SUMMARY
Xanthomonas oryzae pv. oryzae, the causal agent of bacterial blight of rice, produces siderophores only under iron‐limiting conditions. We screened 15 400 mTn5‐induced mutants of X. oryzae pv. oryzae and isolated 27 mutants that produced siderophores even under iron‐replete conditions. We found that the mTn5 insertions in 25 of these mutants were in or close to six genes. Mutants with insertions in five of these genes [colS, XOO1806 (a conserved hypothetical protein), acnB, prpR and prpB] exhibited a deficiency for growth on iron‐limiting medium and a decrease in virulence. Insertions in a sixth gene, XOO0007 (a conserved hypothetical protein), were found to affect the ability to grow on iron‐limiting medium, but did not affect the virulence. Targeted gene disruptants for colR (encoding the predicted cognate regulatory protein for ColS) also exhibited a deficiency for growth on iron‐limiting medium and a decrease in virulence. colR and colS mutants were defective in the elicitation of hypersensitive response symptoms on the nonhost tomato. In addition, colR and colS mutants induced a rice basal defence response, suggesting that they are compromised in the suppression of host innate immunity. Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis demonstrated that a functional ColRS system is required for the optimal expression of several genes encoding components of the type 3 secretion system (T3SS) of X. oryzae pv. oryzae. Our results demonstrate the role of several novel genes, including colR/colS, in the promotion of growth on iron‐limiting medium and the virulence of X. oryzae pv. oryzae.
INTRODUCTION
Xanthomonas oryzae pv. oryzae, the causal agent of bacterial blight of rice, uses a diverse array of virulence functions: surface polysaccharides; adhesins; a type 2 secretion system and its secreted proteins, which include enzymes involved in the degradation of rice cell walls; a type 3 secretion system (T3SS) and its secreted effectors, which function inside rice cells to suppress host innate immunity and up‐regulate the expression of host susceptibility factors; a diffusible signalling factor (DSF)‐mediated cell–cell signalling system; iron uptake functions; a phytase‐like protein, etc. (Buttner and Bonas, 2010; Nino‐Liu et al., 2006).
Several regulatory functions are also required for the virulence of X. oryzae pv. oryzae. One of these regulatory functions is HrpG, a transcriptional activator of the OmpR family of two‐component systems, which controls the expression of the genes for T3SS and its secreted proteins. The HrpG protein exerts its effects by up‐regulating the expression of another transcriptional activator, called HrpX (Buttner and Bonas, 2010). The PhoPQ system is required for virulence and controls the expression of the hrpG (hypersensitive response and pathogenicity G) gene (Lee et al., 2008). In addition to the PhoPQ system, the H‐NS protein XrvA and Trh (a member of the GntR regulator family) also regulate the expression of hrpG (Feng et al., 2009; Tsuge et al., 2006). The PhoPQ system and another two‐component system, RaxRH, are required for Ax21 (a peptide that induces rice XA21‐mediated immunity) activity (2008, 2009). The rpfF (regulation of pathogenicity factor F) gene‐mediated DSF system has been shown to promote the virulence of X. oryzae pv. oryzae by facilitating iron uptake (Chatterjee and Sonti, 2002).
Xanthomonas oryzae pv. oryzae encodes an xss (Xanthomonas siderophore synthesis) operon, which is required for the biosynthesis of a vibrioferrin‐type siderophore, and a feoABC operon‐encoded Feo system for ferrous iron uptake (Pandey and Sonti, 2010). Siderophore‐deficient mutants and feoB mutants grow poorly in iron‐limiting conditions, but only the feoB mutant is virulence deficient (Pandey and Sonti, 2010). Xanthomonas oryzae pv. oryzae produces siderophores only in iron‐limiting conditions. A mutation in the fur (ferric uptake regulator) gene causes the constitutive production of siderophores (Subramoni and Sonti, 2005). In order to identify additional regulatory functions that influence iron uptake in X. oryzae pv. oryzae, we screened 15 400 mTn5‐induced mutants for the ability to produce siderophores even on iron‐replete medium. Such mutants, which we refer to as siderophore overproducers, or sop, mutants, may possess mutations in either regulatory functions or certain iron uptake processes whose suboptimal functioning results in siderophore overproduction as a compensatory measure. Using thermal asymmetric interlaced polymerase chain reaction (TAIL PCR) and, subsequently, PCR with gene‐specific primers, we were able to localize the insertions in 25/27 sop mutants to be in/close to six genes. One of the affected genes is colS, which is the sensor component of the two‐component system, ColRS. Targeted mutagenesis of the colR and colS genes, accompanied by complementation analysis, demonstrates that this two‐component system is required for virulence on rice, an ability to grow on iron‐limiting medium and optimal functioning of T3SS.
RESULTS
Isolation of X. oryzae pv. oryzae sop mutants
An X. oryzae pv. oryzae mTn5 library was generated (see Experimental procedures) using in vivo transposon mutagenesis of strain BXO43 (our laboratory wild‐type strain). A total of 15 400 mTn5 mutants were screened on peptone–sucrose agar‐chrome azurol sulphonate (PSA‐CAS) plates. After repeated screening, 27 sop mutants that exhibited consistent phenotypes of siderophore overproduction (Fig. 1A), without exhibiting apparent growth defects, were selected for further characterization.
Figure 1.
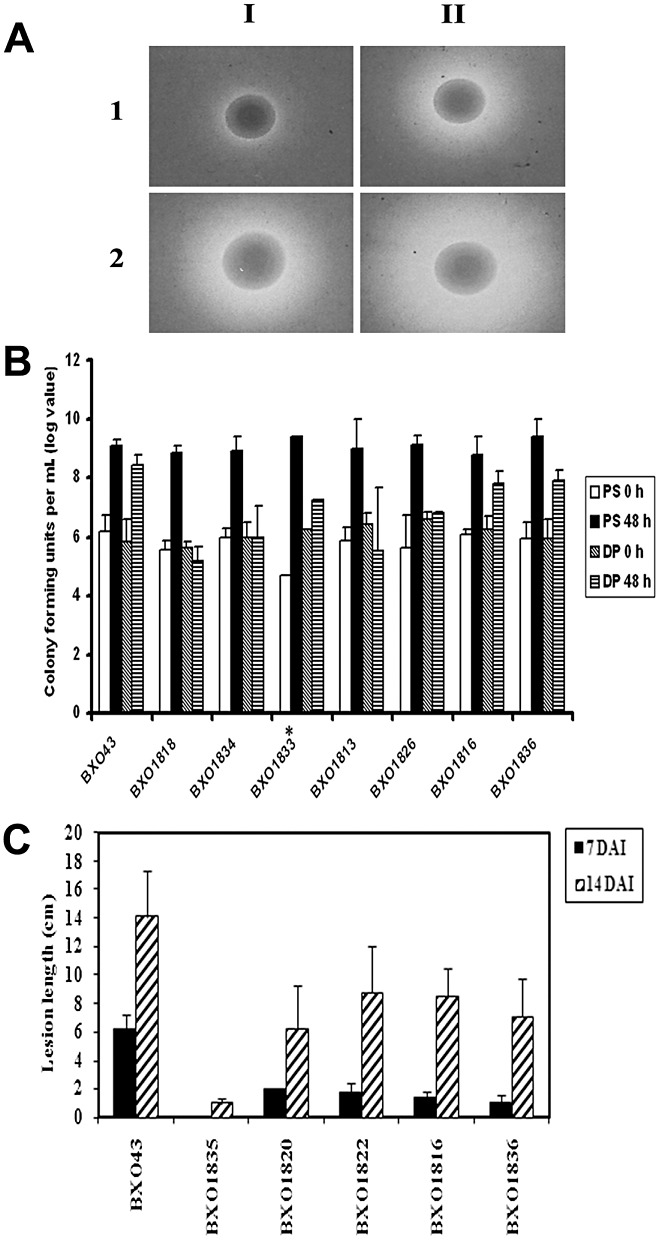
Siderophore phenotype, growth on iron‐limiting medium and virulence properties of siderophore overproducer (sop) mutants of Xanthomonas oryzae pv. oryzae. (A) Siderophore production phenotype of X. oryzae pv. oryzae strains: 1, BXO43 (wild‐type strain); 2, BXO1816 (a representative sop mutant). The strains were grown on: I, peptone–sucrose agar plates containing chrome azurol sulphonate (PSA‐CAS), considered as an iron‐replete condition; II, PSA‐CAS + 40 µm 2,2′‐dipyridyl, an iron chelator (PSA‐CAS + DP), considered as an iron‐limiting condition. Siderophore production is indicated by the presence of an extended halo around the colony. BXO43 produces siderophore only under iron‐limiting conditions, whereas BXO1816 produces siderophore in iron‐replete as well as iron‐limiting conditions. (B) Xanthomonas oryzae pv. oryzae sop mutants are deficient for growth on iron‐limiting medium. Cell numbers were estimated in X. oryzae pv. oryzae cultures grown in peptone–sucrose (PS) and PS + 140 µm DP at 0 h and after 48 h of growth. Data are the mean log colony‐forming unit (cfu) values ± standard deviation from three independent experiments. Strains are as indicated on the x axis: BXO43, BXO1818 (colS1::mTn5), BXO1834 (XOO1806‐2::mTn5), BXO1833 (acnB2::mTn5), BXO1813 (zxx301::mTn5, a mutation in the XOO0007–XOO0008 intergenic region), BXO1826 (XOO0007‐6::mTn5), BXO1816 (prpR1::mTn5) and BXO1836 (prpB1::mTn5). Student's two‐tailed t‐test for independent means was performed and the growth of each of the mutants on iron‐limiting medium was significantly different (P < 0.05) from the wild‐type strain. The asterisk indicates that the cell numbers given for this strain are expressed as the mean of the values obtained from two experiments. (C) Virulence phenotypes of X. oryzae pv. oryzae sop mutants. Inoculations were performed on leaves of 40‐day‐old glasshouse‐grown plants of the susceptible rice cultivar Taichung Native‐1 (TN‐1). Lesion lengths were measured 7 and 14 days after inoculation (DAI). The mean and standard deviation of 15 replicate measurements are given. The results from one experiment are presented. Similar results were obtained in two other independent experiments. Student's t‐test showed that the values for the mutants were significantly different (P < 0.05) from that of the wild‐type strain at both 7 and 14 days. Strains are: BXO43, BXO1835 (colS4::mTn5), BXO1820 (XOO1806‐1::mTn5), BXO1822 (acnB1::mTn5), BXO1816 and BXO1836.
Identification of mutated genes
Using mTn5‐specific as well as arbitrary primers (Table S1, see Supporting Information), TAIL PCR was performed to amplify the flanking regions of mTn5 insertions, and the PCR products thus obtained were sequenced. The expected locations of the transposon insertions in the genome were obtained by performing blast searches with the X. oryzae pv. oryzae KACC10331 genome sequence (Lee et al., 2005). Subsequently, locus‐specific primers were designed to confirm that the insertions were indeed at the expected locations. The locations of insertions in 25 mutants were confirmed by a combination of locus‐ and transposon‐specific primers, and these were found to be in or close to different open reading frames (ORFs). For these 25 mutants, the complete amino acid sequences of X. oryzae pv. oryzae ORFs that carried the mTn5 insertion in/close to the genes were analysed to determine homology with previously characterized proteins in sequence databases and to detect conserved domains (Table 1).
Table 1.
Summary of sequence analyses of open reading frames (ORFs) disrupted in Xanthomonas oryzae pv. oryzae siderophore overproducer (sop) mutants.
| ORF (amino acids)* | No. of mTn5 insertions in/close to gene; phenotypes of mutants† | Predicted function | Presence of conserved domain‡ |
|---|---|---|---|
| XOO0007 (397) | 15; Vir+, Lir‐ | Unknown | Putative Zn‐dependent protease, contains TPR (tetratrico tetrapeptide repeats), 5.2e‐03§ |
| XOO0891 (591) | 1; Vir‐, Lir‐ | PrpR; propionate catabolism regulatory protein | PrpR_N superfamily; propionate catabolism activator, 4.24e‐45§ |
| XOO0892 (298) | 1; Vir‐, Lir‐ | PrpB; 2‐methyl isocitrate lyase | PEP phosphonomutase and related enzymes, 4e‐85§ |
| XOO1207 (450) | 4; Vir‐, Lir‐ | ColS; two‐component system sensor protein | Histidine kinase A |
| (dimerization/phosphoacceptor domain), e‐13§ | |||
| Histidine kinase‐like ATPases, 2.2e‐10§ | |||
| XOO1806 (289) | 2; Vir‐, Lir‐ | Unknown | Orn/DAP/Arg decarboxylases family 2 signature 2; partial, 0.0¶ |
| XOO2862 (863) | 2; Vir‐, Lir‐ | AcnB; aconitate hydratase | AcnB, aconitase B catalytic domain, 0e+00§ |
| Aconitase B swivel domain, 5.88e‐54§ |
Size of the ORF in amino acids.
Vir+ and Vir‐ indicate proficiency and deficiency for virulence, respectively. Lir‐ indicates deficiency for growth on low‐iron medium.
The E‐value obtained from the conserved domain analysis.
Using the National Center for Biotechnology Information (NCBI) Conserved Domain Search program.
Using the EBI InterProScan program.
Fifteen of the 25 sop mutants carried insertions either in a gene annotated as XOO0007, which encodes for a conserved hypothetical protein, or in the intergenic region between this gene and the adjacent tonB gene (XOO0008). Other genes mutated in sop mutants included: colS (XOO1207), which encodes a putative two‐component sensor protein; acnB (XOO2862), which encodes a putative aconitase (Acn); prpB (XOO0892), which encodes a putative methyl isocitrate lyase involved in propionate catabolism; prpR (XOO0891), which encodes a putative regulator of prp genes; and XOO1806, which encodes a conserved hypothetical protein. Southern analysis was performed on 10 sop mutants and all were found to have single‐copy insertions of mTn5 (data not shown). These 10 mutants included the following: two insertions each in the colS, acnB, XOO0007 and XOO1806 genes, and one insertion each in the prpB and prpR genes.
Siderophore overproduction by sop mutants is suppressed by iron supplementation
In order to determine whether the siderophore overproduction phenotype is caused by a defect in sensing the levels of intracellular iron, the secretion of siderophores was monitored on PSA‐CAS plates supplemented with 30 µm FeSO4. All of the sop mutants stopped secreting siderophores under these conditions.
Evaluation of X. oryzae pv. oryzae sop mutants for virulence and growth on iron‐limiting medium
The insertions in the XOO0007 gene are concentrated in the 3′ region of the ORF and two insertions are in the intergenic region between the XOO0007 and XOO0008 (tonB) genes (Fig. S1, see Supporting Information). These mutants are defective for growth on iron‐limiting medium (data are shown for two mutants, BXO1813 and BXO1826, in Fig. 1B), but retain wild‐type levels of virulence (data not shown). The strains carrying insertions in the XOO1806 gene are defective for growth on iron‐limiting medium (data shown only for BXO1834 in Fig. 1B) and exhibit reduced virulence (data shown only for BXO1820 in Fig. 1C).
The sop mutants (BXO1822 and BXO1833) with insertions in the X. oryzae pv. oryzae homologue of acnB (XOO2862) exhibit a virulence deficiency (data shown only for BXO1822 in Fig. 1C) and a growth defect on iron‐limiting medium (data shown only for BXO1833 in Fig. 1B). The two sop mutants (BXO1816 and BXO1836) with insertions in the prpR (XOO0891) and prpB (XOO0892) genes exhibit a deficiency for virulence (Fig. 1C) and for growth on iron‐limiting medium (Fig. 1B).
Four sop mutants (BXO1818, BXO1823, BXO1824 and BXO1835) carried insertions in the colS gene (Fig. 2). The predicted protein product of colS is 450 amino acids in length and exhibits significant similarity to the sensor component of bacterial two‐component regulatory systems. In particular, it exhibits 37% identity and 55% similarity (across a 264‐amino‐acid segment in the C‐terminal region) to ColS, the sensor component of the Pseudomonas fluorescens WCS365 two‐component system that promotes the colonization of the roots of several plants, including potato, tomato, radish and wheat (Dekkers et al., 1998). The ColS protein of X. oryzae pv. oryzae exhibits homology (80% identity and 87% similarity) to the predicted products of the XCC3106, XC_1050 and XAC3249 genes of Xanthomonas campestris pv. campestris strains ATCC33913 and 8004, and Xanthomonas citri ssp. citri strain 306, respectively.
Figure 2.
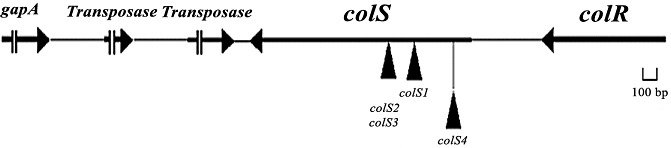
Location of mTn5 insertions and gene organization in the Xanthomonas oryzae pv. oryzae genomic region containing the colS gene. The filled triangles represent the locations of mTn5 insertions. The arrows indicate the transcriptional orientations of the genes. The bold lines indicate open reading frames (ORFs) and the lighter lines indicate intergenic regions. The colS2 (BXO1823) and colS3 (BXO1824) mutants carry the mTn5 insertion at the same site in the colS gene. Immediately downstream of colS are ORFs coding for putative transposase genes, followed by gapA encoding for glyceraldehyde‐3‐phosphate dehydrogenase. These genes are located in the opposite transcriptional orientation with respect to colS. The gene organization given here is as specified by Lee et al. (2005) in the genome of X. oryzae pv. oryzae strain KACC10331.
All four colS mutants (BXO1818, BXO1823, BXO1824 and BXO1835) isolated in this study exhibit virulence deficiency. The data for BXO1835 are shown in Fig. 1C, but similar results were obtained with the other three mutants. These mutants grow as well as the wild‐type strain on peptone–sucrose (PS) medium, but show severe growth deficiency on iron‐limiting medium (Fig. 1B). Data for the BXO1818 strain are shown (Fig. 1B), but similar results were obtained with the other three mutants.
A colR mutant exhibits a Sop phenotype and is deficient for virulence and growth on iron‐limiting medium
The colR gene (XOO1208) is present upstream of the colS gene (Fig. 2), with an intergenic interval of 430 bp in the genome of X. oryzae pv. oryzae strain KACC10331 (Lee et al., 2005). An ISXo1 element is present in this intergenic region in the genome of X. oryzae pv. oryzae strain PXO99A (Salzberg et al., 2008). The predicted protein product of colR is 246 amino acids in length and exhibits significant similarity to the regulator components of bacterial two‐component systems. In particular, it exhibits 58% identity and 75% similarity throughout the length of the protein to ColR, the regulatory component of the P. fluorescens WCS365 two‐component system. The ColR protein of X. oryzae pv. oryzae also exhibits homology (99% identity) to the products of the XCC3107, XC_1049 and XAC3250 genes of X. campestris pv. campestris strains ATCC33913 and 8004, and X. citri ssp. citri strain 306, respectively.
In order to ascertain the phenotypes associated with a colR mutation, an X. oryzae pv. oryzae colR mutant (strain BXO1843) was generated by homologous integration of a recombinant pK18mob vector containing an internal fragment of the colR gene, as described in Experimental procedures. An additional X. oryzae pv. oryzae colS mutant (BXO1844) was generated using a similar strategy. The BXO1843 (colR mutant) and BXO1844 (colS mutant) strains exhibit a Sop phenotype (Fig. 3A). The complementing plasmids carrying the colR gene (pAP24) and colS gene (pAP25) were introduced into the BXO1843 and BXO1844 strains, respectively, to create the complemented strains, BXO2243 (ColR+) and BXO2241 (ColS+). The BXO2243 and BXO2241 strains do not produce siderophores on PSA‐CAS medium (Fig. 3A).
Figure 3.
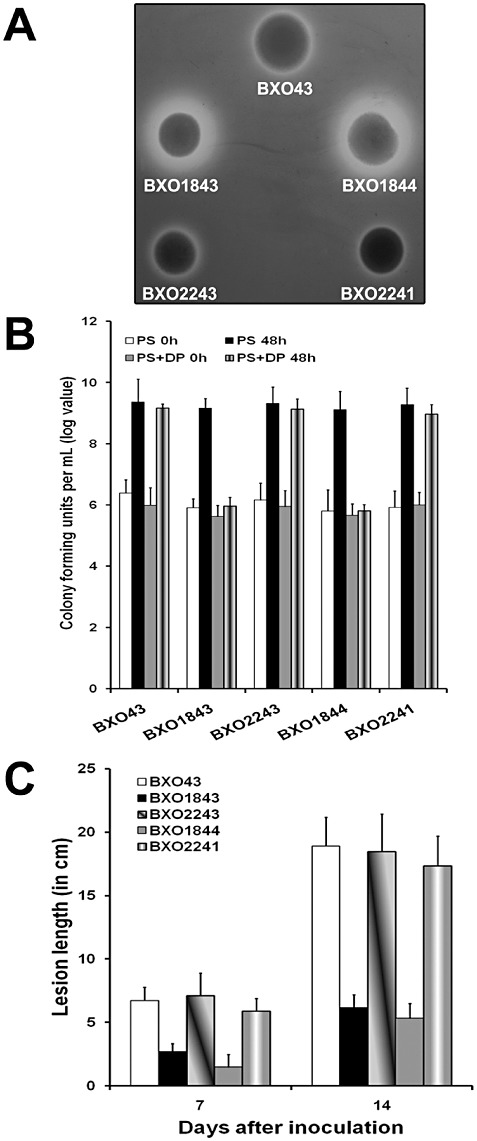
Siderophore production, virulence and growth phenotype of a colR mutant of Xanthomonas oryzae pv. oryzae. Xanthomonas oryzae pv. oryzae colR and colS mutants were constructed by integration of suicide plasmid pK18mob. (A) Siderophore production is assayed as the presence of a halo around colonies growing on peptone–sucrose agar (PSA) medium containing chrome azurol sulphonate (CAS). Strains: BXO43 (wild‐type strain), BXO1843 (colR1::pK18mob), BXO1844 (colS5::pK18mob), complemented strain BXO2243 (BXO1843/pAP24, ColR+) and complemented strain BXO2241 (BXO1844/pAP25, ColS+). The pAP24 and pAP25 plasmids carry the cloned X. oryzae pv. oryzae colR and colS genes, respectively. A colR mutant overproduces siderophores. Wild‐type levels of siderophore are restored by the addition of pAP24. (B) A colR mutant is deficient for growth on iron‐limiting medium. Cell numbers were estimated in X. oryzae pv. oryzae cultures grown in peptone–sucrose (PS) medium and PS + 140 µm 2,2′‐dipyridyl (DP) at 0 h and after 48 h of growth. Data are the mean log cfu values ± standard deviation from three independent experiments. (C) A colR mutant is severely virulence deficient. Inoculations were performed on leaves of susceptible rice cultivar Taichung Native‐1 (TN‐1), as described in Experimental procedures. Lesion lengths were measured 7 and 14 days after inoculation. The mean and standard deviation of 15 replicate measurements are given. The results from one experiment are presented. Similar results were obtained in independent experiments.
In PS medium, no significant difference in growth yield was observed between the wild‐type strain BXO43 and the BXO1843 (colR mutant) and BXO1844 (colS mutant) strains. The BXO1843 and BXO1844 strains exhibit a deficiency for growth when cultured in iron‐limiting conditions (Fig. 3B). However, the complemented strains, BXO2243 (ColR+) and BXO2241 (ColS+), exhibit a level of cell density similar to that observed for BXO43 under these conditions. These results indicate that the ColRS system of X. oryzae pv. oryzae is required for growth under iron limitation.
The virulence phenotypes of BXO43 (wild‐type strain), BXO1843 (colR mutant), BXO2243 (ColR+), BXO1844 (colS mutant) and BXO2241 (ColS+) were determined by inoculating rice leaves and assessing lesion lengths at 7 and 14 days after inoculation. At both time points, the BXO1843 and BXO1844 strains showed significantly smaller lesions when compared with those produced by the BXO43, BXO2243 and BXO2241 strains (Fig. 3C). However, we observed that the BXO1843 and BXO1844 strains produced slightly elongated lesions at the 14‐day time point. Bacteria were re‐isolated from the leading edge of the lesion in rice leaves. These bacteria were found to exhibit the wild‐type phenotype with respect to siderophore production and virulence (data not shown). In addition, these strains were susceptible to kanamycin (Km), indicating that they were ColR+ (BXO1845) and ColS+ (BXO1846) revertants that arose after loss of the integrated plasmid as a result of homologous recombination events involving internally duplicated sequences of the colR and colS genes, respectively. It is relevant to note that reversions were not observed with any of the colS::mTn5 insertion mutants, providing additional evidence that ColR+ and ColS+ revertants observed following inoculation with BXO1843 and BXO1844 are caused by homologous recombination events that result in the loss of integrated plasmid. Taken together, these results indicate that the ColRS system is essential for the virulence of X. oryzae pv. oryzae on rice.
Effect of colR and colS mutations on the expression of genes encoding iron uptake functions
Xanthomonas oryzae pv. oryzae encodes an xss operon that is required for the biosynthesis of siderophores, as well as an feoABC operon‐encoded Feo system for ferrous iron uptake (Pandey and Sonti, 2010). The effect of colR and colS mutations on the expression of the feoB (encodes the ferrous iron transporter) and xssE (encoded in the xss operon) genes was assessed using real‐time quantitative PCR, as described in Experimental procedures. When compared with the wild‐type strain BXO43, expression of the feoB gene was reduced significantly (P < 0.05) in the BXO1843 (colR mutant) and BXO1844 (colS mutant) strains. However, expression of the xssE gene was enhanced significantly (P < 0.05) in the colR/colS mutants (Table 2). In the complemented strains BXO2243 (ColR+) and BXO2241 (ColS+), the expression level of the feoB and xssE genes was comparable with that of BXO43 (Table 2). These results indicate that the X. oryzae pv. oryzae ColRS system is required for optimal expression of the feoB gene.
Table 2.
Effect of Xanthomonas oryzae pv. oryzae colR and colS mutations on expression of xssE and feoB genes.
| Strain | Fold expression change ± SD* | |
|---|---|---|
| xssE | feoB | |
| BXO1843 (colR mutant) | 3.13 ± 0.37† | 0.38 ± 0.20† |
| BXO2243 (complemented colR mutant) | 1.04 ± 0.32‡ | 1.42 ± 0.46‡ |
| BXO1844 (colS mutant) | 2.19 ± 0.73† | 0.47 ± 0.25† |
| BXO2241 (complemented colS mutant) | 0.85 ± 0.21‡ | 1.31 ± 0.49‡ |
The fold expression change (mutant/wild type) (complemented mutant/wild type) was calculated using 2−ΔΔ Ct as described in Experimental procedures. Mean ± standard deviation (SD) of fold expression change for each gene is presented from three biological replicates.
Indicates that the fold expression changes are significantly different (two‐tailed t‐test, P < 0.05) with respect to the wild‐type strain.
Indicates that the fold expression changes are not significantly different (two‐tailed t‐test, P < 0.05) with respect to the wild‐type strain.
colR and colS mutants are defective in the elicitation of the hypersensitive response (HR) and expression of certain hrp genes
The role of the X. oryzae pv. oryzae colR and colS genes in the elicitation of HR symptoms was evaluated by infiltrating bacterial suspensions in leaves of tomato plants, as described in Experimental procedures. Wild‐type strain BXO43 elicited HR symptoms in tomato leaves. The BXO1843 (colR mutant) and BXO1844 (colS mutant) strains exhibited substantially reduced HR. The complemented strains BXO2243 (ColR+) and BXO2241 (ColS+) regained the ability to elicit wild‐type levels of HR (Fig. 4). As expected, BXO2012, a strain that is T3SS deficient [mutated for the hrpB6 (hrcN) gene; Jha et al. (2007)] did not elicit an HR (Fig. 4).
Figure 4.

colR and colS mutants of Xanthomonas oryzae pv. oryzae are deficient in the induction of a hypersensitive response (HR) on tomato. The leaves of tomato plants were infiltrated with bacterial suspensions as described in Experimental procedures. HR symptoms were observed 16 h after infiltration. Wild‐type strain BXO43 and the complemented strains BXO2243 (ColR+) and BXO2241 (ColS+) induced HR symptoms on tomato leaves, but BXO1843 (colS mutant), BX01844 (colR mutant) and BXO2012 (hrpB6::bla; a type 3 secretion system‐deficient mutant) did not.
The HR phenotype associated with the colR and colS mutants suggests a role for the ColRS system in the expression of hrp genes. In X. oryzae pv. oryzae, the hrp gene cluster consists of more than 20 genes representing six operons (hrpA–hrpF) (Lee et al., 2005; Ochiai et al., 2005; Oku et al., 2004; Zhu et al., 2000). In order to determine the role of the ColRS system in the regulation of hrp genes, the expression profile of one gene from each of these operons was assessed using real‐time quantitative PCR in colR and colS mutants. These genes were as follows: hrcC (hrpA operon), hrcT (hrpB operon), hrcU (hrpC operon), hrcR (hrpD operon), hrpE1 (hrpE operon) and hrpF (hrpF operon). We found that, when compared with BXO43, the expression of hrcU, hrcR, hrpE1 and hrpF genes was decreased significantly (P < 0.05) in the BXO1843 (colR mutant) and BXO1844 (colS mutant) strains. In the complemented strains BXO2243 (ColR+) and BXO2241 (ColS+), the expression level of the hrcU, hrcR, hrpE1 and hrpF genes was comparable with that of BXO43 (Fig. 5A,B). These results indicate that the X. oryzae pv. oryzae ColRS system is required for the optimal expression of the hrcU, hrcR, hrpE1 and hrpF genes. However, disruption of the colR and colS genes did not affect the expression of the hrcC and hrcT genes, which are encoded in the hrpA and hrpB operons (Fig. 5A,B).
Figure 5.
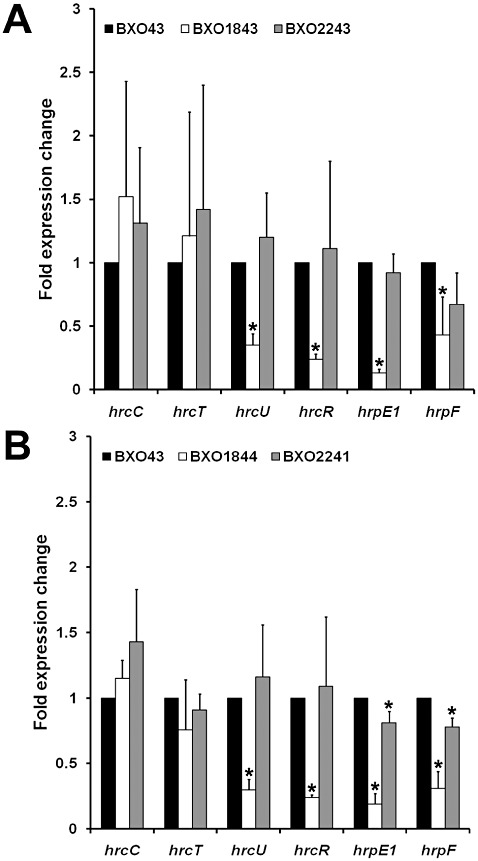
The colR and colS mutants of Xanthomonas oryzae pv. oryzae exhibit reduced expression of hrcU, hrcR, hrpE1 and hrpF genes. In comparison with the wild‐type strain (BXO43), the fold expression change of hrp genes was measured in BXO1843 (colR mutant) and the complemented strain BXO2243 (ColR+) (A) and BXO1844 (colS mutant) and the complemented strain BXO2241 (ColS+) (B). Real‐time quantitative polymerase chain reaction analysis was performed for hrp genes with cDNA synthesized using total RNA extracted from hrp‐inducing XOM2 medium‐grown cultures. The hrp genes analysed are indicated on the x axis. The 16S rRNA gene was used as an endogenous control. The fold expression change (mutant/wild type) (complemented mutant/wild type) was calculated using 2−ΔΔ Ct. The mean ± standard deviation of the fold expression change for each gene is presented from three biological repeats. Asterisks indicate that the fold expression change values are significantly different (two‐tailed t‐test, P < 0.05) with respect to the wild‐type strain.
Xanthomonas oryzae pv. oryzae colR and colS mutants are inducers of rice basal defence responses
The above data suggest that the expression of some T3SS genes is down‐regulated in colR and colS mutants. In an earlier study, we have demonstrated that a T3SS‐deficient mutant (BXO2012) of X. oryzae pv. oryzae induces rice basal defence responses, seen as callose deposition (Jha et al., 2007). Therefore, we assessed the ability of colR and colS mutants to induce callose deposition. As expected, minimal callose deposits were observed in rice leaves infiltrated with BXO43 (Fig. 6A‐a,6B). When compared with BXO43, the BXO1843 (colR mutant) and BXO1844 (colS mutant) strains induced significantly (P < 0.001) larger amounts of callose deposition (Fig. 6A‐b,d,6B). The numbers of callose deposits in the leaves infiltrated with complemented strains BXO2243 (ColR+) and BXO2241 (ColS+) were comparable with those observed in leaves infiltrated with BXO43 (Fig. 6A‐c,e,6B). Taken together, these data indicate that the colR/colS mutants are inducers of rice basal defence responses. However, the numbers of callose deposits induced by either BXO1843 or BXO1844 are lower than those induced by a T3SS‐deficient mutant (BXO2012; Fig. 6A‐f,6B).
Figure 6.
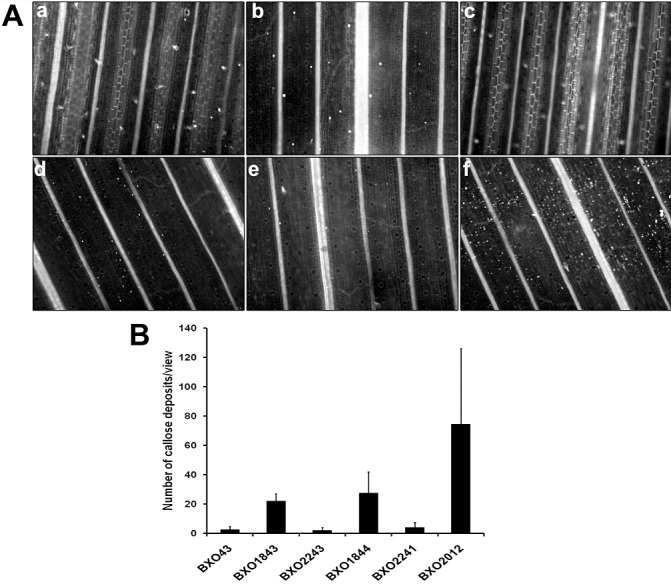
colR and colS mutants of Xanthomonas oryzae pv. oryzae induce callose deposition in rice leaves. (A) Representative images of rice leaves infiltrated with BXO43 (wild‐type strain) (a), BXO1843 (colR mutant) (b), BXO2243 (ColR+, complemented strain) (c), BXO1844 (colS mutant) (d), BXO2241 (ColS+, complemented strain) (e) and BXO2012 (type 3 secretion system‐deficient mutant) (f), and examined under a fluorescence microscope after staining with aniline blue. Each image represents a leaf area of approximately 0.56 µm2. (B) Quantification of callose deposits per field of view (∼ 0.56 µm2) after infiltration of rice leaves with X. oryzae pv. oryzae strains. The numbers of callose deposits obtained with the BXO1843 and BXO1844 strains were significantly (two‐tailed t‐test, P < 0.001) higher than those observed for BXO43. Data were collected from at least three leaves in each experiment, and three different viewing areas from the infiltrated region of each leaf. Similar results were obtained in four independent experiments.
DISCUSSION
We have carried out a genome‐wide screen to identify X. oryzae pv. oryzae mutants that exhibit siderophore production under iron‐replete conditions. We have called them ‘siderophore overproducer’ (sop) mutants. Of the 27 mutants isolated, the mTn5 insertions in 25 mutants were localized to be in or close to six genes. The siderophore overproduction phenotype of all of the sop mutants can be reversed by iron supplementation in the medium, indicating that this phenotype is not caused by a defect in sensing the levels of intracellular iron. This is in contrast with the previously identified X. oryzae pv. oryzae fur regulatory mutant, which fails to shut off siderophore production, even on supplementation with extracellular iron (Subramoni and Sonti, 2005).
Four sop mutants carried insertions in the colS gene, which codes for a putative X. oryzae pv. oryzae two‐component sensor protein. Targeted mutagenesis of colR, the cognate regulator, also results in a Sop phenotype. The X. oryzae pv. oryzae colR and colS mutants are deficient for growth in iron‐limiting conditions, but exhibit no growth deficiency in PS medium. This suggests that the ColRS system of X. oryzae pv. oryzae may be required for the optimal expression of genes involved in some aspects of iron uptake/metabolism. Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis indicates that the expression of the feoB gene is down‐regulated in colR and colS mutants. It is quite possible that the growth deficiency of the colR and colS mutants on iron‐limiting medium is caused by reduced expression of the feoB gene. The overexpression of siderophores in colR and colS mutants, as evident from the phenotype on CAS medium and enhanced expression of the xssE gene, is likely to be a compensatory measure for the reduced expression of feoB.
The X. oryzae pv. oryzae colR/colS mutants are defective in the elicitation of an HR on the nonhost plant, tomato. When compared with the wild‐type, the colR/colS mutants elicit enhanced levels of callose deposition, which is an indicator of the induction of rice innate immunity. Both phenotypes are indicative of a deficiency in T3SS of X. oryzae pv. oryzae, and suggest that there may be reduced expression of hrp/hrc genes in colR/colS mutants. The hrp genes of xanthomonads have been reported to be organized into six operons, namely the hrpC/D/E/F/A/B operons. We examined the effect of the mutation of colR and colS genes of X. oryzae pv. oryzae on the expression of one gene from each of these operons. Mutational inactivation of either colR or colS genes leads to reduced expression of hrcU, hrcR, hrpE1 and hrpF genes, which are encoded within the hrpC/D/E/F operons, but does not affect the expression of the hrcC and hrcT genes, which are encoded in the hrpA/B operons. This suggests that the expression of the hrpC/D/E/F operons, but not expression of the hrpA/B operons, is under the control of the ColRS system of X. oryzae pv. oryzae. In a previous study, Zhang et al. (2008) have shown that the expression of hrpC/E operons is affected in colR/colS mutants of X. campestris pv. campestris. In addition, Yan and Wang (2011) have shown that the expression of hrpD6 and hpaF genes is affected in colR/colS mutants of X. citri ssp. citri.
Mutations in the colR/colS orthologues of X. citri ssp. citri affect biofilm formation, T3SS activity, catalase activity and lipopolysaccharide (LPS) production (Yan and Wang, 2011). These mutants also exhibit enhanced susceptibility to various environmental stresses, such as hydrogen peroxide, phenol and copper. These phenotypes may be attributable to defects in catalase activity and LPS production. Enhanced susceptibility to environmental stresses, such as cadmium, sodium chloride, phenol and antibiotics, has also been reported in colR/colS mutants of X. campestris pv. campestris (Zhang et al., 2008). Mutations in the colR/colS genes of X. campestris pv. campestris and X. citri ssp. citri (Qian et al., 2008; Yan and Wang, 2011; Zhang et al., 2008) cause virulence deficiency. These earlier reports and the results of this study demonstrate that the ColRS system is required for virulence of xanthomonads, because, either directly or indirectly, it serves as a master regulator of a number of virulence‐associated functions.
The ColRS system of the plant growth‐promoting P. fluorescens bacterial strain WCS365 has been shown to be important for root colonization (Dekkers et al., 1998). The promoter of the colS gene of P. putida has been shown to be rhizosphere induced (Ramos‐Gonzalez et al., 2005), indicating its importance for root colonization. In P. putida, the ColRS system promotes tolerance to phenol, resistance to multiple metals and accumulation of Tn4652 events that promote the utilization of phenol under starvation conditions (Horak et al., 2004; Hu and Zhao, 2007; Kivistik et al., 2006). It has been suggested that tolerance to phenol and resistance to metals are promoted by an effect on membrane functionality. The observed deficiencies in the expression of the genes involved in iron transport and T3SS suggest that, in xanthomonads, as well as in pseudomonads, the ColRS system is involved in the regulation of expression of proteins that function in membranes.
Insertions in the colS gene were found in only four of the 25 characterized sop mutants, with the remaining mutants having Tn5 insertions in/close to five other genes. Fifteen sop mutants contained insertions in the XOO0007 gene, which encodes a conserved hypothetical protein and is immediately upstream of the tonB gene. Mutations in this gene affect growth on iron‐limiting medium, but do not cause virulence deficiency. The TonB protein is essential for siderophore‐mediated iron uptake (Postle and Kadner, 2003), and it is possible that insertions in the XOO0007 gene affect siderophore‐mediated iron uptake either because the XOO0007 protein functions in this process or because of a polar effect of an insertion in this gene on tonB expression. Mutations in the tonB gene of X. campestris pv. campestris could be isolated only on ferrous sulphate‐supplemented medium (Wiggerich et al., 1997). The absence of tonB insertions amongst the mutants isolated in our screen may be a result of the fact that, when performing the screen, bacterial cells were grown on medium that was not supplemented with ferrous sulphate. It is possible that tonB expression is driven by two promoters, one of which is upstream of XOO0007, whereas the other is in the 154‐bp intergenic region between XOO0007 and tonB. Therefore, insertions in XOO0007 may only result in reduced expression of tonB. A defect in siderophore‐mediated iron uptake has been shown previously to affect growth on iron‐limiting medium without causing any virulence deficiency (Pandey and Sonti, 2010).
The other sop mutants carried insertions in the following genes: two each in the acnB (XOO2862; encodes an Acn) and XOO1806 (encodes a conserved hypothetical protein) genes; one each in the prpR and prpB genes. The prpR and prpB genes are predicted to encode an activator of genes involved in propionate catabolism and an enzyme involved in propionate catabolism, respectively (Palacios and Escalante‐Semerena, 2004). All of these mutants exhibit a deficiency for virulence and growth on iron‐limiting medium. Insertions in these genes have not been shown previously to be involved in the promotion of virulence in any xanthomonad. Although polar effects cannot be ruled out without complementation data, a scrutiny of the downstream genes indicates that these have also not been shown to be involved in the virulence of xanthomonads.
AcnB has also been shown to function in the post‐transcriptional regulation of gene expression. Studies in Escherichia coli indicate that the regulatory and enzymatic functions of AcnB are mutually exclusive. Under low‐iron conditions, the dimeric form of AcnB switches to a catalytically inactive monomeric form and binds to target mRNAs to regulate their expression (Tang et al., 2005). It is possible that, in X. oryzae pv. oryzae, the correct expression of genes regulated by AcnB is required for the promotion of growth on iron‐limiting medium and virulence.
Although mutations in several novel genes were isolated in our screen, no mutations were isolated in some genes that would be expected to result in siderophore overproduction. These include the fur, feoB and xsuA (which encodes siderophore receptor) genes. The fur mutants grow very poorly on PS medium (Subramoni and Sonti, 2005) and were probably excluded from our screen, because we only picked mutants that did not show an obvious growth defect. Although feoB mutants grow better than fur mutants in laboratory medium, they exhibit a growth defect on medium that is not supplemented with exogenous iron (Pandey and Sonti, 2010). This explains the absence of feoB insertions, which have been shown to overproduce siderophores, amongst the sop mutants. The mTn5 element that was used for mutagenesis in this study causes polar mutations. An insertion in the xsuA gene would be polar on downstream xss genes that are required for siderophore production. Therefore, a mTn5 insertion in the xsuA gene would result in a loss of siderophore production and not siderophore overproduction.
In summary, the results presented here indicate that colR and colS mutants of X. oryzae pv. oryzae exhibit reduced levels of expression of hrp genes and the feoB gene, together with concomitant deficiencies in T3SS activity and growth on iron‐limiting medium. The virulence deficiency associated with the colR and colS mutants could be a result of defects in either T3SS functioning or iron uptake. The acnB, prpR, prpB and XOO1806 mutants exhibit deficiencies for virulence and growth on iron‐limiting medium. It is possible that these mutants could also be affected in certain other functions, in addition to iron uptake/metabolism, that affect virulence. Further studies are needed to understand how these genes, as well as colR/colS, affect virulence and iron uptake/metabolism in X. oryzae pv. oryzae.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids, primers and culture media
The bacterial strains and plasmids used are listed in Table 3. The primers used in this work are listed in Table S1. Xanthomonas oryzae pv. oryzae strains were grown at 28 °C in PS medium, and E. coli strains were grown at 37 °C in Luria Bertoni (LB) medium, as described previously (Pandey and Sonti, 2010). The concentrations of antibiotics used in this study were as follows: rifampicin (Rf), 50 µg/mL; ampicillin (Ap), 100 µg/mL; spectinomycin (Sp), 50 µg/mL; Km, 50 µg/mL for E. coli and 15 µg/mL for X. oryzae pv. oryzae. In addition, cycloheximide (80 µg/mL) was used to reduce fungal contamination. 2,2′‐Dipyridyl (DP), an iron chelator, was prepared as a 10‐mm stock solution and used at the concentrations indicated for each experiment.
Table 3.
Bacterial strains and plasmids used in this study.
| Strain/plasmid | Relevant characteristics* | Reference/source |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | F′/endA1 hsdR17(rk‐ mk+)supE44 thi‐1 recA1 gyrA relA1φ80dlacZΔM15Δ(lacZYA‐argF)U169 | Invitrogen |
| S17‐1 | RP4‐2Tc::Mu‐Km::Tn7 pro hsdR recA Tra+, used as a mobilizing strain | Simon et al. (1983) |
| TP0051 | λ pir derivative of S17‐1, pUT::mTn5gusA40, Apr, Spr | Wilson et al. (1995) |
| Plasmids | ||
| pK18mob | pUC18 derivative, Mob+; Kmr, does not replicate in X. oryzae pv. oryzae | Schafer et al. (1994) |
| pHM1 | Broad host range cosmid vector (∼13.3 kb); Spr | Innes et al. (1988) |
| pSS2 | pK18mob + 374‐bp internal fragment of colR cloned into SmaI site of pK18mob | This work |
| pSS3 | pK18mob + 739‐bp internal fragment of colS cloned into SmaI site of pK18mob | This work |
| pAP24 | pHM1 + 853‐bp fragment containing full‐length colR cloned into HindIII and SacI sites of pHM1 | This work |
| pAP25 | pHM1 + 1462‐bp fragment containing full‐length colS cloned into HindIII and SacI sites of pHM1 | This work |
| Xanthomonas oryzae pv. oryzae strains | ||
| BXO1 | Wild‐type; Indian isolate | Laboratory collection |
| BXO43 | rif‐2; derivative of BXO1 | Laboratory collection |
| BXO2012 | hrpB6::bla rif‐2; HR‐, Apr, derived from BXO43 | Jha et al. (2007) |
| BXO1843 | colR1::pK18 mob rif‐2; Kmr, derived from BXO43 | This work |
| BXO1844 | colS5::pK18 mob rif‐2; Kmr, derived from BXO43 | This work |
| BXO1845† | colR+rif‐2; Kms, revertant of BXO1843 | This work |
| BXO1846† | colS+rif‐2, Kms, revertant of BXO1844 | This work |
| BXO2241 | colS5::pK18mob rif‐2/pAP25; ColS+, Kmr, Spr | This work |
| BXO2243 | colR1::pK18mob rif‐2/pAP24; ColR+, Kmr, Spr | This work |
| sop mutants‡ | ||
| BXO1818 | colS1::mTn5 rif‐2 | This work |
| BXO1823 | colS2::mTn5 rif‐2 | This work |
| BXO1824 | colS3::mTn5 rif‐2 | This work |
| BXO1835 | colS4::mTn5 rif‐2 | This work |
| BXO1820 | XOO1806‐1::mTn5 rif‐2 | This work |
| BXO1834 | XOO1806‐2::mTn5 rif‐2 | This work |
| BXO1822 | acnB1::mTn5 rif‐2 | This work |
| BXO1833 | acnB2::mTn5 rif‐2 | This work |
| BXO1805 | XOO0007‐1::mTn5 rif‐2 | This work |
| BXO1807 | XOO0007‐2::mTn5 rif‐2 | This work |
| BXO1812 | XOO0007‐3::mTn5 rif‐2 | This work |
| BXO1813 | zxx301::mTn5 rif‐2; an insertion in XOO0007–XOO0008 intergenic region | This work |
| BXO1814 | XOO0007‐4::mTn5 rif‐2 | This work |
| BXO1815 | zxx302::mTn5 rif‐2; an insertion in XOO0007–XOO0008 intergenic region | This work |
| BXO1821 | XOO0007‐5::mTn5 rif‐2 | This work |
| BXO1826 | XOO0007‐6::mTn5 rif‐2 | This work |
| BXO1827 | XOO0007‐7::mTn5 rif‐2 | This work |
| BXO1830 | XOO0007‐8::mTn5 rif‐2 | This work |
| BXO1831 | XOO0007‐9::mTn5 rif‐2 | This work |
| BXO1832 | XOO0007‐10::mTn5 rif‐2 | This work |
| BXO1837 | XOO0007‐11::mTn5 rif‐2 | This work |
| BXO1839 | XOO0007‐12::mTn5 rif‐2 | This work |
| BXO1840 | XOO0007‐13::mTn5 rif‐2 | This work |
| BXO1816 | prpR1::mTn5 rif‐2 | This work |
| BXO1836 | prpB1::mTn5 rif‐2 | This work |
rif‐2 indicates a mutation that confers resistance to rifampicin. HR‐ indicates deficiency for hypersensitive response. Apr, Spr and Kmr indicate resistance to ampicillin, spectinomycin and kanamycin, respectively. Kms indicates kanamycin sensitivity. zxx indicates mutations that are in the intergenic region between XOO0007 and XOO0008 (tonB). Gene numbers are as specified by Lee et al. (2005).
These strains were isolated as virulence‐proficient revertants from rice leaves inoculated with X. oryzae pv. oryzae strains BXO1843 and BXO1844.
sop indicates siderophore overproduction phenotype.
Generation of mTn5 mutant library of X. oryzae pv. oryzae
In vivo mutagenesis was performed by setting up biparental matings between donor E. coli (TP0051) containing the transposon mTn5gusA40 (Sp resistance; transcription fusions with gus gene) on a suicide plasmid (pUT::Tn5gusA40) and recipient X. oryzae pv. oryzae strain BXO43. Biparental matings were performed as described by Dharmapuri and Sonti (1999). Briefly, late log‐phase cultures of X. oryzae pv. oryzae were grown in 200 mL of PS at 28 °C. Cells were pelleted, washed and resuspended in 2 mL of sterile distilled water; 40 µL of this cell suspension was spotted onto Hybond N Plus nylon membrane (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, UK) that was overlaid on nutrient agar (Difco Laboratories, Detroit, MI, USA) and incubated at 28 °C for 2 h. An overnight culture of donor E. coli strain TP0051 was pelleted, washed and resuspended in water; 5 µL of this cell suspension [density of 107 colony‐forming units (cfu)/mL] was mixed with recipient cells on the nylon membrane. The plates were incubated at 28 °C for 48 h, after which the cells were scraped from the plate and resuspended in 100 µL of sterile water. The cell suspension was plated on PSA plates containing Rf, Sp and cycloheximide. The plates were incubated at 28 °C and exconjugants were found to grow by 6–7 days. The X. oryzae pv. oryzae mTn5 mutants were then patched onto PSA + Ap and PSA + Sp plates separately to eliminate any Apr colonies which might have arisen as a result of integration of pUT::Tn5gusA40. Only Spr, Aps colonies of X. oryzae pv. oryzae were selected for further study.
Screening for X. oryzae pv. oryzae mutants that overproduce siderophores
PSA plates supplemented with chrome azurol sulphonate (CAS) dye were prepared for the detection of siderophores, as described previously (Chatterjee and Sonti, 2002). Under iron‐limiting conditions (caused by the addition of DP), siderophore production is visible as a yellow–orange zone around the bacterial colony. PSA‐CAS plates are apparently rich in iron as wild‐type X. oryzae pv. oryzae does not secrete siderophores in this condition. Mutants were patched with a toothpick on PSA‐CAS plates and the siderophore production phenotype was assessed after 24 h of incubation at 28 °C. Three rounds of screening were performed with selected sop mutants following the same protocol to eliminate mutants with inconsistent phenotypes.
Southern hybridization
Genomic DNA was isolated from X. oryzae pv. oryzae strains by the protocol described previously (Leach et al., 1990). Restriction digestion of DNA and α‐32P‐dATP labelling were performed as described by Sambrook et al. (1989). Genomic DNA was digested with KpnI (NEB, Beverly, NE, USA) because it does not cleave within the mTn5 sequence (Wilson et al., 1995). Southern hybridization was performed according to the protocol described by Yashitola et al. (1997). The plasmid pUT::Tn5gusA40 isolated from E. coli strain TP0051 was used as a probe.
TAIL PCR
TAIL PCR was performed according to the protocol described by Liu and Whittier (1995). The primers used for TAIL PCR are indicated in Table S1. Briefly, X. oryzae pv. oryzae colonies were picked with a toothpick, resuspended in 100 µL of sterile water and lysis was carried out at 94 °C for 15 min. Primary PCR was performed with 0.2 µm mTn5‐specific primer and 3 µm arbitrary primer (AD). Secondary and tertiary PCRs were performed with a 10‐fold lower concentration of deoxynucleoside triphosphates (dNTPs). PCR cycling conditions were similar to those described by Liu and Whittier (1995). The annealing temperatures for both primary and secondary PCRs were kept at 55 °C. For the tertiary reaction, the annealing temperature was 36 °C. The secondary and tertiary PCR samples were analysed on a 1.5% agarose gel, and a band shift corresponding to a difference in the product sizes of secondary and tertiary PCRs (150 bp with gus side primers and 100 bp with spec side primers of the mTn5 element) was taken as an indication of the presence of a specific product.
DNA sequencing and analysis
DNA sequencing of PCR‐amplified fragments was performed using the primers directed outwardly from the gus and spec genes of the mTn5 element (Wilson et al., 1995). The sequencing reactions and detection were performed using an ABI Prism Automated Sequencer 3730 (Perkin Elmer, Foster City, CA, USA). Homology and conserved domain searches were performed in the National Center for Biotechnology Information (NCBI) database using the blast algorithm (Altschul et al., 1990).
Generation and complementation of colR and colS mutants
The colR and colS genes were each disrupted by integration of the suicide vector pK18mob (Schafer et al., 1994) into chromosomal DNA via homologous recombination. The internal fragments of colR (374 bp) and colS (739 bp) were generated by PCR using genomic DNA of X. oryzae pv. oryzae strain BXO43 as a template with the following respective primer sets: colRIP1/colRIP2 and colSF1/colSR1 (Table S1). These fragments were cloned into pK18mob digested with SmaI, and the plasmids obtained were designated as pSS2 and pSS3, respectively. These constructs were then transferred from E. coli strain S17‐1 (Simon et al., 1983) to X. oryzae pv. oryzae via biparental mating, and colR (BXO1843) and colS (BXO1844) mutants were selected on Km‐containing medium. PCR was performed with the flanking primers (colRPF1/colRPR1 and colSGF1/colSGR1) in combination with vector‐specific primers (M13F and M13R), and the fragments obtained were sequenced to confirm the disruption of colR and colS genes in the mutants.
For complementation, 853‐ and 1462‐bp DNA fragments containing full‐length colR and colS genes, respectively, were amplified by PCR using genomic DNA of X. oryzae pv. oryzae and the primer sets ColRCF/ColRCR and ColSCF/ColSCR. The amplified fragments were individually cloned as HindIII‐SacI fragments into the broad host range vector pHM1 (Innes et al., 1988) to create the recombinant plasmids: pAP24 (for colR) and pAP25 (for colS). The pAP24 and pAP25 plasmids were introduced into the colR (BXO1843) and colS (BXO1844) mutants, respectively, to obtain the complemented strains: BXO2243 and BXO2241.
Assays for virulence, HR and callose deposition
Xanthomonas oryzae pv. oryzae strains were grown to saturation and inoculated on 30–40‐day‐old glasshouse‐grown rice plants of the highly susceptible cultivar Taichung Native (TN‐1). Inoculation was performed by dipping scissors into bacterial cultures and clipping the tips of rice leaves (Kauffman et al., 1973). Lesion lengths were measured 7 and 14 days after inoculation and expressed as the mean lesion length ± standard deviation. For HR assays, leaves of tomato plants were infiltrated with X. oryzae pv. oryzae strains (108 cfu/mL) using a blunt‐ended plastic syringe. HR symptoms (browning of the infiltrated area) were observed 16 h after infiltration. For callose assays, rice leaves were infiltrated with X. oryzae pv. oryzae strains (108 cfu/mL) using a blunt‐ended plastic syringe. After 16 h, the leaves were cleared with 100% ethanol, rehydrated with 50% ethanol and stained with aniline blue. The leaf samples were then mounted on a slide in 50% glycerol, and analysed by an Axio Imager Z1 (Carl Zeiss, Oberkochen, Germany) microscope using a 4′,6‐diamidino‐2‐phenylindole (DAPI) fluorescence filter.
Re‐isolation of bacteria from rice leaves and revertant identification
Infected leaves were surface sterilized by dipping in 1% (v/v) sodium hypochlorite (Loba Chemie, Mumbai, India) for 1 min and washing three times in distilled water. The leaves were cut at the leading edge of the lesion and dipped in 1 mL of sterile water for 5 min. Bacteria that exuded from the cut edges of the leaves were isolated by plating for individual colonies on PSA. These colonies were patched on PSA + Km plates and the ColR+ (BXO1845) and ColS+ (BXO1846) revertants were identified as Km‐sensitive colonies. The Km‐sensitive colonies were patched on PSA‐CAS plates to assess the ability to produce siderophores. In addition, revertants were checked for their ability to cause disease and to grow on iron‐limiting medium.
Growth experiments
Xanthomonas oryzae pv. oryzae strains were grown to log‐phase in PS medium at 28 °C to obtain pre‐inoculum. For each strain, 3 mL PS and 3 mL PS + 140 µm DP were inoculated with 0.1% pre‐inoculum. Cultures were dilution plated on PSA plates immediately (0 h) and 48 h after inoculation to determine cfu/mL. The bacterial count was expressed as the log value of cfu/mL.
RNA isolation and quantitative RT‐PCR
To investigate the effect of colR/colS genes on the expression of hrp, xssE and feoB genes, real‐time quantitative PCR was carried out. Xanthomonas oryzae pv. oryzae strains were grown in PS medium to an optical density at 600 nm (OD600) of 1.0. These cultures were used to isolate RNA and to examine the expression of xssE and feoB genes; in contrast, for hrp genes, cultures incubated in hrp‐inducing XOM2 medium (Tsuge et al., 2002) were used for total RNA isolation. For incubation in XOM2 medium, PS‐grown cultures were centrifuged and the pellets were washed three times with MilliQ water (Millipore, Billerica, MA, USA), and then transferred to XOM2 medium. Cultures were further incubated for 18 h, and total RNA from each of the samples was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The total RNA concentration was determined using a NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA).
RNA samples were treated with RNase‐free DNaseI (NEB) to remove any DNA contamination. A total of 1 µg of RNA was reverse transcribed by SuperScript III reverse transcriptase enzyme (Invitrogen) using 50 ng of random hexamers, according to the supplier's instructions. One microlitre of the 1:10 diluted cDNA was subjected to real‐time quantitative PCR using SYBR GreenER qPCR supermix (Invitrogen) and gene‐specific primers designed to amplify 100–155‐bp fragments from each gene of interest and the gene encoding for 16S ribosomal RNA. The primers (Table S1) were designed to amplify the fragments from hrcC, hrcT, hrcU, hrcR, hrpE1, hrpF, xssE and feoB genes of X. oryzae pv. oryzae. After 10 min of initial denaturation at 95 °C, the samples were subjected to cycling parameters of 95 °C for 15 s, 58 °C for 30 s and 72 °C for 30 s (40 cycles) using a 7900 HT sequence detection system (Applied Biosystems, Foster City, CA, USA). The relative expression of hrp genes was calculated using the 2−ΔΔ Ct method (Livak and Schmittgen, 2001) with 16S rRNA as an endogenous control.
Supporting information
Fig. S1 Location of mTn5 insertions and gene organization in the Xanthomonas oryzae pv. oryzae genomic region containing the XOO0007 gene. The filled triangles represent the locations of the mTn5 insertions. The arrows indicate the transcriptional orientations of the genes. The bold lines indicate open reading frames (ORFs) and the lighter lines indicate intergenic regions. Several sop mutants carry an mTn5 insertion at the same site in the XOO0007 gene. These include XOO0007‐4 (BXO1814), XOO0007‐6 (BXO1826) and XOO0007‐7 (BXO1827); XOO0007‐1 (BXO1805) and XOO0007‐9 (BXO1831); and XOO0007‐8 (BXO1830) and XOO0007‐11 (BXO1837). The zxx301 (BXO1813) and zxx302 (BXO1815) insertions are in the intergenic region between the XOO0007 and tonB genes, which are in the same transcriptional orientation. The gene organization given here is as indicated for the genome of X. oryzae pv. oryzae strain KACC10331 (Lee et al., 2005).
Table S1 List of oligonucleotide primers used in this study.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
AP was supported by research fellowships from the Council of Scientific and Industrial Research (CSIR) and Department of Biotechnology, Government of India. This work was supported, in part, by a National Bioscience Award for Career Development to RVS from the Department of Biotechnology, Government of India.
REFERENCES
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Buttner, D. and Bonas, U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. [DOI] [PubMed] [Google Scholar]
- Chatterjee, S. and Sonti, R.V. (2002) rpfF mutants of Xanthomonas oryzae pv. oryzae are deficient for virulence and growth under low iron conditions. Mol. Plant–Microbe Interact. 15, 463–471. [DOI] [PubMed] [Google Scholar]
- Dekkers, L.C. , Bloemendaal, C.J. , de Weger, L.A. , Wijffelman, C.A. , Spaink, H.P. and Lugtenberg, B.J. (1998) A two‐component system plays an important role in the root‐colonizing ability of Pseudomonas fluorescens strain WCS365. Mol. Plant–Microbe Interact. 11, 45–56. [DOI] [PubMed] [Google Scholar]
- Dharmapuri, S. and Sonti, R.V. (1999) A transposon insertion in the gumG homologue of Xanthomonas oryzae pv. oryzae causes loss of extracellular polysaccharide production and virulence. FEMS Microbiol. Lett. 179, 53–59. [DOI] [PubMed] [Google Scholar]
- Feng, J.X. , Song, Z.Z. , Duan, C.J. , Zhao, S. , Wu, Y.Q. , Wang, C. , Dow, J.M. and Tang, J.L. (2009). The H‐NS‐like protein‐encoding gene xrvA of Xanthomonas oryzae pv. oryzae regulates virulence in rice. Microbiology, 155, 3033–3044. [DOI] [PubMed] [Google Scholar]
- Horak, R. , Ilves, H. , Pruunsild, P. , Kuljus, M. and Kivisaar, M. (2004) The ColR‐ColS two‐component signal transduction system is involved in regulation of Tn4652 transposition in Pseudomonas putida under starvation conditions. Mol. Microbiol. 54, 795–807. [DOI] [PubMed] [Google Scholar]
- Hu, N. and Zhao, B. (2007) Key genes involved in heavy‐metal resistance in Pseudomonas putida CD2. FEMS Microbiol. Lett. 267, 17–22. [DOI] [PubMed] [Google Scholar]
- Innes, R.W. , Hirose, M.A. and Kuempel, P.L. (1988) Induction of nitrogen‐fixing nodules on clover requires only 32 kilobase pairs of DNA from the Rhizobium trifolii symbiosis plasmid. J. Bacteriol. 170, 3793–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, G. , Rajeshwari, R. and Sonti, R.V. (2007) Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. Mol. Plant–Microbe Interact. 20, 31–40. [DOI] [PubMed] [Google Scholar]
- Kauffman, H.E. , Reddy, A.P.K. , Hsieh, S.P.Y. and Merca, S.D. (1973) An improved technique for evaluation of resistance of rice varieties to Xanthomonas oryzae . Plant Dis. Rep. 57, 537–541. [Google Scholar]
- Kivistik, P.A. , Putrins, M. , Puvi, K. , Ilves, H. , Kivisaar, M. and Horak, R. (2006) The ColRS two‐component system regulates membrane functions and protects Pseudomonas putida against phenol. J. Bacteriol. 188, 8109–8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, J.E. , White, F.F. , Rhoads, M.L. and Leung, H. (1990) A repetitive DNA sequence differentiates Xanthomonas campestris pv. oryzae from other pathovars of X. campestris . Mol. Plant–Microbe Interact. 3, 238–246. [Google Scholar]
- Lee, B.M. , Park, Y.J. , Park, D.S. , Kang, H.W. , Kim, J.G. , Song, E.S. , Park, I.C. , Yoon, U.H. , Hahn, J.H. , Koo, B.S. , Lee, G.B. , Kim, H. , Park, H.S. , Yoon, K.O. , Kim, J.H. , Jung, C.H. , Koh, N.H. , Seo, J.S. and Go, S.J. (2005) The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.W. , Jeong, K.S. , Han, S.W. , Lee, S.E. , Phee, B.K. , Hahn, T.R. and Ronald, P. (2008) The Xanthomonas oryzae pv. oryzae PhoPQ two‐component system is required for AvrXA21 activity, hrpG expression, and virulence. J. Bacteriol. 190, 2183–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.W. , Han, S.W. , Sririyanum, M. , Park, C.J. , Seo, Y.S. and Ronald, P.C. (2009) A type I‐secreted, sulfated peptide triggers XA21‐mediated innate immunity. Science, 326, 850–853. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G. and Whittier, R.F. (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert and fragments from P1 and YAC clones for chromosome walking. Genomics, 25, 674–681. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCt method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mahalingam, R. and Fedoroff, N. (2001) Screening insertion libraries for mutations in many genes simultaneously using DNA microarrays. Proc. Natl. Acad. Sci. USA, 98, 7420–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nino‐Liu, D.O. , Ronald, P.C. and Bogdanove, A.J. (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. [DOI] [PubMed] [Google Scholar]
- Ochiai, H. , Inoue, Y. , Takeya, M. , Sasaki, A. and Kaku, H. (2005) Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jpn. Agric. Res. Q. 39, 275–287. [Google Scholar]
- Oku, T. , Tanaka, K. , Iwamoto, M. , Inoue, Y. , Ochiai, H. , Kaku, H. , Tsuge, S. and Tsuno, K. (2004) Structural conservation of the hrp gene cluster in Xanthomonas oryzae pv. oryzae . J. Gen. Plant Pathol. 70, 159–167. [Google Scholar]
- Palacios, S. and Escalante‐Semerena, J.C. (2004) 2‐Methylcitrate‐dependent activation of the propionate catabolic operon (prpBCDE) of Salmonella enterica by the PrpR protein. Microbiology, 150, 3877–3887. [DOI] [PubMed] [Google Scholar]
- Pandey, A. and Sonti, R.V. (2010) Role of the FeoB protein and siderophore in promoting virulence of Xanthomonas oryzae pv. oryzae on rice. J. Bacteriol. 192, 3187–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postle, K. and Kadner, R.J. (2003) Touch and go: tying TonB to transport. Mol. Microbiol. 49, 869–882. [DOI] [PubMed] [Google Scholar]
- Qian, W. , Han, Z.J. , Tao, J. and He, C.Z. (2008) Genome‐scale mutagenesis and phenotypic characterization of two‐component signal transduction systems in Xanthomonas campestris pv. campestris ATCC 33913. Mol. Plant–Microbe Interact. 21, 1128–1138. [DOI] [PubMed] [Google Scholar]
- Ramos‐Gonzalez, M.I. , Campos, M.J. and Ramos, J.L. (2005) Analysis of Pseudomonas putida KT2440 gene expression in the maize rhizosphere: in vitro technology capture and identification of root activated promoters. J. Bacteriol. 187, 4033–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg, S.L. , Sommer, D.D. , Schatz, M.C. , Phillippy, A.M. , Rabinowicz, P.D. , Tsuge, S. , Furutani, A. , Ochiai, H. , Delcher, A.L. , Kelley, D. , Madupu, R. , Puiu, D. , Radune, D. , Shumway, M. , Trapnell, C. , Aparna, G. , Jha, G. , Pandey, A. , Patil, P.B. , Ishihara, H. , Meyer, D.F. , Szurek, B. , Verdier, V. , Koebnik, R. , Dow, J.M. , Ryan, R.P. , Hirata, H. , Tsuyumu, S. , Lee, S.W. , Seo, Y.S. , Sriariyanum, M. , Ronald, P.C. , Sonti, R.V. , Van Sluys, M.A. , Leach, J.E. , White, F.F. and Bogdanove, A.J. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A . BMC Genomics, 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schafer, A. , Tauch, A. , Jager, W. , Kalinowski, J. , Thierbach, G. and Puhler, A. (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene, 145, 69–73. [DOI] [PubMed] [Google Scholar]
- Simon, R. , Priefer, U. and Puhler, A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotech. 1, 784–791. [Google Scholar]
- Subramoni, S. and Sonti, R.V. (2005) Growth deficiency of a Xanthomonas oryzae pv. oryzae fur mutant in rice leaves is rescued by ascorbic acid supplementation. Mol. Plant–Microbe Interact. 18, 644–651. [DOI] [PubMed] [Google Scholar]
- Sun, Q. , Wu, W. , Qian, W. , Hu, J. , Fang, R. and He, C. (2003) High‐quality mutant libraries of Xanthomonas oryzae pv. oryzae and X. campestris pv. campestris generated by an efficient transposon mutagenesis system. FEMS Microbiol. Lett. 226, 145–150. [DOI] [PubMed] [Google Scholar]
- Tang, Y. , Guest, J.R. , Artymiyk, P.J. and Green, J. (2005) Switching aconitase B between catalytic and regulatory modes involves iron‐dependent dimer formation. Mol. Microbiol. 56, 1149–1158. [DOI] [PubMed] [Google Scholar]
- Tsuge, S. , Furutani, A. , Fukunaka, R. , Oku, T. , Tsuno, K. , Ochiai, H. , Inoue, Y. , Kaku, H. and Kubo, Y. (2002) Expression of Xanthomonas oryzae pv. oryzae hrp genes in XOM2, a novel synthetic medium. J. Gen. Plant Pathol. 68, 363–371. [Google Scholar]
- Tsuge, S. , Nakayama, T. , Terashima, S. , Ochiai, H. , Furutani, A. , Oku, T. , Tsuno, K. , Kubo, Y. and Kaku, H. (2006) Gene involved in transcriptional activation of the hrp regulatory gene hrpG in Xanthomonas oryzae pv. oryzae . J. Bacteriol. 188, 4158–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggerich, H.G. , Klauke, B. , Köplin, R. , Priefer, U.B. and Pühler, A. (1997) Unusual structure of the tonB‐exb DNA region of Xanthomonas campestris pv. campestris: tonB, exbB, and exbD1 are essential for ferric iron uptake, but exbD2 is not. J. Bacteriol. 179, 7103–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K.J. , Sessitch, A. , Corbo, J.C. , Giller, K.E. , Akkermans, A.D.L. and Jefferson, R.A. (1995) beta‐Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram‐negative bacteria. Microbiology, 141, 1691–1705. [DOI] [PubMed] [Google Scholar]
- Yan, Q. and Wang, N. (2011) The ColR/ColS two‐component system plays multiple roles in the pathogenicity of the citrus canker pathogen Xanthomonas citri subsp. citri . J. Bacteriol. 193, 1590–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashitola, J. , Krishnaveni, D. , Reddy, A.P.K. and Sonti, R.V. (1997) Genetic diversity within the population of Xanthomonas oryzae pv. oryzae in India. Phytopathology, 87, 760–765. [DOI] [PubMed] [Google Scholar]
- Zhang, S.S. , He, Y.Q. , Xu, L.M. , Chen, B.W. , Jiang, B.L. , Liao, J. , Cao, J.R. , Liu, D. , Huang, Y.Q. , Liang, X.X. , Tang, D.J. , Lu, G.T. and Tang, J.L. (2008) A putative colR (XC1049)‐colS (XC1050) two‐component signal transduction system of Xanthomonas campestris positively regulates hrpC and hrpE operons and is involved in virulence, the hypersensitive response and tolerance to various stresses. Res. Microbiol. 159, 569–578. [DOI] [PubMed] [Google Scholar]
- Zhu, W. , MaGbanua, M.M. and White, F.F. (2000) Identification of two novel hrp‐associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae . J. Bacteriol. 182, 1844–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Location of mTn5 insertions and gene organization in the Xanthomonas oryzae pv. oryzae genomic region containing the XOO0007 gene. The filled triangles represent the locations of the mTn5 insertions. The arrows indicate the transcriptional orientations of the genes. The bold lines indicate open reading frames (ORFs) and the lighter lines indicate intergenic regions. Several sop mutants carry an mTn5 insertion at the same site in the XOO0007 gene. These include XOO0007‐4 (BXO1814), XOO0007‐6 (BXO1826) and XOO0007‐7 (BXO1827); XOO0007‐1 (BXO1805) and XOO0007‐9 (BXO1831); and XOO0007‐8 (BXO1830) and XOO0007‐11 (BXO1837). The zxx301 (BXO1813) and zxx302 (BXO1815) insertions are in the intergenic region between the XOO0007 and tonB genes, which are in the same transcriptional orientation. The gene organization given here is as indicated for the genome of X. oryzae pv. oryzae strain KACC10331 (Lee et al., 2005).
Table S1 List of oligonucleotide primers used in this study.
Supporting info item
Supporting info item


