuum infiltrated with a suspension of A. tumefaciens GV2260 [optical density at 600 nm (OD600) = 0.1]. Photographs were taken 4 days after infection. (B) WT AC+ and hp1 mutant plants were vacuum infiltrated with a suspension of A. tumefaciens GV2260 (OD600 = 0.00001). (C) WT AC+ and hp1 mutant plants were vacuum infiltrated with a suspension of A. tumefaciens EHA105 (OD600 = 0.00001). Bacterial growth was determined at 0, 2 and 4 days post‐inoculation (dpi). Each data point consists of at least six samples. Error bars indicate standard deviation. The statistical significance of the difference was confirmed by Student's t‐test (**P < 0.01). Similar results were obtained in at least two independent experiments.
Agrobacterium‐mediated transient transformation in hp1 leaves is more efficient than in WT AC+ leaves
The efficiency of Agrobacterium‐mediated transient transformation has been widely used to evaluate the susceptibility of plants to Agrobacterium infection (2007, (2008; Wroblewski et al., 2005; Zipfel et al., 2006). We next sought to determine whether hp1 affects the efficiency of transient transformation mediated by nontumorigenic A. tumefaciens GV2260 strain. Agrobacterium harbouring the binary vector pBISN1 expressing a β ‐glucuronidase (GUS) gene (Nam et al., 1999) was vacuum infiltrated into WT AC+ and hp1 leaves, and the transformation efficiency was determined by the GUS activity derived from the transformed leaf tissue. The GUS activity assay was conducted 2 days after Agrobacterium infiltration. As shown in Fig. 3A, GUS activity, indicated by the intensity of 5‐bromo‐4‐chloro‐3‐indolyl glucuronide (X‐Gluc) staining of leaves, was significantly greater in hp1 mutant leaves than in WT AC+ leaves. GUS activity was also quantified. GUS activity in hp1 leaves infiltrated with Agrobacterium was about 70% greater than that in WT AC+ leaves with the same treatment (Fig. 3B). Together, our data suggest that hp1 plants are more susceptible to Agrobacterium infection, and this susceptibility facilitates the efficiency of transient transformation mediated by the nontumorigenic A. tumefaciens strain.
Figure 3.
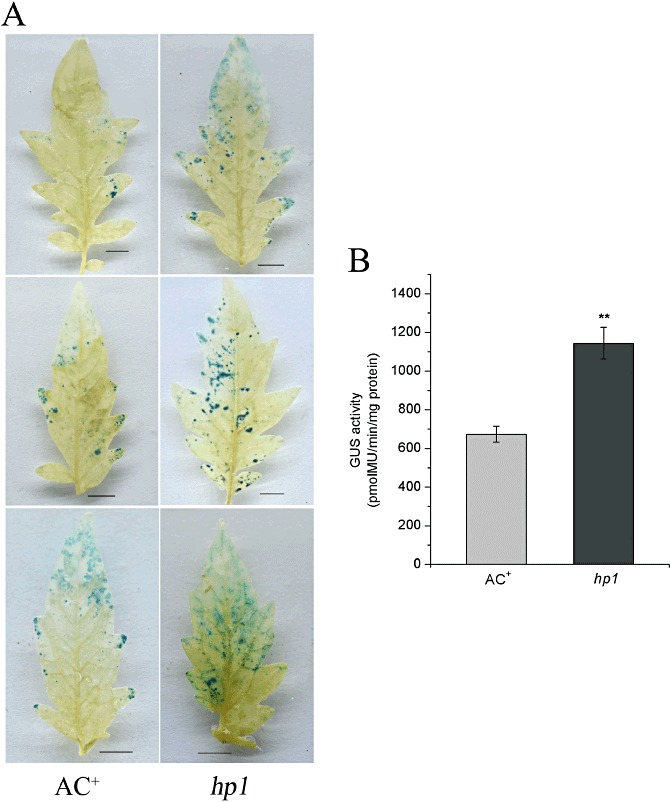
Agrobacterium‐mediated transient transformation in the leaves of wild‐type (WT) AC+ and hp1 mutant plants. (A) Randomly selected representative leaves of WT AC+ and hp1 mutant plants stained with 5‐bromo‐4‐chloro‐3‐indolyl glucuronide (X‐Gluc) at 2 days after infiltration with A. tumefaciens GV2260 expressing the β ‐glucuronidase (GUS) gene at an inoculum of OD600 = 0.1 (optical density at 600 nm). Bars, 1 cm. (B) Quantification of GUS activity in WT AC+ and hp1 mutant leaves. The GUS activity of infected leaves was measured by recording the fluorescence of 4‐methylumbelliferone (MU) at 2 days post‐inoculation (dpi). The data presented are the means with standard deviations of three independent experiments. The statistical significance of the difference was confirmed by Student's t‐test (**P < 0.01).
HP1/DDB1 is required for the induction of the SlPR1a1 gene by A. tumefaciens
PR genes play a significant role in plant defence responses and have been widely used as molecular markers for the defence reaction in plant–pathogen interactions (Durrant and Dong, 2004). The infiltration of nontumorigenic Agrobacterium into Nicotiana benthamiana or Arabidopsis leaves triggers strong defence responses, which are characterized by the induction of the PR‐1 gene at late time courses after infiltration (Pitzschke et al., 2009; Pruss et al., 2008). Significantly, the PR‐1‐characterized host responses are sufficient to confer resistance to subsequent infection with other pathogens, such as tobacco mosaic virus (Pruss et al., 2008). Thus, it is possible that the susceptibility of the hp1 mutant to Agrobacterium infection is a result of the abolishment of defence reactions, such as PR‐1 gene induction. To test this hypothesis, we sought to determine the tomato PR‐1 (SlPR1a1) mRNA expression pattern in A. tumefaciens‐inoculated AC+ and hp1 mutant plants by quantitative reverse transcriptase‐polymerase chain reaction (RT‐PCR) at different time points. As shown in Fig. 4A, A. tumefaciens treatment induced SlPR1a1 expression in WT AC+ plants at 2 dpi, and SlPR1a1 induction was maintained over the next 2 days. The induction pattern was similar to that reported for the induction of PR‐1 by Agrobacterium in tobacco (Pruss et al., 2008). In contrast, the induction of SlPR1a1 expression in hp1 mutant plants was completely abolished throughout all time points examined. It is also notable that, even prior to Agrobacterium infection, the basal level expression of SlPR1a1 was lower in WT AC+ than in the hp1 mutant (Fig. 4A). These results indicate that HP1/DDB1 is essential for the induction of the SlPR1a1 gene by Agrobacterium. They also suggest a correlation between HP1/DDB1‐dependent SlPR1a1 induction and resistance to Agrobacterium.
Figure 4.
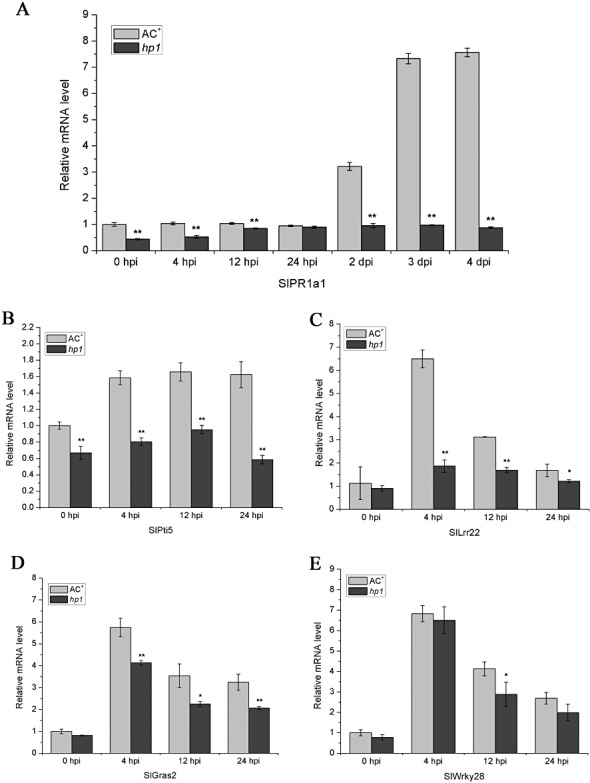
Effect of HP1/DDB1 on the induction of SlPR1a1 (A), SlPti5 (B), SlLrr22 (C), SlGras2 (D) and SlWrky28 (E) by Agrobacterium tumefaciens. Six‐week‐old wild‐type (WT) AC+ and hp1 mutant plants were vacuum infiltrated with A. tumefaciens GV2260 [optical density at 600 nm (OD600) = 0.1]. Total RNA was extracted at the indicated time points for quantitative reverse transriptase‐polymerase chain reaction (RT‐PCR). SlUBI3 was used as an internal control. Data represent the mean ± standard deviation of three independent experiments. The statistical significance of the difference was confirmed by Student's t‐test (*P < 0.05; **P < 0.01). hpi, hours post‐inoculation.
HP1/DDB1 is required for several PTI marker genes induced by A. tumefaciens
Recent publications have demonstrated that plants express PR‐1 and other defence‐related genes in response to Agrobacterium infection through the MAPK‐mediated PTI pathway (Djamei et al., 2007; Pitzschke et al., 2009). We next verified whether the PTI signalling pathways were compromised in the hp1 mutant by examining the induction pattern of several PTI marker genes by Agrobacterium. To this end, we took advantage of several PTI marker genes characterized in tomato, including SlPti5, SlGras2, SlLrr22 and SlWrky28 (Nguyen et al., 2010). To monitor gene induction by Agrobacterium, WT AC+ and hp1 mutant plants were vacuum infiltrated with a bacterial suspension of A. tumefaciens GV2260 at a concentration of 108 cfu/mL. The mRNA abundance of the PTI marker genes was determined by quantitative RT‐PCR at 0, 4, 12 and 24 hpi. As shown in Fig. 4B, E, all four PTI marker genes were rapidly induced by Agrobacterium infection in WT AC+ plant leaves. However, the expression of these markers in hp1 mutant leaves showed different patterns in response to Agrobacterium challenge: SlPti5 and SlLrr22 were not induced; SlGras2 and SlWrky28 were induced, but the induction level was significantly lower than that in WT AC+ plant leaves (Fig. 4B, E). These results suggest that HP1/DDB1 plays a significant role in certain PTI signalling pathways.
SlPR1a1 expression and enhanced resistance to A. tumefaciens by exogenous SA is abolished in the hp1 mutant
It is thought that PR‐1 gene induction by bacteria is mainly regulated through the SA signalling pathway (Durrant and Dong, 2004). Recent studies have also suggested that SA‐mediated defence signalling plays a major role in the resistance to Agrobacterium in N. benthamiana (Anand et al., 2008). In particular, exogenous application of SA can induce PR expression and activate defence responses to bacteria, including Agrobacterium (Anand et al., 2008). Therefore, it was logical to determine whether HP1/DDB1 functions through interaction with SA signalling in the resistance to Agrobacterium by examining whether SA‐regulated defence signalling and resistance to Agrobacterium were compromised in the hp1 mutant. First, we tested the induction of the SlPR1a1 gene by exogenous SA in hp1 mutant plants. As shown in Fig. 5A, exogenous application of SA on WT AC+ leaves strongly induced SlPR1a1 expression, whereas this induction by SA was completely abolished in hp1 mutant leaves. Next, we assessed the SA‐induced resistance to Agrobacterium in the hp1 mutant. We pretreated leaves of WT AC+ and hp1 mutant plants with SA solution and tested their response to A. tumefaciens. Again, to differentiate the subtle effect on Agrobacterium growth in tomato leaves, we inoculated WT AC+ and hp1 mutant plant leaves with A. tumefaciens GV2260 using a low inoculum of 104 cfu/mL. The results showed that, in WT AC+ leaves, pretreatment with SA enhanced significantly the resistance to Agrobacterium growth, as manifested by about five‐fold fewer bacterial populations in SA‐pretreated leaves relative to mock‐treated leaves, suggesting that SA can activate resistance to Agrobacterium. However, in the case of the hp1 mutant, there was no significant difference in Agrobacterium populations in SA‐pretreated and mock‐treated leaves, suggesting that the SA‐triggered resistance was abolished (Fig. 5B). Taken together, our results suggest that HP1/DDB1 is required for SA‐regulated SlPR1a1 induction and resistance to Agrobacterium infection in tomato.
Figure 5.
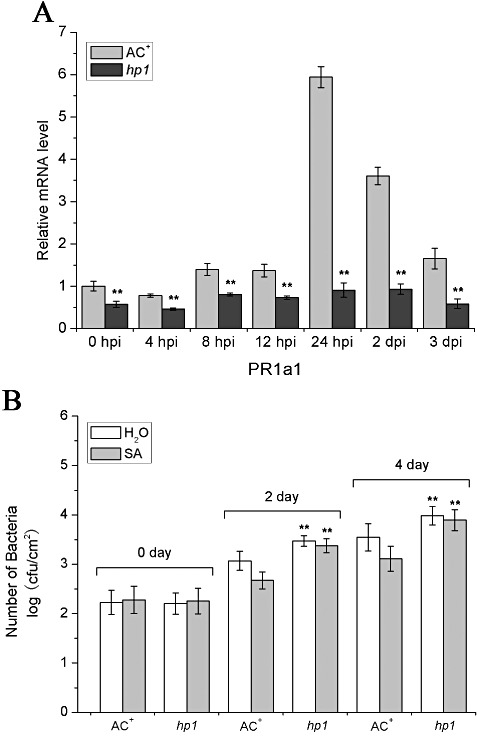
Influence of salicylic acid (SA) treatment on SlPR1a1 expression and the susceptibility of the hp1 mutant to Agrobacterium tumefaciens. (A) SlPR1a1 expression in wild‐type (WT) AC+ and hp1 mutant after SA treatment. Leaf samples sprayed with 0.5 mm SA solution were collected for quantitative reverse transriptase‐polymerase chain reaction (RT‐PCR) analysis at the indicated time points. SlUBI3 was used as an internal control. Data represent the mean ± standard deviation from three independent experiments. The statistical significance of the difference was confirmed by Student's t‐test (**P < 0.01). (B) Bacterial growth in infected leaves of WT AC+ and hp1 mutant plants treated with SA. Leaves were presprayed with 1 mm SA or mock solution, 48 h before inoculation with A. tumefaciens GV2260 at an inoculum of 1 × 104 colony‐forming units (cfu)/mL [optical density at 600 nm (OD600) = 0.00001]. Bacterial growth was determined at 0, 2 and 4 days post‐inoculation (dpi). Each data point consists of at least six samples. Error bars indicate standard deviation. The statistical analysis of the difference between the hp1 mutant and WT AC+ for both buffer and SA treatment was confirmed by Student's t‐test (**P < 0.01). Similar results were obtained in at least two independent experiments.
Functional deficiency of HP1/DDB1 (hp1) affects tumour formation by tumorigenic Agrobacterium
Tumorigenesis is the characteristic pathogenesis of virulent A. tumefaciens strains. However, tumour development during plant–Agrobacterium interactions is determined by both Agrobacterium and the host plant, as it involves the virulence of Agrobacterium and the regulation of tumour formation in the host by plant genes, in which, in particular, the regulation of plant cell division plays a critical role (Anand et al., 2007; Gelvin, 2003; Lee et al., 2009). To further investigate the role of HP1/DDB1 in the tumorigenesis by Agrobacterium, we inoculated the stems of WT AC+ and hp1 mutant plants with virulent A. tumefaciens A348 strain and assessed tumour development. Four weeks after inoculation, tumours had developed on the stems of both WT AC+ and hp1 mutant plants (Fig. 6A). However, the sizes of the tumours on the stems of hp1 mutant plants were significantly smaller than those on WT AC+ plants. We also quantified the biomass of tumours formed on the stems by measuring the fresh and dry weights of the tumours. As shown in Fig. 6B,C, the average fresh and dry weights of the tumours formed on hp1 mutant stems were 57 mg and 5.4 mg, respectively, which accounted for only 70% fresh weight and 50% dry weight of tumours developed in WT AC+ stems. We speculate that the weaker tumorigenesis in the hp1 mutant is a result of the defect of cell division caused by hp1 mutation (Caspi et al., 2008). Further experiments on the tumour cell division rate are needed to verify this hypothesis. Thus, our results show the complexity of tumour formation during plant–Agrobacterium interactions, in which cell division plays a critical role.
Figure 6.
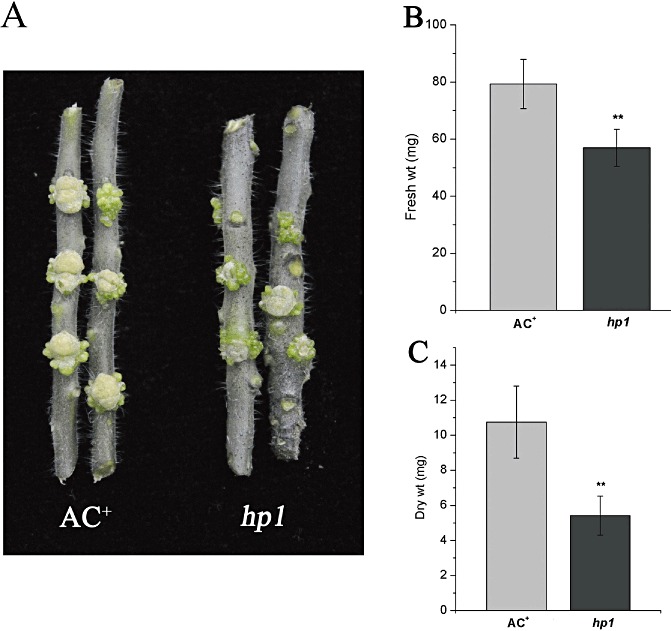
In planta tumorigenesis assays in wild‐type (WT) AC+ and hp1 mutant plants. (A) Tumours formed on stems of WT AC+ and hp1 mutant plants inoculated with the tumorigenic strain Agrobacterium tumefaciens A348. The photographs of the stems with tumours were taken 4 weeks after inoculation. (B, C) Quantification of the biomass of the tumours. The fresh and dry weights of the tumours developed on the stems of WT AC+ and hp1 mutant plants were measured 4 weeks after inoculation. The experiments were repeated twice with at least six samples. Error bars indicate standard deviation. The statistical significance of the difference was confirmed by Student's t‐test (**P < 0.01).
DISCUSSION
The interplay between Agrobacterium infection and plant immunity is very complicated. In general, it is thought that plants can recognize PAMPs from Agrobacterium and mount basal level defence—PTI (Zipfel et al., 2006). However, virulent Agrobacterium has evolved to overcome basal defence by hijacking the PTI signalling pathway to transfer T‐DNA into host chromatin and, consequently, to cause tumour formation (Djamei et al., 2007; Pitzschke et al., 2009; Zaltsman et al., 2010). Disarmed Agrobacterium strains without tumour‐triggering gene expression no longer cause tumorigenesis, but retain the ability to transfer T‐DNA into the plant chromosome. Thus, these nontumorigenic strains have been widely used as vectors to transfer genes of interest into the plant chromosome and, subsequently, to express the encoding proteins, either via stable transformation of the transgenic plants or transient transformation in the plant leaves (Gelvin, 2003). However, nontumorigenic Agrobacterium normally does not cause typical disease symptoms or proliferate to a great extent in leaves (Zipfel et al., 2006). In this study, we found that a tomato DDB1‐deficient mutant (high pigment‐1, hp1), which was originally identified as a spontaneous mutation with enhanced pigmentation, was hypersusceptible to nontumorigenic Agrobacterium infection. The majority of excised hp1 cotyledons infected with A. tumefaciens GV2260 died without developing callus (Fig. 1), suggesting that hp1 compromises the resistance to Agrobacterium. When inoculated with A. tumefaciens GV2260 by infiltration into leaves, the hp1 mutant moderately supported Agrobacterium growth and developed disease symptoms (Fig. 2). A consistently similar result was obtained when WT AC+ and hp1 mutant plant leaves were inoculated with another nontumorigenic A. tumefaciens strain EHA105 (Fig. 2C). Unfortunately, we were unable to test whether complementation of the hp1 mutant could restore the resistance phenotype. It was difficult to obtain restored transgenic plants overexpressing DDB1 driven by the 35S promoter in the hp1 mutant background (Y. Liu and J. Giovannoni, unpublished data), probably because the function of HP1/DDB1 is strictly controlled by the rate‐limiting expression level. We are currently conducting complementation experiments using tomato DDB1 as native promoter. Once the transgenic lines become available, it would be interesting to determine whether overexpression of this gene contributes to the increased level of resistance to Agrobacterium.
It is notable that there is a discrepancy between our results and data from a previous publication, in which it was shown that A. tumefaciens did not multiply after infiltration (Pruss et al., 2008). These contradictory observations could be caused by the different inoculation titres used in the experiments. A high inoculation titre, which was used by Pruss and colleagues, may saturate Agrobacterium growth and affect bacterial propagation because of a lack of nutrients in the harsh conditions of plant tissue. Interestingly, the Arabidopsis efr2 mutant is susceptible to Agrobacterium, as manifested by the increased transformation efficiency and chlorosis symptoms after infiltration with A. tumefaciens GV3101 strain, but does not support Agrobacterium growth in leaves (Zipfel et al., 2006). EFR recognizes the PAMP EF‐Tu from Agrobacterium and mounts a basal defence against Agrobacterium infection (Zipfel et al., 2006). Thus, from our observations, it seems that the hp1 mutation has a greater severity of effect than efr2 on PTI‐mediated resistance to Agrobacterium. As EFR is the PRR receptor to one particular PAMP EF‐fu from Agrobacterium, functional abrogation of one particular PTI pathway may only partially affect the resistance because of the redundancy of PTI pathways. Given the fact that plants belonging to the Solanum family, including tomato, do not contain a functional EFR gene (Zipfel et al., 2006), our results imply that HP1/DDB1 might function as an important signalling component essential for many PTI pathways other than the EF‐Tu/EFR pathway. Supporting the effect of hp1 on the PTI signalling triggered by Agrobacterium, several PTI marker genes exhibited less response to A. tumefaciens. Among the four PTI marker genes examined, SlPti5 and SlLrr22 were not induced, and SlGras2 and SlWaky28 showed reduced induction in hp1 mutant leaves after infiltration with A. tumefaciens GV2260 strain (Fig. 4B, E). Based on the fact that SlPti5, at least, is induced by multiple PAMPs, including flg22 and chitin (Nguyen et al., 2010), it is likely that hp1 mutation affects several PTI pathways, probably by compromising a common factor essential for PTI signalling.
The PR‐1 gene is a well‐known marker gene for defence reactions. In this study, we showed a correlation between the abolishment of SlPR1a1 expression and loss of resistance to Agrobacterium in the hp1 mutant (Fig. 5). Significantly, it has been demonstrated that the Escherichia coli DH5α strain can also induce PR‐1 expression in tobacco, even though it is nonpathogenic on plants, suggesting that PR‐1 induction by Agrobacterium is triggered by the perception of PAMPs (Pruss et al., 2008). A working model for PAMP‐triggered PR‐1 activation can be proposed on the basis of recent publications (Chinchilla et al., 2007; Djamei et al., 2007; Lu et al., 2010; Pitzschke et al., 2009; Zhang et al., 2010; Zipfel et al., 2006): PAMPs from Agrobacterium are first perceived by PRRs; PRRs coordinate with other factors, such as BAK1 and BIK1, to activate the MAPK cascade (Chinchilla et al., 2007; Lu et al., 2010; Zhang et al., 2010), triggering MAPK‐dependent activation of the VIP1 transcription factor, which, in turn, shuttles into the nucleus, where it indirectly activates PR‐1 expression (Djamei et al., 2007; Pitzschke et al., 2009). Thus, any step involved in PR‐1 induction could be a potential target of HP1/DDB1 for interference with defence signalling.
It is thought that the PR‐1 gene is mainly regulated through SA, an important component of the complex plant defence signalling network against pathogens (Durrant and Dong, 2004). Increasing evidence has indicated that SA is also involved in PAMP‐triggered basal defence signalling (Chen et al., 2009; Halim et al., 2009; Liu et al., 2010). However, at least in tomato, it remains unknown how the PAMP‐activated MAPK–VIP pathway and SA pathway coordinate to activate defence responses, such as the induction of the PR‐1 gene. It is unclear whether SA functions in a MAPK‐dependent manner or SA functions parallel to the MAPK–VIP pathway after recognition of PAMPs by PRRs. In this study, we investigated the functional relationship between SA and the hp1 mutation in terms of SlPR1a1 expression and resistance to Agrobacterium infection. We found that the exogenous application of SA on tomato leaves can induce SlPR1a1 expression and confer enhanced resistance to nontumorigenic A. tumefaciens GV2260. However, the hp1 mutant was insensitive to SA treatment, as revealed by the abolishment of SlPR1a1 induction and attenuation of the resistance to A. tumefaciens GV2260 (Fig. 5). Thus, our results indicate that HP1/DDB1 functions downstream of SA in defence signalling leading to SlPR1a1 induction. They also suggest that the deficiency of SA‐regulated defence signalling in the hp1 mutant is caused by interference in SA signal transduction, rather than by a defect in SA biosynthesis. However, we cannot rule out the possibility that the SA‐independent signalling pathway also contributes to HP1/DDB1‐dependent resistance, as we were unable to test the critical requirement of SA for SlPR1a1 induction or resistance to Agrobacterium because of a lack of the NahG transgenic line of WT AC+ which inhibits SA accumulation.
Consistent with the results of the Agrobacterium growth and disease symptom assay, the efficiency of Agrobacterium‐mediated transient expression in hp1 leaves was higher than that in WT AC+ leaves (Fig. 3), again indicating that the hp1 mutant is susceptible to Agrobacterium infection. However, the hp1 mutant did not show enhanced tumorigenesis by tumorigenic A. tumefaciens A348 strain with regard to the size and biomass of the tumours formed in stems (Fig. 6). This is probably because of the role of HP1/DDB1 in the regulation of cell growth. In the hp1 mutant plant, cell growth is reduced significantly compared with that in the WT AC+ plant, probably as a result of the decreased cell division rate in the hp1 mutant (Caspi et al., 2008). Thus, although hp1 facilitates Agrobacterium‐mediated transformation, the cell growth rate of the tumour may be restricted and, consequently, result in smaller sized tumours.
HP1/DDB1 is an important component of the CUL4‐mediated ubiquitin E3 ligase complex, which has multiple functions in plastid division (Liu et al., 2004; Wang et al., 2008), photomorphogenesis (Chen et al., 2010) and stress responses (Lee et al., 2010). Moreover, recent studies have demonstrated that HP1/DDB1 is also involved in the regulation of gene expression by epigenetic modifications (Higa et al., 2006; Pazhouhandeh et al., 2011; Zhao et al., 2010). As described above, PR‐1‐associated resistance is activated through the MAP3–VIP1 pathway and is also regulated through SA signalling. HP1/DDB1 probably contributes to the defence response via interaction with MAP3–VIP1 signalling, SA signalling, or both. We speculate that HP1/DDB1 might regulate the factor(s) downstream of SA in defence signalling, leading to SlPR1a1 expression, either through ubiquitination of the protein for degradation or via histone and/or DNA methylation of the corresponding gene for expression repression. Further experiments, such as the identification of the CUL4–HP1/DDB1 E3 substrate(s), are needed to verify this hypothesis.
EXPERIMENTAL PROCEDURES
Plant material and tomato cotyledon transformation
Tomato plants [wild‐type AC+ (Ailsa Craig) and hp1 mutant] were grown in the glasshouse under standard conditions (26 °C day, 18 °C night; 16 h light, 8 h dark). The light intensity was monitored by a luxmeter (TES‐1334A). Agrobacterium tumefaciens GV2260, carrying the binary vector pBI121, was grown at 28 °C in Luria–Bertani (LB) medium containing rifampicin (50 µg/mL) and kanamycin (50 µg/mL). Tomato cotyledon transformation was carried out according to the method described by Fillatti et al. (1987). The efficiency of callus induction was expressed as the quotient of the number of cotyledons having callus divided by the total number of cotyledons used for transformation in each plate.
Leaf infection with Agrobacterium tumefaciens
Agrobacterium tumefaciens GV2260 or EHA105 strain was propagated at 28 °C in LB medium with 50 µg/mL rifampicin. For leaf inoculation, overnight cultures were subcultured into fresh medium (LB, acetosyringone at 20 µm and appropriate antibiotics), and grown at 28 °C to an optical density at 600 nm (OD600) of 1–1.8. Cells were harvested by centrifugation, resuspended in buffer (10 mm MgCl2, 10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES), pH 5.6, and 0.006% Silwet L‐77) to OD600 = 0.00001 (approximately 1 × 104 cfu/mL). Three six‐week‐old tomato plants were vacuum infiltrated, and well‐expanded leaves were collected for analysis at 0, 2 and 4 dpi. To assess bacterial populations, eight 0.2826‐cm2 leaf discs were punched and ground in 1 mL of 10 mm MgCl2, diluted and plated on LB medium containing appropriate antibiotics. The number of bacteria (cfu/cm2) was counted 2 days after the plates had been kept at 28 °C. In the case of expression assessments of HP1, SlPR1a1, SlPti5, SlGras2, SlWrky28 and SlLrr22, the inoculum of A. tumefaciens GV2260 at OD600 = 0.1 (approximately 1 × 108 cfu/mL) was used.
Agrobacterium tumefaciens‐mediated transient transformation of tomato leaves
The media and culture conditions for induction and infection are detailed in Anand et al. (2007). Briefly, overnight cultures of A. tumefaciens GV2260 carrying the binary vector pBISN1 were washed with distilled water, and induced on agro‐induction medium supplemented with acetosyringone (150 µg/mL) at room temperature (24 °C) for 14–16 h. The induced cultures were washed with sterile distilled water and resuspended in buffer (10 mm MgCl2, 10 mm MES, pH 5.6, and 0.006% Silwet L‐77) to OD600 = 0.1 (approximately 1 × 108 cfu/mL). Six‐week‐old plants were vacuum infiltrated, and well‐expanded leaves were washed with water and immediately stained with X‐Gluc staining solution [0.1 m sodium phosphate buffer, pH 7.0, 0.1% Triton X‐100, 1 mm potassium ferricyanide, 1 mm potassium ferrocyanide, 10 mm ethylenediaminetetraacetic acid (EDTA), 200 mL/L methanol and 2 mm X‐Gluc] for 1 day at 37 °C in the dark. Stained leaves were placed in 95% ethanol and incubated at 37 °C until the leaves were cleared of chlorophyll, with the ethanol being replaced as necessary until the clearing was complete.
GUS activity was analysed using fluorometric assays (Jefferson et al., 1987). Protein extracts were prepared by grinding five to eight leaf discs from the Agrobacterium‐infected WT AC+ or hp1 mutant plants in a microcentrifuge tube containing GUS extraction buffer [50 mm sodium phosphate buffer, 1% sodium dodecylsulphate (SDS), 10 mL/L, 10 mm EDTA, 200 mL/L methanol, 0.1% Triton X‐100, 0.1% β‐mercaptoethanol], and two aliquots were assayed for each of the extracts to determine the protein concentration and GUS activity, following the method described previously (Jefferson et al., 1987). The protein concentration of plant extracts was determined spectrophotometrically using the Gene Quant pro (Amersham Biosciences, Piscataway, NJ, USA) based on the Bradford method (Bradford, 1976). The fluorescence of 4‐methylumbelliferone was measured with a Fluoroskan ascent FL2‐6 (Thermo Electron Corporation, Waltham, MA, USA).
Quantitative RT‐PCR assay for PTI marker gene expression
Six‐week‐old tomato plants were vacuum infiltrated with A. tumefaciens GV2260 in buffer (10 mm MgCl2, 10 mm MES, pH 5.6, and 0.006% Silwet L‐77) at an inoculum of OD600 = 0.1, or with buffer only as the mock inoculation. Leaf tissue was harvested at different time points after inoculation for RNA isolation. The 0‐hpi sample was harvested immediately prior to vacuum infiltration. Total RNAs were extracted using Trizol reagent according to the protocol provided by the manufacturer (Invitrogen, Carlsbad, CA, USA; http://www.Invitrogen.com/), and treated with DNaseI (TaKaRa, Dalian, Liaoning, China; http://www.takara‐bio.com).
Primers for real‐time RT‐PCR were designed for SlPR1a1 (SGN‐U577839; SlPR1a1‐F, 5′‐TGCTGGTGCTGTGAAGATGTG‐3′; SlPR1a1‐R, 5′‐CAGACTTTACCTGGAGCACACG‐3′), SlPti5 (SGN‐U571539; SlPti5‐F, 5′‐ATTCGCGATTCGGCTAGACATGGT‐3′; SlPti5‐R, 5′‐AGTAGTGCCTTAGCACCTCGCATT‐3′), SlLrr22 (SGN‐U585837; SlLrr22‐F, 5′‐AAGATTGGAGGTTGCCATTGGAGC‐3′; SlLrr22‐R, 5′‐ATCGCGATGAATGATCGGTGGAGT‐3′), SlGras2 (SGN‐U567396; SlGras2‐F, 5′‐TAATCCAAGGGATGAGCTTCT‐3′; SlGras2‐R, 5′‐CCACCAACGTGACCACCTT‐3′), SlWrky28 (SGN‐U586086; SlWrky28‐F, 5′‐ACAGATGCAGCTACCTCATCCTCA‐3′; SlWrky28‐R, 5′‐GTGCTCAAAGCCTCATGGTTCTTG‐3′), SlUBI3 (GenBank accession no. X58253; SlUBI3‐F, 5′‐AGGTTGATGACACTGGAAAGGTT‐3′; SlUBI3‐R, 5′‐AATCGCCTCCAGCCTTGTTGTA‐3′). Real‐time PCR was performed using an SsoFast EvaGreen Supermix (Bio‐Rad catalogue no. 172‐5203, Hercules, CA, USA). Each sample was amplified in triplicate and all PCRs were performed on an Applied Biosystems StepOne Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA; http://www.appliedbiosystems.com.cn/). Dissociation curve analysis was performed at the end of each run to ensure that unique products were amplified. The tomato SlUBI3 gene was used as a reference. The RT‐PCR conditions were as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 20 s. The expression level was normalized to the SlUBI3 control, and relative expression values were determined against the buffer‐treated sample or WT AC+ sample using the 2−ΔΔCt method. To confirm the specificity of the PCR, PCR products were verified on a 1% agarose gel for the accurate amplification product size. A pairwise Student's t‐test was performed to obtain the P values indicated in the figures.
SA treatment
SA treatment assay was performed according to Wu and Yang (2010). Briefly, plants were sprayed with a solution of SA containing 0.006% Silwet L‐77. SA solution at a concentration of 0.5 mm was used to determine SlPR1a1 expression, and a 1‐mm solution was used to induce resistance to A. tumefaciens GV2260. Control plants were sprayed with water containing the same concentration of Silwet L‐77. Plants were covered with clear transparent plastic foil for 4 h to retain moisture.
In planta tumour assay
Tumorigenic A. tumefaciens A348 strain containing the octopine type Ti plasmid (pTiA6) was cultured as described above in transient transformation methods. Stems of WT Ailsa Craig or hp1 mutant plants were inoculated by slight injury to the stem, using a needle dipped in A. tumefaciens A348 suspension culture (OD600 = 0.1). Tumours on shoots were scored after 4 weeks.
ACKNOWLEDGEMENTS
We thank Dr Kirankumar Mysore for kindly providing the GUS construct and A. tumefaciens A348 strain. This work was supported by the National Science Fund for Distinguished Young Scholars (No. 30825030), the National Science and Technology Key Project of China (Nos. 2009ZX08001‐011B, 2009ZX08009‐072B and 2011CB100401) and the University of Idaho Startup Funding for F. Xiao.
REFERENCES
- Anand, A. , Vaghchhipawala, Z. , Ryu, C.M. , Kang, L. , Wang, K. , del‐Pozo, O. , Martin, G.B. and Mysore, K.S. (2007) Identification and characterization of plant genes involved in Agrobacterium‐mediated plant transformation by virus‐induced gene silencing. Mol. Plant–Microbe Interact. 20, 41–52. [DOI] [PubMed] [Google Scholar]
- Anand, A. , Uppalapati, S.R. , Ryu, C.M. , Allen, S.N. , Kang, L. , Tang, Y. and Mysore, K.S. (2008) Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens . Plant Physiol. 146, 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers, S. , Li, T. , Yi, X. , MacCoss, M.J. , Moon, R.T. and Zheng, N. (2006) Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature, 443, 590–593. [DOI] [PubMed] [Google Scholar]
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.L. , Gomez‐Gomez, L. , Boller, T. , Ausubel, F.M. and Sheen, J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M. (2005) Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6, 973–979. [DOI] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Braun, S. , Garcia, J.F. , Rowley, M. , Rougemaille, M. , Shankar, S. and Madhani, H.D. (2011) The Cul4‐Ddb1Cdt2 ubiquitin ligase inhibits invasion of a boundary‐associated antisilencing factor into heterochromatin. Cell, 144, 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi, N. , Levin, I. , Chamovitz, D.A. and Reuveni, M. (2008) A mutation in the tomato DDB1 gene affects cell and chloroplast compartment size and CDT1 transcript. Plant Signal. Behav. 3, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore, R.C. , Havens, C.G. , Manning, A.L. , Li, J.M. , Flynn, R.L. , Tse, A. , Jin, J. , Dyson, N.J. , Walter, J.C. and Zou, L. (2010) CRL4(Cdt2)‐mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell, 40, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Xue, L. , Chintamanani, S. , Germain, H. , Lin, H. , Cui, H. , Cai, R. , Zuo, J. , Tang, X. , Li, X. , Guo, H. and Zhou, J.M. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3‐LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis . Plant Cell, 21, 2527–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Huang, X. , Gusmaroli, G. , Terzaghi, W. , Lau, O.S. , Yanagawa, Y. , Zhang, Y. , Li, J. , Lee, J.H. , Zhu, D. and Deng, X.W. (2010) Arabidopsis CULLIN4‐damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1‐SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell, 22, 108–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla, D. , Zipfel, C. , Robatzek, S. , Kemmerling, B. , Nurnberger, T. , Jones, J.D. , Felix, G. and Boller, T. (2007) A flagellin‐induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature, 448, 497–500. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D.G. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Djamei, A. , Pitzschke, A. , Nakagami, H. , Rajh, I. and Hirt, H. (2007) Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science, 318, 453–456. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E. and Dong, X. (2004) Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Fillatti, J.J. , Kiser, J. , Rose, R. and Comai, L. (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat. Biotechnol. 5, 726–730. [Google Scholar]
- Gelvin, S.B. (2003) Agrobacterium‐mediated plant transformation: the biology behind the ‘gene‐jockeying’ tool. Microbiol. Mol. Biol. Rev. 67, 16–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim, V.A. , Altmann, S. , Ellinger, D. , Eschen‐Lippold, L. , Miersch, O. , Scheel, D. and Rosahl, S. (2009) PAMP‐induced defense responses in potato require both salicylic acid and jasmonic acid. Plant J. 57, 230–242. [DOI] [PubMed] [Google Scholar]
- Higa, L.A. , Wu, M. , Ye, T. , Kobayashi, R. , Sun, H. and Zhang, H. (2006) CUL4–DDB1 ubiquitin ligase interacts with multiple WD40‐repeat proteins and regulates histone methylation. Nat. Cell Biol. 8, 1277–1283. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Jefferson, R.A. , Kavanagh, T.A. and Bevan, M.W. (1987) GUS fusions: β‐glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J. , Arias, E.E. , Chen, J. , Harper, J.W. and Walter, J.C. (2006) A family of diverse Cul4‐Ddb1‐interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell, 22, 709–721. [DOI] [PubMed] [Google Scholar]
- Kendrick, R.E. , Kerckhoffs, L.H.J. , Van Tuinen, A. and Koornneef, M. (1997) Photomorphogenic mutants of tomato. Plant Cell Environ. 20, 746–751. [PubMed] [Google Scholar]
- Lee, C.W. , Efetova, M. , Engelmann, J.C. , Kramell, R. , Wasternack, C. , Ludwig‐Muller, J. , Hedrich, R. and Deeken, R. (2009) Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana . Plant Cell, 21, 2948–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H. , Yoon, H.J. , Terzaghi, W. , Martinez, C. , Dai, M. , Li, J. , Byun, M.O. and Deng, X.W. (2010) DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4‐based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell, 22, 1716–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Chen, X. , Garbutt, K.C. , Zhou, P. and Zheng, N. (2006) Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell, 124, 105–117. [DOI] [PubMed] [Google Scholar]
- Lieberman, M. , Segev, O. , Gilboa, N. , Lalazar, A. and Levin, I. (2004) The tomato homolog of the gene encoding UV DAMAGED DNA BINDING protein 1 (DDB1) underlined as the gene that causes the high pigment‐1 mutant phenotype. Theor. Appl. Genet. 108, 1574–1581. [DOI] [PubMed] [Google Scholar]
- Liu, P.P. , Yang, Y. , Pichersky, E. and Klessig, D.F. (2010) Altering expression of benzoic acid/salicylic acid carboxyl methyltransferase 1 compromises systemic acquired resistance and PAMP‐triggered immunity in Arabidopsis . Mol. Plant–Microbe Interact. 23, 82–90. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Roof, S. , Ye, Z. , Barry, C. , van Tuinen, A. , Vrebalov, J. , Bowler, C. and Giovannoni, J. (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc. Natl. Acad. Sci. USA, 101, 9897–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, D. , Wu, S. , Gao, X. , Zhang, Y. , Shan, L. and He, P. (2010) A receptor‐like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA, 107, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli, A.C. , Fenzi, F. , Ciliento, R. , Alfano, F. and Bowler, C. (1999) Phenotype of the tomato high pigment‐2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell, 11, 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, J. , Mysore, K.S. , Zheng, C. , Knue, M.K. , Matthysse, A.G. and Gelvin, S.B. (1999) Identification of T‐DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium . Mol. Gen. Genet. 261, 429–438. [DOI] [PubMed] [Google Scholar]
- Nguyen, H.P. , Chakravarthy, S. , Velasquez, A.C. , McLane, H.L. , Zeng, L. , Nakayashiki, H. , Park, D.H. , Collmer, A. and Martin, G. (2010) Methods to study PAMP‐triggered immunity using tomato and Nicotiana benthamiana . Mol. Plant–Microbe Interact. 23, 991–999. [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh, M. , Molinier, J. , Berr, A. and Genschik, P. (2011) MSI4/FVE interacts with CUL4–DDB1 and a PRC2‐like complex to control epigenetic regulation of flowering time in Arabidopsis . Proc. Natl. Acad. Sci. USA, 108, 3430–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski, M.D. and Deshaies, R.J. (2005) Function and regulation of cullin‐RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20. [DOI] [PubMed] [Google Scholar]
- Pitzschke, A. , Djamei, A. , Teige, M. and Hirt, H. (2009) VIP1 response elements mediate mitogen‐activated protein kinase 3‐induced stress gene expression. Proc. Natl. Acad. Sci. USA, 106, 18 414–18 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss, G.J. , Nester, E.W. and Vance, V. (2008) Infiltration with Agrobacterium tumefaciens induces host defense and development‐dependent responses in the infiltrated zone. Mol. Plant–Microbe Interact. 21, 1528–1538. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Liu, J. , Feng, Y. , Niu, X. , Giovannoni, J. and Liu, Y. (2008) Altered plastid levels and potential for improved fruit nutrient content by downregulation of the tomato DDB1‐interacting protein CUL4. Plant J. 55, 89–103. [DOI] [PubMed] [Google Scholar]
- Wroblewski, T. , Tomczak, A. and Michelmore, R. (2005) Optimization of Agrobacterium‐mediated transient assays of gene expression in lettuce, tomato and Arabidopsis . Plant Biotechnol. J. 3, 259–273. [DOI] [PubMed] [Google Scholar]
- Wu, L. and Yang, H.Q. (2010) CRYPTOCHROME 1 is implicated in promoting R protein‐mediated plant resistance to Pseudomonas syringae in Arabidopsis . Mol. Plant, 3, 539–548. [DOI] [PubMed] [Google Scholar]
- Yen, H.C. , Shelton, B.A. , Howard, L.R. , Lee, S. , Vrebalov, J. and Giovannoni, J.J. (1997) The tomato high‐pigment (hp) locus maps to chromosome 2 and influences plastome copy number and fruit quality. Theor. Appl. Genet. 95, 1069–1079. [Google Scholar]
- Zaltsman, A. , Krichevsky, A. , Loyter, A. and Citovsky, V. (2010) Agrobacterium induces expression of a host F‐box protein required for tumorigenicity. Cell Host Microbe, 7, 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Li, W. , Xiang, T. , Liu, Z. , Laluk, K. , Ding, X. , Zou, Y. , Gao, M. , Zhang, X. , Chen, S. , Mengiste, T. , Zhang, Y. and Zhou, J.M. (2010) Receptor‐like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe, 7, 290–301. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Shen, Y. , Yang, S. , Wang, J. , Hu, Q. , Wang, Y. and He, Q. (2010) Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa . J. Biol. Chem. 285, 4355–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D. , Boller, T. and Felix, G. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]


