SUMMARY
The ascomycete Sclerotinia sclerotiorum is a necrotrophic plant pathogen with an extremely broad host range. It causes stem rot in Camelina sativa, a crucifer with great potential as an alternative oilseed crop. Lignification is a common phenomenon in the expression of resistance against necrotrophs, but the molecular mechanisms underlying this defence response are poorly understood. We present histochemical, gene expression and biochemical data investigating the role of monolignols in the resistance of C. sativa to S. sclerotiorum. Comparative studies with resistant and susceptible lines of C. sativa revealed substantial differences in constitutive transcript levels and gene regulation patterns for members of the gene family encoding cinnamoyl‐CoA reductase (CCR), the first enzyme specifically committed to the synthesis of lignin monomers. These differences were associated with anatomical and metabolic factors. While the induction of CsCCR2 expression after inoculation with S. sclerotiorum was associated with the deposition of lignin mainly derived from guaiacyl monomers, high constitutive levels of CsCCR4 paralleled a high syringyl lignin content in healthy stems of resistant plants. The results provide evidence that plant cell wall strengthening plays a role in the resistance of C. sativa to S. sclerotiorum, and that both constitutive and inducible defence mechanisms contribute to reduced symptom development in resistant germplasm. This study provides the first characterization of quantitative resistance in C. sativa to S. sclerotiorum.
INTRODUCTION
Sclerotinia sclerotiorum (Lib.) de Bary is a ubiquitous necrotrophic plant pathogen with an extremely broad host range, causing disease in at least 408 species belonging to 75 families (Boland and Hall, 1994; Bolton et al., 2006). Among its hosts is Camelina sativa (L.) Crantz, a member of the Brassicaceae family, that is attracting interest as an alternative oilseed crop.
The phenylpropanoid pathway is a crucial component of a plant's defence repertoire against abiotic and biotic stress factors (Dixon et al., 2002). This pathway produces various compounds belonging to a wide range of structural classes and with numerous biological functions. Such compounds include UV protectants and pigments, phytoalexins and phytoanticipins, as well as salicylic acid, a crucial signalling molecule in the interaction between plants and microorganisms. Furthermore, certain phenylpropanoid compounds are polymerized to form defensive barriers, such as lignin (Durbin et al., 2000; Gayoso et al., 2010; Thomma et al., 2001).
Lignin, a major component of secondary cell walls, is an amorphous phenolic heteropolymer resulting from the oxidative polymerization of at least two units of the cinnamyl alcohols (monolignols) p‐coumaryl, coniferyl and sinapyl alcohol, forming p‐hydroxyphenyl (H), guaiacyl (G) and syringyl (S) lignins, respectively. Lignification is a terminal process of highly specialized plant cells capable of forming secondary cell walls, such as xylem and phloem cells, their neighbouring fibre cells and sclereids (Ros Barcelo, 1997). Lignin appears to be a fundamental cell wall component of all vascular plants. Its composition varies across plant lineages and tissue. Gymnosperms generally contain H and mainly G lignin units, whereas, in angiosperms, lignin primarily consists of G and S units and traces of H lignin (Baucher et al., 2003; Boerjan et al., 2003). Within the vascular tissue of angiosperms, such as Arabidopsis thaliana, G lignin predominates in the walls of xylem vessels, whereas fibre and parenchyma cells contain a mixture of G and S lignin, with the latter predominating in heavily lignified parenchyma cells (Chapple et al., 1992; Fergus and Goring, 1970).
There is strong evidence that lignification is a common phenomenon in the expression of disease resistance in plants. Lignin synthesis is induced in response to mechanical damage or wounding, and many plants respond to invading pathogens with the deposition of lignin and lignin‐like material (Boudet et al., 1995; Dixon and Paiva, 1995; Nicholson and Hammerschmidt, 1992; Vance et al., 1980). It is generally thought that lignin plays an important role in the modification of the mechanical properties of cell walls by increasing cell wall rigidity to limit the diffusion of toxins from the pathogen to the host, and of nutrients from the host to the pathogen, and to limit polysaccharide degradation by exogenous enzymes (Ride, 1978; Weng and Chapple, 2010). Lignification of cell walls has been shown to enhance the resistance of many herbaceous and woody plants to fungal attack (Dushnicky et al., 1998; Hammerschmidt and Kuć, 1982; Southerton and Deverall, 1990). Although little is known about the chemical nature, temporal and spatial regulation of deposition or molecular mechanisms involved in gene activation with regard to disease‐induced lignins, it appears that stresses, such as pathogen attack, can change lignin composition with regard to monomer ratios (Gayoso et al., 2010; Hammerschmidt et al., 1985; Pomar et al., 2004; Ros Barcelo, 1997; Southerton and Deverall, 1990).
Despite the substantial impact of S. sclerotiorum on agricultural production, the molecular mechanisms of host resistance to this pathogen have received little attention and remain poorly understood. The evidence for a role of lignification in resistance against S. sclerotiorum is mainly of a histochemical nature (Green et al., 1998; Rodríguez et al., 2004). In a study comparing transgenic carrots over‐expressing different pathogenesis‐related (PR) proteins for their susceptibility to Botrytis cinerea and S. sclerotiorum, the highest levels of resistance were seen in transgenic lines over‐expressing the peroxidase POC1. Petioles of these plants had higher constitutive levels of lignin relative to control plants, and lignification was enhanced after inoculation with S. sclerotiorum in transgenic plants, but not in wild‐type plants (Wally et al., 2008). However, studies conducted on soybean provided evidence that lignification does not play a role in the resistance of this particular host to S. sclerotiorum (Calla et al., 2009; Peltier et al., 2009). Based on the results of gene expression profiling in soybean stems shortly after inoculation with S. sclerotiorum, Calla et al. (2009) proposed that substrates from the lignin pathway are diverted to the synthesis of isoflavonoids and anthocyanins.
Stem rot‐resistant germplasm of C. sativa has been identified recently (C. Eynck et al., unpublished data). The current study reports the histochemical, biochemical and gene expression data investigating the role of monolignol biosynthesis in the resistance of C. sativa to S. sclerotiorum. This study provides the first characterization of quantitative resistance in C. sativa to S. sclerotiorum and contributes to the understanding of stem rot resistance in crucifers.
RESULTS
Disease development
Necrotic lesions were first observed at 48 h post‐inoculation (hpi) on unwounded stems of C. sativa lines 36011 (stem rot‐resistant) and 36012 (stem rot‐susceptible) inoculated with ascospores of S. sclerotiorum at the early flowering stage. At 72 hpi, lesions were significantly (P≤ 0.01) smaller on 36011 than on 36012 plants and, at 96 hpi, 36011 plants showed lesions that were less than half the size of lesions on 36012 plants (Fig. 1A). After 96 hpi, lesions on susceptible plants were characterized by rapid growth. Wilting was first observed at 7 days post‐inoculation (dpi) and most 36012 plants died within 14 dpi. In contrast, disease progression was very slow in 36011 plants; lesions remained confined to the outer cortex of the stem and no wilting occurred (Fig. 2).
Figure 1.
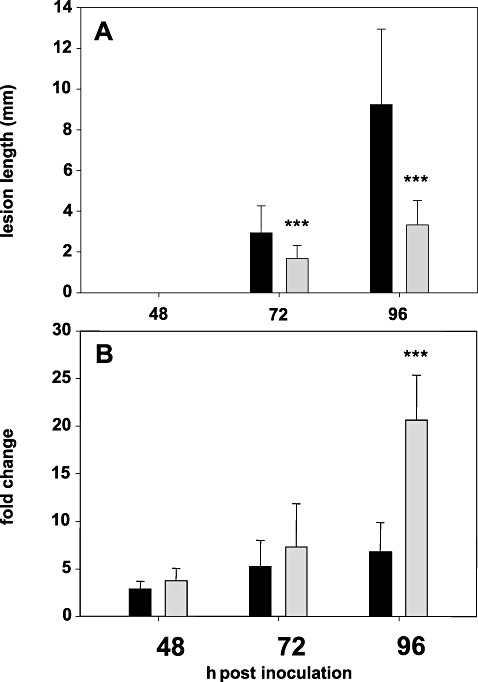
Lesion development and CsCCR2 gene expression in Camelina sativa lines 36011 (grey bars) and 36012 (black bars) at 48–96 h post‐inoculation (hpi). (A) Lesion development. (B) Changes in transcript level relative to mock‐inoculated plants. Columns and bars represent the means and standard deviations of four biological replicates. Asterisks denote a significant difference at P≤ 0.01.
Figure 2.

Typical stem rot symptoms on resistant Camelina sativa line 36011 (A) and susceptible line 36012 (B) at 14 days post‐inoculation (dpi).
Histochemical studies
Stem lignification was studied by bright field microscopy after treatment with phloroglucinol‐HCl and the Mäule reagent, and by confocal laser scanning microscopy.
Synthesis of phenolic compounds, indicated by autofluorescence after illumination with blue light, occurred in inoculated plants of the resistant line in response to infection (Fig. 3A–F). At 3 dpi, accumulation of fluorescent material was observed in cells adjacent to the site of pathogen ingress. In addition, walls of xylem vessels and of cells of the outer layers of the interfascicular tissue in proximity to the infection site exhibited much stronger autofluorescence in inoculated than in mock‐inoculated plants (Fig. 3B). There was a steady increase in the amount of deposited fluorescent material, which formed a lens‐shaped barrier zone around colonized tissue in resistant plants (Fig. 3D, F). Similarly, walls of xylem vessels and of cells of the outer layers of the interfascicular tissue continued to show stronger autofluorescence when compared with mock‐inoculated plants (Fig. 3D, F). There was no autofluorescence observed in cell walls along the border of the developing lesion or the interfascicular tissue in inoculated susceptible plants. Only a slight increase in autofluorescence was observed in the walls of xylem vessels (Fig. 3G–L). Stems of inoculated susceptible plants showed massive colonization by the fungus, detected as clusters of circle‐like cross‐sections of fungal hyphae in peripheral stem tissue (arrowheads, Fig. 3H, J, L; Fig. S1, see Supporting Information).
Figure 3.
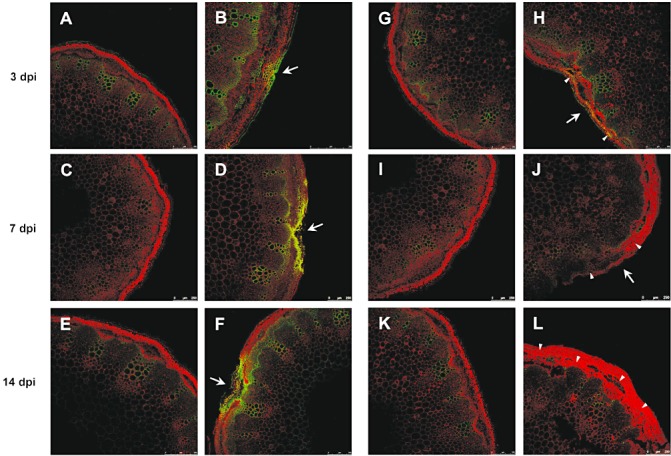
Reaction of Camelina sativa lines 36011 (resistant) and 36012 (susceptible) to infection by Sclerotinia sclerotiorum at 3, 7 and 14 days post‐inoculation (dpi). Line 36011: (A, C, E) mock‐inoculated plants; (B, D, F) inoculated plants; deposition of autofluorescent material. Line 36012: (H, J, L) mock‐inoculated plants; (G, I, K) inoculated plants; massive colonization by the fungus (arrowheads). Arrows indicate point of inoculation.
The Mäule reagent is a histochemical stain used to qualitatively determine lignin monomer composition. Lignified tissues stain red if they contain S units and yellow to brown if only G units are present (Goujon et al., 2003a). Lignin monomer composition typical for angiosperms, i.e. G lignin predominating in walls of xylem vessels and S lignin appearing as the main constituent of the walls of interfascicular fibres, was observed in mock‐inoculated plants (Fig. 4E, G). In inoculated plants, at 7 dpi, stronger staining for S lignin in the walls of xylem parenchyma (Fig. 4F, arrows) and in interfascicular fibre cells was observed than in mock‐inoculated plants. Further, in inoculated resistant plants, a distinct deposition of G lignin, visible as orange to brown staining, was observed in the walls of xylem vessels and cell walls of the outer layers of the interfascicular tissue and xylem parenchyma (Fig. 4F, arrowheads). No clear differences with regard to lignin deposition were observed in the susceptible line between mock‐inoculated and inoculated plants (Fig. 4G, H).
Figure 4.
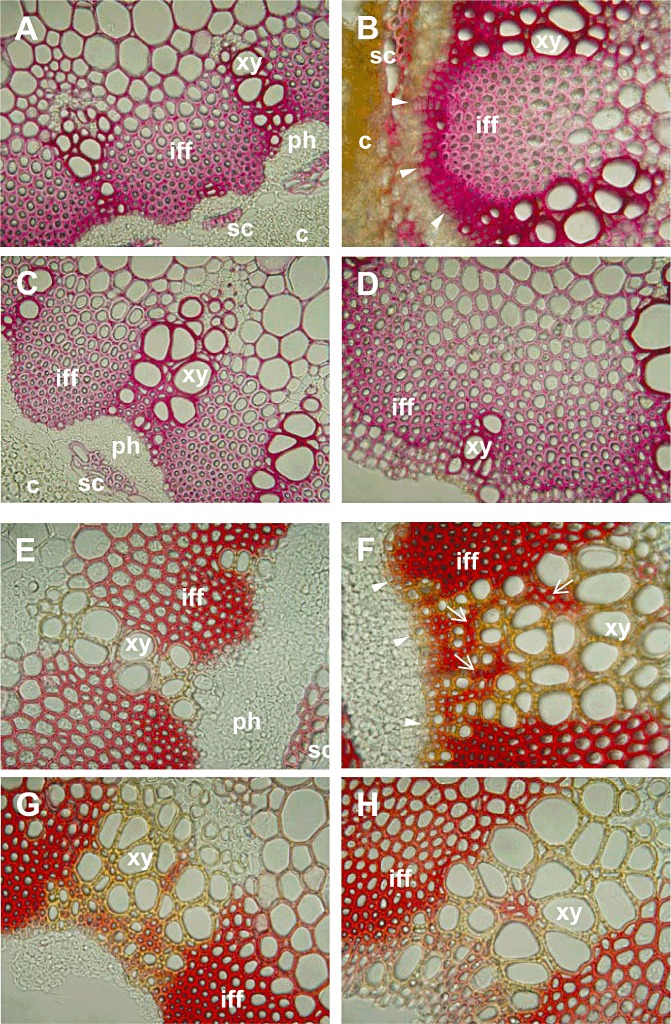
Detection of lignin in Camelina sativa lines 36011 (resistant) and 36012 (susceptible) at 7 days post‐inoculation (dpi). (A–D) Phloroglucinol‐HCl staining (guaiacyl lignin). Line 36011: (A) mock‐inoculated; (B) inoculated; guaiacyl (G) lignin particularly in xylem vessel walls and outer cell layers of interfascicular fibres (arrowheads). Line 36012: (C) mock‐inoculated; (D) inoculated. (E–H) Mäule staining (S lignin indicated by red reaction; G lignin indicated by yellow to brown reaction). Line 36011: (E) mock‐inoculated; (F) inoculated; S lignin in interfascicular tissue and xylem parenchyma cell walls (arrows); G lignin in xylem vessel walls and outer cell layers of interfascicular tissue and xylem parenchyma (arrowheads). Line 36012: (G) mock‐inoculated; (H) inoculated: no differences to mock inoculation. c, cortex; iff, interfascicular fibres; ph, phloem; sc, sclereids; xy, xylem. Magnification, 40×.
The Wiesner test revealed pink‐stained xylem vessels and parenchyma cell walls (Fig. 4A–D). At 7 dpi, inoculated resistant plants exhibited strong staining of vessel and parenchyma cell walls, indicating the deposition of lignin (Fig. 4B). This reaction was extremely prominent in the outer layers of xylem parenchyma and interfascicular tissue (Fig. 4B, arrowheads). In contrast, stem sections of inoculated susceptible plants, vessel elements and cell walls of interfascicular and xylem parenchyma showed only a slightly stronger staining for lignin than the corresponding tissues of healthy plants (Fig. 4C, D).
Gene expression
In order to elucidate the mechanism(s) underlying the differences observed between the lines during the course of the histochemical studies, the expression profiles of 21 genes involved in lignin biosynthesis were analysed by real‐time reverse transcriptase‐polymerase chain reaction (qRT‐PCR) in mock‐inoculated plants and plants inoculated with S. sclerotiorum. Analyses were performed from 12 to 96 hpi using peripheral stem tissue comprising the inoculation site. The Actin2 and CsNIS genes were chosen as reference genes (Table 1).
Table 1.
Monolignol biosynthesis candidate genes in Camelina sativa and reference genes used in this study.
| Gene | Corresponding locus in Arabidopsis thaliana | Enzyme |
|---|---|---|
| CsPAL1 | At2g37040 | Phenylalanine ammonia‐lyase |
| CsPAL2 | At3g53260 | Phenylalanine ammonia‐lyase |
| CsC4H | At2g30490 | Cinnamate‐4‐hydroxylase |
| Cs4CL1 | At1g51680 | 4‐Coumarate CoA ligase |
| CsC3H | At2g40890 | Coumaroyl‐CoA‐3‐hydroxylase |
| CsHCT | At5g48930 | Hydroxycinnamoyl:CoA shikimate/quinate hydroxycinnamoyl transferase |
| CsF5H | At4g36220 | Ferulate‐5‐hydroxylase |
| CsCCOMT1 | At4g26220 | Caffeoyl CoA O‐methyltransferase |
| CsCCOMT2 | At1g24735 | Caffeoyl CoA O‐methyltransferase |
| CsCCR1 | At1g15950 | Cinnamoyl‐CoA reductase |
| CsCCR2 | At1g80820 | Cinnamoyl‐CoA reductase |
| CsCCR3 | At2G02400 | Cinnamoyl‐CoA reductase |
| CsCCR4 | At5g58490 | Cinnamoyl‐CoA reductase |
| CsCCR5 | At2g33590 | Cinnamoyl‐CoA reductase |
| CsCCR6 | At1g76470 | Cinnamoyl‐CoA reductase |
| CsCCR7 | At2g23910 | Cinnamoyl‐CoA reductase |
| CsCCR8 | At2g33600 | Cinnamoyl‐CoA reductase |
| CsCCR9 | At4g30470 | Cinnamoyl‐CoA reductase |
| CsCCR10 | At5g14700 | Cinnamoyl‐CoA reductase |
| CsCAD4 | At3g19450 | Cinnamyl alcohol dehydrogenase |
| CsCAD5 | At4g34230 | Cinnamyl alcohol dehydrogenase |
| CsACT2 | At5g09810 | Actin2 |
| CsNIS | At4g26410 | Unknown function |
The changes in gene expression levels following inoculation with S. sclerotiorum in lines 36011 and 36012 are shown in Table 2. In both lines, the differences in transcript levels between mock‐inoculated and inoculated tissues were most pronounced from 48 to 96 hpi, and the number of differentially expressed genes was highest at 96 hpi. Overall, the transcripts of 12 genes (CsPAL1, CsPAL2, Cs4CL1, CsC3H, CsHCT, CsF5H, CsCCOMT1, CsCCOMT2, CsCCR1, CsCCR3, CsCAD4, CsCAD5) were down‐regulated and the transcripts of six genes (CsCCR2, CsCCR5, CsCCR6, CsCCR8, CsCCR9, CsCCR10) were up‐regulated in both lines at these later time points. In most cases, changes in transcript abundance appeared to be moderate. However, transcript levels of genes down‐regulated in both lines decreased equally or to a significantly greater extent in susceptible plants than in plants showing resistance. Correspondingly, the up‐regulation of genes occurred to the same or a significantly greater extent in resistant plants than in plants of the susceptible line. Members of gene families reacted either in a similar manner (e.g. CsPAL1 and CsPAL2, CsCCOMT1 and CsCCOMT2) or differently (e.g. CsCCR1–CsCCR10, CsCAD4 and CsCAD5) to infection.
Table 2.
Regulation of transcripts (fold change relative to mock‐inoculated plants) in Camelina sativa lines 36011 (resistant) and 36012 (susceptible) inoculated with Sclerotinia sclerotiorum at 12–96 h post‐inoculation (hpi).
| Gene | 12 hpi | 24 hpi | 48 hpi | 72 hpi | 96 hpi | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 36011 | 36012 | 36011 | 36012 | 36011 | 36012 | 36011 | 36012 | 36011 | 36012 | |
| CsPAL1 | 1.2 | 0.9*** | 1.0 | 0.7 | 0.8 | 0.5 | 0.3 | 0.3 | 0.6 | 0.4* |
| CsPAL2 | 1.3 | 1.1* | 0.9 | 0.8 | 0.9 | 0.7 | 0.5 | 0.4 | 0.9 | 0.5*** |
| CsC4H | 1.6 | 1.1 | 1.0 | 0.9 | 1.1 | 0.8 | 0.8 | 0.6 | 1.2 | 0.8** |
| Cs4CL1 | 1.2 | 0.9*** | 1.2 | 0.9 | 0.8 | 0.7 | 0.5 | 0.4 | 0.6 | 0.4 |
| CsC3H | 1.2 | 0.9 | 1.3 | 0.9 | 0.8 | 0.6 | 0.4 | 0.4 | 0.6 | 0.5 |
| CsHCT | 1.3 | 0.9*** | 1.3 | 1.0 | 0.9 | 0.7 | 0.5 | 0.4* | 0.6 | 0.4** |
| CsF5H | 1.3 | 0.9*** | 1.3 | 0.9*** | 1.0 | 0.9 | 0.6 | 0.4 | 0.6 | 0.3*** |
| CsCCOMT1 | 1.3 | 1.4 | 1.2 | 0.9 | 0.7 | 0.7 | 0.7 | 0.5 | 0.7 | 0.5 |
| CsCCOMT2 | 1.6 | 1.0*** | 1.0 | 0.9 | 0.7 | 0.6 | 0.5 | 0.4 | 0.7 | 0.4* |
| CsCCR1 | 1.1 | 1.0 | 1.0 | 0.8 | 0.6 | 0.6 | 0.5 | 0.3*** | 0.7 | 0.3 |
| CsCCR2 | 1.5 | 1.6 | 1.4 | 0.6*** | 3.7 | 2.9 | 7.3 | 5.2 | 20.6 | 6.8*** |
| CsCCR3 | 1.2 | 0.7*** | 1.1 | 0.9*** | 0.7 | 0.5 | 0.5 | 0.4 | 0.6 | 0.5 |
| CsCCR4 | 1.5 | 1.4 | 1.3 | 0.8*** | 0.8 | 0.7 | 0.9 | 0.5 | 1.4 | 0.6*** |
| CsCCR5 | 2.1 | 1.9 | 1.1 | 1.1 | 2.1 | 1.9 | 1.6 | 2.0 | 2.2 | 2.2 |
| CsCCR6 | 0.7 | 1.3 | 1.1 | 1.4 | 7.3 | 6.6 | 8.9 | 4.9 | 4.8 | 5.2 |
| CsCCR7 | 1.0 | 1.1 | 0.7 | 0.8 | 0.8 | 0.6 | 1.4 | 1.0 | 1.1 | 0.6** |
| CsCCR8 | 1.3 | 1.3 | 1.2 | 1.0 | 1.3 | 1.4 | 2.0 | 1.5 | 2.2 | 2.1 |
| CsCCR9 | 1.3 | 1.1 | 1.1 | 1.0 | 1.9 | 1.4 | 1.5 | 1.6 | 1.5 | 1.4 |
| CsCCR10 | 1.5 | 1.8 | 1.3 | 0.8 | 2.4 | 1.8** | 2.6 | 1.9 | 2.9 | 1.6 |
| CsCAD4 | 1.4 | 1.4 | 1.1 | 1.0 | 0.8 | 0.8 | 0.8 | 0.9 | 1.0 | 0.8 |
| CsCAD5 | 1.2 | 1.0 | 1.3 | 0.9*** | 0.9 | 0.7 | 0.7 | 0.6 | 1.0 | 0.6** |
Asterisks indicate statistically significantly different means between the two lines at *P < 0.1, **P < 0.05 and ***P < 0.01.
All genes that were up‐regulated on challenge with the stem rot fungus belonged to the gene family coding for cinnamoyl‐CoA reductase (CCR), the first enzyme of the phenylpropanoid pathway specifically committed to the synthesis of lignin monomers. Transcript levels of seven CCR genes, including CsCCR2 and the paralogues CsCCR4–CsCCR10, were elevated either only in 36011 or in both lines following inoculation. The highest pathogen‐induced change in the transcript ratio was observed for CsCCR2. This gene exhibited elevated expression levels in both lines starting at 48 hpi. Although no clear differences in transcript abundance between 36011 and 36012 were detected at 48 and 72 hpi, expression of CsCCR2 was significantly more strongly up‐regulated in resistant plants at 96 hpi (P≤ 0.01). CsCCR2 transcripts were 20.6‐fold more abundant in inoculated than in mock‐inoculated stem tissue of resistant plants. In the susceptible line, this gene was up‐regulated only 6.8‐fold. This difference in gene regulation in response to pathogen attack at 96 hpi coincided with differences in symptom development (Fig. 1).
qRT‐PCR analysis of the constitutive transcript levels of CsCCR1, CsCCR2, CsCCR3 and CsCCR5–CsCCR10 in stem tissue revealed slight differences in expression among these genes (Fig. 5). The highest relative expression compared with the reference genes in both lines was found for CsCCR7, followed by CsCCR6. No statistically significant differences in constitutive transcript abundance were observed between the lines. In contrast, the constitutive transcript level of CsCCR4, which showed similar expression levels to other CsCCR genes in susceptible 36012, was, on average, 11.6 times higher in resistant line 36011. On inoculation with S. sclerotiorum, CsCCR4 was down‐regulated in susceptible plants, which led to a relative expression level almost 30 times lower than the corresponding transcript level in inoculated resistant plants (Fig. 6). The expression of CsCCR4 did not exhibit tissue specificity; as observed in peripheral stem tissue, expression in the leaves of resistant plants was also about 12 times higher than in the corresponding tissues of susceptible plants. However, transcript levels in leaves were considerably lower than in stem tissue (data not shown), a result that likewise suggests the involvement of CsCCR4 in stem lignification.
Figure 5.
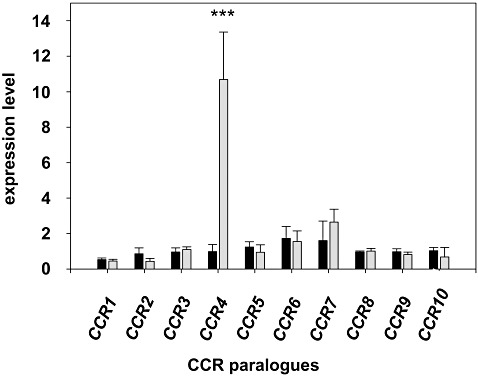
Constitutive expression levels of cinnamoyl‐CoA reductase genes CsCCR1–CsCCR10 in peripheral stem tissue of Camelina sativa resistant line 36011 (grey bars) and susceptible line 36012 (black bars) at 96 h post‐inoculation (hpi). Asterisks denote a significant difference at P≤ 0.01.
Figure 6.
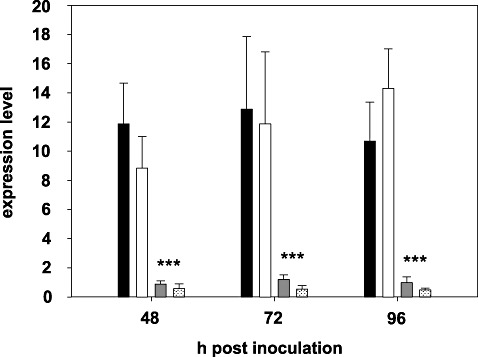
Transcript levels of cinnamoyl‐CoA reductase gene CsCCR4 in peripheral stem tissue of mock‐inoculated and inoculated plants of Camelina sativa lines 36011 (resistant) and 36012 (susceptible) at 48–96 h post‐inoculation (hpi). Line 36011: mock‐inoculated, black bars; inoculated, white bars. Line 36012: mock‐inoculated, dark grey bars; inoculated, light grey bars. Expression levels are given as means and standard deviations of four biological replicates. Asterisks denote a significant difference at P≤ 0.01.
Lignin monomer composition
After the detection of substantial differences in the constitutive transcript levels of CsCCR4 between resistant and susceptible plants, lignin monomer yield and composition in whole stem samples of both lines were compared.
Following the removal of soluble and cell wall‐bound phenolic compounds, stem samples were subjected to thioacidolysis, and the recovered thioethylated monomers were analysed by gas chromatography. In this approach, the uncondensed arylglycerol‐β‐aryl ether‐linked H, G and S monomers of lignin are cleaved from the polymer and their abundances can be determined (Robinson and Mansfield, 2009; Rolando et al., 1992). The total yield and relative proportion of the monomers closely reflect the amount and type of lignin units involved in labile ether bonds. Monomers were detected as the thioethylated products of p‐hydroxycinnamyl aldehydes (Fig. 7, peaks G3 and S3) and the erythro‐ and threo‐ isomers corresponding to each p‐hydroxycinnamyl alcohol (Fig. 7, peaks G1, G2 and S1, S2) (Rolando et al., 1992). Discrete measurements of each form were only possible for the G‐ and S‐derived monomers, as the H‐derived monomers were present only at trace levels in all samples.
Figure 7.
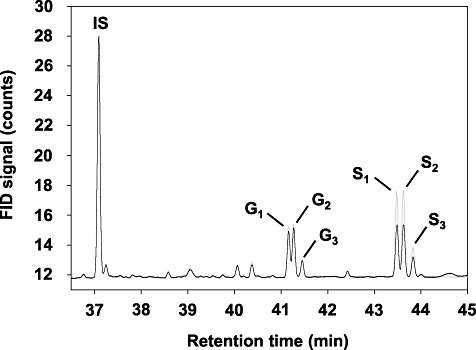
Thioacidolysis products of stem tissue of Camelina sativa line 36011 (resistant, grey line) and 36012 (susceptible, black line). G1, erythro‐guaiacyl‐CHR‐CHR‐CH2R; G2, threo‐guaiacyl‐CHR‐CHR‐CH2R; G3, guaiacyl‐CH2‐CHR‐CHR2; S1, erythro‐syringyl‐CHR‐CHR‐CH2R; S2, threo‐syringyl‐CHR‐CHR‐CH2R; S3, syringyl‐CH2‐CHR‐CHR2; FID, flame ionization detection; IS, internal standard.
Lignin subunit ratios and total monomer yields differed significantly between the lines (P≤ 0.01) (Table 3, Fig. 7). Although total G lignin content was equal in both lines, S lignin content in the resistant line was increased by 40% when compared with that in the susceptible line (5.86 vs. 4.19 peak area/mg dry weight). Correspondingly, the S/G ratio in the resistant line (1.7) was substantially higher than that in the susceptible line (1.2).
Table 3.
Lignin monomer composition in whole stems of Camelina sativa lines 36011 (resistant) and 36012 (susceptible) at early flowering (BBCH 63).
| Line | Lignin monomer composition | |||||
|---|---|---|---|---|---|---|
| G + S* | G* | G (%) | S* | S (%) | S/G ratio | |
| 36011 | 9.26 (±0.38) b | 3.4 (±0.16) a | 36.7 | 5.86 (±0.23) b | 63.3 | 1.7 |
| 36012 | 7.69 (±0.28) a | 3.5 (±0.17) a | 45.5 | 4.19 (±0.12) a | 54.5 | 1.2 |
G and S, guaiacyl and syringyl lignin monomers, respectively.
Mean (±standard deviation) values based on five samples measured in triplicate.
Values in the same column followed by the same letter do not differ at P≤ 0.01.
Quantity is given in peak area/mg dry weight.
As it is generally seen that the induction of expression of lignin biosynthetic genes leads to an increase in lignin content which is not proportional to the increase in transcript level, we analysed peripheral stem tissue of mock‐inoculated and pathogen‐inoculated plants of both lines in order to detect potential changes in lignin monomer yield and monomer ratios in response to an infection with S. sclerotiorum. Tissue was harvested at 7 dpi, subjected to thioacidolysis and analysed by gas chromatography. Plants of the resistant line showed an increase in lignin content after inoculation with S. sclerotiorum when compared with mock‐inoculated plants. Interestingly, this increase was almost exclusively caused by an increase in G lignin subunits (2.93 vs. 2.13 peak area/mg dry weight). Accordingly, the S/G ratio was lower in pathogen‐inoculated resistant plants than in mock‐inoculated plants (0.9 and 1.2, respectively). Only a slight increase in G lignin was observed in the peripheral stem tissue of pathogen‐inoculated susceptible plants (Table 4).
Table 4.
Lignin monomer composition in peripheral stem tissue of Camelina sativa lines 36011 (resistant) and 36012 (susceptible) inoculated with Sclerotinia sclerotiorum (7 days post‐inoculation) compared with mock‐inoculated plants.
| Line | Lignin monomer composition | |||||
|---|---|---|---|---|---|---|
| G + S* | G* | G (%) | S* | S (%) | S/G ratio | |
| 36011 mi | 4.66 (±0.04) | 2.13 (±0.02) | 45.7 | 2.53 (±0.02) | 54.3 | 1.2 |
| 36011 in | 5.52 (±0.04) | 2.93 (±0.03) | 53.1 | 2.59 (±0.02) | 46.9 | 0.9 |
| 36012 mi | 4.09 (±0.07) | 1.64 (±0.02) | 40.1 | 2.45 (±0.05) | 59.9 | 1.5 |
| 36012 in | 3.98 (±0.08) | 1.67 (±0.04) | 42.1 | 2.29 (±0.05) | 57.7 | 1.4 |
G and S, guaiacyl and syringyl lignin monomers, respectively.
Mean (±standard deviation) values based on one biological replicate analysed in triplicate.
Quantity is given in peak area/mg dry weight.
in, inoculated; mi, mock‐inoculated.
DISCUSSION
Sclerotinia sclerotiorum is a soil‐borne necrotrophic pathogen that causes stem rot in C. sativa. The pathogen causes the rapid maceration of host tissue through the concerted action of oxalic acid and cell wall‐degrading enzymes (Hegedus et al., 2008). In a recent study, we identified germplasm of C. sativa exhibiting a high level of resistance to S. sclerotiorum (C. Eynck et al., unpublished data). As in Brassica napus (Garg et al., 2010; Zhao et al., 2009), the resistance phenotype of C. sativa manifests itself in the restriction of lesion progression rather than in the prevention of infection. The results presented here and in previous work (C. Eynck et al., unpublished data) show that, on inoculation at early flowering, lesions remain confined to the stem cortex in resistant plants. The absence of wilt or other symptoms in plant parts above the inoculation site suggests that the fungus is not able to colonize xylem vessels or to effectively release pathogenicity factors, such as toxins or hydrolytic enzymes, into the vascular system of resistant plants.
Histochemical analyses revealed that resistant plants react to infection with the rapid deposition of lignin in cells in the immediate vicinity of the infection site. This reaction was most prominent in the outer layers of xylem parenchyma and interfascicular tissue observed using the Wiesner test. Although it has been more recently disputed, the Wiesner test has been considered to be specific for G lignin, giving weak or no staining in the presence of S units (e.g. Anterola and Lewis, 2002; Sarkanen and Ludwig, 1971). In our study, xylem vessel walls, which are known to have a high content of G lignin in angiosperms (Chapple et al., 1992; Fergus and Goring, 1970), showed much stronger staining than interfascicular fibres treated with phloroglucinol‐HCl. However, staining with the Mäule reagent indicated an abundance of S lignin units in the interfascicular tissue. These observations suggest that the Wiesner reagent indeed shows stronger affinity for G rather than S lignin units, even if it is not absolutely specific for coniferaldehyde end groups. The results of our histochemical analyses therefore indicate that induced lignification in stems of C. sativa as a resistance response to infection by S. sclerotiorum mainly involves the de novo synthesis of G lignin units.
Differences in monolignol composition between lignin formed during the normal development of healthy plants and defence‐related lignin in infected plants have been described for a number of host–pathogen interactions (reviewed by Vance et al., 1980). However, our study is the first to report this phenomenon for plants infected by S. sclerotiorum. All three lignin subunit types have been shown to be specifically increased depending on the pathosystem under investigation (Gayoso et al., 2010; Hammerschmidt et al., 1985; Hawkins and Boudet, 2003; Lange et al., 1995; Menden et al., 2007).
G lignin monomers are monomethoxylated and thus offer more extensive possibilities of cross‐linking compared with S monomer units, which are dimethoxylated (Baucher et al., 2003; Boudet et al., 1995). Increased deposition of the more condensed G lignin in line 36011 after inoculation with S. sclerotiorum may present an effective measure to prevent pathogen ingress into the vascular system, particularly as the main lignin type present in xylem vessel walls is G lignin.
In order to further elucidate the potential role of monolignol biosynthesis in the resistance of C. sativa to S. sclerotiorum, the changes in expression of 21 genes homologous to A. thaliana genes known to be involved in monolignol biosynthesis were analysed. These genes cover each of the enzymatic steps in the pathway, except for the enzyme caffeic acid O‐methyltransferase (COMT), because of the lack of a corresponding homologue in the existing expressed sequence tag (EST) collection.
The conversion of cinnamoyl‐CoA thioesters to the corresponding cinnamaldehydes by CCR is considered as the first step in the general phenylpropanoid pathway specifically committed to the synthesis of monolignols. The activity of CCRs in plants is generally low, indicating that this enzymatic step may be an important control point regulating the carbon flux towards lignins (Chabannes et al., 2001; Lacombe et al., 1997; Piquemal et al., 1998). Transgenic approaches in tobacco, A. thaliana and poplar have demonstrated that CCR activity can profoundly affect the amount and composition of lignin deposited in plant cell walls (Boerjan et al., 2003; Boudet et al., 2003). An in silico analysis of the A. thaliana genome sequence database revealed 11 annotated CCR homologues (Costa et al., 2003). Of these, only the isoforms AtCCR1 and AtCCR2 had been identified and shown to be differentially expressed during development and in response to infection (Lauvergeat et al., 2001). Although AtCCR1 appeared to be involved in constitutive lignification, the expression level of AtCCR2 increased strongly during the incompatible interaction with Xanthomonas campestris pv. campestris, and the enzyme was therefore assumed to be involved in the biosynthesis of phenolics, whose accumulation may lead to resistance. Prior to the identification of CCRs in A. thaliana, Pichon et al. (1998) identified two CCR isoforms in maize (Zea mays) (ZmCCR1 and ZmCCR2) which, likewise, showed differential expression patterns, with ZmCCR1 being predominantly associated with lignification during development. Interestingly, the ectopic lignification in cellulose‐deficient eli1‐1 mutants of A. thaliana involved, among other defence‐related responses, the induction of expression of AtCCR2, but not AtCCR1 (Caño‐Delgado et al., 2003).
We investigated the changes in the expression of 10 CCR genes with a putative function in lignification on inoculation with S. sclerotiorum in both resistant and susceptible lines of C. sativa. As in A. thaliana and maize, CsCCR2 mRNA levels increased in both lines after inoculation with S. sclerotiorum, whereas CsCCR1 expression was not induced. In addition to CsCCR2, the transcript levels of seven additional CsCCRs were elevated in response to infection. Of these, two isoforms were up‐regulated only in resistant line 36011. CsCCR3, like CSCCR1, did not show an increase in expression level on pathogen challenge. These results corroborate the suggestion made earlier (Costa et al., 2003; Lauvergeat et al., 2001) that the CCR gene family may be divided into two groups, one involved in defence responses and the other in developmental lignification. In C. sativa, as in A. thaliana (Kawasaki et al., 2006), these two groups cannot be distinguished on the basis of sequence similarity (data not shown). Of all the CCR homologues that showed up‐regulation after inoculation with S. sclerotiorum, CsCCR2 not only exhibited the strongest induction overall, but also showed a significantly stronger increase in expression in resistant than in susceptible plants. The resistance phenotype of 36011 may therefore be partly associated with gene expression changes of CsCCR2, and thus timely activation of monolignol biosynthesis, prior to the onset of rapid fungal growth, as observed in the susceptible line 36012. Two possible roles of CCR2 expression in the stress response have been suggested (Boudet et al., 2004). First, the synthesis of defence lignins that help to prevent the progression of the pathogen, and/or, second, the synthesis of other defence‐related compounds. It has been reported that coniferaldehyde and coniferyl alcohol, both immediate precursors of G lignin, may function as phytoalexins in the incompatible interaction between flax and Melampspora lini (Keen and Littlefield, 1979).
Purified CCR1 and CCR2 enzymes from a number of species, including poplar (Sarni et al., 1984) and A. thaliana (Lauvergeat et al., 2001), have been shown to be active towards 4‐coumaroyl‐CoA, caffeoyl‐CoA, feruloyl‐CoA, 5‐hydroxyferuloyl‐CoA and sinapoyl‐CoA. However, the highest efficiency for both enzymes was observed with feruloyl‐CoA (Lauvergeat et al., 2001; Luderitz and Grisebach, 1981; Ma, 2007; Patten et al., 2005; Sarni et al., 1984), suggesting the preferential biosynthesis of G lignin. Likewise, the primary phenotype of tobacco plants silenced for CCR was a decrease in total lignin content and an increased S/G ratio, primarily caused by a decrease in extractable G units (O'Connell et al., 2002; Piquemal et al., 1998; Ralph et al., 1998). Hence, not only the results of our histochemical analysis, but also the strong induction of CsCCR2 expression in the resistant line, suggest that increased synthesis of mostly G lignin is part of the resistance response of C. sativa to S. sclerotiorum. This hypothesis is further supported by the results of the chemical analysis. As revealed by thioacidolysis, total lignin content increased 7 days after inoculation with the pathogen in resistant plants only, and this increase was caused almost exclusively by an increase in G units.
Lignification may not only be an induced mechanism in the resistance of C. sativa to S. sclerotiorum, but may also play an important role as a constitutive resistance component. We observed a constitutive transcript level of CsCCR4 that was, on average, about 12 times higher in the stem tissue of the resistant line than in that of the susceptible line. To our knowledge, this is the most distinctive difference in constitutive gene expression for lignin biosynthesis genes between lines of the same species reported to date. In the present study, high CsCCR4 mRNA levels were associated with higher lignin content in whole stem samples of line 36011. Similarly, comparing two wheat cultivars, Ma (2007) reported that high TaCCR1 mRNA levels were associated with both high CCR enzyme activity and a higher lignin content and stem rigidity in a cultivar showing lodging resistance. Strikingly, our data revealed that the difference in constitutive lignin content was caused exclusively by an increased amount of lignin derived from S monomers. This result suggests stronger lignification of tissue rich in S lignin in the resistant line, such as parenchyma cells in the interfascicular tissue (Chapple et al., 1992; Fergus and Goring, 1970).
The latter results lead to the tantalizing hypothesis that the activity of CCR isoform CsCCR4 may be specifically committed to the conversion of sinapoyl‐CoA and, consequently, specific for S lignin biosynthesis. Currently, there is debate about whether the monolignol biosynthetic pathway represents a ‘metabolic grid’ leading to both G and S units, or whether metabolic channelling allows for independent pathways to G and S lignin. The notion of independent pathways is more consistent with recent findings on enzyme specificity and the results of the modification of lignin biosynthetic enzyme activities through transgenic approaches (Dixon et al., 2001). Two enzymes have been identified so far that are considered to be specific for the synthesis of sinapyl alcohol, namely ferulate 5‐hydroxylase (F5H) (Osakabe et al., 1999) and COMT (Li et al., 2000). Arabidopsis thaliana plants over‐expressing F5H under the C4H promoter have been reported to accumulate high levels of S lignin and to show increased resistance to nematode infection (Wuyts et al., 2006). A transgenic approach is currently underway to further elucidate the specificity of CsCCR4 activity.
Lignification is known to be one of a plethora of mechanisms for the prevention of successful colonization by pathogens. However, this study describes, for the first time, the association of monolignol biosynthesis with the resistance of C. sativa to S. sclerotiorum. The results presented provide a strong indication for a role of lignification, both as an induced and constitutive component of resistance, halting the spread of S. sclerotiorum following infection. Although the induced deposition of lignin as a response to pathogen attack appears to mainly involve the de novo synthesis of G lignin monomers, the higher lignin content in healthy plants of the resistant line compared with that in healthy plants of the susceptible line is exclusively based on an increased amount of S lignin. Further work will indicate whether the differences in expression of CsCCR4 and CsCCR2, and the corresponding differences in lignin content and monomer composition, in healthy vs. inoculated plants are indeed crucial for the mounting of an effective defence against S. sclerotiorum in resistant plants of C. sativa. In conclusion, this study has provided new insights into aspects of plant defence against S. sclerotiorum and contributes to an improved general understanding of stem rot resistance in crucifers.
EXPERIMENTAL PROCEDURES
Fungal isolate and inoculum production
Clone 321 of S. sclerotiorum was used, as it is one of the most prevalent aggressive clones in western Canada (Kohli et al., 1995). Inoculum was prepared as described previously (C. Eynck et al., unpublished data). Briefly, sclerotia, produced from fungal mycelium mixed with sterilized hull‐less barley, were incubated on moist sand in darkness at 15 °C for 4–5 weeks and then transferred to daylight under the same temperature conditions to induce apothecia formation. Harvest of ascospores was performed using a vacuum filter; 10‐µL droplets of ascospore solution (2 × 104 viable spores/mL) were applied to senescent petals of B. napus cv. Westar, a canola variety highly susceptible to sclerotinia (Zhao et al., 2009). Inoculated petals were incubated in a growth room at room temperature for 24 h.
Plant material, growth conditions and inoculation procedures
Camelina sativa line 36011, derived by inbreeding from stem rot‐resistant accession CS5, and line 36012, derived by inbreeding from stem rot‐susceptible accession CS17 (C. Eynck et al., unpublished data), were used in the study.
For all experiments described herein, seeds were sown in 10‐cm‐diameter pots containing a soil‐less mixture (Stringam, 1971) amended with controlled release fertilizer (15‐9‐12 Osmocote PLUS, Scotts Fertilizer Company, Marysville, OH, USA). Plants were grown in a growth chamber programmed for 20 °C/18 °C day/night temperatures and 16 h day/8 h night fluorescent illumination (250 µmol/m2/s light).
Plant inoculations were performed at early flowering (BBCH 63, 30% of flowers open; Martinelli and Galasso, 2011). For gene expression analyses, the upper part of the main stem was inoculated at three sites (every other internode) with infested petals. Petals were secured to the stem with strips of Parafilm™. Plants used for the histochemical and lignin analyses were inoculated at a single site on the upper part of the main stem. In all experiments, control plants were treated with petals inoculated with a drop of deionized water (mock inoculation).
Histochemical analysis
Ten inoculated and 10 mock‐inoculated plants were sampled at 7 dpi. Internodes bearing the inoculation site as well as the corresponding internodes of mock‐inoculated plants were excised and preserved in a solution of acetic acid, formalin and ethanol (AFE; 5 : 5 : 90, v/v/v). Cross‐sections (one to two cell layers thick) were made with a razor blade, treated with histochemical reagents and immediately examined under bright field using a Zeiss Axiovert 100 microscope (Zeiss, Mississauga, ON, Canada) equipped with a Sony DSC‐F717 digital camera (Sony, Toronto, ON, Canada). Histochemical stains included phloroglucinol‐HCl (Wiesner test) and the Mäule reagent. Following the protocol established by Jensen (1962) for the Wiesner test, sections were immersed in 1% phloroglucinol solution in 96% ethanol for 2 min. Excess phloroglucinol solution was rinsed off with 32% HCl and sections were mounted in a mixture of glycerol and 32% HCl (1 : 1, v/v). Mäule staining was performed according to the protocol established by Chapple et al. (1992). Sections were treated with 0.5% KMnO4 for 10 min, rinsed with water, treated with 10% HCl for 5 min, rinsed again with water and mounted in concentrated NH4OH for examination.
In a second experiment, stem segments of inoculated and mock‐inoculated plants were sampled at 3, 7 and 14 dpi, dehydrated in an alcohol series and embedded in paraffin using an MVPI tissue processor (Instrumentation Laboratory, Bedford, MA, USA) and a Tissue Tek embedding unit (Miles Laboratories, Elkhart, IN, USA) following a modified protocol according to the manufacturer's manual. Cross‐sections of 20 µm were cut on a rotary microtome (MICROM D6900, Microm International GmbH, Walldorf, Germany), expanded and gently heat fixed onto microscope slides. After the removal of paraffin, staining was performed with 1% acid fuchsin in deionized water dissolved 1 : 10 in lactophenol. After rinsing with lactophenol, samples were mounted in drops of lactophenol and immediately examined using a Leica TCS SP2 confocal laser scanning microscope (CLSM, Leica, Richmond Hill, ON, Canada). Digital images were acquired by two‐channel analysis with ensuing drafting of an overlay (488 nm for excitation/500–530 nm for emission and 543 nm for excitation/560–650 nm for emission). This technique allowed for the detection of phenolic polymers which autofluoresce after excitation with blue light. Acid fuchsin was used as a counter stain to depict nonlignified stem structures. The histochemical experiments were conducted twice.
Lignin biosynthesis
Enzymes for lignin biosynthesis are encoded by multiple gene families, but not every member of a gene family is involved in or contributes equally to lignin biosynthesis (Bi et al., 2011). Therefore, the identification of the most appropriate targets is the first step towards further characterization. A list of candidate lignin biosynthesis genes was established on the basis of previously published gene expression analysis data (Chen et al., 2000; Ehlting et al., 1999; Kiedrowski et al., 1992; Raes et al., 2003; Sibout et al., 2003; Tronchet et al., 2010), mutant analyses (Chapple et al., 1992; 2002a, 2002b; Goujon et al., 2003b; Jones et al., 2001) and the commonly accepted model of the monolignol biosynthesis pathway (e.g. Bhuiyan et al., 2009) (Table 1).
Sequences of candidate genes from A. thaliana were compared with a C. sativa EST library using blastn (Basic Local Alignment Search Tool), and the orthologues with the highest scores were chosen for gene expression analyses. Identical C. sativa sequences were homologous to A. thaliana 4CL1 and 4CL2, and therefore only one 4CL gene was analysed. All members of the CCR gene family were included in the analyses, except for CsCCR11 (At1g68540), because it does not have a putative role in lignin biosynthesis (http://www.arabidopsis.org). A homologue to the A. thaliana COMT gene could not be identified in the C. sativa EST collection.
cDNA synthesis and qRT‐PCR
Tissue was harvested as described by Zhao et al. (2009). At 12, 24, 48, 72 and 96 hpi, peripheral stem tissue of inoculated and mock‐inoculated plants was excised with a razor blade from 10 mm above to 10 mm below the inoculation site and 1 mm deep. For each line, treatment and time point, the tissue of six plants (three inoculation sites per plant) was pooled. The experiment was conducted three times for all time points and an additional replicate was obtained for time points 48, 72 and 96 hpi. In an additional experiment, whole leaves from healthy plants of both lines were sampled just before flowering (BBCH 55; Martinelli and Galasso, 2011) at the fifth internode below the apical meristem in order to elucidate whether the expression of CsCCR4 exhibits tissue specificity.
Total RNA from up to 1 g of ground tissue was isolated using the AMBION Totally RNA kit (AMBION, Austin, TX, USA) and the concentration was measured with an Ultrospec 3000 spectrophotometer (Amersham Pharmacia Biotech Inc., Uppsala, Sweden). Fifty micrograms of total RNA were purified and treated with RNase‐free DNase using the plant RNeasy kit (Qiagen, Mississauga, ON, Canada). The final concentration was determined using a NanoDrop Spectrophotometer (NanoDrop Technologies, Montchanin, DE, USA) and RNA integrity was confirmed using an Agilent 2100 Bioanalyser (Agilent Technologies Inc., Mississauga, ON, Canada). One microgram of total RNA was subjected to cDNA synthesis (Superscript™ III reverse transcriptase, Invitrogen, Burlington, ON, Canada) following the manufacturer's protocol. qPCR assays were performed in a C1000 Thermal Cycler (Bio‐Rad Laboratories, Hercules, CA, USA) in a 20‐µL reaction mixture composed of 3 µL (7.5 ng) cDNA, 1 × Platinum SYBR Green qPCR SuperMIX‐UDG (Invitrogen) and 0.2 µm of each gene‐specific primer. PCR conditions were chosen according to the manufacturer's recommendations: 50 °C for 2 min, 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. Quantitative analysis was performed using the CFX96 Real‐Time PCR Detection System (Bio‐Rad Laboratories). The amplification specificity of primers was confirmed by identification of a single peak in the melting curve analysis. Primers were designed using Invitrogen's OligoPerfect Designer (http://www.tools.invitrogen.com) and the following criteria: an amplicon size of 80–230 bp, a primer size of 20–24 bp and a GC content of 40%–60% (Table S1, see Supporting Information). All designed primers had efficiencies equal to or greater than 90% when tested using a five‐fold dilution series of the template starting with 30 ng. Actin2 and the C. sativa homologue of the A. thaliana gene At4g26410 were used as endogenous reference genes. At4g26410 has been shown to be extremely stable under biotic stress conditions in A. thaliana (Czechowski et al., 2005), and the corresponding C. sativa homologue was shown to be unresponsive to S. sclerotiorum infection in preliminary experiments, herein named CsNIS (Not Induced by Sclerotinia infection). Expression ratios in samples of inoculated plants compared with mock‐inoculated plants were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001) with data corrected for PCR efficiency and normalized against both reference genes. In order to compare data from different PCR runs, an internal calibrator was used in every run. Data were obtained from all biological replicates and each measurement was performed in triplicate.
Analysis of lignin monomer composition (thioacidolysis)
To compare the total yield and proportion of monomers in whole stem samples of healthy plants, five samples of line 36011 and line 36012 at early flowering were analysed by thioacidolysis. Each sample consisted of internodes harvested from three plants. For the analysis of lignin monomer composition in inoculated and mock‐inoculated plants, thioacidolysis was performed on peripheral stem tissue. Tissue was excised according to the procedure described for gene expression analysis. Samples were taken at 7 dpi and pooled from 30 inoculation sites for each treatment and line. All samples were extracted and analysed in triplicate.
Soluble phenolic acids and cell wall‐bound phenolic compounds were extracted as described by Eynck et al. (2009). Subsequently, samples consisting of 10 mg of ground, oven‐dried extract‐free cell wall residue material were subjected to a modified thioacidolysis procedure developed by Robinson and Mansfield (2009) with further minor modifications.
Each sample was transferred to a 5‐mL glass Wheaton vial with a Teflon‐lined screw cap and 1 mL of the freshly prepared thioacidolysis reagent [2.5% BF3 etherate, 10% ethanethiol, in recently distilled dioxane (v/v)] was added to each vial. Nitrogen gas was used to blank the reaction prior to sealing. Vials were incubated in a dry heating block at 100 °C for 4 h with hourly gentle agitation. To stop the reaction, samples were placed at −20 °C for 5 min. Subsequently, 200 µL of tetracosane (5 mg/mL methylene chloride) was added as an internal standard. Further, 300 µL of 0.4 m sodium bicarbonate was added to adjust the pH of the reaction to between pH 3 and 4. Extraction of the reaction products from the aqueous mixture was achieved by the addition of 2 mL of water and 1 mL of methylene chloride. Each sample was vortexed and allowed to settle for about 5 min. After phase separation, 1.5 mL of the lower organic phase containing the lignin breakdown products was removed, filtered and cleared of residual water by passing through a Pasteur pipette packed with a paper plug and about 2.5 cm of granular anhydrous Na2SO4. The cleared filtrates were evaporated to complete dryness in a dry heating block at 65 °C overnight and resuspended in 2 mL of methylene chloride. Derivatization of samples was performed by combining 100 µL of sample with 100 µL of pyridine and 400 µL of N,O‐bis(trimethylsilyl)acetamide. An incubation of at least 2 h at 25 °C was required before 5 µL of sample was analysed by gas chromatography.
Gas chromatography
Gas chromatography was conducted on a Hewlett Packard 6890 instrument (Agilent Technologies, Santa Clara, CA, USA), equipped with an autosampler, splitless injector, flame ionization detector (FID) and a 30‐m HP5 ms 0.32‐mm internal diameter capillary column. Five microlitres were injected and separated using helium as a carrier gas at a flow rate of 1 mL/min. Inlet and detector temperatures were set to 250 °C and the oven profile set‐up was as follows: 130 °C initial temperature, 3 min hold, ramp temperature 3 °C/min for 40 min to end in a final temperature of 250 °C, 5 min hold and then cool. The identification of peaks was consistent with Rolando et al. (1992) and Robinson and Mansfield (2009).
Statistical analysis
Statistical analysis was conducted using the software StatGraphics (Statpoint Technologies Inc., Warrenton, VA, USA). Differences among means were tested with Fisher's least significance difference test and significance was determined at P≤ 0.01, 0.05 and 0.1.
Supporting information
Fig. S1 Camelina sativa line 36012, 14 days after inoculation with Sclerotinia sclerotiorum. Fungal hyphae appear green after staining with WGA Alexa Fluor 488 and plant structures appear red after staining with Congo Red.
Table S1 Sequences of primers used for real‐time reverse transcriptase‐polymerase chain reaction (qRT‐PCR).
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
The authors thank J. A. Nettleton for technical assistance. Funding from the Canadian Food Inspection Agency (Project AAFC0703) and the Saskatchewan Agriculture Development Fund (Project 20080118) is gratefully acknowledged.
Reproduced with the permission of the Minister of Agriculture and Agri‐Food Canada.
REFERENCES
- Anterola, A.M. and Lewis, N.G. (2002) Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry, 61, 221–294. [DOI] [PubMed] [Google Scholar]
- Baucher, M. , Halpin, C. , Petit‐Conil, M. and Boerjan, W. (2003) Lignin: genetic engineering and impact on pulping. Crit. Rev. Biochem. Mol. Biol. 38, 305–350. [DOI] [PubMed] [Google Scholar]
- Bhuiyan, N.H. , Selvaraj, G. , Wei, Y. and King, J. (2009) Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, C. , Chen, F. , Jackson, L. and Gill, B.S. (2011) Expression of lignin biosynthetic genes in wheat during development and upon infection by fungal pathogens. Plant Mol. Biol. Rep. 29, 149–161. [Google Scholar]
- Boerjan, W. , Ralph, J. and Baucher, M. (2003) Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. [DOI] [PubMed] [Google Scholar]
- Boland, G.J. and Hall, R. (1994) Index of plant hosts of Sclerotinia sclerotiorum . Can. J. Plant Pathol. 16, 93–108. [Google Scholar]
- Bolton, M.D. , Thomma, B.P.H. and Nelson, B.D. (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. [DOI] [PubMed] [Google Scholar]
- Boudet, A.M. , Lapierre, C. and Grima‐Pettenati, J. (1995) Biochemistry and molecular biology of lignification. New Phytol. 129, 203–236. [DOI] [PubMed] [Google Scholar]
- Boudet, A.M. , Kajita, S. , Grima‐Pettenati, J. and Goffner, D. (2003) Lignin and lignocellulosics: a better control of synthesis for new and improved uses. Trends Plant Sci. 8, 576–581. [DOI] [PubMed] [Google Scholar]
- Boudet, A.M. , Hawkins, S. and Rochange, S. (2004) The polymorphisms of the genes/enzymes involved in the last two reductive steps of monolignol synthesis: what is the functional significance? C. R. Biol. 327, 837–845. [DOI] [PubMed] [Google Scholar]
- Calla, B. , Vuong, T. , Radwan, O. , Hartman, G.L. and Clough, S.J. (2009) Gene expression profiling soybean stem tissue early response to Sclerotinia sclerotiorum and in silico mapping in relation to resistance markers. Plant Genome, 2, 149–166. [Google Scholar]
- Caño‐Delgado, A. , Penfield, S. , Smith, C. , Catley, M. and Bevan, M. (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana . Plant J. 34, 351–362. [DOI] [PubMed] [Google Scholar]
- Chabannes, M. , Barakate, A. , Lapierre, C. , Marita, J.M. , Ralph, J. , Pean, M. , Danoun, S. , Halpin, C. , Grima‐Pettenati, J. and Boudet, A.‐M. (2001) Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down‐regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 28, 257–270. [DOI] [PubMed] [Google Scholar]
- Chapple, C.C.S. , Vogt, T. , Ellis, B.E. and Sommerville, C.R. (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell, 4, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Meyermans, H. , Burggraeve, B. , De Rycke, R.M. , Inoue, K. , De Vleesschauwer, V. , Steenackers, M. , Van Montagu, M.C. , Engler, G.J. and Boerjan, W.A. (2000) Cell‐specific and conditional expression of caffeoyl‐coenzyme A‐3‐O‐methyltransferase in poplar. Plant Physiol. 123, 853–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, M.A. , Collins, R.E. , Anterola, A.M. , Cochrane, F.C. , Davin, L.B. and Lewis, N.G. (2003) An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof. Phytochemistry, 64, 1097–1112. [DOI] [PubMed] [Google Scholar]
- Czechowski, T. , Stitt, M. , Altmann, T. , Udvardi, M.K. and Scheible, W.‐R. (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R.A. and Paiva, N.L. (1995) Stress‐induced phenylpropanoid metabolism. Plant Cell, 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R.A. , Chen, F. , Guo, D. and Parvathi, K. (2001) The biosynthesis of monolignols: a ‘metabolic grid’ or independent pathways to guaiacyl and syringyl units? Phytochemistry, 57, 1069–1084. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A. , Achnine, L. , Kota, P. , Liu, C.J. , Reddy, M.S.S. and Wang, L. (2002) The phenylpropanoid pathway and plant defence—a genomics perspective. Mol. Plant Pathol. 3, 371–390. [DOI] [PubMed] [Google Scholar]
- Durbin, M.L. , McCaig, B. and Clegg, M.T. (2000) Molecular evolution of the chalcone synthase multigene family in the morning glory genome. Plant Mol. Biol. 42, 79–92. [PubMed] [Google Scholar]
- Dushnicky, L.G. , Ballance, G.M. , Summer, M.J. and MacGregor, A.W. (1998) The role of lignification as a resistance mechanism in wheat to a toxin‐producing isolate of Pyrenophora tritici‐repentis . Can. J. Plant Pathol. 20, 35–47. [Google Scholar]
- Ehlting, J. , Büttner, D. , Wang, Q. , Douglas, C.J. , Somssich, I.E. and Kombrink, E. (1999) Three 4‐coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionary divergent classes in angiosperms. Plant J. 19, 9–20. [DOI] [PubMed] [Google Scholar]
- Eynck, C. , Koopmann, B. , Karlovsky, P. and von Tiedemann, A. (2009) Internal resistance in winter oilseed rape inhibits systemic spread of the vascular pathogen Verticillium longisporum . Phytopathology, 99, 802–811. [DOI] [PubMed] [Google Scholar]
- Fergus, B.J. and Goring, D.A.I. (1970) The location of guaiacyl and syringyl lignins in birch xylem tissue. Holzforschung, 24, 113–117. [Google Scholar]
- Franke, R. , Hemm, M.R. , Denault, J.W. , Ruegger, M.O. , Humphreys, J.M. and Chapple, C. (2002a) Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J. 30, 47–59. [DOI] [PubMed] [Google Scholar]
- Franke, R. , Humphreys, J.M. , Hemm, M.R. , Denault, J.W. , Ruegger, M.O. , Cusumano, J.C. and Chapple, C. (2002b) The Arabidopsis REF8 gene encodes the 3‐hydroxylase of phenylpropanoid metabolism. Plant J. 30, 33–45. [DOI] [PubMed] [Google Scholar]
- Garg, H. , Atri, C. , Sandhu, P.S. , Kaur, B. , Renton, M. , Banga, S.K. , Singh, H. , Singh, C. , Barbetti, M.J. and Banga, S.S. (2010) High level of resistance to Sclerotinia sclerotiorum in introgression lines derived from hybridization between wild crucifers and the crop Brassica species B. napus and B. juncea . Field Crops Res. 117, 51–58. [Google Scholar]
- Gayoso, C. , Pomar, F. , Novo‐Uzal, E. , Merino, F. and Martínez de Ilárduya, O. (2010) The Ve‐mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 10, 232–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujon, T. , Sibout, R. , Eudes, A. , MacKay, J. and Jouanin, L. (2003a) Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana . Plant Physiol. Biochem. 41, 677–687. [Google Scholar]
- Goujon, T. , Sibout, R. , Pollet, B. , Maba, B. , Nussaume, L. , Bechtold, N. , Lu, F. , Ralph, J. , Mila, I. , Barrière, Y. , Lapierre, C. and Jouanin, L. (2003b) A new Arabidopsis thaliana mutant deficient in the expression of O‐methyltransferase impacts lignins and sinapoyl esters. Plant Mol. Biol. 51, 973–989. [DOI] [PubMed] [Google Scholar]
- Green, S. , Gaunt, R.E. , Harvey, I.C. and Bourdot, G.W. (1998) Histopathology of Ranunculus acris infected by a mycoherbicide, Sclerotinia sclerotiorum . Australas. Plant Pathol. 27, 73–79. [Google Scholar]
- Hammerschmidt, R. and Kuć, J. (1982) Lignification as a mechanism for induced systemic resistance in cucumber. Physiol. Plant Pathol. 20, 61–71. [Google Scholar]
- Hammerschmidt, R. , Bonnen, A.M. , Bergstrom, G.C. and Baker, K.K. (1985) Association of epidermal lignification with non‐host resistance of cucurbits to fungi. Can. J. Bot. 63, 2393–2398. [Google Scholar]
- Hawkins, S.W. and Boudet, A.M. (2003) Defense lignin and hydroxycinnamyl alcohol dehydrogenase isoforms activities in wounded Eucalyptus gunnii . For. Pathol. 33, 91–104. [Google Scholar]
- Hegedus, D.D. , Li, R. , Buchwaldt, L. , Parkin, I. , Whitwill, S. , Coutu, C. , Bekkaoui, D. and Rimmer, S.R. (2008) Brassica napus possesses an expanded set of polygalacturonase inhibitor protein genes that are differentially regulated in response to Sclerotinia sclerotiorum infection, wounding and defense hormone treatment. Planta, 228, 241–253. [DOI] [PubMed] [Google Scholar]
- Jensen W.A. (ed.) (1962) Botanical Histochemistry: Principles and Practice. San Francisco, CA: Freeman and Co. [Google Scholar]
- Jones, L. , Ennos, A.R. and Turner, S.R. (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin‐deficient mutant of Arabidopsis . Plant J. 26, 205–216. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T. , Koita, H. , Nakatsubo, T. , Hasegawa, K. , Wakabayashi, K. , Takahashi, H. , Umemura, K. , Umezawa, T. and Shimamoto, K. (2006) Cinnamoyl‐CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defence signalling in rice. Proc. Natl. Acad. Sci. USA, 103, 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen, N.T. and Littlefield, L.J. (1979) The possible association of phytoalexins with resistance gene expression in flax to Melampspora lini . Physiol. Plant Pathol. 14, 265–280. [Google Scholar]
- Kiedrowski, S. , Kawalleck, P. , Hahlbrock, K. , Somssich, I.E. and Dangl, J.L. (1992) Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis RPM1 disease resistance locus. EMBO J. 11, 4677–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli, Y. , Brunner, L.J. , Yoell, H. , Milgroom, M.G. , Anderson, J.B. , Morrall, R.A.A. and Kohn, L.M. (1995) Clonal dispersal and spatial mixing in populations of the plant pathogenic fungus Sclerotinia sclerotiorum . Mol. Ecol. 4, 69–77. [Google Scholar]
- Lacombe, E. , Hawkins, S. , Van Doorsselaere, J. , Piquemal, J. , Goffner, D. , Poeydomenge, O. , Boudet, A.‐M. and Grima‐Pettenati, J. (1997) Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant J. 11, 429–441. [DOI] [PubMed] [Google Scholar]
- Lange, B.M. , Lapierre, C. and Sandermann, H. Jr (1995) Elicitor‐induced spruce stress lignin (structural similarity to early developmental lignins). Plant Physiol. 108, 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvergeat, V. , Lacomme, C. , Lacombe, E. , Lasserre, E. , Roby, D. and Grima‐Pettenati, J. (2001) Two cinnamoyl‐CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry, 57, 1187–1195. [DOI] [PubMed] [Google Scholar]
- Li, L. , Popko, J.L. , Umezawa, T. and Chiang, V.L. (2000) 5‐Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J. Biol. Chem. 275, 6537–6545. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2–ΔΔ CT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luderitz, T. and Grisebach, H. (1981) Enzymic synthesis of lignin precursors. Comparison of cinnamoyl‐CoA reductase and cinnamoyl alcohol:NADP+ dehydrogenase from spruce (Picea abies L.) and soybean (Glycine max L.). Eur. J. Biochem. 119, 115–124. [DOI] [PubMed] [Google Scholar]
- Ma, Q.‐H. (2007) Characterization of a cinnamoyl‐CoA reductase that is associated with stem development in wheat. J. Exp. Bot. 58, 2011–2021. [DOI] [PubMed] [Google Scholar]
- Martinelli, T. and Galasso, I. (2011) Phenological growth stages of Camelina sativa according to the extended BBCH scale. Ann. Appl. Biol. 158, 87–94. [Google Scholar]
- Menden, B. , Kohlhoff, M. and Moerschbacher, B. (2007) Wheat cells accumulate a syringyl‐rich lignin during the hypersensitive resistance response. Phytochemistry, 68, 513–520. [DOI] [PubMed] [Google Scholar]
- Nicholson, R.L. and Hammerschmidt, R. (1992) Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30, 369–389. [Google Scholar]
- O'Connell, A. , Holt, K. , Piquemal, J. , Grima‐Pettenati, J. , Boudet, A. , Pollet, B. , Lapierre, C. , Petit‐Conil, M. , Schuch, W. and Halpin, C. (2002) Improved paper pulp from plants with suppressed cinnamoyl‐CoA reductase or cinnamyl alcohol dehydrogenase. Transgenic Res. 11, 495–503. [DOI] [PubMed] [Google Scholar]
- Osakabe, K. , Tsao, C.C. , Li, L. , Popko, J.L. , Umezawa, T. , Carraway, D.T. , Smeltzer, R.H. , Joshi, C.P. and Chiang, V.L. (1999) Coniferyl aldehyde 5‐hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. USA, 96, 8955–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten, G.S. , Cardenas, C.L. , Cochrane, F.C. , Laskar, D.D. , Bedgar, D.L. , Davin, L. and Lewis, N.G. (2005) Reassessment of effects on lignification and vascular development in the irx4 Arabidopsis mutant. Phytochemistry, 66, 2092–2107. [DOI] [PubMed] [Google Scholar]
- Peltier, A.J. , Hatfield, R.D. and Grau, C.R. (2009) Soybean stem lignin concentration relates to resistance to Sclerotinia sclerotiorum . Plant Dis. 93, 149–154. [DOI] [PubMed] [Google Scholar]
- Pichon, M. , Courbou, I. , Beckert, M. , Boudet, A.‐M. and Grima‐Pettenati, J. (1998) Cloning and characterization of two maize cDNAs encoding Cinnamoyl‐CoA Reductase (CCR) and differential expression of the corresponding genes. Plant Mol. Biol. 38, 671–676. [DOI] [PubMed] [Google Scholar]
- Piquemal, J. , Lapierre, C. , Myton, K. , O'Connell, A. , Schuch, W. , Grima‐Pettenati, J. and Boudet, A.‐M. (1998) Down‐regulation of cinnamoyl‐CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J. 13, 71–83. [Google Scholar]
- Pomar, F. , Novo, M. , Bernal, M.A. , Merino, F. and Ros Barceló, A. (2004) Changes in stem lignins (monomer composition and crosslinking) and peroxidase are related with the maintenance of leaf photosynthetic integrity during Verticillium wilt in Capsicum annuum . New Phytol. 163, 111–123. [DOI] [PubMed] [Google Scholar]
- Raes, J. , Rohde, A. , Christensen, J.H. , Van de Peer, Y. and Boerjan, W. (2003) Genome‐wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133, 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph, J. , Hatfield, R.D. , Piquemal, J. , Yahiaoui, N. , Pean, M. , Lapierre, C. and Boudet, A.M. (1998) NMR characterization of altered lignins extracted from tobacco plants down‐regulated for lignification enzymes cinnamyl‐alcohol dehydrogenase and cinnamoyl‐CoA reductase. Proc. Natl. Acad. Sci. USA, 95, 12 803–12 808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ride, J.P. (1978) The role of cell wall alterations in resistance to fungi. Ann. Appl. Biol. 89, 302–306. [Google Scholar]
- Robinson, A.R. and Mansfield, S.D. (2009) Rapid analysis of poplar lignin monomer composition by a streamlined thioacidolysis procedure and near‐infrared reflectance‐based prediction modeling. Plant J. 58, 706–714. [DOI] [PubMed] [Google Scholar]
- Rodríguez, M.A. , Venedikian, N. , Bazzalo, M.E. and Godeas, A. (2004) Histopathology of Sclerotinia sclerotiorum attack on flower parts of Helianthus annuus heads in tolerant and susceptible varieties. Mycopathologia, 157, 291–302. [DOI] [PubMed] [Google Scholar]
- Rolando, C. , Monties, B. and LaPierre, C. (1992) Thioacidolysis In: Methods in Lignin Chemistry (Lin S. and Dence C., eds), pp. 334–349. Berlin: Springer‐Verlag. [Google Scholar]
- Ros Barcelo, A. (1997) Lignification in plant cell walls. Int. Rev. Cytol. 176, 87–132. [PubMed] [Google Scholar]
- Sarkanen, K.V. and Ludwig, C.H. (1971) Definition and nomenclature In: Lignins. Occurrence, Formation, Structure and Reactions (Sarkanen K.V. and Ludwig C.H., eds), pp. 1–41. New York: Wiley Interscience. [Google Scholar]
- Sarni, R. , Grand, C. and Boudet, A.M. (1984) Purification and properties of cinnamoyl‐CoA reductase and cinnamyl alcohol dehydrogenase from poplar stems (Populus ×euramericana). Eur. J. Biochem. 139, 259–265. [DOI] [PubMed] [Google Scholar]
- Sibout, R. , Eudes, A. , Pollet, B. , Goujon, T. , Mila, I. , Granier, F. , Séguin, A. , Lapierre, C. and Jouanin, L. (2003) Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol. 132, 848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southerton, S.G. and Deverall, B.J. (1990) Histochemical and chemical evidence for lignin accumulation during the expression of resistance to leaf rust fungi in wheat. Physiol. Mol. Plant Pathol. 36, 483–494. [Google Scholar]
- Stringam, G.R. (1971) Genetics of four hypocotyls mutants in Brassica campestris L. J. Hered. 62, 248–250. [Google Scholar]
- Thomma, B.P.H.J. , Penninckx, I.A. , Broekaert, W.F. and Cammue, B.P.A. (2001) The complexity of disease signaling in Arabidopsis . Curr. Opin. Immunol. 13, 63–68. [DOI] [PubMed] [Google Scholar]
- Tronchet, M. , Balague, C. , Kroj, T. , Jouanin, L. and Roby, D. (2010) Cinnamyl alcohol dehydrogenase‐C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 11, 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance, C.P. , Kirk, T.K. and Sherwood, R.T. (1980) Lignification as a mechanism of disease resistance. Annu. Rev. Phytopathol. 18, 259–288. [Google Scholar]
- Wally, O. , Jayaraj, J. and Punja, Z. (2008) Comparative resistance to foliar fungal pathogens in transgenic carrot plants expressing genes encoding for chitinase, β‐1,3‐glucanase and peroxidase. Eur. J. Plant Pathol. 123, 331–342. [Google Scholar]
- Weng, J.‐K. and Chapple, C. (2010) The origin and evolution of lignin biosynthesis. New Phytol. 187, 273–285. [DOI] [PubMed] [Google Scholar]
- Wuyts, N. , Lognay, G. , Swennen, R. and De Waele, D. (2006) Nematode infection and reproduction in transgenic and mutant Arabidopsis and tobacco with altered phenylpropanoid metabolism. J. Exp. Bot. 57, 2825–2835. [DOI] [PubMed] [Google Scholar]
- Zhao, J. , Buchwaldt, L. , Rimmer, S.R. , Sharpe, A. , McGregor, L. , Bekkaoui, D. and Hegedus, D.D. (2009) Patterns of differential gene expression in Brassica napus cultivars infected with Sclerotinia sclerotiorum . Mol. Plant Pathol. 10, 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Camelina sativa line 36012, 14 days after inoculation with Sclerotinia sclerotiorum. Fungal hyphae appear green after staining with WGA Alexa Fluor 488 and plant structures appear red after staining with Congo Red.
Table S1 Sequences of primers used for real‐time reverse transcriptase‐polymerase chain reaction (qRT‐PCR).
Supporting info item
Supporting info item


