Abstract
Purpose:
The relationships of genetic variation in the vitamin D pathway with circulating 25-hydroxyvitamin D3 [25(OH)D] levels and survival remain largely unknown for patients with metastatic colorectal cancer (mCRC).
Methods:
Among 535 patients participating in a randomized trial of chemotherapy for mCRC, we prospectively measured baseline plasma 25(OH)D and examined 124 tagging single nucleotide polymorphisms (SNPs) within seven genes in the vitamin D pathway, including 5 SNPs associated with circulating 25(OH)D levels in previous genome-wide association studies (GWAS). We evaluated whether these SNPs were associated with plasma 25(OH)D levels and patient outcome (overall survival, time to progression, and tumor response), using linear, logistic, and Cox proportional hazards regression.
Results:
We observed a significant association between 25(OH)D levels and an additive genetic risk score determined by the 5 GWAS-identified SNPs (P=0.0009). We did not observe any direct association between 25(OH)D-associated SNPs, individually or as a genetic risk score, and patient outcome. However, we found a significant interaction between 25(OH)D levels and rs12785878 genotype in DHCR7 on overall survival (Pinteraction=0.02).
Conclusion:
Germline genetic variation in the vitamin D pathway informs baseline 25(OH)D levels among patients with mCRC. The association between 25(OH)D levels and overall survival may vary by DHCR7 genotype.
Keywords: 25-hydroxyvitamin D3, single nucleotide polymorphisms, metastatic colorectal cancer, survival
Introduction
Vitamin D is hypothesized to play an important role in colorectal carcinogenesis. Vitamin D receptors (VDR) and 1-α-hydroxylase, which converts 25-hydroxyvitamin D3 [25(OH)D] into 1,25-dihydroxyvitamin D3 [1,25(OH)2D], are expressed in colon cancer cells (1–3). By binding to VDR, 1,25(OH)2D induces differentiation and apoptosis (4–6), and inhibits proliferation (7), angiogenesis (8, 9), and metastasis (10) of colon cancer.
The major sources of vitamin D in humans are intake from foods and dietary supplements and exposure to ultraviolet B rays. In addition to environmental factors, twin and family studies indicate that genetics contributes substantially to variation in circulating 25(OH)D levels, with heritability estimated between 29% and 80% (11–14). Genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) that are associated with circulating 25(OH)D levels in or near four gene regions: (1) CYP2R1 (cytochrome P450, family 2, subfamily R, member 1), encoding vitamin D 25-hydroxylase, which converts vitamin D3 into 25(OH)D; (2) CYP24A1, encoding 24-hydroxylase, which degrades 25(OH)D and 1,25(OH)2D; (3) DHCR7/NADSYN1 (7-dehydrocholesterol reductase/nicotinamide adenine dinucleotide synthetase 1), which removes 7-dehydrocholesterol from the synthetic vitamin D pathway; and (4) GC, encoding vitamin D binding protein (15, 16). Among these SNPs, rs12785878 and rs3829251 in DHCR7/NADSYN1 and rs2282679 in GC are located in introns, whereas rs10741657 and rs6013897 are proximal to CYP2R1 and CYP24A1, respectively.
Because the GWAS data above were obtained from healthy individuals, the role of these vitamin D-related genetic variants is unknown for patients with metastatic colorectal cancer (mCRC), a population with a high prevalence of vitamin D deficiency (17). In this study, we investigated whether the SNPs above, as well as tagging SNPs from genes encoding factors with a biological role in vitamin D metabolism, were associated with baseline plasma 25(OH)D levels and treatment outcome among a cohort of patients with mCRC enrolled in a large, completed, phase III cooperative group clinical trial of palliative chemotherapy. In a previous analysis of the same study cohort, we did not detect an overall association between 25(OH)D levels and patient outcome, though higher 25(OH)D levels were associated with improved overall survival among patients receiving infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX) (17).
Methods
Study population
Patients included in this study were drawn from the North Central Cancer Treatment Group (NCCTG) trial N9741, a phase III cooperative group trial of chemotherapy for patients with previously untreated mCRC. NCCTG is now a part of the Alliance for Clinical Trials in Oncology. Between October 1998 and April 2001, patients were enrolled and randomized to receive irinotecan, bolus fluorouracil, and leucovorin (IFL); FOLFOX; or irinotecan and oxaliplatin (IROX). Full details of the trial have been described elsewhere (18). Briefly, enrolled patients were required to have histologically proven unresectable colorectal adenocarcinoma, Eastern Cooperative Oncology Group (ECOG) performance status ≤ 2, and adequate renal, liver, and bone marrow function. Exclusion criteria included previous treatment for advanced disease, symptomatic peripheral neuropathy or central nervous system metastases, uncontrolled or severe comorbid illnesses, and ≥ 3 loose stools per day. The protocol was approved by the institutional review board of each participating institution. Patients provided informed consent and were given the option of inclusion in a biomarker companion study for future research.
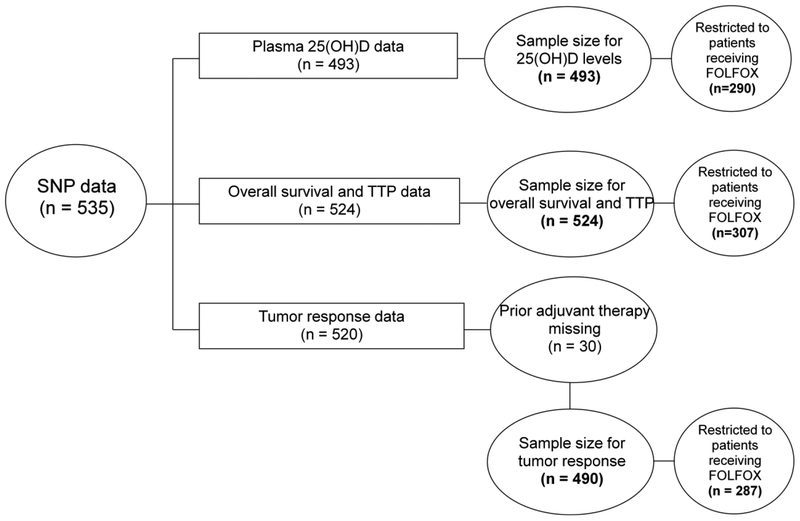
In total, 1,379 patients were enrolled in N9741 after the incorporation of the amendment to collect blood samples. Among those who provided blood samples, SNP data were available for 535 patients, of which 493 had 25(OH)D levels and 524 had survival information. Figure 1 illustrates the derivation of the final sample size. In a previous study, we did not observe any appreciable difference in baseline characteristics between the overall cohort of patients enrolled in N9741 and the subset participating in the biomarker companion study (19). Further, patients who did and did not provide blood samples had similar overall survival (median survival times: 18.1 and 17.0 months, respectively) (20).
Figure 1. Derivation of sample size.
25(OH)D 25-hydroxyvitamin D3, FOLFOX fluorouracil, leucovorin, oxaliplatin, SNP single-nucleotide polymorphism, TTP time to progression
Assessment of patient outcome
Death and disease progression were assessed among all patients, whereas objective response to chemotherapy was assessed among patients with evaluable disease. Overall survival was calculated from study entry to death or last contact. Time to progression (TTP) was calculated from study entry to disease progression or last disease assessment. Progression and response criteria have been described elsewhere (18).
Genotyping of SNPs in the vitamin D pathway
A total of 124 tagging SNPs were selected from seven gene regions in the vitamin D pathway: CYP2R1, CYP24A1, CYP27B1, DHCR7/NADSYN1, GC, RXRA (retinoid X receptor alpha), VDR. Tag SNPs were selected within each gene ± 2kb using data from the HapMap Project Phase I/II and III, with an r2 cutoff of 0.8. Five SNPs previously identified as being associated with 25(OH)D levels (Tier 1 SNPs) were forced in: rs1993116 (r2=1 with rs10741657), rs6013897, rs12785878, rs11234027 (r2=1 with rs3829251), and rs2282679 (15, 16). SNP genotyping was performed using the Sequenom platform, with DNA from unrelated HapMap participants and CEPH (Centre d’Etude du Polymorphisme Humain) families serving as positive controls and Mendelian controls, respectively (21). Polymerase chain reaction (PCR) and extension primers were designed using Sequenom Assay Design software. PCR reactions, shrimp alkaline phosphatase digestion, and extension reactions were performed according to Sequenom’s standard protocol with one exception: a linear adjustment to the PCR primer concentrations was made to standardize mass spectrometer peak heights. Any SNP or individual that had a success rate <85% was removed from further analysis. The Pedstats and MERLIN software packages were also used to identify and remove unlikely genotypes and genotypes that produced Mendelian errors. Invariant SNPs were also removed. The remaining SNPs were tested for Hardy−Weinberg equilibrium among unrelated individuals, using the software HWSIM. SNPs that passed this test were analyzed in Haploview, and several more SNPs were excluded as having been tagged by other genotyped SNPs.
Plasma 25(OH)D assessment
Blood samples were collected at study entry and sent to the Mayo Central Laboratory for Clinical Trials (Rochester, MN). To measure 25(OH)D, plasma samples were sent by overnight delivery to Heartland Assays (Ames, IA) for radioimmunoassay (22). Masked quality control samples were interspersed among the samples, and all laboratory personnel were blinded to patient outcome. The mean intra-assay coefficient of variation was 8%.
Statistical Analyses
We estimated the association between tagging SNPs and 25(OH)D levels, using linear regression either unadjusted or adjusted for age, sex, race/ethnicity, and season of blood collection. Genotypes were coded as 0, 1, or 2 to reflect the number of copies of the allele (additive models). We considered the possibility that a combination of Tier 1 SNPs may be associated with patient outcome. Therefore, we calculated an additive genetic risk score by summing the number of risk alleles (i.e., associated with lower 25(OH)D levels in GWAS) across the 5 Tier 1 SNPs, yielding a possible range of 0–10 alleles. In sensitivity analyses, we calculated a weighted genetic risk score that weighed each risk allele using the coefficient from unadjusted linear regression, and the findings remained unchanged (data not shown). A P value of <0.05 was considered statistically significant for Tier 1 SNPs and the genetic risk score. For the other tagging SNPs (Tier 2 discovery SNPs; Supplementary Table 1), a false discovery rate (FDR) of <0.05 was considered statistically significant to correct for multiple comparisons (23).
Cox proportional hazards regression (24) was used to examine the association of Tier 1 SNPs, the genetic risk score, and any significant Tier 2 SNP with overall survival and TTP. Logistic regression was used to estimate odds ratios (ORs) for tumor response. In multivariable models, we adjusted for age, sex, race/ethnicity, ECOG performance status, number of metastatic sites, and treatment arm. Tests of interaction between 25(OH)D levels and 25(OH)D-associated SNPs on overall survival were performed by entering their product in the model, evaluated by a likelihood ratio test. Our previous data demonstrated that higher 25(OH)D levels were associated with improved overall survival among patients receiving FOLFOX (17). Thus, we repeated the analyses above among patients receiving FOLFOX. All analyses were based on the study database frozen on 6/25/2013 and were performed with SAS software, version 9.4 (SAS Institute, Cary, NC), and all P values are two sided. Data collection and statistical analyses were conducted by the Alliance Statistics and Data Center. Data quality was ensured by review of data by the Alliance Statistics and Data Center following Alliance policies.
Results
Patient characteristics
Baseline patient characteristics of 524 patients are presented in Table 1. The mean age was 60 years (standard deviation, 11 years), with 59% males and 41% females. The mean plasma 25(OH)D level was 21.0 ng/mL (standard deviation, 10.2 ng/mL). Fifty percent of patients were vitamin D deficient (<20 ng/mL) and 32% were vitamin D insufficient (20 − <30 ng/mL). Only 11% of patients had 25(OH)D levels ≥33 ng/mL, the threshold previously shown to be associated with a potential beneficial effect on both colorectal cancer (CRC) incidence and survival (25,26). The median follow-up time among living patients was 9.2 years (90th percentiles: 10.7 years), until final data lock. During the follow-up, 85% of patients had progressed and 95% had died. Among 490 patients evaluable for response, 50% had a confirmed tumor response.
Table 1.
Baseline characteristics of patients with metastatic colorectal cancera
| Characteristic | No. | % |
|---|---|---|
| 25(OH)D, ng/mL, mean (SD) | 21.0(10.2) | |
| Age, years, mean (SD) | 60(11) | |
| Sex | ||
| Female | 215 | 41.0 |
| Male | 309 | 59.0 |
| Race/ethnicity | ||
| White | 450 | 85.9 |
| Black | 38 | 7.3 |
| Other | 31 | 5.9 |
| Unknown/missing | 5 | 0.9 |
| ECOG performance statusb | ||
| 0–1 | 500 | 95.4 |
| 2 | 24 | 4.6 |
| No. of metastatic sites, median (range) | 2(1–4) | |
| Liver-only metastasis | ||
| Yes | 127 | 24.2 |
| No | 397 | 75.8 |
| One metastatic site, not including liver | ||
| Yes | 38 | 7.3 |
| No | 486 | 92.7 |
| Multiple metastatic sites, not including liver | ||
| Yes | 62 | 11.8 |
| No | 462 | 88.2 |
| Prior adjuvant therapy | ||
| Yes | 78 | 14.9 |
| No | 415 | 79.2 |
| Missing | 31 | 5.9 |
| Treatment arm | ||
| IFL | 114 | 21.8 |
| FOLFOX | 307 | 58.6 |
| IROX | 103 | 19.7 |
| Season of blood collection | ||
| Summer (June, July, August) | 135 | 27.4 |
| Fall (September, October, November) | 78 | 15.8 |
| Winter (December, January, February) | 133 | 27.0 |
| Spring (March, April, May) | 147 | 29.8 |
| Missing | 31 | 5.9 |
| Geographic region of registering sitec | ||
| Midwestern US | 228 | 43.5 |
| Northeastern US and Canada | 114 | 21.8 |
| Southern US and Puerto Rico | 85 | 16.2 |
| Western US | 56 | 10.7 |
| Missing | 41 | 7.8 |
25(OH)D 25-hydroxyvitamin D3, ECOG Eastern Cooperative Oncology Group, FOLFOX fluorouracil, leucovorin, oxaliplatin, IFL irinotecan, bolus fluorouracil, leucovorin, IROX irinotecan, oxaliplatin, SD standard deviation
Baseline information available for 524 patients
Grade 0 = fully active, able to carry on all pre-disease performance without restriction; grade 1 = restricted in physically strenuous activity but ambulatory and able to carry out work of a light or sedentary nature, e.g., light house work, office work; grade 2 = ambulatory and capable of all selfcare but unable to carry out any work activities, up and about more than 50% of waking hours
Defined according to the US Census Bureau: Midwest = IL, IN, IA, KS, MI, MN, MO, NE, ND, OH, SD, WI; Northeast = CT, ME, MA, NH, NJ, NY, PA, RI, VT; South = AL, DE, FL, GA, KY, LA, MD, NC, OK, SC, TN, TX, VA, WV; West = AZ, CA, CO, HI, ID, MT, NV, NM, OR, UT, WA, WY
Association between genetic variation in the vitamin D pathway and plasma 25(OH)D levels
We examined the association between SNPs, individually or as a genetic risk score, and plasma 25(OH)D levels. In unadjusted models, four Tier 1 SNPs (rs1993116, rs12785878, rs11234027, rs6013897) and the genetic risk score were associated with 25(OH)D levels (all P<0.05; Table 2). After adjustment for covariates, rs1993116 in CYP2R1 and the genetic risk score remained associated with 25(OH)D levels (P=0.005 and 0.0009, respectively). In the multivariable model, each one-allele increase in the genetic risk score was associated with a decrease of 0.93 ng/mL in 25(OH)D level. The score remained associated with 25(OH)D levels among patients receiving FOLFOX (P=0.003; data not shown). Three Tier 2 SNPs tended to be associated with 25(OH)D levels with an FDR <0.15 in either unadjusted or multivariable model (Table 3), but none of them reached the predefined FDR for statistical significance, so we did not explore them in further analyses.
Table 2.
Association between 25(OH)D-associated SNPs in GWAS (Tier 1 SNPs), an additive genetic risk score, and plasma 25(OH)D levels among patients with metastatic colorectal cancer
| Genetic varianta | Position (nearest gene) | Major/minor allele | MAF | Risk allele | Unadjusted | Multivariableb | ||
|---|---|---|---|---|---|---|---|---|
| Coefficient | P | Coefficient | P | |||||
| rsl993116 | CYP2R1 | C/T | 0.38 | С | −2.31 | 0.0004 | −1.76 | 0.005 |
| rs6013897 | CYP24A1 | T/A | 0.23 | A | −1.89 | 0.02 | −1.39 | 0.06 |
| rsl2785878 | DHCR7 | T/G | 0.32 | G | −1.38 | 0.04 | −0.38 | 0.58 |
| rsl1234027 | NADSYN1 | G/A | 0.19 | A | −1.76 | 0.03 | −1.12 | 0.15 |
| rs2282679 | GC | A/C | 0.26 | С | −0.96 | 0.19 | −1.13 | 0.11 |
| Genetic risk score | - | - | - | - | −1.31 | <0.0001 | −0.93 | 0.0009 |
25(OH)D 25-hydroxyvitamin D3, GWAS genome-wide association studies, MAF minor allele frequency, SNP single-nucleotide polymorphism
Additive effect of a risk allele (i.e., associated with lower 25(OH)D levels) on 25(OH)D levels (continuous in ng/mL)
Adjusted for age (continuous), sex (female, male), race/ethnicity (white, black, other, unknown/missing), and season of blood collection (summer, fall, winter, spring)
Table 3.
Three tagging SNPs in the vitamin D pathway (Tier 2 discovery SNPs) marginally associated with plasma 25(OH)D levels among patients with metastatic colorectal cancera
| SNPb | Position (nearest gene) | Major/minor allele | MAF | Unadjusted | Multivariablec | ||
|---|---|---|---|---|---|---|---|
| Coefficient | FDR | Coefficient | FDR | ||||
| rs7041 | GC | G/T | 0.46 | −1.91 | 0.13 | −1.32 | 0.40 |
| rsl045570 | RXRA | G/T | 0.17 | 2.66 | 0.13 | 2.59 | 0.07 |
| rs4842196 | RXRA | A/C | 0.30 | 0.98 | 0.61 | 2.29 | 0.07 |
25(OH)D 25-hydroxyvitamin D3, FDR false discovery rate, MAF minor allele frequency, SNP single-nucleotide polymorphism
SNPs associated with 25(OH)D levels with an FDR <0.15 in either unadjusted or multivariable model, none of which reached the predefined FDR for statistical significance
Additive effect of a minor allele on 25(OH)D levels (continuous in ng/mL)
Adjusted for age (continuous), sex (female, male), race/ethnicity (white, black, other, unknown/missing), and season of blood collection (summer, fall, winter, spring)
Association between 25(OH)D-associated SNPs and patient outcome
We did not detect any direct association between individual Tier 1 SNPs or the genetic risk score and overall survival, TTP, or tumor response overall or among patients receiving FOLFOX (Table 4 and Supplementary Table 2). Overall, for each one-allele increase in the genetic risk score, the multivariable hazard ratios (HRs) were 1.01 [95% confidence interval (CI), 0.95 to 1.07; P=0.79] for death and 0.99 (95% CI, 0.93 to 1.05; P=0.76) for disease progression, and the multivariable OR for tumor response was 1.03 (95% CI, 0.92 to 1.16; P=0.57) (Table 4).
Table 4.
Association between 25(OH)D-associated SNPs in GWAS (Tier 1 SNPs), an additive genetic risk score, and survival among patients with metastatic colorectal cancer
| Outcomea | Unadjusted | Multivariableb | ||
|---|---|---|---|---|
| HR or OR (95% CI) | P | HR or OR (95% CI) | P | |
| Overall survival | ||||
| rsl993116 | 1.03 (0.91–1.17) | 0.64 | 0.99 (0.87–1.14) | 0.94 |
| rs6013897 | 0.96 (0.83–1.12) | 0.64 | 0.94 (0.80–1.11) | 0.47 |
| rsl2785878 | 1.05 (0.92–1.20) | 0.46 | 1.00 (0.86–1.16) | 0.99 |
| rsl1234027 | 1.05 (0.89–1.23) | 0.57 | 1.05 (0.89–1.25) | 0.54 |
| rs2282679 | 1.07 (0.92–1.24) | 0.36 | 1.06 ((Ш–1.24) | 0.42 |
| Genetic risk score | 1.03 (0.97–1.08) | 0.36 | 1.01 (0.95–1.07) | 0.79 |
| Time to progression | ||||
| rsl993116 | 0.91 (0.80–1.04) | 0.16 | 0.89(0.77–1.02) | 0.11 |
| rs6013897 | 0.92 (0.78–1.09) | 0.36 | 0.90 (0.75–1.07) | 0.23 |
| rsl2785878 | 1.11 (0.97–1.28) | 0.14 | 1.09 (0.93–1.27) | 0.27 |
| rsl1234027 | 1.06 (0.90–1.25) | 0.47 | 1.03 (0.87–1.23) | 0.73 |
| rs2282679 | 1.06 (0.91–1.23) | 0.47 | 1.05 (0.89–1.23) | 0.56 |
| Genetic risk score | 1.01 (0.95–1.07) | 0.82 | 0.99 (0.93–1.05) | 0.76 |
| Tumor response | ||||
| rsl993116 | 1.05 (0.82–1.35) | 0.68 | 1.04 (0.80–1.35) | 0.78 |
| rs6013897 | 1.04 (0.78–1.39) | 0.78 | 1.09 (0.80–1.50) | 0.58 |
| rsl2785878 | 0.95 (0.73–1.22) | 0.68 | 1.09 (0.82–1.44) | 0.56 |
| rsl1234027 | 0.94 (0.69–1.28) | 0.70 | 0.98 (0.70–1.36) | 0.90 |
| rs2282679 | 1.11 (0.84–1.46) | 0.45 | 1.04 (0.78–1.40) | 0.77 |
| Genetic risk score | 1.01 (0.91–1.13) | 0.83 | 1.03 (0.92–1.16) | 0.57 |
25(OH)D 25-hydroxyvitamin D3, CI confidence interval, ECOG Eastern Cooperative Oncology Group, FOLFOX fluorouracil, leucovorin, oxaliplatin, GWAS genome-wide association studies, HR hazard ratio, IFL irinotecan, bolus fluorouracil, leucovorin, IROX irinotecan, oxaliplatin, OR odds ratio, SNP single-nucleotide polymorphism
Additive effect of a risk allele (i.e., associated with lower 25(OH)D levels) on patient outcome
Adjusted for age (continuous), sex (female, male), race/ethnicity (white, black, other, unknown/missing), ECOG performance status (0–1,2), number of metastatic sites (continuous), and treatment arm (IFL, FOLFOX, IROX)
Association between plasma 25(OH)D levels and patient outcome, stratified by 25(OH)D-associated SNPs
We examined the association between plasma 25(OH)D levels and overall survival, stratified by Tier 1 SNPs and the genetic risk score. We found that the association varied by rs12785878 genotype in DHCR7 (Table 5). For each 1-ng/mL increase in 25(OH)D level, the multivariable HR for death was 0.99 (95% CI, 0.97–1.00), 1.01 (95% CI, 0.99–1.02), and 1.02 (95% CI, 0.98–1.07) among patients with the TT, TG, and GG genotype at rs12785878, respectively (Pinteraction=0.02).
Table 5.
Association between plasma 25(OH)D levels and overall survival among patients with metastatic colorectal cancer, stratified by rs12785878 genotype in DHCR7
| rsl2785878 | No. patients | Multivariable hazard ratio (95% confidence interval)a | pinteraction |
|---|---|---|---|
| TT | 232 | 0.99 (0.97–1.00) | |
| TG | 205 | 1.01 (0.99–1.02) | 0.02 |
| GG | 55 | 1.02 (0.98–1.07) |
25(OH)D 25-hydroxyvitamin D3, CI confidence interval, ECOG Eastern Cooperative Oncology Group, FOLFOX fluorouracil, leucovorin, oxaliplatin, IFL irinotecan, bolus fluorouracil, leucovorin, IROX irinotecan, oxaliplatin
Effect of each 1-ng/mL increase in 25(OH)D level on overall survival, adjusted for age (continuous), sex (female, male), race/ethnicity (white, black, other, unknown/missing), ECOG performance status (0–1, 2), number of metastatic sites (continuous), treatment arm (IFL, FOLFOX, IROX), and season of blood collection (summer, autumn, winter, spring)
Discussion
In this cohort of patients with previously untreated mCRC, baseline plasma 25(OH)D levels were associated with rs1993116 in CYP2R1 as well as with an additive genetic risk score determined by the 5 SNPs associated with 25(OH)D levels in previous GWAS. We did not observe any direct association between 25(OH)D-associated SNPs and patient outcome, including overall survival, TTP, and tumor response, individually or as a genetic risk score. However, we detected an interaction between 25(OH)D levels and rs12785878 genotype in DHCR7 on overall survival.
We observed an association between 25(OH)D levels and rs1993116 in CYP2R1. CYP2R1 is a microsomal enzyme that catalyzes C-25 hydroxylation of vitamin D3 into 25(OH)D in the liver and other organs (27). A previous study suggested that rs12794714 in CYP2R1 (r2=0.46 with rs1993116) could affect CRC susceptibility in African Americans (28). Although the other Tier 1 SNPs were not individually associated with 25(OH)D levels, the additive genetic risk score was associated with the levels. One potential explanation is that our study had limited statistical power to detect a small effect of individual alleles on 25(OH)D levels but was able to see an effect from the combined contribution of the 5 Tier 1 SNPs. Another potential explanation is that the SNPs identified in GWAS of healthy participants may not contribute substantially to 25(OH)D levels among patients with mCRC, a population with a high prevalence of vitamin D deficiency (17).
Our findings do not support any direct association between 25(OH)D-associated SNPs and patient outcome. The null association could be due to limited statistical power, yet alternative explanations are possible. First, vitamin D may have limited impact on patient outcome once CRC has metastasized, although this is disputed by a recent large analysis of plasma 25(OH)D levels and survival among patients with mCRC enrolled in a phase III trial, CALGB/SWOG 80405 (29). However, some preclinical data suggest that VDR expression is decreased in less-differentiated colon cancer cell lines (30), which may result in loss of response of colon tumor cells to vitamin D actions. Second, the GWAS-identified SNPs collectively explain only ~5% of the variance in 25(OH)D levels (31). Third, our genetic risk score assumes that each included risk allele would be associated with worse patient outcome given their association with lower 25(OH)D levels. If this assumption is invalid, summing the number of risk alleles would underestimate the true association.
Many prospective epidemiological studies have investigated the association between vitamin D status and CRC survival, most of which found that higher vitamin D levels were associated with improved CRC survival (32). However, knowledge gaps exist with respect to the interactions between vitamin D and genetic variation (33). A prospective cohort study in Scotland reported an interaction between plasma 25(OH)D levels and VDR genotype in relation to survival of non-metastatic CRC (34). In the current study, the association between 25(OH)D levels and overall survival varied according to rs12785878 genotype in DHCR7, which encodes the ultimate enzyme that converts 7-dehydrocholesterol into cholesterol. The enzyme is an important regulatory switch between cholesterol and vitamin D synthesis, since both processes require 7-dehydrocholesterol as the substrate (35). The SNP rs12785878 is located in an intron and does not modify the amino acid sequence of the protein. However, rs12800438, a SNP in high linkage disequilibrium (r2=0.96) with rs12785878, has been found to be associated with DHCR7 mRNA levels in the liver (36), where cholesterol and 25(OH)D are mainly produced. Therefore, the differential association between 25(OH)D levels and overall survival by rs12785878 genotype can potentially be explained by the differential expression of DHCR7 in the liver. These data support the biological role of vitamin D in CRC survival and may inform which patients will benefit, or potentially experience harm, from vitamin D supplementation. To confirm this finding, functional studies are warranted to examine DHCR7 as a regulator of vitamin D activity in relation to CRC survival.
A prospective study nested within a randomized chemotherapy trial, such as that in our analysis, has several advantages. First, all patients had histologically proven mCRC at study entry, limiting the impact of heterogeneity by disease stage. Second, patient treatment and follow-up were standardized, with regular examinations to prospectively record the date and nature of cancer progression. Detailed information on prognostic factors was prospectively collected at study entry. In addition, using germline genetic variants as proxies for vitamin D status limits bias due to non-genetic confounding and reverse causation, because these variants cannot be influenced by environmental, lifestyle or disease-related factors operating later in life.
Several limitations should be considered. First, our study participants are predominantly individuals of European descent, limiting the generalizability of our findings. However, the GWAS that identified 25(OH)D-associated SNPs were conducted in populations of European descent, so the underlying genetic association may not hold in other populations. Moreover, a racially homogeneous population is advantageous as it reduces the potential for confounding by population substructure. Second, if these SNPs are correlated with other loci that affect patient outcome, our results would be confounded (37). Third, we did not examine rs8018720 in SEC23A and rs10745742 in AMDHD1, which were associated with serum 25(OH)D levels in a recent European GWAS (38), but we will include them in future studies.
In conclusion, germline genetic variation in the vitamin D pathway informs baseline 25(OH)D levels among patients with mCRC. Our findings do not support any direct association between 25(OH)D-associated SNPs and patient outcome, though one SNP in DHCR7 modifies the association between 25(OH)D levels and overall survival. This may be due to the fact that our study had a relatively small sample size, and/or that these SNPs account for only a small proportion of the variance in 25(OH)D levels. Future studies examining the association between vitamin D-related genetic variants and patient outcome in larger populations are currently underway.
Supplementary Material
Grant Support:
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821 and U10CA180882 (to the Alliance for Clinical Trials in Oncology), U10CA180790, U10CA180820 (ECOG-ACRIN), U10CA180838, U10CA180867, U10CA180888 (SWOG), and U24CA196171. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Charles S. Fuchs declares consulting for Agios, Bain Capital, Bayer, Celgene, Dicerna, Eli Lilly, Entrinsic Health, Five Prime Therapeutics, Genentech, Gilead Sciences, KEW, Merck, Merrimack Pharmaceuticals, Pfizer, Sanofi, Taiho, and Unum Therapeutics. He also serves as a Director for CytomX Therapeutics and owns unexercised stock options for CytomX Therapeutics and Entrinsic Health. Other authors declare that they have no conflict of interest.
ClinicalTrials.gov Identifier: NCT00003594 (https://clinicaltrials.gov/ct2/show/NCT00003594)
References
- 1.Meggouh F, Lointier P, Saez S. (1991) Sex steroid and 1,25-dihydroxyvitamin D3 receptors in human colorectal adenocarcinoma and normal mucosa. Cancer research. 51: 1227–33. [PubMed] [Google Scholar]
- 2.Vandewalle B, Adenis A, Hornez L, Revillion F, Lefebvre J. (1994) 1,25-dihydroxyvitamin D3 receptors in normal and malignant human colorectal tissues. Cancer letters. 86: 67–73. [DOI] [PubMed] [Google Scholar]
- 3.Zehnder D, Bland R, Williams MC, et al. (2001) Extrarenal expression of 25-hydroxyvitamin d(3)-1 alpha-hydroxylase. The Journal of clinical endocrinology and metabolism. 86: 888–94. [DOI] [PubMed] [Google Scholar]
- 4.Vandewalle B, Wattez N, Lefebvre J. (1995) Effects of vitamin D3 derivatives on growth, differentiation and apoptosis in tumoral colonic HT 29 cells: possible implication of intracellular calcium. Cancer letters. 97: 99–106. [DOI] [PubMed] [Google Scholar]
- 5.Diaz GD, Paraskeva C, Thomas MG, Binderup L, Hague A. (2000) Apoptosis is induced by the active metabolite of vitamin D3 and its analogue EB1089 in colorectal adenoma and carcinoma cells: possible implications for prevention and therapy. Cancer research. 60: 2304–12. [PubMed] [Google Scholar]
- 6.Palmer HG, Gonzalez-Sancho JM, Espada J, et al. (2001) Vitamin D(3) promotes the differentiation of colon carcinoma cells by the induction of E-cadherin and the inhibition of beta-catenin signaling. The Journal of cell biology. 154: 369–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scaglione-Sewell BA, Bissonnette M, Skarosi S, Abraham C, Brasitus TA. (2000) A vitamin D3 analog induces a G1-phase arrest in CaCo-2 cells by inhibiting cdk2 and cdk6: roles of cyclin E, p21Waf1, and p27Kip1. Endocrinology. 141: 3931–9. [DOI] [PubMed] [Google Scholar]
- 8.Iseki K, Tatsuta M, Uehara H, et al. (1999) Inhibition of angiogenesis as a mechanism for inhibition by 1alpha-hydroxyvitamin D3 and 1,25-dihydroxyvitamin D3 of colon carcinogenesis induced by azoxymethane in Wistar rats. International journal of cancer. Journal international du cancer. 81: 730–3. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Garcia NI, Palmer HG, Garcia M, et al. (2005) 1alpha, 25-Dihydroxyvitamin D3 regulates the expression of Id1 and Id2 genes and the angiogenic phenotype of human colon carcinoma cells. Oncogene. 24: 6533–44. [DOI] [PubMed] [Google Scholar]
- 10.Evans SR, Shchepotin EI, Young H, Rochon J, Uskokovic M, Shchepotin IB. (2000) 1,25-dihydroxyvitamin D3 synthetic analogs inhibit spontaneous metastases in a 1,2-dimethylhydrazine-induced colon carcinogenesis model. Int J Oncol. 16: 1249–54. [DOI] [PubMed] [Google Scholar]
- 11.Wjst M, Altmuller J, Braig C, Bahnweg M, Andre E. (2007) A genome-wide linkage scan for 25-OH-D(3) and 1,25-(OH)2-D3 serum levels in asthma families. The Journal of steroid biochemistry and molecular biology. 103: 799–802. [DOI] [PubMed] [Google Scholar]
- 12.Orton SM, Morris AP, Herrera BM, et al. (2008) Evidence for genetic regulation of vitamin D status in twins with multiple sclerosis. The American journal of clinical nutrition. 88: 441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shea MK, Benjamin EJ, Dupuis J, et al. (2009) Genetic and non-genetic correlates of vitamins K and D. European journal of clinical nutrition. 63: 458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter D, De Lange M, Snieder H, et al. (2001) Genetic contribution to bone metabolism, calcium excretion, and vitamin D and parathyroid hormone regulation. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 16: 371–8. [DOI] [PubMed] [Google Scholar]
- 15.Ahn J, Yu K, Stolzenberg-Solomon R, et al. (2010) Genome-wide association study of circulating vitamin D levels. Human molecular genetics. 19: 2739–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang TJ, Zhang F, Richards JB, et al. (2010) Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 376: 180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng K, Sargent DJ, Goldberg RM, et al. (2011) Vitamin D status in patients with stage IV colorectal cancer: findings from Intergroup trial N9741. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 29: 1599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldberg RM, Sargent DJ, Morton RF, et al. (2004) A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 22: 23–30. [DOI] [PubMed] [Google Scholar]
- 19.Meyerhardt JA, Sloan JA, Sargent DJ, et al. (2005) Associations between plasma insulin-like growth factor proteins and C-peptide and quality of life in patients with metastatic colorectal cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 14: 1402–10. [DOI] [PubMed] [Google Scholar]
- 20.McLeod HL, Sargent DJ, Marsh S, et al. (2010) Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 28: 3227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutz JE, Manning WA, Province MA, McLeod HL. (2011) Genomewide analysis of inherited variation associated with phosphorylation of PI3K/AKT/mTOR signaling proteins. PloS one. 6: e24873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollis BW. (1997) Quantitation of 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D by radioimmunoassay using radioiodinated tracers. Methods in enzymology. 282: 174–86. [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. Series B (Methodological). 289–300. [Google Scholar]
- 24.Cox DR. (1992) Regression models and life-tables. Breakthroughs in statistics: Springer; pp. 527–41. [Google Scholar]
- 25.Gorham ED, Garland CF, Garland FC, et al. (2007) Optimal vitamin D status for colorectal cancer prevention: a quantitative meta analysis. American journal of preventive medicine. 32: 210–6. [DOI] [PubMed] [Google Scholar]
- 26.Ng K, Meyerhardt JA, Wu K, et al. (2008) Circulating 25-hydroxyvitamin d levels and survival in patients with colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 26: 2984–91. [DOI] [PubMed] [Google Scholar]
- 27.Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW. (2004) Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proceedings of the National Academy of Sciences of the United States of America. 101: 7711–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pibiri F, Kittles RA, Sandler RS, et al. (2014) Genetic variation in vitamin D-related genes and risk of colorectal cancer in African Americans. Cancer causes & control : CCC 25: 561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng K, Venook AP, Sato K, et al. (2015) Vitamin D status and survival of metastatic colorectal cancer patients: Results from CALGB/SWOG 80405 (Alliance). American Society of Clinical Oncology. [Google Scholar]
- 30.Shabahang M, Buras RR, Davoodi F, Schumaker LM, Nauta RJ, Evans SR. (1993) 1,25-Dihydroxyvitamin D3 receptor as a marker of human colon carcinoma cell line differentiation and growth inhibition. Cancer research. 53: 3712–8. [PubMed] [Google Scholar]
- 31.Hiraki LT, Major JM, Chen C, et al. (2013) Exploring the genetic architecture of circulating 25-hydroxyvitamin D. Genetic epidemiology. 37: 92–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morales-Oyarvide V, Meyerhardt JA, Ng K. (2016) Vitamin D and Physical Activity in Patients With Colorectal Cancer: Epidemiological Evidence and Therapeutic Implications. Cancer J. 22: 223–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mondul AM, Weinstein SJ, Layne TM, Albanes D. (2017) Vitamin D and Cancer Risk and Mortality: State of the Science, Gaps, and Challenges. Epidemiol Rev. 39: 28–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zgaga L, Theodoratou E, Farrington SM, et al. (2014) Plasma vitamin D concentration influences survival outcome after a diagnosis of colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 32: 2430–9. [DOI] [PubMed] [Google Scholar]
- 35.Prabhu AV, Luu W, Sharpe LJ, Brown AJ. (2016) Cholesterol-mediated Degradation of 7-Dehydrocholesterol Reductase Switches the Balance from Cholesterol to Vitamin D Synthesis. The Journal of biological chemistry. 291: 8363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strawbridge RJ, Deleskog A, McLeod O, et al. (2014) A serum 25-hydroxyvitamin D concentration-associated genetic variant in DHCR7 interacts with type 2 diabetes status to influence subclinical atherosclerosis (measured by carotid intima-media thickness). Diabetologia. 57: 1159–72. [DOI] [PubMed] [Google Scholar]
- 37.Sheehan NA, Didelez V, Burton PR, Tobin MD. (2008) Mendelian randomisation and causal inference in observational epidemiology. PLoS medicine. 5: e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang X, O’Reilly PF, Aschard H, et al. (2018) Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nature communications. 9: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.