Summary
The incidence and severity of light leaf spot epidemics caused by the ascomycete fungus Pyrenopeziza brassicae on UK oilseed rape crops are increasing. The disease is currently controlled by a combination of host resistance, cultural practices and fungicide applications. We report decreases in sensitivity of modern UK P. brassicae isolates to the azole (imidazole and triazole) class of fungicides. By cloning and sequencing the P. brassicae CYP51 (PbCYP51) gene, encoding the azole target sterol 14α‐demethylase, we identified two non‐synonymous mutations encoding substitutions G460S and S508T associated with reduced azole sensitivity. We confirmed the impact of the encoded PbCYP51 changes on azole sensitivity and protein activity by heterologous expression in a Saccharomyces cerevisiae mutant YUG37:erg11 carrying a controllable promoter of native CYP51 expression. In addition, we identified insertions in the predicted regulatory regions of PbCYP51 in isolates with reduced azole sensitivity. The presence of these insertions was associated with enhanced transcription of PbCYP51 in response to subinhibitory concentrations of the azole fungicide tebuconazole. Genetic analysis of in vitro crosses of sensitive and resistant isolates confirmed the impact of PbCYP51 alterations in coding and regulatory sequences on a reduced sensitivity phenotype, as well as identifying a second major gene at another locus contributing to resistance in some isolates. The least sensitive field isolates carry combinations of upstream insertions and non‐synonymous mutations, suggesting that PbCYP51 evolution is ongoing and the progressive decline in azole sensitivity of UK P. brassicae populations will continue. The implications for the future control of light leaf spot are discussed.
Keywords: demethylation inhibitors, fungicide resistance, light leaf spot, P450, target site mutation
Introduction
Pyrenopeziza brassicae B. Sutton & Rawlinson (anamorph: Cylindrosporium concentricum Grev.) is an ascomycete fungus causing light leaf spot on Brassica species. Historically, light leaf spot has been problematic on oilseed rape in Scotland and northern England. However, in 2012, over 92% of winter oilseed rape crops in the UK were affected by light leaf spot in all regions surveyed (Crop Monitor, 2012; http://www.cropmonitor.co.uk). Disease incidence was highest in south‐west England, which was previously considered to be at lower risk than northern England (Welham et al., 2004). The long‐term (2001–2010) mean incidence of light leaf spot in the UK was 11% of sampled plants, but, in 2012, infections were reported on 35% of plants.
The control of P. brassicae is normally reliant on a combination of cultural practices, cultivar resistance and foliar fungicides (Latunde‐Dada et al., 2007). Boys et al. (2007) reported that the efficacy of cultivar resistance is often reduced when varieties are deployed on a wide scale. As oilseed rape cultivation has increased over recent years and there are no cultivars completely resistant to the disease, the importance of fungicides for the control of light leaf spot remains high. The azole (imidazole and triazole) fungicides are the most effective active ingredients against light leaf spot, although the methyl benzimidazole carbamate (MBC) carbendazim is also widely used (Home Grown Cereals Authority (HGCA), 2003). Previous reports have described resistance to MBCs in P. brassicae (Sutherland et al., 1994), and recently we have shown that the resistance‐conferring β‐tubulin substitutions E198A and L240F are widespread in UK P. brassicae populations (Carter et al., 2013). Therefore, control of this disease on oilseed rape now relies on the effectiveness of the azole component of fungicide sprays. Azole fungicides have been widely used to manage fungal pathogens of both animals and plants for over 30 years (Sheehan et al., 1999). They bind to the cytochrome P450 sterol 14α‐demethylase (CYP51, syn. ERG11), an essential enzyme in the biosynthesis of sterols (Parks et al., 1995). Of the many sterols identified in fungi, ergosterol is the most common and is required for fungal growth (Weete et al., 2010). Reduced sensitivity to azoles has been reported previously in some Scottish P. brassicae isolates (Burnett, 2003), and there is anecdotal evidence of reduced azole efficacy in other locations. The molecular and genetic basis of reduced azole sensitivity in P. brassicae has not been investigated previously.
Studies of filamentous fungi have reported three predominant mechanisms of azole resistance: mutations in the CYP51 gene, overexpression of CYP51 and enhanced fungicide efflux. Enhanced fungicide efflux, mediated by the overexpression of efflux pumps [e.g. adenosine triphosphate binding cassette (ABC) transporters, major facilitator proteins], has frequently been associated with resistance in the human pathogenic yeasts, for example Candida albicans (Prasad et al., 1995; Sanglard et al., 1997). In contrast, enhanced efflux has rarely been reported in field isolates of plant‐pathogenic fungi. For example, genes encoding ABC transporters have been identified and their capacity to export azoles has been shown by heterologous expression and targeted knockout studies in Botrytis cinerea, Zymoseptoria tritici (synonym: Mycosphaerella graminicola), Magnaporthe oryzae and Penicillum digitatum (De Waard et al., 2006). Yet, to date, only in B. cinerea has enhanced efflux to multiple fungicides been shown to reduce the performance of fungicides in the field (Kretschmer et al., 2009). Overexpression of CYP51 has been associated with reduced azole sensitivity in the plant pathogens Sclerotinia homoeocarpa (Hulvey et al., 2012), Z. tritici (Cools et al., 2012), Pe. digitatum (Hamamoto et al., 2000), Monilinia fructicola (Luo and Schnabel, 2008) and Cercospora beticola (Bolton et al., 2012; Nikou et al., 2009). In some species, CYP51 promoter modifications have been correlated with increased CYP51 expression, suggesting that these sequence changes operate as transcriptional enhancers (Hamamoto et al., 2000; Sun et al., 2013).
In plant‐pathogenic fungi, CYP51 mutations are frequently associated with azole‐resistant phenotypes. To date, these have been best studied in the wheat pathogen Z. tritici (Cools et al., 2010, 2011; Fraaije et al., 2007; Leroux et al., 2007; Leroux and Walker, 2011). Common CYP51 substitutions in azole‐resistant strains of Z. tritici include V136A, Y137F, A379G, I381V, Y459D, Y461H, Y461S, ΔY459/G460 and S524T (Cools and Fraaije, 2013). Substitutions at equivalent residues, as well as some unique alterations, have been identified in other plant‐pathogenic fungi, including Blumeria graminis f. sp. hordei and tritici (Wyand and Brown, 2005), Eryisphe necator (Delye et al., 1997), Puccinia triticina (Stammler et al., 2009) and Mycosphaerella fijiensis (Cañas‐Gutiérrez et al., 2009). Biochemical studies of CYP51 from the human pathogen C. albicans have demonstrated substitutions identified in clinical strains that confer reduced sensitivity to azoles by reducing binding affinity (e.g. G464S), which may also reduce enzyme activity (Kelly et al., 1999). More recently, expression in Saccharomyces cerevisiae has demonstrated that some CYP51 substitutions in Z. tritici (e.g. D134G) may be compensatory, enhancing the activity of proteins carrying substitutions that alone reduce enzyme activity (Cools et al., 2011).
In this article, we report the molecular characterization of P. brassicae isolates with reduced sensitivity to azole fungicides using a combination of classical genetics and candidate gene approaches. The inheritance of azole resistance is complex, conferred by one or two genes, with contributions from minor loci. We sequenced the P. brassicae CYP51 (PbCYP51) gene and identified amino acid substitutions, G460S and S508T, correlated with reduced azole sensitivity. Heterologous expression in S. cerevisiae showed that these substitutions reduced fungicide sensitivity. In addition, by sequencing upstream of the predicted PbCYP51 translation start codon, we identified sequence insertions in less sensitive isolates which were associated with increased induced PbCYP51 expression. The least‐sensitive field isolates carried combinations of PbCYP51 upstream insertions and non‐synonymous mutations. The implications of these genetic changes for the future evolution of azole sensitivity in UK P. brassicae populations are discussed.
Results
PbCYP51 changes are associated with decreased azole sensitivity in P. brassicae
The ratios between the 50% effective concentration (EC50) values of the most‐ and least‐sensitive field isolates of P. brassicae in vitro were approximately 57‐, 102‐, 119‐, 277‐ and 34‐fold to prochloraz, flusilazole, tebuconazole, prothioconazole and metconazole, respectively (Table S1, see Supporting Information). Four P. brassicae isolates obtained before 1996 were significantly more sensitive to tebuconazole (Kolmogorov–Smirnov two‐sample test, P = 0.05), metconazole (P = 0.035), flusilazole (P = 0.002), prochloraz (P = 0.006) and prothioconazole (P = 0.004) than isolates obtained from 2003 onwards (Table S1). There was positive cross‐resistance between the five azole fungicides (Fig. S1, see Supporting Information). Cross‐resistance occurred between the imidazole prochloraz and triazoles (prothioconazole, tebuconazole, metconazole, flusilazole), as well as between the triazoles.
Sequencing of PbCYP51 identified two non‐synonymous mutations (Table 1, Table S1), at codon position 460 (G460S) in isolates Pb7, PbUK062 and PbUK071, and at position 508 (S508T) in isolates BS9053 and Pb1. Predicted PbCYP51 amino acid sequences were otherwise identical in all isolates. Subsequent screening of the full collection of isolates by restriction fragment length polymorphism (RFLP) detected substitution G460S in an additional six isolates and S508T in another three. The mean resistance factors for isolates carrying substitution G460S alone were between 7 (prochloraz) and 12 (tebuconazole/prothioconazole). Isolates carrying substitution S508T alone had resistance factors between 7 (prothioconazole/prochloraz) and 17 (flusilazole) (Table 1).
Table 1.
Azole sensitivities of Pyrenopeziza brassicae isolates carrying particular PbCYP51 variants
| PbCYP51 variant | Fungicide | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tebuconazole | Metconazole | Flusilazole | Prothioconazole | Prochloraz | ||||||
| Mean EC50 (Range) (μg/mL) | RFa | Mean EC50 (Range) (μg/mL) | RF | Mean EC50 (Range) (μg/mL) | RF | Mean EC50 (Range) (μg/mL) | RF | Mean EC50 (Range) (μg/mL) | RF | |
|
Wild‐type (n b = 14) |
0.04 (0.01–0.15) |
1 |
0.02 (0.004–0.09) |
1 |
0.07 (0.01–0.20) |
1 |
0.14 (0.02–0.79) |
1 |
0.03 (0.01–0.08) |
1 |
|
G460S (n = 8) |
0.49 (0.23–0.79) |
12 |
0.15 (0.10–0.22) |
8 |
0.68 (0.22–2.09) |
10 |
1.63 (0.38–3.00) |
12 |
0.20 (0.87–0.31) |
7 |
|
S508T (n = 4) |
0.33 (0.28–0.49) |
8 |
0.16 (0.11–0.20) |
8 |
1.12 (0.56–2.00) |
16 |
0.93 (0.30–3.02) |
7 |
0.22 (0.18–0.33) |
7 |
|
46‐bp insert (n = 1) |
0.40 | 10 | 0.15 | 8 | 1.14 | 16 | 3.17 | 22 | 0.31 | 10 |
|
151‐bp insert (n = 1) |
0.57 | 14 | 0.27 | 14 | 1.59 | 23 | 3.77 | 27 | 0.36 | 12 |
|
232‐bp insert (n = 3) |
0.44 (0.39–0.49) |
11 |
0.16 (0.14–0.19) |
8 |
0.91 (0.79–1.10) |
13 |
4.04 (3.40–4.50) |
29 |
0.24 (0.12–0.55) |
8 |
|
S508T/151‐bp insert (n = 1) |
1.21 | 30 | 0.34 | 17 | 3.10 | 44 | 5.55 | 39 | 0.41 | 14 |
Resistance factors (RF) were calculated as the ratio of the EC50 to the mean EC50 values of isolates obtained between 1993 and 1995. Mean EC50 values were calculated on a logarithmic scale and back‐transformed.
Number of isolates with the CYP51 variant tested.
Amplification of a 663‐bp region upstream of the predicted translation start codon of PbCYP51 identified a 232‐bp sequence insertion in isolates C1E and C1D, a 151‐bp insertion in isolates WC6 and I3 and a 46‐bp insertion in isolate C5A (Table S1). The 232‐bp sequence insertion upstream of PbCYP51 correlated with a mean 11‐fold decrease in tebuconazole sensitivity, 29‐fold reduction in prothioconazole sensitivity, 13‐fold reduction in flusilazole sensitivity, eight‐fold reduction in metconazole sensitivity and eight‐fold reduction in prochloraz sensitivity (Table 1), and a decrease in sensitivity to the unrelated fungicide cycloheximide (data not shown). Isolates with the 46‐ and 150‐bp insertions in PbCYP51 also showed reduced sensitivity to all the azoles tested (Tables 1 and S1). Isolate I3, with both the 150‐bp insertion and S508T, was the least sensitive of all isolates to all triazoles tested (Table 1). There was no change in the cycloheximide sensitivity of isolates carrying the 46‐ and 150‐bp insertions (data not shown).
Saccharomyces cerevisiae YUG37::erg11 transformants expressing PbCYP51 variants are less sensitive to azoles
To verify the effect of the identified nucleotide changes on azole sensitivity, wild‐type (WT) and mutant PbCYP51 genes were expressed in S. cerevisiae YUG37::erg11. Transformants expressing WT PbCYP51 protein were able to grow on SD GAL + RAF medium (synthetic dropout medium with galactose and raffinose) amended with doxycycline, demonstrating that WT PbCYP51 can complement native S. cerevisiae YUG37::erg11 CYP51 function (Fig. 1). Transformation of S. cerevisiae YUG37::erg11 with the constructs pYES‐PbCYP51S508T and pYES‐PbCYP51G460S/S508T did not affect the capacity of PbCYP51 to complement the orthologous gene in S. cerevisiae YUG37::erg11 (Fig. 1). However, PbCYP51 with substitution G460S did not complement S. cerevisiae CYP51; it was therefore not possible to determine the effect of G460S on azole sensitivity. Sensitivity testing of S. cerevisiae YUG37::erg11 expressing PbCYP51/S508T showed reduced sensitivity to all azoles tested. The combination of G460S and S508T further decreased sensitivity (Table 2). The introduction of PbCYP51 substitutions did not affect the sensitivity to cycloheximide (Table 2).
Figure 1.
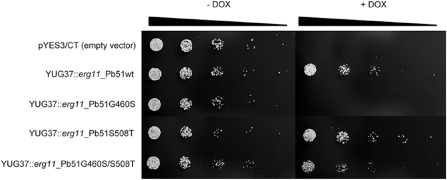
Complementation of Saccharomyces cerevisiae strain YUG37:erg11 with wild‐type (Pb51wt) and mutated variants of the Pyrenopeziza brassicae CYP51 gene. Growth in the absence (–DOX) and presence (+DOX) of doxycycline. Bar indicates five, five‐fold dilutions of cell suspensions starting at 1 × 106 cells/mL.
Table 2.
Azole sensitivities of S accharomyces cerevisiae YUG37::erg11 transformants
| Transformant | Introduced amino acid substitution(s) | Tebuconazole | Metconazole | Prochloraz | Prothioconazole | Flusilazole | Cycloheximide | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC50 a (μg/mL) | RFb | EC50 (μg/mL) | RF | EC50 (μg/mL) | RF | EC50 (μg/mL) | RF | EC50 (μg/mL) | RF | EC50 (μg/mL) | RF | ||
| YUG37::erg11_Pb51wt | None | 0.005 | 1 | 0.001 | 1 | 0.003 | 1 | 0.018 | 1 | 0.005 | 1 | 0.046 | 1 |
| YUG37::erg11_Pb51S508T | S508T | 0.036 | 8 | 0.007 | 7 | 0.018 | 6 | 0.067 | 4 | 0.061 | 11 | 0.071 | 2 |
| YUG37::erg11_Pb51G460S/S508T | G460S and S508T | 0.16 | 35 | 0.028 | 26 | 0.034 | 12 | 0.15 | 9 | 0.11 | 21 | 0.056 | 1 |
| Least significant ratioc | 2.1 | 4.5 | 12.5 | 1.1 | 4.0 | 1.7 | |||||||
EC50 values are means of results from two independent replicates calculated on a logarithmic scale and back‐transformed.
Resistance factors (RF) were calculated as the ratio of EC50 to the EC50 of transformants expressing wild‐type PbCYP51.
Back‐transformed values differing by a ratio greater than this are significantly different at P = 0.05.
PbCYP51 upstream insertions are associated with induced overexpression
The shortest of the inserts found upstream of the predicted translation start codon (46 bp) was also found within the 151‐ and 233‐bp inserts (Fig. 2A). blast searches revealed no significant sequence similarity with sequences on the National Center for Biotechnology Information (NCBI) database (data not shown). After 6 days of growth, in the absence of fungicide treatment and with cultures in the linear growth phase, there were no significant differences between PbCYP51 expression of the calibrator isolate E2C and isolates E2D, PbUK073, WC6 and C1E (Fig. 2B). There was a six‐fold increase in constitutive PbCYP51 expression in isolate C5A. Treatment with tebuconazole increased the quantity of PbCYP51 transcripts in all isolates. In isolates with upstream inserts (WC6, C1E and C5A), the ratio between PbCYP51 transcription with and without azole induction was 4–50‐fold higher than the ratio in isolates without the insert (E2C, E2D, PbUK073).
Figure 2.
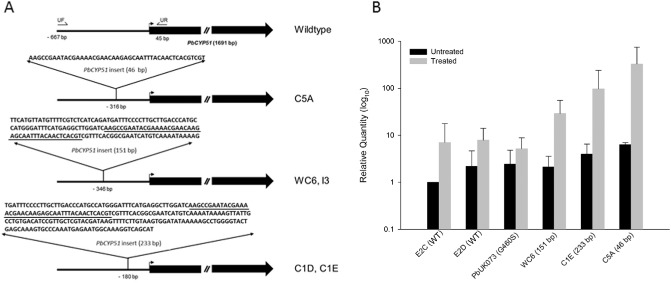
(A) Schematic diagram of PbCYP51 upstream insertions. UF indicates the position of primer CYP51upstreamF and UR indicates the position of primer CYP51upstreamR. Underlining indicates the location of the 46‐bp duplicated sequence within the 151 and 233‐bp inserts. Arrow indicates translation start site. (B) Expression of PbCYP51 in Pyrenopeziza brassicae isolates measured as the mean relative quantity compared with the sample E2C (untreated). Error bars denote 95% confidence level. PbCYP51 genotypes are given in parentheses.
Genetic control of reduced azole sensitivity in P. brassicae differs between isolates
Ascospore progeny were obtained from eight crosses between P. brassicae isolates with different PbCYP51 alleles and/or tebuconazole sensitivities (Table S3, see Supporting Information). Cumulative probability distributions for observed and expected progeny log EC50 values were generated for four crosses (Fig. 3A,C,E,G) and the progeny genotypes at PbCYP51 were determined (Fig. 3B,D,F,H). The tebuconazole sensitivities of progeny from cross PbFr054 (WT) × C1E (232‐bp insert) did not follow a bimodal distribution. There was evidence for the segregation of a second unlinked major gene conferring reduced sensitivity (Fig. 3A). The 232‐bp PbCYP51 upstream insert partially, but not fully, co‐segregated with reduced tebuconazole sensitivity, with three individuals carrying this insert categorized within the sensitive phenotypes (Fig. 3B). Both features of the data suggest that an additional gene contributes to reduced azole sensitivity in C1E. Cumulative probability distributions of the cross between C1E and E2C (data not shown) supported this finding. The phenotype distribution and genotype segregation of progeny from the crosses between PbUK062 (G460S) and E2D (WT) (Fig. 3E,F) and between PbFr054 (WT) and I3 (150 bp + S508T) (Fig. 3C,D) were consistent with single major genes accounting for the difference between the isolates. All progeny with EC50 values greater than 0.5 μg/mL had both the PbCYP51 upstream insert and the amino acid change S508T (Fig. 3D). In progeny labelled as ‘mixture’, both WT and mutant PbCYP51 promoter sequences were detected, suggesting mixed isolate cultures (Fig. 3D). Progeny from cross E2D (WT) × PbUK062 (G460S) also followed a bimodal sensitivity distribution (Fig. 3E), with G460S only detected in less sensitive progeny (Fig. 3F). Therefore, a single major gene difference accounts for the reduced sensitivity to azoles in P. brassicae isolates PbUK062 and I3. In contrast, there was evidence that two major loci contribute to differences in azole sensitivity between isolates PbUK071 (G460S) and E2A (Fig. 3G).
Figure 3.
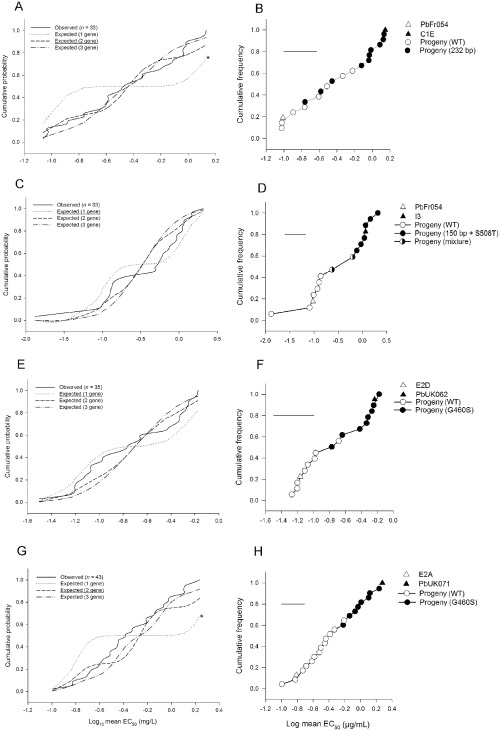
(A, C, E, G) Probability distributions of observed 50% effective concentrations (EC50) to tebuconazole against expected distributions based on the differences being caused by one, two or three genes of equal additive effect. (B, D, F, H) Distribution of parental alleles in relation to EC50 . Crosses are: (A, B) PbFr054 × C1E [single‐gene hypothesis (Kolmogorov–Smirnoff one‐sample test, D = 0.33, P < 0.01), two‐gene hypothesis (D = 0.13, P > 0.2) and three‐gene hypothesis (D = 0.11, P > 0.2)]; (C, D) PbFr054 × I3 [single‐gene hypothesis (Kolmogorov–Smirnoff one‐sample test, D = 0.15, P > 0.2), two‐gene hypothesis (D = 0.21, P > 0.1) and three‐gene hypothesis (D = 0.21, P > 0.1)]; (E, F) E2D × PbUK062 [single‐gene hypothesis (Kolmogorov–Smirnoff one‐sample test, D = 0.18, P > 0.2), two‐gene hypothesis (D = 0.15, P > 0.2) and three‐gene hypothesis (D = 0.19, P > 0.15); (G, H) E2A × PbUK071 [single‐gene hypothesis (Kolmogorov–Smirnoff one‐sample test, D = 0.42, P < 0.05), two‐gene hypothesis (D = 0.2, P > 0.05) and three‐gene hypothesis (D = 0.14, P > 0.2)]. The most parsimonious inference is underlined. *indicates significant difference between observed and expected distributions. Data are adjusted means from two independent replicates. Parental EC50 was determined in the same experiments as the progeny. Error bar denotes two estimated standard errors of an isolate mean, representing approximate 95% confidence limits for a single isolate.
Discussion
Azole fungicides currently offer satisfactory control of P. brassicae in the field. However, there have been reports of a decline in their effectiveness (Burnett, 2003). In this study, we report modifications in the coding and predicted regulatory regions of the PbCYP51 gene of modern P. brassicae isolates with reduced azole sensitivity, and show that changes in other (unknown) genes can contribute to decreases in sensitivity. The accumulation of these changes in a single isolate is associated with the greatest reductions in sensitivity. We propose that the increased frequency of these variants in the UK P. brassicae populations has caused the observed reductions in the efficacy of azole fungicides in the field.
The changes identified in PbCYP51 encode amino acid substitutions G460S and S508T. Substitution S508T is equivalent to the Z. tritici CYP51 (MgCYP51) substitution S524T which, when expressed in S. cerevisiae strain YUG37::erg11, also confers reduced sensitivity to all azoles (Cools et al., 2011). In Z. tritici, S524T is only found in combination with other MgCYP51 changes, but, in P. brassicae, S508T was found alone. PbCYP51 substitution G460S was also associated with reduced azole sensitivity in P. brassicae. However, variants with this substitution alone did not complement S. cerevisiae strain YUG37::erg11. The equivalent substitution in C. albicans, G464S, has been repeatedly implicated in resistance to clinical azoles (Morio et al., 2010; Sanglard et al., 1998). In C. albicans, G464S perturbs the haem environment of CYP51, which results in decreased binding affinity for azoles, and also reduces enzyme activity (Kelly et al., 1999). The failure of PbCYP51 with G460S to complement yeast CYP51 suggests similar alterations in functionality, although it is unclear whether a reduction in PbCYP51 activity reduces P. brassicae isolate fitness. In C. albicans, in vitro growth was unaffected by the replacement of WT CYP51 with a G464S variant, suggesting that this substitution incurs no substantial fitness cost (Sasse et al., 2012). The expression of PbCYP51 with G460S/S508T combined complemented yeast CYP51 and reduced the sensitivity to all azoles tested by 9–35‐fold, much more than S508T alone. Therefore, PbCYP51 variants with multiple substitutions can cause further reductions in azole sensitivity without reducing enzyme activity. In Z. tritici, the accumulation of substitutions in MgCYP51 causes stepwise reductions in azole sensitivity, with particular combinations of alterations required to maintain optimum enzyme activity (Cools and Fraaije, 2013; Cools et al., 2010, 2011; Leroux et al., 2007). Although P. brassicae isolates with the PbCYP51 variant G460S/S508T were not isolated in this study, the cloning and sequencing of polymerase chain reaction (PCR) products from P. brassicae population samples indicated that this variant was present in P. brassicae populations (H. E. Carter et al., Rothamsted Research, Harpenden, UK, unpublished results).
Some P. brassicae isolates (WC6, C1D, C1E and C5A) have the same PbCYP51 coding sequence as sensitive isolates, but are up to 55‐fold less sensitive to azoles. In these isolates, sequencing upstream of the translation start site identified sequence duplications in the predicted regulatory regions of PbCYP51. We measured PbCYP51 transcription in the presence of subinhibitory concentrations of tebuconazole in these isolates, in isolates E2C and E2D with WT PbCYP51 and in isolate PbUK073 with substitution G460S. Transcription was significantly greater in isolates with upstream sequence insertions. Increases in expression caused by alterations in the predicted regulatory regions of CYP51 are most often constitutive. For example, the insertion of a recently defined 194‐bp transposable element (Sun et al., 2013) in CYP51A (Ghosoph et al., 2007; Hamamoto et al., 2000) or CYP51B (Sun et al., 2011) promoter is responsible for constitutive overexpression in imazilil‐resistant Pe. digitatum isolates. Luo and Schnabel (2008) identified a 65‐bp repetitive element in the CYP51 gene of azole‐resistant M. fructicola isolates which correlated with an increase in constitutive expression of MfCYP51. In Z. tritici, constitutive MgCYP51 overexpression is associated with an insertion of a 120‐bp transcriptional enhancer derived from the 5′ end of a gene with unknown function on another chromosome (Cools et al., 2012). Induced expression has been less widely implicated in azole resistance than has increased constitutive expression, although it has been suggested to contribute to resistance in Sc. homoeocarpa (Hulvey et al., 2012) and Ce. beticola (Bolton et al., 2012). In P. brassicae isolates (with the exception of a slight increase in C5A), constitutive PbCYP51 expression was unaffected by upstream sequence insertions, whereas azole‐induced PbCYP51 expression was enhanced substantially. Induced expression was highest in C5A, the isolate with the shortest sequence insert. In addition, the 46‐bp sequence duplicated in isolate C5A was present in the 151‐ and 232‐bp insertions of the other PbCYP51‐overexpressing isolates. Therefore, this 46‐bp region is sufficient to enhance the transcription of PbCYP51 on azole treatment, suggesting that it encompasses a motif or element responsive to sterol depletion.
Crossing experiments indicated differences in the genetics underlying resistance in different P. brassicae isolates and showed that loci other than PbCYP51 were involved. For example, a variant allele at a second locus further reduced azole sensitivity in isolate PbUK071, but was absent from PbUK062. A major gene other than PbCYP51 also reduced sensitivity to azoles in C1E; we have no indication whether this is the same locus as in PbUK071. These other genes could participate directly in resistance by, for example, encoding a protein capable of exporting azole from the cell. The reduced sensitivity of C1E to the unrelated fungicide cycloheximide supports this suggestion (data not shown). The resistance distributions in the offspring of the crosses were broadly symmetrical on a logarithmic scale, which does not suggest substantial epistasis between PbCYP51 and the other loci. Isolate I3, which carries a combination of a 151‐bp upstream insert and substitution S508T, was the least sensitive isolate tested. Progeny from the cross PbFr054 × I3 followed a bimodal distribution of tebuconazole sensitivities, with the range between that of the parental isolates, indicating that a single locus was responsible for the difference in resistance. It would be of interest to compare the offspring distribution of crosses between I3 and isolates with only PbCYP51 changes and crosses between I3 and PbUK071 or C1E, and between PbUK071 and C1E. The progeny of crosses involving loci other than PbCYP51 would be predicted to include phenotypes with resistance beyond the range of either parent.
The most resistant P. brassicae isolate (I3) studied here has a combination of CYP51 promoter insertion with target protein variation. This combination of mechanisms is consistent with the evolution of decreased azole sensitivity in other pathogens. Together with genetic evidence for mechanisms controlled by variation at other loci, it suggests that continued selection will be effective in increasing resistance in populations to levels that could hinder disease control. In some fungi (e.g. Z. tritici; Fraaije et al., 2007), resistance to different azoles is negatively correlated, allowing the substitution of azole fungicides or possibly the use of mixtures to reduce the practical impact of resistance. In this study, the strong positive cross‐resistance between azoles suggests that the use of diverse azole chemistries will not be an effective strategy to maintain disease control. In the short term, molecular diagnostics to rapidly detect resistant isolates will be useful. However, this study indicates that light leaf spot control with azole fungicides is likely to become difficult, and resistance breeding and alternative management techniques should be developed for the future control of P. brassicae.
Experimental Procedures
Collection of P. brassicae isolates
The P. brassicae isolates used in this study are listed in Table S1. Isolates were obtained by incubating infected Brassica species leaf material at 4 °C for 3–4 days. Acervuli were suspended in sterile water to obtain asexual spore suspensions. Aliquots of spore suspensions were spread onto 1% malt extract agar (MEA; Oxoid, Basingstoke, Hampshire, UK) plates amended with 100 mg/L penicillin and 200 mg/L streptomycin sulphate. Plates were incubated for 2–3 weeks at 15 °C. Single spore isolates were obtained by subculturing individual colonies onto new 1% MEA plates. Additional isolates were obtained from the oilseed rape genetic improvement network (OREGIN) isolate collection (Latunde‐Dada et al., 2006)
In vitro fungicide sensitivity testing
Fungicide sensitivity assays were modified from Fraaije et al. (2007). Flat‐bottomed microtitre plates (Costar, Corning, New York, NY, USA) were filled with aliquots of 2 × potato dextrose broth (PDB; Oxoid) amended with decreasing concentrations of technical grade prothioconazole (120, 40, 13.33, 4.44, 1.48, 0.494, 0.165, 0.055, 0.0183, 0.006 and 0.002 μg/mL), tebuconazole, flusilazole, metconazole (30, 10, 3.33, 1.11, 0.37, 0.12, 0.041, 0.014, 0.0046, 0.0015 and 0.00051 μg/mL) or prochloraz (2.5, 1.25, 0.625, 0.312, 0.156, 0.0782, 0.0390, 0.0195, 0.00977, 0.00488 and 0.00244 μg/mL) from stock solutions (10 mg/mL) in acetone. A 100‐μL aliquot of spore suspension (1 × 106 spores/mL) was added to each well. Plates were incubated at 15 °C for 11 days. Fungal growth was measured by absorbance at 630 nm using a FLUOstar OPTIMA microplate reader (BMG Labtech, Offenburg, Germany). Fungicide sensitivities for each isolate were calculated as 50% effective concentrations (EC50) using a dose–response relationship (four‐parameter fit).
DNA extraction and PbCYP51 sequencing
Macerated P. brassicae mycelium was added to 5‐cm Petri dishes containing 5 mL of PDB. After 21 days of growth at 15 °C, mycelia were collected by vacuum filtration, freeze–dried and ground using a FastPrep FP120 machine (Savants Instruments, Holbrook, NY, USA) at 4 m/s for 25 s. DNA was extracted as described previously by Fraaije et al., (1999). A 314‐bp fragment of PbCYP51 was amplified from azole‐sensitive isolate PbFr002 using primer pair F4/R4 (Table S2, see Supporting Information). PCR was carried out in a Biometra T3000 thermocycler (Biotron GmbH, Göttingen, Germany) in 30‐μL reaction volumes containing 60 ng genomic fungal template DNA, 0.5 μm of each primer, 200 μm of each deoxynucleoside triphosphate (dNTP), 1.5 mm MgCl2, 1 × reaction buffer and 1.5 units Red Hot DNA polymerase (ABgene, Epsom, Surrey, UK). The reaction conditions were as follows: 98 °C initial denaturation for 30 s, 40 cycles at 98 °C for 10 s, 55 °C for 30 s and 72 °C for 1 min, with a final extension at 72 °C for 5 min. The upstream PbCYP51 sequence was amplified with the Universal GenomeWalker kit (Clontech, Palo Alto, CA, USA). Genomic DNA was digested and ligated to GenomeWalker adaptors according to the manufacturer's protocol. Primary and secondary PCRs were carried out in 50‐μL reaction volumes containing 2.5 units Easy‐A High Fidelity PCR Cloning Enzyme (Stratagene, La Jolla, CA, USA), 1 × Easy‐A reaction buffer, 0.5 μm adapter primers (AP1/AP2, Stratagene), 0.5 μm gene‐specific primer (CYPgspUP/CYPnestedUP; Table S2), 200 μm of each dNTP and 1 μL of library DNA. The thermocycling parameters were as recommended by the manufacturer's protocol. The downstream PbCYP51 sequence was amplified using primers CYPgspDOWN/CYPnestedDOWN (Table S2). PCR was carried out using Red Hot DNA polymerase. The annealing temperature was 60 °C and the extension time was 1 min. PCR products were visualized by separation on ethidium bromide‐stained 1% (w/v) agarose gels run in 1 × Tris‐borate–ethylenediaminetetraacetic acid (EDTA) buffer, followed by exposure to UV light. PCR products were purified using the Wizard SV gel and PCR clean‐up system (Promega, Madison, WI, USA), according to the manufacturer's protocol. Purified PCR products were ligated to the pGEM‐Easy vector (Promega) and transformed into JM109 high efficiency competent Escherichia coli cells (Promega). Purified plasmid DNA was sequenced by MWG Eurofins Operon (Ebersberg, Germany) using the primers M13uni(‐21) and M13rev(‐29). The complete PbCYP51 was amplified from 17 isolates of different azole sensitivities using PbCYP51F and PbCYP51R (Table S2). PCRs were carried out using Phusion high‐fidelity DNA polymerase in 30‐μL reaction volumes. The annealing temperature was 56 °C and the extension time was 2 min. Purified PCR products were sequenced using CF1, CF2 and CR2 (Table S2). Sequences were assembled and translated in Vector NTI. The predicted protein sequences were aligned using MUSCLE (Edgar, 2004) and amino acid substitutions were noted.
PCR‐RFLP
A 1244‐bp PbCYP51 fragment encompassing PbCYP51 codons 460 and 508 was amplified using CYP51expressionF1/CYP51R (Table S2). PCR was carried out using Phusion high‐fidelity DNA polymerase. The annealing temperature was 55 °C and the extension time was 1.5 min. To detect G460S, approximately 10 ng of PCR product were digested with 2 units of TspRI (CASTG) and 1 × NE buffer 4 in a total volume of 10 μL at 65 °C for 2 h. For the detection of S508T, approximately 100 ng of PCR product were digested with 1 unit of BssSI (CACGAG) and 1 × NE buffer 3 in a total volume of 10 μL at 37 °C for 2 h. Restriction fragments were separated by electrophoresis on 1% (w/v) agarose gels.
RNA extraction and cDNA synthesis
Total RNA was extracted from freeze–dried mycelia of P. brassicae isolates with TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. A subsequent overnight incubation of extracts in 4 m lithium chloride was used to further purify RNA. Five micrograms of total RNA were reverse transcribed with oligo(dT)20 using the SuperScript III First Strand Synthesis System (Invitrogen), according to the supplier's instructions.
Heterologous expression of PbCYP51
The full‐length coding sequence of PbCYP51 was amplified from a 1:10 dilution of synthesized cDNA from isolate E2D using primers CYP51KpnI and CYP51EcoRI (Table S2), which incorporated KpnI and EcoRI restriction sites, respectively, to enable cloning in yeast expression vector pYES2/CT (Invitrogen). PCRs were carried out with EasyA High Fidelity PCR cloning enzyme at an annealing temperature of 55 °C and an extension time of 2 min. The purified PCR product was cloned into the pGEM‐Easy vector to create pGEM‐PbCYP51wt and an internal KpnI restriction site was removed using the QuikChange II site‐directed mutagenesis kit (Stratagene), according to the supplier's instructions. The pGEM‐PbCYP51 construct was digested with KpnI and EcoRI (New England Biolabs, Ipswich, MA, USA), and PbCYP51 was cloned into yeast expression vector pYES2/CT, creating plasmid pYES‐PbCYP51, which was transformed into S. cerevisiae strain YUG37:erg11 (MATa ura3‐52 trp1‐63 LEU2::tTA tetO‐CYC1::ERG11) (Revankar et al., 2004) using the S. cerevisiae EasyComp transformation kit (Invitrogen), according to the manufacturer's instructions. Transformants were plated out onto SD minimal medium containing 1.6 g/L dropout medium supplement without uracil (Sigma, St Louis, MO, USA), 2 g/L yeast nitrogen base without amino acids (Difco, Detroit, MI, US), 2% galactose and 2% raffinose with and without 3 μg/mL doxycycline.
Site‐directed mutagenesis
The PbCYP51 substitutions G460S and S508T were introduced alone and in combination into the pYES‐PbCYP51 plasmid by site‐directed mutagenesis with primers SDMG460S and SDMS508T using the QuikChange II site‐directed mutagenesis kit. All pYES‐PbCYP51 constructs were sequenced to confirm the correct introduction of mutations and to ensure the validity of sequences. Saccharomyces cerevisiae YUG37::ERG11 was transformed with pYES‐PbCYP51 constructs and grown on SD dropout medium as described previously.
Complementation analysis of S. cerevisiae YUG37:erg11 transformants
Saccharomyces cerevisiae YUG37:erg11 transformants were grown in 20 mL of liquid SD medium for 24 h with shaking at 250 rpm at 30 °C. Cell suspensions were adjusted to 1 × 106 cells/mL. Decreasing concentrations of cell suspensions (5 μL of five five‐fold dilutions) were inoculated onto SD GAL + RAF agar plates with and without 3 μg/mL doxycycline. The plates were incubated at 20 °C and 30 °C, and photographed after 72 h. The pYES2/CT vector was used as a negative control.
Fungicide sensitivity testing of S. cerevisiae YUG37:erg11 transformants
Saccharomyces cerevisiae YUG37:erg11 transformants were grown for 24 h with shaking at 250 rpm at 30 °C in 20 mL SD GAL + RAF liquid medium. Cell concentrations were adjusted to 1 × 106 cells/mL and 100‐μL aliquots were added to wells of flat‐bottomed microtitre plates. SD GAL + RAF medium with doxycycline (6 μg/mL) was amended with 0, 2.82 × 10−5, 8.47 × 10−5, 0.000254, 0.000762, 0.00229, 0.00686, 0.0206, 0.0617, 0.185, 0.556, 1.67 or 5 μg/mL flusilazole, tebuconazole, prothioconazole, metconazole, prochloraz and cycloheximide, and 100 μL were added to the wells of microtitre plates. The plates were incubated for 5 days at 20 °C. Fungicide sensitivities were determined as EC50 values for replicate transformants, and resistance factors were calculated as the ratio of the mean EC50 values to those of S. cerevisiae YUG37:erg11 transformants expressing WT PbCYP51.
Sequencing predicted PbCYP51 regulatory regions
Sequences immediately upstream of the predicted PbCYP51 coding sequence were amplified using the primer pair CYP51upstreamF/CYP51upstreamR in all P. brassicae isolates listed in Table S1 and in the progeny of crosses PbFr054 × C1E and PbFr054 × I3. PCR was carried out using Phusion high‐fidelity DNA polymerase. The annealing temperature was 57 °C and the extension time was 1 min. PCR products that differed from the PCR product size of the reference isolate PbFr002 were sequenced using CYP51upstreamR.
Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR)
Three‐millilitre aliquots of P. brassicae spore suspensions (1 × 106 spores/mL) were added to 6 mL of PDB in 55‐mm Petri dishes. After 4 days, tebuconazole was added to cultures at the approximate EC50 for each isolate (0.5 μg/mL for WC6, PbUK073, C1E and C5A, and 0.05 μg/mL for E2C and E2D). Cultures were incubated for a further 48 h before mycelia were harvested by centrifugation at 4 °C, snap frozen in liquid nitrogen and freeze–dried for 24 h. For each isolate, an untreated culture grown under the same conditions was also harvested. The experiment was repeated three times. Quantitative RT‐PCRs were carried out using SYBR Green Jumpstart Taq Ready Mix (Sigma). The 20‐μL reaction mix contained 5 μL of cDNA of a 1:10 dilution, 0.25 μm of each primer and 10 μL of 2 × SYBR Green Jumpstart Taq Ready mix (Sigma). The thermocycling parameters were 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 36 s. A melt curve was performed following thermocycling. Reactions were carried out in triplicate using the ABI 7500 real‐time PCR system (Applied Biosystems, Foster City, CA, USA). The endogenous controls were P. brassicae β‐tubulin (Table S2) and P. brassicae glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) (Singh, 1998). Data were analysed using the supplied 7500 SDS software. Mean relative transcript abundances of three biological replicates were calculated relative to the untreated control (E2C) using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Crossing of P. brassicae isolates
Mating types of P. brassicae isolates were determined using primers Mt3, PbM‐2 and PbM‐1–3 (Table S2; Foster et al., 2002). PCR was carried out using 1.25 U OneTaq DNA Polymerase (New England Biolabs), 1 × OneTaq Standard Reaction Buffer, 200 μm each dNTP, 0.1 μm PbM‐1‐3 and M2, and 0.2 μm Mt3. Thermocycling parameters were 94 °C initial denaturation for 30 s, 30 cycles at 94 °C for 30 s, 56 °C for 30 s and 68 °C for 1 min, with a final extension at 68 °C for 5 min. Sexual crosses were established by inoculating equal volumes (40 μL) of spore suspension (1 × 106 spores/mL) of isolates of opposite mating types onto 1% MEA plates. Plates were incubated in the dark at 15 °C for 12–20 weeks. After apothecia had formed, the Petri dish lids were filled with 1% MEA, the plates were inverted and individual germinating ascospores were subcultured. To confirm sexual recombination, progeny were genotyped using mating type, β‐tubulin and CYP51 sequence polymorphisms. Progeny were tested for in vitro sensitivity to carbendazim as described previously. Adjusted mean log EC50 values were determined using analysis of variance (ANOVA). Cumulative probability distributions for observed and expected progeny log EC50 values were generated by ranking progeny log EC50 values and dividing by the total number of observations. Expected distributions under the hypotheses that one, two or three independent and additive loci were segregating in the cross were generated using the corresponding Mendelian expectations for segregation in haploid crosses (1:1, 1:2:1, 1:3:3:1) and the observed standard error of the mean for an isolate EC50. One‐sample Kolmogorov–Smirnov tests were used to test the differences between the observed and expected cumulative probability distributions.
Supporting information
Fig. S1 Patterns of cross‐resistance.
Table S1 Characteristics of Pyrenopeziza brassicae isolates.
Table S2 Primers used in this study.
Table S3 Characteristics of parent isolates used in crossing experiments.
Acknowledgements
This work was carried out as part of a CASE PhD studentship sponsored by the Biotechnology and Biological Sciences Research Council (BBSRC) and Bayer CropScience. The authors would like to thank James Fountaine, Neil Havis, Peter Gladders, James Townsend, John Lucas and Olu Latunde‐Dada for providing infected leaf material/isolates. Rothamsted Research receives grant‐aided support from the BBSRC.
References
- Bolton, M.D. , Birla, K. , Rivera‐Varas, V. , Rudolph, K.D. and Secor, G.A. (2012) Characterization of CbCyp51 from field isolates of Cercospora beticola . Phytopathology, 102, 298–305. [DOI] [PubMed] [Google Scholar]
- Boys, E.F. , Roques, S.E. , Ashby, A.M. , Evans, N. , Latunde‐Dada, A.O. , Thomas, J.E. , West, J.S. and Fitt, B.D.L. (2007) Resistance to infection by stealth: Brassica napus (winter oilseed rape) and Pyrenopeziza brassicae (light leaf spot) in Europe. Eur. J. Plant Pathol. 118, 307–321. [Google Scholar]
- Burnett, F.J. (2003) Light Leaf Spot (Pyrenopeziza brassicae) in Oilseed Rape: Extent of Triazole Fungicide Resistance in Scotland; Fungicide Strategies , HGCA Project Report OS63e. Kenilworth, Warwickshire: Home Grown Cereals ; Authority. [Google Scholar]
- Cañas‐Gutiérrez, G.P. , Angarita‐Velásquez, M.J. , Restrepo‐Flórez, J.M. , Rodríguez, P. , Moreno, C.X. and Arango, R. (2009) Analysis of the CYP51 gene and encoded protein in propiconazole‐resistant isolates of Mycosphaerella fijiensis . Pest Manag. Sci. 65, 892–899. [DOI] [PubMed] [Google Scholar]
- Carter, H.E. , Cools, H.J. , West, J.S. , Shaw, M.W. and Fraaije, B.A. (2013) Detection and molecular characterisation of methyl benzimidazole carbamate resistant Pyrenopeziza brassicae isolates. Pest Manag. Sci. 69, 1040–1048. [DOI] [PubMed] [Google Scholar]
- Cools, H.J. and Fraaije, B.A. (2013) Mini‐review: update on mechanisms of azole resistance in Mycosphaerella graminicola and implications for future control. Pest Manag. Sci. 69, 150–155. [DOI] [PubMed] [Google Scholar]
- Cools, H.J. , Parker, J.E. , Kelly, D.E. , Lucas, J.A. , Fraaije, B.A. and Kelly, S.L. (2010) Heterologous expression of mutated eburicol 14α‐demethylase (CYP51) proteins of Mycosphaerella graminicola to assess effects on azole fungicide sensitivity and intrinsic protein function. Appl. Environ. Microbiol. 76, 2866–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools, H.J. , Mullins, J.G.L. , Fraaije, B.A. , Parker, J.E. , Kelly, D.E. , Lucas, J.A. and Kelly, S.L. (2011) Impact of recently emerged sterol 14α‐demethylase (CYP51) variants of Mycosphaerella graminicola on azole fungicide sensitivity. Appl. Environ. Microbiol. 77, 3830–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools, H.J. , Bayon, C. , Atkins, S. , Lucas, J.A. and Fraaije, B.A. (2012) Over‐expression of the sterol 14α‐demethylase gene (MgCYP51) in Mycosphaerella graminicola isolates confers a novel azole fungicide sensitivity phenotype. Pest Manag. Sci. 68, 1034–1040. [DOI] [PubMed] [Google Scholar]
- Crop Monitor (2012) Home Grown Cereals Authority (HGCA). Kenilworth, Warwickshire: HGCA. [Google Scholar]
- De Waard, M.A. , Andrade, A.C. , Hayashi, K. , Schoonbeek, H.J. , Stergiopoulos, I. and Zwiers, L.H. (2006) Impact of fungal drug transporters on fungicide sensitivity, multidrug resistance and virulence. Pest Manag. Sci. 62, 195–207. [DOI] [PubMed] [Google Scholar]
- Delye, C. , Laigret, F. and Corio‐Costet, M.F. (1997) A mutation in the sterol 14α‐demethylase gene in Uncinular necator that correlates with resistance to sterol biosynthesis inhibitors. Appl. Environ. Microbiol. 63, 2966–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, S.J. , Ashby, A.M. and Fitt, B.D.L. (2002) Improved PCR‐based assays for pre‐symptomatic diagnosis of light leaf spot and determination of mating type of Pyrenopeziza brassicae on winter oilseed rape. Eur. J. Plant Pathol. 108, 379–383. [Google Scholar]
- Fraaije, B.A. , Lovell, D.J. , Rohel, E.A. and Hollomon, D.W. (1999) Rapid detection and diagnosis of Septoria tritici epidemics in wheat using a polymerase chain reaction PicoGreen assay. J. Appl. Microbiol. 86, 701–708. [Google Scholar]
- Fraaije, B.A. , Cools, H.J. , Kim, S.‐H. , Motteram, J. , Clark, W.S. and Lucas, J.A. (2007) A novel substitution I381V in the sterol 14α‐demethylase (CYP51) of Mycosphaerella graminicola is differentially selected by azole fungicides. Mol. Plant Pathol. 8, 245–254. [DOI] [PubMed] [Google Scholar]
- Ghosoph, J.M. , Schmidt, L.S. , Margosan, D.A. and Smilanick, J.L. (2007) Imazalil resistance linked to a unique insertion sequence in the PdCYP51 promoter region of Penicillium digitatum . Postharvest Biol. Technol. 44, 9–18. [Google Scholar]
- Hamamoto, H. , Hasegawa, K. , Nakaune, R. , Lee, Y.J. , Makizumi, Y. , Akutsu, K. and Hibi, T. (2000) Tandem repeat of a transcriptional enhancer upstream of the sterol 14 alpha‐demethylase gene (CYP51) in Penicillium digitatum . Appl. Environ. Microbiol. 66, 3421–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Home Grown Cereals Authority (HGCA) (2003) Light Leaf Spot Control in Winter Oilseed Rape , Topic Sheet 75. Kenilworth, Warwickshire: Home Grown Cereals ; Authority. [Google Scholar]
- Hulvey, J. , Popko, J.T. , Sang, H. , Berg, A. and Jung, G. (2012) Overexpression of ShCYP51B and ShatrD in Sclerotinia homoeocarpa isolates exhibiting practical field resistance to a demethylation inhibitor fungicide. Appl. Environ. Microbiol. 78, 6674–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, S.L. , Lamb, D.C. , Loeffler, J. , Einsele, H. and Kelly, D.E. (1999) The G464S amino acid substitution in Candida albicans sterol 14 alpha‐demethylase causes fluconazole resistance in the clinic through reduced affinity. Biochem. Biophys. Res. Commun. 262, 174–179. [DOI] [PubMed] [Google Scholar]
- Kretschmer, M. , Leroch, M. , Mosbach, A. , Walker, A.‐S. , Fillinger, S. , Mernke, D. , Schoonbeek, H.‐J. , Pradier, J.‐M. , Leroux, P. , De Waard, M.A. and Hahn, M. (2009) Fungicide‐driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea . PLoS Pathol. 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latunde‐Dada, A.O. , Hornby, P.E. , Castells‐Brooke, N. , Evans, N. , Fitt, B.D.L. and King, G.J. (2006) OREGIN collection of oilseed rape fungal pathogen isolates managed by a relational database accessible to stakeholders via the Internet. IOBC/WPRS Bull. 29, 225–227. [Google Scholar]
- Latunde‐Dada, A.O. , West, J.S. , Measures, S. , Huang, Y.‐J. , Pirie, E. and Fitt, B.D.L. (2007) New methods to understand quantitative resistance to Leptosphaeria maculans and Pyrenopeziza brassicae in oilseed rape. In: Proceedings of the 12th International Rapeseed Congress: Sustainable Development in Cruciferous Oilseed Crops Production . Wuhan, China, March 26–30, p. 310 Princeton: Science Press, USA Inc. [Google Scholar]
- Leroux, P. and Walker, A.S. (2011) Multiple mechanisms account for resistance to sterol 14 alpha‐demethylation inhibitors in field isolates of Mycosphaerella graminicola . Pest Manag. Sci. 67, 44–59. [DOI] [PubMed] [Google Scholar]
- Leroux, P. , Albertini, C. , Gautier, A. , Gredt, M. and Walker, A.S. (2007) Mutations in the cyp51 gene correlated with changes in sensitivity to sterol 14α‐demethylation inhibitors in field isolates of Mycosphaerella graminicola . Pest Manag. Sci. 63, 688–699. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2–ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luo, C.X. and Schnabel, G. (2008) The cytochrome p450 lanosterol 14 alpha‐demethylase gene is a demethylation inhibitor fungicide resistance determinant in Monilinia fructicola field isolates from Georgia. Appl. Environ. Microbiol. 74, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morio, F. , Loge, C. , Besse, B. , Hennequin, C. and Le Pape, P. (2010) Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole‐susceptible and azole‐resistant clinical isolates: new substitutions and a review of the literature. Diagn. Microbiol. Infect. Dis. 66, 373–384. [DOI] [PubMed] [Google Scholar]
- Nikou, D. , Malandrakis, A. , Konstantakaki, M. , Vontas, J. , Markoglou, A. and Ziogas, B. (2009) Molecular characterization and detection of overexpressed C‐14 alpha‐demethylase‐based DMI resistance in Cercospora beticola field isolates. Pestic. Biochem. Physiol. 95, 18–27. [Google Scholar]
- Parks, L. , Smith, S. and Crowley, J. (1995) Biochemical and physiological effects of sterol alterations in yeast—a review. Lipids, 30, 227–230. [DOI] [PubMed] [Google Scholar]
- Prasad, R. , Dewergifosse, P. , Goffeau, A. and Balzi, E. (1995) Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27, 320–329. [DOI] [PubMed] [Google Scholar]
- Revankar, S.G. , Fu, J. , Rinaldi, M.G. , Kelly, S.L. , Kelly, D.E. , Lamb, D.C. , Keller, S.M. and Wickes, B.L. (2004) Cloning and characterization of the lanosterol 14α‐demethylase (ERG11) gene in Cryptococcus neoformans . Biochem. Biophys. Res. Commun. 324, 719–728. [DOI] [PubMed] [Google Scholar]
- Sanglard, D. , Ischer, F. , Monod, M. and Bille, J. (1997) Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology, 143, 405–416. [DOI] [PubMed] [Google Scholar]
- Sanglard, D. , Ischer, F. , Koymans, L. and Bille, J. (1998) Amino acid substitutions in the cytochrome P‐450 lanosterol 14α‐demethylase (CYP51A1) from azole‐resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42, 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse, C. , Dunkel, N. , Schafer, T. , Schneider, S. , Dierolf, F. , Ohlsen, K. and Morschhauser, J. (2012) The stepwise acquisition of fluconazole resistance mutations causes a gradual loss of fitness in Candida albicans . Mol. Microbiol. 86, 539–556. [DOI] [PubMed] [Google Scholar]
- Sheehan, D.J. , Hitchcock, C.A. and Sibley, C.M. (1999) Current and emerging azole antifungal agents. Clin. Microbiol. Rev. 12, 40–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, G. (1998) Molecular analysis of sexual morphogenesis in the light leaf spot pathogen, Pyrenopeziza brassicae . PhD Thesis, University of Cambridge.
- Stammler, G. , Cordero, J. , Koch, A. , Semar, M. and Schlehuber, S. (2009) Role of the Y134F mutation in cyp51 and overexpression of cyp51 in the sensitivity response of Puccinia triticina to epoxiconazole. Crop Prot. 28, 891–897. [Google Scholar]
- Sun, X. , Xu, Q. , Ruan, R. , Zhang, T. , Zhu, C. and Li, H. (2013) PdMLE1, a specific and active transposon acts as a promoter and confers Penicillium digitatum with DMI resistance. Environ. Microbiol. Rep. 5, 135–142. [DOI] [PubMed] [Google Scholar]
- Sun, X.P. , Wang, J.Y. , Feng, D. , Ma, Z.H. and Li, H.Y. (2011) PdCYP51B, a new putative sterol 14 alpha‐demethylase gene of Penicillium digitatum involved in resistance to imazalil and other fungicides inhibiting ergosterol synthesis. Appl. Microbiol. Biotechnol. 91, 1107–1119. [DOI] [PubMed] [Google Scholar]
- Sutherland, K.G. , Griffin‐Walker, V. and Oxley, S.J.P. (1994) Brighton Crop Protection Conference: Pests and Diseases. Alton, Hants: British Crop Protection Council. [Google Scholar]
- Weete, J.D. , Abril, M. and Blackwell, M. (2010) Phylogenetic distribution of fungal sterols. Plos ONE, 5, e10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welham, S.J. , Turner, J.A. , Gladders, P. , Fitt, B.D.L. , Evans, N. and Baierl, A. (2004) Predicting light leaf spot (Pyrenopeziza brassicae) risk on winter oilseed rape (Brassica napus) in England and Wales, using survey, weather and crop information. Plant Pathol. 53, 713–724. [Google Scholar]
- Wyand, R.A. and Brown, J.K.M. (2005) Sequence variation in the CYP51 gene of Blumeria graminis associated with resistance to sterol demethylase inhibiting fungicides. Fungal Genet. Biol. 42, 726–735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Patterns of cross‐resistance.
Table S1 Characteristics of Pyrenopeziza brassicae isolates.
Table S2 Primers used in this study.
Table S3 Characteristics of parent isolates used in crossing experiments.


