Summary
Expansins are cell wall loosening agents, known for their endogenous function in cell wall extensibility. The Arabidopsis expansin‐like A2 (EXLA2) gene was identified by its down‐regulation in response to infection by the necrotrophic pathogen Botrytis cinerea, and by the reduced susceptibility of an exla2 mutant to the same pathogen. The exla2 mutant was equally susceptible to Pseudomonas syringae pv. tomato, but was more resistant to the necrotrophic fungus Alternaria brassicicola, when compared with the wild‐type or with transgenic, ectopic EXLA2‐overexpressing lines. The exla2 mutants also enhanced tolerance to the phytoprostane‐A 1. This suggests that the absence or down‐regulation of EXLA2 leads to increased resistance to B. cinerea in a CORONATINE INSENSITIVE 1 (COI1)‐dependent manner, and this down‐regulation can be achieved by phytoprostane‐A 1 treatment. EXLA2 is induced significantly by salinity and cold, and by the exogenous application of abscisic acid. The exla2 mutant also showed hypersensitivity towards increased salt and cold, and this hypersensitivity required a functional abscisic acid pathway. The differential temporal expression of EXLA2 and the phenotypes in transgenic plants with altered expression of EXLA2 indicate that plant cell wall structure is an important player during Arabidopsis developmental stages. Our results indicate that EXLA2 appears to be important in response to various biotic and abiotic stresses, particularly in the pathogenesis of necrotrophic pathogens and in the tolerance to abiotic stress.
Introduction
The plant cell wall (CW) forms a barrier against pathogen attack and interconnects cells. It plays a wide array of distinct, sometimes opposite, roles. For example, the CW determines cell structure, provides resistance to mechanical stress and protects against pathogens. Yet, it must be permeable to signal molecule trafficking (Levy et al., 2002). Expansins are plant CW‐remodelling proteins that are mainly involved in the pH‐dependent extension of plant CWs, so‐called acid growth (McQueen‐Mason et al., 1992). This is determined by the activity of an H+‐ATPase localized in the plasma membrane to pump protons into the CW, making the pH 4.5–5.5, and causing the CW to relax (Cosgrove, 2000). Expansins are also involved in cell enlargement and CW modifications induced by plant hormones, including abscisic acid (ABA) (Zhao et al., 2012), auxin (McQueen‐Mason et al., 1992), brassinosteroids (BLs; Sun et al., 2005), gibberellins (GAs; Cho and Kende, 1997), cytokinins (CKs; Downes and Crowell, 1998) and the classical defence hormones salicylate (SA), jasmonate (JA) and ethylene (ET; Cho and Cosgrove, 2002). Expansins are multigene families found in all phyla of the kingdom Plantae (Lee et al., 2001). Expansin genes are classified into four families: α‐ and β‐expansins (EXPA and EXPB, respectively), expansin‐like (EXLA) and expansin‐like related (EXLB). In the Arabidopsis genome, 26 α‐ and six β‐expansin genes were identified (Cosgrove, 2000; Kim et al., 2009). Three expansin‐like genes were found in Arabidopsis: EXLA1, EXLA2 and EXLA3 (Cosgrove, 2000; Kende et al., 2004). The mode of action of expansins, causing loosening and extension of the CW, is still not clear. It has been hypothesized that they might disrupt noncovalent bonding between cellulose microfibrils and matrix glucans on the CW; others have proposed that they might be involved in the disturbance of hydrogen bonding between wall polysaccharides (Cosgrove, 2000). No hydrolytic or enzymatic activity has been found on the plant CW.
Recently, ABA has been reported as a regulator of plant defence against necrotrophic pathogens (Adie et al., 2007; Audenaert et al., 2002; Laluk et al., 2011). Despite the high susceptibility to the oomycete Pythium irregulare and the fungus A. brassicicola, ABA‐impaired mutants show increased resistance to Botrytis cinerea (Adie et al., 2007). The transcriptome data also confirm that ABA affects JA biosynthesis and JA‐dependent genes in response to P. irregulare. JA is a hormone which regulates plant maturation and pollen development, and is involved in the response to a variety of stresses. JA is produced from the polyunsaturated fatty acid α‐linolenic acid after a series of enzymatic conversions leading to 12‐oxo‐phytodienoic acid (OPDA) (Mueller, 1997). OPDA undergoes three cycles of β‐oxidation by OPDA reductase3 (OPR3) yielding JA. On stress, OPDA and JA regulate downstream gene expression via the CORONATINE INSENSITIVE 1 (COI1)‐dependent pathway. The cyclopentenone OPDA also acts independently of the JA/COI1 pathway to alter gene expression (Ribot et al., 2008). Although the JA‐insensitive mutant coi1 is highly susceptible to necrotrophic pathogens, the opr3 mutant is highly resistant (AbuQamar et al., 2006; Chehab et al., 2011; Stintzi et al., 2001). The cyclopentenone phytoprostane (PP) is also derived from α‐linolenic acid, but formed via the nonenzymatic, free radical‐catalysed pathway (Mueller, 1997). OPDA and PP are structurally similar to JA, but contain a reactive unsaturated carbonyl structure in the cyclo‐ring. The phytoprostane‐A1 (PPA1) induces glutathione‐S‐transferase (GST), increases phytoalexin biosynthesis and triggers the expression of the genes involved in primary and secondary metabolism (Thoma et al., 2003). Arabidopsis seedlings treated with PPA1 result in root growth inhibition that is mediated through COI1, but independent of JA (Mueller et al., 2008; Stotz et al., 2013). However, the regulation of PPA1‐responsive genes is dependent on TGA transcription factors.
Microarray‐based and functional analyses demonstrate an involvement of expansins in the early stages of symbiosis of the arbuscular mycorrhizal fungus Glomus intraradices on tomato (Solanum lycopersicum; Dermatsev et al., 2010). In addition, the legume sweetclover (Melilotus alba) expansin gene, MaEXP1, is up‐regulated during the development of nitrogen‐fixing nodules (Giordano and Hirsch, 2004). Expansins are also induced under drought. The temperature‐tolerant grass and Agrostis scabra induce the expansin‐like gene, AsEXPL1, after exposure to heat (Xu et al., 2007). Similarly, the resurrection plant, Craterostigma plantagineum, expresses α‐expansin in leaves during dehydration (Jones and McQueen‐Mason, 2004). In Arabidopsis, the accumulation of ABA and the expression of EXPA3, EXPA4, EXPA8, EXPA10 and EXLB1 genes correlate with drought acclimation (Harb et al., 2010). ABA and indole acetic acid (IAA) induce expansin activity to enhance coleoptile growth in wheat on drought stress (Zhao et al., 2012). In response to cold stress, all three AtEXLA genes are up‐regulated (Lee et al., 2005), which confirms that expansins are potentially involved in the programming of plant development under stress.
The role of Arabidopsis expansin‐like genes in defence has not been studied in any detail. We show here that EXLA2 is involved in defence against necrotrophic fungi and in tolerance to abiotic stresses. The induction of EXLA2 is dependent on ABA responses, but its expression is suppressed by exogenous PPA1 application (Mueller et al., 2008). Mutations in EXLA2 show increased resistance to B. cinerea and Alternaria brassicicola, elevated levels of tolerance to the cyclopentenone PPA1 and reduced tolerance to increased salt or cold. EXLA2 influences plant growth and development, including flowering time, morphology and plant size. Overall, this is the first report of an expansin‐like gene in Arabidopsis that links plant development and defence.
Results
EXLA2 gene encodes for an expansin‐like A2 protein
The analyses of EXLA2 cDNA (798 bp; Fig. S1, see Supporting Information) predicted an open reading frame of 265 amino acids corresponding to the coding sequence (Fig. S2, see Supporting Information). blast searches against the National Center for Biotechnology Information (NCBI) and The Arabidopsis Information Resource (TAIR) databases using the EXLA2 sequence predicted this protein to be a member of the expansin‐like family (Fig. S2; Kende et al., 2004). EXLA2 encodes a 28.6‐kDa protein (isoelectric point, 8.25).
Consistent with other expansins, the sequence of the EXLA2 protein has two conserved domains: (i) a glycoside hydrolase‐like family 45; and (ii) a group‐2 grass pollen allergen, preceded by a 21‐amino‐acid signal peptide (Fig. S2). We aligned our EXLA2 amino acid sequence with expansin‐like proteins in other plant species. Protein sequences contain conserved cysteine (C) residues in the N‐terminal region and conserved tryptophan (W) residues in the C‐terminal region, and an ambiguous signal peptide (Fig. S2). In addition, all expansin‐like proteins have one to three NXT/S motifs, which may be N‐linked glycosylation sites (Lee and Kende, 2002). Expansin‐like proteins have an additional tryptophan residue in the C‐terminal region and two additional cysteine residues in the N‐ and C‐terminal regions. All expansin‐like proteins have a unique conserved motif (CDRC) at the N‐terminus of domain 1, and an extension of 17 amino acids in domain 2, unlike expansin proteins (Fig. S2; Lee et al., 2001; Sampedro and Cosgrove, 2005). Expansins loosen CWs via a nonenzymatic mechanism by inducing the slippage of cellulose microfibrils in the plant CW (Cosgrove, 2000); EXLA2 may have a different mechanism of action from that of other expansins.
Expression of EXLA2 during development, defence and following hormone treatment
Because of its altered expression in B. cinerea‐infected Arabidopsis plants (AbuQamar et al., 2006), we studied the role of the EXLA2 gene (At4g38400) in plant defence. We showed that the EXLA2 gene is expressed at low basal levels in healthy wild‐type leaves (Fig. 1A). At 24 h post‐inoculation (hpi) with B. cinerea, however, the levels of EXLA2 transcript decreased by at least five‐fold when compared with noninoculated wild‐type plants. The transcripts of EXLA2 were strongly reduced between 24 and 48 hpi with B. cinerea (Fig. 1A). In addition, none of the virulence or avirulence strains of Pseudomonas syringae pv. tomato (Pst) (PstDC3000 or PstDC3000AvrRPM1) used altered EXLA2 gene expression compared with mock‐inoculated leaves (Fig. S3A, see Supporting Information). This suggests that EXLA2 may have a function in plant resistance specific to necrotrophic pathogens. We also checked the expression of EXLA2 in response to hormones, abiotic stresses and developmental stages (Fig. 1B–D).
Figure 1.
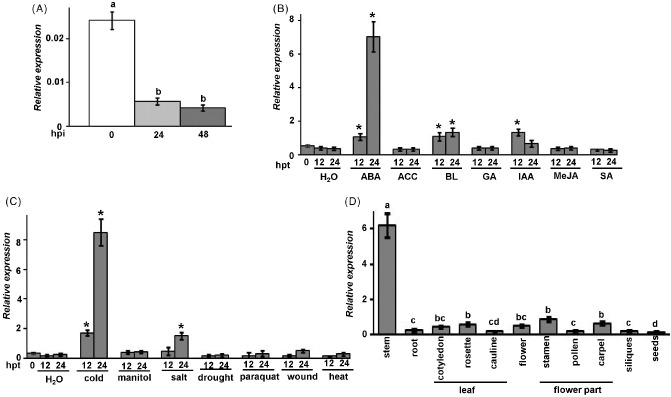
Expansin‐like A2 (EXLA2) gene expression in response to stresses and development. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) amplification of EXLA2 relative to Arabidopsis Actin2 gene, in response to Botrytis cinerea (A), hormone treatments (ABA, abscisic acid; ACC, aminocyclopropane‐1‐carboxylic acid; BL, brassinosteroid; GA, gibberellin; IAA, indole acetic acid; MeJA, methyl jasmonate; SA, salicylic acid) (B), abiotic stresses (C) and plant tissues (D). hpi/hpt, hours post‐inoculation/treatment.
Phytohormones play a major role in plant resistance to necrotrophic fungi (Robert‐Seilaniantz et al., 2007). The exogenous application of ABA induced significantly the expression of EXLA2, and BL and IAA induced slight expression. The expression of EXLA2 was unaltered on treatment with aminocyclopropane‐1‐carboxylic acid (ACC; a natural precursor of ET), GA, methyl jasmonate (MeJA) or SA at 12 h post‐treatment (hpt; Fig. 1B). At 24 hpt with IAA or BL, EXLA2 exhibited marginally basal or induced levels of EXLA2 expression, whereas plants sprayed with the ABA hormone consistently expressed EXLA2, suggesting that EXLA2 is highly induced on treatment with ABA. Because ABA is also a major regulator of abiotic stress responses (Fujita et al., 2006), we determined the expression of EXLA2 in response to cold, osmotic, salt, drought, oxidative, wounding and heating stresses. EXLA2 was up‐regulated to both cold and salt (Fig. 1C), which indicates that plant responses to pathogen infection and cold/salinity stress can regulate antagonistically EXLA2 expression.
We also studied the tissue‐specific expression of EXLA2 during different stages of plant development. In comparison with all other tissues, EXLA2 transcripts were more abundant in the stem than in other tissues (Fig. 1D). Root, leaf and flower tissues showed moderate to low levels of EXLA2 expression. The lowest level of basal expression was found in seeds (Fig. 1D). The predominant expression of EXLA2 in stem tissues suggests a potential function of EXLA2 during plant development.
Mutations in EXLA2 enhance resistance to necrotrophic fungi
To correlate the expression of EXLA2 with its function in plant defence, the responses of EXLA2 mutants, overexpression lines and wild‐type plants to different plant pathogens were examined. Only one T‐DNA insertion allele of the EXLA2 gene (SALK_147678; exla2) was available (Fig. 2A, B; Sessions et al., 2002). We generated a reduced EXLA2 gene expression line (EXLA2 RNAi #4; Fig. 2B) using the 3′‐EXLA2 gene‐specific region lacking sequences shared with the paralogues (Fig. S1). We also selected two Arabidopsis overexpression (35S:EXLA2) lines #2 and #5 that constitutively express EXLA2 at a higher level (Fig. S3B).
Figure 2.
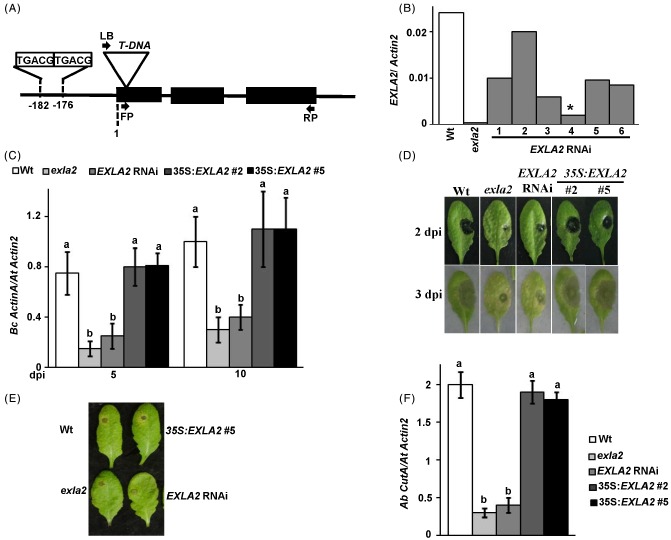
Reduced expansin‐like A2 (EXLA2) transcript levels enhance resistance to necrotrophic pathogens. (A) Genomic organization of the EXLA2 gene and the exla2 insertion allele. (B) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of exla2 T‐DNA insertion and EXLA2 RNAi mutant plants with lower constitutive EXLA2 expression. (C) Fungal growth in plants after spray inoculation with Botrytis cinerea. (D) Disease symptoms in leaves after drop inoculation with B. cinerea. dpi, days post‐inoculation. Disease symptoms (E) and fungal growth (F) in leaves after drop inoculation with Alternaria brassicicola. Wt, wild‐type.
The exla2 and EXLA2 RNAi #4 mutant lines and the transgenic 35S:EXLA2 plants were tested for susceptibility to B. cinerea as described previously (AbuQamar et al., 2006). At 7 dpi of spray‐inoculated exla2 and EXLA2 RNAi mutant plants, B. cinerea infection showed no expansion, with only 3%–6% of the plants showing complete damage; however, almost 20% of the wild‐type plants showed complete maceration (Fig. S4A, see Supporting Information). The EXLA2‐overexpressing plants developed comparable disease symptoms and tissue damage to the wild‐type plants. At 14 dpi with B. cinerea, the majority of the wild‐type plants and overexpression lines had rotted off completely, whereas only approximately 30% of both mutant plants showed complete decay (Fig. S4A). These results were confirmed by determining the fungal growth in the wild‐type and transgenic plants. At 5 and 10 dpi, wild‐type plants exhibited more fungal biomass than the mutants, as assessed by accumulation on B. cinerea of ActinA relative to AtActin2 (Benito et al., 1998; Fig. 2C). The fungal growth on the 35S:EXLA2 #2 and #5 plants was indistinguishable from that on the wild‐type at this level of inoculation. The same results were obtained when all genotypes were drop inoculated with a high concentration of B. cinerea spores (5 μL of 5 × 105 spores/mL). At 2 and 3 dpi, all leaves of different drop‐inoculated genotypes showed relatively increased susceptibility towards B. cinerea, yet both exla2 and EXLA2 RNAi mutant leaves exhibited reduced susceptibility to this pathogen when compared with the wild‐type or EXLA2‐overexpressing transgenic lines (Fig. 2D). This can be seen by the lesser extent of necrotic spots, chlorosis and tissue maceration in both mutants.
We also inoculated plants with A. brassicicola, another necrotrophic fungal pathogen. We evaluated the disease symptoms on leaves that had been drop inoculated with A. brassicicola at 5 dpi. The inoculation of wild‐type plants with fungal spores produced disease lesions that were surrounded by limited chlorosis around the inoculation site, indicating that the wild‐type plants were resistant to the fungal pathogen (Fig. 2E; Zheng et al., 2006). In both exla2 mutant and EXLA2 RNAi plants, inoculation of the fungal pathogen resulted in decreased disease symptoms characterized by no chlorotic lesions surrounding the inoculation spot (Fig. 2E). The EXLA2‐overexpressing (line #5) plants showed the same level of chlorosis on their leaves surrounding the site of inoculation as that on wild‐type leaves. We also measured fungal growth in inoculated plants using quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of the A. brassicicola cutinaseA gene (Brouwer et al., 2003). The exla2 and EXLA2 RNAi mutant plants showed a clearly reduced A. brassicicola cutinaseA gene transcript level relative to wild‐type plants, indicating reduced fungal growth in these mutants (Fig. 2F). Both symptom development and growth of the fungus confirmed that the EXLA2 mutant plants were more resistant to A. brassicicola. Statistically, no significant increase in pathogen growth after A. brassicicola inoculation was observed in transgenic 35S:EXLA2 relative to the wild‐type plants, with disease lesions that did not expand significantly after 5 dpi (Fig. 2E, F). These results suggest a common host response strategy against these pathogens.
By contrast, there was no difference in the bacterial growth of Pst in any of the EXLA2 mutants, 35S:EXLA2 overexpression lines or wild‐type plants when infiltrated with either the virulent (PstDC3000) or avirulent (PstDC3000AvrRpm1) strains of the bacterial pathogen (Fig. S4B, C). This suggests that there is limited or no role of EXLA2 in plant defence to the bacterial infection used in this study.
Mutations in EXLA2 enhance tolerance to PPA 1 and oxidative stress
We also tested the EXLA2 genotypes with the cyclopentenones PPA1 and OPDA (Mueller et al., 2008), and the oxidative stress inducer paraquat (AbuQamar et al., 2009; Mueller et al., 2008; Stotz et al., 2013; Taki et al., 2005). We first tested whether PPA1 or OPDA affects the root growth of the wild‐type and exla2 mutant plants. On control medium without any of the oxylipins, OPDA or PPA1, the roots of exla2 mutants were relatively longer (123%) than wild‐type roots (Fig. 3A, top, B). We compared the root growth inhibition of exla2 T‐DNA insertion and EXLA2 RNAi mutants with wild‐type seedlings on treatment with 75 μm PPA1. It was clearly demonstrated that the inhibition of root growth in these mutants in response to PPA1 treatment was less than that observed in wild‐type roots, and was almost similar to that of the wild‐type control grown on Murashige–Skoog (MS) medium alone (Fig. 3A, middle, B). Similarly, the coi1 mutant did not show a significant inhibition in root length when treated with 25 μm PPA1 (Stotz et al., 2013). The dosage differences of PPA1 might be attributed to the slight inhibition in root growth in exla2 in this study. The addition of 75 μm of OPDA inhibited strongly root growth in all seedlings. The inhibition of root growth in the EXLA2 mutants in the presence of OPDA was similar to that observed in the wild‐type (Fig. 3A, bottom, B). In response to these cyclopentenones, 35S:EXLA2‐overexpressing transgenic plants showed a similar root growth inhibition response to the wild‐type seedlings (Fig. 3B). Collectively, we found that root growth was inhibited by OPDA in all plants, but the exla2 mutants were fairly tolerant to PPA1 treatment. This indicates that mutations in EXLA2 enhanced tolerance to PPA1, but not to OPDA.
Figure 3.
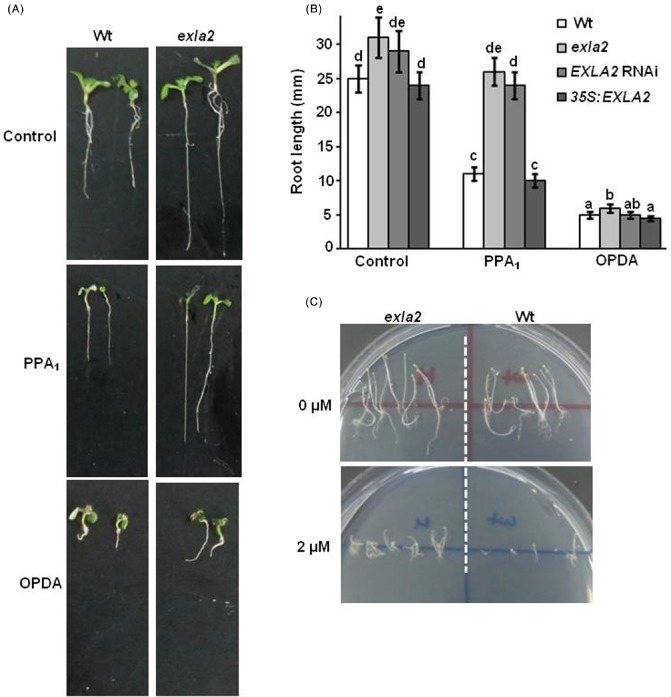
Mutations in expansin‐like A2 (EXLA2) enhance tolerance to phytoprostane‐A1 (PPA 1) and oxidative stress. Germination of seeds and survival (A) and root growth of seedlings (B) grown on 12‐oxo‐phytodienoic acid (OPDA)‐ or PPA 1‐containing medium (75 μm). (C) Germination and seedling survival of seeds plated on paraquat‐containing medium. Wt, wild‐type.
In addition, we found that the exla2 mutant plants enhanced tolerance to paraquat (Fig. 3C). In general, the increased tolerance to oxidative stress and PPA1, and resistance to B. cinerea, suggest that EXLA2 regulates plant responses to oxidative stress, reactive oxylipins and defence against necrotrophic fungi.
Identification of the pathways regulating EXLA2 expression
To further examine which pathways are involved in the reduced expression of EXLA2 in B. cinerea‐infected plants, the expression of EXLA2 was quantified in mutants impaired in JA, ET, SA, ABA and phytoalexin responses. Therefore, defective mutants of various defence response pathways were tested, including the ET response (ethylene‐insensitive 2, ein2), JA signalling (coi1), camalexin accumulation (phytoalexin‐deficient2, pad2), ABA response (ABA‐insensitive 1‐1, abi1‐1) and SA accumulation plants expressing the bacterial salicylate hydroxylase nahG gene, nahG, in response to B. cinerea (Fig. 4A, B). These mutants have been shown previously to exhibit increased susceptibility to B. cinerea (AbuQamar et al., 2006; Ferrari et al., 2003; Gosti et al., 1999; Thomma et al., 1999; van Wees et al., 2003). As in the wild‐type plants, the basal levels of EXLA2 transcripts in all impaired mutants were similar (Fig. 4A). Although only coi1 mutant plants showed basal levels of EXLA2 expression after inoculation with B. cinerea; the other infected mutants and wild‐type plants exhibited reduced levels of EXLA2 transcripts. This indicates that EXLA2 requires COI1 for its repressed expression during the disease response and that EXLA2 is regulated via COI1.
Figure 4.
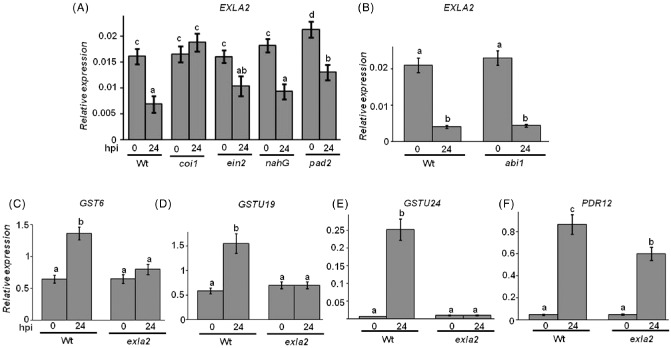
Expression of expansin‐like A2 (EXLA2) in defence response pathways and of oxylipin‐regulated genes during Botrytis cinerea infection. Botrytis cinerea suppressed EXLA2 expression in Arabidopsis coi1, ein2, nahG and pad2 mutants (A) and the abi1 mutant (B). Expression of oxylipin‐regulated genes GST6 (C), GSTU19 (D) GSTU24 (E) and PDR12 (F) in exla2 mutant plants. Gene definitions are given in the text. Wt, wild‐type.
Down‐regulation of EXLA2 alters the expression of cyclopentenone‐regulated genes in response to B. cinerea
JA, OPDA and PPA1 accumulate on pathogen infection (Mueller et al., 2008; Stotz et al., 2013; Thoma et al., 2003). The role of JA in plant resistance to B. cinerea via the COI1‐dependent pathway has been well documented (Block et al., 2005; Kazan and Manners, 2008; Méndez‐Bravo et al., 2011; Yan et al., 2009). In contrast, the role of the electrophilic oxylipins, OPDA and PPA1, in plant defence is still poorly known.
In order to link EXLA2 function in defence to specific pathway(s), we assessed the response of B. cinerea‐infected tissues to molecular markers of different signalling pathways. The increased transcript levels of EXLA2 in coi1 and the responses of exla2 to B. cinerea suggest that EXLA2 may function through COI1. Therefore, we tested the expression of JA/COI1‐regulated genes in both the wild‐type and exla2 mutant plants infected with B. cinerea. At 24 hpi with B. cinerea, the transcript levels of MBP ( myrosinase binding protein), HPL (hydroperoxide lyase) and PDF1.2 (plant defensin 1.2) were induced significantly in both wild‐type and exla2 mutant plants (Fig. S5A–C, see Supporting Information). These B. cinerea‐induced genes are COI1 dependent and activated by JA and/or OPDA (Stintzi et al., 2001; Taki et al., 2005). This suggests that the down‐regulation of EXLA2 does not require these oxylipins, and hence coi1 disrupts this repression after pathogen infection.
To verify which of the oxylipins modulates B. cinerea‐regulated gene expression, we assessed the expression profile of oxylipin‐regulated genes to B. cinerea in the exla2 mutant (AbuQamar et al., 2006; Mueller et al., 2008; Stotz et al., 2013; Windram et al., 2012). Using qRT‐PCR, we analysed the expression of a number of genes that were responsive or known to be up‐regulated by PPs or OPDA. Among these are genes related to detoxification, such as GSTs (GST6, GSTU19 and GSTU24) (Mueller et al., 2008; Stotz et al., 2013). All three genes were up‐regulated in response to B. cinerea in the wild‐type (Fig. 4C–E). No induction of GST6, GSTU19 or GST24 was observed in the exla2 mutant after infection, suggesting that the regulation of these genes is affected by the down‐regulation of EXLA2 to the necrotrophic fungus B. cinerea. PDR12 (pleiotropic drug resistance 12) is also known for its strong up‐regulation in response to PPs; yet, its induction was significantly lower in exla2 relative to the wild‐type. Previously, GST6, GSTU19 and PDR12 exhibited a strong induction by PPs (Mueller et al., 2008). However, UDP‐glucuronyl/glycosyltransferase (UGT73B2), ABC transporter (multidrug resistance‐associated protein 1, MRP1) and TOLB‐like (TOLB protein‐related) were induced in response to B. cinerea in both the mutant and wild‐type seedlings (Fig. S5D, E). Previous reports have shown that electrophilic oxylipins accumulate in plants during pathogen infection (Block et al., 2005; Thoma et al., 2003). All the tested genes were either responsive or up‐regulated by PPs or OPDA, but only slightly or not induced by JA (Mueller et al., 2008; Stotz et al., 2013), which indicates that B. cinerea affects the cyclopentenones. Overall, our data suggest that there is a common regulation between electrophilic oxylipins and B. cinerea, and that EXLA2 plays a major role in this regulation.
Arabidopsis PR‐1 (pathogenesis related‐1) and PR‐4 are markers of the SA and ET signalling pathways, respectively. As expected, the transcript levels of PR‐1 and PR‐4 were induced significantly at 24 hpi with B. cinerea in both wild‐type and exla2 mutant plants (Fig. S5C). Together, the lack of alteration in the repression of EXLA2 expression in nahG and ein2 mutants or in the induction of PR‐1 and PR‐4 in the wild‐type or exla2 mutant suggests that neither the SA nor the ET signalling pathway modulates EXLA2 expression in response to B. cinerea.
EXLA2 mutation causes hypersensitivity to NaCl and cold, mediated by ABA
To study the function of the EXLA2 gene, plants of all EXLA2 genotypes were assayed for their responses to hormones and abiotic stress. We tested the germination responses of the exla2 mutant on medium supplemented with ABA. Although, exla2 plants showed enhanced growth on MS medium lacking ABA relative to the wild‐type, the germination of seedlings of this mutant showed hypersensitivity to medium supplemented with ABA (Fig. 5A). This was measured by a reduction in root growth in the exla2 mutant, when compared with the wild‐type plants, as the concentration of ABA increased (Fig. 5B). Not only was seed germination altered in exla2, but also the roots of exla2 seedlings were sensitive to ABA (Fig. 5C, D). We also checked the germination of seeds using other hormones. The germination on medium supplemented with SA, MeJA, IAA, GA or ACC was not affected by the exla2 mutation, limiting the role of EXLA2 to particular hormone responses (Fig. S6, see Supporting Information).
Figure 5.
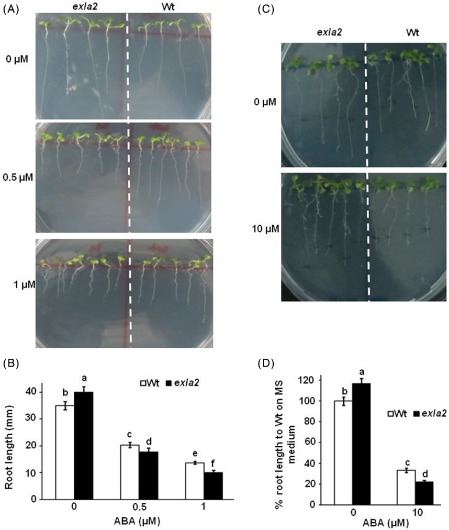
Abscisic acid (ABA) responses of expansin‐like A2 (exla2) mutant and wild‐type (Wt) seedlings. (A) Germination and seedling survival of seeds plated on ABA‐containing medium. (B) Root length measurements of seedlings growing on ABA. (C) Sensitivity of root growth in seedlings after transfer to ABA‐containing medium. (D) Percentage of reduction in root growth after transfer to ABA‐containing medium.
The exla2 mutant plants were also tested for the sensitivity of seed germination and seedling root growth to increased salinity. Mutant plants of exla2 displayed significantly reduced seed germination and growth on medium containing high salt concentrations compared with the corresponding Col‐0 wild‐type plants (Fig. 6A, B). On normal MS medium, roots of exla2 plants were longer and healthier than those of wild‐type plants (Fig. 6A, B). However, on MS medium supplemented with 50 mm NaCl, both the wild‐type and exla2 plants germinated fully, but the exla2 plants showed reduced radicle growth after germination. This retardation in root growth after germination continued at 100 mm NaCl; the germination of exla2 seeds was mostly inhibited at 150 mm NaCl (Fig. 6A, B). To test seedling sensitivity to NaCl, seedlings were pre‐germinated on MS medium and then transferred to medium containing 150 mm NaCl. The root growth of exla2 was reduced dramatically relative to that of wild‐type plants (Fig. S7A, B, see Supporting Information).
Figure 6.
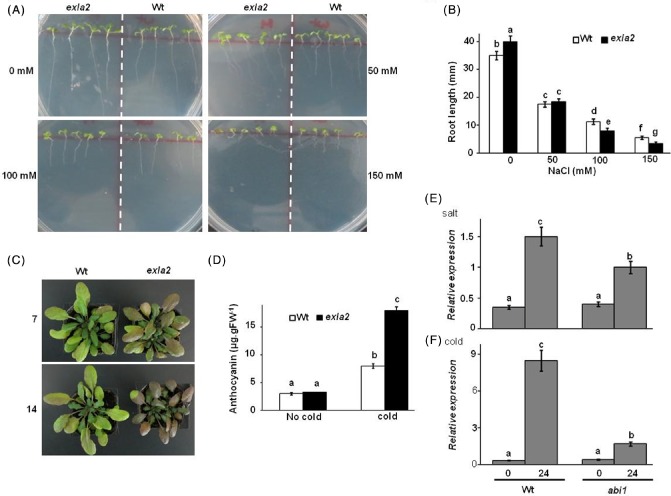
Expansin‐like A2 (EXLA2) mutation is sensitive to salt and cold. Seed germination and seedling survival (A) and seedling root length measurements (B) on NaCl‐supplemented medium. Seedling sensitivity (C) and anthocyanin content in leaves at 7 days post‐treatment (dpt) (D) with cold stress. EXLA2 expression in abi1 mutant in response to salt (E) and cold (F). Wt, wild‐type.
In order to investigate whether impaired gene expression of EXLA2 also extended to cold stress, 2‐week‐old, soil‐grown exla2 mutant and wild‐type plants were exposed to cold stress at 4 °C in the light. In contrast with the wild‐type, the growth of exla2 mutant plants was reduced dramatically (Fig. 6C, top panel); moreover, the exla2 mutant accumulated anthocyanin in leaves after 1 week of cold treatment, but less accumulation was observed in wild‐type leaves (Fig. 6D). In all experiments, the 35S:EXLA2‐overexpressing transgenic lines showed similar phenotypes to the wild‐type plants, indicating that EXLA2 is required, but not sufficient, for Arabidopsis tolerance to abiotic stress. Because ABA is known to regulate abiotic stress, we determined the expression of ABA‐induced EXLA2 in the mutant of the negative regulator of ABA signalling, ABI1 (Gosti et al., 1999). The ABA‐insensitive abi1‐1 showed reduced induction of EXLA2 expression in response to salt and cold relative to wild‐type plants (Fig. 6E, F). The induction of EXLA2 gene was weaker after cold stress than after salt stress in abi1‐1 mutants. In summary, ABA positively regulates EXLA2 expression to abiotic stress.
EXLA2 is essential for normal plant growth
The expression of expansins correlates with plant growth and development (Cho and Kende, 1997; Wu et al., 1996). Our expression profiling data confirmed the constitutive expression of EXLA2 at different developmental stages of Arabidopsis (Fig. 1D). In order to determine the effects of the transgenic EXLA2 lines, the phenotypes of all genotypes of EXLA2 were monitored from germination to seed maturation. Indeed, we did not distinguish any abnormal phenotype in the leaf shape in EXLA2 mutant plants; however, altered vegetative growth under normal growth conditions was observed. During the stages of vegetative growth, we noticed larger and greater numbers of rosette leaves in exla2 and EXLA2 RNAi plants relative to the wild‐type (Fig. 7A; Fig. S9A, see Supporting Information). Interestingly, transgenic plants overexpressing EXLA2 were smaller in size (Fig. 7A), but showed no difference in the total number of leaves when compared with the wild‐type (Fig. S9A). Clearly, exla2 showed the tallest plants, and the plant height was reduced gradually in EXLA2 RNAi, followed by the wild‐type and, finally, 35S: EXLA2 transgenic plants, which were the shortest of all (Fig. 7C). Therefore, plant height is strongly associated with the levels of expression of EXLA2 (Fig. 2B; Fig. S1B).
Figure 7.
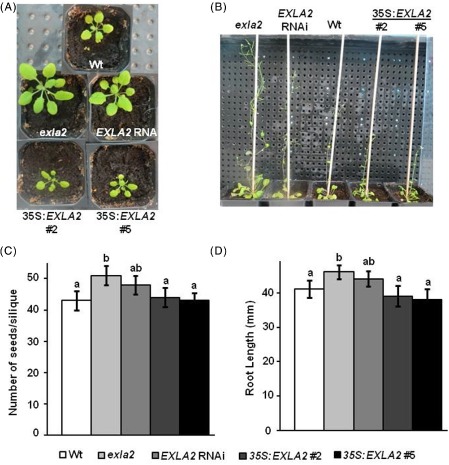
Phenotypic analysis of expansin‐like A2 (EXLA2) mutant and overexpressing transgenic plants. Vegetative growth at 14 days post‐germination (dpg) (A), plant size at 40 dpg (B), seed number per silique (C) and root elongation measurement (D). Wt, wild‐type.
We also checked whether altered levels of EXLA2 transcript showed differences in the time of flowering (Yang and Karlson, 2012). Except for exla2, all plants with different EXLA2 expression levels initiated primary fluorescence at 24 days post‐germination (dpg), which was similar to that in the wild‐type; however, seedlings of exla2 mutant alleles started to flower 3 days earlier (Table 1). This suggests that the down‐regulation of EXLA2 has an effect on the early time of flowering.
Table 1.
Analysis of flowering time in EXLA2 genotypes
| Genotype | Flowering time (dpg) | Sample size (n) |
|---|---|---|
| Wt | 23.8 ± 1.01 | 12 |
| exla2 | 21.2 ± 0.90 | 18 |
| EXLA2 RNAi | 22.6 ± 1.0 | 14 |
| 35S:EXLA2 #2 | 24.3 ± 0.82 | 15 |
| 35S:EXLA2 #5 | 24.5 ± 0.62 | 15 |
dpg, days post‐germination; Wt, wild‐type.
Siliques of exla2 allele mutant plants were longer than those of the wild‐type (Fig. S9B, C) and showed an increased seed number when compared with the wild‐type or any other transgenic plants (Fig. 7C). We also measured the root length under aseptic conditions. Ten days after planting the seeds, an increase in root length was observed on MS plates in the mutant allele, whereas the EXLA2 RNAi root system did not increase significantly (Fig. 7D); this was correlated with the relatively increased levels of EXLA2 gene expression in the RNAi line (Fig. 2B). The 35S:EXLA2 seedlings did not show an alteration in root elongation (Fig. 7D).
It should also be mentioned that EXLA2 expression correlates significantly with the expression of several genes associated with plant development (Fig. S8, see Supporting Information). As a result, EXLA2 is required for normal plant growth throughout the plant developmental stages.
Discussion
Here, we have described the molecular and genetic function of the EXLA2 gene in plant defence against necrotrophic pathogens and abiotic stress. Expansin and expansin‐like (collectively expansins) are CW‐loosening proteins that increase the extensibility of the CW (McQueen‐Mason et al., 1992). With no known enzymatic activity, expansins regulate germination, fruit ripening and pollination, and may mediate defence responses in planta. Several genetic studies have shown that structures of the CW play an important role in plant morphology and growth, tolerance to abiotic stresses and pathogenesis to necrotrophs, including B. cinerea. We have identified mutations in EXLA2 that enhance disease resistance to B. cinerea and A. brassicicola. Mutations altered in plant CW or cutin/cuticle structure, e.g. att1 (aberrant induction of type III genes), bdg (bodyguard), lcr (lacerata), rwa2 (reduced wall acetylation 2) and sma4 (symptoms to multiple avr genotypes 4), exhibit high resistance to B. cinerea (Kurdyukov et al., 2006; Manabe et al., 2011; Tang et al., 2007; Wellesen et al., 2001; Xiao et al., 2004). In tomato, double suppression of both polygalacturonase and LeEXP1 decreases susceptibility to B. cinerea in ripening fruit (Cantu et al., 2008). The down‐regulation of EXLA2 protects Arabidopsis by limiting pathogen invasion. Therefore, EXLA2 may function in defence against necrotrophic fungi. We hypothesize that the greater the repression of EXLA2, the greater the resistance towards necrotrophic pathogens; yet, the mode of action in the Arabidopsis–B. cinerea interaction is unknown.
In plants, OPDA and PP are cyclopentenone signalling molecules that are synthesized enzymatically and nonenzymatically from α‐linolenic acid (Creelman and Mullet, 1997; Durand et al., 2009; Mosblech et al., 2009; Mueller, 1997). The cyclopentenone oxylipins, OPDA and phytoprostane (i.e. PPA1), are biologically active via the action of reactive oxygen species (ROS) on pathogen infection (Sattler et al., 2006). In addition, PP activates the expression of stress response genes, leading to enhanced protection from subsequent oxidative stress (Loeffler et al., 2005; Thoma et al., 2003). In our study, mutants impaired in EXLA2 expression were tolerant to oxidative stress and PPA1 (Fig. 3). Yet, little is known about OPDA and/or PPA1 as electrophilic oxylipins in response to B. cinerea. We examined whether these cyclopentenones altered the expression of EXLA2. Microarray studies have revealed that the down‐regulation of EXLA2 is associated with PPA1 (Mueller et al., 2008). We hypothesize that the accumulation of cyclopentenones on B. cinerea infection leads to the repression of EXLA2. In tomato (Solanum lycopersicum), OPDA and PPA1 accumulate on infection with pathogens, including B. cinerea (Thoma et al., 2003). Notably, EXLA2 contains a duplication of the specific binding sequence of the TGA motif in its promoter (Fig. 2A). We speculate that there might be a transcriptional regulator that binds to this motif, and thereby regulates the expression of the EXLA2 gene during B. cinerea infection. Thus, further investigation is required to test this hypothesis. We confirmed that the major responses of exla2 seedlings are dependent (at least partly) on cyclopentenones by analysing the expression of oxylipin‐regulated genes (AbuQamar et al., 2006; Mueller et al., 2008; Stotz et al., 2013) in response to B. cinerea. It has been reported that 38% of the genes are co‐regulated by the cyclopentenone PPA1 and B. cinerea (Mueller et al., 2008). The representative set of genes related to detoxification, GST6, GSTU19 and GSTU24, and the PDR12 gene encoding the ABC transporter protein (Mueller et al., 2008; Stotz et al., 2013), were strongly up‐regulated in response to B. cinerea; yet their induction was altered in the exla2 mutant. Although the regulation of UGT73B2, MRP1 and TOLB‐like was not affected by B. cinerea infection in the EXLA2‐impaired mutant, all of the mentioned genes were also up‐regulated by the cyclopentenones OPDA and PPA1, but not by the cyclopentanone JA (Fig. 4; Mueller et al., 2008). Cyclopentenone oxylipins accumulate in plants during pathogen infection (Block et al., 2005; Thoma et al., 2003). Our data illustrate that these genes regulated by cyclopentenones are not properly expressed after B. cinerea infection in exla2. Altogether, the overlap of the responses of the exla2 mutant to B. cinerea, oxidative stress and the cyclopentenone PPA1 leads to common regulation between electrophilic oxylipins and B. cinerea that is associated with EXLA2.
We also demonstrated that EXLA2 was regulated via COI1. Our finding of a basal level of EXLA2 expression in the coi1 mutant, compared with the wild‐type (Fig. 4A), on exposure to B. cinerea confirms that EXLA2 also requires COI1 for repressed expression during the disease response. Both the coi1 and exla2 mutants showed increased tolerance to PPA1 treatment (Fig. 3; Stotz et al., 2013), but opposite responses to B. cinerea (Fig. 2; AbuQamar et al., 2006). The differences in disease response might be a result of the regulation of JA and/or its conjugates in the coi1 mutant, but not in exla2; however, further investigation is required. Mutant plants defective in OPR3 and EXLA2 displayed increased resistance to B. cinerea and A. brassicicola (Stintzi et al., 2001; Fig. 2), suggesting that electrophilic oxylipins regulate COI1‐dependent gene expression in response to necrotrophic pathogens (Chehab et al., 2011; Ribot et al., 2008; Stintzi et al., 2001). In general, our findings confirm that cyclopentenone oxylipins mediate resistance to B. cinerea through the JA‐independent COI1 signalling pathway. In addition, the EXLA2 repressed expression in nahG and ein2 mutants, similar to the wild‐type, and the induction of PR‐1 and PR‐4 in both wild‐type and exla2, on exposure to B. cinerea, illustrate that EXLA2 expression is not modulated by SA or ET signalling pathways in response to B. cinerea. Altogether, the down‐regulation of EXLA2 and its specificity to cyclopentenone and B. cinerea suggest that JA‐independent COI1 signalling regulates the plant response to B. cinerea.
Phytohormones act individually or may crosstalk on pathogen infection. In response to plant necrotrophic fungi, ABA may act as a positive or negative regulator. In tomato, ABA signalling mutants, abi1, are resistant to necrotrophic pathogens (Audenaert et al., 2002), but susceptible to these pathogens in Arabidopsis (Gosti et al., 1999). Arabidopsis mutants impaired in ABA biosynthesis and insensitive to ABA show an increased susceptibility to P. irregulare and A. brassicicola (Mauch‐Mani and Mauch, 2005), but are more resistant to B. cinerea (Adie et al., 2007). We conclude a limited effect of ABA in disease resistance mediated by EXLA2.
We argue that the EXLA2 gene has a discrete function in plant development and response. The induction of EXLA2 and the sensitivity of its mutants to cold and salt confirm other data of the up‐regulation of AtEXLA2 in Arabidopsis and Populus species on exposure to abiotic stress (Janz et al., 2012; Lee et al., 2005). In addition, the high basal expression of EXLA2 in stem tissues (Fig. 1), the salt‐induced EXLA2 homologue in developing xylem (Janz et al., 2012) and its up‐regulation during CW regeneration of cotton protoplasts (Yang et al., 2008) display an involvement of EXLA2 in CW modification and metabolism. Altogether, this shows the importance of EXLA2 in adaptive responses to abiotic stresses, and that expansin‐like genes may play a role in plant growth and the stress response (Lee et al., 2005). In most cases, the silencing of expansin genes leads to growth inhibition, whereas excessive ectopic expression leads to abnormal growth (Quiroz‐Castañeda and Folch‐Mallol, 2011). AtEXP10 antisense lines were smaller in plant size and rosettes, whereas overexpressing lines were larger (Cho and Cosgrove, 2000). By contrast, the mutation of EXLA2 showed larger plants, and transgenic EXLA2‐overexpressing plants were smaller (Fig. 7). Although exla2 plants exhibited early flowering, long siliques and an increased number of seeds per silique, EXLA2 RNAi plants were not significantly different from those of the wild‐type or ectopic EXLA2‐overexpressing transgenic lines. We believe that the ‘leaky’ EXLA2 expression of the RNAi lines indicates that a null expression of EXLA2 is required for these phenotypes.
Overall, EXLA2 has the potential to serve in plant development and defence through the regulation of endogenous signal molecules and/or pathogen‐derived effectors. Future investigations into the identification of pathogen‐suppressed EXLA2 gene expression and the relationship with membrane‐associated microbe pattern (MAMP)‐triggered defence will help to explain the functions of EXLA2 in innate immunity against B. cinerea.
Experimental Procedures
Plant growth, pathogen cultures and disease assays
The Arabidopsis wild‐type plants, mutants and transgenic overexpression lines used were in the Columbia background. Plants were grown in soil under fluorescent light (150 μE/m2/s; 12 h light/12 h dark) at 23 ± 2 °C and 60% relative humidity. The plant growth conditions and assays have been described previously (AbuQamar et al., 2006).
The culture of B. cinerea strain BO5‐10 and A. brassicicola strain MUCL20297, spore collection, plant inoculation and disease assays were performed on whole plants or detached leaves as described previously (AbuQamar et al., 2006; Zheng et al., 2006). Bacterial culture and disease assays were performed as described by Zheng et al. (2006).
Determination of fungal growth in inoculated plants
Botrytis cinerea and A. brassicicola growth in inoculated plants was determined on the basis of the levels of expression of B. cinerea ActinA and A. brassicicola cutinaseA, respectively, using qRT‐PCR (Benito et al., 1998; van Wees et al., 2003). The relative expression of B. cinerea ActinA and A. brassicicola cutinaseA to that of AtActin2 expression was determined as described previously (Bluhm and Woloshuk, 2005). Three technical replicates of the qRT‐PCR assay were used for each sample from three biological replicates.
Generation of transgenic lines and identification of the EXLA2 mutant allele
We used blast searches against the NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and TAIR (http://www.arabidopsis.org/) databases to predict the protein sequence of EXLA2. exla2 (SALK_147678; stock number N647678) was obtained from the Nottingham Arabidopsis Stock Centre (NASC, Nottingham, UK) (Sessions et al., 2002). The T‐DNA insertion in the exla2 mutant was confirmed by PCR using a T‐DNA‐specific primer (LBa1, 5′‐TGGTTCACGTAGTGGGCCATCG‐3′) and an EXLA2‐specific primer (RP, 5′‐AGAAACTCAAAACCAAAAATCAGTG‐3′). Homozygous exla2 mutant plants were identified by PCR using a pair of primers corresponding to sequences flanking the T‐DNA insertion (LP, 5′‐GGCTATCAGTACAGCATGTTATGTG‐3′; RP).
The EXLA2 cDNA (clone name RAFL04‐09‐M02; accession number AF378855) was obtained from the RIKEN BioResource Center (Ibaraki, Japan) (Seki et al., 1998, 2002). To generate overexpression plants, full‐length EXLA2 cDNA was cloned after the cauliflower mosaic virus 35S promoter into a modified version of the binary vector pCAMBIA 1200, transformed into Agrobacterium strain GV3101, and transformed into plants by Arabidopsis floral dip transformation (Clough and Bent, 1998). Transgenic plants were selected on MS medium supplemented with hygromycin, and overexpressing lines were identified by qRT‐PCR using the full‐length EXLA2 cDNA.
To generate an EXLA2 RNAi construct, 250 bp from the 3′ end (3 bp) and the 3′ untranslated region (247 bp) of EXLA2 were amplified by PCR with the primers EXLA2 RNAi‐LP (5′‐GCACTAGTCCATGGGATCACATCTGGAACTGA‐3′; SpeI and NcoI sites are shown in italic) and EXLA2 RNAi‐RP (5′‐CGGGATCCGGCGCGCCCAAAAAAACTGTAATGTC‐3′; BamHI and AscI sites are shown in italic), and cloned into the RNAi vector pGSA1165 (http://www.chromdb.org/rnai/order_vectors.html).
Morphological analysis of transgenic lines
Plants were grown at 23 °C up to 60 dpg for morphological analyses. For statistical analyses, 20 different plants were measured and photographed. Root elongation tests and measurements were carried out according to Yang and Karlson (2012).
Sensitivity treatments
All seeds were surface sterilized and plated on MS medium with 2% (w/v) sucrose and 0.8% (w/v) agar, pH 5.7 (Zheng et al., 2006). Sensitivity assays were performed using MS medium supplemented with different concentrations of chemicals. Ten‐day‐old seedlings grown in vitro were treated with 75 μm OPDA or PPA1 (Cayman Europe, Tallinn, Estonia), as described previously (Mueller et al., 2008); 50, 100 and 200 mm NaCl; 2 and 4 μm IAA; 0.5 and 1 μm ABA; 25, 50 and 100 μm MeJA, 10 μm GA; 25, 50 and 100 μm SA; or 2 μm paraquat. For paraquat, seedlings were grown in the dark. For cold treatment, seedlings were grown in soil for 2 weeks and then subjected to cold acclimation at 4 °C for another 14 days (Dong et al., 2006). Anthocyanin measurements were carried out and quantified photometrically by the absorbance at 535 nm (Teng et al., 2005).
Gene expression treatments
Gene expression analyses were determined as described previously (Laluk et al., 2011; Veronese et al., 2006; Zheng et al., 2006). Plants (4 weeks old) grown on soil were sprayed with 3 × 105 B. cinerea spores/mL, 100 μm SA, 100 μm MeJA, 100 μm ACC, 100 μm IAA, 100 μm ABA, 2 μm BL, 100 μm GA, 150 mm NaCl, 300 mm mannitol or 100 μm paraquat (methyl viologen).
Seedlings on soil were cold treated with a continuous temperature of 4 °C on crushed ice in a cold chamber. Wounding was performed by pressing approximately 60% of the leaf surface area using serrated forceps.
RNA extraction and expression analysis
RNA extraction, cDNA synthesis and qRT‐PCR expression analyses were performed as described previously (Dhawan et al., 2009). RT‐PCR and qRT‐PCR were performed using gene‐specific primers, with Arabidopsis Actin2 as an endogenous reference for normalization. A minimum of three technical replicates was used for each sample with a minimum of two biological replicates for qRT‐PCR. Expression levels were calculated by the comparative cycle threshold method, and normalization to the control was performed as described previously (Bluhm and Woloshuk, 2005). The primers used are listed in Table S1 (see Supporting Information).
Statistical analysis
All experiments were repeated at least three times with similar results. Analysis of variance and Duncan's multiple range test were performed to determine the statistical significance (SAS Institute, 1999). Mean values followed by asterisks or letters are significantly different from the corresponding control, or different from each other, respectively (P = 0.05).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Actin2 (At3G18780), PR‐1 (At2g14610), EXLA1 (At3g45970), EXLA3 (At3g45960); PR‐4 (At3g04720); ABI1 (At4G26080); CYP81D11 (At3g28740), UGT73B2 (At4g34135), GST6 (At2g47730), GSTU24 (At1g17170), GSTU19 (At1g78380), MRP1 (At1g30400), PDR12 (At1g15520), HSP70 (At3g12580), HSP17.6 (At2g29500), OPR1 (At1g76680), OPR3 (At2g06050), ELI3‐2 (At4g37990), AOX3 (At1g32350), TOLB (At4g01870), MBP (At3g16460), HPL (At4g15440), PHO1;H10 (At1g69480), GST6 (At2g47730).
Supporting information
Fig. S1 Multiple sequence alignment of expansin‐like A2 (EXLA2) and closely related Arabidopsis cDNA.
Fig. S2 Multiple sequence alignment of expansin‐like A2 (EXLA2) and closely related Arabidopsis and other EXLA2 proteins in plant species.
Fig. S3 Expression of expansin‐like A2 (EXLA2) on exposure to Pseudomonas syringae and identification of AtEXLA2‐overexpressing plants.
Fig. S4 Plant responses of altered expansin‐like A2 (EXLA2) expression on exposure to Botrytis cinerea and Pseudomonas syringae.
Fig. S5 Expression of oxylipin‐responsive and defence‐related genes during Botrytis cinerea infection.
Fig. S6 Germination and growth responses of the expansin‐like A2 (exla2) mutant are not altered on exposure to salicylic acid (SA), methyl jasmonate (MeJA), indole acetic acid (IAA), gibberellin (GA) or aminocyclopropane‐1‐carboxylic acid (ACC).
Fig. S7 Sensitivity of Arabidopsis expansin‐like A2 (EXLA2) mutant to salt.
Fig. S8 Expansin‐like A2 (EXLA2) expression at different developmental stages in relation to other genes.
Fig. S9 Additional morphological analyses of expansin‐like A2 (EXLA2) mutant and overexpressing transgenic plants.
Table S1 List of primers (sequence 5′ to 3′) used in this study.
Acknowledgements
We thank Professor Tesfaye Mengiste for providing us with the binary vector and the strains of Pseudomonas syringae. We also thank Mr Noushad Karuvantevida for technical assistance in the qRT‐PCR analysis. This project was funded by the UAEU (FOS/MRG‐03/11) and the UAEU‐NRF [27/11/2] to SAQ.
The authors have no conflicts of interest to declare.
References
- AbuQamar, S. , Chen, X. , Dhawan, R. , Bluhm, B. , Salmeron, J. , Lam, S. , Dietrich, R.A. and Mengiste, T. (2006) Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 48, 28–44. [DOI] [PubMed] [Google Scholar]
- AbuQamar, S. , Luo, H. , Laluk, K. , Mickelbart, M. and Mengiste, T. (2009) Cross‐talk between biotic and abiotic stress responses is mediated by the tomato AIM1 transcription factor. Plant J. 58, 347–360. [DOI] [PubMed] [Google Scholar]
- Adie, B.A. , Pérez‐Pérez, J. , Pérez‐Pérez, M.M. , Godoy, M. , Sánchez‐Serrano, J.J. , Schmelz, E.A. and Solano, R. (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell, 19, 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert, K. , De Meyer, G.B. and Hofte, M.M. (2002) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid‐dependent signaling mechanisms. Plant Physiol. 128, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito, E.P. , ten Have, A. , van 't Klooster, J.W. and van Kan, J.A.L. (1998) Fungal and plant gene expression during synchronized infection of tomato leaves by Botrytis cinerea . Eur. J. Plant Pathol. 104, 207–220. [Google Scholar]
- Block, A. , Schmelz, E. , Jones, J.B. and Klee, H.J. (2005) Coronatine and salicylic acid: the battle between Arabidopsis and Pseudomonas for phytohormone control. Mol. Plant Pathol. 6, 79–83. [DOI] [PubMed] [Google Scholar]
- Bluhm, B.H. and Woloshuk, C.P. (2005) Amylopectin induces fumonisin B1 production by Fusarium verticillioides during colonization of maize kernels. Mol. Plant–Microbe Interact. 18, 1333–1339. [DOI] [PubMed] [Google Scholar]
- Brouwer, M. , Lievens, B. , Van Hemelrijck, W. , Van den Ackerveken, G. , Cammue, B.P. and Thomma, B.P. (2003) Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real‐time fluorescence PCR. FEMS Microbiol. Lett. 228, 241–248. [DOI] [PubMed] [Google Scholar]
- Cantu, D. , Vicente, A.R. , Greve, L.C. , Dewey, F.M. , Bennett, A.B. , Labavitch, J.M. and Powell, A.L.T. (2008) The intersection between cell wall disassembly, ripening and fruit susceptibility to B. cinerea . Proc. Natl. Acad. Sci. USA, 105, 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab, E.W. , Kim, S. , Savchenko, T. , Kliebenstein, D. , Dehesh, K. and Braam, J. (2011) Intronic T‐DNA insertion renders Arabidopsis opr3 a conditional jasmonic acid‐producing mutant. Plant Physiol. 156, 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.T. and Cosgrove, D.J. (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Aridopsis thaliana . Proc. Natl. Acad. Sci. USA, 97, 9783–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.T. and Cosgrove, D.J. (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell, 14, 3237–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.T. and Kende, H. (1997) Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell, 9, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2000) Loosening of plant cell walls by expansins. Nature, 407, 321–326. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A. and Mullet, J.E. (1997) Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. [DOI] [PubMed] [Google Scholar]
- Dermatsev, V. , Weingarten‐Baror, C. , Resnick, N. , Gadkar, V. , Wininger, S. , Kolotilin, I. , Mayzlish‐Gati, E. , Zilberstein, A. , Koltai, H. and Kapulnik, Y. (2010) Microarray analysis and functional tests suggest the involvement of expansins in the early stages of symbiosis of the arbuscular mycorrhizal fungus Glomus intraradices on tomato (Solanum lycopersicum). Mol. Plant Pathol. 11, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan, R. , Luo, H. , Foerster, A. , AbuQamar, S. , Du, H.‐N. , Briggs, S. , Scheid, O. and Mengsite, T. (2009) HISTONE MONOUBIQUITINATION 1 interacts with a subunit of the mediator complex and regulates defense responses against necrotrophic fungal pathogens in Arabidopsis. Plant Cell, 21, 1000–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C.‐H. , Hu, X. , Tang, W. , Zheng, X. , Kim, Y.S. , Lee, B.‐H. and Zhu, J.‐K. (2006) A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 26, 9533–9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes, B.P. and Crowell, D.N. (1998) Cytokinin regulates the expression of a soybean β‐expansin gene by a post‐transcriptional mechanism. Plant Mol. Biol. 37, 437–444. [DOI] [PubMed] [Google Scholar]
- Durand, T. , Bultel‐Poncé, V. , Guy, A. , Berger, S. , Mueller, M.J. and Galano, J.‐M. (2009) New bioactive oxylipins formed by non‐enzymatic free‐radical‐catalyzed pathways: the phytoprostanes. Lipids, 44, 875–888. [DOI] [PubMed] [Google Scholar]
- Ferrari, S. , Plotnikova, J.M. , De Lorenzo, G. and Ausubel, F.M. (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4 . Plant J. 35, 193–205. [DOI] [PubMed] [Google Scholar]
- Fujita, M. , Fujita, Y. , Noutoshi, Y. , Takahashi, F. , Narusaka, Y. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. [DOI] [PubMed] [Google Scholar]
- Giordano, W. and Hirsch, A.M. (2004) The expression of MaEXP1, a Melilotus alba expansin gene, is upregulated during the sweetclover–Sinorhizobium meliloti interaction. Mol. Plant–Microbe Interact. 17, 613–622. [DOI] [PubMed] [Google Scholar]
- Gosti, F. , Beaudoin, N. , Serizet, C. , Webb, A.A. , Vartanian, N. and Giraudat, J. (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell, 11, 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb, A. , Krishnan, A. , Madana, M.R. , Ambavaram, M.M.R. and Pereira, A. (2010) Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 154, 1254–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz, D. , Lautner, S. , Wildhagen, H. , Behnke, K. , Schnitzler, J. , P., Rennenberg, H. , Fromm, J. and Polle, A. (2012) Salt stress induces the formation of a novel type of ‘pressure wood’ in two Populus species. New Phytol. 194, 129–141. [DOI] [PubMed] [Google Scholar]
- Jones, L. and McQueen‐Mason, S.A. (2004) Role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum . FEBS Lett. 559, 61–65. [DOI] [PubMed] [Google Scholar]
- Kazan, K. and Manners, J.M. (2008) Jasmonate signaling: toward an integrated view. Plant Physiol. 146, 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende, H. , Bradford, K. , Brummell, D. , Cho, H.T. , Cosgrove, D. , Fleming, A. , Gehring, C. , Lee, Y. , McQueen‐Mason, S. , Rose, J. and Voesenek, L. (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol. Biol. 55, 311–314. [DOI] [PubMed] [Google Scholar]
- Kim, E.S. , Lee, H.J. , Bang, W.G. , Choi, I.G. and Kim, K.H. (2009) Functional characterization of a bacterial expansin from Bacillus subtilis for enhanced enzymatic hydrolysis of cellulose. Biotechnol. Bioeng. 102, 1342–1353. [DOI] [PubMed] [Google Scholar]
- Kurdyukov, S. , Faust, A. , Nawrath, C. , Bar, S. , Voisin, D. , Efremova, N. , Franke, R. , Schreiber, L. , Saedler, H. , Metraux, J.P. and Yephremov, A. (2006) The epidermis‐specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis . Plant Cell, 18, 321–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk, K. , AbuQamar, S. and Mengiste, T. (2011) The Arabidopsis mitochondrial localized pentatricopeptide repeat protein PGN functions in defense against necrotrophic fungi and abiotic stress tolerance. Plant Physiol. 156, 2053–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.‐H. , Henderson, D.A. and Zhu, J.‐K. (2005) The Arabidopsis cold‐responsive transcriptome and its regulation by ICE1. Plant Cell, 17, 3155–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. and Kende, H. (2002) Expression of α‐expansin and expansin‐like genes in deepwater rice. Plant Physiol. 130, 1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. , Choi, D. and Kende, H. (2001) Expansins: ever‐expanding numbers and functions. Curr. Opin. Plant Biol. 4, 527–532. [DOI] [PubMed] [Google Scholar]
- Levy, I. , Shani, Z. and Shoseyov, O. (2002) Modification of polysaccharides and plant cell wall by endo‐1,4‐beta‐glucanase and cellulose‐binding domains. Biomol. Eng. 19, 17–30. [DOI] [PubMed] [Google Scholar]
- Loeffler, C. , Berger, S. , Guy, A. , Durand, T. , Bringmann, G. , Dreyer, M. , von Rad, U. , Durner, J. and Mueller, M.J. (2005) B1‐phytoprostanes trigger plant defense and detoxification responses. Plant Physiol. 137, 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe, Y. , Nafisi, M. , Verhertbruggen, Y. , Orfila, C. , Gille, S. , Rautengarten, C. , Cherk, C. , Marcus, S.E. , Somerville, S. , Pauly, M. , Knox, J.P. , Sakuragi, Y. and Scheller, H.V. (2011) Loss‐of‐function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea . Plant Physiol. 155, 1068–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch‐Mani, B. and Mauch, F. (2005) The role of abscisic acid in plant–pathogen interactions. Curr. Opin. Plant Biol. 8, 409–414. [DOI] [PubMed] [Google Scholar]
- McQueen‐Mason, S. , Durachko, D.M. and Cosgrove, D.J. (1992) Two endogenous proteins that induce wall extension. Plant Cell, 4, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez‐Bravo, A. , Calderón‐Vázquez, C. , Ibarra‐Laclette, E. , Raya‐González, J. , Ramírez‐Chávez, E. , Molina‐Torres, J. , Guevara‐García, A.A. , López‐Bucio, J. and Herrera‐Estrella, L. (2011) Alkamides activate jasmonic acid biosynthesis and signaling pathways and confer resistance to Botrytis cinerea in Arabidopsis thaliana . Plos ONE, 6, e27251. doi: 10.1371/journal.pone.0027251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosblech, A. , Feussner, I. and Heilmann, I. (2009) Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 47, 511–517. [DOI] [PubMed] [Google Scholar]
- Mueller, M.J. (1997) Enzymes involved in jasmonic acid biosynthesis. Physiol. Plant. 100, 653–663. [Google Scholar]
- Mueller, S. , Hilbert, B. , Dueckershoff, K. , Roitsch, T. , Krischke, M. , Mueller, M.J. and Berger, S. (2008) General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell, 20, 768–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroz‐Castañeda, R.E. and Folch‐Mallol, J.L. (2011) Plant cell wall degrading and remodeling proteins: current perspectives. Biotechnol. Appl. 28, 205–215. [Google Scholar]
- Ribot, C. , Zimmerli, C. , Farmer, E.E. , Reymond, P. and Poirier, Y. (2008) Induction of the Arabidopsis PHO1;H10 gene by 12‐oxo‐phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1‐dependent pathway. Plant Physiol. 147, 696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Navarro, L. , Bari, R. and Jones, J.D. (2007) Pathological hormone imbalances. Curr. Opin. Plant Biol. 10, 372–379. [DOI] [PubMed] [Google Scholar]
- Sampedro, J. and Cosgrove, D.J. (2005) The expansin superfamily. Genome Biol. 6, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute (1999) The SAS system for windows In: Release 8.0 SAS Institute. Cary, NC. [Google Scholar]
- Sattler, S.E. , Mene‐Saffrane, L. , Farmer, E.E. , Krischke, M. , Mueller, M.J. and DellaPenna, D. (2006) Nonenzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol‐deficient mutants. Plant Cell, 18, 3706–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M. , Carninci, P. , Nishiyama, Y. , Hayashizaki, Y. and Shinozki, K. (1998) High‐efficiency cloning of Arabidopsis full length cDNA by biotinylated CAP trapper. Plant J. 15, 707–720. [DOI] [PubMed] [Google Scholar]
- Seki, M. , Narusaka, M. , Kamiya, A. , Ishida, J. , Satou, M. , Tetsuya, S. , Nakajima, M. , Enju, A. , Akiyama, K. , Oono, Y. , Muramatsu, M. , Hayashizaki, Y. , Kawai, J. , Carninci, P. , Itoh, M. , Ishii, Y. , Arakawa, T. , Shibata, K. , Shinagawa, A. and Shinozaki, K. (2002) Functional annotation of a full‐length Arabidopsis cDNA collection. Science, 296, 141–145. [DOI] [PubMed] [Google Scholar]
- Sessions, A. , Burke, E. , Presting, G. , Aux, G. , McElver, J. , Patton, D. , Dietrich, B. , Ho, P. , Bacwaden, J. , Ko, C. , Clarke, J.D. , Cotton, D. , Bullis, D. , Snell, J. , Miguel, T. , Hutchison, D. , Kimmerly, B. , Mitzel, T. , Katagiri, F. , Glazebrook, J. , Law, M. and Goffa, S.A. (2002) A high‐throughput Arabidopsis reverse genetics system. Plant Cell, 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi, A. , Weber, H. , Reymond, P. , Browse, J. and Farmer, E.E. (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc. Natl. Acad. Sci. USA, 98, 12 837–12 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz, H.U. , Mueller, S. , Zoeller, M. , Mueller, M.J. and Berger, S. (2013) TGA transcription factors and jasmonate‐independent COI1 signalling regulate specific plant responses to reactive oxylipins. J. Exp. Bot. 64, 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Veerabomma, S. , Abdel‐Mageed, H.A. , Fokar, M. , Asami, T. , Yoshida, S. and Allen, R.D. (2005) Brassinosteroid regulates fiber development on cultured cotton ovules. Plant Cell Physiol. 46, 1384–1391. [DOI] [PubMed] [Google Scholar]
- Taki, N. , Sasaki‐Sekimoto, Y. , Obayashi, T. , Kikuta, A. , Kobayashi, K. , Ainai, T. , Yagi, K. , Sakurai, N. , Suzuki, H. , Masuda, T. , Takamiya, K. , Shibata, D. , Kobayashi, Y. and Ohta, H. (2005) 12‐Oxo‐phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound‐induced gene expression in Arabidopsis . Plant Physiol. 139, 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, D. , Simonich, M.T. and Innes, R.W. (2007) Mutations in LACS2, a long‐chain acyl‐coenzyme A synthetase, enhance susceptibility to avirulent Pseudomonas syringae but confer resistance to Botrytis cinerea in Arabidopsis. Plant Physiol. 144, 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S. , Keurentjes, J. , Bentsink, L. , Koornneef, M. and Smeekens, J. (2005) Sucrose‐specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma, I. , Loeffler, C. , Sinha, A.K. , Gupta, M. , Krischke, M. , Steffan, B. , Roitsch, T. and Mueller, M.J. (2003) Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J. 34, 363–375. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P. , Eggermont, K. , Tierens, K.F. and Broekaert, W.F. (1999) Requirement of functional ethylene‐insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea . Plant Physiol. 121, 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese, P. , Nakagami, H. , Bluhm, B. , AbuQamar, S. , Chen, X. , Salmeron, J. , Dietrich, R.A. , Hirt, H. and Mengiste, T. (2006) The membrane‐anchored BOTRYTIS INDUCED KINASE1 has distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell, 18, 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees, S.C. , Chang, H.S. , Zhu, T. and Glazebrook, J. (2003) Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 132, 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellesen, K. , Durst, F. , Pinot, F. , Benveniste, I. , Nettesheim, K. , Wisman, E. , Steiner‐Lange, S. , Saedler, H. and Yephremov, A. (2001) Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid omega‐hydroxylation in development. Proc. Natl. Acad. Sci. USA, 98, 9694–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windram, O. , Madhou, P. , McHattie, S. , Hill, C. , Hickman, R. , Cooke, E. , Jenkins, D.J. , Penfold, C.A. , Baxter, L. , Breeze, E. , Kiddle, S.J. , Rhodes, J. , Atwell, S. , Kliebenstein, D.J. , Kim, Y.S. , Stegle, O. , Borgwardt, K. , Zhang, C. , Tabrett, A. , Legaie, R. , Moore, J. , Finkenstadt, B. , Wild, D.L. , Mead, A. , Rand, D. , Beynon, J. , Ott, S. , Buchanan‐Wollaston, V. and Denby, K.J. (2012) Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high‐resolution temporal transcriptomic analysis. Plant Cell, 24, 3530–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Sharp, R.E. , Durachko, D.M. and Cosgrove, D.J. (1996) Growth maintenance of the maize primary root at low water potentials involves increases in cell wall extension properties, expansin activity and wall susceptibility to expansins. Plant Physiol. 111, 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, F. , Goodwin, S.M. , Xiao, Y. , Sun, Z. , Baker, D. , Tang, X. , Jenks, M.A. and Zhou, J.M. (2004) Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 23, 2903–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Tian, J. , Belanger, F.C. and Huang, B. (2007) Identification and characterization of an expansin gene AsEXP1 associated with heat tolerance in C3 Agrostis grass species. J. Exp. Bot. 58, 3789–3796. [DOI] [PubMed] [Google Scholar]
- Yan, J. , Zhang, C. , Gu, M. , Bai, Z. , Zhang, W. , Qi, T. , Cheng, Z. , Peng, W. , Luo, H. , Nan, F. , Wang, Z. and Xie, D. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell, 21, 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Tu, L. , Zhu, L. , Fu, L. , Min, L. and Zhang, X. (2008) Expression profile analysis of genes involved in cell wall regeneration during protoplast culture in cotton by suppression subtractive hybridization and macroarray. J. Exp. Bot. 59, 3661–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. and Karlson, D. (2012) Effects of mutations in the Arabidopsis Cold Shock Domain Protein 3 (AtCSP3) gene on leaf cell expansion. J. Exp. Bot. 63, 4861–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M.R. , Han, Y.Y. , Feng, Y.N. , Li, F. and Wang, W. (2012) Expansins are involved in cell growth mediated by abscisic acid and indole‐3‐acetic acid under drought stress in wheat. Plant Cell Rep. 31, 671–685. [DOI] [PubMed] [Google Scholar]
- Zheng, Z. , AbuQamar, S. , Chen, Z. and Mengiste, T. (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Multiple sequence alignment of expansin‐like A2 (EXLA2) and closely related Arabidopsis cDNA.
Fig. S2 Multiple sequence alignment of expansin‐like A2 (EXLA2) and closely related Arabidopsis and other EXLA2 proteins in plant species.
Fig. S3 Expression of expansin‐like A2 (EXLA2) on exposure to Pseudomonas syringae and identification of AtEXLA2‐overexpressing plants.
Fig. S4 Plant responses of altered expansin‐like A2 (EXLA2) expression on exposure to Botrytis cinerea and Pseudomonas syringae.
Fig. S5 Expression of oxylipin‐responsive and defence‐related genes during Botrytis cinerea infection.
Fig. S6 Germination and growth responses of the expansin‐like A2 (exla2) mutant are not altered on exposure to salicylic acid (SA), methyl jasmonate (MeJA), indole acetic acid (IAA), gibberellin (GA) or aminocyclopropane‐1‐carboxylic acid (ACC).
Fig. S7 Sensitivity of Arabidopsis expansin‐like A2 (EXLA2) mutant to salt.
Fig. S8 Expansin‐like A2 (EXLA2) expression at different developmental stages in relation to other genes.
Fig. S9 Additional morphological analyses of expansin‐like A2 (EXLA2) mutant and overexpressing transgenic plants.
Table S1 List of primers (sequence 5′ to 3′) used in this study.


