Abstract
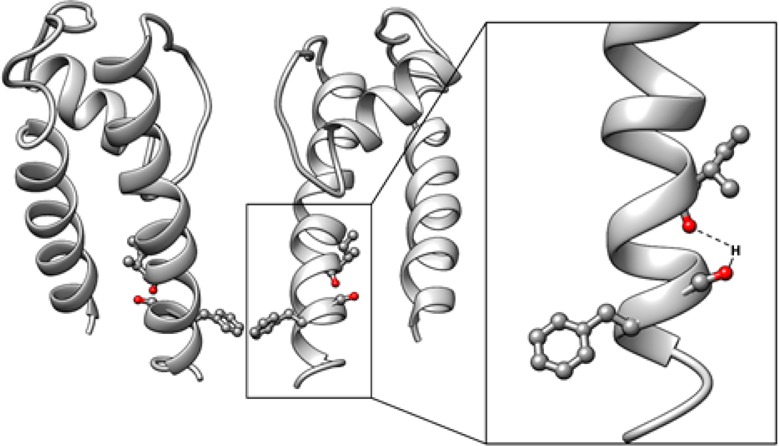
Gating of ion channels is based on structural transitions between open and closed states. To uncover the chemical basis of individual gates, we performed a comparative experimental and computational analysis between two K+ channels, KcvS and KcvNTS. These small viral encoded K+ channel proteins, with a monomer size of only 82 amino acids, resemble the pore module of all complex K+ channels in terms of structure and function. Even though both proteins share about 90% amino acid sequence identity, they exhibit different open probabilities with ca. 90% in KcvNTS and 40% in KcvS. Single channel analysis, mutational studies and molecular dynamics simulations show that the difference in open probability is caused by one long closed state in KcvS. This state is structurally created in the tetrameric channel by a transient, Ser mediated, intrahelical hydrogen bond. The resulting kink in the inner transmembrane domain swings the aromatic rings from downstream Phes in the cavity of the channel, which blocks ion flux. The frequent occurrence of Ser or Thr based helical kinks in membrane proteins suggests that a similar mechanism could also occur in the gating of other ion channels.
Introduction
Ion channels fluctuate stochastically between conductive “open” states and nonconductive “closed” states.1 This process of gating determines the flux of ions across membranes and is therefore a crucial parameter for the regulation of channel activity in the physiological context. Many physical factors like voltage or mechanical stretch and chemical factors like ligands or signaling molecules can modulate the stability of individual closed or open states and as a consequence regulate ion fluxes across membranes. The recent availability of high-resolution structures of channel proteins makes it possible to correlate experimentally measured gating events from single channel recordings with dynamic structural phenomena.2,3 Well documented correlations between channel gating and structural modifications are known from the model K+ channel KcsA. Two gates have been identified in this channel. One gate is located at the inner bundle crossing and is activated by H+. The second gate is in the selectivity filter. Additional structure/function studies have uncovered some electrostatic interactions among amino acid (AA) side chains, which are presumably responsible for the stochastic switching between open and closed states of the channel. For the H+ dependent gating in the KcsA channel a complex network of inter- and intrasubunit salt bridges and hydrogen bonds were identified near the bundle crossing; these are stabilized or destabilized according to pH.3 In the case of the KcsA filter gate a network of AA side chains surrounding the selectivity filter together with the ions in the filter and water behind the filter affect the stability of the conformational states of the pore. A key role in this network is played by a single carboxyl-carboxylate interaction between a Glu (E71) in the pore helix and an Asp (D80) in the signature sequence. Substitution of E71 with Ala (E71A) led to a complete disappearance of inactivation and a constitutively open channel at low pH.2
Here we use small chlorovirus encoded K+ channels to search for additional structural motives for ion channel gating. These channels, which represent the pore module of complex K+ channels,4 have the advantage of being truly minimal because they consist of two transmembrane domains, a pore loop, short N and C termini and small extracellular loops (Figure 1a). Like other K+ channels they form functional tetramers and have many functional properties present in more complex K+ channels. These properties include K+ selectivity, a susceptibility to blockers and, important for this study, stochastic gating between defined open and closed states.5,6 By using K+ channel genes from chloroviruses isolated from different environments we have created a large library of structural orthologs, which exhibit distinct functional differences.7,8 In the present study we analyzed two small K+ channel orthologs, which differ in the presence or absence of a long-lived closed state, even though they have high AA identity. Comparative analysis of the two channels identified a Phe in the inner transmembrane helix as a gate, which can obstruct the ion pathway in the cavity. The stochastic insertion and removal of the hydrophobic barrier is achieved by the formation of H-bonds between an adjacent Ser with upstream-positioned AAs in the transmembrane helix. Hence stochastic gating of this long-lived closed state presumably reflects the formation of a kink in the inner transmembrane helix. This in turn causes an orientation of the Phe in the ionic pathway where it functions as a barrier of ion flux.
Figure 1.
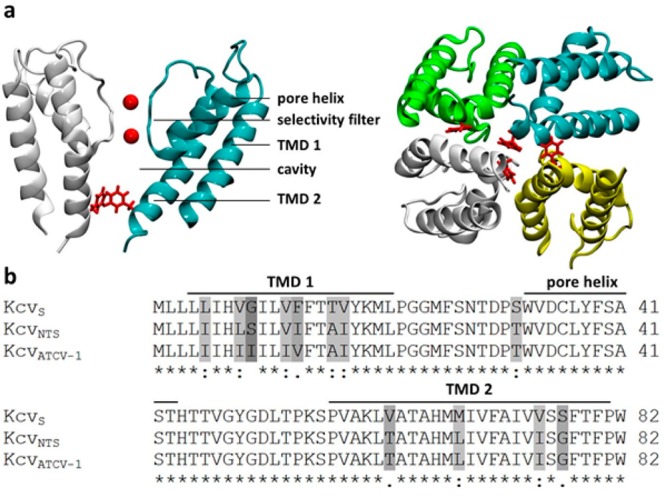
Chlorovirus encoded K+ channels. (a) Cartoon representations of a snapshot of KcvS taken from MD simulation at 80 ns of the production run. Left: Side view of the channel (two opposing monomer units). Right: view from bottom to top of the full tetrameric channel. Potassium ions are shown as red spheres, and F78 as explicit side chains in red. (b) Sequence alignment of three viral encoded K+ channels. The position of the transmembrane domains TMD1 and TMD2 as well as the pore helix are indicated by bars. The sequences are 86% identical; amino acid differences are highlighted in gray.
Results and Discussion
Two Similar Channels with Different Open Probabilities
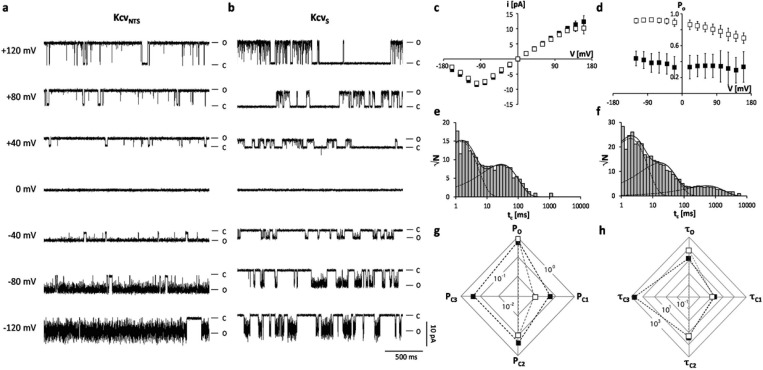
The viral encoded proteins KcvNTS and KcvS are small K+ channels with only 82 AAs per monomer. The two proteins differ in 11 AAs of which 8 are conservative and 3 are semiconservative exchanges (Figure 1b). The difference between KcvNTS and a third channel KcvATCV-1, which was analyzed previously,9 is only four AAs. When reconstituted in planar lipid bilayers, KcvNTS and KcvS generated channel fluctuations with a similar unitary conductance and the same characteristic i/V relation (Figure 2a–c). The latter is characterized by an Ohmic conductance at positive voltages and a negative slope conductance at negative voltages. The decrease in channel amplitude at negative voltages is caused by a rapid flickering of the channel, which results in unresolved channel openings (Figure 2a,b).6 These recordings imply that the AA deviations between the two channels have no large impact on the unitary conductance and on fast gating, which is presumably dominated by a selectivity filter gate.6
Figure 2.
The difference in open probability of two similar K+ channels originates from a long closed time, which is absent in KcvNTS and present in KcvS. (a,b) Characteristic single channel fluctuations of KcvNTS and KcvS at different voltages in a planar lipid bilayer. The closed (C) and open (O) levels are indicated along the current traces. (c) Mean single channel i/V relations (±sd) of KcvNTS (open squares) and KcvS (filled squares) from 6 and 9 independent recordings, respectively. (d) Mean open probabilities (±sd) of the two channels from 6 and 9 independent recordings. Exemplary closed dwell times at +120 mV for KcvNTS (e) and KcvS (f). The data in e can be fitted with two exponentials while the data in f require a third exponential. The long-lived closed state in f is absent in e. The multiple kinetic variables of the two channels including the probabilities (P) for occupying the open state (O) and the closed states (C1–C3) (g) as well as mean lifetimes (in ms) of the open state (τo) and of the three closed states (τc1–τc3) (h) are presented in two-dimensional radar plots. The probabilities of occupancy and mean lifetimes were calculated from three independent 5 min recordings. The symbols in d, g and h correspond to those in c.
In spite of the sequence similarity, the two channels differed significantly in their open probability (Po) (Figure 2a,b,d). While KcvNTS showed robust high Po values of 0.84 ± 0.08 over the entire voltage window, KcvS exhibited a much lower open probability of only 0.35 ± 0.04. Comparative analysis of the open and closed dwell times showed that the two channels differed mainly in one distinct parameter namely a long closed time τC3, which was absent in KcvNTS (Figure 2e,g) but present and frequently occurred in KcvS (Figure 2f,g). The two channels also exhibited a difference in the mean open dwell times, which are at +120 mV 79 ± 19 ms and 15 ± 3 ms for KcvNTS and KcvS, respectively (Figure S1, Table S1). This difference is caused by variable probabilities/rate constants with which the channel fluctuates between the open state and the first, short closed state (C1). The associated closing frequency has however only a marginal effect on Po. The mean lifetime τC1 of the first closed state (1.3 ± 0.2 ms and 1.8 ± 0.8 ms at +120 mV for KcvNTS and KcvS, respective) is so short that it hardly affects Po (Figure 2g,h, Table S1). The low open probability of KcvS is therfore primarily the result of the long lasting closed events (τC3) with a mean lifetime of 480 ± 145 ms at +120 mV (Figure 2h, Table S1).
From these data we conclude that the high sequence similarity and the fact that the two channels differ in one defined closed state make their comparative analysis an ideal system for studying the molecular basis of a channel gate. Since the different AAs are distributed over the two transmembrane domains (TMDs) (Figure 1b) we first tackled the location of the gate, which is responsible for the long closed time. To this end, chimeras were generated, in which we swapped the two TMDs (Figure 3a). Figure 3b shows that both chimeras produced channel activity with the same unitary conductance (Figure 3c) but with distinct kinetics.
Figure 3.
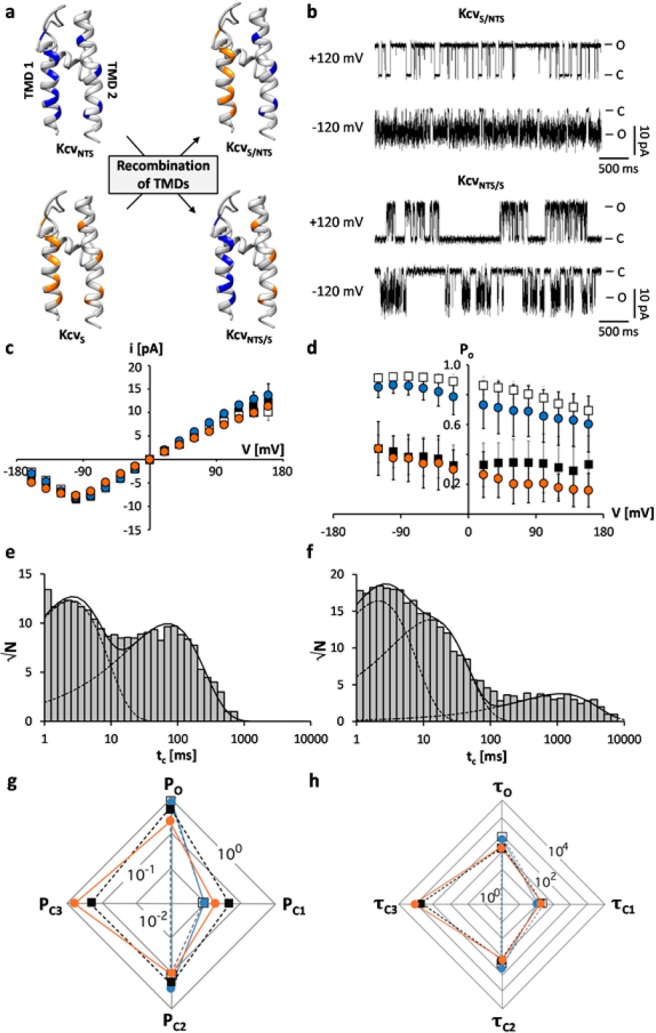
The KcvS long closed time is related to TMD2. (a) Chimeras were constructed by swapping transmembrane domains between KcvNTS (in blue) and KcvS (orange), for illustration purposes only. (b) Characteristic single channel fluctuations of chimeras KcvS/NTS and KcvNTS/S at ±120 mV in planar lipid bilayers. (c) Mean single channel i/V relations (±sd) of KcvNTS (open squares), KcvS (filled squares), KcvS/NTS (blue circles, n = 9) and KcvNTS/S (orange circles, n = 3). (d) Mean open probabilities (±sd) of wt channels and chimeras (n = 9 and n = 3 for KcvS/NTS and KcvNTS/S, respectively). Exemplary closed dwell times at +120 mV for KcvS/NTS (e) and KcvNTS/S (f). The data in e can be fitted with two exponentials while the data in f require a third exponential. Radar plots for probabilities (P) of the wt channels and the chimeras for occupying the open state (O) and the closed states (C1–C3) (g) and of mean lifetimes (in ms) of the open state (τo) and of the three closed states (τc1–τc3) (h). The probabilities of occupancy and mean lifetimes were calculated from three independent 5 min recordings. The symbols in d, g and h correspond to those in c.
It appears that the exchange of the two TMDs led to a reverse gating characteristics: KcvNTS/S now shows the long closed state and low Po whereas KcvS/NTS acquires a high Po and only short closings (Figure 3b–h, Table S1). From the results of these experiments we conclude that the structural information, which is responsible for the long closed time, is located in TMD2. Also TMD1 must have some impact on channel gating, but this contribution is not relevant for the long closed time.
Ser77 and Phe78 Are Crucial for Long-Lived Closed State
There are only four AA differences between the TMD2 of KcvNTS and KcvS. To identify the relevant AA in TMD2 for gating we mutated all 4 candidates in KcvNTS into the respective residue of KcvS. A functional analysis of the mutants showed that mutations T62V, L68M, and I75V in KcvNTS had no impact on gating (Figure S2). Only the mutual exchange of S and G at position 77 in KcvNTS and KcvS completely inverted the gating properties of the two channels: the KcvS S77G mutant acquired the high open probability of KcvNTS and the open probability of KcvNTS G77S decreased to that of KcvS (Figure 4a,b,d). The impact of the mutations on Po can be explained by the disappearance of the long closed time in KcvS S77G and its appearance in KcvNTS G77S (Figure 4e–h, Table S1).
Figure 4.
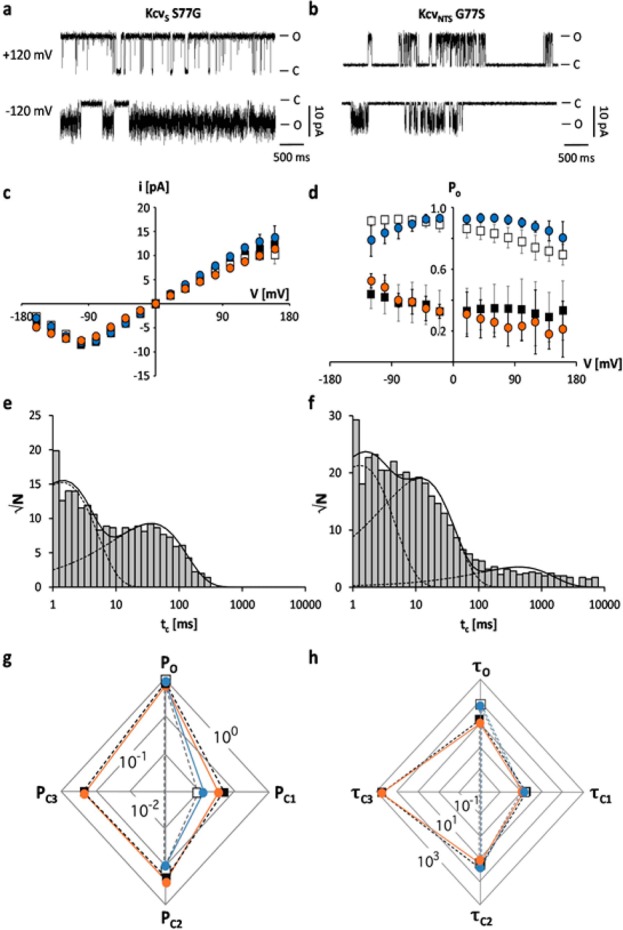
The difference of KcvNTS and KcvS in position 77 is responsible for the absence or presence of a long closed time. Characteristic single channel fluctuations of mutant KcvS S77G (a) and KcvNTS G77S (b) at ±120 mV in planar lipid bilayers. (c) Mean single channel i/V relations (±sd) of KcvNTS (open squares), KcvS (filled squares), KcvNTS G77S (orange circles, n = 5) and KcvS S77G (blue circles, n = 3). (d) Mean open probabilities (±sd) of wt channels and mutants (n = 5 and n = 3 for KcvNTS G77S and KcvS S77G, respectively). Exemplary closed dwell times at +120 mV for KcvS S77G (e) and KcvNTS G77S (f). The data in e can be fitted with two exponentials while the data in f again require a third exponential. Radar plots for probabilities (P) of the wt channels and mutants for occupying the open state (O) and the closed states (C1–C3) (g) and for mean lifetimes (in ms) of the open state (τo) and of the three closed states (τc1–τc3) (h). The probabilities of occupancy and mean lifetimes were calculated from three independent 5 min recordings. The symbols in d, g and h correspond to those in c.
We reasoned that due to their small size neither the AA Gly nor Ser at position 77 could be a gate, which could by itself obstruct the flow of ions. Scrutiny of the channel sequences however revealed that the next AA downstream of position 77 was the aromatic AA Phe in both channels (F78). We predicted that the Phe aromatic side chain was positioned in or out of the ion pathway, depending on the nature of the adjacent AA. To test if F78 had any influence on the gating of the two channels we mutated it to an Ala in KcvS. The data show that removal of the aromatic side chain in the KcvS channel resulted in an increase in open probability and the disappearance of the long closed state C3 (Figure 5a,d,e,g,h, Table S1). To further test the hypothesis that F78 is a gate in combination with the adjacent AA we constructed the double mutant KcvNTS G77S F78A. A functional test of this mutant showed that replacement of the aromatic AA neutralized the effect of the G77S mutation (Figure 5b,d,f–h, Table S1), which alone caused a decrease in Po (Figure 4b,d,f–h, Table S1). The KcvNTS G77S F78A mutant had the same high open probability as KcvNTS and the dwell time histogram showed no long closed times (Figure 5d,f–h).
Figure 5.
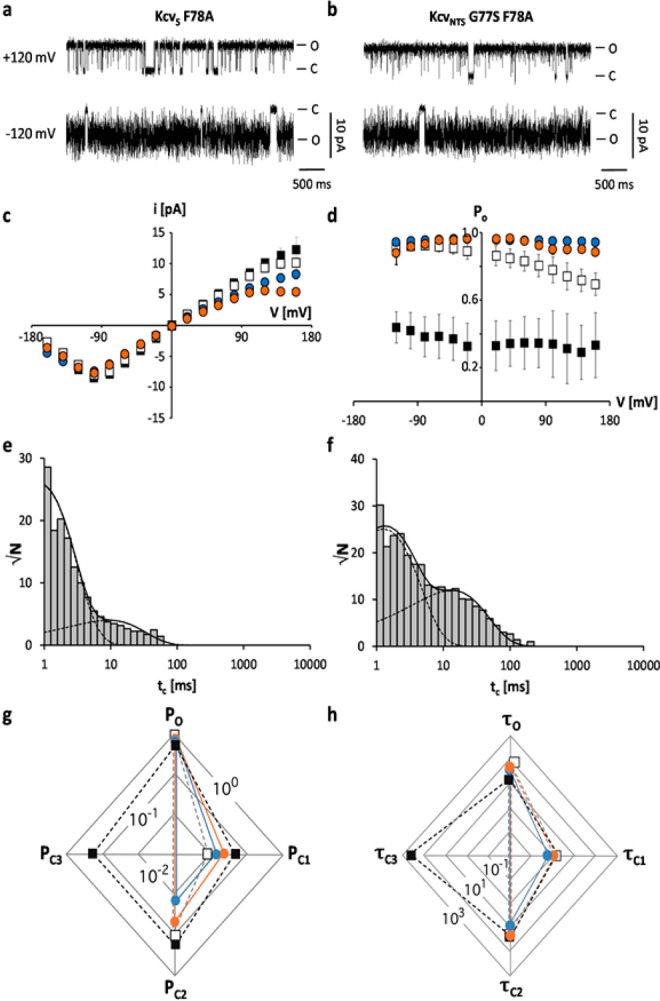
The aromatic AA F78 functions in combination with the upstream AA as a gate for the long closed time. Characteristic single channel fluctuations of mutant KcvS F78A (a) and KcvNTS G77S F78A (b) at ±120 mV in planar lipid bilayers. (c) Mean single channel i/V relations (±sd) of KcvNTS (open squares), KcvS (filled squares), KcvS F78A (orange circles, n = 4) and KcvNTS G77S F78A (blue circles, n = 7). (d) Mean open probabilities (±sd) of wt channels and mutants (n = 4 and n = 7 for KcvS F78A and KcvNTS G77S F78A, respectively). Exemplary closed dwell times at +120 mV for KcvS F78A (e) and KcvNTS G77S F78A (f). The data in e and f can now be fitted with two exponentials. Radar plots for probabilities (P) of the wt channels and mutants for occupying the open state (O) and the closed states (C1–C3) (g) and for mean lifetimes (in ms) of the open state (τo) and of the three closed states (τc1–τc3) (h). The mean lifetimes were calculated from three independent 5 min recordings. The symbols in d, g and h correspond to those in c.
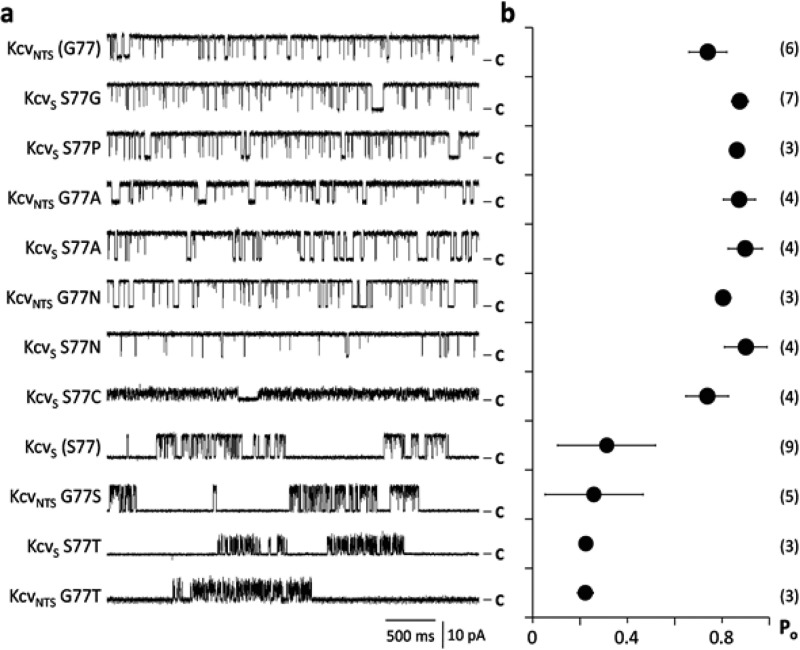
To understand the nature of the gate, i.e., whether the aromatic rings form a barrier by π:π interactions or whether the side chains are only steric hindrances we replaced F78 in KcvS with either Leu or Tyr. An analysis of mutant channel activity showed that the substitution of F78 with Leu—a large, nonaromatic and hydrophobic amino acid—caused a Po value, which was significantly lower than that in the F78A mutant and even lower than in the KcvS wt channel. By analyzing the single channel traces we detected the long lasting closed state, which was eliminated in the F78A mutant. The lower Po of KcvS F78L compared to that of KcvS was the result of a higher probability of occupancy and a longer mean lifetime of the third, long lasting closed state (Figure S4, Table S1). This closed state must be caused by the concerted action of Leu with the upstream Ser because a F78L substitution alone in KcvNTS had no effect on gating (Table S1). F78 in KcvS was also substituted with the polar aromatic Tyr. This caused a significant increase in Po (Figure S4, Table S1). The results of these experiments establish that F78 is a gate, which is responsible for the long closed events in KcvS. It obstructs the flow of ions by creating a hydrophobic barrier in the cavity of the channel. The dynamics of this gate are apparently determined by the adjacent AAs Gly in KcvNTS and Ser in KcvS. It was previously proposed in other channels that the flexible AA Gly could create a hinge in the inner TMD of Kir channels and that this hinge might be essential for gating.10,11 In analogy to this model it may be possible that a flexible G77 favors the removal of F78 from the ion pathway while the small Ser does not. To test the relevance of a potential Gly hinge for gating in KcvNTS and KcvS we mutated it into several other small AAs namely Ala, Asn, Ser, Pro and Thr. However, the results of these experiments do not support the hypothesis of a Gly hinge; they show that G77 in KcvNTS can be replaced by other AAs like Ala, which provide no flexibility in an α-helix, while still preserving the high open probability (Figure 6a,b). The results of these experiments suggest that Gly is not essential for the high open probability in KcvNTS but that Ser is important for the low open probability in KcvS. To test this hypothesis we replaced S77 in KcvS by a number of alternative AAs and measured the function of the mutant channels. The data in Figure 6a,b show that S77 can be replaced in KcvS by different AAs and that these changes result in an increase in open probability. The only replacement of S77, which conserves the low open probability of KcvS, is S77T (Figure 6a,b). In agreement, only G77S and G77T in KcvNTS caused a low, KcvS-like open probability. These experimental results refute the hypothesis that a Gly mediated hinge plays a crucial role in gating. Instead the data support a gating mechanism, which depends on the OH group of either Ser or Thr.
Figure 6.
The AA S77 is responsible for the gate with the long closed time. (a) Characteristic single channel fluctuations at +120 mV of KcvS and KcvNTS and mutants in which the position 77 in both channels was exchanged with an AA with a different flavor. (b) Mean open probabilities (±sd) of wt channels and mutants. The number of independent measurements is shown in brackets. Data in b correspond to the constructs on the left in a. Only AAs Ser and Thr in position 77 generate a low open probability with a long lasting closed time.
A scrutiny of the i/V relations of all the mutants indicated that most AA exchanges have either no or only minor effects on the unitary conductance of the channels (Figure 3c, 4c, 5c, S2–S4). In other cases, like in the KcvS mutants, in which F78 was replaced by other AAs, the mutation lowered the apparent unitary conductance of the channel (Figure 5a,c). Since there was no apparent correlation between a mutation on gating and on conductance, we concluded that both processes were independent. The orientation of the F78 side chain in the pathway is apparently not effected by the current through the open channel and vice versa.
Since Ser has no apparent interactive partner within TMD1 (Figure 1) we conclude that the relevant gating entities must be located solely within TMD2. This is consistent with the fact that KcvATCV-1 also has a high open probability9 in spite of some sequence variability with KcvNTS in TMD1 (Figure 1). Potential interacting partners of S77 in TMD2 could be the side chains of S76 or T79. If one of these AAs were involved in the formation of a hydrogen bond with S77, we would expect that substitution with Ala in KcvS would lead to a significant increase in Po and the disappearance of the long closed events. However, the functional tests showed that both mutants, KcvS S76A and KcvS T79A, still exhibited the typical gating of the wt channel (Figure S3). Therefore, S76 and T79 are not part of the gating mechanism. What remains as an explanation for the gating mechanism, in which S77 plays the crucial role, are interactions of the S77 side chain with the peptide backbone. In this context, it is interesting to note that Ser and Thr were found as important structural AAs in α-helixes.12,13 They can form H-bonds with a carbonyl oxygen in the preceding helix turn and such an intrahelical interaction can generate a kink in the α-helix. We reasoned that such a transient Ser interaction in the TMD2 of KcvS could generate a kink in TMD2 in a stochastic manner and that this structural modulation could affect the positioning of F78 in the ion pathway. This hypothesis is consistent with the experimental data. We found that S77 can be functionally replaced in KcvS by Thr but not by Cys (Figure 6a). While Ser and Thr are able to form an intrahelical H-bond, Cys cannot reach the required side chain conformation because of the steric hindrance between the thiol group and the carbonyl oxygen of the AA three positions upstream inside the α-helix.14
Molecular Dynamics Simulations Disclose Intrahelical Hydrogen Bond
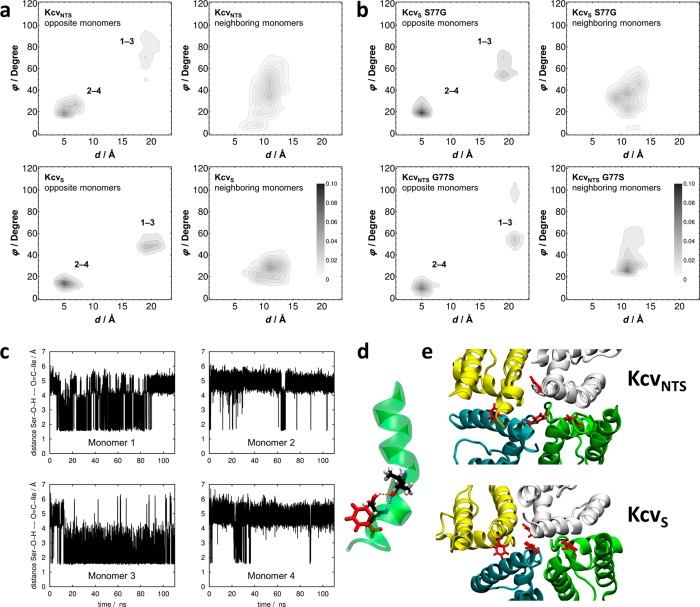
To quantify the structural and dynamical implications of differing sequences between KcvS and KcvNTS on the putative gate in the absence of experimental structural information, we developed a computational model of the two variants. They were examined by molecular dynamics (MD) simulations to determine the angle φ between normal vectors placed on the centers of the F78 phenyl rings along with the respective distance d between phenyl centers (see Figure S5). Distributions of these coordinate pairs were averaged over neighboring and opposite monomers for both channel variants, as shown in Figure 7a. KcvS shows a densely populated area of opposite monomer residues with an angle less than 20° and around 3–8 Å distance, and another area with less population at around 50–60° and more than 18 Å distance. The area with the smaller angle/distance pairs can be identified with π-stack geometries. KcvNTS has similar areas of likely monomer distances at 3–8 and about 20 Å, but with on average larger angles between the phenyl residues. The angles of the area at around 3−8 Å distance fall in the range between 20° and 40°, implying much weaker π:π interactions. The angle of the area around 20 Å is also much larger with angles between 60° and 100°. Angle/distance pairs between phenyl rings in neighboring monomers of KcvS occur with high probability at 8–13 Å and 20°–40°, whereas in the KcvNTS channel a substantially less focused area is found covering a wider angle range. In summary, the results indicate more pronounced structural rigidity for KcvS over KcvNTS, combined with a slightly larger propensity of the KcvS variant to form π-stack pairs between opposite monomers that could potentially block the translocation pathway. This structural and dynamical observation could be correlated with the functional differences between both variants expressed by the larger open probability of KcvNTS.
Figure 7.
Dynamics of TMD2 in KcvS/KcvNTS and mutants. (a) Distribution of distance-angle pairs characterizing π-stack geometries in KcvNTS (top row) and KcvS (bottom row) averaged over all F78 pairs in opposite (left column) and neighboring (right column) monomers. The geometric criteria are shown Figure S5. Numbers in the left panel denote different monomers. (b) Analogous data for KcvS S77G and KcvNTS G77S mutants, demonstrating the transfer of π-stack characteristics upon mutating the key residues. (c) Time series of the distances between the carbonyl oxygen of I73 and the hydroxyl hydrogen of S77 for all four monomers. (d) Schematic representation of a hydrogen bond formed between the amide oxygen of I73 and the hydroxyl hydrogen of S77 within monomer 3 of KcvS supplemented by orientation of F78 (snapshot taken at 80 ns). (e) Snapshots of F78 (red) in tetramer of KcvNTS (top) and KcvS (bottom) representing a wider and a narrow translocation pathway corresponding to statistics shown in a.
MD simulations were also used to test the hypothesis that the functional gate hinges upon possible hydrogen bond interactions between S77 and the helix backbone at the I73 position in KcvS (Figure 7d). We therefore calculated the distances between the S77 hydroxyl hydrogens and I73 backbone amide oxygens as a measure for H-bonding patterns for all monomers, as depicted in Figure 7c. Two distinct states were identified, one of which with a smaller distance at about 2 Å corresponded to an H-bond. Remarkably, oppositely placed and neighboring residues behaved differently, at least on the observed simulation time scale. Monomers 1 and 3 (opposite) show predominantly the hydrogen bond, whereas in the other pair of opposite monomers (2 and 4), it occurs much less frequently. This inherent asymmetry is likely inherited from the KirBac1.1 template structure that can be interpreted as a “dimer of dimers” pairwise asymmetric state, which also explains the 1–3/2–4 difference found for the π-stack statistics.
Deeper analysis of π-stack statistics for separate monomer units (Figure 7c) revealed that monomers 2 and 4 (broken H-bonds) were more likely to form π-stack geometries (not shown). In other words, the formation of hydrogen bonds within a monomer is correlated with the phenyl ring motion by bending them away from the translocation pathway, allowing oppositely placed phenyl groups to interact more strongly, thereby blocking ion translocation as a signature of a closed channel.
In order to test the hypothesis that our geometric π-stack definitions represent valid descriptors that correlate with functional states, we designed two computational control experiments. Following the experimental strategy, we created KcvS S77G and KcvNTS G77S mutants based on the respective last frames of the KcvS and KcvNTS runs. This inverts the experimental behavior, and—as hoped for—we could also reproduce the angle/distance distributions of KcvS by KcvNTS G77S and those of KcvNTS by KcvS S77G (see Figure 7b). Hence, we conclude that our π-stack descriptor (angle/distance pairs) indeed correlates with open/closed state propensities even in the absence of a functionally open, conductive simulation system.
Absence or Presence of Ser77 Alters Orientation of C-Terminal Part of TMD2
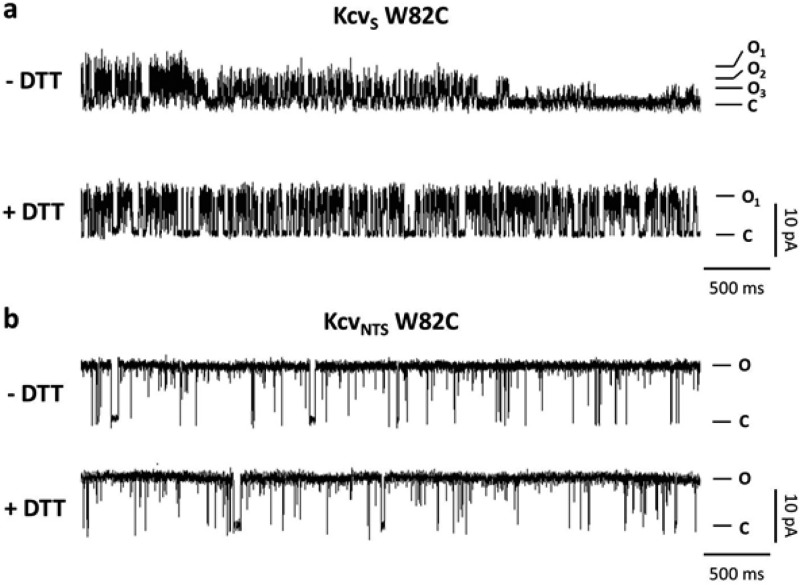
The experimental and computational data agree with the view that the gating differences between the two channels are determined by the presence or absence of a kink in TMD2. The modeling data further suggest that this could result in an altered orientation of the TMD2 downstream of F78. In order to test the hypothesis that a helix kink has an effect on the orientation of the last part of TMD2 we experimentally created mutants in which the AA W82 in KcvNTS and KcvS was replaced with a Cys. We reasoned that a different orientation of the TMD helixes in the mutant channels should affect the propensity of forming sulfhydryl bridges between the terminal Cys. The functional data of the mutant channels strongly supports this hypothesis. The exemplary current traces of the KcvS W82C mutant show that this channel has a reduced unitary conductance with different conductance levels (Figure 8a). The low unitary conductance was partially rescued by adding DTT. This finding suggests that the terminal Cys forms sulfhydryl bridges, which lowers the conductance at the cytosolic exit of this mutant channel. The KcvNTS W82C mutant on the other hand functions like the wt KcvNTS channel and does not respond to DTT (Figure 8b). The terminal Cys must be oriented in the KcvNTS W82C mutant in such a way that it is unable to form sulfhydryl bridges.
Figure 8.
The two channels KcvS and KcvNTS have a different orientation of the C-terminus. Characteristic single channel fluctuations at +120 mV of KcvS W82C (a) and KcvNTS W82C (b) in absence (−DTT) and presence of 5 mM DTT (+DTT) in bath solution. The prevailing open (O) and closed (C) levels are indicated along the traces.
Conclusion
Ion channels typically exhibit open and closed states with different lifetimes in single channel recordings. It is generally believed that each of these closed dwell times reflects the dynamics of a distinct gate. In the present study we causally linked one long-lived closed state in the KcvS channel to a simple electrostatic interaction in the channel protein. The presence of a Ser (or Thr) in the inner TMD of the KcvS channel promotes formation of an intrahelical H-bond and a consequent kink in this helix. The kink, which according to the data in Figure 8 also generates a twist in the lower part of the helix, has by itself no impact on gating. The channel only acquires a long-lived closed state when the AA downstream of the critical Ser has an aromatic or a long hydrophobic side chain. The computational data suggest the following scenario: the formation of hydrogen bonds within a transmembrane domain moves the phenyl ring of this domain out of the cavity giving room for a movement of the adjacent phenyl groups toward the pore center. These structural changes in the ns time window trigger a long-lived conformation, which closes the ion pathway. Hence the closing of the pore reflects a close interplay between all four monomers.
A similar gate, in which hydrophobic aromatic or aliphatic side chains serve as an obstruction of ion flow, has previously been proposed for other ion channels including complex K+ channels.15 From the crystal structure of KirBac channels it was deduced that the aromatic side chain of a Phe within the intracellular half of the inner helix could project into the channel pore and close it. Opening of the channels was presumably promoted by an upstream Gly kink in the inner helix with a simultaneous retraction of the aromatic ring from the ion pathway.16−19 Since many Kir channels have a hydrophobic aromatic Phe or aliphatic Leu at this position,20 it was proposed that they may constitute a general gate in the Kir family. In the case of Kir6.2 and Kir1.1 it was already experimentally confirmed that the respective Phe and Leu are involved in a pH sensitive gating of these channels.20,21
The present data further underscore the relevance of Phe and Leu in gating. But different from Kir channels their function as a gate is not connected to a flexible Gly hinge. Both the KcvS and the KcvNTS channels have a Phe in the same position in the inner helix. This Phe serves as a gate only in combination with an adjacent Ser. Placing a Gly in this position, which occurs in many Kcv channels, does not affect gating. The Phe side chain only becomes relevant as a gate in combination with an upstream AA like Ser or Thr, which is able to form an intrahelical H-bond. In this scenario Phe serves as the door and Ser as the hinge of the door in the KcvS channel.
The detailed analysis of single channel data shows that the Ser/Phe gate at the entry to the Kcvs channel functions autonomously. The presence or absence of this gate has no appreciable impact on the two remaining closed dwell times. This is different from other studies in which channel gating was examined in the context of an individual gate. In the case of the KcsA channel it was reported that any modification of the filter gate or of the pH sensitive gate always resulted in a complex modulation of several open and/or closed dwell times.2,3 In Kir6.2 channels, removal of the critical Phe at the inner gate affects the stability of both open and closed states.20 This is consistent with the idea that the dynamics of the Ser/Phe gate in KcvS is governed only by a single conformational alteration. The filter gate or the pH sensitive gate in KcsA in contrast may involve a network of electrostatic interactions, which include more than one defined conformational state.
While the MD simulation data provided a detailed insight into the structural components of the gate they do not completely describe the dynamics of the gating process due to the simulation time restriction. MD provides structural/dynamical correlates with function, not the complete microscopic picture. However, the MD simulations confirm that the intrahelical H-bond promotes a positioning of the Phe in the ionic pathway; this can generate π:π stacking interactions between aromatic rings from different monomers. Computational mutation tests confirmed the relevance of π:π stacking for understanding the gating transition, in line with the experimental finding that switching S and G at position 77 inverts the open/closed propensities of KcvNTS and KcvS. The results of the simulations suggest that the α-helix can dynamically fluctuate between a straight and bended conformation, correlating with switching H-bond patterns. A similar observation was reported from long MD simulations in which the backbone structure of an α-helix in the pore domain of a voltage gated Na+ channel undergoes dynamic deformation in response to the making and breaking of intrahelical H-bonds.22 All of these events occur in the time window of ns (Figure 7c) or μs22 while the lifetime of the closed state is in the range of hundreds of ms. Hence it is still not possible to directly correlate the lifetime of the closed state with the lifetime of an interaction or a conformational state of the channel protein. We can only exclude the possibility that the respective long-lived close state reflects the short lifetime of π:π stacking between the aromatic rings of neighboring Phe; notably a similar long close time is also observed when the Phe residues are replaced by Leu.
The importance of intrahelical H-bonds and their potential role in channel gating have not been reported previously. However, we predict that similar mechanisms, which are relevant in the simple Kcv channels, could be important in the gating of complex eukaryotic channels. Extensive structural studies have shown that either Ser or Thr induce either alone or in combination with Pro distinct distortions in transmembrane domains of many proteins including ion channels.23−25 Some isolated data already suggest that such Ser or Thr mediated intrahelical H-bonds have an effect on protein function.24,26 Notably, pairs of Ser or Thr together with aromatic or large hydrophobic AAs are also frequently present in the inner TMDs of K+ channels. The BK channel for example has a SFF338–340 motive at the cytosolic end of the pore lining TMD in which Ser and both Phes are involved in channel gating.27,28
Experimental Section
Cloning, Mutagenesis, Protein Expression and Purification
The two viruses encoding the KcvNTS and KcvS channels were originally isolated from alkaline lakes in western Nebraska. For cell-free protein expression the coding sequences of KcvNTS and KcvS as well as their chimeras (KcvNTS/S, KcvS/NTS) were cloned into the pEXP5-CT/TOPO-vector with the pEXP5-CT/TOPO TA Expression Kit (Invitrogen). To express the channel proteins in their native form, a stop-codon was inserted directly upstream of the coding sequence of a 6xHis-tag. The sequences of KcvNTS/S and KcvS/NTS were generated by fusion-PCR.29 All mutations were introduced by site-directed mutagenesis using a protocol based on the method described in.30 The coding regions of all constructs were sequenced. In vitro expression of the channel proteins were performed with the MembraneMax HN Protein Expression Kit (Invitrogen) following the manufacturer’s instructions. The in vitro expression takes place in the presence of nanolipoproteins (NLPs)31 fused with multiple His-tags. This allows the purification of the native channel proteins by metal chelate affinity chromatography. For purification the crude reaction mixture was loaded on a 0.2 mL HisPur Ni-NTA spin column (Thermo Scientific). All steps were performed according to the manufacturer’s instructions, with the exception of the elution step. To elute the NLP/channel conjugates the spin column was washed three times with two resin-bed volumes of 250 mM imidazole.
Planar Lipid Bilayer Experiments
All planar lipid bilayer experiments were performed at room temperature (20–25 °C) with a conventional bilayer setup (IonoVation, Osnabrück Germany). The preparation of the recording chambers was done as described earlier.32 Both chambers were connected with Ag/AgCl electrodes to the head-stage of a patch-clamp amplifier (L/M-EPC-7, List-Medical, Darmstadt). Membrane potentials were referred to the cis compartment. Current traces were filtered at 1 kHz with a 4-pole Bessel filter and digitized with a sampling frequency of 5 kHz by an A/D-converter (LIH 1600, HEKA Elektronik, Lambrecht, Germany). 1,2-Diphytanoyl-sn-glycero-3-phosphocholine (DPhPC, Avanti Polar Lipids, Alabaster, AL, USA) bilayers were formed by the Pseudo painting/air bubble technique33 in symmetrical 100 mM KCl solution (pH 7, buffered with 10 mM HEPES). For reconstitution of channel proteins a small amount (1–3 μL) of the purified NLP/channel-conjugates diluted in 250 mM imidazole solution was added directly below the bilayer in the trans compartment with a bent Hamilton syringe. After successful incorporation of a single channel in the DPhPC bilayer, constant voltages were applied for 1 to 5 min. Voltage was changed in steps of 20 mV between +160 mV and −160 mV.
Data Analysis
Single-channel current traces were recorded with Patchmaster (HEKA) and analyzed with the custom-made program Kielpatch (http://www.zbm.uni-kiel.de/aghansen/software.html). Single channel amplitudes were measured visually. For determination of open probabilities an automated Hinkley jump detector34 was used. Dwell time histograms were fitted with sums of 1 to 3 exponential functions. The number of missed closed times, which were not resolved because of a limited temporal resolution of the recording setup, were estimated from the obtained fitting parameters. The calculated numbers of missed closed-events were used to correct the mean lifetimes of the open state. For this we assumed (i) that the missed closed events are randomly distributed over the open events, and (ii) that the missed closed events have a negligible effect on the total time of the channel in the open state.
For calculating the probabilities of occupancy, we assumed a simple Markov model in which all closed states were directly coupled to the open state with no unobservable closed–closed transitions. If additional information on the state transitions become available in the future, which require a modification of this model topology, algorithms are available to transform equivalent models into each other.35
Homology Modeling
The closed KirBac1.1 structure (PDB code: 1P7B(16)) was chosen as a template for initial homology modeling of the KcvS and KcvNTS geometries, following closely the methodology for developing KcvPBCV-14 and KcvATCV-131 models. The template tetramer was constructed from the KirBac1.1 monomer by applying the provided symmetry operations using VMD 1.9.1.36 Deletion of the cytosolic domain resulted in a truncated KirBac1.1 tertiary structure sharing the same total number of 82 residues. Sequence alignments using CLUSTAL (version 2.1)37 revealed large gaps, which would lead to helix breaks. These were circumvented by restraining residues 10–15 and 75–82 to an α-helical structure during model preparation using Papwoth (version 9.10)38,39 with default settings for optimization cycles.
The energetically most favored model out of ten was identified by its DOPE score and further processed. Hydrogen atoms were added by CHARMM’s HBUILD tool,40 keeping all titratable residues in their standard protonation state at neutral pH; C-terminal residues remained deprotonated and the N-termini residues were protonated; His residues were protonated at their δ-nitrogen. The resulting models were minimized in the presence of two ions (selectivity filter binding sites S2 and S4) and two water molecules (sites S1 and S3) introduced at the filter positions by adapting the available information from the KirBac1.1 structure. PROCHECK was utilized in its online version (http://www.ebi.ac.uk/pdbsum/)41 to assess the model quality, ignoring hydrogen atoms.
Molecular Dynamics Simulations
Again following the methodology for simulating the KcvATCV-1 wild type system32 the KirBac1.1-based homology models of KcvS and KcvNTS were inserted into a pre-equilibrated DMPC bilayer membrane utilizing the CHARMM-GUI tools42 by deletion of lipid molecules colliding with any protein residues within a sharp radius of 0.5 Å of the protein, resulting in a membrane comprising 192 lipid species in total with 90 molecules in the upper and 102 in the lower layer induced by the conic protein shape. 16726 water molecules (TIP3P model)43 were added, removing water molecules inside the membrane. By random exchange with 60 water molecules 31 Cl– and 29 K+ ions were introduced into the bulk solution creating a concentration of approximately 100 mM KCl. With the two K+ ions already located inside the selectivity filter electroneutrality of the entire system was achieved. The initial system dimensions were ca. 90 Å × 90 Å × 115 Å along x, y and z dimensions, with the latter representing the channel axis. Periodic boundary conditions were employed by treating electrostatics with the particle mesh Ewald algorithm44 using a spacing of 0.1 Å between grid points and a real-space short-range cutoff of 12 Å. The temperature was set to 300 K for production runs, controlled by a Langevin thermostat with a damping coefficient of 1 ps–1,45 and a pressure of 1 atm was applied by a Langevin-piston Nose-Hoover barostat.46 The CHARMM22* force field for proteins and CHARMM27 for lipids were applied throughout the simulations47,48 with ion parameters as used earlier.4,32 NAMD (version 2.9) was used for performing simulations.49 After minimization and equilibration with gradually removing restraints over 12.7 ns, production runs were performed over 110 ns for each variant, sampled at each ps for further evaluation. A snapshot of the KcvS structure is shown in Figure 1A. The quality of the MD simulations is demonstrated in Supplementary Figure S6, where the root-mean-square displacements (RMSD) indicate long-time stability. Point mutations to create KcvNTS G77S and KcvS S77G were introduced by the VMD35 “Mutator” plugin on the last frame of the original MD runs of the respective wildtypes and further simulated over 80 ns.
Acknowledgments
This research was supported by the Deutsche Forschungsgemeinschaft SCHR1467/1–1 (IS) the European Research Council NoMAGIC grant (AM, GT) the LOEWE initiative iNAPO (GT), and PGR00139 from MAECI (Ministero affari esteri e cooperazione internazionale), CARIPLO grant 2014–0660 (AM). NSF – EPSCOR Grant EPS-1004094 (JVE) and the COBRE program of the National Center for Research Resources Grant P20-RR15535 (JVE). The IT and Media Center (ITMC) of the TU Dortmund is acknowledged for computational support (SMK).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b01158.
Open and closed dwell times of channel mutants and more details on molecular dynamics simulations (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Hille B.Ion Channels of Excitable Membranes, 3rd ed.; Sinauer Associates Inc.: Sunderland, 2001. [Google Scholar]
- Cordero-Morales J. F.; Cuello L. G.; Zhao Y.; Jogini V.; Cortes D.; Roux B.; Perozo E. Nat. Struct. Mol. Biol. 2006, 13, 311. 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- Thompson A. N.; Posson D. J.; Rarsa P. V.; Nimigean C. M. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 6900. 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayefeh S.; Kloss T.; Kreim M.; Gebhardt M.; Baumeister D.; Hertel B.; Richter C.; Schwalbe H.; Moroni A.; Thiel G.; Kast S. M. Biophys. J. 2009, 96, 485. 10.1016/j.bpj.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G.; Baumeister D.; Schroeder I.; Kast S. M.; Van Etten J. L.; Moroni A. Biochim. Biophys. Acta, Biomembr. 2011, 1808, 580. 10.1016/j.bbamem.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Abenavoli A.; DiFrancesco M.; Schroeder I.; Epimashko S.; Gazzarrini S.; Hansen U. P.; Thiel G.; Moroni A. J. Gen. Physiol. 2009, 134, 869. 10.1085/jgp.200910266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M.; Moroni A.; Gazzarrini S.; DiFrancesco D.; Thiel G.; Severino M.; Van Etten J. L. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 5318. 10.1073/pnas.0307824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S.; Kang M.; Van Etten J. L.; DiFrancesco D.; Thiel G.; Moroni A. J. Biol. Chem. 2004, 279, 28443. 10.1074/jbc.M401184200. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S.; Kang M.; Abenavoli A.; Romani G.; Olivari C.; Gaslini D.; Ferrara G.; Van Etten J. L.; Kreim M.; Kast S. M.; Thiel G.; Moroni A. Biochem. J. 2009, 420, 295. 10.1042/BJ20090095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.; Lee A.; Chen J.; Cadene M.; Chait B. T.; MacKinnon R. Nature 2002, 417, 523. 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- Shrivastava I. H.; Bahar I. Biophys. J. 2006, 90, 3929. 10.1529/biophysj.105.080093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros J. A.; Deupi X.; Olivella M.; Haaksma E. E.; Pardo L. Biophys. J. 2000, 79, 2754. 10.1016/S0006-3495(00)76514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S. E.; Roberts K.; Vaidehi N. J. Mol. Graphics Modell. 2009, 27, 944. 10.1016/j.jmgm.2009.02.004. [DOI] [PubMed] [Google Scholar]
- McGregor M. J.; Islam S. A.; Sternberg M. J. E. J. Mol. Biol. 1987, 198, 295. 10.1016/0022-2836(87)90314-7. [DOI] [PubMed] [Google Scholar]
- Zhou H. X.; McCammon J. A. Trends Biochem. Sci. 2009, 35, 179. 10.1016/j.tibs.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A.; Gulbis J. M.; Antcliff J. F.; Rahman T.; Lowe E. D.; Zimmer J.; Cuthbertson J.; Ashcroft F. M.; Ezaki T.; Doyle D. A. Science 2003, 300, 1922. 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- Kuo A.; Domene C.; Johnson L. N.; Doyle D. A.; Vénien-Bryan C. Structure 2005, 13, 1463. 10.1016/j.str.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Nishida M.; Cadene M.; Chait B.-T.; MacKinnon R. EMBO J. 2007, 26, 4005. 10.1038/sj.emboj.7601828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton G. M.; Altieri S.; Heginbotham L.; Unger V. M.; Morais-Cabral J. H. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 1511. 10.1073/pnas.0711533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A.; Wu J.; Wang R.; Jiang C. Biochim. Biophys. Acta, Biomembr. 2007, 1768, 39. 10.1016/j.bbamem.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Sackin H.; Nanazashvili M.; Palmer L. G.; Krambis M.; Walters D. E. Biophys. J. 2005, 88, 2597. 10.1529/biophysj.104.051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux C.; Vorobyov I.; Allen T. W. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 3454. 10.1073/pnas.1320907111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deupi X.; Olivella M.; Sanz A.; Dölker N.; Campillo M.; Pardo L. J. Struct. Biol. 2010, 169, 116. 10.1016/j.jsb.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Deupi X.; Olivella M.; Govaerts C.; Ballesteros J. A.; Campillo M.; Pardo L. Biophys. J. 2004, 86, 105. 10.1016/S0006-3495(04)74088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea M.; Lugtenburg I.; Mayol E.; Cordomi A.; Deupí X.; Pardo L. BMC Bioinf. 2015, 16, 26. 10.1186/s12859-015-0699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansuk K.; Deupi X.; Torrecillasm I. R.; Jongejan A.; Nijmeijer S.; Bakker R. A.; Pardo L.; Leurs R. Mol. Pharmacol. 2011, 79, 262. 10.1124/mol.110.066068. [DOI] [PubMed] [Google Scholar]
- Strutz-Seebohm N.; Pusch M.; Wolf S.; Stoll R.; Tapken D.; Derwert K.; Attali B.; Seebohm G. Cell. Physiol. Biochem. 2011, 27, 443. 10.1159/000329965. [DOI] [PubMed] [Google Scholar]
- Panaghie G.; Purtell K.; Tai K. K.; Abbott G. W. Biophys. J. 2008, 95, 2759. 10.1529/biophysj.108.133678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R. M.; Hunt H. D.; Ho S. N.; Pullen J. K.; Pease L. R. Gene 1989, 77, 61. 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Papworth C.; Bauer J. C.; Braman J.; Wright D. A. Strategies 1996, 9, 3. 10.1080/08924562.1996.11000299. [DOI] [Google Scholar]
- Katzen F. BioTechniques 2008, 45, 190. 10.2144/000112951. [DOI] [PubMed] [Google Scholar]
- Braun C. J.; Lachnit C.; Becker P.; Henkes L. M.; Arrigoni C.; Kast S. M.; Moroni A.; Thiel G.; Schroeder I. Biochim. Biophys. Acta, Biomembr. 2014, 1838, 1096. 10.1016/j.bbamem.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Braun C.; Baer T.; Moroni A.; Thiel G. J. Neurosci. Methods 2014, 233C, 13. 10.1016/j.jneumeth.2014.05.031. [DOI] [PubMed] [Google Scholar]
- Schultze R.; Draber S. J. Membr. Biol. 1993, 132, 41. 10.1007/BF00233050. [DOI] [PubMed] [Google Scholar]
- Kienker P. Proc. R. Soc. London, Ser. B 1989, 236, 269. 10.1098/rspb.1989.0024. [DOI] [PubMed] [Google Scholar]
- Humphry W.; Dalke A.; Schulten K. J. J. Mol. Graphics 1996, 14, 33. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Larkin M. A.; Blackshields G.; Brown N. P.; Chenna R.; McGettigan P. A.; McWilliam H.; Valentin F.; Wallace I. M.; Wilm A.; Lopez R.; Thompson J. D.; Gibson T. J.; Higgins D. G. Bioinformatics 2007, 23, 2947. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Webb B.; Sali A. Methods Mol. Biol. 2014, 1137, 1. 10.1007/978-1-4939-0366-5_1. [DOI] [PubMed] [Google Scholar]
- Shen M. Y.; Sali A. Protein Sci. 2006, 15, 2507. 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T.; Karplus M. Proteins: Struct., Funct., Genet. 1988, 4, 148. 10.1002/prot.340040208. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A.; MacArthur M. W.; Moss D. S.; Thornton J. M. J. Appl. Crystallogr. 1993, 26, 283. 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Jo S. J. Comput. Chem. 2008, 29, 1859. 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. J. J. Chem. Phys. 1983, 79, 926. 10.1063/1.445869. [DOI] [Google Scholar]
- Darden T.; York D.; Pedersen L. J. Chem. Phys. 1993, 98, 10089. 10.1063/1.464397. [DOI] [Google Scholar]
- Kubo R.; Toda M.; Hashitsume N.. Statistical Physics II: Non equilibrium Statistical Mechanics, 2nd ed.; Springer: Berlin, Heidelberg, 1991. [Google Scholar]
- Feller S. E.; Zhang Y.; Pastor R. W.; Brooks B. R. J. Chem. Phys. 1995, 103, 4613. 10.1063/1.470648. [DOI] [Google Scholar]
- Buck M.; Bouguet-Bonnet S.; Pastor R. W.; MacKerell A. D. Biophys. J. 2006, 90, 36. 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A. D.; Bashford D.; Bellott M.; Dunbrack R. L.; Evanseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; Joseph-McCarthy D.; Kuchnir L.; Kuczera K.; Lau F. T.; Mattos C.; Michnick S.; Ngo T.; Nguyen D. T.; Prodhom B.; Reiher W. E.; Roux B.; Schlenkrich M.; Smith J. C.; Stote R.; Straub J.; Watanabe M.; Wiórkiewicz-Kuczera J.; Yin D.; Karplus M. J. Phys. Chem. B 1998, 102, 3586. 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Phillips J. C.; Braun R. J. Comput. Chem. 2005, 26, 1781. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.