SUMMARY
Streptomyces strains were isolated from scab lesions on potatoes collected from different parts of Norway. Twenty‐eight plant‐pathogenic strains, as tested on seedlings of radish and on potato, were identified on the basis of physiological and molecular criteria. Polymerase chain reaction (PCR) analysis, using species‐specific primers, and sequencing of the 16S rRNA gene identified 14 nonmelanin‐producing strains to S. turgidiscabies. Fourteen melanin‐producing strains were detected with primers specific to S. scabies, but whole‐genome microarray analysis, based on 12 766 probes designed for 8848 predicted open reading frames (ORFs) of S. scabies, showed that the 14 strains were different from S. scabies. They were subsequently identified to be S. europaeiscabiei based on the internal transcribed spacer (ITS) sequences of the rRNA genes. This is the first report of the occurrence of S. turgidiscabies and S. europaeiscabiei in Norway. The putative 762 genes exhibiting the highest sequence differences between strains of S. europaeiscabiei and S. scabies according to microarray analysis were concentrated in relatively few gene ontology (GO) categories, including ‘symbiosis and mutualism through parasitism’, ‘cell death’ and ‘responses to biotic stimulus’, whereas genes related to primary metabolism appeared to be more conserved. Microarray data and 16S rRNA gene phylogeny showed, consistently, that there were two genetically distinguishable groups of S. europaeiscabiei on the basis of differences in 131 genes. The results provide novel information about the genetic variability of S. europaeiscabiei and the gene‐specific variability between the genomes of S. europaeiscabiei and S. scabies. The usefulness of a custom‐designed, whole‐genome oligonucleotide microarray in a survey of bacterial plant pathogens was demonstrated.
INTRODUCTION
Streptomyces species are soil‐dwelling, filamentous, Gram‐positive bacteria belonging to the phylum Actinobacteria (Holt and Bergey, 1994). Only few of the hundreds of described species are plant pathogenic, among which those causing common scab on potato are the most widely studied because of the significant yield losses they cause (Hiltunen et al., 2005; Loria et al., 2006; Wanner, 2009). Common scab‐inducing Streptomyces species can also cause diseases on other root crops, such as radish, carrot, turnip, beet and sweet potato, among others (Locci, 1994). Symptoms of common scab on potato tubers are erumpent, superficial or pitted lesions depending on the pathogen strain and potato cultivar (Lambert and Loria, 1989), but the symptoms are not species specific and cannot be used to identify the pathogen. World‐wide distributed common scab pathogens are represented by Streptomyces scabies, which was the first described species causing common scab on potatoes (Lambert and Loria, 1989). Streptomyces turgidiscabies was found more recently to cause common scab on potato in Japan (Miyajima et al., 1998) and Finland (Kreuze et al., 1999), and later found to be distributed in other parts of the world (Kim et al., 1999; Wanner, 2009). Although these species cause similar common scab symptoms, they can be distinguished by the production of melanin pigment on culture media and the spiral‐like spore chains that are characteristic of S. scabies but not found with S. turgidiscabies.
The plant pathogenicity of Streptomyces species depends on the production of thaxtomin A, a nitrated dipeptide toxin, which causes symptoms of common scab on potato tubers (Healy et al., 2000; Lawrence et al., 1990). Thaxtomin A is synthesized by S. scabies, S. turgidiscabies and S. europaeiscabiei (Bouchek‐Mechiche et al., 2000; King et al., 1989; Miyajima et al., 1998). It induces plant cell hypertrophy in expanding tissues (Fry and Loria, 2002; Scheible et al., 2003). The nec1 and tomA genes are present in a wide range of common scab‐inducing Streptomyces strains, but not in all (Seipke and Loria, 2008; Wanner, 2009). Thus, they are not required for pathogenicity, but contribute to virulence. nec1 encodes a protein inducing necrosis in plant tissue (Bukhalid and Loria, 1997) and is not required for the production of thaxtomin A. tomA encodes a virulence factor homologous to tomatinase, an enzyme belonging to saponinases found in plant‐pathogenic fungi (Kers et al., 2005). Pathogenicity and virulence genes are clustered in a pathogenicity island (PAI) which can be transferred horizontally from pathogens to saprophytic strains of Streptomyces, creating new pathogenic strains (Bukhalid et al., 2002; Healy et al., 1999; Kers et al., 2005). It has been suggested that the direction of horizontal transfer has been from S. scabies to S. turgidiscabies (Bukhalid et al., 2002), but recent results from microarray‐based comparisons of the PAI regions in strains of S. turgidiscabies and S. scabies have revealed that the scenario may be more complicated (Aittamaa et al., 2010). The PAI of S. turgidiscabies consists of two nonoverlapping modules, the first of which is similar to a genomic island in S. scabies, but the other module contains only a short region highly similar to the genome of S. scabies (Huguet‐Tapia et al., 2011). Indeed, also in strain 87.22 of S. scabies, whose genome has been sequenced, the PAI consists of two genomic regions which, however, are much more distant from each other than the PAI regions in S. turgidiscabies (Aittamaa et al., 2010; Lerat et al., 2009). The genes associated with thaxtomin production, such as txtA, txtB, txtC and txtR, are found in the first segment of the PAI, called the ‘toxicogenic region’, whereas the virulence genes, tomA and nec1, are located in the second segment, designated as a ‘colonization region’ (Lerat et al., 2009).
In the recent past, many new plant‐pathogenic Streptomyces species have been described. Previously, common scab‐inducing bacteria that produced melanin were designated to S. scabies, but the introduction of molecular analysis for the characterization of the strains suggested that their genetic variability was wider than expected for a single species. Hence, DNA–DNA hybridization assays (Healy and Lambert, 1991; Paradis et al., 1994), protein profile analysis (Paradis et al., 1994), complementation tests (Lorang et al., 1995) and also host range analysis (Goyer and Beaulieu, 1997) suggested that there could be additional, melanin‐producing species that cause common scab (Goyer et al., 1996). One of these recently detected species is Streptomyces europaeiscabiei, which was first described in France and is frequently found in Europe and also North America (Bouchek‐Mechiche et al., 2000; Flores‐Gonzalez et al., 2008; Wanner, 2009). However, molecular surveys for the common scab pathogens have been conducted only in relatively few countries, including the USA, Canada, Japan, Korea, Finland, Spain and France (Bouchek‐Mechiche et al., 2000; Flores‐Gonzalez et al., 2008; Goyer et al., 1996; Kreuze et al., 1999; Lambert and Loria, 1989; Loria et al., 1997; Miyajima et al., 1998; Park et al., 2003; Wanner, 2009).
Common scab has been known in Norway since the 1920s and an investigation of scab diseases on potato in Norway was carried out in 1967–68 (Jørstad, 1922; Værdal, 1973). However, there is limited knowledge about the Streptomyces species that cause common scab in Norway as no surveys have been performed recently. DNA microarray technology is a powerful tool for obtaining large‐scale genomic information, e.g. comparison of pathogen strains. One or a few genes can be amplified using polymerase chain reaction (PCR), but the whole genome of an organism can be analysed simultaneously using microarray. For diagnostic purposes, taxonomic discrimination of organisms can be performed using a large number of probes simultaneously (Aittamaa et al., 2008). The aim of this study was to identify pathogenic strains of Streptomyces isolated from common scab lesions on potatoes in Norway and to compare genetic differences between the strains using microarray analysis.
RESULTS
Pathogenicity of the bacterial strains
A number of the bacterial strains isolated from potato scab lesions formed colonies phenotypically characteristic of Streptomyces spp. when grown on yeast malt extract (YME) and peptone yeast extract iron (PYI) media (Loria et al., 2001). The strains were tested for pathogenicity by the inoculation of radish seedlings in vitro, as described by Flores‐Gonzalez et al. (2008). Fourteen strains which produced melanin on PYI were found to be pathogenic, as they prevented any growth of the seedlings. Fourteen additional strains caused severe hypertrophy of the seedlings, but did not produce melanin on PYI (Table 1; Fig. S1, see Supporting Information). The other strains (K1, K3 and K4) showed no apparent effect on growth, caused no symptoms on the seedlings and were considered to be nonpathogenic. Three melanin‐producing and four nonmelanin‐producing strains were also tested for pathogenicity on tubers of the scab‐susceptible potato cultivar Blue Congo under controlled conditions in the glasshouse. They all induced similar symptoms of common scab, in contrast with the control strains (K1 and K3), which induced no symptoms (Table 1).
Table 1.
Strains of Streptomyces from Norway characterized in this study.
| Origin of strain | Biological characteristics | Species detection by PCR | ITS analysis | PCR detection of virulence genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | County in Norway | Potato variety | Melanin on PYI | Pathogenicity: radish | Pathogenicity: potato | Universal | S. turgidiscabies | S. scabies | Hpy99I | TxtAB | Nec1 | TomA |
| 3 | Troms | Gullauge | − | + | + | + | + | − | Nt | + | + | + |
| 5 | Troms | Gullauge | − | + | Nt | + | + | − | Nt | + | + | + |
| 6 | Aust‐Agder | N 97‐5‐23 | − | + | + | + | + | − | Nt | + | + | + |
| 7 | Rogaland | Pimpernel | − | + | Nt | + | + | − | Nt | + | + | + |
| 8 | Rogaland | Pimpernel | − | + | Nt | + | + | − | Nt | + | + | + |
| 9 | Vestfold | Opera | + | + | + | + | − | + | − | + | + | + |
| 10 | Vestfold | Opera | + | + | Nt | + | − | + | − | + | + | + |
| 14 | Oppland | Saturna | + | + | Nt | + | − | + | − | + | + | + |
| 15 | Buskerud | Saturna | + | + | Nt | + | − | + | − | + | + | + |
| 17 | Buskerud | Saturna | + | + | Nt | + | − | + | − | + | + | + |
| 18 | Hedmark | Fakse | + | + | + | + | − | + | − | + | + | + |
| 19 | Hedmark | Laila | − | + | + | + | + | − | Nt | + | − | + |
| 21 | Nord‐Trøndelag | Saturna | + | + | Nt | + | − | + | − | + | + | + |
| 22 | Nord‐Trøndelag | Beate | − | + | Nt | + | + | − | Nt | + | − | + |
| 26 | Nord‐Trøndelag | Rutt | + | + | Nt | + | − | + | − | + | − | − |
| 31 | Hedmark | Bintje | + | + | Nt | + | − | + | − | + | − | − |
| 32 | Telemark | Folva | + | + | + | + | − | + | − | + | + | + |
| 33 | Vestfold | Laila | + | + | Nt | + | − | + | − | + | + | + |
| 35 | Akershus | Folva | + | + | Nt | + | − | + | − | + | + | + |
| 36 | Akershus | Folva | − | + | Nt | + | + | − | Nt | + | − | + |
| 37 | Møre og Romsdal | Kerrs Pink | − | + | + | + | + | − | Nt | + | − | + |
| 39 | Møre og Romsdal | Kerrs Pink | − | + | Nt | + | + | − | Nt | + | − | + |
| 40 | Møre og Romsdal | Folva | + | + | Nt | + | − | + | − | + | − | − |
| 43 | Hedmark | Unknown | − | + | Nt | + | + | − | Nt | + | + | + |
| 44 | Østfold | Unknown | − | + | Nt | + | + | − | Nt | + | + | + |
| 46 | Østfold | Brage | − | + | Nt | + | + | − | Nt | + | − | + |
| 48 | Østfold | Brage | − | + | Nt | + | + | − | Nt | + | − | + |
| 50 | Østfold | Laila | + | + | Nt | + | − | + | − | + | − | − |
| K1 | Oppland | Saturna | + | − | − | + | − | − | Nt | − | − | − |
| K3 | Nord‐Trøndelag | Asterix | + | − | − | + | − | − | Nt | − | − | − |
| K4 | Oppland | Unknown | + | − | Nt | + | − | − | Nt | − | − | − |
ITS, internal transcribed spacer; Nt, not tested; PCR, polymerase chain reaction; PYI, peptone yeast extract iron.
Microarray analysis‐based grouping of the bacterial strains
The 28 pathogenic strains (Table 1), putatively classified as Streptomyces species on the basis of morphological criteria, were subjected to comparative genomic hybridization (CGH) using a microarray containing probes specific to genes of S. scabies (Table S1, see Supporting Information) and S. turgidiscabies (Aittamaa et al., 2010), which have been reported to be the causal agents of common scab on potatoes in the neighbouring countries of Finland and Sweden (Aittamaa et al., 2010; Kreuze et al., 1999; Lehtonen et al., 2004; Lindholm et al., 1997). The type strain of S. scabies (ATCC 49173) (Lambert and Loria, 1989) and strain St32 of S. turgidiscabies (Lehtonen et al., 2004) isolated from potato were included for comparison. Clustering of the strains was performed on the basis of the signals from 12 766 probes designed for the 8848 open reading frames (ORFs) of S. scabies predicted initially in the draft genome sequence of S. scabies strain 87.22 (GenBank accession no. FN554889). Data from two independent hybridization experiments showed that the bacterial strains could be placed into two major clades (Fig. 1). Clade I contained the strain St32 of S. turgidiscabies and 14 strains (nos. 3, 5, 6, 7, 8, 19, 22, 36, 37, 39, 43, 44, 46 and 48) isolated in this study. The remaining 14 strains (nos. 9, 10, 14, 15, 17, 18, 21, 26, 31, 32, 33, 35, 40 and 50) forming clade II were those which produced melanin when grown on PYI. However, the type strain of S. scabies (ATCC 49173) was not placed in either of the two clades (Fig. 1), suggesting that the strains in clade II do not belong to S. scabies.
Figure 1.
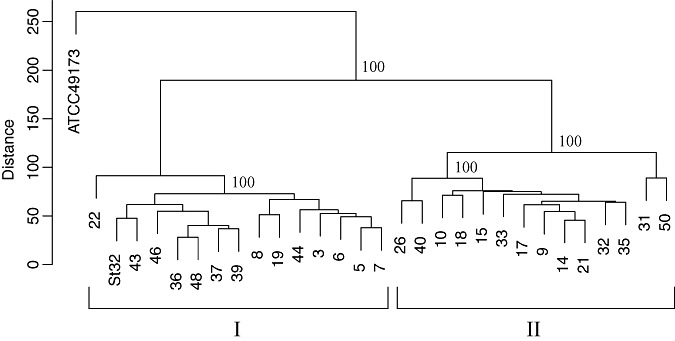
Microarray analysis‐based grouping of the 28 pathogenic Streptomyces strains isolated from potato scab lesions in this study, including S. scabies (type strain ATCC 49173) and S. turgidiscabies (strain St32) for comparison. Clustering was performed on the basis of the signals from 12 766 probes designed for 8848 genes of S. scabies obtained in two independent hybridizations. The signal intensities of the microarray data were preprocessed, the local background signal intensity was subtracted and the signals were subsequently normalized after transformation into the log2 domain. The microarray data were clustered hierarchically using the R package pvclust. Clade I consists of the S. turgidiscabies strain St32 and 14 strains (nos. 3, 5, 6, 7, 8, 19, 22, 36, 37, 39, 43, 44, 46 and 48) isolated in this study. The remaining 14 strains (nos. 9, 10, 14, 15, 17, 18, 21, 26, 31, 32, 33, 35, 40 and 50) form clade II. The type strain of S. scabies (ATCC 49173) is not placed in either clade. Euclidean distances and P values calculated from 1000 bootstraps were used.
Identification of the melanin‐producing pathogenic strains
Because the strains of clade II were not identified to S. turgidiscabies or S. scabies on the basis of microarray analysis, they were also characterized by other methods. The 14 pathogenic strains which did not produce melanin on PYI were detected with the PCR primers specific to the 16S rRNA gene of S. turgidiscabies, whereas the 14 pathogenic strains which produced melanin were detected with the primers for the 16S rRNA gene sequence of S. scabies (Table 1). The three nonpathogenic strains isolated from common scab lesions and included for comparison (K1, K3 and K4) produced melanin on PYI, but were negative when tested with the primers for S. scabies and S. turgidiscabies (Table 1).
Subsequently, the 16S rRNA gene sequences of the 28 pathogenic strains and the three nonpathogenic strains were determined and subjected to phylogenetic analysis. The 14 pathogenic, nonmelanin‐producing strains were clustered with strain St32 and the type strain (ATCC 700248) of S. turgidiscabies (Fig. 2). However, the 14 pathogenic strains which produced melanin were clustered with the type strains of S. scabies (ATCC 49173) and S. europaeiscabiei (CFBP 4497). The nonpathogenic strains were not placed in either cluster (Fig. 3). Strains 31 and 50 clustered in a subgroup of clade II (Fig. 2). Sequence comparisons revealed that, although other strains in clade II contained nucleotides TAGT at positions 261–264 of the 16S rRNA gene (numbering according to S. scabies ATCC49173), strains 31 and 50 had the sequence TAAT, and the strains of clade I contained the sequence TAAC at the same position (data not shown).
Figure 2.
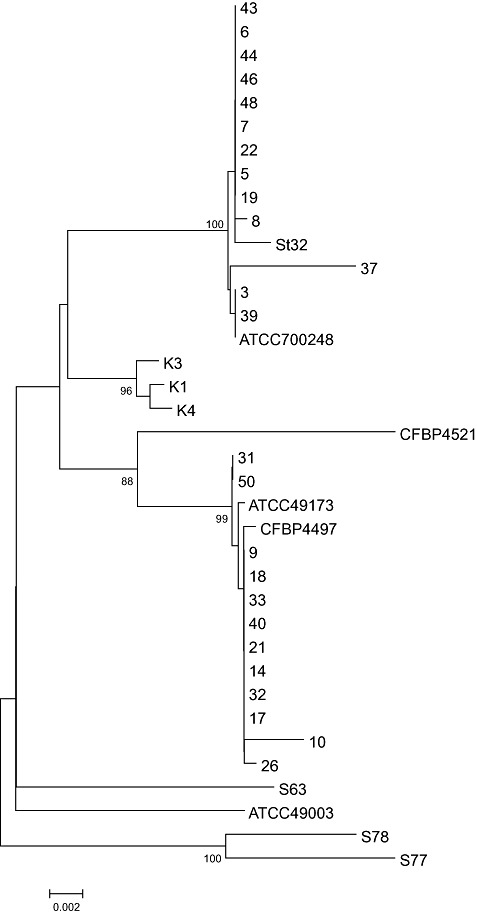
Phylogenetic analysis of Streptomyces strains based on the neighbour‐joining analysis of the 16S rRNA gene sequences, including Norwegian strains of Streptomyces (accession numbers: HQ441807–HQ441834), the type strains of S. scabies (strain ATCC 49173; accession no. AB026199), S. turgidiscabies (strain ATCC 700248; AB026221), S. europaeiscabiei (strain CFBP 4497; AJ007423), S. acidiscabies (strain ATCC49003; AB026220), S. luridiscabiei (strain S63; NR_025155), S. niveiscabiei (strain S78; AF361786), S. puniciscabiei (strain S77; NR_025156), S. stelliscabiei (strain CFBP4521; NR_025294) and S. turgidiscabies (strain St32; EU828541) for comparison. Bootstrap values >90 are shown. The bar indicates a distance of 0.002 substitutions per site.
Figure 3.
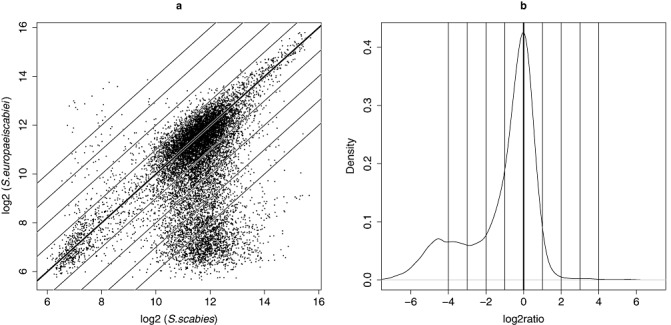
Microarray‐based comparison of Streptomyces scabies (type strain ATCC49173) and S. europaeiscabiei (strain 10). (a) Scatter plot of log intensities of the signals from 12 879 probes, 12 766 of which were designed for the initially predicted 8848 open reading frames (ORFs) of S. scabies (strain 87.22) draft genome sequence (FN554889) and 113 of which were designed for the pathogenicity islands of S. turgidiscabies (strain Car8). (b) Distribution of log ratios between S. europaeiscabiei (strain 10) and Streptomyces scabies (type strain ATCC49173). The mode of distribution at log ratio 0 corresponds to the main diagonal of (a). The small peak at log ratio −4 consists of probe signals indicating divergent/absent genes/genomic regions in S. europaeiscabiei. Diagonal lines in scatterplot (a) correspond to log ratio locations in (b), so that the top‐most line represents log ratio 4 and the bottom‐most line represents log ratio −4.
The contrasting results obtained by CGH and 16S rRNA gene analyses concerning the identity of the 14 melanin‐producing pathogenic strains suggested that these strains might belong to S. europaeiscabiei. The 16S rRNA gene sequences of S. scabies and S. europaeiscabiei are indistinguishable and the species demarcation was based originally only on the distant genomic relatedness evaluated by DNA–DNA hybridization (Bouchek‐Mechiche et al., 2000). However, the two species can also be distinguished on the basis of differences in the sequences of the 16S‐23S ribosomal internal transcribed spacer (ITS) region (Flores‐Gonzalez et al., 2008). The ITS sequence was amplified by PCR from the 14 melanin‐producing pathogenic strains and subjected to restriction analysis with Hpy99I (Flores‐Gonzalez et al., 2008). Amplicons were not restricted by Hpy99I (Table 2), in contrast with the strain ME01‐11h of S. scabies included as a positive control, which indicated that the 14 strains belonged to S. europaeiscabiei.
Table 2.
Primers used for detection and sequencing of genes from Streptomyces strains.
| Primer name† | Primer sequence (5′–3′) | Target gene | Reference | |
|---|---|---|---|---|
| TxtAB1 | F‡ | CCACCAGGACCTGCTCTTC | TxtAB operon | Wanner (2006) |
| TxtAB2 | R | TCGAGTGGACCTCACAGATG | TxtAB operon | Wanner (2006) |
| Nf | F | ATGAGCGCGAACGGAAGCCCCGGA | Nec1 | Bukhalid et al. (1998) |
| Nr | R | GCAGGTCGTCACGAAGGATCG | Nec1 | Bukhalid et al. (1998) |
| Tom3 | F | GAGGCGTTGGTGGAGTTCTA | TomA | Wanner (2006) |
| Tom4 | R | TTGGGGTTGTACTCCTCGTC | TomA | Wanner (2006) |
| 16S‐1F* | F | CATTCACGGAGAGTTTGATCC | 16S rRNA | Bukhalid et al. (2002) |
| 16S 435–455* | F | AGCAGGGAAGAAGCGAAAGT | 16S rRNA | Wanner (2009) |
| 16S 873–893* | F | CCGCAAGGCTAAAACTCAAA | 16S rRNA | Wanner (2006) |
| 16S_1049–1069* | F | AGCTCGTGTCGTGAGATGTTG | 16S rRNA | This study |
| 16S1‐R* | R | AGAAAGGAGGTGATCCAGCC | 16S rRNA | Bukhalid et al. (2002) |
| 16S_455–435* | R | ACTTTCGCTTCTTCCCTGCT | 16S rRNA | Wanner (2006) |
| 16S_893–873* | R | TTTGAGTTTTAGCCTTGCGG | 16S rRNA | Wanner (2006) |
| 16S_1366–1346* | R | AACGTATTCACCGCAGCAAT | 16S rRNA | Wanner (2006) |
| ITS‐L | F | GTCAAGTCATCATGCCCCTT | 16S intergenic region | Flores‐Gonzalez et al. (2008); Song et al. (2004) |
| ITS‐R | R | AAACTTGGCCACAGATGCTC | 16S intergenic region | Flores‐Gonzalez et al. (2008); Song et al. (2004) |
| pA | F | AGAGTTTGATCCTGGCTCAG | Universal (16S rRNA) | Edwards et al. (1989) |
| pH′ | R | AAGGAGGTGATCCAGCCGCA | Universal (16S rRNA) | Edwards et al. (1989) |
| ScabI | F | CAACACTCTCGGGCATCCGA | S. scabies (16S rRNA) | Lehtonen et al. (2004) |
| ScabII | R | TTCGACAGCTCCCTCCTTAC | S. scabies (16S rRNA) | Lehtonen et al. (2004) |
| TurgI | F | CCTCGCATGGGGGTGGGTTA | S. turgidiscabies (16S rRNA) | Lehtonen et al. (2004) |
| TurgII | R | CGACAGCTCCCTCCCCGTAA | S. turgidiscabies (16S rRNA) | Lehtonen et al. (2004) |
Primers marked with an asterisk were for sequencing.
F, forward primer; R, reverse primer.
Genetic differences between S. scabies, S. europaeiscabiei and S. turgidiscabies
Results of hierarchical clustering based on the whole S. scabies genome microarray data and a large number of differentiating probes (Fig. 3) indicated that differences between the genome sequences of S. scabies and S. europaeiscabiei were significant. Standard deviations of signal intensities for the 14 strains of S. europaeiscabiei were calculated using data from two independent hybridization experiments. The plotting pattern (Fig. 4) indicated that the standard deviation of the signal intensities was large in the terminal ends of the genome. Indeed, there were many probes designed based on S. scabies gene sequences that hybridized poorly with the genomic DNA of S. europaeiscabiei.
Figure 4.
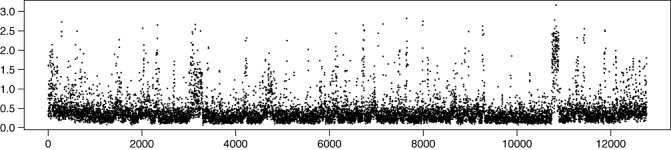
Standard deviations of signal intensities for14 strains of Streptomyces europaeiscabiei observed in microarray analysis in two independent hybridizations. The 12 766 probes designed for 8848 open reading frames (ORFs) of S. scabies probes are ordered according to their location in the genome of S. scabies. The units on the x‐axis are probe indices along the genome sequence (S. scabies strain 87.22; FN554889). Distances in kilobases correspond approximately to the probe index multiplied by 0.7.
The hybridization signal intensity among the strains of S. europaeiscabiei was particularly low for c. 1180 probes (log2 ratio < −3), corresponding to 762 genes (ORFs) of S. scabies (Table S1). These results indicate that the 762 genes possess significant sequence differences between the two species or are found only in S. scabies. Fisher's exact test indicated that these 762 genes (further called ‘divergent genes’) included particularly high or low proportions of the genes of certain gene ontology (GO) categories (Table 3) formed on the basis of the annotation of 6157 of a total of 8746 ORFs (Table S1) predicted in the recently released final genome sequence of S. scabies strain 87.22 [available at the National Center for Biotechnology Information (NCBI) database; https://www.ncbi.nlm.nih.gov/]. The GO categories enriched with the ‘divergent genes’ included, for example, ‘cell death’ (GO:0008219; 13 of the total of 27 genes), ‘symbiosis, encompassing mutualism through parasitism’ (GO:0044403; five of six genes) and ‘response to biotic stimulus’ (GO:0009607; four of five genes). There were many ‘divergent genes’ among transposases (29 of 115 genes; χ 2; P < 2,2E‐16), two‐component system response regulators (seven of 61 genes; χ 2; P < 2,2E‐16) and phage‐related genes (eight of 52 genes; χ 2; P < 2,2E‐16). However, genes related to primary metabolism were under‐represented among the ‘divergent genes’ (Table 3). The probes detecting many ‘divergent genes’ between S. europaeiscabiei and S. scabies were designed for genes that were often found in clusters in the genome sequence of S. scabies. As many as 28 gene clusters defined on this basis contained at least five ‘divergent genes’, and three clusters contained at least 29 ‘divergent genes’. Genes in the clusters belonged to various categories, for example, secondary metabolism, such as polyketide synthases, nonribosomal peptide synthetases, monooxygenases and ABC transporters (Table S1). Many probes designed to genes in the ‘toxicogenic region’ of PAI in S. scabies detected ‘divergent genes’ between S. europaeiscabiei and S. scabies, whereas probes designed for genes in the ‘colonization region’ revealed little genetic variability.
Table 3.
Enrichment analysis (over‐ or under‐representation) in gene ontology (GO) categories of 762 genes showing large sequence variability between the strains of Streptomyces europaeiscabiei and S. scabies, as analysed using probes designed to genes of S. scabies *.
| GO | Name | P value | Representation |
|---|---|---|---|
| GO:0016265 | Death | 4.54E‐08 | Over |
| GO:0008219 | Cell death | 4.54E‐08 | Over |
| GO:0044464 | Cell part | 9.02E‐06 | Under |
| GO:0005737 | Cytoplasm | 2.54E‐05 | Under |
| GO:0044424 | Intracellular part | 2.86E‐05 | Under |
| GO:0009058 | Biosynthetic process | 7.89E‐05 | Under |
| GO:0044419 | Interspecies' interaction between organisms | 2.05E‐05 | Over |
| GO:0051704 | Multi‐organism process | 2.05E‐05 | Over |
| GO:0044403 | Symbiosis, encompassing mutualism through parasitism | 2.05E‐05 | Over |
| GO:0005975 | Carbohydrate metabolic process | 5.35E‐04 | Under |
| GO:0043232 | Intracellular nonmembrane‐bounded organelle | 9.79E‐04 | Under |
| GO:0043228 | Nonmembrane‐bounded organelle | 9.79E‐04 | Under |
| GO:0043229 | Intracellular organelle | 0.001112 | Under |
| GO:0006091 | Generation of precursor metabolites and energy | 0.001133 | Under |
| GO:0005623 | Cell | 0.001578 | Under |
| GO:0005198 | Structural molecule activity | 0.001683 | Under |
| GO:0016020 | Membrane | 0.002413 | Under |
| GO:0005886 | Plasma membrane | 0.002413 | Under |
| GO:0003723 | RNA binding | 0.003294 | Under |
| GO:0009607 | Response to biotic stimulus | 2.11E‐04 | Over |
| GO:0006259 | DNA metabolic process | 2.61E‐04 | Over |
| GO:0044444 | Cytoplasmic part | 0.00396 | Under |
| GO:0030529 | Ribonucleoprotein complex | 0.004842 | Under |
| GO:0005840 | Ribosome | 0.004842 | Under |
| GO:0043226 | Organelle | 0.005546 | Under |
| GO:0019748 | Secondary metabolic process | 0.006092 | Under |
| GO:0044238 | Primary metabolic process | 0.008245 | Under |
| GO:0006412 | Translation | 0.014966 | Under |
| GO:0032991 | Macromolecular complex | 0.01718 | Under |
| GO:0003677 | DNA binding | 0.00135 | Over |
| GO:0008152 | Metabolic process | 0.039758 | Under |
| GO:0009056 | Catabolic process | 0.048668 | Under |
| GO:0006519 | Cellular amino acid and derivative metabolic process | 0.049959 | Under |
| GO:0044281 | Small‐molecule metabolic process | 0.049959 | Under |
The genes corresponding to the GO categories are listed in Table S1.
Analysis of the microarray data revealed that the probes which distinguished strains 31 and 50 from other strains of S. europaeiscabiei (Fig. 1) detected 131 genes. Signals of probes for these genes were much lower or higher for strains 31 and 50 than for the other strains (log2 ratio ≥−3 and log2 ratio ≤−3) (Table S1). The GO category ‘transcription regulator activity’ (GO:0030528) was enriched with 12 genes in strains 31 and 50 (P= 6,6E‐2) (Table S1), with much higher hybridization signal intensity than in the strains forming the main clade of S. europaeiscabiei (Fig. 1).
Comparison of strains using probes for PAI genes
Clustering of the strains on the basis of the signal intensities generated by hybridization with 17 probes specific to four PAI genes (tomA, nec1, txtA and txtB) resulted in four main clusters (Fig. 5). One cluster consisted of 10 strains of S. europaeiscabiei and the type strain of S. scabies (ATCC 49173). The second cluster contained eight Norwegian strains of S. turgidiscabies and the Swedish strain St32 of S. turgidiscabies. The third cluster included six strains of S. turgidiscabies, and a fourth cluster contained four strains of S. europaeiscabiei (Fig. 5).
Figure 5.
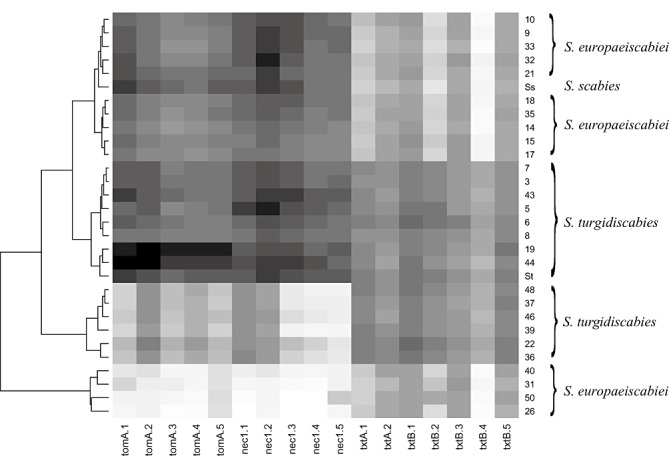
Clustering of the Streptomyces strains based on signal intensities from two independent hybridizations of 17 probes designed for four genes (tomA, nec1, txtA and txtB) of the pathogenicity islands of S. turgidiscabies and used for microarray analysis. Higher signal intensities are indicated with darker colours. Ss, S. scabies strain ATCC 49173; St, S. turgidiscabies strain St32.
Results from CGH showed clearly that the nec1 and tomA genes gave high‐intensity signals in the same 18 strains of S. europaeiscabiei and S. turgidiscabies (strains 3, 5, 6, 7, 8, 9, 10, 14, 15, 17, 18, 19, 21, 32, 33, 35, 43 and 44) (Fig. 5). Consistent with these results, nec1 was amplified by PCR with the same primer pair from all of these strains, except strain 19 (Table 1). Moreover, results from PCR revealed the presence of tomA in six additional strains (strain 22, 36, 37, 39, 46, 48) of S. turgidiscabies which showed low‐intensity signals in CGH.
The signal intensity from probes specific to the txtAB operon was high for all S. turgidiscabies strains, but more variable for the strains of S. europaeiscabiei (Fig. 5).
DISCUSSION
In this study, pathogenic Streptomyces species isolated from common scab lesions on potato in Norway were identified, and the genetic variability of the strains was investigated using microarray‐based comparative genome analysis. The analysis was facilitated by the availability of sequence data from the whole genome of S. scabies and the PAI sequence of S. turgidiscabies, which were utilized to design probes and to verify their target specificity. Microarray analysis revealed two distinct clades of Streptomyces strains, one of which contained 14 Norwegian Streptomyces strains and the Swedish strain of S. turgidiscabies included for comparison, and the other contained the remaining 14 Norwegian pathogenic Streptomyces strains. Sequence analysis of the 16S rRNA genes did not distinguish the Norwegian Streptomyces strains of the second clade from S. scabies, but restriction analysis of the 16S‐23S intergenic spacer region showed that these Norwegian strains could be designated as S. europaeiscabiei. Thus, the whole‐genome‐wide microarray analysis revealed that S. europaeiscabiei and S. scabies can be readily distinguished from each other. This is consistent with the studies of Bouchek‐Mechiche et al. (2000), who concluded that the type strain of S. europaeiscabiei (CFBP4497) can be considered to be a genomic species separated from the phenetic cluster of S. scabies (ATCC49173). However, our data provide a genome‐wide, gene‐level resolution of the differences. The type strain of S. scabies (ATCC 49173) was not placed in either clade, which demonstrates that microarray‐based CGH can be used as a thorough approach to distinguish between species such as S. scabies and S. europaeiscabiei that are identical with regard to their 16S rRNA gene sequences, but possess significant systematic differences in the sequences of many other genes. Furthermore, the results revealed that strains 31 and 50 constitute a genetically distinguishable race of S. europaeiscabiei.
Probes for the thaxtomin synthesis genes (txtAB), used as markers of the ‘toxicogenic region’ of PAI (Lerat et al., 2009), generated strong signals in all S. turgidiscabies strains. Hence, in this respect, the strains tested appeared to be similar to strain Car8 of S. turgidiscabies from Japan (Kers et al., 2005) according to which the probes for PAI were designed (Aittamaa et al., 2010). However, microarray data indicated that the txtAB sequences of S. turgidiscabies are divergent from those in S. scabies and S. europaeiscabiei. Previous studies have shown that the txtAB genes can be rather divergent also among strains of S. scabies (St‐Onge et al., 2008) and S. turgidiscabies (Aittamaa et al., 2010). The txtAB sequences among the strains of S. turgidiscabies characterized from Finland (Aittamaa et al., 2010) show much wider genetic variability than the strains from Norway tested in this study. Results of PCR analysis with the primer pair txtAB1/txtAB2 showed that all tested strains of S. turgidiscabies and S. europaeiscabiei harbour the txtAB genes, and pathogenicity tests on radish and potato also confirmed the pathogenicity of the strains. Thus, the necessary genes for pathogenicity were present, but their sequences varied. However, the negative results of PCR analysis suggested that nec1 was lacking from more than one‐third of the pathogenic strains tested. This is consistent with previous findings showing that nec1 is not required for pathogenicity, but may influence virulence (Flores‐Gonzalez et al., 2008; Kreuze et al., 1999; Wanner, 2006, 2009).
nec1 and tomA, used as markers of the ‘colonization region’ of PAI (Lerat et al., 2009), could be detected in 10 of the 14 S. europaeiscabiei strains by PCR. Microarray analysis revealed high signal intensities with probes for tomA and nec1 in the same 10 strains. However, PCR analysis showed that tomA was present in six additional strains that exhibited only low signals with the tomA‐specific probes in microarray analysis, indicating sequence divergence.
The standard deviation of the hybridization intensities was largest for probes detecting genes at the terminal ends of the genome in S. europaeiscabiei. These results are supported by the observations of Hsiao and Kirby (2007), indicating that the linear chromosome of genus Streptomyces contains a conserved central part and more variable termini. However, there were also other genomic locations in which the standard deviation of the signals was much larger than elsewhere. These areas were located between probes 3000–3300 and 7400–7600, corresponding to 200‐kb genomic regions between the genomic locations 2.30–2.52 Mbp and 8.35–8.54 Mbp in the genome of S. scabies, respectively.
There were 1180 probes for 762 genes that generated the lowest hybridization signal intensities among the strains of S. europaeiscabiei. The corresponding genes of S. scabies represent a large proportion of genes in the GO categories ‘cell death’, ‘symbiosis, encompassing mutualism through parasitism’ and ‘response to biotic stimulus’. ‘Two‐component system response regulators’ were also found to be enriched among the genes differentiating these two species. Thus, the results suggest that S. scabies and S. europaeiscabiei have undergone species‐specific adaptation in their interactions with plants or other microorganisms. Two‐component system response regulators are involved in signalling, and the stimulus–response coupling mechanism allows organisms to sense and respond to changes in various environmental conditions (Stock et al., 2000). This may be needed because Streptomyces species have a complex developmental life cycle, including vegetative mycelium and aerial hyphae, in which spores form on depletion of nutrients (Dworkin et al., 2007). Programmed cell death, in turn, occurs in response to both abiotic and biotic stress, and, in Streptomyces, is accompanied by the presence of enzymes involved in the degradation of cellular macromolecules, regulatory proteins and stress‐induced proteins (Manteca et al., 2006). However, many GO categories related to primary metabolism were under‐represented among the ‘divergent genes’, indicating that they are rather conserved among the Streptomyces species studied.
CGH carried out with gene‐specific probes in a microarray format provides information about thousands of genes simultaneously and is useful for the comprehensive characterization of genetic variability. Results can reveal important information and insights concerning the biology and populations of the pathogen, which can then be used in the management of the disease. To our knowledge, this is the first comparative genomic study of the important potato pathogens S. europaeiscabiei, S. turgidiscabies and S. scabies. This is also the first report of S. turgidiscabies and S. europaeiscabiei in Norway.
EXPERIMENTAL PROCEDURES
Bacterial strains
Streptomyces species were isolated from potato scab lesions from 21 locations in 12 counties of Norway spanning approximately 1400 km from south to north. Scabby potato tubers were surface disinfected in 70% ethanol for 20 s. After several rinses in sterile distilled water, a small piece of potato tissue was cut from under the surface of a single lesion and homogenized in 200 µL of sterile distilled water. An aliquot of 100 µL of the homogenate was plated out on water agar and incubated at 28 °C in the dark. Single colonies, phenotypically characteristic of Streptomyces, were transferred to YME agar (Loria et al., 2001) after approximately 6 days. Subsequent transfer to fresh medium was performed to obtain pure cultures. Streptomyces strains were grown on YME agar plates at 28 °C and maintained on YME agar plugs at −70 °C. The strains were grown on PYI plates to test their ability to produce melanin (Loria et al., 2001).
In addition to the bacterial strains obtained in this study, a number of previously characterized Streptomyces strains were included for comparison. The type strain of S. scabies (ATCC49173), isolated from potato in New York, USA (Lambert and Loria, 1989), was obtained from the American Type Culture Collection (ATCC). Strain 32 of S. turgidiscabies, isolated from potato in northern Sweden (Aittamaa et al., 2010; Lehtonen et al., 2004), was obtained from the collection of the Department of Agricultural Sciences, University of Helsinki, Finland.
Pathogenicity tests
Streptomyces strains were tested for pathogenicity on radish seedlings as described previously by Flores‐Gonzalez et al. (2008). Radish seeds were disinfected in 0.5% sodium hypochlorite solution for 1 min and rinsed several times with sterile distilled water. They were placed on an 8‐day‐old culture of a Streptomyces strain growing on Difco™ oatmeal agar (OMA) (Becton, Dickinson and Company, Baltimore, MD, USA) and incubated at room temperature for 8 days. Seeds grown on OMA plates with three txtAB‐negative Streptomyces strains and on OMA plates without bacteria were used as controls.
A few txtAB‐positive and txtAB‐negative strains were also tested for pathogenicity on potato. Strains K1 and K3 that did not damage radish seedlings were used for inoculation as controls. The Streptomyces strains were grown on YME agar for 2 weeks. P‐Soil (Tjerbo Torvfabrikk, Rakkestad, Norway) and Agra‐perlite (Pull Rhenen, Rhenen, the Netherlands) were autoclaved three times, mixed (1:1, v/v) and placed in pots (5 L). Bacterial cultures of a strain from two plates were mixed thoroughly in the upper layer of the soil–perlite mixture, after which a healthy minituber of the scab‐susceptible cultivar Blue Congo was planted in the pot. A few pots were not inoculated with any bacterial strain and were included as controls. The plants were grown in a glasshouse under controlled conditions at 18–20 °C in natural daylight in July–August. An automated drip‐water system was used for irrigation and the supply of fertilizer. The experiment was carried out with three replicates. The tubers were harvested 9 weeks after planting and the symptoms were recorded.
DNA isolation
Streptomyces strains were grown on YME at 28 °C for 6–8 days and cells were scraped from the plate into a mortar and ground with a pestle in liquid nitrogen. DNA was extracted using the DNeasy Plant Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. Lysis buffer and RNase (included in the kit) were added before the samples were incubated at 65 °C for 1 h. DNA was eluted in 40 µL AE buffer included in the kit. The quality and amount of DNA were checked by agarose gel electrophoresis and a NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Analysis of the 16S rRNA gene and intergenic sequences
The primers developed for the 16S rRNA gene sequences by Lehtonen et al. (2004) were used to detect S. scabies and S. turgidiscabies by PCR according to the authors' instructions. However, S. scabies and S. europaeiscabiei cannot be distinguished on the basis of the 16S rRNA gene sequences. Therefore, the ITS region of the 16S operon was amplified using the primer pair ITS‐R/ITS‐L (Table 1) and the amplicons were subjected to digestion with the restriction enzyme Hpy99I (New England Biolabs, Beverly, MA, USA), as described previously (Flores‐Gonzalez et al., 2008; Song et al., 2004). The type strain of S. scabies ATCC49173 (Hpy99I+, i.e. amplicons digestible with Hpy99I) and three additional strains ME01‐11h (S. scabies; Hpy99I+), ID02‐12 (S. europaeiscabiei; Hpy99I−) and PE07‐1C (Hpy99I−) were used as controls (courtesy of Dr L. Wanner, US Department for Agriculture, Beltsville, MD, USA).
The 16S rRNA genes were amplified for sequence analysis by PCR using the primer pair 16S1‐F/16S1‐R (Table 1) (Bukhalid et al., 2002). The PCR mix (50 µL) contained 0.4 µm of each primer, 0.2 mm deoxynucleoside triphosphates (dNTPs), 0.5 µL Phusion High‐Fidelity DNA polymerase (Finnzymes, Espoo, Finland), 1.5 mm MgCl2, 3% dimethylsulphoxide (DMSO) and 2.5 µL DNA. The PCR cycle consisted of denaturation at 98 °C for 30 s, followed by 35 cycles consisting of denaturation at 98 °C for 10 s, annealing at 54 °C for 30 s, extension at 72 °C for 1 min 15 s and final extension at 72 °C for 7 min. PCR products were analysed on a 1% agarose gel in tris(hydroxymethyl)aminomethane (Tris)/borate/ethylenediaminetetracetic acid (EDTA) (TBE) buffer by electrophoresis. Products of the expected size (∼1.5 kb) were purified from the gel using an E.Z.N.A Gel Extraction Kit according to the manufacturer's instructions (Omega Bio‐tek, Doranville, GA, USA). DNA was eluted in 30 µL of 10 mm Tris‐HCl buffer for sequencing with the primers indicated in Table 1. Sequencing was performed at Macrogen, Inc., Seoul, South Korea. Sequences were assembled using the Staden package (Bonfield et al., 1995).
Base calling and quality assignment of the 16S rRNA gene sequences were performed using pregap4 with the Phred algorithm. Assembly of the sequence from multiple fragments was carried out using gap4. Because all 16S rRNA gene sequences were not complete, the sequence alignment used for phylogenetic analysis was made up of partial 16S rRNA gene sequences with a final multiple alignment length of 1426 nucleotides. The 16S rRNA gene sequences determined in this study were deposited in the NCBI database under the accession numbers HQ441807–HQ441834.
The 16S rRNA sequences of the type strains of S. scabies (ATCC 49173; accession no. AB026199), S. turgidiscabies (strain ATCC 700248; AB026221), S. europaeiscabiei (strain CFBP 4497; AJ007423), S. acidiscabies (strain ATCC 49003; AB026220), S. luridiscabiei (strain S63; NR_025155), S. niveiscabiei (strain S78; AF361786), S. puniciscabiei (strain S77; NR_025156), S. stelliscabiei (strain CFBP4521; NR_025294) and strain St32 (EU828541) of S. turgidiscabies, described from northern Sweden (Lehtonen et al., 2004), were included for comparison. Sequence alignments were performed using Gap4 software (Bonfield et al., 1995). Neighbour‐joining analysis of 16S rRNA gene sequences (Saitou and Nei, 1987) and the construction of phylogenetic trees were performed using mega software (Tamura et al., 2007). The Kimura two‐parameter model (Kimura, 1980) was used for analysis, and a consensus tree was built on the basis of 1000 bootstraps.
Detection of the txtAB operon, Nec1 and TomA by PCR
The txtAB operon encoding thaxtomin synthetase was detected by PCR using primers txtAB1 and txtAB2 (Table 1), as described previously (Wanner, 2006). The PCR cycle included initial DNA denaturation at 94 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 30 s, primer annealing at 46 °C for 30 s, extension at 72 °C for 35 s and final extension at 72 °C for 7 min. The genes nec1 and TomA were detected using the primer pairs Nf/Nr and Tom3/Tom4, respectively (Table 1), as described previously (Bukhalid et al., 1998; Wanner, 2006). The PCR cycle was the same as above, except that the annealing temperatures were 57 °C and 54 °C, respectively. PCR products were separated on a 1% agarose gel run in TBE buffer, and stained with ethidium bromide. Results were documented using GelDoc and Quantity One software (Bio‐Rad, Hercules, CA, USA). The experiment was repeated.
Microarray analysis
Probes were designed on the basis of the whole‐genome draft sequence of S. scabies (strain 87.22) (FN554889) and the PAI of S. turgidiscabies (AY707079, AY707080, AY707081) using OligoArray 2.1 software (Rouillard et al., 2003). The criteria for probe design included a similar melting temperature of approximately 87 °C and a probe length of 40 nucleotides. Poly(T) was added at the 3′ end of the probe to reach the final probe length of 60 nucleotides. A total of 12 766 probes was designed for 8848 ORFs of S. scabies, whereas 113 probes were designed for the PAI of S. turgidiscabies. Probes were synthesized in situ on Agilent 8 × 15 K custom‐designed microarrays (Agilent, Santa Clara, CA, USA). The probes for PAI were synthesized in triplicate on the array.
On the basis of the results of the radish seedling tests, 28 pathogenic Streptomyces strains (Table 2) were selected for CGH and species' identification by microarray analysis. DNA samples (500 ng) were labelled with Cy3 or Cy5 dCTP (GE Healthcare, Amersham, Buckinghamshire, UK) and purified using the E.Z.N.A. Gel Extraction Kit (Omega Bio‐tek) according to the manufacturer's instructions. DNA concentration and incorporation of the label were checked using a NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies) prior to and after labelling. Two DNA samples, from different Streptomyces strains, one labelled with Cy3 and the other with Cy5, were combined (1:1), denatured at 95 °C for 2 min and hybridized on the same subarray. Hybridization was carried out at 65 °C for 16–18 h using reagents from Agilent and the BioPrime Array CGH Genomic labelling system protocol (Invitrogen, Carlsbad, CA, USA). Cy3 and Cy5 were swapped between the samples when the experiment was repeated (Table S2, see Supporting Information).
Analysis of the microarray data
Microarray slides were scanned using a GenePix 4200 AL scanner with a pixel resolution of 5 µm. GenePix Pro 6.0 software was used for image analysis and spot segmentation. All spots from all arrays were visually investigated, and erroneous signals caused, for example, by scratches or dust were manually flagged and ignored in further analysis. The microarray data (signal intensities) were preprocessed and normalized using R software (R Development Core Team, 2008) with the Limma package (Smyth, 2005). Local background signal intensity was subtracted from the signal intensity of each spot using Normexp with an offset value of 50, and signals were transformed into the log2 domain. Signal intensities were subsequently normalized to an S. scabies reference (ATCC49173).). Four independent hybridizations of the reference strain were combined by quantile normalization and calculation of the median for each probe. Normalization of other strains was based on calculating the log ratio against the reference strain and setting the mode (location of the main peak) to zero (Fig. 3). Normalized log intensities were obtained by adding the log intensity value of the reference to the normalized log ratio (note that the log ratio equals the subtraction of log intensities). After normalization, duplicate hybridizations were averaged. Hybridizations of the different samples showed two modes, high and low signal intensity, reflecting the hybridization of a mixture of present and absent genes/probes, as described in Aittamaa et al. (2010).
The microarray data generated in this study were deposited at Gene Expression Omnibus (GEO) according to the Minimum Information About a Microarray Experiment (MIAME), and are available under accession number GSE20487.
The microarray data were clustered hierarchically using the R package pvclust (Suzuki and Shimodaira, 2006). Euclidean distances and P values calculated from 1000 bootstraps were used. Those probes among the total of 12 766 S. scabies probes that differentiated S. europaeiscabiei from S. scabies ATCC49173 were determined using different log ratio thresholds. In addition, a moderated t‐test (Smyth, 2005) was applied to each probe in order to check whether the log intensity differences (log ratios) were statistically significant (Table 4). P values of the t‐test were corrected using the false discovery rate method (Benjamini and Hochberg, 1995) with multiple testing. Standard deviations of the hybridization intensities were calculated for each probe in order to assess the variation within the S. europaeiscabiei cluster (Fig. 4).
Table 4.
The number of probes differentiating the 14 tested strains of Streptomyces europaeiscabiei from the type strain ATCC49173 of S. scabies in microarray analysis.
| Log2 ratio | Probes | ORFs | Max FDR |
|---|---|---|---|
| Minimum > 4 | 7 | 6 | 4.9e‐15 |
| Minimum > 3 | 15 | 9 | 1.2e‐11 |
| Minimum > 2 | 23 | 16 | 1.2e‐11 |
| Minimum > 1 | 50 | 42 | 1.9e‐08 |
| Maximum < −1 | 2785 | 1925 | 5.0e‐05 |
| Maximum < −2 | 1836 | 1225 | 5.8e‐08 |
| Maximum < −3 | 1180 | 792 | 2.0e‐10 |
| Maximum < −4 | 557 | 394 | 1.2e‐12 |
A total of 12 766 probes was designed for the initially predicted 8848 open reading frames (ORFs) of S. scabies (strain 87.22) draft genome sequence (FN554889). Two probes were designed to some of the ORFs. Hence, the number of differentiating probes at a given log2 ratio can be larger than the number of ORFs. Max FDR is the largest false discovery rate value of a probe observed within the group of probes.
To be able to perform enrichment analysis (Fisher's exact test) on the genes that showed high sequence variability between S. scabies and S. europaeiscabiei, a new annotation of the whole S. scabies genome sequence was carried out. We downloaded cDNA sequences (NCBI accession no.: NC_013929) and used the software blast2go (Conesa et al., 2005) for reannotation. blastx (e‐value: ≤1E‐03) in the NCBI‐NR database was then performed to find GO accessions with distributions of cellular component, biological processes and molecular function. Detailed information is given in Table S1.
Supporting information
Fig. S1 Radish seedlings grown on oat meal agar with Streptomyces strains for 9 days. (A) Seeds grown on plates with S. europaeiscabiei (strain 9) did not germinate. (B) Seedlings grown with S. turgidiscabies (strain 8) were stunted and suffered from hypertrophy. (C) Seedlings grown with the Streptomyces strain K1 grew normally without symptoms. (D) Comparison of two seedlings grown with S. turgidiscabies strain 8 with a seedling grown with Streptomyces strain K1 (to the right).
Table S1 The 762 ‘divergent genes’ between Streptomyces europascabiei and S. scabies, and the 131 ‘divergent genes’ between two races of S. europascabiei. ‘Divergent genes’ were detected on the basis of significant differences in microarray signal intensities between S. europascabiei and S. scabies, or between two races of S. europascabiei. Probes were designed according to the gene sequences of S. scabies (strain 87.22) and are listed in a separate folder of the Excel file. Annotation of the whole genome sequence of S. scabies (strain 87.22), downloaded from the National Center for Biotechnology Information (NCBI) sequence database (accession no. NC_013929), is also presented.
Table S2 Experimental design for the hybridization of 30 Streptomyces strains on 32 microarrays. Strains ATCC49173 and St32 were hybridized four times, whereas all other strains were hybridized twice including dye swaps. One hybridization failed with strains 33 and 44 (omitted from the table).
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
Thanks are due to Leslie Wanner, Arne Hermansen and Arild Sletten for materials, valuable support and comments on the manuscript. Financial support from the Research Council of Norway (NFR 185053/I10), the Foundation for Research Levy on Agricultural Products, the Agricultural Agreement Research Fund and Norwegian food industries is gratefully acknowledged.
REFERENCES
- Aittamaa, M. , Somervuo, P. , Laakso, I. , Auvinen, P. and Valkonen, J.P.T. (2010) Microarray‐based comparison of genetic differences between strains of Streptomyces turgidiscabies with focus on the pathogenicity island. Mol. Plant Pathol. 11, 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aittamaa, M. , Somervuo, P. , Pirhonen, M. , Mattinen, L. , Nissinen, R. , Auvinen, P. and Valkonen, J.P.T . (2008)Distinguishing bacterial pathogens of potato using a genome‐wide microarray approach. Mol. Plant Pathol. 9,705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B, 57, 289–300. [Google Scholar]
- Bonfield, J.K. , Smith, K.F. and Staden, R. (1995) A new DNA sequence assembly program. Nucleic Acids Res. 23, 4992–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchek‐Mechiche, K. , Gardan, L. , Normand, P. and Jouan, B. (2000) DNA relatedness among strains of Streptomyces pathogenic to potato in France: description of three new species, S. europaeiscabiei sp nov, and S. stelliscabiei sp. nov associated with common scab, and S. reticuliscabiei sp nov associated with netted scab. Int. J. Syst. Evol. Microbiol. 50, 91–99. [DOI] [PubMed] [Google Scholar]
- Bukhalid, R.A. , Chung, S.Y. and Loria, R. (1998) nec1, a gene conferring a necrogenic phenotype, is conserved in plant‐pathogenic Streptomyces spp., and linked to a transposase pseudogene. Mol Plant–Microbe Interact. 11, 960–967. [DOI] [PubMed] [Google Scholar]
- Bukhalid, R.A. and Loria, R. (1997) Cloning and expression of a gene from Streptomyces scabies encoding a putative pathogenicity factor. J. Bacteriol. 179, 7776–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhalid, R.A. , Takeuchi, T. , Labeda, D. and Loria, R. (2002) Horizontal transfer of the plant virulence gene, nec1, and flanking sequences among genetically distinct Streptomyces strains in the Diastatochromogenes cluster. Appl. Environ. Microbiol. 68, 738–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa, A. , Götz, S. , García‐Gómez, J. , Terol, J. , Talón, M. and Robles, M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Dworkin, M. , Falkow, S. , Rosenberg, E. , Schleifer, K.‐H. and Stackebrandt, E. (2007) The Prokaryotes: Volume 3: Archaea. Bacteria: Firmicutes, Actinomycetes. New York, NY: Springer‐Verlag. [Google Scholar]
- Edwards, U. , Rogall, T. , Blocker, H. , Emde, M. and Bottger, E.C. (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucl. Acids Res . 17, 7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores‐Gonzalez, R. , Velasco, I. and Montes, F. (2008) Detection and characterization of Streptomyces causing potato common scab in Western Europe. Plant Pathol. 57, 162–169. [Google Scholar]
- Fry, B.A. and Loria, R. (2002) Thaxtomin A: evidence for a plant cell wall target. Physiol. Mol. Plant Pathol. 60, 1–8. [Google Scholar]
- Goyer, C. and Beaulieu, C. (1997) Host range of streptomycete strains causing common scab. Plant Dis. 81, 901–904. [DOI] [PubMed] [Google Scholar]
- Goyer, C. , Otrysko, B. and Beaulieu, C. (1996) Taxonomic studies on streptomycetes causing potato common scab: a review. Can. J. Plant Pathol. 18, 107–113. [Google Scholar]
- Healy, F.G. , Bukhalid, R.A. and Loria, R. (1999) Characterization of an insertion sequence element associated with genetically diverse plant pathogenic Streptomyces spp. J. Bacteriol. 181, 1562–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy, F.G. and Lambert, D.H. (1991) Relationships among Streptomyces spp. causing potato scab. Int. J. Syst. Bacteriol. 41, 479–482. [Google Scholar]
- Healy, F.G. , Wach, M. , Krasnoff, S.B. , Gibson, D.M. and Loria, R. (2000) The txtAB genes of the plant pathogen Streptomyces acidiscabies encode a peptide synthetase required for phytotoxin thaxtomin A production and pathogenicity. Mol. Microbiol. 38, 794–804. [DOI] [PubMed] [Google Scholar]
- Hiltunen, L.H. , Weckman, A. , Ylhainen, A. , Rita, H. , Richter, E. and Valkonen, J.P.T. (2005) Responses of potato cultivars to the common scab pathogens, Streptomyces scabies and S. turgidiscabies . Ann. Appl. Biol. 146, 395–403. [Google Scholar]
- Holt, J.G. and Bergey, D.H. (1994) Bergey's Manual of Determinative Bacteriology. Baltimore, MD: Williams & Wilkins. [Google Scholar]
- Hsiao, N. and Kirby, R. (2007) Comparative genomics of Streptomyces avermitilis, Streptomyces cattleya, Streptomyces maritimus and Kitasatospora aureofaciens using a Streptomyces coelicolor microarray system. Antonie Van Leeuwenhoek 93, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet‐Tapia, J.C. , Badger, J.H. , Loria, R. and Pettis, G.S. (2011) Streptomyces turgidiscabies Car8 contains a modular pathogenicity island that shares virulence genes with other actinobacterial plant pathogens. Plasmid, 65, 118–124. [DOI] [PubMed] [Google Scholar]
- Jørstad, I. (1922) Report of agricultural and horticultural plant diseases during 1920–21. I. Cereal crops and vegetables (Beretning om plantesykdommer i land‐og havebruket 1920–21. I. Landbruksvekster og grønnsaker). Report of the Minister of Agriculture 1922. Ministry of Agriculture, Kristiania, Norway.
- Kers, J. , Cameron, K. , Joshi, M. , Bukhalid, R. , Morello, J. , Wach, M. , Gibson, D.M. and Loria, R. (2005) A large, mobile pathogenicity island confers plant pathogenicity on Streptomyces species. Mol. Microbiol. 55, 1025–1033. [DOI] [PubMed] [Google Scholar]
- Kim, Y.S. , Cho, J.M. , Park, D.H. , Lee, H.G. , Kim, J.S. , Seo, S.T. , Shin, K.Y. , Hur, J.H. and Lim, C.K. (1999) Production of thaxtomin A by Korean isolates of Streptomyces turgidiscabies and their involvement in pathogenicity. Plant Pathol. J. 15, 168–171. [Google Scholar]
- Kimura, M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. [DOI] [PubMed] [Google Scholar]
- King, R. , Lawrence, C. , Clark, M. and Calhoun, L. (1989) Isolation and characterization of phytotoxins associated with Streptomyces scabies . J. Chem. Soc. Chem. Commun. 13, 849–850. [Google Scholar]
- Kreuze, J. , Suomalainen, S. , Paulin, L. and Valkonen, J.P.T. (1999) Phylogenetic analysis of 16S rRNA genes and PCR analysis of the nec1 gene from Streptomyces spp. causing common scab, pitted scab, and netted scab in Finland. Phytopathology, 89, 462–469. [DOI] [PubMed] [Google Scholar]
- Lambert, D.H. and Loria, R. (1989) Streptomyces scabies sp. nov., nom. rev. Int. J. Syst. Bacteriol. 39, 387–392. [Google Scholar]
- Lawrence, C.H. , Clark, M.C. and King, R.R. (1990) Induction of common scab symptoms in aseptically cultured potato‐tubers by the vivotoxin, thaxtomin. Phytopathology, 80, 606–608. [Google Scholar]
- Lehtonen, M.J. , Rantala, H. , Kreuze, J.F. , Bang, H. , Kuisma, L. , Koski, P. , Virtanen, E. , Vihlman, K. and Valkonen, J.P.T. (2004) Occurrence and survival of potato scab pathogens (Streptomyces species) on tuber lesions: quick diagnosis based on a PCR‐based assay. Plant Pathol. 53, 280–287. [Google Scholar]
- Lerat, S. , Simao‐Beaunoir, A.M. and Beaulieu, C. (2009) Genetic and physiological determinants of Streptomyces scabies pathogenicity. Mol. Plant Pathol. 10, 579–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm, P. , Kortemaa, H. , Kokkola, M. , Haahtela, K. , SalkinojaSalonen, M. and Valkonen, J.P.T. (1997) Streptomyces spp. isolated from potato scab lesions under Nordic conditions in Finland. Plant Dis. 81, 1317–1322. [DOI] [PubMed] [Google Scholar]
- Locci, R. (1994) Actinomycetes as plant pathogens. Eur. J. Plant Pathol. 100, 179–200. [Google Scholar]
- Lorang, J.M. , Liu, D. , Anderson, N.A. and Schottel, J.L. (1995) Identification of potato scab inducing and suppressive species of Streptomyces . Phytopathology, 85, 261–268. [Google Scholar]
- Loria, R. , Bukhalid, R.A. , Fry, B.A. and King, R.R. (1997) Plant pathogenicity in the genus Streptomyces . Plant Dis. 81, 836–846. [DOI] [PubMed] [Google Scholar]
- Loria, R. , Clark, C.A. , Bukhalid, R. and Fry, B. (2001) Streptomyces In: Laboratory Guide for Identification of Plant Pathogenic Bacteria (Schaad N.W., Jones J.B. and Chun W., eds), pp. 236–249. St. Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Loria, R. , Kers, J. and Joshi, M. (2006) Evolution of plant pathogenicity in Streptomyces . Annu. Rev. Phytopathol. 44, 469–487. [DOI] [PubMed] [Google Scholar]
- Manteca, A. , Mäder, U. , Connolly, B.A. and Sanchez, J. (2006) A proteomic analysis of Streptomyces coelicolor programmed cell death. Proteomics, 6, 6008–6022. [DOI] [PubMed] [Google Scholar]
- Miyajima, K. , Tanaka, F. , Takeuchi, T. and Kuninaga, S. (1998) Streptomyces turgidiscabies sp. nov. Int. J. Syst. Bacteriol. 48, 495–502. [DOI] [PubMed] [Google Scholar]
- Paradis, E. , Goyer, C. , Hodge, N.C. , Hogue, R. , Stall, R.E. and Beaulieu, C. (1994) Fatty‐acid and protein profiles of Streptomyces scabies strains isolated in eastern Canada. Int. J. Syst. Bacteriol. 44, 561–564. [Google Scholar]
- Park, D.H. , Yu, Y.M. , Kim, J.S. , Cho, J.M. , Hur, J.H. and Lim, C.K. (2003) Characterization of streptomycetes causing potato common scab in Korea. Plant Dis. 87, 1290–1296. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2008) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available at http://www.R‐project.org. [Google Scholar]
- Rouillard, J.‐M. , Zuker, M. and Gulari, E. (2003) OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 31, 3057–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. (1987) The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Scheible, W.R. , Fry, B. , Kochevenko, A. , Schindelasch, D. , Zimmerli, L. , Somerville, S. , Loria, R. and Somerville, C.R. (2003) An Arabidopsis mutant resistant to thaxtomin A, a cellulose synthesis inhibitor from Streptomyces species. Plant Cell. 15, 1781–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipke, R.F. and Loria, R. (2008) Streptomyces scabies 87‐22 possesses a functional tomatinase. J. Bacteriol. 190, 7684–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G. (2005) Limma: linear models for microarray data In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor (Gentleman R., Carey V., Dudoit S., Irizarry R. and Huber W., eds), pp. 397–420. New York: Springer. [Google Scholar]
- Song, J. , Lee, S.‐C. , Kang, J.‐W. , Baek, H.‐J. and Suh, J.‐W. (2004) Phylogenetic analysis of Streptomyces spp. isolated from potato scab lesions in Korea on the basis of 16S rRNA gene and 16S‐23S rDNA internally transcribed spacer sequences. Int. J. Syst. Evol. Microbiol. 54, 203–209. [DOI] [PubMed] [Google Scholar]
- Stock, A.M. , Robinson, V.L. and Goudreau, P.N. (2000) Two‐component signal transduction. Annu. Rev. Biochem. 69, 183–215. [DOI] [PubMed] [Google Scholar]
- St‐Onge, R. , Goyer, C. , Coffin, R. and Filion, M. (2008) Genetic diversity of Streptomyces spp. causing common scab of potato in eastern Canada. Syst. Appl. Microbiol. 31, 474–484. [DOI] [PubMed] [Google Scholar]
- Suzuki, R. and Shimodaira, H. (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Dudley, J. , Nei, M. and Kumar, S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Værdal, J. (1973) Utbredelse av skurv på poteter i Norge. Forskning og forsøk i landbruket 24, 483–497. [Google Scholar]
- Wanner, L.A. (2006) A survey of genetic variation in Streptomyces isolates causing potato common scab in the United States. Phytopathology, 96, 1363–1371. [DOI] [PubMed] [Google Scholar]
- Wanner, L.A. (2009) A patchwork of Streptomyces species isolated from potato common scab lesions in North America. Am. J. Potato Res. 86, 247–264. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Radish seedlings grown on oat meal agar with Streptomyces strains for 9 days. (A) Seeds grown on plates with S. europaeiscabiei (strain 9) did not germinate. (B) Seedlings grown with S. turgidiscabies (strain 8) were stunted and suffered from hypertrophy. (C) Seedlings grown with the Streptomyces strain K1 grew normally without symptoms. (D) Comparison of two seedlings grown with S. turgidiscabies strain 8 with a seedling grown with Streptomyces strain K1 (to the right).
Table S1 The 762 ‘divergent genes’ between Streptomyces europascabiei and S. scabies, and the 131 ‘divergent genes’ between two races of S. europascabiei. ‘Divergent genes’ were detected on the basis of significant differences in microarray signal intensities between S. europascabiei and S. scabies, or between two races of S. europascabiei. Probes were designed according to the gene sequences of S. scabies (strain 87.22) and are listed in a separate folder of the Excel file. Annotation of the whole genome sequence of S. scabies (strain 87.22), downloaded from the National Center for Biotechnology Information (NCBI) sequence database (accession no. NC_013929), is also presented.
Table S2 Experimental design for the hybridization of 30 Streptomyces strains on 32 microarrays. Strains ATCC49173 and St32 were hybridized four times, whereas all other strains were hybridized twice including dye swaps. One hybridization failed with strains 33 and 44 (omitted from the table).
Supporting info item
Supporting info item
Supporting info item


