Summary
Viruses commonly modify host endomembranes to facilitate biological processes in the viral life cycle. Infection by viruses belonging to the genus Bymovirus (family Potyviridae) has long been known to induce the formation of large membranous inclusion bodies in host cells, but their assembly and biological roles are still unclear. Immunoelectron microscopy of cells infected with the bymovirus Wheat yellow mosaic virus (WYMV) showed that P1, P2 and P3 are the major viral protein constituents of the membranous inclusions, whereas NIa‐Pro (nuclear inclusion‐a protease) and VPg (viral protein genome‐linked) are probable minor components. P1, P2 and P3 associated with the endoplasmic reticulum (ER), but only P2 was able to rearrange ER and form large aggregate structures. Bioinformatic analyses and chemical experiments showed that P2 is an integral membrane protein and depends on the active secretory pathway to form aggregates of ER membranes. In planta and in vitro assays demonstrated that P2 interacts with P1, P3, NIa‐Pro or VPg and recruits these proteins into the aggregates. In vivo RNA labelling using WYMV‐infected wheat protoplasts showed that the synthesis of viral RNAs occurs in the P2‐associated inclusions. Our results suggest that P2 plays a major role in the formation of membranous compartments that house the genomic replication of WYMV.
Keywords: endoplasmic reticulum, protein localization, rearrange, replication‐associated inclusion bodies, Wheat yellow mosaic virus
Introduction
Virus infections often induce modifications in their host cells. These may include the formation of aggregate or inclusion structures, changes in organelle architecture and alterations to endomembranes. These cellular modifications are usually mediated by viral‐encoded proteins, but, in some cases, they are associated with cellular responses or defences against virus infection (Moshe and Gorovits, 2012). Positive‐strand RNA viruses commonly utilize and modify cell endomembranes to facilitate viral genome replication. Through the activity of viral membrane‐associated proteins, positive‐strand RNA viruses rearrange endoplasmic reticulum (ER) or, depending on the virus, modify peripheral membranes of chloroplasts, mitochondria, endosomes, peroxisomes or vacuoles, to form a variety of membranous compartments, such as inclusions, spherules, vesicles and multivesicular bodies, which serve as viral replication sites and protect the viral genome from host immune responses (den Boon et al., 2010; Laliberte and Sanfacon, 2010; Miller and Krijnse‐Locker, 2008; Verchot, 2011). Moreover, there is growing evidence indicating that many movement proteins of plant viruses are intimately linked with ER or ER‐derived structures (Harries et al., 2010). Thus, ER modification may also be important for intra‐ or intercellular movement of some viruses.
Potyviridae is one of the largest families of plant viruses, many of which cause economically important crop diseases (Adams et al., 2011). Viruses belonging to the family have positive single‐stranded RNA genomes that are encapsidated in flexible filamentous virions. Genome organization is well conserved in the family, but several genera have been recognized on the basis of sequence similarity and the vectors that transmit the respective viruses (Adams et al., 2011). Most viruses in the family have monopartite genomes that encode a single polyprotein, but those in the genus Bymovirus have bipartite genomes. Wheat yellow mosaic virus (WYMV) is a member of the genus Bymovirus (Namba et al., 1998) and is responsible for winter wheat yellow mosaic disease outbreaks in China and Japan (Han et al., 2000; Kühne, 2009). The single polyprotein encoded on RNA1 of bymoviruses is clearly homologous with the central and C‐terminal regions of the polyproteins encoded by monopartite members of the family. WYMV RNA1 (∼7.6 kb) encodes a large polyprotein that is proteolytically processed to generate eight mature proteins, namely P3, 7K, CI (cylindrical inclusion), 14K, VPg (viral protein genome‐linked), NIa‐Pro (nuclear inclusion‐a protease), NIb and CP (coat protein), whereas RNA2 (∼3.5 kb) encodes a polyprotein that is cleaved into two proteins, P1 and P2 (Namba et al., 1998) (see also Fig. 2A). In addition, the presence of a small open reading frame (ORF), termed pipo, which overlaps with the P3 coding region, has been predicted in the WYMV genome (Chung et al., 2008). Detailed functional analysis of these WYMV‐encoded proteins has not been reported, but the functions of many can be inferred from those of their well‐characterized homologues in other members of the family. Bymoviruses are transmitted to roots by a soil‐inhabiting fungal‐like organism, Polymyxa graminis (Plasmodiophorales) (Tamada and Kondo, 2013), whereas the monopartite viruses within the family Potyviridae are transmitted by arthropod vectors (aphids, mites and whiteflies). Helper component protease (HC‐Pro) is required for virus transmission by arthropod vectors (Stenger et al., 2005; Urcuqui‐Inchima et al., 2001; Young et al., 2007), but is absent from bymoviruses, whereas P2 is unique to bymoviruses and is thought to be essential for bymovirus transmission by P. graminis (Adams et al., 2001; Dessens and Meyer, 1996). In the monopartite genera of the family Potyviridae, HC‐Pro and P1 (not homologous to the P1 encoded on the bymovirus RNA2) function as RNA silencing suppressors (Anandalakshmi et al., 1998; Kasschau and Carrington, 1998; Tatineni et al., 2012; Valli et al., 2006; Young et al., 2012). Similar to VPg encoded by viruses belonging to the genus Potyvirus (family Potyviridae), WYMV VPg also targets the nucleus. Moreover, WYMV VPg nucleocytoplasmic shuttling is regulated by both nuclear localization and nuclear export signals, and requires association with the CP (Sun et al., 2013b).
Figure 2.
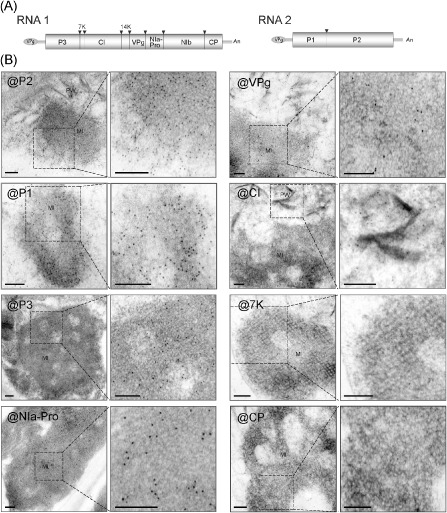
Viral protein constituents of membranous inclusion structures in Wheat yellow mosaic virus (WYMV)‐infected cells. (A) Schematic presentation of the WYMV genome. Boxes represent the open reading frame of the polyproteins that are cleaved by viral proteases into 10 functional proteins. Black triangles indicate proteinase cleavage sites. (B) Electron micrographs showing immunogold labelling of inclusion structures. Ultrathin sections were prepared from leaves of WYMV‐infected wheat and subjected to immunogold labelling using WYMV P1, P2, P3, NIa‐Pro, VPg, CI, 7K and CP antisera; 10 nm gold particle‐conjugated goat antibody against rabbit immunoglobulin G (IgG) was used as the secondary antiserum. The close‐up views of the dashed rectangular areas are also presented (right images). Bars, 0.2 μm.
The cytopathological effects of potyviruses have been widely studied. It has long been known that potyviruses induce morphologically varied types of intracellular inclusion structures in infected cells, including cylindrical inclusions (pinwheels or scrolls), nuclear inclusions, laminated aggregates and proliferated ER (Andrews and Shalla, 1974; Baunoch et al., 1988, 1990; Edwardson, 1966; Edwardson and Christie, 1983; Otulak and Garbaczewska, 2012). Although the biological significance of most of these structures remains unclear, they provide useful diagnostic criteria for some potyviruses (Edwardson, 1966; Edwardson and Christie, 1983; Edwardson et al., 1984). Membranous inclusion bodies have been reported in plant cells infected with bymoviruses (Hibino et al., 1981; Hooper and Wiese, 1972; Huth et al., 1984; Langenberg, 1985), but it is not clear how viruses induce the formation of these structures. In this study, we used immunoelectron microscopy and in planta transient expression of viral proteins in Nicotiana benthamiana to analyse the viral protein constituents and the formation of membrane‐derived inclusions induced by WYMV infection. Our data demonstrate that the membrane‐derived inclusions contain multiple viral proteins and that WYMV P2 probably plays a key role in the formation of these inclusions through its ability to associate with and rearrange ER membranes. Moreover, P2 interacts with and recruits other viral proteins into these inclusions. Finally, we demonstrate the association of the inclusion bodies with WYMV RNA synthesis.
Results
Formation of membranous inclusion bodies in WYMV‐infected cells
Ultrathin sections prepared from symptomatic leaves of WYMV‐infected wheat were examined under an electron microscope. In most of the cells examined, amorphous, crystalline lattice‐like inclusion bodies of various sizes were observed in the cytoplasm (Fig. 1A,B, MI). In some cells, these inclusion bodies were clustered to form large complexes (Fig. 1A). The periphery of the inclusion bodies appeared to be connected with the rough ER (Fig. 1A,B, ER) and membranes were visible in both the interior and exterior regions of these inclusion structures that appeared less dense (Fig. 1C, ER). Sometimes, large inclusion bodies with highly condensed and well‐ordered crystal‐like arrays were observed (Fig. 1D). This pattern may indicate that the structure is in the late stage of inclusion formation. No such inclusions were observed in non‐infected samples (data not shown). These observations suggest that the inclusion bodies may emerge from the rearrangement of ER membranes induced by WYMV infection. The morphology of these membrane‐derived inclusions was similar to those observed previously in cells infected with other bymoviruses (Hooper and Wiese, 1972; Huth et al., 1984; Langenberg, 1985). WYMV particles were not seen inside the membrane‐derived inclusions, but were sometimes observed to aggregate as filamentous bundles in the cytoplasm (Fig. 1A, V). In addition, pinwheel inclusions, similar to those typically seen in potyvirus infections, were also observed (Fig. 1A, PW). No inclusion structures could be recognized within the nuclei of infected cells.
Figure 1.
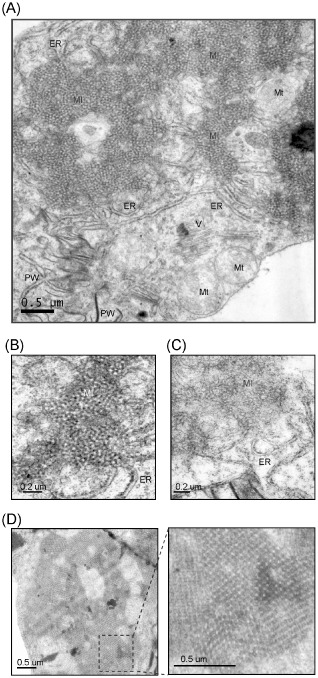
Electron micrographs of the mesophyll cells of Wheat yellow mosaic virus‐infected wheat. Ultrathin sections were stained with uranyl acetate and lead citrate. (A) The presence of membranous inclusion structures in the cytoplasm. ER, endoplasmic reticulum; MI, membranous inclusion; Mt, mitochondria; PW, pinwheel inclusion; V, virus particles. (B, C) More detailed views of membranous inclusion structures showing the connection with the ER at the periphery. (D) An inclusion structure with a highly condensed and well‐ordered crystal‐like array. A close‐up view of the dashed rectangular area is also presented (right image).
WYMV‐induced membranous inclusion bodies contain multiple viral proteins
To determine the viral protein constituents of membrane‐derived inclusions by immunogold labelling, we first attempted to produce antisera to each of the 10 mature proteins encoded by WYMV after their expression in Escherichia coli. Antisera specific for P1, P2, P3, 7K, CI, VPg, NIa‐Pro and CP were obtained and their specificities were confirmed in Western blot assays using protein samples from WYMV‐infected wheat plants (data not shown and see also Figs 3A and 6C). Unfortunately, preparations of 14K and NIb antisera were unsuccessful owing to very low expression of these proteins in E. coli. In Western blotting using WYMV‐infected wheat plants, P3 antiserum reacted with two closely migrating bands (data not shown and see also Fig. 6C). Owing to the unavailability of a specific antiserum, we were unable to confirm that the smaller band is the WYMV P3N‐PIPO protein, as predicted previously (Chung et al., 2008). The estimated molecular weight of WYMV P3N‐PIPO is around 30 kDa, whereas P3 is 36 kDa; therefore, it is possible that the smaller band is P3N‐PIPO. Ultrathin sections were prepared from leaf tissue of WYMV‐infected wheat plants and used for immunogold labelling with the antisera prepared. P1, P2, P3, VPg and NIa‐Pro antisera, but not 7K, CI and CP antisera, labelled the inclusion structures (Fig. 2B). There was much more labelling with P1, P2 and P3 antisera than with VPg and NIa‐Pro antisera (Fig. 2B), suggesting that P1, P2 and P3 may be the major viral protein constituents of these inclusions. However, it is also possible that the reactivity of P1, P2 and P3 antisera is higher than that of VPg and NIa‐Pro antisera. The CI antiserum labelled pinwheel inclusions (Fig. 2B, @CI), consistent with previous reports that the pinwheel inclusions of potyviruses are associated with CI protein accumulation (Arbatova et al., 1998; Hammond, 1998; Rouis et al., 2002).
Figure 3.
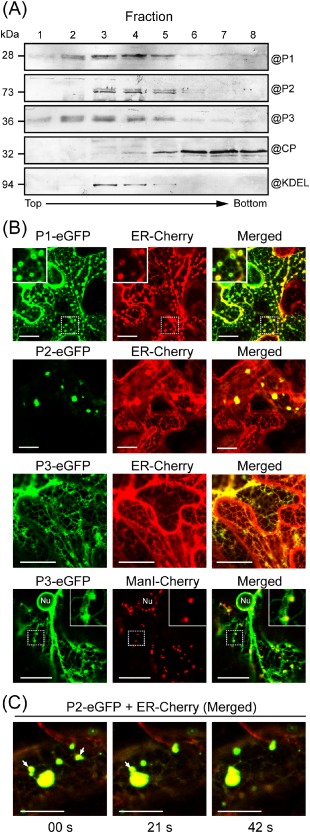
Association of Wheat yellow mosaic virus (WYMV) P1, P2 and P3 with the endoplasmic reticulum (ER). (A) Sucrose gradient fractions of crude membrane extracts from leaves of WYMV‐infected wheat. Fractions were analysed by Western blotting using WYMV P1, P2, P3, CP and KDEL antisera. (B) Epidermal cells of Nicotiana benthamiana transiently co‐expressing P1‐, P2‐ or P3‐enhanced green fluorescent protein (eGFP) and endoplasmic reticulum (ER) luminal marker (ER‐Cherry), or P3‐eGFP and Golgi marker (ManI‐Cherry). Images were derived from a single confocal section. The insets in the top corner are close‐up views of the dashed rectangular areas. Nu, nucleus. (C) Time‐lapse imaging (consecutive images every 21 s) of epidermal cells co‐expressing P2‐eGFP and ER‐Cherry. eGFP and Cherry fluorescent signals were merged. The mobile inclusions are marked with arrows. Fluorescent proteins were observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 20 μm.
Figure 6.
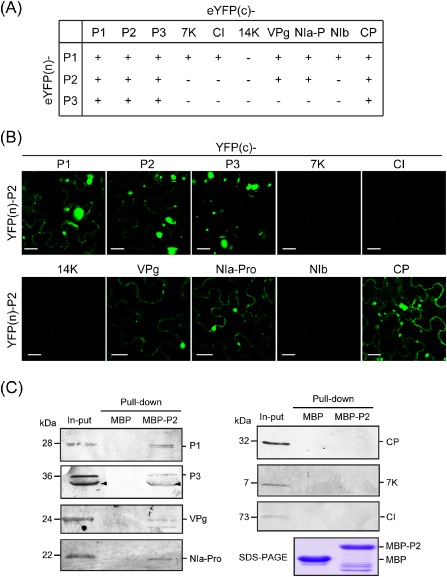
Interaction of Wheat yellow mosaic virus (WYMV) P2 with other viral proteins. (A) A summary of the results of the bimolecular fluorescence complementation (BiFC) assay to examine the interaction of WYMV P1, P2 or P3 with other viral proteins. ‘+’ and ‘–’ indicate interaction and no interaction, respectively. (B) Interaction of WYMV P2 with other viral proteins in BiFC assay. Leaves of Nicotiana benthamiana were infiltrated with mixtures of agrobacteria harbouring the constructs indicated above and on the left side of the images. Yellow fluorescent protein (YFP) fluorescence was observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 20 μm. (C) Interaction of WYMV P2 with other viral proteins in pull‐down assay. Maltose binding protein (MBP)‐P2 and unfused MBP [lower panel, sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE)] were used to pull‐down total proteins extracted from leaves of WYMV‐infected wheat. Pull‐down samples were analysed by Western blotting using P1, P3, NIa‐Pro, VPg, CI, 7K and CP antisera. Arrowheads mark antiserum‐reactive bands possibly from the P3N‐PIPO protein.
WYMV P2 rearranges ER membranes into aggregate structures
Because P1, P2 and P3 seem to be the major viral protein constituents of membrane‐derived inclusion bodies in WYMV‐infected cells (Fig. 2B), we analysed the membrane association of these proteins. Preliminary subcellular fractionation experiments using protein extracts from WYMV‐infected leaves showed that P1, P2 and P3 associated with the crude membrane‐containing fraction (data not shown). The crude membrane fraction was then separated by ultracentrifugation in a continuous sucrose gradient. The gradient was roughly divided into eight fractions and subjected to Western blot analysis using WYMV P1, P2, P3 and CP antisera. KDEL antiserum was used as an ER marker. Western blotting showed peak levels of P2 in fractions 3−5, which coincided with the distribution of the ER marker in the gradient (Fig. 3A). P1 and P3 showed a broader distribution pattern, but were generally associated with the upper portions of the gradient (Fig. 3A). This suggests that P2 associates with ER, and that P1 and P3 may associate with several types of membranous structure in the cell. In contrast, CP was predominantly found in the lower portions of the gradient (Fig. 3A), suggesting that CP does not associate with the membranous structures.
To further examine their subcellular localization in planta, P1, P2 and P3 were fused to the N‐terminus of enhanced green fluorescent protein (P1‐, P2‐ and P3‐eGFP) and transiently expressed in the leaves of N. benthamiana by agroinfiltration. The expression of eGFP in epidermal cells was observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Because WYMV usually infects plants at cool temperatures (Kühne, 2009; Ohto and Naito, 1997), plants were kept at 16 °C after agroinfiltration to simulate natural conditions during WYMV infection. To visualize the association with ER, each of the fusion proteins was simultaneously expressed with an ER luminal marker (ER‐Cherry). P1‐eGFP labelled the tubular ER network, but more strongly accumulated in numerous ring‐like structures located at the vertices of the connecting ER tubules (Fig. 3B). These ring‐like structures were not observed when ER‐red fluorescent protein (ER‐RFP) was expressed alone (data not shown), suggesting that P1 expression mediates the proliferation of ER vertices. P2‐eGFP predominantly localized in numerous large irregular shaped or sometimes small punctate aggregate structures in the cytoplasm. Interestingly, P2‐eGFP overlapped with the ER marker at these structures (Fig. 3B). Consecutive optical cross‐sections (thickness, 2 μm) of the large aggregate structures showed that the red fluorescence consistently overlapped with the green fluorescence (data not shown), showing that these structures are derived from ER membranes. Notably, cortical ER networks were much less visible in the areas surrounding aggregates, suggesting that P2 rearranges tubular ER into aggregates. Time‐lapse imaging demonstrated that some of the aggregates were mobile and merged with other aggregates (Fig. 3C). P3‐eGFP accumulated in the perinuclear and cortical ER and also localized in small mobile vesicular bodies. When P3‐eGFP was co‐expressed with a Golgi‐specific marker (ManI‐Cherry), these small vesicular bodies overlapped with Golgi stacks (Fig. 3B). Thus, unlike P1 and P2, P3 targets both the ER and the Golgi apparatus. Immunogold labelling showed that NIa‐Pro and VPg are also protein constituents of WYMV‐induced membranous inclusions (Fig. 2B). In epidermal cells of N. benthamiana, VPg fused to the N‐terminus of eGFP (VPg‐eGFP) localized exclusively in the nucleus; NIa‐Pro fused to the N‐terminus of eGFP (NIa‐Pro‐eGFP) showed no particular subcellular localization, but was diffusely distributed throughout both the cytoplasm and nucleus; 7K fused to eGFP (7K‐eGFP) localized to the cortical ER network (Fig. S1, see Supporting Information). Thus, among these proteins, only P2 exhibits the ability to rearrange ER into aggregate structures. This may imply that P2 is a key factor for the formation of ER‐derived inclusion structures observed in WYMV‐infected cells.
WYMV P2 and P3 are integral membrane proteins
Because of their association with membranes, the amino acid sequences of P1, P2 and P3 were analysed for the presence of hydrophobic regions. Analysis using the Wimley and White octanol hydrophobicity scale (Wimley and White, 1996) showed that multiple hydrophobic regions exist in the P1, P2 and P3 sequences [Fig. 4A for P2 and Fig. S2 (see Supporting Information) for P1 and P3]. To determine whether P1, P2 and P3 are integral or peripheral membrane proteins, we performed cell fractionation and chemical treatments using protein extracts from the leaf tissue expressing P1‐, P2‐ or P3‐eGFP. Western blotting using a GFP‐specific antiserum showed that P1‐, P2‐ and P3‐eGFP associated with the crude membrane fractions (P30), whereas unfused eGFP associated with the soluble fraction (S30) (Fig. 4C and data not shown for unfused eGFP). Treatment of P30 with 1% Triton X‐100, which solubilizes membrane proteins, resulted in the association of all three fusion proteins with S30. When P30 was treated with 0.1 m Na2CO3 or 4 m urea, which dislodge proteins that are weakly or peripherally associated with membranes, P1 shifted to the soluble fraction, whereas most of P2 and P3 remained in the membrane fraction, although a small amount of P2 became associated with S30 (Fig. 4C). This result suggests that P2 and P3, but not P1, are integral membrane proteins. Because small amounts of P2 were released into P30 by treatment with 0.1 m Na2CO3 or 4 m urea, it is possible that some portions of P2 are peripherally attached onto the membranes. Transmembrane protein prediction programs (TMpred and TopPred II) similarly predicted with high certainties the presence of transmembrane domains in hydrophobic regions of both P2 and P3 (Fig. 4B for P2 and Fig. S2 for P3). Thus, both chemical and computational analyses support the conclusion that P2 and P3 are integral membrane proteins.
Figure 4.
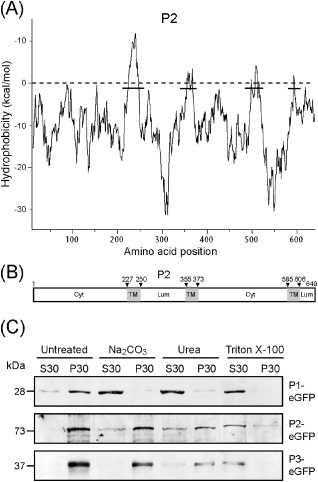
Membrane integration of Wheat yellow mosaic virus P1, P2 and P3. (A) Hydrophobic plot of P2 generated with MPEx according to the Wimley and White octanol hydrophobicity scale. The plot shows the mean values using a window of 19 residues. Lines indicate the predicted hydrophobic regions. (B) A schematic map showing the position of three putative transmembrane domains (TM, grey boxes) in P2 as predicted by TMpred. Arrowheads indicate the amino acid positions of transmembrane domains. Cyt and Lum indicate the cytoplasmic and luminal domains, respectively. (C) Subcellular fractionation and chemical treatments of protein extracts from Nicotiana benthamiana leaves expressing P1‐, P2‐ or P3‐enhanced green fluorescent protein (eGFP). Ultracentrifugation was used to separate soluble (S30) and crude (P30) membrane‐containing fractions. P30 was subjected to chemical treatments and separated again into S30 and P30. Equal amounts of S30 and P30 were analysed by Western blotting using a GFP‐specific antiserum.
Rearrangement of ER membranes by WYMV P2 requires an active secretory pathway
The early secretory pathway has been implicated in the intracellular transport of membrane‐associated proteins that are encoded by various viruses (Andika et al., 2013; Genovés et al., 2010; Laporte et al., 2003; Ribeiro et al., 2009; Vogel et al., 2007; Wei and Wang, 2008). Using chemical and protein inhibitors, we investigated the possible involvement of the secretory pathway in the subcellular localization of P2. Brefeldin A (BFA) is a fungal metabolite that inhibits vesicle trafficking along the secretory pathway (Nebenführ et al., 2002), whereas Sar1[H74L] is a dominant‐negative mutant of the Ras‐like small GTPase (Sar1p) which specifically inhibits coat protein complex II (COPII)‐mediated ER to Golgi transport (Robinson et al., 2007). In leaves expressing P2‐eGFP infiltrated with BFA solution, P2‐eGFP accumulated in the cortical ER network and no aggregate structures were observed, in contrast with the controls infiltrated with buffer plus dimethylsulphoxide (DMSO), where P2‐eGFP localized in aggregate structures (Fig. 5A). Similarly, co‐expression with the Sar1[H74L] mutant led to the accumulation of P2‐eGFP in the ER network and inhibited the formation of aggregate structures (Fig. 5A). These results indicate that the activity of P2 to rearrange ER membranes depends on the active secretory pathway. Because P3 associates with the Golgi apparatus (Fig. 3B), a similar experiment was also performed with P3‐eGFP. The results showed that treatment with BFA or co‐expression with the Sar1[H74L] mutant inhibited the localization of P3‐eGFP to the Golgi stack and resulted in saturated accumulation of P3‐eGFP in the cortical ER network (Fig. 5A), indicating that the transport of P3 from the ER to the Golgi involves the active secretory pathway.
Figure 5.
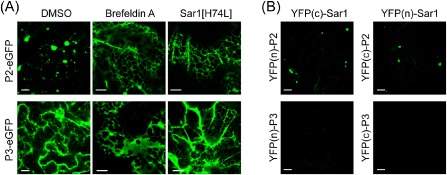
A role for the secretory pathway in the subcellular localization of Wheat yellow mosaic virus P2 and P3. (A) Nicotiana benthamiana leaves expressing P2‐ or P3‐enhanced green fluorescent protein (eGFP) were infiltrated with 10 mg/mL brefeldin A solution, or P2‐ or P3‐eGFP was co‐expressed with Sar1[H74L] in leaves. (B) Bimolecular fluorescence complementation (BiFC) assay to examine the interaction of P2 or P3 with Sar1. Leaves of N. benthamiana plants were infiltrated with mixtures of agrobacteria harbouring the constructs indicated above and on the left side of the images. Fluorescent proteins were observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 10 μm.
Sar1 is one of the components of the COPII transport vesicles that deliver cargo proteins from the ER into their final intracellular destination. Interaction of Sar1 with cargo proteins has been reported in several studies (Barlowe, 2003). We assessed the possibility of the interaction of P2 with Sar1 using a bimolecular fluorescence complementation (BiFC) assay (Hu and Kerppola, 2003). P2 and Sar1 were fused to the N‐terminal or C‐terminal portions of yellow fluorescent protein [YFP(n) or YFP(c)] which had been inserted into the expression cassette of a binary vector. The resulting plasmids were transformed into Agrobacterium and used to co‐infiltrate N. benthamiana leaves. The reconstruction of YFP fluorescence, indicating a protein–protein interaction, was visualized using CLSM at 3 dai. Bright YFP fluorescence was observed in leaves expressing either YFP(n)‐P2 and YFP(c)‐Sar1 or YFP(n)‐Sar1 and YFP(c)‐P2 (Fig. 5B), showing that P2 and Sar1 interact. YFP fluorescence reconstituted by P2 and Sar1 interaction was localized in the aggregate structures in the cytoplasm, showing that the interaction occurs in the aggregate membranes induced by P2. By contrast, no interaction between P3 and Sar1 was observed in the BiFC assay (Fig. 5B). In control experiments, no fluorescence was detected when all fusion constructs were co‐expressed with unfused YFP(n/c) constructs (data not shown).
WYMV P2 recruits other viral proteins into aggregates of ER membranes
The accumulation of multiple WYMV‐encoded proteins in the ER‐derived inclusion bodies may be mediated by interactions among these proteins. First, we employed a BiFC assay to assess the interaction of P1, P2 and P3 with other viral proteins. The results of the assay are summarized in Fig. 6A. P1 interacted with itself and with most of the WYMV‐encoded proteins, except 14K. P2 also interacted with itself and P1, P3, VPg, NIa‐Pro and CP, whereas P3 interacted only with itself, P1, P2 and CP (Figs 6B and S3, see Supporting Information). It is important to note that P1, P2 and P3 interacted with themselves and also with each other, suggesting that an interconnected P1, P2 and P3 heterocomplex might be essential for the formation of the intact ER‐derived inclusion structures observed in WYMV‐infected cells.
In the next experiment, the interaction of P2 with other viral proteins was investigated by pull‐down assay. Maltose binding protein (MBP)‐tagged P2 fusion (MBP‐P2) and unfused MBP proteins were expressed in E. coli (Fig. 6C, bottom panel), and used as bait proteins in a pull‐down assay with a total protein extract from leaves of WYMV‐infected wheat. After pull down, protein samples were subjected to Western blotting using antisera specific to the WYMV‐encoded proteins. P1, P3, VPg and NIa‐Pro, but not CP, 7K and CI, were detected in the protein complexes obtained from the MBP‐P2 pull‐down sample, whereas no viral protein was detected in unfused MBP pull‐down samples (Fig. 6C). P2 and CP interacted in the BiFC assay but did not in the pull‐down assay, but, in other respects, the results of the two assays were in agreement. As P2, P1, P3, VPg and NIa‐Pro, but not CP, were detected in the inclusions (Fig. 2), the results of the pull‐down assay probably reflect the protein constituents of the inclusion bodies.
The YFP fluorescence reconstituted by interaction of P2 with other viral proteins predominantly localized in the aggregate structures (Fig. 6B), suggesting that P2 may recruit these proteins into the aggregates of membranes. To further assess this concept, we carried out co‐expression experiments in which YFP(n)‐P2 and YFP(c)‐P2 were simultaneously expressed with P1‐, P3‐, VPg‐ or NIa‐Pro‐eGFP in N. benthamiana leaves. CLSM observation showed that P1‐, P3‐, VPg‐ and NIa‐Pro‐eGFP primarily localized in the P2‐induced inclusion structures that were visualized through eYFP fluorescence reconstructed by P2 self‐interaction (Fig. 7A). This distribution pattern was substantially different when P1‐, P3‐, VPg‐ or NIa‐Pro‐eGFP was expressed alone, indicating that P2 redirects these proteins into the aggregate membranes by a direct interaction. NIb is the replicase protein of potyviruses (Urcuqui‐Inchima et al., 2001). In BiFC assay, P2 did not interact with NIb (Fig. 6B). Consistent with this, NIb‐eGFP did not co‐localize with the P2‐induced aggregates in the co‐expression experiment (Fig. 7A). Because NIb interacts with P1, it is possible that NIb can be recruited into P2‐induced aggregates through its association with P1. To examine this possibility, NIb‐eGFP was co‐expressed with YFP(n)‐P1 and YFP(c)‐P2. As expected, some portions of NIb‐eGFP co‐localized with the aggregate structures formed by the P1 and P2 interaction (Fig. 7B).
Figure 7.
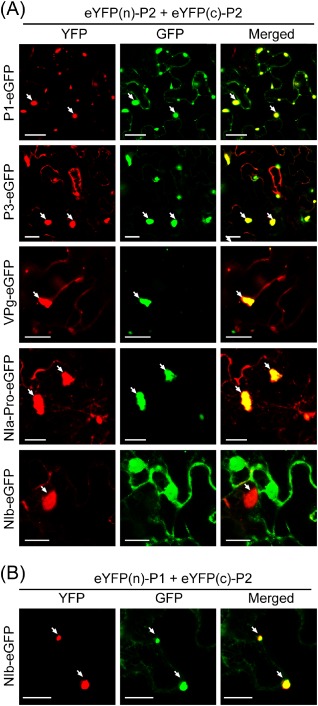
The ability of Wheat yellow mosaic virus P2 to recruit other viral proteins into aggregate structures. (A, B) Epidermal cells of Nicotiana benthamiana simultaneously expressing YFP(n)‐P2, YFP(c)‐P2 and P1, P3, VPg, NIa‐Pro or NIb‐eGFP (A), or YFP(n)‐P1, YFP(c)‐P2 and NIb‐eGFP (B). Fluorescent proteins were observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). GFP, green fluorescent protein; YFP, yellow fluorescent protein. Bars, 20 μm.
Newly synthesized WYMV RNAs co‐localize with P2‐associated granular bodies
We performed in vivo RNA labelling to investigate whether WYMV RNA synthesis occurs in these ER‐derived inclusions. Protoplasts isolated from the young leaves of WYMV‐infected wheat plants were treated with actinomycin D for 1 h to block host DNA transcription, and then incubated with 5‐bromouridine 5′‐triphosphate (BrUTP) for 4 h to incorporate BrUTP into newly synthesized viral RNAs prior to fixing the cells and double immunofluorescence labelling using antisera specific for P2 and BrUTP. Visualization of P2 antiserum using tetramethyl rhodamine isothiocyanate (TRITC) showed that P2 primarily localized in multiple granular bodies distributed throughout the cytoplasm (Fig. 8), a pattern reminiscent of the inclusion bodies observed under an electron microscope (Fig. 1). Visualization of BrUTP‐containing RNA using fluorescein isothiocyanate (FITC) revealed that newly synthesized viral RNAs co‐localized with P2 in these granular bodies (Fig. 8). In a control experiment, when protoplasts were subjected to single immunofluorescence labelling using pre‐immune rabbit or mouse serum, no signal was detected (Fig. 8). In addition, no fluorescence was detected when protoplasts isolated from leaves of non‐infected wheat plants were subjected to a similar RNA labelling procedure (data not shown). This observation suggests that WYMV RNA synthesis occurs in the P2‐associated inclusion bodies.
Figure 8.
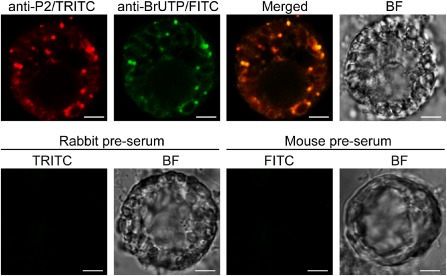
Co‐localization of newly synthesized Wheat yellow mosaic virus (WYMV) RNA with P2‐associated inclusions. Protoplasts isolated from the leaves of WYMV‐infected wheat were treated with actinomycin D prior to labelling with 5‐bromouridine 5′‐triphosphate (BrUTP), and subjected to double immunofluorescence labelling using antisera specific for P2 and BrUTP or single immunofluorescence labelling using pre‐immune rabbit or mouse serum in control experiments. Fluorescence was observed using confocal laser scanning microscopy (CLSM). BF, bright field. FITC, fluorescein isothiocyanate; TRITC, tetramethyl rhodamine isothiocyanate. Bars, 4.5 μm.
Discussion
Large ER‐derived inclusions with crystalline lattice‐like structures are consistently observed in cells infected with bymoviruses (Hibino et al., 1981; Hooper & Wiese, 1972; Huth et al., 1984; Langenberg, 1985; and this study). To our knowledge, similar structures have not been observed in cells infected with viruses of other genera in the family Potyvridae, suggesting that these large membranous inclusion bodies are a distinct cytopathological feature of bymovirus infection. Previously, immunogold labelling of barley cells infected with Barley yellow mosaic virus (BaYMV, genus Bymovirus) showed that P1 and P2 are associated with these membranous inclusion bodies (Schenk et al., 1993). In this study, we performed immunogold labelling using a larger panel of viral protein‐specific antisera. In addition to P1 and P2, we also detected the presence of P3 and, to a lesser extent, NIa‐Pro and VPg, in the membranous inclusions (Fig. 2B). P3 antiserum might also react with P3N‐PIPO protein (see Fig. 6C); therefore, we cannot rule out the possibility that the inclusion bodies contain P3N‐PIPO protein. Because P1, P2 and P3 appear to be the major viral protein constituents of the inclusions, we particularly analysed the properties of these proteins. Although all of these proteins associated with ER, only P2 induced ER‐derived aggregates when expressed alone (Fig. 3B), indicating that P2 has an intrinsic ability to rearrange ER membranes. Moreover, P2 was identified as an integral membrane protein in a chemical treatment experiment (Fig. 4C). When co‐expressed with P2 in transient experiments, P1, P3, NIa‐Pro and VPg relocated into the P2‐formed aggregates of ER membranes (Fig. 7A). This is consistent with the observations that P1, P3, NIa‐Pro and VPg are present in the inclusions (Fig. 2B) and that they interact directly with P2 (Fig. 6). Thus, P2 probably recruits P1, P3, NIa‐Pro and VPg into membranous inclusions through direct binding. Taken together, our results underline a key role for P2 in the formation of ER‐derived inclusion bodies during WYMV infection.
We further observed that the active secretory pathway is required for the ER remodelling activities of P2. Treatment with BFA or co‐expression with the Sar1[H74L] mutant retained P2‐eGFP in the cortical ER and consequently inhibited the formation of P2‐induced aggregates (Fig. 5A). Other studies have also shown that the ER remodelling induced by some viral proteins depends on the secretory pathway. For example, the formation of ER‐derived large aggregate structures by the 37K movement protein of Chinese wheat mosaic virus was disrupted by BFA treatment or overexpression of the Sar1[H74L] mutant (Andika et al., 2013). Another study showed that the secretory pathway was required for ER remodelling induced by Red clover necrotic mosaic virus (RCNMV) p27, a replication protein involved in the formation of RCNMV replication complexes. Accordingly, inhibition of the secretory pathway resulted in reduced RCNMV replication in protoplasts. RCNMV p27 interacts with ADP ribosylation factor 1 (Arf1) and relocates it into ER‐derived aggregate structures (Hyodo et al., 2013). Arf1 is a small GTPase that is involved in the formation of coat protein complex I (COPI) vesicles. It has been suggested that the membrane deformation activity of Arf1 may contribute to the formation of RCNMV replication complexes. RCNMV p27 also recruits Sar1 into p27‐induced aggregates, although no direct interaction has been demonstrated (Hyodo et al., 2013). Intriguingly, in our BiFC assay, WYMV P2 appeared to interact with Sar1 in aggregate structures (Fig. 5B). It would be interesting to investigate whether P2 also interacts with Arf1 and has similar activity to p27 in remodelling the early secretory pathway.
Positive‐strand RNA viruses reorganize host endomembranes to provide scaffolds for the formation of viral replication complexes (Laliberte and Sanfacon, 2010; Miller and Krijnse‐Locker, 2008; Verchot, 2011). Our immunofluorescence labelling experiment using WYMV‐infected wheat protoplasts provides important evidence that viral genome replication occurs in the ER‐derived inclusion bodies (Fig. 8). Oat mosaic virus and Barley mild mosaic virus (both are members of the genus Furovirus) isolates, which have deletions in the C‐terminal region of P2, are still able to replicate in their hosts (Timpe and Kuhne, 1995; Zheng et al., 2002). In a study using infectious cDNA clones, a BaYMV mutant with the entire P2 coding region deleted replicated as efficiently as the wild‐type virus in barley protoplasts; however, when inoculated into barley plants, this mutant showed reduced efficiency in systemic infection, induced milder symptoms and accumulated to a much lower level in upper systemic leaves (You and Shirako, 2010). This result indicates that BaYMV P2 is dispensable for viral replication in infected cells, but facilitates efficient viral systemic movement. Together, these studies seem to indicate that P2 might not be absolutely required for bymovirus replication. In the formation of potyvirus replication complexes, as exemplified by Turnip mosaic virus (TuMV), 6K is an integral membrane protein that has been shown to be the key factor as 6K‐induced vesicles move from the ER to the chloroplast periphery and induce chloroplast membrane invaginations (Restrepo‐Hartwig and Carrington, 1994; Wei et al., 2010; Wei and Wang, 2008). Viral genomic and double‐stranded RNAs, and also viral replicase components (NIa and NIb), localize at the 6K‐induced vesicles that associate with chloroplasts, suggesting that TuMV replication occurs in these compartments (Wei et al., 2010). When fused to eGFP, WYMV 7K, the counterpart of the 6K protein of potyviruses, accumulated in cortical ER, but did not associate with chloroplast membranes (Fig. S1). In our electron microscopy observations, WYMV infection did not induce chloroplast membrane invaginations or any alteration in the morphological structure of chloroplast peripheral membranes. Thus, although 7K does not form invaginations in the chloroplast envelope, it is possible that 7K could constitute the essential membrane anchor of the bymovirus replication complex, as monopartite potyviruses also replicate on the ER (Schaad et al., 1997).
Considering the above‐mentioned observations, the P2‐induced membranous inclusions could be essential for other biological processes in bymovirus infection. In this respect, P2 is comparable with the TGB1 protein (p25) of Potato virus X (PVX). TGB1 organizes the perinuclear X‐body, an inclusion structure thought to be the site of viral replication, by remodelling host actin and ER membranes (Tilsner et al., 2012). However, neither the TGB1‐organized X‐body nor the expression TGB1 protein itself is essential for PVX replication (Batten et al., 2003; Tilsner et al., 2012). Given the multifunctional roles of TGB1, it has been proposed that the ER membrane remodelling activities of TGB1 might relate to viral movement, RNA silencing suppression or viral RNA packaging of PVX (Tilsner et al., 2012). Because P2 is involved in vector transmission, it is possible that the ER‐derived inclusion bodies induced by P2 have a specific function in the efficient transfer of WYMV particles to P. graminis cytoplasm across the vector plasma membranes. It is worth mentioning that the formation of electron‐lucent inclusion bodies induced by infection of Cauliflower mosaic virus is essential for aphid transmission (Khelifa et al., 2007). It is also possible that the interaction of P2 with the secretory pathway may be associated with the transport of virus across the plant cell–vector boundary. We noted that no WYMV particles were observed within the membranous inclusions (Fig. 1), which may suggest that the formation of this structure is not related to virion assembly. Future studies will attempt to resolve the precise roles for the ER‐derived inclusion bodies in the bymovirus life cycle by detailed analyses of the cytopathological effects and characteristics of virus mutants generated by reverse genetics.
Experimental Procedures
Virus and plant materials
WYMV‐infected wheat plants were obtained from a field in Yangzhou City, Jiangsu Province, China (Chen, 1999). Nicotiana benthamiana plants were grown in soil and kept in a growth cabinet at 24 °C or 16 °C with 16 h of daylight.
Plasmid constructs
DNA fragments corresponding to the WYMV‐encoded proteins were amplified by reverse transcription‐polymerase chain reaction (RT‐PCR) using total RNA extracted from leaves of infected wheat. To prepare eGFP fusion constructs, the eGFP fragment. with a SalI site incorporated upstream of the coding region, was first inserted between the BamHI and SmaI sites of the binary vector pBin61 (Voinnet et al., 1998) to produce pBin‐eGFP. DNA fragments were then inserted between the BamHI and SalI sites of pBin‐eGFP. For the BiFC assay, DNA fragments were inserted into pBin‐YFP(n) and pBin‐YFP(c) plasmids (Sun et al., 2013a) using the BamHI and SmaI sites. The Sar1 coding region was amplified from the Sar1‐YFP plasmid (Wei and Wang, 2008). DNA fragments were introduced between the BamHI and NotI sites of pGEX‐6P‐1 (Life Technologies, Carlsbad, CA, USA) and between the BamHI and SalI sites of pMBP‐2CX (New England BioLabs, Ipswich, MA, USA) for the expression of glutathione S‐transferase (GST) and MBP‐tagged recombinant proteins in E. coli, respectively. The fluorescent organelle markers ManI‐Cherry and ER‐Cherry and the plasmid Sar1[H74L] mutant have been described previously (Nelson et al., 2007; Wei and Wang, 2008).
Antisera preparation
GST‐tagged proteins were expressed as described previously (Sun and Suzuki, 2008). For antisera production, purified proteins were diluted in an injection buffer [50 mm Tris‐HCl (pH 8.0), 75 mm NaCl and 1 × protease inhibitor cocktail (Roche, Mannheim, Germany)], and emulsified with Freund's incomplete adjuvant (Difco, Franklin Lakes, NJ, USA). Antigen (1 mg/mL) was subcutaneously injected into New Zealand white rabbits. Antisera were produced by Shengong Biotech Co. Ltd. (Shanghai, China).
Immunogold electron microscopy
The preparation of ultrathin sections of WYMV‐infected wheat leaf tissues and immunogold labelling generally followed the protocol described previously (Sun et al., 2013c; Xiong et al., 2008). For immunogold labelling, the grids were first incubated with antibodies specific for WYMV‐encoded proteins (1:50 dilution) and then with gold (10 nm)‐conjugated goat anti‐rabbit immunoglobulin G (IgG) (1:50) (Sigma‐Aldrich, St. Louis, MO, USA). The grids were examined under a transmission electron microscope H‐7650 (Hitachi, Tokyo, Japan) and photographed with a Gatan 830 CCD camera (Gatan, Pleasanton, CA, USA).
Agrobacterium infiltration and BFA treatment
For Agrobacterium infiltration, plasmid constructs were transformed into Agrobacterium tumefaciens strain C58C1. Bacterial pellets were resuspended in agroinfiltration buffer (10 mm MgCl2, 10 mm 2‐morpholinoethanesulphonic acid and 100 μm acetosyringone) at an optical density at 600 nm (OD600) of unity and used to infiltrate N. benthamiana leaves with syringes. BFA (Sigma‐Aldrich) stocks were dissolved in DMSO and diluted with agroinfiltration buffer to concentrations of 10 μg/mL. The leaves were observed 4 h after infiltration with BFA solution. For the control experiment, leaves were infiltrated with DMSO diluted in agroinfiltration buffer (0.1%).
Western blot analysis
Preparation of protein samples, sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE), electroblotting and immunodetection for Western blot analysis were performed as described previously (Sun and Suzuki, 2008). For the detection of WYMV‐encoded proteins, primary polyclonal antibodies (1:1000) and a secondary polyclonal alkaline phosphatase (AP)‐conjugated goat anti‐rabbit IgG (1:5000) (Sigma) were used. GFP and ER‐associated proteins were detected using primary anti‐GFP (1:10 000) and anti‐KDELR2 (1:2000) monoclonal sera (Zhongshan Jinqiao, Beijing, China), respectively, and a secondary polyclonal AP‐conjugated goat anti‐mouse IgG (1:10 000) (Sigma).
MBP pull‐down assay
Crude protein extracts of bacterial cells expressing MBP‐P2 or unfused MBP were incubated with an amylose‐resin column (New England BioLabs) for 4 h with rotation to immobilize the proteins. Total protein was extracted from leaves of WYMV‐infected wheat with a binding buffer [20 mm Tris‐HCl (pH 7.0), 0.2 m NaCl, 1 mm ethylenediaminetetraacetic acid (EDTA), 0.5% NP‐40 and 1 × protease inhibitor cocktail] and allowed to flow through an amylose‐resin column with immobilized MBP‐P2 or unfused MBP. After washing three times with the binding buffer, the resin was resuspended in SDS‐PAGE sample buffer, denatured by boiling, run on SDS‐PAGE and analysed by Western blotting.
Subcellular fractionation and chemical treatment
For subcellular fractionation, leaves (5 g) were homogenized in 10 mL of grinding buffer [20 mm Tris‐HCl (pH 8.0), 200 mm NaCl, 1 mm EDTA and 1 × protease inhibitor cocktail]. The homogenate was centrifuged twice at 1000 g for 15 min to remove chloroplasts, cell walls and debris. The supernatant was centrifuged at 30 000 g for 30 min to obtain the soluble (S30) and crude membrane (P30) fractions. For sucrose gradient fractions, P30 (0.5 mL) was loaded onto a 5‐mL 10%–60% continuous sucrose gradient (in grinding buffer) and ultracentrifuged at 148 862 g for 2 h. The gradient was divided into 11 density fractions (0.5 mL each) and analysed by Western blotting. For chemical treatments, P30 was resuspended in grinding buffer alone (controls) or containing 0.1 m Na2CO3 (pH 11), 4 m urea or 1% Triton X‐100, and incubated for 30 min on ice. After ultracentrifugation to obtain S30 and P30, samples were analysed by Western blotting using a GFP‐specific antiserum.
Protoplast isolation and in vivo RNA labelling
For the isolation of protoplasts, wheat leaves were cut longitudinally and soaked in an enzyme solution containing 0.6 m mannitol, 10 mm CaCl2 (pH 5.6), 1% cellulase (Onozuka R‐10) and 0.05% macerozyme (both from Yakult Honsha Co., Ltd., Tokyo, Japan), and kept under vacuum for 30 min prior to incubation for 4 h at 25 °C in the dark. Protoplasts were filtered through a 0.64‐mm mesh sieve and collected by centrifugation at 500 g for 10 min. Washing (three times) was performed in a solution containing 0.6 m mannitol and 10 mm CaCl2 (pH 5.6). In vivo RNA labelling and fixing of protoplasts on cover slides were carried out as described previously (Cotton et al., 2009). Immunofluorescence labelling was performed using primary P2 (1:1000) and monoclonal BrUTP (1:500) (Sigma) antisera, and detected with secondary TRITC‐conjugated goat anti‐rabbit and FITC‐conjugated goat anti‐mouse IgGs (1:500) (Zhongshan Jinqiao).
Fluorescence signal imaging
Fluorescent proteins were observed using a TCS SP5 confocal laser scanning microscope (Leica, Solms, Germany). The fluorescence signals were visualized with laser excitation/emission filters of 488/500–508 nm for eGFP, 514/580–600 nm for eYFP, 543/610–655 nm for Cherry and TRITC, and 488/505–550 nm for FITC.
Amino acid sequence analyses
Hydrophobicity plots were generated with MPEx (http://blanco.biomol.uci.edu/mpex/) according to the octanol hydrophobicity scale (Wimley and White, 1996). For the prediction of transmembrane domains, programs TopPred II (Claros and von Heijne, 1994; http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html) and TMpred (Hofmann and Stoffel, 1993; http://www.ch.embnet.org/software/TMPRED_form.html) were used.
Supporting information
Fig. S1 Subcellular localization of Wheat yellow mosaic virus VPg, NIa‐Pro and NIb. Epidermal cells of Nicotiana benthamiana transiently expressing P1‐, P2‐ or P3‐enhanced green fluorescent protein (eGFP). Images were derived from a single confocal section. Fluorescent proteins and 4′,6‐diamidino‐2‐phenylindole (DAPI) staining were observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 20 μm.
Fig. S2 Hydrophobic profile of Wheat yellow mosaic virus P1 and P3. (A) Hydrophobic plot of P1 and P3 generated with MPEx according to the Wimley and White octanol hydrophobicity scale. The plot shows the mean values using a window of 19 residues. Lines indicate the predicted hydrophobic regions. (B) A schematic map showing the position of two putative transmembrane domains (TM, grey boxes) in P3 as predicted by TMpred. Arrowheads indicate the amino acid positions of the transmembrane domains. Cyt and Lum indicate the cytoplasmic and luminal domains, respectively.
Fig. S3 Interaction of Wheat yellow mosaic virus P1 or P3 with other viral proteins in bimolecular fluorescence complementation (BiFC) assay. Leaves of Nicotiana benthamiana plants were infiltrated with mixtures of agrobacteria harbouring the constructs indicated above and on the left side of the images. Fluorescent proteins were observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 20 μm.
Acknowledgements
We are deeply grateful to Taiyun Wei for providing Sar1‐YFP and Sar1[H74L] plasmids, Li Xie for technical assistance and Mike Adams for critical reading of the manuscript. This study was funded by grants from the Project of China Agriculture Research System (CARS‐3‐1), the Ministry of Agriculture of China and the Project of Molecular Mechanism of Plant Defense to Pest and Disease (2012CB722504).
References
- Adams, M. , Antoniw, J. and Mullins, J. (2001) Plant virus transmission by plasmodiophorid fungi is associated with distinctive transmembrane regions of virus‐encoded proteins. Arch. Virol. 146, 1139–1153. [DOI] [PubMed] [Google Scholar]
- Adams, M. , Zerbini, F. , French, R. , Rabenstein, F. , Stenger, D. and Valkonen, J. (2011) Family Potyviridae. In: Virus Taxonomy, 9th Report of the International Committee for Taxonomy of Viruses, (King A.M.Q., Adams M.J., Carstens E.B. and Lefkowitz E.J., eds), 1069–1089. San Diego, CA: Elsevier Academic Press. [Google Scholar]
- Anandalakshmi, R. , Pruss, G.J. , Ge, X. , Marathe, R. , Mallory, A.C. , Smith, T.H. and Vance, V.B. (1998) A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA, 95, 13 079–13 084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andika, I.B. , Zheng, S. , Tan, Z. , Sun, L. , Kondo, H. , Zhou, X. and Chen, J. (2013) Endoplasmic reticulum export and vesicle formation of the movement protein of Chinese wheat mosaic virus are regulated by two transmembrane domains and depend on the secretory pathway. Virology, 435, 493–503. [DOI] [PubMed] [Google Scholar]
- Andrews, J.H. and Shalla, T. (1974) The origin, development and conformation of amorphous inclusion body components in tobacco etch virus‐infected cells. Phytopathology, 64, 234–243. [Google Scholar]
- Arbatova, J. , Lehto, K. , Pehu, E. and Pehu, T. (1998) Localization of the P1 protein of Potato Y potyvirus in association with cytoplasmic inclusion bodies and in the cytoplasm of infected cells. J. Gen. Virol. 79, 2319–2323. [DOI] [PubMed] [Google Scholar]
- Barlowe, C. (2003) Signals for COPII‐dependent export from the ER: what's the ticket out? Trends Cell Biol. 13, 295–300. [DOI] [PubMed] [Google Scholar]
- Batten, J.S. , Yoshinari, S. and Hemenway, C. (2003) Potato virus X: a model system for virus replication, movement and gene expression. Mol. Plant Pathol. 4, 125–131. [DOI] [PubMed] [Google Scholar]
- Baunoch, D. , Das, P. and Hari, V. (1988) Intracellular localization of TEV capsid and inclusion proteins by immunogold labeling. J. Ultrastruct. Mol. Struct. Res. 99, 203–212. [Google Scholar]
- Baunoch, D. , Das, P. and Hari, V. (1990) Potato virus Y helper component protein is associated with amorphous inclusions. J. Gen. Virol. 71, 2479–2482. [DOI] [PubMed] [Google Scholar]
- den Boon, J.A. , Diaz, A. and Ahlquist, P. (2010) Cytoplasmic viral replication complexes. Cell Host Microbe, 8, 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. (1999) Molecular comparisons amongst wheat bymovirus isolates from Asia, North America and Europe. Plant Pathol. 48, 642–647. [Google Scholar]
- Chung, B.Y. , Miller, W.A. , Atkins, J.F. and Firth, A.E. (2008) An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA, 105, 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros, M.G. and von Heijne, G. (1994) TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10, 685–686. [DOI] [PubMed] [Google Scholar]
- Cotton, S. , Grangeon, R. , Thivierge, K. , Mathieu, I. , Ide, C. , Wei, T. Wang, A. and Laliberte, J.F. (2009) Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 83, 10 460–10 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessens, J.T. and Meyer, M. (1996) Identification of structural similarities between putative transmission proteins of Polymyxa and Spongospora transmitted bymoviruses and furoviruses. Virus Genes, 12, 95–99. [DOI] [PubMed] [Google Scholar]
- Edwardson, J. and Christie, R. (1983) Cytoplasmic cylindrical and nucleolar inclusions induced by Potato virus‐A . Phytopathology, 73, 290–293. [Google Scholar]
- Edwardson, J. , Christie, R. and Ko, N. (1984) Potyvirus cylindrical inclusions—Subdivision‐IV. Phytopathology, 74, 1111–1114. [Google Scholar]
- Edwardson, J.R. (1966) Cylindrical inclusions in the cytoplasm of leaf cells infected with tobacco etch virus . Science, 153, 883–884. [DOI] [PubMed] [Google Scholar]
- Genovés, A. , Navarro, J.A. and Pallás, V. (2010) The intra‐ and intercellular movement of Melon necrotic spot virus (MNSV) depends on an active secretory pathway. Mol. Plant–Microbe Interact. 23, 263–272. [DOI] [PubMed] [Google Scholar]
- Hammond, J. (1998) Serological relationships between the cylindrical inclusion proteins of potyviruses. Phytopathology, 88, 965–971. [DOI] [PubMed] [Google Scholar]
- Han, C. , Li, D. , Xing, Y. , Zhu, K. , Tian, Z. , Cai, Z. , Yu, J. and Liu, Y. (2000) Wheat yellow mosaic virus widely occurring in wheat (Triticum aestivum) in China. Plant Dis. 84, 627–630. [DOI] [PubMed] [Google Scholar]
- Harries, P.A. , Schoelz, J.E. and Nelson, R.S. (2010) Intracellular transport of viruses and their components: utilizing the cytoskeleton and membrane highways. Mol Plant–Microbe Interact. 23, 1381–1393. [DOI] [PubMed] [Google Scholar]
- Hibino, H. , Usugi, T. and Saito, Y. (1981) Comparative electron microscopy of inclusions associated with five soil‐borne filamentous viruses of cereals. Ann. Phytopathol. Soc. Jpn. 47, 510–519. [Google Scholar]
- Hofmann, K. and Stoffel, W. (1993) TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe‐Seyler, 374, 166. [Google Scholar]
- Hooper, G. and Wiese, M. (1972) Cytoplasmic inclusions in wheat affected by wheat spindle streak mosaic. Virology, 47, 664–672. [DOI] [PubMed] [Google Scholar]
- Hu, C. and Kerppola, T. (2003) Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth, W. , Lesemann, D.E. and Paul, H.L. (1984) Barley yellow mosaic virus: purification, electron microscopy, serology, and other properties of two types of the virus. J. Phytopathol. 111, 37–54. [Google Scholar]
- Hyodo, K. , Mine, A. , Taniguchi, T. , Kaido, M. , Mise, K. , Taniguchi, H. and Okuno, T. (2013) ADP ribosylation factor 1 plays an essential role in the replication of a plant RNA virus. J. Virol. 87, 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K. and Carrington, J. (1998) A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell, 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Khelifa, M. , Journou, S. , Krishnan, K. , Gargani, D. , Esperandieu, P. , Blanc, S. and Drucker, M. (2007) Electron‐lucent inclusion bodies are structures specialized for aphid transmission of Cauliflower mosaic virus . J. Gen. Virol. 88, 2872–2880. [DOI] [PubMed] [Google Scholar]
- Kühne, T. (2009) Soil‐borne viruses affecting cereals: known for long but still a threat. Virus Res. 141, 174–183. [DOI] [PubMed] [Google Scholar]
- Laliberte, J.F. and Sanfacon, H. (2010) Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 48, 69–91. [DOI] [PubMed] [Google Scholar]
- Langenberg, W.G. (1985) Immunoelectron microscopy of Wheat spindle streak and Soil‐borne wheat mosaic virus doubly infected wheat. J. Ultrastruct. Res. 92, 72–79. [Google Scholar]
- Laporte, C. , Vetter, G. , Loudes, A.M. , Robinson, D.G. , Hillmer, S. , Stussi‐Garaud, C. and Ritzenthaler, C. (2003) Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY‐2 cells. Plant Cell, 15, 2058–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S. and Krijnse‐Locker, J. (2008) Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshe, A. and Gorovits, R. (2012) Virus‐induced aggregates in infected cells. Viruses, 4, 2218–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba, S. , Kashiwazaki, S. , Lu, X. , Tamura, M. and Tsuchizaki, T. (1998) Complete nucleotide sequence of Wheat yellow mosaic bymovirus genomic RNAs. Arch. Virol. 143, 631–643. [DOI] [PubMed] [Google Scholar]
- Nebenführ, A. , Ritzenthaler, C. and Robinson, D.G. (2002) Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol. 130, 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B.K. , Cai, X. and Nebenführ, A. (2007) A multicolored set of in vivo organelle markers for co‐localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ohto, Y. and Naito, S. (1997) Propagation of Wheat yellow mosaic virus in winter wheat under low temperature conditions. Ann. Phytopathol. Soc. Jpn. 63, 361–365. [Google Scholar]
- Otulak, K. and Garbaczewska, G. (2012) Cytopathological Potato virus Y structures during Solanaceous plants infection. Micron, 43, 839–850. [DOI] [PubMed] [Google Scholar]
- Restrepo‐Hartwig, M.A. and Carrington, J.C. (1994) The tobacco etch potyvirus 6‐kilodalton protein is membrane associated and involved in viral replication. J. Virol. 68, 2388–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro, D. , Goldbach, R. and Kormelink, R. (2009) Requirements for ER‐arrest and sequential exit to the Golgi of Tomato spotted wilt virus glycoproteins. Traffic, 10, 664–672. [DOI] [PubMed] [Google Scholar]
- Robinson, D.G. , Herranz, M.‐C. , Bubeck, J. , Pepperkok, R. and Ritzenthaler, C. (2007) Membrane dynamics in the early secretory pathway. Crit. Rev. Plant Sci. 26, 199–225. [Google Scholar]
- Rouis, S. , Ayadi, H. , Bouaziz, S. , Lakhoua, L. and Gargouri, R. (2002) In situ immunocytochemical detection of potyviral proteins in plant cells. Biotech. Histochem. 77, 111–115. [DOI] [PubMed] [Google Scholar]
- Schaad, M.C. , Jensen, P.E. and Carrington, J.C. (1997) Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum‐targeted viral protein. EMBO J. 16, 4049–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, P. , Steinbiß, H.‐H. , Müller, B. and Schmitz, K. (1993) Association of two Barley yellow mosaic virus (RNA 2) encoded proteins with cytoplasmic inclusion bodies revealed by immunogold localisation. Protoplasma, 173, 113–122. [Google Scholar]
- Stenger, D.C. , Hein, G.L. , Gildow, F.E. , Horken, K.M. and French, R. (2005) Plant virus HC‐Pro is a determinant of eriophyid mite transmission. J. Virol. 79, 9054–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. and Suzuki, N. (2008) Intragenic rearrangements of a mycoreovirus induced by the multifunctional protein p29 encoded by the prototypic hypovirus CHV1‐EP713. RNA, 14, 2557–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Andika, I.B. , Kondo, H. and Chen, J. (2013a) Identification of the amino acid residues and domains in the cysteine‐rich protein of Chinese wheat mosaic virus that are important for RNA silencing suppression and subcellular localization. Mol. Plant Pathol. 14, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Bian, J. , Andika, I.B. , Hu, Y. , Sun, B. , Xiang, R. , Kondo, H. and Chen, J. (2013b) Nucleo‐cytoplasmic shuttling of VPg encoded by Wheat yellow mosaic virus requires association with the coat protein. J. Gen. Virol. 94, 2790–2802. [DOI] [PubMed] [Google Scholar]
- Sun, L. , Xie, L. , Andika, I.B. , Tan, Z. and Chen, J. (2013c) Non‐structural protein P6 encoded by Rice black‐streaked dwarf virus is recruited to viral inclusion bodies by binding to the viroplasm matrix protein P9‐1. J. Gen. Virol. 94, 1908–1916. [DOI] [PubMed] [Google Scholar]
- Tamada, T. and Kondo, H. (2013) Biological and genetic diversity of plasmodiophorid‐transmitted viruses and their vectors. J. Gen. Plant Pathol. 79, 307–320. [Google Scholar]
- Tatineni, S. , Qu, F. , Li, R. , Jack Morris, T. and French, R. (2012) Triticum mosaic poacevirus enlists P1 rather than HC‐Pro to suppress RNA silencing‐mediated host defense. Virology, 433, 104–115. [DOI] [PubMed] [Google Scholar]
- Tilsner, J. , Linnik, O. , Wright, K.M. , Bell, K. , Roberts, A.G. , Lacomme, C. , Cruz, S.S. and Oparka, K.J. (2012) The TGB1 movement protein of Potato virus X reorganizes actin and endomembranes into the X‐body, a viral replication factory. Plant Physiol. 158, 1359–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpe, U. and Kuhne, T. (1995) In vitro transcripts of a full‐length cDNA of a naturally deleted RNA2 of Barley mild mosaic virus (BaMMV) replicate in BaMMV‐infected plants. J. Gen. Virol. 76, 2619–2623. [DOI] [PubMed] [Google Scholar]
- Urcuqui‐Inchima, S. , Haenni, A.L. and Bernardi, F. (2001) Potyvirus proteins: a wealth of functions. Virus Res. 74, 157–175. [DOI] [PubMed] [Google Scholar]
- Valli, A. , Martin‐Hernandez, A.M. , Lopez‐Moya, J.J. and Garcia, J.A. (2006) RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro. J. Virol. 80, 10 055–10 063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot, J. (2011) Wrapping membranes around plant virus infection. Curr. Opin Virol. 1, 388–395. [DOI] [PubMed] [Google Scholar]
- Vogel, F. , Hofius, D. and Sonnewald, U. (2007) Intracellular trafficking of Potato leafroll virus movement protein in transgenic Arabidopsis. Traffic, 8, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. , Vain, P. , Angell, S. and Baulcombe, D.C. (1998) Systemic spread of sequence‐specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Wei, T. and Wang, A. (2008) Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI‐ and COPII‐dependent manner. J. Virol. 82, 12 252–12 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. , Huang, T.S. , McNeil, J. , Laliberte, J.F. , Hong, J. , Nelson, R.S. and Wang, A. (2010) Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 84, 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley, W.C. and White, S.H. (1996) Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3, 842–848. [DOI] [PubMed] [Google Scholar]
- Xiong, R. , Wu, J. , Zhou, Y. and Zhou, X. (2008) Identification of a movement protein of the tenuivirus Rice stripe virus . J. Virol. 82, 12 304–12 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, Y. and Shirako, Y. (2010) Bymovirus reverse genetics: requirements for RNA2‐encoded proteins in systemic infection. Mol. Plant Pathol. 11, 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, B.A. , Hein, G.L. , French, R. and Stenger, D.C. (2007) Substitution of conserved cysteine residues in Wheat streak mosaic virus HC‐Pro abolishes virus transmission by the wheat curl mite. Arch. Virol. 152, 2107–2111. [DOI] [PubMed] [Google Scholar]
- Young, B.A. , Stenger, D.C. , Qu, F. , Morris, T.J. , Tatineni, S. and French, R. (2012) Tritimovirus P1 functions as a suppressor of RNA silencing and an enhancer of disease symptoms. Virus Res. 163, 672–677. [DOI] [PubMed] [Google Scholar]
- Zheng, T. , Chen, J. , Chen, J.P. and Adams, M.J. (2002) The complete sequence of Oat mosaic virus and evidence for deletion and duplication in RNA2. Arch. Virol. 147, 635–642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Subcellular localization of Wheat yellow mosaic virus VPg, NIa‐Pro and NIb. Epidermal cells of Nicotiana benthamiana transiently expressing P1‐, P2‐ or P3‐enhanced green fluorescent protein (eGFP). Images were derived from a single confocal section. Fluorescent proteins and 4′,6‐diamidino‐2‐phenylindole (DAPI) staining were observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 20 μm.
Fig. S2 Hydrophobic profile of Wheat yellow mosaic virus P1 and P3. (A) Hydrophobic plot of P1 and P3 generated with MPEx according to the Wimley and White octanol hydrophobicity scale. The plot shows the mean values using a window of 19 residues. Lines indicate the predicted hydrophobic regions. (B) A schematic map showing the position of two putative transmembrane domains (TM, grey boxes) in P3 as predicted by TMpred. Arrowheads indicate the amino acid positions of the transmembrane domains. Cyt and Lum indicate the cytoplasmic and luminal domains, respectively.
Fig. S3 Interaction of Wheat yellow mosaic virus P1 or P3 with other viral proteins in bimolecular fluorescence complementation (BiFC) assay. Leaves of Nicotiana benthamiana plants were infiltrated with mixtures of agrobacteria harbouring the constructs indicated above and on the left side of the images. Fluorescent proteins were observed using confocal laser scanning microscopy (CLSM) at 3 days after inoculation (dai). Bars, 20 μm.


